Abstract Abstract
Based on sequence data of nuclear ITS and plastid matK, trnL-F and psbA-trnH markers, the phylogeny of the subtribes Caraganinae and Chesneyinae in tribe Caraganeae was inferred. The results support the monophyly of each of the subtribes. Within subtribes Caraganinae, Calophaca and Halimodendron are herein transferred into Caragana to ensure its generic monophyly. The subtribe Chesneyinae is composed of four well-supported genera: Chesneya, Chesniella, Gueldenstaedtia and Tibetia. Based on phylogenetic, morphological, distributional and habitat type evidence, the genus Chesneya was divided into three monophyletic sections: Chesneya sect. Chesneya, Chesneya sect. Pulvinatae and Chesneya sect. Spinosae. Chesneya macrantha is herein transferred into Chesniella. Spongiocarpella is polyphyletic and its generic rank is not maintained. The position of Chesneya was incongruent in the nuclear ITS and the plastid trees. A paternal chloroplast capture event via introgression is hypothesized for the origin of Chesneya, which is postulated to have involved the common ancestor of Chesniella (♂) and that of the Gueldenstaedtia – Tibetia (GUT) clade (♀) as the parents.
Keywords: Caragana, Chesneya, Chesniella, chloroplast capture, generic delimitation, phylogeny
Introduction
Caraganeae Ranjbar is a mid-sized tribe in Leguminosae, established by Ranjbar and Karamian (2003) based on five genera: Calophaca Fisch. ex DC., Caragana Fabr., Chesneya Lindl. ex Endl., Gueldenstaedtia Fisch. and Halimodendron Fisch. ex DC., numbers of genera may be altered when treated by different workers (see below). Caraganeae ranges from eastern Europe, central and western Asia to Mongolia, China and the Himalayas, extending northward to Siberia (Lock 2005; Ranjbar et al. 2014). This tribe is diagnosed by the asymmetrical axillary peduncles or pedicels attached to the slightly gibbous calyx and dehiscent pods (except for Halimodendron; Polhill 1981; Ranjbar and Karamian 2003; Ranjbar et al. 2014).
A few recent studies referred to the concept of Caraganeae. Molecular work of Ranjbar et al. (2014) classified Caraganeae into two subtribes: Caraganinae and Chesneyinae Ranjbar, F. Hajmoradi & Waycott. Duan et al. (2015) recognized this tribe based on the genera Calophaca, Caragana and Halimodendron. However, the former was inferred from a limited sampling scheme and few DNA markers, while the latter was subject to the undersampled for Chesneyinae. Hence, the monophyly of this tribe and the division of subtribes need to be further evaluated.
Within the subtribe Caraganinae, the genus Caragana has attracted much attention (Komarov 1908; Moore 1968; Gorbunova 1984; Zhao 1993, 2009; Zhou 1996; Zhang 1997; Sanchir 1999; Sanczir 2000; Hou et al. 2008; Zhang et al. 2009). The infrageneric classifications of Caragana mainly focused on several morphological characters: leaves paripinnate vs. digitate, with four vs. more leaflets, and petioles and rachises caducous vs. persistent. Recent phylogenetic analyses resolved that Caragana was paraphyletic, with Halimodendron and Calophaca embedded in it (Zhang et al. 2009, 2015a; Zhang and Fritsch 2010; Duan et al. 2015). Thus, proposal of a new generic delimitation for Caragana may be possible based on more comprehensive phylogenetic evidence.
The genera Chesneya and Gueldenstaedtia formed a well-supported clade (Sanderson and Wojciechowski 1996), and were treated as the subtribe Chesneyinae (Ranjbar et al. 2014). Within this subtribe, the generic delimitations were controversial, especially concerning the status of Chesniella Boriss. (Borissova 1964), Spongiocarpella Yakovl. et Ulzij. (Yakovlev and Sviazeva 1987), and Tibetia (Ali) H. P. Tsui (Tsui 1979). The former two genera were separated from Chesneya, while Tibetia was a segregate of Gueldenstaedtia and has been revised in several studies (Cui 1998; Zhu 2004; Zhu 2005a, 2005b; Bao and Brach 2010). Zhang et al. (2015b) supported the monophyly of Chesneya and proposed a classification system, but some sections were only weakly supported. Hence, the phylogeny of Chesneyinae and its associated genera needs to be further explored.
We herein employ sequence data from nrDNA ITS and plastid matK, trnL-F and psbA-trnH to a) test the monophyly of Caraganeae and its subtribes; b) estimate the phylogeny of genera in Caraganeae; and c) discuss the taxonomic implications of this phylogeny on the generic and the infrageneric classification of the tribe.
Materials and methods
Taxon sampling
Our sampling was designed largely following the generic demarcations in Flora Reipublicae Popularis Sinicae (Liou 1993; Li 1993; Cui 1998). We included 101 accessions, covering 97 species, containing 39 species of Caraganinae (represented by Calophaca, Halimodendron and all 5 sections of Caragana according to Zhang 1997) and 40 accessions (36 species) of Chesneyinae (including Chesneya, Chesniella, Gueldenstaedtia and Tibetia, tentatively treating Spongiocarpella in Chesneya, which were more comprehensively sampled than previous studies [Ranjbar et al. 2014; Duan et al. 2015; Zhang et al. 2015b]). 82 new sequences were generated in this work.
To better resolve the relationships of subtribes Caraganinae and Chesneyinae, 11 Galegeae species (8 genera) and 5 Hedysareae species (4 genera) were also sampled. Cicer microphyllum Royle ex Bentham, Dalbergia hupeana Hance, Lathyrus latifolius L., Robinia pseudoacacia L., Trifolium repens L. and Wisteria sinensis (Sims) Sweet were selected as outgroups based on previous studies (Wojciechowski et al. 2000, 2004; Wojciechowski 2003). Sequences of 40 accessions (representing 40 species) were downloaded from GenBank (see Suppl. material 1 for details). Most accessions we sampled were collected from the field or herbarium specimens. Onobrychis arenaria DC. was obtained from seedlings germinated from seeds provided by the Royal Botanic Gardens, Kew.
DNA extraction, amplification and sequencing
Total genomic DNAs were extracted from silica-gel dried leaves or herbarium material using the Plant DNA Extraction Kit - AGP965/960 (AutoGen, Holliston, MA, USA) or the DNeasy Plant Mini Kit (Qiagen, Valencia, USA). Polymerase chain reactions (PCR) were prepared in 25µL containing 1.5 mM MgCl2, 0.2 mM of each dNTP, 0.4 mM of each primer, 1 U of Taq polymerase (Bioline, Aberdeen, Scotland, UK), and using 10–50 ng (2.5 µL) template DNAs, following Wen et al. (2007). The PCRs for ITS (primer pair: ITS4 and ITS5a) and psbA-trnH (primer pair: psbA and trnH) were performed according to Stanford et al. (2000) and Hamilton (1999), respectively. The PCR primer pair for trnL-F was “c” and “f” as in Zhu et al. (2013) and Taberlet et al. (1991), and the thermal cycling program followed Soejima and Wen (2006). The barcoding region of the matK marker was amplified and sequenced with the primer pair Kim-3F/Kim-1R (CBOL Plant Working Group 2009; China Plant BOL Group 2011), and the amplification conditions were: 95°C (5min) for DNA pre-denaturation; 94°C (40s), 48°C (40s) and 72°C (100s) for 35 cycles; 72°C (10min) for final extension. PCR products were cleaned using ExoSAP-IT (cat. # 78201, USB Corporation, Cleveland, OH, USA) following the manufacturer’s instruction. Purified products were sequenced from both directions with BigDye 3.1 reagents on an ABI 3730 automated sequencer (Applied Biosystems, Foster City, CA, USA).
Phylogenetic analysis
Sequences were assembled with Geneious 7.1 (http://www.geneious.com/), and aligned using MUSCLE 3.8.31 (Edgar 2004), followed by manual adjustments in Geneious 7.1. Because the chloroplast markers putatively evolve as a single molecule, sequences of the three plastid markers (matK, trnL-F and psbA-trnH) were directly concatenated. Topological discordance was investigated by comparing the ITS and the concatenated plastid trees (as in García et al. 2014). To further determine the compatibility between these two datasets, an (ILD) test and an (AU) test were conducted with PAUP* (Swofford 2003) and CONSEL (Shimodaira and Hasegawa 2001; using site-wise likelihood values estimated by RA×ML; Stamatakis et al. 2008) programs, respectively. The tests retrieved the p values of 0.01 and 0.0001, respectively, suggesting that the incongruence between these two datasets was significant. The ITS and the concatenated plastid sequences were thus analyzed separately.
Phylogenetic analyses were carried out using Bayesian inference (BI; Rannala and Yang 1996; Mau et al. 1999) with MrBayes 3.2.5 (Ronquist and Huelsenbeck 2003; Ronquist et al. 2012). Nucleotide substitution model parameters were determined prior to BI using the corrected (AIC) in jModeltest 2.1.7. (Posada 2008; Darriba et al. 2012). For the ITS dataset, boundaries of the 5.8S region to the ITS1 and the ITS2 regions were determined by comparison with the published 5.8S sequence of Vicia faba L. (Nazar and Wildeman 1981; Yokota et al. 1989), and the sequence substitution models for the ITS1, 5.8S and ITS2 regions were determined separately. Similarly, the models for each of the three plastid markers were estimated for the best-fit models, which were used in the BI analysis for concatenated plastid sequences in a partitioned scheme.
In the BI, the (MCMC) search was run by two replicates for 10,000,000 generations, sampling one tree every 1,000 generations. After the first 2,500,000 generations (2,500 trees) were discarded as burn-in, a 50% majority-rule consensus tree and posterior probabilities were obtained among the remaining trees. Results were checked using the program Tracer 1.5 (Rambaut and Drummond 2007) to ensure that plots of the two runs were converging and the value of the effective sample size for each replicate was above 200. (ML) analyses were conducted using RAxML-MPI v8.2 (Stamatakis 2014) with dataset partition scheme the same as in the BI and the following settings: rapid bootstrap analysis with 1,000 replicates and search for best-scoring ML tree in one program run, starting with a random seed, selecting the GTR model. (LBS), as well as (PP) were labeled on the corresponding branches of the Bayesian trees.
Results
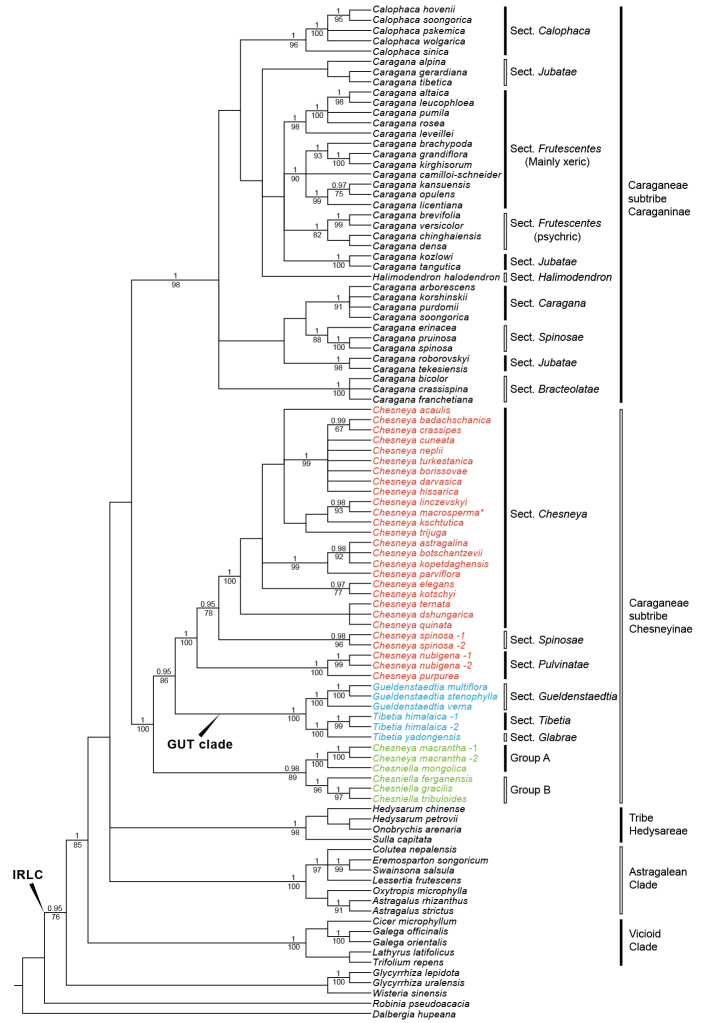
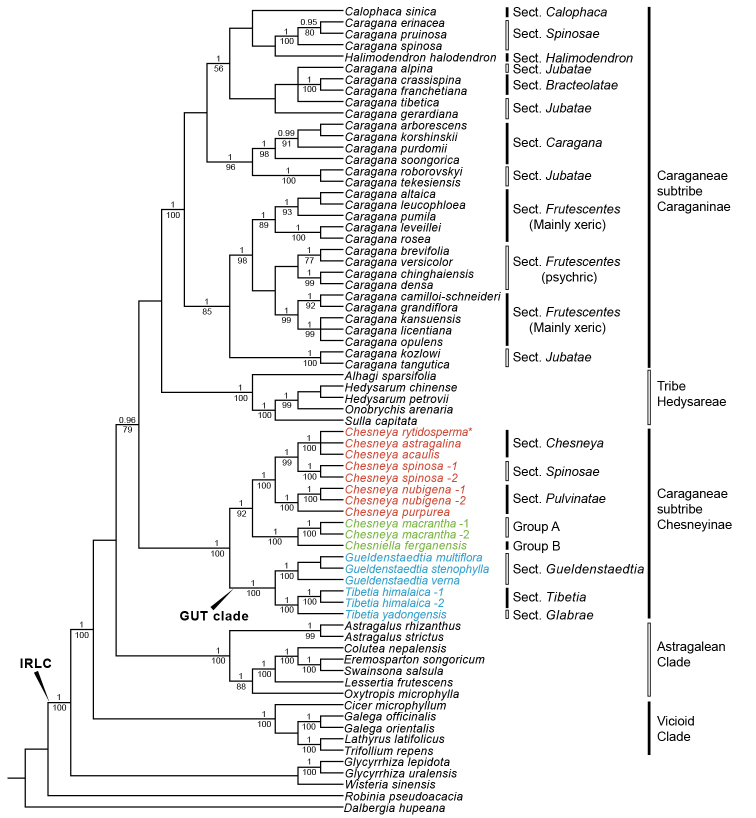
Sequence characteristics are shown in Table 1. Our ML results are basically congruent in topology with the corresponding BI trees, the support values of the former were thus labeled on the corresponding branches of the latter (see legend of Figs 1, 2). Thanks to some extra sequences from GenBank (see Suppl. material 1), especially those of Chesneya and Chesniella, the ITS tree (Fig. 1) was more comprehensively sampled than the plastid tree (Fig. 2), which was of help to increase the general support of the former.
Table 1.
Sequence characteristics with gaps as missing data: alignment length, the number of the constant, variable and potential (Pi) sites, and the best-fit nucleotide substitution model determined by AIC.
| Dataset | Length | Constant | Variable | Pi | Model |
|---|---|---|---|---|---|
| ITS1 | 266 | 81 | 185 | 148 | GTR+I+G |
| 5.8S | 164 | 143 | 21 | 14 | TrNef+I+G |
| ITS2 | 279 | 113 | 166 | 131 | GTR+G |
| matK | 807 | 485 | 322 | 189 | GTR+G |
| trnL-F | 1412 | 921 | 491 | 279 | TVM+I+G |
| psbA-trnH | 793 | 472 | 321 | 175 | TIM1+G |
Figure 1.
Bayesian tree of the nrDNA ITS data, showing relationships of genera in subtribes Caraganinae, Chesneyinae and their close relatives. The labeled sections of Gueldenstaedtia and Tibetia followed Tsui (1979) and Zhu (2005a), respectively. Bayesian posterior probabilities (PP ≥ 0.95) and maximum likelihood bootstrap (LBS ≥ 70%) are given above and below branches, respectively. The asterisk indicates the name of Chesneya macrosperma has not been published, its voucher was storied in LE (details see Zhang et al. 2015b).
Figure 2.
Bayesian tree of the concatenated plastid data of matK, trnL-F and psbA-trnH sequences, showing genera in subtribes Caraganinae, Chesneyinae and their close relatives. The labeled sections of Gueldenstaedtia and Tibetia followed Tsui (1979) and Zhu (2005a), respectively. Bayesian posterior probabilities (PP ≥ 0.95) and maximum likelihood bootstrap (LBS ≥ 70%) are given above and below branches, respectively. The asterisk indicates the type species of Chesneya.
Nuclear data
In the ITS tree (Fig. 1), the Astragalean clade (PP = 1, LBS = 100%; including Astragalus L., Colutea L., Eremosparton Fisch. & C.A.Mey., Lessertia R.Br. ex W.T.Aiton, Oxytropis DC., and Swainsona Salisb.), the Vicioid clade (PP = 1, LBS = 100%; represented by Trifolium, Lathyrus, Cicer and Galega L.), tribe Hedysareae (PP = 1, LBS = 98%), subtribes Caraganinae (PP = 1, LBS = 98%) and Chesneyinae (PP = 1, LBS = 100%) were each strongly supported.
Subtribe Caraganinae contained three genera, within which Calophaca was monophyletic (PP = 1, LBS = 96%), but Calophaca and Halimodendron were embedded within the paraphyletic Caragana. Within subtribe Chesneyinae, Gueldenstaedtia (PP = 1, LBS = 100%) and Tibetia (PP = 1, LBS = 100%) were each monophyletic and together they formed a clade (the GUT clade, shown in blue; PP = 1, LBS = 100%). Two accessions of former Chesneya macrantha Cheng f. ex H.C.Fu constituted a robustly supported branch nested in a monophyletic Chesniella (displayed in green; PP = 0.98, LBS = 89%), while other accessions of Chesneya formed another clade (Chesneya s.s.; shown in red; PP = 1, LBS = 100%; Fig. 1), which contained three well-supported sections (details see Discussion; PP = 1 & LBS = 100%, PP = 0.98 & LBS = 96% and PP = 1 & LBS = 100%, respectively).
Plastid data
Similar to the ITS results, the plastid tree (Fig. 2) also showed the monophyly of both subtribes Caraganinae (PP = 1, LBS = 100%) and Chesneyinae (PP = 1, LBS = 100%). Calophaca and Halimodendron were nested in Caragana in different places from the ITS tree, but such placement was weakly supported. Caragana also showed its paraphyly, with Chesneya sect. Bracteolatae (Kom.) M.L.Zhang (PP = 1, LBS = 100%), Chesneya sect. Caragana Kom. (PP = 1, LBS = 98%), Chesneya sect. Frutescentes (Kom.) Sanchir (PP = 1, LBS = 98%) and Chesneya sect. Spinosae (Kom.) Y.Z.Zhao (PP = 1, LBS = 100%) each strongly supported. Unlike in the ITS tree, Chesneya s.s. and Chesniella were sisters in the plastid tree (PP = 1, LBS = 92%; Fig. 2). As in the ITS tree, the GUT clade (PP = 1, LBS = 100%) contained Gueldenstaedtia (PP = 1, LBS = 100%) and Tibetia (PP = 1, LBS = 100%), with each genus being monophyletic.
Discussion
Caraganeae comprises ca. 100 species distributed in temperate Asia, extending to eastern Europe (Ranjbar and Karamian 2003; Lock 2005). The two subtribes (Caraganinae and Chesneyinae) recognized by Ranjbar et al. (2014) are each well-supported in our analyses. However, our results did not recover a monophyletic Caraganeae (Figs 1, 2). Similarly, the previously expanded delimitation of Hedysareae sensu Lock (2005; also see Cardoso et al. 2013), which included the genera of subtribe Caraganinae and tribe Hedysareae sensu Amirahmadi et al. (2014), is not confirmed herein (Figs 1, 2).
Subtribe Caraganinae is composed of Calophaca, Caragana and Halimodendron (Ranjbar et al. 2014). Morphologically, this subtribe differs from Chesneyinae by several characters, including habit (shrubs vs. perennial herbs or subshrubs), leaf type (paripinnate [except for Calophaca] vs. imparipinnate) and nerve type on wing petals (pinnate vs. palmate except for Chesneya; Lock 2005; Ranjbar et al. 2014; Duan et al. 2015). Caraganinae is also distinct from Hedysareae (as delimited in Amirahmadi et al. 2014 and Duan et al. 2015) based on the following morphological characters: shrubs, rarely small trees; paripinnate, rarely imparipinnate leaves (Calophaca); solitary flowers, or a few flowers in fascicles, rarely forming a raceme; pods cylindric, rarely compressed, glabrous or hairy, with dehiscent and twisted valves (except for Halimodendron; Polhill 1981; Liu et al. 2010b). Caraganinae is also related to the Astragalean clade; yet due to the morphological diversity of the latter, there are few diagnosable features to differentiate the Astragalean clade from Caraganinae, except for the twisted valves of Caraganinae (Calophaca and Caragana).
An expanded generic concept of Caragana
Within Caraganinae, Halimodendron contains only Halimodendron halodendron (Pall.) Druce with its distribution roughly overlapping with that of Calophaca (Lock 2005). This species is morphologically unique in Caraganinae with its inflated pods (Gorshkova 1945; Liu et al. 2010b). Consistent with previous studies (Zhang et al. 2009; Zhang and Fritsch 2010), our results also showed that Halimodendron is nested within Caragana. The phylogenetic evidence hence supports treating Halimodendron as a section within Caragana, i.e., Caragana sect. Halimodenron (Fisch. ex DC.) L.Duan, J.Wen & Zhao Y.Chang. We also resurrect the name Caragana halodendron (Pallas) Dumont de Courset based on Halimodendron halodendron (Figs 1, 2; see Taxonomic Treatment).
Calophaca morphologically resembles Caragana, and it is only distinguished from the latter by its imparipinnate leaves, rachises without thorns, and relatively denser racemes (Borissova 1945; Liu et al. 2010b). Calophaca contains 5–8 species mainly distributed in mountainous areas of central Asia, with one species extending to eastern Europe, and one endemic to northern China (Borissova 1945; Tutin et al. 1968; Yakovlev et al. 1996; Lock 2005; Liu et al. 2010b; Zhang et al. 2015a). The embedded position of Calophaca within Caragana argues that its classification needs to be placed in the broader phylogenetic framework of Caragana, which is supported by our results (Figs 1, 2) and several previous studies (e.g., Zhang et al. 2009, 2010, 2015a, b; Duan et al. 2015). We thus merge Calophaca into Caragana and recognize it at the sectional level as Caragana sect. Calophaca (Fisch. ex DC.) L.Duan, J.Wen & Zhao Y.Chang (see Taxonomic Treatment). The species-level nomenclatural changes will be made in a follow-up paper.
The taxonomy of Caragana has been investigated by various authors (Komarov 1908; Poyarkova 1945; Moore 1968; Sanczir 1979, 2000; Gorbunova 1984; Zhao 1993; Zhou 1996; Zhang 1997; Sanchir 1999; Chang 2008). However, Caragana s.s. as previously circumscribed is clearly paraphylytic (Zhang et al. 2009; Duan et al. 2015). We herein propose the delimitation of Caragana s.l. to ensure the generic monophyly (see Taxonomic Treatment). Caragana as defined now contains taxa of Calophaca, former Caragana s.s. and Halimodendron (Figs 1, 2), which is classified into seven sections: Caragana sect. Bracteolatae M.L.Zhang, Caragana sect. Calophaca, Caragana sect. Caragana, Caragana sect. Frutescentes (Kom.) Sancz., Caragana sect. Halimodenron, Caragana sect. Jabatae (Kom.) Y.Z.Zhao and Caragana sect. Spinosae (Kom.) Y.Z.Zhao. Although Caragana s.l. is morphologically diverse, this genus can be diagnosed by its shrubby habit, saccate, oblique calyx bases, pinnate nerves on the wing petals and twisted, dehiscent pods (except for Caragana holodendron). The expanded concept of Caragana is also supported by cytological evidence (Moore 1968; Chang 1993; Li 1993; Zhou et al. 2002; Chang 2008): most xeric and psychric taxa of Caragana s.l. have the same basic chromosome number (x = 8).
At the sectional level, our ITS tree (Fig. 1) indicated a strongly supported Caragana sect. Calophaca. On the other hand, former Caragana s.s. was divided into five sections mainly based on the combinations of leaf (pinnate or digitate) and petiole/rachis (persistent or caducous) characters (Zhang 1997). Three main sections, Caragana sect. Bracteolatae, Caragana sect. Caragana and Caragana sect. Frutescentes, evolved likely accompanying the rapid uplifts of the (QTP) at around 8 Ma (Zhang et al. 2009). These three sections also largely correspond to psychrophytic, mesophytic and xerophytic habitats, respectively (Zhang and Fritsch 2010). Our analyses supported the monophyly of the three sections, with Caragana sect. Frutescentes only being monophyletic in the plastid tree (also see Zhang et al. 2009; Duan et al. 2015; and see below for an exceptional case in Caragana sect. Frutescentes). Our ITS results failed to resolve a monophyletic Caragana sect. Frutescentes (Fig. 1), but this may be due to insufficient informative sites in the ITS data. Furthermore, we only sampled one series for Caragana sect. Spinosae (Caragana ser. Spinosae Kom.), thus cannot assess its monophyly (Figs 1, 2). Caragana sect. Jabatae was suggested to have experienced a rapid radiation at 3.4–1.8 Ma (Zhang and Fritsch 2010), which may partly explain its poorly resolved relationships in our trees (Figs 1, 2; also see Zhang et al. 2009; Duan et al. 2015).
At the infra-sectional level, Caragana ser. Bracteolatae Kom. and Caragana ser. Spinosae are well-supported by our results (not labeled in the trees). Our results are therefore not completely congruent with Zhang et al. (2009), possibly due to differences in taxon sampling. Interestingly, a strongly supported psychric group is found within the mainly xeric section Caragana sect. Frutescentes (Zhao 2009). This group is represented by Caragana brevifolia Kom., Caragana chinghaiensis Y.X.Liou, Caragana densa Kom. and Caragana versicolor Benth. (in Fig. 1; but weakly supported in the plastid tree). Most species of Caragana sect. Frutescentes range from eastern Europe to northern China, Mongolia and Siberia, however, this above-mentioned psychric group is distributed in the southern edge of northern China, extending to Tibet and its neighboring regions. It may represent a vicariant transitional group of Caragana sect. Bracteolatae, Caragana sect. Jubatae pro parte, Caragana sect. Spinosae pro parte (psychrophytic habitat) and Caragana sect. Frutescentes. Other cases of vicariant distributions have been noted in Caragana, and vicariance was considered as an important biogeographic pattern for this genus. For example, three closely related species in Caragana sect. Caragana, Caragana microphylla Lam., Caragana intermedia Kuang & H.C.Fu and Caragana korshinskii Kom., show non-overlapping to only slightly overlapping distributions in northeast to northwest China (Shue and Hao 1989; Zhang and Wang 1993; Zhang 1998; Chang 2008).
Phylogeny of Chesneyinae
The subtribe Chesneyinae, as established by Ranjbar et al. (2014), was supported to be monophyletic in our trees (Figs 1, 2). Three main clades can be recognized within this subtribe: the GUT clade, Chesneya s.s. and Chesniella (Figs 1, 2).
This subtribe contains ca. 50 species and differs from the Astragalean clade by twisted valves (e.g., in Chesneya), but a few species of Astragalus also have twisted legumes. Taxa of Chesneyinae are distinguished from Hedysareae by their dehiscent pods (Borissova 1945; Yakovlev et al. 1996; Liu et al. 2010a). The genera of Chesneyinae are distributed in central and eastern Asia, Tibet, Mongolia and Siberia, extending to eastern Turkey and Armenia (Fig. 3A; Borissova 1945; Davis 1970; Rechinger 1984; Lock and Schrire 2005; Liu et al. 2010a), which are largely adapted to xerophytic (Chesneya and Chesniella), mesophytic (Gueldenstaedtia) and psychrophytic (Tibetia) habitats, respectively, although some species of Chesneya (see discussion below) and a few of Gueldenstaedtia are psychric taxa. The uplift of the QTP and aridification of the former Tethys region might have driven the origination and divergence of genera in the subtribe Chesneyinae (Wen et al. 2014; Meng et al. 2015; Zhang et al. 2015b).
Figure 3.
Distribution (A) and representative plants (B–H) of genera in Chesneyinae. A red – Chesneya, green – Chesniella, blue – Gueldenstaedtia and yellow – Tibetia B Chesneya acaulis C Chesneya spinosa D Chesneya nubigena E Chesniella macrantha F Chesniella ferganensis G Gueldenstaedtia verna H Tibetia yadongensis.
Topological discordance between ITS and plastid trees in subtribe Chesneyinae
The ITS and plastid topologies are incongruent within Chesneyinae. Chesneya s.s. formed a clade with the GUT clade in the ITS tree (Fig. 1), whereas it was sister to Chesniella in the plastid tree (Fig. 2). Both relationships were well-supported. Various mechanisms have been proposed to explain discordant topologies between gene trees, such as allopolyploidy, hybridization, horizontal gene transfer, incomplete lineage sorting (ILS), different rate of molecular evolution, and chloroplast capture (Degtjareva et al. 2012; García et al. 2014; Yi et al. 2015).
Allopolyploidy can be ruled out for two reasons. First, taxa within Chesneyinae are diploid (Nie et al. 2002; Yang 2002; Sepet et al. 2014), with no evidence of polyploidy in this subtribe and its allied tribes. Second, deep lineages of Chesneyinae basically display a consistent chromosome number (x = 8; Nie et al. 2002; Sepet et al. 2014), with the only exception of Gueldenstaedtia (x = 7; Yang 2002), which has relatively recently diverged (ca. 15.23 Ma; Zhang et al. 2015b).
ILS and chloroplast capture seem more likely mechanisms for the present case (Tsitrone et al. 2003; Deng et al. 2015; Sun et al. 2015). A time-calibrated phylogeny may facilitate the exploration of the likely mechanism. Incomplete lineage sorting, which rarely occurs in deep lineage (Sun et al. 2015), prevails with bifurcation patterns of the shallow lineages of gene trees (especially at the specific level; Xu et al. 2012), and usually takes place in groups with relatively recent diversification times (García et al. 2014). Zhang et al. (2015b) estimated that the main clades of subtribe Chesneyinae split at ca. 28 Ma, which is beyond the time frame supporting ILS of ancestral polymorphisms (as suggested by Xu et al. 2012). On the other hand, biogeographic patterns can also be taken into consideration (Goodman et al. 1999). Given peripatry and parapatry may have been involved in the evolution of Chesneyinae, if ILS occurred, the main clades would hardly be resolved with well-supported dichotomy as presented herein. Hence, although ILS could not be completely excluded in this case, we regarded chloroplast capture as the most likely cause for the discordant position of Chesneya s.s.
Compared to the biparental inheritance of the nuclear genome, plastid DNA of angiosperms is usually uniparentally transmitted, especially maternally (Corriveau and Coleman 1988; McCauley et al. 2007; Wicke et al. 2011). Nevertheless, the plastid DNA of the inverted repeat lacking clade (IRLC; see Figs 1, 2; also as in Lavin et al. 1990; Wojciechowski et al. 2000) in Leguminosae was reported to be inherited paternally or biparentally (Zhang et al. 2003), confirmed by cytoplasmic and phylogenetic studies focusing on Medicago L. (paternal transmission; Schumann and Hancock 1989; Masoud et al. 1990; Havananda et al. 2010) and Wisteria Nutt. (Hu et al. 2005; Trusty et al. 2007). As Chesneya s.s. belongs to IRLC, a paternal inheritance scenario might be the case for the plastid DNA of Chesneya s.s.
We herein hypothesize a chloroplast capture event in the origin of Chesneya s.s. as follows. The common ancestor of Chesniella served as the putative paternal parent of Chesneya s.s. (sister to Chesneya s.s. in the plastid tree; Fig. 2). The maternal parent most likely was the common ancestor of the GUT clade. Their hybrids, with plastid from the paternal parent, may have continuously backcrossed with the maternal parent, and led to Chesneya s.s. inheriting most of the nuclear genome maternally (Fig. 1). Such a chloroplast capture event via introgression likely took place in the Miocene, because the divergence of Chesneya s.s. was dated to be 16.56 Ma and that of Chesniella was estimated as 19.81 Ma (Zhang et al. 2015b).
Analyses of Zhang et al. (2015b) revealed that the divergence of Chesneya and Chesniella most likely occurred around the QTP. Our analysis further indicated the psychric group of Chesneya diverged first in this genus (Chesneya sect. Pulvinatae, see Discussion below). It is probable that the common ancestor of Chesniella adapted to psychrophytic habitats. However, the extant Chesniella is rarely distributed on the QTP. As for the GUT clade, Gueldenstaedtia possesses a unique chromosome number (x = 7; Yang 2002) within the subtribe. Most species of Gueldenstaedtia are adapted to mesophytic habitats of temperate northern and eastern Asia (Fig. 3A), in contrast to the rest of Chesneyinae, which are psychric or xeric taxa. Such a correlation among the variation of chromosome numbers and adaptation to different habitats has also been recorded in other taxa, such as Hedysarum (Tang 2005; Duan et al. 2015), Passiflora (Hansen et al. 2006) and Amaryllidaceae (García et al. 2014). But the mechanisms of these types of adaptation need to be further explored with robust phylogenetic, ecological and biogeographic analyses in our future efforts.
Phylogeny and treatment of Chesneya, Chesniella and Spongiocarpella
Chesneya is the type genus of Chesneyinae, with ca. 35 species (see Fig. 3B–D). This genus has its distribution from the Himalayan region to northwestern China and Mongolia, through central and western Asia, westward to Turkey and Armenia (Fig. 3A; Borissova 1945; Davis 1970; Yakovlev et al. 1996; Lock and Schrire 2005; Fig. 3A). Our results suggest that the formerly circumscribed Chesneya, which contains two well-supported but separated parts: the core Chesneya s.s. and the outlier Chesneya macrantha (Fig. 3E) (as in Li 1993 & Zhu and Larsen 2010), is not monophyletic (Figs 1, 2). Chesneya spinosa P.C.Li (Fig. 3C) of Chesneya s.s. is morphologically similar to Chesneya macrantha (Li 1981). However, Chesneya spinosa is distributed in southern Tibet, while Chesneya macrantha is restricted to the dry lands of Mongolia and northwestern China (Li and Ni 1985; Fu 1989). They occupy psychrophytic and xerophytic habitats, respectively, and are clearly not sister to each other (Figs 1, 2).
Chesneya macrantha is nested within a monophyletic Chesniella according to our ITS tree (Fig. 1), and in the plastid tree, it is sister to the type of Chesniella: Chesniella ferganensis (Korsh.) Boriss. (Borissova 1964; see Fig. 2, 3F). Chesneya macrantha shows some distinct morphologies from the other species in Chesniella, including its pulvinate habit and persistent leaf rachis (Li 1993), but this species generally share distribution areas, xerophytic habitats, and some synapomorphies, such as membranous stipules, hairy standard and ovate leaflets with cuneate apices, with Chesniella (Li and Ni 1985; Fu 1989; Zhu and Larsen 2010). Therefore, the transfer of Chesneya macrantha to Chesniella is supported by morphological, geographic and phylogenetic evidence (see Taxonomic Treatment). On the other hand, Chesneya was thus re-delimited based on the monophyletic Chesneya s.s.
After its establishment by Lindley (1839), Chesneya was divided into Chesneya sect. Macrocarpon Boriss. and Chesneya sect. Microcarpon Boriss. mainly based on pod morphology (Borissova 1945). The latter was segregated as the genus Chesniella by Borissova (1964), and this treatment was followed by Li (1993) and Zhu and Larsen (2010). Zhang et al. (2015b) informally classified Chesneya into five sections without detailed taxonomic treatment. Not all their sections were monophylytic, and the diagnostic characters and distributions of several sections were overlapping to some extent.
The presently demarcated Chesneya was assigned into three strongly supported sections herein (as in the key of Chesneya proposed by Li 1993; details see Figs 1, 2 and Taxonomic Treatment). Chesneya sect. Macrocarpon possesses non-pulvinate habit, reduced stems, truncate or emarginate leaflet apices and caducous petiole and rachis (Borissova 1945). This section is composed of most species of Chesneya, including the type species: Chesneya rytidosperma Jaub. et Spach (see Fig. 2; Borissova 1945; Davis 1970; Rechinger 1984). Chesneya sect. Macrocarpon was thus treated as Chesneya sect. Chesneya (Fig. 3B). Unlike this section, petioles and rachises of Chesneya sect. Pulvinatae M.L.Zhang (Zhang et al., 2015b; see Fig. 3D) are persistent and pubescent. However, most species in Chesneya sect. Pulvinatae have blackened and curved petioles and rachises, while those of one of its species, Chesneya spinosa, are hardened and spiny. Besides, Chesneya spinosa formed a clade separated from Chesneya sect. Pulvinatae. Hence, it is appropriate to segregate this species to form a new monotypic section: Chesneya sect. Spinosae L.Duan, J.Wen & Zhao Y.Chang (see see Fig. 3C and Taxonomic treatment).
The infra-sectional relationships within Chesneya sect. Chesneya are basically unresolved in our ITS trees (Fig. 1), and this section is undersampled in the plastid trees (Fig. 2). As for Chesneya sect. Pulvinatae, two accessions of Chesneya nubigena (D.Don) Ali formed a clade, being sister to Chesneya purpurea P.C.Li (Figs 1, 2). Based on such well-supported tree topologies and several morphological differences, such as smaller leaflets and purple corollae, the specific status of Chesneya purpurea was retained herein (as in Li 1981, 1993).
The xeric Chesneya sect. Chesneya grows on dry slopes or desert margins of northwestern China, Mongolia and central Asia (see Fig. 3B; Borissova 1945; Rechinger 1984; Lock and Simpson 1991; Yakovlev et al. 1996; Zhu and Larsen 2010). This section is morphologically similar to Chesniella (Fig. 3F) and their distributions are more or less overlapping (Borissova 1945; Li, 1993), whereas they are not phylogenetically close to each other (Figs 1, 2). Such a phenomenon may be due to convergent evolution (Degtjareva et al. 2012). Chesneya sect. Spinosae (Fig. 3C) and Chesneya sect. Pulvinatae (Fig. 3D) are restricted to Tibet and adjacent regions, adapting to high-altitude psychrophytic habitats (Ali 1977; Zhu and Larsen 2010). The evolutionary history of Chesneya appears complex, whereas the elevation of the QTP and the subsequent aridifications may have played an important role (Meng et al. 2015; Zhang et al. 2015b), as in former Calophaca (Zhang et al. 2015a), Caragana (Zhang and Fritsch 2010) and Hedysarum (Shue 1985; Duan et al. 2015).
Most previous workers did not accept the generic status of Chesniella, treating it within Chesneya (Borissova 1945; Li 1981; Rechinger 1984; Zhu and Cao 1986; Fu 1987, 1989; Yakovlev 1988; Yakovlev et al. 1991). Nevertheless, Li (1993) and Zhu and Larsen (2010) stated that the former is distinguishable from the latter by non-reduced stems, membranous stipules, obviously smaller calyxes, flowers and pods. With the inclusion of Chesneya macrantha (Fig. 3E), our results justified the monophyly of Chesniella (Figs 1, 2), consistent with Zhang et al. (2015b). Within Chesniella, two well-supported groups were resolved in our ITS tree (Fig. 1). Chesniella macrantha and Chesneya mongolica (Maxim.) Boriss. constituted group A, the group B included Chesniella ferganensis, Chesneya gracilis Boriss. and Chesneya tribuloides (Nevski.) Boriss. The former confined in Mongolia and Inner Mongolia of China, to the contrast, the latter ranged from northwestern China to central Asia, which implied vicariance caused by Altai Mountain may drive the divergence of these two groups. However, due to undersampling and distinct morphology of Chesneya macrantha in Chesniella, the evolution history and infrageneric taxonomy of this genus needs to be further explored.
Yakovlev and Sviazeva (1987) erected Spongiocarpella as a segregate genus from Chesneya in the light of the former’s spongiose legumes. Such treatment was followed by Yakovlev (1988), Fu (1989) and Yakovlev et al. (1996), but was rejected by Li (1993), Zhu (1996), Qian (1998) and Zhu and Larsen (2010). Based on field and herbarium studies, we concur with Zhu (1996) that the sponge-like pericarp is an unstable character. Additionally, several species formerly assigned to Spongiocarpella were represented in our study, including Chesneya nubigena (D.Don) Ali, Chesneya Spinosa and Chesniella macrantha. They did not form a monophyletic group (Figs 1, 2). Thus, our data do not support the generic status of Spongiocarpella (as in Zhu 1996; Zhu and Larsen 2010; Ranjbar et al. 2014; Zhang et al. 2015b).
Monophyly of Gueldenstaedtia and Tibetia
Gueldenstaedtia is a small genus comprised of ca. 10 species and is distinguished from Chesneya by its palmately nerved wing petals (vs. pinnately in Chesneya) and non-twisted pod valves (vs. twisted) (see Fig. 3G; Liu et al. 2010a). This genus ranges from the Sino-Himalayan region to Mongolia and Siberia (Lock and Schrire 2005; see Fig. 3A). It was established by Fischer (1823) and revised by Fedtschenko (1927), Jacot (1927) and Kitagawa (1936). Ali (1962) divided it into Gueldenstaedtia subg. Gueldenstaedtia and Gueldenstaedtia subg. Tibetia Ali, but the latter was elevated to the generic rank by Tsui (1979) based on characters of stems, stipules, styles and seeds (see Fig. 3H). The genus Tibetia was generally accepted in subsequent revisions (Shue 1992; Yakovlev et al. 1996; Cui 1998; Wu 1999; Zhu 2004, 2005a; Bao and Brach 2010), and it is confined to Tibet and the adjacent regions including southern Gansu, southern Qinghai, western Sichuan and northwestern Yunnan of China, northern India, Nepal and Buhtan (Tsui 1979; Grierson and Long 1987; Lock and Schrire 2005; Zhu 2005a; Bao and Brach 2010).
Gueldenstaedtia and Tibetia were each supported to be monophyletic, and the two genera together form the GUT clade (Figs 1, 2). It seems valid to retain the generic status of each genus, which is also supported by karyological studies (Nie et al. 2002; Yang 2002; Zhu 2005b): Gueldenstaedtia (x = 7) vs. Tibetia (x = 8). Within Gueldenstaedtia, three species were sampled (all belonging to Gueldenstaedtia sect. Gueldenstaedtia according to Tsui 1979), but these species were all treated to be Gueldenstaedtia verna (Georgi) Boriss. s.l. by some workers (Yakovlev 1988; Zhu 2004; Bao and Brach 2010). Further work is needed to test the delimitation of Gueldenstaedtia verna s.l.
Within Tibetia, two accessions of Tibetia himalaica (Baker) H.P.Tsui grouped together, which were sister to Tibetia yadongensis H.P.Tsui (Figs 1, 2). The tree topology and the morphological characters (e.g., elongate stem and round or retuse leaflets apex) seem to be consistent with treating Tibetia himalaica as a distinct species (also see Tsui 1979; Cui 1998; Zhu 2005a; Bao and Brach 2010).
Taxonomic treatment
Caragana
Fabr., Enum. Ed. 2. 421. 1763, emend. nov. L.Duan, J.Wen & Zhao Y.Chang
Calophaca Fisch. ex DC., Prod. 2: 270. 1825, syn. nov. Type: Calophaca wolgarica Fisch., Prod. 2: 270. 1825.
Halimodendron Fisch. ex DC., Prod. 2: 269. 1825, syn. nov. Type: Halimodendron halodendron (Pall.) Druce, Rep. Bot. Soc. Exch. Club Brit. Isles 4: 626. 1917.
Type.
Caragana arborescens Lam., Encycl. 1(2): 615. 1785.
Description.
Shrubs, subshrubs or rarely small trees. Stipules caducous or persistent. Leaves paripinnate, rarely imparipinnate (Chesneya sect. Calophaca), 4–27-foliolate; leaflet blades with margin entire. Lax raceme or fascicled flowers axillary, or flowers solitary. Calyx tubular or campanulate, base usually oblique, teeth 5. Corolla yellow, purple, pink or white; standard ovate to suborbicular, clawed or reflexed at margin; wings and keel often auriculate. Stamens diadelphous (9+1). Ovary sessile to stipitate, with ovule 1-many; style filiform. Pod inflated, compressed, cylindric or linear, dehiscent or rarely indehiscent (Chesneya sect. Halimodendron), with twisted or thickened valve.
Distribution and habitat.
This genus contains ca. 100 species, ranging from eastern Europe, Caucasus, western and central Asia, Sino-Himalayan region to Mongolia and Siberia.
Caragana sect. Calophaca
(Fisch. ex DC.) L.Duan, J.Wen & Zhao Y.Chang stat. & comb. nov.
urn:lsid:ipni.org:names:77157989-1
Calophaca Fisch. ex DC., Prod. 2: 270. 1825. Type: Calophaca wolgarica Fisch., Prod. 2: 270. 1825.
Distribution and habitat.
This section includes 5–8 species, distributed in Caucasus, central Asia, northwestern Xinjaing, Innner Mongolia and Shanxi of China.
Caragana sect. Halimodendron
(Fisch .ex DC.) L.Duan, J.Wen & Zhao Y.Chang stat. & comb. nov.
urn:lsid:ipni.org:names:77157990-1
Halimodendron Fisch. ex DC., Prod. 2: 269. 1825. Type: Halimodendron halodendron (Pall.) Druce, Rep. Bot. Soc. Exch. Club Brit. Isles 4: 626. 1917.
Type.
Caragana halodendron (Pallas) Dumont de Courset, Bot. Cult. 3: 513. 1802.
Distribution and habitat.
This section is monotypic and distributes in Caucasus, northeastern Turkey, northern Iran, northern Afghanistan, northern Pakistan, central Asia, western Mongolia, Shanxi and Xinjiang of China.
Key to the sections of Caragana
| 1 | Leaves imparipinnate; ovary sessile | Caragana sect. Calophaca |
| – | Leaves paripinnate; ovary subsessile or stipitate | 2 |
| 2 | Racemose; pedicel non-articulate; pods inflated, indehiscent, valve thickened; seeds few | Caragana sect. Halimodendron |
| – | 2–5 flowers in fascicles, or solitary flower; pedicel articulate; pods compressed, cylindric or linear, dehiscent, valve twisted; seeds many | 3 |
| 3 | Petiole and rachis always caducous; leaves pinnate | Caragana sect. Caragana |
| – | Petiole and rachis persistent, usually spinelike; leaves pinnate or digitate | 4 |
| 4 | Leave digitate | Caragana sect. Frutescentes |
| – | Leave pinnate or partly digitate | 5 |
| 5 | Leave digitate or pinnate with 4 leaflets on short branchlets, leave pinnate on long branchlets | Caragana sect. Spinosae |
| – | Leaves pinnate | 6 |
| 6 | Petiole and rachis persistent | Caragana sect. Jubatae |
| – | Petiole and rachis persistent on long branchlets, caducous on short branchlets | Caragana sect. Bracteolatae |
Chesneya
Lindl. ex Endl., Gen.: 1275. 1840.
Spongiocarpella Yakovlev & N.Ulziykhutag, Bot. Zhur. 17(2): 249. 1987. syn. nov. Type: Spongiocarpella nubigena (D.Don) Yakovl., Bot. Zhur. 17(2): 249. 1987, based on Chesneya nubigena (D.Don) Ali. (see blow)
Type.
Chesneya rytidosperma Jaub. et Spach, Ill. Pl. Orient. 1(5): 93. 1842.
Chesneya sect. Chesneya
Chesneya sect. Macrocarpon Boriss., Fl. U.S.S.R. 11: 280. 1945. syn. nov. Type: Chesneya rytidosperma Jaub. et Spach, Ill. Pl. Orient. 1(5): 93. 1842.
Description, distribution and habitat.
This section includes the majority of Chesneya species. It can be diagnosed by reduced stems and caducous petiole and rachis. It contains ca. 20 xeric species, ranging from desert and dry slope of northwestern China and western Tibet to central and western Asia and Caucasus.
Chesneya sect. Pulvinatae
M.L.Zhang, Biochem. Syst. Ecol. 63: 89. 2015.
Spongiocarpella Yakovlev & N. Ulziykhutag, Bot. Zhur. 17(2): 249. 1987. Type: Spongiocarpella nubigena (D.Don) Yakovl., Bot. Zhur. 17(2): 249. 1987.
Type.
Chesneya nubigena (D.Don) Ali, Scientist (Karachi) iii: 4. 1959.
Description, distribution and habitat.
This psychric section is composed of Chesneya nubigena, Chesneya polystichoides (Hand.-Mazz.) Ali and Chesneya purpurea. It differs from other sections by blackened, curved and non-spiny petiole and rachis, distributed on high-altitude slope in eastern Himalayas and southern and eastern Tibet.
Chesneya sect. Spinosae
L.Duan, J.Wen & Zhao Y.Chang sect. nov.
urn:lsid:ipni.org:names:77157991-1
Type.
Chesneya spinosa P.C.Li, Acta Phytotax. Sin. 19(2): 236. 1981.
Description, distribution and habitat.
This monotypic section is recognized by its hardened-spiny petiole and rachis. It is restricted in high-altitude psychrophytic rocky slope in southern Tibet.
Key to the sections of Chesneya
| 1 | Plant non-pulvinate, petiole and rachis caducous, leaflet apices truncate or emarginate | Chesneya sect. Chesneya |
| – | Plant pulvinate, petiole and rachis persistent, leaflet apices acute | 2 |
| 2 | Petiole and rachis hardened and spiny, leaflet apices with short spines | Chesneya sect. Spinosae |
| – | Petiole and rachis blackened and curved, leaflet apices without short spines | Chesneya sect. Pulvinatae |
Chesniella macrantha
(Cheng f. ex H.C.Fu) L.Duan, J.Wen & Zhao Y.Chang comb. nov.
urn:lsid:ipni.org:names:77157988-1
Chesneya macrantha Cheng f. ex H.C.Fu, Fl. Intramongol. 3: 291. 1977.
Note.
Information of the type specimen was not included in its protolog, which was recorded in Acta Phytotax. Sin. 19(2): 237. 1981: China. Inner Mongolia: Baganmao, 29 May 1931, T.N.Liou 2146 (holotype: PE!).
Specimens examined.
CHINA. Ningxia: Mt. Helan, 1200m, May 15 1923, R.C.Ching 108 (US); Inner Mongolia: Alasan Left Banner, Xiazi valley, 24 Apr 2009, Z.Y.Chang et al. 2009054 (WUK); Mt. Yabulai, Agui temple, 1300m, Apr 26 2008, L.R.Xu 2008008 (WUK); Xinjiang: Qomul, 43° 05.330’N, 93° 42.030’E, 1311m, 6 Jun 2004, Z.Y.Chang et al. 2004516 (WUK).
Distribution and habitat.
Dry slopes in Mongolia and Inner Mongolia, Ningxia and Xinjiang of China.
Supplementary Material
Acknowledgement
We are grateful to the curators of the following herbaria for providing leaf samples, seeds or DNA samples: CS, F, K, TURP, US, WUK and XJBI. We thank Matthew Kweskin, Mingqin Zhou, and Jianqiang Zhang for lab assistance; Natasha Crump for Russian literature translation; Robin Everly for literature search; and Martin F. Wojciechowski, Ying Feng, Shimin Duan, Irina Illarionova and Roman Ufimov for checking voucher information. The Laboratories of Analytical Biology of the National Museum of Natural History (Smithsonian Institution), the Natural Science Foundation of China (Project no. 30270106, 30870155) and the China Scholarship Council are gratefully acknowledged for financial support of the study.
Citation
Duan L, Yang X, Liu P, Johnson G, Wen J, Chang Z (2016) A molecular phylogeny of Caraganeae (Leguminosae, Papilionoideae) reveals insights into new generic and infrageneric delimitations. PhytoKeys 70: 111–137. doi: 10.3897/phytokeys.70.9641
Supplementary materials
Voucher information and GenBank accession numbers
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Lei Duan, Xue Yang, Peiliang Liu, Gabriel Johnson, Jun Wen, Zhaoyang Chang
Data type: Multi-records
Explanation note: Data are arranged in the order: taxon name, locality, collector(s), collection number and herbarium, GenBank accession numbers for ITS, matK, trnL-trnF, psbA-trnH. Newly generated sequences are indicated by an asterisk (*); missing sequences are indicated by a dash (–).
References
- Ali SI. (1959) Chesneya nubigena. Scientist (Karachi: ) 3: 4. [Google Scholar]
- Ali SI. (1962) A taxonomic revision of the genus Gueldenstaedtia Fisch. Candollea 18: 137–159. [Google Scholar]
- Ali SI. (1977) Gueldenstaedtia. In: Nasir E, Ali SI. (Eds) Flora of West Pakistan. University of Karachi, Karachi, 100: 214. [Google Scholar]
- Amirahmadi A, Osaloo SK, Moein F, Kaveh A, Maassoumi AA. (2014) Molecular systematics of the tribe Hedysareae (Fabaceae) based on nrDNA ITS and plastid trnL-F and matK sequences. Plant Systematics and Evolution 300: 729–747. doi: 10.1007/s00606-013-0916-5 [Google Scholar]
- Bao BJ, Brach AR. (2010) Gueldenstaedtia and Tibetia. In: Wu ZY, Hong DY, Raven PH. (Eds) Flora of China, Vol. 10 Science Press, Beijing: & Missouri Botanical Garden Press, St Louis, 506–509. [Google Scholar]
- Borissova AG. (1945) Calophaca, Chesneya and Gueldenstaedtia. In: Komarov VL. (Ed.) Flora of U.S.S.R., Vol. 11 Israel Program for Scientific Translantions, Jerusalem, 275–288. [Google Scholar]
- Borissova AG. (1964) De genere Chesneya Lindl. et genere novo Chesniella Boriss. (Familia Leguminosae) notula. Novosti Sistematiki Nizshikh Rastenii, 178–190.
- Cardoso D, Pennington RT, de Queiroz LP, Boatwright JS, Van Wyk BE, wojciechowski MF, Lavin M. (2013) Reconstructing the deep-branching relationships of the papilionoid legume. South African Journal of Botany 89: 58–75. doi: 10.1016/j.sajb.2013.05.001 [Google Scholar]
- CBOL Plant Working Group (2009) A DNA barcode for land plants. Proceedings of the National Academy of Sciences of the United States of America 106: 12794–12797. doi: 10.1073/pnas.0905845106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CW. (1993) Calophaca. In: Fu KT. (Ed.) Flora Reipublicae Popularis Sinicae, vol. 42(1). Science Press, Beijing, 67–71. [Google Scholar]
- Chang ZY. (2008) A taxonomical study of Caragana Fabr. from China. Ph.D. Thesis, Northeast Forestry University, China. [Google Scholar]
- Cheng F. (1977) Chesneya marantha. In: Fu HC. (Ed.) Flora Chinae Intramongolicae, Tom. 3 Inner Mongolian People’s Publishing House, Huhhot, 291. [Google Scholar]
- China Plant BOL Group (2011) Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proceedings of the National Academy of Sciences of the United States of America 108: 19641–19646. doi: 10.1073/pnas.1104551108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corriveau JL, Coleman AW. (1988) Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. American Journal of Botany 75: 1443–1458. doi: 10.2307/2444695 [Google Scholar]
- Cui HB. (1998) Gueldenstaedtia and Tibetia. In: Cui HB. (Ed.) Flora Reipublicae Popularis Sinicae, vol. 42(2). Science Press, Beijing, 146–163. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. doi: 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis PH. (1970) Flora of Turkey and the East Aegean Islands, vol. 3 Edingburgh University Press, Edingburgh, 47–48. [Google Scholar]
- Degtjareva GV, Valiejo-Roman CM, Samigullin TH, Guara-Requena M, Sokoloff DD. (2012) Phylogenetics of Anthyllis (Leguminosae: Papilionoideae: Loteae): partial incongruence between nuclear and plastid markers, a long branch problem and implications for morphological evolution. Molecular Phylogenetics and Evolution 62: 693–707. doi: 10.1016/j.ympev.2011.11.010 [DOI] [PubMed] [Google Scholar]
- Deng T, Nie ZL, Drew BT, Volis S, Kim C, Xiang CL, Zhang JW, Wang YH, Sun H. (2015) Does the Arcto-Tertiary biogeographic hypothesis explain the disjunct distribution of northern hemisphere herbaceous plants? the case of Meehania (Lamiaceae). PLoS ONE 10(2). doi: 10.1371/journal.pone.0117171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druce GC. (1917) Halimodendron halodendron. (Report) Botanical Society and Exchange Club of the British Isles 4: 626. [Google Scholar]
- Duan L, Wen J, Yang X, Liu PL, Arslan E, Ertuğrul K, Chang ZY. (2015) Phylogeny of Hedysarum and tribe Hedysareae (Leguminosae: Papilionoideae) inferred from sequence data of ITS, matK, trnL-F and psbA-trnH. Taxon 64: 49–64. doi: 10.12705/641.26 [Google Scholar]
- Dumont de Courset GLM. (1802) Caragana halodendrum. Le Botaniste Cultivateur 3: 513. [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. doi: 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison NW, Liston A, Steiner JJ, Williams WM, Taylor NL. (2006) Molecular phylogenetics of the clover genus (Trifolium-Leguminosae). Molecular Phylogenetics and Evolution 39: 688–705. doi: 10.1016/j.ympev.2006.01.004 [DOI] [PubMed] [Google Scholar]
- Fabricius PC. (1763) Caragana. Enumeratio Methodica Plantarum, ed 2 Helmstad Co., Paris, 421. [Google Scholar]
- Fedtschenko BA. (1927) Revision préliminaire du genre “Güldenstaedtia” Fisch. Nuovo Giornale Botanico Italiano 2(34): 1435–1442. [Google Scholar]
- Fischer FE. (1823) Gueldenstaedtia. Mémoires de la Société des Naturalistes de Moscou 6: 170. [Google Scholar]
- Fischer FE. (1825) Caophaca and Halimodendron. Prodromus Systematis Naturalis Regni Vegetabilis. Treuttel & Würtz, Paris, 269–270. [Google Scholar]
- Fu HC. (1987) Chesneya. In: Liou YX. (Ed.) Flora in Desertis Reipublicae Populorum Sinarum, Tom. 2 Science Press, Beijing, 230–232. [Google Scholar]
- Fu HC. (1989) Chesneya, Gueldenstaedtia and Spongiocarpella. In: Ma YC. (Ed.) Flora Intramongolica, vol. 3 Typis Intramongolicae Popularis, Huhhot, 238–245. [Google Scholar]
- Gao T, Sun Z, Yao H, Song J, Zhu Y, Ma X, Chen S. (2011) Identification of Fabaceae plants using the DNA barcode matK. Planta Medica 77: 92–94. doi: 10.1055/s-0030-1250050 [DOI] [PubMed] [Google Scholar]
- García N, Meerow AW, Soltis DE, Soltis PS. (2014) Testing deep reticulate evolution in Amaryllidaceae tribe Hippeastreae (Asparagales) with ITS and chloroplast sequence data. Systematic Botany 39: 75–89. doi: 10.1600/036364414X678099 [Google Scholar]
- Goodman SJ, Barton NH, Swanson G, Abernethy K, Pemberton JM. (1999) Introgression through rare hybridization: a genetic study of a hybrid zone between red and sika deer (genus Cervus) in Argyll, Scotland. Genetics 152: 355–371. http://www.genetics.org/content/152/1/355.short [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova N. (1984) De Generis Caragana Lam. (Fabaceae) Notae Systematicae. Novosti Sistematiki Vysshikh Rastenii 21: 92–101. [Google Scholar]
- Gorshkova SG. (1945) Halimodendron. In: Komarov VL. (Ed.) Flora of U.S.S.R. Israel Program for Scientific Translantions, Jerusalem, 242–244. [Google Scholar]
- Grierson AJC, Long DG. (1987) Gueldenstaedtia. In: Grierson AJC, Long DG. (Eds) Flora of Buhtan, vol. 1(3). Royal Botanic Garden, Edinburgh, 722–723. [Google Scholar]
- Hamilton M. (1999) Four primer pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Molecular Ecology 8: 521–523. [PubMed] [Google Scholar]
- Hansen AK, Gilbert LE, Simpson BB, Downie SR, Cervi AC, Jansen RK. (2006) Phylogenetic relationships and chromosome number evolution in Passiflora. Systematic Botany 31: 138–150. doi: 10.1600/036364406775971769 [Google Scholar]
- Havananda T, Brummer EC, Maureira-Butler IJ, Doyle JJ. (2010) Relationships among diploid members of the Medicago sativa (Fabaceae) species complex based on chloroplast and mitochondrial DNA Sequences. Systematic Botany 35: 140–150. doi: 10.1600/036364410790862506 [Google Scholar]
- Hou X, Liu JE, Zhao YZ. (2008) Molecular phylogeny of Caragana (Fabaceae) in China. Journal of Systematics and Evolution 46: 600–607. doi: 10.3724/SP.J.1002.2008.07071 [Google Scholar]
- Hu JM, Lavin M, Wojciechowski MF, Sanderson MJ. (2000) Phylogenetic systematics of the tribe Millettieae (Leguminosae) based on chloroplast trnK/matK sequences and its implications for evolutionary patterns in Papilionoideae. American Journal of Botany 87: 418–430. doi: 10.2307/2656638 [PubMed] [Google Scholar]
- Hu YF, Zhang Q, Sodmergen (2005) Potential cytoplasmic inheritance in Wisteria sinensis and Robinia pseudoacacia (Leguminosae). Plant and Cell Physiology 46: 1029–1035. doi: 10.1093/pcp/pci110 [DOI] [PubMed] [Google Scholar]
- Jacot AP. (1927) The genus Gueldenstaedtia (Leguminosae). Journal of the North China Branch of the Royal Asiatic Society 58: 85–126. [Google Scholar]
- Jaubert HF, Spach É. (1842) Chesneya rytidosperma. Illustrationes Plantarum Orientalium 1(5): 93. [Google Scholar]
- Kitagawa M. (1936) Amblytropis. In: Nakai T, Honda M, Satake Y, Kitagawa M. (Eds) Report of the first scientific expedition to Manchouko, sect. 4. Under the leadership of Shingeyasu Tokunaga, Japan, 87. [Google Scholar]
- Komarov VL. (1908) Generis Caraganae Monographia. Acta Horti Petropolitani 29: 177–388. [Google Scholar]
- Lanarck J-B. (1785) Caragana arborescens. Encyclopédie Méthodique, Botanique 1(2): 615. [Google Scholar]
- Lavin M, Doyle JJ, Palmer JD. (1990) Evolutionary significance of the loss of the chloroplast-DNA inverted repeat in the Leguminosae subfamily Papilionoideae. Evolution 44: 390–402. doi: 10.2307/2409416 [DOI] [PubMed] [Google Scholar]
- Li PC. (1981) Note on Chinese Chesneya. Acta phytotaxonomica Sinica 19(2): 235–237. http://www.jse.ac.cn/wenzhang/FL19-2-11.pdf [Google Scholar]
- Li PC. (1993) Chesneya, Chesniella and Halimodendron. In: Fu KT. (Ed.) Flora Reipublicae Popularis Sinicae, vol. 42(1). Science Press, Beijing, 71–78. [Google Scholar]
- Li PC, Ni CC. (1985) Chesneya and Gueldenstaedtia. In: Wu CY. (Ed.) Flora Xizangica, vol. 2 Science Press, Beijing, 789–797. [Google Scholar]
- Lindley J. (1839) Chesneya. In: Endlicher S. (Ed.) Genera Plantarum Secundum Ordines Naturales Disposita. UD Fr. Beck University Bibliopolam, Vindobonae, 1275. [Google Scholar]
- Liou YX. (1993) Caragana. In: Fu KT. (Ed.) Flora Reipublicae Popularis Sinicae, vol. 42(1). Science Press, Beijing, 13–67. [Google Scholar]
- Liu YX, Chang ZY, Yakovlev GP. (2010a) Caragana. In: Wu ZY, Hong DY, Raven PH. (Eds) Flora of China, vol. 10 Science Press, Beijing: & Missouri Botanical Garden Press, St Louis, 528–545. [Google Scholar]
- Liu YX, Xu LR, Chang ZY, Zhu XY, Sun H, Yakovlev GP, Choi BH, Larsen K, Bartholomew B. (2010b) Tribe Hedysareae. In: Wu ZY, Hong DY, Raven PH. (Eds) Flora of China, vol. 10 Science Press, Beijing: & Missouri Botanical Garden Press, St Louis, 512–545. [Google Scholar]
- Lock JM. (2005) Hedysareae. In: Lewis G, Schrire B, Mackinder B, Lock M. (Eds) Legumes of the World. Royal Botanic Garden, Kew, 489–495. [Google Scholar]
- Lock JM, Schrire BD. (2005) Galegeae. In: Lewis G, Schrire B, Mackinder B, Lock M. (Eds) Legumes of the World. Royal Botanic Garden, Kew, 475–487. [Google Scholar]
- Lock JM, Simpson K. (1991) Legumes of West Asia: a checklist. Royal Botanic Gardens, Kew, 133–134. [Google Scholar]
- Masoud SA, Johnson LB, Sorensen EL. (1990) High transmission of paternal plastid DNA in alfalfa plants demonstrated by restriction fragment polymorphic analysis. Theoretical and Applied Genetics 79: 49–55. doi: 10.1007/BF00223786 [DOI] [PubMed] [Google Scholar]
- Mau B, Newton MA, Larget B. (1999) Bayesian phylogenetic inference via Markov chain Monte Carlo methods. Biometrics 55: 1–12. doi: 10.1111/j.0006-341X.1999.00001.x [DOI] [PubMed] [Google Scholar]
- McCauley DE, Sundby AK, Bailey MF, Welch ME. (2007) Inheritance of chloroplast DNA is not strictly maternal in Silene vulgaris (Caryophyllaceae) evidence from experimental crosses and natural populations. American Journal of Botany 94: 1333–1337. doi: 10.3732/ajb.94.8.1333 [DOI] [PubMed] [Google Scholar]
- Meng HH, Gao XY, Huang JF, Zhang ML. (2015) Plant phylogeography in arid Northwest China: Retrospectives and perspectives. Journal of Systematics and Evolution 53: 33–46. doi: 10.1111/jse.12088 [Google Scholar]
- Moore RJ. (1968) Chromosome number and phylogeny in Caragana (Leguminosae). Canadian Journal of Botany 46: 1513–1522. doi: 10.1139/b68-209 [Google Scholar]
- Nazar RN, Wildeman AG. (1981) Altered features in the secondary structure of Vicia faba 5.8S rRNA. Nucleic Acids Research 9: 5345–5358. doi: 10.1093/nar/9.20.5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie ZL, Gu ZJ, Sun H. (2002) Cytological study of Tibetia (Fabaceae) in the Hengduan Mountains region, China. Journal of Plant Research 115: 17–22. doi: 10.1007/s102650200003 [DOI] [PubMed] [Google Scholar]
- Osaloo SK, Kazemi NM, Maassoumi AA, Pouyani ER. (2006) Phylogenetic status of Oreophysa microphylla (Fabaceae-Galegeae) based on nrDNA (ITS region) and cpDNA (trnL intron/trnL-trnF intergenic spacer) sequences. Rostaniha 7: 177–188. http://en.journals.sid.ir/ViewPaper.aspx?ID=93558 [Google Scholar]
- Osaloo SK, Maassoumi AA, Murakami N. (2003) Molecular systematics of the genus Astragalus L. (Fabaceae): Phylogenetic analyses of nuclear ribosomal DNA internal transcribed spacers and chloroplast gene ndhF sequences. Plant Systematics and Evolution 242: 1–32. doi: 10.1007/s00606-003-0014-1 [Google Scholar]
- Polhill RM. (1981) Carmichaelieae, Galegeae and Hedysareae. In: Polhill RM, Raven PH. (Eds) Advances in Legume Systematics, part. 1 Royal Botanic Garden, Kew, 357–370. [Google Scholar]
- Posada D. (2008) jModelTest: Phylogenetic model averaging. Molecular Biology and Evolution 25: 1253–1256. doi: 10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- Poyarkova AI. (1945) Caragana. In: Komarov VL. (Ed.) Flora of U.S.S.R., Vol. 11 Israel Program for Scientific Translantions, Jerusalem, 244–275. [Google Scholar]
- Qian ZG. (1998) A revision of the genus Chesneya from East Himalayas. Acta Botanica Yunnanica 20(4): 399–402. http://en.cnki.com.cn/Article_en/CJFDTOTAL-YOKE804.003.htm [Google Scholar]
- Rambaut A, Drummond AJ. (2007) Tracer v1.5. http://beast.bio.ed.ac.uk/Tracer
- Ranjbar M, Hajmoradi F, Waycott M, Van Dijk KJ. (2014) A phylogeny of the tribe Caraganeae (Fabaceae) based on DNA sequence data from ITS. Feddes Repertorium 125: 1–7. doi: 10.1002/fedr.201400051 [Google Scholar]
- Ranjbar M, Karamian R. (2003) Caraganeae, a new tribe with notes on the genus Chesneya Lindl. ex Endl. (Fabaceae) from flora of Iran. Thaiszia Journal of Botany 13: 67–75. http://www.bzupjs.sk/thaiszia/index.html [Google Scholar]
- Rannala B, Yang Z. (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. Journal of Molecular Evolution 43: 304–311. doi: 10.1007/BF02338839 [DOI] [PubMed] [Google Scholar]
- Rechinger KH. (1984) Flora Iranica, No. 147. Akademische Drucku, Verlagsanstalt, Graz, 88–100. [Google Scholar]
- Riahi M, Zarre S, Maassoumi AA, Osaloo SK, Wojciechowski MF. (2011) Towards a phylogeny for Astragalus section Caprini (Fabaceae) and its allies based on nuclear and plastid DNA sequences. Plant Systematics and Evolution 293: 119–133. doi: 10.1007/s00606-011-0417-3 [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. doi: 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. doi: 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchir C. (1999) System of the genus Caragana Lam. (Fabaceae). Acta Scientiarum Naturalium Universitatis NeiMongol 30: 501–512. http://en.cnki.com.cn/Article_en/CJFDTOTAL-NMGX904.018.htm [Google Scholar]
- Sanczir C. (1979) Genus Caragana Lam. Systematic, geography and economic significance in study on flora and vegetation of P. R. Mongolia, vol. 1 Academic Press, Ulan-Bator, 232–233. [Google Scholar]
- Sanczir C. (2000) Systema generis Caragana Lam. (Fabaceae). Novosti Sistematiki Vysshikh Rastenii 32: 76–90. [Google Scholar]
- Sanderson MJ, Wojciechowski MF. (1996) Diversification rates in a temperate legume clade: Are there ‘’so many species’’ of Astragalus (Fabaceae)? American Journal of Botany 83: 1488–1502. doi: 10.2307/2446103 [Google Scholar]
- Schumann CM, Hancock JF. (1989) Paternal inheritance of plastids in Medicago sativa. Theoretical and Applied Genetics 78: 863–866. doi: 10.1007/BF00266672 [DOI] [PubMed] [Google Scholar]
- Sepet H, Özdemir C, Bozdağ B. (2011) Cytological study the genus Chesneya Lindl. (Fabaceae) in Turkey. Caryologia 64: 184–188. doi: 10.1080/00087114.2002.10589782 [Google Scholar]
- Shimodaira H, Hasegawa M. (2001) CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17: 1246–1247. doi: 10.1093/bioinformatics/17.12.1246 [DOI] [PubMed] [Google Scholar]
- Shue LZ. (1985) The ecological differentiation of the Hedysarum L. and geographical distribution in China. Acta Botanica Boreali-Occidentalia Sinica 4: 275–285. http://en.cnki.com.cn/Article_en/CJFDTotal-DNYX198504004.htm [Google Scholar]
- Shue LZ. (1992) Gueldenstaedtia and Tibetia. In: Fu KT. (Ed.) Flora Loess-Plateaus Sinicae, vol. 2 China Forestry Publishing House, Beijing, 378–381. [Google Scholar]
- Shue LZ, Hao XY. (1989) Studies in the taxonomy and their floristic geography of Caragana Fabr. (Leguminosae) in Loess Plateau and Qinling mountains. Acta Botanica Boreali-Occidentalia Sinica 9: 92–101. http://en.cnki.com.cn/Article_en/CJFDTOTAL-DNYX198902005.htm [Google Scholar]
- Soejima A, Wen J. (2006) Phylogenetic analysis of the grape family (Vitaceae) based on three chloroplast markers. American Journal of Botany 93: 278–287. doi: 10.3732/ajb.93.2.278 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. doi: 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. (2008) A rapid bootstrap algorithm for the RAxML Web servers. Systematic Biology 57: 758–771. doi: 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- Stanford AM, Harden R, Parks CR. (2000) Phylogeny and biogeography of Juglans (Juglandaceae) based on matK and ITS sequence data. American Journal of Botany 87: 872–882. doi: 10.2307/2656895 [PubMed] [Google Scholar]
- Sun M, Soltis DE, Soltis PS, Zhu XY, Burleigh JG, Chen ZD. (2015) Deep phylogenetic incongruence in the angiosperm clade Rosidae. Molecular Phylogenetics and Evolution 83: 156–166. doi: 10.1016/j.ympev.2014.11.003 [DOI] [PubMed] [Google Scholar]
- Swofford DL. (2003) PAUP*. Phylogenetic analysis using parsimony (* and other methods), version 4.0. Sinauer, Sunderland, Massachusetts. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology 17: 1105–1109. doi: 10.1007/BF00037152 [DOI] [PubMed] [Google Scholar]
- Tang HL. (2005) Studies on systematics of the genus Hedysarum L. from China. M.S. Thesis, Northwest A&F University, China. [Google Scholar]
- Trusty JL, Johnson KJ, Lockaby BG, Goertzen LR. (2007) Bi-parental cytoplasmic DNA inheritance in Wisteria (Fabaceae): evidence from a natural experiment. Plant and Cell Physiology 48: 662–665. doi: 10.1093/pcp/pcm036 [DOI] [PubMed] [Google Scholar]
- Tsitrone A, Kirkpatrick M, Levin DA. (2003) A model for chloroplast capture. Evolution 57: 1776–1782. doi: 10.1111/j.0014-3820.2003.tb00585.x [DOI] [PubMed] [Google Scholar]
- Tsui HP. (1979) Revisio Gueldenstaestiae Sinensis et Genus Novum affine Eae - Tibetia (Ali) H.P.Tsui. Bulletin of Botanical Laboratory of North-Eastern Forestry Institute 5: 31–55. [Google Scholar]
- Tutin TG, Heywood VH, Burges NA, Moore DM, Valentine DH, Walters SM, Webb DA. (1968) Flora Europaea, vol. 2 Cambridge University Press, Cambridge, 108. [Google Scholar]
- Wen J, Nie ZL, Soejima A, Meng Y. (2007) Phylogeny of Vitaceae based on the nuclear GAI1 gene sequences. Canadian Journal of Botany 85: 731–745. doi: 10.1139/B07-071 [Google Scholar]
- Wen J, Zhang JQ, Nie ZL, Zhong Y, Sun H. (2014) Evolutionary diversifications of plants on the Qinghai-Tibetan Plateau. Frontiers in Genetics 5: . doi: 10.3389/fgene.2014.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, Schneeweiss GM, de Pamphilis CW, Müller KF, Quandt D. (2011) The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Molecular Biology 76: 273–297. doi: 10.1007/s11103-011-9762-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowski MF. (2003) Reconstructing the phylogeny of legumes (Leguminosae): An early 21st century perspective. In: Klitgaard BB, Bruneau A. (Eds) Advances in legume systematics, part 10 Royal Botanic Gardens, Kew, 5–35. http://www.public.asu.edu/~mfwojci/pdfs/WojciechowskiALS10.pdf [Google Scholar]
- Wojciechowski MF, Sanderson MJ, Steele SP, Liston A. (2000) Molecular phylogeny of the “Temperate Herbaceous Tribes” of Papilionoid legumes: a supertree approach. In: Herendeen PS, Bruneau A. (Eds) Advances in legume systematics, part 9 Royal Botanic Gardens, Kew, 277–298. http://www.public.asu.edu/~mfwojci/pdfs/WojoALS2000.pdf [Google Scholar]
- Wojciechowski MF, Lavin M, Sanderson MJ. (2004) A phylogeny of legumes (Leguminosae) based on analyses of the plastid matK gene resolves many well-supported subclades within the family. American Journal of Botany 91: 1846–1862. doi: 10.3732/ajb.91.11.1846 [DOI] [PubMed] [Google Scholar]
- Wu ZL. (1999) Gueldenstaedtia & Tibetia. In: Liu SW. (Ed.) Flora Qinghaiica, vol. 2 Qinghai People’s Publishing House, Xining, 230–233. [Google Scholar]
- Xu B, Wu N, Gao XF, Zhang LB. (2012) Analysis of DNA sequences of six chloroplast and nuclear genes suggests incongruence, introgression, and incomplete lineage sorting in the evolution of Lespedeza (Fabaceae). Molecular Phylogenetics and Evolution 62: 346–358. doi: 10.1016/j.ympev.2011.10.007 [DOI] [PubMed] [Google Scholar]
- Yakovlev GP. (1988) Plantae Asiae Centralis, Fasciculus 8a Chemico-Pharmaceutic Acad., St. Petersburg, 44–45. [Google Scholar]
- Yakovlev GP, Sviazeva OA. (1987) On the taxonomy of the new genus Spongiocarpella (Fabaceae). Botanicheskii Zhurnal 72: 249–260. [Google Scholar]
- Yakovlev GP, Sytin AK, Roskov YR. (1996) Legumes of Northern Eurasia: a checklist. Royal Botanic Gardens, Kew, 280–284, 293, 361. [Google Scholar]
- Yang DK. (2002) The karyotype analysis of Gueldenstaedtia from Shandong. Guihaia 22: 349–351. http://www.cqvip.com/qk/90987x/200204/6597629.html [Google Scholar]
- Yi TS, Jin GH, Wen J. (2015) Chloroplast capture and intra- and inter-continental biogeographic diversification in the Asian-New World disjunct plant genus Osmorhiza (Apiaceae). Molecular Phylogenetics and Evolution 85: 10–21. doi: 10.1016/j.ympev.2014.09.028 [DOI] [PubMed] [Google Scholar]
- Yokota Y, Kawata T, Iida Y, Kato A, Tanifuji S. (1989) Nucleotide sequences of the 5.8S rRNA gene and internal transcribed spacer regions in carrot and broad bean ribosomal DNA. Journal of Molecular Evolution 29: 294–301. doi: 10.1007/BF02103617 [DOI] [PubMed] [Google Scholar]
- Zhang ML. (1997) A reconstructing phylogeny in Caragana (Fabaceae). Acta Botanica Yunnanica 19: 331–341. http://journal.kib.ac.cn/ynzwyj/CN/article/downloadArticleFile.do?attachType=PDF&id=1326 [Google Scholar]
- Zhang ML. (1998) A preliminary analytic biogeography in Caragana (Fabaceae). Acta Botanica Yunnanica 20: 1–11. http://en.cnki.com.cn/Article_en/CJFDTOTAL-YOKE801.000.htm [Google Scholar]
- Zhang ML, Wang CR. (1993) Phenetic, cladistic and biogeographic studies on the genus Caragana in Qinling mountain and the Loess Plateau. Acta Botanica Yunnanica 15: 332–338. http://en.cnki.com.cn/Article_en/CJFDTOTAL-YOKE199304002.htm [Google Scholar]
- Zhang ML, Fritsch PW. (2010) Evolutionary response of Caragana (Fabaceae) to Qinghai-Tibetan Plateau uplift and Asian interior aridification. Plant Systematics and Evolution 288: 191–199. doi: 10.1007/s00606-010-0324-z [Google Scholar]
- Zhang ML, Fritsch PW, Cruz BC. (2009) Phylogeny of Caragana (Fabaceae) based on DNA sequence data from rbcL, trnS-trnG, and ITS. Molecular Phylogenetics and Evolution 50: 547–559. doi: 10.1016/j.ympev.2008.12.001 [DOI] [PubMed] [Google Scholar]
- Zhang ML, Wen ZB, Fritsch PW, Sanderson SC. (2015a) Spatiotemporal evolution of Calophaca (Fabaceae) reveals multiple dispersals in central Asian mountains. PLoS ONE 10(4). doi: 10.1371/journal.pone.0123228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ML, Wen ZB, Hao XL, Byalt VV, Sukhorukov AP, Sanderson SC. (2015b) Taxonomy, phylogenetics and biogeography of Chesneya (Fabaceae), evidenced from data of three sequences, ITS, trnS-trnG, and rbcL. Biochemical Systematics and Ecology 63: 80–89. doi: 10.1016/j.bse.2015.09.017 [Google Scholar]
- Zhao YZ. (1993) Taxonomical study of the genus Caragana from China. Acta Scientiarum Naturalium Universitatis NeiMongol 24: 631–653. http://www.cqvip.com/qk/92088x/199306/1127239.html [Google Scholar]
- Zhao YZ. (2009) Classification and its floristic geography of Caragana Fabr. in the world. Inner Mongolia University Press, Hohhot, 14–91. [Google Scholar]
- Zhou DW. (1996) Study on taxonomy of the genus Caragana Fabricii. Journal of Northeast Mormal University 4: 69–76. http://en.cnki.com.cn/Article_en/CJFDTotal-DBSZ604.019.htm [Google Scholar]
- Zhou QX, Yang YP, Zhang ML. (2002) Karyotypes of fourteen species in Caragana. Bulletin of Botanical Research 22(4): 492–498. http://www.cnki.com.cn/Article/CJFDTotal-MBZW200204022.htm [Google Scholar]
- Zhu WD, Nie ZL, Wen J, Sun H. (2013) Molecular phylogeny and biogeography of Astilbe (Saxifragaceae) in Asia and eastern North America. Botanical Journal of the Linnean Society 171: 377–394. doi: 10.1111/j.1095-8339.2012.01318.x [Google Scholar]
- Zhu XY. (1996) Note on the genus Chesneya Lindl. ex Endl. (Fabaceae). Acta phytotaxonomica Sinica 34(5): 558–562. http://www.jse.ac.cn/wenzhang/f940114.pdf [Google Scholar]
- Zhu XY. (2004) A revision of the genus Gueldenstaedtia (Fabaceae). Annales Botanici Fennici 41: 283–291. http://www.annbot.net/PDF/anbf41/anbf41-283.pdf [Google Scholar]
- Zhu XY. (2005a) A taxonomic revision of Tibetia (Leguminosae; Papilionoideae; Galegeae). Botanical Journal of the Linnean Society 148: 475–488. doi: 10.1111/j.1095-8339.2005.00414.x [Google Scholar]
- Zhu XY. (2005b) Pollen and seed morphology of Gueldenstaedtia Fisch. & Tibetia (Ali) H.P.Tsui (Leguminosae) – with a special reference to their taxonomic significance. Nordic Journal Botany 23: 373–384. doi: 10.1111/j.1756-1051.2003.tb00408.x [Google Scholar]
- Zhu XY, Cao R. (1986) The recently discovered species and the revision of some species of Chesneya in China. Acta Sientiarum Naturalim Universitatis Intramongolicae 17: 757–761. http://www.cnki.com.cn/Article/CJFDTotal-NMGX198604022.htm [Google Scholar]
- Zhu XY, Larsen K. (2010) Calophaca and Chesneya. In: Wu ZY, Hong DY, Raven PH. (Eds) Flora of China, vol. 10 Science Press, Beijing: & Missouri Botanical Garden Press, St Louis, 500–503, 527–528. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Voucher information and GenBank accession numbers
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Lei Duan, Xue Yang, Peiliang Liu, Gabriel Johnson, Jun Wen, Zhaoyang Chang
Data type: Multi-records
Explanation note: Data are arranged in the order: taxon name, locality, collector(s), collection number and herbarium, GenBank accession numbers for ITS, matK, trnL-trnF, psbA-trnH. Newly generated sequences are indicated by an asterisk (*); missing sequences are indicated by a dash (–).