Abstract
Lincosamides, streptogramins, phenicols, and pleuromutilins (LSPPs) represent four structurally different classes of antimicrobial agents that inhibit bacterial protein synthesis by binding to particular sites on the 50S ribosomal subunit of the ribosomes. Members of all four classes are used for different purposes in human and veterinary medicine in various countries worldwide. Bacteria have developed ways and means to escape the inhibitory effects of LSPP antimicrobial agents by enzymatic inactivation, active export, or modification of the target sites of the agents. This review provides a comprehensive overview of the mode of action of LSPP antimicrobial agents as well as of the mutations and resistance genes known to confer resistance to these agents in various bacteria of human and animal origin.
LSPP antimicrobials bind to target sites on bacterial 50S ribosomal subunits and inhibit protein synthesis. Bacterial resistance mechanisms to LSPPs include enzymatic inactivation, active export, and target site modification.
For more than 70 years, antimicrobial agents have been indispensable for the control of bacterial infections in human and veterinary medicine. They inhibit bacteria mainly by interfering with cell-wall synthesis, nucleic acid synthesis, or protein synthesis. Their efficacy is, however, hampered by the continuous development of resistance not only by the target bacteria but also by members of the physiological microbiota in both humans and animals. Lincosamides, streptogramins, phenicols, and pleuromutilins (LSPPs) represent four classes of antimicrobial agents that inhibit protein synthesis by interacting with the 50S subunit of bacterial ribosomes. Bacterial resistance to LSPP antimicrobial agents can be a result of the acquisition of endogenous mutations or horizontally transmitted resistance genes (Schwarz et al. 2006). The mechanisms known so far, associated with resistance to LSPP antimicrobial agents, commonly fall into three categories: enzymatic inactivation, active efflux, and/or structural changes at the ribosomal target site. Many of the resistance genes known so far that confer LSPP resistance are located on mobile genetic elements, which facilitate their dissemination across strain, species, and even genus boundaries. The understanding of the mode of action of antimicrobial agents has in the past led to the development of derivatives of known antimicrobial agents, designed to escape preexisting bacterial resistance mechanisms (Schwarz and Kehrenberg 2006). As such, the knowledge of not only the mode of action but also of the mode of resistance is a key parameter in the understanding of the complex interaction of antimicrobial agents and bacteria.
In the present review, we summarize the current knowledge of the mode of action of LSPP antimicrobial agents and provide an overview of the genetic basis of resistance to these agents in bacteria of human and animal origin.
USE OF LSPP ANTIMICROBIAL AGENTS IN HUMAN AND VETERINARY MEDICINE
Lincosamides
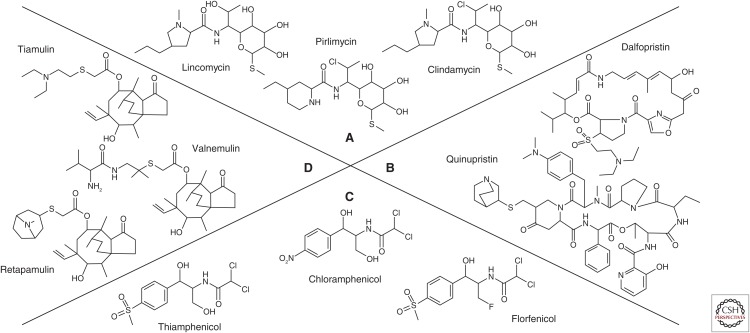
Lincosamides consist of three components: an amino acid (aa) (l-proline substituted by a 4′-alkyl chain) and a sugar (lincosamine), connected by an amide bond (Fig. 1A) (Bryskier 2005a). The first lincosamide, lincomycin, was isolated in 1962 from Streptomyces lincolnensis ssp. lincolnensis found in a soil sample from Lincoln, NE (MacLeod et al. 1964; Bryskier 2005a). In 1967, lincomycin was licensed in the United States for the treatment of infections caused by Gram-positive bacteria. Although approved for use in human medicine, lincomycin is rarely used nowadays. In veterinary medicine, lincomycin is approved for use in various infections in swine, dogs, and cats. In combination with spectinomycin, it is not only approved for ruminants, pigs, and poultry (Giguère 2013) but also for dogs, cats, and carrier pigeons.
Figure 1.
Structural formulas of the lincosamides, streptogramins, phenicols, and pleuromutilins (LSPP) antimicrobial agents. (A) The lincosamides lincomycin, clindamycin, and pirlimycin, (B) the streptogramin A dalfopristin and the streptogramin B quinupristin, (C) the phenicols chloramphenicol, thiamphenicol, and florfenicol, and (D) the pleuromutilins tiamulin, valnemulin, and retapamulin.
As lincomycin has only a limited spectrum of activity, various chemical modifications were introduced to improve the pharmacokinetics of lincomycin and to expand its antibacterial spectrum. The 7-chloro-7-deoxylincomycin derivative, clindamycin, proved to be the most effective. Clindamycin was approved by the Food and Drug Administration (FDA) in 1970 in the United States. The antimicrobial spectrum of clindamycin includes staphylococci, group A and B streptococci, Streptococcus pneumoniae, most anaerobic bacteria, and Chlamydia trachomatis (Smieja 1998). Moreover, clindamycin also shows activity against several protozoa, such as Plasmodium spp. and Toxoplasma spp. (Bryskier 2005a). However, it shows little, if no, activity against most aerobic Gram-negative bacilli, Nocardia spp., Mycobacterium spp., as well as Enterococcus faecalis and Enterococcus faecium (Giguère 2013). In veterinary medicine, clindamycin must not be used in food-producing animals, but may be used in dogs and cats under the Animal Medicinal Drug Use Clarification Act of 1994 (AMDUCA) in the United States and under similar regulations in other countries. Clindamycin is often used in human medicine for the treatment of infections caused by anaerobic bacteria. Because of its activity against anaerobes, it can lead to disruption of the intestinal microbiota and Clostridium difficile overgrowth causing diarrhea and colitis (Gerding et al. 1995). A similar situation has been observed in horses in which lincosamide administration can cause C. difficile–associated disease (CDAD), that is, a severe to fatal enterocolitis (Diab et al. 2013; Giguère 2013). It should be noted that lincosamides are also highly toxic to rabbits, guinea pigs, and hamsters, where toxins produced by Clostridium perfringens and other clostridia have been implicated in lincomycin- and clindamycin-induced enteritis (Morris 1995).
A new lincosamide, pirlimycin, was approved in 2000 in the United States and in 2001 in the European Union (EU). Pirlimycin represents a cis-4-ethyl-l-picecolic acid amide of clindamycin (Ahonkhai et al. 1982). It is exclusively approved for veterinary applications, that is, as an intramammary infusion for the control of staphylococci and streptococci associated with bovine subclinical mastitis.
Streptogramins
Streptogramins (pristinamycin, virginiamycin, mikamycin, and quinupristin–dalfopristin) consist of two structurally different components, A and B (Fig. 1B). The A components, such as pristinamycin IIA, virginiamycin M, mikamycin A, or dalfopristin, are polyunsaturated macrolactones. The B components, such as pristinamycin IB, virginiamycin S, mikamycin B, or quinupristin, are cyclic hexadepsipeptides (Allignet et al. 1996; Giguère 2013).
Pristinamycin was identified in culture filtrates of Streptomyces pristinaespiralis in 1962, virginiamycin in Streptomyces virginiae in 1955, and mikamycin in Streptomyces mitakaensis in 1956 (Vazquez 1966). Natural mixtures of streptogramins have been available for clinical use in Europe since the mid-1950s. Streptogramins cannot cross the outer membrane of most Gram-negative bacteria and are primarily effective against Gram-positive bacteria (Wright 2007). They have been used topically or orally in the treatment of skin, bone, and respiratory infections, mainly caused by staphylococci (Allignet et al. 1996). The first semisynthetic injectable streptogramin compound, quinupristin–dalfopristin, was approved in 1999. Quinupristin–dalfopristin represents one of the few potential antimicrobial agents for the treatment of infections in humans caused by multiresistant E. faecium, especially in cases of vancomycin- and/or linezolid-resistant isolates and also for multiresistant staphylococci, including methicillin-resistant Staphylococcus aureus (MRSA) (see who.int/iris/bitstream/10665/43765/1/9789241595742_eng.pdf). Therefore, it is considered as a last resort drug for human use (see ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2011/11/WC500118230.pdf).
Virginiamycin has been developed largely as a growth promoter (Giguère 2013) but its use for that purpose was phased out in the EU by the end of 2005. Since January 1, 2006, antimicrobial agents are not allowed to be used as growth promoters in the EU anymore. Streptogramins are currently not approved for therapeutic purposes in veterinary medicine in the EU, but virginiamycin may be used for specific indications in swine and horses in North America (Giguère 2013).
Phenicols
Chloramphenicol was isolated from Streptomyces venezuelae in 1947. It was the first phenicol antimicrobial agent and also the first natural product found to contain a nitro group (Fig. 1C). Because of the relative simplicity of its structure, chloramphenicol has been produced synthetically since 1950 (Schwarz et al. 2004). Thiamphenicol is a derivative of chloramphenicol, in which the p-nitro group has been replaced by a sulfomethyl group. Florfenicol is a fluorinated derivative of thiamphenicol in which the hydroxyl group at C3 has been replaced with fluorine (Dowling 2013).
Based on its activity against a wide range of Gram-positive and Gram-negative bacteria, chloramphenicol was initially considered a promising antimicrobial agent (Shaw 1983). However, serious adverse effects have been observed since the mid-1960s. These included a dose-unrelated irreversible aplastic anemia, a dose-related reversible bone marrow suppression, or the Gray syndrome in neonates and infants. In addition, hypersensitivity to chloramphenicol has been observed occasionally (Schwarz et al. 2004). Based on these adverse effects and the availability of less toxic antimicrobial agents with a similar spectrum of activity, chloramphenicol is now used in human medicine only for the treatment of a small number of life-threatening infections or for topical applications, for example, in eye infections. In veterinary medicine, chloramphenicol is still used in pets and non-food-producing animals. It was banned from use in food-producing animals in the EU in 1994 and in many other countries soon thereafter.
Thiamphenicol shows lower antimicrobial activity than chloramphenicol and, therefore, has rarely been used in human and veterinary medicine. Both, thiamphenicol and florfenicol do not cause dose-unrelated irreversible aplastic anemia, but may cause dose-dependent bone marrow suppression in animals (Dowling 2013). Florfenicol is exclusively approved for use in food-producing animals. Since 1995, florfenicol has been approved in numerous countries for the treatment of respiratory disease, pododermatitis, and keratoconjunctivitis in cattle, swine respiratory disease, air sacculitis in broiler chickens, but also for various diseases in fish. The application of florfenicol in horses is not recommended (Dowling 2013).
Pleuromutilins
Pleuromutilins are diterpene antimicrobial agents (Fig. 1D). They were discovered as natural antimicrobial agents in 1950/1951 (Bryskier 2005b; Novak and Shlaes 2010). The first pleuromutilin was found in the basidiomycete Pleurotus mutilus, later renamed as Clitopilus scyphoides (Bryskier 2005b). Tiamulin and valnemulin are semisynthetic pleuromutilins. Tiamulin was the first pleuromutilin to be approved for veterinary use in 1979, followed by valnemulin in 1999 (Novak and Shlaes 2010). Both tiamulin and valnemulin are exclusively used in veterinary medicine, mainly for the control of bacterial infections in pigs and poultry. They are particularly active against anaerobic bacteria, Mycoplasma spp., and selected Gram-positive and Gram-negative bacteria (Giguère 2013). In swine, tiamulin is commonly used for the treatment of swine dysentery caused by Brachyspira hyodysenteriae, swine pneumonia caused by Actinobacillus pleuropneumoniae and Mycoplasma hyodysenteriae, colonic spirochaetosis caused by Brachyspira pilosicoli, and proliferative enteropathy caused by Lawsonia intracellularis. In poultry, both pleuromutilins are commonly used for the treatment of Mycoplasma gallisepticum infections. Pleuromutilins are not approved for use in ruminants and should also not be administered to horses (Giguère 2013).
In 2007, the first pleuromutilin antimicrobial agent, retapamulin, was approved for use in humans (Novak and Shlaes 2010). However, the use of retapamulin is limited to topical treatment of impetigo caused by methicillin-susceptible S. aureus (MSSA) or Streptococcus pyogenes in patients of 9 months or older.
MODE OF ACTION OF LSPP ANTIMICROBIAL AGENTS
Early studies of the mechanisms of action of various antimicrobial agents relied on their effects in various functional assays. The interpretations of these studies were influenced by the lack of knowledge of their binding sites, and whether or not the effect assayed for was a major/minor or a direct/indirect effect. Details and summaries of these studies for translational inhibitors have been reviewed by Wilson (2009). After solution of the bacterial ribosome structure in the year 2000 (Ban et al. 2000), the unraveling of mechanisms of action for protein synthesis inhibitors increased tremendously because the binding sites of the antimicrobial agents could now be determined. The ribosome crystals were coincubated with the antimicrobial agents and thereafter the detailed position of binding could be determined for each antimicrobial agent. From the position in the ribosomes, the overall mode of action could then be deduced.
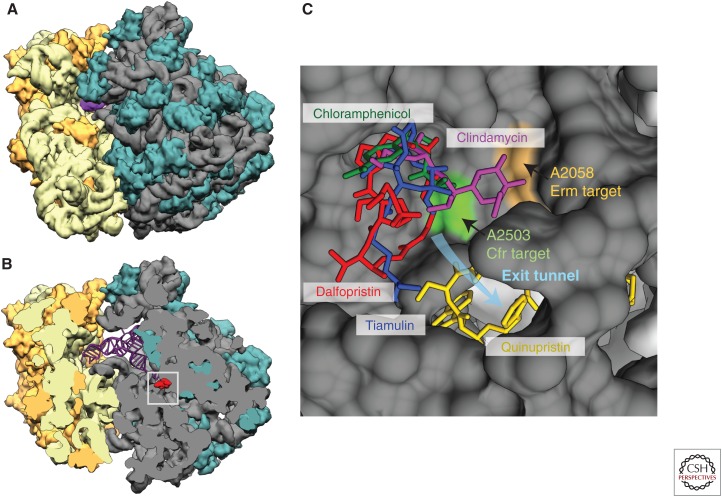
LSPP antimicrobial agents have very diverse chemical structures as shown by the examples presented in Figure 1. Despite their differences, they have all been found to bind at the peptidyltransferase center (PTC) of bacterial ribosomes. The ribosome structures with the bound antimicrobial agents have been obtained from various bacteria (Escherichia coli, Deinococcus radiodurans, Thermus thermophilus, and S. aureus) and one archaeon Haloarcula marismortui, and they show some minor differences in antibiotic binding. Details and summaries of these structures have been described by Wilson (2011, 2014) and Eyal et al. (2015). It is possible to transfer the binding positions from one structure to another to compare their binding sites as presented in Figure 2, which shows binding of one example from each antimicrobial class placed together. It is thus obvious that LSPP antimicrobial agents have overlapping binding sites and that each of them occupies specific sites in the ribosome in the area where the ribosome extends the nascent peptide chain. The binding site is in the bottom of the cleft of the 50S ribosomal subunit where the 3′-ends of aminoacyl-tRNA and peptidyl-tRNA are positioned for peptide transfer. The site is composed exclusively of RNA, and mainly RNA from the central part of domain V of 23S RNA (Nissen et al. 2000), and it is highly conserved in all bacteria. The same site in the ribosome also binds the oxazolidinones including linezolid.
Figure 2.
Lincosamides, streptogramins, phenicols, and pleuromutilins (LSPP) binding to the ribosome. (A) A model of the Escherichia coli bacterial ribosome with the two ribosomal subunits (based on RCSB Protein Data Bank [PDB] 4V9D; see rcsb.org, last accessed August 9, 2015), 30S with RNA in light yellow and proteins in darker yellow, 50S with RNA in grey and proteins in greenish. The magenta is the anticodon tip of tRNA in P-site. (B) A cut-view of the 50S subunit from A with a streptogramin A molecule (dalfopristin from PDB 4U26) in red to mark the peptidyltransferase center (PTC) area. The square marks the approximate area shown as a blow-up in C. (C) The overlapping binding sites of lincosamides, streptogramins, phenicols, and pleuromutilins exemplified by clindamycin (in pink) from PDB 4V7T, quinupristin (in yellow), and dalfopristin (in red) from PDB 4U26, chloramphenicol (in green) from PDB 4V7V, and tiamulin (in blue) from PDB 1XBP, respectively, and all placed in the E. coli structure. All PDB structures are from rcsb.org/pdb/home/home.do (Berman et al. 2000). The colored nucleotide bases indicate the positions methylated by Erm and Cfr methyltransferases. Nucleotides “in front” of the antimicrobial agents have been removed to be able to see the agents in their binding site.
Streptogramin B antimicrobial agents bind at a site adjacent to the PTC in the beginning of the nascent peptide exit tunnel (Fig. 2) and overlap the binding site of macrolides (reviewed in Poehlsgaard and Douthwaite 2005). The environment of the PTC seems to facilitate binding of a range of antimicrobial agents, which in this way interfere with the peptide transfer process. They can thus either disturb the positioning of aminoacyl-tRNA or peptidyl-tRNA for peptide transfer or directly block some movements required during peptide transfer. The overall mechanism of action is the inhibition of peptide transfer by sterically blocking the transfer process. The exact effect depends on the access of the antimicrobial agents to the PTC (during the initiation or the nascent chain elongation), “on” and “off” rates of each agent, and their binding to the ribosomal A- and/or P-site.
The phenicols are small molecules and at least one of them, chloramphenicol, seems to have an alternative binding site in archael ribosomes as discussed by Dunkle et al. (2010), but still with its active site in the A-site of the PTC. The structural data on phenicol binding to the ribosome have been obtained with chloramphenicol, but florfenicol and thiamphenicol probably bind similarly. Lincosamides bind adjacent to and a bit overlapping with chloramphenicol and also at the A-site of PTC (Fig. 2) (Dunkle et al. 2010). The streptogramins appear naturally in pairs and bind together and work synergistically. The A component binds in PTC and the B component binds right beside it at the entrance of the tunnel (Fig. 2). The binding and effect on translation has recently been revisited by Noeske et al. (2014). All pleuromutilins have a conserved tricyclic core that fits nicely in a “cave” in the A-site PTC of the ribosomes (Davidovich et al. 2007). They vary by extensions pointing toward the P-site and occupying different positions (Davidovich et al. 2007).
The older literature focused on the effect of various assays such as the puromycin reaction, fMet-tRNA binding, A- and P-site tRNA binding, translocation, etc.; however, some of these assays were under unnatural conditions and sometimes contradicting results were obtained. Nowadays, focus is more on defining the binding sites and using this knowledge: (1) to develop derivatives that bind more strongly to the ribosome and/or that bind despite target site modifications providing resistance; (2) to avoid other resistance mechanisms; and (3) to improve solubility and pharmacokinetics of the antimicrobial agents without compromising their effect.
PREVALENCE OF RESISTANCE TO LSPP ANTIMICROBIAL AGENTS
To classify a bacterial isolate as resistant or susceptible to an antimicrobial agent, clinical breakpoints are necessary. The Clinical and Laboratory Standards Institute (CLSI) has published a wide range of clinical breakpoints applicable to bacteria from humans and from animals (CLSI 2015, 2016). Among the lincosamides, no clinical breakpoints are available for lincomycin, whereas those for pirlimycin are applicable only to S. aureus and certain streptococcal species from bovine mastitis. For clindamycin, clinical breakpoints are only available for human staphylococci, streptococci, and anaerobes as well as for canine staphylococci and β-hemolytic streptococci. For the streptogramin combination quinupristin–dalfopristin, only clinical breakpoints for staphylococci, streptococci, and enterococci are available. Among the pleuromutilins, the only clinical breakpoint available is for tiamulin and applicable to porcine A. pleuropneumoniae. In contrast, clinical breakpoints for chloramphenicol are available for a wide variety of Gram-positive and Gram-negative bacteria of human origin, whereas those for florfenicol are only applicable to bovine and porcine respiratory tract pathogens. In addition to CLSI, the European Committee on Antimicrobial Suceptibility Testing (EUCAST) provides slightly divergent clinical breakpoints (see eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_6.0_Breakpoint_table.pdf). Although there are numerous national resistance monitoring programs, those in human medicine often cover only pathogens from specific infections, mainly food-borne diarrheal diseases, whereas those in veterinary medicine usually monitor indicator bacteria and commensal bacteria, and only a very limited number of pathogenic bacteria (Silley et al. 2012).
Table 1 shows some examples of resistance prevalences for LSPP antimicrobial agents among pathogenic bacteria from the national resistance monitoring programs DANMAP (see danmap.org), GERM-Vet (see www.bvl.bund.de/DE/09_Untersuchungen/untersuchungen_node.html;jsessionid=76F5E0F5BC375CA81F9CEF5BE44991E1.2_cid322), NARMS (see cdc.gov/narms/reports), NORM/NORM-VET (see vetinst.no/eng/Publications/NORM-NORM-VET-Report), as well as some long-term surveillance studies (Portis et al. 2012; Lindeman et al. 2013).
Table 1.
Resistance rates to LSPP antimicrobial agents from selected national resistance monitoring programs and surveillance studies
| LSPP antimicrobial agent | Bacteria | Origin | Year | Isolates tested | Resistant isolates (%) | MIC (mg/L) | Country | Referencesa |
|---|---|---|---|---|---|---|---|---|
| Clindamycin | Staphylococcus aureus (MSSA) | Human—blood culture | 2013 | 1155 | 1.5 | ≥1 | Norway | NORM/NORM-VET |
| S. aureus (MRSA) | Human | 2013 | 1528 | 18.9 | ≥1 | Norway | NORM/NORM-VET | |
| S. aureus (MSSA) | Human— bacteraemia | 2014 | 381 | 8.4 | ≥1 | Denmark | DANMAP | |
| S. aureus (MRSA) | Human | 2014 | 1932 | 33.0 | ≥1 | Denmark | DANMAP | |
| Staphylococcus pseudintermedius | Dog—clinical | 2013 | 201 | 18.0 | ≥0.5 | Norway | NORM/NORM-VET | |
| S. pseudintermedius | Dog—skin infections | 2011 | 54 | 38.9 | ≥4 | Germany | GERM-Vet | |
| Pirlimycin | S. aureus | Cattle—mastitis | 2009 | 210 | 1.4 | ≥4 | Germany | GERM-Vet |
| S. aureus | Cattle—mastitis | 2010 | 342 | 3.0 | ≥4 | United States, Canada | Lindeman et al. 2013 | |
| Streptococcus dysgalactiae | Cattle—mastitis | 2009 | 158 | 17.7 | ≥4 | Germany | GERM-Vet | |
| S. dysgalactiae | Cattle—mastitis | 2010 | 257 | 7.0 | ≥4 | United States, Canada | Lindeman et al. 2013 | |
| Streptococcus uberis | Cattle—mastitis | 2009 | 289 | 27.0 | ≥4 | Germany | GERM-Vet | |
| S. uberis | Cattle—mastitis | 2010 | 289 | 25.0 | ≥4 | United States, Canada | Lindeman et al. 2013 | |
| Quinupristin–dalfopristin | S. aureus | Poultry—clinical | 2011 | 43 | 27.9 | ≥2 | Germany | GERM-Vet |
| S. aureus | Horse—clinical | 2011 | 33 | 0.0 | ≥2 | Germany | GERM-Vet | |
| S. pseudintermedius | Dog—skin infections | 2011 | 54 | 0.0 | ≥2 | Germany | GERM-Vet | |
| Enterococcus faecium | Broiler meat | 2014 | 177 | 2.8 | ≥8 | Denmark | DANMAP | |
| E. faecium | Beef | 2014 | 56 | 0.0 | ≥8 | Denmark | DANMAP | |
| E. faecium | Pork | 2014 | 23 | 0.0 | ≥8 | Denmark | DANMAP | |
| Tiamulin | Actinobacillus pleuropneumoniae | Swine—respiratory tract | 2012 | 41 | 2.4 | ≥32 | Germany | GERM-Vet |
| Chloramphenicol | Nontyphoidal Salmonella | Human | 2013 | 2178 | 3.9 | ≥32 | United States | NARMS |
| Salmonella Typhi | Human | 2013 | 279 | 9.3 | ≥32 | United States | NARMS | |
| Escherichia coli O157 | Human | 2013 | 177 | 2.8 | ≥32 | United States | NARMS | |
| E. coli | Turkey—clinical | 2012 | 159 | 18.9 | ≥32 | Germany | GERM-Vet | |
| S. pseudintermedius | Dog—skin infections | 2011 | 54 | 24.1 | ≥32 | Germany | GERM-Vet | |
| Florfenicol | Pasteurella multocida | Cattle—respiratory tract | 2012 | 77 | 1.3 | ≥8 | Germany | GERM-Vet |
| P. multocida | Cattle—respiratory tract | 2009 | 328 | 11.6 | ≥8 | United States, Canada | Portis et al. 2012 | |
| Mannheimia haemolytica | Cattle—respiratory tract | 2009 | 304 | 8.6 | ≥ 8 | United States, Canada | Portis et al. 2012 | |
| Bordetella bronchiseptica | Swine—respiratory tract | 2012 | 90 | 2.2 | ≥ 8 | Germany | GERM-Vet | |
| A. pleuropneumoniae | Swine—respiratory tract | 2012 | 41 | 0.0 | ≥ 8 | Germany | GERM-Vet |
TSPP, Lincosamides, streptogramins, phenicols, and pleuromutilins.
aWebsites of the national resistance monitoring programs are given in the section “Prevalence of Resistance to LSPP Antimicrobial Agents.”
MECHANISMS OF RESISTANCE
In general, antimicrobial resistance can be based on two different mechanisms: the acquisition of mutations or resistance genes. Resistance-mediating mutations usually occur in genes or regions that represent the target site of the antimicrobial agents and prevent efficient binding of the antimicrobial agents to these sites. However, mutations may also enhance expression of efflux genes or alter the substrate spectrum of transporters and thereby cause resistance or decreased susceptibility. Resistance genes may confer antimicrobial resistance by either enzymatic inactivation, active efflux, or modifications at the target sites of the antimicrobial agents. Although mutations occur spontaneously and are vertically transferred during division of a cell that harbors the mutation, resistance genes are often associated with mobile genetic elements that are disseminated vertically during cell division, but also horizontally by the gene transfer processes.
Ribosomal Mutations Associated with Resistance to LSPP Antimicrobial Agents
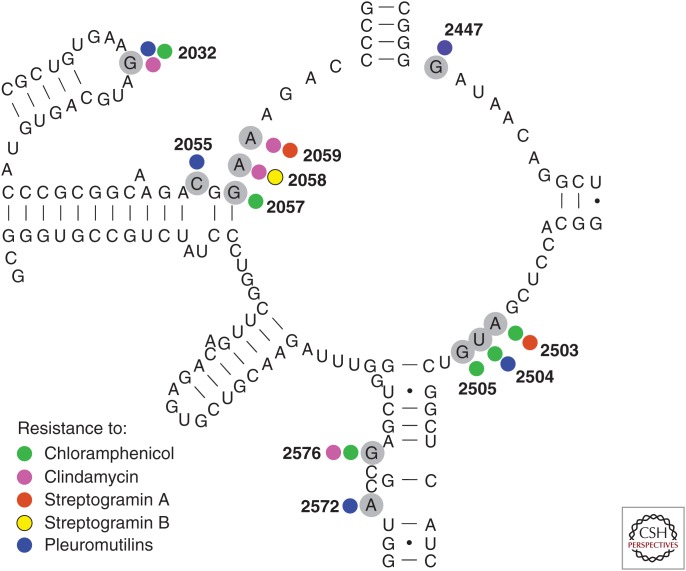
All LSPP antimicrobial agents bind to a well-conserved area of the ribosome consisting mainly of RNA. Although the RNA sequence in the area is conserved and therefore not expected to allow many mutations, a number of mutations in domain V of 23S RNA causing resistance has been observed. It is also noteworthy that most bacteria have multiple 23S rRNA (ribosomal RNA) copies meaning that recombinations have to occur to obtain a full effect of such mutations. It has been shown that, for example, S. aureus under antimicrobial pressure will increase the number of mutated ribosomal RNA operons over time (Besier et al. 2008). Mutations occur constantly but maintaining 23S RNA resistance mutations depends on the number of copies of 23S RNA genes, how dominant the mutation is, and how “expensive” it is to contain a mutated RNA nucleotide in the exact position. In addition, it differs from species to species which mutations show up, and sometimes it is hard to prove that the resistance effect is because of the mutations observed as other mutations may contribute to or be the main effector. In addition, the identification of resistance mutations in clinical or animal isolates might be complicated by the lack of a wild-type comparator or the bacterial species might be hard to assay for resistance. The published literature thus contains more- or less-proven examples of resistance mutations, and only the most proven and solid data has been included here. All 23S RNA mutations causing resistance to the LPPS antimicrobial agents have been found in the part of 23S RNA shown in the secondary structure model shown in Figure 3. This region encompasses the RNA located at the PTC and contains the nucleotides known to be involved in binding of antimicrobial agents to the PTC.
Figure 3.
A secondary structure model of the peptidyl transferase loop of domain V of 23S rRNA (Escherichia coli sequence and numbering) with nucleotides providing antibiotic resistance marked with gray circles. Data are all from bacteria except the streptogramin A data that come from an archae (Porse and Garrett 1999). The bacterial data are from the following studies: Douthwaite (1992), Vester and Douthwaite (2001), Miller et al. (2008), Long et al. (2009, 2010), and Li et al. (2011), and references herein. The smaller circles indicate resistance to the various antibiotics and with the same color code as in Figure 2.
As can be seen, some of the RNA mutations cause resistance to multiple antimicrobial agents and some also to agents not mentioned in this review. Especially the adenine residues at positions 2058 and 2059 are well-studied nucleotides for macrolide and lincosamide resistance (Vester and Douthwaite 2001). Antibiotics with partly overlapping binding sites may also bind to some of the same nucleotides and, thus, cross-resistance can be seen. However, the predictions of such a cross-resistance do not follow an easy recognizable pattern (Long et al. 2010) and, furthermore, multiple mutations can show enhanced effects (Douthwaite 1992; Long et al. 2010).
In addition to 23S rRNA, there are also a few ribosomal proteins located close to PTC, including L3 and L4 (Nissen et al. 2000), in which mutations have been correlated with antimicrobial resistance. L3 mutations associated with tiamulin resistance have been summarized by Klitgaard et al. (2015), and most data relate to the aa 144–151 region of L3 (E. coli numbering), which is also the region closest to PTC. The mutations have been found in E. coli, Staphylococcus spp., and Brachyspira spp., and especially mutations at positions 149–150 have been proven to confer pleuromutilin resistance (see Klitgaard et al. 2015, and references therein). The same L3 region contains mutations associated with linezolid resistance and also examples of cross-resistance to linezolid and tiamulin (Klitgaard et al. 2015). Ribosomal protein L22 is located further down the tunnel, and a six aa insertion is reported to confer resistance to streptogramin B compounds in S. pneumoniae (Cattoir et al. 2007). As for the 23S RNA mutations, it is a cost-benefit matter whether a resistance-mediating mutation in a ribosomal protein “survives,” and it is probably often followed by other “helper mutations” (also called compensatory mutations) as seen by Gentry et al. (2007). Increasing genome-sequencing capability and decreasing costs should facilitate more investigations on these contexts in the near future. In the current situation, investigators are often looking for what others have seen and might, therefore, ignore other positions that may be important for resistance.
Genes Conferring Resistance to LSPP Antimicrobial Agents by Enzymatic Inactivation
Enzymatic Inactivation of Lincosamides
Lincosamides are commonly inactivated by lincosamide nucleotidyltransferases, and their genes are found in various organisms. The known lincosamide nucleotidyltransferases show highest activity, as measured by the corresponding minimum inhibitory concentrations (MICs), against lincomycin, moderate activity against pirlimycin, and lowest activity against clindamycin (Lüthje and Schwarz 2006; Zhao et al. 2014).
The gene lnu(A) encodes a lincosamide nucleotidyltransferase of 161 aa (Brisson-Noël and Courvalin 1986). This gene is often located on small plasmids that usually contain only the lnu(A) gene and a plasmid replication gene. Several different types of lnu(A)-carrying plasmids have been described, many of them in S. aureus and coagulase-negative staphylococci (CoNS) of animal origin (Loeza-Lara et al. 2004; Lüthje and Schwarz 2007b; Lozano et al. 2012a).
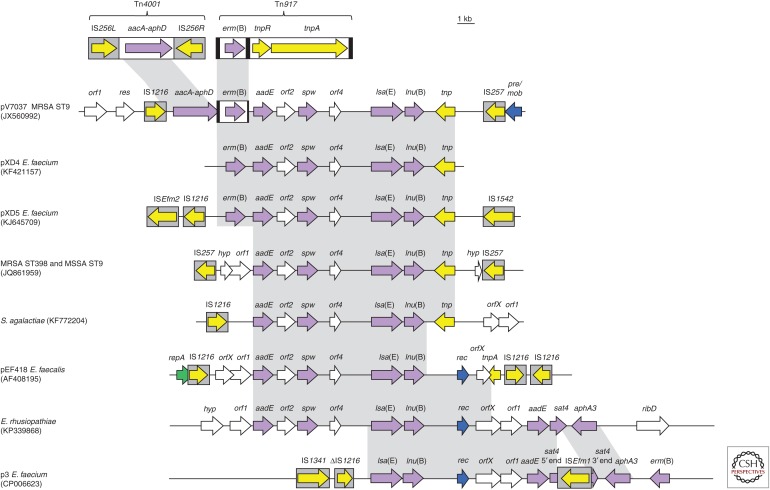
The gene lnu(B) codes for a lincosamide nucleotidyltransferase of 267 aa and was first described in E. faecium of human origin (Bozdogan et al. 1999), and later identified in porcine S. dysgalactiae ssp. equisimilis (Lüthje and Schwarz 2007b). Recently, it has been detected as part of multiresistance gene clusters on plasmids or in the chromosomal DNA of S. aureus, Staphylococcus hyicus, E. faecium, E. faecalis, Streptococcus agalactiae, and Erysipelothrix rhusiopathiae of human and animal origin (Fig. 4) (Lozano et al. 2012a; Li et al. 2013, 2014b; Montilla et al. 2014; Silva et al. 2014; Wendlandt et al. 2014, 2015a; Zhang et al. 2015a).
Figure 4.
lsa(E)- and lnu(B)-carrying multiresistance gene clusters. Schematic presentation of the structural variability among lsa(E) and lnu(B)-carrying multiresistance gene clusters found on plasmids or in the chromosomal DNA of Enterococcus faecalis, E. faecium, Streptococcus agalactiae, Erysipelothrix rhusiopathiae, MRSA (methicillin-resistant Staphylococcus aureus), and MSSA (methicillin-susceptible S. aureus). All genes are indicated by arrows with the arrowhead showing the direction of transcription. Insertion sequences are presented as gray boxes with the arrow inside indicating the transposase gene. All antimicrobial resistance genes are depicted as violet arrows. aacA-aphD, gentamicin/kanamycin/tobramycin resistance; aadE, streptomycin resistance; aphA3, kanamycin/neomycin resistance; erm(B), MLSB resistance; lnu(B), lincosamide resistance; lsa(E), lincosamide/pleuromutilin/streptogramin A resistance; spw, spectinomycin resistance; sat4, streptothricin resistance. Genes involved in transposition are shown in yellow, whereas the genes involved in plasmid replication and plasmid recombination/mobilization are shown in green and blue, respectively. Genes with other functions are displayed in white. The Δ symbol indicates a truncated gene. A 1-kb scale is shown in the upper right corner. The gray-shaded regions show >99% sequence identity.
The gene lnu(C) was first found in a human S. agalactiae isolate and codes for a 164-aa nucleotidyltransferase that inactivates lincomycin and clindamycin (Achard et al. 2005). It has also been discovered in the swine pathogen Haemophilus parasuis (Chen et al. 2010). The gene lnu(D) was detected in the chromosomal DNA of a Streptococcus uberis isolate from a case of bovine mastitis and codes for a nucleotidyltransferase of 164 aa (Petinaki et al. 2008). The gene lnu(E), which codes for a 173 aa protein, was found on a plasmid in Streptococcus suis in which it was interrupted by the integration of an ISEnfa5-cfr-ISEnfa5 segment (Zhao et al. 2014). This gene was de novo synthesized, and after cloning and expression in S. aureus RN4220, shown to confer resistance to lincomycin. The gene lnu(F), originally referred to as linF codes for a 273-aa nucleotidyltransferase and was first identified on a plasmid from an E. coli isolate of human origin. Further analysis showed that the lnu(F) gene was part of a gene cassette located in a class 1 integron (Heir et al. 2004). A lnu(F)-related gene, originally reported as linG, which also encodes a 273-aa nucleotidyltransferase that shows 93% identity to Lnu(F), has been found as part of a gene cassette in a class 1 integron from a Salmonella enterica serovar Stanley isolate of human origin (Levings et al. 2006). Finally, a gene, described as linAN2, has been detected on the mobilizable 11-kb transposon NBU2 in human clinical isolates of Bacteroides fragilis and Bacteroides thetaiotaomicron. It codes for a nucleotidyltransferase of 171 aa, which shares only about 50% identity to Lnu(A) proteins (Wang et al. 2000).
From an evolutionary point of view, at least two groups of lincosamide nucleotidyltransferases can be differentiated. One group includes the proteins Lnu(A), Lnu(C), Lnu(D), and Lnu(E) that harbor a conserved domain at their amino terminus, which shows similarity to the aminoglycoside nucleotidyltransferase ANT(2″)-Ia (Petinaki et al. 2008). This observation points toward a divergent evolution from a common ancestor. The other group includes the proteins Lnu(B) and Lnu(F) with sequence similarity to the β-subunit of DNA polymerases. It is thus likely that they have developed from nucleotide polymerases, which are ubiquitous in bacteria (Morar et al. 2009).
Enzymatic Inactivation of Streptogramins
Inactivation of streptogramin A antimicrobial agents is commonly mediated by acetyltransferases. In staphylococci, three genes, vat(A) (Allignet et al. 1993), vat(B) (Allignet and El Solh 1995), or vat(C) (Allignet et al. 1998), which code for acetyltransferases of 219, 212, or 212 aa, respectively, have been described. These three vat genes have been identified on plasmids of different sizes, which occasionally also harbor additional resistance genes. In contrast to staphylococci from human sources, vat genes have rarely been detected in animal staphylococci. Solely, the vat(B) gene has been identified in two Staphylococcus xylosus isolates of poultry origin (Aarestrup et al. 2000). In enterococci, the plasmid-borne vat(D) gene from E. faecium, initially described as satA, was the first vat gene detected in this genus (Rende-Fournier et al. 1993). In another study, it was shown that the vat(D) gene, which codes for a 209-aa protein, is linked to the macrolide–lincosamide–streptogramin B (MLSB)-resistance gene erm(B) and that both genes are cotransferred (Hammerum et al. 2001). The vat(E) gene, coding for an acetyltransferase of 214 aa and originally published as satG, was first identified in an E. faecium isolate from a sewage treatment plant in Germany (Werner and Witte 1999). A survey of streptogramin-resistant E. faecium from retail meat samples revealed some variability in the vat(E) genes. However, no correlation was seen between the number of aa substitutions and the MICs of quinupristin–dalfopristin (Simjee et al. 2001). Besides Gram-positive bacteria, a novel type of vat gene, vat(F), which codes for a 221-aa protein, was detected in the chromosomal DNA of Yersinia enterocolitica (Seoane and García Lobo 2000). A vat(H) gene coding for a 216-aa acetyltransferase was detected together with the vga(D) gene on a plasmid in an E. faecium isolate of human origin (Jung et al. 2010). The origin of the VAT enzymes remains to be elucidated. Homologs and orthologs of the Vat enzymes, which are present in clinically resistant bacteria, are widely distributed in chromosomes of numerous environmental bacterial species, including species, such as Y. enterocolitica, which are intrinsically resistant to streptogramins (Wright 2007). It should be noted that streptogramin A–acetylating Vat proteins are related to CatB chloramphenicol acetyltransferases in their sizes, aa sequence, presumable active center, and tertiary structure (Murray and Shaw 1997), which points toward a common evolutionary origin.
So far, two genes, vgb(A) and vgb(B), have been described in staphylococci, which code for streptogramin B–specific lactone hydrolases of 298 and 295 aa, respectively (Allignet et al. 1988, 1998). Both genes are plasmid-borne. Plasmid pIP1714, which carries vgb(B), also harbors vat(C). This plasmid was isolated from a Staphylococcus cohnii ssp. cohnii found in the environment of a hospital where pristinamycin was extensively used (Allignet et al. 1998). There is little information about vgb genes in staphylococci from animals. The same two vat(B)-positive S. xylosus from poultry also carried a vgb(B) gene (Aarestrup et al. 2000). Orthologous and homologous vgb genes are present in the genomes of many environmental bacteria, including Streptomyces coelicolor and other Streptomyces spp., which belong to the same genus as the streptogramin producers (Wright 2007). Such genes may have served as precursors from which the vgb genes found in pathogenic bacteria have evolved.
Enzymatic Inactivation of Phenicols
Nonfluorinated phenicols, such as chloramphenicol and thiamphenicol, are commonly inactivated by chloramphenicol O-acetyltransferases (CATs) (Shaw 1983; Schwarz et al. 2004). All CATs transfer an acetyl group from a donor molecule (e.g., acetyl-CoA) to the hydroxyl group at C3 of the phenicol molecule. This acetyl group is then shifted to the hydroxyl group at C1, and the hydroxyl group at C3 is available again for a second acetylation step. Neither mono- nor diacetylated phenicol molecules have antimicrobial activity (Murray and Shaw 1997). None of the CAT enzymes are able to inactivate florfenicol, because the hydroxyl group at C3 is replaced by fluorine and cannot act as an acceptor site for acetyl groups. There are two types of CATs, both of which have a trimeric structure composed of three identical monomers, and the respective cat gene codes for the monomer (Murray and Shaw 1997). The sizes of the classical CAT monomers vary between 207 and 238 aa, whereas CATs of the second type vary in their sizes between 209 and 219 aa (Schwarz et al. 2004).
The classical CATs, encoded by catA genes (Schwarz et al. 2004; Roberts and Schwarz 2009), represent a highly diverse group of enzymes whose members show an overall identity of approximately 44%. These enzymes have been detected in Gram-positive and Gram-negative aerobic and anaerobic bacteria and can be subdivided into at least 22 different groups when using a threshold value of ≥80% aa identity to define a group (Schwarz et al. 2004). The genes catA1, catA2, and catA3 (also known as catI, catII, and catIII) are exclusively found in Gram-negative bacteria and are expressed constitutively. The CatA3 (CATIII) enzyme was the first to be crystallized, and the analysis of its crystal structure provided insight into the folding of the CAT monomers and helped to identify the aa that were important for the structure and function of the CAT enzyme (Leslie et al. 1988). Another three groups of classical CATs were named according to the plasmids (pC221, pC223/pSCS7, and pC194), on which their genes were first detected. Although initially found in staphylococci, the corresponding cat genes have in the meantime been identified in a number of Gram-positive genera. The expression of these cat genes is inducible by chloramphenicol and is regulated by translational attenuation. The regulatory region comprises a reading frame for a short peptide and two inverted repeated sequences, and is located immediately upstream of the respective cat gene (Lovett 1990). Closely related CATP and CATD proteins were first identified in the Gram-positive anaerobe Clostridium spp. where they are located on transposons (Lyras and Rood 2000). However, these genes have also been identified in Gram-negative Neisseria meningitidis (Galimand et al. 1998; Shultz et al. 2003). Both genes are expressed constitutively. The remaining 15 groups of classical CATs are represented by individual enzymes whose genes have so far been detected in only a single species of either Gram-positive or Gram-negative bacteria (Schwarz et al. 2004).
The second type of CAT enzymes, encoded by catB genes (Murray and Shaw 1997; Schwarz et al. 2004; Roberts and Schwarz 2009), is only distantly related to the classical CATs. Their members are structurally similar to acetyltransferases involved in streptogramin A resistance (Murray and Shaw 1997). In general, these CAT enzymes confer lower MICs to chloramphenicol than the classical CATs. Hence, it was speculated that members of this second type of CAT might have a physiological role other than chloramphenicol resistance in their host bacteria (Murray and Shaw 1997). Using the same threshold value as for the classical CATs, at least five different groups can be distinguished, although all enzymes have approximately 77% identity with each other (Schwarz et al. 2004). Many of these cat genes are part of gene cassettes in class 1 and class 2 integrons in Gram-negative bacteria (Recchia and Hall 1995), whereas others have been identified on transposons.
In addition to CATs, inactivation of chloramphenicol by O-phosphorylation has been observed in the chloramphenicol producer S. venezuelae and is believed to contribute to self-defense of the host (Mosher et al. 1995). It is not known whether fluorinated chloramphenicol analogues can be inactivated by phosphorylation. Moreover, a chloramphenicol hydrolase gene has been found in the chloramphenicol producer S. venezuelae and considered to contribute to self-defense of the host (Mosher et al. 1990). Moreover, the gene estDL136 from a soil metagenome library was found to specify a hydrolase, which, when cloned in E. coli, inactivated both chloramphenicol and florfenicol (Tao et al. 2012).
Enzymatic Inactivation of Pleuromutilins
To the best of our knowledge, no pleuromutilin-inactivating enzymes have been described so far.
Genes Conferring Resistance to LSPP Antimicrobial Agents by Active Efflux
Active efflux can be based on multidrug transporters or specific transporters. In the following sections, multidrug transporters and specific transporters are described whose substrate spectrum includes one or more of the LSPP antimicrobial agents.
Resistance to LSPP Antimicrobial Agents by Multidrug Transporters
Usually, multidrug transporters confer an increase in the MICs of their substrates, but not necessarily to levels that are indicative of clinical resistance. Multidrug transporter systems assigned to the resistance/nodulation/cell division (RND) family have been reported to export phenicols from the bacterial cell. They include the AcrAB-TolC system in E. coli (McMurray et al. 1994), the MexAB-OprM and MexCD-OprJ systems in P. aeruginosa (Paulsen et al. 1996), as well as the OqxAB system in Enterobacteriaceae (Hansen et al. 2007), among others. Initially, intrinsic resistance of Enterobacteriaceae and other Gram-negative bacteria to macrolides, lincosamides, and streptogramins was considered to be based on the relative impermeability of the outer membrane to these compounds (Leclercq and Courvalin 1991). However, the observation that E. coli strains, such as CS1562 or AS19, are hypersusceptible to these antimicrobial agents caused by a deficiency of the TolC porin, suggests the involvement of RND pumps, which use TolC as an outer membrane component, in intrinsic resistance to macrolides, lincosamides, and streptogramins.
In Gram-positive bacteria, several 12-transmembrane segments (TMS) multidrug transporters of the major facilitator superfamily (MFS), such as Blt and Bmr proteins from Bacillus subtilis and NorA from S. aureus, have been reported to have a substrate spectrum that includes chloramphenicol (Paulsen et al. 1996). Another two closely related 12-TMS multidrug efflux proteins, MdfA and Cmr, which are able to export chloramphenicol, have been identified in E. coli (Nilsen et al. 1996; Edgar and Bibi 1997). Overexpression of the chromosomal multidrug exporter MdeA of S. aureus conferred a 16-fold increase in the MIC of virginiamycin, while no significant increase in the MICs of other LSPP antimicrobial agents tested was seen (Huang et al. 2004). In addition, the gene lmr(B) from B. subtilis coding for a 479-aa MFS protein (Kumano et al. 2003; Murata et al. 2003) was shown to confer resistance to multiple antimicrobial agents, including lincomycin, when overexpressed. The Lmr(P) protein of Lactococcus lactis specifies a 408-aa multidrug transporter of the MFS, which confers resistance to lincosamides, macrolides, streptogramins, and tetracyclines (Putman et al. 2001; Poelarends et al. 2002). A similar substrate spectrum was determined for the ATP-binding cassette (ABC) transporter encoded by the gene lmr(A) from L. lactis when expressed in a hypersusceptible E. coli strain (Putman et al. 2000; Poelarends et al. 2002). The 480-aa MFS protein LmrS from S. aureus has been reported to confer 4- to 16-fold increases in the MICs of numerous antimicrobial agents (including lincomycin, chloramphenicol, florfenicol, and linezolid) and other substances when expressed in E. coli (Floyd et al. 2010).
Active Efflux of Lincosamides by Specific Exporters
The 492-aa ABC transporter Lsa(B) conferred, in contrast to other Lsa proteins, only elevated MICs of lincosamides, which, however, are below the clinical breakpoint for resistance (Kehrenberg et al. 2004). The lsa(B) gene was detected in close proximity to the multiresistance gene cfr on plasmids in Staphylococcus sciuri and Staphylococcus warneri (Kehrenberg et al. 2004, 2007). The lmrA gene of the lincomycin producer S. lincolnensis specifies an MFS protein of 481 aa, which exports lincomycin and it is believed to be part of a self-defense system of this organism (Peschke et al. 1995). A gene, designated lmrB, which codes for a 481-aa MFS exporter in Corynebacterium glutamicum, was shown to confer resistance to lincosamides but not to other antimicrobial agents tested (Kim et al. 2001). This Lmr(B) protein is only distantly related to the Lmr(B) protein in B. subtilis described above.
Active Efflux of Lincosamides, Pleuromutilins, and Streptogramin A Antimicrobial Agents by Specific Exporters
During recent years, several ABC transporters have been identified in staphylococci, streptococci, and enterococci, which confer combined resistance to lincosamides, pleuromutilins, and streptogramin A antimicrobial agents. A recent review provided detailed information about most of the corresponding multiresistance genes, such as vga(A), vga(A)V, vga(A)LC, vga(C), vga(E), vga(E)V, lsa(A), lsa(C), lsa(E), eat(A)v, and sal(A) (Wendlandt et al. 2015b).
The ABC transporters specified by the genes vga(A) (Allignet et al. 1992), vga(A)V (Haroche et al. 2000), vga(A)LC (Novotná and Janata 2006; Gentry et al. 2008), vga(B) (Allignet and El Solh 1997), vga(C) (Kadlec and Schwarz 2009), vga(E) (Schwendener and Perreten 2011), and vga(E)V (Li et al. 2014a) show sizes of 522, 524, 522, 552, 522, 524, and 524 aa, respectively. All of these can export streptogramin A antimicrobial agents, whereas the Vga(A), Vga(C), and Vga(E) proteins also export lincosamides and pleuromutilins. The vga(A) genes are most widespread and can be located on the 5.5-kb transposon Tn5406 (Haroche et al. 2002). Studies on clinical S. aureus isolates from France identified Tn5406 inserted into the chromosomal att554 site, a site that resembles the integration site in a type III SCCmec cassette and on plasmids. The vga(A) genes can also be located on plasmids, ranging from small plasmids of 5.7 kb that harbor only the vga(A) gene (Kadlec et al. 2010) to large plasmids of 25–46 kb that carry additional resistance genes (Allignet and El Solh 1999; Weiß et al. 2014). The vga(C) gene was initially identified on a 14-kb multiresistance plasmid from a porcine livestock–associated (LA)-MRSA isolate (Kadlec and Schwarz 2009) but has also been identified on a small plasmid of 5.3 kb (Kadlec et al. 2010).
The vga(E) was shown to be part of the 11.5-kb transposon Tn6133, which was detected initially in a porcine LA-MRSA from Switzerland (Schwendener and Perreten 2011) and soon thereafter in LA-MRSA from cattle, chickens, and turkeys in Germany (Hauschild et al. 2012; Monecke et al. 2013). In 2014, a variant of the Vga(E) protein was described, which shared only 86% aa identity with the original Vga(E) protein (Li et al. 2014a). The Vga(E) variant also confers pleuromutilin–lincosamide–streptogramin A resistance, and the corresponding gene vga(E)V was located on a 5.6-kb plasmid in porcine Staphylococcus simulans and S. cohnii isolates. The vga(D) gene codes for a 525-aa protein and was found in E. faecium of human origin (Jung et al. 2010). Vga(D) was the first ABC transporter in E. faecium, which conferred resistance to streptogramin A antimicrobial agents. However, it is not known whether this ABC transporter also exports lincosamides and pleuromutilins.
The species-specific chromosomal gene lsa(A) from E. faecalis codes for an ABC transporter of 496 aa, which is believed to play a role in the intrinsic resistance of E. faecalis to lincosamides and streptogramins. Disruption of the lsa(A) gene was associated with an at least 40-fold decrease in MICs of quinupristin–dalfopristin, clindamycin, and dalfopristin, whereas complementation of the disruption mutant with an intact lsa(A) resulted in restoration of the MICs to wild-type levels (Singh et al. 2002). Another study reported that Lsa(A) not only confers resistance to lincosamide and streptogramin A, but also to pleuromutilins (Malbruny et al. 2011). The same resistance pattern is seen for the lsa(C) gene from S. agalactiae that codes for an ABC transporter of 492 aa (Malbruny et al. 2011). The first lsa gene in staphylococci, lsa(E), which mediates combined resistance to lincosamides, pleuromutilins, and streptogramin A, has been described in human MRSA ST398 and MSSA ST9 (Wendlandt et al. 2013b). This gene codes for an ABC transporter protein of 494 aa and is part of a multiresistance gene cluster that is most likely of enterococcal origin (Fig. 4) (Wendlandt et al. 2013b). Several variants of this multiresistance gene cluster have been identified in MRSA ST398 of human origin and in MRSA/MSSA ST9 of human and pig origin in Europe and Asia (Lozano et al. 2012a; Li et al. 2013; Wendlandt et al. 2013a, 2014, 2015b). Moreover, as summarized in Figure 4, variants of the lsa(E)-containing multiresistance gene cluster have also been found in E. faecalis and E. faecium of human and swine origin in China (Li et al. 2014b; Si et al. 2015), in S. agalactiae of human origin in Argentina (Montilla et al. 2014), and also in E. rhusiopathiae of swine origin in China (Zhang et al. 2015a).
Although E. faecalis is intrinsically resistant to lincosamides, pleuromutilins, and streptogramin A by production of the ABC transporter Lsa(A), E. faecium is naturally susceptible. E. faecium harbors a gene eat(A) for an ABC transporter of 500 aa that shows only 66% aa identity to Lsa(A) and has no function in antimicrobial resistance. Despite this, an aa substitution (Thr450Ile) was found in the Eat(A) protein of isolates that were resistant to lincosamides, pleuromutilins, and streptogramin A antimicrobial agents. Single-nucleotide replacement and transfer of the mutated gene eat(A)V into a susceptible E. faecium isolate confirmed a role of this mutation in resistance to lincosamides, pleuromutilins, and streptogramin A (Isnard et al. 2013).
The gene sal(A) from S. sciuri codes for an ABC transporter of 541 aa, which was reported to confer resistance to streptogramin A and lincosamides (Hot et al. 2014). The sal(A) gene has exclusively been found in the chromosomal DNA of S. sciuri isolates inserted between the two housekeeping genes, iscS and mnmA, of the staphylococcal core genome. A recent study confirmed that Sal(A) also confers pleuromutilin resistance (Wendlandt et al. 2015a).
Active Efflux of Macrolides and Streptogramin B by Specific Exporters
The gene msr(A) codes for a 488-aa ABC transporter protein that confers resistance to macrolides and streptogramin B (Ross et al. 1990) and is often found together with the gene mph(C) that codes for a macrolide phosphotransferase (Lüthje and Schwarz 2006). These two genes have been detected among CoNS and MRSA CC398 from humans in Germany and Spain, respectively (Gatermann et al. 2007; Lozano et al. 2012b), and on plasmids of the S. aureus clone USA300 (Kennedy et al. 2010). The 33-kb plasmid pMS97 has been reported to carry msr(A) and mph(C) together with the MLSB-resistance gene erm(Y) (Matsuoka et al. 2003). In veterinary medicine, the msr(A) gene has been detected in S. aureus isolates of poultry (Nawaz et al. 2000) and dog origin (Lüthje and Schwarz 2007b), in canine S. pseudintermedius (Lüthje and Schwarz 2007b), and in various species of CoNS from cases of bovine mastitis (Lüthje and Schwarz 2006). Moreover, Jaglic et al. (2012) identified the gene msr(A) in not-further-specified CoNS isolates from turkeys, pigs, and cattle.
The species-specific and chromosomally located gene msr(C) of E. faecium codes for a 492-aa ABC transporter. Functional deletion of msr(C) resulted in a two- to eightfold decrease in MICs of the macrolides erythromycin, azithromycin, and tylosin, as well as of the streptogramin B quinupristin. This E. faecium–specific gene shows only 53% identity to msr(A) (Singh et al. 2001). The msr(D) from S. pyogenes (Gay and Stephens 2001) has been reported to confer macrolide and ketolide, but not streptogramin resistance (Poole 2005). The msr(E) gene found in various Gram-negative bacteria is also known to be involved in the efflux of macrolides (Kadlec et al. 2011), but it is not known whether this gene also confers resistance to streptogramin B antimicrobial agents.
Active Efflux of Phenicols by Specific Exporters
Specific exporters that transport only chloramphenicol or chloramphenicol and florfenicol out of the bacterial cell are mainly members of the MFS efflux proteins (Paulsen et al. 1996; Poole 2005). They commonly show 10 to 14 TMS (Butaye et al. 2003). Based on an 80% threshold value, at least 11 genetic groups of specific phenicol exporters can be distinguished, including four groups from soil and environmental bacteria (Schwarz et al. 2004; Roberts and Schwarz 2009). The cmr and cmx genes found in Corynebacterium spp. are located on plasmids, whereas the cmx gene is associated with transposon Tn5564 (Schwarz et al. 2004). The genes cmr and cmrA from Rhodococcus spp. are also found on plasmids, with the cmrA being part of transposon Tn5561 (Nagy et al. 1997). The S. venezuelae cmlv gene is believed to play a role in self-defense of the chloramphenicol producer (Mosher et al. 1990).
In clinically important bacteria, such as S. enterica, E. coli, Klebsiella pneumoniae, or Pseudomonas aeruginosa, several closely related cmlA genes have been identified on gene cassettes. Unlike other cassette-borne genes, cmlA is inducibly expressed via translational attenuation (Stokes and Hall 1991; Recchia and Hall 1995). The chloramphenicol exporter CmlB1, which shares 74%–77% identity with CmlA proteins, was identified on a plasmid from Bordetella bronchiseptica (Kadlec et al. 2007), a bacterium involved in respiratory tract infections in swine. The cmlB1 gene is also inducibly expressed via translational attenuation. Neither CmlA nor CmlB1 proteins can efficiently export florfenicol from the bacterial cell, and bacteria carrying the corresponding genes are classified as florfenicol susceptible (Schwarz et al. 2004).
In contrast to the aforementioned genes, the floR gene codes for a MFS protein that can export both chloramphenicol and florfenicol (Schwarz et al. 2004). The floR gene (also referred to as flo or pp-flo) can be found in the chromosomal DNA of multiresistant S. enterica serovars, including Typhimurium DT104 and Newport, Vibrio cholerae, E. coli, B. bronchiseptica, and Acinetobacter baumannii, or on plasmids of E. coli, K. pneumoniae, Pasteurella multocida, Pasteurella trehalosi, A. pleuropneumoniae, and Stenothrophomonas maltophilia (Schwarz et al. 2004). Recently, the floR gene has been identified as part of the chromosomally located integrative and conjugative element ICEPmu1 from P. multocida, which harbors a total of 12 antimicrobial resistance genes (Michael et al. 2012). A new floR gene variant, floRv, whose product showed only 84%–92% aa identity to the so-far known FloR proteins, has recently been identifed in a multidrug resistance genomic island of a porcine S. maltophilia isolate (He et al. 2015).
There are also a few chloramphenicol/florfenicol exporters in Gram-positive bacteria. The gene fexA is part of transposon Tn558 and was first identified on a plasmid from Staphylococcus lentus (Kehrenberg and Schwarz 2005). Expression of fexA is inducible with either chloramphenicol or florfenicol. A translational attenuator, similar to those of cat genes from Staphylococcus spp. and Bacillus pumilus, is located immediately upstream of the fexA gene (Kehrenberg and Schwarz 2004). A fexA gene variant, fexAv, which conferred only chloramphenicol resistance, was detected in a canine S. pseudintermedius (Gómez-Sanz et al. 2013). In comparison to FexA, FexAv showed two aa substitutions Gly33Ala and Ala37Val. Both substitutions appear to be important for substrate recognition as site-directed mutagenesis to the original fexA gene, restored the chloramphenicol/florfenicol resistance phenotype. In 2010, a novel chloramphenicol/florfenicol resistance gene, designated pexA, was identified from an Alaskan soil sample (Lang et al. 2010). The gene fexB, which also confers resistance to chloramphenicol and florfenicol, was found on nonconjugative plasmids of E. faecium and Enterococcus hirae (Liu et al. 2012a).
Active Efflux of Phenicols and Oxazolidinones by a Specific Exporter
Most recently, a novel gene, designated optrA and coding for an ABC transporter of 655 aa, has been identified on a conjugative plasmid in E. faecalis (Wang et al. 2015). In contrast to the MFS transporters described above, the optrA gene confers resistance, not only to chloramphenicol and florfenicol, but also to the oxazolidinones linezolid and tedizolid. The optrA gene was shown to be functionally active in E. faecalis, E. faecium, and S. aureus. A first survey conducted in China revealed that the optrA gene was more frequently found in enterococci of animal than of human origin (Wang et al. 2015). In a study including 1159 enterococcal isolates from five hospitals in China, the optrA gene was detected at a prevalence of 2.9%, and a distinct increase in optrA carriers was seen from 2010 to 2014 (Cai et al. 2015a). The analysis of the genetic environment of optrA in E. faecalis revealed a substantial heterogeneity (He et al. 2016). Besides in isolates from China, the optrA gene has also been identified in two clinical E. faecalis isolates from Italy (Brenciani et al. 2016). Finally, the analysis of 50 porcine staphylococci from China identified the optrA gene in a single S. sciuri isolate where it was located together with the genes cfr, fexA, aadD, ble, and aacA-aphD on a 60.5-kb nonconjugative multiresistance plasmid (Li et al. 2016).
Genes Conferring Resistance to LSPP Antimicrobial Agents by Target Site Modification
Target Site Modifications that Confer Resistance to Macrolides, Lincosamides, and Streptogramin B Antimicrobial Agents
Combined resistance to macrolides, lincosamides, and streptogramin B antimicrobial agents is commonly mediated by rRNA methylases that target the adenine residue at position 2058 in the domain V of the 23S rRNA. The adenine residue at position 2058 (Fig. 2) is located in the overlapping binding region of these three classes of antimicrobial agents. Methylation of this residue prevents macrolides, lincosamides, and streptogramin B antimicrobial agents from binding to their ribosomal target sites (reviewed in Weisblum 1995a; Roberts 2008). The corresponding erm genes, which code for these methylases, are often located on plasmids, transposons, or integrative and conjugative elements, which facilitate their dissemination across strain, species, and sometimes even genus boundaries. Based on a threshold value of <80% aa identity (Roberts et al. 1999), 46 classes of rRNA methylases are currently distinguished (see faculty.washington.edu/marilynr/ermweb1.pdf). Many erm genes are inducibly expressed via translational attenuation with 14- and 15-membered macrolides acting as inducers (Weisblum 1995b). Lincosamides and streptogramins, but also 16-membered macrolides usually do not act as inducers. Studies on erm(A) and erm(C) genes in staphylococci showed that constitutive expression can develop rapidly in the presence of such noninducers and is commonly a result of deletions, duplications, or point mutations in the translational attenuator (Schmitz et al. 2002a,b; Lüthje and Schwarz 2007a). Detailed information on the erm genes and their occurrence in the various bacteria can be seen in a review on macrolides and ketolides (Fyfe et al. 2016).
Target Site Modifications that Confer Resistance to Phenicols, Lincosamides, Oxazolidinones, Pleuromutilins, and Streptogramin A Antimicrobial Agents
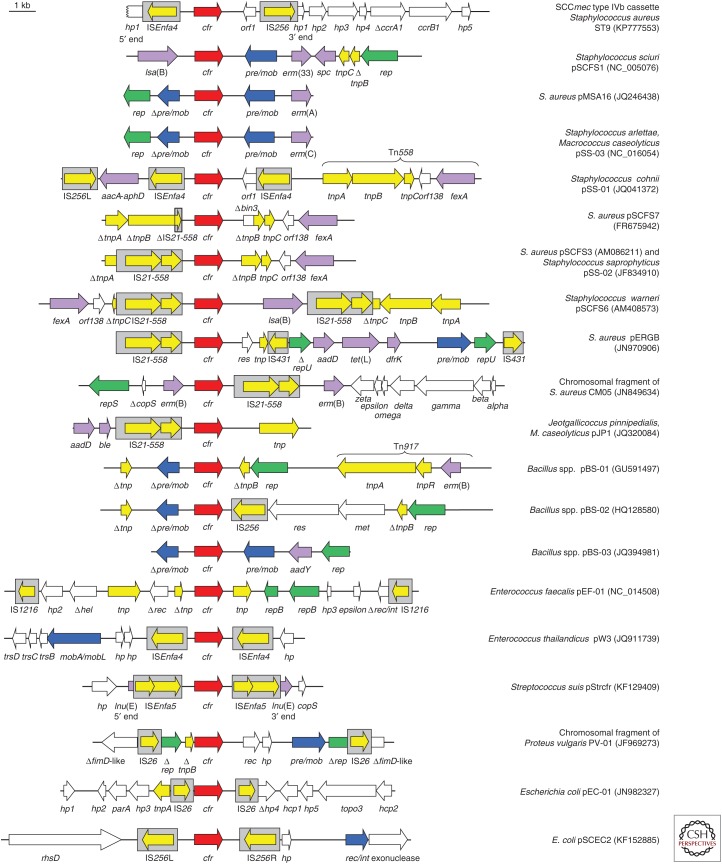
The gene cfr was the first gene that conferred combined resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A (PhLOPSA) antimicrobial agents. This gene was identified on a plasmid in a bovine S. sciuri and initially referred to as a chloramphenicol/florfenicol resistance gene (Schwarz et al. 2000). The clarification of the resistance mechanism, however, showed that cfr is a multiresistance gene, which confers resistance to the aforementioned classes of antimicrobial agents (Kehrenberg et al. 2005; Long et al. 2006). In addition, Cfr also provides decreased susceptibility to some large 16-membered macrolides, such as spiramycin and josamycin (Smith and Mankin 2008). The gene cfr encodes an rRNA methylase that targets the adenine residue at position 2503 in the domain V of the 23S rRNA (Fig. 2). This target site is located in the overlapping binding site of the PhLOPSA agents and the additional Cfr-mediated methylation is believed to prevent these antimicrobial agents from binding to the ribosome. Further studies identified the cfr gene on a number of plasmids in S. aureus, Staphylococcus hyicus, and various CoNS species from pigs, cattle, horses, chickens, and ducks (Kehrenberg and Schwarz 2006; Kehrenberg et al. 2007, 2009; Wang et al. 2012d,e, 2013a; He et al. 2014). The gene cfr was also detected in S. aureus and CoNS from infections in humans (Toh et al. 2007; Mendes et al. 2008, 2013; Bonilla et al. 2010; Shore et al. 2010; Gopegui et al. 2012; Locke et al. 2012; Cui et al. 2013; LaMarre et al. 2013; Feßler et al. 2014; Bender et al. 2015, Cai et al. 2015b). Most recently, the cfr gene was found to be integrated in a type IVb SCCmec cassette of an MRSA CC9 isolate of swine origin (Fig. 5) (Li et al. 2015).
Figure 5.
Schematic presentation of the structural variability in the regions surrounding the cfr gene on plasmids or in the chromosomal DNA of various Gram-positive and Gram-negative bacteria. All genes are indicated by arrows with the arrowhead showing the direction of transcription. Insertion sequences are presented as gray boxes with the arrow inside indicating the transposase gene. The cfr gene is shown as a red arrow, whereas all other antimicrobial resistance genes are depicted as violet arrows. Genes involved in transposition are shown in yellow, whereas the genes involved in plasmid replication and plasmid recombination/mobilization are shown in green and blue, respectively. Genes with other functions are displayed in white. The Δ symbol indicates a truncated gene. A 1-kb scale is shown in the upper left corner. aadD, kanamycin/neomycin resistance; aadY, streptomycin resistance; aacA-aphD, gentamicin/kanamycin/tobramycin resistance; ble, bleomycin resistance; dfrK, trimethoprim resistance, erm(A), erm(B), erm(C), erm(33), MLSB resistance; fexA, chloramphenicol/florfenicol resistance; lsa(B), elevated minimum inhibitory concentrations (MICs) of lincosamides; spc, spectinomycin resistance; tet(L), tetracycline resistance.
Screening studies conducted in China revealed the presence of the gene cfr also in other Gram-positive bacteria such as Bacillus spp. (Dai et al. 2010; Zhang et al. 2011; Wang et al. 2012b), Enterococcus spp. (Liu et al. 2012b, 2013, 2014), Macrococcus caseolyticus and Jeotgalicoccus pinnipedialis (Wang et al. 2012c), and S. suis (Wang et al. 2013b). In most of these cases, the cfr gene was located on plasmids of variable sizes. Moreover, the cfr gene was also found in a small number of Gram-negative bacteria, for example, in the chromosomal DNA of a Proteus vulgaris isolate (Wang et al. 2011) or on plasmids in E. coli (Fig. 5) (Wang et al. 2012a; Zhang et al. 2014, 2015b).
A comparative analysis of the plasmids on which the cfr gene was found in the different bacteria revealed a high structural variability in the regions surrounding the cfr gene (Shen et al. 2013). Moreover, numerous insertion sequences, such as IS21-558, IS256, IS1216, ISEnfa4, ISEnfa5, and IS26, were found in close proximity to the cfr gene (Fig. 5). These insertion sequences may play a role in the transfer of cfr between different plasmids but also in the chromosomal integration of cfr-carrying segments. In addition, many cfr-carrying plasmids harbor additional resistance genes, which enable the coselection and persistence of the cfr gene under a selection pressure imposed by non-PhLOPSA antimicrobial agents.
Recently, a cfr-like gene, whose product showed only 75% aa identity to the original Cfr protein from S. sciuri, was detected in C. difficile, and claimed to be responsible for linezolid resistance (Marín et al. 2015; Schwarz and Wang 2015). In a different study, this gene, meanwhile designated cfr(B), has been shown to confer multiple antimicrobial resistance by the same mechanism as the original cfr gene (Hansen and Vester 2015). Most recently, the cfr(B) gene was also detected in E. faecium recovered from human specimens in the United States (Deshpande et al. 2015).
CONCLUDING REMARKS
For a number of years, fewer and fewer new antimicrobial agents have been approved for use in human and veterinary medicine despite rising trends in antimicrobial resistance in bacterial pathogens of human, veterinary, and zoonotic relevance. This is particularly true for veterinary medicine as all new antimicrobial agents approved during the last 25 years represent derivatives of already known substances. For the development of antimicrobial agents with improved binding to the ribosome, the exact mode of action of the antimicrobial agents, but also the knowledge about their binding sites, are indispensable prerequisites. Moreover, the knowledge about resistance-mediating mutations and the mechanisms specified by resistance genes are important aspects that need to be taken into account when developing new antimicrobial agents that may even overcome existing resistance mechanisms. Florfenicol is a good example to illustrate how knowledge about the most common phenicol resistance mechanism, namely, the enzymatic inactivation of chloramphenicol by chloramphenicol acetyltransferases, was used to generate an antimicrobial agent that was resistant to inactivation by the most widespread resistance mechanism and, in addition, did not show the adverse side effects of the parental substance. However, bacteria are versatile organisms that are able to quickly develop or acquire new resistance mechanisms. In the case of the synthetic and new agent florfenicol, the first phenicol exporters and target site–modifying enzymes, that conferred florfenicol resistance, were identified only a few years after the introduction of florfenicol into use in animals (Kim and Aoki 1996; Arcangioli et al. 1999; Schwarz et al. 2000).
The introduction of any new antimicrobial agent into clinical use will create a new selection pressure under which bacteria develop and/or acquire sooner or later new resistance genes or resistance-mediating mutations. This will reduce the efficacy of the antimicrobial agents and create a demand for newer and better antimicrobial agents. Changes in the current practice of how we use antimicrobial agents not only in human and veterinary medicine, but also in horticulture and aquaculture, are unavoidable, and good antimicrobial stewardship practice should be adopted by everyone involved in antimicrobial use and application (Prescott 2014). The future will show whether new technologies, such as whole-genome sequencing, which have become widely available during the past few years, can successfully be used for the identification of new bacterial targets that serve for the development of future antimicrobial agents for specific bacterial pathogens and disease conditions (Prescott 2014).
ACKNOWLEDGMENTS
We thank our technical staff, Kerstin Meyer, Roswitha Becker, Vivian Hensel, Ute Beermann, Marita Meurer, and Regina Ronge for their invaluable help and support and our colleagues and cooperation partners for the fruitful collaboration in the various research projects that formed the basis of this article. Work in our laboratories is supported by the German Federal Ministry of Education and Research (BMBF) through the German Aerospace Center (DLR) Grant number 01KI1313D (RESET II) and Grant number 01KI1301D (MedVet-Staph II), respectively, as well as by the German Research Foundation (DFG) Grant numbers SCHW382/10-1 and SCHW382/10-2; Grants from the National Basic Research Program of China (2013CB127200) and the National Natural Science Foundation of China (31370046); and a Grant from the Danish National Research Foundation (12-125943).
Footnotes
Editors: Lynn L. Silver and Karen Bush
Additional Perspectives on Antibiotics and Antibiotic Resistance available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Aarestrup FM, Agersø Y, Ahrens P, Jørgensen JC, Madsen M, Jensen LB. 2000. Antimicrobial susceptibility and presence of resistance genes in staphylococci from poultry. Vet Microbiol 74: 353–364. [DOI] [PubMed] [Google Scholar]
- Achard A, Villers C, Pichereau V, Leclercq R. 2005. New lnu(C) gene conferring resistance to lincomycin by nucleotidylation in Streptococcus agalactiae UCN36. Antimicrob Agents Chemother 49: 2716–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahonkhai VI, Cherubin CE, Shulman MA, Jhagroo M, Bancroft U. 1982. In vitro activity of U-57930E, a new clindamycin analog, against aerobic Gram-positive bacteria. Antimicrob Agents Chemother 21: 902–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allignet J, El Solh N. 1995. Diversity among the Gram-positive acetyltransferases inactivating streptogramin A and structurally related compounds and characterization of a new staphylococcal determinant, vatB. Antimicrob Agents Chemother 39: 2027–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allignet J, El Solh N. 1997. Characterization of a new staphylococcal gene, vgaB, encoding a putative ABC transporter conferring resistance to streptogramin A and related compounds. Gene 202: 133–138. [DOI] [PubMed] [Google Scholar]
- Allignet J, El Solh N. 1999. Comparative analysis of staphylococcal plasmids carrying three streptogramin-resistance genes: vat-vgb-vga. Plasmid 42: 134–138. [DOI] [PubMed] [Google Scholar]
- Allignet J, Loncle V, Mazodier P, El Solh N. 1988. Nucleotide sequence of a staphylococcal plasmid gene, vgb, encoding a hydrolase inactivating the B components of virginiamycin-like antibiotics. Plasmid 20: 271–275. [DOI] [PubMed] [Google Scholar]
- Allignet J, Loncle V, El Solh N. 1992. Sequence of a staphylococcal plasmid gene, vga, encoding a putative ATP-binding protein involved in resistance to virginiamycin A-like antibiotics. Gene 117: 45–51. [DOI] [PubMed] [Google Scholar]
- Allignet J, Loncle V, Simenel C, Delepierre M, el Solh N. 1993. Sequence of a staphylococcal gene, vat, encoding an acetyltransferase inactivating the A-type compounds of virginiamycin-like antibiotics. Gene 130: 91–98. [DOI] [PubMed] [Google Scholar]
- Allignet J, Aubert S, Morvan A, El Solh N. 1996. Distribution of genes encoding resistance to streptogramin A and related compounds among staphylococci resistant to these antibiotics. Antimicrob Agents Chemother 40: 2523–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allignet J, Liassine N, El Solh N. 1998. Characterization of a staphylococcal plasmid related to pUB110 and carrying two novel genes, vatC and vgbB, encoding resistance to streptogramins A and B and similar antibiotics. Antimicrob Agents Chemother 42: 1794–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangioli MA, Leroy-Sétrin S, Martel JL, Chaslus-Dancla E. 1999. A new chloramphenicol and florfenicol resistance gene flanked by two integron structures in Salmonella typhimurium DT104. FEMS Microbiol Lett 174: 327–332. [DOI] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. [DOI] [PubMed] [Google Scholar]
- Bender J, Strommenger B, Steglich M, Zimmermann O, Fenner I, Lensing C, Dagwadordsch U, Kekulé AS, Werner G, Layer F. 2015. Linezolid resistance in clinical isolates of Staphylococcus epidermidis from German hospitals and characterization of two cfr-carrying plasmids. J Antimicrob Chemother 70: 1630–1638. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The protein data bank. Nucl Acids Res 28: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besier S, Ludwig A, Zander J, Brade V, Wichelhaus TA. 2008. Linezolid resistance in Staphylococcus aureus: Gene dosage effect, stability, fitness costs, and cross-resistances. Antimicrob Agents Chemother 52: 1570–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla H, Huband MD, Seidel J, Schmidt H, Lescoe M, McCurdy SP, Lemmon MM, Brennan LA, Tait-Kamradt A, Puzniak L, et al. 2010. Multicity outbreak of linezolid-resistant Staphylococcus epidermidis associated with clonal spread of a cfr-containing strain. Clin Infect Dis 51: 796–800. [DOI] [PubMed] [Google Scholar]
- Bozdogan B, Berrezouga L, Kuo MS, Yurek DA, Farley KA, Stockman BJ, Leclercq R. 1999. A new resistance gene, linB, conferring resistance to lincosamides by nucleotidylation in Enterococcus faecium HM1025. Antimicrob Agents Chemother 43: 925–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenciani A, Morroni G, Vincenzi C, Manso E, Mingoia M, Giovanetti E, Varaldo PE. 2016. Detection in Italy of two clinical Enterococcus faecium isolates carrying both the oxazolidinone and phenicol resistance gene optrA and a silent multiresistance gene cfr. J Antimicrob Chemother 71: 307–313. [DOI] [PubMed] [Google Scholar]
- Brisson-Noël A, Courvalin P. 1986. Nucleotide sequence of gene linA encoding resistance to lincosamides in Staphylococcus haemolyticus. Gene 43: 247–253. [DOI] [PubMed] [Google Scholar]
- Bryskier A. 2005a. Lincosamines. In Antimicrobial agents: Antibacterials antifungals (ed. Bryskier A), pp. 592–603. ASM, Washington, DC. [Google Scholar]
- Bryskier A. 2005b. Mutilins. In Antimicrobial agents: Antibacterials antifungals (ed. Bryskier A), pp. 1239–1241. ASM, Washington, DC. [Google Scholar]
- Butaye P, Cloeckaert A, Schwarz S. 2003. Mobile genes coding for efflux-mediated antimicrobial resistance in Gram-positive and Gram-negative bacteria. Int J Antimicrob Agents 22: 205–210. [DOI] [PubMed] [Google Scholar]
- Cai J, Wang Y, Schwarz S, Lv H, Li Y, Liao K, Yu S, Zhao K, Gu D, Wang X, et al. 2015a. Enterococcal isolates carrying the novel oxazolidinone resistance gene optrA from hospitals in Zhejiang, Guangdong, and Henan, China, 2010–2014. Clin Microbiol Infect 21: 1095.e1–1095.e4. [DOI] [PubMed] [Google Scholar]
- Cai JC, Hu YY, Chen GX, Zhou HW, Zhang R. 2015b. Dissemination of the same cfr-carrying plasmid among methicillin-resistant Staphylococcus aureus and coagulase-negative staphylococcal isolates in China. Antimicrob Agents Chemother 59: 3669–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoir V, Merabet L, Legrand P, Soussy CJ, Leclercq R. 2007. Emergence of a Streptococcus pneumoniae isolate resistant to streptogramins by mutation in ribosomal protein L22 during pristinamycin therapy of pneumococcal pneumonia. J Antimicrob Chemother 59: 1010–1012. [DOI] [PubMed] [Google Scholar]
- Chen LP, Cai XW, Wang XR, Zhou XL, Wu DF, Xu XJ, Chen HC. 2010. Characterization of plasmid-mediated lincosamide resistance in a field isolate of Haemophilus parasuis. J Antimicrob Chemother 65: 2256–2258. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). 2015. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals, 3rd ed CLSI supplement VET01S, CLSI, Wayne, PA. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). 2016. Performance standards for antimicrobial susceptibility testing, 26th ed CLSI supplement M100S, CLSI, Wayne, PA. [Google Scholar]
- Cui L, Wang Y, Li Y, He T, Schwarz S, Ding Y, Shen J, Lv Y. 2013. Cfr-mediated linezolid-resistance among methicillin-resistant coagulase-negative staphylococci from infections of humans. PLoS ONE 8: e57096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Wu CM, Wang MG, Wang Y, Wang Y, Huang SY, Xia LN, Li BB, Shen JZ. 2010. First report of the multidrug resistance gene cfr and the phenicol resistance gene fexA in a Bacillus strain from swine feces. Antimicrob Agents Chemother 54: 3953–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Bashan A, Auerbach-Nevo T, Yaggie RD, Gontarek RR, Yonath A. 2007. Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity. Proc Natl Acad Sci 104: 4291–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande L, Ashcraft D, Kahn H, Pankey G, Jones R, Farrell D, Mendes R. 2015. Detection of a new cfr-like gene, cfr(B), in Enterococcus faecium recovered from human specimens in the United States: Report from The SENTRY Antimicrobial Surveillance Program. Antimicrob Agents Chemother 59: 6256–6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diab SS, Songer G, Uzal FA. 2013. Clostridium difficile infection in horses: A review. Vet Microbiol 167: 42–49. [DOI] [PubMed] [Google Scholar]
- Douthwaite S. 1992. Functional interactions within 23S rRNA involving the peptidyltransferase center. J Bacteriol 174: 1333–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling PM. 2013. Chloramphenicol, thiamphenicol, and florfenicol. In Antimicrobial therapy in veterinary medicine, 5th ed (ed. Giguère S, Prescott JF, Dowling PM), pp. 269–277. John Wiley, Hoboken, NJ. [Google Scholar]
- Dunkle JA, Xiong L, Mankin AS, Cate JH. 2010. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc Natl Acad Sci 107: 17152–17157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Bibi E. 1997. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J Bacteriol 179: 2274–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyal Z, Matzov D, Krupkin M, Wekselman I, Paukner S, Zimmerman E, Rozenberg H, Bashan A, Yonath A. 2015. Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus. Proc Natl Acad Sci 112: E5805–E5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feßler AT, Calvo N, Gutiérrez N, Muñoz Bellido JL, Fajardo M, Garduño E, Monecke S, Ehricht R, Kadlec K, Schwarz S. 2014. Cfr-mediated linezolid resistance in methicillin-resistant Staphylococcus aureus and Staphylococcus haemolyticus associated with clinical infections in humans: Two case reports. J Antimicrob Chemother 69: 268–270. [DOI] [PubMed] [Google Scholar]
- Floyd JL, Smith KP, Kumar SH, Floyd JT, Varela MF. 2010. LmrS is a multidrug efflux pump of the major facilitator superfamily from Staphylococcus aureus. Antimicrob Agents Chemother 54: 5406–5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Fyfe C, Grossman TH, Kerstein K, Sutcliffe J. 2016. Resistance to macrolide antibiotics in public health pathogens. Cold Spring Harb Perspect Med 10.1101/cshperspect.a025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimand M, Gerbaud G, Guibourdenche M, Riou JY, Courvalin P. 1998. High-level chloramphenicol resistance in Neisseria meningitidis. N Engl J Med 339: 868–874. [DOI] [PubMed] [Google Scholar]
- Gatermann SG, Koschinski T, Friedrich S. 2007. Distribution and expression of macrolide resistance genes in coagulase-negative staphylococci. Clin Microbiol Infect 13: 777–781. [DOI] [PubMed] [Google Scholar]
- Gay K, Stephens DS. 2001. Structure and dissemination of a chromosomal insertion element encoding macrolide efflux in Streptococcus pneumoniae. J Infect Dis 184: 56–65. [DOI] [PubMed] [Google Scholar]
- Gentry DR, Rittenhouse SF, McCloskey L, Holmes DJ. 2007. Stepwise exposure of Staphylococcus aureus to pleuromutilins is associated with stepwise acquisition of mutations in rplC and minimally affects susceptibility to retapamulin. Antimicrob Agents Chemother 51: 2048–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry DR, McCloskey L, Gwynn MN, Rittenhouse SF, Scangarella N, Shawar R, Holmes DJ. 2008. Genetic characterization of Vga ABC proteins conferring reduced susceptibility of Staphylococcus aureus to pleuromutilins. Antimicrob Agents Chemother 52: 4507–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerding DN, Johnson S, Peterson LR, Mulligan ME, Silva J Jr. 1995. Clostridium difficile-associated diarrhea and colitis. Infect Control Hosp Epidemiol 16: 459–477. [DOI] [PubMed] [Google Scholar]
- Giguère S. 2013. Lincosamides, pleuromutilins, and streptogramins. In Antimicrobial therapy in veterinary medicine, 5th ed (ed. Giguère S, Prescott JF, Dowling PM), pp. 199–210. Wiley, Hoboken, NJ. [Google Scholar]
- Gómez-Sanz E, Kadlec K, Feßler AT, Zarazaga M, Torres C, Schwarz S. 2013. A novel fexA variant from a canine Staphylococcus pseudintermedius isolate that does not confer florfenicol resistance. Antimicrob Agents Chemother 57: 5763–5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopegui ER, Juan C, Zamorano L, Pérez JL, Oliver A. 2012. Transferable multidrug resistance plasmid carrying cfr associated with tet(L), ant(4′)-Ia, and dfrK genes from a clinical methicillin-resistant Staphylococcus aureus ST125 strain. Antimicrob Agents Chemother 56: 2139–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerum AM, Flannagan SE, Clewell DB, Jensen LB. 2001. Indication of transposition of a mobile DNA element containing the vat(D) and erm(B) genes in Enterococcus faecium. Antimicrob Agents Chemother 45: 3223–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen LH, Vester B. 2015. A cfr-like gene from Clostridium difficile confers multiple antibiotic resistance by the same mechanism as the cfr gene. Antimicrob Agents Chemother 59: 5841–5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen LH, Jensen LB, Sørensen HI, Sørensen SJ. 2007. Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J Antimicrob Chemother 60: 145–147. [DOI] [PubMed] [Google Scholar]
- Haroche J, Allignet J, Buchrieser C, El Solh N. 2000. Characterization of a variant of vga(A) conferring resistance to streptogramin A and related compounds. Antimicrob Agents Chemother 44: 2271–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroche J, Allignet J, El Solh N. 2002. Tn5406, a new staphylococcal transposon conferring resistance to streptogramin A and related compounds including dalfopristin. Antimicrob Agents Chemother 46: 2337–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauschild T, Feßler AT, Kadlec K, Billerbeck C, Schwarz S. 2012. Detection of the novel vga(E) gene in methicillin-resistant Staphylococcus aureus CC398 isolates from cattle and poultry. J Antimicrob Chemother 67: 503–504. [DOI] [PubMed] [Google Scholar]
- He T, Wang Y, Schwarz S, Zhao Q, Shen J, Wu C. 2014. Genetic environment of the multi-resistance gene cfr in methicillin-resistant coagulase-negative staphylococci from chickens, ducks, and pigs in China. Int J Med Microbiol 304: 257–261. [DOI] [PubMed] [Google Scholar]
- He T, Shen J, Schwarz S, Wu C, Wang Y. 2015. Characterization of a genomic island in Stenotrophomonas maltophilia that carries a novel floR gene variant. J Antimicrob Chemother 70: 1031–1036. [DOI] [PubMed] [Google Scholar]
- He T, Shen Y, Schwarz S, Cai J, Lv Y, Li J, Feßler AT, Zhang R, Wu C, Shen J, et al. 2016. Genetic environment of the transferable oxazolidinone/phenicol resistance gene optrA in Enterococcus faecalis isolates of human and animal origin. J Antimicrob Chemother 10.1093/jac/dkw016. [DOI] [PubMed] [Google Scholar]
- Heir E, Lindstedt BA, Leegaard TM, Gjernes E, Kapperud G. 2004. Prevalence and characterization of integrons in blood culture Enterobacteriaceae and gastrointestinal Escherichia coli in Norway and reporting of a novel class 1 integron–located lincosamide resistance gene. Ann Clin Microbiol Antimicrob 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hot C, Berthet N, Chesneau O. 2014. Characterization of sal(A), a novel gene responsible for lincosamide and streptogramin A resistance in Staphylococcus sciuri. Antimicrob Agents Chemother 58: 3335–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, O’Toole PW, Shen W, Amrine-Madsen H, Jiang X, Lobo N, Palmer LM, Voelker L, Fan F, Gwynn MN, et al. 2004. Novel chromosomally encoded multidrug efflux transporter MdeA in Staphylococcus aureus. Antimicrob Agents Chemother 48: 909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isnard C, Malbruny B, Leclercq R, Cattoir V. 2013. Genetic basis for in vitro and in vivo resistance to lincosamides, streptogramins A, and pleuromutilins (LSAP phenotype) in Enterococcus faecium. Antimicrob Agents Chemother 57: 4463–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglic Z, Vlkova H, Bardon J, Michu E, Cervinkova D, Babak V. 2012. Distribution, characterization and genetic bases of erythromycin resistance in staphylococci and enterococci originating from livestock. Zoonoses Public Health 59: 202–211. [DOI] [PubMed] [Google Scholar]
- Jung YH, Shin ES, Kim O, Yoo JS, Lee KM, Yoo JI, Chung GT, Lee YS. 2010. Characterization of two newly identified genes, vgaD and vatG, conferring resistance to streptogramin A in Enterococcus faecium. Antimicrob Agents Chemother 54: 4744–4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec K, Schwarz S. 2009. Identification of a novel ABC transporter gene, vga(C), located on a multiresistance plasmid from a porcine methicillin-resistant Staphylococcus aureus ST398 strain. Antimicrob Agents Chemother 53: 3589–3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec K, Kehrenberg C, Schwarz S. 2007. Efflux-mediated resistance to florfenicol and/or chloramphenicol in Bordetella bronchiseptica: Identification of a novel chloramphenicol exporter. J Antimicrob Chemother 59: 191–196. [DOI] [PubMed] [Google Scholar]
- Kadlec K, Pomba CF, Couto N, Schwarz S. 2010. Small plasmids carrying vga(A) or vga(C) genes mediate resistance to lincosamides, pleuromutilins and streptogramin A antibiotics in methicillin-resistant Staphylococcus aureus ST398 from swine. J Antimicrob Chemother 65: 2692–2693. [DOI] [PubMed] [Google Scholar]
- Kadlec K, Brenner Michael G, Sweeney MT, Brzuszkiewicz E, Liesegang H, Daniel R, Watts JL, Schwarz S. 2011. Molecular basis of macrolide, triamilide, and lincosamide resistance in Pasteurella multocida from bovine respiratory disease. Antimicrob Agents Chemother 55: 2475–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrenberg C, Schwarz S. 2004. fexA, a novel Staphylococcus lentus gene encoding resistance to florfenicol and chloramphenicol. Antimicrob Agents Chemother 48: 615–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrenberg C, Schwarz S. 2005. Florfenicol–chloramphenicol exporter gene fexA is part of the novel transposon Tn558. Antimicrob Agents Chemother 49: 813–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrenberg C, Schwarz S. 2006. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob Agents Chemother 50: 1156–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrenberg C, Ojo KK, Schwarz S. 2004. Nucleotide sequence and organization of the multiresistance plasmid pSCFS1 from Staphylococcus sciuri. J Antimicrob Chemother 54: 936–939. [DOI] [PubMed] [Google Scholar]
- Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. 2005. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: Methylation of 23S ribosomal RNA at A2503. Mol Microbiol 57: 1064–1073. [DOI] [PubMed] [Google Scholar]
- Kehrenberg C, Aarestrup FM, Schwarz S. 2007. IS21-558 insertion sequences are involved in the mobility of the multiresistance gene cfr. Antimicrob Agents Chemother 51: 483–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrenberg C, Cuny C, Strommenger B, Schwarz S, Witte W. 2009. Methicillin-resistant and -susceptible Staphylococcus aureus strains of clonal lineages ST398 and ST9 from swine carry the multidrug resistance gene cfr. Antimicrob Agents Chemother 53: 779–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy AD, Porcella SF, Martens C, Whitney AR, Braughton KR, Chen L, Craig CT, Tenover FC, Kreiswirth BN, Musser JM, et al. 2010. Complete nucleotide sequence analysis of plasmids in strains of Staphylococcus aureus clone USA300 reveals a high level of identity among isolates with closely related core genome sequences. J Clin Microbiol 48: 4504–4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EH, Aoki T. 1996. Sequence analysis of the florfenicol resistance gene encoded in the transferable R-plasmid of a fish pathogen, Pasteurella piscicida. Microbiol Immunol 40: 665–669. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kim Y, Lee MS, Lee HS. 2001. Gene lmrB of Corynebacterium glutamicum confers efflux-mediated resistance to lincomycin. Mol Cells 12: 112–116. [PubMed] [Google Scholar]
- Klitgaard RN, Ntokou E, Norgaard K, Biltoft D, Hansen LH, Traedholm NM, Kongsted J, Vester B. 2015. Mutations in the bacterial ribosomal protein L3 and their association with antibiotic resistance. Antimicrob Agents Chemother 59: 3518–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano M, Fujita M, Nakamura K, Murata M, Ohki R, Yamane K. 2003. Lincomycin resistance mutations in two regions immediately downstream of the –10 region of lmr promoter cause overexpression of a putative multidrug efflux pump in Bacillus subtilis mutants. Antimicrob Agents Chemother 47: 432–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMarre J, Mendes RE, Szal T, Schwarz S, Jones RN, Mankin AS. 2013. The genetic environment of the cfr gene and the presence of other mechanisms account for the very high linezolid resistance of Staphylococcus epidermidis isolate 426-3147L. Antimicrob Agents Chemother 57: 1173–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang KS, Anderson JM, Schwarz S, Williamson L, Handelsman J, Singer RS. 2010. Novel florfenicol and chloramphenicol resistance gene discovered in Alaskan soil by using functional metagenomics. Appl Environ Microbiol 76: 5321–5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq R, Courvalin P. 1991. Intrinsic and unusual resistance to macrolide, lincosamide, and streptogramin antibiotics in bacteria. Antimicrob Agents Chemother 35: 1273–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AGW, Moody PCE, Shaw WV. 1988. Structure of chloramphenicol acetyltransferase at 1.75 Å resolution. Proc Natl Acad Sci 85: 4133–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levings RS, Hall RM, Lightfoot D, Djordjevic SP. 2006. linG, a new integron-associated gene cassette encoding a lincosamide nucleotidyltransferase. Antimicrob Agents Chemother 50: 3514–3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BB, Wu CM, Wang Y, Shen JZ. 2011. Single and dual mutations at positions 2058, 2503 and 2504 of 23S rRNA and their relationship to resistance to antibiotics that target the large ribosomal subunit. J Antimicrob Chemother 66: 1983–1986. [DOI] [PubMed] [Google Scholar]
- Li B, Wendlandt S, Yao J, Liu Y, Zhang Q, Shi Z, Wei J, Shao D, Schwarz S, Wang S, et al. 2013. Detection and new genetic environment of the pleuromutilin-lincosamide-streptogramin A resistance gene lsa(E) in methicillin-resistant Staphylococcus aureus of swine origin. J Antimicrob Chemother 68: 1251–1255. [DOI] [PubMed] [Google Scholar]
- Li J, Li B, Wendlandt S, Schwarz S, Wang Y, Wu C, Ma Z, Shen J. 2014a. Identification of a novel vga(E) gene variant that confers resistance to pleuromutilins, lincosamides and streptogramin A antibiotics in staphylococci of porcine origin. J Antimicrob Chemother 69: 919–923. [DOI] [PubMed] [Google Scholar]
- Li XS, Dong WC, Wang XM, Hu GZ, Wang YB, Cai BY, Wu CM, Wang Y, Du XD. 2014b. Presence and genetic environment of pleuromutilin–lincosamide–streptogramin A resistance gene lsa(E) in enterococci of human and swine origin. J Antimicrob Chemother 69: 1424–1426. [DOI] [PubMed] [Google Scholar]
- Li D, Wu C, Wang Y, Fan R, Schwarz S, Zhang S. 2015. Identification of multiresistance gene cfr in methicillin-resistant Staphylococcus aureus from pigs: Plasmid location and integration into a staphylococcal cassette chromosome mec complex. Antimicrob Agents Chemother 59: 3641–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Wang Y, Schwarz S, Cai J, Fan R, Li J, Feßler AT, Zhang R, Wu C, Shen J. 2016. Co-location of the oxazolidinone resistance genes optrA and cfr on a multi-resistance plasmid from Staphylococcus sciuri. J Antimicrobial Chemother 10.1093/jac/dkw040. [DOI] [PubMed] [Google Scholar]
- Lindeman CJ, Portis E, Johansen L, Mullins LM, Stoltman GA. 2013. Susceptibility to antimicrobial agents among bovine mastitis pathogens isolated from North American dairy cattle, 2002–2010. J Vet Diagn Invest 25: 581–591. [DOI] [PubMed] [Google Scholar]
- Liu H, Wang Y, Wu C, Schwarz S, Shen Z, Jeon B, Ding S, Zhang Q, Shen J. 2012a. A novel phenicol exporter gene, fexB, found in enterococci of animal origin. J Antimicrob Chemother 67: 322–325. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang Y, Wu C, Shen Z, Schwarz S, Du XD, Dai L, Zhang W, Zhang Q, Shen J. 2012b. First report of the multidrug resistance gene cfr in Enterococcus faecalis of animal origin. Antimicrob Agents Chemother 56: 1650–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang Y, Schwarz S, Li Y, Shen Z, Zhang Q, Wu C, Shen J. 2013. Transferable multiresistance plasmids carrying cfr in Enterococcus spp. from swine and farm environment. Antimicrob Agents Chemother 57: 42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang Y, Dai L, Wu C, Shen J. 2014. First report of multiresistance gene cfr in Enterococcus species casseliflavus and gallinarum of swine origin. Vet Microbiol 170: 352–357. [DOI] [PubMed] [Google Scholar]
- Locke JB, Rahawi S, LaMarre J, Mankin AS, Shaw KJ. 2012. Genetic environment and stability of cfr in methicillin-resistant Staphylococcus aureus CM05. Antimicrob Agents Chemother 56: 332–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeza-Lara PD, Soto-Huipe M, Baizabal-Aguirre VM, Ochoa-Zarzosa A, Valdez-Alarcón JJ, Cano-Camacho H, López-Meza JE. 2004. pBMSa1, a plasmid from a dairy cow isolate of Staphylococcus aureus, encodes a lincomycin resistance determinant and replicates by the rolling-circle mechanism. Plasmid 52: 48–56. [DOI] [PubMed] [Google Scholar]
- Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. 2006. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob Agents Chemother 50: 2500–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long KS, Poehlsgaard J, Hansen LH, Hobbie SN, Bottger EC, Vester B. 2009. Single 23S rRNA mutations at the ribosomal peptidyl transferase centre confer resistance to valnemulin and other antibiotics in Mycobacterium smegmatis by perturbation of the drug binding pocket. Mol Microbiol 71: 1218–1227. [DOI] [PubMed] [Google Scholar]
- Long KS, Munck C, Andersen TM, Schaub MA, Hobbie SN, Böttger EC, Vester B. 2010. Mutations in 23S rRNA at the peptidyl transferase center and their relationship to linezolid binding and cross-resistance. Antimicrob Agents Chemother 54: 4705–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett PS. 1990. Translational attenuation as the regulator of inducible cat genes. J Bacteriol 172: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano C, Aspiroz C, Sáenz Y, Ruiz-García M, Royo-García G, Gómez-Sanz E, Ruiz-Larrea F, Zarazaga M, Torres C. 2012a. Genetic environment and location of the lnu(A) and lnu(B) genes in methicillin-resistant Staphylococcus aureus and other staphylococci of animal and human origin. J Antimicrob Chemother 67: 2804–2808. [DOI] [PubMed] [Google Scholar]
- Lozano C, Rezusta A, Gómez P, Gómez-Sanz E, Báez N, Martin-Saco G, Zarazaga M, Torres C. 2012b. High prevalence of spa types associated with the clonal lineage CC398 among tetracycline-resistant methicillin-resistant Staphylococcus aureus strains in a Spanish hospital. J Antimicrob Chemother 67: 330–334. [DOI] [PubMed] [Google Scholar]
- Lüthje P, Schwarz S. 2006. Antimicrobial resistance of coagulase-negative staphylococci from bovine subclinical mastitis with particular reference to macrolide-lincosamide resistance phenotypes and genotypes. J Antimicrob Chemother 57: 966–969. [DOI] [PubMed] [Google Scholar]
- Lüthje P, Schwarz S. 2007a. Molecular analysis of constitutively expressed erm(C) genes selected in vitro in the presence of the non-inducers pirlimycin, spiramycin and tylosin. J Antimicrob Chemother 59: 97–101. [DOI] [PubMed] [Google Scholar]
- Lüthje P, Schwarz S. 2007b. Molecular basis of resistance to macrolides and lincosamides among staphylococci and streptococci from various animal sources collected in the resistance monitoring program BfT-GermVet. Int J Antimicrob Agents 29: 528–535. [DOI] [PubMed] [Google Scholar]
- Lyras D, Rood JI. 2000. Transposition of Tn4451 and Tn4453 involves a circular intermediate that forms a promoter for the large resolvase, TnpX. Mol Microbiol 38: 588–601. [DOI] [PubMed] [Google Scholar]
- MacLeod AJ, Ross HB, Ozere RL, Digout G, van Rooyen CE. 1964. Lincomycin: A new antibiotic active against staphylococci and other Gram-positive cocci: Clinical and laboratory studies. Can Med Assoc J 91: 1056–1060. [PMC free article] [PubMed] [Google Scholar]
- Malbruny B, Werno AM, Murdoch DR, Leclercq R, Cattoir V. 2011. Cross-resistance to lincosamides, streptogramins A, and pleuromutilins due to the lsa(C) gene in Streptococcus agalactiae UCN70. Antimicrob Agents Chemother 55: 1470–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín M, Martín A, Alcalá L, Cercenado E, Iglesias C, Reigadas E, Bouza E. 2015. Clostridium difficile with high linezolid MICs harbor the multiresistance gene cfr. Antimicrob Agents Chemother 59: 586–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka M, Inoue M, Endo Y, Nakajima Y. 2003. Characteristic expression of three genes, msr(A), mph(C) and erm(Y), that confer resistance to macrolide antibiotics on Staphylococcus aureus. FEMS Microbiol Lett 220: 287–293. [DOI] [PubMed] [Google Scholar]
- McMurray LM, George AM, Levy SB. 1994. Active efflux of chloramphenicol in susceptible Escherichia coli strains and in multiple-antibiotic-resistant (Mar) mutants. Antimicrob Agents Chemother 38: 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes RE, Deshpande LM, Castanheira M, DiPersio J, Saubolle MA, Jones RN. 2008. First report of cfr-mediated resistance to linezolid in human staphylococcal clinical isolates recovered in the United States. Antimicrob Agents Chemother 52: 2244–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes RE, Deshpande LM, Bonilla HF, Schwarz S, Huband MD, Jones RN, Quinn JP. 2013. Dissemination of a pSCFS3-like cfr-carrying plasmid in Staphylococcus aureus and Staphylococcus epidermidis clinical isolates recovered from hospitals in Ohio. Antimicrob Agents Chemother 57: 2923–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael GB, Kadlec K, Sweeney MT, Brzuszkiewicz E, Liesegang H, Daniel R, Murray RW, Watts JL, Schwarz S. 2012. ICEPmu1, an integrative conjugative element (ICE) of Pasteurella multocida: Analysis of the regions that comprise 12 antimicrobial resistance genes. J Antimicrob Chemother 67: 84–90. [DOI] [PubMed] [Google Scholar]
- Miller K, Dunsmore CJ, Fishwick CW, Chopra I. 2008. Linezolid and tiamulin cross-resistance in Staphylococcus aureus mediated by point mutations in the peptidyl transferase center. Antimicrob Agents Chemother 52: 1737–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monecke S, Ruppelt A, Wendlandt S, Schwarz S, Slickers P, Ehricht R, Jäckel SC. 2013. Genotyping of Staphylococcus aureus isolates from diseased poultry. Vet Microbiol 162: 806–812. [DOI] [PubMed] [Google Scholar]
- Montilla A, Zavala A, Cáceres Cáceres R, Cittadini R, Vay C, Gutkind G, Famiglietti A, Bonofiglio L, Mollerach M. 2014. Genetic environment of the lnu(B) gene in a Streptococcus agalactiae clinical isolate. Antimicrob Agents Chemother 58: 5636–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morar M, Bhullar K, Hughes DW, Junop M, Wright GD. 2009. Structure and mechanism of the lincosamide antibiotic adenylyltransferase LinB. Structure 17: 1649–1659. [DOI] [PubMed] [Google Scholar]
- Morris TH. 1995. Antibiotic therapeutics in laboratory animals. Lab Anim 29: 16–36. [DOI] [PubMed] [Google Scholar]
- Mosher RH, Ranade NP, Schrempf H, Vining LC. 1990. Chloramphenicol resistance in Streptomyces: Cloning and characterization of a chloramphenicol hydrolase gene from Streptomyces venezuelae. J Gen Microbiol 136: 293–301. [DOI] [PubMed] [Google Scholar]
- Mosher RH, Camp DJ, Yang K, Brown MP, Shaw WV, Vining LC. 1995. Inactivation of chloramphenicol by O-phosphorylation. J Biol Chem 27: 27000–27006. [DOI] [PubMed] [Google Scholar]
- Murata M, Ohno S, Kumano M, Yamamane K, Ohki R. 2003. Multidrug resistant phenotype of Bacillus subtilis spontaneous mutants isolated in the presence of puromycin and lincomycin. Can J Microbiol 49: 71–77. [DOI] [PubMed] [Google Scholar]
- Murray IA, Shaw WV. 1997. O-Acetyltransferases for chloramphenicol and other natural products. Antimicrob Agents Chemother 41: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy I, Schoofs G, Vanderleyden J, De Mot R. 1997. Transposition of the ISv21-related element ISv1415 in Rhodococcus erythropolis. J Bacteriol 179: 4635–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz MS, Khan SA, Khan AA, Khambaty FM, Cerniglia CE. 2000. Comparative molecular analysis of erythromycin-resistance determinants in staphylococcal isolates of poultry and human origin. Mol Cell Probes 14: 311–319. [DOI] [PubMed] [Google Scholar]
- Nilsen IW, Bakke I, Vader A, Olsvik O, El-Gewely MR. 1996. Isolation of cmr, a novel chloramphenicol resistance gene encoding a putative efflux pump. J Bacteriol 178: 3188–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930. [DOI] [PubMed] [Google Scholar]
- Noeske J, Huang J, Olivier NB, Giacobbe RA, Zambrowski M, Cate JH. 2014. Synergy of streptogramin antibiotics occurs independently of their effects on translation. Antimicrob Agents Chemother 58: 5269–5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak R, Shlaes DM. 2010. The pleuromutilin antibiotics: A new class for human use. Curr Opin Investig Drugs 11: 182–191. [PubMed] [Google Scholar]
- Novotná G, Janata J. 2006. A new evolutionary variant of the streptogramin A resistance protein, Vga(A)LC, from Staphylococcus haemolyticus with shifted substrate specificity towards lincosamides. Antimicrob Agents Chemother 50: 4070–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen IT, Brown MH, Skurray RA. 1996. Proton-dependent multidrug efflux systems. Microbiol Rev 60: 575–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschke U, Schmidt H, Zhang HZ, Piepersberg W. 1995. Molecular characterization of the lincomycin-production gene cluster of Streptomyces lincolnensis 78-11. Mol Microbiol 16: 1137–1156. [DOI] [PubMed] [Google Scholar]
- Petinaki E, Guérin-Faublée V, Pichereau V, Villers C, Achard A, Malbruny B, Leclercq R. 2008. Lincomycin resistance gene lnu(D) in Streptococcus uberis. Antimicrob Agents Chemother 52: 626–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poehlsgaard J, Douthwaite S. 2005. The bacterial ribosome as a target for antibiotics. Nat Rev Microbiol 3: 870–881. [DOI] [PubMed] [Google Scholar]
- Poelarends G, Mazurkiewicz P, Konings W. 2002. Multidrug transporters and antibiotic resistance in Lactococcus lactis. Biochim Biophys Acta 1555: 1–7. [DOI] [PubMed] [Google Scholar]
- Poole K. 2005. Efflux-mediated antimicrobial resistance. J Antimicrob Chemother 56: 20–51. [DOI] [PubMed] [Google Scholar]
- Porse BT, Garrett RA. 1999. Sites of interaction of streptogramin A and B antibiotics in the peptidyl transferase loop of 23 S rRNA and the synergism of their inhibitory mechanisms. J Mol Biol 286: 375–387. [DOI] [PubMed] [Google Scholar]
- Portis E, Lindeman C, Johansen L, Stoltman G. 2012. A ten-year (2000–2009) study of antimicrobial susceptibility of bacteria that cause bovine respiratory disease complex—Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni—in the United States and Canada. J Vet Diagn Invest 24: 932–944. [DOI] [PubMed] [Google Scholar]
- Prescott JF. 2014. The resistance tsunami, antimicrobial stewardship, and the golden age of microbiology. Vet Microbiol 171: 273–278. [DOI] [PubMed] [Google Scholar]
- Putman M, van Veen HW, Degener JE, Konings WN. 2000. Antibiotic resistance: Era of the multidrug pump. Mol Microbiol 36: 772–774. [DOI] [PubMed] [Google Scholar]
- Putman M, van Veen HW, Degener JE, Konings WN. 2001. The lactococcal secondary multidrug transporter LmrP confers resistance to lincosamides, macrolides, streptogramins and tetracyclines. Microbiology 147: 2873–2880. [DOI] [PubMed] [Google Scholar]
- Recchia GD, Hall RM. 1995. Gene cassettes: A new class of mobile element. Microbiology 141: 3015–3027. [DOI] [PubMed] [Google Scholar]
- Rende-Fournier R, Leclercq R, Galimand M, Duval J, Courvalin P. 1993. Identification of the satA gene encoding a streptogramin A acetyltransferase in Enterococcus faecium BM4145. Antimicrob Agents Chemother 37: 2119–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MC. 2008. Update on macrolide–lincosamide–streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol Lett 282: 147–159. [DOI] [PubMed] [Google Scholar]
- Roberts MC, Schwarz S. 2009. Tetracycline and chloramphenicol resistance mechanisms. In Antimicrobial drug resistance, Vol. 1, Mechanisms of drug resistance (ed. Mayers DJ, et al. ), pp. 183–193. Springer, New York. [Google Scholar]
- Roberts MC, Sutcliffe J, Courvalin P, Jensen LB, Rood J, Seppala H. 1999. Nomenclature for macrolide and macrolide–lincosamide–streptogramin B resistance determinants. Antimicrob Agents Chemother 43: 2823–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JI, Eady EA, Cove JH, Cunliffe WJ, Baumberg S, Wootton JC. 1990. Inducible erythromycin resistance in staphylococci is encoded by a member of the ATP-binding transport super-gene family. Mol Microbiol 4: 1207–1214. [DOI] [PubMed] [Google Scholar]
- Schmitz F-J, Petridou J, Astfalk N, Köhrer K, Scheuring S, Schwarz S. 2002a. Molecular analysis of constitutively expressed erm(C) genes selected in vitro by incubation in the presence of the noninducers quinupristin, telithromycin, or ABT-773. Microb Drug Resist 8: 171–177. [DOI] [PubMed] [Google Scholar]
- Schmitz F-J, Petridou J, Jagusch H, Astfalk N, Scheuring S, Schwarz S. 2002b. Molecular characterization of ketolide-resistant erm(A)-carrying Staphylococcus aureus isolates selected in vitro by telithromycin, ABT-773, quinupristin and clindamycin. J Antimicrob Chemother 49: 611–617. [DOI] [PubMed] [Google Scholar]
- Schwarz S, Kehrenberg C. 2006. Old dogs that learn new tricks: Modified antimicrobial agents that escape pre-existing resistance mechanisms. Int J Med Microbiol 296: 45–49. [DOI] [PubMed] [Google Scholar]
- Schwarz S, Wang Y. 2015. Nomenclature and functionality of the so-called cfr gene from Clostridium difficile. Antimicrob Agents Chemother 59: 2476–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S, Werckenthin C, Kehrenberg C. 2000. Identification of a plasmid-borne chloramphenicol–florfenicol resistance gene in Staphylococcus sciuri. Antimicrob Agents Chemother 44: 2530–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A. 2004. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol Rev 28: 519–542. [DOI] [PubMed] [Google Scholar]
- Schwarz S, Cloeckaert A, Roberts MC. 2006. Mechanisms and spread of bacterial resistance to antimicrobial agents. In Antimicrobial resistance in bacteria of animal origin (ed. Aarestrup FM), pp. 73–98. ASM, Washington, DC. [Google Scholar]
- Schwendener S, Perreten V. 2011. New transposon Tn6133 in methicillin-resistant Staphylococcus aureus ST398 contains vga(E), a novel streptogramin A, pleuromutilin, and lincosamide resistance gene. Antimicrob Agents Chemother 55: 4900–4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane A, García Lobo JM. 2000. Identification of a streptogramin A acetyltransferase gene in the chromosome of Yersinia enterocolitica. Antimicrob Agents Chemother 44: 905–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw WV. 1983. Chloramphenicol acetyltransferase, enzymology and molecular biology. Crit Rev Biochem 14: 1–46. [DOI] [PubMed] [Google Scholar]
- Shen J, Wang Y, Schwarz S. 2013. Presence and dissemination of the multiresistance gene cfr in Gram-positive and Gram-negative bacteria. J Antimicrob Chemother 68: 1697–1706. [DOI] [PubMed] [Google Scholar]
- Shore AC, Brennan OM, Ehricht R, Monecke S, Schwarz S, Slickers P, Coleman DC. 2010. Identification and characterization of the multidrug resistance gene cfr in a Panton-Valentine leukocidin-positive sequence type 8 methicillin-resistant Staphylococcus aureus IVa (USA300) isolate. Antimicrob Agents Chemother 54: 4978–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz TR, Tapsall JW, White PA, Ryan CS, Lyras D, Rood JI. 2003. Chloramphenicol-resistant Neisseria meningitidis containing catP isolated in Australia. J Antimicrob Chemother 52: 856–859. [DOI] [PubMed] [Google Scholar]
- Si H, Zhang WJ, Chu S, Wang XM, Dai L, Hua X, Dong Z, Schwarz S, Liu S. 2015. Novel plasmid-borne multidrug resistance gene cluster including lsa(E) from a linezolid-resistant Enterococcus faecium isolate of swine origin. Antimicrob Agents Chemother 59: 7113–7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silley P, Simjee S, Schwarz S. 2012. Surveillance and monitoring of antimicrobial resistance and antibiotic consumption in humans and animals. Rev Sci Tech 31: 105–120. [DOI] [PubMed] [Google Scholar]
- Silva NC, Guimarães FF, Manzi MP, Júnior AF, Gómez-Sanz E, Gómez P, Langoni H, Rall VL, Torres C. 2014. Methicillin-resistant Staphylococcus aureus of lineage ST398 as cause of mastitis in cows. Lett Appl Microbiol 59: 665–669. [DOI] [PubMed] [Google Scholar]
- Simjee S, McDermott PF, Wagner DD, White DG. 2001. Variation within the vat(E) allele of Enterococcus faecium isolates from retail poultry samples. Antimicrob Agents Chemother 45: 2931–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KV, Malathum K, Murray BE. 2001. Disruption of an Enterococcus faecium species-specific gene, a homologue of acquired macrolide resistance genes of staphylococci, is associated with an increase in macrolide susceptibility. Antimicrob Agents Chemother 45: 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KV, Weinstock GM, Murray BE. 2002. An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob Agents Chemother 46: 1845–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smieja M. 1998. Current indications for the use of clindamycin: A critical review. Can J Infect Dis 9: 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LK, Mankin AS. 2008. Transcriptional and translational control of the mlr operon, which confers resistance to seven classes of protein synthesis inhibitors. Antimicrob Agents Chemother 52: 1703–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes HW, Hall RM. 1991. Sequence analysis of the inducible chloramphenicol resistance determinant in the Tn1696 integron suggests regulation by translational attenuation. Plasmid 26: 10–19. [DOI] [PubMed] [Google Scholar]
- Tao W, Lee MH, Wu J, Kim H, Kim JC, Chung E, Hwang EC, Lee SW. 2012. Inactivation of chloramphenicol and florfenicol by a novel chloramphenicol hydrolase. Appl Environ Microbiol 78: 6295–6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh SM, Xiong L, Arias CA, Villegas MV, Lolans K, Quinn J, Mankin AS. 2007. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol Microbiol 64: 1506–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez D. 1966. Studies on the mode of action of the streptogramin antibiotics. J Gen Microbiol 42: 93–106. [DOI] [PubMed] [Google Scholar]
- Vester B, Douthwaite S. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob Agents Chemother 45: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Shoemaker NB, Wang G-R, Salyers AA. 2000. Characterization of a Bacteroides mobilizable transposon, NBU2, which carries a functional lincomycin resistance gene. J Bacteriol 182: 3559–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang Y, Wu CM, Schwarz S, Shen Z, Zhang W, Zhang Q, Shen JZ. 2011. Detection of the staphylococcal multiresistance gene cfr in Proteus vulgaris of food animal origin. J Antimicrob Chemother 66: 2521–2526. [DOI] [PubMed] [Google Scholar]
- Wang Y, He T, Schwarz S, Zhou D, Shen Z, Wu C, Wang Y, Ma L, Zhang Q, Shen J. 2012a. Detection of the staphylococcal multiresistance gene cfr in Escherichia coli of domestic-animal origin. J Antimicrob Chemother 67: 1094–1098. [DOI] [PubMed] [Google Scholar]
- Wang Y, Schwarz S, Shen Z, Zhang W, Qi J, Liu Y, He T, Shen J, Wu C. 2012b. Co-location of the multiresistance gene cfr and the novel streptomycin resistance gene aadY on a small plasmid in a porcine Bacillus strain. J Antimicrob Chemother 67: 1547–1549. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang Y, Schwarz S, Shen Z, Zhou N, Lin J, Wu C, Shen J. 2012c. Detection of the staphylococcal multiresistance gene cfr in Macrococcus caseolyticus and Jeotgalicoccus pinnipedialis. J Antimicrob Chemother 67: 1824–1827. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang W, Wang J, Wu C, Shen Z, Fu X, Yan Y, Zhang Q, Schwarz S, Shen J. 2012d. Distribution of the multidrug resistance gene cfr in Staphylococcus species isolates from swine farms in China. Antimicrob Agents Chemother 56: 1485–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XM, Zhang WJ, Schwarz S, Yu SY, Liu H, Si W, Zhang RM, Liu S. 2012e. Methicillin-resistant Staphylococcus aureus ST9 from a case of bovine mastitis carries the genes cfr and erm(A) on a small plasmid. J Antimicrob Chemother 67: 1287–1289. [DOI] [PubMed] [Google Scholar]
- Wang Y, He T, Schwarz S, Zhao Q, Shen Z, Wu C, Shen J. 2013a. Multidrug resistance gene cfr in methicillin-resistant coagulase-negative staphylococci from chickens, ducks, and pigs in China. Int J Med Microbiol 303: 84–87. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li D, Song L, Liu Y, He T, Liu H, Wu C, Schwarz S, Shen J. 2013b. First report of the multiresistance gene cfr in Streptococcus suis. Antimicrob Agents Chemother 57: 4061–4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lv Y, Cai J, Schwarz S, Cui L, Hu Z, Zhang R, Li J, Zhao Q, He T, et al. 2015. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J Antimicrob Chemother 70: 2182–2190. [DOI] [PubMed] [Google Scholar]
- Weisblum B. 1995a. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother 39: 577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum B. 1995b. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob Agents Chemother 39: 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiß S, Kadlec K, Feßler AT, Schwarz S. 2014. Complete sequence of a multiresistance plasmid from a methicillin-resistant Staphylococcus epidermidis ST5 isolated in a small animal clinic. J Antimicrob Chemother 69: 847–859. [DOI] [PubMed] [Google Scholar]
- Wendlandt S, Li B, Ma Z, Schwarz S. 2013a. Complete sequence of the multi-resistance plasmid pV7037 from a porcine methicillin-resistant Staphylococcus aureus. Vet Microbiol 166: 650–654. [DOI] [PubMed] [Google Scholar]
- Wendlandt S, Lozano C, Kadlec K, Gómez-Sanz E, Zarazaga M, Torres C, Schwarz S. 2013b. The enterococcal ABC transporter gene lsa(E) confers combined resistance to lincosamides, pleuromutilins and streptogramin A antibiotics in methicillin-susceptible and methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother 68: 473–475. [DOI] [PubMed] [Google Scholar]
- Wendlandt S, Li J, Ho J, Porta MA, Feßler AT, Wang Y, Kadlec K, Monecke S, Ehricht R, Boost M, et al. 2014. Enterococcal multiresistance gene cluster in methicillin-resistant Staphylococcus aureus from various origins and geographical locations. J Antimicrob Chemother 69: 2573–2575. [DOI] [PubMed] [Google Scholar]
- Wendlandt S, Kadlec K, Feßler AT, Schwarz S. 2015a. Identification of ABC transporter genes conferring combined pleuromutilin–lincosamide–streptogramin A resistance in bovine methicillin-resistant Staphylococcus aureus and coagulase-negative staphylococci. Vet Microbiol 177: 353–358. [DOI] [PubMed] [Google Scholar]
- Wendlandt S, Shen J, Kadlec K, Wang Y, Li B, Zhang WJ, Feßler AT, Wu C, Schwarz S. 2015b. Multidrug resistance genes in staphylococci from animals that confer resistance to critically and highly important antimicrobial agents in human medicine. Trends Microbiol 23: 44–54. [DOI] [PubMed] [Google Scholar]
- Werner G, Witte W. 1999. Characterization of a new enterococcal gene, satG, encoding a putative acetyltransferase conferring resistance to streptogramin A compounds. Antimicrob Agents Chemother 43: 1813–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN. 2009. The A–Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol 44: 393–433. [DOI] [PubMed] [Google Scholar]
- Wilson DN. 2011. On the specificity of antibiotics targeting the large ribosomal subunit. Ann NY Acad Sci 1241: 1–16. [DOI] [PubMed] [Google Scholar]
- Wilson DN. 2014. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol 12: 35–48. [DOI] [PubMed] [Google Scholar]
- Wright GW. 2007. The antibiotic resistome: The nexus of chemical and genetic diversity. Nature Rev Microbiol 5: 175–186. [DOI] [PubMed] [Google Scholar]
- Zhang WJ, Wu CM, Wang Y, Shen ZQ, Dai L, Han J, Foley SL, Shen JZ, Zhang Q. 2011. The new genetic environment of cfr on plasmid pBS-02 in a Bacillus strain. J Antimicrob Chemother 66: 1174–1175. [DOI] [PubMed] [Google Scholar]
- Zhang WJ, Xu XR, Schwarz S, Wang XM, Dai L, Zheng HJ, Liu S. 2014. Characterization of the IncA/C plasmid pSCEC2 from Escherichia coli of swine origin that harbours the multiresistance gene cfr. J Antimicrob Chemother 69: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Xu C, Wang H, Lei C, Liu B, Guan Z, Yang C, Yang Y, Peng L. 2015a. Presence and new genetic environment of pleuromutilin–lincosamide–streptogramin A resistance gene lsa(E) in Erysipelothrix rhusiopathiae of swine origin. Vet Microbiol 177: 162–167. [DOI] [PubMed] [Google Scholar]
- Zhang WJ, Wang XM, Dai L, Hua X, Dong Z, Schwarz S, Liu S. 2015b. Novel conjugative plasmid from Escherichia coli of swine origin that coharbors the multiresistance gene cfr and the extended-spectrum-β-lactamase gene blaCTX-M-14b. Antimicrob Agents Chemother 59: 1337–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Wendlandt S, Li H, Li J, Wu C, Shen J, Schwarz S, Wang Y. 2014. Identification of the novel lincosamide resistance gene lnu(E) truncated by ISEnfa5-cfr-ISEnfa5 insertion in Streptococcus suis: De novo synthesis and confirmation of functional activity in Staphylococcus aureus. Antimicrob Agents Chemother 58: 1785–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]