Abstract
Varroa destructor is an obligate ectoparasitic mite and the most important biotic threat currently facing honey bees (Apis mellifera). We used neutral microsatellites to analyze previously unreported fine scale population structure of V. destructor, a species characterized by extreme lack of genetic diversity owing to multiple bottleneck events, haplodiploidy, and primarily brother-sister matings. Our results surprisingly indicate that detectable hierarchical genetic variation exists between apiaries, between colonies within an apiary, and even within colonies. This finding of within-colony parasite diversity provides empirical evidence that the spread of V. destructor is not accomplished solely by vertical transmission but that horizontal transmission (natural or human-mediated) must occur regularly.
Keywords: Varroa destructor, population genetic structure, haplodiploidy, sib mating, inbred
1. Introduction
There is widespread concern about the global decline of pollinators which are integral to the reproduction of nearly 90% of the world’s flowering plant species (National Research Council 2007; Ollerton et al. 2011). Specifically, the precipitous decline in honey bee (Apis mellifera) colonies over the past decades is of particular cause for alarm due to the disproportionate role honey bees play in critical agricultural pollination services (Pettis and Delaplane 2010). Pollination services by animals contribute more than an estimated US $200 billion a year to the world economy (Gallai et al. 2009). In addition, to this large economic value, animal-pollinated crops contribute greatly to human health and nutrition. These crops form approximately one-third of the calories consumed by humans (Klein et al. 2007), and represent close to 10% of agricultural yields (Gallai et al. 2009). Disruptions in pollination services could thus pose a serious and destabilizing risk to the global agriculture enterprise as well as human health and nutrition.
Pathogens and parasites are implicated as one of the primary factors in current honey bee declines (vanEngelsdorp et al. 2009). Of these biotic threats, the ectoparasitic mite Varroa destructor is the most important explanatory factor in colony losses worldwide (Rosenkranz et al. 2010). Originally from Asia, V. destructor has spread quickly wherever it has been introduced, and infested colonies typically do not survive for more than two years without treatment (Boecking and Genersch 2008). V. destructor is also a significant vector for secondary viral infections (Kevan et al. 2006) and may facilitate higher viral replication when coinfection occurs (Nazzi and Conte 2016). Following its introduction to North America in the mid 1980s (Rosenkranz et al. 2010), V. destructor was effectively controlled with acaricides but has now developed resistance to most of these chemicals (Pettis 2004; Gracia-Salinas et al. 2006). Despite the attention being given to management of V. destructor, we still have a surprisingly limited understanding of its basic biology, including the population structure within and among honey bee colonies and apiaries.
Our ability to apply population genetic tools to questions of basic V. destructor biology has been handicapped by a distinctive lack of genetic diversity. This low diversity limits the ability of analyses to resolve differences between populations at multiple levels. Three factors contribute to this low diversity: genetic bottleneck events, haplodiploid sex determination, and a unique life history resulting in a majority of matings occurring between siblings (Rosenkranz et al. 2010).
Neutral microsatellite markers have been developed and used to describe the genetic structure of V. destructor at a global scale by utilizing samples from geographically widespread populations (Evans 2000; Solignac et al. 2003; Solignac et al. 2005), but there is a dearth of in-depth information on the amount of variation found at a fine spatial scale between populations, within populations, and within colonies. How much genetic variation can be expected within a population or colony—or indeed whether we can expect any variation at all - remains unknown. Such questions are directly relevant to understanding parasitism dynamics in mites. This relevant gap in the knowledge of local genetic structure for V. destructor needs to be addressed to deal with important questions such as regional transmission, pathogenicity and evolution.
To help fill this gap, we studied two aspects of fine scale genetic population structure of V. destructor. First, we investigated the partitioning of genetic variation within and between managed colonies. We also examined whether this variation changes over time and if variation is related to geographic spatial structure. Second, we assessed the ability of current microsatellite markers to inform us about mite transmission between honey bee colonies, including distinguishing between horizontal and vertical transmission.
2. Materials and Methods
In the spring and summer of 2011, we obtained mite samples from ten apiaries managed by the University of Georgia Honey Bee Lab around Athens, Georgia, USA. Each apiary consisted of 20 honey bee colonies housed in standard Langstroth hive boxes. We analyzed 589 adult female mites (from a total of 200 colonies and over three time periods: April, July, and September) for microsatellite genotypes. The colonies and apiaries were established at the start of the study. There was no exchange of bees or equipment between these apiaries and no additional bees were added to the study from other sources (details in Supplement).
We extracted total genomic DNA from adult female mites using the DNeasy Blood and Tissue Kit from Qiagen (Valencia, CA, USA. details in Supplement) and amplified ten polymorphic neutral microsatellite loci identified from two previous studies that were useful for population studies and amplify reliably: vj272, vj292, vj294, vj295 (Evans 2000), and VD001, VD112, VD114, VD119, VD126, VD163 (Solignac et al. 2003). We used Qiagen Type-It Microsatellite PCR Kits for the PCR reactions.
We analyzed microsatellite loci for the presence of null alleles, large allele dropout and scoring errors due to stutter peaks using MICRO-CHECKER v.2.2.3 (Van Oosterhout et al. 2004). We assessed departure from Hardy-Weinberg equilibrium (HWE) using ARLEQUIN v.3.5.1.3 (Excoffier and Lischer 2010) using Fisher’s exact tests at each locus/population combination (100 within apiary locus by locus tests and 100 within colony locus by locus tests each employing 1,000,000 Markov chain steps) with a sequential Bonferroni correction (Rice 1989). Most polymorphic pairwise loci/population combinations were not in HWE (Table S1), as was expected based on the high level of inbreeding in V. destructor. This departure of HWE limited the types of population genetic analyses that we could perform. We could not, for example, use Bayesian clustering programs, such as STRUCTURE (Pritchard et al. 2000) due to assumptions of HWE and random mating populations.
We conducted baseline analyses to determine the number of alleles per locus, and allelic range and subsequent derivation of the Garza-Williamson Index. These were calculated in ARLEQUIN to empirically test for genetic evidence of past reduction in population size, or bottleneck events (Garza and Williamson 2001). The index uses a generalized step-wise mutation model specifically designed for microsatellite data. We also employed BOTTLENECK program v.1.2.02 (Piry et al. 1999) to test for reductions in population size.
We calculated overall and pairwise estimates of RST and FST, as well as FIS and FIT in FSTAT and ARLEQUIN to determine genetic differentiation. RST is the preferred parameter when dealing with microsatellite data as it explicitly deals with the stepwise mutation nature of microsatellites (Slatkin 1995). It has become common practice, however, to report both measures (Balloux and Lugon-Moulin 2002). Pairwise FST and RST values were calculated for loci found to be in HWE using GENEPOP v.4.2 (Rousset 2008). The results were recalculated using adjusted allele frequencies calculated by MICRO-CHECKER.
To evaluate the partitioning of genetic variation within and between mite populations (in this case at the apiary, colony, and individual levels), we used one-way analysis of molecular variance (AMOVA) in ARLEQUIN, and determined locus-by-locus significance using 10,000 permutations. We examined changes in haplotype frequencies between the sampling dates using χ2 tests. We used genetic assignment to assign mites to apiaries and to determine detection of first-generation migrants using GENECLASS2 (Piry et al. 2004).
We also examined changes in population structure over time and over geographic space. We ran the same AMOVA analyses over different time periods to assess temporal changes. To evaluate isolation by distance (IBD; i.e. population genetic changes in geographic space with individual mite data pooled by apiary), we used IBDWS v.3.23 (Jensen et al. 2005) to assess the correlation of Euclidean distance between populations and genetic differentiation with Mantel tests. We ran a Mantel test, for pairwise combinations of the genetic similarity (M) and the log transformation of geographic distance for each combination for those values. We ran 10,000 randomizations on FST and RST using both the unadjusted and adjusted allele frequencies (adjusted allele frequencies account for bias resulting from null alleles adjusting the frequencies of the amplified alleles) determined by MICRO-CHECKER.
To compare the allelic diversity we found with that of previously reported diversity (Solignac et al. 2005) at a global scale we used a rarefaction approach to perform a new analysis on the raw data of the global survey (kindly provided by Michel Solignac) and our current study to account for different sampling intensities. This new analysis used the nine microsatellite loci in common between our study and the 2005 data (vj292, vj294, vj295, VD001, VD112, VD114, VD119, VD126 and VD163). Of note is that locus vj272 was not used in the 2005 study and this was the locus in our study that had the greatest allelic diversity. We used ADZE (Szpiech et al. 2008) to perform a rarefaction analysis calculating the number of unique alleles that can be expected for a random sample of each population at different sample sizes. We compared our samples to the previously reported global survey at the continental level. Lastly, we used results from the previously described analyses related to population genetic differentiation and variation to determine if inferences can be made about parasite transmission, including distinction of horizontal vs. vertical transmission.
3. Results
We analyzed 589 V. destructor mites from 200 honey bee colonies arranged in 10 equal-size apiaries at each of 10 microsatellite loci. In our baseline analyses we identified 42 alleles. Numbers of alleles per locus ranged from 2 to 13. The observed heterozygosity for the population across all 10 loci was 0.09, a level consistent with the inbreeding-biased mating system of this mite. Microsatellite scoring analyses detected no evidence for large allele dropout. Seven (VD126, VD112, vj295, vj294, vj292, VD001, and vj272) of the ten loci showed indications of homozygote excess, which is to be expected for a population such as V. destructor that violates panmixia assumptions. However, it could also suggest that null alleles are present. Locus VD001 exhibited a number of alleles with one unit difference; these may be due to a PCR stuttering artifact that can result in scoring errors.
At the apiary level (i.e. pooling the allele frequencies across all colonies within apiaries), 78 out of 91 (86%) polymorphic locus/population combinations were not in HWE (Table S1; P < 0.05; range from P < 0.0001 to P = 0.048). This is not unexpected considering the phylogenetic history and reproductive system of V. destructor and the assumptions built into Hardy-Weinberg equilibrium. At the colony level (i.e. pooling of allele frequencies across all individuals within one colony which was performed on one colony for each apiary), 42 out of 77 (55%) colony samples were not in HWE (Supplement Table S2; P < 0.05; range from P < 0.0001 to P = 0.042). This decrease when looking at individual mites within colony values is most likely due to insufficient statistical power due to the lower mite sample size in individual colonies compared to apiaries (Supplement Tables S1 and S2).
We found a low, but statistically significant, level of population differentiation across all apiaries and loci (global FST = 0.017, P < 0.0001; global weighted RST = 0.096, P < 0.0081). The individual apiaries ranged in FIS value from 0.384 to 0.659 with a statistically significant overall population FIS value of 0.479 (P < 0.0001). Population pairwise FST values at the apiary level are shown in Table 1. The FIT value that measures the deviation from HWE in the total population is 0.488 (P < 0.0001).
Table 1.
Population (apiary) pairwise FST values.
| Apiary | 1 (51) | 2 (49) | 3 (58) | 4 (44) | 5 (30) | 6 (41) | 7 (38) | 8 (28) | 9 (35) |
|---|---|---|---|---|---|---|---|---|---|
| 2 (49) | 0.032* | ||||||||
| 3 (58) | 0.026* | 0.018* | |||||||
| 4 (44) | 0.028* | 0.014* | 0.016* | ||||||
| 5 (30) | 0.018* | 0.270* | 0.025* | 0.014 | |||||
| 6 (41) | 0.008 | 0.013 | 0.000 | 0.010 | 0.012 | ||||
| 7 (38) | 0.026* | 0.038* | 0.024* | 0.035* | 0.022* | 0.005 | |||
| 8 (28) | 0.040* | 0.031* | 0.021* | 0.014 | 0.016* | 0.015 | 0.020* | ||
| 9 (35) | 0.006 | 0.025* | 0.019* | 0.023* | 0.019* | 0.001 | 0.016 | 0.028* | |
| 10 (34) | 0.021* | 0.035* | 0.036* | 0.033* | 0.030* | 0.009 | 0.014 | 0.029* | 0.019* |
Significant differences at the P < 0.05 level are indicated with asterisks. Numbers in parentheses indicate number of mites genotyped.
AMOVA calculations from ARLEQUIN are shown in Table 2. The percentage of variation that can be accounted for within individuals is 51.20%. Genetic assignment procedures using GENECLASS2 were only able to successfully assign 19.7% of the individuals (assignment threshold of 0.05) to the proper apiary and analysis of first generation migrants was therefore not useful, meaning we could not use microsatellites to determine inter-colony transmission. The apiary mean across all loci for the Garza-Williamson Index (GWI) had a fairly tight range over the 10 apiaries (0.36 to 0.41; Supplement Figure S1). A general rule of thumb for the GWI is that a value below 0.7 indicates a population has gone through a bottleneck (Garza and Williamson 2001). None of the loci in any of the apiaries had a GWI greater than 0.67 (Supplement Figure S1). The BOTTLENECK analysis similarly showed a reduction in past population size with 8 out of the 10 apiaries exhibiting heterozygous excess consistent with a recent bottleneck (two-tailed Wilcoxon sign-rank test using a two-phase mutation model for microsatellite data; significant P values ranged from 0.0019 to 0.027; while insignificant values were 0.20 and 0.25).
Table 2.
Results of hierarchical analysis of molecular variance (AMOVA) comparing V. destructor mites from 10 apiaries.
| Source of Variation | d.f. |
Sum of
squares |
Variance
components |
Percentage of
variation |
P-value |
|---|---|---|---|---|---|
| Among groups (between apiaries) | 9 | 27.16 | 0.012 | 1.32 | 0.0020 |
| Among populations within groups (between colonies) |
164 |
226.04 |
0.022 |
2.55 |
0.040 |
| Among individuals within populations (within colonies) |
415 |
515.35 |
0.40 |
44.93 |
<0.00001 |
| Within individuals | 589 | 265.5 | 0.45 | 51.20 | <0.00001 |
| Total | 1177 | 1034.04 | 0.88 | 100.00 |
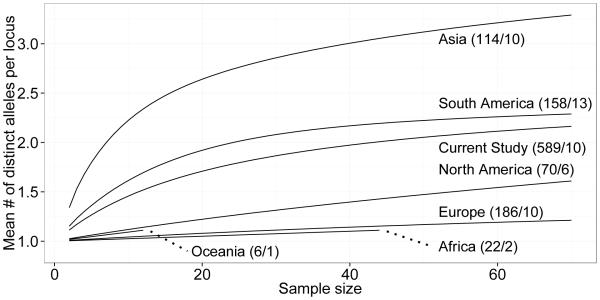
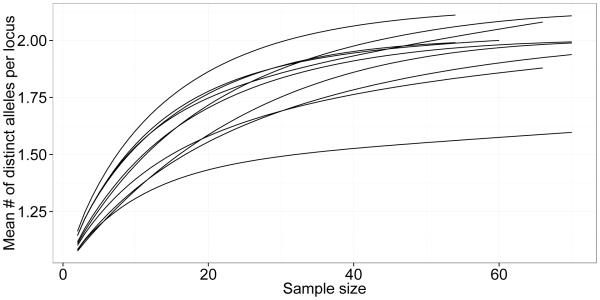
We compared the allelic diversity we found with previously reported V. destructor microsatellite data at the continental scale (Solignac et al. 2005) in a new analysis (Figure 1). As expected, the allelic diversity of V. destructor is highest in Asia (N = 41 unique alleles across the nine loci) where the initial host switches are thought to have occurred. However, the fine scale diversity found in this study is greater than what would be expected from the samples previously taken from North America. We found 27 unique alleles for the nine loci in our ten intensively sampled sites, while the 2005 study (Solignac et al. 2005) found 18 unique alleles in the six North American sites surveyed (three sites in the US and three in Mexico). We expected our samples to contain a subset of the previously reported diversity from North America, but instead we found substantially greater diversity. We also compared the allelic richness between the ten apiaries in this study (Figure 2). The diversity was mostly similar across apiaries (and comparable with the overall rarefaction analysis for the current study in Figure 1) while one apiary (referred to as apiary PS) showed a reduced allelic richness compared with the other apiaries (more in line with the richness identified for North America in Figure 1).
Fig1.
Fine scale allelic diversity (this study) compared to a new analysis of previously reported global allelic diversity (Evans 2000; Solignac et al. 2003; Solignac et al. 2005) using rarefaction to account for different sampling intensities. The estimated number of distinct alleles that can be expected for a random sample of each population is shown for different sample sizes. The number of individuals genotyped for the nine microsatellite loci and the number of sample sites are indicated in the parentheses. Note that Africa and Oceania did not have enough individuals genotyped to perform a full analysis.
Fig 2.
Allelic diversity in ten apiaries using a rarefaction approach. The estimated number of distinct alleles that can be expected for a random sample of each population is shown for different sample sizes. Each line refers to an individual apiary. The outlier with reduced allelic diversity is apiary PS.
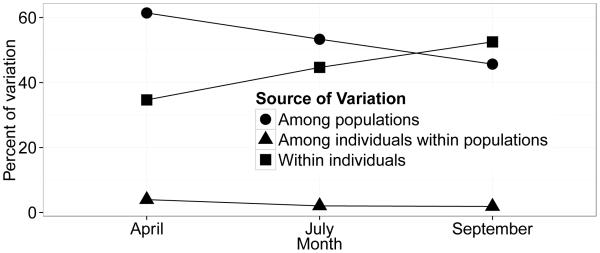
Figure 3 shows the change in variation (AMOVA) over our three sample periods (all individual mite data pooled by time period). Due to the reduced number of samples when pooling time periods we can not analyze AMOVA between apiaries. This shows a modest increase in the amount of total variation accounted for within individuals over time. Finally, we assessed isolation by distance. The Mantel test for IBD showed no significant correlation between the genetic similarity (M) and log-transformed (most appropriate for our two-dimensional spatial data; Rousset 1997) geographic distance between the apiaries (M vs. log dist. P = 0.27).
Fig 3.
AMOVA change in source of population variation over time.
4. Discussion
In this first analysis of the fine scale population structure of V. destructor in a managed apiary setting, we found a surprising amount of genetic diversity given the mite’s legacy of genetic bottlenecks and its inbred reproductive system. Despite these diversity-limiting factors, there is still detectable variation between and within apiaries and colonies, though we did not detect any relationship between geographic and genetic distance. We also documented a modest increase in amount of total variation accounted for within individuals over time (Figure 3), which could be due to de novo generation of diversity or more likely—because of the short time scales in this study—horizontal transmission of mites between colonies.
Genetic variation in Varroa destructor
In some ways, our findings are paradoxical: we found little genetic variation in V. destructor, but given its host-switching and biogeographical history and life cycle, the amount of assignable variation is surprisingly high. Since Apis mellifera is a social insect it is logical calculate variation at the four levels (including within- and between-colony levels) assigned in our AMOVA test (Table 2), which means that direct comparisons to F-statistics (FST, FIS, and FIT) are not appropriate. We found that the AMOVA (Table 2) percentage of variation within populations (colonies, 44.93%) and within individuals (51.20%) to make up the most significant part of the fine scale variation within V. destructor. This indicates a population structure with genetically distinct individuals and more sexual outcrossing than we would expect. The small amount of variation shown among groups (between apiaries, 1.32%) and among populations within groups (between colonies, 2.55%) indicates a genetic similarity between the groups and gene flow between them.
Three elements of V. destructor and honey bee biology and management may contribute to the genetic variation we observed: 1) within-colony mite genetic exchange; and between-colony mite transmission driven by 2) natural bee movement; and 3) human-induced movement. First, one potential explanation of this diversity is within-colony genetic exchange between mites. Most matings in V. destructor occur between siblings. If, however, circumstances encourage more than one foundress mite to enter the same brood cell to lay eggs, outcrossing can occur, enhancing within-colony genetic diversity. This scenario may occur more frequently at points in the annual cycle when the ratio of mites to bee brood is high, as in late summer. Such within-cell-outcrossings would particularly increase genetic diversity if there were mites from multiple sources within the same colony, i.e. if mite transmission between colonies had occurred.
Horizontal transmission of mites between colonies is another possible driver of higher-than-expected mite genetic diversity. Transmission is primarily driven by bee movement between colonies that can occur through two mechanisms: natural or human-mediated movement. Natural movement of bees between colonies occurs through robbing and drifting (Pfeiffer and Crailsheim 1998) . It is well known that honey bees will rob other (usually weaker) colonies, primarily for their stored honey. “Drifting” refers to movement of bees between colonies, for example if a worker honey bee becomes disoriented and enters an alien colony after foraging. Drifting is likely aggravated in human-managed apiaries with high colony densities. Unfortunately, rates of inter-colony movement via robbing and drifting remain poorly characterized. Recent research suggests that robbing and drifting can result in a large amount of mite transfer especially in apiaries that contain a high density of colonies (Frey and Rosenkranz 2014). There remains, however, ambiguity in the published work linking bee drift to mite movement, with evidence that bees from heavily infested colonies do (Kralj and Fuchs 2006) or do not (Goodwin et al. 2006) drift more often than their mite-free counterparts.
Bee movement between colonies is also mediated by humans via beekeeping management. For example, movement of frames of brood (which can contain large mite numbers) and adult bees between colonies is a common beekeeping practice. Beekeepers may do this, for example, to bolster a struggling colony. At a commercial scale, many of the 36,000 US beekeeping operations and their 3 million colonies (USDA 2014) are migratory. These large scale operations move hundreds of colonies at a time on trailers, often over thousands of kilometers a year (Rucker et al. 2012). This movement is driven by crop pollination contracts, as with the almond industry that is wholly dependent on honey bee pollination and requires 1.4 million migratory colonies to descend on one half million acres in California for three weeks every year (National Research Council 2007). Thus if two migratory beekeeping operations were to pollinate crops in adjoining farm properties, this could lead to interactions between bees from two different geographic provenances. This outcome is underscored by the rapidity of the spread of V. destructor in New Zealand, following its recent introduction there (Stevenson et al. 2005).
The intensity of human management also likely explains our inability to detect isolation by distance (IBD) or use genetic assignment criteria in V. destructor. In our study, bees and frames were explicitly not moved in order to control for this confounding factor and to assess the background level of mite transmission in an apicultural setting without human intervention. Because the colonies we used came from a single non-migratory beekeeping operation, prior to the study the typical beekeeping practices used in this operation likely led to a high potential for admixture. There was likely not sufficient time following establishment of our study to evolve differences based on apiary isolation. An additional factor to consider is that IBD may occur in nature only over very large spatial scales in V. destructor, given that honey bee swarms can travel large distances (Schmidt 1995). While beekeepers work to prevent swarming, a certain level of swarming is inevitable. Thus, even if our study had had sufficient time for evolutionary divergence the spatial scale may have been too small to pick up an IBD signal.
The comparison of allelic diversity to previously reported continental level diversity shows more diversity than might be expected (N = 27 unique alleles across nine loci in the current study compared to N = 18 found in the 2005 study for North America; Figure 1). We expected the diversity in our current study to be a subset of that found across the six sites previously surveyed in North America (three sites in the US and three in Mexico). One likely reason for this unexpected result is that the number of mite individuals from North America sampled previously (N = 70 from 6 sites) was too low to fully gauge the diversity of this population compared to the intensive fine scale sampling completed for the current study (N = 589 from 10 sites). Figure 1 shows the continental level rarefaction analysis. The North American population is increasing linearly rather than asymptotically indicating that the previously reported sampling had not exhausted the process of finding unique alleles. Thus, there may be more within-apiary and local allelic diversity than expected and potentially less diversity at larger scales. This is not inconsistent with the recent introduction and rapid spread of V. destructor where the diversity could be haphazardly scattered rather than systematically patterned. Future studies should consider the need to sample some smaller units intensively in order to assess hierarchical partitioning of genetic variability.
Finally, the level of within-colony variation that we found is consistent with horizontal transmission (between unrelated colonies) of V. destructor. If there were solely vertical transmission we would not expect to see this level of diversity. Single transmission events of foundress mites, with subsequent sibling-only matings, would have resulted in much lower levels of variation within individuals. Indeed, the fact that we see such high levels of variation accounted for at the within-individual level lends credence to the idea that transmission between colonies is maintained by multiple sustained events rather than rare single transmission events.
Little is known empirically about the mechanisms and magnitudes of transmission of V. destructor, and this finding adds to anecdotal evidence supporting horizontal transmission (Frey et al. 2011). For example, V. destructor spread very quickly after it was introduced into North America (including to low-density feral populations) (Kraus and Page 1995), implying that there is likely a mechanism beyond vertical transmission by which V. destructor infests new colonies. Unfortunately, the microsatellite markers that exist for V. destructor do not provide enough resolution to use these markers to track this transmission.
Conclusion
We found that existing microsatellite markers in V. destructor display unanticipated levels of diversity, in contrast to the limited diversity we would expect given historical bottlenecks, haplodiploidy, brother-sister matings, and previously reported diversity within North America. This variation may help explain how a species with such low genetic diversity can nevertheless evolve acaricide resistance and spread rapidly wherever it has been introduced. We still know little about the mechanisms and magnitude of V. destructor transmission, which is a key parameter underlying parasite dynamics. While new genetic markers with higher resolution (e.g. SNPs) could contribute to understanding transmission patterns, empirical work must also be done on understanding the extent of bee movement between colonies, whether by human management or naturally via robbing and drifting.
Supplementary Material
Acknowledgements
We thank Nicholas Weaver, Brett Nolan, Ben Rouse, and Cindy Bee for maintaining the colonies and mite collection. We are also grateful to Lucy Anderson, Laila Atalla, Andre Delva, Ashley Ellis, Ian Fried, David Gruenewald, Aaron Hopes, Elsa Lake, Kristen McCrae, Ivan Shoemaker, and Alison Tammany for assistance with DNA extraction and microsatellite scoring. We are grateful to Michel Solignac for providing data needed for rarefaction analyses.
This research was supported by the U.S. Department of Agriculture (AFRI grant No. 2011-67013-30131 to BJB, JCdR, and KSD), the National Institutes of Health (R01-109501-01, to JCdR, BJB, and KSD) and the National Science Foundation (DGE-1444932, to TLD).
References
- Balloux F, Lugon-Moulin N. The estimation of population differentiation with microsatellite markers. Mol Ecol. 2002;11:155–165. doi: 10.1046/j.0962-1083.2001.01436.x. [DOI] [PubMed] [Google Scholar]
- Boecking O, Genersch E. Varroosis – the ongoing crisis in bee keeping. J Consum Prot & Food Saf. 2008;3:221–228. [Google Scholar]
- Evans JD. Microsatellite loci in the honey bee parasitic mite Varroa jacobsoni. Mol Ecol. 2000;9:1436–1438. doi: 10.1046/j.1365-294x.2000.00998-3.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Frey E, Rosenkranz P. Autumn invasion rates of Varroa destructor (Mesostigmata: Varroidae) into honey bee (Hymenoptera: Apidae) colonies and the resulting increase in mite populations. J Econ Entomol. 2014;107:1–8. doi: 10.1603/ec13381. [DOI] [PubMed] [Google Scholar]
- Frey E, Schnell H, Rosenkranz P. Invasion of Varroa destructor mites into mite-free honey bee colonies under the controlled conditions of a military training area. J Apic Res. 2011;50:138–144. [Google Scholar]
- Gallai N, Salles J-M, Settele J, Vaissière BE. Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol Econ. 2009;68:810–821. [Google Scholar]
- Garza JC, Williamson EG. Detection of reduction in population size using data from microsatellite loci. Mol Ecol. 2001;10:305–318. doi: 10.1046/j.1365-294x.2001.01190.x. [DOI] [PubMed] [Google Scholar]
- Goodwin RM, Taylor MA, Mcbrydie HM, Cox HM. Drift of Varroa destructor-infested worker honey bees to neighbouring colonies. J Apic Res. 2006;45:155–156. [Google Scholar]
- Gracia-Salinas MJ, Ferrer-Dufol M, Latorre-Castro E, et al. Detection of fluvalinate resistance in Varroa destructor in Spanish apiaries. J Apic Res. 2006;45:101–105. [Google Scholar]
- Jensen JL, Bohonak AJ, Kelley ST. Isolation by distance, web service. BMC Genet. 2005 doi: 10.1186/1471-2156-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevan PG, Hannan MA, Ostiguy N, Guzman-Novoa E. A summary of the Varroa-virus disease complex in honey bees. Am Bee J. 2006:694–697. [Google Scholar]
- Klein A-M, Vaissière BE, Cane JH, et al. Importance of pollinators in changing landscapes for world crops. Proc R Soc Biol Sci. 2007;274:303–313. doi: 10.1098/rspb.2006.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralj J, Fuchs S. Parasitic Varroa destructor mites influence flight duration and homing ability of infested Apis mellifera foragers. Apidologie. 2006;37:577–587. [Google Scholar]
- Kraus B, Page RE. Effect of Varroa jacobsoni (Mesostigmata: Varroidae) on feral Apis mellifera (Hymenoptera: Apidae) in California. Environ Entomol. 1995;24:1473–1480. [Google Scholar]
- National Research Council . Status of pollinators in North America. The National Academies Press; Washington, D.C: 2007. [Google Scholar]
- Nazzi F, Conte YL. Ecology of Varroa destructor, the Major Ectoparasite of the Western Honey Bee, Apis mellifera. Annu Rev Entomol. 2016;61:417–432. doi: 10.1146/annurev-ento-010715-023731. [DOI] [PubMed] [Google Scholar]
- Ollerton J, Winfree R, Tarrant S. How many flowering plants are pollinated by animals? Oikos. 2011;120:321–326. [Google Scholar]
- Pettis JS. A scientific note on Varroa destructor resistance to coumaphos in the United States. Apidologie. 2004;35:91–92. [Google Scholar]
- Pettis JS, Delaplane KS. Coordinated responses to honey bee decline in the USA. Apidologie. 2010;41:256–263. [Google Scholar]
- Pfeiffer KJ, Crailsheim K. Drifting of honeybees. Insectes Soc. 1998;45:151–167. [Google Scholar]
- Piry S, Alapetite A, Cornuet J-M, et al. GENECLASS2: A Software for Genetic Assignment and First-Generation Migrant Detection. J Hered. 2004;95:536–539. doi: 10.1093/jhered/esh074. [DOI] [PubMed] [Google Scholar]
- Piry S, Luikart G, Cornuet J-M. BOTTLENECK: A computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered. 1999;90:502–503. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- Rosenkranz P, Aumeier P, Ziegelmann B. Biology and control of Varroa destructor. J Invertebr Pathol. 2010;103:S96–S119. doi: 10.1016/j.jip.2009.07.016. [DOI] [PubMed] [Google Scholar]
- Rousset F. GENEPOP’007: A complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Rousset F. Genetic differentiation and estimation of gene flow from F-Statistics under isolation by distance. Genetics. 1997;145:1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rucker RR, Thurman WN, Burgett M. Honey bee pollination markets and the internalization of reciprocal benefits. Am J Agric Econ. 2012;94:956–977. [Google Scholar]
- Schmidt JO. Dispersal distance and direction of reproductive european honey bee swarms (Hymenoptera: Apidae) J Kans Entomol Soc. 1995;68:320–325. [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solignac M, Cornuet J, Vautrin D, et al. The invasive Korea and Japan types of Varroa destructor, ectoparasitic mites of the Western honeybee (Apis mellifera), are two partly isolated clones. Proc R Soc Biol Sci. 2005;272:411–419. doi: 10.1098/rspb.2004.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solignac M, Vautrin D, Pizzo A, et al. Characterization of microsatellite markers for the apicultural pest Varroa destructor (Acari: Varroidae) and its relatives. Mol Ecol Notes. 2003;3:556–559. [Google Scholar]
- Stevenson MA, Benard H, Bolger P, Morris RS. Spatial epidemiology of the Asian honey bee mite (Varroa destructor) in the north island of New Zealand. Prev Vet Med. 2005;71:241–252. doi: 10.1016/j.prevetmed.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Szpiech ZA, Jakobsson M, Rosenberg NA. ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics. 2008;24:2498–2504. doi: 10.1093/bioinformatics/btn478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA . 2012 Census of Agriculture. National Agricultural Statistics Service; Washington, D.C: 2014. [Google Scholar]
- vanEngelsdorp D, Evans JD, Saegerman C, et al. Colony collapse disorder: A descriptive study. PLoS ONE. 2009;4:e6481. doi: 10.1371/journal.pone.0006481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–538. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.