Abstract
Cell lineages, which shape the body architecture and specify cell functions, derive from the integration of a plethora of cell intrinsic and extrinsic signals. These signals trigger a multiplicity of decisions at several levels to modulate the activity of dynamic gene regulatory networks (GRNs), which ensure both general and cell-specific functions within a given lineage, thereby establishing cell fates. Significant knowledge about these events and the involved key drivers comes from homogeneous cell differentiation models. Even a single chemical trigger, such as the morphogen all-trans retinoic acid (RA), can induce the complex network of gene-regulatory decisions that matures a stem/precursor cell to a particular step within a given lineage. Here we have dissected the GRNs involved in the RA-induced neuronal or endodermal cell fate specification by integrating dynamic RXRA binding, chromatin accessibility, epigenetic promoter epigenetic status, and the transcriptional activity inferred from RNA polymerase II mapping and transcription profiling. Our data reveal how RA induces a network of transcription factors (TFs), which direct the temporal organization of cognate GRNs, thereby driving neuronal/endodermal cell fate specification. Modeling signal transduction propagation using the reconstructed GRNs indicated critical TFs for neuronal cell fate specification, which were confirmed by CRISPR/Cas9-mediated genome editing. Overall, this study demonstrates that a systems view of cell fate specification combined with computational signal transduction models provides the necessary insight in cellular plasticity for cell fate engineering. The present integrated approach can be used to monitor the in vitro capacity of (engineered) cells/tissues to establish cell lineages for regenerative medicine.
The life of cells in multicellular organisms is directed by dynamic gene programs, which guide and define lineage progression from pluripotent to differentiated states through series of temporal decisions. Knowledge of these programs and decisions reveals not only how cells acquire physiological functionalities, it also provides key information for therapy, as deviations from this blueprint can lead to disease. Moreover, the possibility to interfere with cell programming by treating stem cells or reprogramming somatic cells may generate specific autologous cell types for regenerative medicine in a personal medicine context.
Cell lineages derive from series of subsequent programming decisions. Cell differentiation models, particularly those where the series of transitions within a lineage is initiated by a single chemical trigger like all-trans retinoic acid (RA), significantly facilitated the study of cell fate acquisition. The use of RA (rather than complex culture conditions) as a defined trigger of regulatory events is essential to elucidate the dynamically regulated “downstream” gene networks. In this context, our study of F9 embryo carcinoma (EC) cells provided a first detailed view of RA-induced gene program diversification through a plethora of regulatory decisions (Mendoza-Parra et al. 2011).
EC cells can differentiate into all three primary germ layers (Soprano et al. 2007). While F9 cells differentiate into primitive endoderm when treated with RA in monolayer, parietal or visceral endodermal differentiation is observed when RA is either complemented with cyclic AMP or when cells are cultured as embryoid bodies in suspension. P19 EC cells differentiate into either skeletal muscle or neuronal cell types upon treatment with dimethlysulfoxide or RA, respectively. Thus, RA can induce cell fate commitment toward two distinct primary germ layers. However, the temporal evolution of the corresponding gene programs and the regulatory mechanisms remained elusive.
RA signaling is initiated by its binding to retinoid receptor heterodimers (RAR/RXR), members of the nuclear receptor (NR) family of ligand-regulated TFs (Laudet and Gronemeyer 2002). Upon ligand binding, RAR/RXR recruits coactivator complexes leading to the transcriptional activation of target genes (TGs) (Gronemeyer et al. 2004; Rosenfeld et al. 2006). The complexity of the RA signaling is largely increased by the expression of three RXR and three RAR isotypes (alpha, beta, and gamma), as each RAR/RXR combination could regulate cognate gene programs (Chiba et al. 1997). Interestingly, particular isotype-selective RAR ligands (Alvarez et al. 2014) induced specific cell fate transitions: F9 cells show similar morphological cell differentiation phenotypes when treated with RA or the RARG-selective ligand BMS961, but not with the RARA-selective ligand BMS753. In contrast, in P19 cells BMS753 and RA induce the same morphological differentiation, while BMS961 has no such effect (Taneja et al. 1996). These observations strongly support a critical role of RAR isotypes in the establishment of different cell fate commitment processes.
Given that RARA/G isotypes are expressed similarly in both EC cells (Supplemental Fig. S1), we reconstructed the dynamics of GRNs that are at the basis of the cell fate decisions in F9 and P19 cells by characterizing common and cell-specific RA-induced gene programs (Supplemental Fig. S2). We subsequently developed a computational signal transduction model that was used to (1) verify the temporal transcriptional coherence of the reconstructed GRN, and (2) predict potential downstream TFs that drive neuronal cell fate commitment. Using CRISPR/dCas9 (D10/N863A) technology, we activated the transcription of several predicted factors and assessed their capacity to induce the acquisition of neuronal identity. Overall, this study provides a detailed view of the complex regulatory wirings that are commonly initiated in both EC model systems but lead to distinct cells fates and which can be engineered for redirecting cell fate decisions.
Results
RA induces both common and cell fate-specific programs in F9 and P19 cells
As RA induces a neuronal cell fate of P19 cells, while driving endodermal differentiation of F9 cells, we first defined common and cell-specific RA-induced programs in these models. We used previously established monolayer cultures (Monzo et al. 2012) for efficient morphological P19 cell differentiation by RA and showed that this process is driven by RARA by using RAR isoform-specific agonists (Supplemental Fig. S3A). Neuronal cell fate commitment was confirmed by the induction of neurogenin 1 (Neurog1) and Neurod1 (Supplemental Fig. S3B). Analysis of the global transcriptome changes during P19 cell differentiation revealed a previously reported progressive increase of differentially expressed genes (DEGs) (Wei et al. 2002). Indeed, after 2 h of RA treatment, only 51 genes showed an induction of ≥1.8-fold, while >1000 were induced after 72 h (Supplemental Fig. S4).
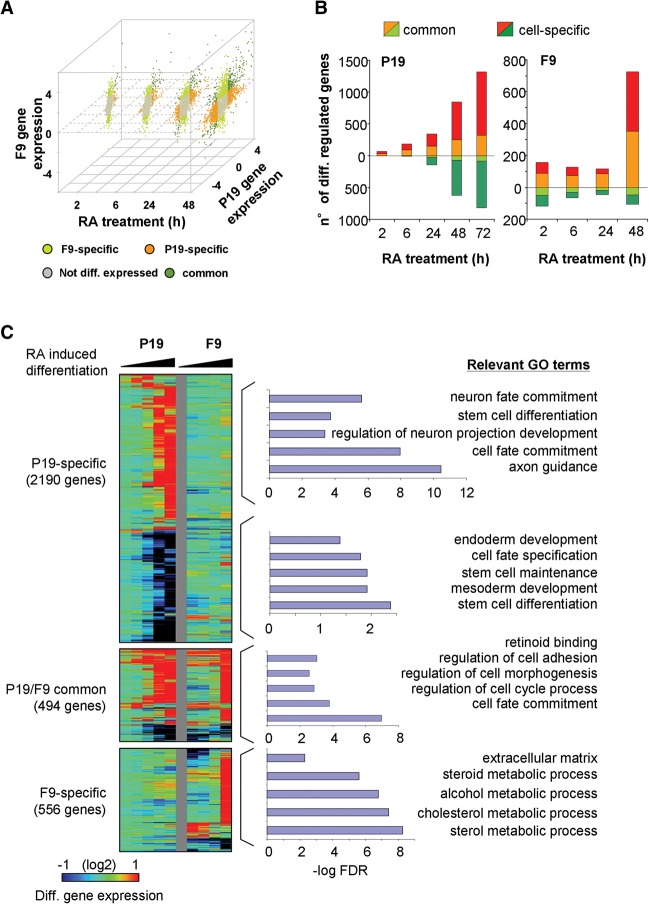
A comparison of the temporal transcriptome changes during endodermal F9 (Mendoza-Parra et al. 2011) and neuronal P19 cell differentiation revealed that >60% of genes are commonly regulated in both cell lines, albeit with different kinetics in some cases (Fig. 1). F9 cells present a higher number of DEGs in the first hours of RA treatment (Fig. 1A), but most of these early responders are also observed in P19 cells at later time points. In keeping with the progressive expression of the differentiated phenotype, divergent cell type-selective gene expression increased toward later time points, such that at 72 h, only <30% of the genes differentially expressed in P19 were similarly regulated in F9 cells (Fig. 1B). Gene Ontology (GO) analysis classified the commonly RA-regulated genes as involved in retinoid binding or cell fate commitment. Among them were classical RA-induced genes (e.g., Rarb, Cyp26a1, or Hoxa1), while pluripotency factors were down-regulated (Fig. 1C). As expected, P19-specific RA-induced genes are enriched for GO terms like neuronal fate commitment, while down-regulated genes are enriched for terms like endoderm or mesoderm development and stem cell maintenance, which are repressed during neuronal cell fate acquisition.
Figure 1.
Common and specific RA-induced differentiation programs characterized in F9 and P19 embryonal carcinoma cells. (A) Scatterplot illustrating transcriptome changes in F9 and P19 EC cells at different time points during RA-induced differentiation. Gene expression levels relative to the undifferentiated state were classified as common, EC-specific, or not differentially expressed, based on a defined fold change threshold (up-regulated genes, fold change > 1.8; down-regulated genes, fold change < 0.5) at a given time point. (B) Differential gene expression levels in both model systems were used for computing the number of differentially regulated genes (y-axis) at various time points covering the first 72 h of RA treatment (x-axis). DEGs were classified as either commonly or cell-specifically expressed. This classification takes into consideration the gene expression response over all evaluated time points, in contrast to A, where a classification per time point is performed. (C) Temporal changes in transcriptional expression in either F9 or P19 EC cells are displayed for common and cell type-specific genes. Relevant GO terms per common or cell type-specific group of genes are displayed.
Chromatin state dynamics during neuronal and endodermal differentiation correlate with gene coexpression patterns
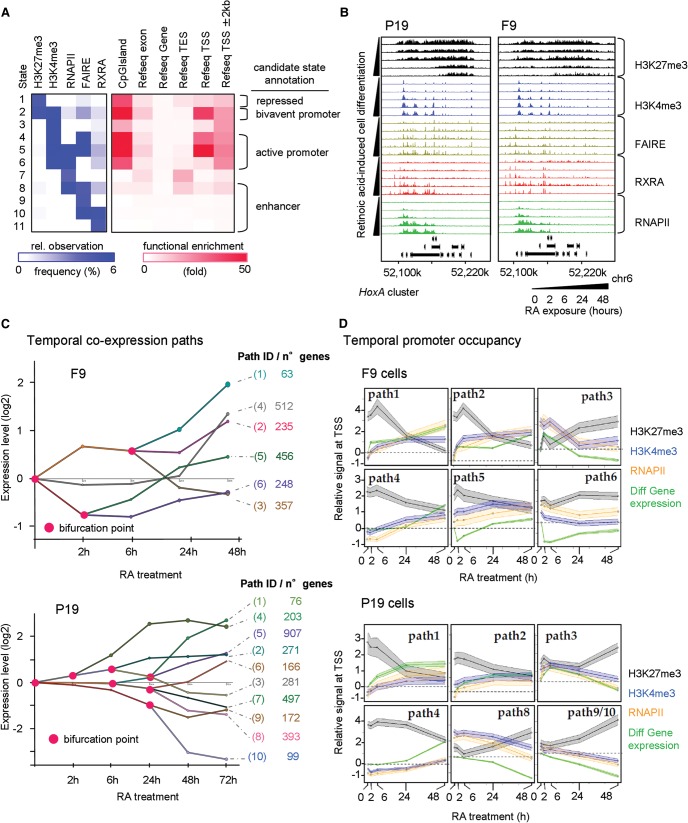
While the above transcriptome profiling revealed the RA-induced changes, an understanding of the corresponding regulatory mechanisms requires additional analyses of the RA-modulated key players and the information on epigenome and chromatin structure changes. To this end, we mapped RXRA binding sites to identify cognate TGs and complemented this readout with the characterization of epigenetic marks indicative for active and repressed transcription, open chromatin regions, and RNAPII binding at regulated genes. Our combinatorial analysis of the generated data sets demonstrated the existence of genomic regions preferentially enriched for repressive marks (H3K27me3), bivalent/poised (H3K27me3 and H3K4me3), or active promoter regions (H3K4me3 and/or RNAPII), but also for candidate enhancer regions where open chromatin sites co-occurred with RXRA binding (Fig. 2A).
Figure 2.
Multiparametric view of retinoid-induced cell fate transitions. (A) Chromatin state analysis performed over all profiled factors at all time points in P19 and F9 cells. Based on the predicted states resulting from a combination of all studied factors (left panel, relative observation frequency); four major candidate states were inferred: repressed, bivalent or active promoter, and enhancer-related states. This classification is supported by their functional enrichment levels associated with the described genomic annotations (right panel). (B) The HoxA cluster at Chromosome 6 displaying temporal changes in the enrichment of H3K27me3 and H3K4me3, the chromatin accessibility status (FAIRE-seq), the recruitment of the RXRA, and the transcriptional activity revealed by the profiling of the RNAPII. (C) Stratification of the temporal transcriptome profiling during RA-induced F9 (upper panel) or P19 cell differentiation (lower panel) in gene coexpression paths, accompanied by relevant bifurcation points (pink circles). Numbers of genes composing each of the coexpression paths are displayed (right). (D) Dynamics of promoter chromatin states during RA-induced F9 (upper panel) or P19 cell differentiation (lower panel). Gene promoters of the coexpression paths displayed in C are analyzed for temporal enrichment of (1) the repressive histone modification mark H3K27me3 (black), (2) the active histone modification mark H3K4me3 (blue), and (3) RNAPII (orange). Changes of mRNA levels relative to the noninduced condition are also displayed (“Diff Gene expression”; green). The y-axis corresponds to the average relative enrichment level derived from Epimetheus normalization (Supplemental Fig. S5). The shaded area corresponds to a 95% confidence interval.
An example of the temporal connection between these various regulatory events is the HoxA cluster, where the progressive loss of the repressive H3K27me3 mark during RA-induced differentiation both in P19 and F9 cells correlates with a gain in FAIRE, RXRA, and RNAPII enrichment patterns (Fig. 2B). These progressive changes of chromatin accessibility/TF association and gain of marks for active transcription with concomitant loss of “repressive” marks correlated with the collinear mechanism for transcription activation of Hox genes, previously described in other systems (Kashyap et al. 2011; Montavon and Duboule 2013).
To evaluate the coherence between epigenetic status and transcriptional activity, temporal transcriptomes were analyzed in the context of gene coexpression paths with the Dynamic Regulatory Events Miner (DREM) (Ernst et al. 2007). This analysis gave rise to a total of six coexpression paths (Mendoza-Parra et al. 2011) for the endodermal differentiation and 10 coexpression paths for the neuronal cell fate acquisition (Fig. 2C).
Assuming that genes with similar temporal expression patterns share common temporal alterations of epigenome and RNAPII recruitment patterns, we assessed the enrichment of H3K27me3, H3K4me3, and/or RNAPII at the promoter regions of genes differentially expressed in both model systems and displayed it in a coexpression path context. To accurately define temporal enrichment patterns, we first normalized the ChIP-seq profiles using a novel two-step normalization procedure (Supplemental Methods; Supplemental Fig. S5).
We observed in general a positive correlation between the temporal evolution of gene coexpression paths and normalized H3K4me3 and RNAPII enrichment patterns at promoter regions of concerned genes, while a negative correlation was seen with the repressive H3K27me3 mark (Fig. 2D). Given the presence of both common and endodermal (F9)/neurogenesis (P19)-specific gene programming in each path, we analyzed these programs separately (Supplemental Fig. S6). As expected, the evolution of the chromatin states of gene promoters from the common program was highly similar in F9 and P19, while the states of fate-specific programs showed significant temporal divergence. In coexpression paths with a similar epigenetic landscape in both cell lines (path1 in F9; path1, 2, and 4 in P19), RA induction led to a temporal increase in the ratios of “active” over “repressive” chromatin in a F9/P19-specific manner, coinciding with increased gene expression.
In contrast, genes of other paths showed already in the noninduced state distinct epigenetic and/or RNAPII association characteristics (paths2, 4, and 5 in F9; path3 and partially path1 in P19 cells). Paths composed of genes gradually repressed during differentiation in an endodermal (F9)/neurogenesis (P19)-specific manner frequently gained in “repressive” chromatin (path9/10 of P19 cells; path6 in F9 cells). Importantly, the temporal evolution of specific genes fully reflected the global promoter characteristics within these paths, as for the commonly regulated Rarb or Pou5f1 and the P19-specific Neurog1 or Tal2 gene promoters (Supplemental Fig. S7).
Altogether, these data support the concept that RA-induced common and fate-specific temporal changes in gene programming closely correlate with changes in the ratios of “active” and “repressive” chromatin marks at the cognate promoter regions.
Dissection of common and divergent target gene programming in neuronal and endodermal lineage-committed cells by RAR isotype-specific ligands
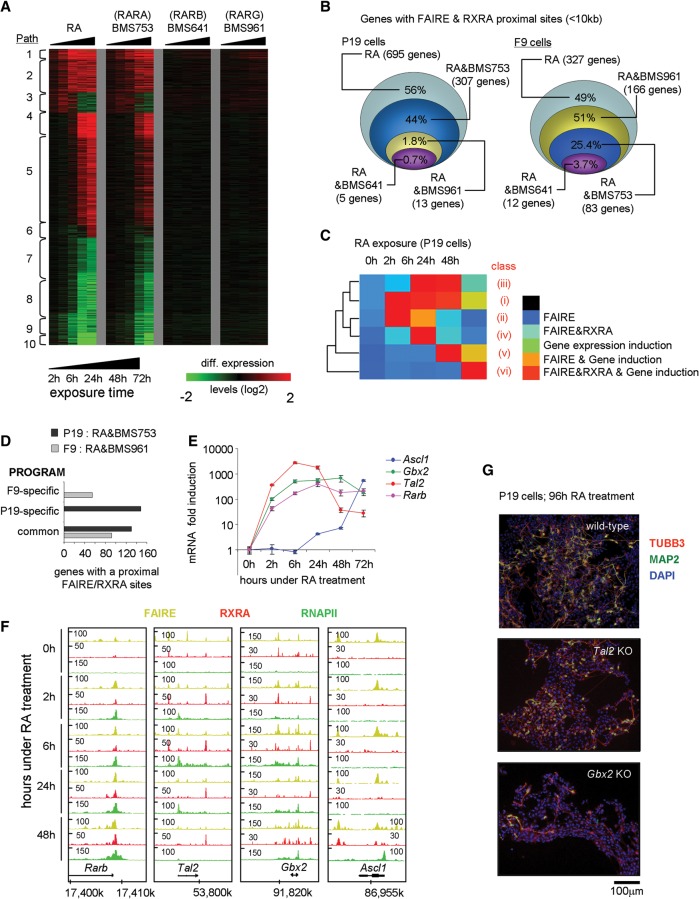
To identify core GRNs for the cell fate transitions, we established P19 transcriptomes after treatment with RAR subtype-specific agonists. Gene coexpression paths were nearly identical for the RARA-specific agonist (BMS753) and RA (Fig. 3A), in keeping with the common induction of a neuronal fate (Supplemental Fig. S3). No such effect was seen with RARB or RARG-specific agonists (Fig. 3A; Supplemental Figs. S8, S9). In F9 cells, both RA and the RARG agonist (BMS961) induced endodermal differentiation, as revealed by corresponding gene expression changes (Supplemental Fig. S8; Mendoza-Parra et al. 2011). Despite the similar response kinetics of RA and BMS753, the RARA agonist did not regulate the same number of genes as RA, suggesting that only a fraction of the RA responsive genes in P19 cells is required for phenotypic differentiation. Apparently, the BMS753-regulon corresponds to a minimal regulatory network, but the regulatory input of RA is more complex and extends beyond known differentiation features.
Figure 3.
Different RAR subtypes induce chromatin alteration in RA-responsive genes of P19 and F9 cells. (A) Heat map illustrating the transcriptional responses of genes comprising the 10 coexpression paths characterized in P19 cells during RA-induced differentiation or in the presence of the indicated RAR isotype-specific agonists. (B) DEGs during RA-induced differentiation in P19 or F9 cells that present FAIRE and RXRA binding in proximity (<10 kb from the TSS) are compared with their corresponding transcriptional response in the presence of RAR isotype-specific agonists. (C) Heat map illustrating temporal SOTA classification of P19 genes positive for RXRA binding, and/or display altered chromatin structure (FAIRE-seq), and/or are induced in response to RA. This classification gave rise to the identification of six classes of genes with different temporal induction patterns (Supplemental Fig. S5). (D) Number of DEGs F9 or P19 cells commonly regulated by RA and BMS753 or RA and BMS961 and presenting a proximal FAIRE and RXRA binding site, stratified for the cell-specific (P19, F9) and common programs. (E) RT-qPCR revealing the temporal RA-induced mRNA expression profiles of bona fide RA target genes. (F) FAIRE-seq, RXRA, and RNAPII ChIP-seq profiles for the factors assessed in E. Rarb, Gbx2, and Tal2 are early responding genes, while Ascl1 gets significantly induced only after 24 h of RA induction. (G) Immunofluorescence micrograph of wild-type and CRISPR/Cas9-inactivated Tal2 or Gbx2 P19 cells after 96 h of RA treatment. Cells were stained for the neuronal markers TUBB3 (red) and MAP2 (green); nuclei were stained with DAPI (blue). Gbx2-inactivated cells present a lower frequency of double-stained TUBB3/MAP2 cells and shorter axon-like extension than Tal2-inactivated or wild-type cells.
To reveal the direct RAR-RXR heterodimer TGs, we compared the proximal binding of RXRA (<10 kb distance) and the co-occurrence of open chromatin regions with RA or RAR subtype-specific agonist-regulated genes. From 695 RA-induced genes with FAIRE and RXRA sites in proximity, 44% responded to BMS753 but <2% to BMS961 or BMS641 (Fig. 3B). A similar analysis for F9 cells showed that from 327 RA–up-regulated genes displaying FAIRE and RXRA sites in proximity, about half (166 genes) responded also to the RARG-specific agonist BMS961, while significantly less (∼25% and <4%) responded to RARA or RARB agonists, corroborating our previous findings (Mendoza-Parra and Gronemeyer 2013). Together, our results provide a complete gene regulatory framework accounting for the observations (Taneja et al. 1996) that RARA triggers neuronal differentiation of P19, while RARG induces endodermal differentiation of F9 cells.
To link the appearance of FAIRE and RXRA sites to transcription activation, we classified genes according to their temporal induction during RA or BMS753 treatment and proximal FAIRE and RXRA co-occurrence (SOTA, self-organization tree algorithm) (Fig. 3C; Supplemental Fig. S10). This methodology classified the transcriptional activation of P19 genes in six temporal patterns. Importantly, each class of the RA-induced P19 RXRA target genes contains a great number of genes that are equally induced in F9 cells, irrespective of the divergent cell fate acquisition (Fig. 3D; Supplemental Fig. S10). Among those are not only early induced prototypical TGs, like Rarb (Fig. 3E,F), Foxa1 (Tan et al. 2010; Mendoza-Parra et al. 2011), and Hoxa1, but also late-induced direct TGs, such as Pbx1, Pbx2, Cdh2, Sox6, and Sox11 (Supplemental Fig. S10). This shows that, despite significantly advanced divergent differentiation, RA still continues to induce an identical subset of TGs irrespective of endodermal or neuronal differentiation.
As expected, the P19-specific direct RXRA targets comprise factors involved in neurogenesis, mostly expressed at late time points during differentiation (Ascl1 [Fig. 3E,F; Voronova et al. 2011; Huang et al. 2012, 2015]; Gata3 [Martinez-Monedero et al. 2008]). Interestingly, however, the expression of some P19-specific TGs was already affected during the first hours of RA-treatment, among them, the TFs Gbx2 (Bouillet et al. 1995; Inoue et al. 2012; Nakayama et al. 2013), and Tal2, which is essential for midbrain neurogenesis (Achim et al. 2013) and contains an intronic RA response element (Kobayashi et al. 2014, 2015). We identified two additional RXRA binding sites proximal to Tal2—a constitutive RXRA binding site ∼3 kb downstream from the coding region and a second site upstream of the transcription start site (TSS) (∼5 kb), which is similarly occupied in the absence of ligand but persists only until 6 h after initiating RA treatment (Fig. 3F).
To evaluate the importance of TAL2 and GBX2 for RA-induced neuronal commitment, we used CRISPR/Cas9-mediated gene inactivation (Supplemental Fig. S11A). Tal2-gene inactivation did not impair the expression of other neuronal-specific factors like ASCL1, NEUROD1, POU3F4, or NEUROG1 (Supplemental Fig. S11B). In contrary, Gbx2-inactivation reduced their expression severely, suggesting that GBX2 rather than TAL2 is a critical mediator of RA-induced neuronal commitment. This has been further supported by immunohistochemical analysis of the neuron-specific tubulin, beta 3 class III (TUBB3) and the microtubule-associated factor MAP2 (Fig. 3G; Supplemental Fig. S9), as RA induction of Gbx2-inactivated cells resulted in dramatically reduced numbers of TUBB3 and MAP2-stained cells, concomitantly with a major reduction of axonal extensions.
A network of TFs drives cell fate lineage decisions
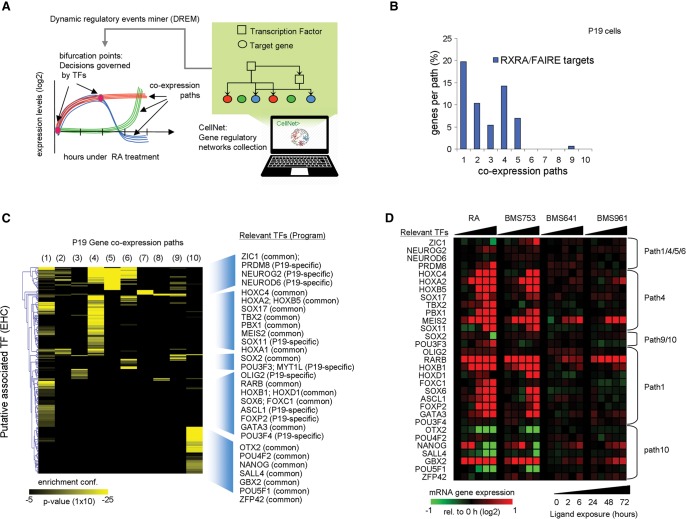
The above integrative approach identified direct RXRA TGs, several of which are TFs. Conceptually, these genes could initiate TF-guided signal transduction cascades, ultimately generating the differentiated phenotype. To identify TFs relevant for the RA-induced neuronal fate of P19 cells, we established DREM-predicted coexpression paths (Fig. 2C). DREM evaluates the enrichment of coexpression paths for TGs associated with given TFs retrieved from TF-TG collections (Fig. 4A). Indeed, correlating RXRA binding/FAIRE site annotations with DREM-based gene coexpression analysis revealed the presence of RA target genes in the early path1-5, compliant with the inductive role of RXR-RAR heterodimers (Fig. 4B).
Figure 4.
Inferring relevant TF-TG relationships during RA-induced neuronal cell fate transition. (A) Scheme of the strategy applied by DREM to integrate static TF-TG regulatory relationships with the temporal gene coexpression paths. In addition, the information provided by the CellNet collection and the RXRA binding/FAIRE site information have been integrated. (B) Bar graph depicting the fraction of RXRA and FAIRE TGs in each coexpression path assessed in P19 cells during the RA-induced differentiation. (C) TFs’ enrichment per coexpression path in P19 cells as predicted by DREM (hypergeometric distribution probability). The heat map illustrates the enrichment confidence per coexpression path for each of the most confident TF associations (CellNet database) further classified by Euclidean hierarchical clustering (EHC). On the right, the identity of relevant TFs per cluster and their implication in common and endodermal (F9)-/neuronal (P19)-specific gene programs is displayed. (D) mRNA expression levels of relevant TFs displayed in C assessed with either RA or RAR-specific agonists.
To identify additional relevant TFs, we reconstructed the RA-induced TF-TG networks involved in neuronal (P19) and endodermal (F9) differentiation by integrating the GRN interactions that constitute CellNet (Morris et al. 2014) into the DREM analysis (Fig. 4A). We identified multiple TFs associated with several coexpression paths but also path-specific TFs (Fig. 4C). Several of them were differentially expressed upon exposure to RA or RARA-specific agonists, supporting a direct implication in the predicted bifurcation (Fig. 4D). The negatively regulated coexpression path 10 associated with the self-renewal and pluripotency factors NANOG, POU5F1, ZFP42, SOX2, or SALL4 or with GBX2 and OTX2, TFs expressed very early during neuroectoderm development (Millet et al. 1999). Note that RA induction of GBX2 negatively regulates the expression of OTX2 in the anterior brain (Li and Joyner 2001; Inoue et al. 2012), corroborating their inverse expression patterns (Fig. 4D). Similarly, the early induced path1 is enriched for homeobox TF (HOXB1, HOXD1)-TGs but also for targets of ASCL1, OLIG2, and POU3F4, which are specifically expressed in neural tissues and, moreover, impose a neuronal fate on MEFs (Vierbuchen et al. 2010). Furthermore, the intermediate to late-induced path4 is enriched for MEIS2, PBX1, TBX2, or HOXA1, the latter being essential for neuronal commitment of mouse embryonic stem cells (Martinez-Ceballos and Gudas 2008). Integrating the CellNet TF-TG regulatory network information into the endodermal differentiation model (F9) revealed a set of TFs specifically involved in endodermal gene programming (“F9-specific”) (Supplemental Fig. S12). However, we found a surprisingly large number of TFs that are commonly involved in both RA-induced endodermal and neuronal differentiation. A comparison of the GRNs inferred from these analyses is provided below.
Generation of comprehensive RA-driven signal transduction networks for neuronal and endodermal cell fates
To provide a comparative view of the signal transduction cascades driving the differential cell fates induced by RA in F9 and P19 cells, we integrated the CellNet TF-TG relationships (Morris et al. 2014), complemented by direct RA target and DREM analysis data, resulting in the reconstruction of a comprehensive GRN (2981 nodes, 44,931 edges) (Fig. 5A; Supplemental File S1). Two major nodes (blue squares) represent the initial RXRA/RAR signal interpreter in P19 or F9 cells. Each of them is associated with its direct targets of the common or fate-specific programs.
Figure 5.
Temporal signal propagation in RA-induced GRNs for neuronal and endodermal cell fate decisions. (A) Structure of the reconstructed GRN displaying genes that are selectively or commonly regulated during neuronal and endodermal cell differentiation. For illustration purposes, all edges were removed; arrows indicate the direct regulation of each of these programs by TFs that are bona fide direct RA responsive genes (blue squares; black arrows). Gene expression changes are illustrated as heat maps. (B) Signal transduction model aiming at evaluating the coherence between the reconstructed GRN and the temporal gene expression changes. The starting node where the initial cue activates the signal transduction is depicted, as well as the downstream node interconnections required for its propagation. The temporal transcriptional state for each node is defined as 1, 0, or −1 (up-regulated, nonresponsive, or down-regulated, respectively). The model excludes signal cascade progression branches (illustrated by crosses) when (1) the state of a node remains nonresponsive; (2) the directionality of the TF-TG relationship is opposite to the temporal signal flux; or (3) the TF-TG relationships are not part of the main signal transduction propagation branches. (C) Temporal transcriptional evolution of the reconstructed GRNs in P19 or F9 RA-induced cell differentiation. Note that common programs dominate at early time points, while the neuronal/endodermal programs take over at late time points. (D) Fraction of transduced nodes per regulatory program for both model systems (F9-specific, common, P19-specific), as assessed by the signal transduction model. As illustrated in C, the common gene regulatory program is activated early (>80% in both cell lines after 2 h of RA treatment), while the cell fate-specific program is set up progressively (∼60% of specific programs in either of the model systems after 48 h of RA). (E) Responsiveness of common and neuronal-/endodermal-specific GRNs described in A to agonists selective for the three RAR isotypes. (F) Fraction of reconstituted gene regulatory programs (GRPs) (after 72 h of RA treatment) in both model systems when either the RA or RAR-specific agonists-derived transcriptomes are used for modeling signal transduction propagation.
As CellNet was established using different cell types, it comprises also TF-TG interactions that are irrelevant for RA-dependent gene regulation. To exclude such interactions, we developed a computational approach that evaluates the coherence of the TF-TG relationships with the temporal evolution of transcription activation (Fig. 5B). Specifically, all interconnections from nodes not differentially expressed or originating from nodes not related to the initial cue were excluded, reducing the reconstructed GRN to 1931 nodes and 11,625 edges. The temporal evolution of common and fate-specific networks is evident from the superposition of RA-dependent gene expression patterns at the first four time points of the reconstructed GRN (Fig. 5C; Supplemental File S1) and from the increasing fraction of transduced nodes for each lineage-specific program (Fig. 5D).
The reconstructed network reveals also the RAR isotype-selective induction of endodermal or neuronal fates. Indeed, the RARA-specific agonist BMS753 fully recapitulates the neurogenic RA-response of P19 in both common and P19-specific gene regulatory programs, while only a minor fraction of this program is regulated in F9 (Fig. 5E,F; Supplemental File S1). Similarly, the RARG-specific BMS961 activates endodermal programming as RA in F9 but remains as ineffective in P19 as the RARB-agonist BMS641 in both cell fate programs. Further reduction by applying topological criteria generated a network (80 nodes, 626 edges) (Supplemental Fig. S17) with major nodes distributed in four subnetworks: two implicated in cell differentiation (pluripotency, HOX factors) and two neuronal/endodermal regulatory programs.
In summary, the reconstructed GRN reconstitutes a scenario in which cascades of TF-driven common and specific regulatory programs are responsible for acquisition of endodermal and neuronal fates. Thus, cell fate specification is predefined by a given cellular context even when the same chemical trigger is used for program initiation.
Identification of “master regulators” from a hierarchical analysis of the GRN
The reconstructed GRN for neuronal/endodermal fates reveals common and cell fate-specific factors, which instruct the two RA-induced differentiation programs. The neurogenic GRN contains several known neuronal TFs, but the majority of these are activated late. To identify early key TFs (“master regulators”) critical for cell fate commitment, we simulated the capacity of each of the 1087 nodes of the P19-specific program to propagate the transcriptional regulatory cascade toward the latest time point, corresponding to the ultimate biological readout (Fig. 6A; Supplemental Fig. S13). This analysis predicted less than 75 nodes as master regulators of the neurogenic program (Fig. 6B; Supplemental Fig. S13). Among them, several known neuronal TFs, like NEUROD1, NEUROG2, POU3F2, or MYT1L, reconstitute <20% of the P19 program, while other “early” factors, like ASCL1 (Huang et al. 2012, 2015), NR2F2 (Zhou et al. 2015), or NR4A2 (Park et al. 2006) reconstitute >60% (Fig. 6B). Importantly, this analysis identified additional TFs (e.g., GBX2, TAL2, TSHZ1, DMRT1, LHX2) with the capacity to reconstitute >50% of the P19-specific program. Moreover, the reconstructed GRN revealed direct and indirect links between many of these factors and connection to the neuronal factors ASCL1, NEUROG1, NEUROG2, and/or POU3F2 (Fig. 6C).
Figure 6.
Predicting master regulators of neurogenesis by modeling signal transduction propagation. (A) Scheme of the signal transduction propagation model initiated at a downstream layer in the reconstructed GRN. (B) 1087 nodes comprising the P19-specific GRP (x-axis) ranked according to their performance in reconstituting the ultimate level of the P19-specific program (y-axis). Previously known neuronal factors are depicted in association with their position in the ranking (gray). Less characterized factors with significant signal propagation performance toward the final level are in blue. (C) Transcriptional regulatory relationships among the newly predicted factors in B are depicted in the context of their interconnections with relevant neuronal markers. Their relative temporal transcriptional response under RA-driven conditions is indicated (color coded). (D) Immunofluorescence micrographs illustrating the presence of the neuronal markers TUBB3 (red) and MAP2 (green), in P19 cells after CRISPR/dCas9 (D10/N863A)-mediated transcription activation of Tal2, Gbx2, Lhx2, or Dmrt1 treated with the RARG-specific agonist BMS961 or vehicle. (E) Immunofluorescence micrographs revealing the presence of the neuronal markers TUBB3 (red) and MAP2, SOX1, or Nestin (NES; green) in F9 cells after CRISPR/dCas9 (D10/N863A)-mediated transcription activation of Tal2, Lhx2, and Dmrt1 treated with BMS961 and the RARB-specific BMS641. In the right panel, a mock-CRISPR/dCas9 (D10/N863A) transfection assay (no guide RNA) in F9 cells under identical treatment conditions is displayed.
To evaluate the relevance of predicted TF-TG relationships, we used the CRISPR/dCas9 transcription activation strategy to induce expression of endogenous factors (Konermann et al. 2015). Specifically, we used guide RNAs to target the Tal2, Gbx2, Lhx2, or Dmrt1 promoters for VP64-mediated transcription activation. To study if the common regulatory program is required for efficient cell fate specification, we performed the activation assays in the presence or absence of the RARG-specific agonist BMS961. This ligand does not induce neuronal differentiation of P19 cells (Supplemental Fig. S9) but activates components of the common program. Tal2 activation (>200-fold in the presence of BMS961) resulted in induced mRNA expression of Gbx2 (greater than sevenfold), Lhx2 (>3.5-fold), and of the neuronal factors Pou3f2, Neurog2, and Neurog1 (>3.5-fold). Similarly, Gbx2, Lhx2, or Dmrt1 activation resulted in increased expression of known neuronal factors (Supplemental Fig. S14). The BMS961-enhanced response of most neuronal factors supported our hypothesis that the common program is required for/supports the fate-selective programs. In all cases, the engineered activation of these factors (Tal2, Gbx2, Lhx2, or Dmrt1) induced the response of the above neuronal markers and led to a positive immunostaining for the neuronal markers TUBB3 and MAP2 (Fig. 6D).
To ultimately demonstrate the potential of the identified neurogenic key factors to impose a neurogenic fate onto a differently committed cell, we used the CRISPR/dCas9 (D10/N863A) strategy to induce in F9 cells the expression of known neurogenic TFs and master regulators predicted by our transcription propagation approach. As illustrated in Supplemental Figure S15A, inefficient CRISPR/dCas9 (D10/N863A)-mediated activation of neuronal factors in F9 cells was observed in the absence of retinoids. We therefore hypothesized that the activation of the common gene program is required for efficient CRISPR/dCas9 (D10/N863A)-mediated induction of these factors in F9 cells. Indeed, exposing CRISPR/dCas9 (D10/N863A)-transfected cells to ATRA (Supplemental Fig. S15A) or RAR subtype-specific agonists (Supplemental Fig. S16A) resulted in dramatically increased expression of the neurogenic factors. This is also supported by the presence (ATRA, BMS753) and absence (EtOH) of morphological changes in CRISPR/dCas9 (D10/N863A)-transfected cells (Supplemental Fig. S15B). Together, this suggested that activation of a subset of the RA-induced program(s) is required for optimal CRISPR/dCas9 (D10/N863A)-mediated transcription activation, possibly due to modulation of promoter accessibility. Using this combinatorial approach, induction of neurogenesis-specific genes was seen upon CRISPR/dCas9 (D10/N863A)-mediated activation of cognate genes for both known neurogenic factors (ASCL1, NEUROG2, POU3F2, MYT1L, OLIG2) (Supplemental Fig. S16A) and the new ones predicted in the present study (TAL2, LHX2, DMRT1) (Supplemental Fig. S16B). In all cases, F9 neuronal transdifferentiation was confirmed by immunostaining for TUBB3, SOX1, Nestin, and MAP2 (Fig. 6E). Together, these results demonstrated that the use of signal propagation models from reconstructed GRNs identifies novel (and confirms known) key TFs involved in cell fate acquisition.
The EC GRNs are relevant for mouse embryonic stem cell differentiation
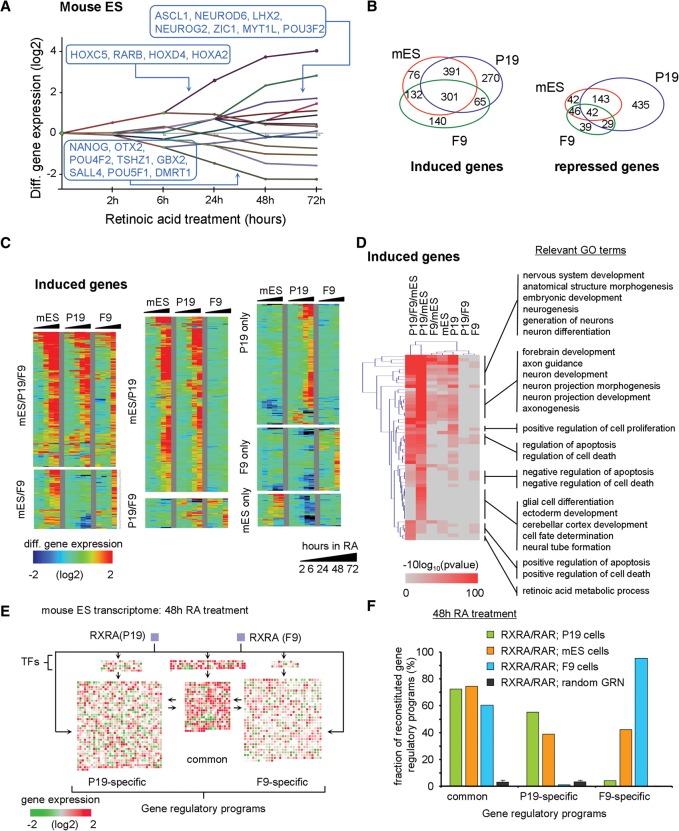
To explore the relevance of our observations and networks for RA-driven mouse ESC differentiation, we have analyzed publicly available temporal studies (GSE30176 [Lin et al. 2011]; GSE34279 [Gaertner et al. 2012]). Reconstruction of its dynamic regulatory map resulted in 14 coexpression paths (Fig. 7A). The integration of the CellNet TF-TG collection predicted several self-renewal TFs enriched in the most down-regulated group of genes, as well as factors like OTX2, GBX2, TSHZ1, or DMRT1, identified here as relevant components of the RA-induced neuronal differentiation. Other coexpression paths are also enriched for components identified in the P19 model, revealing major similarities.
Figure 7.
Relevance of the inferred GRP in EC cells in comparison to the mouse ES model system. (A) Dynamic regulatory map reconstructed from publicly available temporal transcriptome data of RA-treated mES cells. (B) Venn diagram illustrating the number of DEGs shared with either P19 or F9 cells during the RA-induced program (all time points included). (C) Temporal mRNA gene expression levels (heat map; induced genes) associated with each of the cell model systems and displayed based on the classification in B (for repressed genes, see Supplemental Fig. S18). (D) GO analysis of induced genes displayed in B. (E) Genes expressed in mouse ES cells after 48 h of RA treatment revealing common and F9-/P19-specific programs and color-coded according to their expression levels relative to the noninduced state. Genes composing all three GRPs are regulated in ES cells, despite the expected neuronal cell fate commitment. (F) Fraction of reconstituted GRPs in all three cell systems (after 48 h of RA treatment). Note that in mouse ES cells, both the P19- and F9-specific programs are induced at a level of ∼40%; this contrasts with the much more specific neuronal and endodermal programs in P19 or F9 cells, respectively.
Comparing RA-regulated genes in mouse ES and EC cells revealed that >75% of these genes are commonly up-regulated in ES and P19 cells; about half of those are also induced in F9 cells (Fig. 7B,C). Similarly, >65% of the genes repressed in ES are also repressed in P19 cells, again supporting a similar response to RA (Fig. 7B; Supplemental Fig. S18). Despite these similarities, each of the systems contained sets of additional DEGs. GO analysis for each of the observed sets of common P19 and ES up-regulated genes retrieved neuronal fate-related terms, while up-regulated genes shared by all three systems were specifically enriched for RA metabolic processes (Fig. 7D).
Unexpectedly, the transcriptional response of ES cells contributed significantly to both the common and the specific (P19/neuronal; F9 endodermal) gene regulatory programs (GRPs) (Fig. 7E,F), corroborating earlier reports of nonhomogeneous RA-induced differentiation of mouse ES cells (Sartore et al. 2011). Indeed, improved differentiation protocols involve complex cocktails of factors to increase the yield and purity of neuronal precursors (Ying et al. 2003; Abranches et al. 2009).
Discussion
Cell fate transitions are fundamental for the genesis of multicellular organisms, and aberrations from this body plan can generate pathologies. One such process is neurogenesis, a highly complex phenomenon that involves a plethora of instructive signals, including cell-to-cell communication and extrinsic chemical signals, which during organogenesis generate regionally organized cells with diverse functionality.
Interestingly, the blueprint of neurogenesis, which includes the principal architecture of the brain, is already encoded within neuronal stem cells. Indeed, 3D cultures of cerebral organoids have been developed from ES or iPS cells (Lancaster et al. 2013). Notably, neurogenesis occurs also in the adult mammalian brain (Eriksson et al. 1998; Ming and Song 2011), and the plasticity of cell fates in adult tissues prompted critical reflection about concepts of stemness, cell differentiation, and regeneration (Sanchez Alvarado and Yamanaka 2014). However, while some key TFs can be sufficient for cell reprogramming (Weintraub et al. 1989; Zhou et al. 2008; Ieda et al. 2010; Sekiya and Suzuki 2011), our knowledge about the temporal evolution and regulation of gene networks, which specify cell fates and plasticity, has remained fragmentary. Therefore, we have initiated a study to define the temporal regulation of gene programs that are initiated by a single compound, the morphogen all-trans retinoic acid, in P19 cells, which are committed to undergo neuronal differentiation. The involvement of RA in the developing nervous system and the adult brain, including its role in regeneration, is well-documented (Vergara et al. 2005). We have compared these programs with those responsible for RA-induced endodermal differentiation of F9 cells (Mendoza-Parra et al. 2011) and defined common and cell-specific programs, as well as subnetworks initiated by nodes critical for lineage identity. The results of this analysis were used to instruct cells adapting a neuronal fate by a combination of subtype-specific retinoids and CRISPR/dCas-mediated activation of endogenous genes.
RA induces modular gene programs in committed EC cells
A comparison of RA-induced neuronal and endodermal GRNs revealed common, endodermal-, and neuronal-specific programs; most of the well-known RA-targets (e.g., Rarb, Hox genes) belong to the common program. The specific programs can be activated by RARA (neuronal) and RARG (endodermal)-selective retinoids (Alvarez et al. 2014), which both activate the common program (Fig. 5E). Given that RA regulates multiple embryonic (e.g., limb development) and cell physiological (e.g., differentiation, apoptosis) phenomena in different compartments (e.g., hematopoietic system, skin) at different developmental stages (e.g., embryogenesis, organogenesis, adult homeostasis), the overall RA-program is likely composed of common and specific modules. Thus, genes supporting stemness (Sox2, Nanog, Myc) are commonly repressed in both EC cell lines, as differentiated cells lose pluripotency. The coordinately regulated Hox genes may provide spatiotemporal information to the neuronal and endodermal progeny; for example, the self-organizing capacity observed for ES/iPS cell-derived cerebral organoids (Lancaster et al. 2013) may be linked to the ability of Hox genes to define the body plan.
We noted that the common program does not operate in isolation, as it enables CRISPR-activated key genes (Fig. 6D) to induce neuronal differentiation. This indicates intimate links between the cell fate-specific and common programs, which may be of importance for identifying conditions that support/improve the efficiency/functionality of engineered ES/iPS cells for regenerative purposes. It is likely that similar scenarios exist for other nuclear receptors/TFs with similar pleiotropic action as retinoid receptors. It would be interesting to compare in this respect the common and specific gene programs induced by retinoids and vitamin D during hematopoiesis.
The molecular origin of the divergent cell-specific gene programs in P19 and F9 cells remains elusive. While it is clear that different RAR isotypes trigger neurogenic (P19, RARA) and endodermal (F9, RARG) differentiation, we have so far not been able to identify RAR subtype-selective pioneer principles (Zaret and Carroll 2011). Thus, it is unlikely that an RAR subtype-specific gene-regulatory event drives lineage specification; rather, it appears that P19 and F9 cells are already committed. This is supported by the differential epigenetic makeup of P19- and F9-specific genes. In general, activated P19-specific genes lose repressive H3K27me3 marks (with or without gaining H3K4me3 marks) in P19 but not in F9 cells, and vice versa (see Supplemental Fig. S6). Genes that became repressed in one EC cell line showed generally increased levels of H3K27me3 with or without loss of H3K4me3; no such effect was seen in the other EC cell line. However, genes of the common program showed similar epigenetic changes, irrespective of the epigenetic status of genes from the neuronal-/endodermal-specific program.
Notably, the commitment of P19 and F9 cells to their respective lineage was not irreversible, as we could transdifferentiate F9 cells into neurons by activating the common RA-induced program together with the CRISPR/Cas9-mediated induction of endogenous F9 genes that were identified as master regulators of the neuronal program using our novel signal propagation approach (Fig. 6). Notably, activation of the common program was requisite for transdifferentiation.
The RA-regulated programs of ES and EC cells share common and divergent features
A comparative analysis of the gene programs initiated by RA in P19, F9, and ES cells (Lin et al. 2011; Gaertner et al. 2012) yielded the initially surprising result that the ES program was a composite of both EC cells rather than a mimic of the neurogenic P19 program (Supplemental Fig. S17). However, this result reflects that (1) only a fraction of ES cells develop into neurons, (2) sophisticated ES culture conditions are required for efficient differentiation in vitro (Studer 2014), and (3) exogenous RA addresses simultaneously all accessible developmental programs in ES cells, including endodermal ones, thus justifying our choice of committed P19 cells for defining the neurogenic GRN.
A novel in vitro signal propagation approach to identify master regulators
Validation of the RA-dependent neuronal GRN in P19 revealed unexpected results. For example, inactivation of the early induced Tal2 had no obvious consequences on neurogenesis (Fig. 3G), while inactivation of the similarly expressed Gbx2 strongly impaired neurogenesis. However, even though not required, CRISPR-mediated activation of endogenous Tal2 was sufficient to drive neurogenesis (together with the common program), as did the activation of Gbx2 (see Fig. 6E). Thus, the program is composed of both necessary and sufficient actors, including significant functional redundancy.
One of the questions that derives from the present definition of the neuronal network refers to its plasticity in supporting transdifferentiation. Fibroblasts can be converted to electrophysiologically responsive, marker-positive neurons by exogenously expressed ASCL1, POU3F2, and MYT1L (Wapinski et al. 2013); similar results were obtained by overexpressing two neurogenins in human iPS cells (Busskamp et al. 2014). All these factors are activated rather late in the RA-induced GRN following complex regulatory events (Fig. 6C). This suggests two scenarios: (1) either the complex history of temporally organized gene regulatory events is necessary, as it generates a spatiotemporal “memory” for the development, functional specification, and structural organization of all the cells that constitute a functional CNS, and the transdifferentiation experiments reveal only a testable fraction of this scenario; or (2) the cellular plasticity allows for virtually any cell fate conversion given the correct set of conditions and factors is provided (see also Sanchez Alvarado and Yamanaka 2014). Validating these scenarios experimentally requires blueprints of the developmental programs driving differentiation of CNS compartments and cell types in vivo and an assessment of how this program can be recapitulated in the structures of cerebral organoids.
The value of reconstructing networks
We demonstrate here that by reconstructing the cellular network corresponding to induced cell fate transitions, it is possible to infer relevant factors, their interdependency, and hierarchical position. Particularly useful was the approach to validate nodes and connectivities that were imported from heterologous settings by monitoring their temporal coherence with the current expression data and confirming the functional relevance of predicted key factors by CRISPR-based approaches. By evaluating the potential of a factor to generate the final nodes of the network, we identified several known (e.g., NR4A2, ASCL1, NR2F2) and novel (TAL2, GBX2, LHX2, DMRT1) key factors involved in retinoid-induced neurogenesis (see Fig. 6B). Note that identification of DMRT1 as a potential neuronal differentiation factor previously involved enormous transcriptome profiling efforts (Yamamizu et al. 2013).
Modeling temporal signal propagation in reconstructed GRNs is a general approach to reveal transcriptional interconnection and identify master regulators in any system. Indeed, for validating the corresponding Cytoscape plugin, we applied it to diverse phenomena, including differentiation, reprogramming, and tumorigenesis, supporting its general utility (MA Mendoza-Parra, PE Cholley, J Moehlin, M Lieb, and H Gronemeyer, unpubl.). We thus believe that the comprehensive approach described here is not limited to understanding the molecular circuits underlying physiological and, when altered, pathological cell fate transition. It provides, moreover, a comprehensive way to monitor the ability of stem, reprogrammed, or transdifferentiated cells to properly adopt a desired cell fate.
Methods
Cell culture
F9 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 4.5 g/L glucose; P19 cells were grown in DMEM supplemented with 1 g/L glucose, 5% FCS, and 5% delipidated FCS. Both media contained 40 µg/mL Gentamicin. F9 or P19 EC cells were cultured in monolayer on gelatin-coated culture plates (0.1%). For cell differentiation assays, RA was added to plates to a final concentration of 1 µM for different exposure times. For treatment with RAR subtype-specific agonists, cells were incubated with BMS961 (RARG-specific; 0.1 µM), BMS753 (RARA-specific; 1 µM), and/or BMS641 (RARB-specific; 0.1 µM).
RT-qPCR and transcriptomics
Total RNA was extracted from EC cells treated with either RA or RAR-specific agonists, using the GenElute Mammalian Total RNA Miniprep kit (Sigma). Two micrograms of the extracted RNA were used for reverse transcription (AMV-RTase, Roche; Oligo[dT], New England Biolabs; 1 h at 42°C and 10 min at 94°C). Transcribed cDNA was diluted 10-fold and used for real-time quantitative PCR (Roche LC480) (primers, Supplemental Methods).
For transcriptomics analysis, AffymetrixGeneChip Mouse Gene 1.0 ST arrays were used (Supplemental Methods). For comparing transcriptomes, we normalized all raw CELL files with the Affymetrix software Expression Console.
Chromatin immunoprecipitation assays
ChIP assays were performed according to standard procedures (Supplemental Methods). All ChIP and FAIRE assays were validated using positive and negative controls. ChIP validation assays were performed by quantitative real-time PCR using the Qiagen Quantitect kit.
Massive parallel sequencing and quality control
qPCR-validated ChIPs were quantified (Qubit dsDNA HS kit; Invitrogen); multiplexed sequencing libraries were prepared from 10 ng of the ChIPed material (Supplemental Methods).
Sequence-aligned files were qualified for enrichment using the NGS-QC Generator (Mendoza-Parra et al. 2013b). Briefly, this methodology computes enrichment quality descriptors discretized in a scale ranging from “AAA” (Best) to “DDD” (worst). Based on this quantitative method, all ChIP-seq and FAIRE data sets described in this study presented quality grades higher than “CCC”; integrative studies were thus performed exclusively with high-quality data sets.
Enrichment pattern detection and intensity profile normalization
Relevant binding sites in all ChIP-seq and FAIRE-seq data sets were identified with MeDiChISeq (Mendoza-Parra et al. 2013a); multiprofile comparisons were done after quantile normalization (Supplemental Methods; Mendoza-Parra et al. 2012).
Dynamic regulatory maps and RA-driven GRN reconstruction
We reconstructed GRNs by combining several layers of information. First, we identified direct TGs as those containing (1) a proximal RXRA and FAIRE enrichment event (<10 kb distance), and (2) responding to both RA and the corresponding BMS-specific agonist. Downstream regulatory processes were reconstructed by integrating the TF-TG collection of CellNet (Cahan et al. 2014; Kim and Scholer 2014) in the RA-regulated EC GRPs deduced by DREM (Supplemental Methods).
The integration in Cytoscape (version 2.8) of the RXRA-direct targets per cell type with the downstream regulatory networks assessed from the DREM/CellNet approach generated a GRN composed of 2981 nodes and 44,931 edges, organized in common or EC-specific regulated programs. GRN complexity was reduced by applying topological metrics (Yu et al. 2007; Chin et al. 2014). The ultimate reduced GRN was composed of 80 nodes and 626 edges, with a ranking color code (heat map) displaying the hub importance metrics (Supplemental Fig. S17). The organization of reduced GRN and its visualization were performed with the Cytoscape package Cerebral (Supplemental File S1; Barsky et al. 2007).
Modeling signal transduction progression in reconstructed GRNs
To validate the relevance of the TF-TGs relationships composing the reconstructed F9/P19 GRN, we developed a computational framework for modeling signal propagation within the network. It takes as initial information: (1) the topology of the reconstructed network in which the TF-TG directionality is essential; (2) the temporal transcriptional information associated with each of the nodes composing the network; and (3) the node from which the signal transduction is initiated, (starting node) to follow the temporal evolution of signal(s) until the ultimate time points of the experimental data set (final nodes). In this context, the signal propagation model evaluates in the first round the transcriptional response at the first time point (e.g., 2 h of RA treatment) of the TGs associated with the starting node. In the second round, the model defines starting nodes, initially defined by the user as well as those with a differential transcriptional behavior in the first round. In this manner, the second round evaluates the interconnections (edges) between the newly defined starting nodes and their corresponding targets by evaluating their transcriptional behavior at the second time point (e.g., 6 h of RA treatment). Such analysis over all available transcriptional time points reveals the coherence between the TF-TGs relationships and the temporal transcriptional information. Finally, the number of retrieved nodes at the end of the signal transduction model is compared with the expected user-provided list of final nodes. The signal propagation was performed multiple times using a randomized network as a control.
The GRN reduction (Fig. 5), the prediction of factors driving the neuronal program (Fig. 6), as well as the evaluation over mouse ES data sets (Fig. 7) have been performed using an in-house R script (Supplemental File S2); a Cytoscape plugin is in preparation.
Targeted gene knockouts with the CRISPR/Cas9 system
Cells were transfected with pairs of double-nickase plasmids encoding the Cas9D10A mutation and a 20-nt guide RNA (Santa Cruz Biotech). Single cell-derived cultures were treated with ATRA, and loss-of-expression from the targeted genes was validated by qPCR relative to control cultures (Supplemental Methods).
CRISPR/dCas9 (D10/N863A) transcriptional activation and immunohistochemical staining
EC cells were transfected with CRISPR/dCas9 (D10/N863A) activation plasmids (Santa Cruz Biotech) using lipofection and treated with ATRA, RAR-specific agonists, or ethanol, complemented with antibiotics. Six days later, cells were fixed, permeabilized, and immunostained as specified (Supplemental Methods).
Data access
Affymetrix microarrays and Illumina platform ChIP-seq and FAIRE-seq data described in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE68291.
Supplementary Material
Acknowledgments
We thank all the members of our laboratory and the IGBMC sequencing and microarray platform for discussion. Special thanks go to Anna Podlesny for sharing expertise in ICC assays. This study was supported by AVIESAN-ITMO Cancer, the Ligue National Contre le Cancer, the Institut National du Cancer (INCa), and the Agence Nationale de la Recherche (ANRT-07-PCVI-0031-01, ANR-10-LABX-0030-INRT, and ANR-10-IDEX-0002-02).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.208926.116.
References
- Abranches E, Silva M, Pradier L, Schulz H, Hummel O, Henrique D, Bekman E. 2009. Neural differentiation of embryonic stem cells in vitro: a road map to neurogenesis in the embryo. PLoS One 4: e6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achim K, Peltopuro P, Lahti L, Tsai HH, Zachariah A, Astrand M, Salminen M, Rowitch D, Partanen J. 2013. The role of Tal2 and Tal1 in the differentiation of midbrain GABAergic neuron precursors. Biol Open 2: 990–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez R, Vaz B, Gronemeyer H, de Lera AR. 2014. Functions, therapeutic applications, and synthesis of retinoids and carotenoids. Chem Rev 114: 1–125. [DOI] [PubMed] [Google Scholar]
- Barsky A, Gardy JL, Hancock RE, Munzner T. 2007. Cerebral: a Cytoscape plugin for layout of and interaction with biological networks using subcellular localization annotation. Bioinformatics 23: 1040–1042. [DOI] [PubMed] [Google Scholar]
- Bouillet P, Chazaud C, Oulad-Abdelghani M, Dolle P, Chambon P. 1995. Sequence and expression pattern of the Stra7 (Gbx-2) homeobox-containing gene induced by retinoic acid in P19 embryonal carcinoma cells. Dev Dyn 204: 372–382. [DOI] [PubMed] [Google Scholar]
- Busskamp V, Lewis NE, Guye P, Ng AH, Shipman SL, Byrne SM, Sanjana NE, Murn J, Li Y, Li S, et al. 2014. Rapid neurogenesis through transcriptional activation in human stem cells. Mol Syst Biol 10: 760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P, Li H, Morris SA, Lummertz da Rocha E, Daley GQ, Collins JJ. 2014. CellNet: network biology applied to stem cell engineering. Cell 158: 903–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba H, Clifford J, Metzger D, Chambon P. 1997. Specific and redundant functions of retinoid X Receptor/Retinoic acid receptor heterodimers in differentiation, proliferation, and apoptosis of F9 embryonal carcinoma cells. J Cell Biol 139: 735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. 2014. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 8 (Suppl 4): S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson PS, Perfilieva E, Bjork-Eriksson T, Alborn AM, Nordborg C, Peterson DA, Gage FH. 1998. Neurogenesis in the adult human hippocampus. Nat Med 4: 1313–1317. [DOI] [PubMed] [Google Scholar]
- Ernst J, Vainas O, Harbison CT, Simon I, Bar-Joseph Z. 2007. Reconstructing dynamic regulatory maps. Mol Syst Biol 3: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertner B, Johnston J, Chen K, Wallaschek N, Paulson A, Garruss AS, Gaudenz K, De Kumar B, Krumlauf R, Zeitlinger J. 2012. Poised RNA polymerase II changes over developmental time and prepares genes for future expression. Cell Rep 2: 1670–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronemeyer H, Gustafsson JA, Laudet V. 2004. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov 3: 950–964. [DOI] [PubMed] [Google Scholar]
- Huang HS, Turner DL, Thompson RC, Uhler MD. 2012. Ascl1-induced neuronal differentiation of P19 cells requires expression of a specific inhibitor protein of cyclic AMP-dependent protein kinase. J Neurochem 120: 667–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HS, Redmond TM, Kubish GM, Gupta S, Thompson RC, Turner DL, Uhler MD. 2015. Transcriptional regulatory events initiated by Ascl1 and Neurog2 during neuronal differentiation of P19 embryonic carcinoma cells. J Mol Neurosci 55: 648–705. [DOI] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. 2010. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142: 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue F, Kurokawa D, Takahashi M, Aizawa S. 2012. Gbx2 directly restricts Otx2 expression to forebrain and midbrain, competing with class III POU factors. Mol Cell Biol 32: 2618–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap V, Gudas LJ, Brenet F, Funk P, Viale A, Scandura JM. 2011. Epigenomic reorganization of the clustered Hox genes in embryonic stem cells induced by retinoic acid. J Biol Chem 286: 3250–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KP, Scholer HR. 2014. CellNet—where your cells are standing. Cell 158: 699–701. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Komori R, Ishida K, Kino K, Tanuma S, Miyazawa H. 2014. Tal2 expression is induced by all-trans retinoic acid in P19 cells prior to acquisition of neural fate. Sci Rep 4: 4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Suzuki M, Morikawa M, Kino K, Tanuma SI, Miyazawa H. 2015. Transcriptional regulation of Tal2 gene by all-trans retinoic acid (atRA) in P19 cells. Biol Pharm Bull 38: 248–256. [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. 2015. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517: 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. 2013. Cerebral organoids model human brain development and microcephaly. Nature 501: 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudet V, Gronemeyer H. 2002. The nuclear receptor factsbook. Academic Press, San Diego. [Google Scholar]
- Li JY, Joyner AL. 2001. Otx2 and Gbx2 are required for refinement and not induction of mid-hindbrain gene expression. Development 128: 4979–4991. [DOI] [PubMed] [Google Scholar]
- Lin C, Garrett AS, De Kumar B, Smith ER, Gogol M, Seidel C, Krumlauf R, Shilatifard A. 2011. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC). Genes Dev 25: 1486–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Ceballos E, Gudas LJ. 2008. Hoxa1 is required for the retinoic acid-induced differentiation of embryonic stem cells into neurons. J Neurosci Res 86: 2809–2819. [DOI] [PubMed] [Google Scholar]
- Martinez-Monedero R, Yi E, Oshima K, Glowatzki E, Edge AS. 2008. Differentiation of inner ear stem cells to functional sensory neurons. Dev Neurobiol 68: 669–684. [DOI] [PubMed] [Google Scholar]
- Mendoza-Parra MA, Gronemeyer H. 2013. Genome-wide studies of nuclear receptors in cell fate decisions. Semin Cell Dev Biol 24: 706–715. [DOI] [PubMed] [Google Scholar]
- Mendoza-Parra MA, Walia M, Sankar M, Gronemeyer H. 2011. Dissecting the retinoid-induced differentiation of F9 embryonal stem cells by integrative genomics. Mol Syst Biol 7: 538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Parra MA, Sankar M, Walia M, Gronemeyer H. 2012. POLYPHEMUS: R package for comparative analysis of RNA polymerase II ChIP-seq profiles by non-linear normalization. Nucleic Acids Res 40: e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Parra MA, Nowicka M, Van Gool W, Gronemeyer H. 2013a. Characterising ChIP-seq binding patterns by model-based peak shape deconvolution. BMC Genomics 14: 834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Parra MA, Van Gool W, Mohamed Saleem MA, Ceschin DG, Gronemeyer H. 2013b. A quality control system for profiles obtained by ChIP sequencing. Nucleic Acids Res 41: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet S, Campbell K, Epstein DJ, Losos K, Harris E, Joyner AL. 1999. A role for Gbx2 in repression of Otx2 and positioning the mid/hindbrain organizer. Nature 401: 161–164. [DOI] [PubMed] [Google Scholar]
- Ming GL, Song H. 2011. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron 70: 687–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon T, Duboule D. 2013. Chromatin organization and global regulation of Hox gene clusters. Philos Trans R Soc Lond B Biol Sci 368: 20120367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzo HJ, Park TI, Montgomery JM, Faull RL, Dragunow M, Curtis MA. 2012. A method for generating high-yield enriched neuronal cultures from P19 embryonal carcinoma cells. J Neurosci Methods 204: 87–103. [DOI] [PubMed] [Google Scholar]
- Morris SA, Cahan P, Li H, Zhao AM, San Roman AK, Shivdasani RA, Collins JJ, Daley GQ. 2014. Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell 158: 889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama Y, Kikuta H, Kanai M, Yoshikawa K, Kawamura A, Kobayashi K, Wang Z, Khan A, Kawakami K, Yamasu K. 2013. Gbx2 functions as a transcriptional repressor to regulate the specification and morphogenesis of the mid-hindbrain junction in a dosage- and stage-dependent manner. Mech Dev 130: 532–552. [DOI] [PubMed] [Google Scholar]
- Park CH, Kang JS, Shin YH, Chang MY, Chung S, Koh HC, Zhu MH, Oh SB, Lee YS, Panagiotakos G, et al. 2006. Acquisition of in vitro and in vivo functionality of Nurr1-induced dopamine neurons. FASEB J 20: 2553–2555. [DOI] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK. 2006. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev 20: 1405–1428. [DOI] [PubMed] [Google Scholar]
- Sanchez Alvarado A, Yamanaka S. 2014. Rethinking differentiation: stem cells, regeneration, and plasticity. Cell 157: 110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartore RC, Campos PB, Trujillo CA, Ramalho BL, Negraes PD, Paulsen BS, Meletti T, Costa ES, Chicaybam L, Bonamino MH, et al. 2011. Retinoic acid-treated pluripotent stem cells undergoing neurogenesis present increased aneuploidy and micronuclei formation. PLoS One 6: e20667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya S, Suzuki A. 2011. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 475: 390–393. [DOI] [PubMed] [Google Scholar]
- Soprano DR, Teets BW, Soprano KJ. 2007. Role of retinoic acid in the differentiation of embryonal carcinoma and embryonic stem cells. Vitam Horm 75: 69–95. [DOI] [PubMed] [Google Scholar]
- Studer L. 2014. The nervous system. In Essentials of stem cell biology (ed. Lanza R, Atala A), pp. 163–184. Academic Press, Amsterdam. [Google Scholar]
- Tan Y, Xie Z, Ding M, Wang Z, Yu Q, Meng L, Zhu H, Huang X, Yu L, Meng X, et al. 2010. Increased levels of FoxA1 transcription factor in pluripotent P19 embryonal carcinoma cells stimulate neural differentiation. Stem Cells Dev 19: 1365–1374. [DOI] [PubMed] [Google Scholar]
- Taneja R, Roy B, Plassat JL, Zusi CF, Ostrowski J, Reczek PR, Chambon P. 1996. Cell-type and promoter-context dependent retinoic acid receptor (RAR) redundancies for RARβ2 and Hoxa-1 activation in F9 and P19 cells can be artefactually generated by gene knockouts. Proc Natl Acad Sci 93: 6197–6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara MN, Arsenijevic Y, Del Rio-Tsonis K. 2005. CNS regeneration: a morphogen's tale. J Neurobiol 64: 491–507. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. 2010. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463: 1035–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronova A, Fischer A, Ryan T, Al Madhoun A, Skerjanc IS. 2011. Ascl1/Mash1 is a novel target of Gli2 during Gli2-induced neurogenesis in P19 EC cells. PLoS One 6: e19174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski OL, Vierbuchen T, Qu K, Lee QY, Chanda S, Fuentes DR, Giresi PG, Ng YH, Marro S, Neff NF, et al. 2013. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 155: 621–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Harris T, Childs G. 2002. Global gene expression patterns during neural differentiation of P19 embryonic carcinoma cells. Differentiation 70: 204–219. [DOI] [PubMed] [Google Scholar]
- Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, Miller AD. 1989. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci 86: 5434–5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamizu K, Piao Y, Sharov AA, Zsiros V, Yu H, Nakazawa K, Schlessinger D, Ko MS. 2013. Identification of transcription factors for lineage-specific ESC differentiation. Stem Cell Rep 1: 545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Stavridis M, Griffiths D, Li M, Smith A. 2003. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol 21: 183–186. [DOI] [PubMed] [Google Scholar]
- Yu H, Kim PM, Sprecher E, Trifonov V, Gerstein M. 2007. The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput Biol 3: e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25: 2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. 2008. In vivo reprogramming of adult pancreatic exocrine cells to β-cells. Nature 455: 627–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Liu F, Tian M, Xu Z, Liang Q, Wang C, Li J, Liu Z, Tang K, He M, et al. 2015. Transcription factors COUP-TFI and COUP-TFII are required for the production of granule cells in the mouse olfactory bulb. Development 142: 1593–1605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.