Summary
Fast excitatory synaptic signaling in the mammalian brain is mediated by AMPA-type ionotropic glutamate receptors. In neurons, AMPA receptors co-assemble with auxiliary proteins, such as stargazin, which can markedly alter receptor trafficking and gating. Here we used luminescence resonance energy transfer measurements to map distances between the full-length, functional AMPA receptor and stargazin expressed in HEK-293 cells and to determine the ensemble structural changes in the receptor due to stargazin. In addition, we used single molecule fluorescence resonance energy transfer to study the structural and conformational distribution of the receptor, and how this distribution is affected by stargazin. Our nanopositioning data place stargazin below the AMPA receptor ligand-binding domain, where it is well-poised to act as a scaffold to facilitate the long-range conformational selection observations seen in single molecule experiments. These data support a model of stargazin acting to stabilize or select conformational states which favor activation.
Keywords: AMPA Receptors, Transmembrane AMPA Receptor Regulatory Proteins, Stargazin, AMPA Receptor Modulation, Desensitized State
Graphical Abstract
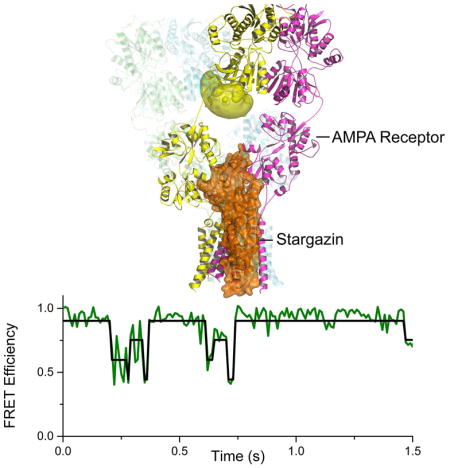
Model of AMPA/Stargazin complex determined by lanthanide resonance energy transfer nanopositioning in live cells (top); single molecule FRET trajectory revealing conformational changes in full-length AMPA receptors (bottom).
Glutamate receptors are the predominant mediators of excitatory synaptic signaling in the central nervous system and play an important role in the regulation of synaptic strength, learning and memory, and in diverse neuropathologies, including epilepsy and ischemia (Dingledine et al., 1999). Based on agonist affinity profiles, glutamate receptors can be subdivided into three subfamilies: α-amino-5-methyl-3-hydroxy-4-isoxazole propionate (AMPA) receptors, N-methyl-D-aspartate (NMDA) receptors, and kainate receptors (Hollmann and Heinemann, 1994). Amongst the three subtypes, AMPA receptors mediate the fast component of excitatory signaling. As such, AMPA receptors and their gating properties are critical in shaping the dynamics of the synaptic transmission. With the discovery of the family of related transmembrane AMPA receptor regulatory proteins (TARPs), it has become increasingly clear that auxiliary proteins modulate many aspects of AMPA receptor function, making a vital contribution to excitatory synaptic- and neuronal–glial signaling. Stargazin, also known as γ2, is the most well-characterized member of the TARP protein family. Initially determined to play important roles in AMPA receptor trafficking (Bats et al., 2007), stargazin and the other TARPs have more recently been established as key contributors to the diversity of signaling attributed to the AMPA receptor, with this diversity arising from the additional TARP-mediated modulation of channel gating (Cho et al., 2007; Tomita et al., 2005). Electrophysiological measurements show that stargazin increases the efficacy and potency of agonists at AMPA receptors, slows glutamate-mediated AMPA receptor deactivation and desensitization, accelerates recovery from desensitization, reduces polyamine block, and enables a superactivation of AMPA receptors (Carbone and Plested, 2016; Cho et al., 2007; Maclean and Bowie, 2011; Tomita et al., 2005). The modulation of AMPA receptor agonist efficacy has been attributed to stargazin increasing closure of the ligand-binding domain of the AMPA receptor (MacLean et al., 2014). However, such a mechanism cannot account for all the effects of TARPs on AMPA receptor gating, and indeed recent work has demonstrated that TARPs also alter the various biophysical properties of AMPA receptors through distinct processes (Dawe et al., 2016). Therefore, a full explanation for how TARPs shape the features of AMPA receptor gating is still largely unknown.
The structure of the AMPA receptor in isolation has been studied extensively under various ligand conditions (Durr et al., 2014; Meyerson et al., 2014; Yelshanskaya et al., 2014). These structures show that the receptor is a tetramer that is organized as a dimer of dimers, with extracellular amino-terminal and ligand-binding domains, transmembrane segments, and an intracellular C-terminal domain. Subunit crossover between the amino-terminal and ligand-binding domains results in different subunit groupings at each level, with the amino-terminal domain comprising A/B and C/D dimers, and the ligand-binding domain comprising A/D and B/C dimers. These structures also show that the ligand-binding domain undergoes a cleft closure upon binding agonists, and this cleft closure conformational change is thought to drive receptor activation (channel opening). In the continued presence of agonists, the dimer interface at the ligand-binding domain decouples, leading to receptor desensitization (Gonzalez et al., 2010). This decoupling within ligand-binding domain dimers is also observed between the amino-terminal domain dimers. The extent of this decoupling is still unclear, as a wide range of decoupling between the dimers has been observed in the reported X-ray and cryo-EM structures (Durr et al., 2014; Meyerson et al., 2014; Yelshanskaya et al., 2014).
Here we advance a model of the AMPA Receptor-stargazin complex by positioning a claudin-based homology model of stargazin through luminescence resonance energy transfer (LRET)-derived distance measurements between the AMPA receptor and the stargazin. This model of the complex in HEK-293 cells is comparable to the recently published cryo-EM structures of the AMPA receptor in complex with stargazin, also expressed in HEK cells (Twomey et al., 2016; Zhao et al., 2016). Furthermore, we have used LRET and single molecule fluorescence resonance energy transfer (smFRET) to examine the extent of amino-terminal domain decoupling associated with desensitization, and thus show the receptor favors less decoupling in the presence of stargazin. This result suggests that stargazin acts as a scaffold to reduce decoupling between subunits and hence stabilizes the active open conformation of the receptor.
Results and Discussion
Mapping stargazin to the AMPA receptor
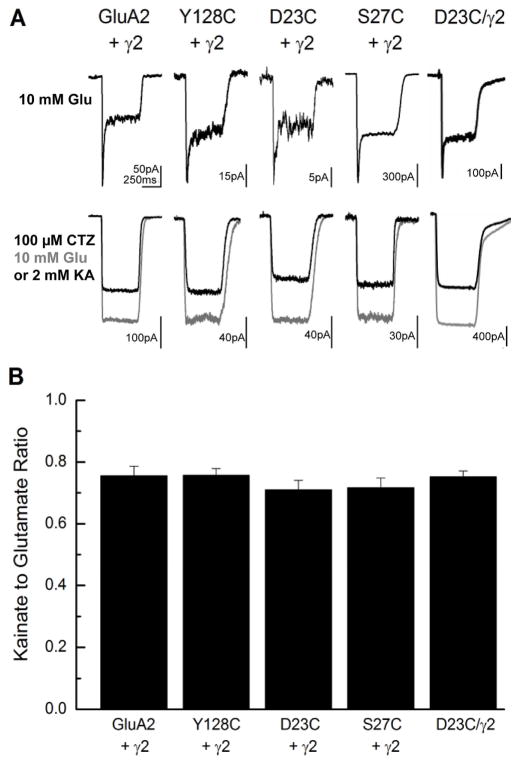
In order to perform the LRET investigations to measure the distance between the AMPA receptor and stargazin we modified the GluA2 subunit by mutating the surface-accessible cysteine residues at positions 89, 196, and 436 to serines. Additionally, we introduced a Factor Xa protease recognition sequence at position 228, allowing us to perform the experiments under near physiological conditions with proteins expressed in HEK-293T cells without the need for purification. This site was chosen as it required minimal substitution from residues TDGD to IDGR. By performing LRET measurements before and after protease cleavage, we were able to quantify the background and determine the specific signal arising from GluA2 receptors (Dolino et al., 2014; Gonzalez et al., 2008; MacLean et al., 2014; Rambhadran et al., 2010, 2011; Sirrieh et al., 2013, 2015). This basal LRET construct will hereafter be referred to as GluA2*. For our mapping studies, the tyrosine at site 128 was further mutated to cysteine, introducing a reactive site for a maleimide derivative of terbium chelate, which served as the donor fluorophore. We confirmed that this construct, GluA2*Y128C, was functional and that stargazin modulation was intact by measuring kainate-to-glutamate ratios electrophysiologically (Figure 1). Kainate is a poor agonist at AMPA receptors but in the presence of stargazin it becomes more efficacious (Tomita et al., 2005). We found that when stargazin was co-transfected, kainate was equally efficacious for both wild type GluA2 as well as GluA2*Y128C (kainate-to-glutamate ratio of 0.76 ± 0.03, n = 4 for wild-type; 0.76 ± 0.02, n = 5 for GluA2*Y128C, p = 0.63). In order to measure the distances between the GluA2 subunit and stargazin, acceptor fluorophores were tagged on stargazin by introducing the unnatural amino acid p-acetyl-L-phenylalanine at various sites on the extracellular domain of stargazin. The keto group of p-acetyl-L-phenylalanine could then be coupled to hydrazide derivatives of either fluorescein or Alexa 555. The background-subtracted LRET lifetime decays of GluA2*Y128C and stargazin tagged with donor and acceptor fluorophores, respectively, are shown in Figure 2a. The donor-only lifetimes for the GluA2*Y128C receptor tagged with terbium chelate in the absence of acceptor labeling is shown in Supplemental Figure S1. Based on the donor-only and sensitized acceptor lifetimes, the distances between site 128 on GluA2 and sites 44, 51, and 61 on stargazin were determined using the Förster equation (Supplemental Table S2). The errors reported in the Table are determined from error propagation from the error in the exponential fits of the data. The error in the absolute distances due to κ2 is approximately 10%. The use of an isotropic atomic donor reduces this error even further, as does the long lifetime of terbium luminescence, which allows for sufficient rotational diffusion of the acceptor (dos Remedios and Moens, 1995).
Figure 1. Stargazin modulation of AMPA receptor mutants used for LRET.
A) Representative whole cell recordings from HEK cells expressing wild type GluA2, GluA2*-Y128C, GluA2*-D23C and GluA2*-S27C in the presence of stargazin (γ2), and tandem GluA2*-D23C/γ2, in response to 10 mM glutamate (Glu) alone (upper panel) and either 10 mM glutamate or 2 mM kainate (KA) in the presence of 100 μM cyclothiazide (CTZ) (lower panel). B) Summary data showing the kainate to glutamate response ratio.
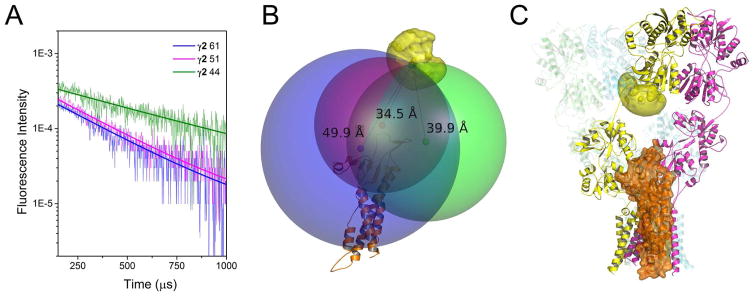
Figure 2. LRET-Nanopositioning system-based model of AMPA receptor-stargazin interaction.
A) LRET lifetimes between GluA2 amino-terminal domain site 128 of the apo AMPA receptor and sites 44 (green), 51 (magenta) and 61 (blue) in the extracellular region of stargazin (γ2) are shown. See also Figure S1. B) Stargazin homology model (orange) with the root mean positions of its fluorophores shown as red, green, and blue hard spheres. The LRET-determined distances to the donor fluorophore (39.9 Å for γ2 site 44, 34.5 Å for γ2 site 51, and 49.9 Å for γ2 site 61) were used to generate the larger spheres. The AMPA receptor structure and stargazin model were positioned to place the terbium fluorophore at the point of intersection of the LRET-radii spheres. C) Our model of the means of interaction between the AMPA receptor and stargazin. See also Figure S3
LRET lifetimes were also obtained between the AMPA receptor and stargazin in the presence of glutamate (Supplemental Figure S1). The distances between site 128 on AMPA receptor and site 44 and 51 on stargazin in the presence of glutamate are 39.0 Å and 37.5 Å. These distances are slightly different from the 39.9 Å and 34.5 Å respectively under apo conditions (Supplemental Table S2), and could indicate local rearrangements between the AMPA receptor and stargazin upon binding glutamate. The LRET data obtained with intact HEK-293 cells do not show dissociation of stargazin from the AMPA receptor in the presence of glutamate as previously reported by Tomita and coworkers (Morimoto-Tomita et al., 2009). However local rearrangements could account for the autoinactivation of AMPA receptors at high glutamate concentrations in the presence of stargazin and might underlie the observed dissociation of stargazin from the AMPA receptors under western blot conditions where membrane preparations were used (Morimoto-Tomita et al., 2009).
Based on the distances obtained from LRET measurements, we used a method similar to the nano-positioning methods previously reported (Kalinin et al., 2012; Muschielok and Michaelis, 2011; Tatulian, 2014). The SWISS-MODEL server (Biasini et al., 2014) was used, which generated a homology model for stargazin based on claudin-19. Then, the approximate root mean positions of the respective fluorophores were calculated as previously described (Kalinin et al., 2012) for both the AMPA receptor structure and our generated stargazin model. Spheres were drawn around the positions of the acceptor fluorophore using the LRET-determined distances as radii (Figure 2b). The sites of intersection of the three spheres were then considered as the likely spatial position of the donor fluorophore on the AMPA receptor. A series of rigid-body translations and rotations were performed to superpose the calculated root mean position of the donor fluorophore with the triangulated points of intersection of the acceptor fluorophore spheres with the additional constraint of maintaining the transmembrane domains of both the AMPA receptor and stargazin parallel, such that they are both are positioned in the membrane. The model that best fit the distance with minimal steric clashes (Figure 2c), positions the extracellular domain of stargazin next to the ligand-binding domain of the AMPA receptor, with a significant portion interacting with the linker regions and placed below the lower lobe of the ligand-binding domain. While the precise location of stargazin is harder to predict based on the LRET distances, placing stargazin close to the B or D ligand-binding domain in the A-B and C-D interfaces provides the most interactions between the transmembrane segments of stargazin and AMPA receptor. Additional poses were also obtained (Supplemental Figure S3); however, these showed minimal interactions at the level of the transmembrane segments of the AMPA receptor and stargazin.
Our model of stargazin in complex with the AMPA receptor is able to account for the previous functional and LRET measurements, which showed the ligand-binding domain of the AMPA receptor being more closed in the presence of stargazin (MacLean et al., 2014). It is also consistent with the prior peptide based mapping of stargazin on the AMPA receptor that showed significant interactions between the extracellular domain of stargazin and the ligand-binding domain of the AMPA receptor (Cais et al., 2014).
While this manuscript was under review, two laboratories published cryo-EM structures of the AMPA receptor-stargazin complex (Twomey et al., 2016; Zhao et al., 2016). Both structures show that stargazin interacts with the B and D subunits. One of the structures shows weak interactions of stargazin with the A and C subunits (Zhao et al., 2016). These data are remarkably consistent with our predicted model. The similarity of our model, generated based on LRET experiments on AMPA receptors co-expressed with stargazin in HEK-293 cells, to the cryo-EM structures of purified protein, with one structure being of a tandem construct between the AMPA receptor and stargazin, speaks to the robustness of the structure of the complex.
Decoupling of amino-terminal domain dimers of the AMPA receptor due to desensitization
Recent structures of the AMPA receptor show decoupling between the amino-terminal domains of the AMPA receptor associated with desensitization (Durr et al., 2014; Meyerson et al., 2014; Yelshanskaya et al., 2014). The extent of decoupling at the amino-terminal domain has been debated, with some structures showing large decoupling and others smaller or no decoupling (Durr et al., 2014; Meyerson et al., 2014; Yelshanskaya et al., 2014). Decoupling, as referred to here, is decoupling between the dimers at the amino-terminal domain, and not decoupling within the dimer as seen in the ligand-binding domain (Gonzalez et al., 2010). Here we used LRET to study the inter-subunit distance between proximal subunits B and D at sites 23 and 27. For these measurements, cysteines were introduced at site 23 or 27 in the GluA2* background. In both GluA2*D23C and GluA2*S27C, stargazin modulation of kainate efficacy was intact (Figure 1; kainate-to-glutamate ratio of 0.71 ± 0.03, n = 4 for GluA2*D23C, p = 0.34 compared to wild-type; 0.72 ± 0.03, n = 4 for GluA2* S27C, p = 0.42 compared to wild-type).
The receptors were tagged with a 1:4 ratio of acceptor:donor, thus ensuring that the majority of the receptors probed have at most one acceptor fluorophore per receptor (Cha et al., 1999). Labeling the same site on the four subunits allows measurement of the inter-subunit distance at the same structural point on the four subunits. Measurements of the desensitized conformations were obtained using saturating glutamate (1 mM) and measurements of the open conformation were obtained using saturating glutamate as well as cyclothiazide (100 μM). The receptors tagged with donor alone showed no significant changes between these conditions and could be well-represented by a single exponential decay for both sites 23 and 27 (Supplemental Figure S4). The background-subtracted LRET lifetimes for the donor-acceptor tagged receptors also could be represented by a single exponential decay (Figure 3), consistent with the structure of the AMPA receptor that shows the shortest distance for site 23 and site 27 is the inter-subunit distance between subunits B and D. The distances across the other subunits are significantly longer than the R0 = 45 Å of the Terbium-Fluorescein LRET pair, and, consistent with this, they do not significantly contribute to the LRET signal. Based on the donor-only and donor-acceptor lifetimes, the distances were calculated using Förster equation (Supplemental Table S5).
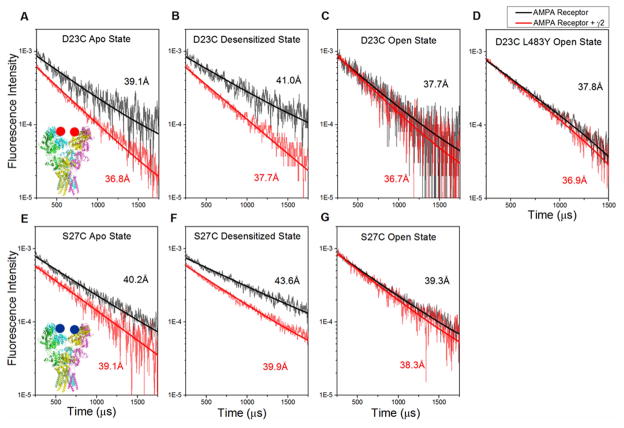
Figure 3. Amino-terminal domain subunits of the AMPA receptor decouple upon desensitization and are stabilized in the presence of stargazin.
LRET lifetimes between the proximal GluA2 subunits at site 23 (Top Panel) and at site 27 (Bottom Panel). Insets show location of sites 23 and 27 on the AMPA receptor. A) and E): Apo or resting condition. B) and F): Desensitized condition with the presence of 1 mM glutamate. C) and G): Open condition with the presence of 1 mM glutamate + 100 μM CTZ. D): GluA2 L483Y non-desensitizing mutant (open conformation) in the presence of 1 mM glutamate. See also Figure S4.
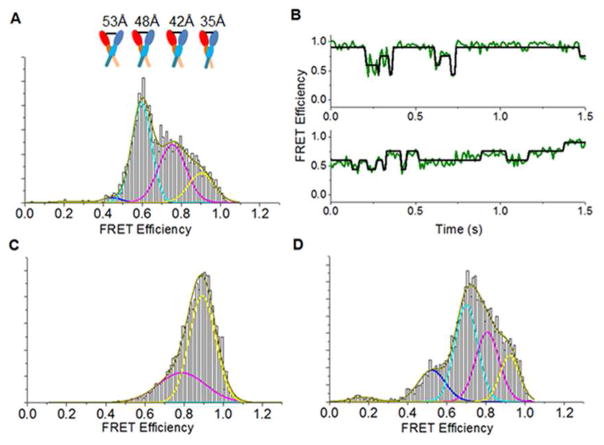
These ensemble measurements show that the average resting conformation and the average open (glutamate-and-cyclothiazide-bound) conformation, both exhibit shorter distances relative to the predominantly desensitized (glutamate-bound) conformation of the receptor. The change in distance between the open state and the desensitized conformation show increases of 3.3 ± 0.1 Å and 4.3 ± 0.04 Å, at sites 23 and 27 respectively. This increase indicates a decoupling between the subunits at the amino-terminal domain upon desensitization. Since a wide range of decoupling has been noted in the X-ray (PDBID: 4U4F) (Durr et al., 2014; Yelshanskaya et al., 2014) and cryo-EM structures (EMD2686, EMD2687, EMD2688) (Meyerson et al., 2014), we studied this decoupling further using smFRET studies. The smFRET histogram for the glutamate-bound, desensitized AMPA receptor labeled at site 23 (GluA2*D23C) is shown in Figure 4a. The data were analyzed using HaMMy software (McKinney et al., 2006) to obtain the states that the protein samples (representative traces shown in Figure 4b). The combined histograms for all the molecules could be fit to the same states observed in the HaMMy analysis. Moreover, analyzing the raw data with wavelet denoising (Dolino et al., 2015; Landes et al., 2011; Ramaswamy et al., 2012) resulted in the same states (Supplemental Figure S6), providing independent confirmation of the existence of these states. The analysis of the smFRET data shows that under desensitized conditions the protein samples varying degrees of decoupling, with stabilization into predominantly four conformational states. Two additional states were also present that, combined, comprise less than 1% of the population and are likely due to background or to denatured protein. The occupancy, FRET efficiency, and corresponding FRET distances of the four significant states are shown in Supplemental Table S7. These data reveal the complete spectrum of states that the amino-terminal domain subunits probe under desensitized conditions. It is interesting to note that the FRET distances correlate well with previously published X-ray and cryo-EM structures (Supplemental Table S7) (Meyerson et al., 2014; Yelshanskaya et al., 2014), suggesting that the desensitized receptor samples each of these structures to varying degrees. We have also studied the receptor under predominantly open conditions, i.e. with both, glutamate and cyclothiazide-bound, using smFRET (Figure 4c). Consistent with the LRET data, the smFRET data show that when bound to both cyclothiazide and glutamate, the receptor occupies predominantly high-efficiency FRET states. The data could be fit to three conformational states, with 98% of the receptor being in the conformations having distances of 36 Å and 41 Å. This suggests a compact receptor with very little decoupling at the amino terminal domain under open channel conditions.
Figure 4. The presence of stargazin promotes a more compact form of the AMPA receptor.
smFRET histograms showing the population distribution of the AMPA receptor labeled at site 23 of the amino-terminal domain under A) desensitized condition (presence of 1mM glutamate) with corresponding representative smFRET traces shown in B, C) open condition (presence of 1mM glutamate + 100 μM CTZ) and D) in tandem with γ2, in the desensitized condition (presence of 1mM glutamate). State 1, yellow peak; State 2, magenta peak; State 3, cyan peak; State 4, blue peak. Each state in A is accompanied by a cartoon representation of the corresponding distance between sites 23 of the B and D subunits of the AMPA receptor.
The sampling of multiple conformations of the AMPA receptor as seen with our smFRET data is also consistent with electrophysiological evidence for multiple kinetic states for a single AMPA receptor functional condition (Jin et al., 2003). Four desensitized kinetic states have been identified based on electophysiological measurements for the GluA1 subtype of AMPA receptors (Robert and Howe, 2003), and other iGluRs also populate multiple desensitized states (Amico-Ruvio and Popescu, 2010; Borschel et al., 2012; Kussius et al., 2009).
Decoupling of amino-terminal domains of the AMPA receptor in the presence of stargazin
LRET measurements were also performed on AMPA receptors co-transfected with stargazin. Comparing the LRET lifetimes at sites 23 and 27 for the AMPA receptor in the absence and presence of stargazin (Figure 3 and Supplemental Table S5), it is evident that the average distances are not altered under open conditions. However, the distances are shorter under resting and desensitized conditions when stargazin is present, with the effect being most dramatic under the desensitized state. Given that the desensitized receptor is the most decoupled, these results suggest that the presence of stargazin reduces the extent of decoupling, implying a mechanism to explain the increased recovery from desensitization.
In order to see the shift in the occupancy of the different states, we obtained smFRET histograms of the AMPA receptor/stargazin tandem construct tagged at site 23 (GluA2*-D23C/γ2) in the desensitized (glutamate-bound) condition, and compared it to our previous data of the AMPA receptor without stargazin. Stargazin modulation of kainate efficacy was intact for the tandem GluA2*D23C/γ2 construct (Figure 1; kainate-to-glutamate ratio of 0.75 ± 0.02, n = 4, p = 0.81 compared to wild-type). The smFRET histograms as well as the individual traces show that similar numbers of states are observed in the desensitized receptor in the presence of stargazin as in its absence (Figure 4d). However, in the presence of stargazin, each of the states observed in the receptor in the absence of stargazin is shifted to higher efficiencies (less decoupling). Thus, although the overall landscape of the receptor in the absence and presence of stargazin is similar, the shorter distance seen in each state suggests a more compact (less decoupled) receptor (Supplemental Table S7) under desensitized conditions.
The overall reduction in decoupling caused by stargazin in each conformational state is consistent between both our LRET and smFRET data, suggesting that stargazin acts as a scaffold, stabilizing a more compact structure of the AMPA receptor. Such stabilization by stargazin of the AMPA receptor would account for the stabilization of the receptor in an open, activated conformation and destabilization of the desensitized conformations, perhaps contributing to the accelerated recovery from desensitization, increased agonist efficacy and potency, and resensitization of the AMPA receptor by stargazin. Our work thus provides insight into the structural basis for the functional consequences of stargazin on the AMPA receptor.
Experimental Procedures
Cloning and mutagenesis
In order to specifically label cysteines on GluA2 using maleimide fluorophore derivatives, the extracellular, surface-exposed non-disulfide bonded cysteines at positions 89, 196, and 436 (numbered according to the GluA2 crystal structure PDB ID: 3KG2) were mutated to serines. Additionally, a Factor Xa protease site (IDGR) was introduced at position 228 with the mutations T228I and D231R. Upon cleavage, the labeled amino-terminal domains dissociate, thus removing the specific AMPA receptor contribution to the LRET signal, and enabling distance measurements in full-length receptors expressed in whole cells without the need for protein solubilization and purification. This background construct, called GluA2*, was further modified to substitute cysteines at specific sites of the amino-terminal domain for fluorophore labeling; the mutants generated were GluA2*-D23C, GluA2*-S27C and GluA2*-Y128C. For measuring AMPA receptor-STG distances, Amber TAG stop codons were introduced at positions 44, 51, and 61 of γ2, and the original stop codon was mutated to TAA. A GluA2*-D23C/stargazin tandem construct was generated by replacing the stop codon with a stretch of GGSGGSGGSG residues followed by the stargazin sequence at the C-terminus (called GluA2*-D23C/γ2). All mutations were introduced using standard site-directed mutagenesis protocols and checked by sequencing.
Functional characterization of mutants
HEK293 tsA201 cells at 40–50% confluency were transfected using jetPrime (PolyPlus) following the manufacturer’s instructions with the relevant GluA2, γ2, and eGFP DNA at a mass ratio of 10:15:1 μg per 10 ml of media. After 10–12 h of incubation cells were re-plated at a low density. 10 μM NBQX was present in the medium during and after transfection. Whole cell patch clamp recordings were performed 24–48 h after transfection using fire-polished borosilicate glass pipettes with 3–5 MΩ resistance, filled with internal solution: 135 mM CsF, 33 mM CsOH, 2 mM MgCl2, 1 mM CaCl2, 11 mM EGTA, and 10 mM HEPES, pH 7.4. The external solution was: 150 mM NaCl, 1 mM CaCl2 and 10 mM HEPES, pH 7.4. Solutions with no added ligand, 10 mM glutamate, 2 mM kainate, and/or 100 μM cyclothiazide were locally applied to lifted cells using a stepper motor system (SF-77B; Warner Instruments) with triple barrel tubing. Recordings were performed using an Axopatch 200B amplifier (Molecular Devices) at −60 mV hold potential, acquired at 10 kHz using pCLAMP10 software (Molecular Devices) and filtered online at 5 kHz.
LRET investigations
HEK 293T cells were transiently transfected with the constructs described above using jetPRIME® Polyplus according to the manufacturer’s guidelines and were maintained in 30 μM NBQX during and after transfection. For LRET measurements, cells were transfected with either GluA2 DNA alone or a GluA2:γ2 μg ratio of 5:15.
In order to incorporate the unnatural amino acid p-acetyl-L-phenylalanine into the γ2 protein during translation, cells were co-transfected with plasmids containing suppressor tRNACUA and the p-acetyl-L-phenylalanyl-tRNA synthetase, along with the GluA2 and γ2 plasmids. After transfection, the medium was supplemented with 500 μM amino acid p-acetyl-L-phenylalanine (AcF) (RSP Amino Acids).
LRET experiments were done 48 h post-transfection. Cells were collected and washed 3–4 times using extracellular buffer containing 145 mM NaCl, 1.8 mM MgCl2, 1 mM CaCl2, 3 mM KCl, 10mM glucose, and 10 mM HEPES, pH 7.4. The washed HEK cells were then labeled with 300 nM donor and 75 nM acceptor fluorophores in 3 ml extracellular buffer, rotating in the dark for 1 h. The donor fluorophore was always terbium chelate (Invitrogen), while acceptor fluorophores were either Alexa 555 hydrazide or fluorescein thiosemicarbazide (ThermoFisher) for cells expressing AcF incorporated γ2, and fluorescein maleimide (ThermoFisher) for cysteine mutants. After labeling, cells were washed and resuspended in 2 ml of the buffer and used for LRET measurements. A cuvette- based LRET system, QuantaMaster model QM3-SS with Fluorescan Software (Photon Technology International) was used for analysis. All samples were excited at 337 nm. Emission was detected at 545 nm for donor-only samples, at 515 nm for fluorescein-labeled samples, and at 565 nm for Alexa 555-labeled samples. To study the effect of γ2 on the AMPA receptor amino terminal domains, distance measurements were collected in the presence of 1 mM glutamate alone as well as with 100 μM cyclothiazide (CTZ). LRET measurements were taken before and after protease cleavage by Factor Xa to enable quantification and subtraction of background fluorescence in order to isolate the specific signal due the AMPA receptor (and stargazin). Distances between the donor and acceptor fluorophores were calculated from LRET lifetime (TDA) and donor-only lifetime (TD) using the Förster equation:
| (Eq. 1) |
where R is the distance between donor and acceptor fluorophore, R0 is the distance yielding half-maximal energy transfer for a given fluorophore pair (65 Å for Terbium-Alexa 555 and 45 Å for Terbium-fluorescein), TD is the measured lifetime of the donor when bound to the protein and without acceptor fluorophore present, and TDA is the lifetime of the donor fluorophore when bound to the protein and transferring energy to the acceptor fluorophore, which we have measured here as the lifetime of the sensitized emission of the acceptor.
Nano Positioning
A homology model of stargazin was constructed using the SWISS-MODEL server (Biasini et al., 2014), which generated a model based on claudin-19. Root mean positions of the fluorophores were calculated using the FRET Positioning and Screening software as previously described (Kalinin et al., 2012). Based on the root mean positions of the acceptor fluorophores on the stargazin model, spheres were drawn in PyMOL centered upon those positions with radii corresponding to the distances derived from the LRET measurements. The sites of intersection of the three spheres were then considered to be the triangulated position where we would expect to locate the root mean position of the donor fluorophore. The structure of the AMPA receptor (PDBID: 3KG2) with the calculated accessible volume and root mean position of Tyr128Cys-Tb chelate underwent a series of rigid-body translations and rotations and visually inspected until a model was found which superposed the triangulated point of intersection with the calculated root mean position of terbium. Additional constraints of maintaining the transmembrane domains in a logical position and minimizing steric clashing were used to generate the models shown.
smFRET Measurements
HEK293T cells were transfected, harvested, and washed as described for the LRET investigations. The cells were then labeled with maleimide derivatives of 300nM Alexa 555 donor and 1.2 μM Alexa 647 (Invitrogen) acceptor fluorophores. After washing, the cells were solubilized in phosphate buffered saline containing 1mM n-dodecyl-β-D-maltoside (DDM) and 0.2mM cholesteryl hemisuccinate (CHS) with protease inhibitor (ThermoScientific). The lysed cells were centrifuged at 100,000×g at 4 °C for 1 h and the supernatant was used for smFRET sample preparation. Slides for smFRET studies were prepared and measurements were taken as described previously (Dolino et al., 2015). The background-corrected signal was used to calculate the FRET efficiency using the equation:
| (Eq. 2) |
where EA is the apparent FRET efficiency, IA is the background-corrected acceptor fluorescence intensity and ID is the background-corrected donor fluorescence intensity. From this FRET efficiency, the distance was determined through the Förster equation,
| (Eq. 3) |
where R is the distance between the dyes, and R0 is the Förster radius. The Förster radius is 51 Å for the Alexa Fluor 555-Alexa Fluor 647 fluorophore pair used for these experiments. Error in FRET efficiencies was set to 0.03 based on measurements under the same conditions performed with a rigid DNA double strand. After processing the data, the traces were further filtered for single molecule verification and excluded if they showed multistep bleaching or exceptionally high background adapted from a normal distribution.
Statistics
For electrophysiological analysis of the mutants, n represents a single cell. The representative traces are an average of 5 to 10 individual traces from a single cell. The statistical significance was calculated using Student’s t test, with p < 0.05 considered significant. For LRET measurements, each sample was scanned 3 times, each scan was an average of 99 sweeps and each sweep comprised 500 pulses. The LRET lifetimes were normalized for comparison and error was calculated using the Error Propagation Calculator developed by Thomas Huber in the Physics Department of Gustavus Adolphus College. smFRET data was obtained from 12 molecules (10282 data points) under open receptor consitions, 16 molecules (2384 data points) under desensitized receptor conditions without stargazin, and 28 molecules (4269 data points) under desensitized receptor conditions with stargazin. All data were analyzed using Origin 9.0 (OriginLab Corp.)
Supplementary Material
Acknowledgments
This study was supported by National Institute of Health Grants R01 GM113212 to V. J., American Heart Association Fellowship 16POST30030007 to S.A.S., the Schissler Foundation Fellowship to D.M.D., and K99NS094761 to D.M.M. The authors declare no competing financial interests.
Footnotes
Author Contributions
S.A.S. and G.L. made the GluA2 mutant constructs and performed the LRET investigations; S.C. performed the smFRET measurements; D.M.D., S.C., C.F. and S.A.S. analyzed the smFRET data; D.M.D. modeled the AMPA-stargazin complex by nano-positioning; and D.M.M. performed the electrophysiology experiments. S.A.S., D.M.D., D.M.M. and V.J. designed the experiments, interpreted the results, and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amico-Ruvio S, Popescu G. Stationary gating of GluN1/GluN2B receptors in intact membrane patches. Biophysical Journal. 2010;98:1160–1169. doi: 10.1016/j.bpj.2009.12.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bats C, Groc L, Choquet D. The Interaction between Stargazin and PSD-95 Regulates AMPA Receptor Surface Trafficking. Neuron. 2007;53:719–734. doi: 10.1016/j.neuron.2007.01.030. [DOI] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic acids research. 2014;42:W252–258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borschel WF, Myers JM, Kasperek EM, Smith TP, Graziane NM, Nowak LM, Popescu GK. Gating reaction mechanism of neuronal NMDA receptors. J Neurophysiol. 2012;108:3105–3115. doi: 10.1152/jn.00551.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cais O, Herguedas B, Krol K, Cull-Candy SG, Farrant M, Greger IH. Mapping the interaction sites between AMPA receptors and TARPs reveals a role for the receptor N-terminal domain in channel gating. Cell reports. 2014;9:728–740. doi: 10.1016/j.celrep.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone AL, Plested AJ. Superactivation of AMPA receptors by auxiliary proteins. Nature communications. 2016;7:10178. doi: 10.1038/ncomms10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha A, Snyder GE, Selvin PR, Bezanilla F. Atomic scale movement of the voltage-sensing region in a potassium channel measured via spectroscopy. Nature. 1999;402:809–813. doi: 10.1038/45552. [DOI] [PubMed] [Google Scholar]
- Cho CH, St-Gelais F, Zhang W, Tomita S, Howe JR. Two families of TARP isoforms that have distinct effects on the kinetic properties of AMPA receptors and synaptic currents. Neuron. 2007;55:890–904. doi: 10.1016/j.neuron.2007.08.024. [DOI] [PubMed] [Google Scholar]
- Dawe GB, Musgaard M, Aurousseau MR, Nayeem N, Green T, Biggin PC, Bowie D. Distinct Structural Pathways Coordinate the Activation of AMPA Receptor-Auxiliary Subunit Complexes. Neuron. 2016;89:1264–1276. doi: 10.1016/j.neuron.2016.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacological Reviews. 1999;51:7–61. [PubMed] [Google Scholar]
- Dolino DM, Cooper D, Ramaswamy S, Jaurich H, Landes CF, Jayaraman V. Structural dynamics of the glycine-binding domain of the N-methyl-D-aspartate receptor. J Biol Chem. 2015;290:797–804. doi: 10.1074/jbc.M114.605436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolino DM, Ramaswamy SS, Jayaraman V. Luminescence resonance energy transfer to study conformational changes in membrane proteins expressed in mammalian cells. Journal of visualized experiments : JoVE, 51895. 2014 doi: 10.3791/51895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Remedios CG, Moens PD. Fluorescence resonance energy transfer spectroscopy is a reliable “ruler” for measuring structural changes in proteins. Dispelling the problem of the unknown orientation factor. Journal of structural biology. 1995;115:175–185. doi: 10.1006/jsbi.1995.1042. [DOI] [PubMed] [Google Scholar]
- Durr KL, Chen L, Stein RA, De Zorzi R, Folea IM, Walz T, McHaourab HS, Gouaux E. Structure and dynamics of AMPA receptor GluA2 in resting, pre-open, and desensitized states. Cell. 2014;158:778–792. doi: 10.1016/j.cell.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez J, Du M, Parameshwaran K, Suppiramaniam V, Jayaraman V. Role of dimer interface in activation and desensitization in AMPA receptors. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9891–9896. doi: 10.1073/pnas.0911854107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez J, Rambhadran A, Du M, Jayaraman V. LRET investigations of conformational changes in the ligand binding domain of a functional AMPA receptor. Biochemistry. 2008;47:10027–10032. doi: 10.1021/bi800690b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollmann M, Heinemann S. Cloned glutamate receptors. Annual Review of Neuroscience. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- Jin R, Banke TG, Mayer ML, Traynelis SF, Gouaux E. Structural basis for partial agonist action at ionotropic glutamate receptors. Nature neuroscience. 2003;6:803–810. doi: 10.1038/nn1091. [DOI] [PubMed] [Google Scholar]
- Kalinin S, Peulen T, Sindbert S, Rothwell PJ, Berger S, Restle T, Goody RS, Gohlke H, Seidel CA. A toolkit and benchmark study for FRET-restrained high-precision structural modeling. Nature methods. 2012;9:1218–1225. doi: 10.1038/nmeth.2222. [DOI] [PubMed] [Google Scholar]
- Kussius CL, Kaur N, Popescu GK. Pregnanolone Sulfate Promotes Desensitization of Activated NMDA Receptors. J Neurosci. 2009;29:6819–6827. doi: 10.1523/JNEUROSCI.0281-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landes CF, Rambhadran A, Taylor JN, Salatan F, Jayaraman V. Structural landscape of isolated agonist-binding domains from single AMPA receptors. Nature chemical biology. 2011;7:168–173. doi: 10.1038/nchembio.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean DM, Bowie D. Transmembrane AMPA receptor regulatory protein regulation of competitive antagonism: a problem of interpretation. The Journal of physiology. 2011;589:5383–5390. doi: 10.1113/jphysiol.2011.219485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean DM, Ramaswamy SS, Du M, Howe JR, Jayaraman V. Stargazin promotes closure of the AMPA receptor ligand-binding domain. The Journal of general physiology. 2014;144:503–512. doi: 10.1085/jgp.201411287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophysical journal. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML, Subramaniam S. Structural mechanism of glutamate receptor activation and desensitization. Nature. 2014;514:328–334. doi: 10.1038/nature13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto-Tomita M, Zhang W, Straub C, Cho CH, Kim KS, Howe JR, Tomita S. Autoinactivation of neuronal AMPA receptors via glutamate-regulated TARP interaction. Neuron. 2009;61:101–112. doi: 10.1016/j.neuron.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschielok A, Michaelis J. Application of the nano-positioning system to the analysis of fluorescence resonance energy transfer networks. The journal of physical chemistry B. 2011;115:11927–11937. doi: 10.1021/jp2060377. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Cooper D, Poddar N, MacLean DM, Rambhadran A, Taylor JN, Uhm H, Landes CF, Jayaraman V. Role of conformational dynamics in alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid (AMPA) receptor partial agonism. The Journal of biological chemistry. 2012;287:43557–43564. doi: 10.1074/jbc.M112.371815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambhadran A, Gonzalez J, Jayaraman V. Subunit arrangement in N-methyl-D-aspartate (NMDA) receptors. J Biol Chem. 2010;285:15296–15301. doi: 10.1074/jbc.M109.085035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambhadran A, Gonzalez J, Jayaraman V. Conformational changes at the agonist binding domain of the N-methyl-D-aspartic acid receptor. The Journal of biological chemistry. 2011;286:16953–16957. doi: 10.1074/jbc.M111.224576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert A, Howe JR. How AMPA receptor desensitization depends on receptor occupancy. J Neurosci. 2003;23:847–858. doi: 10.1523/JNEUROSCI.23-03-00847.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrieh RE, MacLean DM, Jayaraman V. Amino-terminal domain tetramer organization and structural effects of zinc binding in the N-methyl-D-aspartate (NMDA) receptor. The Journal of biological chemistry. 2013;288:22555–22564. doi: 10.1074/jbc.M113.482356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrieh RE, MacLean DM, Jayaraman V. Subtype-dependent N-Methyl-d-aspartate Receptor Amino-terminal Domain Conformations and Modulation by Spermine. The Journal of biological chemistry. 2015;290:12812–12820. doi: 10.1074/jbc.M115.649723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatulian SA. Molecular-scale GPS: positioning a biosensor peptide on RyR. Biophysical journal. 2014;107:2003–2005. doi: 10.1016/j.bpj.2014.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S, Adesnik H, Sekiguchi M, Zhang W, Wada K, Howe JR, Nicoll RA, Bredt DS. Stargazin modulates AMPA receptor gating and trafficking by distinct domains. Nature. 2005;435:1052–1058. doi: 10.1038/nature03624. [DOI] [PubMed] [Google Scholar]
- Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J, Sobolevsky AI. Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy. Science. 2016;353:83–86. doi: 10.1126/science.aaf8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelshanskaya MV, Li M, Sobolevsky AI. Structure of an agonist-bound ionotropic glutamate receptor. Science. 2014;345:1070–1074. doi: 10.1126/science.1256508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chen S, Yoshioka C, Baconguis I, Gouaux E. Architecture of fully occupied GluA2 AMPA receptor-TARP complex elucidated by cryo-EM. Nature. 2016;536:108–111. doi: 10.1038/nature18961. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.