Abstract
Proteins of the intermediate filament (IF) supergene family are ubiquitous structural components that comprise, in a cell type–specific manner, the cytoskeleton proper in animal tissues. All IF proteins show a distinctly organized, extended α-helical conformation prone to form two-stranded coiled coils, which are the basic building blocks of these highly flexible, stress-resistant cytoskeletal filaments. IF proteins are highly charged, thus representing versatile polyampholytes with multiple functions. Taking vimentin, keratins, and the nuclear lamins as our prime examples, we present an overview of their molecular and structural parameters. These, in turn, document the ability of IF proteins to form distinct, highly diverse supramolecular assemblies and biomaterials found, for example, at the inner nuclear membrane, throughout the cytoplasm, and in highly complex extracellular appendages, such as hair and nails, of vertebrate organisms. Ultimately, our aim is to set the stage for a more rational understanding of the immediate effects that missense mutations in IF genes have on cellular functions and for their far-reaching impact on the development of the numerous IF diseases caused by them.
Intermediate filaments constitute flexible and stress-resistant networks in animal cells. They are composed of sequence-related fibrous proteins and represent the major determinant of cell and tissue plasticity.
1. INTRODUCTION
1.1. Discovery and Characterization of Intermediate Filament Proteins
The first intermediate filament (IF) proteins studied were keratins. The name keratin is derived from the Greek word for horn: κερας. They are—like the non-IF proteins myosin, fibrinogen, and collagen—abundant and highly insoluble constituents of metazoan cells and tissues. However, through the action of kinases, cells can promptly solubilize them down to characteristic coiled-coil complexes. In contrast, IF proteins can serve as the base for complex materials needed for various functions of vertebrate organisms. The so-called “hard” or “trichocytic” keratins can form, through a complex series of cellular differentiation steps, appendages such as wool, hair, nail, horn, or the baleen bristles of whales, and they are also needed to build bird feathers (Moll et al. 2008; Greenwold et al. 2014).
Historically, keratins were studied extensively by biochemists because of their prominent occurrence in animals, which enabled their isolation and further investigation by amino acid sequencing and various biophysical techniques. Because of their highly ordered fibrous arrangement in appendages such as hair and wool, they were among the first objects to be analyzed by X-ray diffraction in the early 1930s. William T. Astbury noticed their perplexingly simple diffraction patterns, differing so much from the very complex patterns obtained from crystals of globular proteins. In the course of analyzing more-fibrous proteins, he formulated the general conclusion “…that the x-ray and elastic properties of myosin are closely related to those of keratin” (Astbury 1939). Hence, keratin and myosin were grouped together with two other types of fibrous proteins—epidermin (epidermal keratins) and fibrinogen—as k-m-e-f proteins. Most notably, depending on the physical state of the material, these X-ray diffraction experiments revealed different signals from both hair and muscle, which, respectively, contain keratin and myosin in abundance and in an oriented form. Hence, for hair, a ground state (i.e., a mechanically nonstressed state) called α-keratin can be discriminated from a mechanically stressed state, when the hair is stretched by up to 100%, called β-keratin. Hence, stretching of hair, and very similarly of wool fibers, leads to an α-to-β transformation (Astbury 1933; MacArthur 1943; Kreplak et al. 2004). This transformation is a reversible process, and quite recently it has been simulated with atomic precision (Chou and Buehler 2012). Interestingly, silk and feathers also yielded the β-type diffraction pattern; however, these materials are made from distinctly different non-IF-proteins—fibroin and feather keratin—which are glycine-serine-alanine-rich and glycine-proline-serine-rich proteins, respectively (Astbury 1939; Gregg et al. 1984).
Before these diffraction data could be explained with molecular precision, it needed the definition of the α-helix and the β-pleated sheet as the two principal folds for proteins (Pauling and Corey 1951, 1953). On the basis of these data, both Francis Crick and Linus Pauling, together with Robert Corey, proposed the concept of a coiled-coil structure for keratins, with the nonpolar residues of two aligned α-helices pointing inward toward the axis of the coiled coil (Crick 1952, 1953; Pauling and Corey 1953). Following up these proposals, Fraser and colleagues investigated the tips of porcupine quills by quantitative measurements of X-ray diffraction patterns, using a Fourier transform approach originally formulated by Francis Crick, and thereby evaluated how the coiled-coil structure could manifest within these macrostructures (Fraser et al. 1964a,b). The coiled-coil principle was later directly verified on the basis of sequence information obtained for myosin and tropomyosin (Sodek et al. 1972; McLachlan et al. 1975): If an amino acid sequence supposed to form an α-helix shows heptad repeats (a-b-c-d-e-f-g)n of the kind that the first and the fourth position (i.e., a and d) are hydrophobic, such α-helical segments are prone to form a left-handed coiled-coil superstructure.
The next breakthrough in the elucidation of the biological role of IFs in vertebrates came with their identification as a third major filament system in ultrathin sections of myofibers by electron microscopy (EM). The group of Howard Holtzer detected smooth and relatively uniform filaments that coexisted with actin and myosin filaments in muscle (Ishikawa et al. 1968). The apparent diameter was reported to be in the range of 10 nm and, therefore, “intermediate” between that of actin filaments (6 nm) and myosin filaments (15 nm). This criterion was then used to investigate other tissues, as well as cultured cells, for the presence of 10-nm-type filaments (Goldman 1971; Small and Sobieszek 1977). As filaments of this diameter were detected in nonmuscle cells as well, they were from then on called “intermediate” between actin filaments and microtubules. Both their property to reorganize massively, after dissociation of the microtubules by colchicine, into large “juxtanuclear birefringent caps” consisting of 10-nm filament bundles (Goldman 1971) and their striking insolubility in physiological buffers facilitated their efficient purification in a native state from cultured cells (Starger and Goldman 1977; Starger et al. 1978). In parallel, tissues found to be rich in IF structures were used to devise biochemical procedures to purify the constituent proteins for structural and functional analyses (Steinert et al. 1976). Because of their pronounced insolubility under various ionic conditions, denaturing agents, such as urea and guanidine-hydrochloride, had to be used to solubilize them for further purification and analysis.
A first hint of the molecular relatedness of the various 10-nm filament proteins came from the reaction of a monoclonal antibody that, in immunofluorescence microscopy and immunoblots, recognized proteins constituting astrocyte filaments, neurofilaments, muscle IFs, and “tonofilaments” from epithelial cells and fibroblasts, indicating that they all shared a common sequence epitope (Pruss et al. 1981). In the same year, extensive amino acid sequence data of porcine desmin compared with chicken desmin and porcine lens vimentin were published that revealed that the molecules were closely related by amino acid sequence (Geisler and Weber 1981). Extending this notion further, the same group published the striking sequence similarity between desmin and published wool keratins within the central α-helical segment (Weber and Geisler 1982). Following up on these data, the group of Peter M. Steinert, one of the pioneers in the IF field, described the first full-length amino acid sequence of an epidermal keratin, mouse K10 (Steinert et al. 1983). These data essentially substantiated the molecular and evolutionary relationship of the various proteins from multiple tissues, all forming 10-nm-wide filaments.
2. THE DOMAIN ORGANIZATION: TRIPARTITE FIBROUS PROTEINS
The completed amino acid sequence of desmin allowed researchers to propose a common structural model for IF proteins—that is, a tripartite organization consisting of a central α-helical rod domain of conserved size flanked by non-α-helical end domains (Fig. 1A). Based on the folding principles for a two-stranded α-helical coiled coil, the primary amino acid sequences of IF proteins opened the possibility to propose rather distinct folding models for the α-helical rod (Geisler and Weber 1982; Quax-Jeuken et al. 1983). Using structure prediction according to Chou and Fasman (1978), they further suggested the rod to be substructured into three helices—Ia, Ib, and II. These structure predictions correlated well with data obtained by circular dichroism measurements: The isolated α-helical rod segment of desmin indeed showed 83% α-helical content. Last, but not least, the investigators concluded from chemical-cross-linking data of desmin rods that IF proteins form dimeric coiled coils. This prediction was indeed later verified by several studies on nuclear lamin A, keratins, and vimentin by X-ray crystallography and site-directed spin-labeling electron paramagnetic resonance (Fig. 1B,C) (discussed below in Sec. 6). It was further revealed that two dimers can associate in a half-staggered antiparallel manner to generate an ∼46-nm-long tetrameric complex that is the “brickstone” for further assembly into filaments (Quinlan et al. 1984; Geisler et al. 1992). The model for the tetrameric complex as derived from the structural determination of the vimentin coiled-coil structure indicates that it is, including the modeled end domains, 62 nm long (Chernyatina et al. 2015). Note the very different dimensions of the fibrous IF protein complex as compared with the mode of globular actin association within a filament segment (Fig. 1C).
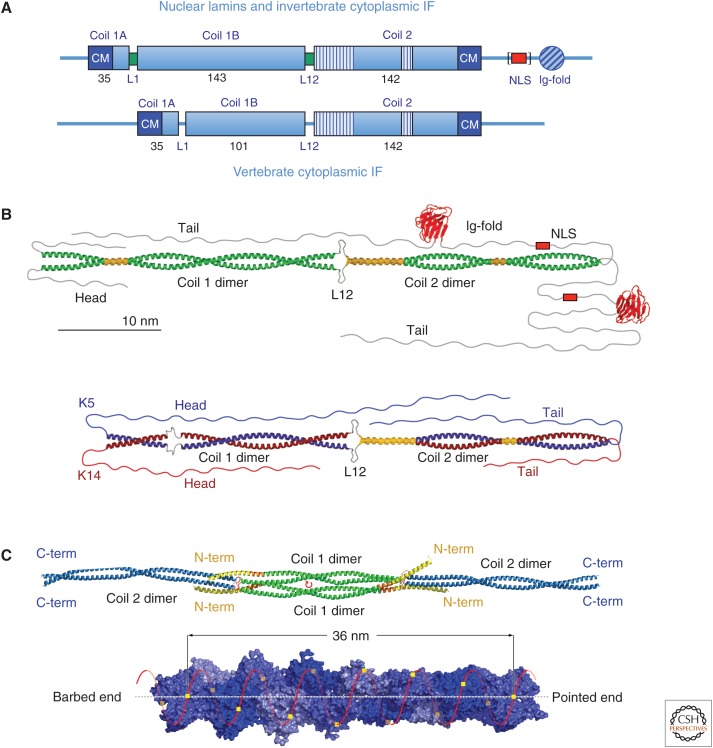
Figure 1.
Structural organization of intermediate filament (IF) proteins. (A) Domain organization of nuclear lamin and invertebrate cytoplasmic (top) and vertebrate cytoplasmic (bottom) IF proteins. Helical segments are designated “coil” and drawn as boxes, with the numbers represent the amino acid length of each segment. The striped parts of coil 2 indicate regions with hendecad repeats instead of the standard heptad repeats, which mediate coiled-coil formation. The red box designates the nuclear localization signal (NLS)—this is found in lamins, but not in invertebrate cytoplasmic IF proteins (hence, it is bracketed in the figure). The “Ig-fold” is a structurally conserved domain of approximately 100 amino acids showing an immunoglobulin-like fold found in nearly all nuclear lamins and most invertebrate cytoplasmic IF proteins. “CM” indicates conserved amino acid sequence motifs that occur at the amino-terminal and carboxy-terminal end of the rod, referred to as IF consensus motif 1 and 2, respectively. L1 and L12 indicate intrinsically disordered linker segments connecting the corresponding helices, although in lamins they can adopt an ordered state (green boxes). (B) Structural model of a lamin A homodimer (top) and a keratin heterodimer formed by K5 and K14 (bottom); the α-helical segments colored yellow represent parallel helices or “paired bundles”—the short paired bundle in the middle of the coil 2 domain has also been referred to as a “stutter.” (Redrawn from Herrmann et al. 2009, with permission from the American Society for Clinical Investigation.) (C) (Top) molecular structure of a vimentin tetramer featuring an antiparallel, half-staggered alignment of two coiled-coil dimers via coil 1 in the A11 mode. The center of symmetry is indicated by a small red circular arrow. (Adapted from Chernyatina et al. 2012). C-term, carboxyl terminal; N-term, amino terminal. (Bottom) A 14-subunit-long actin filament segment showing polar helical packing of 13 subunits in six left-handed turns with an axial increase of 2.75 nm per subunit. (Adapted from Dominguez and Holmes 2011, © Annual Reviews; http://www.annualreviews.org.)
With more sequences becoming available, homology profiles were calculated that strengthened the original suggestion for a tripartite organization of IF proteins as a general principle and gave more insight into the specific structural and molecular properties: The then newly sequenced nuclear lamin A did fit well into this structural paradigm, although its sequence showed a special feature: Helix 1b contained 42 amino acids, or “six heptads,” more than cytoplasmic IF proteins (Conway and Parry 1988). Hence, it was proposed that the human lamin A rod domain harbors 351 amino acids and the human vimentin rod domain 303, the lamin rod being longer by 48 amino acids because of three additional residues in both linkers L1 and L12 (Herrmann and Aebi 2004). In the following years, the originally proposed general plan for the structural organization of IF proteins was confirmed and extended by the identification and sequencing of a plethora of vertebrate and invertebrate IF genes, including the 70 human genes encoding IF proteins (Szeverenyi et al. 2008). However, as there is always an exception to the rule, the single lamin of the nematode Caenorhabditis elegans shows a two-heptad deletion in coil 2 (Karabinos et al. 2003). Sequence comparison with human lamin B2 localized the deletion to the beginning of coil 2 immediately after the “paired bundle” region (Fig. 1A).
3. INTERMEDIATE FILAMENT SEQUENCE-HOMOLOGY CLASSES: MOLECULES UNDER CONSTRUCTION
When amino acid sequences from the various IF proteins from different cell types and animal species were compared, it became evident that IF proteins could readily be grouped, by different investigators, into “type I”–“type V” (Conway and Parry 1988) or sequence homology classes (SHC 1–SHC 5) (Herrmann and Aebi 2000). Interestingly, this sequence-derived classification also reflected their biological function and tissue origin. Hence, keratins are grouped into SHC 1 and SHC 2. This classification, according to the degree of sequence identity, correlates well with the behavior of keratins in two-dimensional gel electrophoresis. Originally, the “acidic” keratins were termed type I (pI 4.9–5.4) and the “basic” keratins type II (pI 6.1–7.8). The numbering started with K1, the most basic and largest keratin, and continued on to K19, the then-smallest acidic keratin (Moll et al. 1982). Notably, all keratin filaments are built from heterodimers of one type I protein and one type II protein (Quinlan et al. 1984; Hatzfeld and Weber 1990; Steinert 1990). Moreover, using two-dimensional gel electrophoresis with different urea concentrations in the first dimension, it was shown that, when keratin pairs were combined in a “promiscuous” manner at the monomeric state in 8 m urea from basic and acidic group members, they still formed stable heterodimers in 5 m urea (Hatzfeld and Franke 1985). These experiments indicated that the attractive forces mediating coiled-coil formation are strong. Accordingly, vimentin was shown by analytical ultracentrifugation to form dimers in 6 m urea. When the urea concentration was further lowered to 5 m, the dimers associated to form ordered tetrameric complexes—a quite unusual feature for proteins (Herrmann et al. 1996).
The human genome harbors 67 keratin genes, 54 of which are functional (Schweizer et al. 2007). Keratins come as “soft” components, such as found in living cells, and as “hard” (or “trichocytic”) components of appendages such as hair and nails. Of these 54 genes, soft keratins are accounted for by 37 genes (17 SHC 1 and 20 SHC 2) and hard keratins by 17 genes (11 SHC 1 and six SHC 2). Hard keratins are generally very rich in cysteines and, in the hair bulb, the keratins come with a plethora of associated proteins (KAPs), represented by 21 different families containing 85 genes (Rogers et al. 2006). These proteins help to stabilize the flexible keratin IFs in three dimensions before they become heavily oxidized. As a result, highly oriented fibrous structures are formed that yield spectacular X-ray diffraction patterns (Kreplak et al. 2004). From a cell-biological point of view, the formation of the various IF-based appendages requires a highly complex series of differentiation steps of epithelial stem cells into tissue specializations or so-called “microfactories” (Fuchs 2009).
4. INTERMEDIATE FILAMENT PROTEINS ARE SELF-ASSEMBLING COMPONENTS
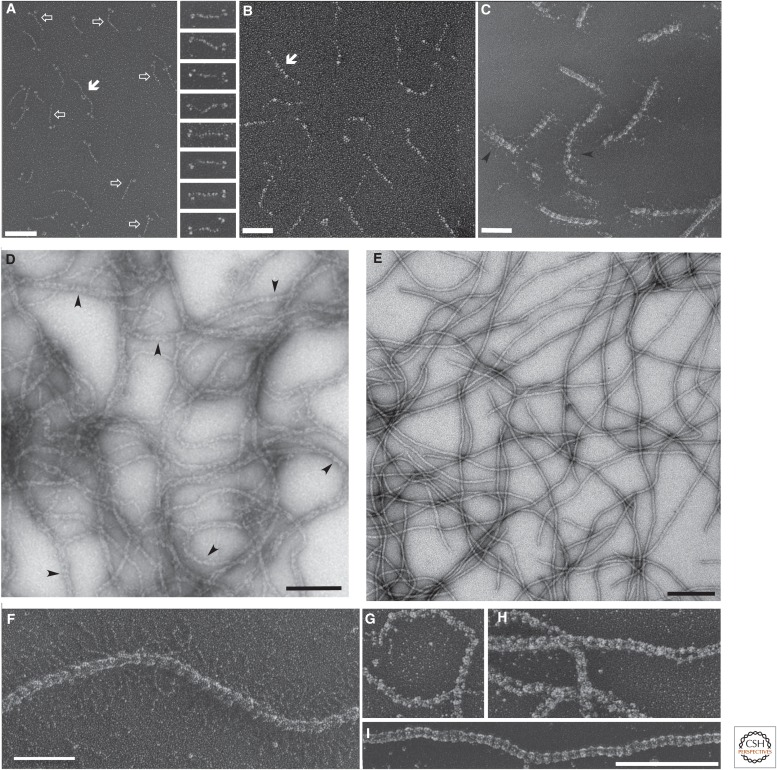
Biochemical purification of the various IF proteins from tissues and cultured cells revealed that they all can be renatured after purification under chaotropic conditions, and nearly all of them would self-assemble into 10-nm-like filaments (Steinert et al. 1976; Renner et al. 1981). Depending on the conditions, IF proteins can be renatured in such a way that they form distinct stable oligomers. Hence, the smallest complexes that can be obtained for lamins in the absence of denaturing agents are dimers (Fig. 2A, left panel). As a fundamental difference to lamins, invertebrate and vertebrate cytoplasmic IF proteins form in the absence of urea at least tetrameric complexes through the association of two dimers in an antiparallel, half-staggered manner. This orientation is immediately manifested for the invertebrate proteins by the location of their globular carboxy-terminal “tail” domains at either end of the tetrameric complexes, as detected by EM (Fig. 2A,B). Simply by manipulating the ionic strength conditions, the tetrameric complexes form larger assemblies, such as minifilaments (Fig. 2C), and eventually long, mature IFs (Fig. 2D–I).
Figure 2.
Electron microscopy of distinct supramolecular assemblies formed by various authentic and recombinant intermediate filament (IF) proteins. (A) Recombinant chicken lamin B2 dimers in 25 mm Tris-HCl (pH 8.5), 150 mm NaCl, 1 mm EGTA, 1 mm dithiothreitol (DTT) (micrograph on the left); and native Ascaris (invertebrate) cytoplasmic IF protein tetramers in 10 mm Tris-HCl (pH 8.0), 1 mm 2-mercaptoethanol (gallery on the right). The open arrows indicate single dimer molecules, and the solid arrow designates a head-to-tail complex of two dimers. Scale bar, 100 nm. (B) Recombinant human vimentin A11 tetramers in 5 mm Tris-HCl (pH 8.4). Scale bar, 50 nm. (C) 10-sec assembly intermediates of recombinant human vimentin at 37°C in 25 mm Tris-HCl (pH 7.5), 50 mm NaCl. Scale bar, 100 nm. (D) Recombinant human lamin A assembled by dialysis into 25 mm MES-NaOH (pH 6.5), 250 mm NaCl. Scale bar, 200 nm. (C,D) Arrowheads indicate prominent “beading” of the filaments. (E) Recombinant human vimentin assembled for 1 h at 37°C in 25 mm Tris-HCl (pH 7.5), 50 mm NaCl. Scale bar, 200 nm. (F) Native neurofilament triplet protein filaments. Note the radial projections emanating from the filament core. Scale bar, 100 nm. (G,H) Full-width filaments assembled from native Ascaris IF protein. (I) For comparison, an IF formed by native chicken gizzard desmin is displayed. Scale bar, 250 nm (also for G and H). Shown are glycerol-sprayed/rotary-metal-shadowed (A–C, F–I) and negatively stained (D,E) specimens. For details of these powerful techniquies, see Aebi and Baschong (2006). (The micrographs are reproduced from A, Heitlinger et al. 1991 [left] and Geisler et al. 1998 [gallery on the right]; B,C, Kreplak et al. 2008; F, Heins and Aebi 1994; G–I, Geisler et al. 1998.)
By standard negative-staining procedures as used for transmission electron microscopy (TEM), cytoplasmic IFs appear smooth and of relatively uniform diameter (Steinert et al. 1976). However, inspection of unstained keratin, desmin, and vimentin IFs by scanning transmission electron microscopy (STEM), which allows determination of their mass-per-length (MPL) characteristics, yielded three “classes” of filaments. Authentic vimentin IFs showed MPL values of 25, 37, and 51 kDa/nm, with corresponding filament diameters of 13.8, 15.1, and 16.2 nm (Steven et al. 1983). Similarly, recombinant vimentin IFs also revealed three MPL “classes” with 38, 47, and 56 kDa/nm and corresponding “full width at half maximum MPL” diameters of 9.8, 10.7, and 11.6 nm (Herrmann et al. 1996). The average value of 10.7 nm compares well with the value of 9.7 nm measured for fixed and negatively stained preparations (Herrmann et al. 1996; Mücke et al. 2004a). In contrast, when assembly was performed by dialysis instead of dilution and, therefore, the ionic conditions were changed slowly rather than instantaneously, more-homogeneous filaments, with an average MPL of 37 kDa/nm corresponding to 29 subunits per filament cross section, were obtained. In summary, IFs have the potential to be strongly “polymorphic,” in contrast to actin filaments. On a related matter, an often-overlooked feature is that microtubules can adopt structures with 13 to 16 protofilaments within one and the same filament and, thus, have the potential to display a considerable polymorphism (Chrétien et al. 1992).
Interestingly, cytoplasmic IFs reveal a characteristic periodic substructure by TEM after glycerol spraying/rotary metal shadowing—a 21.5-nm longitudinal “beading” repeat for vertebrate IFs and one of 24.5 nm for lower-invertebrate IFs (Henderson et al. 1982; Geisler et al. 1998). An explanation for this is that, on addition of glycerol (up to 50%) for spraying and the subsequent drying of the sample in vacuo on mica, the glycerol competes with the filament for water: As a result, a periodic collapse of structural elements along the filament occurs, thereby generating local constrictions of subdomains with respect to the filament axis (Fig. 2F–I). A similar type of beading is seen with negatively stained lamin filaments when they are adsorbed to the glow-discharged EM grids in the absence of glycerol and without prior chemical fixation (Fig. 2D). In this case, the beading repeat is 24.5 nm, just as it is for lower-invertebrate cytoplasmic IFs, as both of them have the longer coil 1B (see Fig. 1A).
Although not examined further here, it is important to note that lamins, too, assemble into 10-nm-type filaments both in vivo in the nuclear envelope of the Xenopus oocyte (B-type lamin) and in vitro after reconstitution of A-type lamins from isolated rat liver nuclei. In the latter case, however, the 10-nm filaments do not represent a stable species—instead they grow laterally into complex fibers and sheets to eventually yield highly ordered paracrystalline arrays consisting of multiple fibers oriented in parallel (Aebi et al. 1986; Ben-Harush et al. 2009). Hence, it is the absence of nuclear envelope transmembrane proteins (see Schirmer et al. 2003) in the “minimal” in vitro assembly system that allows A-type lamins to engage in multiple self-interactions. In agreement with this interpretation, strong overexpression of A-type lamins (e.g., by the baculovirus system) yields extensive paracrystalline fiber formation in transfected cultured Sf9 cells, originating from the moth Spodoptera frugiperda (Klapper et al. 1997).
5. THE EVOLUTION OF IF GENES ACCOMPANIES THE GENERATION OF NEW CELLULAR FUNCTIONS
The finding of a plethora of genes coding for IF proteins in all metazoan species opened the way to deeply investigate the evolutionary details of this multigene family (Erber et al. 1999). These sequences include those of IF proteins from the nematode C. elegans and the freshwater polyp Hydra attenuata (for review, see Herrmann and Aebi 2004). The nematode harbors one lamin and 13 cytoplasmic IF proteins, whereas the polyp expresses only one lamin but no cytoplasmic IF protein (Dodemont et al. 1994; Erber et al. 1999). However, the polyp expresses proteins in its mechanosensitive cilia—nematocilins A and B—whose amino acid sequence and structural organization resemble cytoplasmic IF proteins to some extent (Hwang et al. 2008). Likewise, the sea anemone Nematostella expresses a cytoplasmic IF-like protein—cytovec—which is sequence-related to the Hydra vulgaris nematocilins (Zimek et al. 2012). Indeed, these are interesting examples documenting the rapid evolutionary changes often detected in IF genes. Hence, the remarkable adaptability of the IF structure principle makes these proteins powerful building blocks involved in the development of new cellular shapes and functions. Accordingly, IF-related proteins have been found in protozoa too—a part of the living world still not explored as much as vertebrates with respect to their genomes (Kollmar 2015).
A look at the principal domain organization of lamin A and vimentin illustrates both the common and the specific features of IF proteins: Within the central α-helical “rod” domain, both the nuclear and the cytoplasmic protein harbor a coil 2 segment of identical length and organization (see Fig. 1A). Both contain a linker L12 of similar size between coil 1 and coil 2. Coil 1 is further subdivided into two α-helical segments by linker L1. The first segment, coil 1A, is 35 amino acids long in both the nuclear lamin A and cytoplasmic vimentin, whereas coil 1B shows a striking difference as it is 42 amino acids, or six heptads, longer in lamin A (143 amino acids) compared with vimentin (101 amino acids). Notably, all lower-invertebrate cytoplasmic IF proteins show the longer, lamin-A-like coil 1B. The short coil 1B is first found in urochordates such as Styela plicata (Riemer and Weber 1998). Therefore, as a nuclear “urlamin” is believed to be the first identifiable IF protein in evolution, the further shaping of cytoplasmic IF proteins from lower-invertebrate to higher-invertebrate species can now be studied in detail (Peter and Stick 2015).
The amino-terminal “head” domains differ considerably in size among IF species, and their amino acid sequences indicate typical intrinsically unstructured polypeptide segments. For example, the head domain of keratin K1 is 179 amino acids long, whereas that of the “stem cell marker” nestin is only seven amino acids long. Future studies are required to have a closer look at the structural properties of these short-headed IF proteins compared with bona fide IF proteins (i.e., those that form 10-nm filaments).
The carboxy-terminal tail domains differ in size at least as much as do the head domains. Certain simple epithelial keratins harbor a tail of only a few amino acids—for example, the human keratin 19 has a tail of only 15 residues. In contrast, nestin tails of human and rat are 1314 and 1497 amino acids long, respectively. In this case, the 183-residue longer rat nestin tail carries several insertions within the common sequence. Evolution did obviously “tinker” with some of the tissue-specific IF proteins to generate new functions. For example, the amino acid sequences of rat and human nestin vary considerably: Their rod domains are still 82% identical, but their tail domains are only 55% identical (Dahlstrand et al. 1992). In contrast, human and rat lamin C are 99% identical in their rods and 92% in their tails—values that are close to those found for other principal IF proteins, such as desmin and vimentin, in these two species.
In line with their nuclear localization, lamins contain a “classical” nuclear localization signal—the three principal human lamins contain the sequence Lys-Lys-Arg-Lys in lamin A and Lys-Arg-Lys-Arg in lamins B1 and B2. Furthermore, lamins—and many invertebrate cytoplasmic IF proteins—harbor in the middle of their tail domain an approximately 108-residue conserved sequence element adopting a distinct immunoglobulin-type fold (Fig. 1A; Erber et al. 1999). This domain represents a compact, highly versatile interaction module positioned outside of the filament body and therefore is ideally suited to take part in multiple distinct binding events, both with factors within the nuclear envelope and within the nucleoplasm (Shumaker et al. 2008).
6. COILED-COIL STRUCTURE AND TETRAMER FORMATION
Reassuringly, structural models of IF proteins obtained by structure prediction from their primary amino acid sequences are very similar to the structures determined more recently by protein crystallography and other complementary methods. Nevertheless, some of the key features within the central α-helical rod domain had to await crystallization of suitable IF protein fragments (Fig. 1B).
For example, in preparation for the crystallization of the 35-amino-acid-long coil 1A of human vimentin, analytical ultracentrifugation experiments revealed that this segment did not form a stable dimer (Strelkov et al. 2001; Meier et al. 2009). Even at the high concentration used in the crystallization experiments (typically 10 mg/mL), coil 1A existed as a monomeric α-helical chain in the crystals (Strelkov et al. 2002). Hence, a coiled-coil structure was observed for coil 1A only in the context of the entire coil 1 (Chernyatina et al. 2012). In contrast, coil 1B forms a well-organized left-handed coiled coil, as determined by X-ray crystallography and by site-directed spin labeling electron paramagnetic resonance (SDSL-EPR) analysis (Aziz et al. 2012). Indeed, the entire vimentin coil 1 forms a dimer, even at low protein concentrations, indicating that it represents an independent folding unit (Mücke et al. 2004a).
However, there is a possibility that at the very end of coil 1B, a hendecad (11-residue) repeat segment can form and thereby allow for the elongation of the helical domain by four amino acids into linker L12 (see Fig. 1) (Aziz et al. 2012; Chernyatina et al. 2012). Sequence comparisons indicated that the assumed start sequence of linker L12 is conserved and, thus, probably mediates a unique function: In the four classes of cytoplasmic IF proteins—that is, SHC 1 to SHC 4—it reads Gln-Ala-Gln-Iso, whereas, in lamins, the SHC 5 class, it is Arg-Arg-Lys-His, with little change in homologous amino acids in different species (Herrmann and Aebi 2004).
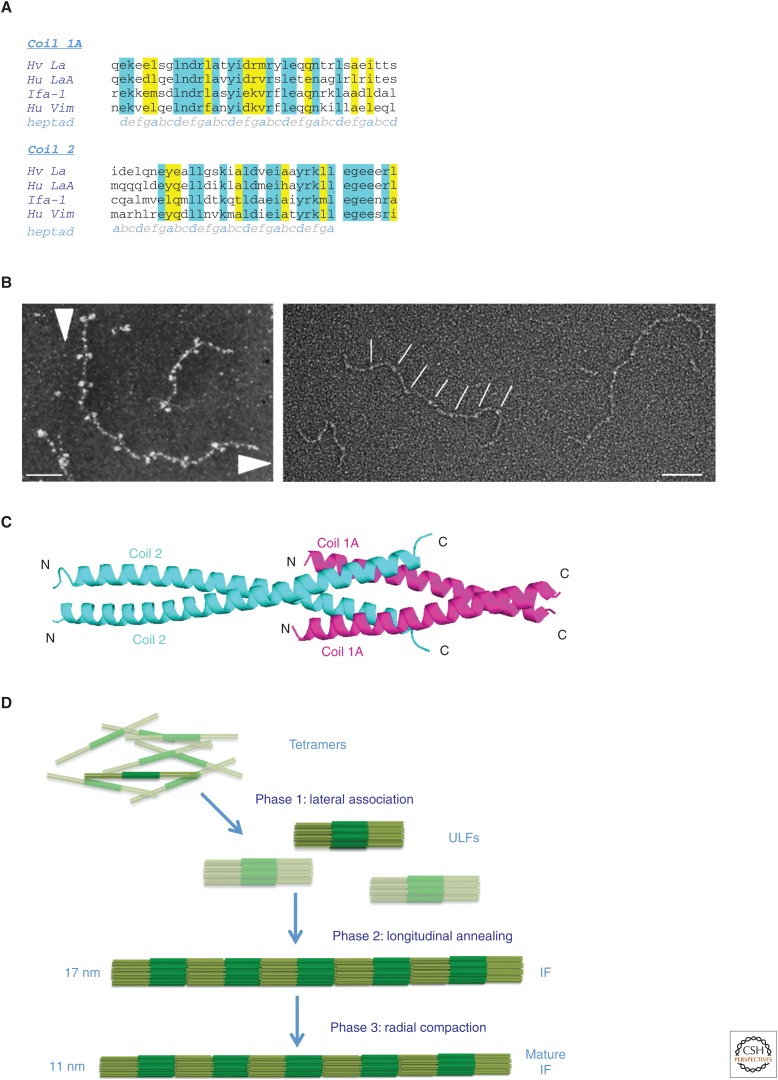
In the first sequence-derived structural models, coil 2 continued for three amino acids beyond the IF consensus motif 2, referred to as TYRKLLEGEE in the one-letter amino acid code (Fig. 3A). In lamins, another glutamic acid followed the consensus motif such that four glutamic acids, apposed to each other on the two chains, would terminate the coiled coil. This high charge density at the end of a coiled coil seemed to be unfavorable, and, therefore, the first atomic structure to be determined was from the last 28 amino acids of coil 2 of vimentin. As it was not known whether this four-heptad peptide would form a coiled coil by itself, 31 residues from the transcription factor GCN4, which had been shown to form a coiled coil (O’Shea et al. 1991), were added at its amino terminus as a “trigger” to force coiled-coil formation of the vimentin segment (Herrmann et al. 2000). The resulting atomic structure clearly revealed that the last amino acid of the coiled coil was the second leucine of the IF consensus motif 2 (Leu404), and that already the first glutamic acid of this motif (Glu405) bent away from the α-helical axis. Also, the atomic structure disclosed that both helices harbor an intrahelical salt bridge between Lys390 and Asp394. In addition, the structure revealed one interhelical salt bridge between Glu396 of the one chain and Arg401 of the other chain, which is assumed to stabilize the coiled coil (Burkhard et al. 2001). Notably, these four amino acids are evolutionarily highly conserved in IF proteins as diverse as human hair keratins and Hydra lamin, indicating they mediate a principal molecular interaction in filament formation (Herrmann et al. 2000).
Figure 3.
Assembly mechanism of intermediate filament (IF) proteins. (A) Sequence comparison of coil 1A containing the IF consensus motif 1 and of the carboxy-terminal end of coil 2 IF consensus harboring motif 2 (see Fig. 1A). Amino acids are shown in the one-letter code. Blue represents identical amino acids, yellow represents three or at least two identical amino acids, and the remaining residues are of very similar character; heptad indicates the position of each amino acid in the coiled-coil heptad pattern (abcdefg). Hv La, Hydra vulgaris lamin; Hu LaA, human lamin A; Ifa-1, cytoplasmic IF protein from C. elegans; Hu Vim, human vimentin. (B) Glycerol spraying/low-angle rotary metal shadowing of lamin polypeptides. (Left) Head-to-tail polymerization of lamin A dimers. The two arrowheads indicate the direction from the carboxy-terminal to the amino-terminal end of the dimer, referring to the two globular domains, which represent the carboxy-terminal tail domains (reprinted from Heitlinger et al. 1991). Scale bar, 50 nm. (Right) The same type of head-to-tail polymers formed by a human lamin A variant that is missing most of its tail (amino acids 417–664). The near-vertical bars point to thickenings at intervals of ∼49 nm representing the site of the head-to-tail overlap of two consecutive dimers. Scale bar, 100 nm. (C) Molecular model of the head-to-tail overlap: The end of coil 2 of one dimer overlaps by ∼3 nm with the beginning of coil 1A of the next dimer. Here, the helices are oriented such that they either form a four-stranded coiled coil or two two-stranded heterocoiled coils, each formed by one coil 1A and one coil 2 chain. Note that all four helices have the same orientation. N, amino terminus; C, carboxyl terminus. (D) Schematic representation of the three-phase assembly model for cytoplasmic IFs—involving, first, lateral association and then longitudinal annealing, followed by a maturation phase that involves radial compaction—as originally proposed by Herrmann and Aebi 1998. The degree of radial compaction—from 17 to 11 nm—is to some extent buffer-dependent (see Herrmann et al. 1996, 1999; Mücke et al. 2004a; Georgakopoulou et al. 2009).
Originally, coil 2 was also subdivided into two helical segments—coil 2A and coil 2B—connected by linker L2. However, as all three subsegments showed the same number of amino acids in all IF proteins investigated (i.e., 19, 8, and 121) and as L2 did not show any “helix-breaker”-type amino acid such as proline, the nature of the folding of coil 2 remained ambiguous. However, it was proposed that linker L2 has the potential to form an α-helix as well (Parry and Fraser 1985; Fraser et al. 1985). In addition, at exactly the same position in coil 2B of all IF protein sequences studied to date, eight and a half heptads downstream from the start of coil 2B, there is a so-called “stutter”—a discontinuity of the heptad repeat pattern that formally corresponds to a four-residue insertion (Parry and Steinert 1995). As the structural appearance of such a stutter in an IF protein coiled-coil dimer was unknown, the atomic structure of a segment covering most of coil 2B of vimentin (amino acids 328–411) was determined by X-ray crystallography. Accordingly, the crystal structure revealed that the two α-helices run parallel for a short distance, corresponding to an 11-residue (hendecad) repeat segment (Strelkov et al. 2002). Corresponding structures for coil 2B were subsequently obtained for lamin A (Strelkov et al. 2004) and a heterodimer formed by keratin K5 and K14 fragments (Lee et al. 2012). In both cases, the coiled-coil structural organization followed the one originally elucidated for vimentin (Strelkov et al. 2002).
Eventually, the atomic structure of the entire coil 2 of human vimentin was solved, showing that this originally defined coil 2A and L2 represent hendecad segments (Nicolet et al. 2010; Chernyatina and Strelkov 2012). Hence, coil 2 is predicted to be 115 amino acids long, with the first 36 amino acids forming hendecad repeats instead of heptad repeats, and another hendecad repeat residing within the second half of coil 2. Taken together, coil 2 adopts a distinct fold in all IF proteins, from keratins to lamins, indicating that this specific structure is a crucial “hub” for IF assembly.
As mentioned above, vimentin dimerizes in 6 m urea, and tetramers are formed in 5 m urea. Further dialysis into low-ionic-strength buffer such as 2 mm Tris-HCl (pH 9) preserves tetramers, as shown by analytical ultracentrifugation. This biophysical behavior is consistently shown by all cytoplasmic IF proteins (Lichtenstern et al. 2012). Chemical cross-linking of entire filaments followed by sequencing of cross-linked peptides revealed the existence of four types of tetrameric orientations (Parry and Steinert 1995). These forms are A11, half-staggered, antiparallel dimers associating via coil 1 (this is also the major form found in soluble tetramers [Mücke et al. 2004a]); A22, half-staggered, antiparallel dimers associating via coil 2 (this association has been found in filaments and in headless vimentin under physiological salt conditions and, thus, coil 2 shows a salt-induced binding site); A12, in-register, antiparallel dimers (this association has been found in mature filaments and might be formed by dimers of neighboring tetramers); and ACN, the longitudinal, “amino terminus to carboxyl terminus,” head-to-tail overlap of two coiled-coil dimers. In lamins, the head-to-tail overlap of several dimers “in tandem” can be directly documented by EM (Fig. 3B). The globular structures observed by rotary metal shadowing techniques represent the tail domain and, thus, serve as a marker for the directionality of the assembled fiber made from dimeric complexes by head-to-tail association (Fig. 3B; left panel). Moreover, note that also the tailless lamin A still assembles and that now the head-to-tail overlap region can be seen directly (Fig. 3B; right panel, white bars). From a large series of such EM images, the length of the overlap has been determined to be in the range of 2–4 nm (Heitlinger et al. 1992; Kapinos et al. 2010). For cytoplasmic IFs, this overlap distance was determined to be 1–2 nm by chemical cross-linking experiments (Parry and Steinert 1995). However, the molecular length of the bidirectional cross-linkers and the accessibility of their substrate—the ε-amino groups of lysines—might influence this value considerably, as cross-linking can “force” more-distant side chains to come closer together. Accordingly, EM showed a highly stringent periodicity in vertebrate cytoplasmic IFs of 43 nm (Henderson et al. 1982) and, together with the calculated length of the coiled-coil dimer of 46 nm, these two values would yield an overlap of 3 nm. From these data, we proposed that this overlap could involve the formation of two heterodimeric coiled-coil segments between helices from the first three heptads of coil 1A and those from the last three heptads of coil 2 (Fig. 3C); alternatively, the four α-helical segments might form a four-stranded coiled coil or lock in laterally (Strelkov et al. 2004; Kapinos et al. 2010).
The exact mode of the A11 organization has been determined jointly by SDSL-EPR, using spin-labeled vimentin filaments and X-ray diffraction analysis of an extended coil 1B fragment of vimentin: The interaction of the two coiled coils is centered around the glutamic acids in position 191 (Fig. 1C) (Aziz et al. 2012; Chernyatina et al. 2012). The total length of a vimentin tetramer without the nonstructured “head” and “tail” domains is 62 nm and is therefore six times longer than the diameter of an IF. In contrast, the globular subunits of an actin filament are arranged in a single helix, with 13 subunits packed in six left-handed turns, showing an axial increase of 2.76 nm per subunit (Fig. 1C). Accordingly, the structure can also be described as a right-handed long-pitch two-start helix (Dominguez and Holmes 2011). Because of this distinct subunit arrangement, the actin filament is polar, and this polarity has a consequence for assembly: Actin filaments show a fast-growing and a slow-growing end (for review, see Pollard 2016). In contrast, because of the antiparallel association of the two dimers in a tetramer as a building block for assembly, IFs are nonpolar.
7. FILAMENT ASSEMBLY: THE UNIT-LENGTH FILAMENT PARADIGM
Different from actin and tubulin, which are globular proteins with nucleotidase activity that is intimately connected to the assembly process, IF proteins are fibrous and lack enzymatic activity. Solely under the influence of monovalent cations, the 62-nm-long A11 tetramers laterally associate into ∼58-nm-long mini-filaments termed unit-length filaments (ULFs) (Herrmann et al. 1996). This process has been termed “phase 1” of IF assembly, and it is so fast that A11 tetramers are all consumed in ULF formation within seconds. ULFs are productive precursors for the formation of long filaments (Fig. 3D). Note that ULFs, because of their half-staggered, antiparallel association of dimers within the tetramer, are made of a central “core” of 32 coil 1 domains and two flanking segments consisting of 16 coil 2 domains each. In phase 2, elongation occurs by longitudinal annealing of ULFs via coil 2, such that filaments containing two, three, and more ULFs are formed. In phase 3, the filament diameter radially compacts by 20%–40%, yielding mature filaments of ∼10 nm in diameter (Herrmann and Aebi 1998; Georgakopoulou et al. 2009). These filaments show a high flexibility, with an apparent persistence length lp of 0.3–1 µm (Mücke et al. 2004b). Notably, various desmin disease mutants that were not able to form proper IFs in vitro were shown to get stuck at one of the three described stages, either staying in the arrested state or aggregating massively into amorphous structures over time (Bär et al. 2005). These desmin aggregates have indeed been discussed as a cause for disease development in the wider context of protein aggregate myopathies (Clemen et al. 2015).
IF proteins are highly insoluble under a wide range of ionic conditions, including standard “physiological” conditions. This property dictates the way that they can be isolated, purified, and crystallized; in addition, it may influence investigations of their assembly and their interaction with other cellular constituents. Hence, a “cocycling” type of assembly–disassembly regime, as successfully used to coisolate, for instance, microtubule-associated proteins (MAPs), is only possible when a denaturation step with chaotropic agents such as urea or sodium dodecyl sulfate or dissolution in low-ionic-strength buffers is applied (Renner et al. 1981; Steinert et al. 1981). Such a “cycling” regime has been documented to work also for plectin, a high-molecular-weight cytolinker protein (∼500 kDa), which copurifies with vimentin when cells are dissolved in buffers containing 1% nonionic detergent and 1.5 m potassium chloride (Herrmann and Wiche 1987; Foisner et al. 1988).
For decades, filament-like “precursors” were observed in various types of keratin preparations analyzed by EM. However, these were, in most cases, met with caution and skepticism, as they were obtained together with various heterogeneous, in many cases somewhat unraveled, filamentous structures. These were of different lengths, depending on the disassembly or reassembly conditions used, and occurred in the presence of phosphate buffer or even 4 m urea (Franke et al. 1982; Eichner et al. 1985). Notably, epidermal keratin IFs showed a distinct subfilamentous organization that could best be described as a partially unraveled, multistranded helix of two to four protofibrils when reconstituted filaments were deposited onto an EM grid and washed with phosphate buffer before negative staining (Aebi et al. 1983). However, it was not clear in what order tetramers would associate laterally and longitudinally to form an IF. The idea of elongated protofibrils serving as IF precursors was attractive as the nuclear IF protein lamin A had been shown to form, under a certain assembly regime, long strands of head-to-tail–associated dimers that eventually would laterally associate in an antiparallel manner to yield filamentous structures, although without reaching a “steady state” in the form of a structurally distinct filament with a uniform 10-nm diameter (Aebi et al. 1986; Heitlinger et al. 1991; Foeger et al. 2006; Ben-Harush et al. 2009).
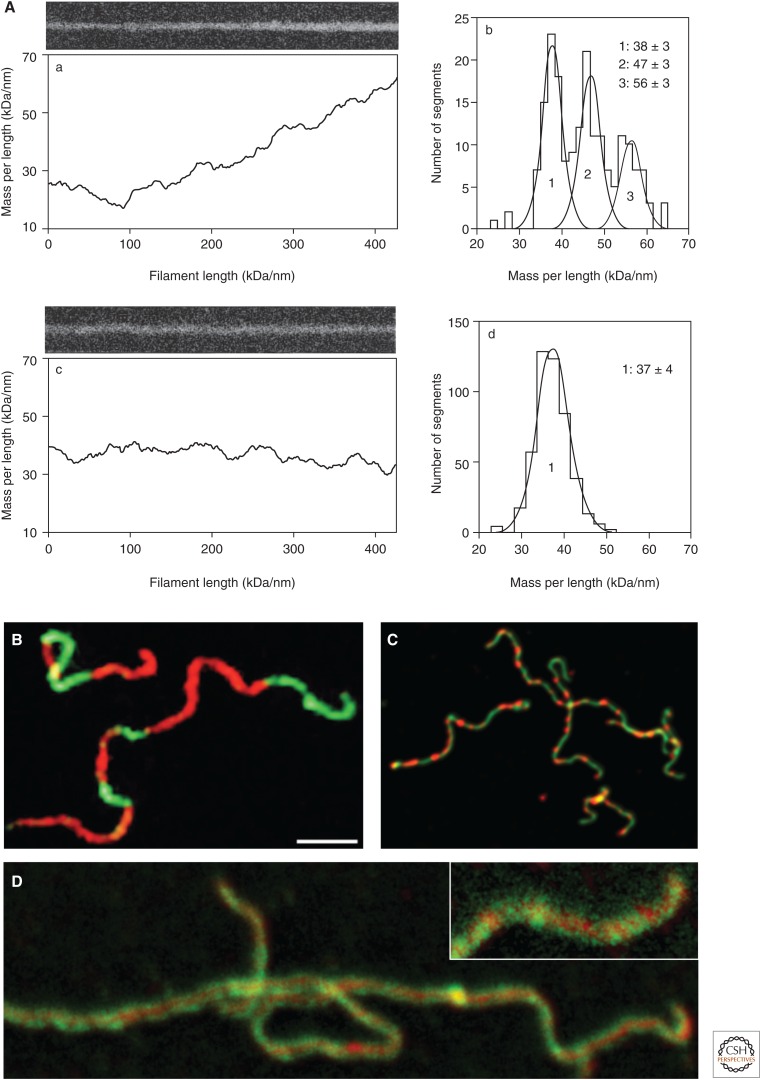
However, although cytoplasmic IFs looked rather uniform, determination of MPL by STEM showed that they were “polymorphic” (i.e., distinct segments of individual filaments contained different numbers of subunits per filament cross section), although this polymorphism was less evident with directly isolated (native) filaments compared with IFs reassembled from renatured subunits (Steven et al. 1982). Hence, the molecular mechanism of assembly was not immediately evident. As it turned out, the mode of assembly very strongly impacts on the homogeneity of the filaments formed. For example, “kick-start” of assembly by the addition of a concentrated salt solution to tetramers to establish “assembly buffer” conditions yields filaments with varying MPLs along one and the same filament (Fig. 4A,a,b). In contrast, assembly of tetramers by dialysis into assembly buffer results in homogeneous filaments in terms of their MPL (Fig. 4A,c,d) (Herrmann et al. 1996).
Figure 4.
Determination of intermediate filament (IF) architecture by complementary experimental methods. (A) Measuring the mass-per-length (MPL) of IFs by scanning transmission electron microscopy (STEM). (Left) Dark-field electron micrographs of unstained freeze-dried single IFs (top) together with the corresponding MPL tracings (bottom). (Right) MPL histograms obtained from measuring several hundred segments along many filaments. Fitting these histograms by multiple Gaussian curves reveals distinct MPL peak values (in kDa/nm). Assembly was performed either by dilution (“kick-start” mode; a,b) or by dialysis (“slow” mode; c,d) of tetramers from low-salt buffer into filament-forming buffer, as described by Herrmann et al. (1996). (B) Red and green fluorescently labeled vimentin were each assembled separately for 1 h before being mixed to trace how assembly continued over the next two days. Inspection of the assembly products in sample chambers confined in height, so that long filaments could swiftly equilibrate on the surface, was performed by total internal reflection fluorescence (TIRF) microscopy. Scale bar, 3 µm. (Reprinted from Winheim et al. 2011.) (C) IFs obtained from cultured human rhabdomyosarcoma-derived (RD) cells in the course of their extraction with 0.2% NP40 were fluorescently labeled with antibodies directed against vimentin (green) and desmin (red). (D) Single vimentin IFs were double-labeled with a mouse monoclonal antibody detecting an epitope at the beginning of coil 2 of the central rod (red) and a rabbit serum detecting the last nine amino acids of the carboxy-terminal tail domain (green). The filaments in C and D are ∼20-µm long; imaging was by stimulated emission depletion (STED) microscopy. The inset clearly depicts the peripheral space occupied by the tips of the carboxy-terminal vimentin tail domains around the filament core. (The micrograph in C was provided by Michaela Hergt [B065, DKFZ]; the micrograph in D was produced by Michaela Hergt [B065, DKFZ] in collaboration with Jasmin Zahn [E190, DKFZ]).
8. ASSEMBLY MECHANISM AND KINETICS
One way to follow the assembly of IFs over time is by turbidometric methods, in which changes of light scattering are followed over time at 300 nm (Steinert and Gullino 1976; Steinert et al. 1976). However, the temporal resolution achieved in these early experiments was not very high because assembly was followed in the minute-to-hour range and because soluble complexes were prepared by “desalting” urea-denatured molecules into assembly buffer using molecular-sieving columns, thus, starting assembly in the column. In any case, it was shown that, at physiological protein concentrations (1–2 mg/mL), IF assembly reached a plateau by ∼2 h, indicating that, by this time, the assembly precursors were used up.
In an attempt to identify IF assembly intermediates, quick-freeze/deep-etch EM was performed on vimentin assemblies that were formed by dialysis of tetramers that had been renatured into a buffer containing 10 mm Tris-acetate buffer (pH 8.5), 50 mm NaCl, and 1 mm MgCl2 (Ip et al. 1985). This revealed 60-nm-long filaments that produced mature IFs when further dialyzed into a buffer of physiological ionic strength and pH (170 mm NaCl, 10 mm imidazole, pH 7.0). However, these studies did not reveal assembly intermediates, and the underlying assumption that tetramers are in the A12 form turned out to be incorrect (see Mücke et al. 2004a).
To follow quantitatively the kinetics of assembly of vimentin, viscometry was performed by a kick-start assembly schedule (Hofmann et al. 1991). In this, tetramers were “forced” to assemble by the addition of a concentrated buffered salt solution, leading to the instantaneous increase of the ionic strength from 5 mm Tris-HCl to 25 mm Tris-HCl plus 160 mm NaCl and a simultaneous drop of the pH value from 8.4 to 7.5. Visual inspection of the mature filament assemblies by EM revealed them to be homogeneous and aggregate-free. With a standard Ostwald capillary viscometer, the time resolution was in the minute range: Typically, measurements were performed at 1 min after initiation of assembly, and then every 5 min. Depending on the ionic conditions and temperature, the increase in relative viscosity was very fast and leveled off by 10 min of assembly, indicating that, on average, the filaments had reached a length larger than the mesh size and thereby had established the properties of a network (Herrmann et al. 1999). Accordingly, the mesh size of vimentin IFs was determined to be 200 nm (at 0.5 mg/mL) and 150 nm (at 2 mg/mL) under standard assembly conditions in a rheometer by small-amplitude oscillatory shear measurements (Schopferer et al. 2009).
The mechanism underlying this network formation is based on the rapid lateral association of A11 tetramers (Fig. 1C) into ULFs (Fig. 2C). The formation and further growth of these “immediate early” assembly products was documented by a time-lapse EM approach. Accordingly, the assembly reaction was quenched after initiation of assembly from one second onward by addition of assembly buffer containing the strong fixative glutaraldehyde at various time points (Herrmann et al. 1996). During the first minute of assembly, electron micrographs also yielded a “background” of fibers representing tetrameric assembly products beyond the ULF state. After one minute, this background had disappeared, indicating the consumption of the ULF-assembly precursors. As measured in a quantitative study, the mean filament length grew from 55 nm at 5 sec to 130 nm by 1 min, indicating that these filaments on average consist of three ULFs (Kirmse et al. 2007). Note that, during longitudinal annealing of two ULFs, the length grows only by 43 nm because of the overlap of the two coil 2 segments in the annealing zone. Hence, a “double” ULF is ∼100-nm long (55 nm plus 43 nm). Every further addition of an ULF yields a 43-nm increase in length.
9. QUANTITATIVE DESCRIPTION AND KINETIC MODELING OF ASSEMBLY
To describe the kinetics of assembly, a suite of different types of complexes that can theoretically assemble from tetramers, such as 8-mers, 12-mers, 16-mers, and further multiples of 4, was investigated. The resulting association constants were applied to the experimentally derived mean length values and compared with a scenario in which tetramers predominantly formed eight-mers and multiples thereof, which eventually yielded the best approximation to the measured assembly data (Kirmse et al. 2007). In a next step, the assembly process was modeled according to the experimentally determined filament length histograms at different time points and by considering geometrical constraints, as well as the diffusion behavior of rod-like aggregates (Portet et al. 2009). This way, it became clear how the measured IF assembly differs from the monomer-addition-type polymerization shown, for example, by actin and tubulin.
With respect to the late stage of IF assembly, it was shown by TIRF microscopy of preassembled IFs that filaments of micrometer length and longer do longitudinally anneal, although it takes days for a significant number of elongation events to occur (Winheim et al. 2011). By this approach, IFs exceeding 10 µm in length were obtained, formed by micrometer-long alternating red-fluorescent and green-fluorescent segments (Fig. 4B). Furthermore, it has directly been demonstrated using fluorescently labeled vimentin that in vitro the exchange of subunits from assembled filaments is rather low (Nöding et al. 2014). The segmental organization of IFs has also been visualized in IFs of cultured cells. In the case of human rhabdomyosarcoma-derived RD cells, which contain both vimentin and the muscle-specific desmin, individual IFs show alternating segments, being either rich in vimentin (green) or in desmin (red) indicating that vimentin and desmin segregate from each other up to the level of ULFs (Fig. 4C) (M Hergt and H Herrmann, unpubl.).
In summary, the work on IF assembly clearly revealed that a “nucleation”-type of reaction, as observed for actin and tubulin, does not hold true for vimentin. Referring to this different assembly mechanism of IFs, it has been mooted by Douglas that the complex evolution in the assembly dynamics of the IFs, as proposed by Portet and colleagues (Portet et al. 2009), is probably more of a norm than an exception (Douglas 2009).
In the case of the cytoplasmic IFs, ULFs represent the basic building blocks or “monomers” of the assembly process. However, in sharp contrast to dynamic polymer situations, on initiation of assembly, growing filaments hardly release subunits such as ULFs, and the equilibrium is very much on the side of the “polymer.” In fact, the magnitude of this exchange reaction was determined to be typically one tetramer per micron of filament per hour, which is equivalent to one out of 200 tetramers per hour (Nöding et al. 2014; Köster et al. 2015). The reason for this behavior might reside in the fact that IF proteins are polyelectrolytes with a negatively charged rod domain and a very basic head domain. As a consequence, the IF is built as a complex network of ionic interactions formed by the acidic rod and the basic head domain, complemented by additional hydrophobic interactions of the head and the rod with neighboring subunits within the filament. Hence, IFs formed in vitro cannot be dissolved by incubation with high concentrations of nonionic detergents and salt (1% NP40, 1.5 m KCl); however, phosphorylation by protein kinases dissolves IFs very effectively, as first shown for vimentin IFs (Inagaki et al. 1987). In cells, additional mechanisms using various posttranslational modifications exist to remodel IF networks dynamically (Snider and Omary 2014).
10. MECHANICAL PROPERTIES OF INTERMEDIATE FILAMENTS AND NETWORK BEHAVIOR
An important organizational feature of IFs is the positioning of the protein tail domain within the filament. By stimulated emission depletion microscopy with domain-specific antibodies, the tips of the protein tail domain appear to form a peripheral “cloud” around the core of the filament (Fig. 4D). Also, a direct demonstration of the tails protruding from the filament axis was shown by EM after glycerol spraying coupled with rotary metal shadowing of filaments assembled from the low-molecular-weight and high-molecular-weight forms of the neurofilament triplet proteins (NF-L, NF-H) formed in vitro. As documented in Figure 2F, the long NF-H tails project radially, thereby giving the neurofilaments a millipede-like appearance (see also Hisanaga and Hirokawa 1988; Heins and Aebi 1994). Furthermore, the complex interaction of the tail domains in filaments made from NF-H, NF-L, the medium-molecular-weight neurofilament protein NF-M, and another neuron-specific IF protein, α-internexin, has recently been investigated by small-angle X-ray scattering experiments, yielding a concept for their complex and precisely tunable function as elements in a composite “bottlebrush-type” network (Kornreich et al. 2015).
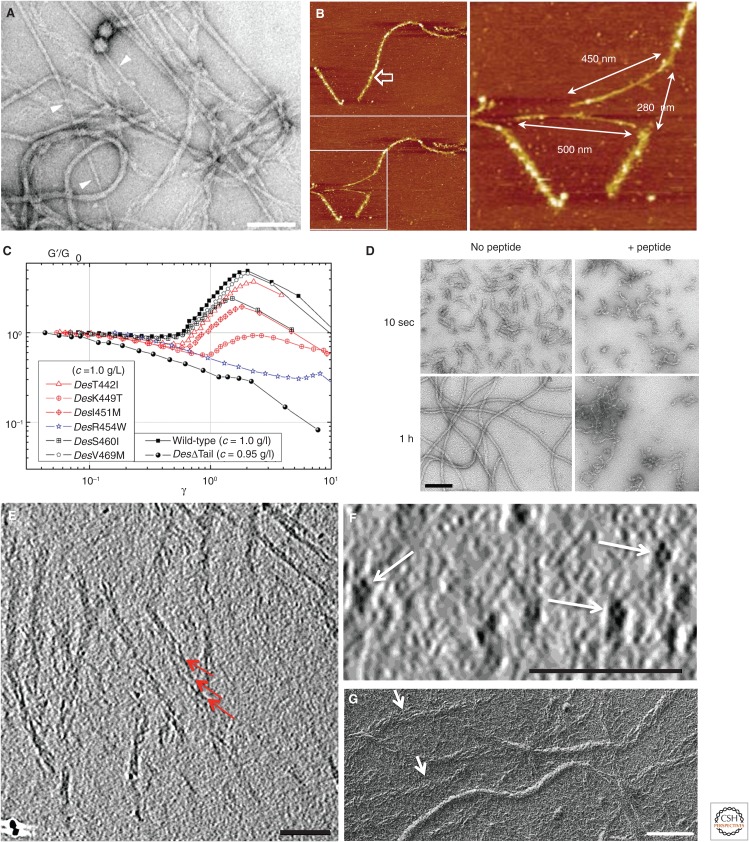
The tail domains are probably of general significance for the unusual resistance of IFs to mechanical stress displayed, for example, when filaments are stretched in bulk on an EM grid (Fig. 5A) or manipulated individually by the tip of an atomic force microscope (AFM) (Fig. 5B). By TEM, stretched IFs show a shrinking of their diameter, typically from 12 to 4 nm (Fig. 5A, arrowheads). A similar shrinking is observed by AFM, in which individual IFs can be stretched by up to 250% before they rupture (Fig. 5B) (Kreplak et al. 2005).
Figure 5.
Mechanical properties of intermediate filaments (IFs). (A) A desmin IF network stretched on a glow-discharged carbon-coated copper grid followed by glutaraldehyde fixation and negative staining for electron microscopy. Scale bar, 100 nm. The arrowheads point to stretched IFs with a diameter of ∼4 nm. For comparison, the average diameter of the unstretched IFs is ∼12 nm. (Reprinted from Kreplak et al. 2008.) (B) Native rat neurofilaments were subjected to pulling by an atomic force microscope (AFM) tip with a lateral force of 30–40 nN and a scanning speed of 40 nm/sec. The upper left panel shows the filament before being pulled; the lower left image documents the breakage of the filament through the action of the AFM cantilever after being operated under high force; the white boxed area is shown enlarged on the right: arrows indicate the length of the stretched filament segments after pulling and breaking, as well as the length of the original filament segment, now appearing as a gap of 280 nm. (Reprinted from Kreplak et al. 2005.) (C) Rheological investigation of the effect of disease-causing point mutations in the human desmin tail domain (symbols in left box) on IF strain stiffening when compared with wild-type and tailless desmin (symbols in right box). The reduced storage modulus G′/G0 is depicted as a function of the strain amplitude γ. (Adapted from Bär et al. 2010.) Note that tailless desmin shows no strain stiffening at all. (D) Inhibition of vimentin filament assembly in the presence of a peptide representing the last 58 amino acids of coil 2, as described in Strelkov et al. 2002. Note that the presence of this peptide effectively prevents longitudinal annealing of unit-length filaments (ULFs), but not their formation by lateral association of tetramers. (E,F) Cryo-electron tomography of unfixed, unstained recombinant human vimentin assembled for 1 h at 37°C under standard IF-forming conditions (see Goldie et al. 2007). In E, a selected x–y slice through a tomographic reconstruction stack is shown. In this view, filaments are either compact or on occasion partially unraveled, thereby exposing individual octameric protofibrils that show a twisted arrangement (red arrows). In F, an x–z slice of the same tomographic reconstruction stack is shown, yielding a prominent packing pattern of cross-sectioned IFs, with four protofibrils per filament (white arrows). Scale bars, 100 nm. (G) Unidirectionally metal-shadowed vimentin IFs were obtained from plunge-frozen/vitrified samples after freeze-drying (see Kirmse et al. 2010). Arrows point to partially unraveled IFs. Scale bar, 100 nm. (E,F, Adapted from Goldie et al. 2007; G, provided by Andreas Hoenger, University of Colorado at Boulder, Boulder, CO.)
Because of their exposure to the filament surface, the IF tails have important roles in the gelation—or network formation—of long filaments (Beck et al. 2010). Although the IF tail domain appears to control filament width (see below), it can be completely “deleted” by recombinant techniques and the resulting tailless proteins still assembled perfectly (Eckelt et al. 1992; Rogers et al. 1995; Herrmann et al. 1996). In contrast, the biomechanical properties of, for example, “tailless” desmin filaments are completely different from those formed by the wild-type protein. Instead of becoming stiffer, like wild-type desmin networks, when being mechanically stressed, tailless desmin filament networks actually become softer (Fig. 5C; compare black circles with black squares) (see also Schopferer et al. 2009). The behavior of filamentous networks to increase in viscoelasticity on being mechanically stressed is also referred to as “strain stiffening” (Janmey et al. 1991). As an important medical consequence, desmin tail mutations causing both cardiomyopathy and skeletal muscle disease show altered strain-stiffening behavior (Fig. 5C) (Bär et al. 2010).
MPL measurements by STEM of tail-truncated vimentin variants made it clear that the first 30 amino acids of the tail domain are directly engaged in the packing of the coiled-coil domains within the filament core. Hence, tailless vimentin contains more tetramers per filament cross section than wild-type vimentin (i.e., 12 compared with eight to nine tetramers) when assembly is performed in conjunction with dialysis (Herrmann et al. 1996).
Despite their high resistance to mechanical stress, IFs are highly dynamic polymers both in vivo and in vitro. Hence, by simply challenging them with peptides (e.g., one representing the last 58 amino acids of coil 2 that harbors the IF consensus motif 2 [see Fig. 1A]), mature filaments will disintegrate in seconds (Strelkov et al. 2002). When added during assembly, these peptides allow the formation of ULF-like structures but completely inhibit their longitudinal annealing (Fig. 5D). A molecular basis for such a drastic effect might lie in the fact that IFs are built from four protofibrillar strands, with appropriate space around and within the individual protofibrils. Hence, the dimeric head-to-tail overlaps within a protofibril can be rather freely accessed by the peptides. The impact of these peptides on cellular organization has also been shown directly in vivo, when, after its microinjection into serum-starved cells, the entire vimentin IF network retracted into the nuclear periphery and the cells showed extensive membrane ruffling (Helfand et al. 2011).
The subfilamentous structure of vimentin IFs is documented in fine detail by cryo-electron tomography of unfixed, unstained specimens. Accordingly, in selected x–y slices through tomographic reconstruction stacks, some filament segments appear flexible and partially unraveled, whereas others look stiffer and compact (Fig. 5E). Similarly, in selected x–z slices of the same tomographic reconstruction stacks, a prominent packing pattern of cross-sectioned vimentin IFs with typically four protofibrils per filament is evident (Fig. 5F; white arrows). When the same unfixed vimentin IF samples were adsorbed to carbon-coated grids, blotted to remove excess liquid, quick-frozen in liquid nitrogen, and unidirectionally metal shadowed, again a mixture of compact and partially unraveled filament segments was revealed by TEM (Fig. 5G) (see also Kirmse et al. 2010). Notably, as described above, brief incubation of keratin IFs with phosphate buffer on adsorption to an EM grid yielded their subfilamentous nature, too (Aebi et al. 1983).
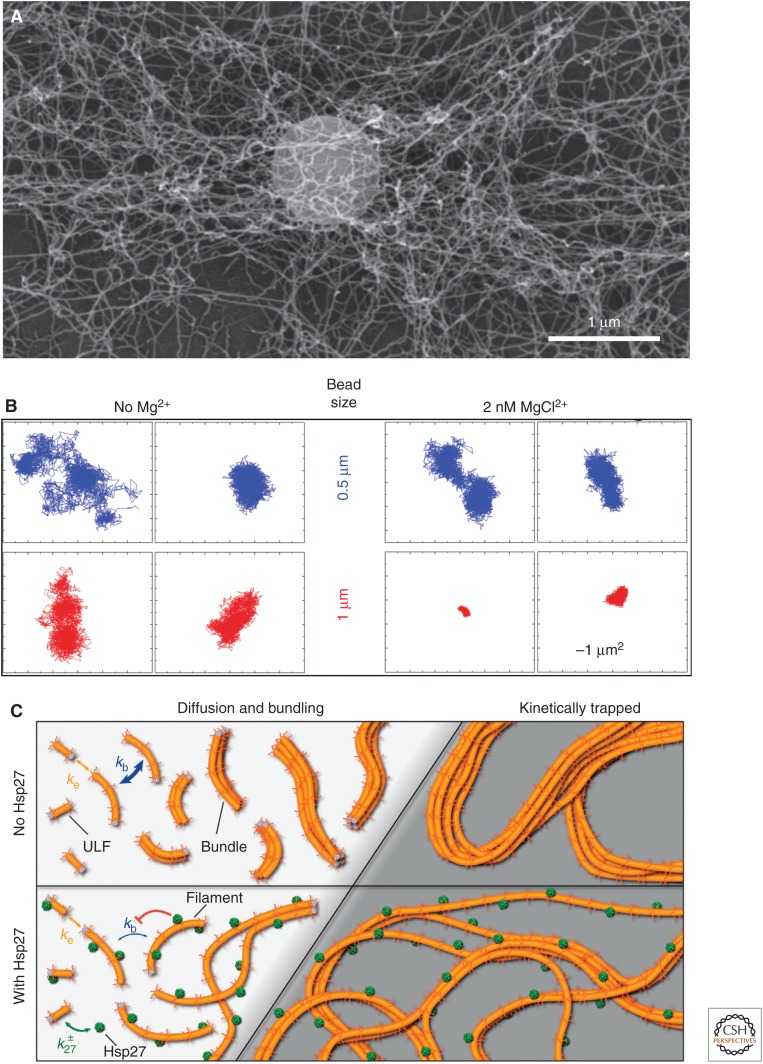
The network’s keratin IF’s form is visualized well by scanning electron microscopy—see, for example, the network with an entrapped microsphere in Figure 6A. The degree of filament–filament interaction is distinctly influenced by the ionic conditions—in particular, by the presence of divalent cations such as Mg2+ and Ca2+ (Hofmann et al. 1991; Köster et al. 2010; Lin et al. 2010; Kayser et al. 2012; Leitner et al. 2012). Hence, depending on the Mg2+ concentration, the mesh size of the IF networks can be varied systematically, as shown by microrheology (Fig. 6B). A “negative” kind of regulation in both bundling and network formation of keratin IFs can be evoked in vitro by the small heat-shock protein Hsp27 (Fig. 6C). Here, the binding of Hsp27 to the keratin K8 tail prevents a “kinetic trapping” reaction because of bundling of short elongating filaments in competition with longitudinal annealing (Kayser et al. 2013). Small heat-shock proteins are also expressed prominently in muscle, and mutations in one of them, αB-crystallin, have been discovered to cause effects in myofibrillar myopathies similar to those caused by desmin mutations, indicating that they are authentic regulators of IF properties in muscle (Clemen et al. 2013).
Figure 6.
Network formation and role of physiological effectors. (A) Scanning electron micrograph of keratin K8/K18 filaments entrapping a 1-µm microsphere, thereby enabling the investigation of network properties by microrheology. (Image courtesy of Ines Martin, University of Ulm, Germany.) (B) Effect of magnesium ions (Mg2+) on vimentin filament networks. Microspheres of different diameter were tracked in the absence (left) and presence (right) of 2 mm MgCl2. The two images for every bead size exemplify the degree of restriction of diffusible space for the corresponding microspheres. Hence, at 2 mm MgCl2, the space is essentially confined for the 1-µm microsphere, whereas the movement of the 0.5-µm microsphere is hardly affected. (Adapted from Köster et al. 2010.) (C) Small heat-shock proteins modulate keratin K8/K18 IF bundling. The presence of Hsp27 impedes the bundling reaction, as the heat-shock protein affects the kinetics of filament formation by interacting with the K8 tail domain. (Reprinted from Kayser et al. 2013.)
Recent studies aimed at addressing the question of how IFs behave in the context of other filament systems, such as actin filaments, have been performed (Jensen et al. 2014). The results show that different cytoskeletal systems influence each other in steric ways; however, a clear concept as to what this means has yet to be formulated. Nevertheless, it has become evident that it is the direct physical interaction between the three major cytoskeletal filament systems that comprises the primary determinants of the mechanical properties of cells and tissues (Huber et al. 2015).
11. CONCLUSION
Research on IF proteins is coming of age. What started with peculiar diffraction patterns, whose beauty and simplicity was the subject of the Ph.D. thesis of one of the greatest biologists of our times—Francis Crick—has now reached a phase in which we investigate the role of these simple coiled-coil proteins in a multitude of biological functions. Indeed, most of these functions are not merely “structural” but often “functional” as well, reflecting that these two terms are strongly coupled. For instance, the functional architecture of the epidermis is highly dependent on the proper keratin network organization. Likewise, the ability of an eagle to generate feathers depends on the intricate interaction of epithelial and mesenchymal cell layers to organize two types of keratins into these complex biomaterials. Now, the role that IFs play in organogenesis and in human disease needs to be explained in molecular terms. Most important in this ongoing endeavor is to explore IF structure, function, and dynamics more systematically in the cellular and tissue context and to consequently complement the emerging findings with corresponding ex vivo and in vitro studies.
ACKNOWLEDGMENTS
H.H. received ongoing support from the German Research Foundation (DFG, HE-1853), the DKFZ-MOST, and the German–Israel Foundation (GIF). U.A. thanks the Swiss National Science Foundation, the Canton Basel-Stadt, and the M.E. Müller Foundation of Switzerland for constant support. Both H.H. and U.A. had the joy to take part in a European Union’s Framework Programme 6 project (STREP) on laminopathies. H.H. wants to thank, in particular, his long-term coworkers on structure and assembly, namely, Monika Mauermann (neé Brettel), Tatjana Wedig, Tanja Tourgaidis (neé Lichtenstern), Dorothee Möller, and Michaela Hergt, for superb assistance. U.A. acknowledges the enthusiastic participation of Ariel Lustig in many of the fundamental studies over the past 20 years. To both of us, the late Markus Häner (Biozentrum Basel) was a wonderful colleague with unbreakable joy for doing experiments, lots of ideas, and incredible technical skills.
Footnotes
Editors: Thomas D. Pollard and Robert D. Goldman
Additional Perspectives on The Cytoskeleton available at www.cshperspectives.org
REFERENCES
*Reference is in this collection.
- Aebi U, Baschong W. 2006. Glycerol spraying/low-angle rotary metal shadowing. In Cell biology—A laboratory handbook, 3rd ed. (ed. Celis JE), pp. 241–246. Elsevier, Amsterdam. [Google Scholar]
- Aebi U, Fowler WE, Rew P, Sun TT. 1983. The fibrillar substructure of keratin filaments unraveled. J Cell Biol 97: 1131–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi U, Cohn J, Buhle L, Gerace L. 1986. The nuclear lamina is a meshwork of intermediate-type filaments. Nature 323: 560–564. [DOI] [PubMed] [Google Scholar]
- Astbury WT. 1933. Some insight information about the properties of the wool fibre. In Fundamentals of fibre structure (Lecture VI), Fundamentals of fibre structure (with an Introduction by Sir William Bragg), pp. 134–179. Oxford University Press/Humphrey Milford, London. [Google Scholar]
- Astbury WT. 1939. X-ray studies of the structure of compounds of biological interest. Annu Rev Biochem 8: 113–133. [Google Scholar]
- Aziz A, Hess JF, Budamagunta MS, Voss JC, Kuzin AP, Huang YJ, Xiao R, Montelione GT, FitzGerald PG, Hunt JF. 2012. The structure of vimentin linker 1 and rod 1B domains characterized by site-directed spin-labeling electron paramagnetic resonance (SDSL-EPR) and X-ray crystallography. J Biol Chem 287: 28349–28361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bär H, Mücke N, Kostareva A, Sjöberg G, Aebi U, Herrmann H. 2005. Severe muscle disease-causing desmin mutations interfere with in vitro filament assembly at distinct stages. Proc Natl Acad Sci 102: 15099–15104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bär H, Schopferer M, Sharma S, Hochstein B, Mücke N, Herrmann H, Willenbacher N. 2010. Mutations in desmin’s carboxy-terminal “tail” domain severely modify filament and network mechanics. J Mol Biol 397: 1188–1198. [DOI] [PubMed] [Google Scholar]
- Beck R, Deek J, Jones JB, Safinya CR. 2010. Gel-expanded to gel-condensed transition in neurofilament networks revealed by direct force measurements. Nat Mater 9: 40–46. [DOI] [PubMed] [Google Scholar]
- Ben-Harush K, Wiesel N, Frenkiel-Krispin D, Moeller D, Soreq E, Aebi U, Herrmann H, Gruenbaum Y, Medalia O. 2009. The supramolecular organization of the C. elegans nuclear lamin filament. J Mol Biol 386: 1392–1402. [DOI] [PubMed] [Google Scholar]
- Burkhard P, Stetefeld J, Strelkov SV. 2001. Coiled coils: A highly versatile protein folding motif. Trends Cell Biol 11: 82–88. [DOI] [PubMed] [Google Scholar]
- Chernyatina AA, Strelkov SV. 2012. Stabilization of vimentin coil2 fragment via an engineered disulfide. J Struct Biol 177: 46–53. [DOI] [PubMed] [Google Scholar]
- Chernyatina AA, Nicolet S, Aebi U, Herrmann H, Strelkov SV. 2012. Atomic structure of the vimentin central α-helical domain and its implications for intermediate filament assembly. Proc Natl Acad Sci 109: 13620–13625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyatina AA, Guzenko D, Strelkov SV. 2015. Intermediate filament structure: The bottom-up approach. Curr Opin Cell Biol 32: 65–72. [DOI] [PubMed] [Google Scholar]
- Chou CC, Buehler MJ. 2012. Structure and mechanical properties of human trichocyte keratin intermediate filament protein. Biomacromolecules 13: 3522–3532. [DOI] [PubMed] [Google Scholar]
- Chou PY, Fasman GD. 1978. Empirical predictions of protein conformation. Annu Rev Biochem 47: 251–276. [DOI] [PubMed] [Google Scholar]
- Chrétien D, Metoz F, Verde F, Karsenti E, Wade RH. 1992. Lattice defects in microtubules: Protofilament numbers vary within individual microtubules. J Cell Biol 117: 1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemen CS, Herrmann H, Strelkov SV, Schröder R. 2013. Desminopathies: Pathology and mechanisms. Acta Neuropathol 125: 47–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemen CS, Stöckigt F, Strucksberg KH, Chevessier F, Winter L, Schütz J, Bauer R, Thorweihe JM, Wenzel D, Schlötzer-Schrehardt U, et al. 2015. The toxic effect of R350P mutant desmin in striated muscle of man and mouse. Acta Neuropathol 129: 297–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JF, Parry DA. 1988. Intermediate filament structure. 3. Analysis of sequence homologies. Int J Biol Macromol 10: 79–98. [Google Scholar]
- Crick FH. 1952. Is α-keratin a coiled coil? Nature 170: 882–883. [DOI] [PubMed] [Google Scholar]
- Crick FH. 1953. The packing of α-helices: Simple coiled-coils. Acta Cryst 6: 689–697. [Google Scholar]
- Dahlstrand J, Zimmerman LB, McKay RD, Lendahl U. 1992. Characterization of the human nestin gene reveals a close evolutionary relationship to neurofilaments. J Cell Sci 103: 589–597. [DOI] [PubMed] [Google Scholar]
- Dodemont H, Riemer D, Ledger N, Weber K. 1994. Eight genes and alternative RNA processing pathways generate an unexpectedly large diversity of cytoplasmic intermediate filament proteins in the nematode Caenorhabditis elegans. EMBO J 13: 2625–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R, Holmes KC. 2011. Actin structure and function. Annu Rev Biophys 40: 169–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas JF. 2009. Theoretical issues relating to thermally reversible gelation by supermolecular fiber formation. Langmuir 25: 8386–8391. [DOI] [PubMed] [Google Scholar]
- Eckelt A, Herrmann H, Franke WW. 1992. Assembly of a tailless mutant of the intermediate filament protein, vimentin, in vitro and in vivo. Eur J Cell Biol 58: 319–330. [PubMed] [Google Scholar]
- Eichner R, Rew P, Engel A, Aebi U. 1985. Human epidermal keratin filaments: Studies on their structure and assembly. Ann N Y Acad Sci 455: 381–402. [DOI] [PubMed] [Google Scholar]
- Erber A, Riemer D, Hofemeister H, Bovenschulte M, Stick R, Panopoulou G, Lehrach H, Weber K. 1999. Characterization of the Hydra lamin and its gene: A molecular phylogeny of metazoan lamins. J Mol Evol 49: 260–271. [DOI] [PubMed] [Google Scholar]
- Foeger N, Wiesel N, Lotsch D, Mücke N, Kreplak L, Aebi U, Gruenbaum Y, Herrmann H. 2006. Solubility properties and specific assembly pathways of the B-type lamin from Caenorhabditis elegans. J Struct Biol 155: 340–350. [DOI] [PubMed] [Google Scholar]
- Foisner R, Leichtfried FE, Herrmann H, Small JV, Lawson D, Wiche G. 1988. Cytoskeleton-associated plectin: In situ localization, in vitro reconstitution, and binding to immobilized intermediate filament proteins. J Cell Biol 106: 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke WW, Schiller DL, Grund C. 1982. Protofilamentous and annular structures as intermediates during reconstitution of cytokeratin filaments in vitro. Biologie Cellulaire 46: 257–268. [Google Scholar]
- Fraser RD, MacRae TP, Miller A. 1964a. The Fourier transform of the coiled-coil model for α-keratin. Acta Cryst 17: 813–816. [DOI] [PubMed] [Google Scholar]
- Fraser RD, MacRae TP, Miller A. 1964b. The coiled-coil model of α-keratin structure. J Mol Biol 10: 147–156. [DOI] [PubMed] [Google Scholar]
- Fraser RD, MacRae TP, Suzuki E, Parry DA. 1985. Intermediate filament structure. 2. Molecular interactions in the filament. Int J Biol Macromol 7: 258–274. [Google Scholar]
- Fuchs E. 2009. The tortoise and the hair: Slow-cycling cells in the stem cell race. Cell 137: 811–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler N, Weber K. 1981. Comparison of the proteins of two immunologically distinct intermediate-sized filaments by amino acid sequence analysis: Desmin and vimentin. Proc Natl Acad Sci 78: 4120–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler N, Weber K. 1982. The amino acid sequence of chicken muscle desmin provides a common structural model for intermediate filament proteins. EMBO J 1: 1649–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler N, Schünemann J, Weber K. 1992. Chemical cross-linking indicates a staggered and antiparallel protofilament of desmin intermediate filaments and characterizes one higher-level complex between protofilaments. Eur J Biochem 206: 841–852. [DOI] [PubMed] [Google Scholar]
- Geisler N, Schünemann J, Weber K, Häner M, Aebi U. 1998. Assembly and architecture of invertebrate cytoplasmic intermediate filaments reconcile features of vertebrate cytoplasmic and nuclear lamin-type intermediate filaments. J Mol Biol 282: 601–617. [DOI] [PubMed] [Google Scholar]
- Georgakopoulou SD, Möller D, Sachs N, Herrmann H, Aebi U. 2009. Near-UV circular dichroism reveals structural transitions of vimentin subunits during intermediate filament assembly. J Mol Biol 386: 544–553. [DOI] [PubMed] [Google Scholar]
- Goldie KN, Wedig T, Mitra A, Aebi U, Herrmann H, Hoenger A. 2007. Dissecting the 3-D structure of vimentin intermediate filaments by cryo-electron tomography. J Struct Biol 158: 378–385. [DOI] [PubMed] [Google Scholar]
- Goldman RD. 1971. The role of three cytoplasmic fibers in BHK-21 cell motility. I. Microtubules and the effects of colchicine. J Cell Biol 51: 752–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwold MJ, Bao W, Jarvis ED, Hu H, Li C, Gilbert MT, Zhang G, Sawyer RH. 2014. Dynamic evolution of the alpha (α) and beta (β) keratins has accompanied integument diversification and the adaptation of birds into novel lifestyles. BMC Evol Biol 14: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg K, Wilton SD, Parry DAD, Rogers GE. 1984. A comparison of genomic coding sequences for feather and scale keratins: Structural and evolutionary implications. EMBO J 3: 175–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzfeld M, Franke WW. 1985. Pair formation and promiscuity of cytokeratins: Formation in vitro of heterotypic complexes and intermediate-sized filaments by homologous and heterologous recombinations of purified polypeptides. J Cell Biol 101: 1826–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzfeld M, Weber K. 1990. The coiled coil of in vitro assembled keratin filaments is a heterodimer of type I and II keratins: Use of site-specific mutagenesis and recombinant protein expression. J Cell Biol 110: 1199–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heins S, Aebi U. 1994. Making heads and tails of intermediate filament assembly, dynamics and networks. Curr Opin Cell Biol 6: 25–33. [DOI] [PubMed] [Google Scholar]
- Heitlinger E, Peter M, Häner M, Lustig A, Aebi U, Nigg EA. 1991. Expression of chicken lamin B2 in Escherichia coli: Characterization of its structure, assembly, and molecular interactions. J Cell Biol 113: 485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitlinger E, Peter M, Lustig A, Villiger W, Nigg EA, Aebi U. 1992. The role of the head and tail domain in lamin structure and assembly: Analysis of bacterially expressed chicken lamin A and truncated B2 lamins. J Struct Biol 108: 74–89. [DOI] [PubMed] [Google Scholar]
- Helfand BT, Mendez MG, Murthy SN, Shumaker DK, Grin B, Mahammad S, Aebi U, Wedig T, Wu YI, Hahn KM, et al. 2011. Vimentin organization modulates the formation of lamellipodia. Mol Biol Cell 22: 1274–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson D, Geisler N, Weber K. 1982. A periodic ultrastructure in intermediate filaments. J Mol Biol 155: 173–176. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Aebi U. 1998. Mechanism of intermediate filament assembly. Curr Opin Struct Biol 8: 177–185. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Aebi U. 2000. Intermediate filaments and their associates: Multi-talented structural elements specifying cytoarchitecture and cytodynamics. Curr Opin Cell Biol 12: 79–90. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Aebi U. 2004. Intermediate filaments: Molecular structure, assembly mechanism, and integration into functionally distinct intracellular scaffolds. Annu Rev Biochem 73: 749–789. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Wiche G. 1987. Plectin and IFAP-300K are homologous proteins binding to microtubule-associated proteins 1 and 2 and to the 240 kDa subunit of spectrin. J Biol Chem 262: 1320–1325. [PubMed] [Google Scholar]
- Herrmann H, Häner M, Brettel M, Müller S, Goldie K, Fedtke B, Lustig A, Franke WW, Aebi U. 1996. Structure and assembly properties of the intermediate filament protein vimentin: The role of its head, rod and tail domains. J Mol Biol 264: 933–953. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Häner M, Brettel M, Ku NO, Aebi U. 1999. Characterization of distinct early assembly units of different intermediate filament proteins. J Mol Biol 286: 1403–1420. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Strelkov SV, Feja B, Rogers KR, Brettel M, Lustig A, Häner M, Parry DAD, Steinert PM, Burkhard P, et al. 2000. The intermediate filament protein consensus motif of helix 2B: Its atomic structure and contribution to assembly. J Mol Biol 298: 817–832. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Strelkov SV, Burkhard P, Aebi U. 2009. Intermediate filaments: Primary determinants of cell architecture and plasticity. J Clin Invest 119: 1772–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisanaga S, Hirokawa N. 1988. Structure of the peripheral domains of neurofilaments revealed by low angle rotary shadowing. J Mol Biol 202: 297–305. [DOI] [PubMed] [Google Scholar]
- Hofmann I, Herrmann H, Franke WW. 1991. Assembly and structure of calcium-induced thick vimentin filaments. Eur J Cell Biol 56: 328–341. [PubMed] [Google Scholar]
- Huber F, Boire A, López MP, Koenderink GH. 2015. Cytoskeletal crosstalk: When three different personalities team up. Curr Opin Cell Biol 32: 39–47. [DOI] [PubMed] [Google Scholar]
- Hwang JS, Takaku Y, Chapman J, Ikeo K, David CN, Gojobori T. 2008. Cilium evolution: Identification of a novel protein, nematocilin, in the mechanosensory cilium of Hydra nematocytes. Mol Biol Evol 25: 2009–2017. [DOI] [PubMed] [Google Scholar]
- Inagaki M, Nishi Y, Nishizawa K, Matsuyama M, Sato C. 1987. Site-specific phosphorylation induces disassembly of vimentin filaments in vitro. Nature 328: 649–652. [DOI] [PubMed] [Google Scholar]
- Ip W, Hartzer MK, Pang YY, Robson RM. 1985. Assembly of vimentin in vitro and its implications concerning the structure of intermediate filaments. J Mol Biol 183: 365–375. [DOI] [PubMed] [Google Scholar]
- Ishikawa A, Bischoff R, Holtzer H. 1968. Mitosis and intermediate-sized filaments in developing skeletal muscle. J Cell Biol 38: 538–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey PA, Euteneuer U, Traub P, Schliwa M. 1991. Viscoelastic properties of vimentin compared with other filamentous biopolymer networks. J Cell Biol 113: 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MH, Morris EJ, Goldman RD, Weitz DA. 2014. Emergent properties of composite semiflexible biopolymer networks. Bioarchitecture 4: 138–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapinos LE, Schumacher J, Mücke N, Machaidze G, Burkhard P, Aebi U, Strelkov SV, Herrmann H. 2010. Characterization of the head-to-tail overlap complexes formed by human lamin A, B1 and B2 “half-minilamin” dimers. J Mol Biol 396: 719–731. [DOI] [PubMed] [Google Scholar]
- Karabinos A, Schünemann J, Meyer M, Aebi U, Weber K. 2003. The single nuclear lamin of Caenorhabditis elegans forms in vitro stable intermediate filaments and paracrystals with a reduced axial periodicity. J Mol Biol 325: 241–247. [DOI] [PubMed] [Google Scholar]
- Kayser J, Grabmayr H, Harasim M, Herrmann H, Bausch AB. 2012. Assembly kinetics determine the structure of keratin networks. Soft Matter 8: 8873. [Google Scholar]
- Kayser J, Haslbeck M, Dempfle L, Krause M, Grashoff C, Buchner J, Herrmann H, Bausch AR. 2013. The small heat shock protein Hsp27 affects assembly dynamics and structure of keratin intermediate filament networks. Biophys J 105: 1778–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmse R, Portet S, Mücke N, Aebi U, Herrmann H, Langowski J. 2007. The kinetics of intermediate filament assembly: Evidence for unit-length and filament end-to-end annealing as a mechanism for longitudinal growth of filaments. J Biol Chem 282: 18563–18572. [DOI] [PubMed] [Google Scholar]
- Kirmse R, Bouchet-Marquis C, Page C, Hoenger A. 2010. Three-dimensional cryo-electron microscopy on intermediate filaments. Methods Cell Biol 96: 565–589. [DOI] [PubMed] [Google Scholar]
- Klapper M, Exner K, Kempf A, Gehrig C, Stuurman N, Fisher PA, Krohne G. 1997. Assembly of A- and B-type lamins studied in vivo with the baculovirus system. J Cell Sci 110: 2519–2532. [DOI] [PubMed] [Google Scholar]
- Kollmar M. 2015. Polyphyly of nuclear lamin genes indicates an early eukaryontic origin of the metazoan-type intermediate filament proteins. Sci Rep 5: 10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornreich M, Malka-Gibor E, Laser-Azogui A, Doron O, Herrmann H, Beck R. 2015. Composite bottlebrush mechanics: α-internexin fine-tunes neurofilament network properties. Soft Matter 11: 5839–5849. [DOI] [PubMed] [Google Scholar]
- Köster S, Lin YC, Herrmann H, Weitz DA. 2010. Nanomechanics of vimentin intermediate filament networks. Soft Matter 6: 1910–1914. [Google Scholar]
- Köster S, Weitz DA, Goldman RD, Aebi U, Herrmann H. 2015. Intermediate filament mechanics in vitro and in the cell: From coiled coils to filaments, fibers and networks. Curr Opin Cell Biol 32: 82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreplak L, Doucet J, Dumas P, Briki F. 2004. New aspects of the α-helix to β-sheet transition in stretched hard α-keratin fibers. Biophys J 87: 640–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreplak L, Bär H, Leterrier JF, Herrmann H, Aebi U. 2005. Exploring the mechanical behavior of single intermediate filaments. J Mol Biol 354: 569–577. [DOI] [PubMed] [Google Scholar]
- Kreplak L, Richter K, Aebi U, Herrmann H. 2008. Electron microscopy of intermediate filaments: Teaming up with atomic force and confocal laser scanning microscopy. Methods Cell Biol 88: 273–297. [DOI] [PubMed] [Google Scholar]
- Lee CH, Kim MS, Chung BM, Leahy DJ, Coulombe PA. 2012. Structural basis for heteromeric assembly and perinuclear organization of keratin filaments. Nat Struct Mol Biol 19: 707–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner A, Paust T, Marti O, Walther P, Herrmann H, Beil M. 2012. Properties of intermediate filament networks assembled from keratin 8 and 18 in the presence of Mg²+. Biophys J 103: 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstern T, Mücke N, Aebi U, Mauermann M, Herrmann H. 2012. Complex formation and kinetics of filament assembly exhibited by the simple epithelial keratins K8 and K18. J Struct Biol 177: 54–62. [DOI] [PubMed] [Google Scholar]
- Lin YC, Yao NY, Broedersz CP, Herrmann H, Mackintosh FC, Weitz DA. 2010. Origins of elasticity in intermediate filament networks. Phys Rev Lett 104: 058101. [DOI] [PubMed] [Google Scholar]
- MacArthur I. 1943. Structure of α-keratin. Nature 152: 38–41. [Google Scholar]
- McLachlan AD, Stewart M, Smillie LB. 1975. Sequence repeats in α-tropomyosin. J Mol Biol 98: 281–291. [DOI] [PubMed] [Google Scholar]
- Meier M, Padilla GP, Herrmann H, Wedig T, Hergt M, Patel TR, Stetefeld J, Aebi U, Burkhard P. 2009. Vimentin coil 1A-A molecular switch involved in the initiation of filament elongation. J Mol Biol 390: 245–261. [DOI] [PubMed] [Google Scholar]
- Moll R, Franke WW, Schiller DL, Geiger B, Krepler R. 1982. The catalog of human cytokeratins: Patterns of expression in normal epithelia, tumors and cultured cells. Cell 31: 11–24. [DOI] [PubMed] [Google Scholar]
- Moll R, Divo M, Langbein L. 2008. The human keratins: Biology and pathology. Histochem Cell Biol 129: 705–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mücke N, Wedig T, Bürer A, Marekov LN, Steinert PM, Langowski J, Aebi U, Herrmann H. 2004a. Molecular and biophysical characterization of assembly-starter units of human vimentin. J Mol Biol 340: 97–114. [DOI] [PubMed] [Google Scholar]
- Mücke N, Kreplak L, Kirmse R, Wedig T, Herrmann H, Aebi U, Langowski J. 2004b. Assessing the flexibility of intermediate filaments by atomic force microscopy. J Mol Biol 335: 1241–1250. [DOI] [PubMed] [Google Scholar]
- Nicolet S, Herrmann H, Aebi U, Strelkov SV. 2010. Atomic structure of vimentin coil 2. J Struct Biol 170: 369–376. [DOI] [PubMed] [Google Scholar]
- Nöding B, Herrmann H, Köster S. 2014. Direct observation of subunit exchange along mature vimentin intermediate filaments. Biophys J 107: 2923–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea EK, Klemm JD, Kim PS, Alber T. 1991. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254: 539–544. [DOI] [PubMed] [Google Scholar]
- Parry DA, Fraser RD. 1985. Intermediate filament structure: 1. Analysis of IF protein sequence data. Int J Biol Macromol 7: 203–213. [Google Scholar]
- Parry DA, Steinert PM. 1995. Intermediate filament structure, pp. 1–183. R.G. Landes, Austin, TX. [Google Scholar]
- Pauling L, Corey RB. 1951. The pleated sheet, a new layer configuration of polypeptide chains. Proc Natl Acad Sci 37: 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauling L, Corey RB. 1953. Compound helical configurations of polypeptide chains: Structure of proteins of the α-keratin type. Nature 171: 59–61. [DOI] [PubMed] [Google Scholar]
- Peter A, Stick R. 2015. Evolutionary aspects in intermediate filament proteins. Curr Opin Cell Biol 32: 48–55. [DOI] [PubMed] [Google Scholar]
- *.Pollard T. 2016. Actin and actin-binding proteins. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portet S, Mücke N, Kirmse R, Langowski J, Beil M, Herrmann H. 2009. Vimentin intermediate filament formation: In vitro measurement and mathematical modeling of the filament length distribution during assembly. Langmuir 25: 8817–8823. [DOI] [PubMed] [Google Scholar]
- Pruss RM, Mirsky R, Raff MC, Thorpe R, Dowding AJ, Anderton BH. 1981. All classes of intermediate filaments share a common antigenic determinant defined by a monoclonal antibody. Cell 27: 419–428. [DOI] [PubMed] [Google Scholar]
- Quax-Jeuken YE, Quax WJ, Bloemendal H. 1983. Primary and secondary structure of hamster vimentin predicted from the nucleotide sequence. Proc Natl Acad Sci 80: 3548–3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan RA, Cohlberg JA, Schiller DL, Hatzfeld M, Franke WW. 1984. Heterotypic tetramer (A2D2) complexes of non-epidermal keratins isolated from cytoskeletons of rat hepatocytes and hepatoma cells. J Mol Biol 178: 365–388. [DOI] [PubMed] [Google Scholar]
- Renner W, Franke WW, Schmid E, Geisler N, Weber K, Mandelkow E. 1981. Reconstitution of intermediate-sized filaments from denatured monomeric vimentin. J Mol Biol 149: 285–306. [DOI] [PubMed] [Google Scholar]
- Riemer D, Weber K. 1998. Common and variant properties of intermediate filament proteins from lower chordates and vertebrates; two proteins from the tunicate Styela and the identification of a type III homologue. J Cell Sci 111: 2967–2975. [DOI] [PubMed] [Google Scholar]
- Rogers KR, Eckelt A, Nimmrich V, Janssen KP, Schliwa M, Herrmann H, Franke WW. 1995. Truncation mutagenesis of the non-α-helical carboxyterminal tail domain of vimentin reveals contributions to cellular localization but not to filament assembly. Eur J Cell Biol 66: 136–150. [PubMed] [Google Scholar]
- Rogers MA, Langbein L, Praetzel-Wunder S, Winter H, Schweizer J. 2006. Human hair keratin-associated proteins (KAPs). Int Rev Cytol 251: 209–263. [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Florens L, Guan T, Yates JR III, Gerace L. 2003. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 301: 1380–1382. [DOI] [PubMed] [Google Scholar]
- Schopferer M, Bär H, Hochstein B, Sharma S, Mücke N, Herrmann H, Willenbacher N. 2009. Desmin and vimentin intermediate filament networks: Their viscoelastic properties investigated by mechanical rheometry. J Mol Biol 388: 133–143. [DOI] [PubMed] [Google Scholar]
- Schweizer J, Langbein L, Rogers MA, Winter H. 2007. Hair follicle–specific keratins and their diseases. Exp Cell Res 313: 2010–2020. [DOI] [PubMed] [Google Scholar]
- Shumaker DK, Solimando L, Sengupta K, Shimi T, Adam SA, Grunwald A, Strelkov SV, Aebi U, Cardoso MC, Goldman RD. 2008. The highly conserved nuclear lamin Ig-fold binds to PCNA: Its role in DNA replication. J Cell Biol 181: 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small JV, Sobieszek A. 1977. Studies on the function and composition of the 10-nm (100-Å) filaments of vertebrate smooth muscle. J Cell Sci 23: 243–268. [DOI] [PubMed] [Google Scholar]
- Snider NT, Omary MB. 2014. Post-translational modifications of intermediate filament proteins: Mechanisms and functions. Nat Rev Mol Cell Biol 15: 163–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodek J, Hodges RS, Smillie LB, Jurasek L. 1972. Amino-acid sequence of rabbit skeletal tropomyosin and its coiled-coil structure. Proc Natl Acad Sci 69: 3800–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starger JM, Goldman RD. 1977. Isolation and preliminary characterization of 10-nm filaments from baby hamster (BHK-21) cells. Proc Natl Acad Sci 6: 2422–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starger JM, Brown WE, Goldman AE, Goldman RD. 1978. Biochemical and immunological analysis of rapidly purified 10-nm filaments from baby hamster kidney (BHK-21) cells. J Cell Biol 78: 93–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert PM. 1990. The two-chain coiled-coil molecule of native epidermal keratin intermediate filaments is a type I-type II heterodimer. J Biol Chem 265: 8766–8774. [PubMed] [Google Scholar]
- Steinert PM, Gullino MI. 1976. Bovine epidermal keratin filament assembly in vitro. Biochem Biophys Res Commun 70: 221–227. [DOI] [PubMed] [Google Scholar]
- Steinert PM, Idler WW, Zimmerman SB. 1976. Self-assembly of bovine epidermal keratin filaments in vitro. J Mol Biol 108: 547–567. [DOI] [PubMed] [Google Scholar]
- Steinert PM, Idler WW, Cabral F, Gottesman MM, Goldman RD. 1981. In vitro assembly of homopolymer and copolymer filaments from intermediate filament subunits of muscle and fibroblastic cells. Proc Natl Acad Sci 78: 3692–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert PM, Rice RH, Roop DR, Trus BL, Steven AC. 1983. Complete amino acid sequence of a mouse epidermal subunit and implications for the structure of intermediate filaments. Nature 302: 794–800. [DOI] [PubMed] [Google Scholar]
- Steven AC, Wall J, Hainfeld J, Steinert PM. 1982. Structure of fibroblastic intermediate filaments: Analysis by scanning transmission electron microscopy. Proc Natl Acad Sci 79: 3101–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steven AC, Hainfeld JF, Trus BL, Wall JS, Steinert PM. 1983. The distribution of mass in heteropolymer intermediate filaments assembled in vitro. Stem analysis of vimentin/desmin and bovine epidermal keratin. J Biol Chem 258: 8323–8329. [PubMed] [Google Scholar]
- Strelkov SV, Herrmann H, Geisler N, Lustig A, Ivaninskii S, Zimbelmann R, Burkhard P, Aebi U. 2001. Divide-and-conquer crystallographic approach towards an atomic structure of intermediate filaments. J Mol Biol 306: 773–781. [DOI] [PubMed] [Google Scholar]
- Strelkov SV, Herrmann H, Geisler N, Wedig T, Zimbelmann R, Aebi U, Burkhard P. 2002. Conserved segments 1A and 2B of the intermediate filament dimer: Their atomic structures and role in filament assembly. EMBO J 21: 1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strelkov SV, Schumacher J, Burkhard P, Aebi U, Herrmann H. 2004. Crystal structure of the human lamin A coil 2B dimer: Implications for the head-to-tail association of nuclear lamins. J Mol Biol 343: 1067–1080. [DOI] [PubMed] [Google Scholar]
- Szeverenyi I, Cassidy AJ, Chung CW, Lee BT, Common JE, Ogg SC, Chen H, Sim SY, Goh WL, Ng KW, et al. 2008. The Human Intermediate Filament Database: Comprehensive information on a gene family involved in many human diseases. Hum Mutat 29: 351–360. [DOI] [PubMed] [Google Scholar]
- Weber K, Geisler N. 1982. The structural relation between intermediate filament proteins in living cells and the α-keratins of sheep wool. EMBO J 1: 1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winheim S, Hieb AR, Silbermann M, Surmann EM, Wedig T, Herrmann H, Langowski J, Mücke N. 2011. Deconstructing the late phase of vimentin assembly by total internal reflection fluorescence microscopy (TIRFM). PLoS ONE 6: e19202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimek A, Thiering S, Weber K, Magin TM. 2012. Genes encoding cytoplasmic intermediate filament proteins of vertebrates revisited: Identification of a cytoplasmic intermediate filament protein in the sea anemone Nematostella. Eur J Cell Biol 91: 809–817. [DOI] [PubMed] [Google Scholar]