Abstract
STUDY QUESTION
In comparison to in vivo development, how do different conditions of in vitro culture (‘one step’ versus ‘sequential medium’) impact DNA methylation and hydroxymethylation in preimplantation embryos?
SUMMARY ANSWER
Using rabbit as a model, we show that DNA methylation and hydroxymethylation are both affected by in vitro culture of preimplantation embryos and the effect observed depends on the culture medium used.
WHAT IS KNOWN ALREADY
Correct regulation of DNA methylation is essential for embryonic development and DNA hydroxymethylation appears more and more to be a key player. Modifications of the environment of early embryos are known to have long term effects on adult phenotypes and health; these probably rely on epigenetic alterations.
STUDY DESIGN SIZE, DURATION
The study design we used is both cross sectional (control versus treatment) and longitudinal (time-course). Each individual in vivo experiment used embryos flushed from the donor at the 2-, 4-, 8-, 16- or morula stage. Each stage was analyzed in at least two independent experiments. Each individual in vitro experiment used embryos flushed from donors at the 1-cell stage (19 h post-coïtum) which were then cultured in parallel in the two tested media until the 2-, 4-, 8- 16-cell or morula stages. Each stage was analyzed in at least three independent experiments. In both the in vivo and in vitro experiments, 4-cell stage embryos were always included as an internal reference.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Immunofluorescence with antibodies specific for 5-methylcytosine (5meC) and 5-hydroxymethylcytosine (5hmeC) was used to quantify DNA methylation and hydroxymethylation levels in preimplantation embryos. We assessed the expression of DNA methyltransferases (DNMT), of ten eleven translocation (TET) dioxigenases and of two endogenous retroviral sequences (ERV) using RT-qPCR, since the expression of endogenous retroviral sequences is known to be regulated by DNA methylation. Three repeats were first done for all stages; then three additional repetitions were performed for those stages showing differences or tendencies toward differences between the different conditions in the first round of quantification.
MAIN RESULTS AND THE ROLE OF CHANCE
The kinetics of DNA methylation and hydroxymethylation were modified in in vitro cultured embryos, and the observed differences depended on the type of medium used. These differences were statistically significant. In addition, the expression of TET1 and TET2 was significantly reduced in post-embryonic genome activation (EGA) embryos after in vitro culture in both tested conditions. Finally, the expression of two retroviral sequences was analyzed and found to be significantly affected by in vitro culture.
LIMITATIONS REASONS FOR CAUTION
Our study remains mostly descriptive as no direct link can be established between the epigenetic changes observed and the expression changes in both effectors and targets of the studied epigenetic modifications. The results we obtained suggest that gene expression could be affected on a large scale, but this remains to be confirmed.
WIDER IMPLICATIONS OF THE FINDINGS
Our results are in agreement with the literature, showing that DNA methylation is sensitive to in vitro culture. As we observed an effect of both tested culture conditions on the tested epigenetic marks and on gene expression, we cannot conclude which medium is potentially closest to in vivo conditions. However, as the observed effects are different, additional studies may provide more information and potential recommendations for the use of culture media in assisted reproductive technology.
STUDY FUNDING/COMPETING INTEREST(S)
This work was supported by an ‘AMP diagnostic prénatal et diagnostic génétique’ 2012 grant from the French Agence de la Biomédecine. This study was performed within the framework of ANR LABEX ‘REVIVE’ (ANR-10-LABX-73). Authors are members of RGB-Net (TD 1101) and Epiconcept (FA 1201) COST actions. The authors declare that there is no competing interest.
Keywords: preimplantation embryo, DNA methylation, DNA hydroxymethylation, embryo culture media, DNA methyltransferases (DNMT), ten eleven translocation (TET), endogenous retrovirus
Introduction
As the number of children born from assisted reproductive technologies has been increasing over the years, the question of safety of ART procedures, including in vitro culture, has become a prevalent issue for public health. Indeed, the recommendations to reduce multiple pregnancies after IVF and ICSI suggest an increase in the duration of in vitro culture before selecting the embryo to be transferred.
Two main in vitro culture strategies are currently used. ‘Sequential’ media are designed to mimic the environmental changes that occur in vivo when the embryo progresses in the oviduct to the uterus. ‘One-step’ media, on the other hand, are designed to contain all that the embryo needs for all stages of preimplantation development, arguing that brutal environmental changes are detrimental to the embryo, which needs to adapt (Watkins et al., 2008a). An additional advantage is that they are compatible with uninterrupted culture associated with the use of time-lapse technology. However, it remains impossible to this day to say which approach is superior (Mantikou et al., 2013; Swain et al., 2016), especially since studies in the human are made difficult by numerous confounding effects. Studies on animal models have shown that the environment surrounding oogenesis and the first steps of embryonic development influences the phenotype and can have effects during the animal's whole life (Fleming et al., 2015), thus extending the concept of Developmental Origin of Health and Diseases (DOHaD) to the periconceptional period. These long term effects likely result from environment-induced epigenetic modifications in a sensitive period of intense epigenetic remodeling (Watkins et al., 2008a, b; El Hajj and Haaf, 2013), leading to subsequent changes in gene expression (Burdge et al., 2007).
DNA methylation on cytosines (5-methylcytosine: 5meC) is an essential epigenetic modification that seems to be particularly affected by in vitro culture (Wright et al., 2011) and to have an impact on the health of children born after ART, in particular through misregulation of imprinted genes (Fauque et al., 2007; Choux et al., 2015), leading to rare syndromes such as Beckwith–Wiedemann and Angelman (Manipalviratn et al., 2009; Owen and Segars, 2009; Lazaraviciute et al., 2015). In addition to its importance in imprinting, DNA methylation is involved in the regulation of many developmental processes such as parental imprinting, X-inactivation and control of endogenous retrotransposons (Watkins et al., 2008b; Corry et al., 2009). Methylation at gene promoters is generally associated with transcriptional repression (Klose and Bird, 2006). This mark is submitted to intense remodeling events during embryonic development. In mice, a major demethylation phase occurs during the 1-cell stage and further demethylation is ongoing until the blastocyst stage (Mayer et al., 2000; Li and O'Neill, 2012; Salvaing et al., 2012; Smith et al., 2012; Wang et al., 2014; Guo et al., 2014a). In rabbits, although DNA methylation is submitted to many changes during the 1-cell stage in both parental genomes (Reis e Silva et al., 2011), the real demethylation phase starts later, after the 4-cell stage (Reis e Silva et al., 2012). Demethylation of the human genome appears less clear; while an initial report had described a similar kinetics to that of rabbit embryo (Fulka et al., 2004), a more recent report has shown that demethylation of human embryos is complete by the end of the 2-cell stage (Guo et al., 2014b). Differences in the timing of DNA demethylation between various species could be related to the timing of embryonic genome activation (EGA), which occurs at the 2-cell stage in mouse embryos but at the 4/8-cell stage in human embryos and around the 8-cell stage in rabbit embryos ((Telford et al., 1990) and our own unpublished data).
Active DNA demethylation can be ensured by several indirect processes (review (Hill et al., 2014)) but it is now clearly established that oxidation of 5meC to 5-hydroxymethylcytosine (5hmeC) by enzymes of the ten eleven translocation (TET) family is the first step in the main active demethylation process (Tahiliani et al., 2009; Ito et al., 2010), particularly in early embryos (Iqbal et al., 2011; Wossidlo et al., 2011). However, the dynamics of DNA hydroxymethylation during early development (Li and O'Neill, 2012; Salvaing et al., 2012) suggests that 5hmeC has functions of its own. In particular, it seems important for pluripotence of mouse embryonic stem cells (Tahiliani et al., 2009; Ito et al., 2010; Koh et al., 2011; Dawlaty et al., 2014) and is preferentially associated with transcriptionally active genes (Ficz et al., 2011; Pastor et al., 2011; Wu et al., 2011; Xu et al., 2011; Kubiura et al., 2012; Mellén et al., 2012). Studies reporting a link between 5hmeC and many diseases, in particular cancers (review (Vasanthakumar and Godley, 2015)) and neurologic disorders such as autism (Papale et al., 2015), have also greatly increased over the last few years. Due to its link to DNA demethylation and to these functions, 5hmeC appears as a potential crucial mark during early development.
In the present study, we chose to use both DNA hydroxymethylation and DNA methylation to assess the two types of culture conditions (‘sequential’ versus ‘one-step’), using respectively the G-1™ PLUS/G-2™ PLUS (G1+/G2+) media (Vitrolife) and the global® (Global) medium (LifeGlobal®), which are most commonly used in ART in France, by comparing embryos cultured in these media to in vivo developed embryos. Due to ethical as well as technical considerations, this type of study cannot be conducted using human embryos. We chose to use rabbit embryos as a model as its characteristics are much closer to human embryo than the more traditional mouse model. As already mentioned above, the timing of EGA in rabbit embryos is closer to that of human embryos than mouse, which is apart from most mammalian species. This can lead to very different epigenetic reprogramming timing and mechanisms between mouse and other mammalian species, as we have shown in the case of X-chromosome inactivation (Okamoto et al., 2011). In addition, the metabolism of rabbit embryos is very close to human embryos (Manes and Daniel, 1969; Kane, 1987; Chi et al., 1988; Capmany and Bolton, 1999; Biggers et al., 2000), which is essential for two reasons: first, to ensure that consumption of the medium nutrients are comparable in both species; and second, because there is a direct link between metabolism and DNA methylation, through the production of the methyl donor S-adenosyl methionine (SAM) (Naviaux, 2008).
We analyzed the evolution of DNA hydroxymethylation and methylation using quantitative immunofluorescence in embryos produced in vivo or cultured in either G1+/G2+ or Global media. We then assessed whether the observed differences could be related to changes in the expression of DNA methylation related enzymes (DNA methyltransferases (DNMT) and TET dioxigenases). Finally, because the expression of endogenous retroviral sequences is known to be repressed by DNA methylation (Rowe and Trono, 2011), we examined the impact of in vitro culture on the expression of two endogenous retroviral sequences.
Material and methods
Ethics
Animal care and handling were carried out according to European regulations on animal welfare. ND, VD, NB and JS have the authorization to work with laboratory animals from the departmental veterinary regulatory services (No 78–73, 78–101, 78–95 and 78–137, respectively). This work has been approved by the local ethics committee (Comethea Jouy-en-Josas/AgroParisTech: agreements 12/107 and 14/35 for in vitro analyses and embryo transfer experiments respectively).
Animals
New Zealand White female rabbits were superovulated as follows: five subcutaneous administrations of pFSH (Stimufol®, Merial, France) were performed during the 3 days prior to mating (two 5 µg doses on the first day with a 12-h interval, two 10 μg doses on the second day with a 12-h interval, and one 5 μg dose on the third day), followed 12 hours later by an intravenous administration of 30 IU HCG (Chorulon, MSD Animal Health, USA) at the time of mating (natural mating with New Zealand White male rabbits).
Recovery of rabbit embryos
In vivo developed embryos were collected from oviducts perfused with Dulbecco's Phosphate Buffer Saline (PBS) at the following stages: 1-cell (19 h post-coitum (hpc)), 2-cell (24 hpc), 4-cell (32 hpc), 8-cell (49 hpc), 16-cell (55 hpc), morula (67 hpc) and blastocyst (80, 88 and 96 hpc). Embryos were always collected from a minimum of two female rabbits.
Embryos for the in vitro culture experiments were all collected at the 1-cell stage (19 hpc). They were rinsed and put in culture in 40 µl drops of either preheated and pre-equilibrated G-1™ PLUS (G1+) medium (Vitrolife, Sweden) or global® (Global) medium supplemented with 10% Human Serum Albumin (HSA, LifeGlobal®, Canada) under mineral oil (Sigma-Aldrich, USA) in an incubator at 38°C under 5% CO2 in air. For each experiment, we took care of the following parameters: ensuring that embryos cultured in both media were treated in the same way and minimizing the time the embryos spent out of the incubator. With this aim, we prepared one dish containing three drops of each medium for each stage of fixation. In addition, embryos were always collected from a minimum of three female rabbits and sorted into 6–10 pools depending on the number collected so that each pool contained an equivalent amount of embryos from each rabbit. Embryos cultured in vitro to the 2-, 4- and 8-cell stages were processed at, respectively, 27, 33 and 49 hpc; at 49 hpc, embryos that were to be kept in culture until later stages were transferred to new drops of media, G-2™ PLUS (G2+) for those that were in G1+, and Global+10% HSA, for those that were already in Global medium. Then 16/32-cell stage embryos, morulae and blastocysts were processed respectively at 57, 72 and 98 hpc. All media were prepared the evening before use and put in the incubator for gas equilibration. Embryos used for molecular biology were snap-frozen in dry ice.
Embryo transfer
Pseudopregnancy was induced by intramuscular injection of 0.8 µg of buserelin (0.2 ml of Receptal, Intervet, Angers France) with a 24–28 h delay (asynchronisation) in comparison to the protocol used for embryos donor rabbits. The embryos were transferred surgically into the oviducts of the pseudopregnant rabbits by laparotomy under general gas (isoflurane) anesthesia as performed routinely in the laboratory. A few minutes before surgery, recipients received one intramuscular injection of ketamine (Imalgen 1000), one intravenous injection of meloxicam (Metacam, Boehringer Ingelheim, Alcyon) and one subcutaneous injection of enrofloxacine (Alcyon). At 24.5 days after transfer (D28 embryonic age), females were euthanized, and implantations and fetuses were counted.
Immunostaining
In vivo collected embryos were incubated in 5 mg/ml Pronase (Sigma-Aldrich, USA) at room temperature (RT) to remove the zona pellucida and mucin coat. All embryos were fixed and treated as previously described (Reis e Silva et al., 2012). For technical reasons, we could not process in vivo developed embryos in the same experiments as in vitro cultured ones. However, in all experiments we processed, at the same time, a minimum of three different stages of either in vivo developed or embryos cultured in both media. The 4-cell stage was always included as an internal reference. Briefly, embryos were fixed in 4% paraformaldehyde (PAF, Sigma-Aldrich, USA) in PBS overnight at 4°C and permeabilized with 0.5% Triton X-100 (Sigma-Aldrich, USA) in PBS for 1 h on a heating plate at 25°C. DNA denaturation was performed using 2 M HCl for 1 h on a heating plate at 25°C. After a 1 h incubation in PBS containing 2% BSA at room temperature, incubation in the primary antibody, either anti-5methylcytosine (5meC 1:500; BI-MECY, Eurogentec, Belgium) or anti-5hydroxymethylcytosine, (5hmeC 1:500; #39769 Active Motif, Belgium) was performed overnight at 4°C. Embryos were incubated the next day in the secondary antibody, either FITC-conjugated anti-mouse (for 5meC) or AlexaFluor488-conjugated anti-rabbit (for 5hmeC) (1:200 dilution; respectively #715-095-151 and #715-545-152, Jackson ImmunoResearch, USA), for 1 h at room temperature. DNA was counterstained using 2 µM Ethidium Homodimer-2 (EthD2, Invitrogen, USA) for 30 min at 37°C. The embryos were mounted in Citifluor (AF1, BioValley, France).
Signal quantifications
The embryos were observed using a Carl Zeiss (Germany) AxioObserver Zl fluorescence microscope equipped with the ApoTome slider (MIMA2 Platform, INRA). The samples were observed for the area containing the nuclei with a 63x Plan-Neofluar oil objective (NA 1.3) and digital optical sections were collected using a Z-series acquisition feature every 0.5 µm. Quantitative analyses of DNA methylation levels and total DNA contents were estimated by quantifying fluorescence signals using Image-J software (National Institute of Health, Bethesda, Maryland, USA) as follows: (i) the z-range was determined for each nucleus and a summing z-projection was realized; (ii) the area of each nucleus was outlined manually and the mean fluorescence intensity was measured for both 5hmeC/5meC and EthD-2 images; (iii) the mean background was determined for each nucleus in its immediate vicinity and subtracted from the measured signal; (iv) the mean fluorescence signal obtained was multiplied by the corresponding nucleus area to obtain the total fluorescence signal; (v) the total fluorescence intensities were normalized by the acquisition time of the corresponding signal; and (vi) the total fluorescence measured for 5hmeC/5meC in each nucleus was corrected by the corresponding measure of EthD-2. Each stage was present in at least two (in vivo developed embryos) or three (in vitro produced embryos) independent experiments, except for the 4-cell stage which was always included. Data from each replicate were normalized by the median of the 4-cell early stage of the same replicate. For embryos at the 4- and 8-cell stages, we calculated the median of all DNA content measures on embryos from one experimental set (all stages pooled together): those with a DNA content inferior to the median were considered to be in the early phases of the cell cycle (G1 and early S phase) while those with a higher DNA content were considered to be in the later phases of the cell cycle (late S and G2).
RNA isolation, reverse transcription and qPCR
Total RNA was isolated from batches of embryos (n = 20 for all stages except blastocyst stage n = 10) using PicoPur Arcturus (Excilone, France) with a DNase I (Qiagen, Germany) treatment as recommended by the supplier. To control the quality of ‘Isolation-RT’ procedure a posteriori, we proceeded as described in Peynot et al. (2015). The reverse transcription reaction was performed on the same number of ‘equivalent embryos’ at each stage by the Superscript III enzyme and primed by random hexamers (300 ng) (Lifetechnologies, France) following the supplier's protocol (25°C for 5 min, 50°C for 60 min and 70°C for 15 min).
qPCR conditions
A pilot experiment was conducted to determine the optimal conditions of use of the primers and to define the number of ‘equivalent embryos’ required for PCR for each embryonic stage and gene (Table I). The primers used for the endogenous reference genes (Hprt1 and Ywhaz) have been published by Mamo et al. (2008) and the Luciferase primers have been previously published Bui et al. (2009). Primers for the genes of interest were designed using Primers Express software (Applied Biosystems, USA). The ERVA and ERVB primers were defined from expressed sequence tags (ESTs) identified from a cDNA library corresponding to genes expressed at the morula stage. Details for primers are reported in Table II. qPCR reactions were performed on a Step One Plus machine (Applied Biosystems, USA) in 25 µl volume containing 12.5 µl of Sybr Green PCR Master Mix (Applied Biosystems, USA), primers and cDNAs. The thermal cycle profile was: 10 min at 95°C then 40 cycles with 15 s at 95°C and 60 s at 60°C except for Ywhaz and Hprt1 (68°C). Dissociation curves were obtained after each PCR run to ensure that a single PCR product had been amplified. The specificity of amplicons was verified by sequence alignment on the rabbit genome.
Table I.
Number of ‘equivalent embryos’ used for each qPCR reaction.
| Gene | Embryonic stages | |
|---|---|---|
| 2PN to morula | Blastocyst | |
| Dnmt1 | 0.2 | 0.2 |
| Dnmt3A | 0.2 | 0.05 |
| Dnmt3B | 0.5 | 0.05 |
| Tet1 | 0.5 | 0.05 |
| Tet2 | 0.5 | 0.05 |
| Tet3 | 0.5 | 0.5 |
| ERVA | 0.2 | 0.2 |
| ERVB | 0.2 | 0.2 |
| Luc, Hprt1 and Ywhaz | 0.2 | 0.2 |
Table II.
Primers and PCR features.
| Gene | Accession number | Primer sequence (nM) | Size (bp) | Annealing temperature (°C) |
|---|---|---|---|---|
| Hprt1 (Mamo et al., 2008) | ENSOCUG00000003186 | F: ACGTCGAGGACTTGGAAAGGGTGTT (200 nM) | 96 | 68 |
| R: GGCCTCCCATCTCCTTCATCACATC (200 nM) | ||||
| Ywhaz (Mamo et al., 2008) | ENSOCUG00000000734 | F: GGTCTGGCCCTTAACTTCTCTGTGTTCTA (200 nM) | 142 | 68 |
| R: GCGTGCTGTCTTTGTATGATTCTTCACTT (200 nM) | ||||
| Luciferase (Bui et al., 2009) | M15077 | F: AGAGATACGCCCTGGTTCCT (100 nM) | 259 | 60 |
| R: ATAAATAACGCGCCCAACAC (100 nM) | ||||
| H2afz | ENSOCUG00000001888 | F: AGAGCCGGCTGCCAGTTCC (200 nM) | 85 | 60 |
| R: CAGTCGCGCCCACACGTCC (200 nM) | ||||
| TET1 | ENSOCUT00000007207 | F: AGAAGCCATCCGTCCTTGTG (300 nM) | 101 | 60 |
| R: TTTGTTGCCAATATCTGCCTTTAA (300 nM) | ||||
| TET2 | ENSOCUT00000007098 | F: GTGCTCACGCCCACAGAGA (300 nM) | 124 | 60 |
| R: GGCAGAACGTGGAGCTGCT (300 nM) | ||||
| TET3 | ENSOCUT00000030004 | F: AACCAGGTGACCAATGAAGAAATAG (300 nM) | 175 | 60 |
| R: AGCGATTGTCTTCCTTGGTCAG (300 nM) | ||||
| Dnmt1 | ENSOCUT00000008429 | F: CATCGACACCGGCCTCA (200 nM) | 102 | 60 |
| R: CCATTGACACCGCCTTCCT (200 nM) | ||||
| Dnmt3A | ENSOCUT00000030812 | F: CTGTCCCAGCTGAAAAGAGGAA (200 nM) | 100 | 60 |
| R: TCCACCTGGATGCCCAAGT (200 nM) | ||||
| Dnmt3B | ENSOCUT00000017835 | F: GTAGGCGGCCCATTCGA (200 nM) | 100 | 60 |
| R: GAAGCGACGTACTTTTCCACCTT (200 nM) | ||||
| ERVA | CU 465723 | F: GGGTGTCCAATGACGGGTAAG (200 nM) | 151 | 60 |
| R: GCCGGGAAGTCTCCATTCA (200 nM) | ||||
| ERVB | CU 465570 | F: TCCTTCGCATCTGGATTGTCA (200 nM) | 119 | 60 |
| R: ACAAACACCACAGGCAAAATAACAC (200 nM) |
F, forward primer; R, reverse primer.
For each qPCR, the efficiency was determined by serial dilutions of iPS cells cDNA (Osteil et al., 2013, iPS cells kindly provided by Pierre Savatier). For each sample, PCR was performed in triplicate.
The results presented in this study were initially obtained from three biological repeats, when a difference between two conditions was found significant or close to significance, three additional repeats were performed to verify the differences.
For each PCR, starting from experimentally obtained Ct values, we inferred the Ct corresponding to one embryo for each gene and each stage, using the standard curve. Then data analysis was performed using the qBase Plus software (Biogazelle, Belgium) (Hellemans et al., 2007). Ywhaz and Hprt1 were used to normalize the results according to the GeNorm procedure. A ΔΔCt method taking into account the efficiency of each qPCR was used to determine the relative normalized expression level of each gene of interest.
Statistical analysis
For 5meC and 5hmeC quantification, statistical analyses were performed using the pairwise.perm.t.test (with 10 000 permutations) from the RVAideMemoire package in the R software (R Development Core Team, 2010). Statistical analysis of RT-qPCR data was performed using non-parametric tests in the R software: K-Sample-Fisher Pitman Permutation test from the ‘coin’ package and non-parametric relative contrast effect (nparcomp) from the ‘nparcomp’ package.
Results
Development of rabbit embryos in human culture media
In order to confirm that rabbit embryos could provide a valuable model to study the effect of media commonly used in ART procedures, we first followed their development in those media. For each experiment, about 60 embryos were collected at the 1-cell stage from two female rabbits and placed in either G1+ or Global medium. In order to mimic the procedures used in ART, we transferred the embryos to a new dish at the 8-cell stage either to change the medium (G2+ instead of G1+) or to refresh the medium (Global). Based on our expertize of the timeline of rabbit embryos development in other commonly used media, we chose to assess development at given time points corresponding to key preimplantation stages (Reis e Silva et al., 2012 and our unpublished data).
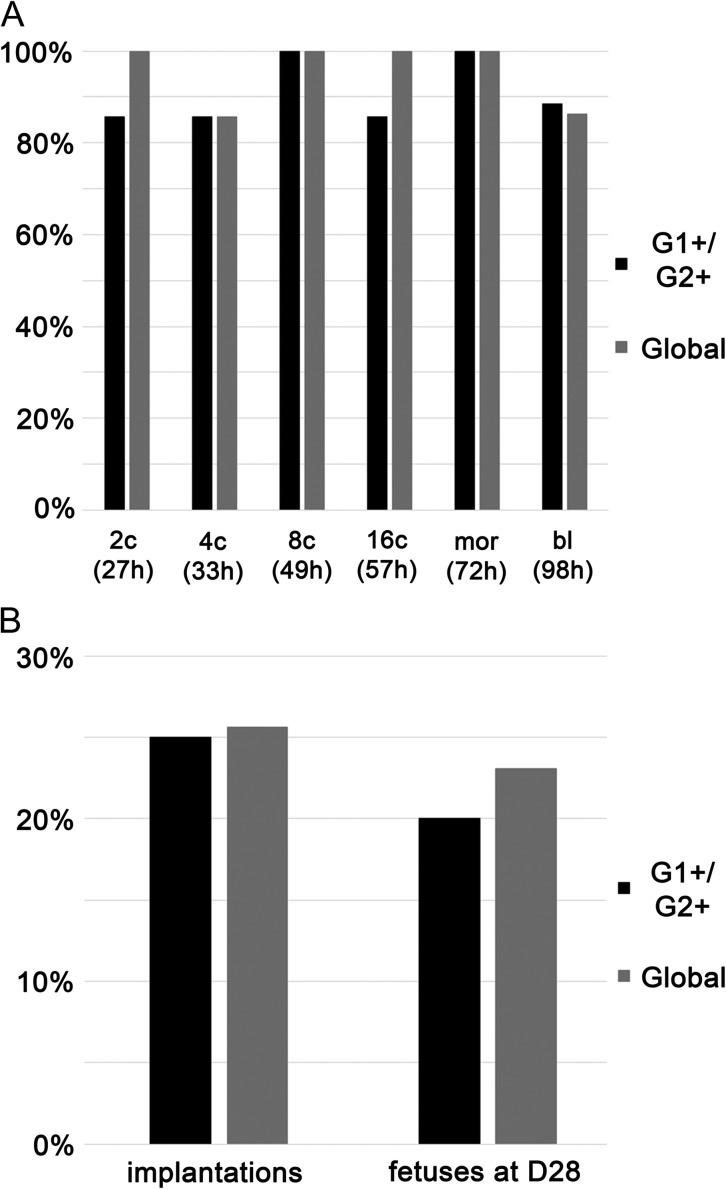
As can be seen in Fig. 1A, at all chosen time points, we observed that over 80% of embryos were at the expected developmental stage, showing that rabbit embryo development in media designed for human embryos follows the same kinetics with the same development rates as that previously observed in other commonly used media. Those time points were used for the rest of the experiments.
Figure 1.
Development of rabbit embryos after culture in G1+/G2+ and Global media. Panel A: preimplantation development. Percentage of embryos collected at the 1-cell stage that had reached a given stage (2c: 2-cell; 4c: 4-cell; 8c: 8-cell; 16c: 16-cell; mor: morula; bl: blastocyst) at a given time point (expressed in hours post-coïtum). Time points were chosen accordingly to our expertize of New Zealand White rabbit in vitro development. Embryos from four different rabbits (obtained in two independent experiments) were separated in dishes (one dish per time point) so as to minimize the time spent out of the incubator. At the 8-cell stage, i.e. the time of medium change, and at the blastocyst stage, end point of the experiments, the development of embryos from all dishes was assessed. A total of 60 embryos (30 for each medium) were followed. Panel B: development after embryonic transfer. Recipient female rabbits were sacrificed at Day 28 and the percentages of embryos that had properly implanted (implantation) and developed to the proper stage (fetuses at day 28) are shown on the graphic. totals of 60 embryos (G1+/G2+) and 39 embryos (Global) were transferred respectively into three and two recipient females and analyzed at Day 28.
Rabbit development to term after embryo culture in human culture media
To determine whether in vitro culture of rabbit embryos was compatible with their further development, 60 and 39 embryos cultured in G1+/G2+ and Global respectively were transferred into three and two recipient females after 73 h of culture (blastocyst stage Day 4). At day 28 (embryonic age), 15 and 10 implantations were observed and 12 (20%) and 9 (23%) morphologically normal fetuses were obtained for G1+/G2+ and Global respectively (Fig. 1B). These developmental rates were well above those commonly obtained after transfer of blastocysts in this species (Jin et al., 2000, and our unpublished data).
Effect of ART culture media on DNA hydroxymethylation in rabbit embryos
In order to assess the effect of in vitro culture in G1+/G2+ and Global media on DNA hydroxymethylation, we used immunofluorescence staining with an antibody specific for 5hmeC and quantified the signal obtained.
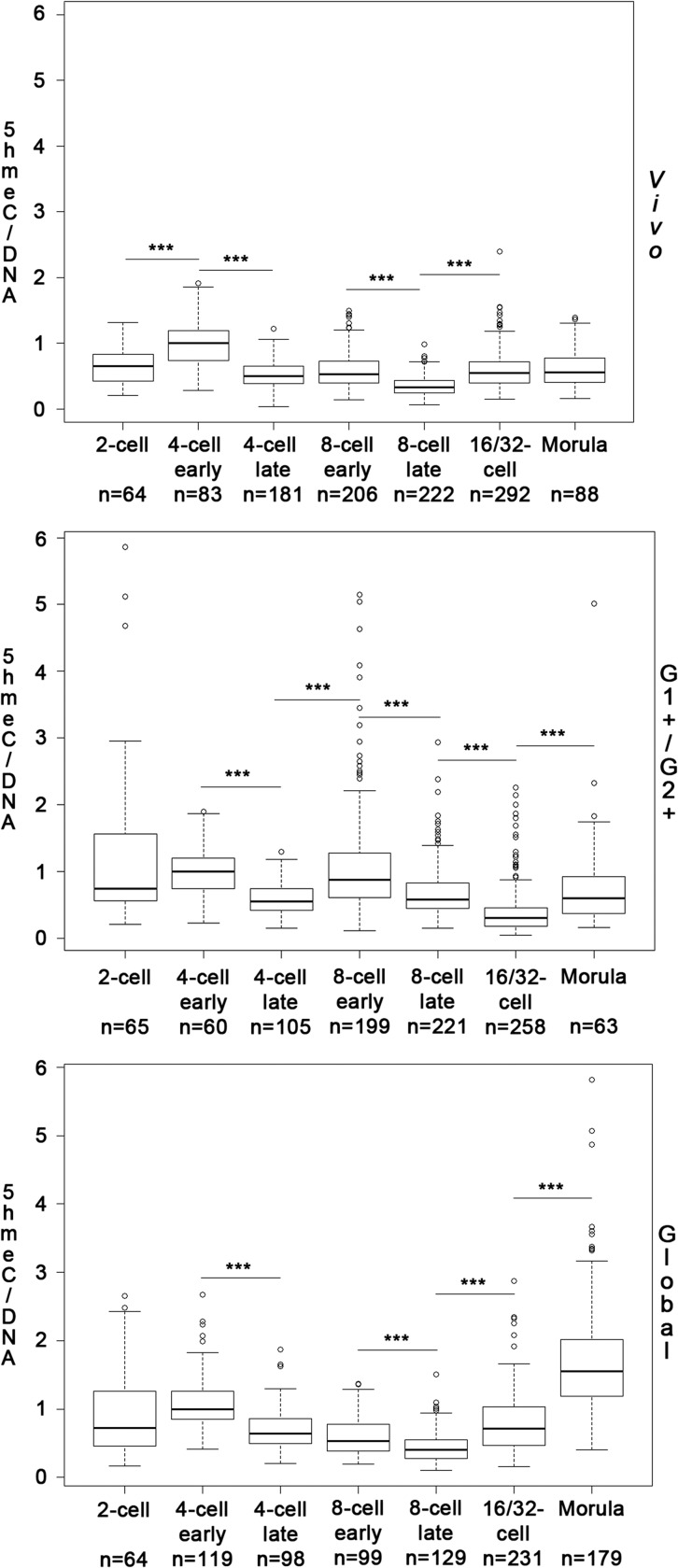
Figure 2 shows the quantification of DNA hydroxymethylation in embryos collected in vivo and cultured in the two in vitro conditions. As DNA content doubles during the cell cycle, we separated nuclei of 4- and 8-cell embryos into ‘early’ and ‘late’ categories based on their DNA content. This separation allowed us to assess the evolution of DNA hydroxymethylation during the cell cycle. We did not separate the 2-cell embryos because we had very few embryos in the ‘late’ pool for in vivo developed embryos. Moreover, as we did not observe differences in DNA hydroxymethylation levels between the 16- and 32-cell stage embryos, we decided to pool them. Finally, all results were normalized by the median of the 4-cell early stage.
Figure 2.
Quantification of DNA hydroxymethylation during rabbit preimplantation development in different conditions. Boxplots showing the 5hmeC/DNA (EthD2) ratios obtained after quantification of immunofluorescence images in embryos developed in vivo (Vivo) or cultured in G1+/G2+ (G1+/G2+) or in Global (Global) medium and normalization by the median of the 4-cell early stage for each condition. The number of nuclei analyzed for each developmental stage is indicated below the corresponding stage. Significant differences between two consecutive stages are indicated on the graphic (*P < 0.01; **P < 0.005; ***P < 0.001).
In in vivo developed embryos, the DNA hydroxymethylation levels significantly increased between the 2- and 4-cell stages. It started decreasing during the 4- and 8-cell stage; no difference was observed between the 4-cell late and 8-cell early stages. At the 16/32-cell stage, DNA hydroxymethylation increased again to reach about the level it had been in 2-cell embryos; it then remained stable at the morula stage. Interestingly, DNA hydroxymethylation behaved differently in the two in vitro conditions we compared, and appeared to be much more variable, especially at the later stages. In embryos cultured in Global medium, it followed a similar pattern to that in in vivo collected embryos until the 16/32-cell stage, even if the decrease observed at the 4-cell stage was slightly lower (Supplementary data, Fig. S1). The main difference was observed at the morula stage, where the level of DNA hydroxymethylation significantly increased and became higher than it was at the 2-cell stage. By contrast, in embryos cultured in G1+/G2+ media, differences appeared much earlier. The DNA hydroxymethylation level decreased during the 4- and the 8-cell stages to a roughly similar extent to that observed in vivo, but surprisingly there was a significant increase between the 4-cell late and the 8-cell early stages, leading to higher hydroxymethylation levels in 8-cell early but also 8-cell late embryos (Supplementary data, Fig. S1). Finally, DNA hydroxymethylation decreased again between the 8-cell late and 16/32-cell stages, where it was lower than in the other two conditions, but increased between the 16/32-cell and morula stages. However, like in in vivo developed embryos, the final level of DNA hydroxymethylation was close to the level observed in 2-cell embryos. In conclusion, the kinetics of DNA hydroxymethylation in embryos cultured in the Global medium appeared equivalent to that observed in in vivo collected embryos until the 16/32-cell stage, but its final level at the morula stage was much higher. In contrast, embryos cultured in G1+/G2+ media showed different kinetics, especially at the time of EGA (8/16-cell stages), but the final level at the morula stage was similar to that observed in in vivo collected embryos.
Effect of ART culture media on DNA methylation in rabbit embryos
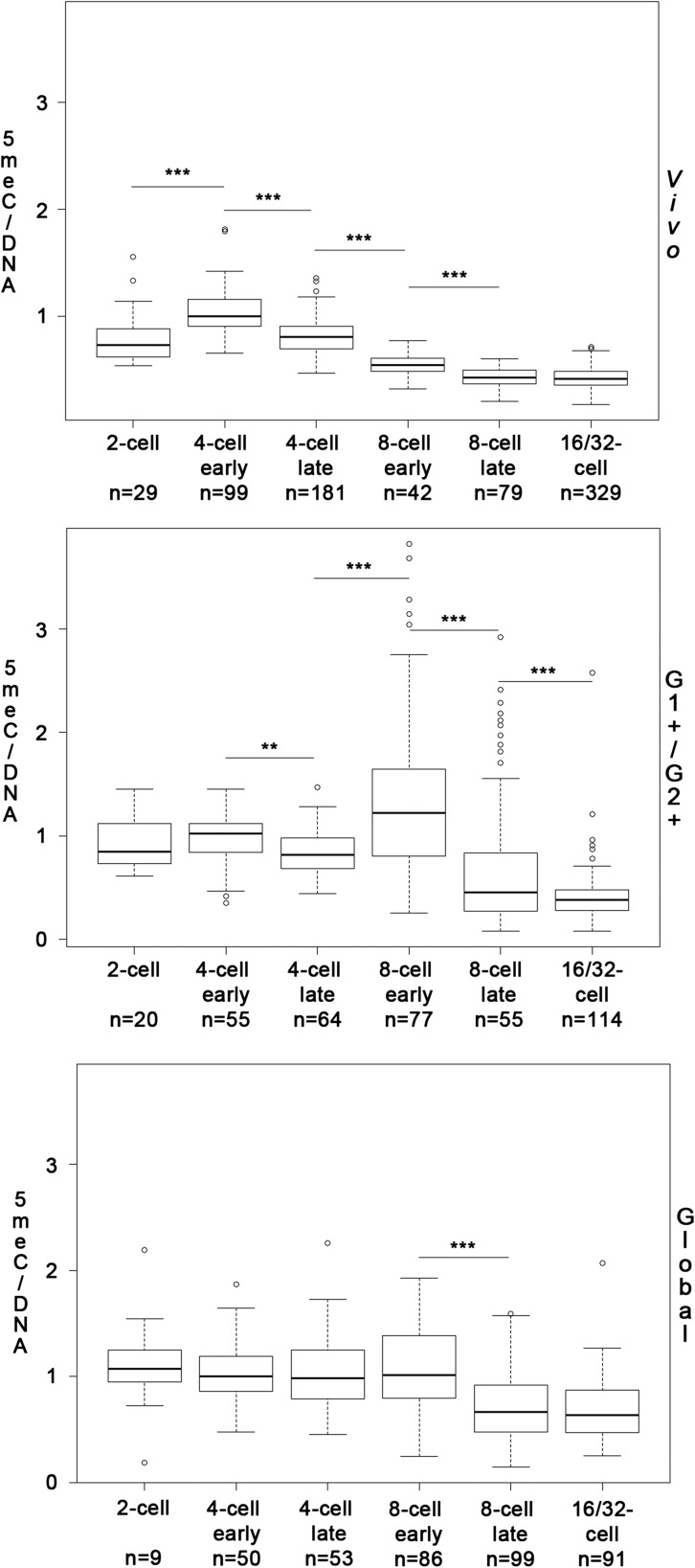
We had previously shown, using a rabbit specific medium, that in vitro culture has an impact on DNA methylation of rabbit embryos. We thus decided to analyze DNA methylation in G1+/G2+ and Global media focusing on the stages where the main differences were observed in our previous study, i.e. between the 2- and the 16/32-cell stages. For comparison, we reused the data previously obtained on in vivo collected embryos, (Reis e Silva et al., 2012), although we separated the 4- and 8-cell stages embryos into ‘early’ and ‘late’ categories, as described in the methods.
As shown in Fig. 3, we observed that DNA methylation followed a kinetic that is similar to that of DNA hydroxymethylation in in vivo collected embryos: increase between the 2- and 4-cell early stages, decrease during the 4-cell stage and the 8-cell stage. There are however two main differences: unlike DNA hydroxymethylation, DNA methylation significantly decreased between the 4-cell late and 8-cell early stages, and there was no increase between the 8- and 16/32-cell stages. Embryos cultured in vitro in G1+/G2+ and Global media both showed very different kinetics (Fig. 3). No DNA demethylation was observed for embryos cultured in Global medium until the 8-cell stage. In fact, the only demethylation phase seemed to occur during the 8-cell stage. Compared to in vivo developed embryos, DNA methylation relative to early 4-cell stage was thus higher in Global whatever the stage (Supplementary data, Fig. S2). In contrast, in embryos cultured in G1+/G2+ media, demethylation occured both during the 4- and 8-cell stages, and between the 8-cell late and 16/32-cell stages but a sharp increase in DNA methylation was observed between the 4-cell late and 8-cell early stages. It is important to note that at the 8-cell stage DNA methylation levels appeared much more variable after in vitro culture than in in vivo developed embryos (Fig. 3).
Figure 3.
Quantification of DNA methylation during rabbit preimplantation development in different conditions. Boxplots showing the 5meC/DNA (EthD2) ratios obtained after quantification of immunofluorescence images in embryos developed in vivo (Vivo) or cultured in G1+/G2+ (G1+/G2+) or in Global (Global) medium and normalization by the median of the 4-cell early stage for each condition. The number of nuclei analyzed for each developmental stage is indicated below the corresponding stage. Significant differences between two consecutive stages are indicated (*P < 0.01; **P < 0.005; ***P < 0.001).
Expression of DNA methylation related enzymes after in vitro culture
As we observed differences in both DNA methylation and hydroxymethylation dynamics between in vivo developed embryos and in vitro cultured ones, we wondered whether these changes could be associated with changes in the expression of genes encoding the related enzymes: DNMTs and/or TET genes.
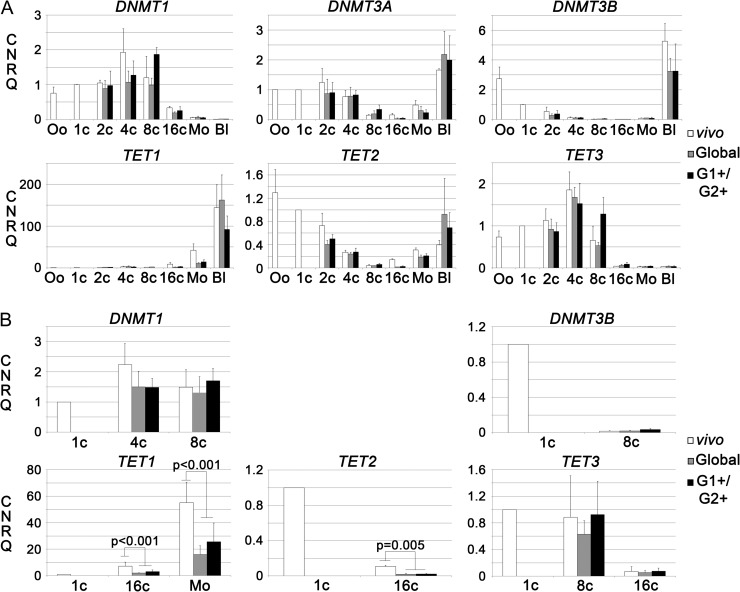
We assessed the expression of DNMT1, DNMT3A, DNMT3B, TET1, TET2 and TET3 by RT-qPCR in preimplantation embryos after in vitro culture in either G1+/G2+ or Global medium and compared the results to those obtained for in vivo developed embryos. We first performed three independent repeats for all stages (Fig. 4A). Our results did not show any modifications of DNMT3A at any of the analyzed stages. On the other hand, we were able to detect differences in the expression of the five other enzymes at some stages that were either statistically significant or nearly significant. In order to verify those differences, we performed three more repeats for the stages at which we had observed differences (Fig. 4B). We could confirm that the expression of TET1 is significantly decreased after in vitro culture in both G1+/G2+ and Global media at the 16-cell (P = 0.01) and morula (P = 0.003) stages. Similarly, the expression of TET2 was significantly decreased at the 16-cell stage (P = 0.006) after culture in both media.
Figure 4.
Expression of DNA methylation related genes during rabbit preimplantation development in different conditions. Panel A: expression of DNMT1, DNMT3A, DNMT3B, TET1, TET2 and TET3 in oocytes (Oo) and at the 1-cell (1c), 2-cell (2c), 4-cell (4c), 8-cell (8c), 16-cell (16c), morula (Mo) and blastocyst (Bl) stages. White bars: in vivo developed embryos: oocytes and 1-cell embryos are common to all conditions as in vitro cultured embryos were collected at the 1-cell stage; grey bars: embryos cultured in Global medium; black bars: embryos cultured in G1+/G2+ media. Each bar represents the mean of three independent experiments with standard deviations and expression levels normalized by the expression level at the 1-cell stage. Panel B: Focus on the expression of DNMT1, DNMT3B, TET1, TET2 and TET3 at certain developmental stages: when a difference detected in the first set of experiments presented in panel A was significant or close to significance, three additional repeats of the points of interest were performed in order to verify the differences. Where significant differences were confirmed, the corresponding P value is indicated on the graphic.
Thus, while the expression of DNMTs was not altered by in vitro culture in G1+/G2+ and Global media, the embryonic expression of TET1 and TET2 was significantly reduced at later stages.
Effect of in vitro culture on the expression of retrovirus sequences
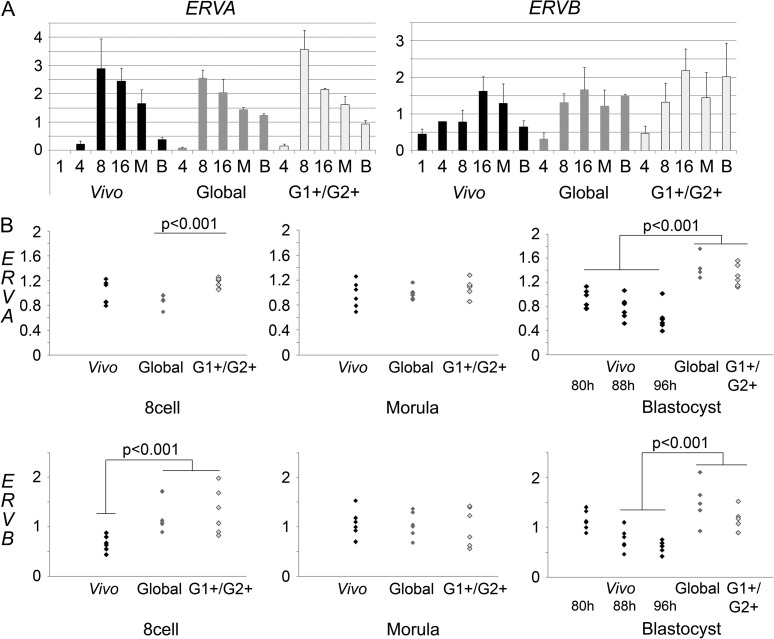
We then wanted to assess whether the different culture conditions could have an impact on gene expression. In order to test this hypothesis, we chose to analyze the expression of two endogenous retroviral sequences (ERVs), belonging to two different families of retroviruses, thereafter referenced as ERVA and ERVB. Indeed, ERVs are known to be regulated by DNA methylation (Rowe and Trono, 2011) and their expression could thus be affected by modifications of this mark after in vitro culture. Moreover, we had previously identified those ERVs as transiently expressed at the time of EGA during development (belonging to the ‘LHL’ cluster as published in (Léandri et al., 2009)). We first analyzed the expression of ERVA and ERVB in three independent experiments (Fig. 5A), and performed three additional experiments for stages where we could detect a significant or close to significant difference between the various conditions (Fig. 5B). We confirm that both ERVA and ERVB are strongly expressed at the time of EGA (Fig. 5A). ERVA expression had already reached its peak at the 8-cell stage, and subsequently decreased until the blastocyst stage (96 hpc), where it was barely detectable. ERVB expression peaked later, between the 8-cell and morula stages, and decreased at the blastocyst stage. At first glance, the global dynamics of ERVA retrovirus did not seem to be affected by in vitro culture: its expression increased strongly between the 4- and 8-cell stages and decreased gradually between the 8-cell and blastocyst stages (Fig. 5A). However, in vitro culture affected the levels of ERVA expression at certain stages. At the 8-cell stage, ERVA expression in embryos cultured in G1+/G2+ was statistically higher than in embryos cultured in Global, although there was no statistical difference between either in vitro condition and the in vivo one (Fig. 5B). Moreover, ERVA expression levels at the blastocyst stage (96 hpc) remained higher in in vitro cultured embryos (P < 0.01) (Fig. 5B). As development is slightly slower in in vitro than in vivo, and because ERVA expression decreases between the morula and blastocyst stages, we wanted to check whether the observed difference was due to this developmental delay. Thus we compared ERVA expression in in vitro developed blastocysts at 96 hpc to its expression in in vivo developed blastocysts at earlier time points: i.e. 80 and 88 hpc. We confirmed that ERVA expression decreases during the blastocyst stage in vivo; however, this expression was still significantly higher in in vitro produced blastocysts at 96 hpc than in the earliest in vivo produced blastocysts (80 hpc) (P < 0.01). Thus, the higher expression observed in vitro was not due to an induced developmental delay (Fig. 5B). The expression pattern of ERVB also seemed to be affected by in vitro culture (Fig. 5A). At the 8-cell stage, ERVB expression was higher in in vitro produced embryos than in in vivo produced ones (Fig. 5B). In 96 hpc blastocysts, we also observed a higher ERVB expression after in vitro culture (P < 0.01). But ERVB expression significantly decreased during the blastocyst stage in vivo and we could not detect a significant difference between 80 hpc in vivo produced blastocysts and 96 hpc in vitro produced ones so that we cannot rule out the possibility that the observed difference for ERVB at that stage was due to a developmental delay after in vitro culture (Fig. 5B).
Figure 5.
Expression of two endogenous retroviral sequences (ERVs) during rabbit preimplantation development in different conditions Panel A: expression kinetics of ERVA and ERVB in embryos developed in vivo (Vivo: black bars), cultured in G1+/G2+ (G1+/G2+: light grey bars) or in Global (Global: dark grey bars) medium. Each bar represents the mean of three independent experiments with standard deviations. 1: 1-cell embryos; 4: 4-cell embryos; 8: 8-cell embryos; 16: 16-cell embryos; M: morula; B: Blastocysts. Panel B: comparison of the expression levels of ERVA and ERVB in the three different conditions at the 8-cell, morula and blastocyst stages: when a difference between two conditions of development in the first set of experiments presented in panel A was significant or close to significance, three additional repeats of the points of interest were performed in order to verify the differences. For in vivo developed blastocysts (black dots), three different time points were analyzed: 80, 88 and 96 hpc and compared with in vitro cultured blastocysts (Global medium: dark grey dots; G1+/G2+ media: light grey dots) at 96 hpc; significant differences are displayed on the graphic. Each dot represents an independent measure.
In conclusion, in vitro culture affects the expression of both tested ERVs, albeit not at the same developmental stages.
Discussion
Using two different media, the ‘sequential’ G1+/G2+ and the ‘one-step’ Global, we showed that in vitro culture impacts the evolution of DNA methylation and hydroxymethylation and that the effect observed is dependent on the medium used. In addition, in vitro culture alters the expression of TET1 and TET2, two genes encoding proteins involved in DNA demethylation, but not the expression of DNMTs. Finally, we showed that the developmentally regulated expression of two endogenous retroviral sequences is altered after in vitro culture, suggesting that the effects we observed on global DNA methylation and hydroxymethylation may have an impact on gene expression.
DNA methylation and hydroxymethylation dynamics in vivo and the effect of in vitro culture
We had previously shown that the in vivo DNA methylation level decreases after the 4-cell stage in rabbit embryos and covers several cell cycles (Reis e Silva et al., 2012). In the present study, we refined this analysis by separating embryos at the 4- and 8-cell stage depending on their DNA content and thus on their position in the cell cycle. This allowed us to show that DNA methylation decrease starts during the 4-stage. In addition, the drop we observe between the end of 4-cell late and 8-cell early stages shows that an active demethylation mechanism is at play.
By contrast, DNA hydroxymethylation also decreases during the 4- and 8-cell stages but does not change between the 4- and 8-cell stages, suggesting passive loss of this mark as in mouse embryo (Inoue and Zhang, 2011). However, as active demethylation, and the consequent formation of new 5hmeC, takes place simultaneously, DNA hydroxymethylation loss cannot only be passive and 5hmeC conversion to 5-formylcytosine (5fC) and 5-carboxylcytosine (5-caC) probably also takes place.
We also showed that DNA methylation and hydroxymethylation kinetics in embryos cultured in vitro are altered. It is also interesting to note that the levels of both marks appear much more variable after in vitro culture (Figs 2 and 3); this variability probably reveals both interindividual and interblastomere heterogeneity in the response to suboptimal environmental conditions. As the rabbits used in this study are not isogenic, genetic differences between same stage embryos could be responsible for inter-individual differences, while the absence of gap junctions during cleavage stages and the resulting metabolic heterogeneity between early blastomeres (Brison et al., 2014) could contribute to inter-blastomere variability.
Indeed, DNA methylation increases between the 4-cell late and 8-cell early stages after culture in G1+/G2+, while no decrease is observed until the 8-cell stage after culture in Global medium. This suggests an increase of DNA methylation activity in G1+/G2+, and an increase of DNA methylation and/or a decrease of DNA demethylation activity in Global media. Yet, at these stages, we did not observe any change in the expression of DNMTs or TETs. Thus, the differences we observe should be explained either by changes in proteins levels or by modifications of their activity. This remains to be determined.
In embryos cultured in G1+/G2+ media, DNA hydroxymethylation follows the DNA methylation increase between the 4- and 8-cell stages, which is consistent since DNA hydroxymethylation can only be generated through the oxidation of methylated DNA (Szwagierczak et al., 2010; Ficz et al., 2011). However, as mentioned above, the stability of DNA methylation levels in embryos cultured in Global medium has no apparent effect on the kinetics of DNA hydroxymethylation. This suggests that the activity of TET enzymes in both media may be different at this stage. Assessing TET enzymes activities is all the more crucial as we observed an increase of DNA hydroxymethylation between the 16/32-cell and morula stages after in vitro culture in both conditions, surprisingly correlated with a decrease in TET1 and TET2 expression. Alternatively, as TET enzymes cannot only catalyze the conversion of 5meC to 5hmeC but also the conversion of 5hmeC to 5fC and 5caC, we may hypothesize that the latter reactions require higher concentrations of TET enzymes.
The effect of in vitro culture and its implications regarding the tested media
Thus, both in vitro culture conditions have different impacts on DNA methylation and hydroxymethylation dynamics. In rabbit embryos cultured in G1+/G2+ media but not in Global medium, both DNA methylation and hydroxymethylation seem perturbed around the time of EGA, which also corresponds to the time of medium change between G1+ and G2+ (8-cell stage). This timing corresponds to the one recommended by Vitrolife for the medium change in human embryos. Our results however suggest that the G1+ medium may not be entirely adapted to development when the embryo is ‘preparing’ for EGA. This cannot be explained by the lack of carbohydrates, as the concentrations of glucose, lactate, pyruvate and citrate are very similar in both G1+ and Global, but may be related to the absence of essential amino acids in G1+ medium (Morbeck et al., 2014), while those are present in rabbit oviductal fluid (Leese et al., 1979). Conversely, the Global medium appears adapted to the EGA developmental period but may not be suited to the later developmental stages (from 16/32-cell).
We show here that the expression of both endogenous retroviral sequences we tested is affected by in vitro culture at critical points of development: around EGA and at the blastocyst stage. Interestingly, expression of these ERV genes is under the control of promoters located in their Long Terminal Repeats (LTR); while the ERV sequences we quantified are found as about 40 and 60 copies in the rabbit genome respectively, their LTRs are scattered throughout the genome and may regulate many endogenous genes (Peaston et al., 2004). The effects we observe for these two sequences may thus reflect a deregulation of many other genes. Moreover, differences in DNA methylation may affect the expression of other genes independently of these LTRs.
In conclusion, additional studies will need to be conducted to identify gene expression perturbations on a broader scale and to determine their relationship with epigenetic perturbations due to the culture conditions. One of our aims in this study was to provide new insight to help the choice of culture media for human ART: sequential or ‘one-step’ media. Unexpectedly, neither in vitro culture condition appeared closer to in vivo development. Further analyses at later stages of rabbit embryo development should however provide useful data to help with this difficult choice. Especially, remethylation occurs differently in the embryonic and extraembryonic lineages of peri-implantation blastocysts (Salvaing et al., submitted) and could thus differentially maintain, amplify or erase the early epigenetic differences we observe here. It will be important to perform those analyses separately in embryonic and extraembryonic tissues just after this remethylation step but also in the perinatal and postnatal periods to decipher whether the early alterations we observe have long term effects.
Supplementary data
Supplementary data are available at http://humrep.oxfordjournals.org/.
Acknowledgements
The authors would like to thank Luc Jouneau for his help with statistical analyses. The authors are indebted to members of Unité Commune d'Expérimentation Animale (UCEA) responsible for our rabbit facility. They are grateful to Eugénie Canon, Vincent Brochard, Catherine Archilla and Linda Maulny for excellent technical assistance, to Anne Jeandin for participating in the early phases of this work, and to Geneviève Jolivet for providing primer sequences for rabbit DNMT. They also thank Pierre Adenot and the MIMA2 facility for access to the Apotome microscopy and the Région Ile-de-France for funding this system.
Authors’ roles
J.S., V.D., N.P., N.D. and N.B. designed the experiments. J.S., N.P., M.N.B., S.V., E.P., C.B. and N.D. performed the experiments. J.S., M.N.B., N.P., S.V. and V.D. analyzed the results. J.S., N.P. and V.D. wrote the article. All authors have critically revised the article.
Funding
This work was supported by an ‘AMP diagnostic prénatal et diagnostic génétique’ 2012 grant from the French Agence de la Biomédecine.This study was performed within the framework of ANR LABEX ‘REVIVE’ (ANR-10-LABX-73). Authors are members of RGB-Net (TD 1101) and Epiconcept (FA 1201) COST actions.
Conflict of interest
The authors declare that they have no conflict of interests.
References
- Biggers JD, McGinnis LK, Raffin M. Amino acids and preimplantation development of the mouse in protein-free potassium simplex optimized medium. Biol Reprod 2000;63:281–293. [DOI] [PubMed] [Google Scholar]
- Brison DR, Sturmey RG, Leese HJ. Metabolic heterogeneity during preimplantation development: the missing link. Hum Reprod Update 2014;20:632–640. [DOI] [PubMed] [Google Scholar]
- Bui LC, Evsikov A V, Khan DR, Archilla C, Peynot N, Hénaut A, Bourhis D Le, Vignon X, Renard JP, Duranthon V. Retrotransposon expression as a defining event of genome reprogramming in fertilized and cloned bovine embryos. Reproduction 2009;138:289–299. [DOI] [PubMed] [Google Scholar]
- Burdge GC, Hanson MA, Slater-Jefferies JL, Lillycrop KA. Epigenetic regulation of transcription: a mechanism for inducing variations in phenotype (fetal programming) by differences in nutrition during early life. Br J Nutr 2007;97:1036–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capmany G, Bolton VN. Synthetic profiles of polypeptides of human oocytes and normal and abnormal preimplantation embryos. J Reprod Fertil 1999;117:125–133. [DOI] [PubMed] [Google Scholar]
- Chi MM, Manchester JK, Yang VC, Curato AD, Strickler RC, Lowry OH. Contrast in levels of metabolic enzymes in human and mouse ova. Biol Reprod 1988;39:295–307. [DOI] [PubMed] [Google Scholar]
- Choux C, Carmignac V, Bruno C, Sagot P, Vaiman D, Fauque P. The placenta: phenotypic and epigenetic modifications induced by Assisted Reproductive Technologies throughout pregnancy. Clin Epigenetics 2015;7:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry GN, Tanasijevic B, Barry ER, Krueger W, Rasmussen TP. Epigenetic regulatory mechanisms during preimplantation development. Birth Defects Res C Embryo Today 2009;87:297–313. [DOI] [PubMed] [Google Scholar]
- Dawlaty MM, Breiling A, Le T, Barrasa MI, Raddatz G, Gao Q, Powell BE, Cheng AW, Faull KF, Lyko F et al. . Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev Cell 2014;29:102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauque P, Jouannet P, Lesaffre C, Ripoche M-A, Dandolo L, Vaiman D, Jammes H. Assisted Reproductive Technology affects developmental kinetics, H19 Imprinting Control Region methylation and H19 gene expression in individual mouse embryos. BMC Dev Biol 2007;7:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore Ta, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 2011;473:398––402. [DOI] [PubMed] [Google Scholar]
- Fleming TP, Velazquez MA, Eckert JJ. Embryos, DOHaD and David Barker. J Dev Orig Health Dis 2015;6:377–383. [DOI] [PubMed] [Google Scholar]
- Fulka H, Mrazek M, Tepla O, Fulka J. DNA methylation pattern in human zygotes and developing embryos. Reproduction 2004;128:703–708. [DOI] [PubMed] [Google Scholar]
- Guo F, Li X, Liang D, Li T, Zhu P, Guo H, Wu X, Wen L, Gu T-P, Hu B et al. . Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell 2014. a;15:447–458. [DOI] [PubMed] [Google Scholar]
- Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, Yan J, Ren X, Lin S, Li J et al. . The DNA methylation landscape of human early embryos. Nature 2014. b;511:606––610. [DOI] [PubMed] [Google Scholar]
- EI Hajj N, Haaf T. Epigenetic disturbances in in vitro cultured gametes and embryos: implications for human assisted reproduction. Fertil Steril 2013;99:632–641. [DOI] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, Paepe A De, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 2007;8:R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill PWS, Amouroux R, Hajkova P. DNA demethylation, Tet proteins and 5-hydroxymethylcytosine in epigenetic reprogramming: An emerging complex story. Genomics 2014;104:324–333. [DOI] [PubMed] [Google Scholar]
- Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science 2011;334:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Jin S, Pfeifer GP, Szabó PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. PNAS 2011;108:3642–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010;466:1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin DI, Kim DK, Im KS, Choi WS. Successful pregnancy after transfer of rabbit blastocysts grown in vitro from single-cell zygotes. Theriogenology 2000;54:1109–1116. [DOI] [PubMed] [Google Scholar]
- Kane MT. Minimal nutrient requirements for culture of one-cell rabbit embryos. Biol Reprod 1987;37:775–778. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 2006;31:89–97. [DOI] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G et al. . Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 2011;8:200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubiura M, Okano M, Kimura H, Kawamura F, Tada M. Chromosome-wide regulation of euchromatin-specific 5mC to 5hmC conversion in mouse ES cells and female human somatic cells. Chromosome Res 2012;20:837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaraviciute G, Kauser M, Bhattacharya S, Haggarty P. A systematic review and meta-analysis of DNA methylation levels and imprinting disorders in children conceived by IVF/ICSI compared with children conceived spontaneously. Hum Reprod Update 2015;21:555–557. [DOI] [PubMed] [Google Scholar]
- Léandri RD, Archilla C, Bui LC, Peynot N, Liu Z, Cabau C, Chastellier A, Renard JP, Duranthon V. Revealing the dynamics of gene expression during embryonic genome activation and first differentiation in the rabbit embryo with a dedicated array screening. Physiol Genomics 2009;36:98–113. [DOI] [PubMed] [Google Scholar]
- Leese HJ, Aldridge S, Jeffries KS. The movement of amino acids into rabbit oviductal fluid. J Reprod Fertil 1979;56:623–626. [DOI] [PubMed] [Google Scholar]
- Li Y, O'Neill C. Persistence of cytosine methylation of DNA following fertilisation in the mouse. PLoS One 2012;7:e30687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamo S, Gal AB, Polgar Z, Dinnyes A. Expression profiles of the pluripotency marker gene POU5F1 and validation of reference genes in rabbit oocytes and preimplantation stage embryos. BMC Mol Biol 2008;9:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manes C, Daniel JC. Quantitative and qualitative aspects of protein synthesis in the preimplantation rabbit embryo. Exp Cell Res 1969;55:261–268. [DOI] [PubMed] [Google Scholar]
- Manipalviratn S, DeCherney A, Segars J. Imprinting disorders and assisted reproductive technology. Fertil Steril 2009;91:417–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantikou E, Youssef MA, Van Wely M, van der Veen F, Al-Inany HG, Repping S, Mastenbroek S. Embryo culture media and IVF/ICSI success rates: a systematic review. Hum Reprod Update 2013;19:210–220. [DOI] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature 2000;403:501–502. [DOI] [PubMed] [Google Scholar]
- Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 2012;151:1417–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morbeck DE, Krisher RL, Herrick JR, Baumann NA, Matern D, Moyer T. Composition of commercial media used for human embryo culture. Fertil Steril 2014;102:759–766, e9. [DOI] [PubMed] [Google Scholar]
- Naviaux RK. Mitochondrial control of epigenetics. Cancer Biol Ther 2008;7:1191–1193. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Patrat C, Thépot D, Peynot N, Fauque P, Daniel N, Diabangouaya P, Wolf J-P, Renard J-P, Duranthon V et al. . Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature 2011;472:370–374. [DOI] [PubMed] [Google Scholar]
- Osteil P, Tapponnier Y, Markossian S, Godet M, Schmaltz-Panneau B, Jouneau L, Cabau C, Joly T, Blachère T, Gócza E et al. . Induced pluripotent stem cells derived from rabbits exhibit some characteristics of naïve pluripotency. Biol Open 2013;2:613–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen CM, Segars JHJ. Imprinting disorders and assisted reproductive technology. Semin Reprod Med 2009;27:417–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papale L, Zhang Q, Li S, Kailei C, Sündüz K, Alisch R. Genome-wide disruption of 5-hydroxymethylcytosine in a mouse model of autism. Hum Mol Genet 2015;24(24):7121–7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P et al. . Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 2011;473:394–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB, Harbor B. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell 2004;7:597–606. [DOI] [PubMed] [Google Scholar]
- Peynot N, Duranthon V, Khan DR. Gene expression analysis in early embryos through reverse transcription quantitative PCR (RT-qPCR). Methods Mol Biol 2015;1222:181–196. [DOI] [PubMed] [Google Scholar]
- R Development Core Team R: A Language and Environment for Statistical Computing Vienna, Austria: R Foundation for Statistical Computing, 2010. [Google Scholar]
- Reis e Silva A, Adenot P, Daniel N, Archilla C, Peynot N, Lucci CM, Beaujean N, Duranthon V. Dynamics of DNA methylation levels in maternal and paternal rabbit genomes after fertilization. Epigenetics 2011;6:1–7. [DOI] [PubMed] [Google Scholar]
- Reis e Silva A, Fleurot R, Daniel N, Lucci M, Beaujean N, Duranthon V. Alteration of DNA demethylation dynamics by in vitro culture conditions in rabbit pre-implantation embryos. Epigenetics 2012;7:1–7. [DOI] [PubMed] [Google Scholar]
- Rowe HM, Trono D. Dynamic control of endogenous retroviruses during development. Virology 2011;411:273–287. [DOI] [PubMed] [Google Scholar]
- Salvaing J, Aguirre-Lavin T, Boulesteix C, Lehmann G, Debey P, Beaujean N. 5-Methylcytosine and 5-Hydroxymethylcytosine Spatiotemporal Profiles in the Mouse Zygote PLoS One 2012;7:e38156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 2012;484:339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain JE, Carrell D, Cobo A, Meseguer M, Rubio C, Smith GD. Optimizing the culture environment and embryo manipulation to help maintain embryo developmental potential. Fertil Steril 2016;105:571–587. [DOI] [PubMed] [Google Scholar]
- Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res 2010;38:e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L et al. . Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009;324:930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford NA, Watson AJ, Schultz GA. Transition from maternal to embryonic control in early mammalian development: a comparison of several species. Mol Reprod Dev 1990;26:90–100. [DOI] [PubMed] [Google Scholar]
- Vasanthakumar A, Godley L. 5-hydroxymethylcytosine in cancer: significance in diagnosis and therapy. Cancer Genet 2015;208:167–177. [DOI] [PubMed] [Google Scholar]
- Wang L, Zhang J, Duan J, Gao X, Zhu W, Lu X, Yang L, Zhang J, Li G, Ci W et al. . Programming and inheritance of parental DNA methylomes in mammals. Cell 2014;157:979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins AJ, Papenbrock T, Fleming TP. The preimplantation embryo: handle with care. Semin Reprod Med 2008. a;26:175–185. [DOI] [PubMed] [Google Scholar]
- Watkins AJ, Ursell E, Panton R, Papenbrock T, Hollis L, Cunningham C, Wilkins A, Perry VH, Sheth B, Kwong WY et al. . Adaptive responses by mouse early embryos to maternal diet protect fetal growth but predispose to adult onset disease. Biol Reprod 2008. b;78:299–306. [DOI] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun 2011;2:241. [DOI] [PubMed] [Google Scholar]
- Wright K, Brown L, Brown G, Casson P, Brown S. Microarray assessment of methylation in individual mouse blastocyst stage embryos shows that in vitro culture may have widespread genomic effects. Hum Reprod Oxford Engl 2011;26:2576–2585. [DOI] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 2011;473:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S et al. . Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell 2011;42:451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.