Martin et al. report that biallelic mutations in NCAPD2, NCAPH, or NCAPD3, encoding subunits of condensin complexes, cause microcephaly. Frequent anaphase chromatin bridge formation observed in apical neural progenitors during neurogenesis are the consequence of failed sister chromatid disentanglement during chromosome compaction.
Keywords: condensin, microcephaly, neurodevelopment, decatenation
Abstract
Compaction of chromosomes is essential for accurate segregation of the genome during mitosis. In vertebrates, two condensin complexes ensure timely chromosome condensation, sister chromatid disentanglement, and maintenance of mitotic chromosome structure. Here, we report that biallelic mutations in NCAPD2, NCAPH, or NCAPD3, encoding subunits of these complexes, cause microcephaly. In addition, hypomorphic Ncaph2 mice have significantly reduced brain size, with frequent anaphase chromatin bridge formation observed in apical neural progenitors during neurogenesis. Such DNA bridges also arise in condensin-deficient patient cells, where they are the consequence of failed sister chromatid disentanglement during chromosome compaction. This results in chromosome segregation errors, leading to micronucleus formation and increased aneuploidy in daughter cells. These findings establish “condensinopathies” as microcephalic disorders, with decatenation failure as an additional disease mechanism for microcephaly, implicating mitotic chromosome condensation as a key process ensuring mammalian cerebral cortex size.
Faithful and accurate segregation of the genome requires chromosome compaction and resolution of sister chromatids. At mitosis, chromosome condensation occurs through the large-scale reorganization of chromatin fibers and ensures the movement of chromosomes to daughter cells without DNA entanglement or entrapment by the cleavage furrow. In metazoans, mitotic chromosome assembly is mediated foremost through the activity of two protein complexes, condensin I and II (Hirano 2012). Condensin I and II complexes share the same overall arrangement with two core ATPases—structural maintenance of chromosomes 2 (SMC2) and SMC4—but differ in their three associated non-SMC subunits. In humans, the non-SMC subunits of condensin I are NCAPD2, NCAPG, and NCAPH (also known as Kleisin-γ), with the respective counterparts in the condensin II complex being NCAPD3, NCAPG2, and NCAPH2 (Kleisin-β) (Hirano and Mitchison 1994; Hirano et al. 1997).
Despite structural similarities, condensin I and condensin II have distinct functions, with condensin II localized in the nucleus during interphase (Hirota et al. 2004) and required for early axial shortening of chromosomes in prophase (Shintomi and Hirano 2011). Cytoplasmic condensin I does not gain access to the chromosomes until after nuclear envelope breakdown (Hirota et al. 2004), when it contributes to the lateral compaction of metaphase chromosomes (Shintomi and Hirano 2011). In addition, condensins are required for the removal of catenations between replicated chromatids that arise during DNA replication by facilitating topoisomerase II in actively resolving entangled sister chromatids (Baxter et al. 2011; Charbin et al. 2014).
In autosomal recessive primary microcephaly, brain size is markedly reduced, resulting in a cranial volume comparable with that of human ancestors several million years ago (Woods et al. 2005). This condition has therefore been extensively studied to identify factors that regulate brain growth and neurogenesis. A similar reduction in cerebral cortex size is also seen in patients with microcephalic primordial dwarfism (MPD), where, unlike primary microcephaly, significant short stature is also evident (Klingseisen and Jackson 2011). The identification of disease genes common to both disorders (O'Driscoll et al. 2003; Bond et al. 2005; Al-Dosari et al. 2010; Kalay et al. 2011; Qvist et al. 2011; Martin et al. 2014) has established that primary microcephaly and MPD represent a phenotype spectrum in which a disproportionate reduction in brain volume relative to body size is seen (Verloes et al. 2009). Disease genes identified in these microcephalic disorders encode proteins integral to cell cycle progression (Klingseisen and Jackson 2011). Many encode centrosomal proteins essential for efficient mitotic spindle formation (Bond et al. 2002, 2005; Griffith et al. 2008; Nicholas et al. 2010), and, consequently, mutations result in perturbed cleavage plane orientation of neural progenitors (Fish et al. 2006; Gruber et al. 2011; Lancaster et al. 2013; Insolera et al. 2014). Therefore, altered symmetric/asymmetric cell division dynamics in such neural stem cells has emerged as the major developmental mechanism for microcephaly (Thornton and Woods 2009). Here we report the identification of mutations in condensin complex genes and demonstrate that these arise from a different mitotic mechanism, which establishes chromosome compaction and decatenation as critical processes ensuring normal mammalian brain growth.
Results
Mutations in condensin subunits cause microcephaly
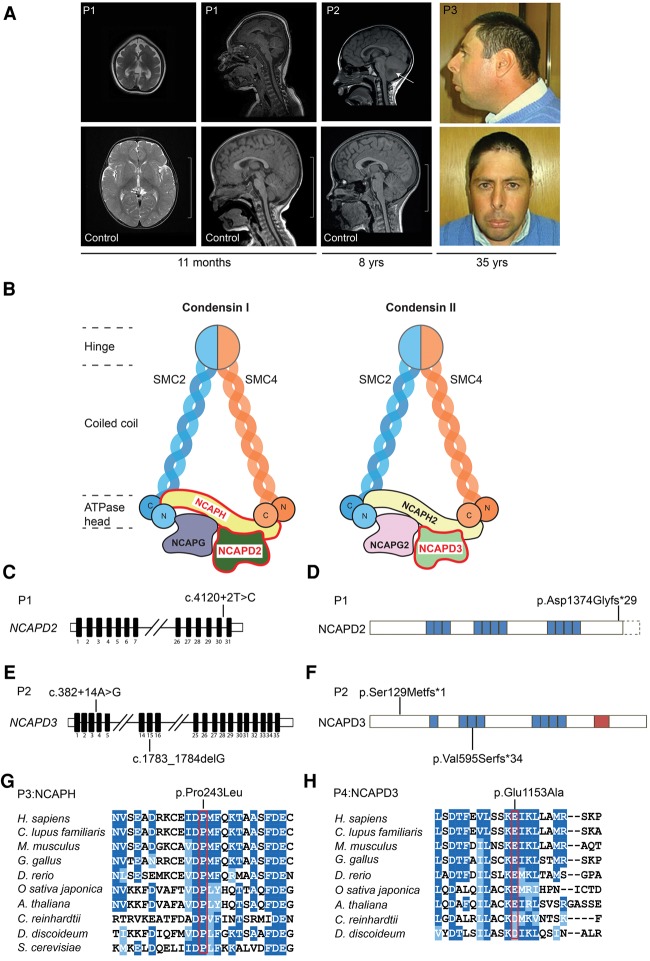
To identify novel microcephaly genes, whole-exome sequencing (WES) was performed on a cohort of patients with extreme microcephaly (occipitofrontal circumference [OFC] less than −4 SD) due to reduced cerebral cortex volume with simplified gyri but without other structural abnormalities (Fig. 1A). In patient P1, a homozygous essential splice site mutation (c.4120+2T>C) in NCAPD2 was detected, encoding one of the three non-SMC subunits of condensin I (Fig. 1B–D). In keeping with a pathogenic variant for a rare recessive disorder, it was present in a heterozygous state in both parents and was not reported in the Exome Aggregation Consortium (ExAC) database (http://exac.broadinstitute.org). To identify further mutations, we examined exome data from a further 94 families from our cohort. While no additional deleterious variants in NCAPD2 were seen, in patient P2, compound heterozygous variants were observed in its paralog, NCAPD3, encoding the corresponding subunit in the condensin II complex (Fig. 1E,F). These comprised a frameshift mutation (c.1783_1784delG) and an intronic substitution (c.382+14A>G) predicted by in silico analysis (Alamut Visual, Interactive Biosoftware) to create a new splice donor site within intron 3.
Figure 1.
Mutations in condensin I and II subunits cause microcephaly. (A) Reduced cerebral cortex size is seen in individuals with mutations in condensin subunits. (Left) Axial (T2-weighted) and sagittal (T1) MRI (magnetic resonance imaging) images from patient P1 at 11 mo of age and patient P2, sagittal (T1), aged 8 yr and 8 mo, compared with age-matched controls. (Arrow) Low-signal area within the cerebellum corresponding to the site of medulloblastoma. Bar, 10 cm. (Left) Photographs demonstrating a reduced OFC and sloping forehead in patient P3. (B) Schematic of condensin I and II complexes, with subunits containing patient mutations highlighted by red-edged boxes. Both condensins contain five subunits, sharing the same two core SMC subunits (SMC2 and SMC4), which dimerize at the hinge domain. Each SMC subunit is composed of two coiled-coil domains extending from the hinge domain and forming an anti-parallel coil that ends in an ATPase head domain. The two complexes then differ in the three non-SMC subunits: A kleisin protein (NCAPH in condensin I and NCAPH2 in condensin II) bridges the ATPase head domains of the two SMC subunits, and two HEAT repeat proteins (NCAPG and NCAPD2 in condensin I and NCAPG2 and NCAPD3 in condensin II) associate with the complex predominantly through interactions with the kleisin subunit. (C–F) Schematics of NCAPD2 and NCAPD3 genes and encoded proteins depicting mutations identified in patients P1 and P2. A homozygous essential splice site mutation is present in NCAPD2 in patient P1 (C) and is predicted to alter the C terminus of the protein (D). Compound heterozygous mutations in patient P2 (E) and their predicted protein consequences (F). (Black) Coding exons; (white boxes) untranslated regions; (blue boxes) HEAT repeats; (red) coiled-coil regions (Ono et al. 2003). (G,H) Missense mutations in patients P3 and P4 disrupt highly conserved amino acid residues. (G) The codon disrupted by the NCAPH Pro243Leu substitution is conserved in eukaryotes. (H) Amino acid 1153 is strongly conserved in condensin II NCAPD3 subunits across animal kingdoms. The alignment of species with complete condensin II complexes demonstrates that many metozoa and plants retain a glutamate at the mutated site in NCAPD3 in patient P4, while, in Amoebazoa (Dictyostelium discoideum) and green algae (Chlamydomonas reinhardtii), a physiochemically similar aspartate residue is present. Not all model organisms are shown, as some eukaryotes such as fungi and yeast possess only a single condensin complex (Hirano 2012), and use of condensin complexes in invertebrates varies (Savvidou et al. 2005). Residues that are dark blue are >60% identical, and those that are light blue are similar.
Having identified mutations in subunits of condensin I and II, we therefore proceeded to screen all eight genes that encode proteins in these two complexes using a custom AmpliSeq multiplex PCR panel and ion–proton semiconductor sequencer. Sequencing of 198 patients with microcephaly phenotypes ranging from primary microcephaly to microcephalic dwarfism identified a third patient (patient P3, OFC −4.2 SD) (Fig. 1A; Supplemental Table S1) with a homozygous missense variant in NCAPH (c.728C>T, p.Pro243Leu), encoding the kleisin subunit of the condensin I complex. This substitution was at an evolutionary conserved residue (Fig. 1G, conserved to Saccharomyces cerevisiae) and was predicted to have a deleterious effect by in silico analysis (Polyphen-2, MutationTaster, and MutationAssessor). We then extended our search to The Deciphering Developmental Disorders Study (The Deciphering Developmental Disorders Study 2015), representing a wide range of developmental disorders (n = 1000 exomes). This identified a fourth microcephalic patient (patient P4, OFC −2.7 SD) (Supplemental Table S1) with a homozygous missense mutation in the condensin II gene NCAPD3 (c.3458T>G, p.Glu1153Ala), resulting in a deleterious amino acid substitution at a residue highly conserved in condensin II orthologs (Fig. 1H). All variants identified were confirmed by capillary sequencing, and all parents were established to be heterozygous carriers, consistent with autosomal recessive inheritance (Table 1). None of the variants were reported in the large-scale control data set ExAC.
Table 1.
Gene mutations in condensin I and II microcephaly patients
The severity of microcephaly correlated with mutation type, ranging from −2.7 to −11.9 SD (Supplemental Table S1; Supplemental Fig. S1A), and was most severe in patients P1 and P2, in whom frameshift/splice-disrupting variants were found. Missense mutations in patients P3 and P4 were accompanied by milder microcephaly. Although less severely affected, stature was also significantly reduced in patients P1 and P2, with height within normal population limits in patients P3 and P4. No distinctive facial features or associated malformations were apparent (Supplemental Fig. S1B), and, apart from intellectual disability, comorbidity was limited except for patient P2, who died of a malignant anaplastic medulloblastoma at 11 yr (Fig. 1A; Supplemental Table S1).
Condensin mutations impair chromosome structural integrity
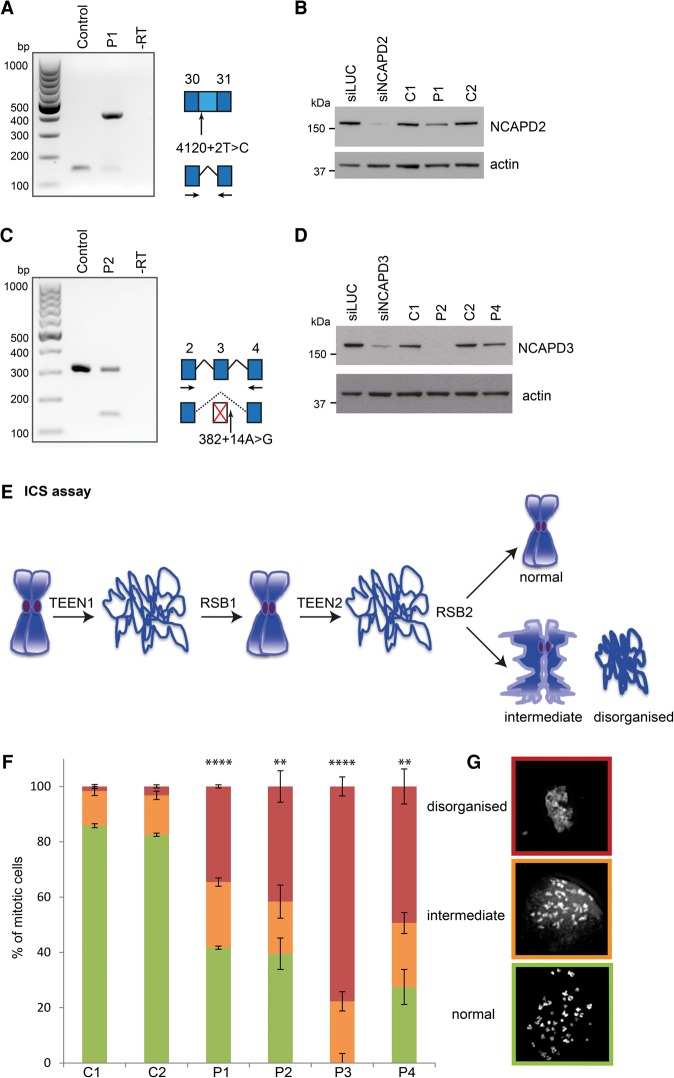
To confirm the pathogenic nature of the identified mutations, we established primary fibroblast lines from all four patients and assessed the effect of these mutations on transcript splicing and protein expression by RT–PCR and immunoblotting, respectively. The nucleotide substitution c.4120+2T>C in NCAPD2 abolished the exon 31 consensus splice donor site, and RT–PCR demonstrated that this resulted in retention of intron 30, with residual wild-type transcript only just detectable (Fig. 2A). The incorporation of intron 30 was confirmed by capillary sequencing (data not shown) and results in a transcript encoding an additional 29 amino acids before a premature termination codon (PTC) that led to omission of the most C-terminal 28 amino acids encoded by exon 31. Furthermore, a significant reduction in NCAPD2 protein was seen in patient primary fibroblasts (P = 0.0086) (Fig. 2B; Supplemental Fig. S2A), presumably resulting from decreased stability of the mutated protein (given the C-terminal location of the mutation, nonsense-mediated decay [NMD] is not expected).
Figure 2.
Mutations impair canonical condensin function in mitotic chromosome compaction. (A,B) The c.4120+2T>C mutation results in retention of intron 30 in NCAPD2 transcripts and reduced NCAPD2 protein levels. (A) RT–PCR using primers in exons 30 and 31 (arrows in the schematic) demonstrated altered splicing of NCAPD2 in patient P1 fibroblasts. The larger 413-base-pair (bp) PCR product corresponded to a transcript containing intron 30, confirmed by subcloning and Sanger sequencing. This transcript encodes an alternative C terminus comprising an additional 29 amino acids before a PTC with consequent loss of 28 amino acids encoded by exon 31. (B) NCAPD2 protein levels are reduced in P1 fibroblasts. Immunoblot with anti-NCAPD2 antibody. (Left two lanes) RNAi of NCAPD2 in RPE1 cells confirms the specificity of the NCAPD2 antibody. (Remaining lanes) Patient P1 and control primary fibroblasts. NCAPD2 protein mobility is not expected to be altered in patient P1, as the mutant protein has a molecular weight similar to that of wild type. (Bottom panel) Loading control: The blot was probed with anti-actin antibody. (C,D) The NCAPD3 c.382+14A>G mutation leads to skipping of exon 3, which, in combination with c.1783_1784delG frameshift mutation in trans, results in markedly reduced NCAPD3 protein. (C) Alternative splicing of NCAPD3 in patient P2 fibroblasts, detected by RT–PCR using primers in exon 2 and exon 4 (arrows, schematic). The wild-type transcript represented by a PCR product of 317 bp is substantially reduced in patient P2 cells, while the smaller PCR fragment of 154 bp is detectable only in patient P2 and corresponds to a transcript in which exon 3 had been skipped (confirmed by subcloning and sequencing). This results in an out-of-frame transcript, which is expected to be subject to NMD. (D) Immunoblotting of NCAPD3 establishes that protein levels are markedly reduced in patient P2. (Left two lanes) RNAi of NCAPD3 in RPE1 cells demonstrating specificity of the NCAPD3 antibody. (Remaining lanes) Patient and control primary fibroblasts. NCAPD3 protein is substantially depleted in patient P2, whereas protein levels are unaffected by the p.Glu1153Ala missense mutation in patient P4 fibroblasts. (Bottom panel) Loading control: The blot was probed with anti-actin antibody. (E) Schematic of intrinsic chromosome structure (ICS) assay. Sequential TEEN decompaction and refolding in RSB buffer was used to establish the structural integrity of mitotic chromosomes. (F,G) Patient mutations lead to compromised ICS. (F) Quantification of abnormal chromosome refolding after ICS assay in C1 and C2 and patient P1–P4 cell fibroblasts (experiments = 3; n > 30 mitoses per sample per experiment). Error bars indicate SEM. Two-tailed t-test, (**) P ≤ 0.01; (****) P ≤ 0.0001; proportion of “disorganized” chromosomes versus C2. (G) Representative images of chromosome morphology from ICS assay. Chromosomes were visualized with DAPI following two sequential rounds of decompaction (TEEN) and refolding (RSB). Patient and control primary fibroblast chromosomes were scored as either reforming the original chromosome morphology (“normal”), partially reforming starting chromosome morphology (“intermediate”), or complete loss of chromosome compaction (“disorganized”).
Heterozygous 1783_1784delG and 382+14A>G variants in NCAPD3 were predicted to introduce PTC after exon 3 and in exon 15, respectively, leading to NMD of the resulting transcripts. Consistent with this, substantially reduced wild-type NCAPD3 transcript levels were seen in patient P2 fibroblasts (Fig. 2C). A transcript resulting from the alternative donor splice site generated by the c.382+14A>G substitution was not detected. However, an additional transcript was observed that deleted exon 3, likely arising from nonsense-associated altered splicing (Wang et al. 2002). This transcript was consequently out of frame, resulting in premature termination (p.Ser74Alafs*3). A low level of normally spliced transcript was detectable that likely accounts for the low levels of residual NCAPD3 protein observed on immunoblotting (Fig. 2D; Supplemental Fig. S2B). We therefore concluded that mutations in both patient P1 and P2 led to substantial effects on the transcript and proteins of the affected condensin subunits. However, we expect these to result in significant loss of function rather than being functionally null, given the essential nature of condensins in vertebrates (Hudson et al. 2003; Smith et al. 2004; Nishide and Hirano 2014; Houlard et al. 2015). In contrast, amino acid substitutions in patients P3 and P4 in NCAPH (p.Pro243Leu) (Supplemental Fig. S2C,D) and NCAPD3 (p.Glu1153Ala) (Fig. 2D), respectively, did not affect cellular protein levels. Although they are outside annotated domains, the substitutions are at highly conserved residues (Fig. 1G,H) and so are likely to affect condensin complex function.
As both condensin I and II have established roles in ensuring axial and radial stability of compacted chromatin (Hudson et al. 2003; Ono et al. 2003; Hirota et al. 2004; Green et al. 2012), we next assessed the impact of patient P1–P4 mutations on condensin function in mitotic chromosome integrity. We used an established intrinsic chromosome structure (ICS) assay (Fig. 2E) that has been used previously to demonstrate structural integrity defects in SMC2, NCAPH, and NCAPD3 conditionally null DT40 cells (Hudson et al. 2003; Green et al. 2012). This is a qualitative assay that assesses whether chromosomes are able to recover higher-order structure after unfolding chromatin in low-salt TEEN buffer (which increases DNA–DNA repulsion through cation depletion). In this in vitro assay, mitotic chromosomes from all patient cell lines failed to efficiently recover their normal condensed appearance after two rounds of unfolding/refolding, in marked contrast to two control primary fibroblast cell lines (C1 = 1.7% ± 0.7%; C2 = 3.2% ± 0.6%; P1 = 34.6% ± 0.6%, P < 0.0001; P2 = 41.6% ± 5.7%, P = 0.0026; P3 = 77.7% ± 3.5%, P < 0.0001; P4 = 49.4% ± 6.3%, P = 0.0019) (Fig. 2F,G). Morphologically, structurally disrupted patient chromosomes resembled those reported previously in DT40 cells lacking NCAPH and NCAPD3 (Fig. 2G; Green et al. 2012). We therefore concluded that mutations in all four patients can disrupt condensin-dependent mitotic chromosome integrity, providing functional supporting evidence for their pathogenicity.
Chromatin bridges in Ncaph2 apical neural progenitors lead to microcephaly
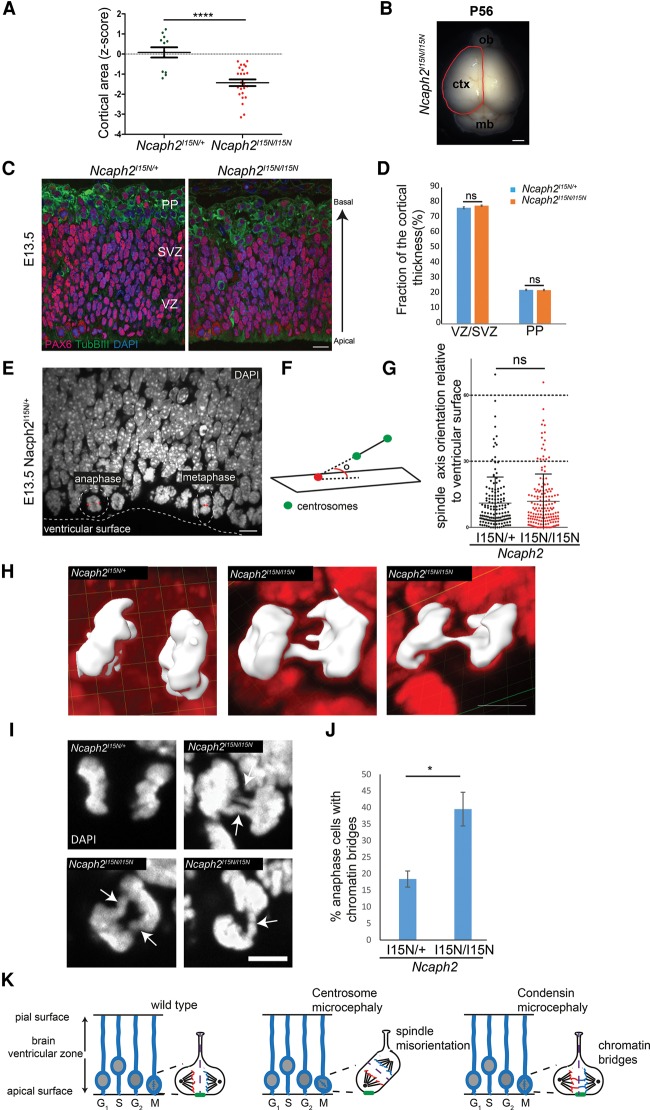
To extend our investigations into a developmental system, we next examined a published hypomorphic loss-of-function Ncaph2 condensin II mouse mutant (Gosling et al. 2007) with a single missense mutation (p.Ile15Asn) in which brain size and neurodevelopment had not been assessed previously. Measurement of brain size at 8 wk postnatally demonstrated significantly reduced brain weight (Ncaph2I15N/+, z = 0.06 ± 0.2; Ncaph2I15N/I15N, z = −0.6 ± 0.15; P = 0.01) and reduced cortical surface area (Ncaph2I15N/+, z = 0.08 ± 0.26; Ncaph2I15N/I15N, z = −1.4 ± 0.16; P = 0.0001) in comparison with Ncaph2I15N/+ littermate controls (Fig. 3A,B; Supplemental Fig. S3A). Reduced body weight was also evident in Ncaph2I15N/I15N mice (Ncaph2I15N/+, z = 0.12 ± 0.23; Ncaph2I15N/I15N, z = −1.45 ± 0.3; P = 0.0003) (Supplemental Fig. S3B). Having confirmed that mutation of a non-SMC condensin subunit also caused microcephaly in mice, we then investigated how neurogenesis was perturbed during embryonic development. In the embryonic day 13.5 (E13.5) Ncaph2I15N/I15N brain, a 4% ± 1.6% reduction in neocortical thickness was seen, while the relative thickness of the ventricular zone (VZ)/subventricular zone (SVZ; PAX6-positive cells, Ncaph2I15N/+ = 76.5% ± 0.7%; Ncaph2I15N/I15N = 78% ± 0.4%; P = 0.12) and the neuronal preplate (PP) layer (Tub βIII-positive, Ncaph2I15N/+ = 22.4% ± 0.4%; Ncaph2I15N/I15N = 22.3% ± 0.4%; P = 0.83) was unchanged (Fig. 3C,D).
Figure 3.
Impaired chromosome segregation instead of altered neural progenitor cleavage plane accounts for reduced cortical size in Ncaph2I15N/I15N mice. (A,B) Cerebral cortex surface area is reduced in Ncaph2I15N/I15N mice at 8 wk. (A) Quantification of cortical area expressed as a Z-score at 8 wk of age. n > 12 mice per genotype. Error bars indicate SEM. Two-tailed t-test, (****) P ≤ 0.0001 versus littermate Ncaph2I15N/+ mice. (B) Cortical area measurement. The red line indicates the measured region of the dorsal cerebral cortex. Bar, 1.5 mm. Z-score is defined as the standard deviations from the mean of age- and sex-matched Ncaph2I15N/+ mice at 8 wk. (C,D) The relative proportion of progenitor cells and neurons is retained during neurogenesis in Ncaph2I15N/I15N mice. (C) Representative images of neurogenesis in E13.5 Ncaph2I15N/+ and Ncaph2I15N/I15N coronal sections immunostained for PAX6 (radial glia; red), TUBβIII (neurons; green), and DAPI (DNA; blue). (D) Quantification of proportions of TUBβIII- and PAX6-positive cells per radial unit, determined by measurement of the thickness of the PP and SVZ/VZ, respectively. Measurements made at four regions per coronal section for two coronal sections at equivalent rostro–caudal positions per brain. n = 10 brains analyzed for Ncaph2I15N/+; n = 9 brains analyzed for Ncaph2I15N/I15N. Error bars indicate SEM. (ns) Nonsignificant with an unpaired t-test. Bar, 10 µm. (E–G) Cleavage plane orientation is not affected in E13.5 Ncaph2I15N/I15N mice. (E) Representative image of mitotic apical progenitors illustrating the normal orientation of the mitotic spindle parallel to the ventricular surface. E13.5 coronal sections and nuclei (DAPI; white) are shown. (Dashed line) Ventricular surface; (dashed circles) mitotic cells; (double-sided arrows) the axis of the mitotic spindle and position of mitotic centrosomes (identified by Aurora A immunostaining). Bar, 10 µm. (F) Schematic outlining measurement of the mitotic spindle axis, defined as the angle between the spindle line vector and the reference plane for the ventricular surface. Green circles indicate centrosomes, and red circles point to where the spindle line vector intersects the plane of the ventricular surface. (G) Quantification of mitotic spindle orientation in apical neural progenitors at metaphase and anaphase in E13.5 neuroepithelium. Data for Ncaph2I15N/+ mice (black circles; mean angle 11°; n = 150 cells from three brains) and Ncaph2I15N/I15N mice (red circles; mean angle 12°; n = 159 cells from three brains) are shown. Error bars indicate SD. P = 0.56 with an unpaired t-test. (H–J) Chromosome segregation is impaired in E13.5 Ncaph2I15N/I15N apical neural progenitors. (H,I) Representative images of chromatin bridges in anaphase apical progenitor cells. Three-dimensional reconstructions (DAPI stain in red; anaphase cell DNA highlighted in white; bar, 4 µm) (H) and two-dimensional DAPI-stained confocal sections (bar, 5 µm) (I) from E13.5 coronal sections. (J) Quantification of chromatin bridges observed in apical progenitors of E13.5 Ncaph2I15N/+ and Ncaph2I15N/I15N mice. Significantly increased numbers of mid and late anaphase cells with chromatin bridges are observed in Ncaph2I15N/I15N mice (n = 46 cells from three brains) compared with Ncaph2I15N/+ mice (n = 72 cells from three brains). Error bars indicate SEM. Two-tailed t-test, P = 0.02 versus Ncaph2I15N/+ mice. (K) Model: During early neurogenesis, neural stem cells divide symmetrically to enlarge the pool of neural progenitors. Mitosis occurs at the ventricular surface, with the axis of the mitotic spindle parallel to this surface. This ensures that the cleavage furrow is perpendicular and that, consequently, basal fate determinants (indicated by green bar) segregate equally between daughter cells, ensuring retention of stem cell identity. (Middle panel) The mitotic spindle fails to align when genes encoding centrosomal proteins are mutated (ASPM, CDK5RAP2, MCPH1, and CPAP), leading to oblique cell cleavage events that result in premature asymmetric cell division. Daughter neuron cells are produced, and expansion of the neural stem cell pool is decreased, consequently reducing the total number of neurons. (Right panel) In contrast, condensin mutations lead to chromatin bridges during neurogenesis, with unaltered spindle orientation. Such chromosome segregation errors would be expected to reduce cell proliferation and survival, depleting the neuronal number generated during neurogenesis.
We also measured the mitotic spindle orientation of apical progenitors, which has been found previously to be perturbed in microcephaly models (Lizarraga et al. 2010; Gruber et al. 2011; Lancaster et al. 2013). As two-dimensional analysis of the spindle axis can lead to systematic measurement errors (Juschke et al. 2014), metaphase and anaphase apical progenitors were imaged by spinning-disc confocal microscope, and axis orientation was determined in three dimensions. The angle between the line bisecting both spindle poles (mitotic centrosomal marker, Aurora A) and the plane of the ventricular surface was measured in E13.5 brain sections (Fig. 3E,F; Supplemental Fig. S4A,B), demonstrating that the mitotic spindle position in Ncaph2I15N/I15N neuroepithelia was indistinguishable from controls (30°–60° bin, Ncaph2I15N/+ = 7% ± 1.4%, Ncaph2I15N/I15N = 10.8% ± 5.9%, P = 0.57; 60°–90° bin, Ncaph2I15N/+ = 0.8% ± 0.8%, Ncaph2I15N/I15N = 0.6% ± 0.6%, P = 0.83) (Fig. 3G; Supplemental Fig. S4C). This therefore indicated that a different process underlies condensin-associated microcephaly, and, upon further characterization, we observed frequent chromatin bridges during anaphase in Ncaph2I15N/I15N apical progenitors (Fig. 3H–J). Such bridges were at a significantly higher frequency than heterozygous controls (Ncaph2I15N/I15N = 39.5% ± 5.5%; Ncaph2I15N/+ = 18.4% ± 2.4%; P = 0.02) (Fig. 3J). As DNA bridges are known to lead to chromosome segregation errors and impaired cell survival, this provided an alternative explanation for condensin microcephaly (Fig. 3K, model). To investigate this mechanism in more detail, we next employed a cell biological approach using the patient and mouse cell lines available to us.
Decatenation failure leads to DNA bridges in condensin-deficient cells
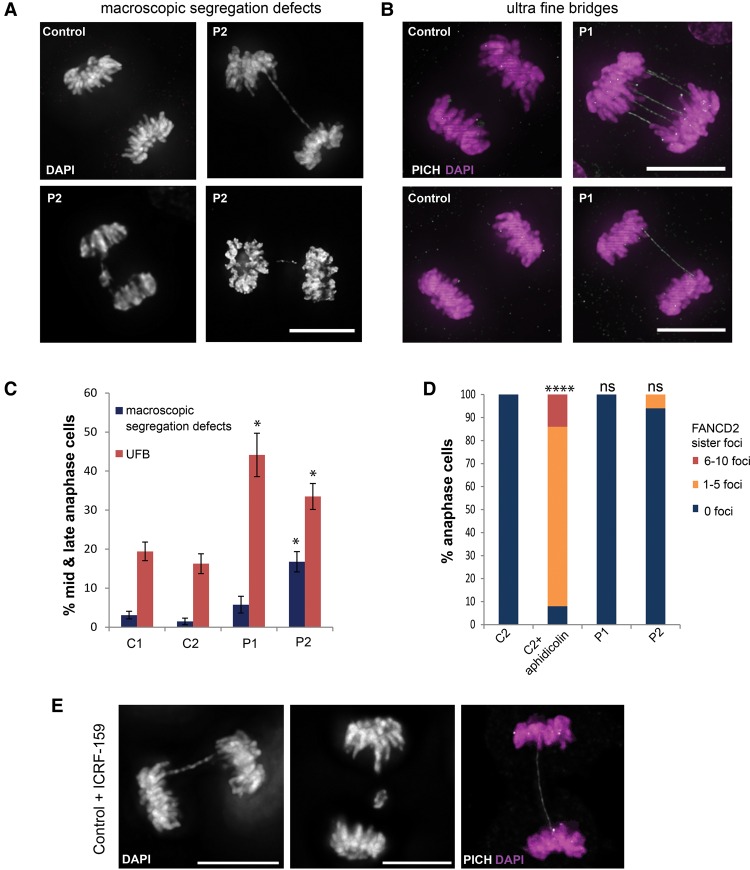
First, to characterize the cause of such DNA bridges, we examined mitosis in condensin-deficient patient cells. Analysis of both patient P1 and P2 fibroblasts, representing mutations in condensin I and II, respectively, demonstrated DNA bridges, confirming that such mitotic abnormalities also occurred in condensin microcephaly patients. In patient P2 (NCAPD3) fibroblasts, elevated levels of chromatin bridges and lagging chromatin/chromosomes were observed during mitosis, evident in 16.7% of anaphases (C1 = 3.1% ± 1%; P2 = 16.7% ± 2.6%, P = 0.003) (Fig. 4A,C). Such chromosome segregation errors have been reported previously in NCAPD3-deficient cells (Ono et al. 2004; Gerlich et al. 2006), but, consistent with patient P2 cells retaining residual condensin II function, we observed that this occurred to a lesser extent than in conditionally null NCAPD3 DT40 cells, in which 80% of anaphase and telophase cells had prominent chromatin bridges (Green et al. 2012). A level of segregation defects similar to that present in patient P2 was also seen in Ncaph2I15N/I15 mouse embryonic fibroblasts (MEFs), significantly higher than heterozygote littermate-derived control lines (Ncaph2I15N/+ #1 = 6.3% ± 0.7%, Ncaph2I15N/I15N#1 = 17.9% ± 1.7%, P = 0.0013; Ncaph2I15N/I15N#2 = 13% ± 0.4%, P = 0.0001) (Supplemental Fig. S5A).
Figure 4.
Condensin patient mutations lead to decatenation failure at mitosis. (A–C) Chromosome segregation is impaired in patient P1 (NCAPD2) and P2 (NCAPD3) primary fibroblasts. Representative images of chromatin bridges and lagging chromosomes/chromosome fragments were detected with DAPI stain (macroscopic chromosome segregation defects) (A), and ultrafine DNA bridges (UFBs) were detected by the presence of PLK1-interacting checkpoint helicase (PICH) and absence of DAPI stain (B). Bars, 10 µm. (C) Quantification of macroscopic chromosome segregation defects (defined as DAPI-positive chromatin bridges, lagging chromosomes, or chromosome fragments) and UFBs, scored in anaphase C1 and C2 and patient P1 and P2 fibroblasts. Increased numbers of mid and late anaphase cells with UFBs are seen in NCAPD2 patient P1 and NCAPD3 patient P2 cell fibroblasts (experiments ≥ 3; n > 50 anaphases per sample per experiment), and patient P2 fibroblasts also had elevated macroscopic chromosome segregation defects compared with C1 and C2 fibroblasts (experiments ≥ 3, n > 100). Error bars indicate SEM. Two-tailed t-test, (*) P ≤ 0.05 versus C1. (D) UFBs in condensin patient cells do not arise from late replicating intermediates. Quantification of the number of FANCD2 sister foci during anaphase in C2 and patient P1 and P2 fibroblasts. C2 fibroblasts treated with 0.1 µM aphidicolin for 16 h to induce late replicating intermediates through replication stress were also included as a positive control. n = 50 cells; experiment = 1. (E) Chromosome decatenation failure due to topoisomerase II α inhibition in control fibroblasts results in chromosome segregation defects identical to those observed in condensin patient cells. Representative images of bulky chromatin bridges and lagging chromatin detected with DAPI stain and UFBs detected by PICH stain. Bars, 10 µm. C2 fibroblasts were treated with 5 µM ICRF-159 for 24 h.
In patient P1 cells, a significantly increased frequency of substantial DNA bridges was not evident on DAPI staining. However finer chromatin bridges have been observed in condensin I-deficient cells (Green et al. 2012), and loss of both condensin complexes has been shown previously to increase the number of ultrafine DNA bridges (UFBs) (Lukas et al. 2011). To sensitively detect UFBs, we performed PICH (PLK1-interacting checkpoint helicase) immunostaining and, using this methodology, observed a substantial increase in persisting UFBs in both condensin I and condensin II cells in mid to late anaphase (C1 = 19.4% ± 2.4%; P1 = 44.1% ± 5.6%, P = 0.02; P2 = 33.5% ± 3.3%, P = 0.04) (Fig. 4B,C).
We next considered the origin of the observed DNA bridges. UFBs are believed to originate either from unresolved sister chromatid entanglements as dsDNA catenanes (Chan et al. 2007) or at regions of incomplete DNA replication, termed late replicating intermediates (LRIs) (Chan et al. 2009). The latter can be induced by replication stress and is associated with paired FANCD2 foci that flank LRI UFBs. Such sister FANCD2 foci were not evident in patient P1 and P2 cells (Fig. 4D), indicating that observed UFBs were more likely due to dsDNA catenenes. Furthermore, UFBs, chromatin bridges, and lagging chromosomes/chromatin identical to those observed in patient cells were also observed in control fibroblasts in which cellular decatenation activity was inhibited by treatment with low levels of the topoisomerase IIα inhibitor ICRF-159 (Fig. 4E).
Micronuclei and aneuploidy are consequences of impaired chromosome segregation
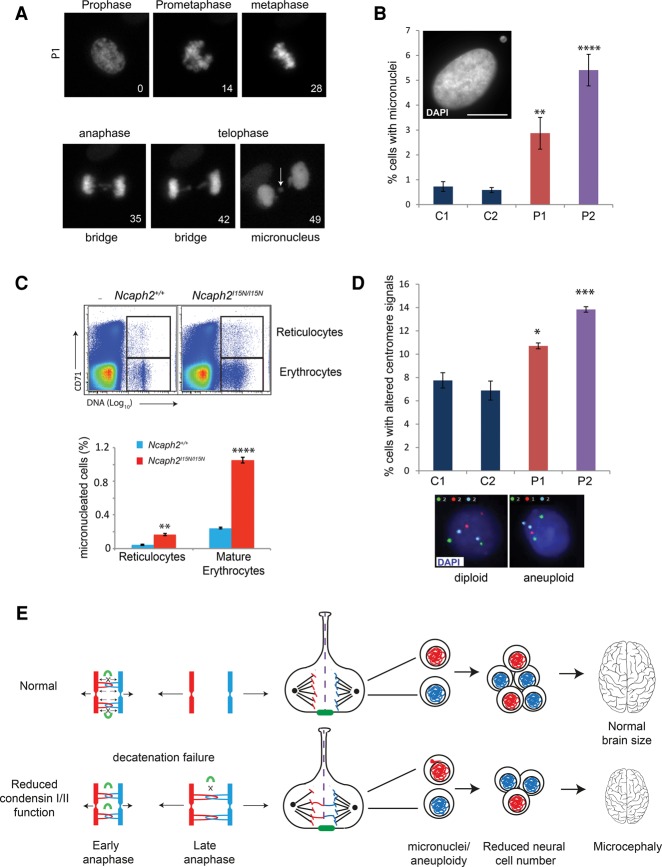
To establish how reduced cellularity arises in condensin patients, we examined the post-mitotic consequences of decatenation failure by live imaging of patient fibroblasts transiently expressing RFP-H2B (Fig. 5A). This confirmed a significant increase of anaphase cells with chromatin bridges in patient P2 fibroblasts, consistent with the previous analysis of fixed patient cells (C1 = 6.1%; C2 = 2.4%; P2 = 14.6%, P = 0.0056) (Supplemental Fig. S5C). Furthermore, some of the chromatin bridges were seen to lead to micronucleus formation at later time points (Fig. 5A). Analysis of fixed DAPI-stained interphase cells confirmed increased levels of such micronuclei in both patient P1 and P2 fibroblast lines (C1 = 0.7% ± 0.2%; P1 = 2.9% ± 0.6%, P = 0.009; P2 = 5.4% ± 0.6%, P = 0.0001) (Fig. 5B). In patient P3 and P4, microcephaly without significant short stature was observed. In keeping with this milder clinical phenotype restricted to the CNS, increased micronucleus formation was less pronounced in patient P3 fibroblasts and was not detected in patient P4 fibroblasts (P3 = 1.3% ± 0.3%, 0.4% ± 0.2%, P = ns). An in vivo erythrocyte micronucleus assay demonstrated that micronucleated cells were significantly more frequent in homozygous mutant mice compared with wild-type control animals (Ncaph2I15N/I15N reticulocytes = 0.168% ± 0.01%, P = 0.005; Ncaph2I15N/I15N mature erythrocytes = 1.5% ± 0.04%, P = 0.01) (Fig. 5C). Interphase FISH with chromosome-specific α-satellite probes (Fig. 5D) demonstrated a significant increase of patient P1 and P2 primary fibroblast cells with altered numbers of centromere signals compared with controls, consistent with a low level of increased aneuploidy in both cell lines (C1 = 7.8% ± 0.7%, P1 = 10.7% ± 0.3%, P = 0.03; P2 = 13.8% ± 0.2%, P = 0.0006). Taken together, we concluded from these cellular experiments that decatentation failure leads to the formation of micronuclei and aneuploidy in condensin-deficient patient cells.
Figure 5.
Decatenation failure results in micronuclei and aneuploidy in condensin mouse and patient cells. (A) Live-imaging stills of chromatin bridge and subsequent micronucleus formation in a patient P1 fibroblast cell. DNA visualized by transient expression of RFP-tagged histone 2B. Time is shown in minutes from nuclear envelope breakdown. The arrow indicates the position of the micronucleus. (B) NCAPD2 patient P1 and NCAPD3 patient P2 fibroblasts have significantly elevated levels of micronuclei. The percentage of micronucleus-positive cells was quantified (experiments = 5, n > 500 cells). Error bars indicate SEM. Two-tailed t-test, (**) P ≤ 0.01; (****) P ≤ 0.0001 versus C1. (Inset) Representative picture of a patient P2 fibroblast cell with a micronucleus (shown in the top left corner). Bar, 10 µm. (C) Elevated frequency of micronuclei in peripheral blood of Ncaph2I15N/I15N mice. (Top panel) Representative FACS plots of CD71 (reticuloctyte marker) versus propidium iodide (DNA content) of Ncaph2I15N/I15N and Ncaph2+/+ adult mice demonstrating gating strategy. Gated cells have subdiploid DNA content (micronuclei). (Top gate) Micronuclei in CD71+ve reticulocytes. (Bottom gate) Micronuclei in mature erythrocytes. In contrast to the usual linear scale for DNA content FACS plots, the DNA axis is presented on a log10 scale, with fully nucleated cells off the scale, and micronucleated cells appearing to the right of enucleated cells within the gates indicated by the boxed regions. (Right panel) Quantification of flow cytometry analysis of micronuclei in the peripheral blood reticulocytes and mature erythrocytes of Ncaph2I15N/I15N and Ncaph2+/+ adult mice. N = 3. Error bars indicate SEM. Two-tailed t-test, (**) P ≤ 0.01; (****) P ≤ 0.0001 versus Ncaph2+/+ mice. (D) Levels of aneuploidy are significantly increased in NCAPD2 patient P1 and NCAPD3 patient P2 fibroblasts. (Top panel) Quantification of the percentage of cells with numerical chromosome imbalances detected by interphase FISH. Bars indicate the average of two sets of probes detecting chromosome 3, 7, and 18 and 4, 10, and 17, respectively. n > 700 cells; experiments ≥ 4. Error bars indicate SEM. Two-tailed t-test, (*) P ≤ 0.05; (***) P ≤ 0.001 versus C1. (Bottom panel) Representative images of interphase FISH of chromosomes 4 (green), 10 (red), and 17 (aqua) in primary fibroblasts. (E) Model: Decatenation failure in condensin-deficient neural stem cells reduces brain size through a reduction in cell number generated during neurogenesis. Impaired mitotic chromosome compaction in condensin-deficient neural stem cells decreases tension in catenated DNA, reducing topoisomerase IIα DNA strand exchange activity (topoisomerase IIα enzyme indicated by green curved line). This delays decatenation of DNA between sister chromatids, leading to increased chromosome segregation errors and subsequent aneuploidy/micronucleus formation. Anueploidy in condensin-deficient neural stem cells is likely to reduce cell number by impaired proliferation potential (Gogendeau et al. 2015) and/or reduced cell survival (Marthiens et al. 2013; Nishide and Hirano 2014) during the restricted developmental window of neurogenesis.
Discussion
Human germline mutations in condensin protein subunits
Here we report that mutations in genes encoding subunits of condensins cause microcephaly, establishing that disrupted chromosome condensation can result in reduced brain size. This provides a further example of where aggregate analysis of genes encoding macromolecular complexes has permitted identification of rare human disease genes (Crow et al. 2006; Bicknell et al. 2011; Mannini et al. 2013). Using a similar strategy, somatic mutations in condensins have also been associated with several cancer types (Leiserson et al. 2015). This and the fatal brain tumor in patient P2 raise the possibility that germline condensin mutations are also cancer-predisposing.
Consistent with the cell-essential nature of condensins due to their requirement for cell division (Strunnikov et al. 1995; Hagstrom et al. 2002; Hudson et al. 2003; Ono et al. 2003; Oliveira et al. 2005; Siddiqui et al. 2006), the patient mutations reported here are all functionally hypomorphic. The implicit developmental viability of these human mutations therefore has utility for providing further insights into the roles of these essential protein complexes at the organism level.
Both condensin I and condensin II are required to resolve UFBs formed by dsDNA catenanes
In addition to identifying three new human disease genes, we established decatenation failure as a novel pathogenic mechanism for microcephaly—mitotic in origin and distinct from mechanisms associated with previously identified disease genes (Fig. 3K, model). This is in contrast to the disrupted transcriptional regulation that underlies “cohesinopathies” (Dorsett 2011), which result from mutations in the paralogous cohesin complex (Musio et al. 2006; Deardorff et al. 2007, 2012). The identification of mutations in both condensin I and condensin II complexes supports a mitotic basis, as both are required for mitotic chromosome integrity (Hudson et al. 2003; Ono et al. 2003; Hirota et al. 2004) and colocalize only at mitosis on compacting chromatin (Hirota et al. 2004). Furthermore, mitotic chromosome integrity is impaired in patient cell lines irrespective of which condensin is affected.
Our results indicate that decatenation failure is likely to be the direct consequence of such impaired chromosome compaction. Substantial chromatin bridging has been described in condensin II-null cells, and finer DAPI-positive bridges occur in condensin I knockout cells (Green et al. 2012). While condensin II patient cells have similar macroscopic chromatin bridges, albeit at lower rates than reported previously, such chromatin bridges were not increased in condensin I cells. This led us to assess whether DNA bridges not visualized by DAPI might be present. PICH immunostaining permitted the detection of UFBs that have not been reported previously in cells deficient for either condensin I or condensin II. Such single nonchromatinized DNA fibers point to failed decatenation as the origin of the bridges (Baumann et al. 2007; Chan et al. 2007; Ke et al. 2011) rather than these arising directly from disordered chromosome structure. Mechanistic studies in yeast have demonstrated that condensins promote decatenation (D'Ambrosio et al. 2008; Baxter et al. 2011; Charbin et al. 2014), and it has been proposed that condensin-mediated chromosome compaction increases tension in catenated DNA that remains between sister chromatids such that it provides directionality to topoisomerase II-mediated strand exchanges toward chromatid decatenation (Fig. 5E, model; Cuylen and Haering 2011).
Studies of replication stress-associated UFBs at fragile sites have implicated condensins in promoting their resolution (Minocherhomji et al. 2015). However, rather than solely dealing with LRI UFBs, we establish here that both condensin complexes are also required to resolve dsDNA catenanes that occur in unperturbed cells, as UFBs were not flanked by FANCD2, a marker of LRI UFBs. Additionally, we observed a differential requirement for condensin I and II in UFB resolution, with condensin II specifically needed for decatenation of centromeric UFBs (Supplemental Fig. S5B), while condensin I UFBs arise at distributed sites that are not predominantly centromeric or ribosomal DNA (data not shown). Such differences could be accounted for by differing localization on mitotic chromosomes (Ono et al. 2003, 2004) and/or their differential roles in axial and lateral chromosome compaction (Shintomi and Hirano 2011). Therefore, the study of hypomorphic mammalian cell lines establishes that both condensins act in the resolution of dsDNA catenanes, suppressing UFB formation during the unperturbed cell cycle.
Decatenation failure as a cause of microcephaly
The model currently favored for the majority of primary microcephaly genes is that altered mitotic spindle position leads to an altered cleavage plane at cytokinesis and, consequently, perturbed symmetric/asymmetric fate decisions (Fig. 3K). In contrast to centrosomal models of microcephaly, spindle orientation is not altered in condensin-deficient apical progenitors (neural stem cells). Furthermore, normal proportions of PAX6 progenitors and TUJ1-positive neurons are maintained in the developing neuroepithelium, consistent with unperturbed symmetric/asymmetric cell fate choice. Therefore, while microcephaly associated with condensin mutations has a mitotic origin, it arises by a different mechanism that originates from DNA bridges in dividing apical neural progenitor cells as a result of decatenation failure.
Macroscopic chromatin bridges are an established cause of chromosome segregation failure (Hoffelder et al. 2004). Consistent with this, we observed lagging chromosome fragments by live imaging in condensin II patient cells and, consequently, micronuclei in the subsequent interphase. Micronuclei also arose in condensin I patient cells, suggesting that UFBs may also prevent timely chromosome segregation, a conclusion supported by findings in PICH knockout DT40 cells (Nielsen et al. 2015). Therefore, despite the differing mitotic roles of the two condensin complexes, missegregation of chromatin likely explains the shared phenotype and disease presentation. Furthermore, the high frequency of UFBs in Blooms syndrome (Chan et al. 2007) would also predict that microcephaly in this disorder occurs by the same mechanism.
Notably, the protein encoded by the previously identified primary microcephaly gene MCPH1 is a regulator of chromosome condensation that inhibits condensin II (Trimborn et al. 2006; Yamashita et al. 2011) and interacts with both NCAPD3 and NCAPG2 subunits (Wood et al. 2008; Yamashita et al. 2011). This raises the possibility that similar mechanisms could contribute to reduced brain size in MCPH1 patients. However, MCPH1 has also been reported to function in centrosomal Chk1-dependent checkpoint signaling (Tibelius et al. 2009) that, when impaired in Mcph1−/− mice, results in altered mitotic spindle orientation and increased asymmetric cell division in neural stem cells (Gruber et al. 2011). Nonetheless, given that mutations in condensins are now also found to cause microcephaly, further investigation of the MCPH1–condensin interaction is warranted.
We observed that chromosome segregation failure leads to aneuploidy in patient cells—itself an established cause of microcephaly (Hanks et al. 2004; Marthiens et al. 2013). Aneuploidy has been shown to impair proliferation potential and cell survival, with chromosome instability models in yeast and mammals establishing that chromosome segregation failure leads to cell cycle delay and/or arrest (Santaguida and Amon 2015), processes that would act to reduce neuronal cell number during the rapid cell proliferation of neurogenesis. DNA damage is also likely to occur at persisting chromatin bridges (Janssen et al. 2011; Maciejowski et al. 2015) and micronuclei (Crasta et al. 2012), which may well impair cell viability, increasing the already high levels of apoptosis that occurs during brain development. Diminished cell survival would also result in reduced brain size and is consistent with the increased cell death seen in Nestin-cre condensin I and II knockouts (Nishide and Hirano 2014). Therefore, we propose that impaired decatenation with subsequent chromosome segregation failure results in reduced cell proliferation and increasing cell death, providing a straightforward explanation for microcephaly associated with hypomorphic condensin mutations. While the restricted developmental window for neurogenesis provides one explanation for the disproportionate effect on brain size, there is also an elevated number of chromatin bridges in condensin-deficient neural stem cells (Fig. 3J, Supplemental Fig. S5A; Nishide and Hirano 2014) that could also explain why this organ is more severely affected.
In summary, through the identification of mutations in multiple condensin subunits, we show that “condensinopathies” result in microcephaly due to impaired DNA decatenation, establishing this as a novel disease mechanism and chromosome compaction as a key process ensuring mammalian cortical size.
Materials and methods
Research subjects
Genomic DNA from the affected individuals and family members was extracted from peripheral blood by standard methods or obtained from saliva samples using Oragene collection kits according to the manufacturer's instructions. Informed consent was obtained from all participating families, and the studies were approved by the ethics review board at the Scottish Multicentre Research Ethics Committee (04:MRE00/19) and the University of Texas Southwestern Medical Center (STU112010-149). Parents provided written consent for the publication of photographs of the patients. For growth measurements, Z-scores (standard deviations from population mean for age and sex) were calculated using LMSgrowth based on British 1990 growth reference data (Freeman et al. 1995).
Growth was impaired prenatally, with low birth weight reported in three of the patients (P1, P2, and P3). No distinctive facial features aside from a sloping forehead were evident (patients P1 and P3). Although seizures and lower limb hypertonia were described in one case (patient P4), no malformations were reported in any of the patients, and no consistent facial phenotype was apparent between the four cases. Three patients had moderate intellectual impairment, with autistic-like behaviors being reported in patient P1; however, development was described as normal in patient P2. Of note, this latter patient developed a highly malignant anaplastic medulloblastoma (World Health Organization grade IV) at 8 yr of age that was unresponsive to treatment, resulting in death at 11 yr. No comorbidities were present in the other three patients, with one adult patient now aged 42 (patient P3). Malignancy was not reported in any of the heterozygous carriers identified in this study.
Animal studies
All mouse studies were conducted under guidance and approval issued by the Medical Research Council in Responsibility in the Use of Animals for Medical Research (July 1993) and Home Office Project License. The Ncaph2I15N/I15N mouse was identified from an ENU screen as having an unusual, isolated T-lymphocyte developmental defect that altered circulating T-cell numbers with increased levels of the activation/memory marker CD44 but was reported as otherwise healthy and fertile and without other phenotypes (Gosling et al. 2007). The Ncaph2I15N/I15N mouse line was maintained on a C57BL6/J background.
Exome sequencing, AmpliSeq resequencing, and Sanger sequencing
Exome sequencing and variant filtering were performed as described previously (Martin et al. 2014; Murray et al. 2015) for patients P1 and P2. Exome sequencing for patient P4 (DDDP261277) was performed by The Deciphering Developmental Disorders Study as described (Wright et al. 2014). Ion AmpliSeq multiplex PCR (Life Technologies) was used for cohort NGS (next-generation sequencing) resequencing as described previously (Murray et al. 2015). Briefly, primers were designed to amplify coding exons of all condensin I and II genes with 25 base pairs (bp) of flanking intronic sequence. PCR amplification performed in one-sixth reaction volumes for primer pools using 5 ng of patient DNA. After pooling patient amplicons, Ion Xpress barcoding was performed, and sequencing was done on the Ion Proton sequencing platform (Life Technologies) using an Ion P1 200 sequencing kit. For variant calling, Torrent Suite software was used, filtering as for exome analysis. All variants were confirmed by capillary sequencing of PCR products amplified from the relevant condensin gene (primer sequences in Supplemental Table S2A) on an ABI 3730 capillary sequencer. Sequence traces were analyzed using Mutation Surveyor software (SoftGenetics, Inc.).
Sequence alignment and secondary structure prediction
Sequences were obtained from Ensembl release 78 and Ensembl Genomes release 25 (except for Chlamydomonas) (from Phytozome 10.3; http://phytozome.jgi.doe.gov/pz/portal.html). Protein sequences were aligned using CLUSTALW (CLUSTAL 2.1). Since not all eukaryotes have condensin II complexes, homologs were selected from the species listed in Figure 1 as described in Hirano (2012). Amino acid similarity was scored using Blossom 45. The locations of NCAPH domains were determined based on the above alignment and secondary structure predictions made by the JPred server (http://www.compbio.dundee.ac.uk/jpred4).
RT–PCR
Total RNA was isolated from human primary fibroblast cell lines using the RNeasy kit (Qiagen) according to the manufacturer's instructions. After DNase I treatment (Qiagen), cDNA was generated using random oligomer primers and AMV RT (Roche). For RT–PCR primers, see Supplemental Table S2B.
Cell culture
Dermal fibroblasts were obtained by skin punch biopsy and cultured in amnioMAX C-100 complete medium (Life Technologies). MEFs were prepared from individual E13.5 embryos after removing the head and abdominal cavities. Each embryo was minced and maintained in DMEM, 10% FCS, 0.1 mM β-mercaptoethanol, 100 U/mL penicillin, and 100 µg/mL streptomycin. All fibroblast cultures were maintained in a 37°C incubator with 5% CO2 and 3% O2. hTERT RPE-1 cells were obtained from the American Type Culture Collection (CRL-4000) and cultured in DMEM:F12, 10% FCS, 0.26% sodium bicarbonate, 100 U/mL penicillin, and 100 µg/mL streptomycin. All cell lines were routinely tested for mycoplasma. siRNA oligonucleotides were transfected into hTERT RPE1 cells using RNAiMAX (Life Technologies) according to the manufacturer's instructions. For siRNA oligonucleotide sequences, see Supplemental Table S2C. pRFP-H2B was transfected into primary fibroblast cells by electroporation with the Neon transfection system (Thermo Fischer Scientific) according to the manufacturer's instructions. Where indicated, cells were treated with 0.3 µM aphidicolin (Sigma-Aldrich) or 5 µM ICRF-159 (Sigma-Aldrich) for 24 h.
Flow cytometry
Peripheral blood micronucleus assays were performed using ∼50 µL of blood collected into 200 µL of heparin solution. Of this mixture, 180 µL was pipetted into 2 mL of methanol, which had been prechilled for 24 h at −80°C. Samples were stored for at least 24 h at −80°C, washed in PBS, and resuspended in 50 µL of PBS. Twenty microliters was transferred to a new tube containing 69 µL of PBS, 1 µL of ant-CD71 antibody (Biolegend, RI7217), and 10 µL of 10 mg/mL RNase A solution and incubated for 1 h on ice. PBS (500 µL) containing 1.25 µg/mL propidium iodide solution was added immediately prior to analysis. FACS data were acquired on a BD FACSAria II cell sorter and processed using FlowJo software (Treestar).
Chromosome analysis
Metaphases and nuclei were isolated in hypotonic buffer (0.25% KCl, 1% Na3C6H5O7), fixed with methanol:acetic acid (3:1), and dropped onto slides. Metaphase chromosomes were visualized with DAPI staining. Interphase FISH used chromosome enumeration probes labeled in Spectrum orange/green/aqua (Abbot Molecular) prepared in CEP hybridization buffer (Abbot Molecular). Slides were scanned on the GSL-120 automated slide scanner and analyzed using Cytovision version 7.3.1 (Leica Biosystems).
ICS assay
Primary fibroblasts were grown on poly-L-lysine-coated (Sigma-Aldrich) coverslips and blocked with 200 ng/µL nocodazole overnight. Cells were incubated in 75 mM KCl buffer for 5 min followed by TEEN buffer (0.5 mM triethanolamine-HCl at pH 8.5, 0.1 mM Na EDTA, 12.5 mM NaCl) for 30 min and RSB (10 mM Tris-HCl at pH 7.4, 210 mM NaCl, 5 mM MgCl2) for 10 min. The TEEN–RSB cycle was then repeated, cells were fixed in methanol:acetic acid (3:1) for 30 min, and coverslips were mounted in DAPI.
Immunofluorescence and microscopy
Human dermal fibroblasts and MEFs were grown on untreated coverslips (Becton Dickinson). Cells were fixed in 4% PFA (TAAB) in PHEM (25 mM Hepes-NaOH at pH 6.8, 100 mM EGTA, 60 mM PIPES, 2 mM MgCl2) for 15 min. Following PFA fixation, cells were permeabilized by treatment in 0.2% Triton X-100 in PHEM for 2 min. Fixed cells were blocked in PBS/1% bovine serum albumin (Sigma-Aldrich) and incubated with anti-PICH antibody (Ke et al. 2011) or FANCD2 (Santa Cruz Biotechnology, sc-20022) for 30 min followed by anti-rabbit IgG Alexa fluor 568-linked (Life Technologies A11036) or anti-mouse IgG Alexa fluor 488-linked (Life Technologies, A11029) and DAPI. For UFB scoring, mid to late anaphase cells were selected on the criteria that tips of segregating sister chromatids were between 12 and 20 µm apart and that no nuclear envelope reformation was visible. Epifluorescent images were acquired using an Imager A1 fluorescence microscope (Zeiss) controlling a Photometrics Coolsnap HQ2 CCD camera, and a piezoelectrically driven objective mount was used to control movement in the Z-dimension. Images were captured using a 100× plan-apo (1.4 NA) objective at 0.2-µm Z-sections and subsequently deconvolved using Volocity software (PerkinElmer). For live imaging, primary fibroblast cells were seeded onto glass-bottom plates (Greiner Bio One), and imaging was acquired in a closed chamber system with Leibovitz L-15 medium (Gibco). TRITC image data sets were collected with a Axiovert 200 fluorescence microscope system (Zeiss) heated to 37°C and equipped with a 40× plan NeoFluor (0.75 NA) objective. Image capture was performed using Metamorph (Molecular Devices). Images with Z optical spacing of 2 µm were recorded every 10 min for a 24-h period.
Western blotting
Cells were lysed in 50 mM Tris-HCl (pH 8), 280 mM NaCl, 0.5% NP40, 0.2 mM EDTA, 0.2 mM EGTA, and 10% glycerol supplemented with protease inhibitor tablet (Roche). Protein samples were run on a 4%–12% NuPAGE Bis-Tris precast gel (Life Technologies) followed by immunoblotting using anti-NCAPD3 (Bethyl Laboratories, A300-604A), NCAPD2 (Bethyl Laboratories, A300-601A), NCAPH (Bethyl Laboratories, A300-603A), NCAPH2 (Bethyl Laboratories, A302-276A), NCAPG2 (Bethyl Laboratories, A300-605A), and actin (Sigma, A2066).
Mouse genotyping, immunofluorescence, and microscopy
For mouse genotyping, ear clips were boiled in 50 µL of 25 mM NaOH and 0.2 mM EDTA for 30 min at 95°C, cooled, neutralized with 50 µL of 40 mM Tris base, and used directly in PCR reactions. For genotyping of embryos, tails were treated with DirectPCR lysis reagent (Viagen) according to the manufacturer's instructions. All genotyping PCRs were performed using the primer pair in Supplemental Table S2D and were digested by BccI prior to electrophoresis.
E13.5 embryos and postnatal mice (postnatal days 50–58) were dissected in ice-cold PBS. The E13.5 embryos were fixed for 5 h in 4% paraformaldehyde/PBS, and postnatal mouse brains were dissected and fixed overnight in 4% formaldehyde/PBS. For cryosectioning, the samples were cryoprotected through a sucrose/PBS series (5%, 10%, 15%, and 20%), embedded in OCT compound (Cellpath), and stored at −80°C. All cryostat sections were cut at 12 µm. Immunofluorescence staining was performed by permeabilization with 0.2% Triton X-100/PBS for 15 min, blocking in 1% BSA/PBS, and primary incubation overnight. Primary antibodies used were anti-TUBβIII (Covance, MMS-435P), PAX6 (Covance, PRB-178P), and Aurora A (BD Transduction Laboratories, 610939). Secondary antibodies were incubated for 2 h, and all immunofluorescence was costained with DAPI. Imaging was performed using dragonfly multimodal imaging platform in three-dimensional fast confocal mode using the 405-, 488-, and 561-nm lasers and a iXon Ultra 888 EMCCD camera (Andor). Images were captured using a 100× plan-apo (1.4 NA) or 60× plan-apo (1.4 NA) objective at 0.2-µm Z-sections.
Three-dimensional measurements of spindle orientations
Analysis was restricted to cells in metaphase and anaphase, as the mitotic spindle stops rotating along the apico–basal axis from metaphase onward (Haydar et al. 2003). Two assumptions were made to measure mitotic spindle orientations in three dimensions. First, the spindle axis was defined by a straight line through two points defined by the XYZ coordinates of the two centrosomes (Aurora A signal). Second, the ventricular surface was defined using three points to establish a three-dimensional plane. These points were manually annotated for each three-dimensional image using OMERO (5.1.4). Mitotic spindle orientation was calculated using a custom-written graphic user interface software developed using Matlab (R2015a). This calculated the normal vector to the plane representing the ventricular surface and then the cosine angle, “A,” between this normal vector and the spindle line vector. The mitotic spindle orientation angle was thus 90° A (i.e., the complementary acute angle). Z-axis calibrations were accounted for when defining the points’ spatial locations. Three-dimensional reconstructions were rendered using Imaris software (Andor).
Statistical analysis
All data are shown as averages, with variance as either SEM or SD. Statistical analysis was performed using Prism (Graphpad Software, Inc.). For all quantitative measurements, normal distribution was assumed, with t-tests performed unpaired and two-sided unless otherwise stated. For categorical data, Χ2-test was used. Data collection and analysis were not performed blind, as the different conditions were clearly recognizable by the experimenter. No statistical methods were used to predetermine sample sizes, which were determined empirically from previous experimental experience with similar assays and/or from sizes generally employed in the field.
Supplementary Material
Acknowledgments
We thank the families, J. Tolmie, and other clinicians for their involvement and participation; the Potentials Foundation; the Walking with Giants Foundation; W. Bickmore and M. Reijns for comments on the manuscript; J. Marsh, C. Ponting, Matt Hurles and Phil Jones for helpful discussions; H. Yu (Howard Hughes Medical Institute) for his kind gift of the PICH antibody; E. Freyer for assistance with FACS analysis; D. Read for technical assistance; the MRC Institute of Genetics and Molecular Medicine (IGMM) core sequencing service and imaging facilities; Western General Hospital Biological Research Facility staff for animal husbandry; and Oxford Gene Technology and Edinburgh Clinical Research Facility for next-generation sequencing. This work was supported by funding from the Medical Research Council, the Lister Institute for Preventative Medicine, and the European Research Council (ERC; 281847 to A.P.J.); a Biotechnology and Biological Sciences Research Council grant (BB/K017632/1 to P.V); a Sir Henry Dale Fellowship (grant 102560/Z/13/Z to A.J.W.); Medical Research Scotland (to L.S.B.); the Potentials Foundation (to C.A.W.); and the Indian Council of Medical Research (BMS 54/2/2013 to S.R.P). The Deciphering Developmental Disorders Study presents independent research commissioned by the Health Innovation Challenge Fund (grant no. HICF-1009-003), a parallel funding partnership between the Wellcome Trust and the Department of Health, and the Wellcome Trust Sanger Institute (grant no. WT098051). The views expressed here are those of the authors and not necessarily those of the Wellcome Trust or the Department of Health. The study has UK Research Ethics Committee approval (10/H0305/83) granted by the Cambridge South Research Ethics Committee, and GEN/284/12 granted by the Republic of Ireland. We acknowledge the support of the National Institute for Health Research through the Comprehensive Clinical Research Network.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.286351.116.
References
- Al-Dosari MS, Shaheen R, Colak D, Alkuraya FS. 2010. Novel CENPJ mutation causes Seckel syndrome. J Med Genet 47: 411–414. [DOI] [PubMed] [Google Scholar]
- Baumann C, Korner R, Hofmann K, Nigg EA. 2007. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell 128: 101–114. [DOI] [PubMed] [Google Scholar]
- Baxter J, Sen N, Martinez VL, De Carandini ME, Schvartzman JB, Diffley JF, Aragon L. 2011. Positive supercoiling of mitotic DNA drives decatenation by topoisomerase II in eukaryotes. Science 331: 1328–1332. [DOI] [PubMed] [Google Scholar]
- Bicknell LS, Bongers EM, Leitch A, Brown S, Schoots J, Harley ME, Aftimos S, Al-Aama JY, Bober M, Brown PA, et al. 2011. Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat Genet 43: 356–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J, Roberts E, Mochida GH, Hampshire DJ, Scott S, Askham JM, Springell K, Mahadevan M, Crow YJ, Markham AF, et al. 2002. ASPM is a major determinant of cerebral cortical size. Nat Genet 32: 316–320. [DOI] [PubMed] [Google Scholar]
- Bond J, Roberts E, Springell K, Lizarraga SB, Scott S, Higgins J, Hampshire DJ, Morrison EE, Leal GF, Silva EO, et al. 2005. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet 37: 353–355. [DOI] [PubMed] [Google Scholar]
- Chan KL, North PS, Hickson ID. 2007. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J 26: 3397–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID. 2009. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol 11: 753–760. [DOI] [PubMed] [Google Scholar]
- Charbin A, Bouchoux C, Uhlmann F. 2014. Condensin aids sister chromatid decatenation by topoisomerase II. Nucleic Acids Res 42: 340–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. 2012. DNA breaks and chromosome pulverization from errors in mitosis. Nature 482: 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, et al. 2006. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat Genet 38: 910–916. [DOI] [PubMed] [Google Scholar]
- Cuylen S, Haering CH. 2011. Deciphering condensin action during chromosome segregation. Trends Cell Biol 21: 552–559. [DOI] [PubMed] [Google Scholar]
- D'Ambrosio C, Kelly G, Shirahige K, Uhlmann F. 2008. Condensin-dependent rDNA decatenation introduces a temporal pattern to chromosome segregation. Curr Biol 18: 1084–1089. [DOI] [PubMed] [Google Scholar]
- Deardorff MA, Kaur M, Yaeger D, Rampuria A, Korolev S, Pie J, Gil-Rodriguez C, Arnedo M, Loeys B, Kline AD, et al. 2007. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of Cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet 80: 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff MA, Wilde JJ, Albrecht M, Dickinson E, Tennstedt S, Braunholz D, Monnich M, Yan Y, Xu W, Gil-Rodriguez MC, et al. 2012. RAD21 mutations cause a human cohesinopathy. Am J Hum Genet 90: 1014–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Deciphering Developmental Disorders Study. 2015. Large-scale discovery of novel genetic causes of developmental disorders. Nature 519: 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. 2011. Cohesin: genomic insights into controlling gene transcription and development. Curr Opin Genet Dev 21: 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish JL, Kosodo Y, Enard W, Paabo S, Huttner WB. 2006. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci 103: 10438–10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JV, Cole TJ, Chinn S, Jones PR, White EM, Preece MA. 1995. Cross sectional stature and weight reference curves for the UK, 1990. Arch Dis Child 73: 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D, Hirota T, Koch B, Peters JM, Ellenberg J. 2006. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr Biol 16: 333–344. [DOI] [PubMed] [Google Scholar]
- Gogendeau D, Siudeja K, Gambarotto D, Pennetier C, Bardin AJ, Basto R. 2015. Aneuploidy causes premature differentiation of neural and intestinal stem cells. Nat Commun 6: 8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosling KM, Makaroff LE, Theodoratos A, Kim YH, Whittle B, Rui L, Wu H, Hong NA, Kennedy GC, Fritz JA, et al. 2007. A mutation in a chromosome condensin II subunit, kleisin β, specifically disrupts T cell development. Proc Natl Acad Sci 104: 12445–12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green LC, Kalitsis P, Chang TM, Cipetic M, Kim JH, Marshall O, Turnbull L, Whitchurch CB, Vagnarelli P, Samejima K, et al. 2012. Contrasting roles of condensin I and condensin II in mitotic chromosome formation. J Cell Sci 125: 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith E, Walker S, Martin CA, Vagnarelli P, Stiff T, Vernay B, Al Sanna N, Saggar A, Hamel B, Earnshaw WC, et al. 2008. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat Genet 40: 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber R, Zhou Z, Sukchev M, Joerss T, Frappart PO, Wang ZQ. 2011. MCPH1 regulates the neuroprogenitor division mode by coupling the centrosomal cycle with mitotic entry through the Chk1–Cdc25 pathway. Nat Cell Biol 13: 1325–1334. [DOI] [PubMed] [Google Scholar]
- Hagstrom KA, Holmes VF, Cozzarelli NR, Meyer BJ. 2002. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev 16: 729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks S, Coleman K, Reid S, Plaja A, Firth H, Fitzpatrick D, Kidd A, Mehes K, Nash R, Robin N, et al. 2004. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet 36: 1159–1161. [DOI] [PubMed] [Google Scholar]
- Haydar TF, Ang E Jr, Rakic P. 2003. Mitotic spindle rotation and mode of cell division in the developing telencephalon. Proc Natl Acad Sci 100: 2890–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. 2012. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev 26: 1659–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ. 1994. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell 79: 449–458. [DOI] [PubMed] [Google Scholar]
- Hirano T, Kobayashi R, Hirano M. 1997. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell 89: 511–521. [DOI] [PubMed] [Google Scholar]
- Hirota T, Gerlich D, Koch B, Ellenberg J, Peters JM. 2004. Distinct functions of condensin I and II in mitotic chromosome assembly. J Cell Sci 117(Pt 26): 6435–6445. [DOI] [PubMed] [Google Scholar]
- Hoffelder DR, Luo L, Burke NA, Watkins SC, Gollin SM, Saunders WS. 2004. Resolution of anaphase bridges in cancer cells. Chromosoma 112: 389–397. [DOI] [PubMed] [Google Scholar]
- Houlard M, Godwin J, Metson J, Lee J, Hirano T, Nasmyth K. 2015. Condensin confers the longitudinal rigidity of chromosomes. Nat Cell Biol 17: 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson DF, Vagnarelli P, Gassmann R, Earnshaw WC. 2003. Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev Cell 5: 323–336. [DOI] [PubMed] [Google Scholar]
- Insolera R, Bazzi H, Shao W, Anderson KV, Shi SH. 2014. Cortical neurogenesis in the absence of centrioles. Nat Neurosci 17: 1528–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. 2011. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science 333: 1895–1898. [DOI] [PubMed] [Google Scholar]
- Juschke C, Xie Y, Postiglione MP, Knoblich JA. 2014. Analysis and modeling of mitotic spindle orientations in three dimensions. Proc Natl Acad Sci 111: 1014–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalay E, Yigit G, Aslan Y, Brown KE, Pohl E, Bicknell LS, Kayserili H, Li Y, Tuysuz B, Nurnberg G, et al. 2011. CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat Genet 43: 23–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y, Huh JW, Warrington R, Li B, Wu N, Leng M, Zhang J, Ball HL, Yu H. 2011. PICH and BLM limit histone association with anaphase centromeric DNA threads and promote their resolution. EMBO J 30: 3309–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingseisen A, Jackson AP. 2011. Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev 25: 2011–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. 2013. Cerebral organoids model human brain development and microcephaly. Nature 501: 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson MD, Vandin F, Wu HT, Dobson JR, Eldridge JV, Thomas JL, Papoutsaki A, Kim Y, Niu B, McLellan M, et al. 2015. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet 47: 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizarraga SB, Margossian SP, Harris MH, Campagna DR, Han AP, Blevins S, Mudbhary R, Barker JE, Walsh CA, Fleming MD. 2010. Cdk5rap2 regulates centrosome function and chromosome segregation in neuronal progenitors. Development 137: 1907–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, Grofte M, Chan KL, Hickson ID, Bartek J, et al. 2011. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol 13: 243–253. [DOI] [PubMed] [Google Scholar]
- Maciejowski J, Li Y, Bosco N, Campbell PJ, de Lange T. 2015. Chromothripsis and kataegis induced by telomere crisis. Cell 163: 1641–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannini L, Cucco F, Quarantotti V, Krantz ID, Musio A. 2013. Mutation spectrum and genotype-phenotype correlation in Cornelia de Lange syndrome. Hum Mutat 34: 1589–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthiens V, Rujano MA, Pennetier C, Tessier S, Paul-Gilloteaux P, Basto R. 2013. Centrosome amplification causes microcephaly. Nat Cell Biol 15: 731–740. [DOI] [PubMed] [Google Scholar]
- Martin CA, Ahmad I, Klingseisen A, Hussain MS, Bicknell LS, Leitch A, Nurnberg G, Toliat MR, Murray JE, Hunt D, et al. 2014. Mutations in PLK4, encoding a master regulator of centriole biogenesis, cause microcephaly, growth failure and retinopathy. Nat Genet 46: 1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minocherhomji S, Ying S, Bjerregaard VA, Bursomanno S, Aleliunaite A, Wu W, Mankouri HW, Shen H, Liu Y, Hickson ID. 2015. Replication stress activates DNA repair synthesis in mitosis. Nature 528: 286–290. [DOI] [PubMed] [Google Scholar]
- Murray JE, van der Burg M, IJspeert H, Carroll P, Wu Q, Ochi T, Leitch A, Miller ES, Kysela B, Jawad A, et al. 2015. Mutations in the NHEJ component XRCC4 cause primordial dwarfism. Am J Hum Genet 96: 412–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, Vezzoni P, Larizza L. 2006. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet 38: 528–530. [DOI] [PubMed] [Google Scholar]
- Nicholas AK, Khurshid M, Desir J, Carvalho OP, Cox JJ, Thornton G, Kausar R, Ansar M, Ahmad W, Verloes A, et al. 2010. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat Genet 42: 1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen CF, Huttner D, Bizard AH, Hirano S, Li TN, Palmai-Pallag T, Bjerregaard VA, Liu Y, Nigg EA, Wang LH, et al. 2015. PICH promotes sister chromatid disjunction and co-operates with topoisomerase II in mitosis. Nat Commun 6: 8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishide K, Hirano T. 2014. Overlapping and non-overlapping functions of condensins I and II in neural stem cell divisions. PLoS Genet 10: e1004847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. 2003. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet 33: 497–501. [DOI] [PubMed] [Google Scholar]
- Oliveira RA, Coelho PA, Sunkel CE. 2005. The condensin I subunit Barren/CAP-H is essential for the structural integrity of centromeric heterochromatin during mitosis. Mol Cell Biol 25: 8971–8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T. 2003. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 115: 109–121. [DOI] [PubMed] [Google Scholar]
- Ono T, Fang Y, Spector DL, Hirano T. 2004. Spatial and temporal regulation of condensins I and II in mitotic chromosome assembly in human cells. Mol Biol Cell 15: 3296–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qvist P, Huertas P, Jimeno S, Nyegaard M, Hassan MJ, Jackson SP, Borglum AD. 2011. CtIP mutations cause Seckel and Jawad syndromes. PLoS Genet 7: e1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Amon A. 2015. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Mol Cell Biol 16: 473–485. [DOI] [PubMed] [Google Scholar]
- Savvidou E, Cobbe N, Steffensen S, Cotterill S, Heck MM. 2005. Drosophila CAP-D2 is required for condensin complex stability and resolution of sister chromatids. J Cell Sci 118: 2529–2543. [DOI] [PubMed] [Google Scholar]
- Shintomi K, Hirano T. 2011. The relative ratio of condensin I to II determines chromosome shapes. Genes Dev 25: 1464–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui NU, Rusyniak S, Hasenkampf CA, Riggs CD. 2006. Disruption of the Arabidopsis SMC4 gene, AtCAP-C, compromises gametogenesis and embryogenesis. Planta 223: 990–997. [DOI] [PubMed] [Google Scholar]
- Smith ED, Xu Y, Tomson BN, Leung CG, Fujiwara Y, Orkin SH, Crispino JD. 2004. More than blood, a novel gene required for mammalian postimplantation development. Mol Cell Biol 24: 1168–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov AV, Hogan E, Koshland D. 1995. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev 9: 587–599. [DOI] [PubMed] [Google Scholar]
- Thornton GK, Woods CG. 2009. Primary microcephaly: do all roads lead to Rome? Trends Genet 25: 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibelius A, Marhold J, Zentgraf H, Heilig CE, Neitzel H, Ducommun B, Rauch A, Ho AD, Bartek J, Kramer A. 2009. Microcephalin and pericentrin regulate mitotic entry via centrosome-associated Chk1. J Cell Biol 185: 1149–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimborn M, Schindler D, Neitzel H, Hirano T. 2006. Misregulated chromosome condensation in MCPH1 primary microcephaly is mediated by condensin II. Cell Cycle 5: 322–326. [DOI] [PubMed] [Google Scholar]
- Verloes A, Drunat S, Gressens P, Passemard S. 2009. Primary autosomal recessive microcephalies and Seckel syndrome spectrum disorders. In GeneReviews (ed. Pagon RA, et al. ), http://www.ncbi.nlm.nih.gov/books/NBK9587 University of Washington, Seattle. [Google Scholar]
- Wang J, Chang YF, Hamilton JI, Wilkinson MF. 2002. Nonsense-associated altered splicing: a frame-dependent response distinct from nonsense-mediated decay. Mol Cell 10: 951–957. [DOI] [PubMed] [Google Scholar]
- Wood JL, Liang Y, Li K, Chen J. 2008. Microcephalin/MCPH1 associates with the Condensin II complex to function in homologous recombination repair. J Biol Chem 283: 29586–29592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods CG, Bond J, Enard W. 2005. Autosomal recessive primary microcephaly (MCPH): a review of clinical, molecular, and evolutionary findings. Am J Hum Genet 76: 717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CF, Fitzgerald TW, Jones WD, Clayton S, McRae JF, van Kogelenberg M, King DA, Ambridge K, Barrett DM, Bayzetinova T, et al. 2014. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet 385: 1305–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita D, Shintomi K, Ono T, Gavvovidis I, Schindler D, Neitzel H, Trimborn M, Hirano T. 2011. MCPH1 regulates chromosome condensation and shaping as a composite modulator of condensin II. J Cell Biol 194: 841–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.