Abstract
Lipopeptides are natural product antibiotics that consist of a peptide core with a lipid tail with a diverse array of target organisms and mechanisms of action. Daptomycin (DAP) is an example of these compounds with specific activity against Gram-positive organisms. DAP has become increasingly important to combat infections caused by Gram-positive bacteria because of the presence of multidrug resistance in these organisms, particularly in methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE). However, emergence of resistance to DAP during therapy is a well-described phenomenon that threatens the clinical use of this antibiotic, limiting further the therapeutic options against both MRSA and VRE. This work will review the historical aspects of the development of DAP, as well as the current knowledge on its mechanism of action and pathways to resistance in a clinically relevant context.
The antibiotic daptomycin is used to combat challenging infections (e.g., those caused by MRSA and VRE). It has a unique and potent mechanism of action, but emerging resistance threatens its clinical use.
Lipopeptides refer to a diverse class of compounds that share the general structure of a peptide core attached to a lipid tail, and are produced by a variety of environmental microorganisms including soil bacteria and fungi. This class of compounds possesses a wide therapeutic potential as evidenced by drugs that are currently in clinical use as antimicrobials, including the polymixins (polymixin B and colistin), echinocandins (caspofungin, micafungin, and anidulafungin), and daptomycin. The first isolation of a lipopeptide antibiotic occurred in 1953 with the discovery of amphomycin (Heinemann et al. 1953). However, this compound (and related molecules) was not further developed as a result, in part, of complex chemical structure and, in some cases, concerns of toxicity. Of note, there has recently been a resurgence of interest in compounds similar to amphomycin, driven by increasing rates of antimicrobial resistance to more traditional therapeutic agents.
Daptomycin (DAP), a lipopeptide antibiotic with in vitro bactericidal activity against Gram-positive bacteria, received approval by the Food and Drug Administration (FDA) in 2003 for soft-tissue infections and in 2006 for Staphylococcus aureus bacteremia and right-sided endocarditis. DAP has become a front-line agent in the treatment of challenging infections caused by both methicillin-resistant S. aureus (MRSA) and vancomycin-resistant Enterococcus faecium (VRE) (Munita et al. 2015). Despite its increasing role in the treatment of serious infections by these organisms, details of the precise mechanism of action and the mechanisms by which bacteria develop resistance are incompletely understood. Here, we will provide a brief overview of the structure and synthesis of DAP, explore what is known about its mechanism of action, and discuss the genetic changes associated with DAP nonsusceptibility (hereafter referred to as daptomycin resistance [DAP-R]) in S. aureus and the enterococci.
HISTORY, STRUCTURE, AND SYNTHESIS OF DAPTOMYCIN
After the discovery of amphomycin, a variety of lipopeptides with antimicrobial properties were identified over the next decade, including crystallomycin (Lomakina and Brazhnikova 1959), aspertocin (Shay et al. 1960), glumamycin (Shibata et al. 1962), laspertomycin, and tsushimycin (Naganawa et al. 1968; Shoji et al. 1968). Further development of these compounds for study and use was limited by several factors, including (1) the heterogeneous mixture of related molecules isolated from the fermentation of source organisms, (2) the complex chemistry needed to manipulate isolated compounds, and (3) a lack of understanding of the genetics behind their production. By the late 1980s, several important breakthroughs would allow DAP to make the journey from drug discovery to the bedside.
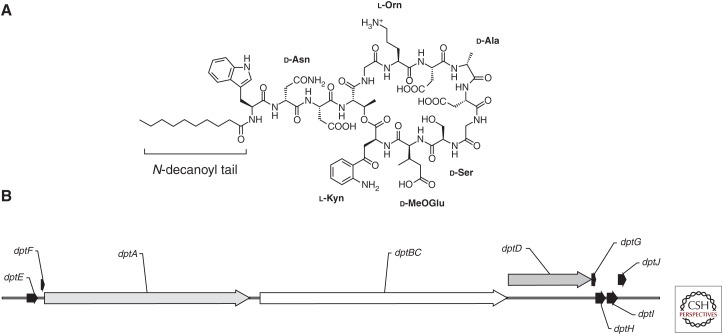
DAP began its journey as a molecule identified as A21987C, a group of lipopeptides produced by an isolate of Streptomyces roseosporus collected from the soil of the slopes of Mount Ararat in Turkey (Eisenstein et al. 2010). It consists of a 13-amino-acid depsipeptide, which harbors a cyclic decapeptide core with three extra-cyclic amino acids attached to an amino-terminal fatty acid tail (Fig. 1A). A distinctive feature of the lipopeptides is the diverse nature of the peptide core. In the case of DAP, the core contains a variety of noncanonical amino acids (kynurenine, ornithine, and 3-methylglutamic acid) and l-enantiomers (d-alanine and d-serine) (Debono et al. 1987). Several of these residues, in particular, kynurenine (of which the carboxyl group is the site of cyclization) and 3-methylglutamic acid have been shown, via substitution, to be important in the antimicrobial activity of the molecule with altered peptides displaying an increase of up to five times the minimum inhibitory concentration (MIC) (Grünewald et al. 2004). Further, six acidic residues in the DAP peptide ring are conserved across other calcium-dependent antimicrobial lipopeptides, highlighting the importance of this inorganic ion in both the mechanism of action and resistance (see below) (Hojati et al. 2002). The fatty acid tail also plays an important role in the activity of the compound, particularly in regard to toxicity. A21987C was found by high-performance liquid chromatography (HPLC) to be a mix of three main constituents differing only in the lipid moiety (with chain lengths of 11, 12, and 13 carbons) at the amino terminus (Debono et al. 1988). It was noted that longer chain lengths correlated with increasing toxicity; however, batch fermentation and subsequent separation was a laborious and difficult task with inefficient yield. The discovery that a penicillin deacylase produced by Actinoplanes utahensis (Debono et al. 1988) could remove the lipid tail opened the door for further characterization of the molecule. Using this technique, a semisynthetic derivative of A21987C with an n-decanoyl tail, named daptomycin, was found to balance antimicrobial activity with toxicity in a mouse model. Large-scale biosynthesis of DAP was achieved by feeding a controlled amount of decanoic acid to cultures of S. roseosporus (Huber et al. 1988).
Figure 1.
Structure of daptomycin and organization of the daptomycin biosynthesis gene cluster in Streptomyces filamentosus. (A) Chemical structure of daptomycin (DAP) with noncanonical amino acids and N-decanoyl fatty acid tail labeled. l-Kyn, l-Kynurenine; l-Orn, l-Ornithine; d-MeOGlu, d-3-methylglutamic acid. (B) Organization of the DAP biosynthesis gene cluster (see text for details). (Sequence information from NCBI database, accession number AY787762.1.)
Lipopeptides, similar to many other natural product antimicrobials, are produced by nonribosomal peptide synthetases (NRPS). These large enzymatic complexes work in an assembly-line fashion to generate a specific peptide sequence. At their core is a series of three enzymatic activities (condensation, adenylation, and thiolation [CAT]) that perform a function analogous to ribosomal polypeptide synthesis, with amino acid specificity determined by the binding characteristics of each adenylation domain rather than an mRNA codon (Marahiel et al. 1997; Fischbach and Walsh 2006). The adenylation domain uses energy from adenosine triphosphate (ATP) to form an aminoacyladenosine monophosphate (AMP) intermediary from its cognate amino acid. Next, the AMP is displaced by the formation of a thioester bond coupling the amino acid to the thiolation domain carrier protein. The condensation domain then catalyzes the addition of the growing peptide chain to the amino acid monomer via an amide linkage, resulting in the passage of the nascent chain from one thiolation domain to the next module in the complex, wherein the process is repeated. Additional enzymes augment this core synthesis machinery, allowing modifications such as the incorporation of d-amino acids and allowing for the cyclic structure of DAP. In S. roseosporus, this machinery is organized into three multimodular subunits, DptA, DptBC, and DptD (Fig. 1B), which are responsible for the synthesis, modification, and cyclization of the 13 amino acid core (Baltz 2009). Two genes, dptE and dptF, located directly upstream of the primary peptide synthesis cluster, show similarity to acyl-CoA ligase and acyl carrier proteins, and are thus predicted to be involved in the addition of fatty acids to the amino-terminal end of DAP (Mchenney et al. 1998; Miao et al. 2005). Downstream are four accessory genes, one of which encodes a protein that shares identity with those known to metabolize tryptophan (a needed step for the synthesis of kynurenine). Another of the accessory genes is predicted to encode a glutamate methyltransferase presumably involved in the production of 3-methylglutarate (Miao et al. 2005).
The understanding of the genetic organization of the DAP NRPS machinery has opened the pathway to further drug modification and discovery. Although the dpt locus is transcribed as a single long mRNA, splitting the DptA, DptBC, and DptD submodules by deletion and subsequent reintroduction into different chromosomal locations (under control of a constitutive erm promoter) was not shown to adversely affect the production of DAP (Coëffet-Le Gal et al. 2006). Further, substitution of various CAT domains between lipopeptide synthesis clusters of different Streptomyces species has allowed the creation of altered peptide cores to screen for desired characteristics, such as increased activity in the presence of surfactant (Nguyen et al. 2006, 2010). Continued experimentation with novel arrangements of NRPS modules may offer further insights into DAP and may lead to discovery of novel compounds with improved activities.
MECHANISM OF ACTION
DAP shares structural similarities with a group of molecules produced by the mammalian innate immune system known as cationic antimicrobial peptides (CAMPs), specifically the human cathelicidin LL-37. These effectors of the innate immune response possess a wide spectrum of activity against bacteria, fungi, and some encapsulated viruses, and are thought to exert their effect by binding to and disrupting membrane integrity (Bals and Wilson 2003). The structural similarities between DAP and CAMPs have led investigators to postulate that they may share a common mechanism of membrane disruption, as DAP is known to bind the Gram-positive bacterial membrane and initiate a series of events that lead to cell death (Straus and Hancock 2006). Although the precise mechanism of action remains to be fully elucidated, there are at least two important interactions required for DAP to exert its bactericidal effect.
First is the interaction between DAP and calcium. Nuclear magnetic resonance (NMR) data of DAP in solution suggests that DAP complexes with calcium in a 1:1 molar ratio to form small (14–16 molecules) DAP micelles that may aid in antimicrobial delivery to the bacterial membrane (Scott et al. 2007). Changes in the NMR signal of the tryptophan at position 1 and the kynurenine at position 13 on the addition of calcium were thought to indicate a possible role for these residues in calcium binding or oligomerization of the molecule (Ho et al. 2008). Other divalent cations, such as magnesium, can induce micelle formation at higher concentrations (2.5:1 ratio), but result in decreased antimicrobial activity as evidenced by an increase in MICs by 64-fold (Ho et al. 2008).
The second important interaction takes place between DAP and the anionic phospholipid phosphatidylglycerol (PG). Once in proximity to the bacterial membrane, DAP undergoes a structural transition to insert into the cell membrane (Jung et al. 2004). This process appears to be dependent on the presence of PG in the target membrane (Muraih et al. 2011) and is facilitated by calcium ions, which decrease the DAP concentration needed for membrane insertion by ∼50-fold (Chen et al. 2014). Indeed, the presence of PG is an important mediator of DAP aggregation on model membranes. Using excimer fluorescence, excitation of DAP-perylene conjugants was seen in PG-containing membranes, but was absent from those made exclusively of phosphatidylcholine (Muraih et al. 2012). Thus, the first key steps of DAP membrane insertion and oligomerization rely on both calcium and PG as crucial mediators.
The dependence on specific phospholipids for the mechanism of action of DAP may also explain the antimicrobial spectrum of this drug, as it has potent activity against Gram-positive organisms, but none against Gram-negative organisms. This effect seems to be independent of the permeability barrier of the outer membrane (OM) of Gram-negative bacteria, as Escherichia coli protoplasts in which the OM was removed showed a fourfold reduction in MICs to vancomycin (a large glycopeptide antibiotic that would otherwise be excluded from the periplasmic space), but no change in DAP MIC (Randall et al. 2013). This observation has led some to suggest that Gram-negative bacteria are devoid of phospholipids that may interact with DAP. Indeed, the membrane lipid composition of E. coli includes 80% of phosphatidylethanolamine (PE) and only 15% PG, as compared with S. aureus, which lacks PE and contains 58% PG and 42% cardiolipin (CL) (Epand et al. 2007). Thus, the differences of phospholipid content may explain the lack of activity of DAP against Gram-negative bacteria.
The series of events that occur after DAP gains access to the membrane are less clear. Early investigations into the mechanism of action of DAP observed that cell-wall synthesis was inhibited by a decreased intracellular pool of UDP-N-acetylmuramyl-pentapeptide in Bacillus megaterium (Mengin-Lecreulx et al. 1990), which the investigators attributed to inhibition of the enzymes involved in the formation of UDP-N-acetylglucosamine. This deficit was, however, subsequently found to be caused by impaired active transport of the amino acids required for murein synthesis, an effect associated with dissipation of the membrane electrochemical gradient (Allen et al. 1991). Analysis of major metabolic pathways and macromolecules showed DAP had little effect on the synthesis of DNA, RNA, or proteins. In contrast, cell envelope metabolism was consistently altered, with radiolabeled acetate incorporation into lipids decreased by 50% and lipoteichoic acid synthesis reduced by 93%. Moreover, both enterococci and Bacillus species displayed important morphologic changes (elongation) with a relative increase in sidewall synthesis (Canepari et al. 1990) on exposure to DAP. The finding that serial washes with ethylenediaminetetraacetic acid (EDTA) was unable to remove DAP from bacterial membranes (Canepari et al. 1990) suggested that the cell membrane was the site of action, an observation that fit well with the amphipathic nature of the DAP molecule. Further, DAP is able to exert its bactericidal action against S. aureus in stationary phase, under conditions in which active metabolism is quenched and without requiring lysis of the target cell (Mascio et al. 2007; Cotroneo et al. 2008), consistent with disruption of the bacterial cell membrane, rather than inhibition of cell-wall or teichoic acid synthesis.
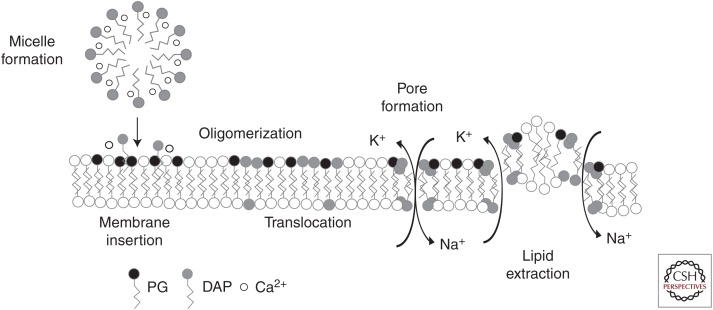
There are currently two proposed mechanisms of oligomeric DAP action (Fig. 2). One hypothesis, originating from the observed correlation of membrane depolarization and cell death in S. aureus, proposes that aggregates of DAP form an oligomeric pore like structure in the membrane, which results in ion leakage and subsequent dissipation of the membrane potential (Silverman et al. 2003). Experimental support for this hypothesis is derived from several studies. Initial stoichiometric calculations using Forester resonance energy transfer showed that ∼7–8 DAP subunits associate in a PG-dependent manner for each oligomeric complex (Muraih and Palmer 2012; Zhang et al. 2014a). Further, the introduction of DAP into the outer leaflet induces a local membrane stress that increases levels of lipid flip-flop, an exchange of lipids between the inner and outer membrane leaflets (Jung et al. 2004), including the transition of DAP from the outer to inner leaflet. In the presence of PG, DAP associates into two oligomers of four units each opposite each other on the membrane, bending the membrane and establishing a pore like structure (Zhang et al. 2014a). Using model liposomes, exposure to DAP was found to make the membrane permeable to small cations such as sodium and potassium, and less so to anions or larger organic acids, suggesting that an influx of sodium ions abolished the membrane potential and served as the effector of DAP action (Zhang et al. 2014b). Interestingly, the presence of another phospholipid (PL), CL, in liposomes containing PG served to inhibit the translocation of DAP from the outer leaflet to the inner one, resulting in tetrameric complexes on the outer surface only (Zhang et al. 2014a). As we will discuss below, alterations of enzymes involved in PL metabolism are a common feature of resistance to DAP in some bacteria, consistent with the important role of PL metabolism in its mechanism of action.
Figure 2.
Proposed mechanisms for the action of daptomycin. In solution, daptomycin (DAP) complexes with calcium to form small micelles, and subsequent membrane insertion is dependent on both the presence of calcium and phosphatidylglycerol (PG). Once inserted, DAP oligomerizes and transitions to the inner membrane leaflet. These complexes then align on opposite sides of the membrane to form a pore channel permeable to small cations, or disrupt membrane integrity by extracting lipids and leading to transient ion leakage.
A second hypothesis centers on a newly described phenomenon termed the lipid extracting effect. Using giant unilamellar vesicles (GUV), Chen et al. (2014) observed that DAP insertion into the membrane results in an initial expansion of vesicle surface area. As DAP concentrations continue to increase, there is a rapid aggregation of lipid on the membrane surface, while at the same time the overall surface area of the vesicle decreases, implying that the lipid clusters are extracted and “released” from the vesicle membrane. Interestingly, this phenomenon is dependent on both calcium and PG, and displays a threshold concentration of DAP required to initiate the membrane changes, which the investigators postulate may correlate with MIC values in bacterial isolates. Further, the extraction of lipids results in the formation of transient water pores, which could theoretically explain the ion leakage observed experimentally (Gurtovenko and Vattulainen 2007). This effect may also explain the observations of Pogliano et al. (2012) in Bacillus subtilis showing that DAP binding to the membrane near the cell septum induced a patchy aggregate of lipid, altering cell morphology to a bent “L” shape and mislocalizing the essential cell division protein DivIVA. Indeed, abnormal septation and thickened cell walls are common features of DAP-R bacteria, and may be because of recognition of altered lipid membranes as signals for new peptidoglycan synthesis away from the septum.
It is important to note that the two hypotheses are not mutually exclusive because both pore formation and lipid extraction may be playing a role once DAP makes contact with the bacterial membrane and could explain the broad effects of the antibiotic in bacterial permeabilization, cell division, and metabolism.
DAPTOMYCIN RESISTANCE
DAP-R in S. aureus and the enterococci has been well documented and it is a serious concern for the treatment of serious infections caused by these organisms (Bayer et al. 2013; Miller et al. 2014). Given the clinical burden of disease that these organisms represent, an understanding of the mechanisms by which they subvert the DAP “attack” is likely to provide novel insights into the manner that bacteria protect their cell membrane and adapt to the antimicrobial challenge. Detailed analyses of both DAP-R laboratory and clinical isolates have revealed several common pathways associated with resistance, namely, alteration of regulatory systems responsible for the bacterial cell envelope stress response, as well as enzymes involved in phospholipid metabolism and membrane homeostasis. Despite the genetic similarities, the mechanisms by which these changes drive DAP-R seem quite varied and are adapted to the biology of each organism, a fascinating feature of bacterial evolution. Thus, we will discuss each relevant species separately.
DAP-R IN Staphylococcus aureus
S. aureus use several strategies to circumvent the DAP effect, the most common appears to involve the alteration of the cell-surface charge (Fig. 3A). Indeed, S. aureus seems to primarily respond to the DAP attack by producing a more positive overall cell-surface charge, presumably to prevent the positively charged DAP–calcium insertion by electrostatic repulsion. This phenotype is classically associated with mutations in mprF (multiple peptide resistance factor), which encodes a bifunctional enzyme that contains a carboxy-terminal cytoplasmic tail responsible for lysinylation of PG and an amino-terminal domain, which consists of eight transmembrane domains. The amino-terminal domain encodes a “flippase” activity, which is responsible for the translocation of lysyl-PG (LPG) from the inner to the outer membrane. A central domain of four transmembrane helices seems to assist with both lysinylation and flippase activities (Ernst et al. 2009).
Figure 3.
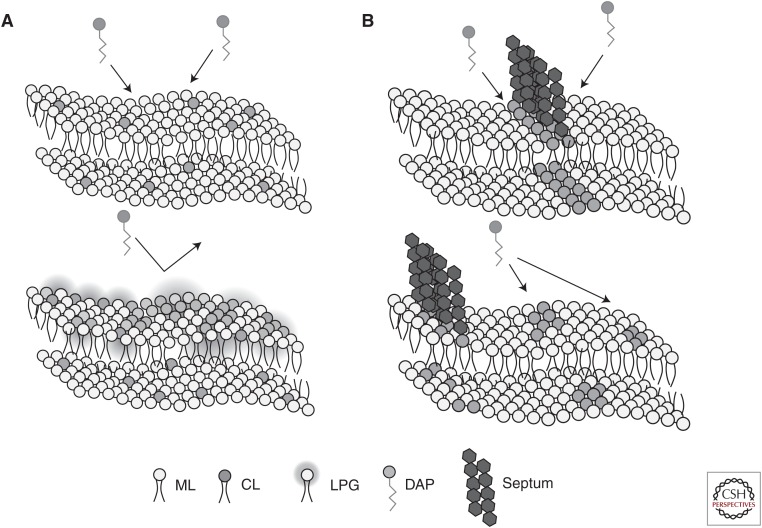
Strategies for resisting daptomycin membrane attack. (A) Repulsion: In Staphylococcus aureus and Enterococcus faecium, changes in cell-surface charge and membrane phospholipid content block daptomycin (DAP) membrane association and oligomerization. (B) Diversion: In E. faecalis sensitive to DAP cardiolipin (CL) clusters at the division septum. In resistant isolates, redistribution of CL microdomains “traps” DAP away from the septum. ML, membrane lipid; LPG, lysylphosphatidylglycerol.
In DAP-R S. aureus, a number of mprF mutations have been described that result in amino acid changes clustering in the central bifunctional region that overall confer a “gain-of-function” of the enzyme (Bayer et al. 2015). Thus, the net result is an increased synthesis and expression of positively charged LPG on the outer membrane. Strong evidence for the role of mprF in DAP-R are studies in which expression of mprF with DAP-R associated mutations (but not wild-type mprF) in trans could restore elevated DAP MICs to strains of S. aureus in which mprF had been deleted from the chromosome (Yang et al. 2013). Moreover, inhibition of MprF protein synthesis in DAP-R strains harboring gain-of-function mutations by antisense RNA (directed against mprF transcripts) was able to reverse DAP-R in vitro (Rubio et al. 2011).
An alternative pathway for DAP-R in S. aureus that results in an increase of cell-surface charge is the overexpression of the dlt operon (Yang et al. 2009; Cafiso et al. 2014). This operon produces the machinery responsible for attaching the positively charged amino acid alanine to cell-wall teichoic acid (WTA), leading to an increase in positive cell-surface charge in a manner similar to increased LPG synthesis. Up-regulation of WTA synthesis (as observed by increased transcription of tagA) and the dlt operon were also associated with increased cell-wall mass, another common phenotype observed in DAP-R staphylococci (Bertsche et al. 2011, 2013). However, despite the strong association between mutations in mprF and increased expression of dlt with increases in net positive cell-surface charge, these changes do not seem to correlate with changes in DAP MICs in all strains (Mishra et al. 2014). Indeed, an in vitro study generated DAP-R isolates with alterations in both mprF and dlt, but the net positive charge of the DAP-R mutants was less than the parent strain (Mishra et al. 2009). Thus, additional characteristics must also play a role in mediating DAP-R in staphylococci.
A second mechanism associated with DAP-R in staphylococci is the alteration of membrane phospholipid composition, which is postulated to either decrease the amount of PG available at the membrane interface or to change the fluidity of the membrane, thus interfering with DAP binding and subsequent oligomerization. Interestingly, by analyzing the action of a membrane active antimicrobial peptide on GUVs, it was found that increases in LPG were not associated with decreased peptide binding (as might be expected in a charge repulsion mechanism) but rather with inhibition of intravesicular dye leakage after binding takes place, consistent with a membrane integrity protective effect (Kilelee et al. 2010). Further, as discussed above, other phospholipid species, such as CL, may also play a protective role in preventing DAP translocation once inserted in the membrane (Zhang et al. 2014a). The enzyme responsible for cardiolipin synthesis, cardiolipin synthase, joins two molecules of PG to make CL (Short and White 1972). Thus, it is tempting to speculate that mutations producing changes in enzyme function may play a role in DAP-R by altering the ratio of PG to CL in the cell membrane. Indeed, genomic analysis of 33 DAP-R strains indicated that, among others, mutations in pgsA (which encodes an enzyme involved in PG synthesis) and cls2 (cardiolipin synthase) were associated with DAP-R (Peleg et al. 2012). Additionally, membrane fluidity (which is highly dependent on PL and fatty acid composition) may also be an important factor that influences the DAP-R phenotype in certain strains. (Jones et al. 2008; Mishra et al. 2011). Interestingly, membranes of DAP-R clinical isolates are more fluid, whereas laboratory isolates tend to have more rigid membranes (Mishra et al. 2009), suggesting that DAP requires an optimal membrane order for insertion and oligomerization and perturbations of this order to either side may be protective. Along these lines, changes (both increase and decrease) in the production of staphyloxanthin, the carotenoid responsible for the golden color of S. aureus, was associated with DAP-R and was postulated to be a result of its influence on membrane fluidity (Mishra and Bayer 2013).
Global regulatory changes in genes modulating cell envelope stress and maintenance in S. aureus have also been associated with development of DAP-R (Utaida et al. 2003; Rose et al. 2012). Interestingly, DAP challenge induces important changes in global gene expression. These genomic pathways are similar to those associated with resistance to other antibiotics such as vancomycin and seem to affect the expression of the cell-wall “stimulon.” Two important two-component regulatory systems (TCS) have been involved in DAP-R, namely, VraSR and YycFG (Muthaiyan et al. 2008; Mehta et al. 2012). Of note, DAP was also found to induce a group of genes that was previously associated with exposure to carbonyl cyanide m-chlorophenylhydrazone, a proton ionophore, reflecting its ability to disrupt the membrane and induce ion leakage (Muthaiyan et al. 2008).
In general, TCS consist of a membrane-bound sensor histidine kinase (HK) responsible for detecting a particular stimulus or cellular perturbation, and a DNA-binding response regulatory (RR) that alters transcription of target genes (Dubrac et al. 2008). Mutations in these proteins can lead to altered expression of the system’s regulon, profoundly affecting membrane homeostasis. The essential TCS YycFG (also known as WalKR) is involved in the control of peptidoglycan biosynthesis in S. aureus, mainly through the regulation of expression of two major autolysins, LytM and AltA (Dubrac and Msadek 2004). The genes encoding this system are clustered with two other genes, yycHI, that are “accessory” to the function of YycFG. Both YycH and YycI are amino-terminal transmembrane proteins with extracellular carboxy-terminal domains that in B. subtilis have been shown to repress the activity of the YycG HK (Szurmant et al. 2007). In nondividing cells, the entire YycFGHI complex remains in the peripheral cell wall, presumably in an inactive state. However, under growth conditions, YycG is recruited to the site of septal formation, whereas YycH and YycI remain in the periphery (Fukushima et al. 2011).
Using an inducible promoter to control YycFG expression, Dubrac et al. (2007) showed that low levels of YycFG expression were associated with decreased peptidoglycan turnover, increased cross-linking, and increased glycan chain length. Interestingly, low levels of YycFG were also associated with increased resistance to lysis by the detergent Triton X-100. By varying the temperature of model lipid membranes, it was shown that the activity of the HK YycG was impacted by membrane fluidity, with the system turned off under highly fluid conditions (Türck and Bierbaum 2012). The investigators suggested a mechanism by which YycG senses changes in membrane fluidity and responds by adjusting cell-wall cross-linking to compensate for stresses caused by osmotic pressure. Of note, in DAP-R isolates, several mutations in yycFG affecting multiple domains of both YycG HK and YycF RR (Friedman et al. 2006; Howden et al. 2011) have been described. Additionally, mutations in the accessory genes have also been noted. For example, a mutation resulting in a frameshift and truncation of ∼10% of the accessory protein YycH (which in B. subtilis is associated with regulating YycF signaling) was associated with DAP-R (Szurmant et al. 2005; Mwangi et al. 2007). Given that the DAP-R phenotype displays some similarities to the YycFG-deficient phenotype (e.g., thickened cell walls, increased membrane fluidity, and resistance to membrane disruption), it is tempting to speculate that the observed changes in YycFG impair the functioning of the operon, down-regulating cell-wall homeostasis to survive the DAP-mediated attack.
The VraSR TCS is orthologous to the LiaSR system of B. subtilis and enterococci (discussed below) and is conserved across the low G+C bacteria (Jordan et al. 2006). It is up-regulated by both vancomycin and DAP exposure, and is associated with cell-wall biosynthesis via transcription of pbp2 (penicillin binding protein 2), tagA (WTA synthesis), prsA (a chaperone), and murZ (UDP-N-acetylglucosamine enolpyruvyl transferase), among others (Kuroda et al. 2003; Mwangi et al. 2007; Camargo et al. 2008). Structural studies have shown that on activation by phosphorylation, the VraR RR undergoes a conformational change allowing for dimerization and a subsequent increase in its binding affinity for target DNA (Leonard et al. 2013). Experimental evidence supports a role for this system in DAP-R, as deletion of the vraSR operon from a DAP-R strain of S. aureus resulted in a DAP-sensitive (DAP-S) phenotype, which could be reversed by supplying the genes in trans (Mehta et al. 2012). Additional mutations associated with the DAP-R phenotype include genes encoding the RNA polymerase subunits rpoB and rpoC (Friedman et al. 2006; Peleg et al. 2012). A mutation in rpoB, resulting in the amino acid change A621E, was associated with increased expression of the dlt operon and correlated with an increase in positive cell-surface charge, whereas RpoB mutations A621E and A477D were both linked to activation of cell-wall biosynthesis and increased cell-wall thickness (Cui et al. 2010; Bæk et al. 2015).
The pathway to DAP-R also results in significant cellular metabolic shifts. Analysis of six strain pairs of S. aureus under normal growth conditions and DAP exposure revealed that there is a decrease in activity of the tricarboxylic acid (TCA) cycle and, instead, carbon sources are redirected into the pentose phosphate pathway (Gaupp et al. 2015). This is corroborated by prior work that had shown levels of succinate dehydrogenase, an enzyme involved in the TCA cycle, were lower in a DAP-R strain when compared with its DAP-S counterpart (Fischer et al. 2011). Additionally, mutations noted upstream of acetyl-CoA synthetase in DAP-R isolates (Friedman et al. 2006) may affect the production of acetyl-CoA, which is involved in lipid synthesis and may also feed into the TCA cycle. Redirection of the flow of metabolites results in the formation of larger pools of amino sugar precursors, which can be used for peptidoglycan, teichoic acid, and nucleotide synthesis (Gaupp et al. 2015). Thus, a metabolic shift primes DAP-R isolates to build larger stores of cell envelope precursors allowing them to weather the storm of DAP-induced membrane stress.
DAP-R IN ENTEROCOCCI
The introduction of DAP provided clinicians with an agent that possessed in vitro bactericidal activity against enterococci, and it quickly became a front-line antibiotic for recalcitrant VRE infections, despite the lack of FDA approval for this indication. Even in early development of DAP, it was noted that longer acyl chain lengths (13–14 carbons) tended to improve activity against enterococci, but with the trade-off of increased toxicity (Debono et al. 1988). Thus, DAP (with its n-decanoyl fatty acyl side chain) is less potent against enterococci, a fact that is reflected in the clinical breakpoints, which are fourfold higher for enterococci compared with S. aureus (4 µg/mL vs. 1 µg/mL). Similar to what has been discussed in staphylococci, development of DAP-R in enterococci seems to affect two important groups of genes, namely, those controlling the cell membrane stress response and phospholipid metabolism. Despite the genetic similarities, the two clinically relevant species, Enterococcus faecalis and E. faecium, seem to display distinctive phenotypic differences in their response to DAP challenge and, thus, we will discuss them separately.
Daptomycin Resistance in E. Faecalis
The genetic bases of DAP-R in E. faecalis were mapped using whole-genome sequencing of both in vitro and clinical isolates that had developed resistance in the presence of the drug (Arias et al. 2011; Palmer et al. 2011). Using a strain pair from a patient with E. faecalis bacteremia who failed DAP therapy, Arias et al. (2011) mapped the genetic changes to genes encoding the LiaFSR system (a conserved TCS associated with DAP-R in B. subtilis) and two enzymes involved in phospholipid metabolism, cardiolipin synthase (Cls), and a glycerophosphoryl diester phosphodiesterase (GdpD). Phenotypic changes associated with the DAP-R phenotype included increased thickness of the cell-wall and abnormal septations. Additionally, the DAP-R derivative was found to have a decrease in the proportion of PG and increased rigidity of the cell membrane (Mishra et al. 2012). However, in contrast to both S. aureus and E. faecium, a distinct characteristic of DAP-R E. faecalis is a rearrangement of cell membrane PL microdomains. Indeed, DAP-S E. faecalis shows prominent concentration of anionic PLs (including CL) at the division septum and in polar areas. Development of DAP resistance markedly changes the architecture of these PL microdomains, moving them away from the division septum, the principle site of DAP action (Tran et al. 2014). This reorganization in E. faecalis seems to be crucial for full expression of the DAP-R phenotype. It is postulated that these PL aggregates may serve as “sink holes” for DAP, diverting the antibiotic away from the vital septal area of the membrane (the diversion hypothesis) (Fig. 3B). Indeed, compelling experimental data suggest that DAP-R E. faecalis strains do not “repel” DAP from the cell surface as shown previously by S. aureus (Tran et al. 2014).
Detailed studies on the molecular basis of the DAP-R phenotype in E. faecalis has identified the LiaFSR system as a major contributor to the adaptive response against DAP and antimicrobial peptide “attack.” This system is conserved across the Firmicutes (VraSR is its ortholog in S. aureus, see above) and has been well-characterized in the model organism B. subtilis (Jordan et al. 2006; Schrecke et al. 2013). The HK LiaS responds to as-yet-unidentified membrane stressors induced by DAP or other membrane active agents. LiaS phosphorylates its cognate RR LiaR, which contains a DNA-binding motif and alters expression of target genes. LiaF serves a regulatory role by inhibiting the activation of LiaR through interactions with LiaS in the absence of membrane stress. The liaFSR operon in E. faecalis consists of only three open reading frames. However, in B. subtilis, an additional three genes, liaG, liaH, and liaI, are targets of LiaR (Wolf et al. 2010) and mediate resistance to antimicrobial peptides via a response that appears to be similar to that described for the phage shock protein (PSP) response of Gram-negative organisms (Brissette et al. 1990; Yamaguchi et al. 2013).
Several lines of experimental evidence point to an activation of the LiaFSR system and its downstream effectors as mediators of DAP-R in E. faecalis. Mutations in the predicted inhibitor LiaF have been associated with increases in DAP MIC, presumably caused by increased activity of the system. A deletion of isoleucine at position 177 of LiaF, (identified in a clinical isolate of E. faecalis) was sufficient to increase the DAP MIC of a susceptible isolate from 1 to 4 µg/mL and resulted in redistribution of membrane phospholipid microdomains (Tran et al. 2014). Further, this same change was noted to abolish the bactericidal action of DAP in vitro (loss of a three log10 decrease in time–kill curve colony counts), despite the MIC being within the “susceptible” range (Munita et al. 2013). In an experimental evolution of a polymorphic population of E. faecalis maintained in continuous culture, changes in the LiaFSR system emerged as the first step in the pathway to DAP resistance (Miller et al. 2013). The most frequently observed mutations involved either insertion or deletion of the isoleucine at position 177 in LiaF (suggesting the importance of this residue for the inhibitory function of LiaF) and appeared after ∼2 weeks as MICs rose into the 3–4 µg/mL range.
Because of the major role of LiaFSR in DAP and antimicrobial resistance, Davlieva et al. (2015) sought to investigate the structural bases of DAP-R associated with mutations in LiaR (which have been commonly identified in clinical isolates of DAP-R enterococci). These studies showed that a substitution of asparagine for aspartate at position 191 of LiaR mimics phosphorylation and changes the oligomeric state of LiaR. Indeed, “wild-type” unphosphorylated LiaR seems to exist as a dimer. When the protein is phosphorylated or harbors mutations that mimic phosphorylation LiaR tetramerizes, increasing the binding affinity for its own and other promoters by 100-fold (Davlieva et al. 2015), resulting in constitutive activation of the LiaFSR system. Furthermore, deletion of the liaR gene results in a “hypersusceptible” phenotype (MICs of 0.047 µg/mL) that is independent of the genetic background into which it is introduced (Reyes et al. 2015). Thus, LiaFSR seems crucial in orchestrating the specific response to a variety of membrane active agents and overexpression of this system results in a membrane protective effect that results in DAP-R.
Once established, LiaFSR mutations allow the accrual of additional genetic changes resulting in the full resistance phenotype (Miller et al. 2013). Mutations in genes affecting membrane phospholipids, particularly cls, have been frequently associated with DAP-R. In E. faecalis, introduction of the altered cls alleles in trans bearing the R218Q substitution or the N77-Q79 deletion were able to confer resistance to the laboratory strain OG1RF (Palmer et al. 2011). Mutations in GdpD had no effect on DAP MICs in isolation, but when introduced along with LiaF mutations, they resulted in a fully resistant phenotype (Arias et al. 2011). Genes in the LiaR regulon bear similarities to the Psp system mediated by liaI and liaH in B. subtilis, although these genes (named liaXYZ) seem to be organized into an independent operon in the E. faecalis genome distant from liaFSR (Miller et al. 2013). Interestingly, point mutations in this group of three genes, specifically a frameshift disrupting the carboxy-terminal end of LiaX and a second frameshift mutation in LiaY have been associated with DAP-R in enterococci both in vitro and in clinical isolates (Palmer et al. 2011; Humphries et al. 2012). Additional mutations in yybT, a cyclic dinucleotide phosphodiesterase predicted to be involved in cell stress and signaling, and gshF, a glutathione synthase, have been described, although their contributions to DAP-R are not well understood (Miller et al. 2013).
Daptomycin Resistance in E. Faecium
Although a number of genetic determinants of DAP-R in E. faecium have been identified, the biochemical bases for their effect on the DAP resistance phenotype are not well understood. Unlike E. faecalis, E. faecium does not display a visible alteration or rearrangement of anionic phospholipids in the membrane, even in isolates with mutations in the LiaFSR system (Tran et al. 2015). Instead, it appears that the impact of mutations in E. faecium results in phenotypic changes that are more akin to those associated with DAP-R in S. aureus (Mishra et al. 2012). Indeed, the overall mechanism for DAP-R in E. faecium appear to involve repulsion of the antibiotic from the cell surface.
Analysis of the genomes of 19 clinical isolates of E. faecium with DAP MICs ranging from 3 to 48 µg/mL revealed that the majority of the strains harbored mutations in either LiaFSR or YycFG and that either pathway can lead to DAP-R (Diaz et al. 2014). In the LiaFSR system, the most common mutation was a W73C change in LiaR accompanied by a T120A substitution in LiaS, suggesting that these changes coevolve during the development of DAP-R (Munita et al. 2012). Four strains also harbored various mutations in LiaF, although these changes did not affect the isoleucine at position 177 as described in E. faecalis. The importance of LiaFSR changes in E. faecium was shown by deletion of the liaR gene from clinical strains harboring mutations in both the LiaFSR and YycFG pathway (Panesso et al. 2015). In both cases, strains developed a “hypersusceptible” phenotype with increased binding of fluorescently labeled DAP to the cell membrane in the absence of liaR. The presence of LiaRS substitution has also been associated with clinical failure of DAP and loss of bactericidal activity of the antibiotic (Munita et al. 2014). Changes in the YycFG pathway are commonly localized to the YycG HK as well as both accessory proteins YycH and YycI (Diaz et al. 2014); however, the role of such mutations in the development of DAP-R remains to be established.
As in both S. aureus and E. faecalis, mutations in cls, the gene encoding cardiolipin synthase, are common in DAP-R E. faecium. They are often found with substitutions in LiaFSR or YycFG and, in this setting, they may contribute to the progression of an isolate from DAP-tolerant to DAP-resistant (Diaz et al. 2014). Exchange of the R218Q cls allele from a DAP-R strain into a susceptible one was not able to increase the MIC, further suggesting that this change alone is not sufficient for the development of a DAP-R phenotype in E. faecium (Tran et al. 2013). Biochemical characterization of Cls proteins from a susceptible and resistant strain pair of E. feacium showed that the R218Q and H215R substitutions mapped to the PLD1 phospholipase catalytic domain resulted in an increase in the Vmax of the enzyme (Davlieva et al. 2013). This is consistent with an enzymatic gain-of-function and may allow for a more rapid depletion of the available PG by shunting this PL to the CL pool during times of membrane stress. Mutations in a pspC-like protein (the above mentioned LiaY), cfa (a cyclooxygenase that catalyzes the addition of a methyl group to unsaturated fatty acids), dlt, and mprF, among others, have been associated with DAP-R E. faecium. However, they appear to be rare in clinical isolates and their role in resistance is currently difficult to assess (Humphries et al. 2012; Tran et al. 2013; Diaz et al. 2014).
CONCLUDING REMARKS
Over the last decade, the increase of multidrug-resistant Gram-positive organisms has brought DAP into the spotlight as a therapeutic option for severe infections. DAP has potent bactericidal activity and a unique mechanism of action, which have made it a useful addition to the clinician’s antibiotic repertoire. As its clinical use continues to increase reports of resistance are becoming more common. To preserve the use of this and other antimicrobial compounds, a deeper understanding of the robust and redundant pathways that mediate the mechanism of resistance may shed light on the biology of bacterial membrane adaptation, including the response to the innate immune system. With continued efforts to unravel the complex networks that mediate DAP-R, additional insights into the coordination of cell envelope synthesis machinery are sure to provide new therapeutic targets to exploit against recalcitrant Gram-positive infections in the future.
Footnotes
Editors: Lynn L. Silver and Karen Bush
Additional Perspectives on Antibiotics and Antibiotic Resistance available at www.perspectivesinmedicine.org
REFERENCES
- Allen NE, Alborn WE Jr, Hobbs JN Jr. 1991. Inhibition of membrane potential-dependent amino acid transport by daptomycin. Antimicrob Agents Chemother 35: 2639–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias CA, Panesso D, McGrath DM, Qin X, Mojica MF, Miller C, Diaz L, Tran TT, Rincon S, Barbu EM, et al. 2011. Genetic basis for in vivo daptomycin resistance in enterococci. N Engl J Med 365: 892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bæk KT, Thøgersen L, Mogenssen RG, Mellergaard M, Thomsen LE, Petersen A, Skov S, Cameron DR, Peleg AY, Frees D. 2015. Stepwise decrease in daptomycin susceptibility in clinical Staphylococcus aureus isolates associated with an initial mutation in rpoB and a compensatory inactivation of the clpX gene. Antimicrob Agents Chemother 59: 6983–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bals R, Wilson JM. 2003. Cathelicidins—A family of multifunctional antimicrobial peptides. Cell Mol Life Sci 60: 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz RH. 2009. Daptomycin: Mechanisms of action and resistance, and biosynthetic engineering. Curr Opin Chem Biol 13: 144–151. [DOI] [PubMed] [Google Scholar]
- Bayer AS, Schneider T, Sahl HG. 2013. Mechanisms of daptomycin resistance in Staphylococcus aureus: Role of the cell membrane and cell wall. Ann NY Acad Sci 1277: 139–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer AS, Mishra NN, Chen L, Kreiswirth BN, Rubio A, Yang SJ. 2015. Frequency and distribution of single-nucleotide polymorphisms within mprF in methicillin-resistant Staphylococcus aureus clinical Isolates and their role in cross-resistance to daptomycin and host defense antimicrobial peptides. Antimicrob Agents Chemother 59: 4930–4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsche U, Weidenmaier C, Kuehner D, Yang SJ, Baur S, Wanner S, Francois P, Schrenzel J, Yeaman MR, Bayer AS. 2011. Correlation of daptomycin resistance in a clinical Staphylococcus aureus strain with increased cell wall teichoic acid production and d-alanylation. Antimicrob Agents Chemother 55: 3922–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsche U, Yang SJ, Kuehner D, Wanner S, Mishra NN, Roth T, Nega M, Schneider A, Mayer C, Grau T, et al. 2013. Increased cell wall teichoic acid production and d-alanylation are common phenotypes among daptomycin-resistant methicillin-resistant Staphylococcus aureus (MRSA) clinical isolates. PLoS ONE 8: e67398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissette JL, Russel M, Weiner L, Model P. 1990. Phage shock protein, a stress protein of Escherichia coli. Proc Natl Acad Sci 87: 862–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafiso V, Bertuccio T, Purrello S, Campanile F, Mammina C, Sartor A, Raglio A, Stefani S. 2014. dltA overexpression: A strain-independent keystone of daptomycin resistance in methicillin-resistant Staphylococcus aureus. Int J Antimicrob Agents 43: 26–31. [DOI] [PubMed] [Google Scholar]
- Camargo IL, Neoh HM, Cui L, Hiramatsu K. 2008. Serial daptomycin selection generates daptomycin-nonsusceptible Staphylococcus aureus strains with a heterogeneous vancomycin-intermediate phenotype. Antimicrob Agents Chemother 52: 4289–4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canepari P, Boaretti M, Lleó MM, Satta G. 1990. Lipoteichoic acid as a new target for activity of antibiotics: Mode of action of daptomycin (LY146032). Antimicrob Agents Chemother 34: 1220–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Sun TL, Sun Y, Huang HW. 2014. Interaction of daptomycin with lipid bilayers: A lipid extracting effect. Biochemistry 53: 5384–5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coëffet-Le Gal MF, Thurston L, Rich P, Miao V, Baltz RH. 2006. Complementation of daptomycin dptA and dptD deletion mutations in trans and production of hybrid lipopeptide antibiotics. Microbiology 152: 2993–3001. [DOI] [PubMed] [Google Scholar]
- Cotroneo N, Harris R, Perlmutter N, Beveridge T, Silverman JA. 2008. Daptomycin exerts bactericidal activity without lysis of Staphylococcus aureus. Antimicrob Agents Chemother 52: 2223–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Isii T, Fukuda M, Ochiai T, Neoh HM, Camargo IL, Watanabe Y, Shoji M, Hishinuma T, Hiramatsu K. 2010. An RpoB mutation confers dual heteroresistance to daptomycin and vancomycin in Staphylococcus aureus. Antimicrob Agents Chemother 54: 5222–5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davlieva M, Zhang W, Arias CA, Shamoo Y. 2013. Biochemical characterization of cardiolipin synthase mutations associated with daptomycin resistance in enterococci. Antimicrob Agents Chemother 57: 289–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davlieva M, Shi Y, Leonard PG, Johnson TA, Zianni MR, Arias CA, Ladbury JE, Shamoo Y. 2015. A variable DNA recognition site organization establishes the LiaR-mediated cell envelope stress response of enterococci to daptomycin. Nucleic Acids Res 43: 4758–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debono M, Barnhart M, Carrell CB, Hoffmann JA, Occolowitz JL, Abbott BJ, Fukuda DS, Hamill RL, Biemann K, Herlihy WC. 1987. A21978C, a complex of new acidic peptide antibiotics: Isolation, chemistry, and mass spectral structure elucidation. J Antibiot (Tokyo) 40: 761–777. [DOI] [PubMed] [Google Scholar]
- Debono M, Abbott BJ, Molloy RM, Fukuda DS, Hunt AH, Daupert VM, Counter FT, Ott JL, Carrell CB, Howard LC, et al. 1988. Enzymatic and chemical modifications of lipopeptide antibiotic A21978C: The synthesis and evaluation of daptomycin (LY146032). J Antibiot (Tokyo) 41: 1093–1105. [DOI] [PubMed] [Google Scholar]
- Diaz L, Tran TT, Munita JM, Miller WR, Rincon S, Carvajal LP, Wollam A, Reyes J, Panesso D, Rojas NL, et al. 2014. Whole-genome analyses of Enterococcus faecium isolates with diverse daptomycin MICs. Antimicrob Agents Chemother 58: 4527–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrac S, Msadek T. 2004. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J Bacteriol 186: 1175–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrac S, Boneca IG, Poupel O, Msadek T. 2007. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J Bacteriol 189: 8257–8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrac S, Bisicchia P, Devine KM, Msadek T. 2008. A matter of life and death: Cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol Microbiol 70: 1307–1322. [DOI] [PubMed] [Google Scholar]
- Eisenstein BI, Oleson FB, Baltz RH. 2010. Daptomycin: From the mountain to the clinic, with essential help from Francis Tally, MD. Clin Infect Dis 50: S10–S15. [DOI] [PubMed] [Google Scholar]
- Epand RF, Savage PB, Epand RM. 2007. Bacterial lipid composition and the antimicrobial efficacy of cationic steroid compounds (ceragenins). Biochim Biophys Acta 1768: 2500–2509. [DOI] [PubMed] [Google Scholar]
- Ernst CM, Staubitz P, Mishra NN, Yang SJ, Hornig G, Kalbacher H, Bayer AS, Kraus D, Peschel A. 2009. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog 5: e1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Walsh CT. 2006. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic, machinery, and mechanisms. Chem Rev 106: 3468–3496. [DOI] [PubMed] [Google Scholar]
- Fischer A, Yang SJ, Bayer AS, Vaezzadeh AR, Herzig S, Stenz L, Girard M, Sakoulas G, Scherl A, Yeaman MR, et al. 2011. Daptomycin resistance mechanisms in clinically derived Staphylococcus aureus strains assessed by a combined transcriptomics and proteomics approach. J Antimicrob Chemother 66: 1696–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L, Alder JD, Silverman JA. 2006. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob Agents Chemother 50: 2137–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima T, Furihata I, Emmins R, Daniel RA, Hoch JA, Szurmant H. 2011. A role for the essential YycG sensor histidine kinase in sensing cell division. Mol Microbiol 79: 503–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaupp R, Lei S, Reed JM, Peisker H, Boyle-Vavra S, Bayer AS, Bischoff M, Herrmann M, Daum RS, Powers R, et al. 2015. Staphylococcus aureus metabolic adaptations during the transition from a daptomycin susceptibility phenotype to a daptomycin nonsusceptibility phenotype. Antimicrob Agents Chemother 59: 4226–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünewald J, Sieber SA, Mahlert C, Linne U, Marahiel MA. 2004. Synthesis and derivatization of daptomycin: A chemoenzymatic route to acidic lipopeptide antibiotics. J Am Chem Soc 126: 17025–17031. [DOI] [PubMed] [Google Scholar]
- Gurtovenko AA, Vattulainen I. 2007. Ion leakage through transient water pores in protein-free lipid membranes driven by transmembrane ionic charge imbalance. Biophys J 92: 1878–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann B, Kaplan MA, Muir RD, Hooper IR. 1953. Amphomycin, a new antibiotic. Antibiot Chemother (Northfield) 3: 1239–1242. [PubMed] [Google Scholar]
- Ho SW, Jung D, Calhoun JR, Lear JD, Okon M, Scott WR, Hancock RE, Straus SK. 2008. Effect of divalent cations on the structure of the antibiotic daptomycin. Eur Biophys J 37: 421–433. [DOI] [PubMed] [Google Scholar]
- Hojati Z, Milne C, Harvey B, Gordon L, Borg M, Flett F, Wilkinson B, Sidebottom PJ, Rudd BA, Hayes MA, et al. 2002. Structure, biosynthetic origin, and engineered biosynthesis of calcium-dependent antibiotics from Streptomyces coelicolor. Chem Biol 9: 1175–1187. [DOI] [PubMed] [Google Scholar]
- Howden BP, McEvoy CR, Allen DL, Chua K, Gao W, Harrison PF, Bell J, Coombs G, Bennett-Wood V, Porter JL, et al. 2011. Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR. PLoS Pathog 7: e1002359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber FM, Pieper RL, Tietz AJ. 1988. The formation of daptomycin by supplying decanoic acid to Streptomyces roseosporus cultures producing the antibiotic complex A21978C. J Biotechnol 7: 283–292. [Google Scholar]
- Humphries RM, Kelesidis T, Tewhey R, Rose WE, Schork N, Nizet V, Sakoulas G. 2012. Genotypic and phenotypic evaluation of the evolution of high-level daptomycin nonsusceptibility in vancomycin-resistant Enterococcus faecium. Antimicrob Agents Chemother 56: 6051–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T, Yeaman MR, Sakoulas G, Yang SJ, Proctor RA, Sahl HG, Schrenzel J, Xiong YQ, Bayer AS. 2008. Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob Agents Chemother 52: 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan S, Junker A, Helmann JD, Mascher T. 2006. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: Identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J Bacteriol 188: 5153–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D, Rozek A, Okon M, Hancock RE. 2004. Structural transitions as determinants of the action of the calcium-dependent antibiotic daptomycin. Chem Biol 11: 949–957. [DOI] [PubMed] [Google Scholar]
- Kilelee E, Pokorny A, Yeaman MR, Bayer AS. 2010. Lysyl-phosphatidylglycerol attenuates membrane perturbation rather than surface association of the cationic antimicrobial peptide 6W-RP-1 in a model membrane system: Implications for daptomycin resistance. Antimicrob Agents Chemother. 54: 4476–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M, Kuroda H, Oshima T, Takeuchi F, Mori H, Hiramatsu K. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol 49: 807–821. [DOI] [PubMed] [Google Scholar]
- Leonard PG, Golemi-Kotra D, Stock AM. 2013. Phosphorylation-dependent conformational changes and domain rearrangements in Staphylococcus aureus VraR activation. Proc Natl Acad Sci 110: 8525–8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakina NN, Brazhnikova MG. 1959. Chemical composition of crystallomycin. Biokhimiia 24: 425–431. [PubMed] [Google Scholar]
- Marahiel MA, Stachelhaus T, Mootz HD. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev 97: 2651–2674. [DOI] [PubMed] [Google Scholar]
- Mascio CT, Alder JD, Silverman JA. 2007. Bactericidal action of daptomycin against stationary-phase and nondividing Staphylococcus aureus cells. Antimicrob Agents Chemother 51: 4255–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mchenney MA, Hosted TJ, Dehoff BS, Rosteck PR Jr, Baltz RH. 1998. Molecular cloning and physical mapping of the daptomycin gene cluster from Streptomyces roseosporus. J Bacteriol 180: 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S, Cuirolo AX, Plata KB, Riosa S, Silverman JA, Rubio A, Rosato RR, Rosato AE. 2012. VraSR two-component regulatory system contributes to mprF-mediated decreased susceptibility to daptomycin in in vivo-selected clinical strains of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 56: 92–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengin-Lecreulx D, Allen NE, Hobbs JN, van Heijenoort J. 1990. Inhibition of peptidoglycan biosynthesis in Bacillus megaterium by daptomycin. FEMS Microbiol Lett 57: 245–248. [DOI] [PubMed] [Google Scholar]
- Miao V, Coëffet-Legal MF, Brian P, Brost R, Penn J, Whiting A, Martin S, Ford R, Parr I, Bouchard M, et al. 2005. Daptomycin biosynthesis in Streptomyces roseosporus: Cloning and analysis of the gene cluster and revision of peptide stereochemistry. Microbiology 151: 1507–1523. [DOI] [PubMed] [Google Scholar]
- Miller C, Kong J, Tran TT, Arias CA, Saxer G, Shamoo Y. 2013. Adaptation of Enterococcus faecalis to daptomycin reveals an ordered progression to resistance. Antimicrob Agents Chemother 57: 5373–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WR, Munita JM, Arias CA. 2014. Mechanisms of antibiotic resistance in enterococci. Expert Rev Anti Infect Ther 12: 1221–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra NN, Bayer AS. 2013. Correlation of cell membrane lipid profiles with daptomycin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 57: 1082–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra NN, Yang SJ, Sawa A, Rubio A, Nast CC, Yeaman MR, Bayer AS. 2009. Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 53: 2312–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra NN, McKinnell J, Yeaman MR, Rubio A, Nast CC, Chen L, Kreiswirth BN, Bayer AS. 2011. In vitro cross-resistance to daptomycin and host defense cationic antimicrobial peptides in clinical methicillin-resistant Staphylococcus aureus isolates. Antimicrob Agents Chemother 55: 4012–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra NN, Bayer AS, Tran TT, Shamoo Y, Mileykovskaya E, Dowhan W, Guan Z, Arias CA. 2012. Daptomycin resistance in enterococci is associated with distinct alterations of cell membrane phospholipid content. PLoS ONE 7: e43958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra NN, Bayer AS, Weidenmaier C, Grau T, Wanner S, Stefani S, Cafiso V, Bertuccio T, Yeaman MR, Nast CC, et al. 2014. Phenotypic and genotypic characterization of daptomycin-resistant methicillin-resistant Staphylococcus aureus strains: Relative roles of mprF and dlt operons. PLoS ONE 9: e107426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munita JM, Panesso D, Diaz L, Tran TT, Reyes J, Wanger A, Murray BE, Arias CA. 2012. Correlation between mutations in liaFSR of Enterococcus faecium and MIC of daptomycin: Revisiting daptomycin breakpoints. Antimicrob Agents Chemother 56: 4354–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munita JM, Tran TT, Diaz L, Panesso D, Reyes J, Murray BE, Arias CA. 2013. A liaF codon deletion abolishes daptomycin bactericidal activity against vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother 57: 2831–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munita JM, Mishra NN, Alvarez D, Tran TT, Diaz L, Panesso D, Reyes J, Murray BE, Adachi JA, Bayer AS, et al. 2014. Failure of high-dose daptomycin for bacteremia caused by daptomycin-susceptible Enterococcus faecium harboring LiaSR substitutions. Clin Infect Dis 59: 1277–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munita JM, Bayer AS, Arias CA. 2015. Evolving resistance among Gram-positive pathogens. Clin Infect Dis. 61: S48–S57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraih JK, Palmer M. 2012. Estimation of the subunit stoichiometry of the membrane-associated daptomycin oligomer by FRET. Biochim Biophys Acta 1818: 1642–1647. [DOI] [PubMed] [Google Scholar]
- Muraih JK, Pearson A, Silverman J, Palmer M. 2011. Oligomerization of daptomycin on membranes. Biochim Biophys Acta 1808: 1154–1160. [DOI] [PubMed] [Google Scholar]
- Muraih JK, Harris J, Taylor SD, Palmer M. 2012. Characterization of daptomycin oligomerization with perylene excimer fluorescence: Stoichiometric binding of phosphatidylglycerol triggers oligomer formation. Biochim Biophys Acta 1818: 673–678. [DOI] [PubMed] [Google Scholar]
- Muthaiyan A, Silverman JA, Jayaswal RK, Wilkinson BJ. 2008. Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrob Agents Chemother 52: 980–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwangi MM, Wu SW, Zhou Y, Sieradzki K, de Lencastre H, Richardson P, Bruce D, Rubin E, Myers E, Siggia ED, et al. 2007. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc Natl Acad Sci 104: 9451–9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganawa H, Hamada M, Maeda K, Okami Y, Takeushi T. 1968. Laspartomycin, a new anti-staphylococcal peptide. J Antibiot (Tokyo) 21: 55–62. [DOI] [PubMed] [Google Scholar]
- Nguyen KT, Ritz D, Gu JQ, Alexander D, Chu M, Miao V, Brian P, Baltz RH. 2006. Combinatorial biosynthesis of novel antibiotics related to daptomycin. Proc Natl Acad Sci 103: 17462–17467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen KT, He X, Alexander DC, Li C, Gu JQ, Mascio C, Van Praagh A, Mortin L, Chu M, Silverman JA, et al. 2010. Genetically engineered lipopeptide antibiotics related to A54145 and daptomycin with improved properties. Antimicrob Agents Chemother 54: 1404–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer KL, Daniel A, Hardy C, Silverman J, Gilmore MS. 2011. Genetic basis for daptomycin resistance in enterococci. Antimicrob Agents Chemother 55: 3345–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panesso D, Reyes J, Gaston EP, Deal M, Londoño A, Nigo M, Munita JM, Miller WR, Shamoo Y, Tran TT, et al. 2015. Deletion of liaR reverses daptomycin resistance in Enterococcus faecium independent of the genetic background. Antimicrob Agents Chemother 59: 7327–7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg AY, Miyakis S, Ward DV, Earl AM, Rubio A, Cameron DR, Pillai S, Moellering RC Jr, Eliopoulos GM. 2012. Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS ONE 7: e28316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J, Pogliano N, Silverman JA. 2012. Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J Bacteriol 194: 4494–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall CP, Mariner KR, Chopra I, O’Neill AJ. 2013. The target of daptomycin is absent from Escherichia coli and other Gram-negative pathogens. Antimicrob Agents Chemother 57: 637–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes J, Panesso D, Tran TT, Mishra NN, Cruz MR, Munita JM, Singh KV, Yeaman MR, Murray BE, Shamoo Y, et al. 2015. A liaR deletion restores susceptibility to daptomycin and antimicrobial peptides in multidrug-resistant Enterococcus faecalis. J Infect Dis 211: 1317–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose WE, Fallon M, Moran JJ, Vanderloo JP. 2012. Vancomycin tolerance in methicillin-resistant Staphylococcus aureus: Influence of vancomycin, daptomycin, and telavancin on differential resistance gene expression. Antimicrob Agents Chemother 56: 4422–4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio A, Conrad M, Haselbeck RJ, Kedar GC, Brown-Driver V, Finn J, Silverman JA. 2011. Regulation of mprF by antisense RNA restores daptomycin susceptibility to daptomycin-resistant isolates of Staphylococcus aureus. Antimicrob Agents Chemother 55: 364–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrecke K, Jordan S, Mascher T. 2013. Stoichiometry and perturbation studies of the LiaFSR system of Bacillus subtilis. Mol Microbiol 87: 769–788. [DOI] [PubMed] [Google Scholar]
- Scott WR, Baek SB, Jung D, Hancock RE, Straus SK. 2007. NMR structural studies of the antibiotic lipopeptide daptomycin in DHPC micelles. Biochim Biophys Acta 1768: 3116–3126. [DOI] [PubMed] [Google Scholar]
- Shay AJ, Adam J, Martin JH, Hausmann WK, Shu P, Bohonos N. 1960. Aspartocin. I: Production, isolation, and characteristics. Antibiot Annu 7: 194–198. [PubMed] [Google Scholar]
- Shibata M, Kanzaki T, Nakazawa K, Inoue M, Hitomi H, Mizuno K, Fujino M, Akira M. 1962. On glumamycin, a new antibiotic. J Antibiot (Tokyo) 15: 1–6. [PubMed] [Google Scholar]
- Shoji JI, Kozuki S, Okamoto S, Sakazaki R, Otsuka H. 1968. Studies on tsushimycin. I: Isolation and characterization of an acidic acylpeptide containing a new fatty acid. J Antibiot (Tokyo) 21: 439–443. [PubMed] [Google Scholar]
- Short SA, White DC. 1972. Biosynthesis of cardiolipin from phosphatidylglycerol in Staphylococcus aureus. J Bacteriol 109: 820–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JA, Perlmutter NG, Shapiro HM. 2003. Correlation of daptomycin bactericidal activity and membrane depolarization in Staphylococcus aureus. Antimicrob Agents Chemother 47: 2538–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus SK, Hancock RE. 2006. Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: Comparison with cationic antimicrobial peptides and lipopeptides. Biochim Biophys Acta 1758: 1215–1223. [DOI] [PubMed] [Google Scholar]
- Szurmant H, Nelson K, Kim EJ, Perego M, Hoch JA. 2005. YycH regulates the activity of the essential YycFG two-component system in Bacillus subtilis. J Bacteriol 187: 5419–5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmant H, Mohan MA, Imus PM, Hoch JA. 2007. YycH and YycI interact to regulate the essential YycFG two-component system in Bacillus subtilis. J Bacteriol 189: 3280–3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TT, Panesso D, Gao H, Roh JH, Munita JM, Reyes J, Diaz L, Lobos EA, Shamoo Y, Mishra NN, et al. 2013. Whole-genome analysis of a daptomycin-susceptible Enterococcus faecium strain and its daptomycin-resistant variant arising during therapy. Antimicrob Agents Chemother 57: 261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TT, Panesso D, Mishra NN, Mileykovskaya E, Guan Z, Munita JM, Reyes J, Diaz L, Weinstock GM, Murray BE, Shamoo Y, et al. 2014. Daptomycin-resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids. mBio 4: e00281–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TT, Munita JM, Arias CA. 2015. Mechanisms of drug resistance: Daptomycin resistance. Ann NY Acad Sci 1354: 32–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Türck M, Bierbaum G. 2012. Purification and activity testing of the full-length YycFGHI proteins of Staphylococcus aureus. PLoS ONE 7: e30403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utaida S, Dunman PM, Macapagal D, Murphy E, Projan SJ, Singh VK, Jayaswal RK, Wilkinson BJ. 2003. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149: 2719–2732. [DOI] [PubMed] [Google Scholar]
- Wolf D, Kalamorz F, Wecke T, Juszczak A, Mäder U, Homuth G, Jordan S, Kirstein J, Hoppert M, Voigt B, et al. 2010. In-depth profiling of the LiaR response of Bacillus subtilis. J Bacteriol 192: 4680–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Reid DA, Rothenberg E, Darwin AJ. 2013. Changes in Psp protein binding partners, localization and behaviour upon activation of the Yersinia enterocolitica phage shock protein response. Mol Microbiol 87: 656–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SJ, Kreiswirth BN, Sakoulas G, Yeaman MR, Xiong YQ, Sawa A, Bayer AS. 2009. Enhanced expression of dltABCD is associated with the development of daptomycin nonsusceptibility in a clinical endocarditis isolate of Staphylococcus aureus. J Infect Dis 200: 1916–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SJ, Mishra NN, Rubio A, Bayer AS. 2013. Causal role of single nucleotide polymorphisms within the mprF gene of Staphylococcus aureus in daptomycin resistance. Antimicrob Agents Chemother 57: 5658–5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Muraih JK, Tishbi N, Herskowitz J, Victor RL, Silverman J, Uwumarenogie S, Taylor SD, Palmer M, Mintzer E. 2014a. Cardiolipin prevents membrane translocation and permeabilization by daptomycin. J Biol Chem 289: 11584–11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Muraih JK, MacCormick B, Silverman J, Palmer M. 2014b. Daptomycin forms cation- and size-selective pores in model membranes. Biochim Biophys Acta 1838: 2425–2430. [DOI] [PubMed] [Google Scholar]