Abstract
Historically, sequencing has been the key technology to assess variation in the genetic code, and has been widely accepted in clinical diagnostics of genetic disease. The advent of next-generation sequencing (NGS) methods increased the size of the analyzed target by several orders of magnitude, while at the same time drastically reducing the cost of sequencing. Current research allows sequencing of germline and tumor whole genomes. However, with the arrival of cutting-edge technology to the clinical diagnostic field, strict regulatory oversight is required to use the advances of the latest research when applied to routine clinical practice. We discuss the differences between sequencing in a research setting and sequencing in a clinical diagnostics setting, as applied to next-generation technology.
Next-generation sequencing is gaining momentum in clinical diagnostics. Strict regulations that govern its use must be established and followed to provide the best possible care of patients and families with genetic disease.
The invention, implementation, and wide adoption of next-generation sequencing (NGS) technology has expanded the horizons of genomics and significantly upgraded the genetic testing toolkit. Research efforts of impressive scale, such as the International Cancer Genome Consortium (ICGC, see icgc.org) and The Cancer Genome Atlas (TCGA, see cancergenome.nih.gov), and the efforts of the University of Cambridge, Genomics England, and Illumina to sequence 10,000 whole genomes of children and adults with rare genetic diseases (Moran 2014), are well underway. Without a doubt, projects of this scale became possible because of the phenomenal power of massively parallel sequencing, commonly referred to as next-generation sequencing.
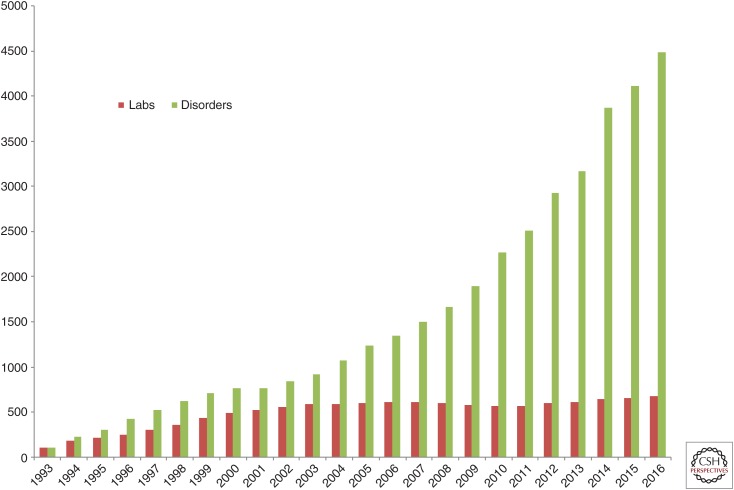
Along with the incredible capabilities afforded in research studies, the horizon of clinical diagnostics has expanded as well. For a number of years, dideoxy-terminator (Sanger) sequencing was the main tool of genetic diagnostics laboratories. A typical Sanger sequencing experiment results in 500–600 base pairs of sequence and covers, on average, a single exon of a gene. In contrast, NGS has enabled inherited disease testing in panels of genes (∼10–200 genes), exomes (∼20,000 genes) and whole human genomes (6 Gbp), diagnostics in solid tumors and infectious diseases, as well as noninvasive prenatal testing. The reasonably accessible cost of NGS instrumentation and consumables, along with the general acceptance of genetic tests in the clinical community and the availability of insurance reimbursement, led to the great increase in the number of genetic service providers. Currently, there are 684 molecular diagnostics laboratories in the GeneTests registry (see genetests.org; Fig. 1). There are close to 500 laboratories registered in the Genetic Testing Registry ([GTR], see ncbi.nlm.nih.gov/gtr); and, of 32,473 molecular tests registered in GTR, 9038 or 27.8% are using NGS (Fig. 2) (Rubinstein et al. 2013).
Figure 1.
Expansion of genetic diagnostic testing. Numbers for 2016 correspond to a partial year as of May 2016. (Courtesy of Genetests.org.)
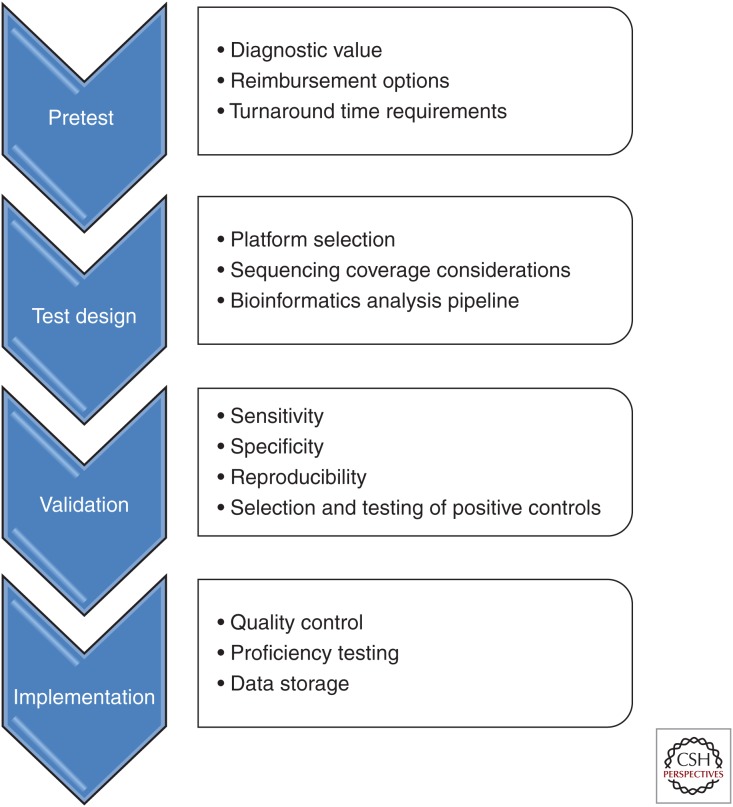
Figure 2.
Development flowchart of an NGS-based clinical diagnostics test.
With the number of NGS-based diagnostic tests and the laboratories providing them constantly growing, the issue of quality control and clinical oversight becomes highly important. Indeed, clinical sequencing provides test results to physicians, diagnoses are made, genetic counseling is given, and patient treatment is prescribed, based on the results of these tests. Additionally, physicians need to be equipped with sufficient knowledge to select the best possible and most relevant genetic test within the complex landscape of institutional and insurance contracts, laboratory providers, and reimbursement options. To make an educated decision, it is necessary that the medical community be familiar with the types of tests available, their sensitivity and specificity, clinical usage and validity, appropriateness for a specific patient/phenotype, as well as the credentials of the laboratory conducting genetic testing.
SEQUENCING PLATFORMS
Multiple NGS platforms are currently available (Glenn 2011; Mardis 2011; Loman et al. 2012; Quail et al. 2012; Ratan et al. 2013). At present, the instrumentation and reagent market is dominated by Illumina, followed by Ion Torrent. Pacific Biosciences provides a powerful technology for long reads, which can be used for specific applications that can benefit from this approach, such as full sequencing of HLA genes (Shiina et al. 2012). The NGS landscape keeps changing, with new players constantly emerging, such as Oxford Nanopore, GnuBio, Genapsys, Genia, etc. In addition, it should be noted that even the most established technology keeps changing at a pace unheard of in previous years. For example, Illumina updates its sequencing chemistry several times a year, providing new opportunities both on existing instruments (HiSeq and MiSeq) as well as pushing technology and throughput on new instruments (X Ten, NextSeq, and MiniSeq). From the standpoint of clinical diagnostics, frequent changes in sequencing technology require constant changes in laboratory protocols and full revalidation of each diagnostic test. Indeed, freezing all of the components of a test for an extended period of time may not be possible, as one has to depend on equipment providers for supplying technology and chemistry. Using the latest updates is necessary to provide the highest quality data at a reasonable price point. Therefore, the validation component of clinical testing becomes a substantial time and financial consideration, requiring significant investment of resources.
TYPES OF CLINICAL NGS-BASED TESTS
Targeted Gene Panels
Targeted gene panels were the first use of NGS in the clinical setting. The idea of simultaneous analysis of multiple genes involved in a certain pathway or for a specific phenotype (such as hypertrophic cardiomyopathy or congenital blindness) took the use of genetic testing to a new level. Various previously developed polymerase chain reaction (PCR)-based strategies (Nilsson et al. 1994; Dahl et al. 2005) provided the technology for target selection from whole genomic DNA. NGS allowed the laboratory to analyze the sequence of all captured fragments massively in parallel (Dahl et al. 2007). On the other hand, parallel synthesis of a high number of oligonucleotides on microarrays provided a powerful strategy for target selection, and this approach was used by multiple groups (Albert et al. 2007; Okou et al. 2007; Porreca et al. 2007). At that time, a typical molecular genetics test was expected to scrutinize 100% of the target nucleotides and detect all of the variations or certify their absence. Although many of the described methods had high target coverage of >90%, none of them could provide 100% analytic sensitivity. Regions that did not attain sufficient sequence coverage had to be augmented by capillary sequencing. Putative pathogenic variants (“mutations”) had to be confirmed by an orthogonal method, usually capillary sequencing, which is considered the gold standard of data quality in the diagnostic setting.
As the size of some targeted panels expands well beyond 1000 exons, the need for supplemental coverage by capillary reads, to provide 100% coverage of every nucleotide in the test, becomes a burden, raising the cost of the test and increasing turnaround time from sample receipt to reporting. In these circumstances, and under the market pressure of faster turnaround times and limited reimbursement rates, some of the test providers might be inclined to use various shortcuts, such as reporting the results without sufficient interpretation or without a confirmation by Sanger sequencing. Generally, this has a negative effect on the quality of the genetic test, and creates an additional level of complexity for ordering physicians. In all cases, a laboratory should make available the test parameters (sensitivity, specificity, etc.) for any molecular test offered in the clinical setting to ordering physicians.
Exome Sequencing
Whole-exome sequencing (WES) inspects all of the exons in the genome for variation compared with a reference, along with the flanking intron/exon boundaries for detection of splicing variants. Covering ∼2% of the genome, exome sequencing provides an efficient and economical approach to the analysis of the protein-coding regions of all ∼20,000 genes. This makes WES a very popular tool for the identification of new Mendelian genes (Bamshad et al. 2011; Gilissen et al. 2012) and for diagnosis of patients with complex phenotypes. In comparison to the initial efforts (Hodges et al. 2007), levels of target coverage have been drastically improved. Modern “augmented” exome designs take the coverage to >99% of the target (e.g., Sureselect Clinical Research Exome, see www.genomics.agilent.com/article.jsp?pageId=2900001&_requestid=24511). With wide availability of commercial kits, WES is becoming a common genetic diagnostics test, and is especially valuable for patients who have an undiagnosed disorder (Adams et al. 2012) in which a specific diagnosis is hard to establish and multiple targeted genetic tests or panels have failed to make a diagnosis previously. It is important to realize the limitations of WES and that “more genes is not always better,” and the level of coverage for each gene should be carefully considered. Indeed, no current WES test can guarantee full coverage of all genes, in contrast to targeted gene panels. Xue et al. (2015) analyzed the conundrum of targeted panel sequencing versus exome sequencing and suggested an efficient algorithm for molecular genetic testing.
Whole-Genome Sequencing
Whole-genome sequencing (WGS) is predicted to be the ultimate diagnostic tool, which examines the genes in their entirety, including deep intronic regions and regulatory regions; in addition, WGS allows detection of genomic rearrangements and better detection of copy number variations. The first NGS sequenced genomes were published in 2008, and despite the tremendous technology improvement, usage of WGS still remains too costly and challenging for routine clinical diagnostic purposes. Analysis of WGS data is also limited by the current level of genetics knowledge and provides only limited advantage over exome sequencing in test sensitivity.
Noninvasive Prenatal Testing
Noninvasive prenatal screening (NIPS) makes use of the cell-free fetal DNA that circulates in the blood of pregnant women. NIPS is being used for detection of certain types of genetic disease very early in pregnancy, thus aiding in pregnancy management. Currently, the NIPS diagnostics space is tangled up in intellectual property disputes among the four companies offering various flavors of this test. Their validated tests are reviewed in Agarwal et al. (2013). The NIPS landscape is changing rapidly as a result of court decisions, legal settlements, and company acquisitions. Because of the intellectual property protection and mutual litigation among the players in this space, the NIPS market remains largely limited in the United States, although not necessarily so in Europe and elsewhere. Lack of wide public availability in the NIPS market may lead to potential issues with appropriate quality-control guidelines (Takoudes and Hamar 2014).
Somatic Cancer Genotyping and Sequencing
NGS is playing an increasingly important role in the diagnosis and management of oncology patients. For example, certain mutations in the KRAS gene correlate with response to anti-epidermal growth factor receptor (EGFR) therapy (Dienstmann et al. 2011). Detection or sequence variation in somatic tissue presents one of the most challenging scenarios for NGS, in part because of the often-limited tissue availability and degraded starting nucleic acid, such as in the case of formalin-fixed, paraffin-embedded (FFPE) samples. Patient samples usually present a heterogeneous mixture of tumor and normal stroma; furthermore, the tumors themselves can be quite heterogeneous. These factors complicate the detection of pathogenic variants, which may represent a small percentage of the sequenced alleles. As a result, the detection level of allele variation must be highly sensitive, with the ability to identify a variant in as few as 2%–5% of the sequencing reads. At this challenging level of detection, it becomes very hard to distinguish the actual mutations from stochastic noise, as this is close to the sensitivity limits of the current sequencing platforms. The usual approach is to increase the depth of read coverage; unfortunately, there is no common standard as to what this depth should be, ranging from 100X to 1000X and beyond. Incorporation of so-called molecular barcodes is another common way to increase the sensitivity of detection in an NGS assay (Kinde et al. 2011). Confirmation of identified variants represents a challenge as well, because commonly used methods like Sanger sequencing cannot effectively detect low allelic fraction. Because of these technical limitations, only a few laboratories perform somatic variant detection by NGS in a clinical setting (Cottrell et al. 2014; Pritchard et al. 2014), although it is a fast-growing field.
CLINICAL DIAGNOSTICS VERSUS RESEARCH SEQUENCING
In trying to distinguish between clinical diagnostic testing and research projects, words “validated” and “regulated” are mentioned most often. Strict regulations govern the space of clinical diagnostics, to certify the clinical and technical validity. It is based on the results of these tests that diagnoses will be established and patients will be treated; thus, the laboratory performing the tests carries a high degree of responsibility. The Centers for Medicare and Medicaid Services (CMS) regulates all laboratory testing performed on humans in the United States through CLIA—the Clinical Laboratory Improvement Amendments (see cms.gov/clia). NGS-based laboratory-developed tests (LDTs) fall under the category of high-complexity testing in CLIA guidelines. In our opinion, the following factors contribute to the clinical acceptance and diagnostic use of NGS-based testing.
Regulatory Requirements for Clinical Laboratories
A laboratory performing clinical tests, where results are issued to an ordering physician, and the results are used for the purpose of diagnosis and/or guiding treatment, is required to carry CLIA certification, the mechanism by which CMS regulates all laboratory testing performed on humans in the United States. Laboratories with CLIA certification follow guidelines regarding personnel qualifications, quality-control programs that include enrollment and participation in proficiency testing programs, maintenance and inspection criteria for the laboratory equipment, etc. The CLIA programs are administered by the states, and individual states may have additional requirements beyond those mandated by CLIA.
Accreditation by the College of American Pathologists (CAP) is a voluntary step, undertaken by most diagnostic laboratories, which serves as a stamp of excellence and a third-party approval of the laboratory practices. In terms of regulatory policy, CAP accreditation represents a more stringent level, compared with CLIA certification, and requires a quality-management program, laboratory safety plan, document control plan, competency assessment program, laboratory director oversight documentation, and specific requirements for a laboratory information system. CAP has issued guidance on the standards for NGS clinical tests (Aziz et al. 2014), which indicate the general direction of regulatory practice, as applied to NGS-based testing.
In addition, some states specifically require that the laboratory performing diagnostic tests on the residents of the state be certified by the state. The burden of this additional certification varies greatly across the states, with the most prominent program being that of the New York State Department of Health (NYSDOH). To conduct testing on New York State residents, not only the laboratory and its general practices, but each individual test offered by the laboratory has to be reviewed by this agency for the laboratory to receive a New York State laboratory license. The need to obtain such certification and the time required to obtain it requires significant laboratory resources for the diagnostic laboratories.
Additionally, guidelines for NGS in the clinical setting are also provided by the American College of Medical Genetics and Genomics (ACMGG) (Rehm et al. 2013). The practical usefulness of these guidelines cannot be overestimated. They are written by the leaders in the NGS diagnostics field, set an excellent example, and provide detailed directions for the genetic diagnostic laboratories.
Finally, the Food and Drug Administration (FDA) has written and is expected to issue guidance, whereby clinical diagnostic tests developed by a laboratory (LDTs) will be treated as devices under the FDA’s current device regulation program, with the laboratories themselves remaining under CLIA and State oversight. This is a highly contentious issue, with many patient and professional groups, as well as the laboratories themselves, believing that the FDA does not have jurisdiction over LDTs and laboratory practices. There is also a possibility that there will be a legislative solution, with Congress enacting laws regarding how diagnostic laboratories will be regulated going forward. (see genomeweb.com/molecular-diagnostics/fda-finalize-ldt-guidance-amid-uncertainty-number-genetic-tests-impacted).
Clinical laboratories will commonly use components that have been sold by the manufacturer “for investigational use” or “research use only” in their LDT, and then the test will be validated according to CAP, CLIA, and state requirements. The close adherence to published standards currently remains the responsibility of the providers, and their oversight to CMS. An additional requirement for FDA test approval would put significant burden on the test providers, slowing down new test development and raising the cost of providing clinical diagnostic tests, which may negatively reflect on the future availability of current and new genetic diagnostic tests.
Preanalytical Handling of Samples
Any laboratory providing clinical diagnostic testing needs to exercise strict control over the accessioning of incoming patient samples. Indeed, accidental switching of samples or even a delay in passing the samples through the testing workflow may result in grave consequences for the patient. A laboratory tracking system should provide information about every stage of the testing process, with samples being tracked from the moment they enter the facility. A critical difference between research and clinical testing, is that a sample for a research study may be allowed to be discarded or replaced by a sample from another subject if there is technical difficulty, whereas in clinical testing every single sample is crucial and represents a separate and individual diagnostic result for which there exists no substitute.
Wet-Bench Test Development and Validation
Clinical tests are developed because there is significant clinical use in performing the test, that is, improved health outcome for the patient. Typically, a laboratory requires multiple peer-reviewed publications implicating a gene/phenotype relationship (or variant/phenotype relationship) before considering the development of a new clinical molecular test. The specific diagnostic requirements will define the scope of the test, and the appropriate choice of technology. Although for some diagnostic conditions it may be sufficient to examine only a set of specific pathogenic variants, or the full sequence of just one or a few genes, for other conditions clinical use and high sensitivity may be met only by examination of an extended gene panel, or even WES. It is the responsibility of clinical experts to define the scope and exclude content, which could give uninterpretable results that may be of questionable value to both the physician and the patient. From this point of view, the principle of “more is better” is generally not followed in the clinical diagnostics setting, while it might be appropriate in a research setting. A good example is presented by the debate over the return of incidental findings in clinical sequencing: ACMG recommended that known and expected pathogenic variants discovered in certain genes (mostly those related to cancer risk or cardiac disease) should be returned to a patient undergoing clinical sequencing regardless of the assay’s indication (Green et al. 2013); however, many diagnostic laboratories offer opt-out policy for their clients, a policy now approved by the ACMGG.
Once the test components are assembled, the test undergoes multiple iterative cycles of improvement and testing, until it matures into a stable workflow. At that point, extensive validation should be performed. The parameters of the validation must be clearly defined up front, and achieved every time the test is run (repeatability). It is equally important to define both what is expected to be covered (depth of sequencing, quality of sequencing) and what is left beyond the scope of the test (genomic regions that are not captured or sequenced sufficiently because of issues, such as the presence of pseudogenes, repeat regions, very high C/G regions, etc.).
Analytic Validity
Analytic validity refers to the accuracy of a genetic test in identifying the presence or absence of sequence variants. Once the test is established, a laboratory would typically run a number of validation samples with known variants that had previously been identified using an alternative method. That method could be Sanger sequencing for nucleotide changes or microarray hybridization for copy number variants. It is not important for the validation scheme to use only pathogenic variants; what is being tested is the power of the newly developed test to detect sequence variation. However, if there are common known pathogenic variants in a specific gene, it is crucial that positive controls for those variants are included in the validation scheme.
Turnaround Time
The turnaround time (TAT) requirement is an example of a metric, which is largely specific to the diagnostic field. A clinical test is typically performed and results reported within a particular timeframe, which is defined by the needs of the physician to guide diagnosis, surveillance, and treatment on one hand, and the technology used in the laboratory on the other hand. Test design should consider TAT requirements and use appropriate methodology to achieve the desired TAT. Failure to deliver results within the promised TAT may be detrimental to the health of a patient, as well as to the reputation of the test provider.
Adequate Data Coverage
Depth of coverage is defined as the number of individual NGS reads covering a given nucleotide. The error rate of an individual NGS read is compensated by the number of multiple overlapping reads, which helps to minimize stochastic errors and also to determine the consensus sequence. The depth of coverage requirement for any sequencing test depends on the type of sequencing technology, read length, assembly algorithm, and other factors (Schatz et al. 2010; Sims et al. 2014). Typical genetic diagnostics tests require minimum coverage of 10–20X, meaning that every nucleotide in the target region is covered by no less than 10 to 20 overlapping reads (Ajay et al. 2011). ACMGG guidelines for NGF (Rehm et al. 2013) recommend performing clinical exome sequencing to 100X coverage overall, so that 90%–95% of the exome is covered to at least 10X depth. The same guidelines suggest alignment of the panel or exome data to the whole genome, to correctly map the reads to the regions of interest, and avoid incorrect mapping to the homologous regions in the genome. For WGS, 30–40X is considered standard. There is no clear guidance on the depth of coverage required for somatic sequencing; typically 100X to 500X is achieved and correlated to the level of minor allele detection. It is common to set the limit of minor allele detection at 5%; going below 2% would require the use of molecular barcodes and specialized bioinformatics solutions. As the depth of coverage is related to the probability of correctly determining the sequence, it is an important parameter of the clinical test, and should be specified in the test description provided to consumers. Similarly, the degree to which target regions are covered constitutes an important performance metric, and sets the diagnostic expectations as it impacts the test sensitivity
Validated and Consistent Computational Data Analysis
Considering the enormous amount of data that NGS creates, computational analysis is an absolutely critical part of the diagnostic workflow. Gigabytes of data coming off the sequencer need to be turned into base calls, reads need to be aligned to the reference sequence and nucleotide changes have to be called. At each step, multiple software tools can be used, their combination being referred to as a bioinformatics pipeline. Certain tools are more common in the bioinformatics space; however, there is no standard or specific preference. Regardless of the software tools used by the laboratory, the resulting pipeline should be robust and consistent, producing identical results from identical input over multiple tests. The pipeline often has to be tuned for sensitivity to detect specific mutations, while also minimizing false-positive findings that would unnecessarily extend the time and cost of analysis because of the need to confirm variants before reporting (see section on Confirmatory Testing below). Because of the wide selection of sequencing platforms and bioinformatics algorithms, and in the absence of a common standard, reference projects like “genome in a bottle” (Zook et al. 2014) can be useful, to establish a benchmark for the bioinformatics pipeline. The regulatory criteria for clinical diagnostic testing clearly apply to the bioinformatics tools as well. Once the bioinformatics pipeline has been validated and put in place, any changes to the programming code have to be documented. With every change to the pipeline, full end-to-end validation must be performed to show that the sensitivity and specificity of the overall test has not been compromised.
Confirmatory Testing
NGS technologies operate on massive amounts of data and the findings are based on statistical approaches and the sensitivity of the bioinformatics analysis. Despite the ever-improving quality of both NGS data and available analytic tools, false-positive and false-negative results are observed, when compared with the gold standard of Sanger sequencing. For this reason, ACMGG guidelines strongly suggest confirmation of NGS-identified and reported variants by an orthogonal method. For nucleotide changes, this could be Sanger sequencing; for copy number variants, this could be microarray hybridization or quantitative PCR. For somatic mutation diagnostics, the sensitivity of the confirmatory method should be adequate, and real-time PCR may be a possible approach. The type of confirmatory testing should be clearly specified by the diagnostic laboratory in the description of a clinical test. The need to confirm pathogenic variants represents an important difference between research and clinical sequencing; indeed, while in the former case the general statistics of mutation is sufficient for a scientific study, in the clinical case, a false-positive result reported to a physician may lead to an incorrect diagnosis and treatment of a patient.
Clinical Utility
The term “clinical utility” refers to the likelihood that the results of the test will lead to an improved health outcome, be it by simply providing an answer to the question “what does this patient have”? or to directing specific treatment to the disorder. A genetic test will be judged by the added value it brings into the clinic. To successfully launch and run a test, and collect payment for it, its clinical usage must be clearly shown. Detecting mutations in individual genes helps to establish the correct diagnosis. This is especially beneficial in the cases of exome/genome sequencing, in which a patient has commonly gone through a long and expensive diagnostic odyssey without resolving the diagnosis. Typically, exome sequencing will result in a 1/4 to 1/3 positive diagnostic rate (Lee et al. 2014; Yang et al. 2014). One WGS study conducted in a small highly selected group of NICU/PICU patients reported a diagnostic rate as high as 57% (Petrikin et al. 2015). Similarly, diagnostic tests leading to identification of somatic mutations in tumors assist in targeting the tumors with specific drugs and the approach is expected to have tremendous value in the treatment of cancer.
Analytical Sensitivity
ACMGG guideline defines analytical sensitivity as the “proportion of biological samples that have a positive test result or known mutation and that are correctly classified as positive.” Analytical sensitivity of a test can be established retroactively by performing the test on a number of samples with previously identified and confirmed disease-causing variants. It is important that the variants represent true pathogenic mutations, confirmed by clinical analysis and/or functional studies. Analytical sensitivity is directly related to the false-negative rate; if analytical sensitivity is high, few variants will be missed, and thus there will be a low false-negative rate. Analytical sensitivity is critical in the context of diagnostic testing, which should aim to miss no important variants. It is hard to determine the sensitivity of a test that is directed at the discovery of novel variants; in this case, the confidence of novel variant discovery can be extrapolated based on the detection rate of previously confirmed findings. The wider and more diverse is the number of correctly identified variants, the higher is the confidence in the discovery of new mutations.
Analytical Specificity
ACMGG guidelines define analytical specificity as “the proportion of biological samples that have a negative test result or no identified mutation (being tested for) and that are correctly classified as negative.” Analytical specificity is related to the false-positive rate; the higher the specificity of the test, the fewer false-positive results will be identified. The false-positive rate greatly depends on the technology, which can produce artifacts looking like legitimate sequence variants. In this context, the need for performing orthogonal confirmation is very clear and should, in principle, eliminate false-positive variants.
Sensitivity and specificity are closely related. Indeed, a test with very high specificity could be tuned to call too many false-positives or “overcall” the variants; under this condition, the specificity would be low, and the number of suspect mutations to be confirmed by an orthogonal technology would be high, and require an extensive effort by the laboratory. In the opposite case, when the sensitivity of detection is tuned low, false-negative results would become a risk. In clinical diagnostics, it is possible to rule out a false-positive result by performing confirmatory tests, but a false-negative result cannot be confirmed and may lead to detrimental consequences in patients’ care. The optimal balance of sensitivity and specificity is difficult to achieve, and requires extensive validation efforts.
Although the concept of analytical sensitivity and specificity is an intrinsic feature of a clinical diagnostic test, these parameters are not typically inspected closely in a research setting. Commonly, because of the number of samples in a research study and lack of a “calibration” dataset, research analytic tools will assume that certain types of mutations are disease-causing, without further analysis and revision of individual variants. For example, a nonsense variant might automatically be deemed “damaging,” whereas in reality this may not necessarily be the case for a particular gene/phenotype relationship. Research sequencing provides valuable gross statistics; clinical diagnostics deals with individual mutations on a patient-by-patient basis, and is the essence of personalized genomics.
Postanalytical Results Handling, Including Report Writing, Review, and Data Storage
Another aspect that distinguishes research NGS from clinical diagnostics is the results report. In a diagnostics laboratory, tests are ordered by a physician, and the final product generated by the laboratory is a clinical report, which comprises and explains the result of testing, provides result interpretation, and is signed by the doctoral-level laboratory director, who must be board-certified in a relevant field. Once the bioinformatics pipeline performs data alignment and nucleotide variant calling, the variants called are reviewed and interpreted by a qualified analyst with expert knowledge of the analyzed genes, who classifies each variant as pathogenic, likely pathogenic, uncertain significance, likely benign, or benign. The result report is then written, often by a certified genetic counselor or geneticist, who provides relevant technical and clinical information with reference to appropriate publications for the physician’s use,
It is absolutely critical that the chain of custody is tracked throughout the steps of the analysis workflow, beginning with sample receipt and accessioning, and ending with the report writing and distribution. Patient information confidentiality should be preserved throughout the testing process, and the result report should be correctly matched back to the identification data on the submitted sample at the end of the test. These steps require complex laboratory information tracking systems, which should be developed in compliance with appropriate regulations of HIPAA (Health Insurance Portability and Accountability Act), CLIA, and CAP. The same regulations cover the policy of sample and data storage.
In a clinical diagnostics laboratory, compliance with regulations should be controlled through a quality-management program. This program oversees documentation of protocol and informatics pipeline modifications, instrument maintenance and calibration, proficiency testing, training, exception log, corrective action, and similar regulatory aspects. Quality management allows the clinical sequencing laboratory to maintain the exceptionally high standards of diagnostic testing, which are required for physicians to provide the best possible care for patients.
CONCLUSION
Next-generation sequencing, taking the DNA-related research to unprecedented heights, is gaining momentum in the clinical molecular genetic diagnostics space. The power and availability of NGS-based tests is guaranteed to bring patient care to a new, highly advanced level. Strict regulations and standards of clinical care must be established and adhered to for NGS tests to provide the highest possible value in the care of patients and families with genetic disease.
ACKNOWLEDGMENTS
The authors thank Robert Daber for useful suggestions, as well as Brandi Kattman and Amar Kamath for providing valuable information.
Footnotes
Editors: W. Richard McCombie, Elaine R. Mardis, James A. Knowles, and John D. McPherson
Additional Perspectives on Next-Generation Sequencing in Medicine available at www.perspectivesinmedicine.org
REFERENCES
- Adams DR, Sincan M, Fuentes Fajardo K, Mullikin JC, Pierson TM, Toro C, Boerkoel CF, Tifft CJ, Gahl WA, et al. 2012. Analysis of DNA sequence variants detected by high-throughput sequencing. Hum Mutat 33: 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal A, Sayres LC, Cho MK, Cook-Deegan R, Chandrasekharan S. 2013. Commercial landscape of noninvasive prenatal testing in the United States. Prenat Diagn 33: 521–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajay SS, Parker SC, Abaan HO, Fajardo KV, Margulies EH. 2011. Accurate and comprehensive sequencing of personal genomes. Genome Res 21: 1498–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, et al. 2007. Direct selection of human genomic loci by microarray hybridization. Nat Methods 4: 903–905. [DOI] [PubMed] [Google Scholar]
- Aziz N, Zhao Q, Bry L, Driscoll DK, Funke B, Gibson JS, Grody WW, Hegde MR, Hoeltge GA, Leonard DG, et al. 2014. College of American Pathologists’ laboratory standards for next-generation sequencing clinical tests. Arch Pathol Lab Med 139: 481–493. [DOI] [PubMed] [Google Scholar]
- Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J. 2011. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet 12: 745–755. [DOI] [PubMed] [Google Scholar]
- Cottrell CE, Al-Kateb H, Bredemeyer AJ, Duncavage EJ, Spencer DH, Abel HJ, Lockwood CM, Hagemann IS, O’Guin SM, Burcea LC, et al. 2014. Validation of a next-generation sequencing assay for clinical molecular oncology. J Mol Diagn 16: 89–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl F, Gullberg M, Stenberg J, Landegren U, Nilsson M. 2005. Multiplex amplification enabled by selective circularization of large sets of genomic DNA fragments. Nucleic Acids Res 33: e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl F, Stenberg J, Fredriksson S, Welch K, Zhang M, Nilsson M, Bicknell D, Bodmer WF, Davis RW, Ji H. 2007. Multigene amplification and massively parallel sequencing for cancer mutation discovery. Proc Natl Acad Sci 104: 9387–9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienstmann R, Vilar E, Tabernero J. Molecular predictors of response to chemotherapy in colorectal cancer. 2011. Cancer J 17: 114–126. [DOI] [PubMed] [Google Scholar]
- Gilissen C, Hoischen A, Brunner HG, Veltman JA. 2012. Disease gene identification strategies for exome sequencing. Eur J Hum Genet 20: 490–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn TC. 2011. Field guide to next-generation DNA sequencers. Mol Ecol Resour 11: 759–769. [DOI] [PubMed] [Google Scholar]
- Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, McGuire AL, Nussbaum RL, O’Daniel JM, Ormond KE, et al. 2013. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 15: 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, Smith SW, Middle CM, Rodesch MJ, Albert TJ, Hannon GJ, et al. 2007. Genome-wide in situ exon capture for selective resequencing. Nat Genet 39: 1522–1527. [DOI] [PubMed] [Google Scholar]
- Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. 2011. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci 108: 9530–9535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-Rivera F, Das K, Toy T, Harry B, Yourshaw M, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. 2014. JAMA 312: 1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ. 2012. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 30: 434–439. [DOI] [PubMed] [Google Scholar]
- Mardis ER. 2011. A decade’s perspective on DNA sequencing technology. Nature 470: 198–203. [DOI] [PubMed] [Google Scholar]
- Moran N. 2014. 10,000 rare-disease genomes sequenced. Nat Biotechnol 32: 7. [Google Scholar]
- Nilsson M, Malmgren H, Samiotaki M, Kwiatkowski M, Chowdhary BP, Landegren U. 1994. Padlock probes: Circularizing oligonucleotides for localized DNA detection. Science 265: 2085–2088. [DOI] [PubMed] [Google Scholar]
- Okou DT, Steinberg KM, Middle C, Cutler DJ, Albert TJ, Zwick ME. 2007. Microarray-based genomic selection for high-throughput resequencing. Nat Methods 4: 907–909. [DOI] [PubMed] [Google Scholar]
- Petrikin JE, Willig LK, Smith LD, Kingsmore SF. 2015. Rapid whole genome sequencing and precision neonatology. Semin Perinatol 39: 623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porreca GJ, Zhang K, Li JB, Xie B, Austin D, Vassallo SL, LeProust EM, Peck BJ, Emig CJ, Dahl F, et al. 2007. Multiplex amplification of large sets of human exons. Nat Methods 4: 31–936. [DOI] [PubMed] [Google Scholar]
- Pritchard CC, Salipante SJ, Koehler K, Smith C, Scroggins S, Wood B, Wu D, Lee MK, Dintzis S, Adey A, et al. 2014. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J Mol Diagn 16: 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y. 2012. A tale of three next generation sequencing platforms: Comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 24: 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratan A, Miller W, Guillory J, Stinson J, Seshagiri S, Schuster SC. 2013. Comparison of sequencing platforms for single nucleotide variant calls in a human sample. PLoS ONE 8: e55089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm HL, Bale SJ, Bayrak-Toydemir P, Berg JS, Brown KK, Deignan JL, Friez MJ, Funke BH, Hegde MR, Lyon E; Working Group of the American College of Medical Genetics, et al. 2013. ACMG clinical laboratory standards for next-generation sequencing. Genet Med 15: 733–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein WS, Maglott DR, Lee JM, Kattman BL, Malheiro AJ, Ovetsky M, Hem V, Gorelenkov V, Song G, Wallin C, et al. 2013. The NIH genetic testing registry: A new, centralized database of genetic tests to enable access to comprehensive information and improve transparency. Nucleic Acids Res 41: D925–D935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz MC, Delcher AL, Salzberg SL. 2010. Assembly of large genomes using second-generation sequencing. Genome Res 20: 1165–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina T, Suzuki S, Ozaki Y, Taira H, Kikkawa E, Shigenari A, Oka A, Umemura T, Joshita S, Takahashi O, et al. 2012. Super high resolution for single molecule-sequence-based typing of classical HLA loci at the 8-digit level using next generation sequencers. Tissue Antigens 80: 305–316. [DOI] [PubMed] [Google Scholar]
- Sims D, Sudbery I, Ilott NE, Heger A, Ponting CP. 2014. Sequencing depth and coverage: Key considerations in genomic analyses. Nat Rev Genet 15: 121–132. [DOI] [PubMed] [Google Scholar]
- Takoudes T, Hamar B. 2014. Performance of non-invasive prenatal testing when fetal cell-free DNA is absent. Ultrasound Obstet Gynecol 45: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Ankala A, Wilcox WR, Hegde MR. 2015. Solving the molecular diagnostic testing conundrum for Mendelian disorders in the era of next-generation sequencing: Single-gene, gene panel, or exome/genome sequencing. Genet Med 17: 444–451. [DOI] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, Ward P, Braxton A, Wang M, Buhay C, et al. 2014. Molecular findings among patients referred for clinical whole-exome sequencing. 2014. JAMA 312: 1870–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zook JM, Chapman B, Wang J, Mittelman D, Hofmann O, Hide W, Salit M. 2014. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotechnol 32: 246–251. [DOI] [PubMed] [Google Scholar]