Abstract
The advancement of microRNA (miRNA) therapies has been hampered by difficulties in delivering miRNA to the injured kidney in a robust and sustainable manner. Using bioluminescence imaging in mice with unilateral ureteral obstruction (UUO), we report that mesenchymal stem cells (MSCs), engineered to overexpress miRNA-let7c (miR-let7c-MSCs), selectively homed to damaged kidneys and upregulated miR-let7c gene expression, compared with nontargeting control (NTC)-MSCs. miR-let7c-MSC therapy attenuated kidney injury and significantly downregulated collagen IVα1, metalloproteinase-9, transforming growth factor (TGF)-β1, and TGF-β type 1 receptor (TGF-βR1) in UUO kidneys, compared with controls. In vitro analysis confirmed that the transfer of miR-let7c from miR-let7c-MSCs occurred via secreted exosomal uptake, visualized in NRK52E cells using cyc3-labeled pre-miRNA-transfected MSCs with/without the exosomal inhibitor, GW4869. The upregulated expression of fibrotic genes in NRK52E cells induced by TGF-β1 was repressed following the addition of isolated exosomes or indirect coculture of miR-let7c-MSCs, compared with NTC-MSCs. Furthermore, the cotransfection of NRK52E cells using the 3′UTR of TGF-βR1 confirmed that miR-let7c attenuates TGF-β1-driven TGF-βR1 gene expression. Taken together, the effective antifibrotic function of engineered MSCs is able to selectively transfer miR-let7c to damaged kidney cells and will pave the way for the use of MSCs for therapeutic delivery of miRNA targeted at kidney disease.
Introduction
The application of microRNAs (miRNAs), small single-stranded noncoding RNAs of 18–24 bases in length that function in the transcriptional and posttranscriptional regulation of target gene expression by repressing the 3′untranslated region (3′UTR) of mRNA,1,2 may provide a novel option to attenuate the progression of kidney disease. miRNAs were originally considered the product of “junk DNA”, however, more than 2000 miRNAs have now been identified,3 which are involved in the regulation of approximately 30% of all mammalian protein encoding genes that have diverse roles in the cell cycle, apoptosis, tissue development, stem cell division, and development of degenerative disease.4,5 In particular, dysregulation of miRNAs has been implicated in diseases associated with kidney homeostasis including polycystic kidney disease, diabetic nephropathy, and kidney cancer (reviewed in ref. 6).
The characterization and roles of miRNAs in both antifibrotic and profibrotic settings of kidney disease have been reported.2,7 We8 and others9 have identified the functional roles of the miR-let7 family members (miR-let7b and miR-let7c) in renal fibrosis resulting from diabetes mellitus, through regulation of transforming growth factor-β (TGF-β) signaling.8 Profiling of human renal proximal tubule (HK2) cell miRNAs demonstrated that miR-let7c was downregulated under fibrotic conditions,9 suggesting that miR-let7 may have clinically relevant therapeutic potential to repair or reverse establish kidney fibrosis. However, the successful delivery of miRNA to the site of injury remains a significant challenge in the field.2
Mesenchymal stem cells (MSCs) have been demonstrated to be a safe and effective delivery vehicle for therapeutic miRNA treatment, due to their ability to specifically target inflammation in neurodegenerative disorders and have the ability to transfer molecules via exosomal trafficking.10 Exosomes can be loaded with miRNAs, which transfer to neighboring cells or to targeted cells, leading to the repression of target gene expression.11,12 A recent study using prion-infected neuronal cells and deep sequencing detected a distinct exosome-specific miRNA signature.13 This novel mechanism of intercellular communication mediated via exosomes was demonstrated using siRNA as a marker to track exocytic and endocytic pathways.14 MSCs secrete microparticles or exosomes enriched with pre-miRNAs,15 and the miRNA expression profile of human MSCs is associated with a high expression of the miR-let7 family16 suggesting that miR-let7 derived from MSCs may play a protective role in tissue injury and disease. These studies suggest that an exosome-based miRNA delivery may provide a clinically relevant gene-therapy strategy for the treatment of fibrotic kidney disease.
MSCs have demonstrated safety in both completed and ongoing clinical trials17,18,19 including kidney transplantation20,21 and exhibit innate therapeutic effects in heart attacks and respiratory disease.22,23 These immunoprivileged cells rapidly home to injured kidneys24 and release cytokines that promote repair through effects on regulatory immune cells25 and alteration of macrophage phenotype.26 They can prevent and/or reverse kidney fibrosis and improve renal function in both experimental models27,28,29 and human patients.20
The current study uses an innovative strategy to construct a miR-let7 delivery system that utilizes genetically engineered MSCs, transduced to overexpress miR-let7c (miR-let7c-MSCs), as a therapeutic tool to target kidney disease. In a mouse model of unilateral ureteral obstruction (UUO), miR-let7c was selectively delivered to the injured kidney following administration of MSC-miR-let7c, where an elevated expression of kidney miR-let7 corresponded with improved kidney structure and reduction of interstitial collagen, compared with delivery of nontargeting control (NTC) MSCs. In vitro analysis using the addition of isolated exosomes or indirect coculture confirmed that miR-let7c-MSCs induced an increased expression of the target miRNA (let7c) in neighboring rat kidney tubular epithelial cells (NRK52E) via exosome delivery. When NRK52E cells were treated with transforming growth factor (TGF)-β1, miR-let7c-MSC coculture inhibited the upregulated expression of collagen types 1α1 and IVα1, α-smooth muscle actin (α-SMA), and TGF-β type 1 receptor (TGF-βR1). These data indicate that MSCs, overexpressing miR-let7c, can be selectively delivered to damaged kidneys to attenuate fibrosis in vivo and reduce TGF-β1-stimulated injury to kidney cells via exosome-mediated delivery of functionally active miR-let7c leading to repression of TGF-βR1 protein expression in vitro.
Results
miR-let7c expression in in vitro and in vivo kidney fibrotic models
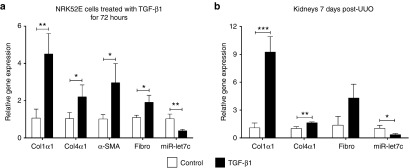
The TGF-β signaling pathway is considered a key mediator in the development of kidney fibrosis. Kidney tubular epithelial cells (NRK52E) were treated with TGF-β1 (5 ng/ml) for 72 hours, and the expression of miR-let7c and related fibrotic genes was determined by qPCR. miR-let7c expression was significantly decreased (P < 0.01) in kidney cells, concurrent with a significant upregulation of collagen type 1α1 (P < 0.01), collagen type IVα1 (P < 0.05), α-SMA (P < 0.05), and fibronectin (P < 0.05) expression levels, in comparison with untreated cells (Figure 1a). Similarly, in the kidneys of mice following UUO, there was a significant upregulation of mRNA expression of collagen Iα1 (P < 0.001) and collagen type IVα1 (P < 0.01), corresponding with a significant decrease in miR-let7c expression (P < 0.05; Figure 1b) in obstructed kidneys relative to contralateral unobstructed kidneys (CUKs). These data indicate a downregulation of miR-let7c in kidney fibrosis in vitro and in vivo consistent with the previous reports on the expression pattern of miR-let7c.9
Figure 1.
miR-let7c and fibrotic gene expression in vitro and in vivo. (a) Kidney tubular epithelial cells (NRK52E) displayed an upregulation of fibrotic markers, including collagen 1α1 (P < 0.01), collagen IVα1 (P < 0.05), α-SMA (P < 0.05), and fibronectin (P < 0.05), in conjunction with a downregulated expression of miR-let7c (P < 0.01) after treatment with TGF-β1. (b) The kidneys of UUO mice showed an upregulated expression of collagen 1α1 (P < 0.001) and collagen IVα1 (P < 0.01) and a reduction of miR-let7c (P < 0.05), compared with CUK kidneys. Results are expressed as the mean ± SEM relative gene expression. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05. col1α1, collagen 1α1; Col4α1, collagen IVα1; Fibro, fibronectin; SMA, smooth muscle actin; UUO, unilateral ureteral obstruction, TGF, transforming growth factor.
Characterization of MSCs engineered to express miR-let7c or NTC
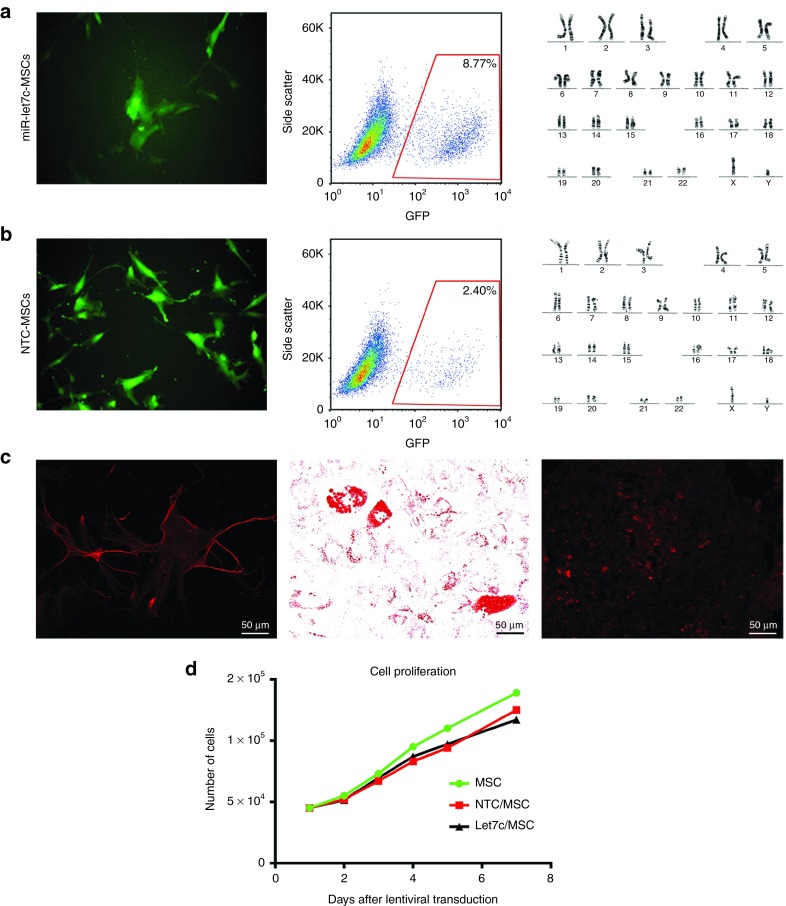
Lentiviral self-inactivating constructs expressing human miR-let7c were used to overexpress miR-let7c in MSCs. MSCs were transduced with miR-let7c (miR-let7c-MSCs) or construct containing a NTC (NTC-MSCs) sequence for control comparison. A turbo green fluorescent protein (GFP) reporter gene was included in the constructs to visualize successful transduction. The transfection of miR-let7c-MSCs showed a high efficiency of GFP expression by immunofluorescence microscopy and fluorescence-activated cell sorting (FACS) analysis (Figure 2a). MSCs transfected with NTC also showed a high efficiency of GFP expression (Figure 2b), with both miR-let7c- and NTC-transfected cells showing a normal karyotype (Figure 2a,b). miR-let7c-MSCs retained a multipotent ability to differentiate into osteocytes, adipocytes, and chondrocytes as shown by positive staining with osteocalcin, fatty acid binding protein-4, and aggrecan, respectively (Figure 2c). Using a proliferation assay, both miR-let7c-MSCs and NTC-MSCs had normal proliferation capabilities (Figure 2d).
Figure 2.
Characterisation of MSCs transduced with miR-let7c (miR-let7c-MSC) or construct containing a NTC (NTC-MSC) sequence. (a) Flow cytometry and fluorescence microscopy for GFP+ expression showed the transfection efficiency of MSCs with miR-let7c, that were karyotypically normal. (b) Successful transduction of MSCs with miR-NTC was demonstrated by fluorescence microscopy and flow cytometry. The transfected cells also displayed karyotypically normality. (c) MSCs displayed multilineage differentiation potential in vitro, differentiating into osteocytes, evidenced by osteocalcin staining (magnification ×200), adipocytes indicated by the presence of lipid droplets stained with Oil Red-O (magnification ×400), and chondrocytes shown by the presence of aggrecan staining (magnification ×200). (d) In addition to displaying a normal morphology, the transduced MSCs, miR-let7c-MSCs displayed a normal proliferative ability. GFP, green fluorescent protein; MSCs, mesenchymal stem cells; NTC, nontargeting control. Bar: b = 100 µm.
MSC exosomal delivery of miR-let7c into kidney cells
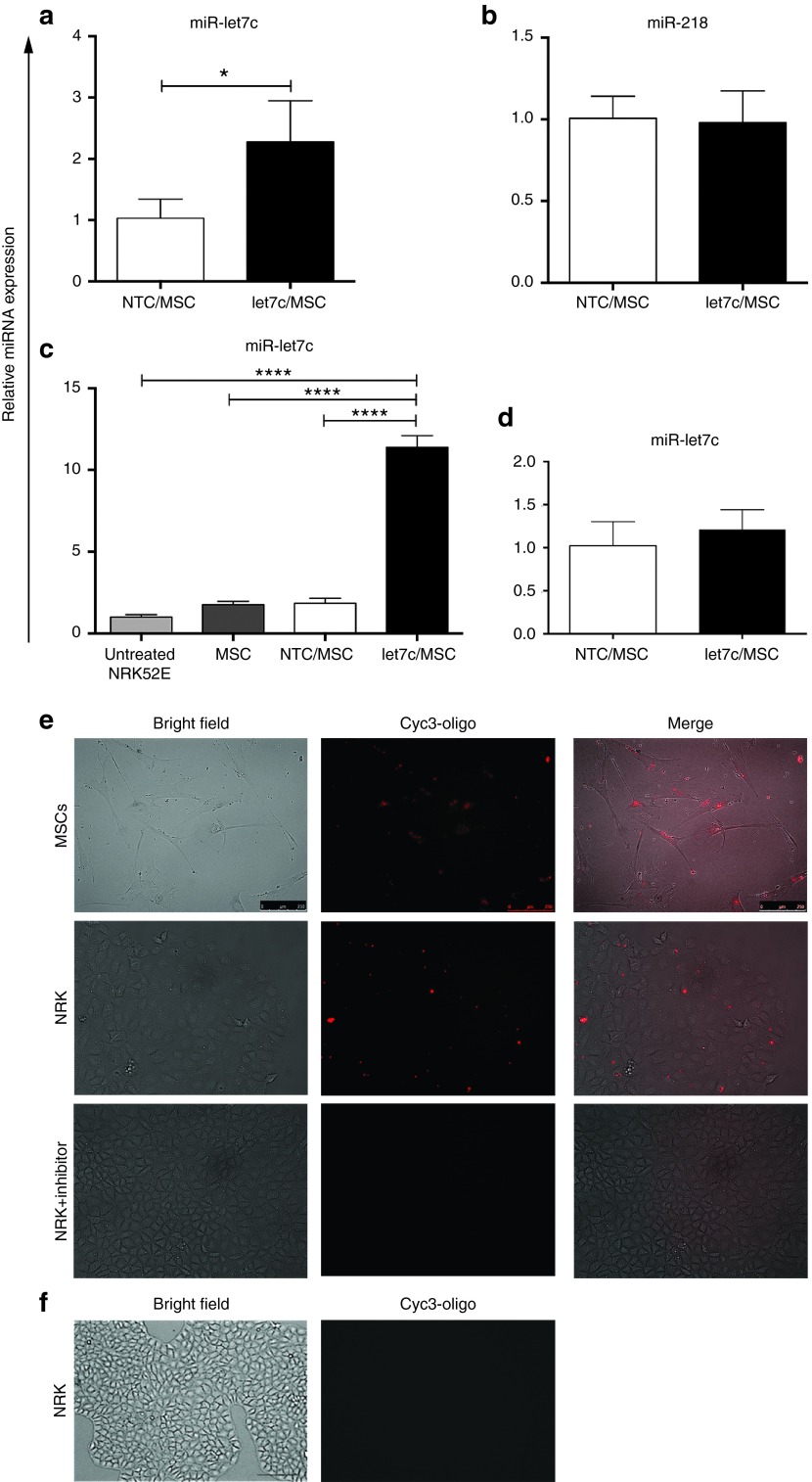
To assess the transfer of miR-let7c expression from MSCs, the exosomes from culture media of modified MSCs were isolated and assessed over 72 hours of culture using a miR-let7c assay. In comparison with isolated exosomes obtained from NTC-MSC culture medium, miR-let7c expression in exosomes from miR-let7c-MSC medium was significantly increased (P < 0.05; Figure 3a), suggesting that miR-let7c expression was increased and exocytosed via MSC-derived exosomes. The expression of miR-let7a and let7d showed no significant change (see Supplementary Figure S2). To avoid the effect of viral transduction of other miRNAs, the endogenous miR-218 from engineered MSCs was assayed as a negative control miRNA, which has previously been identified in MSCs.30 In contrast to miR-let7c, no change of miR-218 was observed in the exosomes isolated from miR-let7c-MSCs, compared with NTC-MSC culture medium (Figure 3b), confirming that endogenous expression of other miRNAs was not affected.
Figure 3.
Exosomal transport and transfer of miR-let7c from MSCs. (a) The exosomes from miR-let7c-MSCs showed a significantly increased expression of let7c in comparison with NTC-MSCs (P < 0.05). In contrast, b shows that miR-218 expression, used as a negative control, was unaltered between the two cell types. (c) Coculture of NRK52E cells with MSCs engineered to overexpress miR-let7c resulted in a significantly increased expression of miR-let7c (P < 0.0001) in the kidney tubular epithelial cells, in comparison with nontransduced MSCs and NTC-MSCs that did not show elevated miR-let7c expression. (d) The addition of an exosome inhibitor, GW4869, was confirmed to inhibit the elevated expression of miR-let7c in NRK52E cells when cocultured with miR-let7c-MSCs and NTC-MSCs. (e) Fluorescence microscopy was used to visualize exosomal transfer of miR-let7c to kidney epithelial cells. The coculture of NRK52E with MSCs transfected with cyc3-oligo showed exosomal transfer of miRNA from the MSCs to the target kidney cells after 3 days. However, the treatment of exosome inhibitor abolished the cyc3-oligo expression in NRK52E cells. (f) Brightfield and immunofluorescence images showing NRK52E cells without cyc3-oligo transfection. Results are expressed as the mean ± SEM relative gene expression. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05. NRK, NRK52E cells; NTC, nontargeting control; MSCs, mesenchymal stem cells. Bar: a = 100 µm.
Next, the endocytosis efficiency of the miR-let7c produced by modified MSCs was examined. An indirect in vitro coculture assay was established to determine if miR-let7c-MSCs could deliver a significantly increased expression of the target miRNA (let7c) to kidney cells via exosomes. A transwell system was used to culture NRK52E kidney cells separately from the MSCs with/without miR-let7c transduction, allowing the MSCs to secrete soluble factors without physical interaction with NRK52E cells. Following 72 hours of coculture, qPCR was performed on NRK52E cells to confirm the successful transfer of let7c miRNA. There was a significant 4.7-fold increase of miR-let7c expression in NRK52E cells cocultured with miR-let7c MSCs, compared with NTC-MSCs (P < 0.0001; Figure 3c). Endogenous miR-let7c expression in NRK52E cocultured with MSCs marginally increased compared with the NRK52E cells without coculture (Figure 3c), suggesting that MSCs have endogenous miR-let7c expression as reported previously.16 To further verify the exosome function in the delivery of miR-let7c, the exosome inhibitor neutral sphingomyelinase (nSMase2), GW4869, was used to treat the cocultured NRK52E cells and miR-let7c-MSCs or NTC-MSCs. There was no difference in miR-let7c expression between the NTC-MSC and miR-let7c-MSC cocultured NRK52E cells (Figure 3d), therefore confirming exosomal transfer of miR-let7c.
To visualize exosome delivery between miR-let7c-MSCs and NRK52E cells, cyc3-labeled pre-miRNA control oligos (cyc3-oligo) were transfected into miR-let7c-MSCs followed by intracellular visualization using immunofluorescence microscopy (Figure 3e). After 72 hours of indirect coculture, cyc3 staining was evident in the cytoplasm of NRK52E cells (Figure 3e). In comparison, the addition of the exosome inhibitor GW4869 abolished the cyc3 staining in NRK52E cells (Figure 3e), suggesting the function of exosome as a cyc3 vehicle. To confirm this intercellular communication, NRK52E cells were also cocultured with MSCs without cyc3-oligo transfection and no cyc3 fluorescent labeling was detected (Figure 3f). These data confirm that miRNA from MSCs can be delivered intercellularly into kidney epithelial cells and provide an important evidence to investigate the antifibrotic effect of miR-let7c-MSCs on NRK52E cells.
MSC expressing miR-let7c reduces α-SMA and collagen in NRK52E cells
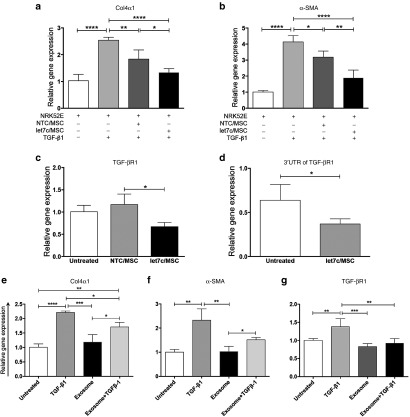
To determine the efficacy of miR-let7c delivered from engineered MSCs, qPCR was used to determine key genes involved in kidney fibrosis in NRK52E cells in response to TGF-β1. NRK52E cells, with or without the addition of TGF-β1, were cocultured using the indirect transwell system (described above) with miR-let7c-MSCs or NTC-MSCs as control comparisons. The addition of TGF-β1 induced a significant increase in collagen IVα1 expression in NRK52E cells compared with all other groups including untreated cells (P < 0.0001). When TGF-β1-treated NRK52E cells were indirectly cocultured with miR-let7c-MSCs, there was a significant decrease in collagen IVα1 expression (P < 0.0001), and compared with NTC-MSCs cocultured with TGF-β1-treated NRK52E cells (P < 0.05; Figure 4a). The coculture of NRK52E cells with NTC-MSCs also showed reduced expression of collagen IVα1 (P < 0.01) in the presence of TGF-β1, confirming our previous report of a protective effect of MSCs in UUO injury.24 The fibrotic marker, α-SMA, was assessed using qPCR and had reduced expression following NRK52E cells cocultured with miR-let7c-MSCs (P < 0.0001; Figure 4b). These data suggest that miR-let7c engineered MSCs had an amplified antifibrotic effect in the presence of TGF-β1, in comparison with NTC-MSCs.
Figure 4.
miR-let7c-MSC regulates fibrotic gene expression in kidney proximal tubular cells through exosomes. (a) The upregulated expression of collagen IVα1 in NRK52E cells, stimulated by the addition of TGF-β1, was significantly reduced following coculture with miR-let7c-MSCs (P < 0.0001), and to a lesser extent with NTC-MSCs (P < 0.01). (b) A downregulated expression of α-SMA was also observed following coculture with miR-let7c-MSCs (P < 0.0001) in the presence of TGF-β1. (c) The coculture of miR-let7c-MSCs also significantly repressed (P < 0.05) the expression of TGF-βR1 gene expression in NRK52E cells. (d) The coculture of miR-let7c-MSCs reduced the 3′UTR TGF-βR1 expression in NRK52E cells (P < 0.05). NRK52E cells were treated with exosomes isolated from the conditioned media of miR-let7c-MSCs cultured for 72 hours in the presence of TGF-β1. The addition of the isolated exosomes reduced the upregulated expression of (e) collagen IV, (f) α-SMA, and (g) TGF-βR1. Results are expressed as the mean ± SEM relative gene expression. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05. Col4α1, collagen IVα1; MSCs, mesenchymal stem cells; NTC, nontargeting control; SMA, smooth muscle actin; TGF-βR1, transforming growth factor beta receptor 1.
TGF-βR1 expression in NRK52E cells was reduced following coculture with MSCs expressing miR-let7c
The gene expression of TGF-βR1, a target gene of miR-let7c, was determined in NRK52E cells cocultured with or without miR-let7c-MSCs or NTC-MSCs (Figure 4c), indicating a reduction of TGF-βR1 by miR-let7c-MSCs (Figure 4c; P < 0.05). miRNAs are known to posttranscriptionally regulate target mRNA by base pairing to partially complementary sites in 3′UTRs preventing translation. For this study, NRK52E cells were transfected with 3′UTR of TGF-βR1 as previously described.9 After 72 hours of coculture, the NRK52E cells were assessed for luciferase reporter assay indicating TGF-βR1 reporter expression. miR-let7c-MSCs repressed luciferase activity for the 3′UTR wild-type constructs (Figure 4d; P < 0.05), as compared with coculture with NRK52E cells alone. NTC-MSCs were also found to have a repressive effect on the 3′UTR constructs (see Supplementary Figure S1). These data suggest that miR-let7c produced from engineered MSCs attenuates TGF-βR1 expression.
Exosomes from miR-let7c-MSC regulate the fibrotic gene expression
To further verify the pivotal role of exosomes as a fibrotic regulator, NRK52E cells were treated with exosomes isolated from miR-let7c-MSC conditioned medium in the presence of TGF-β1 after 72 hours of culture. The addition of exosomes reduced the expression of collagen IVα1 (P < 0.05; Figure 4e), α-SMA (Figure 4f), and TGF-βR1 (P < 0.01; Figure 4g), comparable with untreated NRK52E cells.
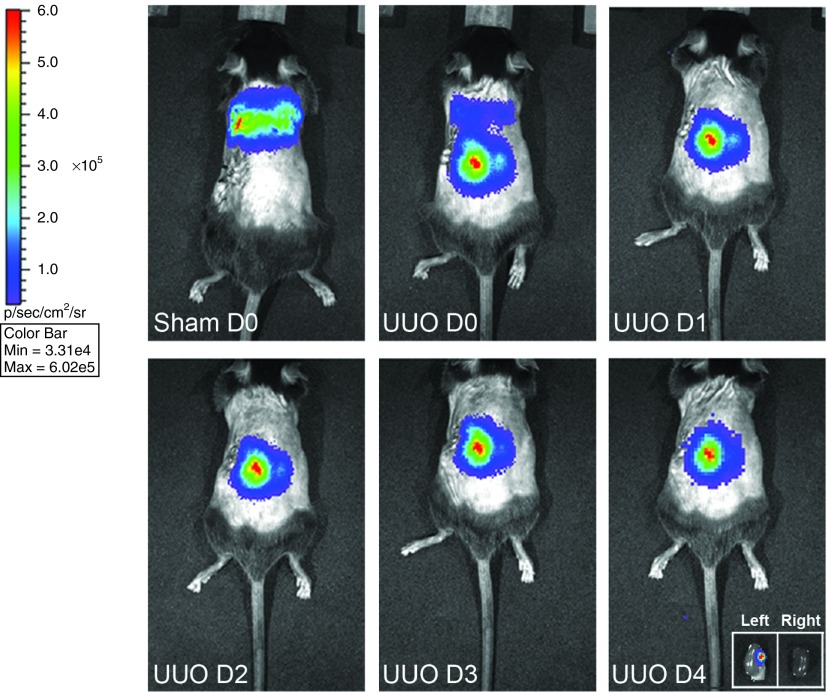
MSCs home to the injured kidney following UUO injury
Bioluminescence imaging was used to trace miR-let7c-MSCs transduced with a bicistronic lentiviral vector encoding firefly luciferase (fluc) and enhanced GFP (eGFP). eGFP+fluc+-miR-let7c-MSCs in vivo in a mouse model of UUO injury. Following intravenous delivery at the time of surgery, eGFP+fluc+-transfected miR-let7c-MSCs homed to the UUO injured kidney by 24 hours where they remained for the 4 days analyzed (Figure 5). Imaging and detection of the excised kidneys confirmed the localization of eGFP+fluc+-miR-let7c-MSCs in the UUO kidney; however, the cells were not evident in the CUKs (Figure 5, inset). In comparison, MSCs administered to sham-operated control mice showed that the cells were transiently localized in the lungs due to an initial entrapment in the pulmonary capillaries (Figure 5), as we have previously reported.23,24,26
Figure 5.
miR-let7c-MSCs migrate to injured UUO kidneys. eGFP+fluc+miR-let7c-MSCs were injected into mice with UUO injury or sham-operated controls at the time of surgery. The bioluminescence fluc signal was examined at day 0 (D0), day 1 (D1), day 2 (D2), day 3 (D3), and day 4 (D4) post injection on the dorsal surface of all animals. In sham-operated control animals, MSCs showed transient localization to the lungs post-MSC injection on D0. In mice with UUO injury, the MSCs homed to the left UUO-injured kidney, but not to the CUK kidney, as early as 1 day post injection and remained until D4. CUK, contralateral unobstructed kidney; D, day; MSCs, mesenchymal stem cells; UUO, unilateral ureteral obstruction.
MSCs deliver miR-let7c to injured kidneys resulting in improved kidney architecture and reduced collagen accumulation in UUO-injured mice
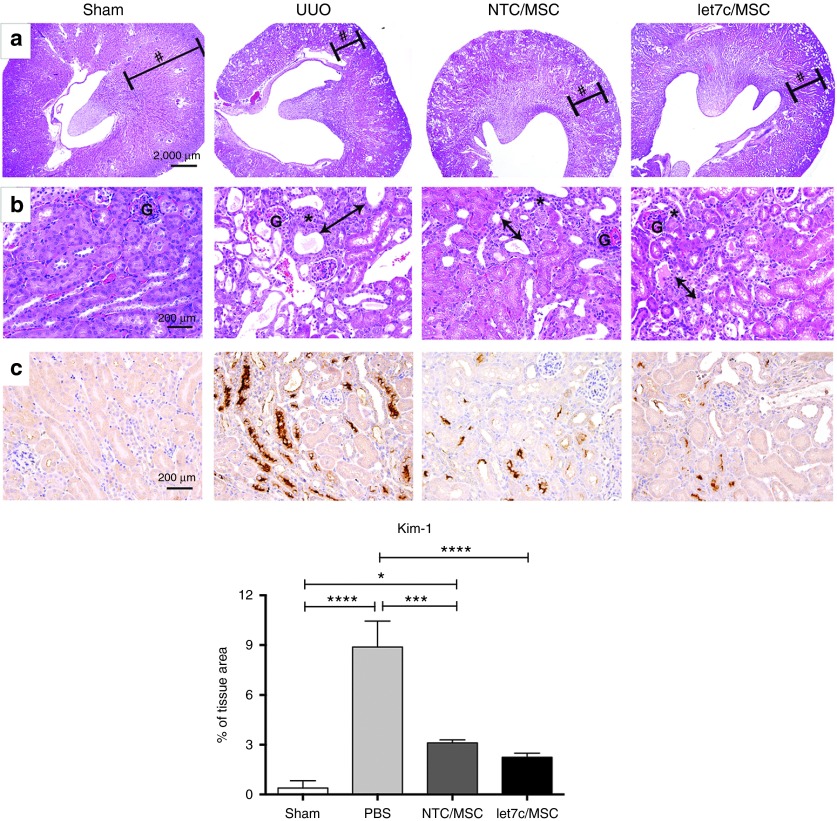
UUO-induced hydronephrosis at 7 days was characterized by the reduction in kidney parenchyma showing dilation of the cortical tubules and interstitial expansion as a result of interstitial inflammatory cell infiltration and collagen accumulation compared with CUKs (Figure 6a,b). At low magnification (Figure 6a), a preservation of kidney architecture was observed in UUO-injured kidneys that received miR-let7c-MSCs or NTC-MSCs. At higher power (Figure 6b), there was reduced tubular dilation, reduced interstitial inflammatory cell infiltration, and greater preservation of intact cortical tubules in UUO kidneys receiving miR-let7c-MSCs compared with vehicle-treated UUO mice. In addition, immunohistochemical staining of kidney injury molecule (Kim)-1 (Figure 6c), a marker of proximal tubular injury, revealed that there was a significant increase in protein localization at 7 days post-UUO compared with the sham-operated control kidneys (P < 0.0001). Both NTC-MSC (P < 0.001) and miR-let7c-MSC (P < 0.0001) treatments resulted in reduced Kim-1 expression; however, in miR-let7c-MSC-treated UUO mice, Kim-1 expression levels were comparable with sham mice confirming the increased protective effects of miR-let7c-MSCs.
Figure 6.
Administration of miR-let7c-MSC improved kidney injury in UUO mice. Representative hematoxylin and eosin stained sections of the CUK control kidney and UUO kidneys at 7 days post injury treated with either PBS vehicle, NTC-MSCs, or miR-let7c overexpressing MSCs. Regardless of treatment, a reduction of the parenchyma (hashtag) was observed in UUO kidneys after 7 days, compared with CUK control kidneys, at (a) low and (b) high magnification. Higher magnification shows that treatment with NTC-MSCs and miR-let7c-MSCs resulted in a decrease in tubular dilation and infiltration of inflammatory cells (asterisk), compared with UUO kidneys receiving vehicle. UUO kidneys administered miR-let7c-MSCs maintained greater structural integrity compared with NTC-MSCs, with increased evidence of intact brush borders in the proximal tubules and a reduction in interstitial matrix expansion (double headed arrow). (c) Representative photomicrographs and semi-quantification of Kim-1 expression in sham and UUO kidneys 7 days post-MSC treatment (magnification ×400) showing a significant reduction in Kim-1 expression in UUO kidneys following injection of miR-let7c-MSCs (P < 0.0001) compared with vehicle or NTC-MSC-injected mice (P < 0.001). CUK, contralateral unobstructed kidney; Kim-1, kidney injury molecule-1; MSCs, mesenchymal stem cells; NTC, nontargeting control; PBS, phosphate-buffered saline. Bar: a = 2000 µm (40×), b = 200 µm (400×).
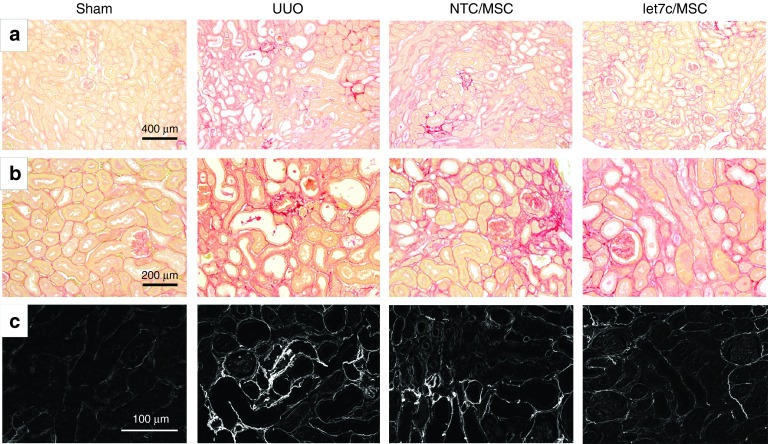
The reduction in accumulation of interstitial collagen was confirmed by assessment of the intensity and distribution of picrosirius red staining that correlated with the presence of interstitial fibrosis following UUO injury. Compared with control CUKs that showed a normal delicate framework of basement membrane collagen, UUO kidneys showed a widespread distribution of interstitial collagen and isolated regions at the arcuate vessels with more prolific, intense staining (Figure 7a,b). Treatment with miR-let7c-MSCs reduced the degree of UUO-induced collagen accumulation in UUO mice (Figure 7a,b), which was confirmed using polarized microscopy (Figure 7c).
Figure 7.
Treatment with miR-let7c-MSCs reduced UUO-induced interstitial fibrosis 7 days post injury. Representative picrosirius red stained kidney sections of the CUK control kidney and UUO kidneys at 7 days post injury receiving vehicle, NTC-MSCs, or miR-let7c overexpressing MSCs. At (a) low power and (b) higher power, 7 days post-UUO injury, there was widespread interstitial fibrosis, with regions of intense cortical red staining. In contrast, treatment with NTC-MSCs reduced the accumulation of interstitial fibrosis, which occurred at a greater extent in UUO kidneys following miR-let7c-MSCs treatment. In c, polarized microscopy was used to show collagen type I and III bundles as depicted by the birefringence (white). Compared with CUK kidneys, UUO kidneys show a significantly upregulated (P < 0.001) collagen accumulation at 7 days (d). When UUO mice received NTC-MSCs, there was a significant reduction in interstitial collagen (P < 0.05), which was further reduced following the administration of miR-let7c-MSCs (P < 0.05). The results shown are mean ±SEM relative to that of the control group, which is expressed as 1. ***P < 0.001, **P < 0.01, *P < 0.05. CUK, contralateral unobstructed kidney; MSCs, mesenchymal stem cells; NTC, nontargeting control; PBS, phosphate-buffered saline; UUO, unilateral ureteral obstruction. Bar: a = 400 µm (200×), b = 200 µm (400×), c = 100 µm (400×).
UUO-induced fibrotic-related gene expression is reduced by miR-let7c-MSC treatment
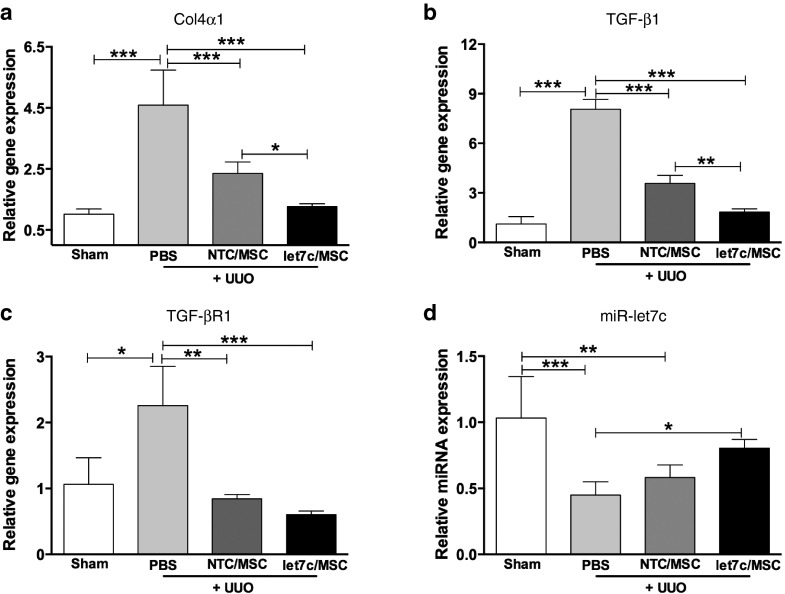
To validate the morphometric analysis, qPCR assessed the relative expression of collagen IVα1, TGF-β1, TGF-βR1, and miR-let7c following 7 days of treatment with/without miR-let7c-MSCs or NTC-MSCs in UUO mice. There was a significant elevation (approximately fourfold) of collagen IVα1 in UUO kidneys treated with vehicle, compared with the sham kidneys (P < 0.001; Figure 8a). The relative expression of collagen IVα1 was significantly reduced following the administration of miR-let7c-MSCs (Figure 8a) compared with UUO mice receiving vehicle (P < 0.001) and NTC-MSCs (P < 0.05), suggesting a potent rescue by miR-let7c-MSCs. Similarly, TGF-β1 expression was significantly elevated in UUO mice receiving vehicle (P < 0.001). Treatment with miR-let7c-MSCs in UUO mice significantly reduced TGF-β1 expression in comparison with UUO mice receiving vehicle (P < 0.001) and NTC-MSCs (P < 0.01; Figure 8b). In addition, TGF-βR1 expression was upregulated in UUO mice, compared with the sham (P < 0.05). Both NTC-MSC and miR-let7c-MSC treatments in UUO mice showed a significant reduction in TGF-βR1 expression (Figure 8c), suggesting a potent repression on TGF-βR1. Meanwhile, the expression of miR-let-7c was significantly decreased in the kidneys of UUO mice receiving vehicle (P < 0.001) and following administration of NTC-MSCs (P < 0.01). However, UUO mice receiving miR-let7c-MSCs had restored miR-let7c expression comparable with UUO mice receiving vehicle (P < 0.05; Figure 8d).
Figure 8.
miR-let7c-MSCs reduced fibrotic gene expression in kidneys from UUO mice. (a) Kidneys of UUO mice showed a significant reduction in collagen IVα1 expression following administration of miR-let7c-MSCs (P < 0.01) and NTC-MSCs (P < 0.01). The collagen IVα1 expression with miR-let7c-MSCs was significantly reduced comparable with NTC-MSC-injected UUO mice (P < 0.05), confirming beneficial effects of miR-let7c-MSCs therapy. (b) TGF-β1 expression was also repressed following treatment with miR-let7c-MSCs (P < 0.001). In comparison, miR-let7c-MSC-injected UUO mice showed a reduction in TGF-β1 (P < 0.01) compared with NTC-MSC-injected UUO mice. (c) TGF-βR1 expression was inhibited following treatment with miR-let7c-MSCs and NTC-MSCs in UUO mice (P < 0.01). (d) miR-let7c expression was significantly reduced in UUO kidneys, compared with sham kidneys (P < 0.01). However, miR-let7c expression was significantly restored following the administration of miR-let7c-MSCs comparable with PBS-injected UUO mice (P < 0.05). Results are expressed as the mean ± SEM relative gene expression. Col4α1, collagen IVα1; MSCs, mesenchymal stem cells; NTC, nontargeting control; TGF-βR1, transforming growth factor receptor 1. ***P < 0.001, **P < 0.01, *P < 0.05.
Discussion
miRNAs have gained increased recognition for their important regulatory role in a variety of biological processes by posttranscriptionally interfering with gene expression. Numerous studies have demonstrated that miRNAs play crucial roles in normal kidney development, tissue homeostasis,31,32 and pathogenesis of kidney disease (reviewed in refs. 2,33) and may therefore represent novel therapeutic targets to attenuate disease progression. However, current methods of miRNA delivery, including through viruses and liposomes, prove unsatisfactory due to safety concerns and low efficiency in vivo, respectively.34 Our data indicate a potential and feasible therapeutic strategy that couples MSCs and miRNAs as an effective, specific, and efficient delivery antifibrotic therapy targeted to kidney injury. Endogenous miRNAs can be transferred by MSCs via microvesicles and exosomes that act as intercellular communicators.35 We now report that MSCs, successfully engineered to overexpress miR-let7c, display normal growth characteristics and are able to deliver miRNAs through exocytosis using both in vitro and in vivo models of renal fibrosis. Using indirect coculture, exosome delivery of miR-let7c produced from miR-let7c-MSCs was able to repress collagen type IVα1 and α-SMA gene expression in tubular epithelial cells that had been stimulated with TGF-β1. Additional assays for the target of miR-let7c, TGF-βR1, also indicated the successful transfer of miR-let7c into NRK52E cells through exosomal delivery.
The increased production of miR-let7c from engineered MSCs provides a delivery vehicle to enable the enhanced ability of MSCs to decrease fibrosis and promote structural repair. An experimental mouse model of UUO was used to demonstrate that MSCs transferred miR-let7c exogenously to damaged kidneys, resulting in an upregulated miR-let7c gene expression and improved kidney structure as evidenced by reduced collagen accumulation and expression of fibrotic genes, compared with unobstructed kidneys. It has been previously reported that genetically engineered MSCs can improve hematopoietic engraftment36 and the targeted delivery of antitumor factors mediated by secretion of growth factors.37 These studies led to several clinical trials using MSCs for the treatment of osteogenesis imperfecta38,39 and Hurler's syndrome.40 Genetically modified MSCs can provide therapeutic biomolecules such as brain-derived neurotrophic factor for the treatment of Huntington's disease41 and duodenal homeobox-1 gene for the treatment of type 1 diabetes.42 The intercellular communication role of exosomes in genetically manipulated MSCs could also be applied for other small RNAs, for example, short hairpin RNA, which has shown a therapeutic protective function in brain.43 However, the lack of a strategy to successfully deliver short hairpin RNA has impeded any clinical advancement. Exocytosis of siRNA into exosomes using genetically modified MSCs may provide an efficient, tissue-specific, and nonimmunogenic delivery technology to broaden the clinical applications for gene therapy.
miRNAs regulate renal fibrosis,44 and several targets have been identified in the regulation of cytogenesis in polycystic kidney disease,45 and fibronectin,44 and collagen46 accumulation in diabetic nephropathy. miR-let7c was recently identified as an effective regulator in kidney fibrosis via regulation of the TGF-β signaling pathway.9,47 We8 and others9 have shown functional roles of the miR-let7 family members, miR-let7b and miR-let7c, in renal fibrosis resulting from diabetes mellitus. Our previous work reports that miR-let7 regulates TGF-βR1 and TGF-β/Smad signaling and may therefore also regulate the activity of matrix proteins.8 TGF-β1 is a classical mediator of renal fibrosis and inflammation.48 Profiling of human proximal tubule kidney-2 (HK2) cell miRNAs demonstrated that miR-let7c was strongly downregulated under fibrotic conditions compared with the levels of other miRNAs.9 miR-let7 is important in the regulation of cell proliferation, including cancer cells and stem cells,49 and is reduced in idiopathic pulmonary fibrosis.50,51
The selective homing of MSCs to the inflammatory site of injury makes specific exosome delivery and transport of miR-let7c using this delivery system particularly attractive. Our results show preferential tracking of MSCs to UUO kidneys following initial entrapment in the lungs, as we have previously reported in other models.52,53 MSCs can themselves elicit kidney repair through paracrine and/or endocrine mechanisms and release trophic growth factors that modulate the immune response and consequently mediate repair.54 Interestingly, endogenous miR-let7 expression has been identified in MSCs.12,55 Our results also indicate that control NTC-MSCs can repress the 3′UTR expression of TGFβ-R1, which is involved in endogenous miR-let7c secretion. Our in vivo studies showed that administration of NTC-MSCs also reduced kidney damage and collagen accumulation, although these renoprotective benefits were further enhanced following miR-let7c-MSC therapy. Importantly, another proinflammatory regulator, metalloproteinase-9 that has been suggested as a target of miR-let7, was also downregulated in the in vivo model after delivery of MSCs.56,57
MSCs may provide novel vehicles to deliver miRNA in a site-specific manner, but have the combined benefits of low immunogenicity.58 Several clinical studies have progressed the use of MSCs as a valuable tool for improving solid organ transplantation without the need for high concentrations of immunosuppressive drugs.59 The application of MSCs in the treatment of kidney disorders has resulted in no adverse side effects in humans.17,60 Indeed, preliminary results have shown promise, with MSC treatment improving graft survival and function, lowering the level of immunosuppression required, decreasing the risk of opportunistic infections, and reducing rejection.20,61
In summary, our studies show that genetic engineering of MSCs using miR-let7c enables effective antifibrotic effects and a high-efficiency delivery of therapeutic miR-let7c. Of particular relevance, this approach allows for the potential of delivering miRNAs, such as miR-let7c, within exosomes where they can be selectively tracked to damaged kidneys to produce a functional response. The targeted delivery of miR-let7 into the injured kidney in chronic and acute kidney damage is likely to profoundly alter the disease and ameliorate the progression to end-stage renal failure. In the long term, this innovative therapeutic approach could be applied to other chronic degenerative diseases that culminate in fibrotic injury.
Materials and Methods
Cell culture. Human bone marrow–derived MSCs (Tulane University, New Orleans, LA) were maintained in Alpha Minimum Essential Medium (Gibco, Scoresby, Vic, Australia) from passage 4 to 10 and supplemented with 16.5% fetal bovine serum (FBS; Atlanta Biologicals), 4 mM l-glutamine (Gibco), and 1% streptomycin sulphate and penicillin (Gibco). The MSCs were characterized for multilineage differentiation potential using a human mesenchymal stem cell functional identification kit (R&D Systems, Minneapolis, MN).
Rat kidney tubular epithelial cells (NRK52E; Sigma ATCC) utilized from passage 34 to 44 were maintained in Dulbecco's modified Eagle medium (Life Technologies, Australia) supplemented with 10% FBS, 4 mM l-glutamine, and 1% streptomycin sulphate and penicillin and cultured with/without TGF-β1 (5 ng/ml; R&D Systems). For coculture studies, NRK52E cells were seeded in 6-well cell culture plates at a density of 80,000 cells/well. Hanging cell culture inserts (1 µm; Millipore, Billerica, MA) were positioned above the NRK52E cells and seeded with human MSCs at a density of 30,000 cells/insert. Cells were cultured using MSC media, as previously described.26
Lentiviral transduction of MSCs and cell sorting. RNA duplexes corresponding to short noncoding oligos labeled with Cy3 (Life Technologies) were transfected in MSCs using lipofectamine2000 (Lipofectamine, Life Technologies) as previously described.8 The transfection efficiency using lipofectamine2000 was approximately 70%, as visualized using fluorescence microscopy. SMARTchoice shMIMIC Lentiviral miRNA expressing pre-miR-let7c and NTC pre-miR were purchased from Thermo Scientific (Waltham, MA). The titration was performed at an multiplicity of infection (MOI) of 20 as per the manufacturer's protocol. To determine the chromosomal and genetic stability of the engineered BM-MSCs, karyotyping was performed (Southern Cross Pathology, Clayton, Australia). MSCs were sorted on GFP expression using flow cytometry to enrich successfully transduced cells (BD Influx Cell Sorter version 7; BD Biosciences) and visualized by fluorescence microscopy (IX81 Olympus, Cellsens Dimension, Center Valley, PA).
Preparation of exosomes. MSCs were tranduced with NTC miRNA or miR-let7c and were maintained in MSC medium using exosome-free FBS. After 4 days of incubation, exosomes were isolated from the supernatants of the MSC cultures using the exosome precipitation solution, ExoQuick, as previously described (System Bioscience, Mountain View, CA). RNA was isolated using a Qiagen RNA extraction kit (Qiagen, Valencia, CA).
RNA extraction and real-time PCR. The RNAs were isolated using Qiagen RNA extraction kit according to the manufacturer's instructions. Gene expression was analyzed by real-time qPCR, performed using the TaqMan system based on real-time detection of accumulated fluorescence (ABI Prism 7500; Perkin-Elmer, Foster City, CA). Fluorescence for each cycle was quantitatively analyzed by an ABI Prism 7500 Sequence Detection System (Perkin-Elmer). Gene expression of the target sequence was normalized in relation to the expression of the endogenous control, β-actin. The dCt method was used to quantitate gene expression and expressed relative to control/untreated samples, which is arbitrarily assigned a value of 1. Each experiment was performed three times with 3–6 replicates per group.
microRNA assay. For miRNA analysis, cDNA synthesis qPCR was performed using TaqMan miRNA assays according to the manufacturer's recommendations (Applied Biosystems, Foster City, CA). Experimental groups were in replicates of six and normalized to U6 snRNA for rat and mouse samples, respectively. Each experiment was conducted three times.
3′UTR luciferase reporter analyses. The 3′UTR luciferase reporter assays were performed as we have previously described.9 NRK52E cells were seeded at a density of 1 × 105 cells per well in 6-well plates 1 day prior to transfection with PRL reporter plasmids (0.5 mg/ml), CMV-galactosidase construct, and miRNA inhibitors using Lipofectamine2000 (Life Technologies) in OptiMEM medium (Life Technologies). 24 hours later, engineered MSCs containing miR-let7c or NTC were seeded on the top of a transwell insert. NRK52E cells were harvested 72 hours post transfection using the Dual-Luciferase reporter assay system (Promega, Madison, WI), and luciferase and pCMV assays were performed according to the manufacturer's recommendations. The 3′-UTR of human TGF-βR1 gene contains an 8-mer (75–81: CUACCUCA) and 7-mer (3889–3895: UACCUCA) let7c binding site conserved across multiple species.
Animal surgery. Male C57BL/6J mice weighing 20–25 g were randomly allocated into the treatment groups of vehicle, NTC MSCs, or miR-let7c MSCs (n = 5/group/timepoint). All surgeries were performed under 2% isoflurane anesthesia (Abbott Australasia, Kurnell, NSW, Australia), administered via inhalation. UUO surgery was performed as previously described,62 whereby the left ureter was visualized via a flank incision and ligated using double tracks with 5.0 surgical silk. The right CUK served as a biological control. After UUO surgery, treatment of phosphate-buffered saline (PBS) vehicle, 1 × 106 NTC MSCs or 1 × 106 miR-let7c MSCs were administered intravenously prior to suturing and recovery. All experiments were approved by the Monash University Animal Ethics Committee, which adheres to the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes.
Bioluminescence imaging. In order to trace MSCs in vivo, a subgroup of animals underwent sham or UUO surgery and were administered 1 × 106 MSCs transduced with a bicistronic lentiviral vector encoding firefly luciferase (fluc) and eGFP.63 The eGFP+fluc+MSCs were FACS sorted and GFP expression was visually confirmed with fluorescence microscopy.26 Using a noninvasive bioluminescent imaging technique, eGFP+fluc+MSCs were administered intravenously and traced in vivo. Mice were anesthetized, injected intraperitoneally with 200 µl of d-luciferin (15 mg/ml in PBS; VivoGlo Luciferin, Promega, San Luis Obispo, CA) on days 0, 1, 2, 3, and 4 post surgery, and imaged using the IVIS 200 system (Xenogen, Alameda, CA). The fluc luminescent signal was captured and analyzed as photons/second/cm2 using Living Image 3.2 software (Xenogen). The localization of MSCs in the kidney was confirmed by imaging the excised CUK and UUO kidneys on day 4.
Histopathology and analysis of kidney collagen content. After 7 days of UUO-induced injury, transverse, paraffin-embedded kidneys sectioned at 4 µm were stained with hematoxylin and eosin to assess changes in kidney structure. Kim-1 immunohistochemical staining was performed with a rat anti-mouse Kim-1 antibody (R&D Systems) using the avidin–biotin complex method. Staining was quantified in 10 nonoverlapping, randomly selected fields of view (n = 3, 3 sections/mouse, ×400 magnification). The area of 3,3′-Diaminobenzidine staining per unit area of tissue was measured using a custom macro in FIJI software, version 2.0.0. Paraffin-embedded kidney sections were stained with picrosirius red to visualize the distribution of interstitial collagen under brightfield and polarized microscopy as described previously.24
Statistical analysis. Values are shown as means ± SEM, unless otherwise specified. GraphPad Prism (GraphPad Software) was used to analyze data by unpaired Student's t-test or by one-way analysis of variance followed by a Tukey's multiple comparison test. P values <0.05 were considered significant.
SUPPLEMENTARY MATERIAL Figure S1. NRK52E cells transfected with the luciferase construct of TGF-βR1 3′UTR were cocultured with NTC-MSCs using a Transwell system for 3 days. Figure S2. Expression of miR-let7c, miR-let7a, and miR-let7d in exosomes from NTC-MSCs and miR-let7c-MSCs.
Acknowledgments
B.W. and B.M.H. acknowledge funding support from Monash University and Kidney Health Australia. J.L.W.-B. is a Senior Research Fellow of the National Health and Medical Research Council of Australia. Work in C.G.'s lab is supported by Science Foundation Ireland Awards 06/IN/.1/B114 and 08/US/B1517. The authors acknowledge technical assistance from Alex Fulcher and Keith Schulze (Monash Micro Imaging, Monash University). The 3′UTR of the human TGF-βR1 was provided by C.G.
Supplementary Material
References
- Chen, K and Rajewsky, N (2007). The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet 8: 93–103. [DOI] [PubMed] [Google Scholar]
- Wang, B and Ricardo, S (2014). Role of microRNA machinery in kidney fibrosis. Clin Exp Pharmacol Physiol 41: 543–550. [DOI] [PubMed] [Google Scholar]
- Kawahara, Y (2014). Human diseases caused by germline and somatic abnormalities in microRNA and microRNA-related genes. Congenit Anom (Kyoto) 54: 12–21. [DOI] [PubMed] [Google Scholar]
- Lewis, BP, Burge, CB and Bartel, DP (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- Hwang, HW and Mendell, JT (2006). MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 94: 776–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran, K, Karolina, DS, Sepramaniam, S, Armugam, A, Wintour, EM, Bertram, JF et al. (2012). Role of microRNAs in kidney homeostasis and disease. Kidney Int 81: 617–627. [DOI] [PubMed] [Google Scholar]
- Patel, V and Noureddine, L (2012). MicroRNAs and fibrosis. Curr Opin Nephrol Hypertens 21: 410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B, Jha, JC, Hagiwara, S, McClelland, AD, Jandeleit-Dahm, K, Thomas, MC et al. (2014). Transforming growth factor-β1-mediated renal fibrosis is dependent on the regulation of transforming growth factor receptor 1 expression by let-7b. Kidney Int 85: 352–361. [DOI] [PubMed] [Google Scholar]
- Brennan, EP, Nolan, KA, Börgeson, E, Gough, OS, McEvoy, CM, Docherty, NG et al.; GENIE Consortium. (2013). Lipoxins attenuate renal fibrosis by inducing let-7c and suppressing TGFβR1. J Am Soc Nephrol 24: 627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, SD, Pollock, K, Kambal, A, Cary, W, Mitchell, GM, Tempkin, J et al. (2012). Genetically engineered mesenchymal stem cells as a proposed therapeutic for Huntington's disease. Mol Neurobiol 45: 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelbrunn, M, Gutiérrez-Vázquez, C, Villarroya-Beltri, C, González, S, Sánchez-Cabo, F, González, MÁ et al. (2011). Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun 2: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi, H, Ekström, K, Bossios, A, Sjöstrand, M, Lee, JJ and Lötvall, JO (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9: 654–659. [DOI] [PubMed] [Google Scholar]
- Bellingham, SA, Coleman, BM and Hill, AF (2012). Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res 40: 10937–10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y and Huang, L (2013). A window onto siRNA delivery. Nat Biotechnol 31: 611–612. [DOI] [PubMed] [Google Scholar]
- Chen, TS, Lai, RC, Lee, MM, Choo, AB, Lee, CN and Lim, SK (2010). Mesenchymal stem cell secretes microparticles enriched in pre-microRNAs. Nucleic Acids Res 38: 215–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, W, Sheng, CT, Tan, B, Lee, QY, Kuznetsov, V, Kiang, LS et al. (2010). Analysis of deep sequencing microRNA expression profile from human embryonic stem cells derived mesenchymal stem cells reveals possible role of let-7 microRNA family in downstream targeting of hepatic nuclear factor 4 alpha. BMC Genomics 11 (suppl. 1): S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders, ME, de Fijter, JW, Roelofs, H, Bajema, IM, de Vries, DK, Schaapherder, AF et al. (2013). Autologous bone marrow-derived mesenchymal stromal cells for the treatment of allograft rejection after renal transplantation: results of a phase I study. Stem Cells Transl Med 2: 107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y, Cao, Y, Li, X, Xu, L, Wang, Z, Liu, P et al. (2014). Cotransplantation of haploidentical hematopoietic and umbilical cord mesenchymal stem cells for severe aplastic anemia: successful engraftment and mild GVHD. Stem Cell Res 12: 132–138. [DOI] [PubMed] [Google Scholar]
- Connick, P, Kolappan, M, Crawley, C, Webber, DJ, Patani, R, Michell, AW et al. (2012). Autologous mesenchymal stem cells for the treatment of secondary progressive multiple sclerosis: an open-label phase 2a proof-of-concept study. Lancet Neurol 11: 150–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, J, Wu, W, Xu, X, Liao, L, Zheng, F, Messinger, S et al. (2012). Induction therapy with autologous mesenchymal stem cells in living-related kidney transplants: a randomized controlled trial. JAMA 307: 1169–1177. [DOI] [PubMed] [Google Scholar]
- Perico, N, Casiraghi, F, Introna, M, Gotti, E, Todeschini, M, Cavinato, RA et al. (2011). Autologous mesenchymal stromal cells and kidney transplantation: a pilot study of safety and clinical feasibility. Clin J Am Soc Nephrol 6: 412–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y and Zhao, RC (2012). The role of chemokines in mesenchymal stem cell homing to myocardium. Stem Cell Rev 8: 243–250. [DOI] [PubMed] [Google Scholar]
- Cardenes, N, Caceres, E, Romagnoli, M and Rojas, M (2013). Mesenchymal stem cells: a promising therapy for the acute respiratory distress syndrome. Respiration 85: 267–278. [DOI] [PubMed] [Google Scholar]
- Huuskes, BM, Wise, AF, Cox, AJ, Lim, EX, Payne, NL, Kelly, DJ et al. (2015). Combination therapy of mesenchymal stem cells and serelaxin effectively attenuates renal fibrosis in obstructive nephropathy. FASEB J 29: 540–553. [DOI] [PubMed] [Google Scholar]
- Spaggiari, GM, Capobianco, A, Abdelrazik, H, Becchetti, F, Mingari, MC and Moretta, L (2008). Mesenchymal stem cells inhibit natural killer-cell proliferation, cytotoxicity, and cytokine production: role of indoleamine 2,3-dioxygenase and prostaglandin E2. Blood 111: 1327–1333. [DOI] [PubMed] [Google Scholar]
- Wise, AF, Williams, TM, Kiewiet, MB, Payne, NL, Siatskas, C, Samuel, CS et al. (2014). Human mesenchymal stem cells alter macrophage phenotype and promote regeneration via homing to the kidney following ischemia-reperfusion injury. Am J Physiol Renal Physiol 306: F1222–F1235. [DOI] [PubMed] [Google Scholar]
- Morigi, M, Introna, M, Imberti, B, Corna, D, Abbate, M, Rota, C et al. (2008). Human bone marrow mesenchymal stem cells accelerate recovery of acute renal injury and prolong survival in mice. Stem Cells 26: 2075–2082. [DOI] [PubMed] [Google Scholar]
- Liu, YL, Wang, YD, Zhuang, F, Xian, SL, Fang, JY, Su, W et al. (2012). Immunosuppression effects of bone marrow mesenchymal stem cells on renal interstitial injury in rats with unilateral ureteral obstruction. Cell Immunol 276: 144–152. [DOI] [PubMed] [Google Scholar]
- Tögel, F, Hu, Z, Weiss, K, Isaac, J, Lange, C and Westenfelder, C (2005). Administered mesenchymal stem cells protect against ischemic acute renal failure through differentiation-independent mechanisms. Am J Physiol Renal Physiol 289: F31–F42. [DOI] [PubMed] [Google Scholar]
- Bengestrate, L, Virtue, S, Campbell, M, Vidal-Puig, A, Hadaschik, D, Hahn, P et al. (2011). Genome-wide profiling of microRNAs in adipose mesenchymal stem cell differentiation and mouse models of obesity. PLoS One 6: e21305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, J and Kreidberg, JA (2013). MicroRNAs in renal development. Pediatr Nephrol 28: 219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L and Qu, L (2013). The function of microRNAs in renal development and pathophysiology. J Genet Genomics 40: 143–152. [DOI] [PubMed] [Google Scholar]
- Schena, FP, Serino, G and Sallustio, F (2014). MicroRNAs in kidney diseases: new promising biomarkers for diagnosis and monitoring. Nephrol Dial Transplant 29: 755–763. [DOI] [PubMed] [Google Scholar]
- Broderick, JA and Zamore, PD (2011). MicroRNA therapeutics. Gene Ther 18: 1104–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings, DJ, Ciaudo, C, Erhardt, M and Voinnet, O (2009). Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol 11: 1143–1149. [DOI] [PubMed] [Google Scholar]
- Nolta, JA, Hanley, MB and Kohn, DB (1994). Sustained human hematopoiesis in immunodeficient mice by cotransplantation of marrow stroma expressing human interleukin-3: analysis of gene transduction of long-lived progenitors. Blood 83: 3041–3051. [PubMed] [Google Scholar]
- Brooke, G, Cook, M, Blair, C, Han, R, Heazlewood, C, Jones, B et al. (2007). Therapeutic applications of mesenchymal stromal cells. Semin Cell Dev Biol 18: 846–858. [DOI] [PubMed] [Google Scholar]
- Horwitz, EM, Gordon, PL, Koo, WK, Marx, JC, Neel, MD, McNall, RY et al. (2002). Isolated allogeneic bone marrow-derived mesenchymal cells engraft and stimulate growth in children with osteogenesis imperfecta: implications for cell therapy of bone. Proc Natl Acad Sci USA 99: 8932–8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz, EM, Prockop, DJ, Fitzpatrick, LA, Koo, WW, Gordon, PL, Neel, M et al. (1999). Transplantability and therapeutic effects of bone marrow-derived mesenchymal cells in children with osteogenesis imperfecta. Nat Med 5: 309–313. [DOI] [PubMed] [Google Scholar]
- Koç, ON, Day, J, Nieder, M, Gerson, SL, Lazarus, HM and Krivit, W (2002). Allogeneic mesenchymal stem cell infusion for treatment of metachromatic leukodystrophy (MLD) and Hurler syndrome (MPS-IH). Bone Marrow Transplant 30: 215–222. [DOI] [PubMed] [Google Scholar]
- Crigler, L, Robey, RC, Asawachaicharn, A, Gaupp, D and Phinney, DG (2006). Human mesenchymal stem cell subpopulations express a variety of neuro-regulatory molecules and promote neuronal cell survival and neuritogenesis. Exp Neurol 198: 54–64. [DOI] [PubMed] [Google Scholar]
- Karnieli, O, Izhar-Prato, Y, Bulvik, S and Efrat, S (2007). Generation of insulin-producing cells from human bone marrow mesenchymal stem cells by genetic manipulation. Stem Cells 25: 2837–2844. [DOI] [PubMed] [Google Scholar]
- Alvarez-Erviti, L, Seow, Y, Yin, H, Betts, C, Lakhal, S and Wood, MJ (2011). Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol 29: 341–345. [DOI] [PubMed] [Google Scholar]
- Wang, Q, Wang, Y, Minto, AW, Wang, J, Shi, Q, Li, X et al. (2008). MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J 22: 4126–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, SO, Masyuk, T, Splinter, P, Banales, JM, Masyuk, A, Stroope, A et al. (2008). MicroRNA15a modulates expression of the cell-cycle regulator Cdc25A and affects hepatic cystogenesis in a rat model of polycystic kidney disease. J Clin Invest 118: 3714–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, M, Zhang, J, Wang, M, Lanting, L, Yuan, H, Rossi, JJ et al. (2007). MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci USA 104: 3432–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, N, Chen, J, Shang, Z, Dou, H, Ji, G, Zou, Q et al. (2013). Piglets cloned from induced pluripotent stem cells. Cell Res 23: 162–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, HY and Chung, AC (2012). TGF-β/Smad signaling in kidney disease. Semin Nephrol 32: 236–243. [DOI] [PubMed] [Google Scholar]
- Johnson, CD, Esquela-Kerscher, A, Stefani, G, Byrom, M, Kelnar, K, Ovcharenko, D et al. (2007). The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 67: 7713–7722. [DOI] [PubMed] [Google Scholar]
- Pandit, KV, Corcoran, D, Yousef, H, Yarlagadda, M, Tzouvelekis, A, Gibson, KF et al. (2010). Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 182: 220–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit, KV, Milosevic, J and Kaminski, N (2011). MicroRNAs in idiopathic pulmonary fibrosis. Transl Res 157: 191–199. [DOI] [PubMed] [Google Scholar]
- Fischer, UM, Harting, MT, Jimenez, F, Monzon-Posadas, WO, Xue, H, Savitz, SI et al. (2009). Pulmonary passage is a major obstacle for intravenous stem cell delivery: the pulmonary first-pass effect. Stem Cells Dev 18: 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, RH, Pulin, AA, Seo, MJ, Kota, DJ, Ylostalo, J, Larson, BL et al. (2009). Intravenous hMSCs improve myocardial infarction in mice because cells embolized in lung are activated to secrete the anti-inflammatory protein TSG-6. Cell Stem Cell 5: 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise, AF and Ricardo, SD (2012). Mesenchymal stem cells in kidney inflammation and repair. Nephrology (Carlton) 17: 1–10. [DOI] [PubMed] [Google Scholar]
- Stoorvogel, W (2012). Functional transfer of microRNA by exosomes. Blood 119: 646–648. [DOI] [PubMed] [Google Scholar]
- Tan, TK, Zheng, G, Hsu, TT, Lee, SR, Zhang, J, Zhao, Y et al. (2013). Matrix metalloproteinase-9 of tubular and macrophage origin contributes to the pathogenesis of renal fibrosis via macrophage recruitment through osteopontin cleavage. Lab Invest 93: 434–449. [DOI] [PubMed] [Google Scholar]
- Ventayol, M, Viñas, JL, Sola, A, Jung, M, Brüne, B, Pi, F et al. (2014). miRNA let-7e targeting MMP9 is involved in adipose-derived stem cell differentiation toward epithelia. Cell Death Dis 5: e1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, JM, Barry, FP, Murphy, JM and Mahon, BP (2005). Mesenchymal stem cells avoid allogeneic rejection. J Inflamm (Lond) 2: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pileggi, A, Xu, X, Tan, J and Ricordi, C (2013). Mesenchymal stromal (stem) cells to improve solid organ transplant outcome: lessons from the initial clinical trials. Curr Opin Organ Transplant 18: 672–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perico, N, Casiraghi, F, Gotti, E, Introna, M, Todeschini, M, Cavinato, RA et al. (2013). Mesenchymal stromal cells and kidney transplantation: pretransplant infusion protects from graft dysfunction while fostering immunoregulation. Transpl Int 26: 867–878. [DOI] [PubMed] [Google Scholar]
- Peng, Y, Ke, M, Xu, L, Liu, L, Chen, X, Xia, W et al. (2013). Donor-derived mesenchymal stem cells combined with low-dose tacrolimus prevent acute rejection after renal transplantation: a clinical pilot study. Transplantation 95: 161–168. [DOI] [PubMed] [Google Scholar]
- Cochrane, AL, Kett, MM, Samuel, CS, Campanale, NV, Anderson, WP, Hume, DA et al. (2005). Renal structural and functional repair in a mouse model of reversal of ureteral obstruction. J Am Soc Nephrol 16: 3623–3630. [DOI] [PubMed] [Google Scholar]
- Payne, NL, Sun, G, McDonald, C, Layton, D, Moussa, L, Emerson-Webber, A et al. (2013). Distinct immunomodulatory and migratory mechanisms underpin the therapeutic potential of human mesenchymal stem cells in autoimmune demyelination. Cell Transplant 22: 1409–1425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.