Abstract
The maternally inherited mitochondrial DNA (mtDNA) D–loop region is widely used for exploring genetic relationships and for investigating the origin of various animal species. Currently, domestic ducks play an important role in animal protein supply. In this study, partial mtDNA D–loop sequences were obtained from 145 samples belonging to six South-East Asian duck populations and commercial duck population. All these populations were closely related to the mallard duck (Anas platyrhynchos), as indicated by their mean overall genetic distance. Sixteen nucleotide substitutions were identified in sequence analyses allowing the distinction of 28 haplotypes. Around 42.76% of the duck sequences were classified as Hap_02, which completely matched with Anas platyrhynchos duck species. The neighbor-joining phylogenetic tree also revealed that South-East Asian duck populations were closely related to Anas platyrhynchos. Network profiles were also traced using the 28 haplotypes. Overall, results showed that those duck populations D-loop haplotypes were shared between several duck breeds from Korea and Bangladesh sub continental regions. Therefore, these results confirmed that South-East Asian domestic duck populations have been domesticated from Anas platyrhynchos duck as the maternal origins.
Keywords: Duck Populations, D-loop Region, Haplotype, Mitochondrial DNA, Phylogeny
INTRODUCTION
Duck is the second most important poultry species for egg, meat, and down feather production in the world. According to the Food and Agriculture Organization of the United Nations (FAO) statistics between 2000 and 2011, the global duck meat production has increased by 1.3 million tons, with the duck meat production in Asia increasing by 3.7% per year during the same period. Asia is the region with the highest duck production accounting for approximately 84% of the total global production in 2011 (FAO, 2014). In Korea, the duck production was 44.6 million in 2000, reaching almost 68.9 million in 2011; in Bangladesh, the duck production increased from 33 to 44 million during the same period.
Two distinct hypotheses have been proposed to explain the ancestry and evolution of the domestic ducks. One hypothesis states that the presently known domestic ducks are mainly originated from the wild duck (wild mallard, Anas boschas) whose domestication process started thousands years ago in Southeast Asia. The other hypothesis suggests that the domestic ducks were originated from the hybridization between the mallard (Anas platyrhynchos) and the Eastern spot-billed duck (Anas p. zonorhyncha) (Cherry and Morris, 2008). Both the Muscovy duck (Cairinamoschata) and A. boschas can interbreed with other Anas species distributed worldwide (Wojcik and Smalec, 2007). Duck species conservation is currently a matter of serious concern due to the uncontrolled breeding, interbreeding, and hybridization of domesticated and natural populations of closely related species all over the world.
In 2012, the Ministry of Finance of Bangladesh reported that 45.12 million of the ducks produced were mostly indigenous types and their crossbreeds (BER, 2012). Among indigenous ducks of Bangladesh, “Nageswari” an egg type native duck breed/variety is locally called “Nagi” (the snake deity) or Deshi Black, due to its head-high snake like posture, has a white breast while the rest of the plumage is black or penciled black, and is believed to have originated in the Sylhet district of Assam, which is now located in Bangladesh (Zaman et al., 2005). Jinding, a popular egg type duck breed in Bangladesh which was imported from China and has been reared by the farmers of duck potential areas since late 20th century. Common indigenous and Deshi white ducks are scatteredly distributed throughout Bangladesh that has been maintained by the farmers as family poultry under traditional husbandry practices (FAO, 2004). Besides, Korean native ducks (KND) are small, similar in appearance to mallards, but cannot fly well. Like most native ducks, KND have a slow growth rate with low feed conversion efficiency (Hong et al., 2012) and a longer rearing period than commercial ducks (CD). Despite these limitations to the commercial production of KND, it has a high consumer demand due to its high meat quality with unique texture and flavor (Kim et al., 2012). In Korea, the National Institute of Animal Science is the main commercial producer of KND and CD. KND conservation has recently become a high-concern issue for the Korean government, which has launched a project in 2014 aiming to recognize the maternal origin, genetic diversity, domestication process, and genetic differentiation among domestic duck breeds.
The D-loop is the major control region of mitochondrial DNA (mtDNA) and its complete or partial sequence is a widely used marker for studies of inter-species variation (Moore and DeFilippis, 1997) and for understanding evolutionary systems (Baker and Marshall, 1997), due to its lack of recombination and 5 to10 times higher nucleotide substitution rates than nuclear DNA markers (Brown et al., 1979). Therefore, D-loop substitutions have been extensively used to characterize the decisive genetic resources in chicken (Hoque et al., 2009; 2011; 2013), ducks (Choi et al., 2014; Jin et al., 2014), and other avian species (Delport et al., 2002).
Several studies using mtDNA D-loop control region genetic variation have been conducted to elucidate the gene flow of domestic and wild ducks and their genetic relationships in several geographical regions (Choi et al., 2014; Jin et al., 2014). The mtDNA cytochrome c oxidase subunit I (COI) genetic variation has been used to discriminate and establish the phylogenetic relationships among Korean dabbling duck species (Jin et al., 2012). The genetic polymorphism and phylogenetic results obtained from D-loop region in five Indonesian native duck breeds suggested that the maternal inheritance patterns of Indonesian native ducks were relatively similar to those of the mallard (A. platyrhynchos) and Eastern spot-billed duck (A. p. zonorhyncha) (Purwantini et al., 2013). Using hyper-variable D-loop sequences Jin et al. (2014) distinguished wild duck breeds from Korean domestic ducks by 26 indels present in partial D-loop sequences. The relationships between black KND and other duck breeds were initially investigated based on partial D-loop sequences (Choi et al., 2014). Results of phylogenetic analyses showed that both black KND and white CD breeds were well differentiated from wild duck breeds, while mallards were not well classified with KND. Thus, the present study aims to investigate the phylogenetic relationships between KND and other South Asian duck breeds using partial D-loop sequences nucleotide substitutions, in order to explore the origin and pattern of genetic diversity among these breeds, as well as to elucidate on conservation breeding programs for native duck breeds.
MATERIALS AND METHODS
Specimen collection and DNA isolation
This study was carried out for two KND populations, a CD breed (Peking), and four Bangladeshi duck populations. Detailed sample information is presented in Table 1. KND blood samples were collected from the wing veins of ducks held in the Poultry Science Division, National Institute of Animal Science (NIAS), Korea, in 3-ml tubes containing ethylenediaminetetraacetic acid (EDTA). A total of 84 blood samples from four duck breeds/varieties were collected from two governmental institutions, the Bangladesh Livestock Research Institute (BLRI) in Dhaka and the Central Duck Breeding Farm (CDBF) in Narayanganj, and from different locations within the Mymensingh, Kishoreganj, and Sherpur districts of Bangladesh (Table 1). Blood samples of all Bangladeshi ducks were preserved on Whatman WB 120205 FTA classic cards (Whatman International Ltd, Maidstone, Kent, UK). Genomic DNA was extracted from all blood samples using the PrimePrep Genomic DNA Isolation from Blood and from Tissue Kits (GeNetBio, Daejeon, Korea), according to the manufacturer’s instructions. DNA concentration of each sample was checked using NanoDrop 2000c UV-Vis Spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA) to confirm the quantity and quality of the extracted DNA. The isolated DNA samples were stored at −20°C and stock genomic DNA was standardized to a maximum of 25 ng/μL for each sample.
Table 1.
Sample information for each of the seven South-East Asian duck populations
| Duck breed/variety | Origin of sample | Location of sample collection site | Sample code | No. of samples |
|---|---|---|---|---|
| Nageswari (Deshi black) | Bangladesh | BLRI, Dhaka; CDBF, Narayanganj; Kishoreganj and Mymensingh districts, Bangladesh | BaB | 36 |
| Deshi white | Bangladesh | BLRI, Dhaka, Bangladesh | BaW | 20 |
| Jinding | Imported from China | CDBF, Narayanganj, Bangladesh | BaJ | 15 |
| Common Indigenous duck | Bangladesh | Sherpur and Mymensingh districts, Bangladesh | BaL | 13 |
| White Korean native duck | South Korea | Yongin, Gyeonggi Province, Korea | WKND | 21 |
| Black Korean native duck | South Korea | NIAS, Korea | KND_N | 20 |
| Commercial (Peking) duck | South Korea | Cherry Valley, Korea | CD | 20 |
| Total | 145 |
Polymerase chain reaction amplification, sequencing and cloning
Polymerase chain reaction (PCR) was used to amplify a partial sequence of the hypervariable D-loop region (about 710 bp long) using the primer set developed by Wu et al. (2011) (Forward, 5′-GTTATTTGGTTATGCATATCGTG-3′; Reverse, 5′-CCATATACGCCAACCGTCTC-3′). PCR reaction mixes included approximately 50 ng of genomic DNA, 10× buffer (containing Tris-HCl [pH 9.0], PCR enhancers, (NH4)2SO4, 20 mM MgCL2), 10 mM dNTPs mixture (2.5 mM of each dATP, dCTP, Dgtp, and dTTP), 10 pM primers, and 1 U Prime Taq (GeNetBio, Korea) in a 20 μL reaction volume. PCR was performed in a My-Genie 96 Thermal Block (Bioneer Co., Daejeon, Korea) using the following profile: pre-denaturation at 95°C for 5 min; 35 cycles of 45 s at 95°C for denaturation, 45 s at 60°C for annealing, and 1 min at 72°C for extension; and a final extension step at 72°C for 7 min. PCR products were visualized under ultraviolet light in 2% agarose gels stained with ethidium bromide. Purification of PCR products was carried out using the PrimePrep PCR purification kit (GeNetBio, Korea) according to the manufacture’s guidelines. The purified PCR products were then sequenced by direct sequencing method. Consensus sequences approximately 606 bp long were retrieved for each individual by assembling sequences from both strands. A total of 27 PCR products from Bangladeshi duck samples, consisting of four breeds/varieties (Nageswari duck: 6; Deshi white duck: 5; Jinding duck: 9; Common Indigenous duck: 9), were cloned into a pGEM-T Easy Vector System (Promega, USA) and transformed using the DH5α competent Escherichia coli strain (Enzynomics, Daejeon, Korea) with the highest transformation efficiency. Collected plasmid DNA was also direct sequenced using the SP6 and T7 universal promoter primers.
Data analysis
All duck D-loop sequences obtained in this study were aligned using ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/) and edited using BioEdit software (Hall, 1999). The nucleotide replacement data was exported to the haplotype assignment software using MEGA ver. 6.06. Gaps in the aligned sequences were excluded from the analysis (Tamura et al., 2013). The number of nucleotide variable sites, number of haplotypes (NHap), nucleotide diversity (Pi), and haplotype diversity (Hd) were calculated using DnaSP ver. 5.1 (Librado and Rozas, 2009). An unrooted neighbor–joining (NJ) tree was produced in MEGA ver. 6.06 to determine the phylogenetic relationships between breeds; tree confidence levels were evaluated by 1,000 bootstrap random replicates and data resampling. Distances between all haplotypes were calculated using the Kimura-2 parameter substitution model. A total of 145 D-loop sequences, 105 from this study and 40 previously published sequences of the KND (Choi et al., 2014), were used in the genetic analysis (Table 1). A total of 158 cloned sequences resulting from the 27 cloned samples from the four Bangladeshi duck breeds were also investigated. In addition, complete D-loop sequences from different domestic duck breeds (Peking, Rongshui, Xilin, Youxian Sheldrake, Longsheng, Jingxi, Linwu, and Youxian) were retrieved from the National Center for Biotechnology Information (NCBI) database and included in the phylogenetic tree (GenBank accession numbers: EU755252.1, EU009397.1 (both Peking duck sequences), KJ833587.1, KJ833586.1, KJ883269.1, KJ739616.1, KJ689447.1, KJ637997.1, and KJ778676.1, respectively). Additionally, NETWORK 4.6.1.0 software was used to construct a median-joining network profile for haplotype differentiation along positional single nucleotide polymorphisms (SNPs) (Bandelt et al., 1999). The partial D-loop haplotype sequences were deposited to GenBank in National Center for Biotechnology Information (NCBI) (GenBank accession numbers: KU845272 ~KU845299).
RESULTS AND DISCUSSION
Polymorphisms and genetic diversity
A total of 145 mtDNA D-loop sequences were obtained: 84 from the Bangladeshi duck populations of Nageswari (BaB), Deshi white (BaW), Jinding (BaJ), and Indigenous (BaL); and 61 from two Korean duck breeds, namely white Korean native duck (WKND), black Korean native duck (KND_N), and one of CD population. The analysis of these 145 duck mtDNA D-loop sequences revealed a total of 16 nucleotide substitutions and one indel (insertion or deletion) (Table 2). Intra-population diversity measures i.e., the number of variable sites (S), the NHap, Hd, and Pi, and a sequenced based neutrality test, Tajima’s D, calculated for each duck population considered in this study are presented in Table 2. Analysis of mtDNA D-loop sequences identified 7 segregating sites in the BaB, one in the BaW, and 4 in the BaJ and BaL populations; KND presented 5, 4, and 7 segregating sites in the WKND, KND_N, and CD populations, respectively. Among the Bangladeshi duck populations, only one indel (nucleotide position 253) was found in the BaB population.
Table 2.
Diversity indices and neutrality test results based on the genetic analysis of mtDNA D-loop sequence fragments for the seven duck populations studied1
| Breed | N | S | Indel | NHap | Pi | K | Hd | Tajima’s D (p>0.10) |
|---|---|---|---|---|---|---|---|---|
| All | 145 | 16 | 1 | 28 | 0.00317 | 1.921 | 0.799 | −0.891 (NS) |
| BaB | 36 | 7 | 1 | 12 | 0.00372 | 2.256 | 0.829 | 0.952 (NS) |
| BaW | 20 | 1 | 0 | 2 | 0.00056 | 0.337 | 0.337 | 0.352 (NS) |
| BaJ | 15 | 4 | 0 | 6 | 0.00255 | 1.543 | 0.886 | 0.815 (NS) |
| BaL | 13 | 4 | 0 | 6 | 0.00237 | 1.436 | 0.833 | 0.387 (NS) |
| WKND | 21 | 5 | 0 | 5 | 0.00195 | 1.181 | 0.633 | −0.453 (NS) |
| KND_N | 20 | 4 | 0 | 3 | 0.00302 | 1.832 | 0.689 | 1.809 (NS) |
| CD | 20 | 7 | 0 | 6 | 0.00372 | 2.253 | 0.789 | 0.466 (NS) |
Number of variable sites (S) number of haplotype (NHap), nucleotide diversity (Pi), and haplotype diversity (Hd), N = number of sequences, number of insertion or deletion (Indel), average number of nucleotide differences (K), Nageswari (BaB), Deshi white (BaW), Jinding (BaJ), Common Indigenous duck (BaL), White Korean native duck (WKND), Black Korean native duck (KND_N), Commercial duck (CD), and not significant (NS).
In general, within-population Hd values were relatively higher for Bangladeshi duck varieties than for Korean duck breeds, except for BaW (Table 2). Tajima’s D test showed non-significant deviations from neutral expectation despite the considerable differences observed in the within population allele frequency distribution. In Bangladeshi duck populations, Pi values ranged between 0.00056 and 0.00372, while values observed in Korean duck populations varied between 0.00195 and 0.00372. The highest Pi value was observed in the BaB and CD populations and the lowest was in the BaW population. Qu et al. (2009) reported that Jinding and Gaoyou ducks had the highest (0.01100) and the lowest (0.00087) mtDNA control region nucleotide polymorphism, respectively, among seven duck breeds in China. Jin et al. (2014) stated that Pi varied between 0.004 and 0.019 among seven species of wild and domestic ducks in Korea. In addition, nucleotide sequences of mtDNA control region showed divergence ranging between 0.0012 and 0.0015 in domestic native Thai ducks. The Pi values obtained for Bangladeshi and KND populations fall within the range depicted in all the above-referred studies, therefore supporting the present findings.
In Bangladeshi duck populations, the highest number of haplotypes (12) was observed in the BaB population that presented 82.9% Hd. BaW, BaJ, and BaL haplotype diversities were 33.7%, 88.6%, and 83.3%, respectively. Korean WKND, KND_N, and CD duck populations presented 5, 3, and 6 haplotypes with a proportional haplotype diversity of 63.3%, 68.9%, and 78.9%, respectively. Hence, the highest haplotype diversity value was observed in BaJ for the Bangladeshi duck populations and in CD for the Korean duck populations. Our results agree with the findings of Leekaew et al. (2008) who reported haplotype diversities varying from 0.700 to 0.800 in Thai native duck populations. Although quite extreme, the haplotype diversity values reported for Chinese domestic duck populations, which varied between 0.324 and 1.000, (Qu et al., 2009), are also in agreement with those found in the present study. Purwantini et al. (2013) suggested that the Indonesian native Magelang duck population, which showed high polymorphism in the mtDNA D-loop region, might have originated from native ducks crossbreeding. Moreover, they found that Indonesian native duck populations’ maternal line originated from Anas platyrhynchos and Anas zonorhyncha. Likewise, previous studies also identified spot-billed and mallard ducks as closely related species (Kulikova et al., 2004).
Haplotype analysis
In this study, the 16 polymorphisms identified in the seven duck populations classified sequences into 28 haplotypes (Table 3), which in turn provided sample identity (Table 4). Among the 28 haplotypes, Hap_02 is the most common containing 62 sequences (42.76%) from both Bangladeshi and Korean duck populations, suggesting that these populations originated from an A. platyrhynchos common ancestor. Haplotype sharing was reported between native Thai and mallard ducks (Leekaew et al., 2008), between Peking and Wild mallard ducks (Qu et al., 2009), and between Chinese domestic duck breeds and mallards, as well as between Spot-billed and domestic ducks (Jin et al., 2014). Our results are, therefore, in agreement with these findings. Hap_01 to Hap_09 were considered to be KND haplotypes, whereas Hap_10 to Hap_28 were only observed in Bangladeshi duck populations. Seven haplotypes (Hap_01, Hap_03, Hap_04, Hap_05, Hap_06, Hap_07, and Hap_08) were presented in 59.02% samples of KND populations and Hap_10 to Hap_28 were found in 52.38% of the Bangladeshi duck populations as a specific haplotype. Most noticeably, the Nageswari duck population had the largest number of unique haplotypes (Hap_10 to Hap_20) and a specific indel, suggesting it is a distinct variety of indigenous Bangladeshi duck.
Table 3.
Polymorphic sites found in the mtDNA D-loop region of the duck populations considered in the present study
| Nucleotide position in mtDNA D-loop region1 | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||||||||
| Polymorphism | InDel | ||||||||||||||||
|
|
|
||||||||||||||||
| Haplotype | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 4 | 6 | 2 |
| 8 | 9 | 0 | 0 | 1 | 2 | 4 | 5 | 5 | 7 | 7 | 9 | 2 | 4 | 3 | 0 | 5 | |
| 2 | 4 | 0 | 2 | 9 | 8 | 3 | 2 | 4 | 2 | 5 | 7 | 6 | 2 | 8 | 8 | 3 | |
|
| |||||||||||||||||
| Hap_01 | C | C | T | T | C | A | A | C | T | C | C | G | C | T | C | T | Δ |
| Hap_02 | - | - | - | - | - | - | - | - | - | - | - | A | - | - | - | - | - |
| Hap_03 | - | - | - | - | - | - | - | - | - | T | - | A | - | - | - | C | - |
| Hap_04 | - | - | - | - | - | - | - | - | - | T | - | A | - | - | - | - | - |
| Hap_05 | - | - | - | - | - | G | - | - | - | - | - | A | T | - | - | - | - |
| Hap_06 | - | - | - | - | T | - | - | - | - | T | - | A | - | - | - | C | - |
| Hap_07 | - | - | - | - | - | G | - | - | - | - | - | A | - | - | - | - | - |
| Hap_08 | - | T | - | - | - | - | - | - | - | - | - | A | - | - | - | - | - |
| Hap_09 | - | - | - | - | - | - | - | T | - | - | - | A | - | - | - | - | - |
| Hap_10 | T | - | - | A | - | - | - | - | - | T | - | A | - | - | - | C | - |
| Hap_11 | T | - | C | - | - | - | - | - | - | - | - | A | - | C | - | - | - |
| Hap_12 | T | - | C | A | - | - | - | - | - | - | - | A | - | C | - | - | - |
| Hap_13 | T | - | - | A | - | - | - | - | - | - | - | A | - | - | - | - | - |
| Hap_14 | - | - | - | - | - | - | - | - | - | T | - | A | - | C | - | C | C |
| Hap_15 | - | - | - | - | - | - | - | - | - | - | - | A | - | C | - | - | - |
| Hap_16 | T | - | C | - | - | - | - | - | - | T | - | A | - | - | - | C | - |
| Hap_17 | - | - | - | A | - | - | - | - | - | - | - | A | - | - | - | - | C |
| Hap_18 | - | - | - | A | - | - | - | - | - | - | - | A | - | C | - | - | - |
| Hap_19 | - | - | C | - | T | - | - | - | - | T | - | A | - | - | - | C | - |
| Hap_20 | - | - | C | - | - | - | - | - | - | - | - | A | - | - | - | - | - |
| Hap_21 | - | - | - | - | - | - | - | - | - | - | - | A | - | - | - | C | - |
| Hap_22 | - | - | - | - | - | - | G | - | - | - | - | A | - | - | - | - | - |
| Hap_23 | - | - | - | A | - | - | - | - | C | - | - | A | - | - | - | - | - |
| Hap_24 | - | - | - | - | - | - | - | - | - | - | - | A | - | - | T | - | - |
| Hap_25 | - | - | - | - | - | - | - | - | C | - | - | A | - | - | - | - | - |
| Hap_26 | - | - | - | - | - | - | - | T | - | - | T | A | - | - | - | - | - |
| Hap_27 | - | - | C | - | - | - | - | T | C | - | - | A | - | - | - | - | - |
| Hap_28 | - | - | - | - | - | - | - | - | - | - | T | A | - | - | - | - | - |
Numbers indicate nucleotide base position in mitochondrial D-loop region;the hyphen (-) indicates that the nucleotide is identical to that in the Hap-1 sequence.
Table 4.
Haplotypes shared amongst Korean and Bangladeshi domestic duck breeds
| Haplotype | No. of samples | Breeds (No. of samples within each breed) |
|---|---|---|
| Hap_01 | 10 | WKND (2), KND_N (5), CD (3) |
| Hap_02 | 62 | WKND (12), KND_N (8), CD (4), BaB (14), BaW (16), BaJ (3), BaL (5) |
| Hap_03 | 5 | WKND (5) |
| Hap_04 | 1 | WKND (1) |
| Hap_05 | 1 | WKND (1) |
| Hap_06 | 15 | KND_N (7), CD (8) |
| Hap_07 | 1 | CD (1) |
| Hap_08 | 3 | CD (3) |
| Hap_9 | 3 | CD (1), BaL (2) |
| Hap_10 | 3 | BaB (3) |
| Hap_11 | 1 | BaB (1) |
| Hap_12 | 3 | BaB (3) |
| Hap_13 | 2 | BaB (2) |
| Hap_14 | 1 | BaB (1) |
| Hap_15 | 2 | BaB (2) |
| Hap_16 | 1 | BaB (1) |
| Hap_17 | 6 | BaB (4), BaJ (2) |
| Hap_18 | 1 | BaB (1) |
| Hap_19 | 1 | BaB (1) |
| Hap_20 | 3 | BaB (3) |
| Hap_21 | 4 | BaW (4) |
| Hap_22 | 3 | BaJ (3) |
| Hap_23 | 3 | BaJ (3) |
| Hap_24 | 2 | BaJ (2) |
| Hap_25 | 4 | BaJ (2), BaL (2) |
| Hap_26 | 2 | BaL (2) |
| Hap_27 | 1 | BaL (1) |
| Hap_28 | 1 | BaL (1) |
Heteroplasmic status of duck mtDNA
Two major patterns, overlapped and ambiguous sequences, appeared in a D-loop sequence fragment of approximately 100 bp in all duck samples. Wu et al. (2011) identified this phenomenon in mtDNA D-loop sequences from domestic mallard ducks blood samples, including 12 samples from Bangladeshi indigenous ducks, amplified using 4 primer pairs. Likewise, we observed heteroplasmic D-loop sequences corresponding to one recognized partial D-loop fragment and to one ambiguous and unidentified sequence. Nuclear mitochondrial (numt) DNA in eukaryotes originates from nuclear DNA aggression on mtDNA and exhibits homologous sequences to mtDNA (Lopez et al., 1994), although presenting different homology degrees and sizes within and between chromosomes (Woischnik and Moraes, 2002). Numts have been found in more than 83 species (Bensasson et al., 2001) and in the human genome their number has been estimated to range from 296 to 621 (Bensasson et al., 2003). Sorenson and Fleischer (1996) reported that nuclear sequences homologous to the mtDNA D-loop region appeared when genomic DNA was extracted from blood samples of diving ducks but not from muscle, feather, and tissue samples. Several recent studies applying cloning sequencing protocols in domestic ducks have been able to retrieve partial D-loop sequences (Leekaew et al., 2008; Jin et al., 2014).
Phylogeny and network profiles
The 28 haplotypes identified in this study, along with the D-loop sequences belonging to different duck breeds retrieved from the NCBI database, were used to construct the NJ phylogenetic tree (Figure 1). Results indicated that Bangladeshi and Korean duck populations originated from the A. platyrhynchos duck breed. According to the phylogenetic tree, Hap_2 was scattered along the tree with other 5 haplotypes all representing the admixture of Bangladeshi and Korean ducks. In addition, eleven haplotypes (Hap_09, Hap_11 to 13, Hap_17, Hap_18, Hap_20, Hap_23, and Hap_26 to Hap_28), which were closely related to the wild duck ancestors, were only found in Bangladeshi duck populations. On the other hand, Hap_01 and Hap-03 to Hap_08, which are unique to CD and KND. The phylogenetic tree also suggested Bangladeshi and Korean ducks were domesticated from A. platyrhynchos. Although the genetic variation in D-loop sequences might not be able to differentiate domestic duck populations, it can separate different species of duck breeds (Choi et al., 2014; Jin et al., 2014). The NJ tree obtained here supports the existence of KND breeds as reported by Choi et al. (2014) and Jin et al. (2014). These authors reported that among seven duck species, four wild species were clearly discriminated from the three domestic duck species. In addition, they reported a relatively close genetic distance between the three domestic ducks, which is in agreement with their similar morphology derived from random crossing A. platyrhynchos and A. p. zonorhyncha (Johnson and Sorenson, 1999). Our median-joining network provided connections between the 28 haplotypes based on SNPs positions (Figure 2) and network analysis provided interesting results including the distinction of Bangladeshi and Korean ducks haplotype groups. In the Bangladeshi duck group, only one haplotype (Hap_09) demonstrated admixture with the Korean duck group through the CD population. Accordingly, the Korean duck group contained four haplotypes from BaB (Hap_10, Hap_14, and Hap_16) and BaW (Hap_21) populations. These haplotype sharing features also existed in other livestock species and might be due to retention of conserved ancestral polymorphisms among the closest species or breeds (Hey, 2006). Due to their geographical distribution, domestic ducks might have the same maternal origin. Our analysis provided informative polymorphisms for discriminating the different duck populations in further analysis.
Figure 1.
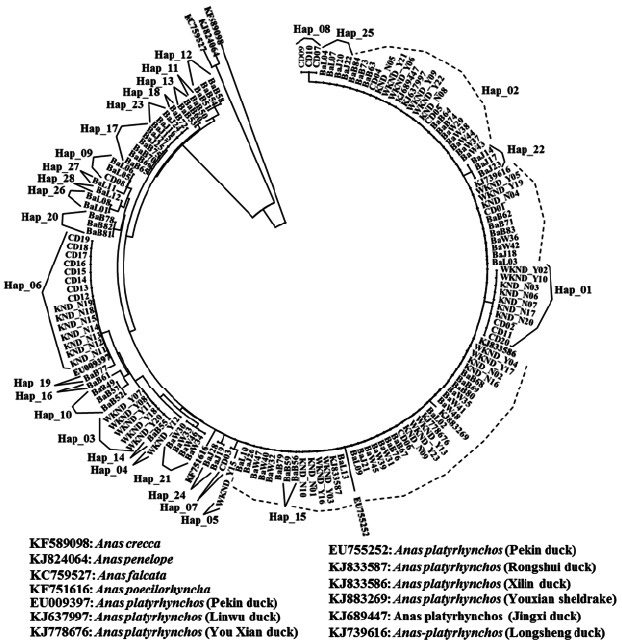
Unrooted Neighbor-Joining (NJ) phylogenetic tree constructed using the D-loop sequences obtained for the Korean and Bangladeshi domestic duck breeds considered in the present study.
Figure 2.
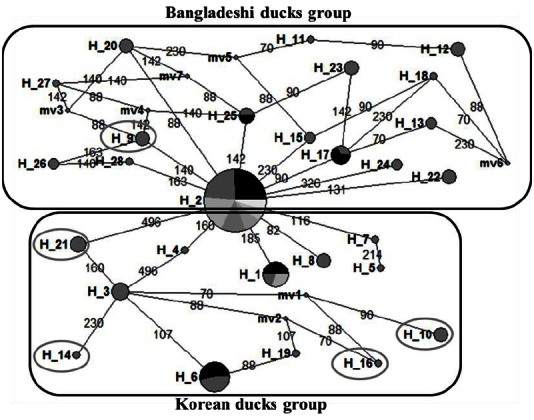
Median-joining network pro les obtained for the duck breeds based on the D-loop region haplotypes. A median vector (mv) is a hypothesized sequence (often ancestral), which is required to connect existing haplotypes within the maximum parsimony network. The oval circles in boxes indicate haplotype sharing with geographically distinct breeds.
In this study, we investigated the polymorphisms of mtDNA D-loop sequences duck populations to identify their origin. Our results revealed that Bangladeshi and Korean ducks were domesticated from an A. platyrhynchos ancestor. Most of the domestic duck populations are not unique and might be inter-crossed throughout their geographical distribution. However, maintaining the valuable native duck populations would be very useful for conservation in a geographical perspective. The polymorphisms identified here for the hypervariable D-loop region could be linked to the identified haplotypes to discriminate the various duck populations. Nevertheless, more molecular studies are required to further improve discrimination between duck breeds.
ACKNOWLEDGMENTS
This study was supported by a grants from the Brain Korea 21 Program (Project No. F16SR28T4002), and the Cooperative Research Program for Agriculture Science & Technology Development (Project No. 010114012015), RDA, Republic of Korea.
Footnotes
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
REFERENCES
- Baker AJ, Marshall HD. Mitochondrial control region sequences as tools for understanding evolution. In: Mindell DP, editor. Avian Molecular Evolution and Systematics. Academic Press; San Diego, USA: 1997. pp. 51–82. [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Bensasson D, Zhang D, Hartl DL, Hewitt GM. Mitochondrial pseudogenes: evolution’s misplaced witnesses. Trends Ecol Evol. 2001;16:314–321. doi: 10.1016/s0169-5347(01)02151-6. [DOI] [PubMed] [Google Scholar]
- Bensasson D, Feldman MW, Petrov DA. Rates of DNA duplication and mitochondrial DNA insertion in the human genome. J Mol Evol. 2003;57:343–354. doi: 10.1007/s00239-003-2485-7. [DOI] [PubMed] [Google Scholar]
- BER (Bangladesh Economic Review) Ministry of Finance, Government of the People’s Republic of Bangladesh. Dhaka, Bangladesh: 2012. pp. 92–93. [Google Scholar]
- Brown WM, George M, Jr, Wilson AC. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci USA. 1979;76:1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry P, Morris TR. Domestic Duck Production: Science and Practice (Paperback ed) CAB International; Wallingford, Oxfordshire, UK: 2008. [Google Scholar]
- Choi NR, Seo DW, Jin SD, Sultana H, Heo KN, Lee JH. Phylogenetic Analysis using mtDNA Sequences in Korean Native Ducks. Kor J Poult Sci. 2014;41:235–240. [Google Scholar]
- Delport W, Ferguson JW, Bloomer P. Characterization and evolution of the mitochondrial DNA control region in hornbills (Bucerotiformes) J Mol Evol. 2002;54:794–806. doi: 10.1007/s00239-001-0083-0. [DOI] [PubMed] [Google Scholar]
- FAO. First Report on the State of the World’s Animal Genetic Resources (AnGR), Bangladesh. 2004. p. 40. [Google Scholar]
- FAO (Food and Agriculture Organization of the United Nations) Global Poultry Trends 2013: Record World Duck Meat Production in 2013. 2014. [Google Scholar]
- Hall T. Bioedit: A biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acid Symp Series. 1999;41:95–98. [Google Scholar]
- Hey J. Recent advances in assessing gene flow between diverging populations and species. Curr Opin Genet Dev. 2006;16:592–596. doi: 10.1016/j.gde.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Hong EC, Choo HJ, Kang BS, Kim CD, Heo KN, Lee MJ, Hwangbo J, Suh OS, Choi HC, Kim HK. Performance of growing period of large-type Korean native ducks. Kor J Poult Sci. 2012;39:143–149. [Google Scholar]
- Hoque MR, Jung CK, Park BK, Choi KD, Lee JH. Genetic variability of mtDNA D-loop region in Korean native chickens. Kor J Poult Sci. 2009;36:323–328. [Google Scholar]
- Hoque MR, Choi NR, Sultana H, Kang BS, Heo KN, Hong SK, Jo C, Lee JH. Phylogenetic analysis of a privately-owned Korean native chicken population using mtDNA D-loop variations. Asian Australas J Anim Sci. 2013;26:157–162. doi: 10.5713/ajas.2012.12459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque MR, Lee SH, Jung KC, Kang BS, Park MN, Lim HK, Choi KD, Lee JH. Discrimination of Korean native chicken populations using SNPs from mtDNA and MHC polymorphisms. Asian Australas J Anim Sci. 2011;24:1637–1643. [Google Scholar]
- Jin SD, Hoque MR, Seo DW, Paek WK, Kang TH, Kim HK, Lee JH. Phylogenetic analysis between domestic and wild duck species in Korea using mtDNA D-loop sequences. Mol Biol Rep. 2014;41:1645–1652. doi: 10.1007/s11033-013-3012-6. [DOI] [PubMed] [Google Scholar]
- Jin SD, Hoque MR, Seo DW, Kim IK, Jo C, Paek WK, Lee JH. Phylogenetic relationships among dabbling duck species in Korea using COI gene variations in mtDNA. J Poult Sci. 2012;49:163–170. [Google Scholar]
- Johnson KP, Sorenson MD. Phylogeny and biogeography of dabbling ducks (Genus: Anas): A comparison of molecular and morphological evidence. Auk. 1999;116:792–805. [Google Scholar]
- Kim HK, Kang BS, Hwangbo J, Kim CD, Heo KN, Choo HJ, Park DS, Suh OS, Hong EC. The study on growth performance and carcass yield of meat-type Korean native ducks. Kor J Poult Sci. 2012;39:45–52. [Google Scholar]
- Kulikova IV, Zhuravlev YN, McCracken KG. Asymmetric hybridization and sex-biased gene flow between eastern Spot-Billed ducks (Anas zonorhyncha) and Mallards (Anas platyrhynchos) in the Russian Far East. Auk. 2004;121:930–949. [Google Scholar]
- Leekaew P, Songserm T, Choothesa A, Boonyaprakob U. A simple method to extract mitochondrial DNA in a non-invasive phylogenetic study of domestic native Thai ducks. Kasetsart J Soc Sci. 2008;42:41–50. [Google Scholar]
- Librado P, Julio R. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lopez JV, Yuhki N, Masuda R, Modi W, O’Brien SJ. Numt, a recent transfer and tandem ampli cation of mitochondrial DNA to the nuclear genome of the domestic cat. J Mol Evol. 1994;39:174–190. doi: 10.1007/BF00163806. [DOI] [PubMed] [Google Scholar]
- Moore WS, DeFilippis VR. The window of taxonomic resolution for phylogenies based on mitochondrial cytochrome b. In: Mindell DP, editor. Avian Molecular Evolution and Systematics. Academic Press; San Diego, CA, USA: 1997. pp. 83–119. [Google Scholar]
- Purwantini D, Yuwanta T, Hartatik T, Ismoyowati Polymorphism of D-loop mitocondrial DNA region and phylogenetic in five Indonesian native duck population. Int J Poult Sci. 2013;12:55–63. [Google Scholar]
- Qu LJ, Lui W, Yang FX, Hou ZC, Zheng JX, Xu GY, Yang N. Origin and domestication history of Peking ducks determined through microsatellite and mitochondrial marker analysis. Sci China, C, Life Sci. 2009;52:1030–1035. doi: 10.1007/s11427-009-0145-x. [DOI] [PubMed] [Google Scholar]
- Sorenson MD, Fleischer RC. Multiple independent transpositions of mitochondrial DNA control region sequences to the nucleus. Proc Natl Acad Sci USA. 1996;93:15239–15243. doi: 10.1073/pnas.93.26.15239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woischnik M, Moraes CT. Pattern of organization of human mitochondrial pseudogenes in the nuclear genome. Genome Res. 2002;12:885–893. doi: 10.1101/gr.227202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik E, Smalec E. Description of the mallard duck (Anas platyrhynchos) karyotype. Folia Biol (Krakow) 2007;55:115–120. doi: 10.3409/173491607781492588. [DOI] [PubMed] [Google Scholar]
- Wu ZY, Zeng SC, Luo YZ, Han JL. A mystery in sequencing the mitochondrial DNA D-loop from blood samples of domestic Mallard duck. J Biol Sci. 2011;11:181–188. [Google Scholar]
- Zaman G, Goswami RN, Aziz A, Nahardeka N, Roy TC, Mahanta JD. Farming system of Nageswari ducks in North-Eastern India (Assam) World’s Poult Sci J. 2005;61:687–693. [Google Scholar]


