Abstract
There is a dynamic interplay between metabolic processes and gene regulation via the remodeling of chromatin. Most chromatin-modifying enzymes use cofactors, which are products of metabolic processes. This article explores the biosynthetic pathways of the cofactors nicotinamide adenine dinucleotide (NAD), acetyl coenzyme A (acetyl-CoA), S-adenosyl methionine (SAM), α-ketoglutarate, and flavin adenine dinucleotide (FAD), and their role in metabolically regulating chromatin processes. A more detailed look at the interaction between chromatin and the metabolic processes of circadian rhythms and aging is described as a paradigm for this emerging interdisciplinary field.
Many cofactors that are produced during metabolic reactions (e.g., acetyl-CoA) influence chromatin-modifying enzymes. Circadian rhythms and aging represent examples of processes that are influenced by these types of interactions.
OVERVIEW
A prominent area in epigenetic research that has emerged in recent years relates to how cellular metabolism regulates various events of chromatin remodeling. Cells sense changes in the environment and translate them into specific modulations of the epigenome through a variety of signaling components, several of which are proteins with histone- and DNA-modifying enzymatic activity. There are now a myriad of residues on DNA and histone tails that can undergo modification at a given time. The enzymes that elicit these modifications rely critically on the availability of phosphate, acetyl, and methyl groups, to mention a few. This constitutes an intriguing link between cellular metabolism and epigenetic control that has previously been largely unappreciated. Although the specificity and degree of change in the levels of cellular metabolites may influence the epigenome, we are unsure of to what degree. However, a number of remarkable studies discussed in this article are revealing a range of responses to the environment.
An example of the intimate connection between metabolism and epigenetic control is epitomized by findings on the FTO (fat mass and obesity-associated) gene. The gene encodes an N6-methyladenosine (m6A) demethylase, an enzyme that controls the levels of RNA methylation (Gerken et al. 2007). Single-nucleotide polymorphisms of the FTO gene, associated with obesity, fundamentally alter its response to food intake and thereby influence the development of obesity. The epigenetic connection came through chromosome capture experiments (described in Dekker and Misteli 2014), showing that certain variants of the FTO gene are uniquely correlated with obesity and physically loop to and interact with IRX3, a homeobox gene located megabases away from FTO. Thus, metabolic dysfunction, such as obesity, manifesting at one gene, directly influences the function of a distant gene that, remarkably, is also implicated in controlling body mass (Smemo et al. 2014).
Chromatin regulation involves enzymes that use cofactors for the reactions that modify DNA or histones. These enzymes either attach small chemical units (i.e., posttranslational modifications or PTMs) or alter nucleosome positioning or composition (i.e., of histone variants). It is assumed that this control depends partly on the variable levels of cellular metabolites acting as enzyme cofactors. For example, acetyltransferases use acetyl-coenzyme A (acetyl-CoA), methyltransferases use S-adenosyl methionine, and kinases use ATP as donors of acetyl, methyl, or phospho groups, respectively; deacetylases can use nicotinamide adenine dinucleotide (NAD), and demethylases can use flavin adenine dinucleotide (FAD) or α-ketoglutarate as coenzymes. In addition, another relevant example relates to remodeler complexes that use ATP for moving, ejecting, or restructuring nucleosomes, as discussed in Becker and Workman (2013).
In this article, we first present a summary of several metabolites that are known to alter the activity of their cognate enzymes and, hence, their potential in regulating chromatin states is discussed. We then present examples of enzymes that directly respond to changes in metabolism, such as the sirtuin enzymes and polyadenosine diphosphate (ADP)-ribose polymerases (PARPs). Finally, we discuss several fields that have been greatly influenced by ideas that altered metabolic signaling could affect epigenetic regulation, such as (1) altered nutrient availability regulating histone and DNA modifications, (2) circadian biological rhythms, and (3) cellular replicative aging and senescence.
1. METABOLITES
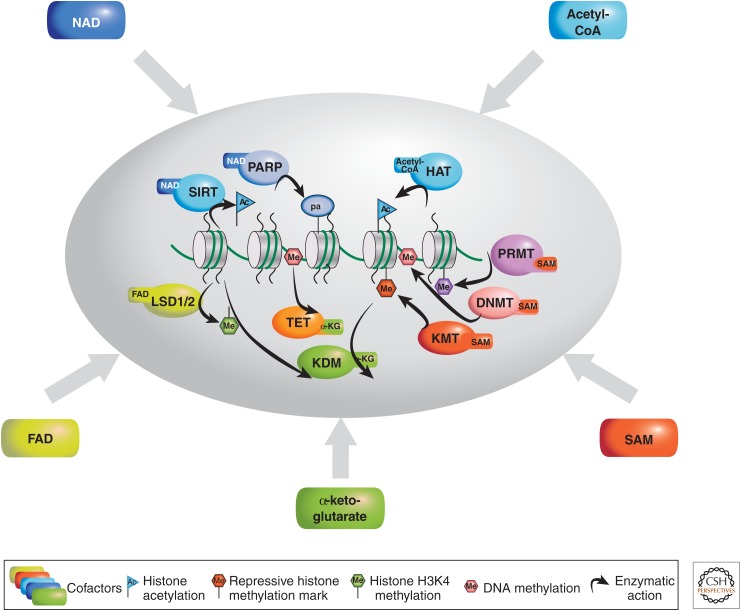
Although it is conceptually conceivable that a variety of metabolites are implicated in various aspects of epigenetic control (Katada et al. 2012; Lu and Thompson 2012), to date solid evidence is scarce. The assumption is that levels of metabolites may change in response to various physiological stimuli, and thereby influence enzymes implicated in chromatin remodeling. In addition, the subcellular concentration of specific metabolites may direct the local activation or inhibition of enzymatic activities. Thus, the presence of “niches” of chromatin-associated metabolites has been hypothesized, which could rationalize the locus-specific modification of otherwise undistinguishable histones and DNA (Katada et al. 2012). The metabolites discussed in this section can be functionally linked to enzymatic activities involved in chromatin remodeling and DNA methylation (Fig. 1).
Figure 1.
The major cofactors involved in enzyme-mediated DNA or histone posttranslational modification (PTM). NAD, nicotinamide adenine dinucleotide; acetyl-CoA, acetyl coenzyme A; SAM, S-adenosyl methionine; FAD, flavin adenine dinucleotide; PARP, poly-ADP-ribose polymerase; HAT, histone acetyltransferase; PRMT, protein arginine methyltransferase; DNMT, DNA methyltransferase; KMT, lysine (K) methyltransferase; KDM, lysine demethylase; TET, ten-eleven translocation protein; LSD, lysine-specific demethylase.
1.1. Acetyl-CoA
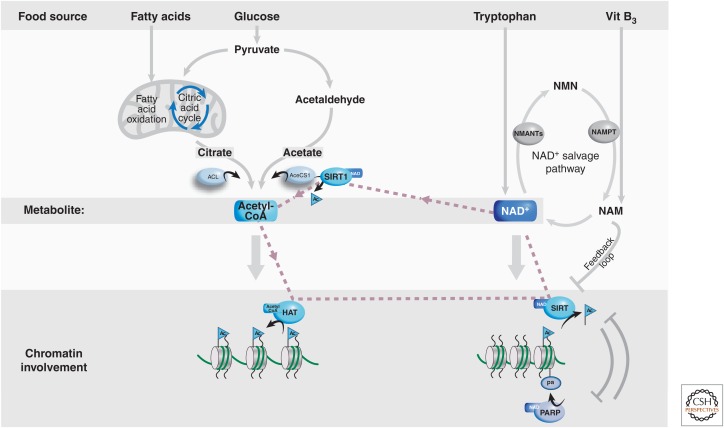
Acetylation of both histone and nonhistone proteins has critical biological consequences (Guan and Xiong 2011). The availability of appropriate levels of acetyl-CoA, the metabolite that provides the acetyl group for the acetylation reaction, could modulate the efficacy and specificity of the reaction. Acetyl-CoA appears to exist in two separate pools in the cell: a mitochondrial and nuclear/cytosolic pool. The mitochondrial pool is derived mainly from the action of the enzyme pyruvate dehydrogenase and fatty acid oxidation (Fig. 2). The nuclear/cytosolic pool of acetyl-CoA, responsible for protein acetylation and fatty acid synthesis, is produced by two enzymes in metazoans: the acetyl-CoA synthetase 1 (AceCS1) and ATP-citrate lyase (ACL) (Albaugh et al. 2011). ACL uses citrate (produced during the tricarboxylic acid [TCA] cycle) as a substrate for the production of acetyl-CoA, whereas AceCS1 uses acetate. In mammals, acetate can be produced by the intestinal flora, alcohol metabolism, prolonged fasting, and class I and II histone deacetylases (HDACs). In Saccharomyces cerevisiae, the homolog of AceCS1 is the major source of acetyl-CoA for histone acetylation (Takahashi et al. 2006). ACL and AceCS1 are present in both the cytosol and nucleus of mammalian cells, and the loss of either leads to reduction in global histone acetylation (Wellen et al. 2009; Ariyannur et al. 2010). In addition, reduction in histone acetylation upon loss of ACL can be rescued by supplementing the cells with acetate, suggesting a critical role for AceCS1 in acetyl-CoA biosynthesis, possibly when cells are shifted from glucose- to acetate-driven metabolism. Overall, these studies thereby indicate that changes in the activity of both ACL and AceCS1 and, thus, in the intracellular levels of acetyl-CoA, can influence histone acetylation and, consequently, chromatin remodeling. Additional work has shown that AceCS1 is itself acetylated (Hallows et al. 2006; Sahar et al. 2014), an event controlled in a cyclic manner by the deacetylase SIRT1 (see Sec. 2.1).
Figure 2.
Biosynthesis pathways of acetyl-CoA and NAD cofactors, and their involvement in chromatin-related processes. Acetyl-CoA is produced via two pathways to metabolize pyruvate, involving the key catalytic action of ACL or AceCS1. Acetyl-CoA is an essential metabolite required for the activity of HATs involved in creating an active chromatin conformation via the acetylation of histones. NAD is produced via the NAD salvage pathway. It is an essential cofactor for the PARP and SIRT enzymes, among other proteins. A link exists between the two pathways, as indicated by the dashed lines, by virtue that the NAD-using enzyme, SIRT1, activates the AceCS1 enzyme via protein deacetylation, which, in turn, produces the metabolite acetyl-CoA.
Finally, an intriguing twist has linked acetyl-CoA with DNA methylation; the levels of DNA methyltransferase 1 (DNMT1) in adipocytes seem to be controlled in part by ACL, which in turn controls adipocyte differentiation (Londoño Gentile et al. 2013).
1.2. NAD
Nicotinamide adenine dinucleotide (NAD) is a key metabolite involved in a large array of cellular metabolic pathways. It is synthesized from tryptophan or two different forms of vitamin B3: nicotinic acid and nicotinamide (NAM) (Fig. 2). The tryptophan pathway provides de novo NAD from protein-based nutrients in all life forms. A switch has occurred during evolution, concerning the synthesis of NAD from vitamin B3, such that lower eukaryotes and invertebrates (including yeast, worms, and flies) use nicotinic acid as the major precursor of NAD, whereas in mammals it is NAM (Magni et al. 2004). The synthesis of NAD in mammals is controlled by an enzymatic loop that is often referred to as the NAD salvage pathway. NAD is metabolized into NAM by NAD-using enzymes. NAM, subsequently, functions as a potent inhibitor of NAD-dependent enzymes, thereby constituting an enzymatic feedback regulatory mechanism (Magni et al. 2004; Nikiforov et al. 2011). NAM is in turn converted into nicotinamide mononucleotide (NMN) by a single, rate-limiting step enzyme: the NAM-phosphoribosyltransferase (NAMPT) (Garten et al. 2009). The regulation of NAMPT is particularly intriguing and will be discussed later in the context of the circadian clock (Sec. 4.4). Finally, a group of three mononucleotide adenylyltransferases convert the NMN to NAD. Changing levels of NAD may then influence the activity of cellular enzymes in two ways, either because NAD is used as a coenzyme, thereby determining the efficacy of the enzymatic activity, or because the enzymes consume NAD to generate NAM (Magni et al. 2004; Nikiforov et al. 2011).
Two groups of enzymes involved in epigenetic control use NAD as coenzyme or are NAD-consumers (Fig. 2). The first is the class III HDACs. This group of seven proteins, commonly called the sirtuins (pronounced “sir-two-ins”) (Finkel et al. 2009), is composed of the mammalian homologs of the yeast Sir2 (silent information regulator 2) (elaborated in Sec. 2.1). The second group of enzymes comprises PARPs, which consume large amounts of NAD to modify proteins by attaching long chains of ADP-ribose (Schreiber et al. 2006; Gibson and Kraus 2012). These two groups of enzymes are discussed in Section 2.2.
1.3. SAM
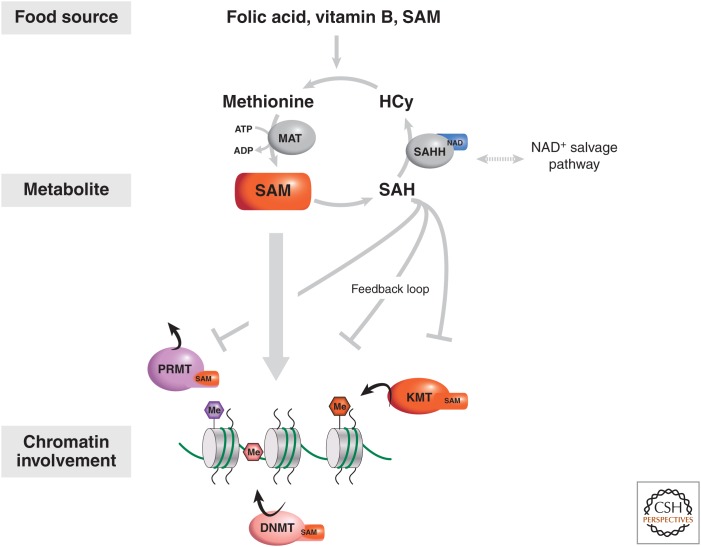
Methylation of both DNA and histones (as well as nonhistone proteins) requires the metabolite S-adenosyl methionine (SAM) as a source of methyl groups. SAM is a common cosubstrate involved in the transfer of methyl groups and is obtained from ATP and methionine by the methionine adenosyltransferases (MATs) enzymes (Fig. 3) (Grillo and Colombatto 2008). The methyl group (CH3) in SAM is reactive and donated to acceptor substrates by trans-methylation. After this reaction, SAM becomes S-adenosyl homocysteine (SAH), which is a potent inhibitor of all methyltransferases. A variety of methyltransferases have been characterized and are involved in epigenetic control (Fig. 1), including the DNA methyltransferases (DNMTs), the peptidylarginine methyltransferases (PRMTs), and the lysine (K) methyltransferases (KMTs) (detailed in Cheng 2014). Because ATP is a source for SAM, the intracellular ATP concentration and sublocalization could influence the availability of SAM for methylation reactions. Furthermore, MATII, one of the MATs responsible for SAM synthesis, serves as a transcriptional corepressor in the oxidative stress response. Specifically, MATII directly interacts with the histone H3 at lysine 9 (H3K9) methyltransferase SETDB1, thereby promoting H3K9 trimethylation and repression of the cyclooxygenase 2 gene (Kera et al. 2013), a prostaglandin synthase involved in the inflammatory response.
Figure 3.
The biosynthesis pathway of S-adenosyl methionine (SAM) and its involvement in chromatin-related processes. SAM is an essential cofactor for PRMT, DNMT, and KMT chromatin-modifying enzymes. This pathway is metabolically influenced by the NAD salvage pathway by virtue that the SAH hydrolase (SAHH) enzyme in its biosynthesis pathway uses NAD. The primary product of SAM metabolism is SAH, which has an inhibitory effect on all the SAM-dependent chromatin-modifying complexes. SAH, S-adenosyl homocysteine; HCy, homocysteine.
Various studies have linked nutrition regimes to the pattern of DNA methylation, indicating that food containing folic acid, vitamin B, and SAM can improve a number of pathophysiological conditions. A revealing example relates to the chemical bisphenol A (BPA), used to make polycarbonate plastic, which was reported as unsafe for human health. Pregnant yellow agouti mice fed with BPA generate a more yellow, unhealthy progeny that correlates with undermethylation of the agouti gene. Feeding pregnant, BPA-treated mothers with methyl-rich foods reverts the negative effect of BPA, and offspring are healthy and brown (Dolinoy et al. 2007). This and additional evidence indicate that diets high in methyl-donating nutrients (such as folic acid, B vitamins, and SAM-e) can alter gene expression, especially during early development when the epigenome is thought to be first established. Conversely, nutritional challenge, such as a high-fat diet regimen, has been shown to induce new oscillations in SAM and SAH levels through a transcriptional and epigenetic reprogramming of the circadian system (Eckel-Mahan et al. 2013), discussed below in Section 4.
1.4. FAD
Riboflavin (vitamin B2) is an essential component for the enzyme cofactors flavin adenine dinucleotide (FAD) and flavin mononucleotide. As with other B vitamins, riboflavin is required for the metabolism of fats, ketone bodies, carbohydrates, and proteins (Fischer and Bacher 2008). Riboflavin is present in many vegetables and meat. During digestion, the various flavoproteins in food are degraded and riboflavin is resorbed. FAD is a redox cofactor used by several enzymes in the control of metabolic pathways (Macheroux et al. 2011). Amine oxidations are widespread reactions in nature. Flavin-dependent amine oxidases are enzymes that catalyze oxidative cleavage of the C–N bond by two-electron reduction of the FAD coenzyme, which produces an imine intermediate that is then hydrolyzed nonenzymatically. Reduced FAD can be reoxidized by molecular oxygen, which generates hydrogen peroxide and thereby makes the enzyme available for a new catalytic cycle (illustrated in Fig. 1 of Shi and Tsukada 2013).
Different classes of demethylases use different metabolites (Hou and Yu 2010). For example, the JMJC family (e.g., Jumonji C) uses a dioxygenase reaction that is dependent on Fe (II) and α-ketoglutarate for demethylating mono-, di-, and trimethylated residues (see Fig. 1 of Shi and Tsukada 2013). In contrast, the demethylases, lysine-specific demethylase 1 and 2 (LSD1 and LSD2), use a FAD-dependent amine oxidation reaction to demethylate mono- and dimethylated substrates (Fig. 1). LSD1, specifically, removes one or two methyl groups from histone H3 at Lys4 via a FAD-dependent reaction (Metzger et al. 2010). LSD1 is inactive in vitro on H3K4me3, consistent with the flavin-catalyzed amine oxidation reaction that requires a lone pair of electrons on the lysine amino group. Thus, depending on the metabolite, different levels of control for H3K4 methylation are exerted.
LSD1 enzymatic activity depends on protein kinase Cα (PKCα)-dependent phosphorylation. PKCα responds both to calcium and to increasing levels of the lipid second messenger diacylglycerol, possibly linking LSD1-mediated demethylation to these signaling pathways (Metzger et al. 2010). Intriguingly, variable phosphorylation of LSD1 appears to control circadian gene expression by inducing direct interaction of LSD1 with the CLOCK:BMAL1 complex (see also Sec. 4.2; Nam et al. 2014). Finally, the balance of methyl groups toward both histone and DNA methylation may be interdependent on reciprocal modifications, as highlighted by the requirement of LSD1 for the maintenance of global DNA methylation (Wang et al. 2009).
2. ENZYMES
The activity of several classes of enzymes may be controlled by changes in metabolite availability or the use of specific metabolites as coenzymes. Here, we discuss examples of enzymes whose role in chromatin regulation has been explored in greater detail.
2.1. SIRT1 and Other Sirtuins
The class III HDACs are composed of a family of mammalian proteins originally identified by similarity to the yeast gene Sir2. In mammals, the sirtuin family is composed of seven members, some of which are mitochondrial (Sirt3, Sirt4, and Sirt5), others that are principally nuclear (Sirt1, Sirt6, and Sirt7), and others are expressed in more than one cellular compartment (Finkel et al. 2009). Sir2 was initially identified as an NAD+-dependent deacetylase and linked to the control of longevity (Imai et al. 2000; Schwer and Verdin 2008; Chalkiadaki and Guarente 2012), although the contribution to life span of Sir2 orthologs in various other organisms is debated (Burnett et al. 2011).
The principal mammalian ortholog of Sir2, SIRT1, has emerged as a key component, linking both histone and nonhistone deacetylation to cellular metabolism. SIRT1, as a class III HDAC, differs from the class I and II deacetylases in that it requires NAD+ as a cofactor for its enzymatic activity, similar to yeast Sir2. SIRT1 breaks down NAD+ during the process of lysine deacetylation, producing O-acetyl-ADP-ribose. During fasting, levels of NAD+ are high, and SIRT1 activity is elevated. However, when energy is in excess, NAD+ is depleted because the rampant flux through the glycolytic cycle promotes the conversion of NAD+ to NADH. It is generally thought that SIRT1 levels do not change (Nakahata et al. 2008) in response to different physiological states, although its variable expression in some specific physiological settings cannot exclude that possibility. Evidence is clear, however, that enzymatic activity is regulated, in large part, by the availability of the enzyme’s cofactor, NAD+ (Finkel et al. 2009).
SIRT1 HDAC activity is mostly directed to K9ac and K14ac of the H3 tail (Chalkiadaki and Guarente 2012). However, increasing evidence indicates that SIRT1 may be a more efficient deacetylase for nonhistone nuclear proteins rather than histones. Indeed, in the dietary fasted state, SIRT1 affects the activity of numerous target proteins, including many involved directly or indirectly in metabolic homeostasis, such as PGC-1α, FOXO, IRS1/2, LXR, HNF-4α, FXR, RAR, TORC2, BMAL1, eNOS, LKB1, AMPK, and SREBP1 (reviewed in Houtkooper et al. 2012). In most cases, the enzymatic deacetylation reaction has been linked to increased activity of a target protein. So, SIRT1-mediated deacetylation of PGC-1α, for instance, activates the protein promoting gluconeogenic gene transcription and the inhibition of glycolytic gene transcription in the liver (Nemoto et al. 2005; Rodgers et al. 2005).
SIRT1 may also contribute to acetylation of histones and nonhistone proteins, indirectly, by regulating the activity of AceCS1 and thereby acetyl-CoA synthesis (Fig. 2). Indeed, the acetylated form of AceCS1 at Lys-661 is inactive, whereas SIRT1-mediated deacetylation activates the enzyme. Interestingly, SIRT1 is the only sirtuin able to deacetylate AceCS1 (Hallows et al. 2006), and this acetylation is cyclic, controlled by the circadian clock (see more in Sec. 4.3), leading to oscillating levels of acetyl-CoA. Because acetyl-CoA is the carbon source for synthesis and elongation of fatty acids, it is of interest that the clock and AceCS1 contribute to the elongation of long fatty acids into very long fatty acids (Sahar et al. 2014). Thus, there is a direct link between NAD+, the coenzyme for SIRT1, and acetyl-CoA synthesis to control the acetylation of both histone and nonhistone proteins (Fig. 2, dashed lines). In summary, the genomic function of SIRT1 appears to result from the combination of a variety of converging molecular functions. The first high-throughput transcriptomic analysis in liver-specific Sirt1-null mice has shown that SIRT1, together with SIRT6, contributes to genomic partitioning of circadian transcription (Masri et al. 2014).
Another sirtuin that may contribute to epigenetic regulation is SIRT3. This enzyme appears to be the major mitochondrial deacetylase in which it targets the local acetyl-CoA synthetase, lecithin-cholesterol aceyltransferase, and 3-hydroxy-3-methylglutaryl-CoA synthase 2, which controls the levels of ketone body production (Shimazu et al. 2010). SIRT3 may also operate as a critical link between circadian metabolism and aging (Peek et al. 2013). This was intimated by the finding that the majority of clock-controlled acetylation relates to mitochondrial proteins, using an unbiased analysis of the circadian acetylome (Masri et al. 2013). In the liver, SIRT3 appears to generate rhythms in the acetylation and activity of oxidative enzymes and respiration. In hematopoietic stem cells, SIRT3 regulates mitochondrial antioxidative stress required to maintain mitochondrial homeostasis during oxidative stress or aging (Peek et al. 2013). Furthermore, it has been observed that, in conditions of critical oxidative stress, the cell responds by resetting the circadian clock, activating antioxidant pathways through BMAL1, heat shock factor 1, and casein kinase II (Tamaru et al. 2013). Interestingly, SIRT1 also participates in a mitochondrial antiaging function by modulating the nuclear-mitochondrial communication. It appears that the decrease in NAD+ levels during aging affects SIRT1 activity and thereby the control of nuclear-encoded mitochondrial genes (discussed further in Sec. 5.3). Hence, the decreased levels in NAD+ might impinge negatively on mitochondrial homeostasis by reducing the activity of sirtuins.
Two other sirtuins that are nuclear and may conceivably participate directly in chromatin remodeling are SIRT6 and SIRT7. Little is known, however, about their deacetylase activity and whether they consume NAD+ with the same affinity and efficacy as SIRT1. It has been proposed that the three nuclear sirtuins may compete for specific NAD+ nuclear pools, thus influencing each other’s enzymatic activities. Intriguingly, free fatty acids are potent endogenous activators of SIRT6 HDAC activity, but not of SIRT1 (Feldman et al. 2013). Thus, endogenous fatty acids could play a role in activating or sensitizing SIRT6, a notion that is particularly appealing when taking into account that the intranuclear concentration and localization of free fatty acids is likely to be dynamic in response to metabolic and nutritional changes.
SIRT6 is unique in its constitutive localization to chromatin (Mostoslavsky et al. 2006). Its genome-wide occupancy is prominent at the transcriptional start site (TSS) of active genomic loci, coinciding with serine 5 phosphorylated RNA polymerase II binding sites (Ram et al. 2011). SIRT6 binding to chromatin has also been reported to be dynamic in response to stimuli, such as tumor necrosis factor α, subsequently, resulting in the alteration of the transcriptional landscape of aging and stress-related genes (Kawahara et al. 2011). SIRT6, specifically, deacetylates H3K9 (Michishita et al. 2008; Kawahara et al. 2009) and H3K56 (Michishita et al. 2009; Yang et al. 2009; Toiber et al. 2013) in a nucleosome-dependent manner (Gil et al. 2013), modulating gene expression and affecting telomere maintenance and genomic stability. Also, SIRT6 interacts with transcription factors, such as NF-κB and HIF1α (Kawahara et al. 2009; Zhong et al. 2010), and contributes to their targeting to gene promoters. A full-scale analysis by high-throughput transcriptomics and metabolomics in the liver has revealed that SIRT6 contributes to genomic partitioning of circadian transcription, which leads to segregated control of cellular metabolism (Masri et al. 2014).
Physiologically, SIRT6 has been implicated in metabolic regulation, as Sirt6−/– mice die at 2–4 wk of age because of a severe accelerated-aging phenotype and hypoglycemia. Lethality is a result of altered rates of glycolysis, glucose uptake, and mitochondrial respiration (Mostoslavsky et al. 2006; Xiao et al. 2010; Zhong et al. 2010). SIRT6 also controls the acetylation state of PGC-1α in a GCN5-dependent manner, which regulates blood glucose levels (Dominy et al. 2010). Liver-specific Sirt6−/– mice develop fatty liver as a result of altered expression of genes involved in fatty acid β-oxidation and triglyceride synthesis (Kim et al. 2010). The SIRT6-targeted deacetylation of H3K9 is an event that has been associated with the down-regulation of cell senescence and apoptotic pathways, further described in Section 5.1.
SIRT7 was first described as being highly concentrated at the level of the nucleolus, involved in ribosomal DNA transcription and interacting with the UBF transcription factor (Grob et al. 2009). This apparently limited function was challenged by the added finding that SIRT7 controls H3K18 acetylation in a NAD-dependent manner, which, in turn, leads to the regulation of genes involved in oncogenic transformation (Barber et al. 2012). Additional findings have confirmed the extranucleolar role of SIRT7. Functional proteomics revealed that SIRT7 interacts with chromatin-remodeling complexes and components of RNA polymerase I machinery (Tsai et al. 2012). Indeed, in the liver, SIRT7 functions at chromatin to suppress endoplasmic reticulum (ER) stress to protect from fatty liver disease. Consistent with this, SIRT7-deficient mice develop chronic hepatosteatosis, in part because SIRT7 acts as a cofactor of MYC-mediated transcriptional repression (Shin et al. 2013).
2.2. PARPs
Poly(ADP-ribosyl)ation is a PTM of proteins, which is catalyzed by PARPs. The founding member of the PARP family, poly(ADP-ribose) polymerase-1 (PARP-1), has been well characterized. The PARP family is composed of at least 18 members, in large part nuclear, which are linked to various genomic functions, including DNA repair and programmed cell death (Schreiber et al. 2006; Gibson and Kraus 2012). PARPs hydrolyze NAD+ and transfer the ADP-ribose moiety to acceptor proteins. Many cellular proteins, including histones, are acceptors of ADP-ribose and can be mono- or poly(ADP-ribosyl)ated. The use of NAD+ by the PARP enzymatic process can be quite massive, thereby reducing the availability of NAD+ for other enzymes; for example, PARP-1 activity depletes the NAD+ pool, thus reducing SIRT1 activity, which causes cell death (Kolthur-Seetharam et al. 2006; Bai et al. 2011). Owing to its role in DNA repair, PARP inhibitors have become attractive drugs against DNA damage-response defective tumors, such as BRCA1/2 mutant cancers, described in Section 1.3.2 of Audia and Campbell (2014). Thus, PARP enzymes may have an indirect role in chromatin remodeling through regulation of sirtuins.
In contrast, PARPs may be directly involved in epigenetic regulation via other mechanisms. For example, PARP-1 may be tethered at CpG regions bound by the insulator protein CTCF, activating PARP’s own auto-ADP-ribosylation. The resulting ADP-ribose polymers then interact in a noncovalent way with DNMT1 to inhibit its methylation activity, ensuring the methylation-free status of CTCF bound promoters (Reale et al. 2005; Guastafierro et al. 2008; Zampieri et al. 2012). This mechanism may ensure that a locus’ promoter region (e.g., of the Dnmt1 housekeeping gene) (Yu et al. 2004) or imprinted control region (e.g., of the Igf2 gene cluster) (Zampieri et al. 2009) are DNA methylation free.
Evidence also exists for PARP-1-mediated poly(ADP-ribosyl)ation of several core histones (at positions K13 of H2A, K30 of H2B, K27 and K37 of H3, and K16 of H4). Inhibitory cross talk with other histone modifications, such as acetylation, has been suggested (Beneke 2012). In particular, acetylated H4K16 appears to inhibit PARP-1-mediated ADP-ribosylation (Messner et al. 2010). Intriguingly, PARP-1 has also been found to be a structural component of chromatin and to modulate the enzymatic activity of various other regulators, such as the histone demethylase KDM5B (Krishnakumar and Kraus 2010). The complexity of poly(ADP-ribosyl)ation and the multitude of enzymes able to elicit this reaction has impeded a full understanding of the pathways through which PARPs may direct specific and functionally relevant chromatin-remodeling events.
3. ALTERED INTERMEDIARY METABOLISM REGULATES EPIGENETIC STATE
A key concept underlying the role that chromatin plays in regulating cell physiology and maintaining homeostasis is that nuclear activity requires coordination with, and response to, the overall cellular metabolic state. However, nuclear processes involving DNA-associated transactions on chromatin are segregated in location and regulation from cytoplasm-based intermediary metabolic enzymes. An emerging idea to reconcile this separation is that alterations in metabolism may directly influence acetyl-CoA, SAM, NAD, and FAD levels in the nucleus, which may then, in turn, alter DNA and histone modifications and hence transcription (Katada et al. 2012). As described below, there is abundant evidence indicating that metabolic state alters histone acetylation, in particular, both during normal growth cycles and as a result of altered metabolic state in disease, such as cancer. However, although histone and DNA methylation can be altered in disease via mutations in enzymes occupying metabolic pathways, the question of whether normal growth cycles actually alter the activity of methylation and demethylation enzymes remains.
3.1. Histone Acetylation Is Regulated by Metabolic State
Initial observations showing the agile histone acetylation response to growth were made in the yeast, S. cerevisiae. A major pathway of acetyl-CoA generation is via the acetyl-CoA synthetases Acs1 and Acs2 (or AceCS1 in mammals), which are enzymes that hydrolyze ATP to catalyze the ligation of acetyl-CoA from acetate and CoA (Fig. 2). Although long considered to be exclusively cytoplasmic, the enzymes also localize to the nucleus (Takahashi et al. 2006). Moreover, impairment of Acs2 globally lowered levels of acetylated histone H3 and H4 tails, and showed remarkably broad decreases in gene expression occurring in specific pathways (Takahashi et al. 2006). These discoveries provided a direct link between enzymes that produce acetyl-CoA to nuclear chromatin regulation and gene expression.
Observations, again in S. cerevisiae, established that the nutrient environment/metabolic state can lead to altered global histone acetylation, namely, that prolonged quiescence during stationary phase leads to decreased histone acetylation (Sandmeier et al. 2002; Ramaswamy et al. 2003). A direct link was shown when elevated glucose levels (the preferred carbon source for yeast) used to stimulate growth were accompanied by the direct metabolic induction of histone acetylation. This acetylation occurred broadly over the genome, catalyzed by the histone acetyltransferases (HATs), Gcn5 and Esa2, in association with their protein complexes, SAGA and picNuA4, respectively, functioning in an untargeted manner (Friis et al. 2009).
A related analysis of intrinsic yeast metabolic cycling provided more insight into histone acetylation fluxes. Yeast metabolic cycling occurs when cells are starved at high cell density and, then, are continuously supplemented with low concentrations of glucose. A synchronous cycle is established in the population, consisting of a growth period accompanied by oxidation, followed by a period of cessation of growth while reductive, building processes occur. Under these conditions, transcription occurs in waves, with up-regulation of growth-related genes followed by induction of stress-related genes, within each respective period (Tu et al. 2005). Interestingly, during the growth portion of the metabolic cycle, a critical metabolite is acetyl-CoA. This transient pulse of acetyl-CoA leads to increased activity of the SAGA-Gcn5 complex and both a transient global increase of acetylation at its substrate sites in histone H3 and a transient targeted increase specifically at growth-related genes (Cai et al. 2011).
These observations of metabolic signaling to directly alter histone acetylation have been extended to mammalian cells. Glucose is the major carbon source for mammalian cells and it can be used by ACL to produce acetyl-CoA (Fig. 2). A small proportion of the ACL enzyme localizes to the nucleus in several mammalian cell types in culture, and small interfering RNA-induced silencing of ACL led to a decrease in global histone acetylation. Indeed, in adipocyte differentiation, ACL is critical for inducing histone acetylation at specific genes and activating transcription (Wellen et al. 2009).
What might be the mechanism by which HATs respond to altered acetyl-CoA concentrations? One possible mechanism is that the binding affinity of the enzymes for acetyl-CoA is in the range of in vivo fluctuations of acetyl-CoA concentrations. Thus, the measured acetyl-CoA concentration during the yeast growth oscillatory cycle (∼3–30 mm) (Cai et al. 2011) bracket the in vitro measured Kd and Km of Gcn5 (∼8.5 and ∼2.5 mm) (Berndsen and Denu 2008). Further speculation is that nuclear localization of the acetyl-CoA-producing enzymes may be induced during growth phases and may directly provide acetyl-CoA to the histone-acetylating enzymes.
Important future questions include determining how far reaching are these mechanisms of increasing activity or driving acetyl-CoA-producing enzymes to localize to the nucleus for directly influencing chromatin modifications. Is this pathway used only to respond to nutrients, or can this pathway be used to respond to cell differentiation (Kaochar and Tu 2012)? In this respect, a new study showed that nutritional challenge, in the form of a high-fat diet in the mouse, leads to a reprogramming of circadian gene expression, using a shift in chromatin remodeling, specifically in H3K9/K14 acetylation. Remarkably, at the metabolic level, it is clear that the shift in histone modifications is paralleled by changes in the levels of NAD+ and acetyl-CoA (Eckel-Mahan et al. 2013).
3.2. Connections of Metabolism to Histone Methylation
The profound changes in acetyl-CoA levels, histone acetylation, and transcription lead to the question of whether levels of metabolites associated with methylation, such as SAM, are similarly altered in the nucleus in response to the nutrient state to then alter DNA and histone methylation (Teperino et al. 2010). A massive response to metabolic change via the up-regulation of histone methylation, as described above for histone acetylation, has not been reported. There are, however, interesting examples, cited below, of altered histone methylation caused by more restricted alterations in metabolic regulation.
The high-energy methyl donor, SAM, is generated from methionine by MAT enzymes in an ATP-consuming reaction. Interestingly, in mammalian cultured immune T cells, MAT has been shown to directly associate with repressed genes, leading to chromatin-localized synthesis of SAM, and fine-tuning of histone methylation and gene down-regulation (Katoh et al. 2011).
Mouse stem cells represent an interesting example of altered histone methylation in response to metabolic adaptation. In general, histone methylation appears to have a critical function in transcriptional regulation and self-renewal in stem cells (Ang et al. 2011; Liang and Zhang 2013). Mouse embryonic stem cells (mESCs) have a specific mode of threonine catabolism, which affects SAM metabolism and H3K4me3, and is crucial for maintaining pluripotency (Shyh-Chang et al. 2013). Culturing proliferating mESCs in threonine-depleted media, therefore, leads to a lowering of SAM and H3K4me3 levels, but not other residue methylation, which slows growth and increases differentiation. However, threonine dependency is specific to mESCs; hence, it is unclear whether histone methylation is similarly influenced by metabolic activities in other stem cell populations, such as human ESCs or adult human stem cells. Furthermore, it remains unknown whether, akin to histone acetylation, histone methylation is a broad energy sensing modification that adapts transcription to the overall metabolic state.
3.3. Oncogenic Mutations in Metabolic Pathways Alter Epigenetic Methylation State
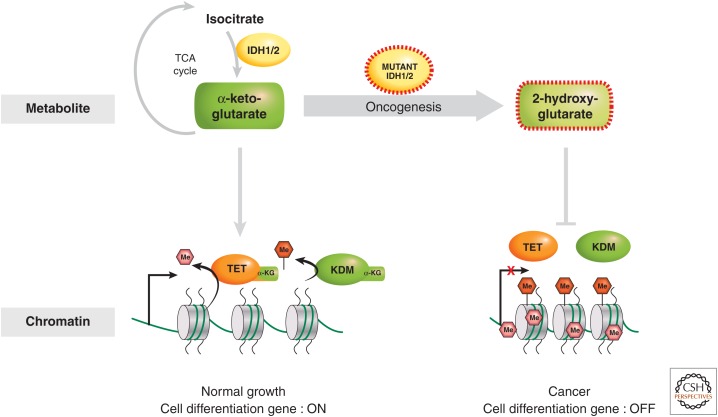
Although it is not clear whether histone methylation responds to normal fluctuations in growth and ranges of nutrient levels, the activity of methyltransferase enzymes can be dramatically altered as a result of mutations in key metabolic pathways. There are leukemic and other malignancies that result from oncogenic gain-of-function mutations in isocitrate dehydrogenase enzymes, IDH1 and IDH2 (Figueroa et al. 2010; Lu et al. 2012; Turcan et al. 2012). Large amounts of 2-hydroxyglutarate are produced from α-ketoglutarate in certain isocitrate dehydrogenase (IDH) mutant enzymes. α-ketoglutarate is a cofactor for DNA ten-eleven translocation (TET) enzymes (which convert 5meC to further oxidized forms, as described in Li and Zhang 2014) and histone lysine demethylases (KDMs); thus, high levels of 2-hydroxyglutarate act to repress the TET and KDM enzymes. Consequently, the IDH1/2 mutations lead to increased levels of DNA methylation and histone lysine methylation and altered transcription (Fig. 4). One model to explain the physiology is that growth-promoting genes have reduced levels of repressive methylation, causing increased transcription, which leads to the oncogenic state. Based on these observations, there may be other cancers that result specifically from mutations or altered activity of metabolic enzymes that change epigenetic regulation.
Figure 4.
Metabolism of the α-ketoglutarate cofactor regulates the activity of the TET and KDM enzymes in normal and cancer cells. The α-ketoglutarate metabolite is produced by the enzymatic action of the IDH proteins. Some cancer driving dominant-negative mutants of IDH cause the accumulation of an aberrant metabolite, 2-hydroxy-glutarate, which blocks TET and KDM activity.
4. THE CIRCADIAN EPIGENOME
There are certain major areas of physiology that are exquisitely reactive to environmental signals and can be gauged by the levels of metabolites and other entities. Circadian rhythms are one area of physiology in which an organism responds to external cycles, including nutrients, via establishing internal cycles. The circadian response largely involves transcriptional regulation and epigenetic mechanisms, as discussed in detail in this section. A second major organismal response to the environment is in the related areas of aging and senescence, discussed in Section 5.
4.1. The Circadian Clock
A wide variety of physiological functions, including sleep–wake cycles, body temperature, hormone secretion, locomotor activity, and feeding behavior depend on the circadian clock—a highly conserved system that enables organisms to adapt to common daily changes, such as the day–night cycle and food availability (Reppert and Weaver 2002; Sahar and Sassone-Corsi 2012). Based on evidence accumulated over several decades, circadian rhythms represent possibly the most prominent area of physiology that has effectively been studied using systems biology.
In mammals, the anatomical structure in the brain that governs circadian rhythms is a small area consisting of approximately 15,000 neurons localized in the anterior hypothalamus, called the suprachiasmatic nucleus (SCN). This “central pacemaker” in the SCN receives signals from the environment and, in turn, coordinates the oscillating activity of peripheral clocks, which are located in almost all tissues (Schibler and Sassone-Corsi 2002). This highly orchestrated network is based on cascades of signaling pathways that lead to the activation of transcriptional programs, involving a remarkable fraction of the genome. Transcriptome studies indicate that ∼10% of all transcripts oscillate and belong to the category of clock-controlled genes (CCGs). This fraction is actually much higher because different tissues express different CCGs (Panda et al. 2002; Masri and Sassone-Corsi 2010). A study covering 14 mouse tissues identified approximately 10,000 of the total almost 25,000 known protein-coding genes as showing circadian oscillations in at least one tissue. The number of common genes showing circadian oscillation in multiple tissues, however, decreases drastically as the number of tissues included in the comparative analysis is increased; that is, only 41 genes displayed circadian oscillation in at least eight out of 14 tissues (Yan et al. 2008).
The capacity for genes to be regulated in a circadian manner appears to depend, in part, on the metabolic state of a given tissue. For example, nutritional challenge in the form of a high-fat diet reprograms the circadian transcriptome in the liver so that normally noncyclic transcriptional and epigenetic regulatory programs become cyclically activated (Eckel-Mahan et al. 2013). Thus, a portion of genes larger than originally thought is intrinsically capable of becoming regulated in a circadian manner under the appropriate metabolic setting.
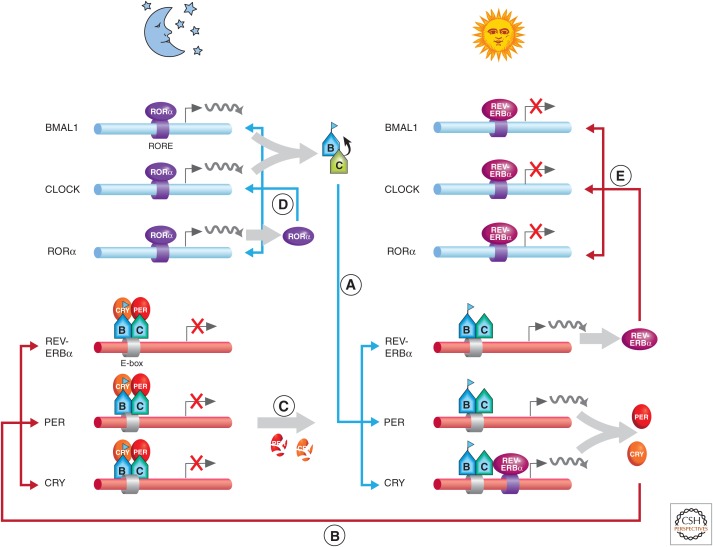
The circadian expression program is governed by interlocking transcription–translation feedback loops, a system that is conceptually conserved among species (Reppert and Weaver 2002). At the heart of the circadian clock molecular network are the core transcription factors CLOCK and BMAL1. These two proteins heterodimerize and direct transcriptional activation of CCGs by binding to E-box sites within their promoters (Fig. 5A). Among these CCGs, CLOCK and BMAL1 also direct transcription of their own repressors, period (PER) and cryptochrome (CRY) family members, creating a tightly self-regulated system. During the day, transcription of PER and CRY is high, leading to protein translation of the circadian repressors and resulting in formation of the inhibitory complex with CLOCK and BMAL1 that abolishes transcription of CCGs (Fig. 5B). The degradation of PER and CRY by nighttime alleviates transcriptional repression and allows CLOCK:BMAL1-mediated transcription to again proceed, establishing an oscillatory rhythm in circadian gene expression (Fig. 5C). An additional level of circadian regulation exists via the orphan nuclear receptors RORα and REV-ERBα, which activate and repress transcription of the Bmal1 gene, respectively (Fig. 5D) (Reppert and Weaver 2002; Sahar and Sassone-Corsi 2012).
Figure 5.
A simplified scheme of the timing of key regulator activity in the molecular circadian clock machinery. (A) A maximum accumulation of CLOCK:BMAL1 heterodimers is achieved by daybreak following nighttime transcription and translation. CLOCK:BMAL1 binding to E-box elements of clock-controlled genes (CCGs) leads to chromatin remodeling and activation of genes. (B) The daytime expression of the repressors, PER and CRY, partially through the acetylated BMAL1 K537 residue, leads to CCG transcriptional repression at night. (C) The nighttime degradation of PER/CRY repression gradually leads to the derepression of CCGs by daybreak. (D) At nighttime, the predominance of RORα binding to the RORE (retinoic acid–related orphan receptor response element) lends to transcription of BMAL1, CLOCK, and RORα. (E) The predominance of REV-ERBα, as a result of the gene’s daytime transcription, has an inhibitory action on BMAL1, CLOCK, and RORα.
The cyclic transcriptional program invokes a similarly cyclic, plastic remodeling of chromatin organization at CCGs, with the need to be strictly timed to ensure the appropriate anticipation of the next cycle of gene expression. The first demonstration that chromatin remodeling is indeed implicated in circadian gene expression came from in vivo studies in SCN neurons; circadian gene transcription was associated with the induction of H3S10 phosphorylation in response to light, via the GABA-B pathway (Crosio et al. 2000). Subsequently, other histone modifications were found to display oscillatory profiles at CCG promoters (Etchegaray et al. 2003; Doi et al. 2006; Ripperger and Schibler 2006; Masri and Sassone-Corsi 2010), and genome-wide profiling has revealed coordinated kinetics of circadian transcription with cyclic histone modifications (Koike et al. 2012; Le Martelot et al. 2012). Chromosome conformation capture 4C technology (described in Dekker and Misteli 2014) has further revealed that circadian genes are physically organized in nuclear interactomes. Remarkably, the formation and disassembly of a given interactome correspond to peaks and troughs of cyclic transcription for the genes constituting the interactome (Aguilar-Arnal et al. 2013).
4.2. CLOCK Is a HAT, and Other Regulators
The search for a specific element that could contribute to circadian chromatin remodeling revealed that CLOCK, the master regulator of circadian transcription, has intrinsic HAT activity (Doi et al. 2006). CLOCK has a MYST (Moz, Ybf2/Sas3, SAS2, Tip60)-type of acetyl-CoA binding domain, although its general structure resembles that of ACTR (acetyltransferase amplified in breast cancer) (Chen et al. 1997). The HAT function of CLOCK is linked to circadian transcriptional activation and preferentially targets H3K9 and K14. In addition, it acetylates its own transcriptional partner, BMAL1, at a unique lysine residue in position 537, an event essential for circadian rhythmicity, described in more detail in Section 4.3 (Fig. 5A) (Hirayama et al. 2007). Another nonhistone target of CLOCK is the glucocorticoid receptor (GR), a finding with intriguing implications because GR-dependent transcription has vast effects on cellular metabolism (Nader et al. 2009).
Several other remodelers have been implicated in circadian chromatin transitions (Fig. 6). Both cAMP response element binding-binding protein (CBP) and p300 are thought to participate in rhythmic histone acetylation and associate with the CLOCK:BMAL1 complex (Curtis et al. 2004). Furthermore, cyclic methylation at H3K27 has been linked to the KMT, EzH2, although the contribution of this specific enzyme to circadian transcription is not established (Etchegaray et al. 2006). Also, a search for components of the PER complex has revealed the HP1γ-Suv39h methyltransferase, suggesting that some of the repressive action of the PER proteins is linked to H3K9 methylation (Duong and Weitz 2014).
Figure 6.
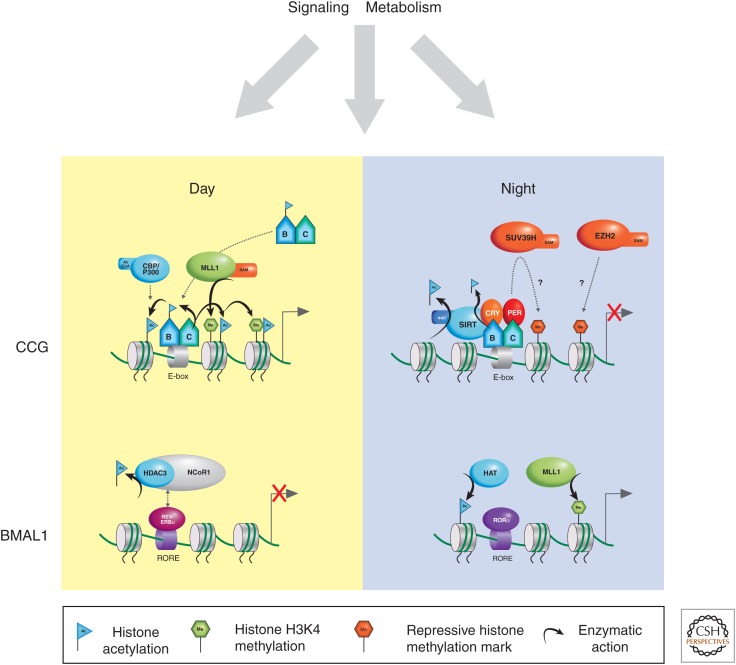
Key chromatin-remodeling factors involved in the activation and repression of CCGs and BMAL1. Recruitment of the CLOCK:BMAL1 heterodimer to CCGs during the day is facilitated by binding to the E-box and via the MLL1 enzyme. A transcriptionally active chromatin conformation is achieved through the combined action of H3K4me3, via the MLL1 lysine methyltransferase enzyme, and the histone acetylating activity of CLOCK, CBP, and P300 (cyan shaded proteins). Repressive chromatin conformations at CCGs may be achieved by the SIRT1-mediated deacetylation of histones at H3K9, K14, and H4K16, and possibly the actions of EZH2-catalyzed H3K27me3 or SUV39H-catalyzed H3K9 methylation. SIRT1 also has an inhibitory effect by deacetylating BMAL1 K537-ac. Silencing of RORE-containing genes, such as BMAL1, by day is, in part, achieved by recruitment to the RORE-bound inhibitor, REV-ERBα, of the NCoR1 complex and associated HDAC3 deacetylase. At night, predominance of RORα-bound ROREs promotes active chromatin structure, typified by histone acetylation and H3K4 methylation.
The critical role of H3K4 methylation in transcriptional activation has pointed to this site as potentially key for circadian gene expression. Remarkably, mono- and dimethylation is not cyclic at CCGs promoters, whereas H3K4 trimethylation cycles (Katada and Sassone-Corsi 2010). H3K4me3 is mediated by MLL1 and favors recruitment of the CLOCK:BMAL1 complex to chromatin to induce subsequent K9/K14 acetylation. Cells lacking the mll1 gene display a drastically impaired circadian cycle (Katada and Sassone-Corsi 2010). Although MLL1 is a central KMT for the circadian system, more than one demethylase has been implicated. First, the histone KDM, Jarid1A, is involved in circadian control, although not by directly counteracting MLL1 nor by demethylating H3K4 (DiTacchio et al. 2011). Rather, Jarid1A is proposed to block HDAC1 to enhance CLOCK:BMAL1-mediated activation, although the precise role of HDAC1 in clock function is not fully established. A second lysine-specific demethylase, LSD1, associates with CLOCK:BMAL1 in a phosphorylation-dependent manner and participates in their recruitment (Nam et al. 2014). As both Jarid1A and LSD1 target H3K4, it is still unclear whether they act on the circadian epigenome in concert or on distinct genomic subdomains or at different times of the circadian cycle.
Further links have been established between circadian control and cellular metabolism, again regarding histone methylation. MLL1 enzymatic activity is modulated by xenobiotic and lipid metabolism (Austenaa et al. 2012). Furthermore, the SAM/SAH ratio, and thus the methylation potential, is critically dependent on the action of the SAH hydrolase (SAHH). The affinity of SAHH for adenosine is affected by the relative amount of NAD and NADH because NAD is an essential coenzyme for the SAHH action (see Fig. 3) (Li et al. 2007). Finally, the levels of SAM and SAH have been found to oscillate in response to nutritional challenge and consequent reprogramming of the circadian clock (Eckel-Mahan et al. 2013).
An intriguing link with metabolism is provided by HDAC3. REV-ERBα, a nuclear receptor that contributes to the clock machinery, appears to act downstream from PPARγ, a key regulator of fat metabolism and adipocyte differentiation. The regulatory function of REV-ERBα is controlled by the nuclear receptor corepressor 1 (NCoR1). This is a corepressor that recruits the HDAC3 histone deacetylase to mediate transcriptional repression of target genes, such as Bmal1. When the NCoR1-HDAC3 association is genetically disrupted in mice, circadian and metabolic defects develop. These mice show a shorter oscillation period, increased energy expenditure, and are resistant to diet-induced obesity. Moreover, there is alteration of cyclic expression in liver lipid metabolism. HDAC3 recruitment to the genome is rhythmic (high during the day and low at night). At these HDAC3-binding sites, REV-ERBα and NCoR1 recruitment are in phase with HDAC3 recruitment, whereas histone acetylation and RNA polymerase II recruitment are antiphasic (Alenghat et al. 2008). Interestingly, genes involved in lipid metabolism in the liver also appear to be the major targets of HDAC3 and REV-ERBα. Depletion of either HDAC3 or REV-ERBα causes fatty liver phenotype, such as increased hepatic lipid and triglyceride content (Alenghat et al. 2008; Feng et al. 2011).
Additional reports show that heme functions as a ligand for REV-ERBα (Raghuram et al. 2007; Yin et al. 2007). Heme binding increases the thermal stability of REV-ERBα and enhances its interaction with the corepressor complex and, hence, is required for its repressor function. Requirement for heme places REV-ERBα in a pivotal position in the regulation of circadian rhythms and metabolism because, importantly, the circadian clock controls cellular heme levels. Aminolevulinate synthase 1 (ALAS1), the rate limiting enzyme in heme biosynthesis, is expressed in a circadian manner and is a specific target gene for the NPAS2 (a paralog of CLOCK)/BMAL1 heterodimer (Yin et al. 2007). Heme, in turn, binds to NPAS2 and inhibits its transactivation ability (Dioum et al. 2002). ALAS1 expression is also regulated by PGC-1α. The metabolic roles attributed to heme are well known; heme is a cofactor for enzymes, such as catalases, peroxidases, and cytochrome P450 enzymes, playing a role in oxygen and drug metabolism (Ponka 1999). Importantly, heme promotes adipocyte differentiation (Chen and London 1981), a function also associated with REV-ERBα and BMAL1. Thus, heme appears to be a key player in the precise coordination of metabolism and circadian function, connecting a transcriptional circadian loop to an enzymatic pathway.
4.3. SIRT1 (and Other Sirtuins) as a Rheostat of the Clock
SIRTs are thought to constitute a functional link between metabolic activity and genome stability and, possibly, aging (Schwer and Verdin 2008; Chalkiadaki and Guarente 2012). SIRT1 promotes cellular resistance to oxidative or radiation-induced stress, promotes mobilization of fat from white adipose tissues, and mediates the metabolism of energy sources in metabolically active tissues (Schwer and Verdin 2008). Thus, the finding that SIRT1 acts as a “rheostat” to modulate CLOCK-mediated acetylase activity and circadian function established an intriguing molecular link (Asher et al. 2008; Nakahata et al. 2008). SIRT1 associates with CLOCK and is recruited to circadian promoters, whereas Sirt1 genetic ablation or pharmacological inhibition of SIRT1 activity leads to significant disturbances in the circadian cycle. Importantly, although the protein levels of SIRT1 do not oscillate, as analyzed in several tissues and under various experimental conditions (Nakahata et al. 2008; Ramsey et al. 2009), its enzymatic activity oscillates in a circadian manner with a peak that corresponds to the lowest level of H3 acetylation at various CCGs (Nakahata et al. 2008; Bellet et al. 2013). This may also be caused by a role of SIRT1 in controlling the stability of some clock proteins, such as PER2 (Asher et al. 2008). Thus, it is possible that SIRT1 contributes to transducing signals originated by cellular metabolites to the circadian machinery. Further evidence comes from the observed circadian oscillation of NAD+ levels in serum-entrained mouse embryonic fibroblasts and mice livers (Nakahata et al. 2009; Ramsey et al. 2009).
Sirtuins, and SIRT1 in particular, establish functional interactions with other proteins implicated in cellular metabolism and signaling (Chalkiadaki and Guarente 2012). At the level of chromatin, the SIRT1 enzymatic activity preferentially targets histone H3K9ac, H3K14ac, and H416ac (Nakahata et al. 2008). In addition, a number of nonhistone proteins are regulated by SIRT1-mediated deacetylation, including p53, FOXO3, PGC-1a and LXR, underlining SIRT1’s pivotal function in cellular control. SIRT1 also deacetylates BMAL1, contributing to its cyclic acetylation levels (Fig. 6A) (Hirayama et al. 2007). An antibody that recognizes BMAL1 acetylated at the unique Lys-537 (K537) has been used in a variety of settings (Hirayama et al. 2007; Nakahata et al. 2008; Chang and Guarente 2013). The BMAL1 K537 antibody shows that acetylation is dependent on CLOCK and highly specific, as shown using a K537R mutant. Using this antibody, it was shown that BMAL1 deacetylation occurs in the presence of both SIRT1 and NAD+ and is inhibited by NAM (Nakahata et al. 2008). Also, BMAL1 acetylation is circadian and acetylation controls the efficient repression of BMAL1 by CRY proteins (Hirayama et al. 2007). These finding were corroborated by quantitative analyses of the BMAL1–CRY interaction by fluorescence polarization and isothermal titration calorimetry (Czarna et al. 2011). Also, comparative brain immunostaining between wild-type and brain-specific Sirt1-deficient mice showed the dependence on SIRT1 for Lys-537 deacetylation of BMAL1 (Chang and Guarente 2013).
An interdependence had been reported between SIRT1 and the nutrient-responsive adenosine monophosphate-activated protein kinase (AMPK) that contributes to the metabolic adaptation to fasting and exercise in skeletal muscles (Cantó et al. 2010). Importantly, AMPK plays a role in the control of circadian rhythms by phosphorylating and inducing the destabilization of CRY1 in a cyclic manner (Lamia et al. 2009). Stimulation of AMPK signaling alters circadian rhythms and mice in which the AMPK pathway is genetically disrupted show alterations in peripheral clocks. Thus, phosphorylation by AMPK enables CRY1 to transduce nutrient signals to circadian clocks in mammalian peripheral organs. How SIRT1, and possibly NAD+ oscillations, may be involved in this control system is still unclear.
A comprehensive analysis of the roles played by SIRT1 and SIRT6 in circadian function by high-throughput transcriptomics in individual liver-specific ablation of these sirtuins showed that SIRT1 and SIRT6 control separate subdomains of the circadian epigenome (Masri et al. 2014). Analysis by metabolomic profiling showed that, whereas SIRT1 controlled metabolites associated with peptides and cofactors, SIRT6 directed the oscillation of lipids and carbohydrates (Masri et al. 2014). This corresponds with the finding that SIRT6 responds more efficiently to long-chain fatty acids rather than NAD (Feldman et al. 2013).
4.4. NAD+, an Oscillating Metabolite
The involvement of SIRT1 in circadian regulation showed a direct link between cyclic rhythms and energy metabolism in the cell (Sahar and Sassone-Corsi 2012). Given that the expression levels of the SIRT1 gene and protein are noncyclic (Nakahata et al. 2008), a major question has been: How can two regulators (CLOCK and SIRT1) that are mostly expressed in a nonoscillatory manner lead to the circadian regulated acetylation of histone H3K9/K14 and BMAL1K537? This apparent discrepancy was solved by subsequent studies, which revealed that NAD+ levels oscillate in a circadian fashion in all cell types analyzed, and it is through the cyclic availability of its own coenzyme that SIRT1 HDAC activity is circadian (Nakahata et al. 2009; Ramsey et al. 2009). The circadian regulation of NAD+ synthesis is itself conceptually remarkable as it links the transcriptional feedback loop of the circadian clock to the NAD+ salvage-dependent enzymatic pathway.
The heavy use of the NAD+ coenzyme by enzymes, such as SIRT1 and PARP-1 (Fig. 1), risks depleting the intracellular stores, which can lead to cell death. Thus, levels of NAD+ need to be controlled, even in the absence of de novo biosynthesis, through nutritional pathways. Production of the NAMPT enzyme is the rate-limiting step within the NAD+ salvage pathway (Nakahata et al. 2009; Ramsey et al. 2009), thus, changes in NAMPT activity directly dictate the levels of intracellular NAD+ (Fig. 2).
The rhythmicity of NAD+ levels, which parallels the antiphasic oscillation in NAM, is governed by the circadian clock, as proven by the abolishment in rhythmicity in cells with a mutation in the circadian machinery (Nakahata et al. 2009). Because SIRT1 associates with and modulates CLOCK:BMAL1 (Nakahata et al. 2008), the oscillation of both SIRT1 activity and NAD+ levels via the NAD+ salvage pathway was suspected to occur via an enzymatic feedback loop. This was shown to, indeed, be the case. First, the regulatory region of the Nampt gene was found to contain two E-box promoter elements that bind CLOCK:BMAL1 and, second, its expression was shown to be controlled by CLOCK:BMAL1 in complex with SIRT1 (Fig. 7). Thus, SIRT1 is present both in the transcriptional regulatory loop of the Nampt gene and in the NAD+ enzymatic salvage pathway. This two-way control results in the circadian expression of the Nampt gene, giving a circadian function to the NAD+ salvage pathway, and thereby regulating the synthesis of NAD+ in a circadian manner.
Figure 7.
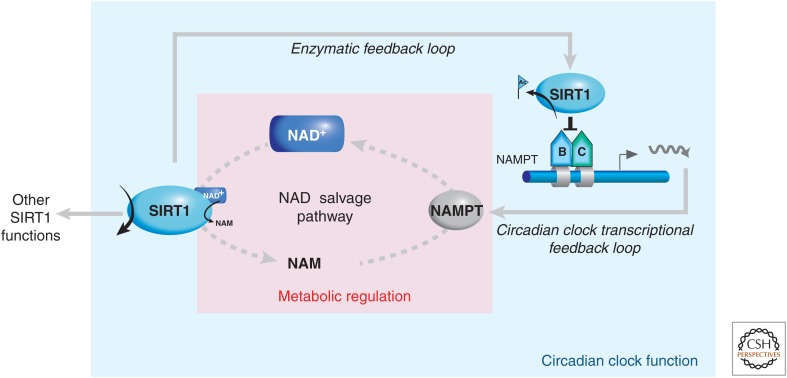
Linking the circadian clock with the NAD+ salvage pathway. The NAMPT enzyme is the limiting factor in the NAD+ salvage pathway; thus, because the NAMPT gene is regulated by the circadian CLOCK:BMAL1 machinery, its product, the NAD+ metabolite, oscillates via this NAMPT transcriptional feedback loop. The HDAC, SIRT1, among other functions, acts as a repressor of the clock machinery by effecting deacetylation of the BMAL1 protein and histones at CCGs (see also Fig. 6). Thus, whereas the enzymatic activity of SIRT1 oscillates in a circadian manner via the circadian controlled supply of its metabolite, NAD+, its activity also constitutes an enzymatic feedback loop for the circadian clock via its repressive activity at the NAMPT gene.
Importantly, the use of a highly specific NAMPT pharmacological inhibitor, FK866, abolishes NAD+ circadian oscillations and, therefore, SIRT1 cyclic activity (Nakahata et al. 2009). This finding is of interest because FK866 is used to control cell death in human cancer tissues. Thus, in addition to revealing a critical enzymatic circadian cycle, these results suggest that a direct molecular coupling exists between the circadian clock, energy metabolism, and cell survival.
The circadian clock is directly implicated in controlling the intracellular levels of critical metabolites, thereby interlocking the transcriptional feedback clock loop with the enzymatic feedback loop of the NAD+ salvage pathway (Fig. 7). This view has been confirmed by recent results, using mice deficient in a NAD+ hydrolase (CD38), in which NAD+ levels become elevated during most of the circadian cycle. These alterations in circadian rhythmicity and CCG expression lead to aberrant metabolism (Sahar et al. 2011). Thus, the oscillation of NAD+ has important consequences on cellular physiology, including changes in chromatin remodeling and other downstream molecular pathways, providing a direct link between circadian control and metabolic regulation via SIRT1, and controlling the cellular levels of its own coenzyme (Fig. 7).
5. THE AGING AND SENESCENT EPIGENOMES AND CONNECTIONS TO METABOLISM
The question of whether epigenetic alterations are a driving feature of cellular aging is important, given the longstanding known connections between life span, chromatin enzymes, and metabolic changes. In particular, the discovery that yeast Sir2, a key protein in heterochromatic silencing and promoting longevity, is an NAD+-dependent HDAC, has suggested that chromatin is involved in aging (Imai et al. 2000; Landry et al. 2000; Lin et al. 2000; Schwer and Verdin 2008; Chalkiadaki and Guarente 2012). Further observations, linking Sirtuin pathways with life span extension by calorie/dietrary restriction, underscored the role of metabolism in regulating longevity. Cellular senescence is a response to environmental stress and cellular damage whereby cells cease to replicate, but are metabolically active. This process is, in fact, physiologically linked to aging, accelerating the process and involving enormous epigenetic alterations. Thus, as discussed below, numerous findings have been made, establishing the involvement of epigenetic regulation in the aging processes.
5.1. Role of Histone Acetylation and Histone Methylation in Longevity
The Sir (silent information regulator) pathway, which includes the Sir2/Sir3/Sir4 proteins in budding yeast, functions to silence gene expression in heterochromatin (detailed in Grunstein and Gasser 2013). Mutations in Sir genes also shorten yeast life span, whereas, conversely, SIR2 gene duplications lengthen life span (Kaeberlein et al. 1999; Lamming et al. 2005). Hst2 is another regulator of life span in yeast (see Table 1 of Seto and Yoshida 2014) and a member of the large family of catalytically related deacetylases, called Sirtuins. Mammalian Sirtuins have many substrates, both nuclear and outside the nucleus. For example, p53 deacetylation by Sirt1 has been linked to cellular senescence (Chua et al. 2005). The large number of substrates has thus called into question whether chromatin is in fact a crucial substrate for Sirtuin action in the aging process and, more generally, whether life span is directly impacted by epigenetic regulation.
Chromatin structure studies over the last few years have, however, confirmed a direct involvement of epigenetics in regulating aging. In yeast, global levels of histone H4K16 acetylation increase in older cells, particularly at heterochromatin regions, such as telomeres and ribosomal DNA. This was corroborated by experiments substituting glutamine at histone H4K16Q genome-wide, to mimic H4K16 acetylation, which led to premature aging via a Sir2-dependent genetic pathway. Also, the constitutive reduction in acetylation near heterochromatin via loss of Sas2, a heterochromatin-directed HAT, lengthened life span (Dang et al. 2009). Taken together, these results confirm chromatin as an aging-relevant Sir2 substrate.
Histone acetylation is typically induced at promoters during gene activation. This modification is able to generally open chromatin structure, as shown directly for H4K16 acetylation on in vitro assembled chromatin (Shogren-Knaak et al. 2006). Thus, the finding that acetylation increases as cells age, and loss of certain acetylation pathways, such as Sas2, extends life span, suggests that chromatin relaxation and opening may negatively impact longevity (Dang et al. 2009). Furthermore, disruptions in chromatin structure occur as a consequence of the age-related decline in overall core histones, which are needed for the continued Asf1 chaperone-assisted chromatin assembly process (Feser et al. 2010), also accelerating aging. Thus, the global reduction of histone levels leads to increased transcription genome-wide and mobilization of transposable elements, which causes genome instability and chromosomal translocations (Hu et al. 2014). In contrast, elevation of histone expression, either by direct up-regulation or by deleting the Hir repressors of histone transcription, delays aging (Feser et al. 2010). These findings emphasize the importance of maintaining the integrity of chromatin structure for appropriate gene expression and genome stability and, hence, enhancing longevity.
Key hypotheses that have followed on from the S. cerevisiae studies are that the modulation of chromatin toward a more relaxed state is relevant to longevity in more complex eukaryotes; there may be a trend toward open chromatin during aging, and these chromatin trends may include alterations in histone methylation as well. In the worm, Caenorhabditis elegans, life span extension was observed following a reduction in components of the trithorax ASH-2 complex, consistent with the latter hypothesis. The ASH-2 complex is orthologous to COMPASS, the complex that methylates H3K4 over promoter TSSs during gene activation. Interestingly, the ASH-2 reduction was unable to extend life span if the reverse key demethylase for H3K4me3 was reduced, strongly implicating the actual need for a reduction in H3K4 methylation to observe a reduction in life span. A gene ontology category, showing altered gene expression with age, includes known life span–regulatory genes, emphasizing the fact that specific genes may have a role in extending life span (Greer et al. 2010). Interestingly, this pathway to extend life span requires an intact germline and it persists through the germline to extend life span in a finite number of subsequent generations (i.e., providing an example of transgenerational memory of epigenetic alterations [Greer et al. 2011]).
Turning to studies in the fly, Drosophila melanogaster, loss of repressive chromatin occurs during aging. The level of histone H3K9me3, a heterochromatic modification associated with pericentric and telomeric locations, decreases at these locations and broadly increases at euchromatin genome-wide. This imbalance in repressive chromatin observed with age is mirrored by similar changes in the localization of HP1, a histone H3K9me3-binding protein, as well as changes in transcription across the genome (Wood et al. 2010). These observations in the yeast, worm, and fly model systems consistently implicate a reduction of heterochromatin and the relaxation of chromatin-mediated transcriptional control during aging, leading to shortened life span.
In the mouse, among the Sirtuins, Sirt6 has a direct relationship to aging via maintaining chromatin integrity (introduced in Sec. 2.1). SIRT6 is nuclear localized and chromatin associated (Masri et al. 2014). Sirt6 gene deletion leads to mice with a short life span and an aging-like degenerative condition because of the impairment of a DNA damage response base-excision repair pathway (Mostoslavsky et al. 2006). In human cultured fibroblasts, the reduction of Sirt6 levels leads to premature senescence (described below in Sec. 5.3), in which cellular replicative life span is dramatically shortened. A key question in all of these correlations with life span is whether chromatin and histones are direct substrates for Sirt6-related rapid aging; SIRT6 was found to specifically target H3K9ac, among numerous major histone acetylation sites, for deacetylation both in vitro and at telomeres in vivo. Further, SIRT6 stabilizes telomeres to antagonize replication-associated dysfunction and chromosome end joining (Michishita et al. 2008). One key SIRT6 pathway, regulating cellular senescence in the mouse, is via NF-κB-regulated genes involved in senescence and apoptosis; these genes are normally maintained in a down-regulated state via Sirt6-dependent deacetylation, but up-regulated on Sirt6 depletion in an NF-κB-dependent manner (Kawahara et al. 2009). Hence, mammalian aging is clearly regulated by histone acetylation, and, consistent with studies in model organisms, low histone acetylation maintains genome integrity, leading to a general model in which compaction of the epigenome and, in particular, the heterochromatic compartment, enhances longevity.
5.2. Calorie Restriction, Life Span Extension, and Epigenetic Regulation
Calorie restriction (CR), an extreme form of altered metabolism, has a beneficial effect on longevity in fungal and most animal models; anecdotal evidence suggests that humans may also benefit from dietary restriction. One pathway to extend life span through lowered nutrients is via the targets of rapamycin (TOR) signaling pathway, which limits protein translation (Kaeberlein et al. 2005). Another well-established CR pathway is via the activation of Sirtuins through elevated NAD+ levels, a cofactor for Sirtuins (Fig. 2) (Cohen et al. 2004). Given the beneficial role of maintaining Sir-2-mediated H4K16 deacetylation for yeast longevity, chromatin is highly likely to be a CR-relevant target.
The ATP-dependent Isw2 complex mediates another CR pathway for life span regulation. In this case, Isw2-dependent nucleosome remodeling represses stress response genes, whereas deletion of Isw2, which partially mimics the effect of CR through the up-regulation of stress response genes, extends life span. This is a conserved life span regulatory pathway in yeast and worms, and reduction of the orthologous complexes also extends life span (Dang et al. 2014).
5.3. Senescence, Premature Aging, and Profound Epigenome Restructuring
Altered chromatin features in aging mammalian cells include large-scale reorganization of cytological heterochromatic structures and dramatic changes in broad epigenomic domains. These alterations involve the nuclear lamina, a protein structural matrix that associates with the interior face of the nuclear membrane. Normally, broad heterochromatic gene-poor genomic regions associate with the lamina, called lamin-associated domains (LADs). They contain relevant repressive chromatin features (e.g., H3K9me3 and HP1 binding) and cognate enzymes map to these large domains (e.g., H3K27me3/EZH2) (Kind and van Steensel 2010). The premature aging syndrome, HGPS (Hutchinson–Guilford progeria syndrome), is caused by dominant negative mutations in lamin A, correlating with reduced levels of heterochromatic H3K9me3 and lowered HP1, the heterochromatin protein that binds to H3K9me3 (Scaffidi and Misteli 2005). Similarly, aged fibroblasts display reconfiguration of nuclear lamin A and a reduction in H3K9me3 marks and HP1, as well as an increase in foci bearing the phosphorylated H2AX variant (indicating DNA damage; see Henikoff and Smith 2014), which are common hallmarks of HGPS (Scaffidi and Misteli 2006). Thus, HGPS is an accelerated aging syndrome caused by dramatic alterations of the nuclear lamina and concomitant reduced heterochromatin.
As mentioned above, cellular senescence is an irreversible cell cycle arrest in response to stress caused by expression of activated oncogenes or long-term replication, both of which are associated with cellular dysfunction and, thus, the cell cycle arrest is protective against cancer (Narita et al. 2003). However, senescence also correlates with aging in primates (Herbig et al. 2006) and, importantly, was recently causally linked to aging in a mouse progerin premature aging model via demonstration that removal of senescent cells ameliorates age-associated tissue pathologies (Baker et al. 2011). Hence, senescence is a provocative state of protection against cancerous malignancy, but also correlates with cellular damage associated with aging. Aberrant large-scale chromatin foci, called senescent-associated heterochromatin foci (SAHF), have been shown to arise during senescence and are enriched in heterochromatin proteins and chromatin changes indicative of DNA damage, such as HMGA (high mobility group AT hook protein; Narita et al. 2003, 2006), H2AXph (S139 phosphorylated histone variant H2AX protein; Sedelnikova et al. 2004), and macroH2A (H2A variant protein with macro domain; Zhang et al. 2005). These acquired SAHFs repress certain genes required for cell proliferation (Narita et al. 2006).
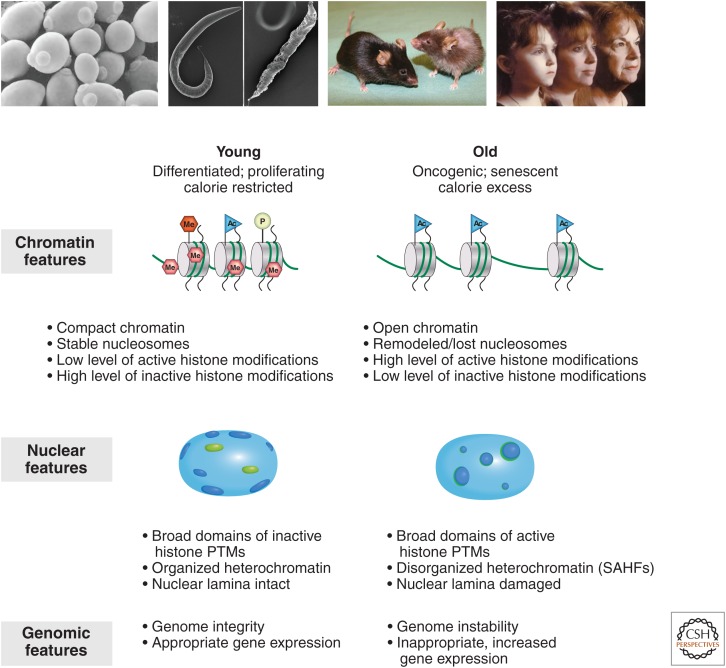
SAHFs have been thought to represent increased heterochromatin, but they do not correlate with obvious changes in heterochromatic H3K9 methylation or H3K27me3. Rather, they show possible higher-order epigenetic changes (Chandra et al. 2012). A reduction of LaminB1 was shown to lead to premature senescence (Shimi et al. 2011; Shah et al. 2013). Interestingly, extremely large domains of reduced heterochromatin develop in senescent cells, which correspond to LADs in proliferating cells, displaying broad increases in H3K4me3 (Shah et al. 2013) and decreases in DNA methylation (Cruickshanks et al. 2013). Hence, senescent cells appear to undergo complex epigenetic restructuring, with focal regions of increased heterochromatin (SAHFs) and broad regions of decreased heterochromatin over LADs (Fig. 8).
Figure 8.
Features of aging and senescent cells. (Top) Aging is a biological inevitability for all eukaryotes, from budding yeast and C. elegans to mice and humans (depicted). The features of senescence and cellular aging are compared between a normal or “young” cell (left) and a diseased or “old” cell (right) at three levels: chromatin, nuclear organization, and integrity of the genome. (Top images [left to right] from chemistryland.com; from Hazreet Gill, Francis Ghandi, and Arjumand Ghazi, University of Pittsburgh School of Medicine, http://www.chp.edu/CHP/ghazilab; courtesy of Christopher Fisher; from victorymedspa.com.)
There are challenging questions surrounding alterations of chromatin during the aging process. Much is known about altered concentration of certain metabolites (such as NAD/NADH ratio) during aging and how these changes affect, for example, Sirtuins; however, virtually nothing is known about how these changes might drive the chromatin alterations genome-wide. It remains to be seen whether other metabolites, such as acetyl-CoA and SAM, alter chromatin during aging, and how this is involved in CR conditions to extend life span. Also, what is the precise relationship between large-scale nuclear structural changes and genome-wide changes in chromatin? And, which age-associated chromatin changes reflect degradative processes and which reflect protective processes?
6. CONCLUDING REMARKS
It is evident from the breadth of subjects discussed in this article that there are many mechanistic and physiological aspects within the broad scope of metabolism and epigenetics. The mechanisms that transduce metabolic signals into epigenetic changes encompass enzymes typified by acetylases and deacetylases, which respond to profound shifts in the cellular milieu via their use of small molecule metabolites as direct cofactors and substrates that alter their enzymatic activity. One key proposal to come from the studies of HATs and HDACs is that many other epigenetic enzymes may also nimbly interact with the environment via their response to changing concentrations of metabolites.
The “circadian epigenome” and the “aging epigenome” represent examples of striking physiological states that are influenced by metabolic changes, which impinge on chromatin. Many more physiological states are altered by metabolic epigenetics, such as numerous types of cancers; many others await elucidation.
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, Danny Reinberg, and Monika Lachner
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
*Reference is also in this subject collection.
- Aguilar-Arnal L, Hakim O, Patel VR, Baldi P, Hager GL, Sassone-Corsi P. 2013. Cycles in spatial and temporal chromosomal organization driven by the circadian clock. Nat Struct Mol Biol 20: 1206–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albaugh BN, Arnold KM, Denu JM. 2011. KAT(ching) metabolism by the tail: Insight into the links between lysine acetyltransferases and metabolism. Chembiochem 12: 290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alenghat T, Meyers K, Mullican SE, Leitner K, Adeniji-Adele A, Avila J, Bućan M, Ahima RS, Kaestner KH, Lazar MA. 2008. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature 456: 997–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang YS, Gaspar-Maia A, Lemischka IR, Bernstein E. 2011. Stem cells and reprogramming: Breaking the epigenetic barrier? Trends Pharmacol Sci 32: 394–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyannur PS, Moffett JR, Madhavarao CN, Arun P, Vishnu N, Jacobowitz DM, Hallows WC, Denu JM, Namboodiri AM. 2010. Nuclear-cytoplasmic localization of acetyl coenzyme a synthetase-1 in the rat brain. J Comp Neurol 518: 2952–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. 2008. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134: 317–328. [DOI] [PubMed] [Google Scholar]
- *.Audia JE, Campbell RM. 2014. Histone modifications and cancer. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austenaa L, Barozzi I, Chronowska A, Termanini A, Ostuni R, Prosperini E, Stewart AF, Testa G, Natoli G. 2012. The histone methyltransferase Wbp7 controls macrophage function through GPI glycolipid anchor synthesis. Immunity 36: 572–585. [DOI] [PubMed] [Google Scholar]
- Bai P, Cantó C, Oudart H, Brunyánszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, et al. 2011. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab 13: 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, Kirkland JL, van Deursen JM. 2011. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479: 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen K, et al. 2012. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 487: 114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Becker PB, Workman JL. 2013. Nucleosome remodeling and epigenetics. Cold Spring Harb Perspect Biol 5: a017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellet MM, Nakahata Y, Boudjelal M, Watts E, Mossakowska DE, Edwards KA, Cervantes M, Astarita G, Loh C, Ellis JL, et al. 2013. Pharmacological modulation of circadian rhythms by synthetic activators of the deacetylase SIRT1. Proc Natl Acad Sci 110: 3333–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beneke S. 2012. Regulation of chromatin structure by poly(ADP-ribosyl)-ation. Front Genet 3: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndsen CE, Denu JM. 2008. Catalysis and substrate selection by histone/protein lysine acetyltransferases. Curr Opin Struct Biol 18: 682–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvári M, Piper MD, Hoddinott M, Sutphin GL, Leko V, McElwee JJ, et al. 2011. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature 477: 482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Sutter BM, Li B, Tu BP. 2011. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell 42: 426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C, Jiang LQ, Deshmukh AS, Mataki C, Coste A, Lagouge M, Zierath JR, Auwerx J. 2010. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab 11: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalkiadaki A, Guarente L. 2012. Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat Rev Endocrinol 8: 287–296. [DOI] [PubMed] [Google Scholar]
- Chandra T, Kirschner K, Thuret JY, Pope BD, Ryba T, Newman S, Ahmed K, Samarajiwa SA, Salama R, Carroll T, et al. 2012. Independence of repressive histone marks and chromatin compaction during senescent heterochromatic layer formation. Mol Cell 47: 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, Guarente L. 2013. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell 153: 1448–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JJ, London IM. 1981. Hemin enhances the differentiation of mouse 3T3 cells to adipocytes. Cell 26: 117–122. [DOI] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. 1997. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90: 569–580. [DOI] [PubMed] [Google Scholar]
- *.Cheng X. 2014. Structural and functional coordination of DNA and histone methylation. Cold Spring Harb Perspect Biol 6: a018747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua KF, Mostoslavsky R, Lombard DB, Pang WW, Saito S, Franco S, Kaushal D, Cheng HL, Fischer MR, Stokes N, et al. 2005. Mammalian SIRT1 limits replicative life span in response to chronic genotoxic stress. Cell Metab 2: 67–76. [DOI] [PubMed] [Google Scholar]
- Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. 2004. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 305: 390–392. [DOI] [PubMed] [Google Scholar]
- Crosio C, Cermakian N, Allis CD, Sassone-Corsi P. 2000. Light induces chromatin modification in cells of the mammalian circadian clock. Nat Neurosci 3: 1241–1247. [DOI] [PubMed] [Google Scholar]
- Cruickshanks HA, McBryan T, Nelson DM, Vanderkraats ND, Shah PP, van Tuyn J, Singh Rai T, Brock C, Donahue G, Dunican DS, et al. 2013. Senescent cells harbour features of the cancer epigenome. Nat Cell Biol 15: 1495–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis AM, Seo SB, Westgate EJ, Rudic RD, Smyth EM, Chakravarti D, FitzGerald GA, McNamara P. 2004. Histone acetyltransferase-dependent chromatin remodeling and the vascular clock. J Biol Chem 279: 7091–7097. [DOI] [PubMed] [Google Scholar]
- Czarna A, Breitkreuz H, Mahrenholz CC, Arens J, Strauss HM, Wolf E. 2011. Quantitative analyses of cryptochrome-mBMAL1 interactions: Mechanistic insights into the transcriptional regulation of the mammalian circadian clock. J Biol Chem 286: 22414–22425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. 2009. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 459: 802–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Sutphin GL, Dorsey JA, Otte GL, Cao K, Perry RM, Wanat JJ, Saviolaki D, Murakami CJ, Tsuchiyama S, et al. 2014. Inactivation of yeast Isw2 chromatin remodeling enzyme mimics longevity effect of calorie restriction via induction of genotoxic stress response. Cell Metab 19: 952–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Dekker J, Misteli T. 2014. Dosage compensation in Drosophila. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA, McKnight SL. 2002. NPAS2: A gas-responsive transcription factor. Science 298: 2385–2387. [DOI] [PubMed] [Google Scholar]
- DiTacchio L, Le HD, Vollmers C, Hatori M, Witcher M, Secombe J, Panda S. 2011. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science 333: 1881–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P. 2006. Circadian regulator CLOCK is a histone acetyltransferase. Cell 125: 497–508. [DOI] [PubMed] [Google Scholar]
- Dolinoy DC, Huang D, Jirtle RL. 2007. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci 104: 13056–13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominy JE Jr, Lee Y, Gerhart-Hines Z, Puigserver P. 2010. Nutrient-dependent regulation of PGC-1α’s acetylation state and metabolic function through the enzymatic activities of Sirt1/GCN5. Biochim Biophys Acta 1804: 1676–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong HA, Weitz CJ. 2014. Temporal orchestration of repressive chromatin modifiers by circadian clock Period complexes. Nat Struct Mol Biol 21: 126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, de Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, Sassone-Corsi P. 2013. Reprogramming of the circadian clock by nutritional challenge. Cell 155: 1464–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray JP, Lee C, Wade PA, Reppert SM. 2003. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421: 177–182. [DOI] [PubMed] [Google Scholar]
- Etchegaray JP, Yang X, DeBruyne JP, Peters AH, Weaver DR, Jenuwein T, Reppert SM. 2006. The polycomb group protein EZH2 is required for mammalian circadian clock function. J Biol Chem 281: 21209–21215. [DOI] [PubMed] [Google Scholar]
- Feldman JL, Baeza J, Denu JM. 2013. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem 288: 31350–31356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. 2011. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331: 1315–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK. 2010. Elevated histone expression promotes life span extension. Mol Cell 39: 724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Deng CX, Mostoslavsky R. 2009. Recent progress in the biology and physiology of sirtuins. Nature 460: 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Bacher A. 2008. Biosynthesis of vitamin B2: Structure and mechanism of riboflavin synthase. Arch Biochem Biophys 474: 252. [DOI] [PubMed] [Google Scholar]
- Friis RM, Wu BP, Reinke SN, Hockman DJ, Sykes BD, Schultz MC. 2009. A glycolytic burst drives glucose induction of global histone acetylation by picNuA4 and SAGA. Nucleic Acids Res 37: 3969–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garten A, Petzold S, Körner A, Imai S, Kiess W. 2009. Nampt: Linking NAD biology, metabolism and cancer. Trends Endocrinol Metab 20: 130–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill LA, et al. 2007. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318: 1469–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, Kraus WL. 2012. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol 13: 411–424. [DOI] [PubMed] [Google Scholar]
- Gil R, Barth S, Kanfi Y, Cohen HY. 2013. SIRT6 exhibits nucleosome-dependent deacetylase activity. Nucleic Acids Res 41: 8537–8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Hauswirth AG, Green EM, Leeman DS, Maro GS, Han S, Banko MR, Gozani O, Brunet A. 2010. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 466: 383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, Benayoun BA, Shi Y, Brunet A. 2011. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 479: 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo MA, Colombatto S. 2008. S-adenosylmethionine and its products. Amino Acids 34: 187–193. [DOI] [PubMed] [Google Scholar]
- Grob A, Roussel P, Wright JE, McStay B, Hernandez-Verdun D, Sirri V. 2009. Involvement of SIRT7 in resumption of rDNA transcription at the exit from mitosis. J Cell Sci 122: 489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Grunstein M, Gasser SM. 2013. Epigenetics in Saccharomyces cerevisiae. Cold Spring Harb Perspect Biol 5: a017491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan KL, Xiong Y. 2011. Regulation of intermediary metabolism by protein acetylation. Trends Biochem Sci 36: 108–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guastafierro T, Cecchinelli B, Zampieri M, Reale A, Riggio G, Sthandier O, Zupi G, Calabrese L, Caiafa P. 2008. CCCTC-binding factor activates PARP-1 affecting DNA methylation machinery. J Biol Chem 283: 21873–21880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows WC, Lee S, Denu JM. 2006. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci 103: 10230–10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Henikoff S, Smith MM. 2014. Histone variants and epigenetics. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM. 2006. Cellular senescence in aging primates. Science 311: 1257. [DOI] [PubMed] [Google Scholar]
- Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, Sassone-Corsi P. 2007. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature 450: 1086–1090. [DOI] [PubMed] [Google Scholar]
- Hou H, Yu H. 2010. Structural insights into histone lysine demethylation. Curr Opin Struct Biol 20: 739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Pirinen E, Auwerx J. 2012. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol 13: 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Chen K, Xia Z, Chavez M, Pal S, Seol JH, Chen CC, Li W, Tyler JK. 2014. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev 28: 396–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 13: 2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. 2005. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310: 1193–1196. [DOI] [PubMed] [Google Scholar]
- Kaochar S, Tu BP. 2012. Gatekeepers of chromatin: Small metabolites elicit big changes in gene expression. Trends Biochem Sci 37: 477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada S, Sassone-Corsi P. 2010. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol 17: 1414–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada S, Imhof A, Sassone-Corsi P. 2012. Connecting threads: Epigenetics and metabolism. Cell 148: 24–28. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Ikura T, Hoshikawa Y, Tashiro S, Ito T, Ohta M, Kera Y, Noda T, Igarashi K. 2011. Methionine adenosyltransferase II serves as a transcriptional corepressor of Maf oncoprotein. Mol Cell 41: 554–566. [DOI] [PubMed] [Google Scholar]
- Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang HY, et al. 2009. SIRT6 links histone H3 lysine 9 deacetylation to NF-κB-dependent gene expression and organismal life span. Cell 136: 62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara TL, Rapicavoli NA, Wu AR, Qu K, Quake SR, Chang HY. 2011. Dynamic chromatin localization of Sirt6 shapes stress- and aging-related transcriptional networks. PLoS Genet 7: e1002153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kera Y, Katoh Y, Ohta M, Matsumoto M, Takano-Yamamoto T, Igarashi K. 2013. Methionine adenosyltransferase II-dependent histone H3K9 methylation at the COX-2 gene locus. J Biol Chem 288: 13592–13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Xiao C, Wang RH, Lahusen T, Xu X, Vassilopoulos A, Vazquez-Ortiz G, Jeong WI, Park O, Ki SH, et al. 2010. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab 12: 224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, van Steensel B. 2010. Genome-nuclear lamina interactions and gene regulation. Curr Opin Cell Biol 22: 320–325. [DOI] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. 2012. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338: 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolthur-Seetharam U, Dantzer F, McBurney MW, de Murcia G, Sassone-Corsi P. 2006. Control of AIF-mediated cell death by the functional interplay of SIRT1 and PARP-1 in response to DNA damage. Cell Cycle 5: 873–877. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. 2010. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell 39: 736–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, et al. 2009. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326: 437–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. 2005. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science 309: 1861–1864. [DOI] [PubMed] [Google Scholar]
- Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. 2000. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci 97: 5807–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Martelot G, Canella D, Symul L, Migliavacca E, Gilardi F, Liechti R, Martin O, Harshman K, Delorenzi M, Desvergne B, et al. 2012. Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol 10: e1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Li E, Zhang Y. 2014. DNA methylation in mammals. Cold Spring Harb Perspect Biol 6: a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QS, Cai S, Borchardt RT, Fang J, Kuczera K, Middaugh CR, Schowen RL. 2007. Comparative kinetics of cofactor association and dissociation for the human and trypanosomal S-adenosylhomocysteine hydrolases. 1. Basic features of the association and dissociation processes. Biochemistry 46: 5798–5809. [DOI] [PubMed] [Google Scholar]
- Liang G, Zhang Y. 2013. Embryonic stem cell and induced pluripotent stem cell: An epigenetic perspective. Cell Res 23: 49–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289: 2126–2128. [DOI] [PubMed] [Google Scholar]
- Londoño Gentile T, Lu C, Lodato PM, Tse S, Olejniczak SH, Witze ES, Thompson CB, Wellen KE. 2013. DNMT1 is regulated by ATP-citrate lyase and maintains methylation patterns during adipocyte differentiation. Mol Cell Biol 33: 3864–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Thompson CB. 2012. Metabolic regulation of epigenetics. Cell Metab 16: 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. 2012. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483: 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macheroux P, Kappes B, Ealick SE. 2011. Flavogenomics—A genomic and structural view of flavin-dependent proteins. FEBS J 278: 2625–2634. [DOI] [PubMed] [Google Scholar]
- Magni G, Amici A, Emanuelli M, Orsomando G, Raffaelli N, Ruggieri S. 2004. Structure and function of nicotinamide mononucleotide adenylyltransferase. Curr Med Chem 11: 873–885. [DOI] [PubMed] [Google Scholar]
- Masri S, Sassone-Corsi P. 2010. Plasticity and specificity of the circadian epigenome. Nat Neurosci 13: 1324–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Patel VR, Eckel-Mahan KL, Peleg S, Forne I, Ladurner AG, Baldi P, Imhof A, Sassone-Corsi P. 2013. Circadian acetylome reveals regulation of mitochondrial metabolic pathways. Proc Natl Acad Sci 110: 3339–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Rigor P, Cervantes M, Ceglia N, Sebastian C, Xiao C, Roqueta-Rivera M, Deng C, Osborne TF, Mostoslavsky R, et al. 2014. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell 158: 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner S, Altmeyer M, Zhao H, Pozivil A, Roschitzki B, Gehrig P, Rutishauser D, Huang D, Caflisch A, Hottiger MO. 2010. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res 38: 6350–6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger E, Imhof A, Patel D, Kahl P, Hoffmeyer K, Friedrichs N, Müller JM, Greschik H, Kirfel J, Ji S, et al. 2010. Phosphorylation of histone H3T6 by PKCβ(I) controls demethylation at histone H3K4. Nature 464: 792–796. [DOI] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, et al. 2008. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452: 492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, Chua KF. 2009. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 8: 2664–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. 2006. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124: 315–329. [DOI] [PubMed] [Google Scholar]
- Nader N, Chrousos GP, Kino T. 2009. Circadian rhythm transcription factor CLOCK regulates the transcriptional activity of the glucocorticoid receptor by acetylating its hinge region lysine cluster: Potential physiological implications. FASEB J 23: 1572–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. 2008. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134: 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. 2009. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324: 654–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam HJ, Boo K, Kim D, Han DH, Choe HK, Kim CR, Sun W, Kim H, Kim K, Lee H, et al. 2014. Phosphorylation of LSD1 by PKCα is crucial for circadian rhythmicity and phase resetting. Mol Cell 53: 791–805. [DOI] [PubMed] [Google Scholar]
- Narita M, Nũnez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113: 703–716. [DOI] [PubMed] [Google Scholar]
- Narita M, Narita M, Krizhanovsky V, Nuñez S, Chicas A, Hearn SA, Myers MP, Lowe SW. 2006. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell 126: 503–514. [DOI] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. 2005. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{α}. J Biol Chem 280: 16456–16460. [DOI] [PubMed] [Google Scholar]
- Nikiforov A, Dölle C, Niere M, Ziegler M. 2011. Pathways and subcellular compartmentation of NAD biosynthesis in human cells: From entry of extracellular precursors to mitochondrial NAD generation. J Biol Chem 286: 21767–21778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. 2002. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109: 307–320. [DOI] [PubMed] [Google Scholar]
- Peek CB, Affinati AH, Ramsey KM, Kuo HY, Yu W, Sena LA, Ilkayeva O, Marcheva B, Kobayashi Y, Omura C, et al. 2013. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science 342: 1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponka P. 1999. Cell biology of heme. Am J Med Sci 318: 241–256. [DOI] [PubMed] [Google Scholar]
- Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. 2007. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBα and REV-ERBβ. Nat Struct Mol Biol 14: 1207–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, Kellis M, Gymrek M, Issner R, Coyne M, et al. 2011. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell 147: 1628–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy V, Williams JS, Robinson KM, Sopko RL, Schultz MC. 2003. Global control of histone modification by the anaphase-promoting complex. Mol Cell Biol 23: 9136–9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, et al. 2009. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324: 651–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reale A, Matteis GD, Galleazzi G, Zampieri M, Caiafa P. 2005. Modulation of DNMT1 activity by ADP-ribose polymers. Oncogene 24: 13–19. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. 2002. Coordination of circadian timing in mammals. Nature 418: 935–941. [DOI] [PubMed] [Google Scholar]
- Ripperger JA, Schibler U. 2006. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet 38: 369–374. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. 2005. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434: 113–118. [DOI] [PubMed] [Google Scholar]
- Sahar S, Sassone-Corsi P. 2012. Circadian rhythms and memory formation: Regulation by chromatin remodeling. Front Mol Neurosci 5: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahar S, Nin V, Barbosa MT, Chini EN, Sassone-Corsi P. 2011. Altered behavioral and metabolic circadian rhythms in mice with disrupted NAD+ oscillation. Aging 3: 794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahar S, Masubuchi S, Eckel-Mahan K, Vollmer S, Galla L, Ceglia N, Masri S, Barth TK, Grimaldi B, Oluyemi O, et al. 2014. Circadian control of fatty acid elongation by SIRT1 protein-mediated deacetylation of acetyl-coenzyme A synthetase 1. J Biol Chem 289: 6091–6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier JJ, French S, Osheim Y, Cheung WL, Gallo CM, Beyer AL, Smith JS. 2002. RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J 21: 4959–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. 2005. Reversal of the cellular phenotype in the premature aging disease Hutchinson–Gilford progeria syndrome. Nat Med 11: 440–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. 2006. Lamin A-dependent nuclear defects in human aging. Science 312: 1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U, Sassone-Corsi P. 2002. A web of circadian pacemakers. Cell 111: 919–922. [DOI] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G. 2006. Poly(ADP-ribose): Novel functions for an old molecule. Nat Rev Mol Cell Biol 7: 517–528. [DOI] [PubMed] [Google Scholar]
- Schwer B, Verdin E. 2008. Conserved metabolic regulatory functions of sirtuins. Cell Metab 7: 104. [DOI] [PubMed] [Google Scholar]
- Sedelnikova OA, Horikawa I, Zimonjic DB, Popescu NC, Bonner WM, Barrett JC. 2004. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat Cell Biol 6: 168–170. [DOI] [PubMed] [Google Scholar]
- *.Seto E, Yoshida M. 2014. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb Perspect Biol 6: a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PP, Donahue G, Otte GL, Capell BC, Nelson DM, Cao K, Aggarwala V, Cruickshanks HA, Rai TS, McBryan T, et al. 2013. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev 27: 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Shi YG, Tsukuda Y. 2013. The discovery of histone demethylases. Cold Spring Harb Perspect Biol 5: a017947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, et al. 2010. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab 12: 654–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Butin-Israeli V, Adam SA, Hamanaka RB, Goldman AE, Lucas CA, Shumaker DK, Kosak ST, Chandel NS, Goldman RD. 2011. The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev 25: 2579–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, He M, Liu Y, Paredes S, Villanova L, Brown K, Qiu X, Nabavi N, Mohrin M, Wojnoonski K, et al. 2013. SIRT7 represses Myc activity to suppress ER stress and prevent fatty liver disease. Cell Rep 5: 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311: 844–847. [DOI] [PubMed] [Google Scholar]
- Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, et al. 2013. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 339: 222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gómez-Marín C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF, et al. 2014. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507: 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. 2006. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol Cell 23: 207–217. [DOI] [PubMed] [Google Scholar]
- Tamaru T, Hattori M, Ninomiya Y, Kawamura G, Varès G, Honda K, Mishra DP, Wang B, Benjamin I, Sassone-Corsi P, et al. 2013. ROS stress resets circadian clocks to coordinate pro-survival signals. PLoS One 8: e82006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teperino R, Schoonjans K, Auwerx J. 2010. Histone methyl transferases and demethylases; can they link metabolism and transcription? Cell Metab 12: 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, Sebastian C, Cosentino C, Martinez-Pastor B, Giacosa S, et al. 2013. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell 51: 454–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YC, Greco TM, Boonmee A, Miteva Y, Cristea IM. 2012. Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol Cell Proteomics 11: 60–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, Kudlicki A, Rowicka M, McKnight SL. 2005. Logic of the yeast metabolic cycle: Temporal compartmentalization of cellular processes. Science 310: 1152–1158. [DOI] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. 2012. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483: 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, et al. 2009. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet 41: 125–129. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. 2009. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324: 1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JG, Hillenmeyer S, Lawrence C, Chang C, Hosier S, Lightfoot W, Mukherjee E, Jiang N, Schorl C, Brodsky AS, et al. 2010. Chromatin remodeling in the aging genome of Drosophila. Aging Cell 9: 971–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Kim HS, Lahusen T, Wang RH, Xu X, Gavrilova O, Jou W, Gius D, Deng CX. 2010. SIRT6 deficiency results in severe hypoglycemia by enhancing both basal and insulin-stimulated glucose uptake in mice. J Biol Chem 285: 36776–36784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Wang H, Liu Y, Shao C. 2008. Analysis of gene regulatory networks in the mammalian circadian rhythm. PLoS Comput Biol 4: e1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Zwaans BM, Eckersdorff M, Lombard DB. 2009. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle 8: 2662–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, et al. 2007. Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science 318: 1786–1789. [DOI] [PubMed] [Google Scholar]
- Yu W, Ginjala V, Pant V, Chernukhin I, Whitehead J, Docquier F, Farrar D, Tavoosidana G, Mukhopadhyay R, Kanduri C, et al. 2004. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat Genet 36: 1105–1110. [DOI] [PubMed] [Google Scholar]
- Zampieri M, Passananti C, Calabrese R, Perilli M, Corbi N, De Cave F, Guastafierro T, Bacalini MG, Reale A, Amicosante G, et al. 2009. Parp1 localizes within the Dnmt1 promoter and protects its unmethylated state by its enzymatic activity. PLoS One 4: e4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampieri M, Guastafierro T, Calabrese R, Ciccarone F, Bacalini MG, Reale A, Perilli M, Passananti C, Caiafa P. 2012. ADP-ribose polymers localized on Ctcf-Parp1-Dnmt1 complex prevent methylation of Ctcf target sites. Biochem J 441: 645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, Erzberger JP, Serebriiskii IG, Canutescu AA, Dunbrack RL, et al. 2005. Formation of macroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell 8: 19–30. [DOI] [PubMed] [Google Scholar]
- Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, et al. 2010. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1α. Cell 140: 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]