Abstract
The long term survival of fecal indicator organisms (FIOs) and human pathogenic microorganisms in sediments is important from a water quality, human health and ecological perspective. Typically, both bacteria and viruses strongly associate with particulate matter present in freshwater, estuarine and marine environments. This association tends to be stronger in finer textured sediments and is strongly influenced by the type and quantity of clay minerals and organic matter present. Binding to particle surfaces promotes the persistence of bacteria in the environment by offering physical and chemical protection from biotic and abiotic stresses. How bacterial and viral viability and pathogenicity is influenced by surface attachment requires further study. Typically, long-term association with surfaces including sediments induces bacteria to enter a viable-but-non-culturable (VBNC) state. Inherent methodological challenges of quantifying VBNC bacteria may lead to the frequent under-reporting of their abundance in sediments. The implications of this in a quantitative risk assessment context remain unclear. Similarly, sediments can harbor significant amounts of enteric viruses, however, the factors regulating their persistence remains poorly understood. Quantification of viruses in sediment remains problematic due to our poor ability to recover intact viral particles from sediment surfaces (typically <10%), our inability to distinguish between infective and damaged (non-infective) viral particles, aggregation of viral particles, and inhibition during qPCR. This suggests that the true viral titre in sediments may be being vastly underestimated. In turn, this is limiting our ability to understand the fate and transport of viruses in sediments. Model systems (e.g., human cell culture) are also lacking for some key viruses, preventing our ability to evaluate the infectivity of viruses recovered from sediments (e.g., norovirus). The release of particle-bound bacteria and viruses into the water column during sediment resuspension also represents a risk to water quality. In conclusion, our poor process level understanding of viral/bacterial-sediment interactions combined with methodological challenges is limiting the accurate source apportionment and quantitative microbial risk assessment for pathogenic organisms associated with sediments in aquatic environments.
Keywords: sediment, viable but non-culturable bacteria, biofilm, fecal indicator organisms, resuspension, survival, virus
Introduction
There are a multitude of bacteria and viruses naturally present within the aquatic environment of which the vast majority are not derived from humans (Rosenwasser et al., 2016). In addition, the majority of bacteria and viruses derived from humans are benign from a human health perspective, however, a small component is considered pathogenic (Filippini and Middelboe, 2007; Lowther et al., 2012). Human pathogenic bacteria and viruses released from point (e.g., wastewater treatment plants) and diffuse sources (e.g., agricultural land) frequently contaminate water bodies downstream and therefore represent a potential risk to human health (e.g., during recreation, contamination of food and drinking water). A large proportion of the pathogenic organisms present in water may also become associated with the sediment, which can be subject to resuspension (Davies et al., 1995; Anderson et al., 2005; Drummond et al., 2014a) and could represent a significant mechanism for delivering pathogens to coastal waters (Yamahara et al., 2007). In coastal and estuarine environments, the survival of fecal indicator organisms (FIOs; indicating the potential presence of pathogenic bacteria and viruses) is positively linked to the concentration of suspended matter in the water column (Howell et al., 1996). In contrast, other factors such as elevated temperatures and exposure to UV radiation tend to have a negative effect on microbial survival in the water column (Chigbu et al., 2005; Kay et al., 2005). Viruses have also been shown to readily adsorb to solid matter in the water (reviewed in Jin and Flury, 2002). Viral particles associated with solids may travel long distances in water, or settle out during transit, where they become more concentrated in the sediments that in the overlying water column. Viral attachment to solid particles may result in permanent inactivation of the virus, however the adsorbed virus particles are often protected from inactivation from stressors (e.g., UV) by the surface they are attached to Schijven and Hassanizadeh (2000) and Chrysikopoulos and Aravantinou (2012). Current monitoring schemes, and the majority of research in this field, typically focuses on quantifying fecally derived bacteria and viruses in the water column, however, numbers within the sediment (including beaches, riverbanks and mudflats) are often orders of magnitude higher (Rao et al., 1986a; Duhamel and Jacquet, 2006; Vignaroli et al., 2013, 2015; Perkins et al., 2014). Greater abundance of pathogenic organisms and viruses in the sediment reservoir is linked to their binding to particle surfaces and enhanced survival within the biofilm matrix (Smith et al., 1978; LaBelle and Gerba, 1980; Danovaro et al., 2008; Pachepsky and Shelton, 2011). Sediments therefore act as a potential reservoir of pathogens and FIOs in aquatic environments that remain undetected until they re-enter the water column due to the action of rainfall, wind, waves, tides recreational boats, and dredging (Howell et al., 1996; Jamieson R. C. et al., 2005) or are accumulated by filter-feeding shellfish destined for human consumption (Lowther et al., 2012; Malham et al., 2014). The survival of human pathogenic bacteria and viruses in rivers and the marine environment is highly species and strain specific (Gerba et al., 1980; Anderson et al., 2005; Byappanahalli et al., 2006). This makes it difficult to generalize about the behavior of pathogenic organisms in a risk assessment context, particularly when they may come from sources which vary both spatially and temporally. Further, these pathogens may have a markedly different viability in aquatic ecosystems compared to non-pathogenic indicator organisms that are frequently used to represent fecal pollution in environmental monitoring (Sinton et al., 2002). Due to the emergence of new pathogens and the need to reduce the economic and social burden of human disease outbreaks, the source-apportionment and transmission of many disease-causing agents is receiving increased attention (Dobrindt et al., 2004; Vignaroli et al., 2013). Within this, there is a clear need to improve our understanding of the behavior, fate and potential mitigation of pathogens associated with sediments as well as the main water body itself. Adequate consideration of pathogens in sediments will enhance our ability to achieve regulatory compliance with legislation associated with protecting bathing and shellfish waters and in the provision of more robust risk assessments (Danovaro et al., 2008; Pachepsky and Shelton, 2011; Malham et al., 2014). Despite significant investment and research into the factors governing bacterial and viral association with sediments, areas such as the factors governing bacterial resuscitation from a viable but non-culturable (VBNC) state, viral quantification in sediments, and resuspension requires additional attention.
Abundance and distribution of fecally derived bacteria and viruses
To effectively determine the human health risk associated with coastal and estuarine sediments, it is important to quantify the size of the pathogen pool. The abundance of FIOs such as Escherichia coli and Enterococcus spp. has been well studied, however, further attention is required for pathogens such as Campylobacter spp., Salmonella spp., E. coli O157:H7 and norovirus, which may cause illness through shellfish consumption or exposure to recreational water (Malham et al., 2014). Previous research has primarily focused on the presence/absence of these microorganisms in sediments, but for an apportionment of risk, a quantitative approach is required (Ramaiah et al., 2005; Setti et al., 2009; Carr et al., 2010; Soares de Lima Grisi and Gorlach-Lira, 2010). The reported number of fecally associated bacteria in coastal and estuarine environment is typically between 0 and 104 colony forming units (CFU) or most probable number (MPN)/100 ml for water and 101 to 106 CFU or MPN/100 g wet weight for sediment (Table 1). Similar trends have been observed in viral abundance in marine and estuarine sediment (Table 1), however, the relative difference in water/sediment abundance cannot be assessed due to the small sample size. Nonetheless, Staggemeier et al. (2015a,b) directly compared the concentrations of adenoviruses in corresponding water and sediment samples derived from freshwater streams, dams, and springs and found that the viral abundance in sediment was significantly higher than in the overlying water. Importantly, they found that adenoviruses may be present in sediment in the absence of the virus in the water column. Anderson et al. (2005) found that sediment had greater spatial variability in bacterial abundance than water, and that populations of enteric organisms can persist in the environment. The high natural variability in the sediment fraction for both bacteria and viruses, has been linked to methodological differences in dissociation from sediment particles which may result in inconsistent enumeration (Anderson et al., 2005; Miura et al., 2011; Pachepsky and Shelton, 2011).
Table 1.
Abundance of fecal bacteria and viruses associated with coastal and estuarine sediments.
| Bacteria | Sediments Range or average | Water column Range or average | References |
|---|---|---|---|
| CFU or MPN 100 g WW−1 | CFU or MPN 100 ml−1 | ||
| Fecal coliforms | 80–200,000 | 8–9400 | Alcântara and Almeida, 1995; Lucena et al., 1996; Bonilla et al., 2007; Abdelzaher et al., 2010; Luna et al., 2010; Vignaroli et al., 2013; Borade et al., 2014. |
| E. coli | 19–100,000 | 0–6700 | Evanson and Ambrose, 2006; Bonilla et al., 2007; Abdelzaher et al., 2010; Stumpf et al., 2010; Borade et al., 2014. |
| Fecal Streptococci | 190–19,000 | 6–240 | Alcântara and Almeida, 1995; Lucena et al., 1996. |
| Enterococcus spp. | 80–136,000 | 0–240 | Evanson and Ambrose, 2006; Bonilla et al., 2007; Abdelzaher et al., 2010; Stumpf et al., 2010; Vignaroli et al., 2013. |
| E. faecalis | ND | 200 | Borade et al., 2014. |
| Clostridium perfringens | 300–1,500,000 | <2–13 | Lucena et al., 1996; Abdelzaher et al., 2010. |
| Staphylococcus aureus | ND | ND | Abdelzaher et al., 2010. |
| Salmonella spp. | ND–262,500 | 600–1500 | Borade et al., 2014. |
| Shigella spp. | ND | 1600 | Borade et al., 2014. |
| Proteus spp. and Klebsiella spp. | 6300–543,700 | 5400–5600 | Borade et al., 2014. |
| Aeromonas spp. | 36,000 | – | Lucena et al., 1996. |
| Vibrio spp.* | 31,300–756,200 | 3000–6600 | Borade et al., 2014. |
| Viruses | Sediments Range or average | Water column Range or average | References |
| PFU or GC 100 g WW−1 | PFU or GC 100 ml−1 | ||
| Enterovirus | 3.3–19.08 (6–75) | ND–160 (ND) | Gerba et al., 1977a; Rao et al., 1984, 1986a; Le Guyader et al., 1994; Alcântara and Almeida, 1995; Lucena et al., 1996; Green and Lewis, 1999; Abdelzaher et al., 2010; Miura et al., 2011.a |
| Norovirus | ND | ND | Abdelzaher et al., 2010. |
| Norovirus GI | ND (24) | ND (0) | Miura et al., 2011; Norman et al., 2013.a |
| Norovirus GII | BDL (ND–6) | ND | Miura et al., 2011; Norman et al., 2013. |
| Rotavirus | 12/4 | 31–265 | Rao et al., 1986a; Alcântara and Almeida, 1995; Green and Lewis, 1999. |
| Hepatitis A virus | (0–87.5) | (0) | Le Guyader et al., 1994; Green and Lewis, 1999; Abdelzaher et al., 2010.a |
| Human adenovirus | 197,000–6,960,000 | 15,700–20,800,000 | Staggemeier et al., 2015a** |
| Human polyomavirus | (Present) | (Present) | Abdelzaher et al., 2010.a |
| Somatic coliphage | ND–240,000 (36) | <1–6 (19) | Alcântara and Almeida, 1995; Lucena et al., 1996; Bonilla et al., 2007.a |
| F+ coliphage | ND–102 (2) | <1–3 (0) | Alcântara and Almeida, 1995; Bonilla et al., 2007.a |
| FRNA-bacteriophage | ND–20 | <1–3 | Alcântara and Almeida, 1995. |
| Bacteroides fragilis bacteriophage | 0–2400 | 0–2640 | Alcântara and Almeida, 1995; Lucena et al., 1996. |
Vibrio spp. are ubiquitous in the marine environment and facultative pathogens.
Human adenovirus detected in freshwater. ND, none detected. –, not analysed; CFU, Colony forming units; MPN, Most probable number; WW, Wet weight. FRNA bacteriophage, Male specific (F) RNA bacteriophage.
Numbers in parenthesis indicate the prevalence of the virus in a separate study (%) BDL, below detection limit (qPCR); PFU, Plaque forming units; GC, Gene Copies.
Pathogens and FIOs also associate with suspended solids (flocs) present in the overlying water column (Rao et al., 1984, 1986a; Jamieson R. et al., 2005). The floc fraction is prone to resuspend easily (Pachepsky et al., 2009a) and is an important but poorly quantified contributor to bacterial loading for water quality monitoring (Malham et al., 2014). However, flocs are ephemeral and prone to break up on disturbance, which provides a technical challenge to enumeration. Numerous studies, have reported a decrease in the number of bacteria and viruses with sediment depth (Obiri-Danso and Jones, 2000; Filippini and Middelboe, 2007; Berthe et al., 2008). Recent research showed a two-log reduction in culturable E. coli from the sediment surface (top 1 cm) to 4 cm in depth (Pachepsky and Shelton, 2011). Generally, the top 2 cm of sediment is considered to have high FIO abundance whereas below 2 cm has significantly lower abundance (Ferguson et al., 1996; Haller and Amedegnato, 2009; Drummond et al., 2014a). Distinct seasonality of bacteria in sediments has been observed, with greater abundance in autumn-winter months compared to spring-summer months (Goyal et al., 1977; Crabill et al., 1999). In contrast, Ishii et al. (2006) found that summer to autumn had greater abundance in soils and winter to spring had the lowest abundance. Meays et al. (2006) noted a distinct diurnal pattern in E. coli abundance in the water column, possibly due to UV light inactivation (Kay et al., 2005; Walters et al., 2013), while the greater stability and protection from stressful conditions could reduce short term changes in abundance. Physio-chemical conditions such as temperature, turbidity, salinity, nutrient and oxygen concentrations and water depth are all important factors controlling the distribution of bacteria (Perkins et al., 2014). The weather, season, disease prevalence in the community; tides and freshwater inputs; time of day; sediment type (sand/mud) and deposition rates; distance from the shore; and predation by, and competition with, the intrinsic microbial community also affects the abundance and distribution of bacteria and viruses (Kirschner et al., 2004; Jamieson R. C. et al., 2005). The complexity of interacting factors that influence pathogen and FIO survival in sediments often restricts direct comparison between studies. Effective surveillance alongside sufficient site/sediment characterization may enable further insights into the influence of the sediment fraction on bathing water quality (Ouattara et al., 2013; Huang et al., 2015). Reports suggest that the number of infectious or culturable pathogens may correlate poorly with the number detected by molecular approaches. Therefore, integrated surveillance schemes using both molecular detection of bacterial/viral genomes by PCR and culture-based methods (e.g., bacterial culture or viral infectivity cell culture tests) may be required (Bae and Schwab, 2008). However, high degrees of inhibition at either the extraction or genome quantification stages suggest that optimization and standardization of molecular methodology in sediments is also required (Miura et al., 2011).
Enteric phages (e.g., F+ RNA coliphages) have been utilized as general markers of fecal pollution. Advantages of this approach includes, target specificity (each phage is typically specific to one host) and their greater environmental persistence in comparison to FIOs; typically 3-fold longer under controlled conditions (Allwood et al., 2003). In addition, source apportionment can be undertaken using different genogroups of F+ RNA coliphages (e.g., I, IV for animal and II and III for human) (Shahrampour et al., 2015). Concentrations of F+ RNA coliphages were between 9 and 20 fold higher in sediments than the overlying water column (Alcântara and Almeida, 1995). Under controlled conditions, F+ RNA coliphages show poor correlation with E. coli, therefore cannot be readily compared to larger historic datasets (usually E. coli or intestinal enterococcus). However, coliphages correlate better with disease incidence and concentrations of pathogens (e.g., norovirus; Doré et al., 2000). Typically, next generation approaches are being used for microbial source tracking (See Section Outlook), however, F+ RNA coliphages still provide a useful indicator of viral culturability.
Sediment characteristics governing bacteria particle interaction
Bacterial adsorption
Bacterial adsorption principally occurs through physicochemical forcing as described by the extended Derjaguin–Landau–Verwey–Overbeek (DVLO) theory (van Loosdrecht et al., 1989). However, a number of factors can increase the rate of association with particle surfaces. Hermansson (1999) showed that a high ionic strength promotes adsorption between particulate matter and bacteria (Jiang et al., 2007). Cao et al. (2011) found that bacterial adsorption to extracellular polymeric substance (EPS) occurs at a greater rate in the presence of sodium ions. Cations reduce the repulsive electrostatic charge of clay particles and allow formation of cation bridges between functional groups of EPS and negatively charged sites of clays. After the long range DVLO interactions have occurred, bacterial cell wall constituents such as extracellular lipopolysaccharides and surface appendages act to increase adsorption by reducing fine scale repulsive forces (e.g., van Loosdrecht et al., 1989; Gilbert et al., 1991). The surface physicochemical properties of particle surfaces therefore influences attachment (Mills and Powelson, 1996; Foppen et al., 2010). A principal factor governing interaction with particles is the surface charge of bacteria. Surface charge can influence binding efficacy to sediment surfaces, as chemical interactions in the electrical double layer dominate effective charge and therefore association between sediment and bacteria. E. coli and other FIOs typically have an overall net negative surface charge due to the prevalence of carboxyl groups within the cell wall and EPS (Foppen and Schijven, 2006), which could result in attraction or repulsion to strongly positively or negatively charged particles respectively. The surface charge of Escherichia coli varies with serotypes suggesting bacteria from different sources could bind differently to sediment (Castro and Tufenkji, 2007; Foppen et al., 2010). Furthermore, Gottenbos et al. (2001) found that bacteria adhered more rapidly to positively charged surfaces but electrostatic interaction impeded bacterial growth after adsorption in pure culture experiments. This interaction decreased the bacterial adenosine triphosphate content and proton motive force upon adhesion (Hong and Brown, 2009) supporting the decreased cell viability identified by van der Mei et al. (2008). Conversely, negatively charged surfaces could promote the opposite, favoring growth of bacteria. Hadjiev et al. (2007) found biofilm attachment is greatest at the maximum surface energy difference between biofilm and material surface. Surface characteristics such as flagellar antigen sites, can vary significantly with species and strain altering the hydrophobicity, electrostatic balance, roughness and surface area parameters of the bacterial surface resulting in markedly different adsorption characteristics to sediment (van Loosdrecht et al., 1987; Stenström, 1989; Bilge et al., 1996; Pachepsky et al., 2009b; Foppen et al., 2010).
Surface characteristics may also be affected by biological aspects such as the metabolic state of the organism. For example, both hydrophobicity and zeta potential (as a measure of wetness) has been shown to be related to the growth rate or phase in E. coli (Allison et al., 1990; Smets et al., 1999). A comparison of 17 E. coli strains, isolated from livestock or water sources, showed an order of magnitude difference in attachment efficacy when binding to quartz sand, with the most efficient stains concurrently possessing the highest number of genes associated with adhesion, toxin production, iron acquisition, or capsular synthesis (Cook et al., 2011). The mineral chemical and surface composition, organic content and particle size affect the propensity of bacterial cells to adhere or release to the particles (Pachepsky et al., 2009b; Hazen and Sverjensky, 2010). Scholl and Harvey (1992) showed that the mineral surface charge controlled initial adhesion of hydrophilic bacteria. Mineralogy and elemental composition often differs between sediment size fractions, with the smaller particles of the clay fraction providing a larger and more reactive surface area for adsorption (Perkins et al., 2014). Most surfaces are coated in reactive groups such as metals, metal oxides and hydroxides and organic material such as proteins through a process known as surface conditioning (Mills and Powelson, 1996). Quartz tends to have greater adhesion when the isoelectric point (pI) of the compound differs greatly from the point of zero charge of quartz. In contrast, a large difference in pI reduces adhesion in clay minerals (Hazen and Sverjensky, 2010). However, surface properties may not modify the microbial viability post initial adhesion (Busscher et al., 1995). The presence of a conditioning film could mask the impact of surface properties by acting as a barrier to chemical and spatial heterogeneity, for example Lorite et al. (2011) showed that a conditioning film reduces film hydrophilicity and roughness of a material surface, which could influence the rate of subsequent film formation. Alternatively, the film could provide a link between the material surface and bacteria (Singh et al., 2011). The importance of roughness is twofold: firstly, it protects the initial bacteria during adhesion from deleterious effects of shear and second, provides greater surface area for adhesion to occur (van Loosdrecht et al., 1989; Stephenson et al., 2013). Singh et al. (2011) identified a threshold of ~20 nm surface roughness where superior protein adsorption substantially decreased attachment rates and biofilm formation by clogging nanoscale pores on the material surface, although whether this influences adhesion of sediment in the field requires further attention.
Fecal coliforms such as E. coli predominantly attach to small particles (< 2 μm), increasing the ease by which they are transported and dispersed in the environment (Muirhead et al., 2006; Goldscheider et al., 2010). Bacterial binding to surfaces, including sediment particles, can be reversible or more permanent (van Loosdrecht et al., 1989; Van Houdt and Michiels, 2005). Fecally derived bacteria are more frequently associated with finer sediments and particles (Chan et al., 1979; Ferguson et al., 1996) than suspended free within the water column (Gerba et al., 1977a; Table 1). Particulate association is important for transport processes: cells attached to larger particles settle to the stream bed, whilst unattached cells, or those attached to small buoyant particles, are likely to be transported further, particularly during storm events (Jamieson, R. C. et al., 2005). Previous research has defined coastal or estuarine sediments as a sink of fecally derived bacteria (Obiri-Danso and Jones, 2000; Deloffre et al., 2005; Berthe et al., 2008; Perkins et al., 2014). Subsequent sediment re-entrainment during storm events, recreational water use, mechanical disturbance and tidal resuspension on mudflats can, therefore, lead to deterioration in microbiological water quality (Crabill et al., 1999).
Survival of bacterial FIOs in water and sediment
Growth and persistence of FIOs and pathogens
The growth of fecally derived bacteria in the environment appears to be restricted mainly to tropical climates or sediments that are subject to intermittent immersion and drying such as riverbank soil, estuaries or coastlines subject to tidal drying and wetting (Table 2) or in the absence of predators (Davies et al., 1995). Maximum decay rates of FIOs in sediments of between −1.1 and −1.3 log10CFU/100 g.sediment./d have been reported (Table 2). Although lower inactivation of FIOs of between −0.011 and −0.138 log10CFU/100 g.sediment./d and persistence of human pathogens has been reported (Davies et al., 1995). Highly variable survival of E. coli in freshwaters has been reported (Table 3) Research suggests sediment associated bacteria exhibit greater survival in marine and river waters compared to free floating bacteria (Roper and Marshall, 1979). For example, the presence of clays resulted in increased E. coli survival from phage attack by ~60% (Roper and Marshall, 1974). Particle-bound fecally derived bacteria are partially shielded from most antimicrobial agents or harmful processes such as UV light that might occur in the water. Schultz-Fademrecht et al. (2008), found a 2–4 log increase in FIOs in a streambed biofilm compared to the overlying water column, possibly due to light inactivation in the water but not the sediment or biofilm. In comparative studies, E. coli survives longer in sediments containing at least 25% clay (< 2 μm) than in those with larger particles (Burton et al., 1987). Sediment particle size plays a role, with coliforms able to survive for between 76 and 83% longer in sediment comprised mainly of clay particles compared with coarser sediments (Howell et al., 1996). Garzio-Hadzick et al. (2010) showed that fine particulates and organic carbon resulted in slower inactivation in streambed sediments. The composition of clay was also found to impact pathogen survival with goethite reducing viability of pathogenic E. coli by 95% compared to other clay types (Cai et al., 2013). The levels of montmorillonite in soil has been associated with reduced occurrence of some human pathogens and greater survival of indigenous bacteria (Filip, 1973); whilst bentonite clays have been shown to inhibit protozoal grazing of Rhizobium in liquid culture (Heijnen et al., 1991) and illite clay antagonizes E. coli by action of Fe2+ ions on the particle surface resulting in loss of outer membrane integrity and therefore viability (Williams et al., 2011; Cai et al., 2013). Future research is necessary to determine whether the elemental/mineral composition of sediment influence the spatial variability of pathogens and fecal indicator bacteria in the environment under representative conditions.
Table 2.
Decay or growth of Fecally derived bacteria in coastal and estuarine sediments.
| Species | Habitat | Temp (°C) | Salinity (PSU)a | Initial inoculation concentration | Decay ratesb (log10 CFU 100 g or ml d−1) | References |
|---|---|---|---|---|---|---|
| E. coli | Fecally contaminated estuarine sediment | 24 | 4–6 | 5 log10 MPN 100 ml−1 | −0.128 (t = 4 days) (grew briefly over first 24 h) | Gerba and McLeod, 1976 |
| Estuarine sediment | 24 | 8–18 | ~5.5 log10 MPN 100 ml−1 | −0.152 (t = 4 days) | ||
| 24 | 8–18 | ~9.5 log10 MPN 100 ml−1 | −0.068 (t = 17 days) | |||
| Coastal swash zone beach sediment | 17 and 23 | 24 | ~6 log10 CFU 100 g−1 | −0.199 (t = 9 days) | Korajkic et al., 2013 | |
| Riverbank soil of a tidally influenced tributary | 25 | 10% seawater | ~3 log10 MPN 100 gDW−1 | Growth after 12 h exceeded the limits of quantification | Solo-Gabriele et al., 2000 | |
| −0.8% initial moisture content | ||||||
| 14% initial moisture content | ~4 log10 MPN 100 gDW−1 | +0.119 (t = 3) | ||||
| 34% initial moisture content | <1 log10 MPN 100 gDW−1 | +0.008 (t = 3) | ||||
| Fecal colifo | ~4 log10 CFU 100 g−1 | −0.018 (t = 28 days) | Davies et al., 1995 | |||
| ~4.8 log10 CFU 100 g−1 | −0.138 (t = 28 days) | |||||
| Fecal streptococcus | ~3.7 log10 CFU 100 g−1 | −0.004 (t = 28 days) | ||||
| ~4 log10 CFU 100 g−1 | −0.011 (t = 28 days) | |||||
| Total Clostridium perfringens | Marine sediment | 22−23 | 34−35 | ~4.4 log10 CFU 100 g−1 | +0.001 (t = 28 days) | |
| ~4.8 log10 CFU 100 g−1 | 0.000 (t = 28 days) | |||||
| C. perfringens spores | ~4.3 log10 CFU 100 g−1 | +0.001 (t = 28 days) | ||||
| ~4.8 log10 CFU 100 g−1 | 0.000 (t = 28 days) | |||||
| Fecal coliformsEnterococcus spp. | Fecally contaminated saltwater sediment | Ambient | ~30 | 5 log10 CFU 100 ml−1 | −1.3 (t = 28 days) | Anderson et al., 2005 |
| 5 log10 CFU 100 ml−1 | −1.1 (t = 28 days) | |||||
| E. coli O157 | Intertidal sand at coastal beaches | 10 | 34 | 8.3 log10 CFU 100 ml−1 | −0.136 (t = 5 days) | Williams et al., 2007 |
| 10 | 34 | 8.3 log10 CFU 100 ml−1 | −0.086 (t = 5 days) | |||
| 10 | 34 | 8.3 log10 CFU 100 ml−1 | −0.110 (t = 5 days) |
PSU, Practical salinity units,
Salinity units not defined in study (assumed %¸)
Bacterial abundance from graphs, normalized per 100 ml or g, and decay rates resolved using the equation from Anderson et al. (2005): r = [ln(Nt)-ln(N0)]/t. Where, Nt is the number of bacteria (log10 CFU 100 ml or g) at time t; N0 is the number of bacteria (log10 CFU 100 ml or g) at time 0, and t, time in days. A negative value denotes a decrease in the number of bacteria, whereas a positive value denotes an increase. The decay rates assume exponential decrease and should be applied with caution in sediments (Davies et al., 1995). DW, Dry weight.
Table 3.
Survival of generic and pathogenic E. coli in water.
| Species | Water source | Factor/Variable investigated | Measure of decline/survival | References | |
|---|---|---|---|---|---|
| E. coli O157 | Sterilized well water (4 sources) | Survival in Different Waters | 2 log decline after 35 days | Geldreich et al., 1992 | |
| E. coli O157 #C4195 and #932 | Portable groundwater source | 5°C | 3.5 log reduction after 70 days | Rice et al., 1992 | |
| E. coli #R1 | 20°C | 5 log reduction after 35 days | |||
| E. coli | Sterile seawater | Rate of die-off under light conditions | Alkan et al., 1995 | ||
| Turbidity | Significant effect | ||||
| Sewage | Significant effect | ||||
| Mixing | Significant effect | ||||
| Temperature | Not-significant effect | ||||
| E. coli K-12 (MC4100) | Filter sterile estuarine water | Organic Matter (Presence/Absence) | Linear regression slopes | Troussellier et al., 1998 | |
| Salinity (Artificial Seawater/Physiological Water) Light (Presence/Absence) | OM+ | OM− | |||
| S – L −+0.050 | −0.005 | ||||
| S + L −−0.006 | −0.020 | ||||
| S – L + −0.060 | −0.110 | ||||
| S + L + −0.120 | −0.100 | ||||
| E coli O157 | Filtered and autoclaved municipal water, in reservoir water, and in water from two recreational lakes | 8°C25°C | 1–2 log drop after 91 days Detection limit reached 49–84 days | Wang and Doyle, 1998 | |
| E. coli O157:H7 (NCTC 12900) | Bottled natural drinking water | Survival in Unsterile mineral water | 3 log reduction after 70 days | Kerr et al., 1999 | |
| Sterile mineral water | 3.5 log reduction after 70 days | ||||
| Sterile distilled water | 4.5 log reduction after 70 days | ||||
| E. coli O157 | River water | Survival in river water | Detection limit reached 27 days | Maule, 2000 | |
| E. coli O157 | Cattle drinking water (2 sources) | Temperature (5 and 15°C) Water source | 1 water source—no difference between temperatures | Rice and Johnson, 2000 | |
| 2 water source—5°C reached detection limit after 8 days | |||||
| 15°C reached detection limit after 4 days | |||||
| E. coli O157 (Environmental) | Cattle water troughs (473) | Water characteristics that encourage survival | Presence/absence 6/473 | LeJeune et al., 2001 | |
| E. coli O157 #3704 Tn5 lux CDABE | Well water from four different sites | Variation in several factors between sites + the presence of different organism | Number of E. coli O157 present reduced by copper, predation by protozoa and in competition with other microorganisms | Artz and Killham, 2002 | |
| E. coli DH5α | Unsterile and sterile groundwater | Influence of microflora | Unsterile T90 2 days | Banning et al., 2002 | |
| Sterile T90 82 days | |||||
| E. coli O157:H7 (NCTC 12900) | River water—with and without feces Sterile distilled water | Difference between temperature and water sources | River water w/o feces—outside <15°C—detection Limit reached after 14 days | McGee et al., 2002 | |
| − inside 15°C—Detection limit reached after >31 days | |||||
| River water w/o Faeces—Outside <15°C—Detection limit reached after 24 days | |||||
| Outside <15°C—Detection limit reached after 17 days | |||||
| Inside 15°C—2.5 log drop after 31 days | |||||
| Escherichia coli Famp (ATCC 700891) | Dechlorinated water | 10°C | D-Value 7.7 Days | Allwood et al., 2003 | |
| 22°C | D-Value 5.7 Days | ||||
| 37°C | D-Value 3.0 Days | ||||
| E. coli O157 #3704 Tn5 lux CDABE and E. coli O157 #3704 | Sterile artificial groundwater | Difference between the strains at 15°C | Both showed a 5-log drop over 70 days | Ritchie et al., 2003 | |
| E. coli O157 | Surface water from lakes and rivers | 6°C | Detection limit reached 32–51 Days | Czajkowska et al., 2005 | |
| 24°C | Detection limit reached 21–32 Days | ||||
| 7 strains of E. coli O157 | Untreated well water | 10°C | 2 strains—1–2 log drop after 56 days | Watterworth et al., 2006 | |
| 2 strains—4 log drop after 56 days | |||||
| 3 strains—detection limit reached <42 days | |||||
| 22°C | 1 strain—6–7 log drop after 56 days | ||||
| 2 strains—detection limit reached <56 days | |||||
| 4 strains—detection limit reached <42 Days | |||||
| E. coli O157 #3704 | Non-sterile: lake Fecally contaminated puddle River Drinking trough | Variation in several factors between sites | T99 12.9 days | Avery et al., 2008 | |
| T99 17.8 days | |||||
| T99 6.0 days | |||||
| T99 6.3 days | |||||
| E. coli O157:H7 | Pond and holding tank water | Difference between water sources | Pond—detection limit reached after 33 days | Suhalim et al., 2008 | |
| Holding tank—detection limit reached after 69 days | |||||
| 6 clinically isolated ETEC strains | Sterile-filtered sea water and freshwater | Induction of VBNC state in water | 2 log drop after 12 weeks | Lothigius et al., 2010 | |
Biofilm formation
Biofilm formation is an important microbial survival strategy in aquatic systems and biofilms are produced when nutrients are abundant (Costerton et al., 1995). Typically, biofilm formation consists of five stages (Van Houdt and Michiels, 2005). The first stage is a reversible association/attachment between the bacterium and the solid surface when brought together by flow of the medium (Figure 1A). This particle association can improve bacterial survival under stressful conditions (Figure 1B). The second stage of biofilm formation is the production of EPS, an important bacterial surface determinant of attachment and fimbriae that anchor the bacterium irreversibly to the solid surface (Junkins and Doyle, 1992; Figure 1C). During the third and fourth stages, the structure of the EPS matrix matures with the addition of macromolecules such as proteins and deoxyribonucleic acid (DNA) (Sutherland, 2001). The fourth stage is distinguished by the alteration of the biofilm to trap and funnel nutrients to those bacteria immobilized in that matrix. The final stage is the steady release of bacteria from the fully mature biofilm, which can occur through shear or sediment resuspension (Figure 1D). It is thought that quorum sensing plays a determinate role in biofilm formation (Costerton et al., 1995) and the response of bacteria to high velocity fluid flow which varies at the transcriptional level (Kim et al., 2016). Further work could elucidate the role of quorum sensing and FIO abundance in sediments.
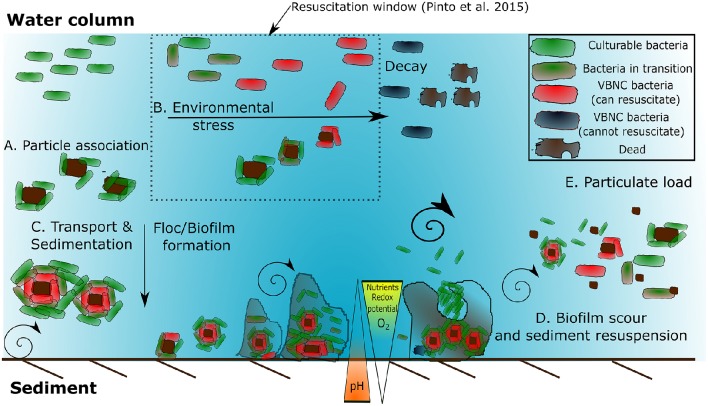
Figure 1.
Factors stimulating bacterial accumulation in the environment, induction to and resuscitation from VBNC state. (A) Bacterial-particle association and bacteria-bacteria association. (B) Environmental stressors such as high/low nutrients, oxygen, redox potential, and oxidative stress induce biofilm formation. (C) Transport and sedimentation provides a downward flux to sediment. As the biofilm grows on the sediment the mass transfer rate is no longer sufficient resulting in localized gradients in electron acceptors and nutrients. This results the induction of VBNC bacteria. (D) High flow events result in shear and can slough the biofilm, reducing the stabilizing effect of the EPS. (E). This can further exacerbate the resuspension of bacteria within the water column leading to increased particulate load (Adapted from Ayrapetyan et al., 2014b; Pinto et al., 2015).
Intertidal mudflats are comprised of very small silt and clay particles deposited when low energy currents and wave action prevails (Stal and de Brouwer, 2003). The stabilization of the sediment is due to a combination of compaction of the sediment during periods of drying (Stal and de Brouwer, 2003) and through the release of EPS by diatoms and bacteria creating stabilizing biofilms (Madsen et al., 1993). Enteric organisms such as E. coli (pathogenic strains), Campylobacter spp., Salmonella spp. and the pathogenic protozoan Cryptosporidium parvum are known for both creating and colonizing existing biofilms in drinking water systems (Wingender and Flemming, 2011). Enterococcus spp. form biofilms in beach sand (Piggot et al., 2012), whilst non-pathogenic E. coli are known to persist or even grow within coastal and estuarine environments, particularly in tropical/subtropical climates (Byappanahalli and Fujioka, 1998) and non-pathogenic Clostridium spp. have been isolated from an estuarine mudflat (Villanueva et al., 2007). At locations where significant fecal contamination occurs, FIOs/pathogens can colonize existing biofilm communities. Enteropathogenic E. coli O157:H7 is known to produce biofilms on a range of solid surfaces such as plastic, steel, wood, plant roots and leaves, facilitating long-term survival in the environment (Cooper et al., 2007). The ability of E. coli O157:H7 to produce biofilms, however, was dependent on the presence of other bacteria (Bauman et al., 2009; Klayman et al., 2009), and it is likely that surface roughness and the age of the biofilm are major determinants for survival (Korber et al., 1997). Biofilms have also been shown to be a reservoir for enteric viruses, suggesting that these entities persist longer in biofilms than in drinking water and wastewater (Skraber et al., 2005, 2009). Biofilms have been shown to provide protection from the surrounding environment, such as from antimicrobial compounds (e.g., chlorine) and UV exposure (Quignon et al., 1997; Ryu and Beuchat, 2005), and can enhance the infectivity of some organisms such as Legionella spp. (Wingender and Flemming, 2011); thus, facilitating persistence of these organisms and viruses. Another important aspect of biofilms is their potential for harboring bacteria in the VBNC state (Bryers, 2000; Schultz-Fademrecht et al., 2008; Wingender and Flemming, 2011). Therefore, quantifying the survival of bacteria in the environment is not a straightforward exercise.
Metabolic activity of fecal bacteria
Fecally derived bacteria are introduced into the aquatic environment through surface run off, wastewater discharge or direct defecation. However, the viability, persistence and metabolic activity within or between indicator species is not constant in the environment (Anderson et al., 2005). For example, the metabolic activity of a bioluminescent strain of E. coli O157:H7 decreased due to exposure to salt water, whilst elevated nutrients boosted its microbial activity (Williams et al., 2007) possibly resulting in growth (Shelton et al., 2014) or reduction in inactivation (Garzio-Hadzick et al., 2010). In most fresh and marine waters, metabolic activity rapidly declines after release from feces, which may result from insufficient carbon source or absence of host factors (Thorn et al., 2011; Li et al., 2014). Knowledge of the physiological state of E. coli is particularly important, as inactive cells (stationary phase), possess greater resistance to environmental stresses such as acidity and anoxia, thereby increasing the probability of survival (Cheville et al., 1996; Saby et al., 1999). Experimental evidence shows that in the log phase, E. coli O157:H7 was more vulnerable to biocides and environmental stress (Arnold and Kaspar, 1995); however, if the environment is suitable for growth, this facilitates rapid resource exploitation and proliferation. Current evidence suggests that E. coli O157:H7 enters a stationary phase after detachment from intestinal margins in ruminants (Poulsen et al., 1995). Subsequently, the bacterium leaves its host in the stationary phase, increasing its chances of survival in the environment. E. coli from cattle feces was shown to be in the VBNC state prior to any environmental exposure (Wu et al., 2009b), suggesting a large fraction of the fecal indicator population may be recalcitrant but non-culturable in agricultural sources when enumerated by conventional microbiological plate counting.
Viable but non-culturable (VBNC) state of fecal indicators and pathogens
VBNC bacteria are defined as cells that are in a state of low metabolic activity, and are therefore viable, but are unable to be cultivated on solid selective microbiological culture media; however, under favorable conditions, VBNC cells may resuscitate and regain the ability to grow on microbiological media. The VBNC state is therefore an important methodological limitation, thus preventing the representative enumeration of bacterial abundance in the environment and clinical settings by microbiological plate count analysis (Oliver, 2010). Under sub-optimal conditions such as starvation, salinity, electron acceptor conditions, temperature or pH bacteria enter a “dormant” state. Return of optimal conditions may result in resuscitation (Oliver, 2005). Therefore, standard water quality monitoring surveys do not adequately represent this sub-population of fecally associated VBNC bacteria/pathogens within the water. Recently, studies have examined VBNC FIOs in sediments and biofilms. These environments tend to be deficient in a growth limiting electron acceptor or nutrient and therefore facilitate a greater proportion of VBNC bacteria than expected in free floating systems (Bryers, 2000; Amel et al., 2008; Lieleg and Ribbeck, 2011). For example, greater numbers of E. coli and Salmonella have been isolated from sediments by molecular methods, than recorded by culturing techniques, indicating that these bacteria could enter the VBNC state in sediments (Amel et al., 2008; Berthe et al., 2008; Luna et al., 2010). In addition, dissolved nucleic acids are more readily extracted than particulate forms which could represent a bias for enumeration (Paul et al., 1991). Vibrio spp. are frequently used as model organisms for VBNC studies and enter and recover from the VBNC state under a variety of different stimuli (see: Oliver and Bockian, 1995; Oliver et al., 1995; Amel et al., 2008; Li et al., 2014; Pinto et al., 2015 for different stimuli). In contrast, studies on sediments are sparse, for example, Amel et al. (2008) found that V. fluvialis entered the VBNC state in sediments and could be resuscitated even after 1 year. Fukushima and Seki (2004) and Randa et al. (2004) challenge the VBNC notion by suggesting that extremely low abundance of suspended V. vulnificus and V. parahaemolyticus in winter months is due to the sediment acting as a microbial reservoir, as opposed to the bacteria entering VNBC. Further, Fukushima and Seki (2004) highlight that the proliferation of Vibrio spp. after a water temperature increase is due to the replication and release of the daughter cells in the sediment or biofilm rather than the resuscitation of cells from the VBNC state in the water column. Lee et al. (2007) found that drinking water pipe material composition was critical in governing the relative proportion of VBNC and culturable bacteria. However, further research is required on methods to enumerate the numbers of fecally associated bacteria entering the VBNC state in sediments (Amel et al., 2008). Delineating resuscitation from growth remains a significant challenge for the use of direct approaches (Ayrapetyan et al., 2014a; Ramamurthy et al., 2014, Table 4). Physiochemical factors governing induction to and resuscitation from VBNC in biofilms requires further attention, particularly on methodologies to sample VBNC bacteria in sediments/biofilms non-destructively.
Table 4.
Comparison of methods to enumerate viable but non-culturable (VBNC) bacteria–suitability for sediment.
| Technique | Direct or indirect | Method aim and calculation required | Advantages | Disadvantages | Suitable for sediment? | References |
|---|---|---|---|---|---|---|
| Nalidixic acid | Indirect | Nalidixic acid is bacteriostatic and inhibits cell division at low concentrations. | Allows differentiation of dividing viable cells from non-dividing VBNC cells. | “Resuscitated” cells would subsequently be inhibited by Nalidixic acid as they start to grow. | No | Ohtomo and Saito, 2001 |
| Need to run in conjunction with direct counts to ensure total counts do not change. | Some bacteria are Nalidixic acid-resistant. | |||||
| VBNC = (live-un-elongated cells under Nalidixic acid treatment) | “Injured” cells may only grow on non-selective media, difficult on sediment samples. | |||||
| BacLight™ | Indirect | Uses two dyes (SYTO9 and Propidium iodide) to stain live cells green and dead cells red. Cells are counted under the microscope. | Membrane integrity is one of the most conservative estimators of viability. Can result in overestimation of viability and VBNC fraction. | Step for disaggregation from sediment required. | No | Hassard et al., 2014 |
| Assumes that dead cells have disrupted membranes. | ||||||
| Cells may form clusters and be difficult to count. | ||||||
| VBNC = (live – culturable) | ||||||
| Fluorescent in-situ hybridisation (FISH) and peptide nucleic acid FISH (PNA-FISH). | Indirect | Oligonucleotide probes hybridise to target DNA/RNA and fluoresce under the microscope. | Relatively straight-forward technique. PNA probes have a superior binding capability than traditional FISH probes. | Need to find a species/strain-specific nucleotide probe. | No | Halpern et al., 2007; Malic et al., 2009 |
| Reliant on microscope quantification | ||||||
| VBNC = (FISH positive – culturable). | Cells may form clusters and be difficult to count. | |||||
| Immunomagnetic separation | Indirect | Antibodies for a specific species or strain are coated onto magnetic beads. | Not a standalone method for VBNC detection, Immunomagnetic separation can be used to isolate the organism of choice from environmental samples in conjunction with a quantification method. | Not 100% specific | Yes | Gwyther et al., 2013 |
| A magnet is used to pull the bacteria-linked beads from an environmental sample. | Step for disaggregation from sediment is required. | |||||
| Adds an extra step into the analysis time. | ||||||
| Relatively straight-forward technique. | Quantification method required. | |||||
| Flow cytometry (FCM) | Indirect | Cells are labeled with nucleic acid stains e.g., BacLight™. The flow cytometer sorts each cell individually, based on fluorescence backscatter which is used to determine abundance of live and dead cells. | FCM can distinguish between reproductively viable, metabolically active, intact and permeabilized cells. | Requires pure cultures or the quantification of entire populations. | Yes | Wallner et al., 1995; Khan et al., 2010 |
| Assumes that dead cells have disrupted membranes. | ||||||
| Rapid in-situ analysis of single cells. | ||||||
| VBNC = (live – culturable) | Difficult to distinguish between bacteria and phages if using environmental samples due to overlap of distributions and signal noise. | |||||
| FCM-FISH has been applied with limited success. | ||||||
| Propidium monoazide—quantitative PCR (PMA-qPCR) | Indirect | PMA binds to DNA in membrane-compromised cells, preventing DNA replication during PCR. | Can target specific species/strain of bacteria. | PMA-qPCR does not in itself distinguish VBNC cells, but enumerates the number gene equivalents from the bacteria with intact membranes. | Yes | Nocker et al., 2007; Gin and Goh, 2013 |
| ΔCTrefers to the difference in qPCR threshold cycles CT between total bacteria and live bacteria | qPCR is quantitative. | Detachment from sediments and particulate matter required as a pretreatment. | ||||
| VBNC = (PMA negative gene equivalents – culturable). | ||||||
| Ethidium monoazide—loop mediated isothermal amplification (EMA-LAMP). | Indirect | EMA binds to DNA in membrane-compromised cells, preventing DNA replication. | Can target specific species/strain of bacteria. | Similar limitations as PMA-qPCR | Yes | Wang et al., 2012, |
| LAMP is quicker than PCR. | ||||||
| VBNC = (EMA negative gene equivalents – culturable). | ||||||
| Reverse transcription-quantitative polymerase chain reaction (RT-qPCR). | Indirect | Quantitative PCR method used to detect expression levels of RNA e.g. rpoS gene mRNA. | Targets RNA expression, which is a proxy for activity in bacteria. | Environmental matrices, particularly sediment, may contain PCR inhibitors which restrict applicability. | Yes | Yaron and Matthews, 2002; Quilliam et al., 2011b; Wingender and Flemming, 2011 |
| Can detect target genes active in VBNC bacteria and compare to levels in culturable bacteria referenced against housekeeping genes. | Difficult to extract RNA. Requires suitable sampling regime and storage. | |||||
| Autoinducers (AI) /resuscitation promotion factors (RPF). | Direct | Synthetic or biologically produced autoinducer 2 (AI2) or RPF to measure culturability of exposed and unexposed population. | Quantifies VBNC bacteria using the same quantification methodology as ‘culturable’ bacteria therefore a representative comparison. | Difficult to distinguish VBNC from additional growth due to autoinducer/RPF. | Yes | Bari et al., 2013; Ayrapetyan et al., 2014a |
| Additional nutrients/cofactors may be required. | ||||||
| VBNC = (AI or RPF culturable)- (normal culturable). | Species specific or broad spectrum RPF can be used depending on requirements. | |||||
| Often species specific. | ||||||
| Theoretical distinction between resuscitation and growth based on growth rates. | ||||||
| Pre-rRNA analysis molecular viability testing. | Indirect | Detects innate synthetic activity of rRNA precursors. | Greater fraction of RNA pool than mRNA so easier to detect. | Possibility for false negatives lack of detection = VBNC cells. | Yes | Cangelosi et al., 2010, |
| Requires a measure of abundance of species of interest. | ||||||
| Direct indicator of growth as pre-rRNAs only formed in growing cells. In dormant cells pre-rRNA levels decline. | Species specific or constitutive precursors can be selected based on required resolution. | |||||
| After nutrient stimulation there is a ratiometric increase in abundance of pre-rRNA from viable cells but not non-viable cells. | Can be used to separate resuscitation from growth, as response time is quicker than that of growth rate of bacteria. | Requires nutrients to stimulate response. | ||||
| Reliant on RT-qPCR for detection so similar limitations at quantification step. | ||||||
| Dilution to extinction—resuscitation potential | Direct | VBNC bacteria can be distinguished from growth by a serial dilution method. Cells are subjected to a log dilution series below 1 CFU/ml of culturable cells. These diluted cells are cultured, if growth is determined then bacteria have resuscitated from VBNC. | Quantitative, utilises the same methodology to determine VBNC as culturable counts therefore directly comparable. | Dilution could inhibit quorum sensing based resuscitation. | Yes | Zhang et al., 2015 |
| VBNC = Resuscitated counts – culturable counts |
Bold indicates the factors used to calculate VBNC.
Indirect approaches such as microscopy combined with live/dead staining, taxon-specific fluorescent in situ hybridization (FISH) and qPCR have all been utilized for measurement of VBNC bacteria in environmental samples by comparing “total” or “live” bacteria with “culturable counts” (Table 4). Indirect methods for VBNC quantification bacteria in environmental water samples are also not appropriate for sediments due to the 3D nature of the matrix, extracellular polymers and blocking of incident light for methods such as BacLight™ staining. Direct methods (utilizing microbiological plate counts) such as the application of resuscitation promotion factors (e.g., autoinducers) have been shown experimentally to be useful for measuring the total bacterial population including the VBNC fraction in water but have yet to be applied to sediment (Atkinson and Williams, 2009; Bari et al., 2013; Ayrapetyan et al., 2014a). The principal issue for these approaches is delineating resuscitation of existing bacteria from growth of daughter bacteria (Ayrapetyan et al., 2014b) and this problem remains with sediment. The phenotypic changes that occur in the VNBC state can be assessed using reverse transcription quantitative PCR (RT-qPCR; Table 4) as alterations to membrane lipid composition, fluidity and a rearrangement of the outer membrane composition have been reported previously (Scherber et al., 2009). Membrane changes in response to stress are modulated via the osmosensor protein EnvZ, which is sensitive to changes in external solute concentration. This cascade is potentially regulated by MzrA, and upregulation increases outer membrane proteins such as ompW (Asakura et al., 2008; Darcan et al., 2009). The porin protein encoded by ompW gene is known to be upregulated by extremes of pH (Wu et al., 2009a), whilst E. coli osmoregulation proteins OmpC/F production are regulated by changes to solute concentration. This is an important survival strategy for coastal and transitional zones, such as estuaries (Rozen and Belkin, 2001). The analysis of pre-ribosomal RNA (pre-rRNA) has received interest recently (Cangelosi et al., 2010). Reported advantages include greater relative abundance of pre-rRNA compared to messenger RNA (mRNA) so response is quicker, which subsequently allows separation of resuscitation from growth (Table 4). The method relies on the ratiometric increase in pre-rRNA levels in bacteria subject to a nutrient-based resuscitation compared to a control in the absence of nutrients; this provides a dormant to non-dormant ratio (Cangelosi et al., 2010). It is still unclear if this approach is valid for sediments.
Viable but non-culturable Pseudomonas spp. exhibited a reduction in nutrient transport, respiration rates and macromolecular synthesis compared to culturable equivalents; however these VBNC cells can still actively divide at a reduced rate (Peneau et al., 2007). Adhesion to the external surface of zooplankton also stimulates fecal enterococci to enter a VBNC state (Signoretto et al., 2004) and this may form a vital part of the transmission pathway (Cellini et al., 2005). Favorable growth conditions and an ideal stoichiometric ratio of carbon to inorganic elements enables recovery from VBNC state, although the resuscitation rate is highly variable depending on species and conditions studied (Arana et al., 2007; Bari et al., 2013; Ayrapetyan et al., 2014a) and may take days to occur (Scherber et al., 2009). Reversion to a culturable state probably involves a resuscitation-promoting or anti-dormancy factor which can cleave peptidoglycan, altering the mechanical properties of the cell wall to facilitate cell division or release lysis products that function as anti-dormancy signals (Ward et al., 2006). Whether VBNC cells are capable of causing infection is poorly understood, and is dependent on the reactivation time, external conditions and if additional vectors/cofactors are required or involved prior to infection. Research into Salmonella has indicated that newly formed VBNC cells do not mount a strong infection response (Passerat et al., 2009) possibly due to lack of suitable resuscitation factors. The resuscitation window is defined as the time or amount of stress a VBNC bacteria can undergo and still resuscitate. If conditions remain unfavorable, then VBNC bacteria go beyond the period where resuscitation can occur, and are considered injured, but may still be viable. Finally, eventual death may occur (Pinto et al., 2015; Figure 1). Zhang et al. (2015) utilized a method known as dilution to extinction (Table 4) and showed that E. coli had significant resuscitation potential after UV treatment, suggesting that routine disinfection induces the VBNC state as opposed to cell death in bacteria. Whether bacterial FIOs and pathogens have “resuscitation potential” which could represent a risk to public health or water quality requires further attention.
The potential for bacteria to enter the VBNC state suggests that sediments may be a greater store of fecally-derived bacteria than previously quantified. Sediments and biofilms provide distinct gradients of nutrients, electron acceptors and pH, whilst protecting from some environmental stressors, such as shear and light (Bryers, 2000). Additional methodological improvements are required to reliably quantify VBNC bacteria in sediment. Gene targets which are expressed and specific to the VBNC response can be used in combination with RT-qPCR quantification, providing a useful approach for VBNC analysis in sediment. This is because probes may be species/strain-specific and are based on the production of messenger RNA (mRNA) or pre-rRNA molecules which are short-lived and can provide high resolution information on temporal gene expression (Yaron and Matthews, 2002; Cangelosi et al., 2010). However, the low extraction efficiency of RNA and downstream qPCR inhibition which is a particular challenge in sediment needs to be overcome (Miura et al., 2011; Carreira et al., 2015).
Fate and behavior of fecally derived viruses in sediments
There has been considerable attention attributed to the fate and transport of viruses in environmental matrixes, such as soil, groundwater and surface water (Schijven and Hassanizadeh, 2000; John and Rose, 2005; Sen and Khilar, 2006). The main factors affecting viral adsorption and persistence in porous media include the type of virus and media, temperature, pH, ionic strength and the presence of organic matter (Jin and Flury, 2002). The dominant mechanisms are well-understood in porous media, however, little information is available on their importance in sediment. Enteric viruses readily adsorb to many types of sediment with reported adsorption rates of between 37 and 100% (Carlson et al., 1968; Gerba et al., 1977b; LaBelle and Gerba, 1979; Gerba et al., 1980; Bitton et al., 1982; Tsai et al., 1983; Johnson et al., 1984). The high adsorption levels in estuarine and marine sediment (Table 5) may be attributed to the high organic content and hydrophobicity of the sediment particles (Chrysikopoulos and Syngouna, 2012). Other factors shown to influence viral adsorption to porous media may have limited impact in sediment due to the production of conditioning films. However, the physico-chemical properties of viral particles and water may play a role in viral adsorption-desorption kinetics in sediment. For instance, Bitton et al. (1982) found complete adsorption (100%) of poliovirus to marine sediment compared to lower adsorption to freshwater sediment (37–45%). LaBelle and Gerba (1979) showed that increased salinity and decreased pH enhance the desorption (5–10%) of echovirus from estuarine sediment, whereas the desorption of other enteric viruses (rotavirus, poliovirus, and coxsachieviruses) was not affected by those changes. Carlson et al. (1968) found that the presence of bivalent cations in solution enhanced viral adsorption to clay, whereas albumin promoted desorption. These results imply that enteric viruses may desorb from sediment when conditions change, for example to heavy rainfalls or tidal changes.
Table 5.
Fecally-derived virus adsorption to sediment.
| Virus type | Sediment type | Adsorption (%) | References |
|---|---|---|---|
| Poliovirus 1 | Marine (99.7% sand, 0.3% clay) | 99% | Bitton et al., 1982 |
| Marine, organic muck | 100% | ||
| Estuarine (20.7% sand, 24.88% clay, 54.4% silt, 3.8% organic matter) | ~100 | LaBelle and Gerba, 1980 | |
| Estuarine | 99.9 | Gerba et al., 1980 | |
| Estuarine (mud and sand) | 99.2–99.98 | Gerba et al., 1977b | |
| Estuarine (99% sand, 1% silt) | 93.4 | Johnson et al., 1984 | |
| Estuarine (52.3% sand, 30.3% silt, 17.4% clay) | 99.8 | ||
| Estuarine (89.3% sand, 6% silt, 4.6% clay) | 98.3 | ||
| Estuarine (37.3% sand, 39.2% silt, 23.5% clay) | 99.9 | ||
| Estuarine (10.1% sand, 48.2% silt, 41.7% clay) | >95 | Tsai et al., 1983 | |
| Estuarine (79.2% sand, 11.8% silt, 9.1% clay) | >95 | ||
| Freshwater (99.6% sand, 0.4% clay) | 37% | Bitton et al., 1982 | |
| Freshwater (99.7% sand, 0.3% clay) | 45% | ||
| Coxsackievirus B1 | Estuarine (99% sand, 1% silt | 64.6 | Johnson et al., 1984 |
| Estuarine (52.3% sand, 30.3% silt, 17.4% clay) | 98.4 | ||
| Estuarine (89.3% sand, 6% silt, 4.6% clay) | 98.6 | ||
| Estuarine (37.3% sand, 39.2% silt, 23.5% clay) | 99.0 | ||
| Coxsackievirus B3 | Estuarine (20.7% sand, 24.88% clay, 54.4% silt, 3.8% organic matter) | 100 | LaBelle and Gerba, 1980 |
| Estuarine | 99.8 | Gerba et al., 1980 | |
| Estuarine (10.1% sand, 48.2% silt, 41.7% clay) | >95 | Tsai et al., 1983 | |
| Estuarine (79.2% sand, 11.8% silt, 9.1% clay) | >95 | ||
| Coxsackievirus B4 | Estuarine | 95 | Gerba et al., 1980 |
| Echovirus 1 | Estuarine (20.7% sand, 24.88% clay, 54.4% silt, 3.8% organic matter) | 90 | LaBelle and Gerba, 1980 |
| Estuarine | 87.0–99.99 | Gerba et al., 1980 | |
| Echovirus 7 | Estuarine | >99.99 | |
| Echovirus 29 | Estuarine | >99.99 | |
| Echovirus 11 | Estuarine (99% sand, 1% silt | 66.6 | Johnson et al., 1984 |
| Estuarine (52.3% sand, 30.3% silt, 17.4% clay) | 98.9 | ||
| Estuarine (89.3% sand, 6% silt, 4.6% clay) | 99.0 | ||
| Estuarine (37.3% sand, 39.2% silt, 23.5% clay) | 99.5 |
Gerba et al. (1980) observed species/strain specific differences in viral adsorption to sediment, suggesting that capsid properties may play an important role in adhesion. The physico-chemical characteristics of viral particles, e.g., pI, hydrophobicity and capsid structures have been shown to play an important role in the adsorption of viruses to porous media. Dowd et al. (1998) highlighted the influence of viral pI on the adsorption rate of viruses, where a smaller pI (3.9–5.3) showed more adsorption than a larger pI (6.6–7.7) despite examined viruses being of similar sizes. Farkas et al. (2015) observed that the adsorption of rotavirus viral surrogates with similar size and pI adsorbed differently to hydrophobic media. Further differences were found in the adsorption of viral surrogates with similar size, zeta potential and hydrophobicity to porous media, suggesting that the composition of viral capsid also affects viral adhesion (Pang et al., 2014; Farkas et al., 2015). Further, Samandoulgou et al. (2015) found that extremes of pH and temperature can change the mechanism of norovirus association with sediment from electrostatic to predominantly hydrophobic, as loss of ordered molecular structure in the protein head results in an increase in hydrophobic attachment sites resulting in greater adsorption of norovirus. Hydrophobic interactions of proteins are enhanced by high salinity thus viral attachment/detachment kinetics in estuarine environments may change rapidly.
From a public health perspective, the inactivation of enteric viruses in sediment is also important. However, most studies focus on the presence/absence and concentration of enteric viruses in sediment and little is known about the inactivation and degradation of viral particles. Viruses in the water column are inactivated at a faster rate than in sediments (Smith et al., 1978; LaBelle and Gerba, 1980; Liew and Gerba, 1980; Rao et al., 1986b), indicating that sediments confer protection for viruses from degradation. The persistence of viruses is largely dependent on sediment and virus type. For instance, coxsachievirus degradation ranged from 0.2 to 2.5 log in three types of sediment in 20 days, whereas poliovirus and echovirus degraded by 0.5–4 log and 2–4 log, respectively (Table 6). As in water, microbial activity enhances the degradation of enteric viruses in the sediment, whereas small changes in temperature and salinity have little effect on inactivation. Inactivating substances, such as enzymes, may also adsorb to particles and thus have no effect on viral degradation (Gerba and Schaiberger, 1975). Interestingly, virus inactivation increased in polluted water even in the absence of microorganisms (LaBelle and Gerba, 1980) probably due to reaction with humics in water. Viral adsorption to sediment particles has also been shown to increase viral thermostability, possibly explaining the recalcitrant nature of enteric viruses in sediments (Liew and Gerba, 1980).
Table 6.
Persistence of Fecally-derived viruses in coastal and estuarine sediments.
| Viral strain | Habitat | Temp (°C) | Salinity (PSU)a | Initial inoculation | Reduction | References |
|---|---|---|---|---|---|---|
| Coxsackievirus B3 Nancy | Estuarine sediment (mud, shell) and seawater | ~7 log10 PFU 100 ml−1 | ~3.8 log in 20 days | Smith et al., 1978 | ||
| Estuarine sediment (sand) and seawater | ~7 log10 PFU 100 ml−1 | ~4 log in 20 days | ||||
| Estuarine sediment (mud, sand) and seawater | ~7 log10 PFU 100 ml−1 | ~2.7 log in 20 days | ||||
| Echovirus 1 Ferouk | Estuarine sediment (sand) and seawater | 30 | 25 | ~7 log10 PFU 100 ml−1 | ~4 log in 10 days | |
| Estuarine sediment (mud, shell) and seawater | ~7 log10 PFU 100 ml−1 | ~4 log in 18 days | ||||
| Estuarine sediment and seawater | 7-8 log10 PFU 100 ml−1 | ~1 log in 6 days | LaBelle and Gerba, 1980 | |||
| Estuarine sediment (mud, sand) and seawater | ~7 log10 PFU 100 ml−1 | ~1.5 log in 20 days | Smith et al., 1978 | |||
| Poliovirus 1 LSc | Estuarine sediment (mud, shell) and seawater | ~7 log10 PFU 100 ml−1 | ~4 log in 14 days | |||
| Estuarine sediment (mud, sand) and seawater | 31 | ~7 log10 PFU 100 ml−1 | ~4 log in 18 days | |||
| Estuarine sediment (sand) and seawater | 25 | ~7 log10 PFU 100 ml−1 | ~4 log in 7 days | |||
| Estuarine sediment and seawater | 31 | 30 | 7–8 log10 PFU 100 ml−1 | ~2.5 log in 7 days | LaBelle and Gerba, 1980 | |
| 33 | 26 | 7–8 log10 PFU 100 ml−1 | ~ 3.3 log in 6 days | |||
| 33 | 27 | 7–8 log10 PFU 100 ml−1 | ~2.8 log in 6 days | |||
| Estuarine sediment and artificial seawater | 4 | ~6 log10 PFU 100 ml−1 | Stable for 33 daysb | Liew and Gerba, 1980 | ||
| 24 | ~6 log10 PFU 100 ml−1 | ~3 log10 over 33 days | ||||
| 37 | ~6 log10 PFU 100 ml−1 | ~2 log in 4 days | ||||
| Estuarine sediment and seawater | 20–25 | 2–20 | 7.8 log10 PFU 100 ml−1 | 7.3 log in 19 days | Rao et al., 1984 | |
| Estuarine suspended solids and seawater | 20–25 | 2–20 | 7.5 log10 PFU 100 ml−1 | 5.8 log in 19 days | ||
| Estuarine “fluffy” sediments and seawater | 20–25 | 2–20 | 7.6 log10 PFU 100 ml−1 | 6.5 log in 19 days | ||
| Simian Rotavirus SA11 | Estuarine sediment and seawater | 20–25 | 2–20 | 7.7 log10 PFU 100 ml−1 | 4.7 log in 19 days | |
| Estuarine suspended solids and seawater | 20–25 | 2–20 | 7.9 log10 PFU 100 ml−1 | 3.9 log in 19 days | ||
| Estuarine “fluffy” sediments and seawater | 20–25 | 2–20 | 7.6 log10 PFU 100 ml−1 | 5.6 log in 19 days |
Salinity assumed PSU = %0.
~Prefix represents viral abundance estimated from graphs.
Viruses may reversibly attach and detach from sediment and re-enter the water column or the sediment-associated viral particles may be transported from polluted to non-polluted waters. Hence, viruses entering the water body from sediment may increase the public health risk. Furthermore, due to water turbulence the viral particles attached to less dense sediment may be easily resuspended. Rao et al. (1986b) implied that solid-associated rotavirus may be transported >5 km (≥3 miles) in estuarine water. Furthermore, sediment-associated viruses may be taken up by shellfish or crustacea that are destined for human consumption. Despite the risks of viral presence in sediment being well-recognized, little is known about the fate of viruses in sediment, and especially the factors which may affect viral adsorption and inactivation in situ e.g., sunlight, chemical contamination and organic matter. This is pertinent when considering the impact of sediment/particle association on the suitability of using viruses as regulatory indicators for drinking waters and bathing/shellfish water quality (Bosch et al., 2008).
Methods for the enumeration of fecally derived viruses from sediments
The identification and quantification of enteric viruses in the environment is challenging mainly due to the lack of reliable methods for accurate quantification and the difficulty in eluting viruses from sediment. The most frequently used methods for quantification of enteric viruses in environmental studies are tissue culture, electron microscopy (EM), enzyme-linked immunosorbent assay (ELISA), flow cytometry and qPCR or RT-qPCR (Weinbauer, 2004; Duhamel and Jacquet, 2006).
Traditional tissue culture approaches involve incubation of virus-containing samples with suitable host cell lines that allow viral replication. The cytopathic effects (host cell damage) can be observed under the light microscope (Dulbecco, 1952; Moce-Llivina et al., 2004). For viruses which do not lyse host cells, a focus-forming assay is used which involves the use of fluorescent antibodies that bind to viral antigens allowing the detection of clusters of infected cells (foci) by fluorescent microscopy (Payne et al., 2006). Nonetheless, culture-based assays can take weeks to perform (Storch, 2000) and often underestimate the number of viruses due to viral aggregation; however, as loss of infectivity is permanent, this provides a useful estimate of infectivity decay rates (Charles et al., 2009). Furthermore, some enteric viruses such as human noroviruses and sapoviruses cannot be maintained in vitro, hence they cannot be quantified by culture. Intact virus particles after incubation with an appropriate dye can be visualized using EM, however this approach cannot reliably distinguish between viral strains or infectious from non-infectious viral particles (Dancho et al., 2012). Tissue culture and EM both require expensive equipment and skilled staff, hence are rarely applied for routine examinations. However, early studies investigating the recovery of enteric viruses from sediment usually applied tissue culture for viral enumeration. In order to detect and quantify sediment-associated viruses using tissue culture or EM, viral particles are eluted from sediment and re-concentrated to reduce sample volume. As shown in Table 7A, the usefulness of different approaches has been evaluated, and recoveries exhibited high variations depending on methodology and sediment/virus type. The best recoveries (>60%) were achieved with the use of casein or beef extract solution as an eluent, followed by polyethylene glycol (PEG) precipitation (Johnson et al., 1984; Lewis et al., 1985).
Table 7.
Enteric virus recoveries from sediment.
| Target virus | Sediment | Elution | Concentration | Quantitation | Mean recovery or range | References |
|---|---|---|---|---|---|---|
| A INDIRECT EXTRACTION: VIRUS ELUTION—CONCENTRATION | ||||||
| Poliovirus 1 | Marine 500 g | 0.25 M glycine, 0.05 M EDTA, pH 11 | 0.06 M AlCl3, pH 3.5 | Culturing | 50% | Gerba et al., 1977b |
| Marine 10 g | 6% beef extract, pH 9 | 8% PEG6000 | Culturing | 6.3–55.8% | Lewis et al., 1985 | |
| Marine 10 g | 4 M urea, 0.05 M lysine, pH 9 | 0.005 M AlCl3, pH 7 | Culturing | 22% | Bitton et al., 1982 | |
| 3% beef extract | pH 3.5–4.5 | 8% | ||||
| 1% purified casein | pH 3.5–4.5 | 14% | ||||
| Estuarine, sewage contaminated 10 g | 0.25 M glycine | None | Culturing | 2.2–3.5% | Tsai et al., 1983 | |
| 0.5% skimmed milk | 0.5–2.7% | |||||
| 0.5% isoelectric casein | 58.8% | |||||
| 3–10% beef extract (paste) | 4.0–9.3% | |||||
| 3–10% beef extract (powder) | 32.7–40.0% | |||||
| 4% nutrient broth | 40–53% | |||||
| Estuarine 10–50 mL | 3% beef extract, 2 M NaNO3, pH 5.5 | 2 M (NH4)2SO4, 0.01 Cat-Floc T pH 3.5 | Culturing | 39–44% | Wait and Sobsey, 1983 | |
| 3% beef extract, 0.25 M glycine, pH 10.50.25 M glycine, 0.05 M EDTA, pH 11 | 0.06 M AlCl3, pH 3.5 | 9.7–18%0–0.1% | ||||
| Freshwater10 g | 6% beef extract, pH 9 | 8% PEG6000 | Culturing | 15.8–76.8% | Lewis et al., 1985 | |
| Freshwater4 types with different sand/silt / clay ratio | 4% nutrient broth, pH 7.5 | None | Culturing | 5.3–10.4% | Johnson et al., 1984 | |
| 4% nutrient broth, pH 9 | 2.0–32.5% | |||||
| 5% beef extract, pH 9 | 0.6–48.9% | |||||
| 0.5% isoelectric casein, pH9 | 0.3–65.3% | |||||
| 0.5% isoelectric casein, 1% crude lecithin, pH9 | 7.0–38.6% | |||||
| 0.5% isoelectric casein, 3% crude lecithin, pH9 | 1.0–25.4% | |||||
| 0.5% isoelectric casein, 1% semi-purified lecithin, pH9 | 0.3–47.9% | |||||
| 0.5% isoelectric casein, 3% semi-purified lecithin, pH9 | 2.6–75.1% | |||||
| 0.5% isoelectric casein, 1% egg lecithin, pH9 | 0.3–56.4% | |||||
| 0.5% isoelectric casein, 3% egg lecithin, pH9 | 0.6–102.6% | |||||
| Silty freshwater 5 g | 0.25 M glycine-NaOH, 0.05 M EDTA | 0.06 M AlCl3, pH 3.5 | qRT-PCR | 1.8% | Miura et al., 2011 | |
| 0.25 M glycine-NaOH, 0.05 M EDTA | 0.1 M MgCl2 | 5.4% | ||||
| 0.1% Laureth-12, 0.01 M Tris, 1 mM EDTA, 0.015% Antifoam Y-30, pH 7.2 | 0.1 M MgCl2 | 0.61% | ||||
| 0.25 M glycine-NaOH, 0.05 M EDTA | 16% PEG6000, 4.7% NaCl | 0% | ||||
| 0.1% Laureth-12, 0.01 M Tris, 1 mM EDTA, 0.015% Antifoam Y-30, pH 7.2 | 16% PEG6000, 4.7% NaCl | 0.18% | ||||
| Sandy freshwater10 g | 4 M urea, 0.05 M lysine, pH 9 | 0.005 M AlCl3, pH 7 | Culturing | 39% | Bitton et al., 1982 | |
| 3% beef extract, pH 9 | pH 3.5–4.5 | 51% | ||||
| 1% purified casein, 0.1% Tween 80 | pH 3.5–4.5 | 59% | ||||
| 1 M trychloroacetate, 1 M glycine | pH 3.5–4.5 | 23% | ||||
| Coxsachievirus B1 | Freshwater4 types with different sand/silt/ clay ratio | 0.5% isoelectric casein, 1% crude lecithin, pH9 | None | Culturing | 12.6–37.8% | Johnson et al., 1984 |
| 0.5% isoelectric casein, 3% crude lecithin, pH9 | 21.8–80.9% | |||||
| 0.5% isoelectric casein, 1% semi-purified lecithin, pH9 | 48.4–61.6% | |||||
| 0.5% isoelectric casein, 3% semi-purified lecithin, pH9 | 54.4–91.5% | |||||
| 0.5% isoelectric casein, 1% egg lecithin, pH9 | 38.6–69.0% | |||||
| 0.5% isoelectric casein, 3% egg lecithin, pH9 | 41.9–73.4% | |||||
| Coxsachievirus B3 | Estuarine, sewage contaminated 10 g | 0.25 M glycine | None | Culturing | 4.3–7.7% | Tsai et al., 1983 |
| 0.5% skim milk | 8.3–9.2% | |||||
| 0.5% isoelectric casein | 18.9% | |||||
| 3–10% beef extract (powder) | 13.0–29.0% | |||||
| 4% nutrient broth | 15.4–25.9% | |||||
| Echovirus 1 | Estuarine 10–50 mL | 3% beef extract, 2 M NaNO3, pH 5.5 | 2 M (NH4)2SO4, 0.01 Cat-Floc T | Culturing | 16–43% | Wait and Sobsey, 1983 |
| 3% beef extract, 0.25 M glycine, pH 10.5 | pH 3.5 | 2.6–4.4% | ||||
| 0.25 M glycine, 0.05 M EDTA, pH 11 | 0.06 M AlCl3, pH 3.5 | 0.1–0.5% | ||||
| Echovirus 11 | Freshwater4 types with different sand/silt/ clay ratio | 0.5% isoelectric casein, 1% crude lecithin, pH9 | None | Culturing | 5.6–42.4% | Johnson et al., 1984 |
| 0.5% isoelectric casein, 3% crude lecithin, pH9 | 33.2–94.7% | |||||
| 0.5% isoelectric casein, 1% semi-purified lecithin, pH9 | 31.6–81.9% | |||||
| 0.5% isoelectric casein, 3% semi-purified lecithin, pH9 | 56.8–78.5% | |||||
| 0.5% isoelectric casein, 1% egg lecithin, pH9 | 66.7–138.4% | |||||
| 0.5% isoelectric casein, 3% egg lecithin, pH9 | 43.3–50.4% | |||||
| Hepatitis A virus | Freshwater | 3% beef extract, 2 M NaNO3, pH 5.5 | 15% PEG6000 | Culturing | 70% | Lewis and Metcalf, 1988 |
| Simian rotavirus 11 | Estuarine10–50 mL | 3% beef extract, 2 M NaNO3, pH 5.5 | 2 M (NH4)2SO4, 0.01 Cat-Floc T | Culturing | 23% | Wait and Sobsey, 1983 |
| 3% beef extract, 0.25 M glycine, pH 10.5 | pH 3.5 | 0% | ||||
| 0.25 M glycine, 0.05 M EDTA, pH 11 | 0.06 M AlCl3, pH 3.5 | 0% | ||||
| Rotavirus WA | Freshwater | 3% beef extract, 2 M NaNO3, pH 5.5 | 15% PEG6000 | Culturing | 70% | Lewis and Metcalf, 1988 |
| Target virus | Sediment | Lysis | Extraction/concentration | Purification | Mean recovery or range | References |
| B DIRECT EXTRACTION OF VIRAL NUCLEIC ACIDS FOLLOWED BY QRT-PCR | ||||||
| Poliovirus 1 | Silty freshwater 5 g | 0.5 N Tris (pH 8), 0.1 M NaCl, 2% SDS, 8 mg skim milk/g sediment | Phenol:chloroform:isoamyl alcohol + isopropanol precipitation | DEAE cellulose column | 0.09% | Miura et al., 2011 |
| 0.5N Tris (pH 8), 0.1 M NaCl, 2% SDS, 8 mg skim milk/g sediment | Phenol:chloroform:isoamyl alcohol + isopropanol precipitation | Oligo(dT) labeled magnetic beads | 0.77% | |||
| 0.1 M EDTA, 0.5 N Tris (pH 8), 0.1 M NaCl, 2% SDS, 8 mg skim milk/g sediment | Phenol:chloroform:isoamyl alcohol + isopropanol precipitation | Oligo(dT) labeled magnetic beads | 11% | |||
| TRIzol reagent | Centrifugation | Oligo(dT) labeled magnetic beads | 0.10% | |||
The ELISA approach involves binding of viral antigens to specific antibodies that are subsequently quantified by adding an enzymatic substrate that produces color changes when bound. This technique has been applied in environmental studies (Fu et al., 1989; Park et al., 2010), and results correlate well with tissue culture findings (Nasser et al., 1995). For many enteric viruses, commercial ELISA kits are available allowing rapid detection, however, the assay may detect degraded viral capsid along with infectious particles. The usefulness of ELISA for sediment samples has not been investigated. The most frequently used methods for viral enumeration are qPCR and RT-qPCR which quantify a small segment of the viral genome of DNA and RNA viruses, respectively. These assays are rapid, sensitive, suitable for all virus types, and can be selective for individual strains (Girones et al., 2010). However standard PCR approaches do not provide any information on the integrity and infectivity of the target virus. When (RT-)qPCR is used, the elution of viral particles is not necessary as nucleic acids can be extracted directly from sediment. Recoveries of viral RNA from sediments range from 0.09 to 11% for direct extraction and RT-qPCR, with improved extraction efficiency when applying indirect elution-concentration approaches (Table 7). There are numerous reports of inhibition of PCR assays by organic matter (e.g., humic acids) often found in environmental samples (Meschke and Sobsey, 1998; Rock et al., 2010) and extraction and enumeration methods strongly influence estimates of viral abundance in sediments (Williamson et al., 2013), which can greatly influence attributed risk in pathogenic strains (Petterson et al., 2015). Recently, methods have been applied to overcome this; for example, Carreira et al. (2015) found that a combination of EDTA in addition to probe sonication and enzymatic pre-treatments resulted in 4.5 fold increase in viral recovery from sediments. Miura et al. (2011) found that a direct extraction method utilizing SDS, EDTA coupled with phenol-chloroform-isoamyl alcohol resulted in an 11% recovery of poliovirus 1 (Table 7). Commercial kits for environmental applications are also available and used to extract viral nucleic acids from various matrixes including biosolids (Ikner et al., 2012), however, their efficiency for sediment has not been evaluated. Comparison of viral abundance in sediments (enterovirus 102.g−1) to the titre which is shed from infected individuals (105–108.g−1) and the high adsorption efficiencies measured in vitro (Table 5) suggests dilution, dispersal, and/or high inactivation in sediments (Melnick and Rennick, 1980; Miura et al., 2011).
Determining viral infectivity is a particular challenge in sediments. Most recently, integrated cell culture (ICC) qPCR/RT-qPCR approaches have been developed (Greening et al., 2002; Fongaro et al., 2013; Ogorzaly et al., 2013). During the assay, cultured viruses are enumerated using qPCR or RT-qPCR, which are more sensitive than microscopy and less affected by viral aggregation. This combined approach allows the accurate quantification of infectious viral particles for strains that can be cultured in vitro within days (Ogorzaly et al., 2013). Viral recoveries may be improved by the combination of traditional indirect extraction followed by ICC-RT-qPCR (Fongaro et al., 2013). An initial assessment of direct nucleic acid extraction followed by RT-qPCR may be useful for rapid evaluation. Also sediments are difficult to isolate and purify viruses without leaking other compounds which also affect quantification. Research on both improving viral recoveries and role of sediment on the persistence of human pathogenic viruses in the environment could further inform modeling viral pathogens, environmental epidemiology and improve risk assessment.
Sediments as a sink/source of fecal bacteria and viruses?
Sediments may accumulate enteric bacteria and viruses and release them back in to the water under specific conditions. Therefore, quantifying the mass balance of fecally derived organisms in an estuary is not a simple task. Inputs of bacteria and viruses will be different for each estuary, depending on the surrounding land use, water use, and hydrological processes such as rainfall and tides. Spatial variation within the estuary itself can also confound the issue (Quilliam et al., 2011a; Perkins et al., 2014), as can the seasonal prevalence of bacteria and viruses (Ishii et al., 2006; He and He, 2008; Siem-Jorgensen et al., 2008). Characterization of individual estuaries is underway (Stapleton et al., 2007; Ouattara et al., 2011; Huang et al., 2015) and data from these surveys are being used in models that monitor the fluxes of FIOs, with the primary aim of predicting beach closures due to poor water quality (Stapleton et al., 2007; He and He, 2008; de Brauwere et al., 2014). The sources of fecally derived bacteria and viruses in the typical mixed estuary include wastewater, agricultural runoff, persistent populations, in situ growth and infrequent deposition events such as animal feces. The vast majority of fecally derived inputs from agriculture are due to livestock farming, although run-off from arable farming may also contribute to the bacterial/viral loading (Cox et al., 2005). Understanding viral pathogen persistence in wastewater treatment works and whether these viruses persist in sediments is in its infancy (Miura et al., 2011; Kitajima et al., 2014). In contrast bacterial persistence has been studied in detail. For example, Ouattara et al. (2011) reported that wastewater inputs of E. coli and intestinal enterococci were 35 and 15 times higher, respectively, than non-point source inputs in the Scheldt Estuary. Weather can also impact the relative contributions of agricultural (diffuse) and wastewater (point source) inputs (Stapleton et al., 2007) further complicating the understanding of pathogen behavior.
Deposition and retention of fecal bacteria in sediments
Settling and deposition of FIOs and pathogens in sediments is a complex process. Laboratory based estimates of settling velocities are 1.17 and 2.4 m/d for small (0.45–10 μm) and large (>10 μm) particles respectively (Auer and Niehaus, 1993). From a modeling perspective, an approximate deposition rate is taken, although reports vary with reported ranges from 2.6 to 25 md−1 (Jamieson, R. et al., 2005). For a review see Pachepsky and Shelton (2011). The settling rate in the field, however, is likely to be lower than these estimates and vary depending on other factors such as turbulence due to waves, wind and tides (Malham et al., 2014). Jamieson, R. et al. (2005) suggested that high bed shear stress limits the exchange between sediments and water column, although in contrast Drummond et al. (2014a) found that deposition of both E. coli and inert fluorescent beads occur rapidly, with 74% of the E. coli in the top 3 cm. Biofilms, vegetation, organic debris and flocs are likely to reduce the deposition and exchange of FIOs and pathogens to the sediment bed (Arnon et al., 2010; Drummond et al., 2014a,b). Arnon et al. (2010) found that the greater flow velocity and sediment particle size increases the mass transfer of particulate and soluble tracers to the sediments and biofilm and that particles preferentially deposit in biofilms as opposed to underlying sediment. Soluble matter is subject to advective and diffuse mass transport between the water column and bed, particulate matter including FIOs are subject to transport, sedimentation, and filtration (Ren and Packman, 2002; Arnon et al., 2010). The dynamic exchange between deposition and resuspension has received increased attention recently.
Release and resuspension of bacteria from sediments
During base flow and in the absence of turbulence, sediment-bound bacteria are unlikely to contribute to the bacterial pathogen abundance in the water column (Pachepsky and Shelton, 2011). Turbulence generated during peak flow results in mixing, an increase in oxygenation, bubble generation, and shear stress, which increases detachment rates from sediment and is dependent on bacterial shape and strain, and biofilm cohesive strength (Gomez-Suarez et al., 2001; Young, 2006; Lemos et al., 2014; Figure 1D). The release/resuspension of bacteria from biofilms within sediments is dependent on the combination of physicochemical forcing (Walter et al., 2013) and biotic factors, such as grazing and quorum sensing (Costerton et al., 1995; Kim et al., 2016) which could impact particulate loading to the water column (Figure 1E).
The release of E. coli from estuarine silts has been linked to rapid decreases in salinity of the water, which can occur in estuarine environments (Weiss, 1951). However, in freshwater systems, the number of E. coli released in successive events are limited to the deposited/proliferated bacteria between events (Shelton et al., 2014) and the sediment depth which is subject to scour (Harvey et al., 2012). The bacterial abundance increases in the water column on the rising curve of the storm hydrograph, due to particulate resuspension under periods of high turbulence (Howlett et al., 2015) often with a delay between the peak in riverflow and the peak in bacterial abundance (Jamieson R. C. et al., 2005; Jamieson R. et al., 2005; Henson et al., 2007). Controlled water release from a reservoir to a stream accounted for a 1–2 log increase in E. coli in the water column, but is dependent on the abundance in the sediment (Drummond et al., 2014a). Similarly, for viruses, desorption of viruses from clay particles can be attributed to reductions in salinity and the addition of organic matter due to rainfall or tides (Gerba and Schaiberger, 1975). In saline environments cations such as Ca2+ form bridges to stabilize bacteria binding to the sediment and also decreases the electrostatic repulsion during the initial stages of adsorption (van Loosdrecht et al., 1989; de Brouwer et al., 2002; Kierek and Watnick, 2003). The input of freshwater into estuarine systems could reduce the efficacy of these bridges, releasing pathogenic bacteria such as V. cholerae from sediment into the water column (Kierek and Watnick, 2003). Guizien et al. (2014) found that bacterial numbers decreased due to grazing and that viral titre in the water column did not significantly increase due to re-settlement of virus-clay complex, suggesting a complex story governing resuspension of enteric microorganisms. The risk associated with the sediment of enteric microorganisms depends on concentration, the ease with which bacteria resuspend or release back into the water column, and the frequency with which this will occur (Cox et al., 2005). Understanding viral resuspension in the environment is reliant on development of suitable methodology to enumerate viruses with accuracy and precision in a reproducible manner.
Outlook
Traditional molecular approaches are useful for absolute quantification of target organisms (e.g., E. coli, Salmonella spp, Enterococcus spp). However, advances in high throughput sequencing (HTS) have been applied to monitor fecal pollution, on a variety of different environmental matrices including wastewater, drinking water, riverine/coastal waters and ground water (Tan et al., 2015). For a more comprehensive review of HTS for assessing water quality see Tan et al. (2015). Targeted sequencing of 16S rRNA for bacteria and 18S rRNA for eukaryote small sub-unit rRNA permits an estimate of diversity and abundance (Henry et al., 2016), although the resolution of the 18S RNA gene as a phylogenetic marker us variable amongst taxa and is often not suitable for the resolution of FIOs belonging to the enterobacteriaceae for example. However, coupled sequencing and flow cytometry approaches can be used for more accurate taxon quantification (Props et al., 2016), although a different quantitative technology would be required for sediments. Bacterial diversity can be readily established in sediments using HTS; however rare sequences (e.g., pathogens) may evade detection, which is dependent on sequencing depth. Therefore, quantitative methods (e.g., qPCR, RT-qPCR) in sediments, remain a critical approach for inferring quantitative change associated with relative abundance HTS datasets.
Host associated genetic markers from bacterial groups such as Bacteroidales have been identified in sediments, which provides useful information for source apportionment (Tan et al., 2015). Genetic fingerprints of 16S rRNA gene or metagenome sequencing can reveal similarities between source (outfall, runoff etc.) and sink (beach sands and sediments) through community analysis (Ervin et al., 2014; Neave et al., 2014). Neave et al. (2014) confirmed the importance of local pollution sources of fecal species, in determining the fecally derived component of sediment associated microbial communities, which have a significant impact on beach sediment quality (e.g., Vignaroli et al., 2013). Sediments provide natural areas of high microbial density, which is of particular concern considered the elevated persistence and accumulation of antimicrobial resistance (AMR). Port et al. (2014) estimated the potential for gene transfer and pathogenicity potential from sediment fecal bacteria from pyrosequencing datasets of sediments. This is pertinent for developing suitable baseline AMR/pathogenicity levels used to inform policy makers on fecally derived hazardous microorganisms/pathogens in sediments. The structuring effect of physiochemical variables such as salinity and sediment porosity on bacterial communities is unsurprising (Hamdan et al., 2013); from a pollution perspective, chemical contamination appears to drive bacterial community structure at least at a local level (Staley et al., 2014), although FIOs appear strongly source dependent.
HTS data has provided novel insights into the dynamics of sediment associated enteric viruses (Paez-Espino et al., 2016). Predictions of viral relative abundance and potential pathogenicity genes can be undertaken with HTS of sediments (Yoshida et al., 2013). However, viral enrichment is often required for the detection of pathogenic components of the virome. Concentration through tangential ultraflow filtration has been applied on dispersed sludges. Following this, immunoprecipitation through antibodies, affinity capture has been applied to isolate pathogenic polioviruses, followed by deep HTS (Furtak et al., 2016), although to our knowledge this approach has yet to be applied for sediments. From an environmental quality perspective, understanding the physiochemical drivers governing the mainitence of the abundance and persistence of viruses and bacterial pathogens in sediments is of principal concern for regulators and requires more attention.
Conclusions
It is anticipated that enteric microorganisms in sediments will continue to be of significant interest for the foreseeable future. It is unlikely that environmental legislation will be widened to cover FIOs/pathogens in sediments; however, understanding the fate of enteric and pathogenic viruses in the environment, including sediments, will be important for implementing potential viral standards. The influence of wastewater treatment works on viral abundance and infectivity, VBNC bacteria, and the role of particulate matter and sediments on source/sinks of these organisms is still unclear. Furthermore, a lack of standardized effective methods for enumerating both VBNC and viruses from environmental matrices (including sediments) has hampered research in these areas. Significant headway on applying correction factors for viral extractions using internal standards has shed light on the problem of poor extraction efficacy and inhibition of molecular methods. However, these workaround methods have yet to be applied to sediment and do not address the fundamental quantification problems. Environmental monitoring should now apply a holistic approach using novel technologies such as “lab on chip” and structural integrity methods which may improve on site diagnostics and should include tests for viral infectivity where this is lacking, all of which are currently not applied to sediments. Further studies are also required to close the loop between conventional plate count and qPCR based methods in sediments particularly regarding the role of VBNC bacteria (if any) on bathing water quality and human/environmental health. An improved process-level understanding is required to consider if FIOs and pathogens from different sources (diffuse or point source derived or from human or animal) have different VBNC or resuscitation potentials. Complete life cycle analysis from terrestrial, fluvial and coastal zones is also required to fully understand the role of sediments in viral transport and infectivity persistence in the environment. Studies which model pathogenic viral abundance to observed FIO numbers have been successfully applied to water and applicability in sediments requires increased attention. Finally, increased acquisition of physiochemical data in addition to routine biological samples will improve our understanding of the fate of viruses, VBNCs and FIOs in sediments and enable the development of suitable environmental risk assessment for microbiological risk of sediments to human health. In conclusion, our poor process-level understanding of viral/bacterial-sediment interactions combined with methodological challenges is limiting accurate source apportionment and quantitative microbial risk assessment for pathogenic organisms associated with sediments in aquatic environments.
Author contributions
FH, CG, and KF wrote the manuscript and undertook data analysis. AA, VJ, SM, JM, DJ, and BC provided technical support and reviewed the manuscript. FH, SM, KF, JM and DJ prepared manuscript for submission. All authors have approved the final version to be published. All authors had substantial contributions to the conception and design of the work.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The authors were commissioned and funded by United Kingdom Water Industry Research Limited to carry out a revised version of this research, which was reported as chapter presented within a technical report. The funding agency did not influence the content of this review.
Acknowledgments
This work was funded by the UK Water Industry Research Limited under funding provision WW11 Coastal and Estuarine Bacteria Loads for Source Apportionment. The research was funded via a Natural Environment Research Council consortium grant under the Macronutrient Cycles Research Programme (NE/J011908/1). The authors would like to thank Dr. Peter Daldorph for advise on the manuscript.
Glossary
| Culturability | The ability of bacteria to form colonies on agar microbiology plates. |
| Viable but non-culturable | Bacteria which can no longer form colonies on agar plates, but are still metabolically active and remain viable. |
| Viable | Refers to all living bacteria |
References
- Abdelzaher A. M., Wright M. E., Ortega C., Solo-Gabriele H. M., Miller G., Elmir S., et al. (2010). Presence of pathogens and indicator microbes at a non-point source subtropical recreational marine beach. Appl. Environ. Microbiol. 76, 724–732. 10.1128/AEM.02127-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcântara F., Almeida M. A. (1995). Virological quality of the Ria de Aveiro: validity of potential microbial indicators. Neth. J. Aquat. Ecol. 29, 419–425. 10.1007/BF02084240 [DOI] [Google Scholar]
- Alkan U., Elliott D. J., Evison L. M. (1995). Survival of enteric bacteria in relation to simulated solar radiation and other environmental factors in marine waters. Water Res. 29, 2071–2080. 10.1016/0043-1354(95)00021-C [DOI] [Google Scholar]
- Allison D. G., Evans D. J., Brown M. R., Gilbert P. (1990). Possible involvement of the division cycle in dispersal of Escherichia coli from biofilms. J. Bacteriol. 172, 1667–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allwood P. B., Malik Y. S., Hedberg C. W., Goyal S. M. (2003). Survival of F-Specific RNA coliphage, feline calicivirus, and Escherichia coli in water : a comparative study. Appl. Environ. Microbiol. 69, 5707–5710. 10.1128/AEM.69.9.5707-5710.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amel B. K. N., Amine B., Amina B. (2008). Survival of Vibrio fluvialis in seawater under starvation conditions. Microbiol. Res. 163, 323–328. 10.1016/j.micres.2006.06.006 [DOI] [PubMed] [Google Scholar]
- Anderson K. L., Whitlock J. E., Harwood V. J. (2005). Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl. Environ. Microbiol. 71, 3041–3048. 10.1128/AEM.71.6.3041-3048.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana I., Orruno M., Perez-Pascual D., Seco C., Muela A., Barcina I. (2007). Inability of Escherichia coli to resuscitate from the viable but nonculturable state. FEMS Microbiol. Ecol. 62, 1–11. 10.1111/j.1574-6941.2007.00362.x [DOI] [PubMed] [Google Scholar]
- Arnold K. W., Kaspar C. W. (1995). Starvation- and stationary-phase-induced acid tolerance in Escherichia coli O157/H7. Appl. Environ. Microbiol. 61, 2037–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon S., Marx L. P., Searcy K. E., Packman A. I. (2010). Effects of overlying velocity, particle size, and biofilm growth on stream–subsurface exchange of particles. Hydrol. Process. 24, 108–114. 10.1002/hyp.7490 [DOI] [Google Scholar]
- Artz R. R., Killham K. (2002). Survival of Escherichia coli O157 : H7 in private drinking water wells : influences of protozoan grazing and elevated copper concentrations. FEMS Microbiol. Lett. 216, 117–122. 10.1111/j.1574-6968.2002.tb11424.x [DOI] [PubMed] [Google Scholar]
- Asakura H., Kawamoto K., Haishima Y., Igimi S., Yamamoto S., Makino S. (2008). Differential expression of the outer membrane protein W (OmpW) stress response in enterohemorrhagic Escherichia coli O157:H7 corresponds to the viable but non-culturable state. Res. Microbiol. 159, 709–717. 10.1016/j.resmic.2008.08.005 [DOI] [PubMed] [Google Scholar]
- Atkinson S., Williams P. (2009). Quorum sensing and social networking in the microbial world. J. R. Soc. Interface 6, 959–978. 10.1098/rsif.2009.0203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer M. T., Niehaus S. L. (1993). Modeling fecal coliform bacteria—1. field and laboratory determination of loss kinetics. Water Res. 27, 693–701. [Google Scholar]
- Avery L. M., Williams A. P., Killham K., Jones D. L. (2008). Survival of Escherichia coli O157:H7 in waters from lakes, rivers, puddles and animal-drinking troughs. Sci. Total Environ. 389, 378–385. 10.1016/j.scitotenv.2007.08.049 [DOI] [PubMed] [Google Scholar]
- Ayrapetyan M., Williams T. C., Oliver J. D. (2014a). Interspecific quorum sensing mediates the resuscitation of viable but non culturable vibrios. Appl. Environ. Microbiol. 80, 2478–2483. 10.1128/AEM.00080-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayrapetyan M., Williams T. C., Oliver J. D. (2014b). Bridging the gap between viable but non-culturable and antibiotic persistent bacteria. Trends Microbiol. 1, 7–13. 10.1016/j.tim.2014.09.004 [DOI] [PubMed] [Google Scholar]
- Bae J., Schwab K. J. (2008). Evaluation of murine norovirus, feline calicivirus, poliovirus, and MS2 as surrogates for human norovirus in a model of viral persistence in surface water and groundwater. Appl. Environ. Microbiol. 74, 477–484. 10.1128/AEM.02095-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banning N., Toze S., Mee B. J. (2002). Escherichia coli survival in groundwater and effluent measured using a combination of propidium iodide and the green fluorescent protein. J. Appl. Microbiol. 93, 69–76. 10.1046/j.1365-2672.2002.01670.x [DOI] [PubMed] [Google Scholar]
- Bari S. M. N., Roky M. K., Mohiuddin M., Kamruzzaman M., Mekalanos J. J., Faruque S. M. (2013). Quorum-sensing autoinducers resuscitate dormant Vibrio cholerae in environmental water samples. Proc. Natl. Acad. Sci. U.S.A. 110, 9926–9931. 10.1073/pnas.1307697110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman W. J., Nocker A., Jones W. L., Camper A. K. (2009). Retention of a model pathogen in a porous media biofilm. Biofouling 25, 229–240. 10.1080/08927010802688566 [DOI] [PubMed] [Google Scholar]
- Berthe T., Touron A., Leloup J., Deloffre J., Petit F. (2008). Faecal-indicator bacteria and sedimentary processes in estuarine mudflats (Seine, France). Mar. Pollut. Bull. 57, 59–67. 10.1016/j.marpolbul.2007.10.013 [DOI] [PubMed] [Google Scholar]
- Bilge S. S., Vary J. C., Dowell S. F., Tarr P. I. (1996). Role of the Escherichia coli O157:H7 O side chain in adherence and analysis of an rfb locus. Infect. Immun. 64, 4795–4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitton G., Chou Y., Farrah S. R. (1982). Techniques for virus detection in aquatic sediments. J. Virol. Methods 4, 1–8. [DOI] [PubMed] [Google Scholar]
- Bonilla T. D., Nowosielski K., Cuvelier M., Hartz A., Green M., Esiobu N., et al. (2007). Prevalence and distribution of fecal indicator organisms in South Florida beach sand and preliminary assessment of health effects associated with beach sand exposure. Mar. Pollut. Bull. 54, 1472–1482. 10.1016/j.marpolbul.2007.04.016 [DOI] [PubMed] [Google Scholar]
- Borade S., Dhawde R., Maloo A., Gajbhiye S. N., Dastager S. G. (2014). Occurrence and seasonal variation in distribution of fecal indicator bacteria in Tapi estuary along the West coast of India. Indian J. Geomarine Sci. 43, 340–347. Available online at: http://drs.nio.org/drs/handle/2264/4508 [Google Scholar]
- Bosch A., Guix S., Sano D., Pinto R. M. (2008). New tools for the study and direct surveillance of viral pathogens in water. Curr. Opin. Biotech. 19, 295–301. 10.1016/j.copbio.2008.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryers (2000). Biofilms II, Process Analysis and Applications, Ecological and Applied Microbiology. New York, NY: Wiley-Liss. [Google Scholar]
- Burton G. A., Gunnison D., Lanza G. R. (1987). Survival of pathogenic bacteria in various fresh-water sediments. Appl. Environ. Microbiol. 53, 633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busscher H. J., Bos R., van der Mei H. C. (1995). Initial microbial adhesion is a determinant for the strength of biofilm adhesion. FEMS Microbiol. Lett. 128, 229–234. [DOI] [PubMed] [Google Scholar]
- Byappanahalli M. N., Richard L., Shively D. A., Sadowsky M. J., Ishii S. (2006). Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ. Microbiol. 8, 504–513. 10.1111/j.1462-2920.2005.00916.x [DOI] [PubMed] [Google Scholar]
- Byappanahalli M. N., Fujioka R. S. (1998). Evidence that tropical soil can support the growth of Escherichia coli. Water Sci. Technol. 38:171–174. 10.1016/S0273-1223(98)00820-8 [DOI] [Google Scholar]
- Cai P., Huang Q., Walker S. L. (2013). Deposition and survival of Escherichia coli O157:H7 on clay minerals in a parallel plate flow system. Environ. Sci. Technol. 47, 1896–1903. 10.1021/es304686a [DOI] [PubMed] [Google Scholar]
- Cangelosi G. A., Weigel K. M., Lefthand-Begay C., Meschke J. S. (2010). Molecular detection of viable bacterial pathogens in water by ratiometric pre-rRNA analysis. Appl. Environ. Microbiol. 76, 960–962. 10.1128/AEM.01810-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Wei X., Cai P., Huang Q., Rong X., Liang W. (2011). Preferential adsorption of extracellular polymeric substances from bacteria on clay minerals and iron oxide. Colloids Surf. B 83, 122–127. 10.1016/j.colsurfb.2010.11.018 [DOI] [PubMed] [Google Scholar]
- Carlson G. F., Jr., Woodard F. E., Wentworth D. F., Sproul O. J. (1968). Virus inactivation on clay particles in natural waters. J. Water Pollut. Control Fed. 40, R89–R106. Available online at: http://www.jstor.org/stable/25036033 [PubMed] [Google Scholar]
- Carr M. R., Wang S. Y., McLean T. I., Flood C. J., Ellender R. D. (2010). Salmonella rarely detected in Mississippi coastal waters and sediment. J. Appl. Microbiol. 109, 2191–2199. 10.1111/j.1365-2672.2010.04851.x [DOI] [PubMed] [Google Scholar]
- Carreira C., Staal M., Middelboe M., Brussaard C. P. D. (2015). Counting viruses and bacteria in photosynthetic microbial mats. Appl. Environ. Microbiol. 81, 2149–2155. 10.1128/AEM.02863-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro F. D., Tufenkji N. (2007). Relevance of nontoxigenic strains as surrogates for Escherichia coli O157: H7 in groundwater contamination potential: role of temperature and cell acclimation time. Environ. Sci. Technol. 41, 4332–4338. 10.1021/es0701558 [DOI] [PubMed] [Google Scholar]
- Cellini L., Di Campli E., Grande R., Di Bartolomeo S., Prenna M., Pasquantonio M. S., et al. (2005). Detection of Helicobacter pylori associated with zooplankton. Aquat. Microb. Ecol. 40, 115–120. 10.3354/ame040115 [DOI] [Google Scholar]
- Chan K. Y., Wong S. H., Mak C. Y. (1979). Effects of bottom sediments on the survival of Enterobacter aerogenes in seawater. Mar. Poll. Bull. 10, 205–210. 10.1016/0025-326X(79)90534-4 [DOI] [Google Scholar]
- Charles K., Shore J., Sellwood J., Laverick M., Hart A., Pedley S. (2009). Assessment of the stability of human viruses and coliphage in groundwater by PCR and infectivity methods. J. Appl. Microbiol. 106, 1827–1837. 10.1111/j.1365-2672.2009.04150.x [DOI] [PubMed] [Google Scholar]
- Cheville A. M., Arnold K. W., Buchrieser C., Cheng C. M., Kaspar C. W. (1996). rpoS regulation of acid, heat, and salt tolerance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 62, 1822–1824. 10.1128/AEM.00863-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chigbu P., Gordon S., Tchounwou P. B. (2005). The seasonality of fecal coliform bacteria pollution and its influence on closures of shellfish harvesting areas in Mississippi Sound. Int. J. Environ. Res. Public Health 2, 362–373. 10.3390/ijerph2005020023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrysikopoulos C. V., Aravantinou A. F. (2012). Virus inactivation in the presence of quartz sand under static and dynamic batch conditions at different temperatures. J. Hazard. Mater 233–234, 148–157. 10.1016/j.jhazmat.2012.07.002 [DOI] [PubMed] [Google Scholar]
- Chrysikopoulos C. V., Syngouna V. I. (2012). Attachment of bacteriophages MS2 and Phi X174 onto kaolinite and montmorillonite: extended-DLVO interactions. Colloids Surf. B 92, 74–83. 10.1016/j.colsurfb.2011.11.028 [DOI] [PubMed] [Google Scholar]
- Cook K. L., Bolster C. H., Ayers K. A., Reynolds D. N. (2011). Escherichia coli diversity in livestock manures and agriculturally impacted stream waters. Curr. Microbiol. 63, 439–449. 10.1007/s00284-011-0002-6 [DOI] [PubMed] [Google Scholar]
- Cooper I. R., Taylor H. D., Hanlon G. W. (2007). Virulence traits associated with verocytotoxigenic Escherichia coli O157 recovered from freshwater biofilms. J. Appl. Microbiol. 102, 1293–1299. 10.1111/j.1365-2672.2006.03178.x [DOI] [PubMed] [Google Scholar]
- Costerton J. W., Lewandowski Z., Caldwell D. E., Korber D. R., Lappin-scott H. M. (1995). Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745. 10.1146/annurev.mi.49.100195.003431 [DOI] [PubMed] [Google Scholar]
- Cox P., Griffith M., Angles M., Deere D., Ferguson C. (2005). Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71, 5929–5934. 10.1128/AEM.71.10.5929-5934.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabill C., Donald R., Snelling J., Foust R., Southam G. (1999). The impact of sediment fecal coliform reservoirs on seasonal water quality in Oak Creek, Arizona. Water Res. 33, 2163–2171. 10.1016/S0043-1354(98)00437-0 [DOI] [Google Scholar]
- Czajkowska D., Witkowska-Gwiazdowska A., Sikorska I., Boszcyzk-Maleszak H., Horoch M. (2005). Survival of Escherichia coli Serotype O157 : H7 in Water and in Bottom-Shore Sediments. Pol. J. Environ. Stud. 14, 423–430. [Google Scholar]
- Dancho B. A., Chen H., Kingsley D. H. (2012). Discrimination between infectious and non-infectious human norovirus using porcine gastric mucin. Int. J. Food Microbiol. 155, 222–226. 10.1016/j.ijfoodmicro.2012.02.010 [DOI] [PubMed] [Google Scholar]
- Danovaro R., Corinaldesi C., Filippini M., Fischer U. R., Gessner M. O., Jacquet S., et al. (2008). Viriobenthos in freshwater and marine sediments: a review. Freshwater Biol. 53, 1186–1213. 10.1111/j.1365-2427.2008.01961.x [DOI] [Google Scholar]
- Darcan C., Ozkanca R., Idil O., Flint K. P. (2009). Viable but Non-Culturable State (VBNC) of Escherichia coli related to EnvZ under the effect of pH, starvation and Osmotic stress in Sea water. Pol. J. Microbiol. 58, 307–317. Available online at: http://www.pjm.microbiology.pl/archive/vol5842009307.pdf [PubMed] [Google Scholar]
- Davies C. M., Long J. A. H., Donald M., Ashbolt N. J. (1995). Survival of fecal microorganisms in marine and fresh-water sediments. Appl. Environ. Microbiol. 61, 1888–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brauwere A., Gourgue O., de Brye B., Servais P., Ouattara N. K., Deleersnijder E. (2014). Integrated modelling of faecal contamination in a densely populated river-sea continuum (Scheldt River and Estuary). Sci. Total Environ. 468, 31–45. 10.1016/j.scitotenv.2013.08.019 [DOI] [PubMed] [Google Scholar]
- de Brouwer J. F. C., Ruddy G. K., Jones T. E. R., Stal L. J. (2002). Sorption of EPS to sediment particles and the effect on the rheology of sediment slurries. Biogeochemistry 61, 57–71. 10.1023/A:1020291728513 [DOI] [Google Scholar]
- Deloffre J., Lafite R., Lesueur P., Lesourd S., Verney R., Guezennec L. (2005). Sedimentary processes on an intertidal mudflat in the upper macrotidal Seine estuary, France. Estuar. Coast. Shelf S. 64, 710–720. 10.1016/j.ecss.2005.04.004 [DOI] [Google Scholar]
- Dobrindt U., Hochhut B., Hentschel U., Hacker J. (2004). Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Micro. 2, 414–424. 10.1038/nrmicro884 [DOI] [PubMed] [Google Scholar]
- Doré W. J., Henshilwood K., Lees D. N. (2000). Evaluation of F-specific RNA bacteriophage as a candidate human enteric virus indicator for bivalve Molluscan shellfish. Appl. Environ. Microbiol. 66, 1280–1285. 10.1128/AEM.66.4.1280-1285.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd S. E., Pillai S. D., Wang S. (1998). Delineating the specific influence of virus isoelectric point and size on virus adsorption and transport through sandy soils. Appl. Environ. Microbiol. 64, 405–410. 10.1016/0043-1354(92)90205-I [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond J. D., Aubeneau A. F., Packman A. I. (2014b). Stochastic modeling of fine particulate organic carbon dynamics in rivers. Water Res. Res. 50, 4341–4356. 10.1002/2013WR014665 [DOI] [Google Scholar]
- Drummond J. D., Davies-Colley R. J., Stott R., Sukias J. P., Nagels J. W., Sharp A., et al. (2014a). Retention and remobilization dynamics of fine particles and microorganisms in pastoral streams. Water Res. 66, 459–472. 10.1016/j.watres.2014.08.025 [DOI] [PubMed] [Google Scholar]
- Duhamel S., Jacquet S. (2006). Flow cytometric analysis of bacteria- and virus-like particles in lake sediments. J. Microbiol. Methods 64, 316–332. 10.1016/j.mimet.2005.05.008 [DOI] [PubMed] [Google Scholar]
- Dulbecco R. (1952). Production of plaques in monolayer tissue cultures by single particles of an animal virus. Proc. Natl. Acad. Sci. U.S.A. 38, 747–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ervin J. S., Van De Werfhorst L. C., Murray J. L. S., Holden P. A. (2014). Microbial source tracking in a coastal California watershed reveals canines as controllable sources of fecal contamination. Environ. Sci. Technol. 48, 9043–9052. 10.1021/es502173s [DOI] [PubMed] [Google Scholar]
- Evanson M., Ambrose R. F. (2006). Sources and growth dynamics of fecal indicator bacteria in a coastal wetland system and potential impacts to adjacent waters. Water Res. 40, 475–486. 10.1016/j.watres.2005.11.027 [DOI] [PubMed] [Google Scholar]
- Farkas K., Varsani A., Pang L. (2015). Adsorption of rotavirus, MS2 bacteriophage and surface-modified silica nanoparticles to hydrophobic matter. Food Environ. Virol. 7, 261–268. 10.1007/s12560-014-9171-3 [DOI] [PubMed] [Google Scholar]
- Ferguson C. M., Coote B. G., Ashbolt N. J., Stevenson I. M. (1996). Relationships between indicators, pathogens and water quality in an estuarine system. Water Res. 30, 2045–2054. 10.1016/0043-1354(96)00079-6 [DOI] [Google Scholar]
- Filip Z. (1973). Clay minerals as a factor influencing biochemical activity of soil microorganisms. Folia Microbiol. 18, 56–74. [DOI] [PubMed] [Google Scholar]
- Filippini M., Middelboe M. (2007). Viral abundance and genome size distribution in the sediment and water column of marine and freshwater ecosystems. FEMS Microbiol. Ecol. 60, 397–410. 10.1128/AEM.00319-06 [DOI] [PubMed] [Google Scholar]
- Fongaro G., Do Nascimento M. A., Rigotto C., Ritterbusch G., da Silva A. D., Esteves P. A., et al. (2013). Evaluation and molecular characterization of human adenovirus in drinking water supplies: viral integrity and viability assays. Virol. J. 10, 1. 10.1186/1743-422X-10-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foppen J. W. A., Schijven J. F. (2006). Evaluation of data from the literature on the transport and survival of Escherichia coli and thermotolerant coliforms in aquifers under saturated conditions. Water Res. 40, 401–426. 10.1016/j.watres.2005.11.018 [DOI] [PubMed] [Google Scholar]
- Foppen J. W., Lutterodt G., Roling W. F. M., Uhlenbrook S. (2010). Towards understanding inter-strain attachment variations of Escherichia coli during transport in saturated quartz sand. Water Res. 44, 1202–1212. 10.1016/j.watres.2009.08.034 [DOI] [PubMed] [Google Scholar]
- Fu Z. F., Hampson D. J., Blackmore D. K. (1989). Detection and survival of group A rotavirus in a piggery. Vet. Rec. 125, 576–578. 10.1136/vr.125.23.576 [DOI] [PubMed] [Google Scholar]
- Fukushima H., Seki R. (2004). Ecology of Vibrio vulnificus and Vibrio parahaemolyticus in brackish environments of the Sada River in Shimane Prefecture, Japan. FEMS Microbiol. Ecol. 48, 221–229. 10.1016/j.femsec.2004.01.009 [DOI] [PubMed] [Google Scholar]
- Furtak V., Roivainen M., Mirochnichenko O., Zagorodnyaya T., Laassri M., Zaidic S. Z., et al. (2016). Environmental surveillance of viruses by tangential flow filtration and metagenomic reconstruction. Eurosurveillance 21, 40–50. 10.2807/1560-7917.ES.2016.21.15.30193 [DOI] [PubMed] [Google Scholar]
- Garzio-Hadzick A., Shelton D. R., Hill R. L., Pachepsky Y. A., Guber A. K., Rowland R. (2010). Survival of manure-borne E. coli in streambed sediment: effects of temperature and sediment properties. Water Res. 44, 2753–2762. 10.1016/j.watres.2010.02.011 [DOI] [PubMed] [Google Scholar]
- Geldreich E. E., Fox K. R., Goodrich J. A., Rice E. W., Clark R. M., Swerdlow D. L. (1992). Searching for a water-supply connection in the Cabool, Missouri sisease outbreak of Escherichia-coli O157-H7. Water Res. 26, 1127–1137. 10.1016/0043-1354(92)90150-3 [DOI] [Google Scholar]
- Gerba C. P., Goyal S. M., Hurst C. J., LaBelle R. L. (1980). Type and strain dependence of Enterovirus adsorption to activated-sludge, soils and estuarine sediments. Water Res. 14, 1197–1198. 10.1016/0043-1354(80)90176-1 [DOI] [Google Scholar]
- Gerba C. P., Goyal S. M., Smith E. M., Melnick J. L. (1977a). Distribution of viral and bacterial pathogens in a coastal canal community. Mar. Poll. Bull. 8, 279–282. 10.1016/0025-326X(77)90170-9 [DOI] [Google Scholar]
- Gerba C. P., McLeod J. S. (1976). Effect of sediments on survival of Escherichia coli in marine waters. Appl. Environ. Microbiol. 32, 114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerba C. P., Schaiberger G. E. (1975). Effect of particulates on virus survival in seawater. J. Water Pollut. Con. F. 47, 93–103. [PubMed] [Google Scholar]
- Gerba C. P., Smith E. M., Melnick J. L. (1977b). Development of a quantitative method for detecting enteroviruses in estuarine sediments. Appl. Environ. Microbiol. 34, 158–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert P., Evans D. J., Evans E., Duguid I. G., Brown M. R. W. (1991). Surface characteristics and adhesion of Escherichia coli and Staphylococcus epidermidis. J. Appl. Bacteriol. 71, 72–77. 10.1111/j.1365-2672.1991.tb04665.x [DOI] [PubMed] [Google Scholar]
- Gin K. Y.-H., Goh S. G. (2013). Modeling the effect of light and salinity on viable but non-culturable (VBNC) Enterococcus. Water Res. 47, 3315–3328. 10.1016/j.watres.2013.03.021 [DOI] [PubMed] [Google Scholar]
- Girones R., Ferrus M. A., Alonso J. L., Rodriguez-Manzano J., Calgua B., de Abreu Corrêa A., et al. (2010). Molecular detection of pathogens in water–the pros and cons of molecular techniques. Water Res. 44, 4325–4339. 10.1016/j.watres.2010.06.030 [DOI] [PubMed] [Google Scholar]
- Goldscheider N., Pronk M., Zopfi J. (2010). New insights into the transport of sediments and microorganisms in karst groundwater by continuous monitoring of particle-size distribution. Geol. Croat. 63, 137–142. 10.4154/gc.2010.10 [DOI] [Google Scholar]
- Gomez-Suarez C., Busscher H. J., van der Mei H. C. (2001). Analysis of bacterial detachment from substratum surfaces by the passage of air-liquid interfaces. Appl. Environ. Microbiol. 67, 2531–2537. 10.1128/AEM.67.6.2531-2537.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottenbos B., Grijpma D. W., van der Mei H. C., Feijen J., Busscher H. J. (2001). Antimicrobial effects of positively charged surfaces on adhering Gram-positive and Gram-negative bacteria. J. Antimicrob. Chemother. 48, 7–13. 10.1093/jac/48.1.7 [DOI] [PubMed] [Google Scholar]
- Goyal S. M., Gerba C. P., Melnick G. L. (1977). Occurrence and distribution of bacterial indicators and pathogens in canal communities along the Texas Coast. Appl. Environ. Microbiol. 43, 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green D. H., Lewis G. D. (1999). Comparative detection of enteric viruses in wastewaters, sediments and oysters by reverse transcription PCR and cell culture. Water Res. 33, 1195–1200. 10.1016/S0043-1354(98)00313-3 [DOI] [Google Scholar]
- Greening G., Hewitt J., Lewis G. (2002). Evaluation of integrated cell culture-PCR (C-PCR) for virological analysis of environmental samples. J. Appl. Microbiol. 93, 745–750. 10.1046/j.1365-2672.2002.01741.x [DOI] [PubMed] [Google Scholar]
- Guizien K., Dupuy C., Ory P., Montanie H., Hartmann H., Chatelain M., et al. (2014). Microorganism dynamics during a rising tide: disentangling effects of resuspension and mixing with offshore waters above an intertidal mudflat. J. Mar. Syst. 129, 178–188. 10.1016/j.jmarsys.2013.05.010 [DOI] [Google Scholar]
- Gwyther C. L., Jones D. L., Golyshin P. N., Edwards-Jones G., McKillen J., McNair, et al. (2013). Bioreduction of sheep carcasses effectively contains and reduces pathogen levels under operational and simulated breakdown conditions. Environ. Sci. Technol. 47, 5267–5275. 10.1021/es400183z [DOI] [PubMed] [Google Scholar]
- Hadjiev D., Dimitrov D., Martinov M., Sire O. (2007). Enhancement of the biofilm formation on polymeric supports by surface conditioning. Enzyme Microb. Tech. 40, 840–848. 10.1016/j.enzmictec.2006.06.022 [DOI] [Google Scholar]
- Haller L., Amedegnato E. (2009). Influence of freshwater sediment characteristics on persistence of fecal indicator bacteria. Water Air Soil Poll. 203, 217–227. 10.1007/s11270-009-0005-0 [DOI] [Google Scholar]
- Halpern M., Landsberg O., Raats D., Rosenberg E. (2007). Culturable and VBNC Vibrio cholerae: interactions with chironomid egg masses and their bacterial population. Microb. Ecol. 53, 285–293. 10.1007/s00248-006-9094-0 [DOI] [PubMed] [Google Scholar]
- Hamdan L. J., Coffin R. B., Sikaroodi M., Greinert J., Treude T., Gillevet P. M. (2013). Ocean currents shape the microbiome of Arctic marine sediments. ISME J. 7, 685–696. 10.1038/ismej.2012.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey J. W., Drummond J. D., Martin R. L., McPhillips L. E., Packman A. I., Jerolmack D. J., et al. (2012). Hydrogeomorphology of the hyporheic zone: stream solute and fine particle interactions with a dynamic streambed. J. Geophys. Res. Biogeosci. 117, G00N11. 10.1029/2012JG002043 [DOI] [Google Scholar]
- Hassard F., Cartmell E., Biddle J., Stephenson T. (2014). Performance of permeable media rotating reactors used for pretreatment of wastewaters. Water Sci. Technol. 69, 1926–1931. 10.2166/wst.2014.105 [DOI] [PubMed] [Google Scholar]
- Hazen R. M., Sverjensky D. A. (2010). Mineral surfaces, geochemical complexities, and the origins of life. Cold Spring Harb. Perspect. Biol. 2, a002162. 10.1101/cshperspect.a002162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L. M., He Z. L. (2008). Water quality prediction of marine recreational beaches receiving watershed baseflow and stormwater runoff in southern California, USA. Water Res. 42, 2563–2573. 10.1016/j.watres.2008.01.002 [DOI] [PubMed] [Google Scholar]
- Heijnen C. E., Hokahin C. H., Vanveen J. A. (1991). Protection of Rhizobium by bentonite clay against predation by flagellates in liquid cultures. FEMS Microbiol. Ecol. 85, 65–72. 10.1111/j.1574-6941.1991.tb01709.x [DOI] [Google Scholar]
- Henry R., Schang C., Coutts S., Kolotelo P., Prosser T., Crosbie N., et al. (2016). Into the deep: evaluation of SourceTracker for assessment of faecal contamination of coastal waters. Water Res. 93, 242–253. 10.1016/j.watres.2016.02.029 [DOI] [PubMed] [Google Scholar]
- Henson S. S., Ahearn D. S., Dahlgren R. A., Nieuwenhuyse E., Van Tate K. W., Fleenor W. E. (2007). Water quality response to a pulsed-flow event on the Mokelumne river, California. River Res. Appl. 23, 185–200. 10.1002/rra.973 [DOI] [Google Scholar]
- Hermansson M. (1999). The DLVO theory in microbial adhesion. Colloids Surf., B 14, 105–119. 10.1016/S0927-7765(99)00029-6 [DOI] [Google Scholar]
- Hong Y., Brown D. G. (2009). Variation in bacterial ATP level and proton motive force due to adhesion to a solid surface. Appl. Environ. Microbiol. 75, 2346–2353. 10.1128/AEM.02671-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell J. M., Coyne M. S., Cornelius P. L. (1996). Effect of sediment particle size and temperature on fecal bacteria mortality rates and the fecal coliform/fecal streptococci ratio. J. Environ. Qual. 25, 1216–1220. 10.2134/jeq1996.00472425002500060007x [DOI] [Google Scholar]
- Howlett E. R., Bowers D. G., Malarkey J., Jago C. F. (2015). Stratification in the presence of an axial convergent front: Causes and implications. Estuar. Coast. Shelf. Sci. 161, 1–10. 10.1016/j.ecss.2015.04.003 [DOI] [Google Scholar]
- Huang G., Falconer R. A., Lin B. (2015). Integrated river and coastal flow, sediment and Escherichia coli modelling for bathing water quality. Water 7, 4752–4777. 10.3390/w7094752 [DOI] [Google Scholar]
- Ikner L. A., Gerba C. P., Bright K. R. (2012). Concentration and recovery of viruses from water: a comprehensive review. Food Environ. Virol. 4, 41–67. 10.1007/s12560-012-9080-2 [DOI] [PubMed] [Google Scholar]
- Ishii S., Ksoll W. B., Hicks R. E., Sadowsky M. J. (2006). Presence and growth of naturalized Escherichia coli in temperate soils from lake superior watersheds. Appl. Environ. Microbiol. 72, 612–621. 10.1128/AEM.72.1.612-621.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson R. C., Joy D. M., Lee H., Kostaschuk R., Gordon R. J. (2005). Resuspension of sediment-associated Escherichia coli in a natural stream. J. Environ. Qual., 34, 581–589. 10.2134/jeq2005.0581 [DOI] [PubMed] [Google Scholar]
- Jamieson R., Joy D. M., Lee H., Kostaschuk R., Gordon R. (2005). Transport and deposition of sediment-associated Escherichia coli in natural streams. Water Res. 39, 2665–2675. 10.1016/j.watres.2005.04.040 [DOI] [PubMed] [Google Scholar]
- Jiang D., Huang Q., Cai P., Rong X., Chen W. (2007). Adsorption of Pseudomonas putida on clay minerals and iron oxide. Colloids Surf. B 54, 217–221. 10.1016/j.colsurfb.2006.10.030 [DOI] [PubMed] [Google Scholar]
- Jin Y., Flury M. (2002). Fate and transport of viruses in porous media. Adv. Agron. 77, 39–102. 10.1016/S0065-2113(02)77013-2 [DOI] [Google Scholar]
- John D. E., Rose J. B. (2005). Review of factors affecting microbial survival in groundwater. Environ. Sci. Technol. 39, 7345–7356. 10.1021/es047995w [DOI] [PubMed] [Google Scholar]
- Johnson R. A., Ellender R. D., Tsai S. C. (1984). Elution of enteric viruses from Mississippi estuarine sediments with lecithin-supplemented eluents. Appl. Environ. Microbiol. 48, 581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junkins A. D., Doyle M. P. (1992). Demonstration of exopolysaccharide production by enterohemorrhagic Escherichia coli. Curr. Microbiol. 25, 9–17. 10.1007/BF01570076 [DOI] [PubMed] [Google Scholar]
- Kay D., Stapleton C. M., Wyer M. D., McDonald A. T., Crowther J., Paul N., et al. (2005). Decay of intestinal enterococci concentrations in high-energy estuarine and coastal waters: towards real-time T90 values for modelling faecal indicators in recreational waters. Water Res. 39, 655–667. 10.1016/j.watres.2004.11.014 [DOI] [PubMed] [Google Scholar]
- Kerr M., Fitzgerald M., Sheridan J. J., McDowell D. A., Blair I. S. (1999). Survival of Escherichia coli O157: H7 in bottled natural mineral water. J. Appl. Microbiol. 87, 833–841. 10.1046/j.1365-2672.1999.00928.x [DOI] [PubMed] [Google Scholar]
- Khan M. M. T., Pyle B. H., Camper A. K. (2010). Specific and rapid enumeration of viable but nonculturable and viable-culturable gram-negative bacteria by using flow cytometry. Appl. Environ. Microbiol. 76, 5088–5096. 10.1128/AEM.02932-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierek K., Watnick P. I. (2003). The Vibrio cholerae O139O-antigen polysaccharide is essential for Ca2+−dependent biofilm development in sea water. Proc. Natl. Acad. Sci. U.S.A. 100, 14357–14362. 10.1073/pnas.2334614100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. K., Ingremeau F., Zhao A., Bassler B. L., Stone H. A. (2016). Local and global consequences of flow on bacterial quorum sensing. Nat. Microbiol. 1, 15005. 10.1038/nmicrobiol.2015.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner A. K. T., Zechmeister T. C., Kavka G. G., Beiwl C., Herzig A., Mach R. L., et al. (2004). Integral strategy for evaluation of Fecal indicator performance in Bird-Influenced Saline Inland waters. Appl. Environ. Microbiol. 70, 7396–7403. 10.1128/AEM.70.12.7396-7403.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Iker B. C., Pepper I. L., Gerba C. P. (2014). Relative abundance and treatment reduction of viruses during wastewater treatment processes - Identification of potential viral indicators viral indicators. Sci. Total Environ. 488–489, 290–296. 10.1016/j.scitotenv.2014.04.087 [DOI] [PubMed] [Google Scholar]
- Klayman B. J., Volden P. A., Stewart P. S., Camper A. K. (2009). Escherichia coli O157:H7 requires colonizing partner to adhere and persist in a capillary flow cell. Environ. Sci. Technol. 43, 2105–2111. 10.1021/es802218q [DOI] [PubMed] [Google Scholar]
- Korajkic A., Wanjugi P., Harwood V. J. (2013). Indigenous microbiota and habitat influence Escherichia coli survival more than sunlight in simulated aquatic environments. Appl. Environ. Microbiol. 79, 5329–5337. 10.1128/AEM.01362-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber D. R., Choi A., Wolfaardt G. M., Ingham S. C., Caldwell D. E. (1997). Substratum topography influences susceptibility of Salmonella enteritidis biofilms to trisodium phosphate. Appl. Environ. Microbiol. 63, 3352–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBelle R. L., Gerba C. P. (1979). Influence of pH, salinity, and organic-matter on the adsorption of enteric viruses to estuarine sediment. Appl. Environ. Microbiol. 38, 93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBelle R. L., Gerba C. P. (1980). Influence of estuarine sediment on virus survival under field conditions. Appl. Environ. Microbiol. 39, 749–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. G., Park S. J. O. O., Kim S. (2007). Influence of pipe materials and VBNC Cells on culturable bacteria in a chlorinated drinking water model system. J. Microbiol. Biotechn. 17, 1558–1562. [PubMed] [Google Scholar]
- Le Guyader F., Dubois E., Menard D., Pommepuy M. (1994). Detection of hepatitis A virus, rotavirus, and enterovirus in naturally contaminated shellfish and sediment by reverse transcription-seminested PCR. Appl. Environ. Microbiol. 60, 3665–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeJeune J. T., Besser T. E., Merrill N. L., Rice D. H., Hancock D. D. (2001). Livestock drinking water microbiology and the factors influencing the quality of drinking water offered to cattle. J. Dairy Sci. 84, 1856–1862. 10.3168/jds.S0022-0302(01)74626-7 [DOI] [PubMed] [Google Scholar]
- Lemos M., Mergulhγo F., Melo L., Simões M. (2014). Food and Bioproducts Processing The effect of shear stress on the formation and removal of Bacillus cereus biofilms. Food Bioprod. Process. 93, 242–248. 10.1016/j.fbp.2014.09.005 [DOI] [Google Scholar]
- Lewis G. D., Loutit M. W., Austin F. J. (1985). A method for detecting human enteroviruses in aquatic sediments. J. Virol. Methods 10, 153–162. [DOI] [PubMed] [Google Scholar]
- Lewis G. D., Metcalf T. G. (1988). Polyethylene glycol precipitation for recovery of pathogenic viruses, including hepatitis A virus and human rotavirus, from oyster, water, and sediment samples. Appl. Environ. Microbiol. 54, 1983–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Mendis N., Triguri H., Oliver J. D., Faucher S. (2014). The importance of VBNC state in human bacterial pathogens. Front. Microbiol. 5:258. 10.3389/fmicb.2014.00258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieleg O., Ribbeck K. (2011). Biological hydrogels as selective diffusion barriers. Trends Cell Biol. 21, 543–551. 10.1016/j.tcb.2011.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew P. F., Gerba C. P. (1980). Thermostabilization of enteroviruses by estuarine sediment. Appl. Environ. Microbiol. 40, 305–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorite G. S., Rodrigues C. M., Souza A. A., De Kranz C., Mizaikoff B., Cotta M. A. (2011). The role of conditioning film formation and surface chemical changes on Xylella fastidiosa adhesion and biofilm evolution. J. Colloid Interface Sci. 359, 289–295. 10.1016/j.jcis.2011.03.066 [DOI] [PubMed] [Google Scholar]
- Lothigius A., Sjöling A., Svennerholm A. M., Bölin I. (2010). Survival and gene expression of enterotoxigenic Escherichia coli during long-term incubation in sea water and freshwater. J. Appl. Microbiol. 108, 1441–1449. 10.1111/j.1365-2672.2009.04548.x [DOI] [PubMed] [Google Scholar]
- Lowther J. A., Gustar N. E., Powell A. L., Hartnell R. E., Lees D. N. (2012). Two-year systematic study to assess norovirus contamination in oysters from commercial harvesting areas in the United Kingdom. Appl. Environ. Microbiol. 78, 5812–5817. 10.1128/AEM.01046-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucena F., Araujo R., Jofre J. (1996). Usefulness of bacteriophages infecting Bacteroides fragilis as index microorganisms of remote faecal pollution. Water Res. 30, 2812–2816. 10.1016/S0043-1354(96)00193-5 [DOI] [Google Scholar]
- Luna G. M., Vignaroli C., Rinaldi C., Pusceddu A., Nicoletti L., Gabellini M., et al. (2010). Extraintestinal Escherichia coli carrying virulence genes in coastal marine sediments. Appl. Environ. Microbiol. 76, 5659–5668. 10.1128/AEM.03138-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen K. N., Nilsson P., Sundback K. (1993). The influence of benthic microalgae on the stability of a subtidal sediment. J. Exp. Mar. Biol Ecol. 170, 159–177. 10.1016/0022-0981(93)90150-M [DOI] [Google Scholar]
- Malham S. K., Rajko-Nenow P., Howlett E., Tuson K. E., Perkins T. L., Pallett D. W., et al. (2014). The interaction of human microbial pathogens, particulate material and nutrients in estuarine environments and their impacts on recreational and shellfish waters. Env. Sci. Process. Impact 16, 2145–2155. 10.1039/C4EM00031E [DOI] [PubMed] [Google Scholar]
- Malic S., Hill K. E., Hayes A., Percival S. L., Thomas D. W., Williams D. W. (2009). Detection and identification of specific bacteria in wound biofilms using peptide nucleic acid fluorescent in situ hybridization (PNA FISH). Microbiology 155, 2603–2611. 10.1099/mic.0.028712-0 [DOI] [PubMed] [Google Scholar]
- Maule A. (2000). Survival of verocytotoxigenic Escherichia coli 0157 in soil, water and on surfaces. Symp. J. Appl. Microbiol. 88, 71S–78S. 10.1111/j.1365-2672.2000.tb05334.x [DOI] [PubMed] [Google Scholar]
- McGee P., Bolton D. J., Sheridan J. J., Earley B., Kelly G., Leonard N. (2002). Survival of Escherichia coli O157:H7 in farm water: its role as a vector in the transmission of the organism within herds. J. Appl. Microbiol. 93, 706–713. 10.1046/j.1365-2672.2002.01752.x [DOI] [PubMed] [Google Scholar]
- Meays C. L., Broersma K., Nordin R., Mazumder A., Samadpour M. (2006). Diurnal variability in concentrations and sources of Escherichia coli in three streams. Can. J. Microbiol. 52, 1130–1135. 10.1139/w06-066 [DOI] [PubMed] [Google Scholar]
- Melnick J. L., Rennick V. (1980). Infectivity titers of enterovirus as found in human stools. J. Med. Virol. 5, 205–220. 10.1002/jmv.1890050305 [DOI] [PubMed] [Google Scholar]
- Meschke J., Sobsey M. (1998). Comparative adsorption of Norwalk virus, poliovirus 1 and F RNA coliphage MS2 to soils suspended in treated wastewater. Water Sci. Technol. 38, 187–189. 10.1016/S0273-1223(98)00823-3 [DOI] [PubMed] [Google Scholar]
- Mills A. L., Powelson D. K. (1996). Bacterial interactions with surfaces in soils, in Bacterial Adhesion Molecular and Ecological Diversity, ed Fletcher M. (New York, NY: Wiley-Liss Inc.), 25–58. [Google Scholar]
- Miura T., Masago Y., Sano D., Omura T. (2011). Development of an effective method for recovery of viral genomic RNA from environmental silty sediments for quantitative molecular detection. Appl. Environ. Microbiol. 77, 3975–3981. 10.1128/AEM.02692-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moce-Llivina L., Lucena F., Jofre J. (2004). Double-layer plaque assay for quantification of enteroviruses. Appl. Environ. Microbiol. 70, 2801–2805. 10.1128/AEM.70.5.2801-2805.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muirhead R. W., Collins R. P., Bremer P. J. (2006). Interaction of Escherichia coli and soil particles in runoff. Appl. Environ. Microbiol. 72, 3406–3411. 10.1128/AEM.72.5.3406-3411.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasser A., Tchorch Y., Fattal B. (1995). Validity of serological methods (ELISA) for detecting infectious viruses in water. Water Sci. Technol. 31, 307–310. [Google Scholar]
- Neave M., Luter H., Padovan A., Townsend S., Schobben X., Gibb K. (2014). Multiple approaches to microbial source tracking in tropical northern Australia. Microbiologyopen 3, 860–874. 10.1002/mbo3.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nocker A., Sossa-Fernandez P., Burr M. D., Camper A. K. (2007). Use of propidium monoazide for live/dead distinction in microbial ecology. Appl. Environ. Microbiol. 73, 5111–5117. 10.1128/AEM.02987-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman S. A., Hobbs R. C., Wuertz S., Melli A., Beckett L. A., Chouicha N., et al. (2013). Fecal pathogen pollution: sources and patterns in water and sediment samples from the upper Cook Inlet, Alaska ecosystem. Env. Sci. Process. Impact 15, 1041–1051. 10.1039/c3em30930d [DOI] [PubMed] [Google Scholar]
- Obiri-Danso K., Jones K. (2000). Intertidal sediments as reservoirs for hippurate negative campylobacters, salmonellae and faecal indicators in three EU recognised bathing waters in North West England. Water Res. 34, 519–527. 10.1016/S0043-1354(99)00146-3 [DOI] [Google Scholar]
- Ogorzaly L., Cauchie H., Penny C., Perrin A., Gantzer C., Bertrand I. (2013). Two-day detection of infectious enteric and non-enteric adenoviruses by improved ICC-qPCR. Appl. Microbiol. Biotechnol. 97, 4159–4166. 10.1007/s00253-013-4782-4 [DOI] [PubMed] [Google Scholar]
- Ohtomo R., Saito M. (2001). Increase in the culturable cell number of Escherichia coli during recovery from saline stress: possible implication for resuscitation from the VBNC state. Microb. Ecol. 42, 208–214. 10.1007/s002480000103 [DOI] [PubMed] [Google Scholar]
- Oliver J. D. (2005). The viable but nonculturable state in bacteria. J. Microbiol. 43, 93–100. [PubMed] [Google Scholar]
- Oliver J. D. (2010). Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol. Rev. 34, 415–425. 10.1111/j.1574-6976.2009.00200.x [DOI] [PubMed] [Google Scholar]
- Oliver J. D., Bockian R. (1995). In-vivo resuscitation, and virulence towards mice, of viable but nonculturable cells of Vibrio vulnificus. Appl. Environ. Microbiol. 61, 2620–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver J. D., Hite F., McDougald D., Andon N. L., Simpson L. M. (1995). Entry into, and resuscitation from, the viable but nonculturable state by Vibrio vulnificus in an estuarine environment. Appl. Environ. Microbiol. 61, 2624–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouattara N. K., de Brauwere A., Billen G., Servais P. (2013). Modelling faecal contamination in the Scheldt drainage network. J. Mar. Syst. 128, 77–88. 10.1016/j.jmarsys.2012.05.004 [DOI] [Google Scholar]
- Ouattara N. K., Passerat J., Servais P. (2011). Faecal contamination of water and sediment in the rivers of the Scheldt drainage network. Environ. Monit. Assess. 183, 243–257. 10.1007/s10661-011-1918-9 [DOI] [PubMed] [Google Scholar]
- Pachepsky Y. A., Guber A. K., Shelton D. R., Hil R. L. (2009a). E. coli re-suspension during an artificial high-flow event in a small first-order creek. Geophys. Res. Abstr. 11, EGU2009–EGU9880. [Google Scholar]
- Pachepsky Y. A., Shelton D. R. (2011). Escherichia coli and fecal coliforms in freshwater and estuarine sediments. Crit. Rev. Env. Sci. Tec. 41, 1067–1110. 10.1080/10643380903392718 [DOI] [Google Scholar]
- Pachepsky Y. A., Yu O., Karns J. S., Shelton D. R., Guber A. K., van Kessel J. S. (2009b). Strain-dependent variations in attachment of E. coli to soil particles of different sizes. Int. Agrophys. 22, 61–66. [Google Scholar]
- Paez-Espino D., Eloe-Fadrosh E. A., Pavlopoulos G. A., Thomas A. D., Huntemann M., Mikhailova N., et al. (2016). Uncovering Earth's virome. Nature 536, 425–430. 10.1038/nature19094 [DOI] [PubMed] [Google Scholar]
- Pang L., Farkas K., Bennett G., Varsani A., Easingwood R., Tilley R., et al. (2014). Mimicking filtration and transport of rotavirus and adenovirus in sand media using DNA-labeled, protein-coated silica nanoparticles. Water Res. 62, 167–179. 10.1016/j.watres.2014.05.055 [DOI] [PubMed] [Google Scholar]
- Park S., Kim E., Yun T., Lee J., Kim C., Seo Y., et al. (2010). Human enteric viruses in groundwater. Food Environ. Virol. 2, 69–73. 10.1007/s12560-010-9029-2 [DOI] [Google Scholar]
- Passerat J., Got P., Dukan S., Monfort P. (2009). Respective roles of culturable and viable-but-nonculturable cells in the heterogeneity of Salmonella enterica serovar typhimurium invasiveness. Appl. Environ. Microbiol. 75, 5179–5185. 10.1128/AEM.00334-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H., Jiang S. C., Rose J. B. (1991). Concentration of viruses and dissolved DNA from aquatic environments by vortex flow filtration. Appl. Environ. Microbiol. 57, 2197–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne A. F., Binduga-Gajewska I., Kauffman E. B., Kramer L. D. (2006). Quantitation of flaviviruses by fluorescent focus assay. J. Virol. Methods 134, 183–189. 10.1016/j.jviromet.2006.01.003 [DOI] [PubMed] [Google Scholar]
- Peneau S., Chassaing D., Carpentier B. (2007). First evidence of division and accumulation of viable but nonculturable Pseudomonas fluorescens cells on surfaces subjected to conditions encountered at meat processing premises. Appl. Environ. Microbiol. 73, 2839–2846. 10.1128/AEM.02267-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins T. L., Clements K., Baas J. H., Jago C. F., Jones D. L., Malham S. K., et al. (2014). Sediment composition influences spatial variation in the abundance of human pathogen indicator bacteria within an estuarine environment. PLoS ONE 9:112951. 10.1371/journal.pone.0112951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petterson S., Grøndahl-Rosado R., Nilsen V., Myrmel M., Robertson L. J. (2015). Variability in the recovery of a virus concentration procedure in water: implications for QMRA. Water Res. 87, 79–86. 10.1016/j.watres.2015.09.006 [DOI] [PubMed] [Google Scholar]
- Piggot A. M., Klaus J. S., Johnson S., Phillips M. C., Solo-Gabriele H. M. (2012). Relationship between enterococcal levels and sediment biofilms at recreational beaches in South Florida. Appl. Environ. Microbiol. 78, 5973–5982. 10.1128/AEM.00603-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Santos M. A., Chambel L. (2015). Thirty years of viable but nonculturable state research: unsolved molecular mechanisms. Crit. Rev. Microbiol. 41, 61–76. 10.3109/1040841X.2013.794127 [DOI] [PubMed] [Google Scholar]
- Port J. A., Cullen A. C., Wallace J. C., Smith M. N., Faustman E. M. (2014). Metagenomic frameworks for monitoring antibiotic resistance in aquatic environments. Environ. Health Perspect. 122, 222–228. 10.1289/ehp.1307009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen L. K., Licht T. R., Rang C., Krogfelt K. A., Molin S. (1995). Physiological-state of Escherichia coli BJ4 growing in the large-intestines of streptomycin-treated mice. J. Bacteriol. 177, 5840–5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Props R., Kerckhof F.-M., Rubbens P., De Vrieze J., Hernandez Sanabria E., Waegeman W., et al. (2016). Absolute quantification of microbial taxon abundances. ISME J. [Epub ahead of print]. 10.1038/ismej.2016.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quignon F., Sardin M., Kiene L., Schwartzbrod L. (1997). Poliovirus-1 inactivation and interaction with biofilm: a pilot-scale study. Appl. Environ. Microbiol. 63, 978–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilliam R. S., Clements K., Duce C., Cottrill S. B., Malham S. K., Jones D. L. (2011a). Spatial variation of waterborne Escherichia coli - implications for routine water quality monitoring. J. Water Health 9, 734–737. 10.2166/wh.2011.057 [DOI] [PubMed] [Google Scholar]
- Quilliam R. S., Williams A. P., Avery L. M., Malham S. K., Jones D. L. (2011b). Unearthing human pathogens at the agricultural-environment interface: a review of current methods for the detection of Escherichia coli O157 in freshwater ecosystems. Agric. Ecosys. Environ. 140, 354–360. 10.1016/j.agee.2011.01.019 [DOI] [Google Scholar]
- Ramaiah N., Kolhe V., Sadhasivan A. (2005). Quantitative analyses of pollution-indicator and pathogenic bacteria in Mumbai waters from ballast water exchange perspective. Environ. Monit. Assess. 104, 295–308. 10.1007/s10661-005-1619-3 [DOI] [PubMed] [Google Scholar]
- Ramamurthy T., Ghosh A., Pazhani G. P., Shinoda S. (2014). Current perspectives on viable but non-culturable (VBNC) Pathogenic bacteria. Front. Public Health, 2:103. 10.3389/fpubh.2014.00103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randa M. A., Polz M. F., Lim E. (2004). Effects of temperature and salinity on Vibrio vulnificus population dynamics as assessed by quantitative PCR. Appl. Environ. Microbiol. 70, 5469–5476. 10.1128/AEM.70.9.5469-5476.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao V. C., Metcalf T. G., Melnick J. L. (1986a). Human viruses in sediments, sludges, and soils. Bull. World Health Organ. 64, 1–13. [PMC free article] [PubMed] [Google Scholar]
- Rao V. C., Metcalf T. G., Melnick J. L. (1986b). Development of a method for concentration of rotavirus and its application to recovery of rotaviruses from estuarine waters. Appl. Environ. Microbiol. 52, 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao V. C., Seidel K. M., Goyal S. M., Metcalf T. G., Melnick J. L. (1984). Isolation of enteroviruses from water, suspended-solids, and sediments from Galveston Bay - survival of poliovirus and rotavirus adsorbed to sediments. Appl. Environ. Microbiol. 48, 404–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Packman A. (2002). Effects of Background Water Composition on Stream–Subsurface Exchange of Submicron Colloids. J. Environ. Eng. 128, 624–634. 10.1061/(ASCE)0733-9372(2002)128:7(624) [DOI] [Google Scholar]
- Rice E. W., Johnson C. H. (2000). Short communication: survival of Escherichia coli O157:H7 in dairy cattle drinking water. J. Dairy Sci. 83, 2021–2023. 10.3168/jds.S0022-0302(00)75081-8 [DOI] [PubMed] [Google Scholar]
- Rice E. W., Johnson C. H., Wild D. K, Reasoner D. J. (1992). Survival of Escherichia coli O157: H7 in drinking water associated with a Waterborne Disease Outbreak of Hemorrhagic Colitis. Lett. Appl. Microbiol. 7, 38–40. 10.1111/j.1472-765X.1992.tb00719.x [DOI] [Google Scholar]
- Ritchie J. M., Campbell G. R., Shepherd J., Beaton Y., Jones D., Killham K., et al. (2003). A stable bioluminescent construct of Escherichia coli O157:H7 for hazard assessments of long-term survival in the environment. Appl. Environ. Microbiol. 69, 3359–3367. 10.1128/AEM.69.6.3359-3367.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock C., Alum A., Abbaszadegan M. (2010). PCR inhibitor levels in concentrates of biosolid samples predicted by a new method based on excitation-emission matrix spectroscopy. Appl. Environ. Microbiol. 76, 8102–8109. 10.1128/AEM.02339-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper M. M., Marshall K. C. (1974). Modification of the interaction between Escherichia coli and Bacteriophage in saline sediment. Microb. Ecol. 1, 1–13. 10.1007/BF02512375 [DOI] [PubMed] [Google Scholar]
- Roper M. M., Marshall K. C. (1979). Effects of salinity on sedimentation and of particulates on survival of bacteria in estuarine habitats. Geomicrobiol. J. 1, 103–116. 10.1080/01490457909377727 [DOI] [Google Scholar]
- Rosenwasser S., Ziv C., Creveld S. G., Van Vardi A. (2016). Virocell metabolism: metabolic innovations during host-virus Interactions in the Ocean. Trends Microbiol. 24, 821–832. 10.1016/j.tim.2016.06.006 [DOI] [PubMed] [Google Scholar]
- Rozen Y., Belkin S. (2001). Survival of enteric bacteria in seawater. FEMS Microbiol. Rev. 25, 513–529. 10.1111/j.1574-6976.2001.tb00589.x [DOI] [PubMed] [Google Scholar]
- Ryu J. H., Beuchat L. R. (2005). Biofilm formation by Escherichia coli O157:H7 on stainless steel: effect of exopolysaccharide and curli production on its resistance to chlorine. Appl. Environ. Microbiol. 71, 247–254. 10.1128/AEM.71.1.247-254.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saby S., Leroy P., Block J. C. (1999). Escherichia coli resistance to chlorine and glutathione synthesis in response to oxygenation and starvation. Appl. Environ. Microbiol. 65, 5600–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samandoulgou I., Hammami R., Morales Rayas R., Fliss I., Jean J. (2015). Stability of secondary and tertiary structures of virus-like particles representing noroviruses: effects of pH, ionic strength, and temperature and implications for adhesion to surfaces. Appl. Environ. Microbiol. 81, 7680–7686. 10.1128/AEM.01278-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherber C. M., Schottel J. L., Aksan A. (2009). Membrane phase behavior of Escherichia coli during desiccation, rehydration, and growth recovery. BBA Biomembranes 1788, 2427–2435. 10.1016/j.bbamem.2009.08.011 [DOI] [PubMed] [Google Scholar]
- Schijven J. F., Hassanizadeh S. M. (2000). Removal of viruses by soil passage: overview of modeling, processes, and parameters. Crit. Rev. Env. Sci. Tec. 30, 49–127. 10.1080/10643380091184174 [DOI] [Google Scholar]
- Scholl M. A., Harvey R. W. (1992). Laboratory investigations on the role of sediment surface and groundwater chemistry in transport of bacteria through a contaminated sandy aquifer. Environ. Sci. Technol. 26, 1410–1417. 10.1021/es00031a020 [DOI] [Google Scholar]
- Schultz-Fademrecht C., Wichern M., Horn H. (2008). The impact of sunlight on inactivation of indicator microorganisms both in river water and benthic biofilms. Water Res. 42, 4771–4779. 10.1016/j.watres.2008.08.022 [DOI] [PubMed] [Google Scholar]
- Sen T. K., Khilar K. C. (2006). Review on subsurface colloids and colloid-associated contaminant transport in saturated porous media. Adv. Colloid Interface Sci. 119, 71–96. 10.1016/j.cis.2005.09.001 [DOI] [PubMed] [Google Scholar]
- Setti I., Rodriguez-Castro A., Pata M. P., Cadarso-Suarez C., Yacoubi B., Bensmael L., et al. (2009). Characteristics and dynamics of Salmonella contamination along the coast of Agadir, Morocco. Appl. Environ. Microbiol. 75, 7700–7709. 10.1128/AEM.01852-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahrampour D., Yavarmanesh M., Habibi Najafi M. B., Mohebbi M. (2015). Application of F+RNA coliphages as source tracking enteric viruses on parsley and leek using RT-PCR. Food Environ. Virol. 7, 381–389. 10.1007/s12560-015-9212-6 [DOI] [PubMed] [Google Scholar]
- Shelton D. R., Pachepsky Y. A., Kiefer L. A., Blaustein R. A., McCarty G. W., Dao T. H. (2014). Response of coliform populations in streambed sediment and water column to changes in nutrient concentrations in water. Water Res. 59, 316–324. 10.1016/j.watres.2014.04.019 [DOI] [PubMed] [Google Scholar]
- Siem-Jorgensen M., Glud R. N., Middelboe M. (2008). Viral dynamics in a coastal sediment: seasonal pattern, controlling factors and relations to the pelagic-benthic coupling. Mar. Biol. Res. 4, 165-U20. 10.1080/17451000801888718 [DOI] [Google Scholar]
- Signoretto C., Burlacchini G., Pruzzo C., Zampini M., Pane L., Franzini G., et al. (2004). Adhesion of Enterococcus faecalis in the nonculturable state to plankton is the main mechanisms responsible for persistence of this bacterium in both lake and saltwater. Appl. Environ. Microbiol. 70, 6892–6896. 10.1128/AEM.70.11.6892-6896.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. V., Vyas V., Patil R., Sharma V., Scopelliti P. E., Bongiorno G., et al. (2011). Quantitative characterization of the influence of the nanoscale morphology of nanostructured surfaces on bacterial adhesion and biofilm formation. PLoS ONE 6:e25029. 10.1371/journal.pone.0025029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinton L. W., Hall C. H., Lynch P. A., Davies-colley R. J. (2002). Sunlight inactivation of fecal indicator bacteria and Bacteriophages from waste stabilization pond effluent in fresh and saline Waters. Appl. Environ. Microbiol. 68, 1122–1131. 10.1128/AEM.68.3.1122-1131.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skraber S., Ogorzaly L., Helmi K., Maul A., Hoffmann L., Cauchie H.-M., et al. (2009). Occurrence and persistence of enteroviruses, noroviruses and F-specific RNA phages in natural wastewater biofilms. Water Res. 43, 4780–4789. 10.1016/j.watres.2009.05.020 [DOI] [PubMed] [Google Scholar]
- Skraber S., Schijven J., Gantzer C., de Roda Husman A. (2005). Pathogenic viruses in drinking-water biofilms: a public health risk?. Biofilms 2, 105–117. 10.1017/S1479050505001833 [DOI] [Google Scholar]
- Smets B. F., Grasso D., Engwall M. A., Machinist B. J. (1999). Surface physicochemical properties of Pseudomonas fluorescens and impact on adhesion and transport through porous media. Colloids Surf. B 14, 121–139. 10.1016/S0927-7765(99)00030-2 [DOI] [Google Scholar]
- Smith E. M., Gerba C. P., Melnick J. L. (1978). Role of sediment in persistence of enteroviruses in estuarine environment. Appl. Environ. Microbiol. 35, 685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares de Lima Grisi T. C., Gorlach-Lira K. (2010). The abundance of some pathogenic bacteria in mangrove habitats of Paraiba do Norte Estuary and crabmeat contamination of mangrove crab Ucides cordatus. Braz. Arch. Biol. Technol. 53, 227–234. 10.1590/S1516-89132010000100028 [DOI] [Google Scholar]
- Solo-Gabriele H. M., Wolfert M. A., Desmarais T. R., Palmer C. J. (2000). Sources of Escherichia coli in a coastal subtropical environment. Appl. Environ. Microbiol. 66, 230–237. 10.1128/AEM.66.1.230-237.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staggemeier R., Bortoluzzi M., da Silva Heck T. M., da Luz R. B., Fabres R. B., Soliman M. C., et al. (2015b). Animal and human enteric viruses in water and sediment samples from dairy farms. Agr. Water Manage. 152, 135–141. 10.1016/j.agwat.2015.01.010 [DOI] [Google Scholar]
- Staggemeier R., Bortoluzzi M., da Silva Heck T. M., Spilki F. R., de Matos Almeida S. E. (2015a). Quantitative vs. conventional PCR for detection of human adenoviruses in water and sediment samples. Rev. Instit. Med. Trop. Sγo Paulo 57, 299–303. 10.1590/S0036-46652015000400005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stal L. J., de Brouwer J. F. C. (2003). Biofilm formation by benthic diatoms and their influence on the stabilization of intertidal mudflats. Berichte-Forschungszentrum Terramare 12, 109–111. [Google Scholar]
- Staley C., Gould T. J., Wang P., Phillips J., Cotner J. B., Sadowsky M. J. (2014). Bacterial community structure is indicative of chemical inputs in the Upper Mississippi River. Front. Microbiol. 5:524. 10.3389/fmicb.2014.00524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton C. M., Wyer M. D., Kay D., Bradford M., Humphrey N., Wilkinson J., et al. (2007). Fate and Transport of Particles in Estuaries. Vol. I, Summary and Conclusions.Bristol: Environment Agency. [Google Scholar]
- Stenström T. A. (1989). Bacterial hydrophobicity, an overall parameter for the measurement of adhesion potential to soil particles. Appl. Environ. Microbiol. 55, 142–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson T., Reid E., Avery L. M., Jefferson B. (2013). Media surface properties and the development of nitrifying biofilms in mixed cultures for wastewater treatment. Proc. Saf. Environ. 91, 321–324. 10.1016/j.psep.2012.07.002 [DOI] [Google Scholar]
- Storch G. A. (2000). Diagnostic virology. Clin. Infect. Dis. 31, 739–751. 10.1086/31401 [DOI] [PubMed] [Google Scholar]
- Stumpf C. H., Piehler M. F., Thompson S., Noble R. T. (2010). Loading of fecal indicator bacteria in North Carolina tidal creek headwaters: Hydrographic patterns and terrestrial runoff relationships. Water Res. 44, 4704–4715. 10.1016/j.watres.2010.07.004 [DOI] [PubMed] [Google Scholar]
- Suhalim R., Huang Y. W., Burtle G. J. (2008). Survival of Escherichia coli O157: H7 in channel catfish pond and holding tank water. Lwt-Food Sci. Technol. 41, 1116–1121. 10.1016/j.lwt.2007.06.023 [DOI] [Google Scholar]
- Sutherland I. W. (2001). The biofilm matrix - an immobilized but dynamic microbial environment. Trends Microbiol. 9, 222–227. 10.1016/S0966-842X(01)02012-1 [DOI] [PubMed] [Google Scholar]
- Tan B., Ng C., Nshimyimana J. P., Loh L. L., Gin K. Y. H., Thompson J. R. (2015). Next-generation sequencing (NGS) for assessment of microbial water quality: current progress, challenges, and future opportunities. Front. Microbiol. 6:1027. 10.3389/fmicb.2015.01027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorn C. E., Quilliam R. S., Williams A. P., Malham S. K., Cooper D., Reynolds B., et al. (2011). Grazing intensity is a poor indicator of waterborne Escherichia coli O157 activity. Anaerobe 17, 330–333. 10.1016/j.anaerobe.2011.04.021 [DOI] [PubMed] [Google Scholar]
- Troussellier M., Bonnefont J. L., Courties C., Derrien A., Dupray E., Gauthier M., et al. (1998). Responses of enteric bacteria to environmental stresses in seawater. Oceanol. Acta 21, 965–981. 10.1016/S0399-1784(99)80019-X [DOI] [Google Scholar]
- Tsai S. C., Ellender R. D., Johnson R. A., Howell F. G. (1983). Elution of viruses from coastal sediments. Appl. Environ. Microbiol. 46, 797–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Mei H. C., Rustema-abbing M., Langworthy D. E., Collias D. I., Mitchell M. D., Bjorkquist, et al. (2008). Adhesion and viability of waterborne pathogens on p-DADMAC coatings. Biotechnol. Bioeng. 99, 165–169. 10.1002/bit.21538 [DOI] [PubMed] [Google Scholar]
- Van Houdt R., Michiels C. W. (2005). Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res. Microbiol. 156, 626–633. 10.1016/j.resmic.2005.02.005 [DOI] [PubMed] [Google Scholar]
- van Loosdrecht M. C. M., Lyklema J., Norde W., Schraa G., Zehnder A. J. B. (1987). Electrophoretic mobility and hydrophobicity as a measure to predict the initial steps of bacterial adhesion. Appl. Environ. Microbiol. 53, 1898–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loosdrecht M. C. M., Lyklema J., Norde W., Zehnder A. J. B. (1989). Bacterial adhesion: a physicochemical approach. Microb. Ecol. 17, 1–15. 10.1007/BF02025589 [DOI] [PubMed] [Google Scholar]
- Vignaroli C., Di Sante L., Magi G., Luna G. M., Di Cesare A., Pasquaroli S., et al. (2015). Adhesion of marine cryptic Escherichia isolates to human intestinal epithelial cells. ISME J. 9, 508–515. 10.1038/ismej.2014.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignaroli C., Luna G. M., Pasquaroli S., Di Cesare A., Petruzzella R., Paroncini, et al. (2013). Epidemic Escherichia coli ST131 and Enterococcus faecium ST17 in coastal marine sediments from an Italian beach. Environ. Sci. Technol. 47, 13772–13780. 10.1021/es4019139 [DOI] [PubMed] [Google Scholar]
- Villanueva L., Navarrete A., Urmeneta J., Geyer R., White D. C., Guerrero R. (2007). Monitoring diel variations of physiological status and bacterial diversity in an estuarine microbial mat: an integrated biomarker analysis. Microb. Ecol. 54, 523–531. 10.1007/s00248-007-9224-3 [DOI] [PubMed] [Google Scholar]
- Wait D. A., Sobsey M. D. (1983). Method for recovery of enteric viruses from estuarine sediments with chaotropic agents. Appl. Environ. Microbiol. 46, 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner G., Erhart R., Amann R. (1995). Flow cytometric analysis of activated sludge with rRNA-targeted probes. Appl. Environ. Microbiol. 61, 1859–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Safari A., Ivankovic A., Casey E. (2013). Detachment characteristics of a mixed culture biofilm using particle size analysis. Chem. Eng. J. 228, 1140–1147. 10.1016/j.cej.2013.05.071 [DOI] [Google Scholar]
- Walters E., Graml M., Behle C., Müller E., Horn H. (2013). Influence of Particle Association and Suspended Solids on UV Inactivation of Fecal Indicator Bacteria in an Urban River. Water Air Soil Poll. 225, 1–9. 10.1007/s11270-013-1822-8 [DOI] [Google Scholar]
- Wang G. D., Doyle M. P. (1998). Survival of enterohemorrhagic Escherichia coli O157: H7 in water. J. Food Protect. 61, 662–667. [DOI] [PubMed] [Google Scholar]
- Wang L., Zhong Q., Li Y. (2012). Ethidium monoazide-loop mediated isothermal amplification for rapid detection of Vibrio parahaemolyticus in viable but non-culturable state. Energy Procedia 17, 1858–1863. 10.1016/j.egypro.2012.02.323 [DOI] [Google Scholar]
- Ward J. M., Keep N. H., Ward J. M., Cohen-gonsaud M., Henderson B. (2006). Wake up! Peptidoglycan lysis and bacterial non-growth states. Trends Microbiol. 14, 271–276. 10.1016/j.tim.2006.04.003 [DOI] [PubMed] [Google Scholar]
- Watterworth L., Rosa B., Schraft H., Topp E., Leung K. T. (2006). Survival of various ERIC-genotypes of Shiga toxin-producing Escherichia coli in well water. Water Air Soil Poll. 177, 367–382. 10.1007/s11270-006-9179-x [DOI] [Google Scholar]
- Weinbauer M. G. (2004). Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28, 127–181. 10.1016/j.femsre.2003.08.001 [DOI] [PubMed] [Google Scholar]
- Weiss C. M. (1951). Adsorption of E. coli on river and estuarine silts. Sewage Ind. Wastes 23, 227–237. [Google Scholar]
- Williams A. P., Avery L. M., Killham K., Jones D. L. (2007). Persistence, dissipation, and activity of Escherichia coli O157:H7 within sand and seawater environments. FEMS Microbiol. Ecol. 60, 24–32. 10.1111/j.1574-6941.2006.00273.x [DOI] [PubMed] [Google Scholar]
- Williams L. B., Metge D. W., Eberl D. D., Harvey R. W., Turner A. G., Prapaipong P., et al. (2011). What makes a natural clay antibacterial? Environ. Sci. Technol. 45, 3768–3773. 10.1021/es1040688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson K. E., Corzo K. A., Drissi C. L., Buckingham J. M., Thompson C. P., Helton R. R. (2013). Estimates of viral abundance in soils are strongly influenced by extraction and enumeration methods. Biol. Fert. Soils 49, 857–869. 10.1007/s00374-013-0780-z [DOI] [Google Scholar]
- Wingender J., Flemming H. C. (2011). Biofilms in drinking water and their role as reservoir for pathogens. Int. J. Hyg. Environ. Heal. 214, 417–423. 10.1016/j.ijheh.2011.05.009 [DOI] [PubMed] [Google Scholar]
- Wu L., Lin X. M., Peng X. X. (2009a). From proteome to genome for functional characterization of pH-dependent outer membrane proteins in Escherichia coli. J. Proteome Res. 8, 1059–1070. 10.1021/pr800818r [DOI] [PubMed] [Google Scholar]
- Wu S., Ueno D., Inoue K., Someya T. (2009b). Direct viable count combined with fluorescence in situ hybridization (DVC-FISH) for specific enumeration of viable Escherichia coli in cow manure. Microbes Environ. 24, 33–38. 10.1264/jsme2.ME08543 [DOI] [PubMed] [Google Scholar]
- Yamahara K. M., Layton B. A., Santoro A. E., Boehm A. B. (2007). Beach sands along the California Coast are diffuse sources of fecal Bacteria to Coastal Waters. Environ. Sci. Technol. 41, 4515–4521. 10.1021/es062822n [DOI] [PubMed] [Google Scholar]
- Yaron S., Matthews K. R. (2002). A reverse transcriptase-polymerase chain reaction assay for detection of viable Escherichia coli O157:H7: investigation of specific target genes. J. Appl. Microbiol. 92, 633–640. 10.1046/j.1365-2672.2002.01563.x [DOI] [PubMed] [Google Scholar]
- Yoshida M., Takaki Y., Eitoku M., Nunoura T., Takai K. (2013). Metagenomic Analysis of Viral Communities in (Hado)Pelagic Sediments. PLoS ONE 8:57271. 10.1371/journal.pone.0057271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K. D. (2006). The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 70, 660–703. 10.1128/MMBR.00001-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Ye C., Lin H., Lv L., Yu X. (2015). UV Disinfection Induces a VBNC State in Escherichia coli and Pseudomonas aeruginosa. Environ. Sci. Technol. 49, 1721–1728. 10.1021/es505211e [DOI] [PubMed] [Google Scholar]