Abstract
Haplotypes are fundamental to fully characterize the diploid genome of an individual, yet methods to directly chart the unique genetic makeup of each parental chromosome are lacking. Here we introduce single-cell DNA template strand sequencing (Strand-seq) as a novel approach to phasing diploid genomes along the entire length of all chromosomes. We demonstrate this by building a complete haplotype for a HapMap individual (NA12878) at high accuracy (concordance 99.3%), without using generational information or statistical inference. By use of this approach, we mapped all meiotic recombination events in a family trio with high resolution (median range ∼14 kb) and phased larger structural variants like deletions, indels, and balanced rearrangements like inversions. Lastly, the single-cell resolution of Strand-seq allowed us to observe loss of heterozygosity regions in a small number of cells, a significant advantage for studies of heterogeneous cell populations, such as cancer cells. We conclude that Strand-seq is a unique and powerful approach to completely phase individual genomes and map inheritance patterns in families, while preserving haplotype differences between single cells.
Diploid organisms, like humans, contain two homologous copies of each chromosome, one inherited from the mother and one from the father. Despite being highly similar, each homologous chromosome harbors a unique set of genetic variants, ranging from single-nucleotide variants (SNVs), insertions, and deletions, to large polymorphic inversions. The collection of genetic variants along a single chromosome is called a haplotype, and the process of assigning variants to corresponding haplotypes is referred to as phasing.
Haplotype-resolved genomes are important in many areas of personalized medicine and genetics, ranging from variant-disease associations (Glusman et al. 2014), mapping regions with loss of heterozygosity (LOH) (Huang et al. 2007), to studies of inheritance patterns in pedigrees and populations (Tewhey et al. 2011). To phase genetic variants (alleles) into haplotypes, both computational and experimental approaches have been developed (Browning and Browning 2011). Currently, massively parallel sequencing provides the most complete set of alleles of an individual. Unfortunately, phasing these variants across the length of a chromosome is currently very challenging unless the parents of the individual are also sequenced (Kitzman et al. 2011; Amini et al. 2014). To overcome this limitation, whole-chromosome sorting (Ma et al. 2010; Fan et al. 2011; Brown et al. 2012) and chromatin capture techniques (Selvaraj et al. 2013) have been developed. However, such techniques are labor- and time-consuming and have not been widely adopted in practice. To overcome these limitations, linked-read sequencing (Zheng et al. 2016) was recently proposed to deliver long-range haplotypes. However, with this method it is not yet possible to phase genetic variants across whole chromosomes.
Here we introduce Strand-seq (Falconer et al. 2012) together with a custom bioinformatics pipeline as a novel, direct approach for haplotyping variants along the entire length of the chromosome. While our approach requires preparation of single-cell libraries, it circumvents the need for generational information and rapidly builds accurate whole-chromosome haplotypes. We directly apply these tools to phase de novo germline variants of an individual and to map parental meiotic recombination events in a family trio. Lastly, we illustrate how the single-cell resolution of our approach allows us to detect changes in the haplotype structure in subpopulations of cells.
Results
Phasing using single-cell template strand sequencing
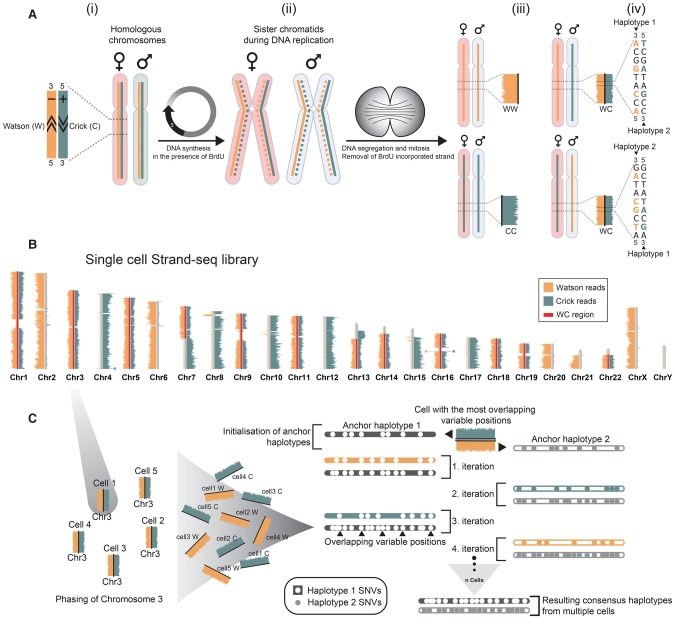
Strand-seq is a single-cell sequencing technique in which only one strand of DNA of each chromosome is sequenced, allowing individual homologs to be distinguished as either Watson (W; reverse strand), or Crick (C; forward strand) based on read alignment to the reference genome (Fig. 1A, i). The principle of Strand-seq is based on template strand identity of sister chromatids generated during DNA replication. During mitosis, each daughter cell inherits one sister chromatid from each parental homolog (Fig. 1A, ii). By sequencing only the original template strand of the inherited chromatids, we can distinguish both homologs in a single cell as two Crick template strands (CC), two Watson templates (WW), or a combination of Watson and Crick templates (WC) (Fig. 1A, iii; Falconer et al. 2012; Hills et al. 2013; Sanders et al. 2016). Consequently, when a cell inherits a chromosome as WC, the parental haplotypes for that chromosome can be readily distinguished (Fig. 1A, iv). This allows the variant alleles found in short sequencing reads of Strand-seq libraries to be phased along entire chromosomes, generating haplotypes that span centromeres, sequence gaps, and regions of homozygosity. By pooling data of multiple Strand-seq libraries from cells that inherited a chromosome as WC, accurate and dense linkage maps of the two parental haplotypes for that chromosome can be achieved.
Figure 1.
Direct whole-chromosome haplotyping using single-cell template strand sequencing (Strand-seq). (A,i) Two homologous chromosomes, one originating from the mother (light red) and one from the father (light blue), are shown. Each homolog is composed of a positive template strand (Crick; teal) and a negative template strand (Watson; orange). (ii) Cells incorporate BrdU during DNA replication, generating hemi-substituted sister chromatids containing one BrdU-negative template strand (solid line) and one BrdU-positive newly synthesized strand (dashed line). (iii) Segregation of sister chromatids in two daughter cells follows the depicted combinations of maternal and paternal template strands. The newly formed DNA strands containing BrdU are selectively removed in daughter cells during library preparation, such that only the original template DNA strands are sequenced. Read density along a chromosome is plotted as horizontal bars. (iv) When daughter cells inherit one Crick and one Watson template strand for a particular chromosome, we can use strand directionality to directly assign all reads to separate haplotypes. (B) Example of a single-cell Strand-seq library, generated from HapMap cell line NA12878. Each chromosome is represented as a vertical ideogram, and the distribution of directional sequencing reads is represented as horizontal lines along each chromosome, with Watson in orange and Crick in teal. WC regions that were selected for haplotype phasing are highlighted by red bars. (C) The custom phasing algorithm StrandPhase processes one chromosome at a time. Cells that inherit one Crick and one Watson template strand for a particular chromosome are selected as input, and the SNVs identified on each template strand are used to derive each single-cell haplotype. In the first iteration, anchor haplotypes are established by pairing single-cell haplotypes exhibiting the highest number of overlapping heterozygous SNVs. This is used to initialize the consensus haplotypes “H1” and “H2,” which are further built upon in subsequent iterations. In the second iteration, the second most-dense single-cell haplotype is considered and compared to both consensus haplotypes, and any new SNVs are added to the consensus haplotype showing the best concordance. With each iteration, the consensus haplotypes are extended until no additional single-cell haplotype can be reliably assigned to the one of the consensus haplotypes.
To evaluate haplotype phasing using Strand-seq, we generated sequencing libraries from an extensively studied HapMap family trio (see Methods, “Raw data production”) (The International HapMap Consortium 2007; The International HapMap 3 Consortium 2010). We selected the child (NA12878) for our initial analysis because this individual was previously phased using parental genotype information and can therefore serve as a reference to assess the validity and precision of our approach. The Strand-seq library for a single NA12878 cell is illustrated in Figure 1B. Within this single cell, reads that aligned to the reference assembly (see Methods, “Raw data processing”) covered ∼5% of the genome, and half of the genome was inherited as WC and thus suitable for phasing (Fig. 1B, red bars). By using SNVs listed in the HapMap reference for NA12878, we phased 77,717 variant alleles in this single cell (1.34% of reference SNVs), with 99.3% of the phased SNVs matching the reference haplotypes. This result illustrates that Strand-seq can be used to rapidly generate highly accurate chromosome-spanning haplotypes from single cells.
Building whole-genome haplotypes from multiple single-cell Strand-seq libraries
In order to build more complete whole-genome haplotypes, Strand-seq data from multiple cells were combined. Each single-cell library samples the genome in a random fashion. By combining Strand-seq data from multiple cells, subsets of phased SNVs can be compiled into a dense consensus haplotype. For this purpose, we developed a Strand-seq phasing algorithm and analysis pipeline called “StrandPhase” (see Methods, “Haplotype data analysis pipeline”; algorithm available at https://github.com/daewoooo/StrandPhase) (Supplemental Fig. S1). Briefly, all WC regions are first identified within each individual cell, and SNVs present on each template strand are phased to build single-cell haplotypes. Then, StrandPhase iteratively adds the phased variants from each single cell into two consensus haplotypes based on the best concordance. Accordingly, our algorithm concatenates haplotype information from multiple single cells, reinforcing and validating the phased variants in a consensus haplotype for each homolog (Fig. 1C).
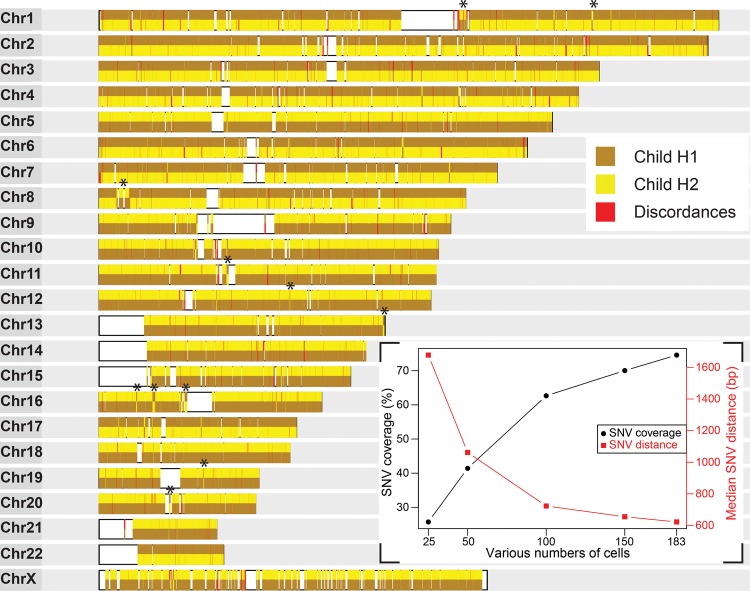
To evaluate the performance of our analysis pipeline, we selected 183 Strand-seq libraries derived from NA12878 based on read depth and coverage distribution (Supplemental Fig. S2). By use of StrandPhase, these data were used to build two consensus haplotypes, each representing a phased parental homolog inherited by the child (NA12878). Across all 183 libraries, the aligned reads covered a total of 2,156,208 SNV positions, representing 74.6% of the variants listed in the HapMap reference (Supplemental Table S1). Of the all identified variants, 1,730,627 SNV alleles were assigned to consensus haplotype 1 (Child H1) and 1,729,512 SNV alleles to consensus haplotype 2 (Child H2) (Supplemental Fig. S3A), yielding a median distance between all phased alleles of 622 bp (1309 bp for heterozygous alleles). As we increased the number of cells analyzed, SNV coverage increased and the distance between subsequent SNVs decreased (Fig. 2, inset), eventually reaching saturation. Next, we compared our haplotypes to the HapMap reference and found 99.3% of our phased SNV alleles concordant with the reported haplotypes (Fig. 2). The long-range information of Strand-seq data generated haplotypes spanning centromeres and reference assembly gaps. In addition to continuous stretches of haplotypes, we also observed smaller haplotype switches (Fig. 2, black asterisks). These switches most likely represent homozygous inversions in these regions (Sanders et al. 2016).
Figure 2.
Accurate and dense whole-genome haplotypes are built from multiple single-cell Strand-seq libraries. Assembled haplotypes of the child derived from 183 Strand-seq libraries. Chromosome ideograms illustrate 151,700 high-confidence (covered in more than one cell) heterozygous SNV positions phased from Strand-seq data and compared with the HapMap reference. The consensus haplotypes determined by Strand-seq are depicted for each chromosome, with each SNV represented by a vertical line and color-coded based on whether it matched the child's reference homolog 1 (brown) or homolog 2 (yellow) listed in the HapMap reference. The contiguous haplotypes extend the whole length of each chromosome, spanning centromeres and reference assembly gaps (white blocks). Discordant alleles that did not match either reference haplotype are shown in red. (Asterisks) Short localized switches in haplotypes that were confirmed as homozygous inversions. (Inset) The percentage of HapMap reference SNVs covered (black line) and the median distance between these SNVs (red line) are plotted for various numbers of single-cell libraries (25, 50, 100, 150), randomly sampled from the entire data set of 183 cells.
Despite the accurate phasing of SNVs spanning every chromosome in the genome, we found 23,782 alleles (0.7%) that were discordant to the HapMap reference. Strikingly, 52.9% of these discordances were observed in more than one cell in our data set, supporting the confidence of our allele phasing (Supplemental Fig. S3B). Because the likelihood of random PCR or sequencing errors occurring at the same genomic position in the same homolog in multiple independent libraries is very low, we propose that discordant phasing at these SNV positions represents errors in the HapMap reference, polymorphic inversions, or somatic mutations in the HapMap cell lines.
To further confirm the specificity of haplotype reconstruction using Strand-seq, we tested haplotyping discordances between Strand-seq and HapMap phasing using publicly available long-read PacBio RNA-seq data from the same NA12878 individual (see Methods, “PacBio and Strand-seq cross-validation”) (The International HapMap Consortium 2007). We cross-referenced the alleles segregating together on each cDNA molecule with both the Strand-seq-derived and HapMap-derived haplotypes. We found nearly perfect concordance (99.2%) of the PacBio data set to our haplotypes, while its concordance to HapMap reference was only 94.7% (Supplemental Table S2). In addition, the same trend was observed in comparison to whole-genome haplotypes reported by Fan et al. (2011; Supplemental Table S3). These results confirm that we can generate accurate haplotypes in the absence of generational (parental or population) information, which represents a major advance in the field.
With the ability to build whole-genome haplotypes, we explored phasing of unique individual variants. Expectedly, trio-based or population-based haplotyping is highly inefficient at phasing variants that occur de novo (Bansal et al. 2011). For example, only one in five de novo variants were phased in recent trio-based whole-genome sequencing (WGS) studies (Francioli et al. 2014; Kloosterman et al. 2015). To investigate the efficiency of haplotype phasing of unique variants within an individual, we applied phasing to 49 previously described and validated germline de novo mutations for NA12878 (Conrad et al. 2011). Of these, 42 were found in our data set and were phased within our consensus haplotypes (Supplemental Table S4). The remaining seven mutations were not covered in our Strand-seq data set. To detect such missing mutations, data from alternative sequencing technologies can be integrated with Strand-seq data, or more Strand-seq libraries can be analyzed to increase the overall genome coverage. A previous study (Conrad et al. 2011) attempted to phase the same alleles but was unsuccessful due to the large distance between each de novo mutation. These results show that Strand-seq can phase both inherited and individual-specific variants, a major advantage for clinical research.
Genome-wide mapping of meiotic recombination breakpoints in a family trio
Having shown that we can build accurate whole-genome haplotypes without the need to sequence family members, we set out to study haplotype inheritance in a family trio. To explore this, we generated Strand-seq libraries for the father (NA12891) and mother (NA12892) of the HapMap child (NA12878). In total, we selected 233 libraries for the father and 267 for the mother, for analysis using our StrandPhase pipeline (Supplemental Table S1). From these data, we captured 82.5% and 72.7% SNVs present in the HapMap reference for the father and mother, respectively, to build whole-genome haplotypes for each parent. We confirmed that phased parental haplotypes agreed with our findings for the child by comparing the heterozygous variants in the child that were homozygous in at least one parent. This allowed us to unambiguously assign the parental origin of 99.7% of the child's heterozygous SNVs and thus predict which homolog was inherited from the maternal lineage versus the paternal lineage (Supplemental Fig. S4). In addition, we were able to assign a parental homolog to the 42 de novo germline mutations identified in the child, with 37 of paternal origin and five of maternal origin. This observation is consistent with previous studies reporting that most de novo mutations in offspring are paternally derived (Francioli et al. 2014; Kloosterman et al. 2015).
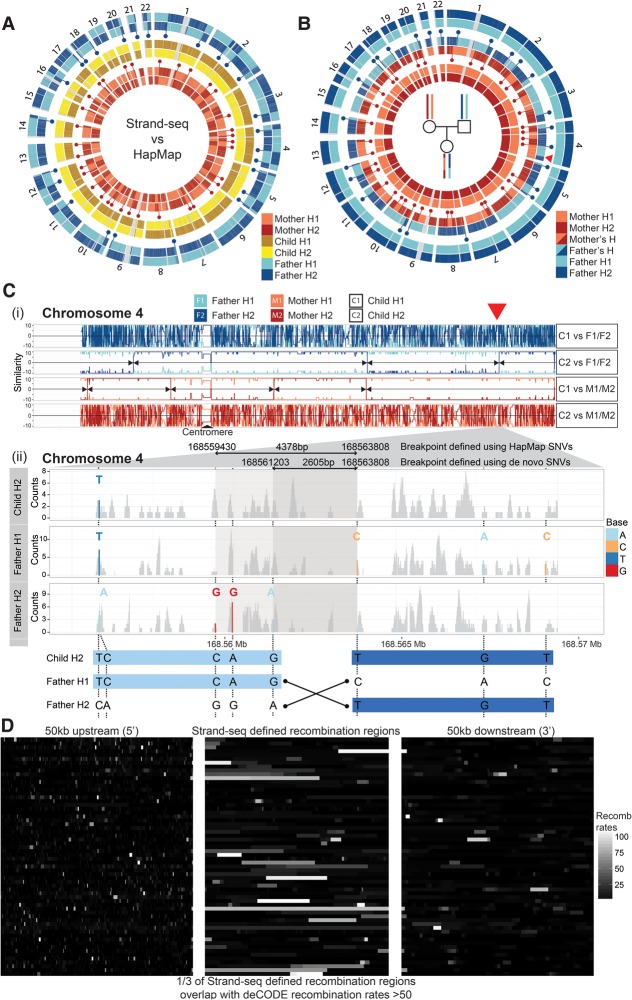
Following the phasing of whole-genome haplotypes for each individual in this family, we explored whether Strand-seq can be used to map individual meiotic recombination events. We compared our assembled haplotypes to those reported in the HapMap reference. Unlike the near complete concordance seen in the child, we observed multiple switches in the parental haplotypes (Fig. 3A, blue and red dots). This is because the methods used to build the HapMap reference relied on the haplotypes of the child to infer the haplotypes of the parents (Duitama et al. 2012). However, the child's genome is composed of recombined germline products, and therefore, the parental haplotypes in the HapMap reference contain a mixture of the parental haplotypes. We infer that the haplotype switches between our data and the HapMap reference data represent the locations of parental meiotic recombination events. Indeed, an independent comparison of our derived consensus haplotypes from the child to those of both parents showed discrete positions where the parental haplotypes inherited by the child had recombined (Fig. 3B, blue and red dots). For instance, the child's paternally derived homolog of Chromosome 1 exhibited two distinct haplotype switches, where the first part of p-arm was most similar to Father H2, the middle matched Father H1 and the last part of q-arm matched Father H2. These haplotype switches represent locations of meiotic recombination in the paternal gamete, resulting in a shuffling of the parental SNV alleles inherited by the child. We observed 38 switches (including two on Chromosome X) in the maternal homologs and 26 on the paternal homologs of the child, consistent with meiotic recombination rate estimates in previous studies (Broman et al. 1998; Lu et al. 2012; Hou et al. 2013; Kirkness et al. 2013).
Figure 3.
Genome-wide mapping of meiotic recombination breakpoints in a family trio. (A) Circular plots of Strand-seq haplotypes (H1 and H2) assembled for a family trio (mother, child, and father) with each pair of homologs compared with the corresponding HapMap reference haplotypes. Only heterozygous SNV positions are plotted along each chromosome. Strand-seq haplotypes for the child (middle circles; yellow and brown) match the HapMap reference along the whole length of the chromosome (see also Fig. 2). Haplotypes from the mother (inner circles; light red and dark red) and father (outer circles; light blue and dark blue) show multiple switches (blue and red dots) between the Strand-seq haplotypes and those listed in the HapMap reference. (B) Comparison of the Strand-seq child's haplotypes to the Strand-seq parental haplotypes, with only the heterozygous SNV positions plotted for each homolog. We compared each of the child's haplotypes independently to both the parental haplotypes. Haplotype switches (blue and red dots) represent sites of meiotic recombination and occur at almost every chromosome, both from the maternal and paternal germline. (Red arrowhead) The switch event illustrated in C. (C,i) Similarity plot for Chromosome 4 depicting pairwise comparison of each child homolog (C1 and C2) with both parental homologs (F1 and F2, or M1 and M2, as indicated) (see Methods, “Mapping meiotic recombination breakpoints”). Lines depict continuous stretches of high (+10) and low (−10) similarity. A high similarity score (e.g., 10) indicated all SNVs were matched between the pairs, whereas a low similarity score (e.g., −10) indicated the homologs were dissimilar. This illustrates that, for this chromosome, C1 was inherited from the father and C2 was inherited from the mother. (Black arrowheads) Locations where the degree of similarity switched between the inherited parental homologs (e.g., from F1 to F2, red arrowhead) and mark locations of meiotic recombination. (ii) Enlarged region of Chromosome 4 showing the homolog-specific BAM files generated for child's homolog (C2) inherited from the father, as well as the corresponding paternal homologs (F1 and F2). Read coverage (gray) was plotted for each BAM file, with heterozygous SNVs highlighted (see legend). By use of these SNVs, the meiotic recombination breakpoint was narrowed to a 2605-bp region (bottom panel). (D) A comparison of the overlap of the meiotic recombination breakpoints predicted in this study to the hotspots reported in the deCODE project. The middle panel illustrates the genomic regions where a meiotic recombination breakpoint was found in our analysis, with each row depicting a distinct recombination event and the shade denoting overlap with the predicted deCODE recombination rates corresponding to these locations (white indicates high levels of recombination; black, low levels of recombination). The left and right panels show 50 kb upstream of and 50 kb downstream from the defined meiotic recombination breakpoint, respectively, again with the shade representing the overlap with deCODE recombination rates. We saw high concordance between our predicted breakpoints and those listed in the deCODE database, where one in three overlapped with deCODE regions predicted to have high (more than 50 standardized units) (Kong et al. 2010) meiotic recombination rates.
To more precisely map these recombination events, we systematically tracked parental haplotype inheritance in the child using a pairwise similarity test (see Methods, “Mapping meiotic recombination breakpoints”) (Supplemental Fig. S5). This allowed us to precisely map recombination breakpoints at locations where similarity of a child haplotype switched, for example, from Father H1 to Father H2 (Fig. 3C, red arrowhead). In total, we mapped all 64 recombination events (Supplemental Fig. S6A) with a resolution as low as 408 bp and a median breakpoint resolution of 14,385 bp (Supplemental Fig. S6B; Supplemental Table S5). The location of recombination events in our study matched almost perfectly to those found in another single-cell phasing study (Fan et al. 2011) be it with a threefold better resolution (see Supplemental Methods; Supplemental Fig. S7A–C). Of interest, we found that one in three of our meiotic recombination locations overlapped with previously identified recombination hotspots (Fig. 3D; Kong et al. 2010).
In addition to meiotic recombination events, which involve reciprocal exchanges of large blocks of homologous chromosomes, we also observed a number of smaller phase switches. For instance, on homolog Child H1 of Chromosome 13, we did not observe any meiotic recombination of the father's homologs. Instead, we localized a short region where the haplotypes exhibited a segmental decrease in similarity to the corresponding paternal haplotype (Supplemental Fig. S8A). Here, we identified four consecutive SNVs that matched homolog Father H1 in a child homolog that otherwise matched homolog Father H2 (Supplemental Fig. S8B). Such a short switch in haplotypes could result from homozygous inversions, from two independent meiotic crossovers in close vicinity, or from a gene conversion event. We examined the template strand directionality of this region and did not find evidence supporting an inversion (Sanders et al. 2016), suggesting this represents either a meiotic or a conversion event. We located 18 additional regions in the child's homologs that exhibited a short haplotype switch involving at least three consecutive heterozygous SNV positions (Supplemental Table S6).
Taken together, our results demonstrate the power of Strand-seq to comprehensively map meiotic recombination breakpoints and predict potential gene conversion events within a family trio. In comparison to the mapping of recombination events using isolated metaphase chromosomes or single sperm cells (Fan et al. 2011; Wang et al. 2012), Strand-seq has the advantage that it avoids genome preamplification and thus reduces PCR sequencing artifacts.
Phasing of structural variants
In addition to SNVs, StrandPhase allows phasing of larger structural variants (SVs), such as deletions and insertions. To phase such variants, we used Strand-seq to split reads into homolog-specific subsets for SV genotyping (Supplemental Fig. S5). Supplemental Figure S9A shows an example of a heterozygous deletion in Father H2 that was inherited by Child H1. Moreover, we propose this technique is able to characterize individual homologs based on the copy number of segmental duplications (Supplemental Fig. S9B, arrowheads). Importantly, balanced rearrangements like inversions that are difficult to detect using current technologies can be reliably mapped and phased using Strand-seq (Sanders et al. 2016). To our knowledge, Strand-seq is the only technique able to simultaneously map and phase heterozygous inversions (Supplemental Fig. S9C–E). To explore the phasing efficacy of larger SVs using our technique, we set out to phase variants previously reported for this family trio. First, the phase of all experimentally validated deletions for NA12878, NA12891, and NA12892, reported by Fan et al. (2011), were confirmed using StrandPhase and matched expected Mendelian inheritance patterns (Supplemental Table S7; Supplemental Fig. S7D). To provide a more comprehensive set of SVs phased by our method, we then phased the heterozygous deletions identified from phase 3 of the 1000 Genomes Project (Sudmant et al. 2015). For this, we selected deletions >1 kb and phased them for NA12878 based on template strand–specific read count information (see Supplemental Methods). Out of 348 selected deletions, 305 matched the phase stated in the 1000 Genomes Project, while eight deletions did not. The remaining 35 deletions could not be reliably assessed because of low coverage in homolog-specific (binary alignment map) BAM files (Supplemental Table S8). In addition to deletions >1 kb, we explored smaller indels as well (see Supplemental Methods). Out of all 302,555 heterozygous short indels, only 68,233 (22.6%) were phased successfully. This low number most likely reflects the genotyping step (see Supplemental Methods), and methods for phasing indels using low coverage single-cell sequencing data need to be improved. However, the concordance of phased indels using Strand-seq in comparison to the 1000 Genomes Project was 97.7%, illustrating high accuracy. Taken together, these results illustrate that our phasing approach can reliably phase different classes of structural variants.
Mapping of regional changes in haplotypes at the single-cell level
Finally, we investigated the potential of Strand-seq to map mosaic recombination events at the single-cell level. For this, we performed a pairwise similarity analysis to compare the consensus haplotypes built for each family member (i.e., H1 and H2) to the single-cell haplotypes of each individual Strand-seq library (see Methods, “Evaluation of single-cell haplotypes”). In total, we identified 44 locations (eight in the mother, 19 in the father, and 17 in the child) where the consensus haplotypes switched in a homolog of a single cell (Supplemental Fig. S10A; Supplemental Table S9). For instance, in one maternal cell, Mother H1 switched to Mother H2 at the centromere of Chromosome 1 (Supplemental Fig. S10B, i). This resulted in one haplotype being converted to the other, thus marking a LOH region within the cell. Notably, this loss was not due to a deletion, since comparable read depths were found for both homologs (data not shown). The observed LOH patterns in these cells suggest that mitotic recombination events might be commonly occurring between homologous chromosomes (Moynahan and Jasin 2010) at a frequency of about 0.06 events per cell (Supplemental Fig. S10C). The possibility to explore LOH events and other genetic rearrangements at the single-cell level using Strand-seq is expected to have many applications in studies of DNA repair and cancer.
Discussion
The results presented here show that Strand-seq, together with StrandPhase, is a novel single-cell haplotyping method that retains linkage information along whole chromosomes. Because Strand-seq does not involve whole-genome amplification (WGA) prior to library preparation, the sequence bias and allelic drop-out introduced by PCR amplification are reduced, allowing extraction of highly accurate phase information from single cells. By compiling SNVs across multiple Strand-seq libraries, we were able to reconstruct whole-genome haplotypes without generational information. Each SNV is independently sampled in multiple single-cell libraries, allowing us to directly cross-validate variant calls made in a sample and to rapidly build highly accurate consensus haplotypes. Highlighting this, our results recapitulate the HapMap Project reference haplotypes without statistical inference, population, or pedigree data, demonstrating the strength of our approach for clinical studies. With the current Strand-seq protocol, around 100 single-cell libraries (with an average genome coverage of ∼2.5% per single-cell library) are sufficient to encompass 60%–70% of the genomic SNVs (Supplemental Fig. S11). In addition to SNVs, we have accurately phased larger SVs, such as deletions and smaller indels, illustrating the utility of Strand-seq for building haplotypes.
An important limitation of our current method is the requirement for BrdU incorporation in dividing cells as the input for Strand-seq, as well as the low genome coverage of single-cell libraries. However, we believe these limitations are mitigated by the possibility to rapidly phase entire chromosomes and track haplotype differences at the single-cell level. Furthermore, incomplete sets of phased alleles obtained by Strand-seq analysis can be augmented by other data, such as short- and long-read WGS technologies. Our analysis shows that the vast majority of nonphased polymorphisms (92.5%) are located near enough to phased variants to be phased using a combination of Strand-seq and regular WGS data (Supplemental Fig. S12). Indeed, we expect that future studies on haplotypes will benefit from the combination of Strand-seq and long-read technologies to assemble complete and chromosome-long haplotypes.
It is also important to note that Strand-seq phasing relies on a reference genome to map directional reads, and therefore, alleles that are not represented in the reference genome, including new duplications, may not be phased. Moreover, balanced rearrangements like inversions cause directional reads to map in opposite directions to the reference genome and are visible as switches in resulting haplotypes (Fig. 2). To overcome this, others have used hybrid phasing approaches based on de novo assembly to improve haplotype accuracy (Pendleton et al. 2015; Mostovoy et al. 2016). To explore how Strand-seq relates to hybrid phasing, we compared our phasing with the large 64-Mb scaffold assembled for Chromosome X by Mostovoy et al. (2016) (for details, see Supplemental Methods; Supplemental Note). The overall concordance between Strand-seq and hybrid phasing for this scaffold was 99.8% (Supplemental Fig. S13). This finding supports that phasing using Strand-seq, despite its dependency on a reference genome assembly, is highly accurate.
Taken together, we propose that Strand-seq is a unique tool to completely phase individual genomes, map meiotic recombination events in family trios, and explore haplotype structure in single cells. By avoiding preamplification, Strand-seq offers unmatched accuracy over other sequencing-based phasing techniques. Moreover, Strand-seq phasing can be combined with mapping of SVs, such as deletions and inversions, which is of major interest for clinical research. As single-cell sequencing becomes more and more accessible, we anticipate that Strand-seq haplotyping will have an important contribution to de novo assembly of haplotype-resolved personal genomes and thereby greatly facilitate studies of genomic variants in human health and disease.
Methods
Raw data production
Cells and cell culture
Epstein-Barr virus (EBV)–transformed B-lymphocyte cell lines GM12878, GM12891, and GM12892 were obtained from the Coriell Institute for Medical Research. The pedigree of all cell lines is UTAH/MORMON from USA, which is part of the International HapMap Project (The International HapMap Consortium 2007; The International HapMap 3 Consortium 2010). Cells were cultured in RPMI 1640 medium (Gibco) supplemented with 15% FBS (Sigma Aldrich) in 37°C at 5% CO2. For Strand-seq, BrdU (40 or 100 µM, final) was added to exponentially growing cells for 24 h.
Single-cell sorting
Cells were harvested, and nuclei were isolated by resuspension in nuclear isolation buffer (100 mM Tris-HCl at pH 7.4, 150 mM NaCl, 1 mM CaCl2, 0.5 mM MgCl2, 0.1% NP-40, 0.2% BSA). In each sample, cells cultured without BrdU were added as an internal control for Hoechst fluorescence. Nuclei were stained with Hoechst-33258 and propidium iodide (PI) by adding both to the isolation buffer at final concentration of 10 µg/mL and incubating on ice for 30 min. Nuclei of cells that underwent a cell division in the presence of BrdU were sorted based on low Hoechst fluorescence (quenched by BrdU in DNA) and PI (gated on G1 phase), using a MoFLo Atrios cell sorter (Beckman Coulter), and deposited into 96-well skirted PCR plates (4Titude) containing 5 µL/well freeze medium (pro-freeze CDM freeze medium [Lonza] containing 15% DMSO).
Library construction
Library preparation was performed using modified versions of a previously described protocol (Falconer et al. 2012). To scale for production on a Bravo automated liquid handling platform (Agilent), the enzymatic reactions were performed in smaller volumes while keeping buffer and enzyme concentrations at the same levels. DNA clean-up steps were performed using AMPure XP paramagnetic beads (Agencourt AMPure, Beckman Coulter). After adapter ligation and 17 PCR cycles, two consecutive AMPure bead clean-ups were performed using a 1.2× bead volume.
Next-generation sequencing
Libraries were pooled for sequencing and 250- to 300-bp size- range fragments were purified using 2% E-gel Agarose EX-gels (Invitrogen). DNA quality was assessed on a high-sensitivity dsDNA kit (Agilent) using the 2100 Bioanalyzer (Agilent), and DNA was quantified on the Qubit 2.0 fluorometer (Life Technologies). For sequencing, clusters were generated on the cBot, and paired-end 100-bp-long or single-end 50-bp-long reads were generated using the HiSeq 2500 sequencing platform (Illumina) following the manufacturer's instructions. For 50-bp- and 100-bp-long reads, 192 and 96 single-cell libraries were pooled together, respectively, and sequenced in one lane of the rapid run flow cell. Each plate included two 10-cell controls and two zero-cell controls.
Raw data processing
The single-cell raw sequencing data were de-multiplexed based on the library-specific barcodes and converted to FASTQ files using Illumina standard software (bcl2fastq, version 1.8.4). The resulting reads were mapped to the human reference genome NCBI36/hg18 using Bowtie 2 aligner (version 2.2.4) (Langmead and Salzberg 2012). After alignment, reads were sorted using SAMtools (version 0.1.19) (Li et al. 2009), and duplicate reads were marked using BamUtil (version 1.0.3). All Strand-seq libraries were prefiltered to avoid haplotype errors arising from low-quality data. For this, we excluded libraries with less than 50 reads/Mb, with >5% level of background reads, and with excessive genomic rearrangements, aneuploidy events, or uneven coverage (Supplemental Fig. S2). BAM files passing our quality criteria served as an input for our haplotyping pipeline.
Haplotype data analysis pipeline
Haplotype analyses were performed using our in-house Perl based scripts (Supplemental Fig. S1). We used aligned BAM files as input files, which were filtered for duplicate reads and low mapping quality reads (mapq < 10) using SAMtools (version 0.1.19) (Li et al. 2009). To build single-cell haplotypes, we first selected chromosomal regions that inherited W and C template strands (WC regions). For this, we scanned the genome of each single cell and counted the number of Crick (forward; “+”) and Watson (reverse; “−”) reads in equally sized regions (default, 1 Mb). Fisher exact tests were used to calculate the probability that a region contained approximately equal numbers of Crick and Watson reads and agreed with the expected 50:50 ratio of a WC region (Sanders et al. 2016). Subsequently only WC regions >5 Mb were selected for further analysis. A list of the selected WC regions analyzed for each individual and single-cell library is available at the StrandPhase repository (available at https://github.com/daewoooo/StrandPhase). Next, we identified SNVs in WC regions by querying variant positions listed in the HapMap reference database (a nonredundant list of SNVs from phase 2 release 22 and from phase 3 release 2) using the SAMtools “mpileup” function (Li et al. 2009). We recorded the specific nucleotide at each variable position separately for the Crick and the Watson template strands, creating low-density haplotypes for every single cell. These partial single-cell haplotypes were then used as the input for the Strand-seq specific phasing algorithm.
To build whole-genome haplotypes, the phasing algorithm StrandPhase analyzed the single-cell haplotypes for a single chromosome at a time. All single-cell haplotypes for every informative chromosome are considered as a separate entity. The first iteration pulled out the pair of single-cell haplotypes that contained the highest density of overlapping heterozygous positions, and set these as the anchor haplotypes. This essentially initialized the two consensus haplotypes, arbitrarily designated “H1” and “H2.” In the next iteration, the single-cell haplotypes containing the highest number of SNV positions overlapping with the anchor haplotypes were selected and compared separately to both H1 and H2. The percentage of mismatches was calculated for each comparison as a missH1 and missH2. Subsequently, the difference between the level of mismatches was calculated as ((missH1 − missH2)/(missH1 + missH2)/2) × 100, and the haplotype showing the highest concordance was added to the corresponding consensus haplotype. Single-cell haplotypes with the degree of difference less than 25 were excluded from the analysis (1.3%–3.7% of single-cell haplotypes were excluded). By iteratively adding additional single-cell haplotypes to H1 and H2, the density of SNVs in each consensus haplotype increased with every additional cell analyzed. Single-cell data that could not be reliably assigned to one of the consensus haplotypes were excluded and reported in a separate file.
PacBio and Strand-seq cross-validation
We incorporated PacBio data using a three-stage approach. First we mapped PacBio reads to the human transcriptome (NCBI36/hg18, Ensembl release 54) using bwasw module implemented in BWA aligner (version 0.7.12.) (Li and Durbin 2010). Second, for every PacBio read, we recorded the specific variant at each position listed in the HapMap reference. Lastly, we added strand information to each allele based on the mapping directionality. To directly compare our haplotypes with the PacBio data set, we selected all PacBio reads that overlapped with at least two heterozygous positions in our Strand-seq haplotypes. We filtered out reads containing SNVs with a base quality less than 20. Next we calculated the percentage of phased PacBio reads that matched the phase we found for our haplotypes to test the level of concordance between these data sets. To assess nonrandom concordance, we randomly shuffled the SNVs between the H1 and H2 Strand-seq haplotypes and counted the number of concordant and discordant reads again. Reshuffling eliminated the concordance between Strand-seq and PacBio data.
PacBio data used for this analysis were downloaded from the SRA database. Accession numbers are SRR1163655 (NA12878), SRR1163657 (NA12891), and SRR1163658 (NA12892) (Tilgner et al. 2014).
Mapping meiotic recombination breakpoints
To map meiotic recombination events with higher resolution, we created homolog-specific BAM files for each family member by merging the phased reads across all single cells into two high-density read files per individual (one representing H1; the other, H2) (Supplemental Fig. S5). During this step, duplicate reads were filtered, and sequencing reads from all single-cell haplotypes were merged together for each consensus haplotype. In order to compare the child with both parents, we temporarily merged the child's homologs with the father's and mother's homologs, respectively, using the SAMtools (Li et al. 2009) “merge” function and performed SNV calling using GATK UnifiedGenotyper (version 3.2-2) (McKenna et al. 2010) with default settings. This identified the heterozygous positions that distinguished the child from each parent, which were used to assign the identity of each of the child's homologs. In order to map meiotic recombination breakpoints at high resolution, we performed a pairwise comparison of each child's homolog to both the maternal and paternal homologs. For this comparison, only parental heterozygous positions covered in the child were considered. Every comparison was encoded as a vector of zeros and ones based on the parental homolog to which child's homolog corresponded (zero, parental homolog 1; one, parental homolog 2). Then a circular binary segmentation algorithm (R package fastseg, minSeg set to 150) (Klambauer et al. 2012) was applied on the binary vectors using a custom R script. Segments <5 Mb were filtered out. Meiotic breakpoints were localized as the end position of one segment and start position of the following segment.
To visualize meiotic breakpoints, we calculated the level of similarity between paired homologs by scanning the chromosome using a 10 k-mer (10 consecutive heterozygous SNVs) long sliding window (moving by one heterozygous position at a time). This allowed us to compare 10 heterozygous SNV positions between the homologs and calculate the degree of similarity in the window. Similarity was calculated as the reverse of Hamming distances with a match score +1 and mismatch penalty −2. Meiotic recombination breakpoints were located as positions where similarity of a single child's homolog abruptly drops and instead matched the other parental homolog. Final mapping and validation of meiotic recombination breakpoints was done by visual confirmation of the haplotype switch.
To look for shorter switches in haplotypes, we used homolog-specific BAM files for each family member, as discussed above. We performed a pairwise comparison of each child's homolog to both maternal and paternal homologs considering only parental heterozygous positions covered in the child. Initially we split each homolog into a smaller region at positions of mapped meiotic recombination events. Then using a 3 k-mer (three consecutive heterozygous SNVs) sliding window (moving by one heterozygous position at a time), we calculated the level of similarity in every window, as mentioned above. Switch event breakpoints were located as positions where similarity of a single child's homolog drops and instead matched the other parental homolog. Lastly, we filtered out regions that overlapped with regional switches in read directionality and with low SNVs of quality (less than 100). A putative gene conversion event was defined as a short region where a single child's homolog corresponding to one parental homolog matched the other homolog instead.
To compare the location of our recombination breakpoint predictions to those listed in the deCODE project, the deCODE recombination hotspot file was downloaded from the UCSC Genome Table Browser database using table browser, HapMap CEU hapmap release 24. We selected deCODE recombination rates overlapping with regions of our meiotic recombination breakpoints. For each meiotic recombination region defined in our data, we looked for overlaps with defined regions of meiotic recombination rates. We repeated this process for regions 50 kb downstream and upstream of Strand-seq–defined meiotic recombination breakpoints.
Evaluation of single-cell haplotypes
To test for haplotype switches at the single-cell level, we performed a pairwise comparison of each single-cell haplotype to both consensus haplotypes for every chromosome. As above, only heterozygous positions between consensus haplotypes and the single cell were considered. For each heterozygous position, the consensus base was called as the highest abundant nucleotide at that position across all cells. We scanned each chromosome by a 3 k-mer (three consecutive heterozygous SNVs) sliding window (moving by one heterozygous position at a time) to systematically compare three heterozygous positions and assess the level of similarity between the single-cell haplotype and the consensus haplotype. For each comparison, the level of similarity was calculated as the reverse of Hamming distances with match score +1 and mismatch penalty −2. We selected putative LOH regions where at least three consecutive heterozygous positions switched in one haplotype of a single cell but not in the other haplotype. We filtered regions <1000 bp to ensure that not all heterozygous positions are part of a single erroneous read but were covered by independent reads. Data visualization was performed using R package ggbio (Yin et al. 2012).
Data access
The StrandPhase software, custom data processing scripts (Perl and R code), and data used in this study are publicly available through GitHub repository (https://github.com/daewoooo/StrandPhase) and can be found in the Supplemental Material. Strand-seq libraries selected for this study have been submitted to the European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena) under accession number PRJEB14185.
Supplementary Material
Acknowledgments
We thank Nancy Halsema, Inge Kazemier, and Karina Hoekstra-Wakker for technical help. Financial support for these studies was provided by a European Research Council Advanced grant (ROOTS-Grant Agreement 294740) to P.M.L.
Author contributions: D.P. performed data analysis and implemented the phasing algorithm. D.P. and A.D.S. wrote the manuscript; N.v.W. cultured cells and prepared Strand-seq libraries. A.D.S., E.F., M.H., and V.G. helped with data analysis and development of bioinformatics approaches. D.C.J.S. and M.R.B. helped with sequencing of Strand-seq libraries. M.R.B., V.G., and P.M.L. designed experiments and helped with writing of the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.209841.116.
References
- Amini S, Pushkarev D, Christiansen L, Kostem E, Royce T, Turk C, Pignatelli N, Adey A, Kitzman JO, Vijayan K, et al. 2014. Haplotype-resolved whole-genome sequencing by contiguity-preserving transposition and combinatorial indexing. Nat Genet 46: 1343–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal V, Tewhey R, Topol EJ, Schork NJ. 2011. The next phase in human genetics. Nat Biotechnol 29: 38–39. [DOI] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. 1998. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63: 861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJB, De Pedro MA, Kysela T, Van Der Henst C, Kim J, De Bolle X, Fuqua C, Brun YV. 2012. Completely phased genome sequencing through chromosome sorting. Proc Natl Acad Sci 109: 3190–3190. [Google Scholar]
- Browning SR, Browning BL. 2011. Haplotype phasing: existing methods and new developments. Nat Rev Genet 12: 703–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Keebler JE, DePristo MA, Lindsay SJ, Zhang Y, Casals F, Idaghdour Y, Hartl CL, Torroja C, Garimella KV, et al. 2011. Variation in genome-wide mutation rates within and between human families. Nat Genet 43: 712–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duitama J, McEwen GK, Huebsch T, Palczewski S, Schulz S, Verstrepen K, Suk EK, Hoehe MR. 2012. Fosmid-based whole genome haplotyping of a HapMap trio child: evaluation of Single Individual Haplotyping techniques. Nucleic Acids Res 40: 2041–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer E, Hills M, Naumann U, Poon SSS, Chavez EA, Sanders AD, Zhao Y, Hirst M, Lansdorp PM. 2012. DNA template strand sequencing of single-cells maps genomic rearrangements at high resolution. Nat Methods 9: 1107–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HC, Wang J, Potanina A, Quake SR. 2011. Whole-genome molecular haplotyping of single cells. Nat Biotechnol 29: 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francioli LC, Menelaou A, Pulit SL, van Dijk F, Palamara PF, Elbers CC, Neerincx PBT, Ye K, Guryev V, Kloosterman WP, et al. 2014. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet 46: 818–825. [DOI] [PubMed] [Google Scholar]
- Glusman G, Cox HC, Roach JC. 2014. Whole-genome haplotyping approaches and genomic medicine. Genome Med 6: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hills M, O'Neill K, Falconer E, Brinkman R, Lansdorp PM. 2013. BAIT: organizing genomes and mapping rearrangements in single cells. Genome Med 5: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Fan W, Yan L, Li R, Lian Y, Huang J, Li J, Xu L, Tang F, Xie XS, et al. 2013. Genome analyses of single human oocytes. Cell 155: 1492–1506. [DOI] [PubMed] [Google Scholar]
- Huang YC, Lee CM, Chen M, Chung MY, Chang YH, Huang WJS, Ho DMT, Pan CC, Wu TT, Yang S. 2007. Haplotypes, loss of heterozygosity, and expression levels of glycine N-methyltransferase in prostate cancer. Clin Cancer Res 13: 1412–1420. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap 3 Consortium. 2010. Integrating common and rare genetic variation in diverse human populations. Nature 467: 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkness EF, Grindberg RV, Yee-Greenbaum J, Marshall CR, Scherer SW, Lasken RS, Venter JC. 2013. Sequencing of isolated sperm cells for direct haplotyping of a human genome. Genome Res 23: 826–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzman JO, Mackenzie AP, Adey A, Hiatt JB, Patwardhan RP, Sudmant PH, Ng SB, Alkan C, Qiu R, Eichler EE, et al. 2011. Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nat Biotechnol 29: 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klambauer G, Schwarzbauer K, Mayr A, Clevert DA, Mitterecker A, Bodenhofer U, Hochreiter S. 2012. cn.MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Res 40: e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Francioli LC, Hormozdiari F, Marschall T, Hehir-kwa JY, Abdellaoui A, Lameijer EW, Moed MH, Koval V, Renkens I, et al. 2015. Characteristics of de novo structural changes in the human genome. Genome Res 25: 792–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Gudbjartsson DF, Masson G, Sigurdsson A, Jonasdottir A, Walters GB, Jonasdottir A, Gylfason A, Kristinsson KT, et al. 2010. Fine-scale recombination rate differences between sexes, populations and individuals. Nature 467: 1099–1103. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Zong C, Fan W, Yang M, Li J, Chapman AR, Zhu P, Hu X, Xu L, Yan L, et al. 2012. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science 338: 1627–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Xiao Y, Huang H, Wang Q, Rao W, Feng Y, Zhang K, Song Q. 2010. Direct determination of molecular haplotypes by chromosome microdissection. Nat Methods 7: 299–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostovoy Y, Levy-Sakin M, Lam J, Lam ET, Hastie AR, Marks P, Lee J, Chu C, Lin C, Džakula Ž, et al. 2016. A hybrid approach for de novo human genome sequence assembly and phasing. Nat Methods 13: 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Jasin M. 2010. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol 11: 196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton M, Sebra R, Pang AW, Ummat A, Franzen O, Rausch T, Stütz AM, Stedman W, Anantharaman T, Hastie A, et al. 2015. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat Methods 12: 780–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders AD, Hills M, Porubský D, Guryev V, Falconer E, Lansdorp PM. 2016. Characterizing polymorphic inversions in human genomes by single-cell sequencing. Genome Res (this issue) 26: 1575–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj S, Dixon JR, Bansal V, Ren B. 2013. Whole-genome haplotype reconstruction using proximity-ligation and shotgun sequencing. Nat Biotechnol 31: 1111–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Hsi-Yang Fritz M, et al. 2015. An integrated map of structural variation in 2,504 human genomes. Nature 526: 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewhey R, Bansal V, Torkamani A, Topol EJ, Schork NJ. 2011. The importance of phase information for human genomics. Nat Rev Genet 12: 215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Grubert F, Sharon D, Snyder MP. 2014. Defining a personal, allele-specific, and single-molecule long-read transcriptome. Proc Natl Acad Sci 111: 9869–9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Fan HC, Behr B, Quake SR. 2012. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell 150: 402–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin T, Cook D, Lawrence M. 2012. ggbio: an R package for extending the grammar of graphics for genomic data. Genome Biol 13: R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GX, Lau BT, Schnall-Levin M, Jarosz M, Bell JM, Hindson CM, Kyriazopoulou-Panagiotopoulou S, Masquelier DA, Merrill L, Terry JM, et al. 2016. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat Biotechnol 34: 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.