Generation of antigen receptor diversity: a double-edged sword
The realization, now more than half a century ago, that B cells can generate antibodies to an astounding variety of chemical structures sparked intense interest in the “generation of diversity question” (reviewed in (1). The correct solution to this puzzle turned out to be both surprising and simple: The exons encoding the antigen binding portions of the receptor (the so-called variable regions) are assembled by chromosomal breakage and rejoining in developing lymphocytes (2). Immunoglobulins and T cell receptors are composed of two polypeptide chains, each of which contributes to the antigen binding domain. The exons encoding the antigen binding domains are assembled from so-called V (variable), D (diversity), and J (joining) gene segments by “cut and paste” DNA rearrangements. This process, termed V(D)J recombination, chooses a pair of segments, introduces double-strand breaks adjacent to each segment, deletes (or, in selected cases, inverts) the intervening DNA, and ligates the segments together (Figure 1). Rearrangements occur in an ordered fashion, with D to J joining proceeding before a V segment is joined to the rearranged DJ segments. This process of combinatorial assembly— choosing one segment of each type from several (sometimes many) possibilities is the fundamental engine driving antigen receptor diversity in mammals. Diversity is tremendously amplified by the characteristic variability at the junctions (loss or gain of small numbers of nucleotides) between the various segments. This process leverages a relatively small investment in germline coding capacity into an almost limitless repertoire of potential antigen binding specificities.
Figure 1. Antigen receptor variable exons are assembled by V(D)J recombination.
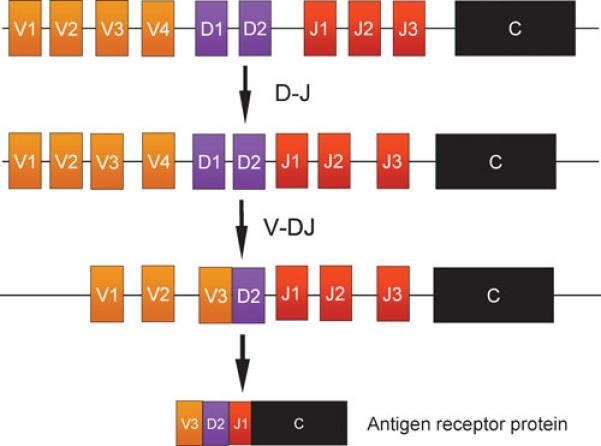
Assembly of a complete variable exon occurs in two steps (in the case of an Ig heavy chain gene or a TCR beta or delta gene), as shown. First, a D and a J segment are chosen from among several possibilities, and are brought together to form a D-J rearrangement. Then a V region is selected and joined with the D-J rearrangement to form a complete VDJ exon. Immunoglobulin light chain genes and TCR alpha and gamma genes rearrange in a single step, involving V-J recombination, as D segments are absent from these loci.
This elegant process does, however, have a potential downside. A system that must break chromosomal DNA several times in order to generate a functional antigen receptor gene-- many millions of times over the lifetime of an organism-- creates significant opportunities for error. The necessity for enforcing a high degree of fidelity in V(D)J recombination has been recognized for decades (reviewed in (3). Aberrant V(D)J recombination events do occur, and they can be life-threatening, underlying the genesis of common lymphoid neoplasms (4-7), as discussed below. Recent genomewide analyses of lymphoid neoplasms have revealed V(D)J recombination-driven oncogenic events, and have intensified interest in the regulatory mechanisms responsible for ensuring fidelity during V(D)J recombination. This chapter reviews basic aspects of V(D)J recombination, mechanisms responsible for aberrant rearrangements, and the types of events uncovered in recent analyses of tumor genomes. Recent advances in understanding mechanisms responsible for safeguarding genomic integrity during V(D)J recombination are also discussed.
The normal V(D)J recombination mechanism
This section briefly overviews the normal mechanism of V(D)J recombination. For more details, see (8, 9). The V(D)J recombinase recognizes conserved DNA sequence elements, termed recombination signal sequences (RSS), located adjacent to each V, D, and J coding segment. RSS consist of conserved heptamer and nonamer elements, separated by 12 or 23 nucleotides of less conserved “spacer” sequence (Figure 2). Efficient recombination only occurs between RSS with different spacer lengths (the “12/23 rule”). Additional restrictions are imparted at some antigen receptor loci by other DNA sequence features (the so-called ‘beyond 12/23 rule”) (10). The RSS are the only DNA segments required to allow V(D)J recombination to occur on artificial substrates, and their relative orientation determines whether the reaction proceeds by inversion or by deletion (Figure 3) (11). An additional outcome, occasionally observed at antigen receptor loci, is formation of a “hybrid joint”, in which a coding segment is joined to an RSS (12) (Figure 3). Hybrid joints do not contribute to antigen receptor diversity, nor do they appear to play a role in oncogenic transformation.
Figure 2. Consensus RSS.
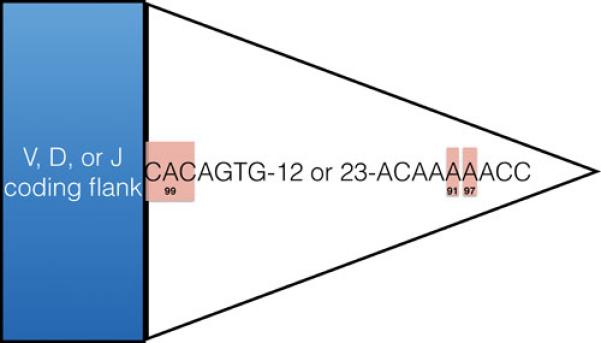
The consensus RSS sequence is shown, with the heptamer abutting the coding flank. The most highly conserved positions of the heptamer and nonamer are shaded in red, with conservation (percent) given below. Sequence conservation data are from (13).
Figure 3. Products of V(D)J recombination.
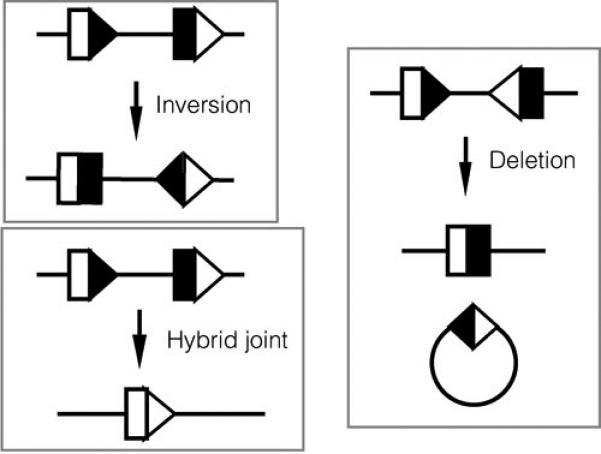
Inversional and deletional recombination are shown in the top portion of the figure. Whether recombination proceeds in a deletional or inversional manner is specified by the relative orientation of the two RSS. Hybrid joint formation is shown at the bottom of the figure, and involves an inappropriate joining of a coding end to a signal end. The reciprocal hybrid joint product, in this case an excised circle, is not shown.
Nucleotide sequences of natural RSS display considerable variability. Those with sequences closest to the consensus support the most efficient recombination (13). The first three nucleotides of the heptamer (closest to the coding flank) show the highest sequence conservation, and are critical for recombination, whereas the remaining heptamer positions are much less important (13) (Figure 2). The nonamer sequence conforms less closely to the consensus, with only a few highly conserved positions (particularly the A/T tract), and the nonamer is dispensable under certain conditions in vitro (13, 14). Spacer length is critical, and can be changed successfully only in increments that preserve the helical spacing of the nonamer and heptamer elements (13). Other nucleotide sequence features can influence recombination efficiency, most notably the sequence of the coding segment adjacent to the heptamer (the coding flank) (14, 15). This reflects a requirement for DNA distortion during DNA cleavage (14-17).
The V(D)J recombinase consists of two lymphoid-specific proteins, RAG1 and RAG2 (18), which work together with non-lymphoid-specific DNA bending factors, HMG1A or HMG1B (19) to carry out DNA cleavage. The RAG1 and RAG2 genes are located quite close to each other in all species examined, and their ORFs are generally encoded in single exons. These observations led to speculation that the V(D)J recombinase may have evolved from an ancestral prokaryotic transposase (20). Indeed, the mechanism of DNA cleavage by RAG1/2 (one step transesterification) (21) is shared with a class of bacterial transposases, and the RAG proteins can catalyze bona fide transposition events (22, 23). Definitive evidence that RAG1/2 indeed evolved from an ancestral transposase remains elusive (24).
The functional anatomy of the RAG1 and RAG2 proteins has been reviewed recently (see (8). Mutational studies have defined the minimally functional regions of both proteins. The “core” region of murine RAG1 is comprised of amino acids 384-1008 of the 1040 amino acid protein (25, 26), and is sufficient to catalyze V(D)J recombination, albeit with some abnormal features (27). Core RAG1 contains elements important for binding to the nonamer as well as amino acids required for catalysis of cleavage. Neither specific DNA binding nor catalytic activities have been attributed to RAG2, leading to the view that RAG1 is the catalytic component of the recombinase, with RAG2 serving as an essential cofactor with some regulatory activities (described below). The essential “core” region of RAG2 historically has been defined as amino acids 1-383 (of 527) (28, 29). Recent work has shown that the minimal region extends only to amino acid 360 (30), closely coinciding with the predicted 6 bladed beta propeller structure (31). This core domain is connected to the C-terminal domain via a flexible acidic hinge. The C-terminus, while dispensable for recombination, is important for optimal recombination (32) and for enforcing the proper order of recombination events in developing lymphocytes (33). In its absence, aberrant recombination events are observed (33-35). The C-terminus is also important for maintaining genomic stability (36-38), as is the acidic hinge (30). Within the C-terminus reside a plant homeodomain (PHD) capable of recognizing histone H3K4 trimethylation (39, 40) and a cell cycle-regulated protein degradation signal (41). These elements are discussed in more detail below.
Efficient cleavage of a DNA substrate requires only RAG1, RAG2, a divalent metal ion, and HMGB1 or HMGB2 (19, 42). Cleavage proceeds via a two step mechanism (Figure 4). First, a nick is introduced between the RSS and the coding flank, then the resulting 3’OH group is used to attack the opposite strand by transesterification, forming a hairpin coding end and a blunt signal end. This second step is similar to transesterification reactions catalyzed by the HIV integrase and by bacterial transposases (21). Whereas nicking can occur independently on either RSS, in the presence of the physiological divalent metal ion (Mg2+), transesterification requires assembly of a synaptic complex including both a 12- and a 23-RSS (43), providing a molecular basis for the 12/23 rule. After cleavage, the four DNA ends remain associated with the RAG proteins in a post-cleavage complex (Figure 5), which retains the signal ends more stably than the coding ends (43-45). This complex is important for the proper rejoining of the broken DNA ends (46-48), and shepherds the ends to the classical nonhomologous end joining (cNHEJ) pathway (44). This function, which prevents access of the ends to the low fidelity, translocation-prone alternative NHEJ joining pathway (49), is hypothesized to be important for maintenance of genomic stability during V(D)J recombination (30, 36), as discussed below. RAG2's C-terminus contributes to the stability of the post-cleavage signal end complex (30, 49). Flexibility of the acidic hinge is also important: mutations (including some nucleotide sequence polymorphisms identified in humans) that reduce the negative charge destabilize the RAG-signal end complex and reduce genomic stability in pre-B cell lines (30).
Figure 4. Biochemistry of cleavage.
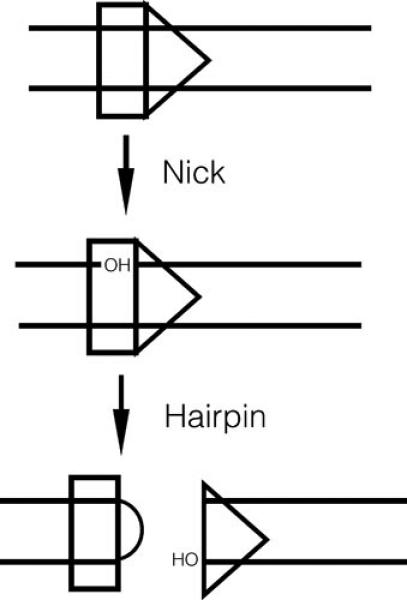
Cleavage occurs at the junction between the heptamer and the adjoining coding flank, and occurs in two steps, as described in the text.
Figure 5. V(D)J recombination overview.
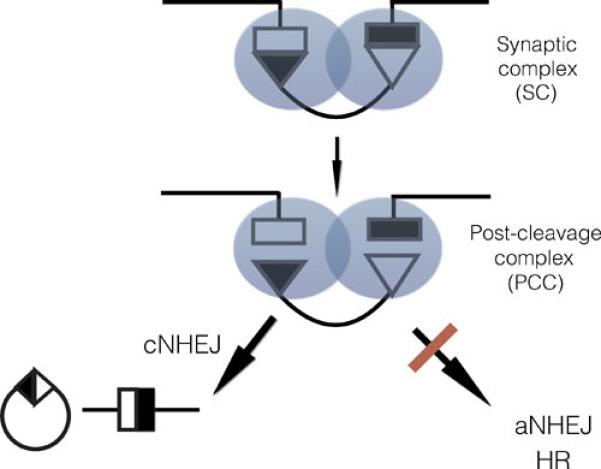
Recombination is thought to be initiated by binding of the RAG proteins to a single 12-RSS (not shown), which then captures the 23-RSS to form a synaptic complex (86). RAG1/2 complexes are shown as shaded circles. Double-strand break formation generates a DNA-protein complex, the post-cleavage complex, which then helps to control the “shepherding” of the broken DNA ends to the classical nonhomologous end joining machinery (left), preventing the ends from accessing other repair mechanisms such as alternative NHEJ or homologous recombination (right).
A characteristic feature of V(D)J recombination is the asymmetric processing of the signal and coding ends. Coding ends are joined with slight variations (small deletions, short insertions), whereas signal ends are generally joined with little or no end processing, so that the majority of the signal joints are perfect heptamer-to-heptamer fusions (50). This asymmetry may be partly explained by the requirement for additional processing of the coding ends, which are covalently sealed. Other factors may also contribute, including the differential stability of the RAG-coding end and RAG-signal end post-cleavage complexes. Hairpin opening occurs through the action of the Artemis endonuclease, which often cuts off-axis, resulting in short, single-stranded extensions which can give rise to palindromic insertions (P nucleotides) at the coding joint. These are never observed in signal joints, which are formed by blunt end-to-end joining. Formation of ends with single stranded extensions may also increase opportunities for loss of nucleotides from the coding ends. Another source of extra nucleotides is provided by TdT, which adds short, nontemplated, GC-rich inserts (N regions) to coding joints (and occasionally to signal joints). The frequent presence of such “microscopic” junctional alterations at coding joints provides a powerful mechanism for amplifying the diversity of antigen binding sites in T cell receptor and immunoglobulin molecules.
RAG-generated DNA ends are normally joined by cNHEJ (reviewed by (51). Inactivation of any of the key components of cNHEJ (e.g. Ku70/80, DNA ligase IV, XRCC4) results in a severe impairment of joining. The few junctions formed under these conditions are often (but not always) abnormal, showing excessive deletions, the frequent presence of microhomologies, and the occasional presence of abnormally long stretches of extra nucleotides. These features have been considered characteristic of alternative joining pathways, collectively termed alternative NHEJ (aNHEJ) (51), although as discussed below they are not always observed. aNHEJ is error-prone in two senses: the tendency toward formation of abnormal junctions, and also an increased propensity for forming gross genomic rearrangements such as chromosome translocations (52-54).
As noted above, the “shepherding” function of the RAG post-cleavage complex prevents the coding and signal ends from accessing aNHEJ. This was demonstrated by the observation that certain RAG mutants allow much higher levels of aNHEJ with artificial substrates in cultured cells, in both the presence and the absence of functional cNHEJ (44, 49). This may be important for preserving genomic stability, as discussed below.
V(D)J recombination errors
As noted above, the rearranging gene strategy that so successfully generates antigen receptor diversity comes with a price: the potential for generating deleterious genomic rearrangements. Indeed, chromosomal rearrangements involving antigen receptor loci were reported in both B and T cell neoplasms shortly after the discovery of V(D)J recombination (4, 55, 56). With the advent of next generation sequencing technologies, the genomic landscapes of these malignancies are being studied with increasingly fine resolution. Aberrant events identified in lymphoid neoplasms include chromosome translocations, relatively small (kilobase to megabase) inversions and deletions (5, 6, 57) and re-insertion of excised fragments bounded by signal ends (58). Recent work highlight the importance of deletions, which affect numerous genes implicated or suspected in tumorigenesis (7, 38, 59, 60) (Mijuskovic et al, submitted). These deletions are RAG-mediated, as they occur between pairs of sequences closely resembling RSS, they often follow the 12/23 rule and reciprocal signal joints have been detected (Mijuskovic et al, submitted). Thus, different types of V(D)J recombination errors play important roles in initiating oncogenic transformation.
Aberrant V(D)J recombination events observed in lymphoid neoplasms fall into two broad conceptual categories: errors in target recognition (Figure 6) and errors in joining (Figure 7). The first type consists of recognition of one authentic RSS and one DNA sequence fortuitously resembling an RSS (termed a “cryptic RSS” or cRSS). Given the relatively small size of RSS sequences, and that recombination does not require strict adherence of this sequence to consensus heptamer/nonamer sequences, it is not surprising that cRSS capable of supporting recombination are present approximately once per kb in random DNA sequence (61). Perhaps the first example of such events was provided by cytogenetic analyses of human lymphoid neoplasms, which revealed chromosome translocations involving authentic RSS at antigen receptor loci and cRSS adjacent to proto-oncogenes (4, 55) (Figure 6a). These events can cause inappropriate expression of the target gene due to, for example, the presence of transcriptional regulatory elements from the antigen receptor loci. Recombination events involving a cRSS/RSS pair can also deregulate oncogenes through amplification, likely through a breakage-fusion-bridge mechanism (62).
Figure 6. V(D)J recombination: recognition errors.
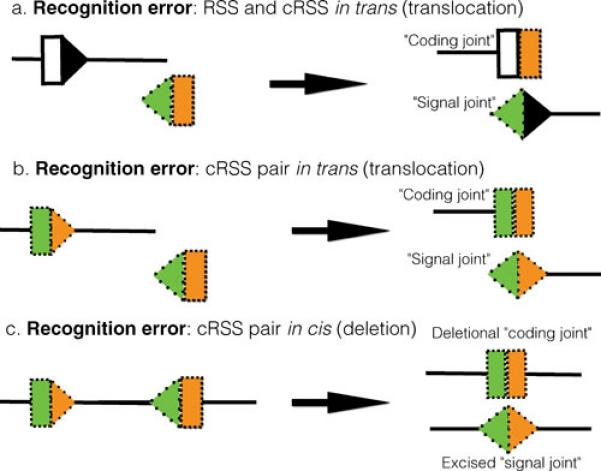
Three types of recognition errors are shown. In (a), recombination occurs between an authentic RSS (black triangle, with its associated coding flank, shown as a white box) and a cRSS (green triangle) with its associated coding flank (orange box), located on a separate DNA molecule. Recombination produces a trans rearrangement, with a pseudo coding joint and a pseudo signal joint. In (b), the recombinase recognizes a pair of cRSS located on separate DNA molecules. These recombine, generating a reciprocal chromosome translocation. The two products bear a pseudo coding joint and a pseudo signal joint. In (c), the recombinase recognizes a pair of cRSS located on the same DNA molecule, and generates a deletion, forming a pseudo coding joint (retained on the chromosome) and an excised circle containing a pseudo signal joint.
Figure 7. V(D)J recombination: joining errors.
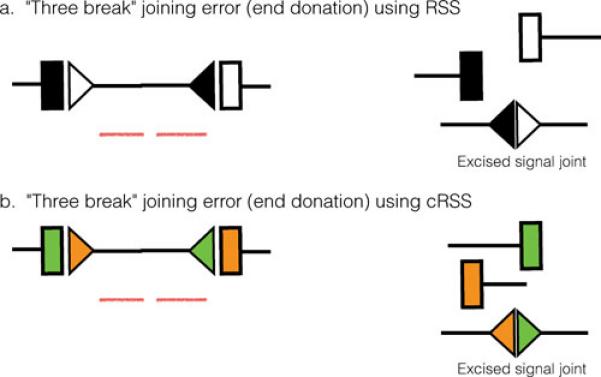
Two versions of a three break event (end donation) are shown. (a) depicts an event occurring between a normal V(D)J recombination event involving authentic RSS and a chromosome break generated by some other means (break in the red DNA molecule). (b) shows a similar event, this time involving a V(D)J recombination event involving a pair of cRSS.
Events also occur between pairs of cRSS. These can occur in trans, generating a chromosome translocation (Figure 6b) as in T-ALL cases involving translocations between TCR gene segments and the SCL locus (3), or in cis, generating a deletional “coding joint” and an excised “signal joint” (Figure 6c). Interestingly, although one might expect events between cRSS to also generate deletional “signal joints” retained in the chromosome, or inversion events, these are rarely observed (7) (Mijuskovic, submitted). Deletional recombination between cRSS pairs generates recurrent deletions at the SIL/SCL locus (63) and in Notch1, Izkf1, PTEN, and other critical genes in lymphoid neoplasms in humans and in mice (5, 7, 38, 57, 59, 60, 64) (Figure 6c). These are now thought to be major drivers of oncogenic transformation in lymphocytes. Another type of target recognition error, less commonly observed, involves RAG-mediated cleavage at non-B form DNA structures. This type of error has been implicated in oncogenic rearrangements joining the Bcl-2 major breakpoint region to an authentic RSS at the immunoglobulin heavy chain locus (65).
Errors in joining involve events that join a RAG-mediated DSB to a broken DNA end created by a non-RAG mediated mechanism. The events observed in lymphoid neoplasms generally involve three DNA breaks, and are referred to as “end donation” (66) or “type 2” events (67). These can involve a pair of breaks made during an apparently normal V(D)J recombination event, which are then mistakenly joined to another break generated by another mechanism (Figure 7a). These can generate chromosome translocations or insertions of signal ended fragments into another chromosomal location (58, 67). Similar events can involve a combination of recognition and joining errors: cleavage at a pair of cRSS followed by joining to a non-RAG mediated DSB (Figure 7b). Although joining errors may involve the normal cNHEJ mechanism, it is thought that such events may be favored by the use of error-prone alternative NHEJ (aNHEJ) mechanisms, (49, 62), which are known to favor formation of translocations (52, 53).
RAG-mediated transposition events cannot be conveniently classified as errors in recognition or in joining, as in this case the initial cleavage event, occurring at a pair of authentic RSS, is followed by RAG-mediated integration of the excised signal-ended fragment at another genomic location. These events, while observed in vitro (22, 23) in artificial systems in cultured cells (68, 69), and at the HPRT locus in human peripheral T cells (70), have not yet been definitively demonstrated in lymphoid neoplasms. It should be noted, however, that certain types of transposition events generate products that would not be recognizable as having been derived from transposition (71).
How is fidelity preserved during V(D)J recombination?
Given the fact that V(D)J recombination occurs in many millions of lymphocytes each day, and that a variety of V(D)J recombination errors that generate oncogenic lesions in lymphoid neoplasms, it seems logical to suppose that mechanisms exist to maintain the fidelity of the process. Perhaps the most basic of these is to ensure that the recombinase is active only in the appropriate target cells, and only during the appropriate developmental stages. Indeed, expression of RAG1 and RAG2 is carefully limited in a cell- and developmental stage-specific fashion. Bypassing these controls by introducing RAG1 and RAG2 transgenes under the control of strong promoters causing constitutive expression during lymphocyte development and in extra-lymphoid tissues results in a spectrum of phenotypes (including lymphopenia, growth retardation, and early death) reminiscent of DNA damage deficiency syndromes (72). An additional temporal control is provided by cell cycle-specific protein degradation of the RAG2 protein, mediated by phosphorylation of a threonine (T490) located in the dispensable C-terminus (41). Disabling this feature (via a T490A mutation) results in accelerated lymphomagenesis in p53-deficient mice (37). Autoubiquitylation of RAG1 may also play a regulatory role (73, 74).
Choice of the joining pathway used to repair RAG-generated breaks also appears to be important in maintaining fidelity. Mice lacking a functional cNHEJ pathway exhibit accelerated lymphomagenesis in the absence of p53 (75, 76), with complex chromosome translocations (mediated by aNHEJ) accompanied by gene amplification (62). Choice of joining pathway appears to involve the RAG post-cleavage complex, as mutations in RAG1 or RAG2 which destabilize the complex allow the ends to be joined by alternative pathways, including homologous recombination and aNHEJ (30, 44, 49). Further work showed that mutations in RAG2's nonessential C-terminus lead to genomic instability, accompanied by chromosomal aberrations (30, 36). To test the hypothesis that the C-terminal mutations caused oncogenic transformation by encouraging joining errors mediated by aNHEJ, lymphomas from two mouse models lacking RAG2's C-terminus were examined by whole genome sequencing. Scant evidence for oncogenic aberrant joining events was observed. Instead, most genomic lesions that could be linked to potentially oncogenic events were deletions between pairs of cRSS (38, 59). These data suggest that interstitial deletions may be more important drivers of RAG-mediated oncogenesis, at least in some systems, than gross chromosomal aberrations, and are in agreement with recent studies of B- and T-ALL in humans (7, 60).
In the case of the RAG2 C-terminal mutants, it is not yet clear whether increased access of RAG-mediated DNA breaks to aNHEJ plays a role in the observed genomic instability. A severe RAG-2 C-terminal truncation which allows high levels of aNHEJ in cultured cells (49) increases access of RAG-mediated breaks to alternative joining mechanisms, as shown by rescue of joining in a Ku80/RAG2 double mutant. This observation supports the idea that the post-cleavage complex enforces pathway choice (38). Recognizing a particular junction as having arisen from aNHEJ in this system is complicated, however, because junctional features considered characteristic of aNHEJ were rarely observed, even in the double mutants (in which junctions must have formed by a cNHEJ-independent process) (38). These data are consistent with previous analysis of rare junctions isolated from Ku80-deficient mice (77). Together, these data indicate that aNHEJ is not always distinguishable on the basis of junction structures, and provide support for the suggestion that aNHEJ may actually consist of several distinct pathways (54, 78), only some of which generate junctional “signatures”. Thus, caution must be observed when inferring the involvement of aNHEJ in aberrant recombination events in cancer genomes by sequence features alone.
As noted above, the 140+ amino acids in RAG2's C-terminus, largely conserved throughout evolution, contain several known or suspected regulatory elements. These may play important roles in maintaining fidelity of V(D)J recombination. Clear evidence implicates the cell cycle-regulated phosphorylation of T490 in suppressing persistence of broken DNA ends through the cell cycle and in suppressing lymphomagenesis. The PHD domain, which recognizes trimethylated histone H3K4, may play a role in limiting recognition of cRSS located outside the antigen receptor loci and/or in downregulating RAG cleavage activity in the absence of this histone modification (79). It should be noted, however, that these potential regulatory activities do not prevent the generation of deletions between cRSS pairs in known or suspected oncogenes and tumor suppressor genes, as these are observed in thymic lymphomas of p53-deficient mice bearing wild-type RAG2 (38, 59) and in T- and B-ALL genomes from patients who presumably bear wild-type RAG alleles (7, 60). Other potential regulatory mechanisms involving the C-terminus include its ability to inhibit RAG-mediated transposition (80, 81) and suppression of bi-allelic cleavage at antigen receptor loci (82).
Clearly, V(D)J recombination fidelity is also enforced by non RAG-specific mechanisms. These include ATM (83, 84), p53 (36, 38, 75, 76), and phosphorylated histone H2AX (85), and other aspects of the DNA damage response (9, 51). It is relatively straightforward to imagine how DNA damage response factors may act to limit errors in joining, such as, for example, limiting the persistence of broken DNA ends or the survival of cells bearing persistent broken ends. How these factors might limit errors in recognition, such as deletions between pairs of cRSS, or the survival of cells bearing such events, is less obvious. Investigating these regulatory mechanisms provides an interesting focus for future research.
Bibliography
- 1.Brandt VL, Roth DB. G.O.D.'s Holy Grail: discovery of the RAG proteins. J Immunol. 2008;180:3–4. doi: 10.4049/jimmunol.180.1.3. [DOI] [PubMed] [Google Scholar]
- 2.Hozumi N, Tonegawa S. Evidence for somatic rearrangement of immunoglobulin genes coding for variable and constant regions. Proc Natl Acad Sci U S A. 1976;73:3628–3632. doi: 10.1073/pnas.73.10.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Onozawa M, Aplan PD. Illegitimate V(D)J recombination involving nonantigen receptor loci in lymphoid malignancy. Genes Chromosomes Cancer. 2012;51:525–535. doi: 10.1002/gcc.21942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsujimoto Y, Gorham J, Cossman J, Jaffe E, Croce CM. The t(14;18) chromosome translocations involved in B-cell neoplasms result from mistakes in VDJ joining. Science. 1985;229:1390–1393. doi: 10.1126/science.3929382. [DOI] [PubMed] [Google Scholar]
- 5.Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, White D, Hughes TP, Le Beau MM, Pui CH, Relling MV, Shurtleff SA, Downing JR. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, Lu C, Chen SC, Wei L, Collins-Underwood JR, Ma J, Roberts KG, Pounds SB, Ulyanov A, Becksfort J, Gupta P, Huether R, Kriwacki RW, Parker M, McGoldrick DJ, Zhao D, Alford D, Espy S, Bobba KC, Song G, Pei D, Cheng C, Roberts S, Barbato MI, Campana D, Coustan-Smith E, Shurtleff SA, Raimondi SC, Kleppe M, Cools J, Shimano KA, Hermiston ML, Doulatov S, Eppert K, Laurenti E, Notta F, Dick JE, Basso G, Hunger SP, Loh ML, Devidas M, Wood B, Winter S, Dunsmore KP, Fulton RS, Fulton LL, Hong X, Harris CC, Dooling DJ, Ochoa K, Johnson KJ, Obenauer JC, Evans WE, Pui CH, Naeve CW, Ley TJ, Mardis ER, Wilson RK, Downing JR, Mullighan CG. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papaemmanuil E, Rapado I, Li Y, Potter NE, Wedge DC, Tubio J, Alexandrov LB, Van Loo P, Cooke SL, Marshall J, Martincorena I, Hinton J, Gundem G, van Delft FW, Nik-Zainal S, Jones DR, Ramakrishna M, Titley I, Stebbings L, Leroy C, Menzies A, Gamble J, Robinson B, Mudie L, Raine K, O'Meara S, Teague JW, Butler AP, Cazzaniga G, Biondi A, Zuna J, Kempski H, Muschen M, Ford AM, Stratton MR, Greaves M, Campbell PJ. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet. 2014;46:116–125. doi: 10.1038/ng.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annu Rev Genet. 2011;45:167–202. doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- 9.Helmink BA, Sleckman BP. The response to and repair of RAG-mediated DNA double-strand breaks. Annual review of immunology. 2012;30:175–202. doi: 10.1146/annurev-immunol-030409-101320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bassing CH, Alt FW, Hughes MM, D'Auteuil M, Wehrly TD, Woodman BB, Gartner F, White JM, Davidson L, Sleckman BP. Recombination signal sequences restrict chromosomal V(D)J recombination beyond the 12/23 rule. Nature. 2000;405:583–586. doi: 10.1038/35014635. [DOI] [PubMed] [Google Scholar]
- 11.Hesse JE, Lieber MR, Gellert M, Mizuuchi K. Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V-(D)-J joining signals. Cell. 1987;49:775–783. doi: 10.1016/0092-8674(87)90615-5. [DOI] [PubMed] [Google Scholar]
- 12.Lewis SM, Hesse JE, Mizuuchi K, Gellert M. Novel strand exchanges in V(D)J recombination. Cell. 1988;55:1099–1107. doi: 10.1016/0092-8674(88)90254-1. [DOI] [PubMed] [Google Scholar]
- 13.Hesse JE, Lieber MR, Mizuuchi K, Gellert M. V(D)J recombination: a functional definition of the joining signals. Genes Dev. 1989;3:1053–1061. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]
- 14.Ramsden DA, McBlane JF, van Gent DC, Gellert M. Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. Embo j. 1996;15:3197–3206. [PMC free article] [PubMed] [Google Scholar]
- 15.Cuomo CA, Mundy CL, Oettinger MA. DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol Cell Biol. 1996;16:5683–5690. doi: 10.1128/mcb.16.10.5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kale SB, Landree MA, Roth DB. Conditional RAG-1 mutants block the hairpin formation step of V(D)J recombination. Mol Cell Biol. 2001;21:459–466. doi: 10.1128/MCB.21.2.459-466.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bischerour J, Lu C, Roth DB, Chalmers R. Base flipping in V(D)J recombination: insights into the mechanism of hairpin formation, the 12/23 rule, and the coordination of double-strand breaks. Mol Cell Biol. 2009;29:5889–5899. doi: 10.1128/MCB.00187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 19.van Gent DC, Hiom K, Paull TT, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. Embo j. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson CB. New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity. 1995;3:531–539. doi: 10.1016/1074-7613(95)90124-8. [DOI] [PubMed] [Google Scholar]
- 21.van Gent DC, Mizuuchi K, Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- 22.Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 23.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 24.Litman GW, Rast JP, Fugmann SD. The origins of vertebrate adaptive immunity. Nat Rev Immunol. 2010;10:543–553. doi: 10.1038/nri2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silver DP, Spanopoulou E, Mulligan RC, Baltimore D. Dispensable sequence motifs in the RAG-1 and RAG-2 genes for plasmid V(D)J recombination. Proc Natl Acad Sci U S A. 1993;90:6100–6104. doi: 10.1073/pnas.90.13.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadofsky MJ, Hesse JE, McBlane JF, Gellert M. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 1994;22:550. doi: 10.1093/nar/22.3.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dudley DD, Sekiguchi J, Zhu C, Sadofsky MJ, Whitlow S, DeVido J, Monroe RJ, Bassing CH, Alt FW. Impaired V(D)J recombination and lymphocyte development in core RAG1-expressing mice. J Exp Med. 2003;198:1439–1450. doi: 10.1084/jem.20030627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuomo CA, Oettinger MA. Analysis of regions of RAG-2 important for V(D)J recombination. Nucleic Acids Res. 1994;22:1810–1814. doi: 10.1093/nar/22.10.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadofsky MJ, Hesse JE, Gellert M. Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res. 1994;22:1805–1809. doi: 10.1093/nar/22.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coussens MA, Wendland RL, Deriano L, Lindsay CR, Arnal SM, Roth DB. RAG2's acidic hinge restricts repair-pathway choice and promotes genomic stability. Cell Rep. 2013;4:870–878. doi: 10.1016/j.celrep.2013.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Callebaut I, Mornon JP. The V(D)J recombination activating protein RAG2 consists of a six-bladed propeller and a PHD fingerlike domain, as revealed by sequence analysis. Cell Mol Life Sci. 1998;54:880–891. doi: 10.1007/s000180050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akamatsu Y, Monroe R, Dudley DD, Elkin SK, Gartner F, Talukder SR, Takahama Y, Alt FW, Bassing CH, Oettinger MA. Deletion of the RAG2 C terminus leads to impaired lymphoid development in mice. Proc Natl Acad Sci U S A. 2003;100:1209–1214. doi: 10.1073/pnas.0237043100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Curry JD, Schlissel MS. RAG2's non-core domain contributes to the ordered regulation of V(D)J recombination. Nucleic Acids Res. 2008;36:5750–5762. doi: 10.1093/nar/gkn553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Talukder SR, Dudley DD, Alt FW, Takahama Y, Akamatsu Y. Increased frequency of aberrant V(D)J recombination products in core RAG-expressing mice. Nucleic Acids Res. 2004;32:4539–4549. doi: 10.1093/nar/gkh778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Curry JD, Schulz D, Guidos CJ, Danska JS, Nutter L, Nussenzweig A, Schlissel MS. Chromosomal reinsertion of broken RSS ends during T cell development. J Exp Med. 2007;204:2293–2303. doi: 10.1084/jem.20070583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deriano L, Chaumeil J, Coussens M, Multani A, Chou Y, Alekseyenko AV, Chang S, Skok JA, Roth DB. The RAG2 C terminus suppresses genomic instability and lymphomagenesis. Nature. 2011;471:119–123. doi: 10.1038/nature09755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L, Reynolds TL, Shan X, Desiderio S. Coupling of V(D)J recombination to the cell cycle suppresses genomic instability and lymphoid tumorigenesis. Immunity. 2011;34:163–174. doi: 10.1016/j.immuni.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gigi V, Lewis S, Shestova O, Mijuskovic M, Deriano L, Meng W, Luning Prak ET, Roth DB. RAG2 mutants alter DSB repair pathway choice in vivo and illuminate the nature of ‘alternative NHEJ’. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, Oettinger MA. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Z, Dordai DI, Lee J, Desiderio S. A conserved degradation signal regulates RAG-2 accumulation during cell division and links V(D)J recombination to the cell cycle. Immunity. 1996;5:575–589. doi: 10.1016/s1074-7613(00)80272-1. [DOI] [PubMed] [Google Scholar]
- 42.van Gent DC, McBlane JF, Ramsden DA, Sadofsky MJ, Hesse JE, Gellert M. Initiation of V(D)J recombinations in a cell-free system by RAG1 and RAG2 proteins. Curr Top Microbiol Immunol. 1996;217:1–10. doi: 10.1007/978-3-642-50140-1_1. [DOI] [PubMed] [Google Scholar]
- 43.Hiom K, Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- 44.Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- 45.Wang G, Dhar K, Swanson PC, Levitus M, Chang Y. Real-time monitoring of RAG-catalyzed DNA cleavage unveils dynamic changes in coding end association with the coding end complex. Nucleic Acids Res. 2012;40:6082–6096. doi: 10.1093/nar/gks255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiu JX, Kale SB, Yarnell Schultz H, Roth DB. Separation-of-function mutants reveal critical roles for RAG2 in both the cleavage and joining steps of V(D)J recombination. Mol Cell. 2001;7:77–87. doi: 10.1016/s1097-2765(01)00156-3. [DOI] [PubMed] [Google Scholar]
- 47.Yarnell Schultz H, Landree MA, Qiu JX, Kale SB, Roth DB. Joining-deficient RAG1 mutants block V(D)J recombination in vivo and hairpin opening in vitro. Mol Cell. 2001;7:65–75. doi: 10.1016/s1097-2765(01)00155-1. [DOI] [PubMed] [Google Scholar]
- 48.Tsai CL, Drejer AH, Schatz DG. Evidence of a critical architectural function for the RAG proteins in end processing, protection, and joining in V(D)J recombination. Genes Dev. 2002;16:1934–1949. doi: 10.1101/gad.984502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA, Shah S, Brandt VL, Meek K, Roth DB. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 50.Lewis S, Gifford A, Baltimore D. DNA elements are asymmetrically joined during the site-specific recombination of kappa immunoglobulin genes. Science. 1985;228:677–685. doi: 10.1126/science.3158075. [DOI] [PubMed] [Google Scholar]
- 51.Deriano L, Roth DB. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu Rev Genet. 2013;47:433–455. doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- 52.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, Rajewsky K, Alt FW. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 53.Simsek D, Brunet E, Wong SY, Katyal S, Gao Y, McKinnon PJ, Lou J, Zhang L, Li J, Rebar EJ, Gregory PD, Holmes MC, Jasin M. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7:e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv Immunol. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- 55.Haluska FG, Finver S, Tsujimoto Y, Croce CM. The t(8; 14) chromosomal translocation occurring in B-cell malignancies results from mistakes in V-D-J joining. Nature. 1986;324:158–161. doi: 10.1038/324158a0. [DOI] [PubMed] [Google Scholar]
- 56.Kagan J, Finan J, Letofsky J, Besa EC, Nowell PC, Croce CM. Alpha-chain locus of the T-cell antigen receptor is involved in the t(10;14) chromosome translocation of T-cell acute lymphocytic leukemia. Proc Natl Acad Sci U S A. 1987;84:4543–4546. doi: 10.1073/pnas.84.13.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haydu JE, De Keersmaecker K, Duff MK, Paietta E, Racevskis J, Wiernik PH, Rowe JM, Ferrando A. An activating intragenic deletion in NOTCH1 in human T-ALL. Blood. 2012;119:5211–5214. doi: 10.1182/blood-2011-10-388504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vanura K, Montpellier B, Le T, Spicuglia S, Navarro JM, Cabaud O, Roulland S, Vachez E, Prinz I, Ferrier P, Marculescu R, Jager U, Nadel B. In vivo reinsertion of excised episomes by the V(D)J recombinase: a potential threat to genomic stability. PLoS Biol. 2007;5:e43. doi: 10.1371/journal.pbio.0050043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mijuskovic M, Brown SM, Tang Z, Lindsay CR, Efstathiadis E, Deriano L, Roth DB. A streamlined method for detecting structural variants in cancer genomes by short read paired-end sequencing. PLoS One. 2012;7:e48314. doi: 10.1371/journal.pone.0048314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mendes RD, Sarmento LM, Cante-Barrett K, Zuurbier L, Buijs-Gladdines JG, Povoa V, Smits WK, Abecasis M, Yunes JA, Sonneveld E, Horstmann MA, Pieters R, Barata JT, Meijerink JP. PTEN micro-deletions in T-cell acute lymphoblastic leukemia are caused by illegitimate RAG-mediated recombination events. Blood. 2014 doi: 10.1182/blood-2014-03-562751. [DOI] [PubMed] [Google Scholar]
- 61.Lewis SM, Agard E, Suh S, Czyzyk L. Cryptic signals and the fidelity of V(D)J joining. Mol Cell Biol. 1997;17:3125–3136. doi: 10.1128/mcb.17.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 63.Aplan PD, Lombardi DP, Ginsberg AM, Cossman J, Bertness VL, Kirsch IR. Disruption of the human SCL locus by “illegitimate” V-(D)-J recombinase activity. Science. 1990;250:1426–1429. doi: 10.1126/science.2255914. [DOI] [PubMed] [Google Scholar]
- 64.Ashworth TD, Pear WS, Chiang MY, Blacklow SC, Mastio J, Xu L, Kelliher M, Kastner P, Chan S, Aster JC. Deletion-based mechanisms of Notch1 activation in T-ALL: key roles for RAG recombinase and a conserved internal translational start site in Notch1. Blood. 2010;116:5455–5464. doi: 10.1182/blood-2010-05-286328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Raghavan SC, Swanson PC, Wu X, Hsieh CL, Lieber MR. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature. 2004;428:88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- 66.Lewis SM. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 67.Marculescu R, Le T, Simon P, Jaeger U, Nadel B. V(D)J-mediated translocations in lymphoid neoplasms: a functional assessment of genomic instability by cryptic sites. J Exp Med. 2002;195:85–98. doi: 10.1084/jem.20011578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chatterji M, Tsai CL, Schatz DG. Mobilization of RAG-generated signal ends by transposition and insertion in vivo. Mol Cell Biol. 2006;26:1558–1568. doi: 10.1128/MCB.26.4.1558-1568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reddy YV, Perkins EJ, Ramsden DA. Genomic instability due to V(D)J recombination-associated transposition. Genes Dev. 2006;20:1575–1582. doi: 10.1101/gad.1432706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Messier TL, O'Neill JP, Hou SM, Nicklas JA, Finette BA. In vivo transposition mediated by V(D)J recombinase in human T lymphocytes. Embo j. 2003;22:1381–1388. doi: 10.1093/emboj/cdg137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roth DB, Craig NL. VDJ recombination: a transposase goes to work. Cell. 1998;94:411–414. doi: 10.1016/s0092-8674(00)81580-9. [DOI] [PubMed] [Google Scholar]
- 72.Barreto V, Marques R, Demengeot J. Early death and severe lymphopenia caused by ubiquitous expression of the Rag1 and Rag2 genes in mice. Eur J Immunol. 2001;31:3763–3772. doi: 10.1002/1521-4141(200112)31:12<3763::aid-immu3763>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 73.Jones JM, Gellert M. Autoubiquitylation of the V(D)J recombinase protein RAG1. Proc Natl Acad Sci U S A. 2003;100:15446–15451. doi: 10.1073/pnas.2637012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yurchenko V, Xue Z, Sadofsky M. The RAG1 N-terminal domain is an E3 ubiquitin ligase. Genes Dev. 2003;17:581–585. doi: 10.1101/gad.1058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Difilippantonio MJ, Zhu J, Chen HT, Meffre E, Nussenzweig MC, Max EE, Ried T, Nussenzweig A. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gao Y, Ferguson DO, Xie W, Manis JP, Sekiguchi J, Frank KM, Chaudhuri J, Horner J, DePinho RA, Alt FW. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature. 2000;404:897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- 77.Bogue MA, Wang C, Zhu C, Roth DB. V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal, and hybrid joint formation. Immunity. 1997;7:37–47. doi: 10.1016/s1074-7613(00)80508-7. [DOI] [PubMed] [Google Scholar]
- 78.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Grundy GJ, Yang W, Gellert M. Autoinhibition of DNA cleavage mediated by RAG1 and RAG2 is overcome by an epigenetic signal in V(D)J recombination. Proc Natl Acad Sci U S A. 2010;107:22487–22492. doi: 10.1073/pnas.1014958107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Elkin SK, Matthews AG, Oettinger MA. The C-terminal portion of RAG2 protects against transposition in vitro. Embo j. 2003;22:1931–1938. doi: 10.1093/emboj/cdg184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Swanson PC, Volkmer D, Wang L. Full-length RAG-2, and not full-length RAG-1, specifically suppresses RAG-mediated transposition but not hybrid joint formation or disintegration. J Biol Chem. 2004;279:4034–4044. doi: 10.1074/jbc.M311100200. [DOI] [PubMed] [Google Scholar]
- 82.Chaumeil J, Micsinai M, Ntziachristos P, Roth DB, Aifantis I, Kluger Y, Deriano L, Skok JA. The RAG2 C-terminus and ATM protect genome integrity by controlling antigen receptor gene cleavage. Nat Commun. 2013;4:2231. doi: 10.1038/ncomms3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bredemeyer AL, Huang CY, Walker LM, Bassing CH, Sleckman BP. Aberrant V(D)J recombination in ataxia telangiectasia mutated-deficient lymphocytes is dependent on nonhomologous DNA end joining. J Immunol. 2008;181:2620–2625. doi: 10.4049/jimmunol.181.4.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mahowald GK, Baron JM, Mahowald MA, Kulkarni S, Bredemeyer AL, Bassing CH, Sleckman BP. Aberrantly resolved RAG-mediated DNA breaks in Atm-deficient lymphocytes target chromosomal breakpoints in cis. Proc Natl Acad Sci U S A. 2009;106:18339–18344. doi: 10.1073/pnas.0902545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, Eckhaus M, Ried T, Bonner WM, Nussenzweig A. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jones JM, Gellert M. Ordered assembly of the V(D)J synaptic complex ensures accurate recombination. Embo j. 2002;21:4162–4171. doi: 10.1093/emboj/cdf394. [DOI] [PMC free article] [PubMed] [Google Scholar]


