Abstract
Amid the complexity of genetic alterations in human cancer, TP53 mutation appears as an almost invariant component, representing by far the most frequent genetic alteration overall. Compared with previous targeted sequencing studies, recent integrated genomics studies offer a less biased view of TP53 mutation patterns, revealing that >20% of mutations occur outside the DNA-binding domain. Among the 12 mutations representing each at least 1% of all mutations, five occur at residues directly involved in specific DNA binding, four affect the tertiary fold of the DNA-binding domain, and three are nonsense mutations, two of them in the carboxyl terminus. Significant mutations also occur in introns, affecting alternative splicing events or generating rearrangements (e.g., in intron 1 in sporadic osteosarcoma). In aggressive cancers, mutation is so common that it may not have prognostic value (all these cancers have impaired p53 function caused by mutation or by other mechanisms). In several other cancers, however, mutation makes a clear difference for prognostication, as, for example, in HER2-enriched breast cancers and in lung adenocarcinoma with EGFR mutations. Thus, the clinical significance of TP53 mutation is dependent on tumor subtype and context. Understanding the clinical impact of mutation will require integrating mutation-specific information (type, frequency, and predicted impact) with data on haplotypes and on loss of heterozygosity.
The clinical significance of TP53 mutations remains elusive in most cancers, but it appears to be tumor- and context-dependent. Combining mutation data with other data (e.g., on haplotypes and loss of heterozygosity) is needed.
In 1989, 10 years after the discovery of the p53 protein, studies by Baker et al. (1989), Nigro et al. (1989), and Takahashi et al. (1989) revealed that the TP53 gene was frequently mutated in many forms of human cancer. More than 25 years later, in the era of genome-wide sequencing of cancer DNA, TP53 is confirmed as the most frequently mutated gene associated with cancer in general. This finding is one of the most surprising lessons from cancer genome sequencing. There is no a priori reason why candidate gene-centered sequencing approaches would have correctly identified one unique gene among 20,000 genes as the most frequently mutated one. Indeed, a comparison of significantly mutated genes (SMGs) detected by whole-exome analysis in 15 types of solid tumors shows that TP53 is the most frequently mutated gene in 11 of them (glioblastoma, clear cell renal cell carcinoma, head and neck squamous cell cancer, serous ovarian carcinoma, non-small-cell lung cancer, small-cell lung cancer, gastric cancer, hepatocellular carcinoma [HCC], cholangiocarcinoma, breast cancer [BC], prostate cancer) (average: 25%–60%), whereas it ranks second to KRAS in pancreatic and colorectal cancer, and third to BRAF and NRAF in melanoma (Watson et al. 2013). In hematopoietic malignancies, TP53 mutations are less frequent (10%–15%) but are among the 10 most frequent SMGs in five of six documented malignancies (the exception is pediatric acute lymphoblastic leukemia [Zhang et al. 2012]).
The first studies on TP53 mutations focused on tumors with allelic deletions at the TP53 locus (17p13.1) and showed that mutations resulting in single amino acid substitutions were frequent on the allele that was retained. This observation was consistent with the theoretical hallmark of a tumor-suppressor gene and Knudsen’s two-hit hypothesis. Soon, however, it was noted that tumors that retained both parental alleles also frequently carried point mutations on one allele, suggesting a form of dominant effect of the mutation. Indeed, independent of the allelic context, nuclear accumulation of mutant p53 protein was detected in a wide spectrum of primary and metastatic lesions, suggesting a selective retention of mutant p53 during tumor development and dissemination (Bartek et al. 1990). In 1991, distinct patterns of somatic TP53 mutations were revealed, respectively, in UV-induced nonmelanoma skin cancers (Brash et al. 1991) and in aflatoxin-related HCC (Bressac et al. 1991). These mutation patterns were consistent with the ones induced by these mutagens in experimental assays, confirming the idea that “carcinogens [can] leave fingerprints” in TP53 sequence as evidence of their etiological role in carcinogenesis (Vogelstein and Kinzler 1992).
In subsequent years, the results literally sparked an industry of sequencing TP53 exons using the Sanger technique of chain-terminating inhibitors in cancer tissues. From a few hundred at the turn of the 1990s, the number of somatic TP53 mutations identified in cancer grew steadily to more than 10,000 by the end of the millennium (Hainaut and Hollstein 2000). Soon, computational resources were developed to compile, retrieve, and compare the already identified somatic mutations. In 1994, two databases collecting and annotating mutations detected by sequencing and published in the peer-reviewed literature were established, and they have been since maintained continuously: the TP53 database at the International Agency for Research on Cancer (IARC), initiated by Monica Hollstein and Curtis Harris (p53.iarc.fr) (Olivier et al. 2002), and the UMD-p53 database, initiated by Thierry Soussi (p53.fr/index.html) (Beroud and Soussi 1998). These databases currently contain more than 30,000 annotated mutations and provide a number of flexible tools to analyze the distribution of mutations across cancers. They have been the basis of several comprehensive reviews discussing the variability and heterogeneity of TP53 mutation patterns with respect to mutagenic processes, cancer etiology, or potential impacts on cancer biology (Greenblatt et al. 1994; Hainaut and Hollstein 2000; Petitjean et al. 2007; Soussi 2014).
In recent years, the development of second-generation sequencing technologies (also called next-generation sequencing [NGS]) has enabled the systematic analysis of cancer exomes and genomes, expanding our understanding of mutation patterns far beyond the knowledge derived from single genes such as TP53. However, despite numerous studies reporting an association between somatic TP53 mutations and poor prognosis and unfavorable treatment outcomes (Olivier et al. 2010; Robles and Harris 2010), the only accepted recommendation for clinical testing to date is for chronic lymphocytic leukemia (CLL), in which somatic TP53 mutation is predictive of poor response to conventional therapies (Stilgenbauer et al. 2014). Thus, the clinical significance of somatic TP53 mutations remains elusive in most forms of cancer. In this review, we revisit our understanding of TP53 mutation patterns in the light of recent data generated by next-generation sequencing, and we discuss how somatic TP53 mutation testing may contribute to inform clinical practice.
TOWARD AN UNBIASED SOMATIC TP53 MUTATION SPECTRUM
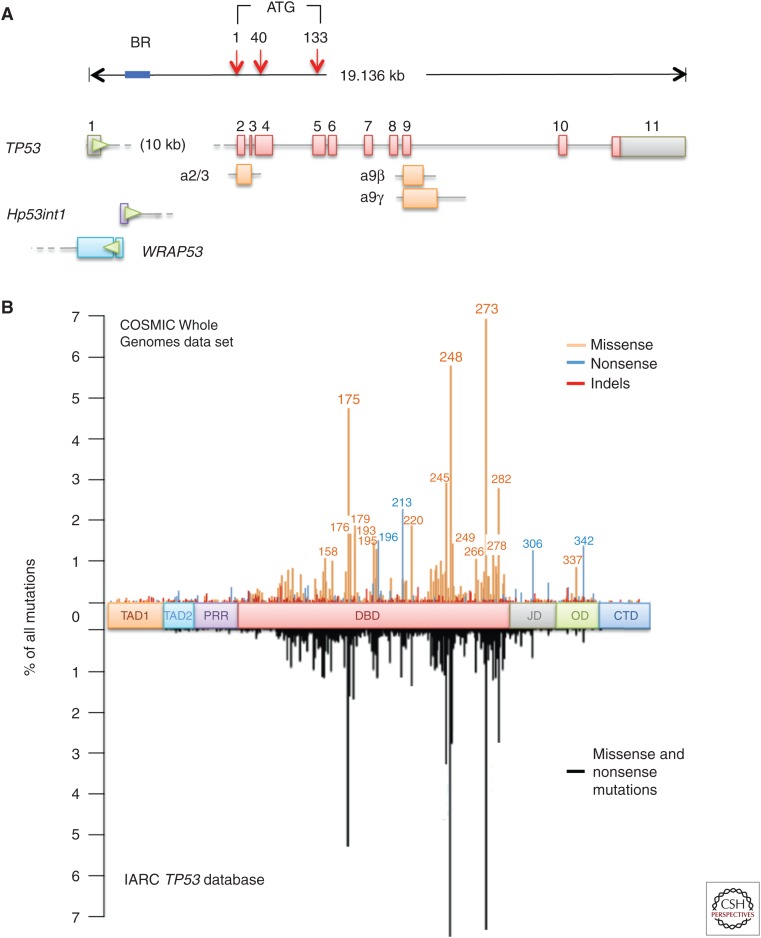
TP53 occupies ∼19.14 kb of genomic DNA on chromosome 17p13.1 and is oriented on the minus strand, antisense to other genes in the neighborhood. It comprises 14 exons, including 10 exons constituting the coding sequence of the canonical, full-length p53 protein of 393 amino acids, one noncoding exon 1, and three alternative exons, exon 2/3, exon 9β, and exon 9γ (exon 9α being the regularly spliced form of exon 9) involved in the synthesis of alternatively spliced transcripts encoding p53 protein isoforms (Fig. 1A). The noncoding exon 1 is separated from the rest of the coding sequence by a highly conserved intron that covers more than half of the gene (10,738 bp). Another gene, WRAP53, is located upstream of the TP53 coding sequence on the opposite (plus) strand. It overlaps with exon 1 and the proximal part of intron 1 and encodes antisense transcripts that are complementary to exon 1 transcripts (Mahmoudi et al. 2009). Two main promoter domains, P1 and P2, have been identified. P1 is the main promoter and is located upstream of exon 1. P2 is located within intron 1 at about 1000 bp of the 3′ end of exon 1 and regulates the expression of an intronic polyadenylated transcript of 1125 b, Hp53int1 (D17S2179E, GenBank: U58658.1) (Reisman et al. 1996). The p53 gene has a complex expression pattern with a single major transcript (encoding the full-length p53 protein) and more than 12 documented transcripts of variable abundance generated by alternative splicing and/or internal promoter usage (a weak promoter has been identified in a region encompassing exons 2–4 [Marcel et al. 2010]). These transcripts encode protein isoforms that differ from canonical p53 by the lack of amino-terminal domains (Δ40p53, lacking residues 1–39; Δ133p53, lacking residues 1–132) or by alternative carboxyl-terminal domains (p53β and p53γ) (Courtois et al. 2004; Khoury and Bourdon 2010; Marcel et al. 2011). Although the precise functions of these isoforms is still poorly understood, there is strong experimental evidence that amino-terminally truncated p53 variants may counteract the suppressor functions of full-length p53 protein (reviewed in Marcel et al. 2011; see also Joruiz and Bourdon 2016).
Figure 1.
TP53 mutation spectrum in human cancer. (A) TP53 locus on chromosome 17p13.1, showing main intron/exon structure (coding exons, red; noncoding exon 1 and 3′ UTR in exon 11, gray) and alternative exons 2/3, 9α, and 9β (orange). The position of Hp53int1 in intron 1 is shown, as well as the region of exon1/intron 1 overlapping with alternative exons 1 of WRAP53. All introns and exons are represented to scale, except intron 1. Blue bar, location of rearrangement breakpoints in osteosarcoma. Red arrows, position of the main (+1) and alternative (+40, +133) protein initiation sites. Green arrowheads, orientation of the coding sequences (TP53 is located on the minus strand of DNA, WRAP53 on the plus strand). (B, top) Codon distribution of mutations (missense, nonsense, and indels) in the coding sequence of TP53, based on mutation data derived from integrated genomic studies compiled in the Whole Genomes Resource (v73) of the COSMIC mutation database (cancer.sanger.ac.uk/cosmic/signatures), showing the position of major hotspots (>2% of all mutations) and mini hotspots (1%–2% of all mutations). (Bottom) Codon distribution of missense and nonsense mutations based on studies using conventional targeted sequencing approaches and compiled from the International Agency for Research on Cancer (IARC) TP53 mutation database (version R.13, 2008 [Olivier et al. 2010]). The IARC distribution is displayed as a mirror image of the COSMIC distribution. TAD1, TAD2, Transcriptional activation domains 1 and 2; PRR, proline-rich region; DBD, DNA-binding domain; JD, junctional domain; OD, oligomerization domain; CTD, carboxy-terminal domain.
After initial studies reported that somatic mutations were clustered over ∼1200 bases in conserved regions of exons 5–8, most studies have sequenced only these regions. Conventional sequencing methods are optimized for the detection of small sequence alterations (single-base mutations, short indels). Larger indels or rearrangements are not readily scored by these techniques. These biases have caused an underrepresentation of somatic mutations occurring outside exons 5–8, many of which are indels. Whole-genome sequencing (WGS) and whole-exome sequencing (WES) mutation data generated by NGS may be, in principle, less biased toward an a priori selection of “hotspot” mutation areas. The COSMIC database maintained at the Sanger Institute (Hinxton, United Kingdom) is the largest public database designed to store and display somatic mutation information relating to cancer. The Whole Genomes Resource (v73) of this database (cancer.sanger.ac.uk/wgs) compiles a total of ∼5000 TP53 mutations detected in WGS/WES projects, corresponding to 1206 different mutation events (as of May 1, 2015). Of those, only 12 mutations are highly recurrent, representing each at least 1% of the entire mutation data set (Table 1). These mutations represent mutational DNA “hotspots.” Of note, 11 of these hotspots occur at CpG sites and constitute a molecular signature of the random deamination of unstable 5-methylcytosine. Altogether, the 12 DNA hotspots represent ∼25%–30% of the total number of mutations. In contrast, 773 mutations are rare events, each occurring only once or twice in the data set, representing altogether ∼20% of the total number of mutations. Because several DNA hotspot mutations fall within the same codons (e.g., 245, 248, or 273), only six “major hotspot” codons comprise each at least 2% of all mutations (175, 213, 245, 248, 273, and 282). Another 13 codons represent “mini hotspots,” comprising each between 1% and 2% of all mutations (158, 176, 179, 193, 195, 196, 220, 249, 266, 278, 306, 337, and 342). Figure 1B shows the distribution of WGS/WES mutations along the p53 coding sequence and compares it with the distribution compiled from the IARC mutation data set in 2010, before the systematic usage of NGS (of note, the latest release of the IARC database [p53.iarc.fr] has included 866 mutations identified by WGS/WES [3% of the data set]). Although these distributions look very similar, the WGS/WES pattern shows that mutations outside exons 5–8 represent 22.5% of the total, almost double that estimated by Sanger sequencing (10%–15%). The higher number of silent mutations detected by NGS explains only 1.5% of this difference. Within the DNA-binding domain of the p53 protein, NGS data show that mutations at codon 213 (80% nonsense, p.R213*) and codon 220 (90% missense, p.Y220C) are hotspots almost as high as the well-defined hotspot codons 245 and 282. NGS data also show a frequently mutated area in the oligomerization domain (residues 326–355, 5% of all mutations). Within this domain, codon 342 (91% nonsense, p.R342*) constitutes a minor hotspot codon. Thus, the Sanger data set appears to be biased toward underrepresentation of mutations outside exons 5–8 and overrepresentation of the mutations known as major hotspots (codons 175, 245, 248, 273, and 282), perhaps because of a tendency of investigators to not fully analyze exon 6 (which is GC-rich and hard to sequence) and to preferentially report mutations detected at codons in exons 5, 7, and 8. Interestingly, the newly scored hotspot codons 213 and 342 contain CpG dinucleotide sequences, as do the standard hotspot codons 175, 245, 248, 273, and 282.
Table 1.
TP53 mutation “hotspots”: 12 mutations representing each of at least 1% of all mutations in the COSMIC Whole Genome Dataset (cancer.sanger.ac.uk/wgs)
| Position (codon) | cDNA/base change | Protein/amino acid change | Type of mutation | % | CpG site | Mutation effect |
|---|---|---|---|---|---|---|
| 175 | c.524G>A | p.R175H | Substitution/missense | 4.0 | Yes | Structural |
| 196 | c.586C>T | p.R196* | Substitution/nonsense | 1.4 | Yes | Structural |
| 213 | c.637C>T | p.R213* | Substitution/nonsense | 2.1 | Yes | Null |
| 220 | c.659A>G | p.Y220C | Substitution/missense | 1.5 | No | Structural |
| 245 | c.733G>A | p.G245S | Substitution/missense | 1.3 | Yes | Structural |
| 248 | c.742C>T | p.R248W | Substitution/missense | 2.1 | Yes | DNA-binding |
| 248 | c.743G>A | p.R248Q | Substitution/missense | 2.6 | Yes | DNA-binding |
| 273 | c.818G>A | p.R273H | Substitution/missense | 2.6 | Yes | DNA-binding |
| 273 | c.817C>T | p.R273C | Substitution/missense | 2.9 | Yes | DNA-binding |
| 282 | c.844C>T | p.R282W | Substitution/missense | 2.2 | Yes | DNA-binding |
| 306 | c.916C>T | p.R306* | Substitution/nonsense | 1.1 | Yes | Carboxy-terminal truncation |
| 342 | c.1024C>T | p.R342* | Substitution/nonsense | 1.3 | Yes | Carboxy-terminal truncation |
Structural, mutation affecting the folding of the DNA-binding domain; null, mutation predicted to impair protein synthesis; DNA-binding, mutation replacing an amino acid making direct and specific contact with DNA; carboxy-terminal truncation, mutation predicted as generating a protein lacking the entire (p.R306*) or part of (p.R342*) carboxy-terminal oligomerization domain.
About 2% of all mutations occur at intron/exon boundaries and affect splicing donor or acceptor sites. Genome-wide splicing mutation analysis has identified TP53 as the gene most commonly affected by splicing mutation in BC (Dorman et al. 2014). However, significant somatic variations occur in TP53 introns at sites other than those implicated in splice junctions, which are not covered in many conventional or exome-sequencing programs and thus are not reported in mutation databases. In BC, mutations in intron 9 colocalizing with alternative exons 9α and 9β have been detected using yeast functional assays (Iggo et al. 2013). These mutations alter the balance between fully spliced and alternatively spliced p53 transcripts, leading to the preferential synthesis of a protein that lacks part of the oligomerization domain. In osteosarcoma, somatic rearrangements in intron 1 have been detected by WGS in ∼20% of the cases (Ribi et al. 2015). The rearrangement breakpoints are located across the entire sequence of intron 1, but most of them cluster in a region of 1.7 kb located immediately upstream of the locus encoding the intronic Hp53int1 transcript, suggesting that breakpoint may be facilitated by open chromatin conformation at this locus (Fig. 1A) (Chen et al. 2014; Ribi et al. 2015). So far, intron 1 rearrangements have not been observed in other sporadic cancers and the reason for the narrow tissue specificity is unknown. The impact of these rearrangements on p53 function remains to be analyzed, but it is noteworthy that an intron 1 rearrangement (455-kb inversion) cosegregates with inherited risk of several cancers in a Li–Fraumeni family. The tumors of these patients show impaired TP53 transcription and loss of heterozygosity (LOH) affecting the wild-type allele (Ribi et al. 2015). Osteosarcoma has been considered as a type of cancer with low rate of TP53 mutations in exons 2–11 (20%), partly balanced by frequent functional inactivation of p53 protein through amplification of MDM2 (Ladanyi et al. 1993). The finding of intron 1 rearrangements in about half of sporadic osteosarcomas leads to a call for reconsidering them as a cancer with a high rate of somatic TP53 mutations.
BEYOND MUTATIONS: IMPACT OF TP53 HAPLOTYPES
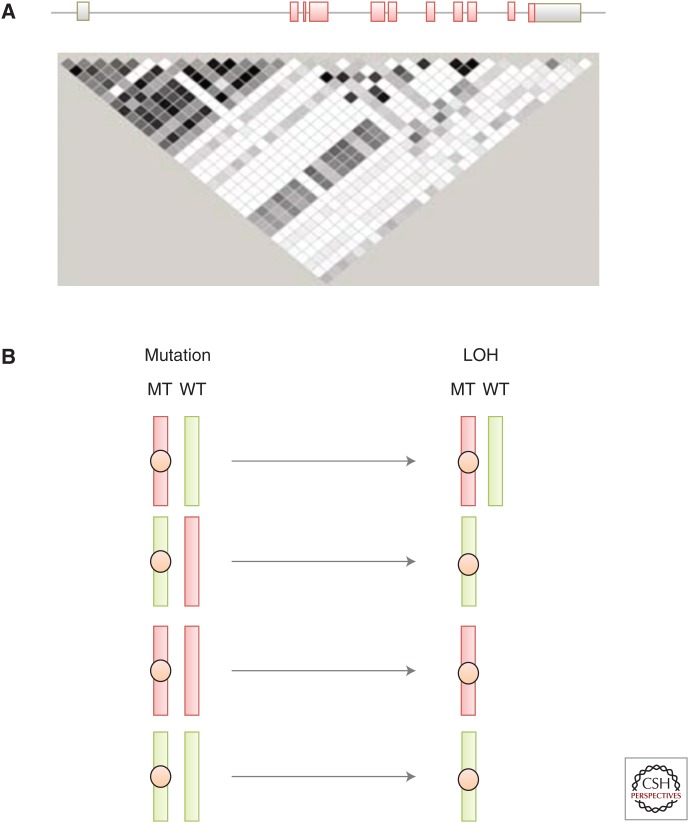
TP53 contains more than 100 validated naturally occurring single-nucleotide polymorphisms (SNPs; see p53.iarc.fr/TP53GeneVariations.aspx) but only a few of them have been studied for their effect on p53 functions. The best characterized exonic SNP is a nonsilent substitution at codon 72 in exon 4 (rs1042522, g.7520197G>C, p.R72P), which is present at minor allele (P) frequency between 10% (Caucasians, Northern Europe) and 50% (Africans, Yoruba), depending on population (Whibley et al. 2009). Several studies have shown that this SNP specifies protein variants with different functional properties in vitro and in experimental in vivo models. The form of p53 with R (arginine) at codon 72 (p53R72) more effectively induces p53-mediated apoptosis than p53P72 (proline), partially through targeting p53 to the mitochondria (Dumont et al. 2003). Meanwhile, p53P72, when compared with p53R72, more efficiently induces cell-cycle arrest and DNA repair (Pim and Banks 2004; Siddique and Sabapathy 2006). The ability of mutant p53 to bind p73, neutralize p73-induced apoptosis, and experimentally transform cells in cooperation with EJ-Ras is enhanced when the mutant protein carries an arginine at position 72 (Marin et al. 2000). In addition, R72 alleles appear to be preferentially mutated and retained in squamous cell carcinomas arising in patients who are R72P germline heterozygotes. Thus, this intragenic polymorphism may act as a modifier of mutant TP53 effect, suggesting that mutations and /or LOH may occur at different rates on different TP53 haplotypes. This hypothesis is supported by results from Mechanic et al. (2007) who analyzed 14 intragenic TP53 SNPs in a case-control study of lung cancer and uncovered an association between several combinations of SNPs and the risk of somatic mutation. The SNPs associated with mutation were located between intron 2 and intron 7, including R72P/rs1042522 (exon 4), rs9895829 (intron 4, c.376-125T>C), rs1625895 (intron 6, c.672+62A>G), and rs12951053 (intron 7, c.782+92T>G), defining the haplotype containing R-T-A-G as more likely to carry a mutation. The reason why this haplotype is preferentially mutated is unknown. In HCC carrying the aflatoxin-induced p.R249S mutation, analysis of 19 SNPs spanning the entire TP53 locus has shown a strong association between mutation and haplotypes that carry a combination of two SNPs (rs17882227 and rs8064946) defining a linkage disequilibrium block extending from upstream of noncoding exon 1 to the first half of intron 1. This domain contains two coding sequences overlapping with TP53 (WRAP53 and Hp53int1) (Ortiz-Cuaran et al. 2013). It is possible that these SNPs may modulate the p53 antisense activity of these transcripts, thus having an impact on p53 expression levels and therefore on the rate at which specific mutant alleles occur and are retained during tumor development. A simple way to rationalize this notion is to consider that different haplotypes express p53 at different levels, defining a continuum of functional p53 responses from “weak” to “strong” TP53 alleles, which may differ for different biological p53 effects (e.g., cell-cycle arrest, apoptosis, control of metabolism, or DNA repair) (Fig. 2). Mutations on a “weak” haplotype may be more likely to require the loss of the “strong” wild-type allele to exert a significant effect. In contrast, mutation on a “strong” haplotype may have disrupting, dominant effects even if a weaker wild-type allele is present.
Figure 2.
Variability and potential impact of TP53 haplotypes. (A) Linkage disequilibrium (LD) plot in the HapMap Panel for Caucasians, using 29 tag SNPs (tSNPs) for typing TP53 haplotypes (Garritano et al. 2010). The degree of LD is indicated by shades from black (strong LD) to white (no LD). This figure shows that most tSNPs in intron 1 and the TP53 promoter are in strong LD, forming a conserved haplotype block. (B) Model for loss of heterozygosity (LOH) at the wild-type (WT) allele depending on the interplay between “strong” (red) and “weak” (green) mutant haplotypes. (Top row) If the mutation (MT) occurs on a “strong” haplotype, its effects may dominate the residual WT “weak” haplotype, making LOH not compulsory. In all other haplotype combinations, LOH is required to eliminate a WT haplotype stronger than or equally strong as the one carrying the mutation.
MUTATION PATTERNS AS SIGNATURES OF MUTAGENIC PROCESSES
TP53 mutations have been extensively used as clues for the etiology of endogenous and exogenous mutagenic processes that operate during carcinogenesis in humans. Multiple independent studies have been conducted by Sanger sequencing of TP53 in tumors selected for their suspected association with a mutagen or mutagenic processes. The spectra produced by the aggregation of these somatic mutations were compared with the mutations experimentally generated by the suspected mutagens in vitro or in vivo systems (Hollstein et al. 1999; Pfeifer 2015). Table 2 summarizes the “molecular signatures” identified in TP53, caused by specific agents or mutagenic processes identified in TP53. This information has been extensively discussed in previous reviews (Greenblatt et al. 1994; Hainaut and Hollstein 2000; Olivier et al. 2004, 2010; Robles and Harris 2010; Pfeifer 2015). Nonmelanoma skin cancer (basal and squamous cell carcinoma) and melanoma show a predominance of G:C>A:T transitions at dipyrimidine sites (mutated base underlined), including about 10% of CC>TT tandem mutations, a type of mutation resulting from UV-light-induced covalent coupling of C=C double bonds at adjacent pyrimidines. These mutations are not caused by other known carcinogens and are almost never observed in tumors of internal organs (Luo et al. 2001). In HCC, a unique transversion at codon 249 (p.R249S; G:C>T:A) is highly prevalent in geographic areas in which the mycotoxin aflatoxin is a widespread contaminant of the food (parts of Africa, Eastern Asia, South America) (reviewed in Gouas et al. 2009). In lung cancer, ∼30% of TP53 mutations are G:C to T:A transversions in smokers, but not in never-smokers. This class of mutation is the main mutagenic effect of polycyclic aromatic hydrocarbons (PAHs) from the tar fraction of cigarette smoke. Exposure of human bronchial cells to the diol epoxide of the PAH benzo[a]pyrene causes DNA damage at the same G:C pairs that are frequently mutated into T:A in lung cancers from smokers (Denissenko et al. 1996; Tretyakova et al. 2002). Recently, a mutational fingerprint of aristolochic acid has been reported in urothelial carcinomas of subjects with a history of exposure to herbal remedies and food products containing seeds of Aristolochia clematitis (Hollstein et al. 2013; Poon et al. 2013). Last, all cancer types harbor at least 20% of G:C>A:T mutations at hypermutable CpG dinucleotides, attributed to the normal cellular event of deamination of 5-methylcytosine to thymine. This process is enhanced by inflammation, resulting in a high prevalence of these mutations in cancers associated with chronic inflammation (Ambs et al. 1999; Cooks et al. 2014). CpG mutations are exceptionally common in a few specific types of cancer including colorectal, gastric, and esophageal cancers (frequently occurring from precursor inflammatory lesions) as well as in brain cancers.
Table 2.
TP53 mutation patterns as a signature of mutational processes
| Base change (underlined) | Type | Mechanism/agent | Promutagenic lesion | Typical TP53 mutations | Cancers | References |
|---|---|---|---|---|---|---|
| CC>TT tandem | Transition | Solar UV | Dipyrimidine dimers | p.S127F, p.Q136*, p.R158C, p.H179Y, p.R196*, p.R248W, p.R278F, p.R282W | Nonmelanoma skin | Brash et al. 1991 |
| G:C>T:A | Transversion | Aflatoxin (AFB1) | AFB1-N7-guanine adducts | p.R249S | Liver (hepatocellular carcinoma) | Bressac et al. 1991 |
| G:C>T:A | Transversion | Tobacco smoke | PAH-N2-guanine adducts | p.V157F, p.R158L, p.G245V, p.G245C, p.R248L, p.R249M, p.R273L, p.D281E, p.E298* | Bronchus and lung, head and neck, esophagus | Denissenko et al. 1996; Tretyakova et al. 2002 |
| A:T>T:A | Transversion | Aristolochic acid | Aristolactam-DNA adducts | p.Q104L, p.N131Y, p.K139*, p.K164*, p.R209*, p.R280S, p.K291* | Bladder/urothelium | Hollstein et al. 2013 |
| G:C>A:T at CpG | Transition | Random deamination | 5- methylcytosine | p.R175S, p.R213*, p.R248Q, p.R248W, p.R282W, p.R306*, p.R342* | All cancers, in particular, colon, stomach, brain | Ambs et al. 1999 |
The introduction of NGS has provided a stepping-stone for a giant leap in our understanding of the signatures of mutational processes in human cancer. In addition to access to large data sets (between a few hundred and >10,000 mutations per tumor genome), NGS data have supported the development of a computational framework that allows deconstructing distinct but overlapping patterns from a set of cancer samples (Alexandrov et al. 2013a,b). Somatic mutations in the genome of any cancer are the result of cumulative mutagenic processes encompassing endogenous and exogenous mechanisms, each operating with different temporal patterns and with different strengths. Often, the cumulative pattern in cancer does not match any of the known single-operative mutational processes. By using algorithms that decipher the minimal set of mutagenic signatures that optimally explain the proportion of each mutation type in each cancer, it is now possible to estimate the contribution of different signatures to each cancer mutation spectrum.
The first NGS studies focusing on mutation patterns were published in Nature in 2010 and reported the patterns of somatic mutations in malignant melanoma (Pleasance et al. 2010a) and small-cell lung carcinoma (Pleasance et al. 2010b). These studies confirmed the widespread impact of UV light and tobacco carcinogens, respectively, in these cancers. Since then, many international consortia have undertaken the sequencing of large numbers of cancer samples, and mutational signature genomic patterns have been identified for many of them (Alexandrov et al. 2013a; Roberts and Gordenin 2014). The COSMIC database website provides a resource documenting up to 30 mutational signatures based on pooled NGS data from 10,952 exomes and 1048 whole genomes across 40 different types of cancers (accessible at cancer.sanger.ac.uk/cosmic/signatures). Several of these signatures confirm and refine mutational processes already described in TP53 (COSMIC signature 1, spontaneous deamination of 5meC; signature 4, tobacco smoke; signature 7, UV light; signature 22, aristolochic acid; and signature 24, aflatoxin).
Surprisingly, there are only a few new mutational signatures informing on exogenous carcinogen exposure in addition to those already detected in TP53. One example is signature 11, which corresponds to a distinct C>T mutation pattern caused by the alkylating agent temozolomide in recurrent glioblastoma multiforme of patients that have undergone this type of chemotherapy. On the other hand, NGS mutational signatures provide new insights into endogenous mutation processes, including mutational signatures caused by the error-prone polymerase η (signature 9, detected in chronic lymphocytic leukemia and malignant B-cell lymphomas), by proofreading defects attributed to polymerase ɛ (signature 10, in subsets of colorectal and uterine cancers), by defective mismatch repair (signatures 6, 15, 30, and 26, in gastric, colorectal, and endometrial cancer), and by defective repair of DNA strand breaks associated with germline and somatic BRCA1 and BRCA2 mutations (signature 3, breast, pancreatic, and ovarian cancers). Several new mutation signatures are still to be assigned to a specific mechanism (signatures 5, 8, 12, 14, 17–19, 21, 23, 25, 28, and 30). For example, esophageal adenocarcinomas carry a very unusual type of mutation, A-to-C mutations at 5′ AA dinucleotides (Pfeifer 2015). There are no known mutational processes, either endogenous or exogenous, that have been shown experimentally to result in these types of mutations. Because these events are highly specific for a tissue traversed by ingested materials, it is plausible that these mutations are caused by exposure to an exogenous mutagen. Furthermore, in addition to the aforementioned exposures to aflatoxins, liver cancer genomes, unlike other cancer genomes, have very diverse patterns of mutations targeting A/T base pairs. Because the liver is the primary organ for metabolism of xenobiotics, it would not be surprising if these A/T-targeted mutations were caused by metabolites of environmental carcinogens.
Some of the most interesting new data provided by NGS is the identification of an endogenous pathway involving cytidine deaminases of the AID/APOBEC family (signatures 2 and 13). Abnormal targeting of cytidine deaminases to random chromosomal locations appears to cause frequent C>T mutations at the cytosine of 5′TpC dinucleotides. The human genome encodes eight APOBEC enzymes (seven of them located at the APOBEC3 gene cluster) that normally serve to restrict viral infection and retrotransposon mobility by deaminating cytosines during the ssDNA stage of their replication stages. These enzymes target TCA/TCT trinucleotides in ssDNA in which they deaminate C to U, which is either directly copied to T or excised by uracil DNA glycosylase, followed by copying of the resulting abasic site into C>T or C>G mutations. Because of the processivity of APOBEC enzymes, these mutations often occur in clusters. AID/APOBEC signatures are present in many forms of cancer, and a germline deletion polymorphism affecting APOBEC3A and APOBEC3B is associated with large numbers of them (Nik-Zainal et al. 2014).
It is now possible to revisit TP53 mutation patterns in the light of these extended and refined mutational signatures. Recent studies have identified APOBEC mutation patterns in TP53 in BC (Lindley 2013) and lung adenocarcinoma (Waters et al. 2015). Although these studies are useful to better understand the sequence and codon context of mutation formation in TP53, the amount of etiological information they reveal is limited compared with multigene studies using NGS.
FUNCTIONAL DIVERSITY OF TP53 MUTATIONS
The spectrum of TP53 mutation in cancer cells and tissues is the result of combined processes acting as filters that select cancer mutations among a broad range of possible mutations. Sequence-context-dependent mutagenesis and DNA repair represent two of these filters. The biological selection of functionally “meaningful” mutations represents a third filter. However, what is a “meaningful” human cancer mutation is still largely unclear.
The lowest common denominator of cancer mutations is loss of transcriptional function (LOF) altering the p53-dependent transcriptional repertoire. Not all mutants are equal in this respect. The use of standardized yeast-based assays to test for LOF at total of 2314 mutants representing all possible substitutions throughout the protein has shown that LOF is virtually complete for frequently occurring mutants (including hotspots) but that some of the rarely occurring mutants retain quasi-wild-type functionality for transactivation (Kato et al. 2003). Most of these rare functional mutants and occur only once or twice in the COSMIC WGS/WES data set (representing <3% of all mutations). They may simply represent passenger mutations with no functional significance, detected only because of their accidental presence in an expanded clone of transformed cells. An interesting parallel exists between these mutants and those occasionally detected in noncancer tissues such as rheumatoid arthritis (RA), a noncancer precursor chronic inflammatory disease characterized by oligoclonal proliferation of nontransformed synoviocytes. RA tissues frequently contain clusters of cell clones with atypical TP53 mutations that include a high proportion of silent mutations (14%), mutations retaining wild-type transactivation capacity (60%), and mutations rarely found in any cancer (70% of them are either absent or reported only once in the WGS/WES COSMIC data set) (Firestein et al. 1997; Yamanishi et al. 2002). Such mutants are unlikely to be selected during carcinogenesis but are carried over as passengers in expanding subclones of RA synoviocytes.
There is compelling experimental evidence that TP53 mutations induce functional changes well beyond the scope of LOF. Many missense p53 mutants are expressed as stable proteins that exert dominant negative (DN) effects by interfering with the product of the remaining wild-type allele. A “prion-like” effect of some mutant p53 over wild type has been shown in vitro, in which the mutant enforces wild-type protein to adopt a denatured, mutant-like conformation (Milner and Medcalf 1991). DN effects may also result from a “sponge” effect of stable mutant p53 protein, absorbing the low-abundance wild-type proteins into oligomers dominated by mutant forms. Aside from DN effects, gain-of-function (GOF) effects have been demonstrated through which mutant p53 exerts a number of prooncogenic biochemical activities. GOF effects have been extensively studied in experimental cell systems and in genetically modified mouse models (Brosh and Rotter 2009; Muller and Vousden 2014). A number of mechanisms have been proposed for mutant p53 GOF, including direct binding to DNA changing (rather than abolishing) gene expression, binding to a large panel of transcription factors enhancing or impairing their function (e.g., binding to the p53 family members p63 and p73), binding to a number of proteins not directly involved in transcription (e.g., binding to the MRE11-RAD50-NSB1 complex, disrupting its function), and regulating microRNA networks (e.g., miR130b, miR155, and miR205) (Brosh and Rotter 2009; Muller and Vousden 2014). Recently, several p53 mutant proteins have been shown to up-regulate a number of chromatin regulatory genes, including the methyltransferases MLL1/KMTA2A and MLL2 (KMT2D) and the acetyltransferase MOZ/KAT6A, resulting in a global increase in histone methylation and acetylation and suggesting that mutant p53 may induce GOF effects through sweeping changes in the epigenetic control of chromatin dynamics (Zhu et al. 2015). The functional consequences of these GOF effects encompass the entire spectrum of the “hallmarks of cancer” (Hanahan and Weinberg 2011; Solomon et al. 2011). Thus, in contrast to LOF, there is no simple standard assay for scoring GOF effects. Furthermore, there is no clear understanding of the structural properties of mutant p53 that support GOF effects. Given the multiplicity of GOF mechanisms, it is unlikely that all effects depend on a single structural feature. In addition, mutants with the most perturbed protein structure are not necessarily those with the strongest GOF effects. In contrast, these effects are expected to be subtle and context-dependent, because they require interactions between mutant p53 and proteins that are themselves expressed in a tissue- and context-dependent manner. The subtlety of GOF effects is illustrated by p.R249S, the mutant caused by aflatoxin-induced mutagenesis in HCC arising in a context of chronic hepatitis B (HB) infection. This mutant is a clear LOF mutant, but is not commonly considered as GOF in most studies. However, its mechanism of action involves the binding of the HB antigen HBx, activating it as a viral oncogene in liver cells (Jiang et al. 2010; Gouas et al. 2012). This effect is a typical context-dependent GOF effect. The binding specificity of p.R249S to HBx may explain the surprisingly narrow spectrum of aflatoxin-induced TP53 mutations in HCC, because other mutants potentially induced by aflatoxin may not bind HBx with the same efficacy as p.R249S, preventing their selection during HB-dependent hepatocarcinogenesis (Denissenko et al. 1998).
CLASSIFYING TP53 MUTATIONS FOR CLINICAL USE
Although a multitude of experimental studies have shown different effects of various mutants on proliferation, apoptosis, or responses to cytotoxic drugs, there is surprisingly little clinical evidence for association between different types of mutations and clinicopathological variables, prognostics, or prediction of therapeutic responses. This does not imply that such associations do not exist. Perhaps the best evidence for clinically relevant differences between different mutations is found in subjects who carry germline TP53 mutations. In a recent report on 322 TP53 mutation carriers from French Li–Fraumeni families, the mean age of tumor onset was statistically different between carriers of missense mutations (23.8 years) and those carrying nonsense mutations or genomic rearrangements (28.5 years). The difference was even larger when comparing hotspot mutations, such as p.R175H or p.R248W (mean age of tumor onset, 15–20 yr) with complete deletion of the TP53 gene (38.5 yr) (Bougeard et al. 2015). Assuming that complete deletion of TP53 represents a “straight” LOF effect, these observations suggest that the more severe clinical phenotype of hotspot mutants is caused by their capacity to impair the wild-type allele (DN effects) and/or by specific GOF effects. It is likely that similar genotype–phenotype correlations occur for somatic mutations.
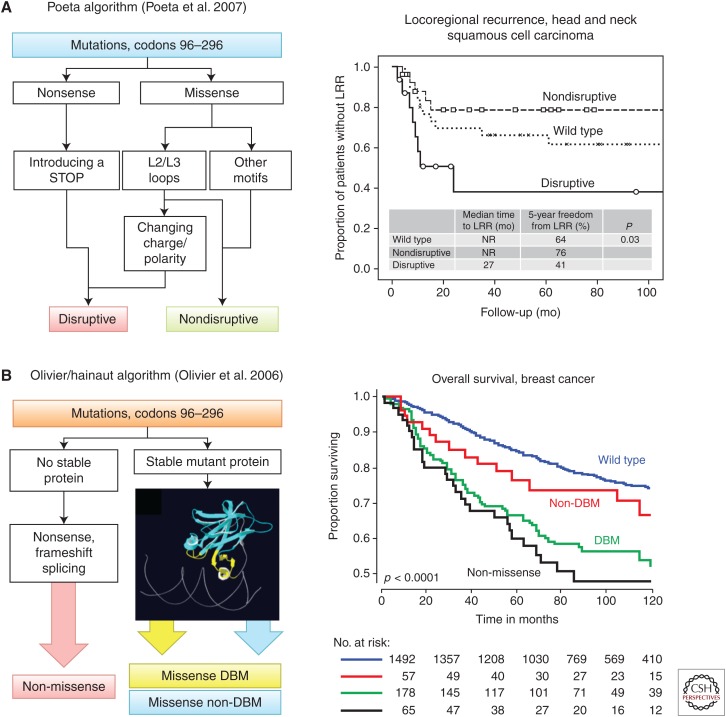
Many clinical studies have tried to subdivide somatic mutations into severity classes based on localization and predicted structural effects. Small clinical cohorts, suboptimal methods used to assess TP53 mutations status, and lack of consistency in how the mutations are classified have hindered a robust correlation with clinical features. Computational approaches using sequence similarities such as SIFT (sorting intolerant from tolerant) generate relatively accurate prediction for loss of transactivation function in vitro (specificity, 90%; sensitivity, 70% [Mathe et al. 2006]) but their clinical usefulness is limited to the identification of rare passenger mutations. Two simple classification algorithms have been used with some success (Fig. 3). The Poeta algorithm classifies mutations into two a priori classes, disruptive and nondisruptive mutations. Disruptive mutations combine predicted-null mutations (nonsense, frameshift, and splice site) and chemically deleterious substitutions in loops forming the DNA-binding surface of the protein (Poeta et al. 2007). Nondisruptive mutations include any other missense mutation. In head and neck squamous cell cancer (HNSqCC), disruptive mutations are associated with decreased overall survival, whereas there is no significant association with nondisruptive mutations. Moreover, disruptive mutations predict locoregional recurrence and HNSqCC cell lines with disruptive mutations are significantly more radioresistant than those expressing nondisruptive mutations (Skinner et al. 2012). The Olivier/Hainaut algorithm specifies three classes of mutations, predicted-null, missense mutation in the DNA-binding motif (DBM), and missense mutation in non-DNA-binding motif (non-DBM) (Olivier et al. 2006). The distinction between DBM and non-DBM mutations is purely topological, based on the observation that interspecies conservation of amino acids in the loops of the DBM is higher than in β-strands and short intervening loops of the non-DBM. The non-DBM is thus predicted to be more tolerant to mutations than the DBM (Mathe et al. 2006). In a large series of 1791 BCs from European patients, the algorithm identified predicted-null mutations as having the strongest association with poor survival, followed by DBM, whereas non-DBM were just marginally worse than wild-type TP53 (Olivier et al. 2006). In the METABRIC BC cohort (1420 patients), no statistical difference was found between the three mutation classes, although DBM mutations appear to have a marginally more severe effect (Silwal-Pandit et al. 2014). Incidentally, the fact that the two different classes of missense mutations appear to have the same (or less) effects as predicted-null mutations argues against DN or GOF effects of missense mutations in BC. When applied to non-small-cell lung cancer (NSCLC), the algorithm does not identify any prognostic value associated with either mutation class. However, in a randomized trial of platinium-based therapy after surgery (525 patients, 221 with mutation; 42%), non-DBM mutations were associated with poor overall survival in the group that received chemotherapy but not in the group treated by surgery only (Ma et al. 2014). This effect was not seen with predicted null or DBM mutations. This observation suggests that non-DBM may exert DN and/or GOF effects that confer resistance to adjuvant therapy. It is interesting to note that non-DBM mutations are frequent in NSCLC of smokers, because of the frequent targeting by tobacco carcinogens of codons encoding residues located in the hydrophobic core of the protein (Le Calvez et al. 2005). The selection of these mutations may contribute to a form of drug resistance in tumor cells.
Figure 3.
TP53 mutation classification algorithms. (A) The Poeta algorithm (left) (Poeta et al. 2007) classifies mutations occurring in the DNA-binding domain in two groups, disruptive (D) and nondisruptive (ND) mutations. D mutations either preclude the synthesis of p53 (nonsense mutations) or cause a structurally important amino acid change in the L2/L3 loops of the DNA-binding domain. Other missense mutations are classified as ND. (Right) D and ND mutations have different prognostic value for locoregional recurrence (LRR) in a cohort of 74 patients with squamous head and neck cancer (Skinner et al. 2012). (B) The Olivier/Hainaut algorithm (left) classifies mutations in three groups: mutations predicting a null p53 protein (non-missense), missense mutations in DNA-binding motifs (missense DBM), and missense mutations outside the DNA-binding motif (missense non-DBM). (Right) These three categories show different prognostic values for overall survival in a cohort of 1794 patients with breast cancer (Olivier et al. 2006).
Overall, these results indicate that the clinical severity of mutations is tumor- and context-dependent. Thus, the same mutations do not carry the same clinical consequences in different cancers. Although it can be assumed that the LOF effects of mutations might be roughly equivalent in different tissue contexts, DN effects are dependent on the retention, expression, and activity of the wild-type allele, and GOF effects may depend on factors that are expressed in a tissue- and context-dependent manner.
TP53 IN THE GENOMIC LANDSCAPE OF CANCER: SEEING THE NEEDLE WITHIN THE HAYSTACK
Until recently, TP53 mutations have been mostly approached as stand-alone events in studies of limited size and statistical power. The development of integrative genomics combining WGS/WES with DNA copy number alterations and studies on mRNA and protein expression provides an unprecedented view of the general genomic contexts in which TP53 mutations occur (Kristensen et al. 2014). In recent months, large consortium studies have combined multidimensional molecular approaches to characterize the landscape of genomic alterations in human cancers. These studies are generating a wealth of information that remains to be fully explored using TP53 mutation as a magnifying lens. Here, we briefly highlight some of the new knowledge that integrative genomics is bringing to our understanding of TP53 mutations in breast and non-small-cell lung cancer.
Breast Cancer (BC)
With mutation in 20%–25% of all cases, TP53 ranks first in a series of seven genes that are mutated in >10% of BCs (including PIK3CA, ERBB2, MYC, FGFR1/ZNF703, GATA3, and CCND1) (Stephens et al. 2012). A global study of the genomic and transcriptomic architecture of breast cancers has identified up to 10 molecular subtypes (integrative clusters) in which TP53 mutations occur at significantly different frequencies (Curtis et al. 2012). A detailed analysis of the TP53 mutation spectrum in the 1420 BCs of the METABRIC cohort has identified mutations in 28.3% of the cases (402 patients), with significant variations across BC subtypes (Silwal-Pandit et al. 2014). Mutations are frequent in basal-like and HER2-enriched tumors (65% and 53.4%, respectively), rare in luminal A and normal-like tumors (9.3% and 11%, respectively), and occur at an intermediate rate in luminal B tumors (24.8%). LOH at the TP53 locus is observed in 80.9% of the mutated tumors, independently of molecular subtype. However, in TP53 wild-type cases, the frequency of LOH varies across subtypes (from 24% in basal-like to 52% in luminal B). The distribution of mutations show striking differences between subtypes, with hotspot mutations dominating the spectrum of basal-like tumors, whereas a more uniform mutation distribution is seen in other subtypes.
When examining the associations between mutation and prognosis, TP53 mutation appears to be strongly correlated with poor overall survival in ER-positive patients but not in ER-negative patients. In fact, the presence of a mutation seems to wipe out the prognostic beneficial effect of ER positivity (Olivier et al. 2006; Silwal-Pandit et al. 2014). This observation suggests that p53 may function as a rate-limiting factor for ER signaling in BC, a conjecture supported by experimental data implicating p53 in estrogen responses in breast cancer cells (Fernandez-Cuesta et al. 2011a,b). Patients with mutant TP53 may thus not respond as well to endocrine breast cancer therapy as patients with wild-type TP53. In a recent study, p53 protein accumulation has been identified as a strong predictor of recurrence in ER-positive BC patients treated by aromatase inhibitor. It remains to be shown whether this accumulation is associated with mutant p53 (Yamamoto et al. 2014).
The association between TP53 mutation and BC prognosis is not uniform across subtypes. Mutations are significantly associated with poor survival in HER2-enriched, luminal B, and normal-like but not in basal-like or luminal A, suggesting a diverse role and impact for TP53 mutations (Fig. 4) (Silwal-Pandit et al. 2014). In basal-like cancers, disruption of the p53 pathway may be an obligate mechanism in virtually all tumors, either by mutation or by other mechanisms. Presence of wild-type TP53 may therefore not imply an intact p53 pathway, explaining the lack of prognostic value of TP53 mutations. In HER2-enriched cancers, mutation may disable the capacity of p53 to operate a safeguard mechanism against oncogene-driven cell proliferation induced by activated HER2. In luminal B and normal-like cancers, disruption of TP53 function is an optional mechanism and the occurrence of a mutation and/or LOH makes a significant difference for tumor growth and progression, thus entailing a clear negative prognostic value. In contrast, in luminal A cancers, mutation is a rare event that may be relatively unimportant. The fact that luminal A mutations rarely occur at hotspots suggest that many of them might be passenger mutations. There is no significant difference in prognostic value between different classes of mutations in any of the subtypes.
Figure 4.
Prognostic value of TP53 mutation in breast and lung cancer driven by oncogenes of the EGF receptor family. (Left) Prognostic value of TP53 mutations in patients with HER2-enriched breast cancer subtype (Silwal-Pandit et al. 2014). (Right) Prognostic value of TP53 mutations in patients with adenocarcinoma of the lung with mutant EGFR (Clinical Lung Cancer Genome Project 2013).
With respect to predictive value of mutations, it has long been suggested that docetaxel may confer a greater therapeutic advantage over anthracyclines in BC with mutated compared with wild-type TP53. This hypothesis has not been substantiated in two large clinical trials, which have confirmed the prognostic value of mutant TP53 but have not provided evidence for a predictive effect of mutation on response to therapy (Bonnefoi et al. 2011; Fernandez-Cuesta et al. 2012).
Non-Small-Cell Lung Cancer (NSCLC)
NSCLC comprises two major histological entities, squamous cell carcinoma (SqCC) and adenocarcinoma (AdC), and several minor histological subtypes including large-cell carcinomas (LCCs). WES reveals TP53 mutations in 81% of SqCC, a frequency at the higher end of those reported in studies using Sanger sequencing (range, 50%–80%), in which TP53 mutation load in SqCC appeared to correlate with smoking history (Le Calvez et al. 2005). TP53 is by far the most frequently mutated gene in SqCC, ahead of MLL2 (20%), PI3KCA (16%), CDKN2A (15%), NFE2L2 (15%), and KEAP1 (12%) (Clinical Lung Cancer Genome Project 2013). There is no obvious difference in the patterns of mutated genes between TP53-mutated and wild-type cases, suggesting that disruption of the p53 pathway is an obligate mechanism in the pathogenesis of SqCC. However, the high frequency precludes using TP53 mutations as a classifier for identifying molecular subtypes or for investigating prognostic and predictive value. In AdC, mutations occur in 46% of the cases, ahead of KRAS (33%), KEAP1 (17%), STK11 (17%), EGFR (14%), NF1 (11%), and BRAF (10%). It is striking to note that TP53 is the only gene that is commonly mutated in both major histological types of NSCLC. In both AdC and SqCC, the pattern of TP53 mutations is characterized by a high rate of transversions occurring at G bases on the nontranscribed strand, a typical signature of mutations induced by tobacco smoke. This pattern is seen in all histological subtypes but less marked in AdC than SqCC. In AdC, this pattern is not detected in EGFR-mutated cases, which mostly develop in never smokers, and in which TP53 mutations occur at a lower frequency than in smoking-related KRAS-mutated cases (30% vs. 60%, respectively). Independent of this association with smoke-related mutation signatures, TP53 mutation does not appear to be associated with any specific molecular feature of AdC.
A multitude of cohort studies has failed to identify a prognostic effect for TP53 mutations on NSCLC survival, even in AdC and after separating TP53 into different classes (see, e.g., Scoccianti et al. 2012; Ma et al. 2014). However, in a comprehensive analysis of 1255 clinically annotated lung cancers by the Clinical Lung Cancer Genome Project (CLCGP), TP53 mutation was found to be associated with a significant negative prognostic value in AdC with mutated EGFR (Fig. 4) (Clinical Lung Cancer Genome Project 2013). In fact, AdC with mutated EGFR seems to have a more favorable prognosis than AdC with other driver oncogene mutations, including KRAS. Although TP53 mutation does not appear to have an effect on the prognosis of AdC with mutated KRAS, it cancels the apparent beneficial effect of EGFR mutation. It should be noted that this beneficial effect is not only attributable to the response of these patients to targeted EGFR therapies. It is also observed in a retrospective series of AdC patients who have been treated with conventional therapies. These observations suggest that wild-type p53 functions as a limiting factor in AdC driven by oncogenic activation of EGFR. There is a striking parallelism between the effect of TP53 mutation in EGFR-mutated lung AdC and in HER2-enriched breast cancer (Fig. 4).
TP53 mutations have been reported as having a predictive effect on the survival of patients treated with adjuvant platinum-based therapy, a standard treatment regimen for stage II–III completely resected NSCLC (Ma et al. 2014). This effect predicts a marginal benefit of adjuvant therapy in patients with wild-type TP53 and marginal detrimental effect in patients with mutated TP53 mutation. There is no effect of TP53 mutation in patients who are treated by surgery alone. As discussed above, this effect is significant only for non-DBM mutations, a class of mutations occurring in regions encoding the hydrophobic core structure of the p53 DNA-binding domain. Mutations in the hydrophobic core domain are frequent in lung cancers of smokers, because of the targeting of codons in this region by carcinogens from tobacco smoke. These mutations may carry GOF effect that, although having no impact on natural tumor development (no prognostic value), accelerates tumor progression or relapse under adjuvant treatment (negative predictive value). This particular situation is a rare illustration of a possible GOF effect of mutant TP53 in a large randomized clinical trial. Presence of a non-DBM mutation appears to be associated with an adverse effect of adjuvant therapy, suggesting that these treatments should be restricted to wild-type TP53 cases. The molecular basis of this effect remains to be identified.
CONCLUDING REMARKS: SOLVING THE TP53 MUTATION EQUATION
The biological and clinical significance of TP53 mutation is the result of a complex equation that combines structural and functional terms. The main structural terms are the mutation itself (position, type, and effect on p53 protein structure), the retention (or not) of a functional wild-type allele (LOH at TP53 locus), and the haplotype structure of both wild-type and mutant alleles, which may be critical in the fine balancing act by which one allele may dominate over the other. Current sequencing strategies are tailored to assess only the first of these terms, and only in an incomplete manner because most studies remain focused on mutations occurring in exons. A recent initiative of researchers in the TP53 mutation field, led by Thierry Soussi, is recommending that the entire sequence of the TP53 gene should be taken into consideration when developing mutational analysis strategies to assess TP53 status. As a next step, it will be important to design algorithms that enable the assignment of haplotypes and LOH from sequencing data, thus providing the technical basis for an assay capable of scoring all the structural terms of the mutant TP53 equation.
The functional terms of the equation are even more complex. Although the major generic term is LOF, DN and GOF represent additional variables that may be extremely context-dependent. There is currently no simple and robust assay to score DN and GOF functions in a clinical setting. This complexity is compounded by the fact that, in many cancers, the p53 pathway may be disrupted by other mechanisms than structural alteration of the TP53 gene, so that tumors that apparently retain intact alleles actually do not have an intact p53 pathway. Many studies have analyzed the transcriptomic patterns of TP53 dysfunction in cancer cells and tissues but so far no robust consensus signature has emerged. Although a number of confirmed p53 target genes exist, their aberrant expression in cancer tissue may not be detectable without an inducer or treatment, or the expression of these genes in cancer tissue may be altered by p53-independent mechanisms. Efforts should be continued to identify surrogate biomarkers of a disrupted p53 pathway. A promising approach is to investigate p53-regulated long and short noncoding RNA networks (Donzelli et al. 2014).
Integrative genomics studies are confirming that somatic TP53 mutation lies at the very center of the patterns of alterations that characterize the genomic landscape of cancer. They also confirm that the biological and clinical significance of TP53 mutation is very context-dependent, with large differences among histological subtypes of the same cancers. We propose to distinguish three patterns of association between TP53 and genomic landscapes, highlighting their contrasting clinical messages.
High Mutation Frequency in Tumors with Profoundly Rearranged Genomes
In many subtypes of cancer (in particular in solid tumors), the mutation frequency is so high that it can be assumed that deregulation of the p53 pathway is an obligate mechanism for carcinogenesis. These cancers often show very disorganized genomes with high overall mutation load, often associated with intense exposure to endogenous or exogenous mutagens. They are poorly responsive to either conventional or pathway-targeted therapies. This is the situation observed in basal-like breast cancer or in SqCC of the lung. The overarching role of TP53 mutation in these cancers might be to promote and maintain metabolic and/or epigenetic programs essential for the expansion of aggressive cancer cells with stem-like phenotypes. In turn, loss of p53 function may promote genomic instability by favoring the occurrence of massive catastrophic genomic rearrangements such as chromothripsis (Rausch et al. 2012). In these tumors, TP53 mutation in itself may not be of much clinical significance for subclassifying cases or stratifying patients. However, a deeper evaluation of all structural terms of the TP53 mutation equation may uncover significant differences in the clinical behavior of cancers with different mutants. Despite having an overall poor prognosis, these tumors with frequent TP53 mutation show a range of clinical behaviors that may depend on factors that modulate the penetrance of TP53 mutation, such as LOH, haplotypes, and alteration of regulators of p53 function. In breast cancer, combining TP53 mutation, LOH, and MDM2 overexpression reveals an additive negative prognostic effect, suggesting that several mechanisms of p53 inactivation may cooperate within the same tumor (Silwal-Pandit et al. 2014).
Intermediate Mutation Frequency in Oncogene-Driven Cancers
Knowledge of TP53 mutation status is proving of more direct relevance in tumors with intermediate TP53 mutation frequencies, such as EGFR-mutated lung AdC and HER2-enriched BC. In these oncogene-driven cancers, TP53 mutation occurs at frequencies between 30% and 50% and is a clear indicator of poor prognosis. In both cases, the driving event is the oncogenic activation of a cell-surface tyrosine kinase receptor of the EGF superfamily and it is plausible that TP53 mutation has similar consequences in both cancers. An interesting hypothesis is that this effect is caused by the involvement of p53 in regulating cell responses to oncogenic signaling through the p14arf–Mdm2 pathway. Inactivation of p53 by mutation may switch off this pathway, thus eliminating a natural safeguard against excessive cell proliferation (Lomazzi et al. 2002). In support of this hypothesis, we have reported that p14arf expression is often down-regulated in lung AdC with mutated EGFR and wild-type TP53, suggesting that this pathway is under strong negative selection pressure in EGFR-mutated lung cancers (Mounawar et al. 2007; Cortot et al. 2014). However, why this effect is seen with EGFR mutation or HER2 enrichment and not with KRAS mutation is not clear. Manipulating the p53 pathway to increase and stabilize wild-type responses may prove a valuable companion approach in the treatment of these cancers, in particular with targeted therapies.
Low Mutation Frequencies in Tumors with Actionable Wild-Type TP53
A broad range of cancers has rare TP53 mutations, in particular at early stages. These cancers include lesions of overall good prognosis such as adenoma of the colon or luminal A breast cancers, in which rare TP53 mutations might just be occasional passengers. Most of these lesions are adequately curated by surgery. However, TP53 mutations are also rare in several cancer types requiring more aggressive treatment, such as melanoma (6%) or osteosarcoma (10%–12%). In these cases, a reassessment of TP53 mutation status is warranted. It is possible that significant structural alterations in TP53 have been missed in conventional studies because they occur in regions of the gene that are not routinely screened. A dramatic example of such a situation is observed in osteosarcoma, in which frameshift alterations/translocations are detected in intron 1 in ∼50% of the cases with no mutation in the coding sequence (Chen et al. 2014; Ribi et al. 2015). Another potential intragenic mechanism of inactivation is over-expression of dominant-negative p53 isoforms, either caused by deregulation of transcription and/or splicing, or by mutations in introns that activate the expression of naturally occurring isoforms (Marcel et al. 2011). For melanoma, the frequent deletion or mutation of the CDKN2A locus, which includes alterations of p14/ARF, may be a contributing factor for the lower mutation prevalence in the TP53 gene itself (Hodis et al. 2012). The fact that many of these tumors have in common the retention of a potentially functional p53 pathway suggests that it may be possible to pharmacologically “resuscitate” a suppressor function. A proof of principle has been given in cutaneous melanoma, in which overexpression of MDM4 promotes the survival of human metastatic melanoma by antagonizing p53 proapoptotic function (Gembarska et al. 2012). Inhibition of the MDM4–p53 interaction restores p53 function in melanoma cells, resulting in increased sensitivity to cytotoxic chemotherapy and to inhibitors of the BRAF (V600E) oncogene, thus identifying MDM4 as a promising target for therapy in melanoma.
In the coming years, TP53 mutation data will continue to grow at an unprecedented, exponential pace because at least the frequently mutated exons of TP53 are included in most current next-generation sequencing panels. During the weeks of redaction of this article (May–July 2015), the COSMIC WGS/WES TP53 mutation data set grew by more than 1000 units. A large concerted effort is needed to mine and annotate this enormous mass of information and to turn it into a source of detailed knowledge for the biologist, the clinician and, ultimately, for the benefit of the patients.
Footnotes
Editors: Guillermina Lozano and Arnold J. Levine
Additional Perspectives on The p53 Protein available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. 2013a. Signatures of mutational processes in human cancer. Nature 500: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. 2013b. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 3: 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambs S, Bennett WP, Merriam WG, Ogunfusika MO, Oser SM, Harrington AM, Shields PG, Felley-Bosco E, Hussain SP, Harris CC. 1999. Relationship between p53 mutations and inducible nitric oxide synthase expression in human colorectal cancer. J Natl Cancer Inst 91: 86–88. [DOI] [PubMed] [Google Scholar]
- Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, vanTuinen P, Ledbetter DH, Barker DF, Nakamura Y, et al. 1989. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science 244: 217–221. [DOI] [PubMed] [Google Scholar]
- Bartek J, Bartkova J, Vojtesek B, Staskova Z, Rejthar A, Kovarik J, Lane DP. 1990. Patterns of expression of the p53 tumour suppressor in human breast tissues and tumours in situ and in vitro. Int J Cancer 46: 839–844. [DOI] [PubMed] [Google Scholar]
- Beroud C, Soussi T. 1998. p53 gene mutation: Software and database. Nucleic Acids Res 26: 200–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefoi H, Piccart M, Bogaerts J, Mauriac L, Fumoleau P, Brain E, Petit T, Rouanet P, Jassem J, Blot E, et al. 2011. TP53 status for prediction of sensitivity to taxane versus non-taxane neoadjuvant chemotherapy in breast cancer (EORTC 10994/BIG 1-00): A randomised phase 3 trial. Lancet Oncol 12: 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougeard G, Renaux-Petel M, Flaman JM, Charbonnier C, Fermey P, Belotti M, Gauthier-Villars M, Stoppa-Lyonnet D, Consolino E, Brugieres L, et al. 2015. Revisiting Li–Fraumeni syndrome from TP53 mutation carriers. J Clin Oncol 33: 2345–2352. [DOI] [PubMed] [Google Scholar]
- Brash DE, Rudolph JA, Simon JA, Lin A, McKenna GJ, Baden HP, Halperin AJ, Ponten J. 1991. A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc Natl Acad Sci 88: 10124–10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressac B, Kew M, Wands J, Ozturk M. 1991. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature 350: 429–431. [DOI] [PubMed] [Google Scholar]
- Brosh R, Rotter V. 2009. When mutants gain new powers: News from the mutant p53 field. Nat Rev Cancer 9: 701–713. [DOI] [PubMed] [Google Scholar]
- Chen X, Bahrami A, Pappo A, Easton J, Dalton J, Hedlund E, Ellison D, Shurtleff S, Wu G, Wei L, et al. 2014. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep 7: 104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical Lung Cancer Genome Project (CLCGP). 2013. A genomics-based classification of human lung tumors. Sci Transl Med 5: 209ra153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooks T, Harris CC, Oren M. 2014. Caught in the cross fire: p53 in inflammation. Carcinogenesis 35: 1680–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortot AB, Younes M, Martel-Planche G, Guibert B, Isaac S, Souquet PJ, Commo F, Girard P, Fouret P, Brambilla E, et al. 2014. Mutation of TP53 and alteration of p14arf expression in EGFR- and KRAS-mutated lung adenocarcinomas. Clin Lung Cancer 15: 124–130. [DOI] [PubMed] [Google Scholar]
- Courtois S, Caron de Fromentel C, Hainaut P. 2004. p53 protein variants: Structural and functional similarities with p63 and p73 isoforms. Oncogene 23: 631–638. [DOI] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. 2012. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486: 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissenko MF, Pao A, Tang M, Pfeifer GP. 1996. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science 274: 430–432. [DOI] [PubMed] [Google Scholar]
- Denissenko MF, Koudriakova TB, Smith L, O’Connor TR, Riggs AD, Pfeifer GP. 1998. The p53 codon 249 mutational hotspot in hepatocellular carcinoma is not related to selective formation or persistence of aflatoxin B1 adducts. Oncogene 17: 3007–3014. [DOI] [PubMed] [Google Scholar]
- Donzelli S, Strano S, Blandino G. 2014. microRNAs: Short non-coding bullets of gain of function mutant p53 proteins. Oncoscience 1: 427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman SN, Viner C, Rogan PK. 2014. Splicing mutation analysis reveals previously unrecognized pathways in lymph node–invasive breast cancer. Sci Rep 4: 7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont P, Leu JI, Della Pietra AC, George DL, Murphy M. 2003. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet 33: 357–365. [DOI] [PubMed] [Google Scholar]
- Fernandez-Cuesta L, Anaganti S, Hainaut P, Olivier M. 2011a. p53 status influences response to tamoxifen but not to fulvestrant in breast cancer cell lines. Int J Cancer 128: 1813–1821. [DOI] [PubMed] [Google Scholar]
- Fernandez-Cuesta L, Anaganti S, Hainaut P, Olivier M. 2011b. Estrogen levels act as a rheostat on p53 levels and modulate p53-dependent responses in breast cancer cell lines. Breast Cancer Res Treat 125: 35–42. [DOI] [PubMed] [Google Scholar]
- Fernandez-Cuesta L, Oakman C, Falagan-Lotsch P, Smoth KS, Quinaux E, Buyse M, Dolci MS, Azambuja ED, Hainaut P, Dell’orto P, et al. 2012. Prognostic and predictive value of TP53 mutations in node-positive breast cancer patients treated with anthracycline- or anthracycline/taxane-based adjuvant therapy: Results from the BIG 02-98 phase III trial. Breast Cancer Res 14: R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein GS, Echeverri F, Yeo M, Zvaifler NJ, Green DR. 1997. Somatic mutations in the p53 tumor suppressor gene in rheumatoid arthritis synovium. Proc Natl Acad Sci 94: 10895–10900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garritano S, Gemignani F, Palmero EI, Olivier M, Martel-Planche G, Le Calvez-Kelm F, Brugieres L, Vargas FR, Brentani RR, Ashton-Prolla P, et al. 2010. Detailed haplotype analysis at the TP53 locus in p.R337H mutation carriers in the population of Southern Brazil: Evidence for a founder effect. Hum Mutat 31: 143–150. [DOI] [PubMed] [Google Scholar]
- Gembarska A, Luciani F, Fedele C, Russell EA, Dewaele M, Villar S, Zwolinska A, Haupt S, de Lange J, Yip D, et al. 2012. MDM4 is a key therapeutic target in cutaneous melanoma. Nat Med 18: 1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouas D, Shi H, Hainaut P. 2009. The aflatoxin-induced TP53 mutation at codon 249 (R249S): Biomarker of exposure, early detection and target for therapy. Cancer Lett 286: 29–37. [DOI] [PubMed] [Google Scholar]
- Gouas DA, Villar S, Ortiz-Cuaran S, Legros P, Ferro G, Kirk GD, Lesi OA, Mendy M, Bah E, Friesen MD, et al. 2012. TP53 R249S mutation, genetic variations in HBX and risk of hepatocellular carcinoma in The Gambia. Carcinogenesis 33: 1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt MS, Bennett WP, Hollstein M, Harris CC. 1994. Mutations in the p53 tumor suppressor gene: Clues to cancer etiology and molecular pathogenesis. Cancer Res 54: 4855–4878. [PubMed] [Google Scholar]
- Hainaut P, Hollstein M. 2000. p53 and human cancer: The first ten thousand mutations. Adv Cancer Res 77: 81–137. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: The next generation. Cell 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al. 2012. A landscape of driver mutations in melanoma. Cell 150: 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollstein M, Hergenhahn M, Yang Q, Bartsch H, Wang ZQ, Hainaut P. 1999. New approaches to understanding p53 gene tumor mutation spectra. Mutat Res 431: 199–209. [DOI] [PubMed] [Google Scholar]
- Hollstein M, Moriya M, Grollman AP, Olivier M. 2013. Analysis of TP53 mutation spectra reveals the fingerprint of the potent environmental carcinogen, aristolochic acid. Mutat Res 753: 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iggo R, Rudewicz J, Monceau E, Sevenet N, Bergh J, Sjoblom T, Bonnefoi H. 2013. Validation of a yeast functional assay for p53 mutations using clonal sequencing. J Pathol 231: 441–448. [DOI] [PubMed] [Google Scholar]
- Jiang W, Wang XW, Unger T, Forgues M, Kim JW, Hussain SP, Bowman E, Spillare EA, Lipsky MM, Meck JM, et al. 2010. Cooperation of tumor-derived HBx mutants and p53–249ser mutant in regulating cell proliferation, anchorage-independent growth and aneuploidy in a telomerase-immortalized normal human hepatocyte-derived cell line. Int J Cancer 127: 1011–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Joruiz SM, Bourdon JC. 2016. p53 isoforms: Key regulators of the cell fate decision. Cold Spring Harb Perspect Med 10.1101/cshperspect.a026039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Han SY, Liu W, Otsuka K, Shibata H, Kanamaru R, Ishioka C. 2003. Understanding the function–structure and function–mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci 100: 8424–8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury MP, Bourdon JC. 2010. The isoforms of the p53 protein. Cold Spring Harb Perspect Biol 2: a000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen VN, Lingjaerde OC, Russnes HG, Vollan HK, Frigessi A, Borresen-Dale AL. 2014. Principles and methods of integrative genomic analyses in cancer. Nat Rev Cancer 14: 299–313. [DOI] [PubMed] [Google Scholar]
- Ladanyi M, Cha C, Lewis R, Jhanwar SC, Huvos AG, Healey JH. 1993. MDM2 gene amplification in metastatic osteosarcoma. Cancer Res 53: 16–18. [PubMed] [Google Scholar]
- Le Calvez F, Mukeria A, Hunt JD, Kelm O, Hung RJ, Taniere P, Brennan P, Boffetta P, Zaridze DG, Hainaut P. 2005. TP53 and KRAS mutation load and types in lung cancers in relation to tobacco smoke: Distinct patterns in never, former, and current smokers. Cancer Res 65: 5076–5083. [DOI] [PubMed] [Google Scholar]
- Lindley RA. 2013. The importance of codon context for understanding the Ig-like somatic hypermutation strand-biased patterns in TP53 mutations in breast cancer. Cancer Genet 206: 222–226. [DOI] [PubMed] [Google Scholar]
- Lomazzi M, Moroni MC, Jensen MR, Frittoli E, Helin K. 2002. Suppression of the p53- or pRB-mediated G1 checkpoint is required for E2F-induced S-phase entry. Nat Genet 31: 190–194. [DOI] [PubMed] [Google Scholar]
- Luo JL, Tong WM, Yoon JH, Hergenhahn M, Koomagi R, Yang Q, Galendo D, Pfeifer GP, Wang ZQ, Hollstein M. 2001. UV-induced DNA damage and mutations in Hupki (human p53 knock-in) mice recapitulate p53 hotspot alterations in sun-exposed human skin. Cancer Res 61: 8158–8163. [PubMed] [Google Scholar]
- Ma X, Rousseau V, Sun H, Lantuejoul S, Filipits M, Pirker R, Popper H, Mendiboure J, Vataire AL, Le Chevalier T, et al. 2014. Significance of TP53 mutations as predictive markers of adjuvant cisplatin-based chemotherapy in completely resected non-small-cell lung cancer. Mol Oncol 8: 555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi S, Henriksson S, Corcoran M, Mendez-Vidal C, Wiman KG, Farnebo M. 2009. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol Cell 33: 462–471. [DOI] [PubMed] [Google Scholar]
- Marcel V, Vijayakumar V, Fernandez-Cuesta L, Hafsi H, Sagne C, Hautefeuille A, Olivier M, Hainaut P. 2010. p53 regulates the transcription of its Δ133p53 isoform through specific response elements contained within the TP53 P2 internal promoter. Oncogene 29: 2691–2700. [DOI] [PubMed] [Google Scholar]
- Marcel V, Dichtel-Danjoy ML, Sagne C, Hafsi H, Ma D, Ortiz-Cuaran S, Olivier M, Hall J, Mollereau B, Hainaut P, et al. 2011. Biological functions of p53 isoforms through evolution: Lessons from animal and cellular models. Cell Death Differ 18: 1815–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin MC, Jost CA, Brooks LA, Irwin MS, O’Nions J, Tidy JA, James N, McGregor JM, Harwood CA, Yulug IG, et al. 2000. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat Genet 25: 47–54. [DOI] [PubMed] [Google Scholar]
- Mathe E, Olivier M, Kato S, Ishioka C, Hainaut P, Tavtigian SV. 2006. Computational approaches for predicting the biological effect of p53 missense mutations: A comparison of three sequence analysis based methods. Nucleic Acids Res 34: 1317–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechanic LE, Bowman ED, Welsh JA, Khan MA, Hagiwara N, Enewold L, Shields PG, Burdette L, Chanock S, Harris CC. 2007. Common genetic variation in TP53 is associated with lung cancer risk and prognosis in African Americans and somatic mutations in lung tumors. Cancer Epidemiol Biomarkers Prev 16: 214–222. [DOI] [PubMed] [Google Scholar]
- Milner J, Medcalf EA. 1991. Cotranslation of activated mutant p53 with wild type drives the wild-type p53 protein into the mutant conformation. Cell 65: 765–774. [DOI] [PubMed] [Google Scholar]
- Mounawar M, Mukeria A, Le Calvez F, Hung RJ, Renard H, Cortot A, Bollart C, Zaridze D, Brennan P, Boffetta P, et al. 2007. Patterns of EGFR, HER2, TP53, and KRAS mutations of p14arf expression in non-small-cell lung cancers in relation to smoking history. Cancer Res 67: 5667–5672. [DOI] [PubMed] [Google Scholar]
- Muller PA, Vousden KH. 2014. Mutant p53 in cancer: New functions and therapeutic opportunities. Cancer Cell 25: 304–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro JM, Baker SJ, Preisinger AC, Jessup JM, Hostetter R, Cleary K, Bigner SH, Davidson N, Baylin S, Devilee P, et al. 1989. Mutations in the p53 gene occur in diverse human tumour types. Nature 342: 705–708. [DOI] [PubMed] [Google Scholar]
- Nik-Zainal S, Wedge DC, Alexandrov LB, Petljak M, Butler AP, Bolli N, Davies HR, Knappskog S, Martin S, Papaemmanuil E, et al. 2014. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat Genet 246: 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. 2002. The IARC TP53 database: New online mutation analysis and recommendations to users. Hum Mutat 19: 607–614. [DOI] [PubMed] [Google Scholar]
- Olivier M, Hussain SP, Caron de Fromentel C, Hainaut P, Harris CC. 2004. TP53 mutation spectra and load: A tool for generating hypotheses on the etiology of cancer. IARC Sci Publ 247–270. [PubMed] [Google Scholar]
- Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, Eyfjord J, Theillet C, Rodriguez C, Lidereau R, Bieche I, et al. 2006. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res 12: 1157–1167. [DOI] [PubMed] [Google Scholar]
- Olivier M, Hollstein M, Hainaut P. 2010. TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2: a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Cuaran S, Cox D, Villar S, Friesen MD, Durand G, Chabrier A, Khuhaprema T, Sangrajrang S, Ognjanovic S, Groopman JD, et al. 2013. Association between TP53 R249S mutation and polymorphisms in TP53 intron 1 in hepatocellular carcinoma. Genes Chromosomes Cancer 52: 912–919. [DOI] [PubMed] [Google Scholar]
- Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M. 2007. TP53 mutations in human cancers: Functional selection and impact on cancer prognosis and outcomes. Oncogene 26: 2157–2165. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP. 2015. How the environment shapes cancer genomes. Curr Opin Oncol 27: 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pim D, Banks L. 2004. p53 polymorphic variants at codon 72 exert different effects on cell cycle progression. Int J Cancer 108: 196–199. [DOI] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, et al. 2010a. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463: 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O’Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. 2010b. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463: 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeta ML, Manola J, Goldwasser MA, Forastiere A, Benoit N, Califano JA, Ridge JA, Goodwin J, Kenady D, Saunders J, et al. 2007. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med 357: 2552–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon SL, Pang ST, McPherson JR, Yu W, Huang KK, Guan P, Weng WH, Siew EY, Liu Y, Heng HL, et al. 2013. Genome-wide mutational signatures of aristolochic acid and its application as a screening tool. Sci Transl Med 5: 197ra101. [DOI] [PubMed] [Google Scholar]
- Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J, Jager N, Remke M, Shih D, Northcott PA, et al. 2012. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 148: 59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisman D, Balint E, Loging WT, Rotter V, Almon E. 1996. A novel transcript encoded within the 10-kb first intron of the human p53 tumor suppressor gene (D17S2179E) is induced during differentiation of myeloid leukemia cells. Genomics 38: 364–370. [DOI] [PubMed] [Google Scholar]
- Ribi S, Baumhoer D, Lee K Edison, Teo AS, Madan B, Zhang K, Kohlmann WK, Yao F, Lee WH, et al. 2015. TP53 intron 1 hotspot rearrangements are specific to sporadic osteosarcoma and can cause Li–Fraumeni syndrome. Oncotarget 6: 7727–7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Gordenin DA. 2014. Hypermutation in human cancer genomes: Footprints and mechanisms. Nat Rev Cancer 14: 786–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles AI, Harris CC. 2010. Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harb Perspect Biol 2: a001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoccianti C, Vesin A, Martel G, Olivier M, Brambilla E, Timsit JF, Tavecchio L, Brambilla C, Field JK, Hainaut P. 2012. Prognostic value of TP53, KRAS and EGFR mutations in nonsmall cell lung cancer: The EUELC cohort. Eur Respir J 40: 177–184. [DOI] [PubMed] [Google Scholar]
- Siddique M, Sabapathy K. 2006. Trp53-dependent DNA-repair is affected by the codon 72 polymorphism. Oncogene 25: 3489–3500. [DOI] [PubMed] [Google Scholar]
- Silwal-Pandit L, Vollan HK, Chin SF, Rueda OM, McKinney S, Osako T, Quigley DA, Kristensen VN, Aparicio S, Borresen-Dale AL, et al. 2014. TP53 mutation spectrum in breast cancer is subtype specific and has distinct prognostic relevance. Clin Cancer Res 20: 3569–3580. [DOI] [PubMed] [Google Scholar]
- Skinner HD, Sandulache VC, Ow TJ, Meyn RE, Yordy JS, Beadle BM, Fitzgerald AL, Giri U, Ang KK, Myers JN. 2012. TP53 disruptive mutations lead to head and neck cancer treatment failure through inhibition of radiation-induced senescence. Clin Cancer Res 18: 290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon H, Madar S, Rotter V. 2011. Mutant p53 gain of function is interwoven into the hallmarks of cancer. J Pathol 225: 475–478. [DOI] [PubMed] [Google Scholar]
- Soussi T. 2014. The TP53 gene network in a postgenomic era. Hum Mutat 35: 641–642. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, et al. 2012. The landscape of cancer genes and mutational processes in breast cancer. Nature 486: 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilgenbauer S, Schnaiter A, Paschka P, Zenz T, Rossi M, Dohner K, Buhler A, Bottcher S, Ritgen M, Kneba M, et al. 2014. Gene mutations and treatment outcome in chronic lymphocytic leukemia: Results from the CLL8 trial. Blood 123: 3247–3254. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Nau MM, Chiba I, Birrer MJ, Rosenberg RK, Vinocour M, Levitt M, Pass H, Gazdar AF, Minna JD. 1989. p53: A frequent target for genetic abnormalities in lung cancer. Science 246: 491–494. [DOI] [PubMed] [Google Scholar]
- Tretyakova N, Matter B, Jones R, Shallop A. 2002. Formation of benzo[a]pyrene diol epoxide-DNA adducts at specific guanines within K-ras and p53 gene sequences: Stable isotope-labeling mass spectrometry approach. Biochemistry 41: 9535–9544. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. 1992. Carcinogens leave fingerprints. Nature 355: 209–210. [DOI] [PubMed] [Google Scholar]
- Waters CE, Saldivar JC, Amin ZA, Schrock MS, Huebner K. 2015. FHIT loss-induced DNA damage creates optimal APOBEC substrates: Insights into APOBEC-mediated mutagenesis. Oncotarget 6: 3409–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson IR, Takahashi K, Futreal PA, Chin L. 2013. Emerging patterns of somatic mutations in cancer. Nat Rev Genet 14: 703–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whibley C, Pharoah PD, Hollstein M. 2009. p53 polymorphisms: Cancer implications. Nat Rev Cancer 9: 95–107. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Hosoda M, Nakano K, Jia S, Hatanaka KC, Takakuwa E, Hatanaka Y, Matsuno Y, Yamashita H. 2014. p53 accumulation is a strong predictor of recurrence in estrogen receptor-positive breast cancer patients treated with aromatase inhibitors. Cancer Sci 105: 81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanishi Y, Boyle DL, Rosengren S, Green DR, Zvaifler NJ, Firestein GS. 2002. Regional analysis of p53 mutations in rheumatoid arthritis synovium. Proc Natl Acad Sci 99: 10025–10030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. 2012. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481: 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Sammons MA, Donahue G, Dou Z, Vedadi M, Getlik M, Barsyte-Lovejoy D, Al-awar R, Katona BW, Shilatifard A, et al. 2015. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature 525: 206–211. [DOI] [PMC free article] [PubMed] [Google Scholar]