Abstract
INTRODUCTION
Blood flow restriction in combination with low-intensity resistance exercise (REFR) increases skeletal muscle size to a similar extent as compared to traditional high-intensity resistance exercise training. However, there are limited data describing the molecular adaptations that occur after REFR.
PURPOSE
To determine whether (hypoxia inducible factor-1 alpha) HIF-1α and REDD1 mRNA is expressed differently in REFR compared to low-intensity resistance exercise with no blood flow restriction (CONTROL). Secondly, to determine if low-intensity resistance exercise is able to induce changes in mRNA expression of several anabolic and catabolic genes as typically seen with high-intensity resistance exercise.
METHODS
Six subjects were studied at baseline and 3h after a bout of leg resistance exercise (20% 1RM) in REFR and CONTROL subjects. Each subject participated in both groups with 3 weeks separating each visit. Muscle biopsy samples were analyzed for mRNA expression using qRT-PCR.
RESULTS
Our primary finding was that there were no differences between CONTROL and REFR for any of the selected genes at 3h post-exercise (P>0.05). However, low-intensity resistance exercise increased HIF-1α, p21, MyoD and muscle RING finger 1 (MuRF1) mRNA expression and decreased REDD1 and Myostatin mRNA expression in both groups (P<0.05).
CONCLUSION
Low-intensity resistance exercise can alter skeletal muscle mRNA expression of several genes associated with muscle growth and remodeling such as REDD1, HIF-1α, MyoD, MuRF1, and Myostatin. Further, the results from REFR and CONTROL were similar indicating that the changes in early post-exercise gene expression were due to the low-intensity resistance exercise bout and not blood flow restriction.
Keywords: mRNA, HIF-1α, REDD1, mTOR, ischemia-reperfusion
Introduction
Recent work has shown that low-intensity resistance exercise with blood flow restriction (REFR) causes muscle hypertrophy (1,2,26,30–32), which can be measured after only two weeks of REFR training (2). In addition, when walking exercise (3×/week at a distance of 1km/day) is combined with blood flow restriction, muscle hypertrophy is also stimulated (1). Thus, it appears that low-intensity muscle contractions when performed under conditions of restricted blood flow have the potential to initiate the necessary cellular signals promoting muscle protein synthesis and cell growth.
The stimulation of muscle growth with very low-intensity resistance exercise (20–50% of 1-RM) and a shortened duration of training (~2 weeks) is of significant interest for understanding the mechanisms regulating human muscle hypertrophy since traditional resistance exercise recommendations for muscle hypertrophy has generally been accepted as being >70% 1-RM, 3×/week for at least 5–12 weeks (18). In an attempt to begin identifying the mechanisms behind REFR induced muscle hypertrophy, we have recently shown that muscle protein synthesis and ribosomal S6 kinase 1 (S6K1) phosphorylation are increased following a single bout of REFR (12). This suggested that mRNA translation was increased following REFR, however, changes in mRNA expression of muscle hypertrophy regulators may also be involved.
A potential mechanism for how REFR promotes muscle hypertrophy may be the subsequent reperfusion upon release of the pressure cuff following the acute muscle hypoxia during the exercise bout. This may promote cell survival and cell growth adaptations within muscle such as the activation of the mammalian target of rapamycin (mTOR), a signaling pathway linked to muscle hypertrophy (23). Previous reports have indicated that hypoxia-inducible factor-1 alpha (HIF-1α) and REDD1 (regulated in development and DNA damage responses) are upregulated following hypoxia (4,13,14,19,27). An increased expression of these proteins have been shown to downregulate the mammalian target of rapamycin (mTOR) signaling pathway (4,7,19,33) However, in our previous publication, mTOR signaling to S6K1 was enhanced following REFR (12). This may have been due to muscle hyperemia-reperfusion (post-exercise when the pressure cuff was released) inhibition of HIF-1α and REDD1. Therefore, it is unknown whether HIF-1α and REDD1 have a regulatory role during the early post-exercise recovery period (3h) following REFR.
Another potential mechanism for acute changes in muscle hypertrophy following REFR may be the upregulation of hypertrophy associated genes in skeletal muscle during post-exercise recovery. Many reports have identified that several mRNAs increase/decrease transiently within 24h following a single bout of resistance exercise in human skeletal muscle (5,6,15,17,21,24,36,37). However, there is no information available identifying changes in mRNA expression following REFR. Our previous report focused on mechanisms supporting translation initiation (12) while neglecting other hypertrophy signals such changes in mRNA expression of genes associated with satellite cell control (p21, Cyclin D1,Insulin-like growth factor-1 (IGF-1), mechano growth factor (MGF), MyoD, Myogenin), cell size (Myostatin), and protein turnover (muscle atrophy F-box (MAFbx), muscle RING finger 1 (MuRF1), mTOR, S6K1).
Therefore, we hypothesized that selected mRNAs associated with muscle growth and remodeling would be altered to a greater extent in the early post-exercise recovery period following REFR as compared to low-intensity resistance exercise without blood flow restriction (CONTROL). The aims of the study were: 1) to determine whether HIF-1α and REDD1 mRNA is expressed differently in REFR compared to low-intensity resistance exercise with no blood flow restriction (CONTROL). 2) to determine if low-intensity resistance exercise is able to induce changes in mRNA expression of several anabolic and catabolic genes as typically seen with high-intensity resistance exercise.
Methods
Subjects
We studied 6 young male subjects in a cross-over design on two separate occasions three weeks apart. All subjects were healthy and physically active, but were not currently engaged in an exercise training program. All subjects gave informed written consent before participating in the study, which was approved by the Institutional Review Board of the University of Texas Medical Branch. Screening of subjects were performed with clinical history, physical exam, and laboratory tests including complete blood count with differential, liver and kidney function tests, coagulation profile, fasting blood glucose and oral glucose tolerance test (OGTT), hepatitis B and C screening, HIV test, TSH, lipid profile, urinalysis, drug screening, and ECG. The subjects’ characteristics are published in Fujita et al. (12). The subjects were initially randomized to a study in which they performed resistance exercise during blood flow restriction (REFR) or a control group in which the subjects performed resistance exercise with no blood flow restriction (CONTROL).
One-repetition maximum (1RM) testing
On two separate occasions (>5 days apart) and more than 5 days prior to conducting the study, each subject was tested for muscle strength by measuring their 1RM on a leg extension machine (Cybex-VR2, Medway, MA) located within the General Clinical Research Center’s (GCRC) Exercise Laboratory. Briefly, 1RM consisted of a single 10-repetition warm-up at 22.5 kg. At this time the exercise supervisor determined a suitable weight for the subject to lift one time. Subsequently, the subject rested for 3 minutes, the weight was increased/decreased and the subject attempted again to lift the assigned weight. This process continued until the subject could no longer lift the weight. The previous completed lift was considered their 1RM. About five days later the subjects repeated this protocol and the higher of the two 1RM values obtained was used to determine the exercise weight (20%) for the resistance exercise portion of this study.
Experimental design
Details to the infusion protocol and tracer methodology previously used can be found in Fujita et al. (12). Each subject was admitted to the GCRC of the University of Texas Medical Branch the day prior to the exercise study. The subjects were then fed a standard dinner, and a snack was given at 2200h. The subjects were studied following an overnight fast under basal conditions and refrained from exercise for 24h prior to study participation. The next morning, a baseline muscle biopsy was obtained from the lateral portion of the vastus lateralis of the leg with the biopsy site between 15 and 25 cm from the mid-patella. The biopsy was performed using a 5 mm Bergström biopsy needle, under sterile procedure and local anesthesia (1% lidocaine). Muscle tissue was immediately blotted and frozen in liquid nitrogen and stored at −80°C until analysis.
Resistance exercise with blood flow restriction (REFR)
After the baseline biopsy, a lower extremity pressure cuff (Kaatsu-Master Mini, Sato Sports Plaza, Tokyo, Japan) was placed around the most proximal portion of each leg. While the subject was seated on a chair, the cuff was pressurized to 120 mmHg for 30 seconds, and then the pressure was released for 10s. This was done four more times while the pressure in the cuff was increased by 20 mmHg with each subsequent re-inflation until 200 mmHg was reached. With the pressure maintained at 200 mmHg, the subjects then performed a set of 30 repetitions of bilateral knee extension exercise at 20% of 1RM, followed by a 30 second rest period. The subjects performed three more sets of 15 repetitions separated by 30 second rest intervals for a total of 4 sets and 75 repetitions. The cuff pressure was maintained at 200 mmHg during the entire 4 to 5 minutes of exercise and the total exercise time was approximately 4 to 5 minutes. Immediately following the fourth set, the pressure cuff was released and the subject rested in bed for 3h. At this time a post-exercise biopsy was sampled from the same incision site but instead the needle was angled away from the baseline biopsy sample area.
Resistance exercise without blood flow restriction (CONTROL)
Three weeks after the first visit, subjects initially assigned to the REFR group repeated the protocol without blood flow restriction (CONTROL) whereas subjects initially assigned to the CONTROL group then completed the blood flow restricted exercise protocol (REFR). The subjects in the CONTROL group performed the identical exercise protocol as the REFR group except that the cuff was not inflated and no pressure was applied to the legs. Therefore, all 6 subjects performed both trials in a randomized, cross-over design.
RNA extraction and cDNA synthesis
Total RNA was isolated by homogenizing 30–40 mg tissue with a homogenizing dispenser (T10 Basic Ultra Turrax, IKA, Wilmington, NC) in a solution containing 1.0 ml of TRI REAGENT and 4 µl of polyacryl carrier (Molecular Research Center, Inc, Cincinnati, Ohio). The RNA was separated into an aqueous phase using 0.2 ml of chloroform and precipitated from the aqueous phase using 0.50 ml of isopropanol. Extracted RNA was washed with 1 ml of 75% ethanol, dried, and then suspended in a known amount (1.5 µl/mg tissue) of nuclease-free water. RNA was quantified spectrophotometrically (BioRad, SmartSpec, Hercules, CA) at a wavelength of 260 nm. RNA concentration was calculated on the basis of total RNA yield. RNA quality was assessed by RNA agarose gel electrophoresis followed by visulation of the 18 and 28S ribosomal RNA bands under ultraviolet light. RNA was DNase-treated using a commercially available kit (DNA-free, Ambion, Austin, TX). One microgram of total RNA was reverse transcribed into cDNA according to the manufacturers’ directions (iScript, BioRad, Hercules, CA). Briefly, a 20 µl reaction mixture was constructed consisting of 1 µg of total RNA, 4 µl of 5× iScript Reaction Mix, 1 µl of iScript Reverse Transcriptase, and a known amount of nuclease-free water. RNA was reverse transcribed into cDNA using a thermocycler (IQ5 Real-Time PCR cycler, BioRad, Hercules, CA) following the following temperature/time protocol: 25°C for 5 min, 42°C for 30 min and 85°C for 5 min. All isolated RNA and cDNA samples were stored at −80°C until further analysis.
PCR primers
Primer pairs were customized using Beacon Designer 2.0 software (Premier Biosoft Int., Palo Alto, CA). Custom-designed primers are located in Table 1. MyoD primer pairs have been used previously (25). All primers were checked for specificity to our genes of interest by conducting a Blast analysis and minimizing primer selection across secondary structures. Primer efficiencies were optimized and PCR product was verified by melt analysis and a single DNA product as identified by a DNA agarose gel.
Table 1.
Human primer sequences used for real-time qRT-PCR.
| Name | Accession No. |
Primer Sequence (5' to 3') | Product size (bp) |
|
|---|---|---|---|---|
| HIF-1α | NM_001530 | Sense | AAGTCTGCAACATGGAAGG | 144 |
| Antisense | ATTTGATGGGTGAGGAATGG | |||
| REDD1 | NM_019058 | Sense | AGAGACGACTGAACTTTTGG | 140 |
| Antisense | CCCGCACACAACTCAATG | |||
| P21 | L25610 | Sense | CAGCATGACAGATTTCTACC | 147 |
| Antisense | CACACAAACTGAGACTAAGG | |||
| Cyclin D1 | NM_053056 | Sense | GAACAAGCTCAAGTGGAACC | 134 |
| Antisense | CACAGAGGGCAACGAAGG | |||
| IGF-1 | NM_000875 | Sense | ATGCGGTGTCCAATAACTAC | 112 |
| Antisense | TTGTTGATGGTGGTCTTCTC | |||
| MGF | U40870 | Sense | GTGGATGAGTGCTGCTTC | 135 |
| Antisense | CTGATACTTCTGGGTCTTGG | |||
| Myogenin | NM_002479 | Sense | TGGAGCTGTATGAGACATCC | 107 |
| Antisense | CTCGTAGCCTGGTGGTTC | |||
| Myostatin | AF019627 | Sense | CCGTCGAGACTCCTACAAC | 150 |
| Antisense | ACACTGTCTTCACATCAATGC | |||
| mTOR | NM_004958 | Sense | TCGTGCCTGTCTGATTCTC | 131 |
| Antisense | GATTCATGCCCTTCTCTTTGG | |||
| S6K1 | NM_003161 | Sense | ACTCAATTTGCCTCCCTACC | 128 |
| Antisense | AGAATGGATGAGCTTGAACTTC | |||
| MuRF1 | NM_032588 | Sense | TGGACTTCTTTACTTTGGATTTAG | 114 |
| Antisense | CTTCCTCTTCCTGATCTTCTTC | |||
| MAFbx | NM_148177 | Sense | ACAGGCATCAAGCTCATTCG | 120 |
| Antisense | ACAAAGGCACAGAAGTTACTCC | |||
| GAPDH | J02642 | Sense | AGGTGAAGGTCGGAGTCAAC | 231 |
| Antisense | GCTCCTGGAAGATGGTGATG |
Semi-quantitative real-time PCR
Determination of relative mRNA expression was performed by real-time RT-PCR using the iQ5 Multicolor Real-Time PCR cycler (Bio-Rad, Hercules, CA). cDNA was diluted (1:8) and analyzed using SYBR Green fluorescence (iQ SYBR Green Supermix, BioRad, Hercules, CA). The reaction vessel contained 12.5 µl iQ SYBR Green Supermix, 0.5–0.7 µl forward and reverse primers, 2.0 µl cDNA and a known amount of sterile water. The total volume of the reaction tube was 25 µl. All samples were run in duplicate. An initial cycle for 5 min at 95°C was used to denature the cDNA. This was followed with 40 PCR cycles consisting of denaturation at 95°C for 20s, and primer annealing and extension at 55°C for 30s. Relative fold changes in mRNA within and baseline differences between groups were determined as described by Livak and Schmittgen (20) after normalizing to GAPDH.
Statistical analysis
All values are expressed as mean ± SEM. Comparisons were performed using a one-way analysis of variance with repeated measures, to determine main effects for group (REFR, CONTROL) and time (Baseline, Post). Post-hoc testing was performed using independent paired t-tests when appropriate. Significance was set at P<0.05.
Results
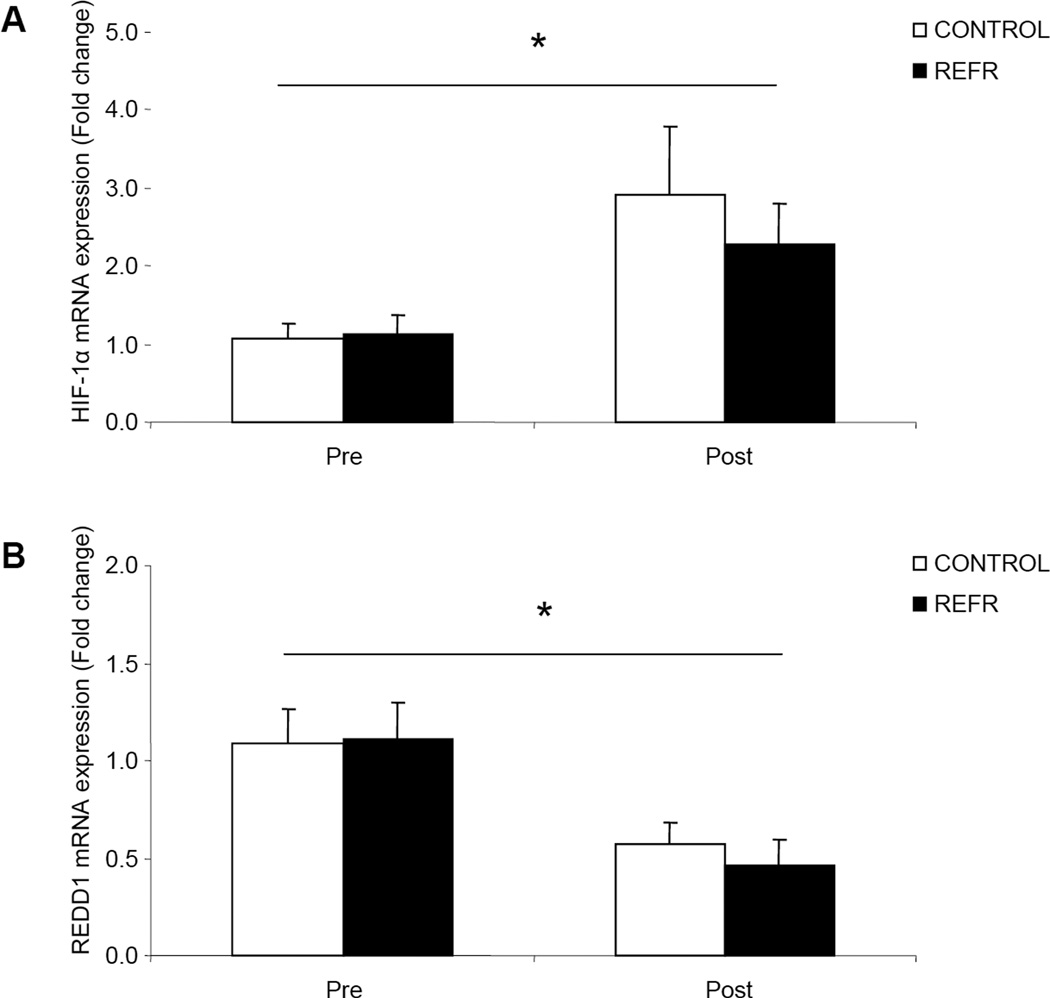
There were no differences in our selected hypoxia related genes between groups at baseline (P>0.05) or after exercise (P>0.05), however, HIF-1α mRNA was significantly increased (Fig. 1A; P<0.05) and REDD1 mRNA was significantly reduced (Fig. 1B; P<0.05) 3h after low-intensity resistance exercise in both groups.
Figure 1.
Results represent A) HIF-1α and B) REDD1 mRNA expression between CONTROL and REFR at baseline and 3h post resistance exercise. Values are mean ± SEM. * indicates main effect for time when CONTROL and REFR are combined (P<0.05).
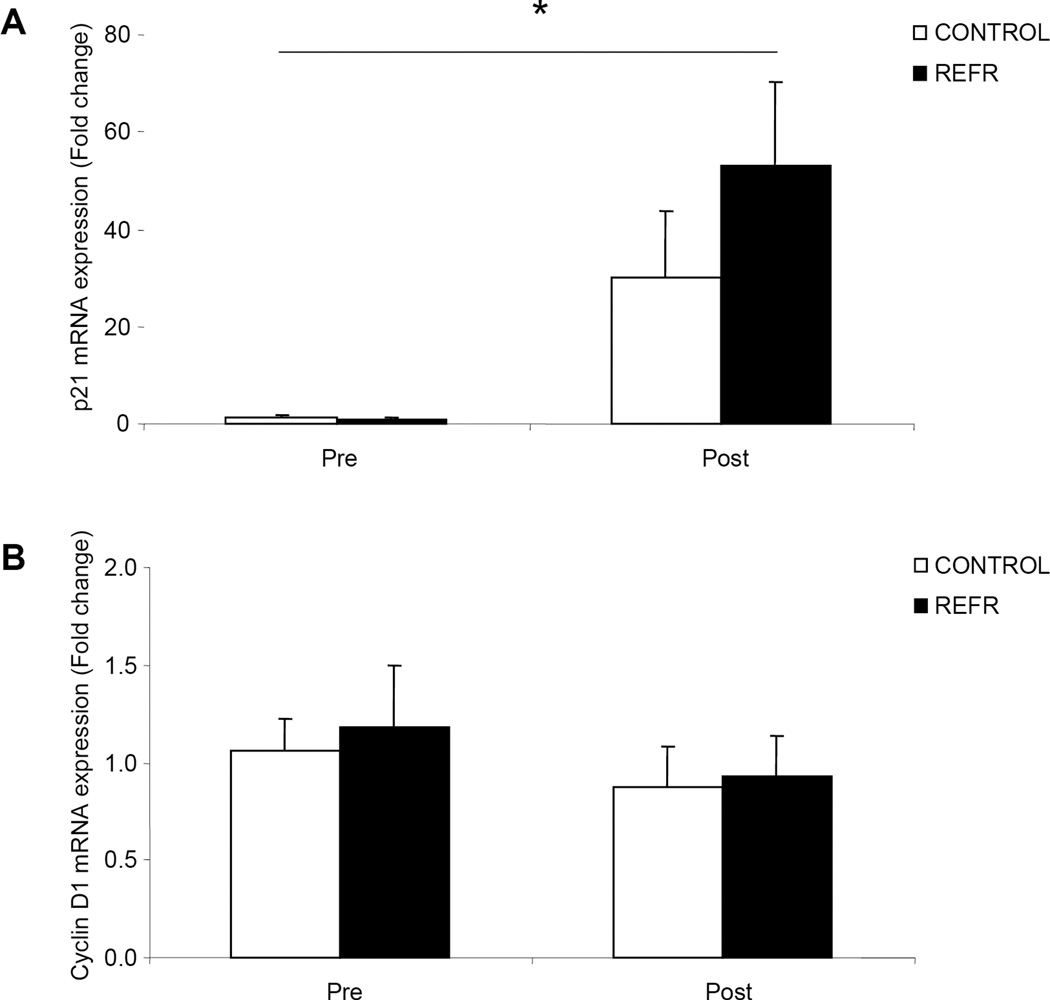
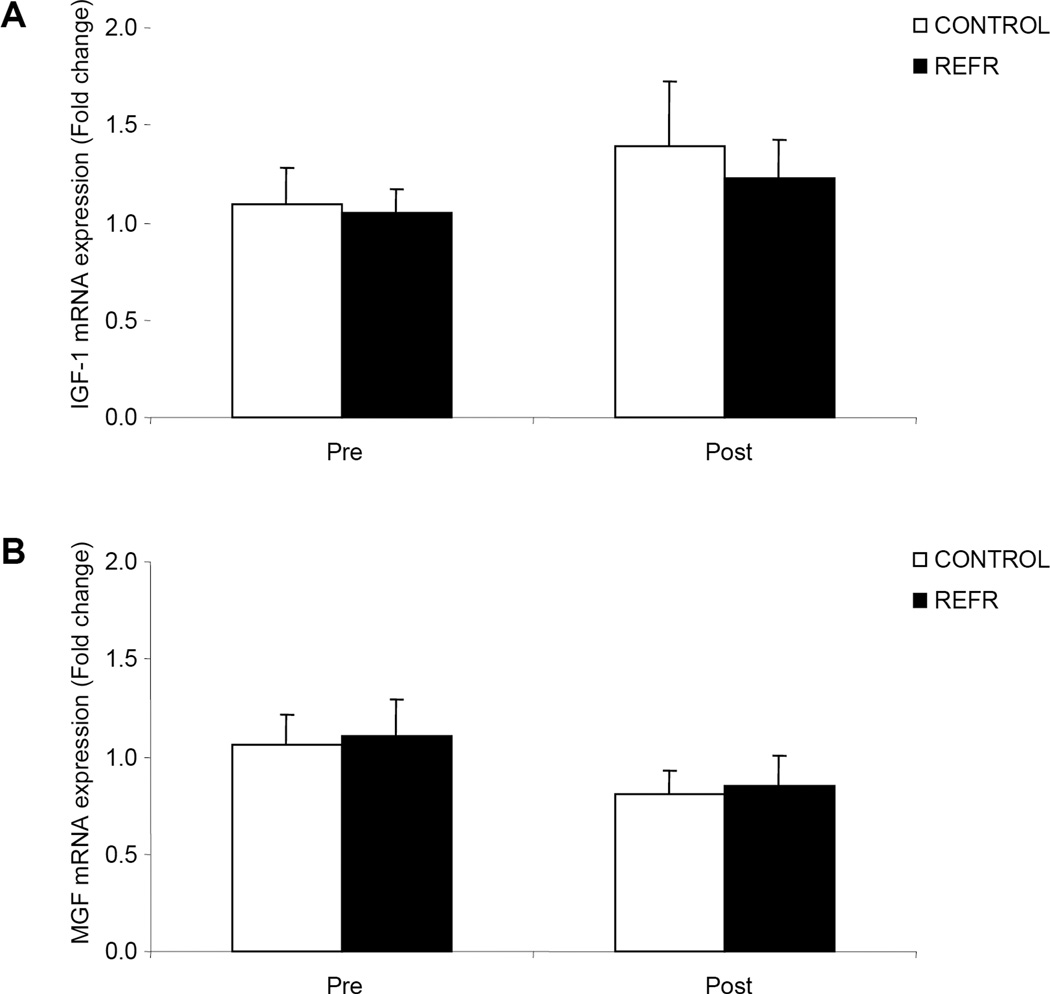
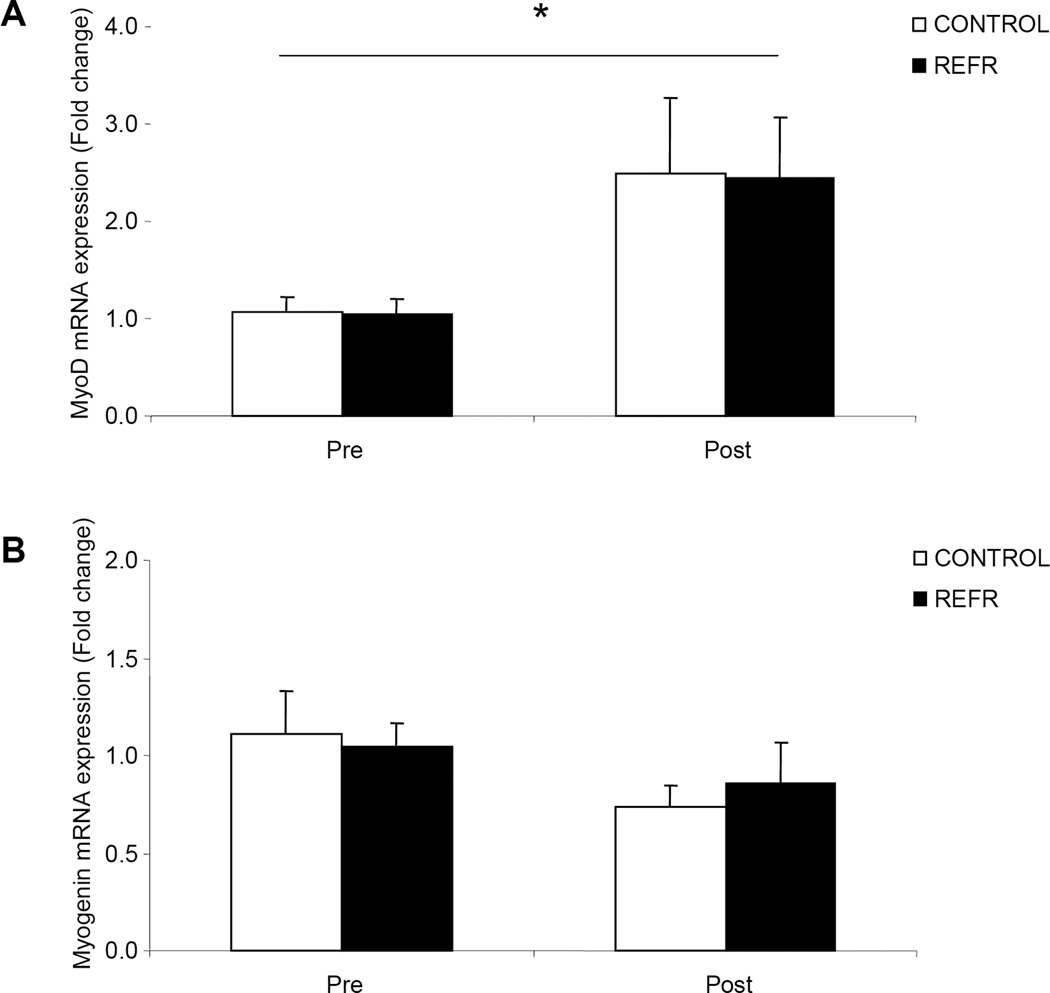
There were no differences in our selected genes associated with satellite cell function between groups at baseline (P>0.05) or after exercise (P>0.05), however, p21 (Fig 2A; P<0.05) increased in both groups. Cyclin D1 was unchanged 3h following an acute bout of low-intensity resistance exercise (Fig. 2B; P>0.05). Further, the mRNA expression of IGF-1 (Fig. 3A) and MGF (Fig.3B) did not change in either group 3h following the low-intensity resistance exercise bout (P>0.05) while MyoD mRNA expression increased in both groups (Fig. 4A; P<0.05). Further, Myogenin mRNA expression (Fig. 4B; P>0.05) was unchanged 3h following the bout of low-intensity resistance exercise.
Figure 2.
Results represent A) p21 and B) Cyclin D1 mRNA expression between CONTROL and REFR at baseline and 3h post resistance exercise. Values are mean ± SEM. * indicates main effect for time when CONTROL and REFR are combined (P<0.05).
Figure 3.
Results represent A) IGF-1 and B) MGF mRNA expression between CONTROL and REFR at baseline and 3h post resistance exercise. Values are mean ± SEM.
Figure 4.
Results represent A) MyoD and B) Myogenin mRNA expression between CONTROL and REFR at baseline and 3h post resistance exercise. Values are mean ± SEM. * indicates main effect for time when CONTROL and REFR are combined (P<0.05).
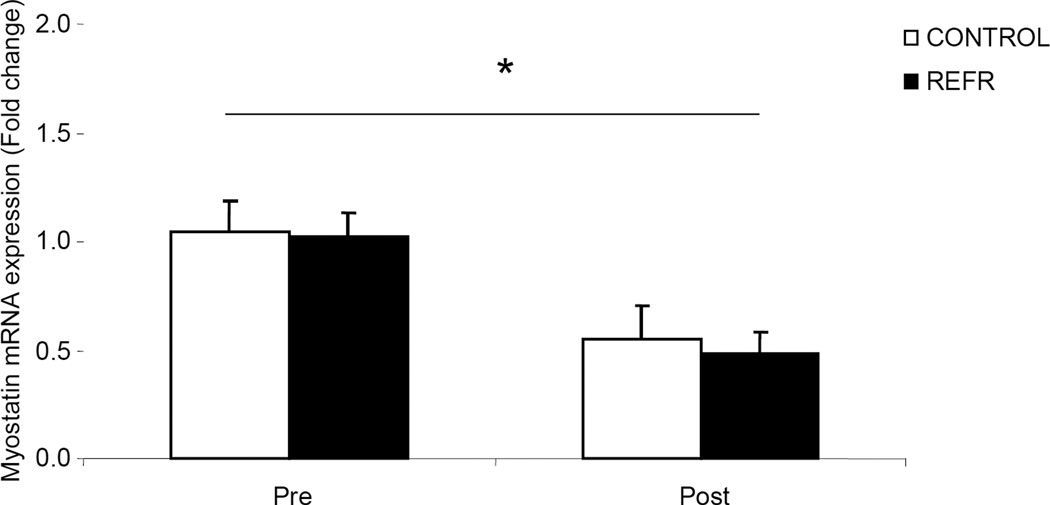
There were no differences in our selected gene involved in the regulation of cell size (Myostatin) between groups at baseline (P>0.05) or after exercise (P>0.05), however, Myostatin mRNA was significantly reduced in both groups 3h after a bout of low-intensity resistance exercise (Fig. 5; P<0.05).
Figure 5.
Results represent Myostatin mRNA expression between CONTROL and REFR at baseline and 3h post resistance exercise. Values are mean ± SEM. * indicates main effect for time when CONTROL and REFR are combined (P<0.05).
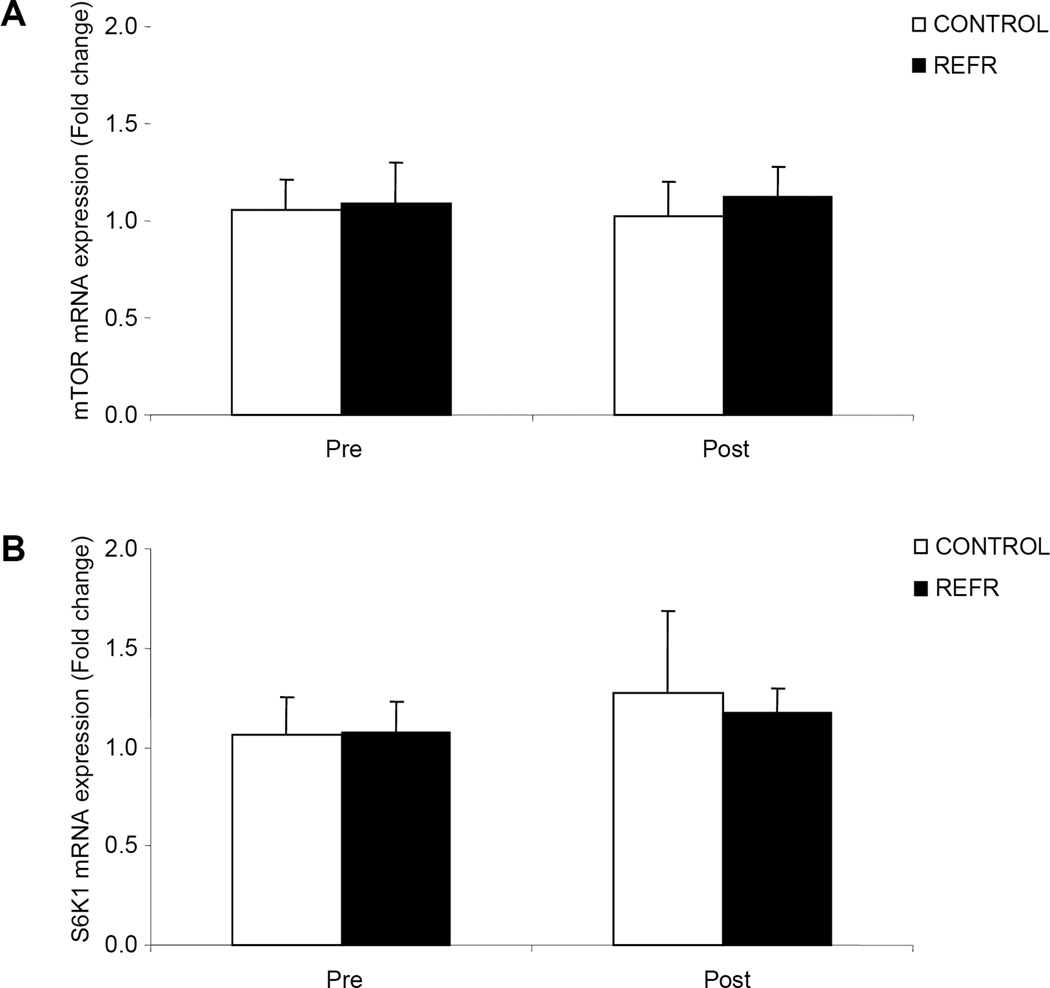
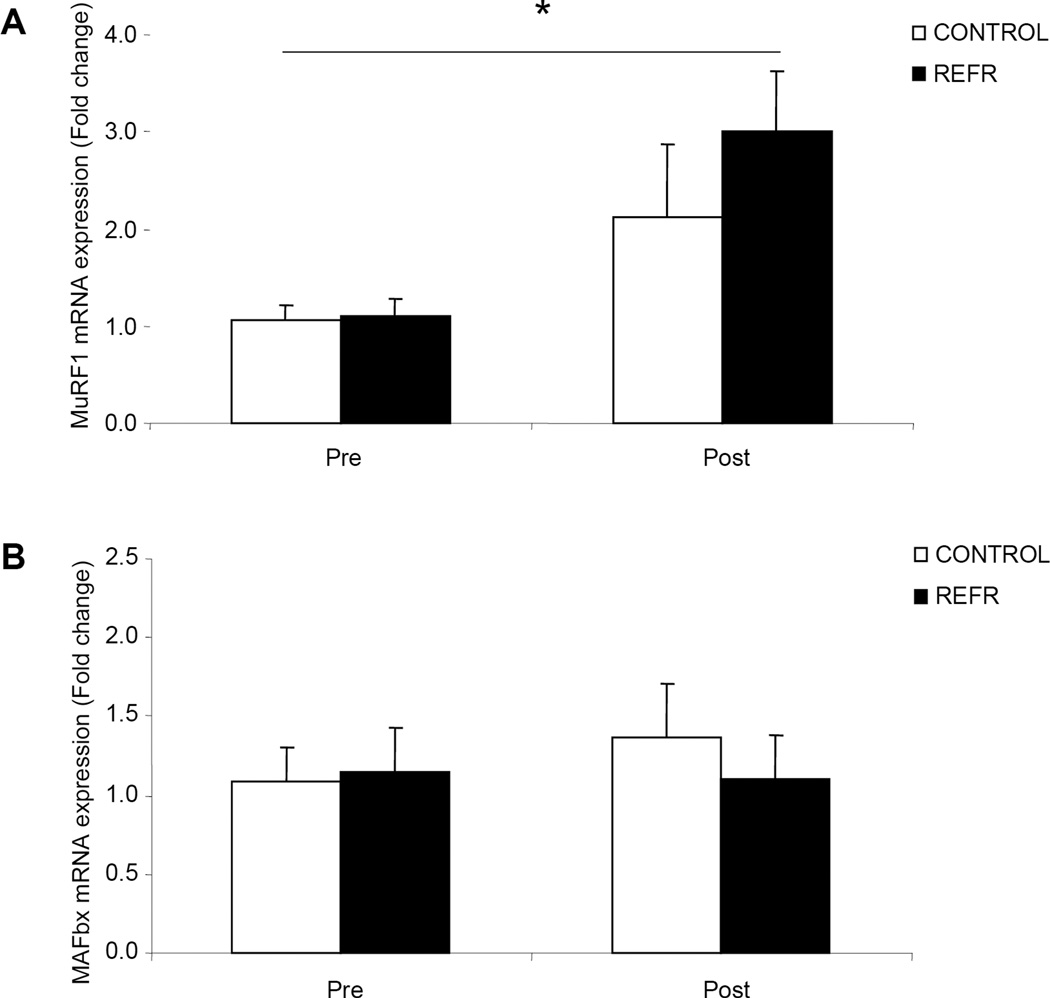
There were no differences in our selected genes involved in muscle protein turnover between groups at baseline (P>0.05) or after exercise (P>0.05). For the genes involved in the regulation of mRNA translation and muscle protein synthesis (mTOR and S6K1) we found no changes in mRNA expression in either group 3h following the bout of low-intensity resistance exercise (Fig. 6; P>0.05). Finally, in our selected genes associated with the regulation of muscle protein breakdown we found an increase in the mRNA expression of MuRF1 (Fig. 7A; P<0.05) and no change in MAFbx (Fig. 7B; P>0.05) for both groups 3h after an acute bout of low-intensity resistance exercise.
Figure 6.
Results represent A) mTOR and B) S6K1 mRNA expression between CONTROL and REFR at baseline and 3h post resistance exercise. Values are mean ± SEM.
Figure 7.
Results represent A) MuRF1 and B) MAFbx mRNA expression between CONTROL and REFR at baseline and 3h post resistance exercise. Values are mean ± SEM. * indicates main effect for time when CONTROL and REFR are combined (P<0.05).
Discussion
The primary finding from our study was that human skeletal muscle gene expression associated with muscle hypertrophy and remodeling is not different when blood flow restriction is performed during a single bout of low-intensity resistance exercise. However, we did detect that HIF-1α, REDD1 and other anabolic and catabolic skeletal muscle mRNAs are altered following a bout of low-intensity resistance exercise (20% of 1RM). The finding that human skeletal muscle REDD1 decreased following resistance exercise is novel and suggests that REDD1 may play a regulatory role in controlling human muscle protein synthesis.
The interaction between low-intensity resistance exercise and blood flow restriction does produce muscle hypertrophy similar to that seen with high-intensity resistance exercise (1,2,26,30–32). The underlying mechanisms for this are still rather unclear. A possible mechanism may be that REFR caused brief ischemia to the muscle during exercise which was followed by significant hyperemia during reperfusion when the cuff was released. Although ischemia/reperfusion was not directly measured in our study, lactate levels were elevated suggesting at least partial ischemia and/or hypoxia during REFR (12). Because hypoxia has been shown to activate the transcription factor HIF-1α, we chose to identify the response of HIF-1α and its downstream target, REDD1, during the 3h recovery period of REFR when the reduced blood flow and hypoxic conditions during exercise have been reversed. We report that HIF-1α and REDD1 mRNA were not different between our groups but rather HIF-1α was elevated and REDD1 decreased as a result of low-intensity resistance exercise alone (Fig. 1A, B). Our data is consistent with other studies which have reported increased HIF-1α mRNA and protein expression in human skeletal muscle after an acute bout of endurance exercise (3,22) and muscle contraction in rats (28) On the other hand, there have been no reports of REDD1 expression in human skeletal muscle following exercise. The relative significance of this finding is perplexing since HIF-1α mRNA expression did not correlate with a corresponding elevation in REDD1. Since the intensity of our resistance exercise protocol more closely resembled that of muscular endurance protocols, an upregulation of HIF-1α may signify recruitment of metabolic genes (rather than an interaction with REDD1) that promote oxygen delivery. Indeed, HIF-1α can directly activate the transcription of several metabolic genes such as VEGF, EPO and GLUT4 (3,28,35). Thus, it would be interesting to determine if HIF-1α is upregulated during high-intensity resistance exercise (e.g. 70% of 1RM) in which aerobic adaptations are less likely to occur. Therefore, additional research is needed to determine HIF-1α involvement following chronic low and high-intensity resistance exercise.
Similarly, REDD1 is induced following cellular stress (7,27,33). Recent reports have determined a connection between REDD1 and the mTOR pathway revealing its role as a repressor of mTOR phosphorylation and protein synthesis (4,7,11,19,33). Therefore, we were interested in determining whether REDD1 mRNA expression was different in REFR because we have previously shown that mTOR signaling to S6K1 post-exercise was greater in REFR as compared to CONTROL (12). We report that there were no differences between CONTROL and REFR. However, we show that REDD1 mRNA expression was significantly reduced in both groups (Fig. 1B) indicating an exercise independent response to REDD1. We believe that this is the first study in human skeletal muscle to show that REDD1 mRNA is altered by exercise. However, it is likely that other factors regulate REDD1 (since HIF-1α mRNA expression did not correlate with REDD1 expression). Further, our finding of reduced REDD1 mRNA expression may prove to be important since a reduction in REDD1 would apparently relieve inhibition on mTOR promoting a stimulation of mTOR signaling, mRNA translation, and muscle cell growth. Although changes in mTOR or S6K1 mRNA were not detected in either of our groups (Fig. 6A and B), it may be that REDD1 plays a permissive role by relieving mTOR inhibition and thereby opening the door to activation from other anabolic regulators. Future work is needed to define the roles of REDD1 or REDD2 in human skeletal muscle
An unexpected, yet, novel finding from this data was that several mRNAs associated with skeletal muscle hypertrophy and atrophy were altered after a single bout of resistance exercise at 20% of 1-RM. Markers of satellite cell activity (p21; Fig. 2A and MyoD; Fig. 4A) were significantly elevated 3h following exposure to low-intensity resistance exercise (P<0.05) while others were unchanged (P>0.05; Cyclin D1; Fig. 2B, IGF-1; Fig. 3A, MGF; Fig. 3B, Myogenin, Fig. 4B). Further, our data suggest a potential increase in muscle protein turnover as indicated by a significant increase in MuRF1 mRNA (Fig. 7A) and a relieved inhibition on cell growth as shown by a significant reduction in Myostatin mRNA (Fig. 5). Previous reports have demonstrated similar changes in these mRNAs following high loads of resistance exercise (6,21,24,25,36), but our data is the first to show that these genes are altered following an acute bout of low-intensity resistance exercise. Therefore, our findings may suggest a level of remodeling (albeit potentially small compared to high intensity resistance exercise) that has occurred in order to reinforce and strengthen the fiber in preparation for future loads. It is also likely that the changes in mRNA expression following low-intensity resistance exercise are temporary (i.e., few hours) whereas high-intensity resistance exercise produces a more sustained upregulation of hypertrophy associated genes (ie., up to 24h or more) as has been previously shown (6,16,21,25,36).
Our data also support the work of others showing that during the early recovery period (following a bout of resistance exercise in humans) that it is primarily enhanced mRNA translation which is responsible for the increase in muscle protein synthesis (10,34). For example, we have shown that REFR (but not low-intensity resistance exercise alone) increases muscle protein synthesis and S6K1 phosphorylation at 3h post-exercise (12), however, no differential effect of REFR was detected on our 13 selected muscle genes in this study. It is likely that a differential response may have been detected at later time points such as 24 hours post-exercise since REFR training clearly induces muscle hypertrophy over time (1,2,12,26,30–32). As pointed out previously (9), mRNA translation seems to be playing a key role in the early stages post-exercise with transcription of muscle genes becoming involved to a greater extent over time with training.
Low-intensity/high repetition is characteristic of muscular endurance and may not be optimal for muscular hypertrophy. This is supported by a study by Takarada et al. which reported that 8 weeks of low-intensity resistance exercise at approximately 20% 1RM did not increase skeletal muscle cross sectional area (32). This has also been demonstrated by others (8), but, in addition, these authors identified an increase in the percentage of type IIAB fibers after 8 weeks of high repetitions. It is quite possible that skeletal muscle hypertrophy at a low exercise intensity may take longer than eight weeks and/or may be a more suitable exercise paradigm for different populations rather than healthy young adults. For instance, Taaffe et al. reported that older subjects performing exercise at an intensity of 40% of 1-RM for 52 weeks, type I fiber significantly increased while type II fibers demonstrated a trend for an increase in size (29). In support of our data, these studies indicate that remodeling of human skeletal muscle can occur with repeated bouts of exercise at a low exercise intensity of 20% of 1RM. Although, it is evident that muscle hypertrophy will be less in comparison to traditional high-intensity resistance exercise training.
In summary, we demonstrate that changes at the mRNA level of various anabolic, catabolic or hypoxia genes are not different following REFR as compared to CONTROL. Further, we present novel data showing that a single bout of low intensity resistance exercise can alter mRNA expression during early post-exercise recovery in human skeletal muscle for genes associated with muscle growth and protein turnover such as REDD1 (↓), HIF-1α (↑), p21 (↑), MyoD (↑), MuRF1 (↑), and Myostatin (↓). We conclude that acute blood flow restriction in combination with resistance exercise does not influence the selected mRNAs differently as compared to when resistance exercise is performed with no blood flow restriction. Future studies are needed to evaluate gene expression at later stages of post-exercise recovery following REFR and in response to REFR training.
Acknowledgments
We wish to thank the nurses and personnel of the General Clinical Research Center of the University of Texas Medical Branch for their help with the conduct of the clinical portion of this study and Dr. Jerson Cadenas for assistance in our original data collection.
This study was supported by, grant R01 AR049877 from the National Institute for Arthritis and Musculoskeletal and Skin Diseases, a research grant from the Japan Kaatsu Training Society, grant S10 RR16650 from the Shared Instrumentation Grant Program, grant P30 AG024832 from the National Institutes on Aging, and grant M01 RR00073 from the General Clinical Research Branch, National Center for Research Resources.
Footnotes
The authors report no conflict of interest or endorsement by ACSM.
References
- 1.Abe T, Kearns CF, Sato Y. Muscle size and strength are increased following walk training with restricted venous blood flow from the leg muscle, Kaatsu-walk training. J Appl Physiol. 2006;100(5):1460–1466. doi: 10.1152/japplphysiol.01267.2005. [DOI] [PubMed] [Google Scholar]
- 2.Abe T, Yasuda T, Midorikawa T, Sato Y, Kearns CF, Inoue K, Koizumi K, Ishii N. Skeletal muscle size and circulating IGF-1 are increased after two weeks of twice daily KAATSU resistance training. Int J Kaatsu Training Res. 2005;1(6–12) [Google Scholar]
- 3.Ameln H, Gustafsson T, Sundberg CJ, Okamoto K, Jansson E, Poellinger L, Makino Y. Physiological activation of hypoxia inducible factor-1 in human skeletal muscle. Faseb J. 2005;19(8):1009–1011. doi: 10.1096/fj.04-2304fje. [DOI] [PubMed] [Google Scholar]
- 4.Arsham AM, Howell JJ, Simon MC. A novel hypoxia-inducible factor-independent hypoxic response regulating mammalian target of rapamycin and its targets. J Biol Chem. 2003;278(32):29655–29660. doi: 10.1074/jbc.M212770200. [DOI] [PubMed] [Google Scholar]
- 5.Bamman MM, Shipp JR, Jiang J, Gower BA, Hunter GR, Goodman A, McLafferty CL, Jr, Urban RJ. Mechanical load increases muscle IGF-I and androgen receptor mRNA concentrations in humans. Am J Physiol Endocrinol Metab. 2001;280(3):E383–E390. doi: 10.1152/ajpendo.2001.280.3.E383. [DOI] [PubMed] [Google Scholar]
- 6.Bickel CS, Slade J, Mahoney E, Haddad F, Dudley GA, Adams GR. Time course of molecular responses of human skeletal muscle to acute bouts of resistance exercise. J Appl Physiol. 2005;98(2):482–488. doi: 10.1152/japplphysiol.00895.2004. [DOI] [PubMed] [Google Scholar]
- 7.Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG., Jr Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18(23):2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campos GE, Luecke TJ, Wendeln HK, Toma K, Hagerman FC, Murray TF, Ragg KE, Ratamess NA, Kraemer WJ, Staron RS. Muscular adaptations in response to three different resistance-training regimens: specificity of repetition maximum training zones. Eur J Appl Physiol. 2002;88(1–2):50–60. doi: 10.1007/s00421-002-0681-6. [DOI] [PubMed] [Google Scholar]
- 9.Carson JA. The regulation of gene expression in hypertrophying skeletal muscle. Exerc Sport Sci Rev. 1997;25(301–320) [PubMed] [Google Scholar]
- 10.Chesley A, MacDougall JD, Tarnopolsky MA, Atkinson SA, Smith K. Changes in human muscle protein synthesis after resistance exercise. J Appl Physiol. 1992;73(4):1383–1388. doi: 10.1152/jappl.1992.73.4.1383. [DOI] [PubMed] [Google Scholar]
- 11.Corradetti MN, Inoki K, Guan KL. The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J Biol Chem. 2005;280(11):9769–9772. doi: 10.1074/jbc.C400557200. [DOI] [PubMed] [Google Scholar]
- 12.Fujita S, Abe T, Drummond MJ, Cadenas JG, Dreyer HC, Sato Y, Volpi E, Rasmussen BB. Blood flow restriction during low-intensity resistance exercise increases S6K1 phosphorylation and muscle protein synthesis. J Appl Physiol. 2007;103(3):903–910. doi: 10.1152/japplphysiol.00195.2007. [DOI] [PubMed] [Google Scholar]
- 13.Jiang BH, Rue E, Wang GL, Roe R, Semenza GL. Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J Biol Chem. 1996;271(30):17771–17778. doi: 10.1074/jbc.271.30.17771. [DOI] [PubMed] [Google Scholar]
- 14.Jiang BH, Semenza GL, Bauer C, Marti HH. Hypoxia-inducible factor 1 levels vary exponentially over a physiologically relevant range of O2 tension. Am J Physiol. 1996;271(4 Pt 1):C1172–C1180. doi: 10.1152/ajpcell.1996.271.4.C1172. [DOI] [PubMed] [Google Scholar]
- 15.Kim JS, Cross JM, Bamman MM. Impact of resistance loading on myostatin expression and cell cycle regulation in young and older men and women. Am J Physiol Endocrinol Metab. 2005;288(6):E1110–E1119. doi: 10.1152/ajpendo.00464.2004. [DOI] [PubMed] [Google Scholar]
- 16.Kim JS, Kosek DJ, Petrella JK, Cross JM, Bamman MM. Resting and load-induced levels of myogenic gene transcripts differ between older adults with demonstrable sarcopenia and young men and women. J Appl Physiol. 2005;99(6):2149–2158. doi: 10.1152/japplphysiol.00513.2005. [DOI] [PubMed] [Google Scholar]
- 17.Kostek MC, Chen YW, Cuthbertson DJ, Shi R, Fedele MJ, Esser KA, Rennie MJ. Gene expression responses over 24h to lengthening and shortening contractions in human muscle: major changes in CSRP3, MUSTN1, SIX1 and FBXO32. Physiol Genomics. 2007 doi: 10.1152/physiolgenomics.00151.2006. [DOI] [PubMed] [Google Scholar]
- 18.Kraemer WJ, Adams K, Cafarelli E, Dudley GA, Dooly C, Feigenbaum MS, Fleck SJ, Franklin B, Fry AC, Hoffman JR, Newton RU, Potteiger J, Stone MH, Ratamess NA, Triplett-McBride T. American College of Sports Medicine position stand. Progression models in resistance training for healthy adults. Med Sci Sports Exerc. 2002;34(2):364–380. doi: 10.1097/00005768-200202000-00027. [DOI] [PubMed] [Google Scholar]
- 19.Liu L, Cash TP, Jones RG, Keith B, Thompson CB, Simon MC. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol Cell. 2006;21(4):521–531. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Louis ES, Raue U, Yang Y, Jemiolo B, Trappe SW. Time Course of Proteolytic, Cytokine, and Myostatin Gene Expression After Acute Exercise in Human Skeletal Muscle. J Appl Physiol. 2007 doi: 10.1152/japplphysiol.00679.2007. [DOI] [PubMed] [Google Scholar]
- 22.Lundby C, Gassmann M, Pilegaard H. Regular endurance training reduces the exercise induced HIF-1alpha and HIF-2alpha mRNA expression in human skeletal muscle in normoxic conditions. Eur J Appl Physiol. 2006;96(4):363–369. doi: 10.1007/s00421-005-0085-5. [DOI] [PubMed] [Google Scholar]
- 23.Moumen A, Patane S, Porras A, Dono R, Maina F. Met acts on Mdm2 via mTOR to signal cell survival during development. Development. 2007;134(7):1443–1451. doi: 10.1242/dev.02820. [DOI] [PubMed] [Google Scholar]
- 24.Psilander N, Damsgaard R, Pilegaard H. Resistance exercise alters MRF and IGF-I mRNA content in human skeletal muscle. J Appl Physiol. 2003;95(3):1038–1044. doi: 10.1152/japplphysiol.00903.2002. [DOI] [PubMed] [Google Scholar]
- 25.Raue U, Slivka D, Jemiolo B, Hollon C, Trappe S. Myogenic gene expression at rest and after a bout of resistance exercise in young (18–30 yr) and old (80–89 yr) women. J Appl Physiol. 2006;101(1):53–59. doi: 10.1152/japplphysiol.01616.2005. [DOI] [PubMed] [Google Scholar]
- 26.Shinohara M, Kouzaki M, Yoshihisa T, Fukunaga T. Efficacy of tourniquet ischemia for strength training with low resistance. Eur J Appl Physiol Occup Physiol. 1998;77(1–2):189–191. doi: 10.1007/s004210050319. [DOI] [PubMed] [Google Scholar]
- 27.Shoshani T, Faerman A, Mett I, Zelin E, Tenne T, Gorodin S, Moshel Y, Elbaz S, Budanov A, Chajut A, Kalinski H, Kamer I, Rozen A, Mor O, Keshet E, Leshkowitz D, Einat P, Skaliter R, Feinstein E. Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Mol Cell Biol. 2002;22(7):2283–2293. doi: 10.1128/MCB.22.7.2283-2293.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silva JL, Giannocco G, Furuya DT, Lima GA, Moraes PA, Nachef S, Bordin S, Britto LR, Nunes MT, Machado UF. NF-kappaB, MEF2A, MEF2D and HIF1-a involvement on insulin- and contraction-induced regulation of GLUT4 gene expression in soleus muscle. Mol Cell Endocrinol. 2005;240(1–2):82–93. doi: 10.1016/j.mce.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 29.Taaffe DR, Pruitt L, Pyka G, Guido D, Marcus R. Comparative effects of high- and low-intensity resistance training on thigh muscle strength, fiber area, and tissue composition in elderly women. Clin Physiol. 1996;16(4):381–392. doi: 10.1111/j.1475-097x.1996.tb00727.x. [DOI] [PubMed] [Google Scholar]
- 30.Takarada Y, Sato Y, Ishii N. Effects of resistance exercise combined with vascular occlusion on muscle function in athletes. Eur J Appl Physiol. 2002;86(4):308–314. doi: 10.1007/s00421-001-0561-5. [DOI] [PubMed] [Google Scholar]
- 31.Takarada Y, Takazawa H, Sato Y, Takebayashi S, Tanaka Y, Ishii N. Effects of resistance exercise combined with moderate vascular occlusion on muscular function in humans. J Appl Physiol. 2000;88(6):2097–2106. doi: 10.1152/jappl.2000.88.6.2097. [DOI] [PubMed] [Google Scholar]
- 32.Takarada Y, Tsuruta T, Ishii N. Cooperative effects of exercise and occlusive stimuli on muscular function in low-intensity resistance exercise with moderate vascular occlusion. Jpn J Physiol. 2004;54(6):585–592. doi: 10.2170/jjphysiol.54.585. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Kubica N, Ellisen LW, Jefferson LS, Kimball SR. Dexamethasone represses signaling through the mammalian target of rapamycin in muscle cells by enhancing expression of REDD1. J Biol Chem. 2006;281(51):39128–39134. doi: 10.1074/jbc.M610023200. [DOI] [PubMed] [Google Scholar]
- 34.Welle S, Bhatt K, Thornton CA. Stimulation of myofibrillar synthesis by exercise is mediated by more efficient translation of mRNA. J Appl Physiol. 1999;86(4):1220–1225. doi: 10.1152/jappl.1999.86.4.1220. [DOI] [PubMed] [Google Scholar]
- 35.Wenger RH, Gassmann M. Oxygen(es) and the hypoxia-inducible factor-1. Biol Chem. 1997;378(7):609–616. [PubMed] [Google Scholar]
- 36.Yang Y, Creer A, Jemiolo B, Trappe S. Time course of myogenic and metabolic gene expression in response to acute exercise in human skeletal muscle. J Appl Physiol. 2005;98(5):1745–1752. doi: 10.1152/japplphysiol.01185.2004. [DOI] [PubMed] [Google Scholar]
- 37.Yang Y, Jemiolo B, Trappe S. Proteolytic mRNA expression in response to acute resistance exercise in human single skeletal muscle fibers. J Appl Physiol. 2006;101(5):1442–1450. doi: 10.1152/japplphysiol.00438.2006. [DOI] [PubMed] [Google Scholar]