Abstract
MicroRNAs (miRNAs) are highly conserved, short non-coding RNAs that regulate gene expression at the posttranscriptional level. Although many miRNAs are identified in muscles and muscle cells, their individual roles are still not fully understood. In the present study, we investigated a muscle highly-expressed miRNA, miR-127-3p, in C2C12 myoblasts and tissues of goats with different muscle phenotypes (Boer vs Wushan black goats). Our results demonstrated that i) miR-127-3p was extensively expressed in tissues of goats; ii) miR-127-3p was higher expressed in muscle, spleen, heart, and skin in the muscular goats (Boer goats) than the control (Wushan black goats). Then we further characterized the dynamical expression of miR-127-3p, MyoD, MyoG, Myf5, Mef2c, and Myosin in the proliferating and differentiating C2C12 myoblasts at day of 0, 1, 3, 5, and 7 in culture mediums. Especially, we found that miR-127-3p was significantly higher expressed in the proliferating than differentiating cells. Our findings suggest that miR-127-3p probably plays roles in the proliferation and differentiation of myoblasts, which further underlies regulation of muscle phenotype in goats.
Keywords: MiR-127-3p, C2C12, Proliferation, Differentiation, Goats
INTRODUCTION
MicroRNAs (miRNAs) are endogenous noncoding small RNAs of approximately 22 nucleotides (nt) in length, which regulate gene expression through the RNA-induced silencing complex (RISC) at posttranscriptional level (Chekulaevac and Filipowicz, 2009). Lines of evidence indicate that miRNAs participate in almost all biological process including cell cycle, development, metabolism, diseases, and so on (Bushati and Cohen, 2007). Each miRNA is predicted to have many targets, and each mRNA may be regulated by more than one miRNA (Lewis et al., 2003; Lim et al., 2005). It has been well known that the myogenic process including myoblast proliferation, withdrawal from the cell cycle, differentiation and fusion into multinuclear myotubes, is not only orchestrated finely by myogenic regulatory factors and myocyte enhancer factor 2 (Singh and Dilworth, 2013), but is regulated by the miRNAs. Recently, three muscle-specific miRNAs, miR-1, miR-133 and miR-206, have been well investigated in skeletal, smooth and cardiac muscles. MiR-1 and miR-133 have been reported to regulate different aspects of skeletal muscle development in vitro and in vivo. MiR-1 promotes myocyte differentiation by repressing expression of histone deacetylase 4 (HDAC4), a negative regulator of differentiation and a repressor of the MEF2 transcription factor (Chen et al., 2006). In C2C12 myoblasts, miR-133a promotes proliferation, in part, by repressing serum response factor. MiR-206 is only expressed in skeletal muscle while miR-1 and miR-133 are expressed in both skeletal and cardiac muscles. MiR-206 also promotes muscle differentiation by the repression of follistatin-like 1 and Utrophin genes expression, and its expression is induced by myoD and myogenin, both of which are critical transcriptional factors for muscle differentiation (Rao et al., 2006). In addition, other myogenic miRNAs have also been identified, including miR-23 (Wilfred et al., 2007), miR-26a (Dey etal., 2012), miR-27b (Crist et al., 2009), miR-29 (Wang et al., 2013), miR-125b (Ge et al., 2011), miR-155 (Seok et al., 2011), miR-181 (Naguibneva et al., 2006), miR-214 (Feng et al., 2011), miR-486 (Dey et al., 2011), miR-208/499 (van Rooij et al., 2009), miR-221/222 (Cardinali et al., 2009), miR-322/424 (Sarkar et al., 2010) and miR-503 (Sarkar et al., 2010). However, those miRNAs identified represent just the tip of the iceberg in the noncoding RNA world. To elucidate the scenario of miRNAs in muscle development, we recently conducted the analysis of miRNA transcriptome in muscle in two sheep groups with different muscle phenotypes by RNA-seq (Zhang et al., 2013). Our results demonstrated that the miR-127-3p was another most highly expressed miRNA in fetal muscle besides miRNA-1, miRNA-133a, and miRNA-378, which suggest miR-127-3p probably is associated with muscle development. MiR-127-3p, locates in chromosome region 14q32.2 (Benetatos et al., 2013), and this locus can transcribe two mature miRNAs (miR-127-3p and miR-127-5p) from the same precursor miRNA. Recent studies show that miR-127 is a critical factor in fetal lung development (Bhaskaran et al., 2009) and may inhibit lung inflammation (Xie et al., 2012). In rat liver cells, down-regulation of miR-127 promotes cell proliferation, while up-regulation of miR-127 inhibits cell proliferation (Pan et al., 2012). These observations suggest the important roles of miR-127 in cell proliferation, differentiation, and development. In addition, miR-127 was recently reported to be widespread expressed in tissues in human and pig (Robertus et al., 2009; Yang et al., 2014). However, little is known about tissue expression profile of miR-127-3p in goat, as well as its dynamical expression patterns in muscle cells during proliferation and differentiation. In this present study, we examined expression of miR-127-3p in skeletal muscle between two goat groups with different muscular phenotypes as well as its dynamical expression during the C2C12 myoblasts proliferation and differentiation.
MATERIALS AND METHODS
Animal
Boer goats (muscular phenotype) and Wushan black goats (the control, less muscle mass) were chosen for investigating the relationship between miR-127-3p and muscle development. We collected seven tissues including heart, liver, spleen, lung, kidney, muscle and skin from three adult goats in each breeds. All samples were dissected and quickly stored in liquid nitrogen for gene expression. All experimental and surgical procedures were approved by the Biological Studies Animal Care and Use Committee, Chongqing, Peoples Republic of China. The goats were housed in one group and were fed according to the nutrient requirements of goat established by the National Research Council in 1985; the feeding was in line with the Instructive Notions with Respect to Caring for Laboratory Animals that was published in 2006 by the Science and Technology Department of China (Approval No. S20072911).
Cell culture
C2C12 myoblasts obtained from the Cell Bank of Chinese Academy of Sciences. C2C12 myoblasts were maintained in growth medium (GM, High Glucose Dulbecco’s Modified Eagle’s Medium, Gibco, Grand Island, NY, USA) containing 1 g/L glucose with with 10% (v/v) fetal bovine serum (FBS, Gibco, Grand Island, NY, USA) at 37°C with 5% CO2. For differentiation, myoblasts were transfered to differentiation medium (DM, Dulbecco’s Modified Eagle Medium supplemented with 2% horse serum (HS, Gibco, USA) at the confluence of 60% to 70%. Replenished with fresh medium daily for 7 days for C2C12 cells in GM and DM, respectively.
RNA extraction
The total RNAs from seven tissues and C2C12 cells were isolated using TriPure Isolation Reagent (Roche, Mannheim, Germany) and High Pure miRNA Isolation Kit (Roche, Germany) according to the manufacturer’s instructions. RNA purity was assessed using a spectrometer (Nanodrop, Wilmington, DE, USA) at ratios of OD260/OD280 between 1.9 and 2.1. RNA integrity was assessed using agarose gel electrophoresis.
Real time-polymerase chain reaction of mRNA and miRNA
The cDNA was synthesized by total RNA using SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA, USA). The expression levels of mRNA were detected using real-time quantitative polymerase chain reaction (PCR) SuperScript III Platinum SYBR Green One-Step qRT-PCR Kit (Invitrogen, USA). The real-time PCR measurements were performed in triplicate on each cDNA sample and analyzed relative quantification results by the 2(−ΔΔCt) method. Gene expression ratios were normalized to glceraldhyde-6-phosphate dehydrogenase (GAPDH) gene in the same sample. The sequences of primers can be found in Supplementary Table S1.
For miRNA, total RNA was used to make cDNA using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). TaqMan MicroRNA Assays consist of miR-127-3p and U6 (Applied Biosystems, USA). TaqMan miRNA Assays were used for reverse transcription (RT) of miR-127-3p (5×), U6 (5×) from 10 ng of total RNA each, according to the manufacturer’s protocol. The quantitative real-time PCR (qRT-PCR) reaction mixture (20 μL) contained 1 μL cDNA, 1 μL TaqMan MicroRNA Assay (20×), 10.0 μL TaqMan Universal PCR Master Mix II, no UNG, 8 μL Nuclease-free water. The cycling conditions were 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, and 60°C for 60 s. The real-time PCR measurements and analysis miRNA were same as mRNA.
Statistical analysis
The significant difference of tissue expression profile in two goat breeds was determined by the Student’s t-test. All data were shown as mean±standard deviation with n = 3.
RESULTS
Tissue expression profile of miR-127-3p in two goat breeds
To assess the conservation of miR-127-3p in mammals, we compared the mature sequences of miR-127-3p in human, mouse, rat, pig, cow, and goat by BLAST (http://www.ncbi.nlm.nih.gov/). We found miR-127-3p is highly conserved among these species (Figure 1A). To examine the tissue specificity of miR-127-3p, we determined miR-127-3p expression of seven tissues in adult Boer and Wushan black goats by real-time qPCR. Our results demonstrated that miR-127-3p was extensively expressed in tissues, unlike the muscle specific expression of miR-1, miR-206, and miR-133 (Rao et al., 2006). Moreover, miR-127-3p was abundantly expressed in muscle, spleen, heart, and skin, but weakly expressed in lung, kidney, and liver in both breeds. Especially, miR-127-3p was significantly higher expressed in muscle, spleen, heart, and skin in Boer goats than that in Wushan black goats (Figure 1B), which suggest that miR-127-3p may be involved in muscle development.
Figure 1.
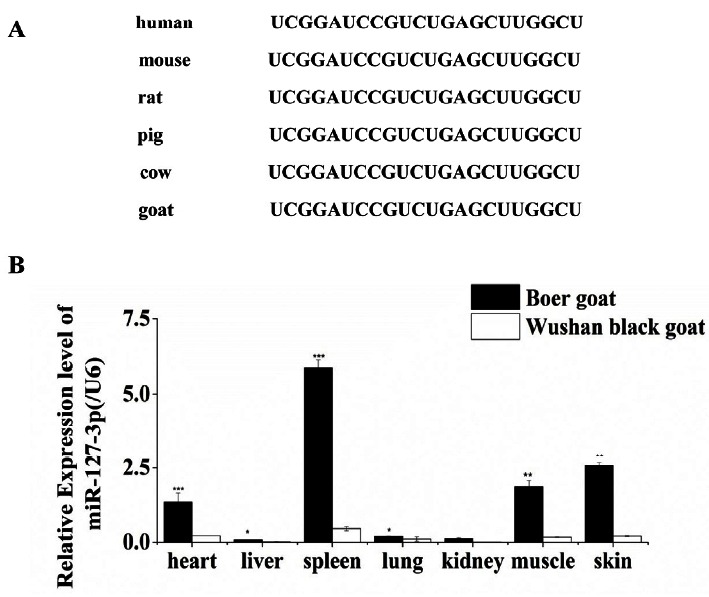
Conservation and tissue expression profile of miR-127-3p. (A) MiR-127-3p is high conserved in mammals. (B) Tissue expression profile of miR-127-3p in adult Boer and Wushan black goats, respectively. The data of miR-127-3p (average±standard error) are normalized to U6 from three independent experiments. (* p<0.1,** p<0.05, and *** p<0.01).
Dynamical expression of miR-127-3p and myogenic marker genes during C2C12 myoblasts proliferation and differentiation
To investigate the expression profiles of miR-127-3p during C2C12 cell proliferation, we determined expression of miR-127-3p and myogenic marker genes (MyoD, MyoG, Myf5, Mef2c, and Myosin) in C2C12 myoblasts in the proliferation medium for 0, 1, 3, 5, and 7 days by real -time RT-PCR. We showed the expression level of miR-127-3p was generally increasing during cell proliferation except a sharp decrease at day 7. While there was a diversity of the expression patterns for the myogenic marker genes including MyoD, MyoG, Myf5, Mef2c, and Myosin during cell proliferation (Figure 2A).
Figure 2.
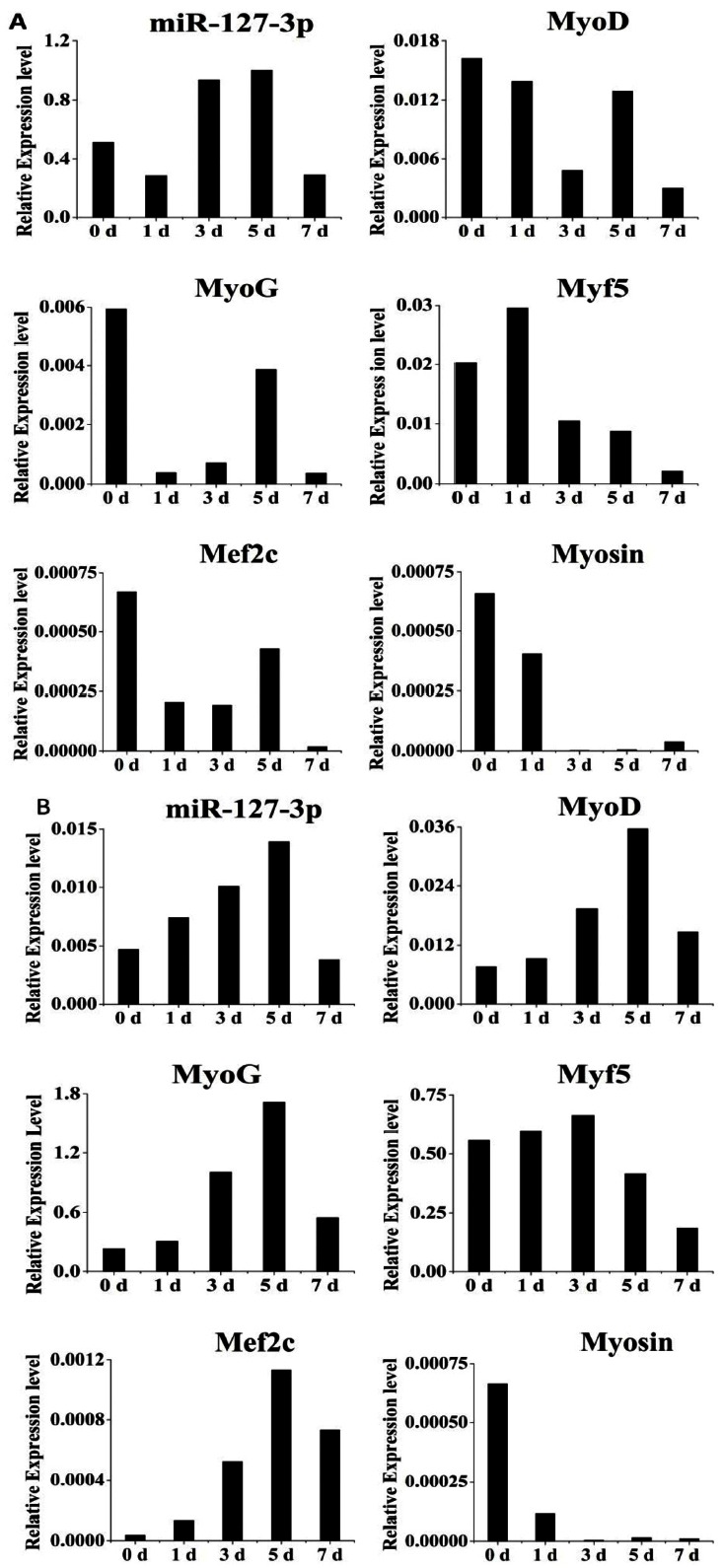
Expression profiles of miR-127-3p and myogenic marker genes during C2C12 myoblasts proliferation and differentiation. C2C12 cells cultured in proliferation medium (A) and differentiation medium (B) for 0, 1, 3, 5, and 7 days were used to determine expression levels of miR-127-3p by real-time polymerase chain reaction. Results are presented as mean relative expression±standard deviation from three independent experiments.
To investigate the dynamical expression of miR-127-3p during C2C12 differentiation, we investigated expression of miR-127-3p and myogenic marker genes above in C2C12 at five time-points by real-time RT-PCR. We found that the expression level of miR-127-3p is increasing in DM during the initial 5 days of differentiation, and then sharply decreasing at day 7 of differentiation (Figure 2B). Interestingly, we found that there were similar expression patterns between miR-127-3p and three myogenic marker genes including MyoD, MyoG, and Mef2c. Whereas the expression profiles of Myf5 and Myosin were various. Especially, we found that miR-127-3p was significantly higher expressed in the proliferating than differentiating cells (p<0.01). It is speculated that specificity of miR-127-3p in muscle cells.
These findings suggest that miR-127-3p is probably associated with muscle development by playing roles in myoblast proliferation and differentiation.
DISCUSSION
MiR-127 was higher expressed in tissues in Tongcheng than in Landrace pigs, but this difference in muscle was confined to the middle and later embryonic period (Yang et al., 2014). Interestingly, in this present study, we demonstrated miR-127-3p was significantly higher expressed in skeletal muscle, spleen, heart, skin, lung, and liver in Boer goats than Wushan black goats (Figure 1B), which is different from the recent study in pig (Yang et al., 2014). A possible reason for this may be the fact that the previous study examined miR-127, which was consist of miR-127-3p and miR-127-5p, while we only examined the miR-127-3p in the present study. In addition, we also found a high conservation of miR-127-3p among human, mouse, rat, pig, cow, and goat (Figure 1A), which suggests this miRNA may play an important role in physiological function in mammals. Generally, our findings preliminarily suggest miR-127-3p is associated with skeletal muscle development and growth in goats. Since miR-127-3p and miR-1/206/133 are high expressed miRNAs in skeletal muscles (~107 reads), as well as miR-1/206/133 play indispensable roles in myogenic cell development process (Anderson et al., 2006; Chen et al., 2006). Based on our findings, we therefore speculate that miR-127-3p may make a difference to myogenic process between different breeds of goats. MiR-1 could strongly enhance myogenesis by increasing expression of myogenic markers, including myogenin, myosin heavy chain, MyoD, MEF2, and skeletal α-actin (Chen et al., 2006). We found that there were obvious differences in expression patterns of myogenic marker genes besides miR-127-3p during C2C12 cells proliferation and differentiation (Figure 2A and 2B). Whether differential expression of miR-127-3p affects these myogenic markers or not remains to be investigated further. Among the myogenic markers, Myf5 and MyoD are mainly involved in controlling myoblast proliferation and early differentiation, which govern myocytes into myogenic program. As terminal differentiation genes, MyoG and MRF4 are necessary to myocytes and fusion into myotubes. The transcriptional activity of MEF2C is regulated during skeletal muscle differentiation. Especially, there were similar expression patterns between miR-127-3p and the three myogenic marker genes including MyoD, MyoG, and Mef2c during differentiation of C2C12 myoblasts (Figure 2B). MiR-127 is a critical factor in fetal lung development (Xie et al., 2012). Thus we consider the mechanism of miR-127-3p regulating skeletal muscle development could be similar to its role in fetal lung development. Besides, miR-127 inhibits proliferation in rat liver cells and in hepatocellular carcinoma. We found miR-127-3p was significantly higher expressed in the proliferating than differentiating cells (p<0.01). Then we speculate that the main function of miR-127-3p in C2C12 cells may be identical to in rat liver cells (Pan et al., 2012), but need to further to study. Taken together, these findings suggest that miR-127-3p is probably associated with muscle development by affecting myoblast proliferation or differentiation. For comprehensively understanding this myogenic miRNA, we think the following issues remain to be elucidated: Since miR-127-3p plays roles in proliferation or differentiation of myoblasts, what are its target genes at certain myogenic stage? What is its regulatory mechanism during myogenesis?
These contribute to well comprehend molecular mechanisms of miR-127-3p in regulation of muscle development and growth, as well as contribute to elucidate the pathogenesis of human muscle disease and gene-targeted therapy.
Supplementary Information
ACKNOWLEDGMENTS
This work was supported by grants from the Chongqing Fundamental Research Fund (cstc2013jcyjA80008; 2013cstc-jbky-00106-zj), and the Chongqing Fund of Agriculture Development (No.15404; No.14413).
Footnotes
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
REFERENCES
- Anderson C, Catoe H, Werner R. MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res. 2006;34:5863–5871. doi: 10.1093/nar/gkl743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetatos L, Hatzimichael E, Londin E, Vartholomatos G, Loher P, Rigoutsos I, Briasoulis E. The microRNAs within the DLK1-DIO3 genomic region involvement in disease pathogenesis. Cell Mol Life Sci. 2013;70:795–814. doi: 10.1007/s00018-012-1080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskaran M, Wang Y, Zhang H, Weng T, Baviskar P, Guo Y, Gou D, Liu L. MicroRNA-127 modulates fetal lung development. Physiol Genomics. 2009;37:268–278. doi: 10.1152/physiolgenomics.90268.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati NA, Cohen SM. MicroRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Cardinali B, Castellani L, Fasanaro P, Basso A, Alema S, Martelli F, Falcone G. Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PLoS ONE. 2009;4:e7607. doi: 10.1371/journal.pone.0007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crist CG, Didier M, Pallafacchina G, Rocancourt D, Cumano A, Conway SJ, Buckingham M. Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc Natl Acad Sci USA. 2009;106:13383–13387. doi: 10.1073/pnas.0900210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey BK, Gagan J, Dutta A. MiR-206 and miR-486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol. 2011;31:203–214. doi: 10.1128/MCB.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey BK, Jeffrey G, Zhen Y, Anindya D. MiR-26a is required for skeletal muscle differentiation and regeneration in mice. Gene Dev. 2012;26:2180–2191. doi: 10.1101/gad.198085.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Cao JH, Li XY, Zhao SH. Inhibition of miR-214 expression represses proliferation and differentiation of C2C12 myoblasts. Cell Biochem Funct. 2011;29:378–383. doi: 10.1002/cbf.1760. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs down-regulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, Cuvellier S, Harel-Bellan A. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8:278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- Pan C, Chen H, Wang L, Yang S, Fu H, Zheng Y, Miao M, Jiao B. Down-regulation of MiR-127 facilitates hepatocyte proliferation during rat liver regeneration. PLoS ONE. 2012;7:e39151. doi: 10.1371/journal.pone.0039151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Rao PK, Kumar RM, Farkhondeh M, Baskervile S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci USA. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertus JL, Harms G, Blokzijl T, Booman M, de Jong D, van Imhoff G, Rosati S, Schuuring E, Kluin P, van den Berg A. Specific expression of miR-17–5p and miR-127 in testicular and central nervous system diffuse large B-cell lymphoma. Mod Pathol. 2009;22:547–555. doi: 10.1038/modpathol.2009.10. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Dey BK, Dutta A. MiR-322/424 and miR-503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol Biol Cell. 2010;21:2138–2149. doi: 10.1091/mbc.E10-01-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok HY, Tatsuguchi M, Callis TE, He A, Pu WT, Wang DZ. MiR-155 inhibits expression of the MEF2A protein to repress skeletal muscle differentiation. J Biol Chem. 2011;286:35339–35346. doi: 10.1074/jbc.M111.273276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Dilworth FJ. Differential modulation of cell cycle progression distinguishes members of the myogenic regulatory factor family of transcription factors. FEBS J. 2013;280:3991–4003. doi: 10.1111/febs.12188. [DOI] [PubMed] [Google Scholar]
- Ge Y, Sun Y, Chen J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. J Cell Biol. 2011;192:69–81. doi: 10.1083/jcb.201007165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ, Jr, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, He HB, Zhang WY, Zhang HX, Bai JB, Liu HZ, Cao JH, Chang KC, Li XY, Zhao SH. MiR-29 targets Akt3 to reduce proliferation and facilitate differentiation of myoblasts in skeletal muscle development. Cell Death Dis. 2013;4:e668. doi: 10.1038/cddis.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfred BR, Wang WX, Nelson PT. Energizing miRNA research: A review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab. 2007;91:209–217. doi: 10.1016/j.ymgme.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Liang J, Liu N, Wang Q, Li Y, Noble PW, Jiang D. MicroRNA-127 inhibits lung inflammation by targeting IgG Fcγ receptor I. J Immunol. 2012;188:2437–2444. doi: 10.4049/jimmunol.1101070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Li Y, Liang R, Zhou R, Ao H, Mu Y, Yang S, Li K, Tang Z. Dynamic expression of microRNA-127 during porcine prenatal and postnatal skeletal muscle development. J Integr Agric. 2014;13:1331–1339. [Google Scholar]
- Zhang S, Zhao F, Wei C, Sheng X, Ren H, Xu L, Lu J, Liu J, Zhang L, Du L. Identification and characterization of the miRNA transcriptome of Ovis aries. PLoS ONE. 2013;8:e58905. doi: 10.1371/journal.pone.0058905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


