SUMMARY
The accumulation of damage caused by oxidative stress has been linked to aging and to the etiology of numerous age-related diseases. The longevity gene, sirtuin 6 (SIRT6), promotes genome stability by facilitating DNA repair, especially under oxidative stress conditions. Here we uncover the mechanism by which SIRT6 is activated by oxidative stress to promote DNA double-strand break (DSB) repair. We show that the stress-activated protein kinase, c-Jun N-terminal kinase (JNK), phosphorylates SIRT6 on serine 10 in response to oxidative stress. This post-translational modification facilitates the mobilization of SIRT6 to DNA damage sites and is required for efficient recruitment of poly (ADP-ribose) polymerase 1 (PARP1) to DNA break sites and for efficient repair of DSBs. Our results demonstrate a post-translational mechanism regulating SIRT6, and they provide the link between oxidative stress signaling and DNA repair pathways that may be critical for hormetic response and longevity assurance.
Graphical Abstract
INTRODUCTION
Reactive oxygen species pose a threat to organismal health and survival by undermining the structural integrity of DNA, lipids, proteins, and other biological macromolecules (Bergamini et al., 2004). The cumulative effect of oxidative stress during an organism’s lifetime has been linked to aging and a number of age-related, degenerative pathologies, including Alzheimer’s disease, amyotrophic lateral sclerosis, Parkinson’s disease, rheumatoid arthritis, and cancer (Schumacker, 2006; Stadtman, 2001; Wiseman and Halliwell, 1996). One of the biggest hazards posed by oxidative stress is the generation of DNA damage and, in particular, DNA double-strand breaks (DSBs). DSBs are an especially cytotoxic lesion since both strands of the DNA back-bone are severed (Lombard et al., 2005). The longevity gene, SIRT6, has been identified as a key factor in stimulating DSB repair (Kaidi et al., 2010; Toiber et al., 2013), particularly in response to oxidative stress (Mao et al., 2011; Van Meter et al., 2011a). DSBs are repaired primarily by two pathways in human cells, homologous recombination (HR) and non-homologous end joining (NHEJ) (Li and Heyer, 2008; Weterings and Chen, 2008), and SIRT6 has been shown to stimulate both of these pathways in response to oxidative stress (Mao et al., 2011).
SIRT6 is a nuclear localized member of the Sirtuin family (Michishita et al., 2005). SIRT6 is required for the maintenance of genomic stability, as SIRT6 deficiency results in severe genomic instability, sensitivity to DNA damage, and premature aging in mice (Mostoslavsky et al., 2006). SIRT6 catalyzes both deacetylation and mono-ADP ribosylation (Gertler and Cohen, 2013), and it also can remove long-chain fatty acyl moieties from lysine residues (Jiang et al., 2013). The first identified deacetylation substrates of SIRT6 are histone H3 lysines 9 and 56 (Michishita et al., 2008; Yang et al., 2009). Deacetylation of these residues plays a role in chromatin regulation upon DNA damage. SIRT6 also is required for mobilization of the DNA-PK catalytic subunit (DNA-PKcs) to chromatin in response to DNA damage (McCord et al., 2009). Non-histone deacetylation substrates of SIRT6 include CtIP (Kaidi et al., 2010), a protein mediating DNA end resection during HR. In addition, SIRT6 is responsible for the recruitment of SNF2H chromatin remodeler to DSBs (Toiber et al., 2013). Known ribosylation substrates of SIRT6 are Kap1 and PARP1. Modification of Kap1 by SIRT6 promotes silencing of L1 retrotransposons (Van Meter et al., 2014). SIRT6-mediated mono-ADP ribosylation of PARP1 on lysine 521 activates PARP1 poly-ADP ribosylation activity and promotes DSB repair under oxidative stress (Mao et al., 2011; Van Meter et al., 2011a).
Despite the significant progress in understanding the role of SIRT6 in maintaining genome stability, little is known about the regulation of SIRT6. Recently, it was reported that Lamin A promotes the catalytic activities of SIRT6 in the context of DNA repair (Ghosh et al., 2015). However, the molecular mechanisms by which SIRT6 is activated in response to oxidative stress remain to be elucidated. Several proteomic screens have identified phosporylation sites on SIRT6 (Dephoure et al., 2008; Miteva and Cristea, 2014; Olsen et al., 2010), and phosphorylation of SIRT6 on Serine 338 was reported to target SIRT6 for degradation (Thirumurthi et al., 2014). However, none of the post-translational modifications have been shown to modulate SIRT6 activity.
Here, we report that SIRT6 is phosphorylated by JNK on Serine 10 in response to oxidative stress. S10 phosporylation is required for the efficient recruitment of SIRT6 to DNA breaks. Furthermore, S10 phosphorylation stimulates SIRT6 mono-ADP ribosylation of PARP1 and promotes rapid recruitment of PARP1 to DNA breaks. Our results define the pathway leading from oxidative stress to activation of DSB repair by SIRT6.
RESULTS
JNK Is Required for SIRT6-Mediated Stimulation of DSB Repair
To gain insight into the mechanism of SIRT6 activation in response to oxidative stress, we conducted a screen using chemical inhibitors of canonical stress response pathways, and we assayed whether SIRT6 was able to stimulate DSB repair in the presence of these inhibitors after exposure to paraquat-induced oxidative stress. For this screen, we used immortalized human dermal fibroblasts cells (HCA2-hTERT) containing a chromosomally integrated reporter of NHEJ (Mao et al., 2011) (Figure S1A). Among the chemical inhibitors screened, only the JNK inhibitor attenuated the ability of SIRT6 to stimulate DSB repair in response to oxidative stress. Importantly, the JNK inhibitor did not affect the levels of SIRT6 protein (Figure 1A). In contrast, other stress-activated protein kinase inhibitors, such as p38, ERK, mTOR, AKT, CDK, and Raf, did not affect the ability of SIRT6 to stimulate DSB repair in response to oxidative stress (Figures S1C–S1H). Silencing JNK expression with JNK-targeting small interfering RNAs (siRNAs) similarly resulted in a failure of SIRT6 to stimulate DSB repair in response to stress (Figure 1B). Using HCA2-hTERT cells containing a chromosomally integrated HR reporter (Figure S1B), we observed that JNK signaling also is required for SIRT6 to stimulate HR in response to oxidative stress (Figure 1C). JNK signaling was similarly required for SIRT6 to induce repair of DSBs in response to stress, as measured by complementary assays, including clearance of γH2AX foci in the presence of the JNK inhibitor or JNK siRNA (Figure 1D) and reduction of mean tail moment in comet assays (Figure 1E).
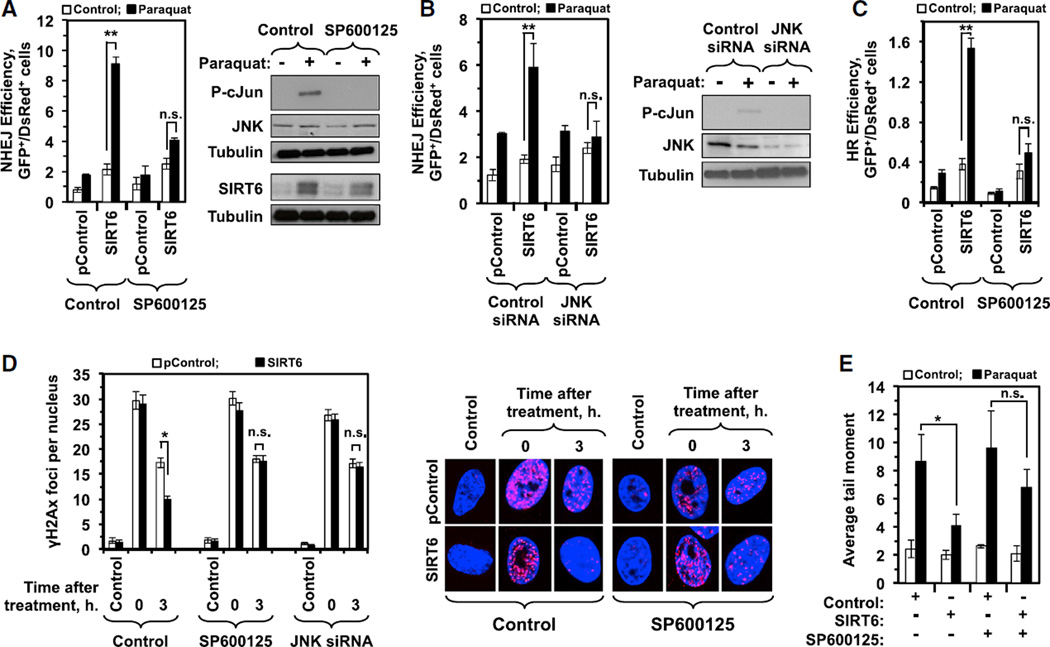
Figure 1. Inhibition of JNK Signaling Abrogates the Ability of SIRT6 to Stimulate DSB Repair in Response to Oxidative Stress.
(A) Immortalized normal diploid human fibroblast cells containing a chromosomally integrated NHEJ reporter cassette (see Figure S1A) were co-transfected with I-SceI and DsRed expression vectors as well as either an SIRT6-encoding plasmid or a control plasmid in the presence or absence of paraquat and a JNK inhibitor (SP600125). SIRT6 expression stimulated NHEJ 2.3-fold relative to control; when cells were pretreated with 1 mM paraquat, SIRT6 expression stimulated NHEJ 9.4-fold relative to control. Pretreating cells with 10 µMJNK inhibitor did not affect the ability of SIRT6 to stimulate NHEJ under basal conditions; when cells were pretreated with both paraquat and JNK inhibitor, however, SIRT6 failed to stimulate NHEJ. Western blots indicate activation of JNK signaling in response to paraquat, as indicated by phosphorylation of c-JUN (p-cJUN); treatment with the JNK inhibitor SP600125 effectively abrogated JNK signaling, but did not affect the paraquat-induced increase in the levels of SIRT6 protein (bottom panel). Error bars indicate SD (n = 6). See also Figure S1.
(B) The requirement of JNK signaling for SIRT6 expression to stimulate NHEJ in response to stress was confirmed using siRNAs. HCA2-hTERT-NHEJ cells were transfected as in (A), but, instead of exposure to a chemical inhibitor, the cells were co-transfected with siRNAs specific to JNK1/2 or a scrambled, control siRNA. SIRT6 expression massively stimulated NHEJ in cells pretreated with paraquat, but failed to do so when the cells also had been transfected with JNK siRNAs. Western blots indicate activation of JNK signaling in response to paraquat, as indicated by phosphorylation of c-JUN (p-cJUN); treatment siRNAs targeting JNK effectively abrogated JNK signaling. Error bars indicate SD (n = 5).
(C) Immortalized normal diploid human fibroblast cells containing a chromosomally integrated HR reporter cassette (see Figure S1B) were co-transfected with I-SceI and DsRed expression vectors as well as either an SIRT6-encoding plasmid or a control plasmid in the presence or absence of paraquat and a JNK inhibitor (SP600125). SIRT6 expression stimulated HR 3.1-fold relative to control; when cells were pretreated with 1 mM paraquat, SIRT6 expression stimulated NHEJ 10.4-fold relative to control. Pretreating cells with 10 µM JNK inhibitor did not affect the ability of SIRT6 to stimulate HR under basal conditions; when cells were pretreated with both paraquat and JNK inhibitor, however, SIRT6 failed to stimulate HR. Error bars indicate SD (n = 4).
(D) SIRT6 expression accelerates the clearance of the DNA DSB marker γH2AX in HCA2-hTERT cells that had been pretreated with 1 mM paraquat for 16 hr. Inhibition of JNK signaling with SP600125 or JNK siRNA abrogates the effect of SIRT6 overexpression. Data represent the average number of γH2AX foci per nucleus. At least 50 nuclei were scored for each time point. Error bars indicate SEM.
(E) Human fibroblasts, transfected with a plasmid encoding either SIRT6 or a control vector, were treated with 1 mM paraquat for 16 hr. Repair was measured 3 hr after the treatment using a comet assay kit (Trevigen) according to the manufacturer’s instructions. Tail moments were determined using CometScore software. One hundred cells were scored for each independent experiment. Error bars indicate SD (n = 3; *p < 0.05 and **p < 0.01). See also Figure S1 for inhibitors of other kinases.
JNK Phosphorylates SIRT6 on Serine 10
JNKs are a family of proline-directed serine/threonine kinases that mediate stress signaling by phosphorylating effector proteins (Davis, 2000); interestingly, multiple studies have implicated JNK signaling in longevity maintenance (Oh et al., 2005; Wang et al., 2003, 2005). We next wanted to test whether JNK directly phosphorylated SIRT6 or whether the requirement for JNK signaling to stimulate SIRT6-mediated DSB repair in response to stress was due to the activation of an intermediate effector protein. We observed that oxidative stress strongly induces interaction between JNK and SIRT6 (Figure 2A). Additionally, when we reconstituted an in vitro system to test whether activated JNK could directly phosphorylate SIRT6, we observed that JNK strongly transferred radiolabel (32P) from [γ-32P]-ATP to SIRT6 under standard kinase reaction conditions (Figure 2B). Interestingly, JNK auto-phosphorylation was reduced in the presence of SIRT6, suggesting that SIRT6 is a preferred target for JNK phosphorylation.
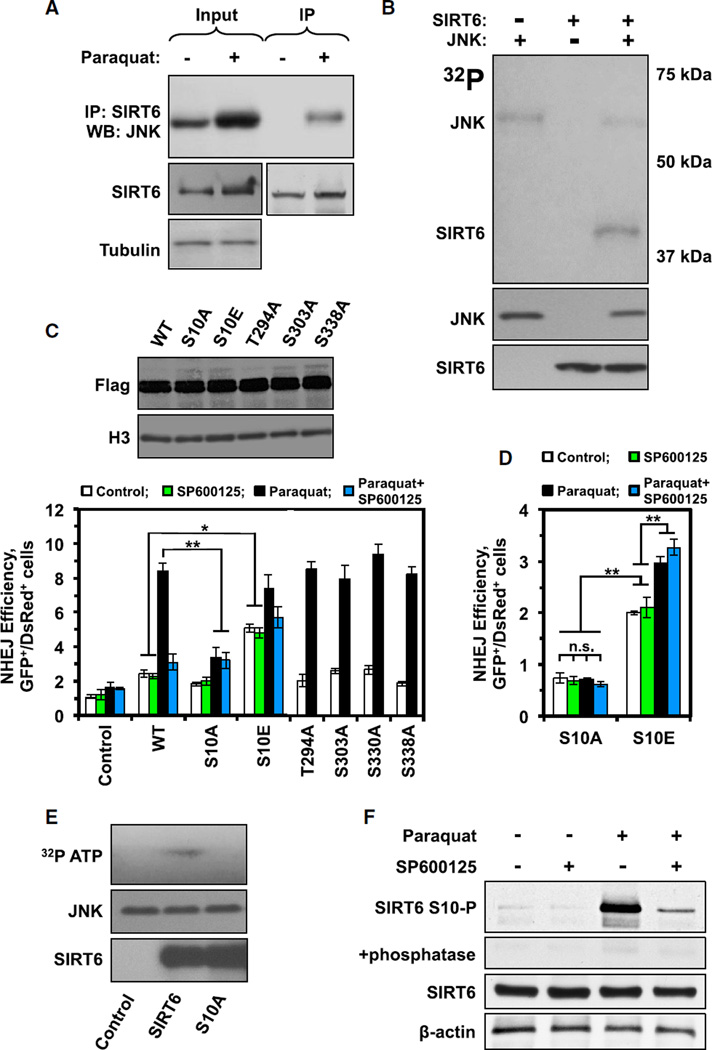
Figure 2. JNK Phosphorylates SIRT6 at Residue S10 to Promote DSB Repair.
(A) CoIP reveals that SIRT6 interacts with JNK in HCA2-hTERT cells only when the cells have been exposed to oxidative stress (1 mM paraquat for 16hr). The experiment was repeated at least four times.
(B) In vitro phosphorylation assay demonstrating that JNK can phosphorylate SIRT6 in vitro. Anisomysin-activated JNK, purified from HEK293 cells, was incubated with BSA and bacterially purified SIRT6 in the presence of 32P-ATP and a kinase reaction buffer. SIRT6 specifically incorporated the radiolabel in these reactions, indicating that it was phosphorylated by JNK. The experiment was repeated three times and a representative gel is shown. See also Figure S2.
(C) SIRT6 plasmids encoding mutations at the indicated putative phosphorylation sites were overexpressed in HCA2-hTERT-NHEJ cells to measure their ability to stimulate NHEJ. SIRT6 T294A, S303A, S330A, and S338A all stimulated NHEJ similarly to WT SIRT6. SIRT6 S10A, however, failed to stimulate NHEJ in response to stress. Expression of an SIRT6 plasmid encoding an S10E phospho-mimetic mutation was able to powerfully stimulate NHEJ in the absence of oxidative stress. The effect of S10E mutation on DNA repair was resistant to JNK inhibition with SP600125. Error bars indicate SD (n = 4). Immunoblot (above) demonstrates that all of the indicated SIRT6 vectors were expressed stably and at comparable levels (*p < 0.05 and **p < 0.01).
(D) NHEJ reporter construct was integrated into SIRT6−/− MEF to measure the SIRT6 S10A and S10E activity in DNA repair in the absence of endogenous SIRT6. Cells expressing SIRT6 S10A mutant showed no stimulation of NHEJ repair in response to paraquat-induced oxidative stress. SIRT6 S10E phospho-mimetic mutant stimulated NHEJ under basal conditions, which could be further stimulated by stress; however, this additional stimulation of NHEJ was not affected by JNK inhibitor SP600125. Error bars indicate SD (n = 3; *p < 0.05 and **p < 0.01).
(E) In vitro phosphorylation assay demonstrates that, while JNK can phosphorylate WT SIRT6, it cannot phosphorylate SIRT6 S10A. Anisomysin-activated JNK, purified fromHEK293 cells, was incubated with BSA and bacterially purified WT SIRT6 of SIRT6 S10A in the presence of 32P-ATP and a kinase reaction buffer. The experiment was repeated three times and a representative gel is shown.
(F) SIRT6 is phosphorylated on S10 in vivo after oxidative stress and the phosphorylation is diminished by JNK inhibitor (SP600125). Custom rabbit polyclonal antibodies (Rb5159 and Rb5160) were generated by immunizing rabbits with YAAGLpSPYADKGKC peptide (see Figure S2D for antibody specificity assays). The hTERT-immortalized human fibroblasts HCA2 were transfected with WT SIRT6-expressing plasmid, then treated with paraquat and/or JNK inhibitor and SIRT6 S10-P, and total SIRT6 levels were assessed by western blot (Rb5159 is pictured; both antibodies gave comparable results). To further confirm the specificity of S10-P antibodies, replicate samples were run and treated with Lambda Protein Phosphatase (LPP) for 1 hr, prior to antibody staining. The experiment was repeated three times and a representative blot is shown.
Several high-throughput proteomic screens had suggested that SIRT6 is subject to extensive phosphorylation at one or more residues, including S10, T294, S303, S330, and S338 (Dephoure et al., 2008; Miteva and Cristea, 2014; Olsen et al., 2010). Consistent with these screens, we observed that SIRT6 is phosphorylated in HCA2-hTERT cells (Figures S2A and S2B). To test if any of these putative SIRT6 phosphorylation sites are important in the context of DSB repair, we constructed five SIRT6 overexpression plasmids wherein these five phosphorylation sites were mutated to alanine residues (S10A, T294A, S303A, S330A, and S338A). When we overexpressed these mutant SIRT6 proteins in our NHEJ reporter assay cells, we observed that SIRT6 T294A, S303A, S330A, and S338A were indistinguishable from wild-type (WT) SIRT6 in their ability to stimulate NHEJ (Figure 2C). Cells overexpressing S10A SIRT6, however, failed to stimulate NHEJ repair in response to oxidative stress. Additionally, cells overexpressing S10E SIRT6, a phospho-mimetic mutant, exhibited heightened DSB repair efficiency, even in the absence of oxidative stress. Overexpression of the S10E mutant also stimulated NHEJ in the presence of a JNK inhibitor, suggesting phosphorylation of SIRT6 is downstream of JNK activation in this pathway (Figure 2C).
Some enhancement of DNA repair by paraquat seen in cells overexpressing SIRT6 S10E and S10A mutants can be explained by endogenous WT SIRT6 protein. To further test this, we generated a mouse SIRT6 knockout (KO) cell line with chromosomally integrated NHEJ reporter, and we repeated the NHEJ assay with SIRT6 S10A and S10E mutants in the absence of the endogenous SIRT6. Under these conditions, S10A mutation completely abolished the ability of SIRT6 to stimulate NHEJ under oxidative stress (Figure 2D). This result indicates that S10 phosphorylation is absolutely required for SIRT6-mediated stimulation of DNA repair. Consistently with our previous results in human fibro-blasts, S10E mutation activated NHEJ even in the absence of paraquat (Figure 2D). Interestingly, paraquat provided additional stimulation of NHEJ, but this additional stimulation was insensitive to the JNK inhibitor. It could be explained by an additional, JNK-independent, post-translational modification of SIRT6 that requires S10 phosphorylation to have an effect or by activation of an SIRT6 downstream factor by oxidative stress.
Consistent with the results of DNA repair assays with SIRT6 phosphorylation site mutants, mass spectrometry of SIRT6 phosphorylated in vitro by JNK revealed S10 phosphorylation, and bioinformatic analysis indicated that residue S10 of SIRT6 conforms to the predictive topology of JNK substrates (Figure S2C). Importantly, recombinant JNK did not phosphorylate S10A SIRT6 in vitro (Figure 2E). To study SIRT6 phosphorylation by JNK in vivo, we generated a phospho-specific SIRT6 S10 antibody. SIRT6 S10 phosphorylation increased upon paraquat treatment and strongly decreased upon the addition of a JNK inhibitor; importantly, treatment with the JNK inhibitor prevented phosphorylation of SIRT6 S10 in response to paraquat treatment (Figure 2F). Treating the samples with lambda protein phosphatase eliminated the band demonstrating the specificity of the antibodies (Figure 2F). Cumulatively, these results suggest that SIRT6 is phosphorylated by JNK at residue S10 and that this modification is required for SIRT6-mediated stimulation of DSB repair in response to oxidative stress.
SIRT6 S10 Phosphorylation Promotes SIRT6 Recruitment to DNA Breaks
We next wanted to gain mechanistic insight into how phosphorylation of SIRT6 by JNK, in response to oxidative insult, resulted in the stimulation of DNA DSB repair. Initial analysis indicated that SIRT6 is enriched at chromatin after oxidative insult (Figure 3A). We therefore hypothesized that phosphorylation of SIRT6, by JNK, may impact the efficiency of SIRT6 recruitment to chromatin. Using laser irradiation-directed confocal microscopy, we observed that SIRT6-GFP is rapidly recruited to DSBs. In the presence of a JNK inhibitor, however, recruitment of SIRT6-GFP to break sites was significantly attenuated (Figure 3B). Consistent with these results, S10A SIRT6-GFP displayed diminished recruitment efficiency to DSBs, relative to WT SIRT6-GFP, while S10E SIRT6-GFP displayed enhanced recruitment efficiency (Figure 3C).
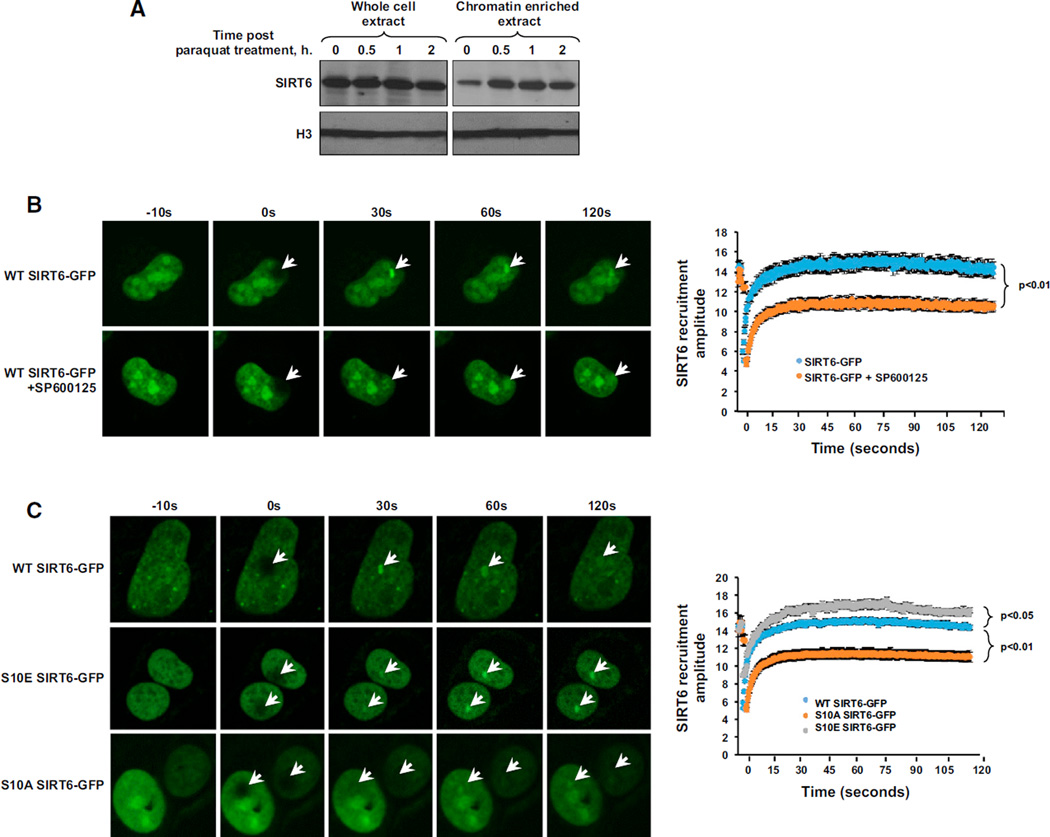
Figure 3. Phosphorylation at SIRT6 Residue S10 Stimulates Recruitment of SIRT6 to DSB Sites.
(A) Chromatin-enriched fractions from WT MEFs indicate that SIRT6 is rapidly recruited to chromatin following exposure of cells to paraquat. Cells were treated with 0.5 mM paraquat, and chromatin-enriched extracts were prepared at the indicated time points (n = 3). A representative blot is shown.
(B) Recruitment of SIRT6-GFP to sites of laser-induced DNA damage was monitored in U2OS cells transfected with SIRT6-GFP in the presence or absence of a JNK inhibitor (SP600125). Cells pretreated with 20 µM JNK inhibitor for 2 hr exhibited severe defects in their ability to recruit SIRT6 to DSB sites. Representative images are shown (n > 8 for each sample). Error bars indicate SEM. See also Figure S3.
(C) Recruitment of WT, S10A, or S10E SIRT6-GFP to sites of laser-induced DNA damage was monitored in U2OS cells. S10E SIRT6-GFP exhibited enhanced recruitment efficiency to sites of DSBs, whereas S10A SIRT6-GFP exhibited diminished recruitment efficiency to DSB sites. Representative images are shown (n > 8 for each sample). Error bars indicate SEM.
We previously showed, using chromatin immunoprecipitation (ChIP) assays, that SIRT6 displays bimodal recruitment kinetics to I-SceI-induced DSBs (Mao et al., 2011). The first rapid wave of recruitment coincides with the maximum I-SceI expression (Mao et al., 2008), while the second wave happens 8 hr later and may be related to post-repair chromatin remodeling to restore the original chromatin state. Consistent with the confocal microscopy results (Figures 3B and 3C), ChIP analysis of SIRT6 recruitment to a site-specific DSB confirmed that JNK was required for the first wave of SIRT6 mobilization to DSB sites under oxidative stress, while the second wave was independent of JNK signaling (Figure S3). Collectively, these results suggest that JNK signaling is required for efficient recruitment of SIRT6 to DSB sites.
SIRT6 S10 Phosphorylation Stimulates SIRT6 Mono-ADP Ribosylation Activity on PARP1
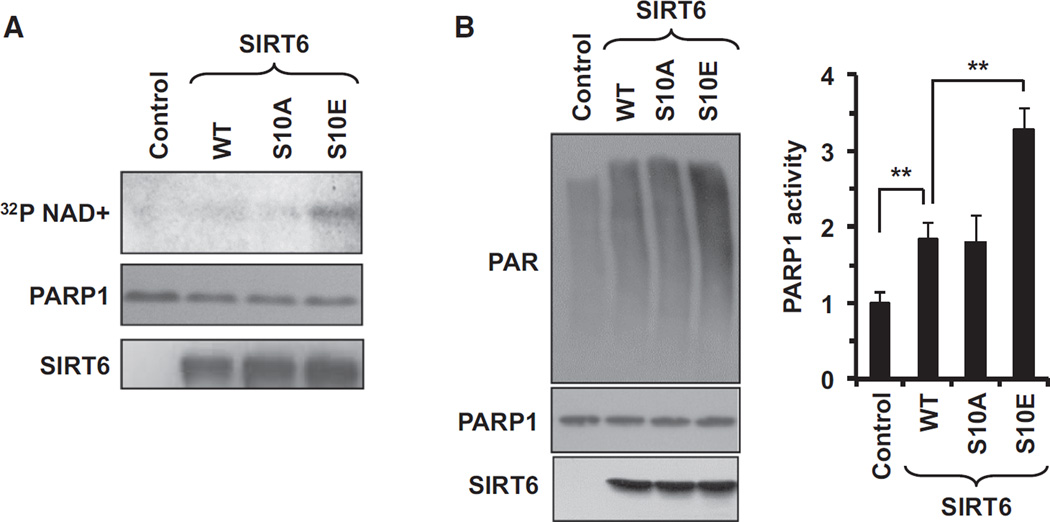
Previous studies had indicated that SIRT6 mono-ADP ribosylation of PARP1 is critical for mediating DSB repair in response to oxidative stress (Mao et al., 2011). It was unclear, however, how the activity of SIRT6 on PARP1 is regulated. In vitro experiments revealed that S10E SIRT6 more robustly mono-ADP ribosylated PARP1 than either WT or S10A SIRT6 (Figure 4A), resulting in higher PARP1 activity (Figure 4B). In this experiment, SIRT6 was purified from E.coli and not phosphorylated, hence, the WT and S10A SIRT6 displayed similar activities. SIRT6 S10A and S10E mutations did not affect SIRT6 deacetylation activity on H3K9ac, consistent with the mono-ADP ribosylation activity playing the primary role in DNA repair function of SIRT6.
Figure 4. Phosphorylation of SIRT6 by JNK Stimulates Its Ability to Activate PARP1.
(A) In vitro mono-ADP ribosylation reaction. Bacterially purified recombinant WT and S10A and S10E SIRT6 proteins were incubated with catalytically inactive recombinant PARP1 (C-terminal truncation, containing only aa 1–655) for 2 hr. S10E SIRT6 was able to more robustly mono-ADP ribosylate the PARP1 substrate than either WT or S10A SIRT6 (n = 3). A representative reaction is shown.
(B) In vitro ribosylation assay demonstrating S10E SIRT6 more robustly stimulates PARP1 activity. Bacterially purified WT, S10A, or S10E SIRT6 was incubated with PARP1. PARP1 activity was measured by quantifying the amount of auto-poly-ADP ribosylation of the protein by immunoblotting with antibodies targeting poly-ADP ribose. The experiment was independently repeated three times; the right panel shows quantification; error bars indicate SD (*p < 0.05 and **p < 0.01).
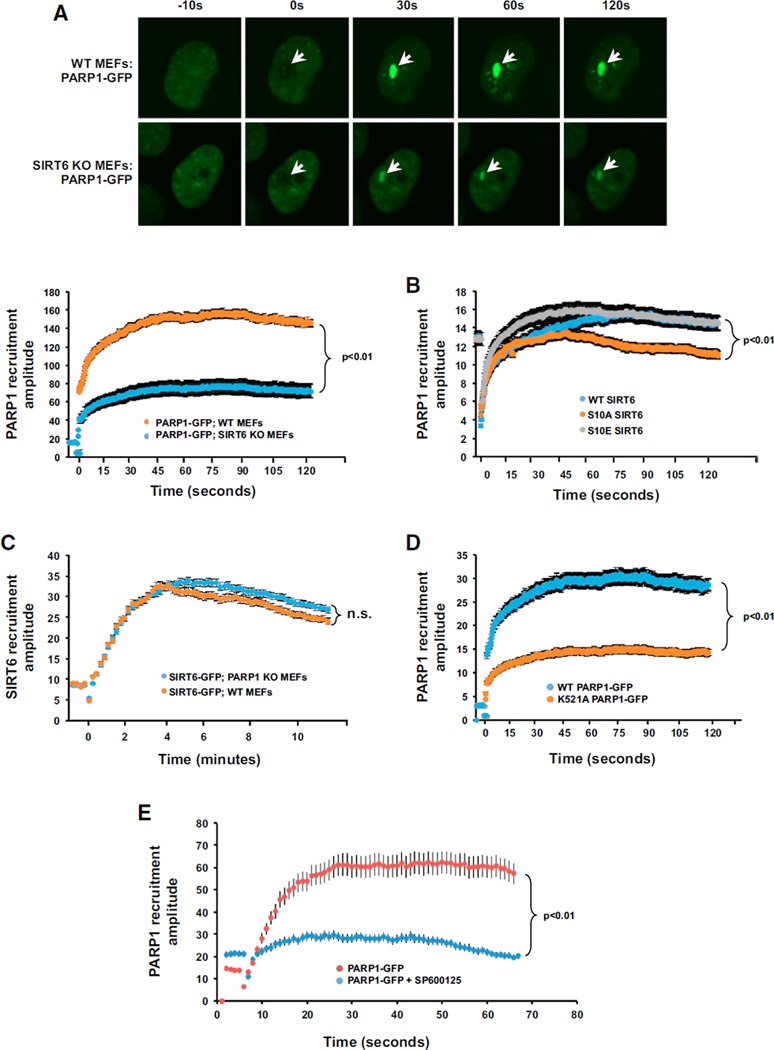
We employed laser irradiation confocal microscopy to dissect the relevance of these observations in a cellular context. Strikingly, we observed that, in the absence of SIRT6, PARP1-GFP was poorly recruited to DSB sites (Figure 5A). However, PARP1 was not required for the efficient recruitment of SIRT6 to DSB sites (Figure 5B), indicating that SIRT6 is upstream of PARP1 in the cascade of protein recruitment to the DNA break site. Consistent with this finding, overexpression of WT or S10E SIRT6, but not S10A SIRT6, was sufficient to stimulate PARP1-GFP recruitment efficiency to DSB sites (Figure 5C). The effect of overexpression of SIRT6 S10 mutations was more profound at the retention stage than the complete absence of SIRT6, which affected initial recruitment (Figure 5A). This could be explained by the presence of endogenous WT SIRT6 molecules reducing the impact of the phosphorylation site mutants. Consistent with this explanation, PARP1 recruitment was severely impaired in the presence of a JNK inhibitor (Figure 5E). SIRT6 mono-ADP ribosylates PARP1 at residue K521, thereby activating PARP1. K521A PARP1-GFP failed to efficiently recruit to DSB sites in WT mouse embryonic fibroblasts (MEFs), suggesting that SIRT6-mediated modification is required for PARP1 recruitment to the DNA breaks (Figure 5D).
Figure 5. Phosphorylation of SIRT6 by JNK Accelerates the Recruitment of PARP1 to DSB Sites.
(A) Recruitment of PARP1-GFP to sites of laser-induced DNA damage was measured in WT and SIRT6 KO MEFs. In the absence of SIRT6, PARP1 exhibited a striking failure to fully recruit to DNA break sites. Representative images are shown (n > 8 for each condition); error bars indicate SEM.
(B) Recruitment of SIRT6-GFP to sites of laser-induced DNA damage was measured in WT and PARP1 KO MEFs. In the absence of PARP1, SIRT6 was able to be fully recruited to DNA break sites. Representative images are shown (n > 8 for each condition). Error bars represent SEM (n.s., not significant).
(C) Recruitment of PARP1-GFP to sites of laser-induced DNA damage in MEFs overexpressing WT, S10A, or S10E SIRT6. Cells overexpressing WT SIRT6 and S10E SIRT6 were able to more robustly recruit PARP1-GFP to sites of DNA damage. By contrast, cells overexpressing S10A SIRT6 failed to stimulate PARP1 recruitment to DNA damage sites. Error bars indicate SEM (n > 8).
(D) Modification of K521A PARP1 is required for the efficient recruitment of PARP1 to DSB sites. Recruitment of PARP-GFP1 or K521A PARP1-GFP to sites of laser-induced DNA damage was measured in MEFs. K521A PARP1-GFP exhibited a failure to efficiently recruit to DNA damage sites. Representative images are shown (n > 8 for each condition). Error bars indicate SEM (n > 8).
(E) JNK inhibition abrogates PARP1 recruitment to DNA damage sites. Recruitment of PARP1-GFP to sites of laser-induced DNA damage was measured in WT MEFs in the presence or absence of a JNK inhibitor. JNK inhibition with SP600125 resulted in failure to recruit PARP1 to DNA damage sites. Error bars indicate SEM (n > 8).
DISCUSSION
Our results indicate that SIRT6 is phosphorylated by JNK specifically in response to oxidative stress at residue S10 and that this modification is necessary for SIRT6 to stimulate DNA DSB repair under stress conditions. Once phosphorylated, SIRT6 is rapidly mobilized to DSB break sites, and it potentiates the activity and recruitment of the apical DNA repair factor, PARP1, which in turn stimulates DNA DSB repair (Figure 6). Our work provides evidence of a post-translational modification stimulating the activity of SIRT6. In summary, our results delineate the pathway connecting oxidative stress response and DSB repair, indicating that S10 phosphorylation plays a central role in stimulating DSB repair in response to oxidative stress.
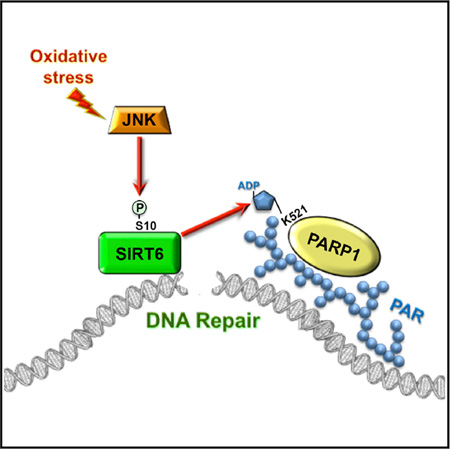
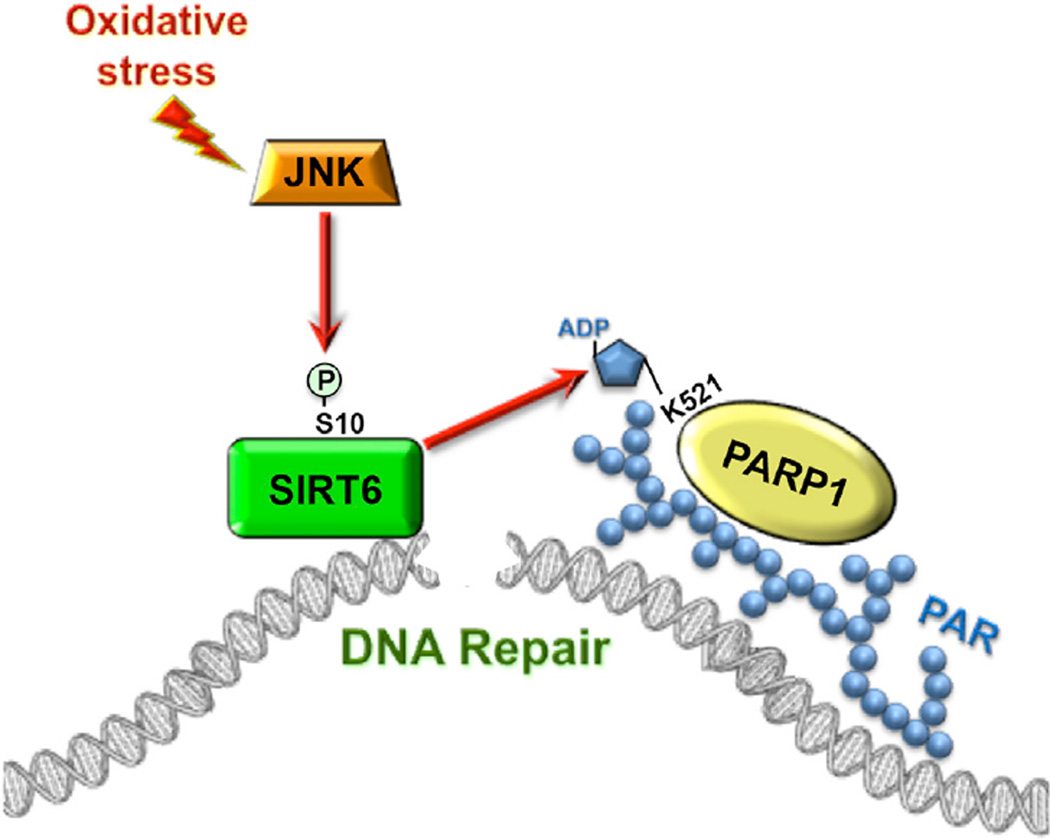
Figure 6. Model for JNK-Mediated Activation of DSB Repair.
Upon oxidative stress JNK phosphorylates SIRT6 on Serine 10. This results in rapid recruitment of SIRT6 to the DSB site and simultaneously stimulates SIRT6 mono-ADP ribosylation of PARP1. PARP1 mono-ADP ribosylation leads to recruitment of PARP1 to DSB site and activates PARP1 poly-ADP ribosylation activity. This sequence of events represents the initial steps in the assembly of repair machinery on a DSB, and it is required for efficient DSB repair under oxidative stress conditions.
Interestingly, SIRT6-mediated stimulation of DSB repair is reminiscent of a hormetic response, wherein mild doses of stress have beneficial effects on an organism by promoting stress and survival pathways. Hormesis has been linked to the lifespan-extending effects of caloric restriction (Masoro, 2006; Merksamer et al., 2013). We hypothesize that the JNK-SIRT6 axis represents a hormetic response pathway, promoting longevity by stimulating DNA DSB repair under moderately stressful conditions.
We demonstrated that SIRT6 recruitment to DNA damage sites is stimulated by oxidative stress. Oxidative damage is one of the more frequent types of damage occurring to DNA during the life course of an organism. It is also hypothesized to be the major type of damage to macromolecules contributing to the aging process. It is possible that the JNK-SIRT6 axis has evolved specifically to modulate the repair of oxidatively damaged DNA. It would be interesting to study whether other physiological types of stress, such as starvation or exercise, may similarly stimulate SIRT6 phosphorylation and/or recruitment to DNA upon DNA damage.
Studies in invertebrates have revealed that mild activation of JNK signaling is longevity promoting, whereas constitutive activation of JNK drives apoptosis, inflammation, and cell death (Wang et al., 2014). It is also interesting to note that JNK has been reported to phosphorylate SIRT1, a gene also linked to longevity maintenance in mammals, thereby stimulating the activity of this sirtuin protein (Ford et al., 2008; Nasrin et al., 2009).
In mammals, several studies have indicated that activation of SIRT6 represents a novel strategy for delaying the onset, and potentially reversing the pathology, of multiple age-related diseases. For example, SIRT6 overexpression protects against diet-induced obesity, has anti-tumorigenic effects, and extends lifespan in mice (Kanfi et al., 2010, 2012; Min et al., 2012; Van Meter et al., 2011b). As such, identifying mechanisms for modulating the activity of SIRT6 is of intense therapeutic and pharmacological interest. Several studies have indicated that caloric restriction and reduction of glucose intake can upregulate SIRT6 expression levels (Kanfi et al., 2008). Our study suggests that the activity of SIRT6 may be modulated post-translationally as well, since phosphorylation of SIRT6 by JNK stimulates the ability of the protein to mediate DNA DSB repair. This finding provides new avenues for developing SIRT6 activators potentially having beneficial effects on health.
EXPERIMENTAL PROCEDURES
Cell Lines and Growth Conditions
HCA2-hTERT, HCA2-hTERT-NHEJ, and HCA2-hTERT-HR cell lines were hTERT-immortalized normal human diploid foreskin fibroblasts. HCA2-hTERT-NHEJ cells contained a single copy of a chromosomally integrated NHEJ reporter cassette (Figure S1A). HCA2-hTERT-HR cells contained a single copy of a chromosomally integrated HR reporter cassette (Figure S1B). MEF cells were isolated from embryos of WT, SIRT6 KO, and PARP1 KO mice of 129 genetic backgrounds. All cell lines were cultured at 37°C in 5% CO2 and 3% O2 in Eagle’s minimal essential medium (EMEM) (Gibco) with 10% fetal bovine serum, 1% penicillin/streptomycin (Gibco), and 1× minimal essential medium (MEM) nonessential amino acids (Gibco).
Transfections
HCA2-hTERT and HCA2-hTERT-NHEJ were transfected with plasmids or siRNA using AMAXA nucleofector with NHDF transfection kit and U20 program. In each transfection two million cells were transfected.
DSB Repair Assays and FACS Analysis
In vivo DSB repair assays were performed as described (Mao et al., 2011). Briefly, cycling cells were co-transfected with 5 µg plasmid encoding I-SceI endonuclease to induce DSBs and 0.1 µg plasmid encoding DsRed to control for transfection efficiency. 4 days after transfection, the numbers of GFP+ and DsRed+ cells were determined by flow cytometry. The ratio between GFP+ and DsRed+ cells was used as a measure of DSB repair efficiency. Fluorescence-activated cell sorting (FACS) analysis was performed on FACS canto machine (BD Biosciences). For each treatment, a minimum of 50,000 cells was analyzed by FACS. Final data analysis was done using FlowJo software.
Plasmids and siRNAs
SIRT6 was cloned into the backbone of pDsRed2-N1 using Titan One Tube RT-PCR kit (Roche, 11939823001). The resulting clones were sequenced and confirmed by comparing them to the NCBI database. All the SIRT6 mutants were generated by site-specific mutagenesis method (Strategene, Quikchange Kit, 200516). The siRNAs against SIRT6 were purchased from QIAGEN. The siRNAs against JNK were purchased from Cell Signaling Technology.
Antibodies
Commercially available antibodies against SIRT6 were purchased from Abcam; ab62738 was used for analyzing endogenous SIRT6 by western blot; ab48192 was used for ChIP; ab62738 was used for co-immunoprecipitation (coIP) and immunostaining.
Custom rabbit polyclonal anti-SIRT6-phosphoS10 (pS10) antibodies were obtained from GenScript. Custom peptide containing pS10 of SIRT6 (YAAGLpSPYADKGKC) was synthesized by GenScript, and it was used for three consecutive immunizations of two rabbits. Final serum was collected and purified against SIRT6-pS10 peptide that was immobilized to agarose beads. Specificity of each anti-SIRT6-pS10 purified antibody (from each rabbit) was gauged by ELISA by comparing the binding of serial dilution of each antibody against pS10-peptide or the non-phosphorylated equivalent (Figure S2D). Excellent specificity for pS10-peptide was found in both samples (as high as 1:2.5 × 105 dilutions showed specificity for pS10 peptide and none for the non-phosphorylated equivalent). For western blot, a dilution of Rb5159 at 1:15,000 was used. For lambda phosphatase treatment, membranes with transferred proteins were incubated with 6 ml buffered solution and 4,000 units phosphatase for 1 hr at 30°C.
H3 and acetylated H3K9 were detected with anti-H3 (Cell Signaling Technology, 9715) and anti-H3K9Ac (Cell Signaling Technology, 9671). Antibodies against JNK were purchased from Santa Cruz Biotechnology and Abcam; sc-137020 was used for coIP experiments, ab64334 was used for western blots, and phospho-JNK was detected using ab4821. PAR was detected with mouse monocolonal antibodies (BD Pharmigen, 550781). Control antibodies used for coIP and ChIP were rabbit anti-human IgG antibody (Abcam, ab938) and GFP antibody (Cell Signaling Technology, 2555). FLAG tag was detected using anti-DYKDDDDK antibodies (Cell Signaling Technology, 2368).
ChIP
Prior to ChIP, HCA2-hTERT-NHEJ cells were treated with 1 mM paraquat and transfected with 5 µg I-SceI expression vector. ChIP was performed as previously described (Mao et al., 2011). To quantify SIRT6 recruitment to the site of I-SceI-induced DSBs, qPCR was performed to amplify the region 7 to 315 nt downstream of the I-SceI site with the primers 5′CCTGAAGATTTGG GGGATTGTGCTTC3′ and 5′CTTGGAAACACCCATGTTGAAATATC3′, using the Platinum Taq DNA polymerase kit (Invitrogen). DNA corresponding to 35,000 cells from IgG and anti-SIRT6-precipitated samples and input equal to 1% of the IPs were used as a template for PCR. The PCR reaction was run in an MJ Research thermocycler with the following steps: one cycle of 95°C for 1 min; 35 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min; and one cycle of 72°C for 5 min. Data from the IPs were normalized to data from the 1% inputs.
CoIP
HCA2-hTERT cells were treated with 1 mM paraquat for 16 hr or a PBS control. IP was performed as previously described (Mao et al., 2011). Anti-SIRT6 antibody was added to the supernatants for overnight incubation at 4°C, followed by incubation with protein A sepharose for 2 hr. Then the beads were spun down for 1 min at 8,000 rpm at 4°C and washed five times with IP lysis buffer. The proteins were eluted with 2× sample buffer (Laemmli buffer:beta-mecaptoethanol = 950:50) by boiling for 10 min and then spun down, and the supernatants were collected and used for western blotting.
Immunofluorescence
For the immunostaining experiments, HCA2-hTERT cells were seeded at 50,000 cells per coverslip and subsequently fixed with 4% paraformaldehyde, washed three times with cold PBS, and permeabilized with 0.25% Triton X-100. Following three PBS washes, the samples were blocked with 1% goat serum for 60 min and then incubated with anti-SIRT6, anti-γH2AX (Abcam, ab22551) overnight. Coverslips were washed with PBS three times and incubated with goat anti-rabbit-FITC or goat anti-mouse-Cy3 for 1 hr in the dark. Then the samples were washed with PBS three times, stained with 1 µg/ml DAPI for 2 min, and washed with PBS. The slides were covered with VECTASHIED mounting media (Vector Laboratories), and they were used for confocal microscopy with a Leica SP5 microscope. A minimum of 50 nuclei was quantified for each sample.
Chemical Inhibitors
The following inhibitors were used at the manufacturer-recommended concentrations: JNK was inhibited using 20 µM SP600125 (Sigma), p38 was inhibited using 100 nM SB202190 (Sigma), ERK was inhibited using 72 nM U0126 (Sigma), CDK1 was inhibited using 500 nM Kenpaullone (Tocris), AKT was inhibited using 83 µM124005 Akt Inhibitor (Millipore), mTOR was inhibited using 200 nM rapamycin (Sigma), and Raf was inhibited using 50 nM PLX4032 (SelleckChem).
In Vitro Phosphorylation Assay
Purified, anisomycin-activated JNK (1 µg, Signal Chem, M34-10G) was incubated with 5 µg bacterially purified recombinant SIRT6 for 2 hr in Kinase Reaction Buffers (Signal Chem, K23-09-05 and K21-09-05) and ATP or [γ-32P]-ATP. Upon the competition of the reaction, proteins were boiled in Laemmli buffer, separated by SDS-PAGE, and exposed to film.
In Vitro Ribosylation Assay
The reactions analyzing mono-ADP ribosylation were performed essentially as described by Liszt et al., (2005). Reactions contained 5 µg hSIRT6 WT or mutant proteins, 5 µg of the truncated PARP1 missing catalytic domain (C-terminal truncation; containing only amino acid [aa] 1–655), or truncated PARP1 harboring K521A, 50 mM Tris$Cl, 150 mM NaCl, 10 mM DTT, 1 µM unlabeled NAD+, and 8 µCi 32P NAD+ (PerkinElmer, NEG023X500UC) in a 50-µl volume. Auto-ribosylation reactions by hSIRT6 and its mutants were carried out at 37°C for 30 min, and ribosylation reactions on catalytically inactive PARP1 by hSIRT6 were kept at 37°C for 2 hr. After reactions were completed, the samples were 2,2,2-trichloroacetic acid (TCA) precipitated to remove unincorporated 32P NAD+. The purified samples were loaded on SDS-PAGE gels, Commassie stained, dried, and subjected to autoradiography. Reactions analyzing activation of PARP1 by human SIRT6 contained 2 µl PARP1 (Sigma, P0996), 5 µg SIRT6WT ormutant proteins, 20 mM Tris$Cl (pH 8.0), 100 mM NaCl, 10 mM MgCl2, 10 µM ZnCl2, 10% glycerol, 300 µM NAD+, 1 mM DTT, and 0.1 µg/ml sonicated salmon sperm DNA in a 50-µl volume. These reactions were performed at 30°C for 30 min. The samples were then analyzed by western blotting using anti-PAR antibody (BD Pharmigen, 550781).
MicroPoint Laser Irradiation and Microscopy
MEF cell lines were used to analyze recruitment of fluorescently labeled DNA repair proteins to laser-induced DNA damage sites. MEFs were transfected using the U-20 program of an Amaxa Nucleofector II electroporation device. Cycling MEFs were harvested 1 day prior to analysis, suspended in NHDF Neucleofector solution, mixed with 1 µg of the relevant plasmid DNA, and transfected. The cells were then plated on 15-mm dishes with thin glass bottoms. Targeted DNA damage was introduced using the MicroPoint Ablation Laser System from Photonic Instruments at 11% laser power as described in Popuri et al., (2012), and fluorescent protein recruitment and retention were monitored. Images were acquired either continuously over the course of 2 min or every 10 s for at least 5 min.
Statistical Analysis
Unless indicated otherwise, the Student’s t test was used to determine if differences between groups were statistically significant. All tests were two-tailed. We set p values of less than 0.05 as a threshold for statistical significance.
Supplementary Material
Highlights.
JNK phosphorylates SIRT6 at residue S10 in response to oxidative stress
SIRT6 S10 phosphorylation is required for the stimulation of DNA break repair
SIRT6 S10 phosphorylation stimulates SIRT6 mono-ADP ribosylation activity on PARP1
In Brief.
Van Meter et al. show that SIRT6 is phosphorylated by JNK on serine 10 in response to oxidative stress. SIRT6 S10 phosphorylation promotes DNA break repair by stimulating SIRT6 and PARP1 recruitment to DNA break sites.
Acknowledgments
This work was supported by the Intramural program of the National Institute on Aging to V.A.B.; grants from the National Institute on Aging to M.V.M., V.G., and A.S.; and Life Extension Foundation to V.G. and A.S. We thank Xiao Tian, Amita Vaidya, and Jorge Azpurua for helpful conversations. We also thank Juliya Ablaeva for technical assistance.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.08.006.
AUTHOR CONTRIBUTIONS
M.V.M. and T.D.M. performed the screens to identify a role for JNK in DSB repair. M.S., M.V.M., and K.B. performed comet assays and γH2AX foci-staining experiments. G.T., M.S., and R.P. generated and characterized SIRT6 S10-P antibody. A.M. and M.V.M. performed irradiation confocal microscopy experiments. B.P.H. and D.A.S. cloned SIRT6 phospho-mutant. M.V.M. performed the in vitro experiments. M.V.M., D.A.S., V.A.B., A.S., and V.G. designed the experiments. M.V.M., V.G., and A.S. wrote the manuscript and analyzed the data with the help of all authors. V.G. and A.S. conceived the project, supervised the project, and acquired funding.
REFERENCES
- Bergamini CM, Gambetti S, Dondi A, Cervellati C. Oxygen, reactive oxygen species and tissue damage. Curr. Pharm. Des. 2004;10:1611–1626. doi: 10.2174/1381612043384664. [DOI] [PubMed] [Google Scholar]
- Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villén J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford J, Ahmed S, Allison S, Jiang M, Milner J. JNK2-dependent regulation of SIRT1 protein stability. Cell Cycle. 2008;7:3091–3097. doi: 10.4161/cc.7.19.6799. [DOI] [PubMed] [Google Scholar]
- Gertler AA, Cohen HY. SIRT6, a protein with many faces. Biogerontology. 2013;14:629–639. doi: 10.1007/s10522-013-9478-8. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Liu B, Wang Y, Hao Q, Zhou Z. Lamin A is an endogenous SIRT6 activator and promotes SIRT6-mediated DNA repair. Cell Rep. 2015;13:1396–1406. doi: 10.1016/j.celrep.2015.10.006. [DOI] [PubMed] [Google Scholar]
- Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, et al. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496:110–113. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329:1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kanfi Y, Shalman R, Peshti V, Pilosof SN, Gozlan YM, Pearson KJ, Lerrer B, Moazed D, Marine JC, de Cabo R, Cohen HY. Regulation of SIRT6 protein levels by nutrient availability. FEBS Lett. 2008;582:543–548. doi: 10.1016/j.febslet.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanfi Y, Peshti V, Gil R, Naiman S, Nahum L, Levin E, Kronfeld-Schor N, Cohen HY. SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell. 2010;9:162–173. doi: 10.1111/j.1474-9726.2009.00544.x. [DOI] [PubMed] [Google Scholar]
- Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, Bar-Joseph Z, Cohen HY. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Mao Z, Bozzella M, Seluanov A, Gorbunova V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst.) 2008;7:1765–1771. doi: 10.1016/j.dnarep.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoro EJ. Role of hormesis in life extension by caloric restriction. Dose Response. 2006;5:163–173. doi: 10.2203/dose-response.06-005.Masoro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord RA, Michishita E, Hong T, Berber E, Boxer LD, Kusumoto R, Guan S, Shi X, Gozani O, Burlingame AL, et al. SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging (Albany, N.Y.) 2009;1:109–121. doi: 10.18632/aging.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merksamer PI, Liu Y, He W, Hirschey MD, Chen D, Verdin E. The sirtuins, oxidative stress and aging: an emerging link. Aging (Albany, N.Y.) 2013;5:144–150. doi: 10.18632/aging.100544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min L, Ji Y, Bakiri L, Qiu Z, Cen J, Chen X, Chen L, Scheuch H, Zheng H, Qin L, et al. Liver cancer initiation is controlled by AP-1 through SIRT6-dependent inhibition of survivin. Nat. Cell Biol. 2012;14:1203–1211. doi: 10.1038/ncb2590. [DOI] [PubMed] [Google Scholar]
- Miteva YV, Cristea IM. A proteomic perspective of Sirtuin 6 (SIRT6) phosphorylation and interactions and their dependence on its catalytic activity. Mol. Cell. Proteomics. 2014;13:168–183. doi: 10.1074/mcp.M113.032847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Nasrin N, Kaushik VK, Fortier E, Wall D, Pearson KJ, de Cabo R, Bordone L. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PLoS ONE. 2009;4:e8414. doi: 10.1371/journal.pone.0008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Svrzikapa N, Jiang F, Davis RJ, Tissenbaum HA. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc. Natl. Acad. Sci. USA. 2005;102:4494–4499. doi: 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- Popuri V, Ramamoorthy M, Tadokoro T, Singh DK, Karmakar P, Croteau DL, Bohr VA. Recruitment and retention dynamics of RECQL5 at DNA double strand break sites. DNA Repair (Amst) 2012;11:624–635. doi: 10.1016/j.dnarep.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacker PT. Reactive oxygen species in cancer cells: live by the sword, die by the sword. Cancer Cell. 2006;10:175–176. doi: 10.1016/j.ccr.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Stadtman ER. Protein oxidation in aging and age-related diseases. Ann. N Y Acad. Sci. 2001;928:22–38. doi: 10.1111/j.1749-6632.2001.tb05632.x. [DOI] [PubMed] [Google Scholar]
- Thirumurthi U, Shen J, Xia W, LaBaff AM, Wei Y, Li CW, Chang WC, Chen CH, Lin HK, Yu D, Hung MC. MDM2-mediated degradation of SIRT6 phosphorylated by AKT1 promotes tumorigenesis and trastuzumab resistance in breast cancer. Sci. Signal. 2014;7:ra71. doi: 10.1126/scisignal.2005076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, Sebastian C, Cosentino C, Martinez-Pastor B, Giacosa S, et al. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol. Cell. 2013;51:454–468. doi: 10.1016/j.molcel.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meter M, Mao Z, Gorbunova V, Seluanov A. Repairing split ends: SIRT6, mono-ADP ribosylation and DNA repair. Aging (Albany, N.Y.) 2011a;3:829–835. doi: 10.18632/aging.100389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meter M, Mao Z, Gorbunova V, Seluanov A. SIRT6 over-expression induces massive apoptosis in cancer cells but not in normal cells. Cell Cycle. 2011b;10:3153–3158. doi: 10.4161/cc.10.18.17435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meter M, Kashyap M, Rezazadeh S, Geneva AJ, Morello TD, Seluanov A, Gorbunova V. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat. Commun. 2014;5:5011. doi: 10.1038/ncomms6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MC, Bohmann D, Jasper H. JNK signaling confers tolerance to oxidative stress and extends lifespan in Drosophila. Dev. Cell. 2003;5:811–816. doi: 10.1016/s1534-5807(03)00323-x. [DOI] [PubMed] [Google Scholar]
- Wang MC, Bohmann D, Jasper H. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell. 2005;121:115–125. doi: 10.1016/j.cell.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Wang L, Karpac J, Jasper H. Promoting longevity by maintaining metabolic and proliferative homeostasis. J. Exp. Biol. 2014;217:109–118. doi: 10.1242/jeb.089920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–124. doi: 10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- Wiseman H, Halliwell B. Damage to DNA by reactive oxygen and nitrogen species: role in inflammatory disease and progression to cancer. Biochem. J. 1996;313:17–29. doi: 10.1042/bj3130017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Zwaans BM, Eckersdorff M, Lombard DB. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle. 2009;8:2662–2663. doi: 10.4161/cc.8.16.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.