Summary
Centrioles coordinate the primary microtubule organizing center of the cell and template the formation of cilia, thereby operating at a nexus of critical cellular functions. Here we use proximity-dependent biotinylation (BioID) to map the centrosome-cilium interface; with 58 bait proteins we generate a protein topology network comprising >7000 interactions. Analysis of interaction profiles coupled with high resolution phenotypic profiling implicates a number of new protein modules in centriole duplication, ciliogenesis and centriolar satellite biogenesis, and highlights extensive interplay between these processes. By monitoring dynamic changes in the centrosome-cilium protein interaction landscape during ciliogenesis, we also identify satellite proteins that support cilia formation. Systematic profiling of proximity interactions combined with functional analysis thus provides a rich resource for better understanding human centrosome and cilia biology. Similar strategies may be applied to other complex biological structures or pathways.
Graphical abstract
We use in vivo proximity-dependent biotinylation(BioID) to generate a protein interaction map of the human centrosome-cilium interface. Avast and functionally rich interaction space is characterized, allowing us to uncover protein modules critical for centrosome and cilium biogenesis. We demonstrate pervasive interplay between centriole duplication, centriolar satellite biogenesis and ciliogenesis, anddiscover pronounced dynamic modulation of the protein interaction landscape during the ciologenesis program. Our work thus provides a rich resource for better understanding human centrosome and cilia biology.
Introduction
The centrosome is composed of a 9-fold symmetric centriole pair surrounded by pericentriolar material, and acts as the primary MT organizing center in mammalian cells. In non-cycling cells centrioles can also template the formation of primary cilia at the plasma membrane (Figure 1A). The mother centriole is specifically decorated with subdistal appendages, required for anchoring MTs, and distal appendages, which effect the docking of the centriole/basal body to the plasma membrane during ciliogenesis (Figure 1A). The distal appendages function with the transition zone (TZ), a membrane-associated ciliary subdomain, to act as a “gate”, controlling transit into and out of the cilium proper (Figure 1A; (Garcia-Gonzalo et al., 2011; Williams et al., 2011)). Centriolar satellites are electron-dense structures that accumulate in the vicinity of centrosomes (Kubo et al., 1999), participate in the MT-dependent trafficking of centrosome and ciliary proteins, and thereby regulate cilia formation and centrosome biogenesis (Tollenaere et al., 2015). Failure to properly regulate centrosome function is linked to aneuploidy and primary microcephaly (Godinho and Pellman, 2014; Sir et al., 2011), and disruption of cilia function can lead to ciliopathies (Reiter et al., 2012).
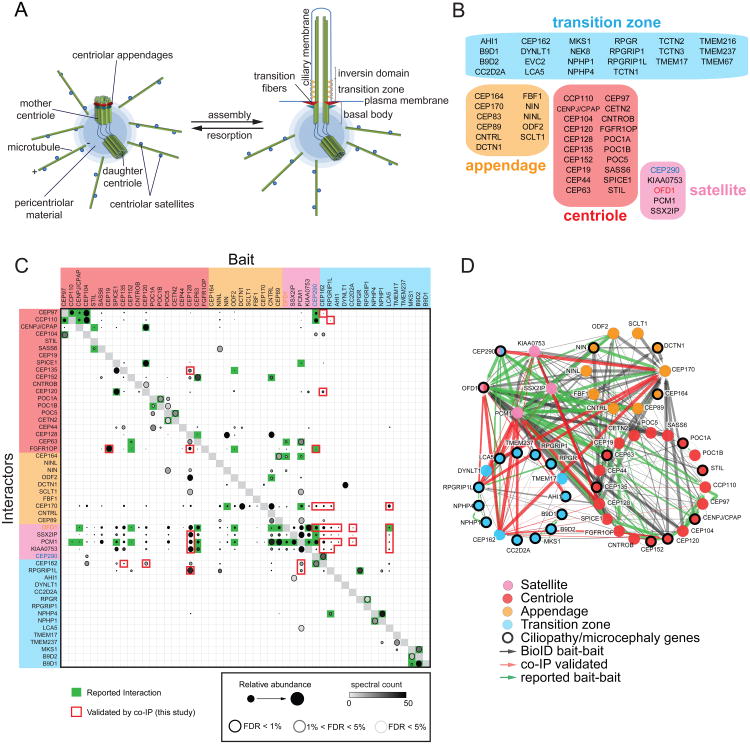
Figure 1. Proximity Mapping Of The Centrosome-Cilium Interface.
A. Schematic representation of the mammalian centrosome and centrosome/basal body-primary cilium interface. Distal and subdistal appendages are indicated in red and blue, respectively.
B. Bait proteins used in this study, grouped and color-coded according to primary localization. Note that CEP290 and OFD1 localize to both centriolar satellites and transition zone/centriole, respectively.
C. Mass spectrometry Dot Plot view of bait-bait interactions. Dot shading (grey – black gradient) indicates total number of spectral counts detected for each prey protein. Dot size indicates relative abundance of prey protein in each BioID analysis. Confidence levels for each bait-bait interaction according to SAINT (significance analysis of interactome; (Teo et al., 2014)) false discovery rate (FDR) are indicated by dot border (light grey <5% FDR; black <1% FDR). Green boxes, previously reported interaction; red box border, BioID bait-bait interaction validated here by co-IP. Four previously reported bait-bait interactions were verified by co-IP as controls (green box highlighted with red border). Baits not interacting with any other bait protein omitted for clarity.
D. Bait-bait PxIs detected in our study. Each node (color-coded circle) represents a unique bait protein associated with the indicated centrosome-cilium substructure. 70 previously reported (green edges), and 206 new (black edges) bait-bait interactions were detected. 30 bait-bait PxIs validated by co-IP highlighted in red. Edge thickness is proportional to peptide counts (maximum number of counts detected in a single MS analysis, or MaxSpec).
Shotgun proteomic, bioinformatic and genomic approaches have identified many cilium and centrosome proteins (Andersen et al., 2003; Keller et al., 2005; Li et al., 2004). However, due to the largely insoluble nature of the centrosome and cilium, generating detailed protein-protein interaction networks for these organelles has remained challenging. The recently developed proximity-dependent biotinylation (BioID) technique can be used to survey protein interactions in living cells (Roux et al., 2012). Briefly, a polypeptide of interest is fused in-frame with a mutant E. coli biotin conjugating enzyme (BirA R118G, or BirA*), which effects the biotinylation of vicinal amine groups on nearby proteins. Following robust cell lysis, biotinylated polypeptides can be affinity purified using streptavidin resin and identified using mass spectrometry (MS). Here, we use BioID to generate a comprehensive in vivo protein proximity map of the human centrosome-cilium interface. A large subset of this network is then subjected to high content screening and functional analyses, to reveal interactors that play critical roles in centrosome and cilia biology.
Results
A proximity interaction (PxI) network at the centrosome-cilum interface
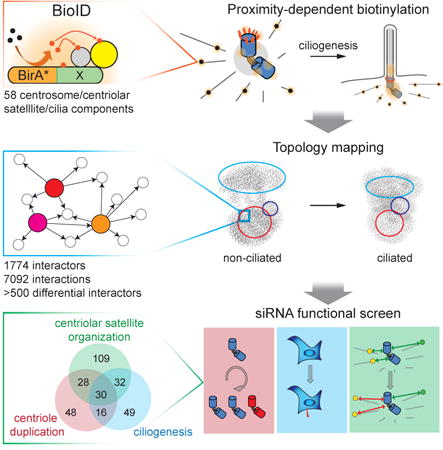
To gain a better understanding of the centrosome and cilium protein interaction landscape, a systematic BioID analysis was conducted on 58 different “bait” polypeptides previously localized to the centriole, centriolar appendages, centriolar satellites or the ciliary transition zone - henceforth referred to as the centrosome-cilium interface (Figures 1B, S1A; see Experimental Procedures). In cycling cells, 4046 high-confidence proximity interactions (PxI) were identified amongst 1405 unique proteins (Table S1; all BioID data available at http://prohits-web.lunenfeld.ca).
To benchmark our interaction dataset, we assembled a high quality reference list of 1554 proteins with previous evidence for centrosome or cilium association (Alves-Cruzeiro et al., 2014; van Dam et al., 2013), hereafter referred to as the centrosome and cilium database (CCDB; Table S1; Experimental Procedures). 25% (336) of our BioID dataset consists of CCDB proteins (Table S1). Our dataset also contains 91 of 144 polypeptides identified in previous proteomic profiling of the centriole (Andersen et al., 2003; Jakobsen et al., 2011), 201 previously reported centrosome/cilium-associated protein-protein interactions, and 55 polypeptides previously linked to ciliopathies or microcephalies (see Table S1). This dataset is thus highly enriched for biologically relevant polypeptides.
213 protein-protein interactions were previously reported between the 58 bait proteins used in our study (Table S1). 70 (30%) of these “bait-bait” interactions (Table S1) are found in our BioID data, amongst 276 detected in total (n.b. 86 bait-bait interactions were detected by BioID in both directions; Figures 1C-D; see Experimental Procedures for further discussion). Using antibodies directed against endogenous proteins to conduct standard co-immunoprecipitation (co-IP) analysis, we validated 30 of these previously unreported bait-bait interactions (Figures 1C-D; Table S1). FLAG IP-MS was also conducted on ten of the BioID bait proteins. 89 interactions were shared between the two methods, representing >40% of all interactors identified in the FLAG IP, and 21% of the BioID interaction space (Figures S1B-C, Table S2). Four of the same bait proteins were also characterized in a recently published BioID study (Firat-Karalar et al., 2014). Despite a number of methodological differences, we detected 35 of the 69 interactors reported in this work, but also uncovered 170 additional interactors for the same set of baits (Figure S1D, Table S2 and Experimental Procedures). BioID thus represents an effective and complementary approach to identify protein-protein interactions at the centrosome-cilium interface.
Proximity mapping identifies new centrosome and centriolar satellite proteins
To better understand the centrosome-cilium protein interaction landscape, a self-organized bait-prey interactome was generated (Figure 2A), in which map location is determined by the number and relative abundance (i.e. total peptide counts) of interactors. As expected, this approach clustered known centrosome-associated functional modules such as the centriolar satellite proteins, and the Augmin/HAUS and prefoldin (PFD) complexes (Figure 2B). Proteins known to work together in centrosome-cilium function were also clustered in this map (Figure 2B), e.g. CEP97-CCP110, SPICE1-CEP120, B9D1-B9D2 and NPHP1-NPHP4.
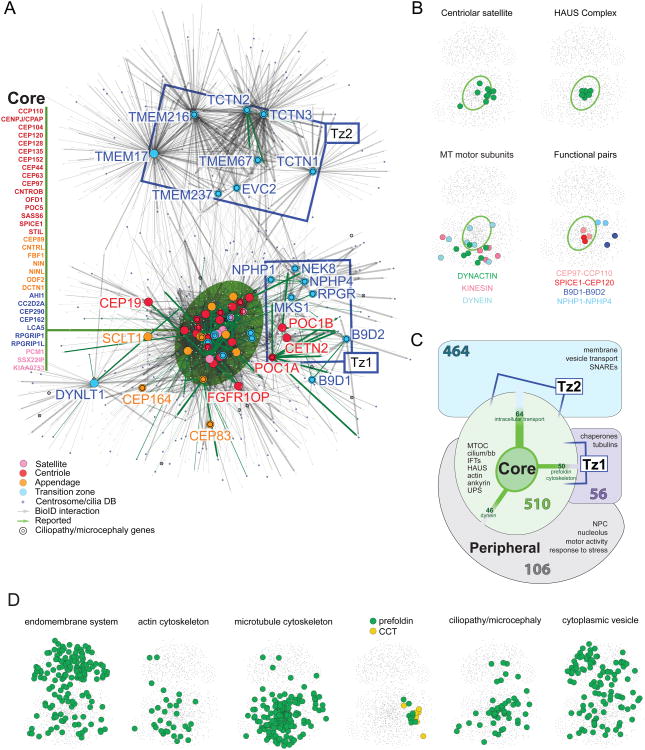
Figure 2. Topology Mapping Of The Centrosome-Cilium Interface.
A. Self-organized, prefuse force-directed topology map (based on peptide count sum of two MS runs) of the centrosome-cilium BioID interactome, consisting of 4046 PxIs amongst 1405 proteins. Bait proteins represented by larger, color-coded nodes; interactors represented by small grey nodes. CCDB proteins are highlighted in blue; proteins linked to ciliopathies or microcephalies highlighted by a black ring. Previously reported PxIs highlighted by green edges; edge thickness proportional to MaxSpec. ‘Core’ component location highlighted by a green ellipse; bait proteins localized to this cluster are listed at left. Ciliary transition zone baits not located in the “core” region cluster into two additional topologically distinct zones (Tz1, Tz2) indicated with blue boxes.
B. Functional module locations in the topology map. Map position of components of the indicated protein group overlaid on a thumbnail of the topology map. Green ellipse indicates “core” location (from Figure 2A). Functional protein groups highlighted in the indicated colors.
C. “Clustergram” schematic of the BioID interactome, depicting the four primary map regions, the number of proteins specific to each cluster, and functional groups of interest relevant to each topological region. Polypeptides shared between groups are indicated by connecting edges, where edge thickness is proportional to the number of shared proteins (cluster-cluster connections with <15 shared interactors not shown; see Table S3 for details).
D. Locations of enriched Gene Ontology (GO) categories or gene groups in the dataset (as indicated) overlaid on the network topology map. See also Table S3.
Standard hierarchical bait-prey clustering was next applied to the dataset (Figure S2). Mapped onto the force-directed layout, these data can be represented as a simplified topological organization of protein groups, or “clustergram” (Figure 2C). A large group of “core” proteins (comprising 32 bait polypeptides from the centriole, appendage, satellite and transition zone, highlighted by a green ellipse) are highly interconnected, and cluster in the center of the map. The 510 additional components of the core cluster (i.e. interacting partners of the 32 bait proteins) include 152 CCDB proteins (Figure 2C; Table S3), and are enriched in e.g. cilium/basal body components, intraflagellar transport (IFT) proteins and WD40-containing polypeptides. Reflecting the connections between centrosome function and intracellular trafficking, this cluster is also enriched in membrane-bound vesicle budding (e.g. dynamin, clathrin) and trafficking (actin and MT cytoskeleton) machineries (Figures 2C, 2D).
Notably, even in non-ciliated cells, the TZ bait proteins were part of an extensive interaction landscape (Figures 2A, 2C, Table S3). Based on their PxI signatures, TZ bait proteins segregated into three distinct subgroups. One group (AHI1, CC2D2A, CEP290, CEP162, LCA5, RPGRIP1 and RPGRIP1L) clusters with core centriole and appendage components, reflecting known associations with the centrosome. A second group, Tz1 (NPHP1, NPHP4, MKS1, RPGR and NEK8), clusters with the centriolar baits POC1A, POC1B and CETN2. The 56 additional members of this cluster are enriched in PFD polypeptides, heat shock proteins and chaperonin containing TCP-1 (CCT) complex components (Figure 2D, Table S3), previously shown to be essential for the folding of actin and tubulin monomers, and required for ciliogenesis (Nachury et al., 2007; Seixas et al., 2010). A third group of TZ baits (Tz2) comprises the membrane-associated TCTN1-3, EVC2, TMEM17, TMEM67, TMEM216 and TMEM237. The 464 Tz2-associated components are enriched (>65%) in membrane polypeptides and vesicle transport machinery (also previously implicated in ciliogenesis; (Lu et al., 2015)). While highly interconnected, this protein group shared relatively few interactions with other protein clusters (Figure 2A; Table S3), likely corresponding to the spatial separation between the centriole apparatus and the plasma membrane in non-ciliated cells. The clustering of NPHP1 and NPHP4 in Tz1, and TMEM67 and TMEM237 in Tz2, and the separation of Tz1 and Tz2 in our network is also consistent with the reported hierarchy of interactions within the transition zone (Garcia-Gonzalo et al., 2011; Williams et al., 2011). Some additional centriole, appendage and TZ bait proteins did not cluster with any of the other groups under these conditions, and are located in a “peripheral” map zone.
152 (∼30%) of the 510 interactors in the core cluster are in the CCDB (Figure 3A). We posited that many of the other proteins mapping to the core cluster are thus likely to be novel centrosome components or regulators. Indeed, when examined with 3D structured-illumination (3D-SIM) and conventional microscopy, 7 of 65 tested candidate proteins (TBC1D31, CCDC66, TEX9, C11orf49, LRRC49, CCDC112 and CCDC18) co-localized with PCM1, a marker of centriolar satellites (Figures 3B, S3A). Consistent with their localization, these proteins were also components of a satellite subnetwork generated by extracting the satellite bait proteins PCM1, SSX2IP, KIAA0753, OFD1 and CEP290 from the primary map (Figure 3C). As a group, the centriolar satellite proteins also exhibited the largest “indegree” (i.e. number of incoming connections) in the PxI network (Figure S3D), and shared a high degree of connectivity with the core module (Figure S3E).
Figure 3. Identification Of New Centrosome/Satellite Components.
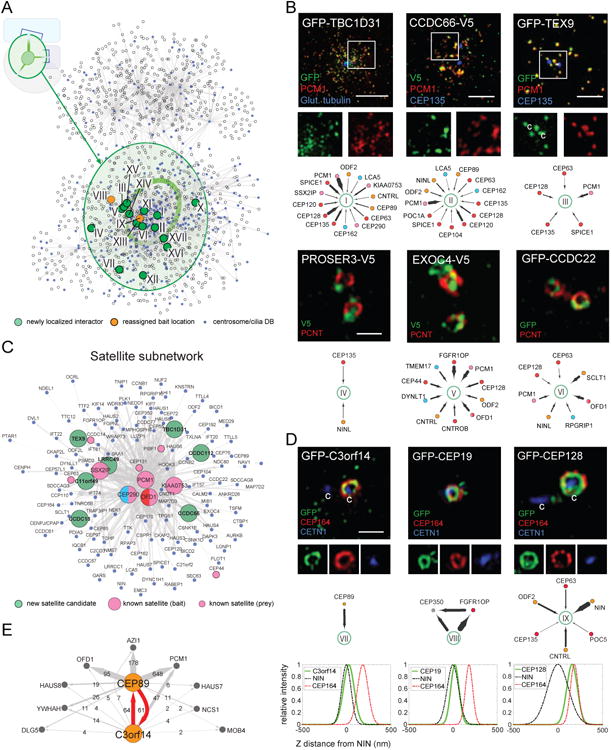
A. Network layout of the BioID dataset, with the”core” region (see Figure 2C) highlighted in green. Newly assigned (green nodes) or re-assigned (orange nodes) centrosome components (labeled with roman numerals) are highlighted. CCDB proteins indicated in blue.
B. (top) 3D-SIM micrographs of RPE-1 cells transiently expressing the indicated proteins, and labelled with the indicated antibodies. Centrioles, where apparent, marked with”c”. Scale bar 2.5μm, insets 1.8×. (bottom) Subnetworks highlighting PxIs of interest, color-coded based on primary localization as in Figure 1B, edge thickness proportional to MaxSpec.
C. Simplified centriolar satellite subnetwork generated from PCM1, SSX2IP, KIAA0753, CEP290 and OFD1 interactomes.
D. 3D-SIM micrographs of RPE-1 cells, as in B. Scale bar 1μm, inset panels 1×, 1×, and 0.8×, from left to right. (middle) Subnetworks highlighting PxIs of interest. (bottom) Intensity profile plots of relative Z axial positioning for the indicated proteins (green fit) with respect to reference markers NIN (subdistal; dashed black fit) and CEP164 (distal; dashed red fit).
E. BioID analysis of C3orf14 and CEP89 (orange nodes); shared PxIs (grey) and peptide counts indicated.
Five other candidate proteins (PROSER3, EXOC4, CCDC22, LUZP1 and KIAA1217) co-localize with PCNT or γ-tubulin (Figures 3D, S3B), suggesting that they are bona fide centrosome components. Two putative MT-associated core proteins were also characterized: KIAA1430, the human ortholog of Drosophila hemingway (Soulavie et al., 2014), localizes to the primary cilium, while MAP7D2 localizes to cytoplasmic MT arrays in RPE-1 cells (Figure S3C).
Our data suggested that the poorly studied C3orf14 polypeptide interacts with the distal appendage protein CEP89 (Figure 3E). The reciprocal interaction (along with nine additional shared PxIs) was confirmed by using C3orf14 as a BioID bait protein (Figure 3E). GFP tagged C3orf14 localized closely to NIN, suggesting that it is a sub-distal centriolar appendage protein (Figure 3D). Finally, CEP19 and CEP128, originally identified as putative mother centriole components (Jakobsen et al., 2011), were found to localize to sub-distal (CEP19) or distal (CEP128) appendages when tagged with GFP (Figure 3D and Table S4). Together, these results demonstrate that proximity mapping can be used to identify novel components of the centrosome-cilium interface.
Functional characterization of the centrosome-cilium interaction landscape reveals extensive interplay between network components
We next used systematic functional analyses to identify centrosome-cilium regulators within the BioID network: 500 proteins, comprising ∼30% of the interactome and selected to encompass all regions of the PxI map, were assessed (Figures 4A, B).
Figure 4. Functional Screening Of Network Components.
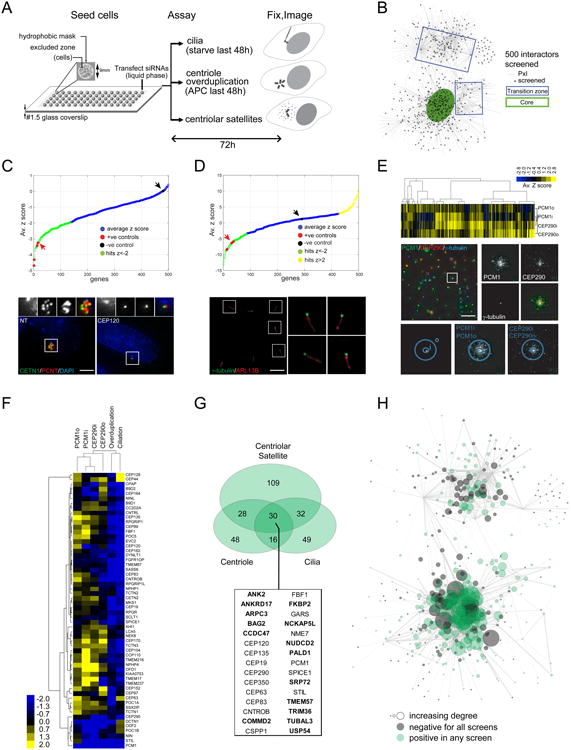
A. Schematic overview of high-throughput microscopy screens (see Experimental Procedures for details).
B. Distribution of the screened network components in the BioID topology map. Screened polypeptides highlighted in black.
C. (top) Z-score distribution of centriole overduplication screen (from Table S5). Positive controls (CEP120, PLK4, STIL, SASS6) highlighted in red; screen hits (Z-score < -2) indicated in green, and negative control (NT, non-targeting siRNA) in black, see arrows. (bottom) Representative micrographs from the screen, showing NT (bottom left panel) or a positive control (CEP120 siRNA; bottom right panel) cell with centriole(s) boxed in white (insets of centriole region in the small panels above). Scale bar 2μm, insets 1.8×.
D. (top) Z-score distribution of cilia screen (from Table S5). Positive controls highlighted in red; hits indicated in green and yellow, negative control highlighted in black (see arrows). (bottom) Representative micrograph of a field of RPE-1 cells from the cilia screen, with γ-tubulin (green)-labelled puncta boxed in white and insets showing examples of primary cilia labelled with anti-ARL13B (red). Scale bar 6μm, insets 3×.
E. (top) Distribution of Z-scores for four parameters measured in the satellite morphology screen, as defined by dilating the centrosomal signal near (“i”) or outside (“o”) the centrosome, for the satellite markers PCM1 and CEP290 (see bottom panels for example, and Experimental Procedures for details; from Table S5). Z-scores were subjected to unsupervised hierarchical clustering and the resulting heat map is shown. (middle) Representative micrograph of a field of control HeLa cells from the satellite screen, with a γ-tubulin (blue)-labelled spot boxed in white, and insets showing typical centrosomal and centriolar satellite patterns for the three channels. Scale bar 40μm, insets 2.5×.
F. Z-scores in each of the three screens for the 58 baits used in this study were subjected to clustering, as in E.
G. Area-proportional Venn diagram of the three screen hit populations. 30 genes that scored in all three screens are listed; genes not in CCDB are indicated in bold.
H. Topology map of the screened subset of network components. Grey nodes were not hits in any screen, green nodes positive in any screen; node size corresponds to indegree (number of incoming connections) for each screened polypeptide.
After a 48h aphidicolin-induced S-phase arrest (which sensitizes cells for defects in centriole duplication;(Balczon et al., 1995)), centriole overduplication in U-2 OS cells was assessed from images collected with automated high-resolution microscopy (Figure 4C; Experimental Procedures). 12 of 15 known centriole duplication factors were hits in this screen (Figure 4C; Table S5). In total, knockdown of 122/500 network components suppressed centriole amplification. 55 of these hits are in CCDB (Tables S1, S5), and the remaining 67 hits included: USP54, a recently identified PLK4 interacting partner (Firat-Karalar et al., 2014); the microcephaly gene WDR62 (Nicholas et al., 2010); the appendage component C3orf14 identified above (see Figure 3D); a number of MT-associated proteins (ANK2, MTUS1, GTSE1, TRIM36, MAP1S); and subunits of the WASH and Arp2/3 complexes (KIAA1033, WASH1 and CCDC53, ARPC3; (Edwards et al., 2014; Rotty et al., 2013)).
Automated microscopy coupled with image analysis of the cilium-specific ARL13B protein was next used to monitor ciliogenesis in siRNA-treated, serum-starved RPE-1 cells (Figure 4D; Experimental Procedures). 19 of 30 human proteins (and 7 of 10 human orthologs of model organism proteins) previously reported to positively regulate ciliation were hits in this assay (Table S5). In total, 86 positive (50 in CCDB) and 41 negative (15 in CCDB) ciliation factors were identified, 62 of which were previously unreported, including the centrosome (KIAA1217) and satellite (CCDC112, TEX9; Figures 3B, S3A-B) proteins identified here.
Finally, we assessed the effects of depleting the same 500 components on PCM1 and CEP290 intensity and distribution (Figures 4A, E). Signal intensity within a 5 (“i”) or 20 (“o”) pixel radius around the centrosome marker was measured for each satellite channel (Figure 4E; Experimental Procedures). In total, knockdown of 199 network components affected either satellite marker, while depletion of 76 components affected both PCM1 and CEP290 parameters (Table S5, Figure 4E). Consistent with published data: (i) knockdown of the MT-anchoring protein SSX2IP affected PCM1 localization (Hori et al., 2015); (ii) known (CCDC14, CEP63 and CEP131; (Firat-Karalar et al., 2014; Staples et al., 2012)) and newly assigned satellite proteins (CCDC112, CCDC18 and CCDC66 (Figures 3B, S3A)) all perturbed satellite intensity; (iii) depletion of MT machinery (ANK2, DCTN1, MAPT, MAP7D1, MAP9, MAP7D3 and MAPRE3) also affected satellite intensity parameters (Table S5). 50% or greater reproducibility was observed using orthogonal silencing triggers in the ciliation and satellite assays (Figure S4A).
Notably, genes that perturbed centriole amplification, ciliation and/or satellite intensity displayed extensive overlap. For example, more than half of the 199 proteins found to modulate satellite parameters also affected centriole duplication (58) or ciliation (62), with 30 of these network components affecting all three screening parameters (Figures 4F-G; Table S5). Network components yielding phenotypes also displayed higher connectivity (Figures 4H, S4B-C), consistent with the notion that increased interactions are indicative of “hubs” in protein networks, and are thus predictive of important functional roles.
Analysis of local proximity profiles reveals novel functional clusters
To further explore the centrosome-cilium structure-function map, we next focused on protein groups sharing similar proximity profiles (Figure 5A, Table S6). One interesting group comprises 17 polypeptides that segregate with the centriole baits CEP120 and SPICE1 (Figure 5A), and includes the centriole duplication and elongation factors CENPJ/CPAP and CEP135. To better understand the role of this module in centriole function, we examined whether knockdown of each of these proteins resulted in phenotypes common to CEP120, SPICE1 or CEP135 (Comartin et al., 2013; Lin et al., 2013): (i) suppression of centriole amplification; (ii) elongation of centrioles; (iii) relocalization of CEP120, or; (iv) glutamylation of cytoplasmic MTs (Figure 5B). Notably, a number of MT-associated polypeptides (ANK2, MTUS1, MAP7, NAP1L1, MAP1S and MAP9) scored in these assays, suggesting that this cluster may regulate centriole duplication and MT organization.
Figure 5. Local Proximity Profiles Identify Functional Clusters: (I) ANK2 Is A Component Of A MT Stability Module That Regulates Centriole Duplication.
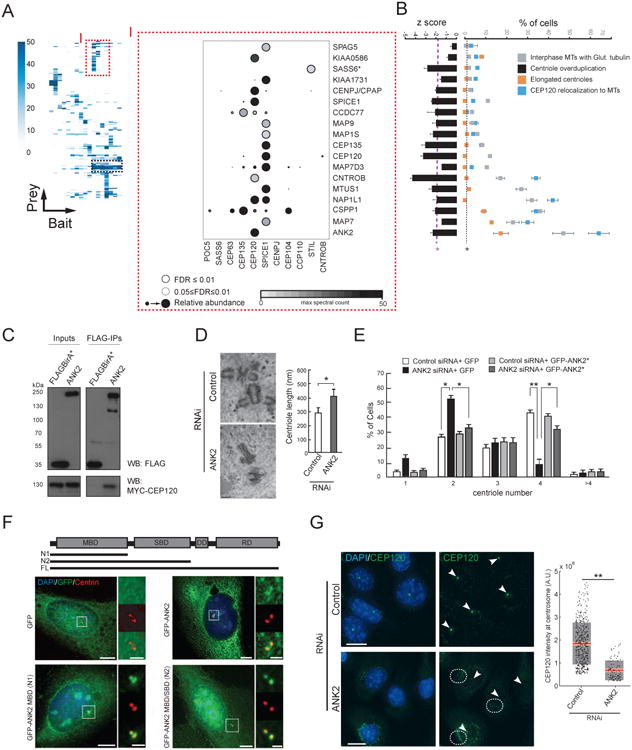
A. (left) Hierarchical clustering reveals modules with similar proximity profiles (dashed boxes). Peptide counts (MaxSpec) indicated for each interactor (from complete cluster map in Table S6). (right) Mass spectrometry DotPlot of selected baits from the region of the CEP120/SPICE1 cluster denoted by “I” (as in Figure 1C, see legend). *SASS6 was not part of this interactor group, and is included here as a control (see B).
B. (left) Interactors (presented in the same order as in A) were profiled for suppression of centriole amplification (see Figure 4A); (right) Qualitative phenotypes related to MT organization (see legend and Experimental Procedures). Dashed purple (overduplication assay) or black (all other assays) lines represent 1.9 and 2 S.D. of the mean of negative control values (*p<0.05, N>100, three replicates), respectively. Negative control values for MT phenotypes were <2% in all cases (not shown).
C. Western blots (as indicated) of input (left) or FLAG IP (right) conducted on lysates from 293 T-REx cells stably expressing FLAGBirA* (tag alone; control) or FLAGBirA*-ANK2, transfected with MYC-CEP120.
D. (left) Electron micrographs of U-2 OS Tet-inducible Myc-PLK4 cells transfected with control (top) or ANK2 (bottom) siRNA for 72 h. At 48 h post-transfection, hydroxyurea and tetracycline were added for 24 h to arrest cells in S-phase and induce centriole overduplication. (right) Average centriole length (±S.D.) in each population (*p=0.03, Student's t-test, N>15). Scale bar 200nm.
E. Effect of ANK2 depletion on centriole number. U-2 OS lines carrying Tet-inducible GFP or the siRNA-resistant GFP-ANK2 (GFP-ANK2*) transgenes were transfected with control or ANK2 siRNA for 72 h. At 48 h post-transfection, tetracycline and hydroxyurea were added to induce ANK2 expression, and arrest cells in S-phase for 24 h. Cells were fixed and labelled with anti-centrin, and the number of centrioles per cell was counted. Bar graph, percent cells with indicated centriole number (N>300, 3 replicates, *p<0.05, **p<0.01).
F. Full length (FL) and truncation constructs of GFP-ANK2 (top) were transfected into U-2 OS cells, and (left) IF microscopy was used to characterize localization at the centriole (boxed in white) with a marker (centrin). (right) Insets from top to bottom: GFP; centrin ; pseudocolor merge. Scale bar 10μm, insets 3×.
G. Representative micrographs (left) of U-2 OS cells treated with control (top) or ANK2 (bottom) siRNA, and labelled with antibodies to endogenous CEP120 (green). White arrowheads denote centrosomal CEP120 puncta. Cytoplasmic regions where CEP120 has relocalized are encircled by white dashes. Scale bar 15μm. (right) Quantification of CEP120 levels at the centrosome (N>300, three replicates, **p<0.01, Student's t-test). Grey region denotes 2 S.D. from the mean (red line) and pink region denotes the 95% confidence interval.
In addition to suppressing centriole amplification, ANK2 knockdown reduced ciliation and perturbed satellite morphology (Figures 4G, S5A-B; Table S5). This protein was thus further analyzed. ANK2 appeared to interact exclusively with CEP120, and this interaction was confirmed by co-IP (Figure 5C). Consistent with the over-duplication assay (above), ANK2-depleted cells overexpressing PLK4 did not over-duplicate centrioles (Figure 5D left panel), and the remaining centrioles were significantly longer (Figure 5D right panel). The duplication phenotype could be rescued with a siRNA-resistant GFP-ANK2 transgene (Figure 5E). N-terminal GFP-tagged ANK2 fragments localized to the centrosome (Figure 5F), but C-terminal truncated fragments did not (Figure S5C), suggesting that the N-terminal MT-binding domain is sufficient to direct ANK2 to the centrosome. During S-phase, CEP120 levels at centrosomes in ANK2-depleted cells were decreased by ∼60% (Figure 5G), with a concomitant redistribution of CEP120 along cytoplasmic MTs (Figures 5G, S5E). ANK2 is thus required for proper CEP120 centrosomal localization.
Notably, upon ANK2 depletion, cytoplasmic MTs were often hyperacetylated (Figures S5E-G), coincident with hyperglutamylation (Figures 5B, S5D) and altered MT-stability (Figure S5G). Defects in MT post-translational modifications were previously observed when CEP120 or SPICE1 were overexpressed (Comartin et al., 2013) or depleted (Figure 5B). These observations are consistent with recent reports implicating ANK2 in MT organization (Stephan et al., 2015), and reveal ANK2 as a key component of a PxI network that controls MT organization and centriole biogenesis.
A second protein cluster (Figure 6A; Table S6) consisted of centrosomal (HAUS6, CEP350, MED4), centriolar satellite (PCM1, SSX2IP, KIAA0753, OFD1, PIBF1, AZI1 and CEP72) and two uncharacterized (WRAP73, CCDC138) proteins. This cluster was also highly enriched in known and candidate regulators of ciliation (Figure 6A), and GFP-tagged (WRAP73, MED4) or endogenously localized (HAUS6) proteins displayed overlap with the satellite marker PCM1 (Figure 6B). MED4 and HAUS6 were previously reported as centrosomal proteins (Lambert et al., 2015; Lawo et al., 2009) but not known to associate with or perturb satellites, or to affect ciliation (Table S5). Since WRAP73 was uncharacterized in this context, we examined it further.
Figure 6. Local Proximity Profiles Identify Functional Clusters: (II) WRAP73 Is Required for Ciliogenesis.
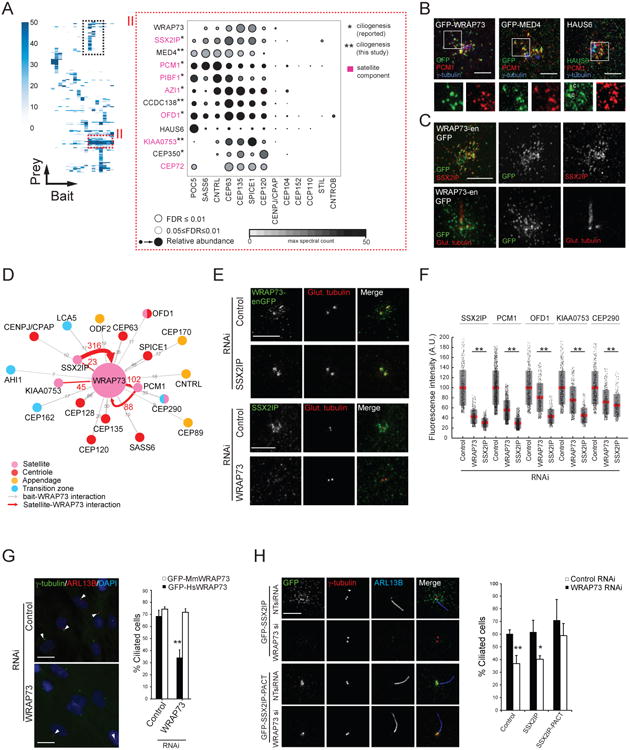
A. (left) Hierarchical clustering reveals a second interactor group with similar proximity profiles (boxed regions; see Figure 5A). (right) Centriolar satellite enriched cluster “II” represented as a Dot Plot of spectral counts. Known satellite components labelled in pink. *previously associated with ciliogenesis; **candidate ciliogenesis regulators.
B. 3D-SIM micrographs of RPE-1 cells transiently expressing the indicated proteins (except HAUS6, which was endogenously detected), and labelled with the indicated antibodies. Centrioles, where apparent, are marked with “c”. Scale bar 2.5μm. Insets 1.8×.
C. 3D-SIM micrographs of a non-ciliated (top) or ciliated (bottom) RPE-1-WRAP73-GFP knock-in cell (denoted WRAP73-enGFP) labelled with the indicated antibodies. Scale bar 3μm.
D. WRAP73 BioID subnetwork. Interactors (along with associated peptide counts) indicated according to legend.
E. (top) 3D-SIM images of control (NT siRNA) or SSX2IP-depleted RPE-1-WRAP73-GFP knock-in cells (WRAP73-enGFP) labelled with anti-GFP and anti-poly-glutamylated tubulin. Scale bar 2μm. (bottom) IF analysis of control (NT siRNA) and WRAP73-depleted RPE-1 cells labelled with the indicated antibodies. Scale bar 5μm.
F. Mean fluorescence intensity of the indicated proteins (top) in a region surrounding the centrosome (see “i” parameter in 4E) in WRAP73- and SSX2IP-depleted cells. Grey region denotes 2 S.D. from the mean (red line), pink region denotes 95% confidence interval. **p<0.01 by Student's t-test, N>400.
G. (left) IF analysis of control and WRAP73-depleted RPE-1 cells labelled with DAPI and the indicated antibodies. Arrowheads indicate ciliated cells. Scale bar 20μm. (right) Percentage ciliated cells (N>100 cells per replicate, three replicates) in serum-starved RPE-1 cells stably expressing GFP-tagged human or mouse WRAP73; **p<0.01 by Student's t-test.
H. (left) IF analysis (as indicated) of control (NT siRNA) and WRAP73-depleted RPE-1 cells stably expressing either GFP-SSX2IP or GFP-SSX2IP-PACT. Scale bar 5μm. (right) Percentage ciliated cells (N>100 cells per replicate, three replicates) in serum-starved RPE-1 (control) or RPE-1 cells stably expressing GFP-SSX2IP or GFP-SSX2IP-PACT, treated with the indicated siRNA; *p<0.05, **p<0.01 by Student's t-test.
CRISPR genome editing was used to generate a WRAP73-GFP knock-in cell line. WRAP73-GFP localized with SSX2IP in non-ciliated cells (Figure 6C) and like SSX2IP, at or near basal bodies in ciliated cells ((Klinger et al., 2014); Figure 6C). WRAP73 displayed reciprocal PxIs with the satellite proteins PCM1, KIAA0753 and SSX2IP (Figure 6D), and WRAP73-SSX2IP, -CEP135 and -SPICE1 interactions were confirmed by co-IP (Figure S6A). The satellite portion of WRAP73 was nocodazole-sensitive, but a small centriolar pool remained (Figure S6B), and WRAP73 and SSX2IP were dependent on each other for localization to centriolar satellites (Figures 6D-E). WRAP73 or SSX2IP knockdown lead to lower levels of the satellite proteins PCM1, KIAA0753, CEP290 and OFD1 in the vicinity of the centrosome (Figure 6F). Like several other members of this cluster, ciliogenesis was also perturbed following siRNA-mediated depletion of WRAP73 (Figures 6F and 6A). This phenotype was verified with a rescue by the siRNA-resistant mouse GFP-WRAP73 transgene (Figure 6G) and by CRISPR-mediated ablation of the WRAP73 locus in RPE-1 cells (Figures S6C-G).
We next examined two specific steps in the ciliogenesis program: WRAP73 depletion did not affect CEP164 localization (Figure S6H) and CCP110 persisted on centrioles (Figure S6I). Low ciliation levels could not be rescued by CCP110 co-depletion, indicating that WRAP73 function is not restricted to CCP110 removal (Figures S6J-K). Similar to SSX2IP knockdown (Klinger et al., 2014), there was a ∼30% reduction in RAB8A recruitment to the cilium (Figures S6L-M). To test whether the WRAP73 ciliogenesis defect was due to a loss of SSX2IP, GFP-SSX2IP was fused to the PACT domain of AKAP450 to target it to the centrosome. This fusion protein localized correctly even upon WRAP73 depletion (Figure 6H). Importantly, once SSX2IP localization to the centrosome was reconstituted, there was no significant loss in ciliation (p=0.251; Figures 6H). Together, these results indicate that WRAP73 functions in partnership with SSX2IP, and is a new component of a centriolar satellite module that affects ciliogenesis.
A number of other interesting local proximity profiles were observed. For example, centriole baits displayed novel PxIs with members of known complexes (TPGS1-LRRC49 (Janke et al., 2005); most HAUS, CCT and PFD subunits), or functional units (CEP97-CCP110-CEP104; SIPA1L1-3; TNR6A-B; DVL2-3; Table S6). Individual protein profiles were also instructive: e.g. CCDC18, a novel satellite protein identified here (Figures 3A, S3A), affected satellite morphology (Table S5) and displayed a proximity profile similar to the satellite protein CCDC14 ((Firat-Karalar et al., 2014); Table S6), suggesting that they may partner. Further examination of the numerous other local proximity groups present in our dataset is thus predicted to yield important biological insights.
Dynamic modulation of the proximity interaction landscape during ciliogenesis
To begin to understand how the protein interaction landscape is altered in response to ciliogenesis, we performed a second round of BioID on 40 bait proteins (the appendage, satellite and TZ polypeptides) under ciliated conditions (Figure 7A, see Experimental Procedures). A net loss of ∼46% of CCP110 and ∼35% of CEP97 peptides in the ciliated interactome was observed (Figure S7A), consistent with their removal from mother centrioles during ciliogenesis (Spektor et al., 2007). The ciliated BioID map consists of 1355 proteins and 2910 PxIs (compared to 1236 polypeptides and 2862 PxIs in the non-ciliated dataset for the same set of baits). The non-ciliated and ciliated interactomes share 1024 network components, while 328 proteins were observed only under ciliating conditions.
Figure 7. Modulation Of The Proximity Interaction Landscape During Ciliogenesis.
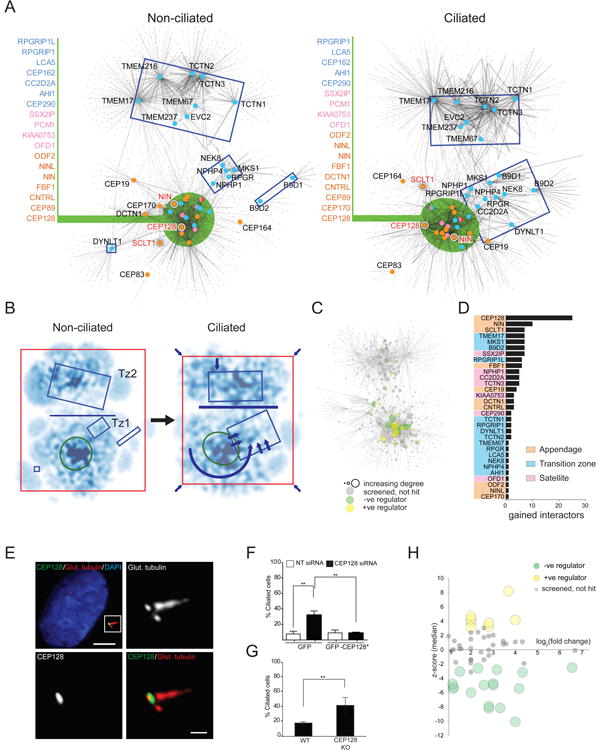
A. Self-organized prefuse force directed BioID-based topology network (as in Figure 2) of appendage, centriolar satellite and transition zone bait proteins in cells maintained under standard (non-ciliated; left) or serum-starved (ciliated; right) culture conditions.
B. Kernel density representation of non-ciliated and ciliated topology maps, with darker regions representing higher node density. Rectangles highlight specific bait-containing regions, as in A. Horizontal blue line highlights the interaction interface between Tz2 and the remainder of the topology map; arrows depict TZ re-organization and network contraction.
C. Distribution of cilia screen hits (negative regulators in green; positive regulators in yellow; not scoring grey; not tested white) overlaid on the ciliated topology map. Node diameter proportional to indegree.
D. Number of gained cilia screen hits in serum-starved 293 T-REx cells, ranked by bait. Baits with no such interactors omitted for clarity.
E. CEP128 localization (boxed in white) in serum-starved RPE-1 cells labeled with the indicated antibodies. Scale bar 5μm, inset 0.5μm.
F. Cycling RPE-1 cells stably expressing GFP or siRNA-resistant GFP-CEP128 (GFP-CEP128*) transfected with control (NT siRNA) or CEP128 siRNA for 72 h. Percentage ciliated cells; N>200 cells per replicate, three replicates, **p<0.01 by Student's t-test.
G. Percent ciliated cells in cycling control or CEP128 knockout RPE-1 cells; **p<0.01 by Student's t-test, N>100, three replicates.
H. Median z-scores of ciliation efficiency versus log2 fold-change peptide count for each of 57 CEP128 interactors that were enriched by ciliogenesis induction. Significant z-scores labelled in yellow or green, as indicated.
Topological force-directed maps of the non-ciliated versus ciliated interactomes (Figure 7A) revealed two key changes: (i) a >40% net gain of PxIs shared between the Tz1, Tz2 and central core groups (Figure 7B, Table S5), attributable in large part to increased PxIs with cytoskeletal and membrane trafficking components. These protein groups are thus mapped much closer to each other than in non-ciliated conditions; (ii) the Tz1 module is re-organized, gaining a number of PxIs that shift it closer to the interface between the core and Tz2 (Figure 7B).
To evaluate the functional significance of the changes in the interaction landscape in response to ciliation, we conducted functional screens on 35 novel interactors found only in the ciliated state (Figure 7C; Table S5). Notably, six of these proteins were identified as candidate ciliation regulators (1 positive, 5 negative), while 12 and 11 components scored in the centriole duplication and satellite morphology screens, respectively. Overall, 23 of the newly screened proteins scored in at least one of our functional assays (Table S5). After filtering for proteins that scored in the ciliation screen, the three bait polypeptides that gained the highest number of PxIs during ciliation were the appendage proteins CEP128, NIN, and SCLT1 (Figure 7D). SCLT1 and NIN play essential roles in ciliogenesis (Figure 4D; (Graser et al., 2007; Tanos et al., 2013)), and our functional screen suggested that CEP128 is a new negative regulator of ciliation (Table S5; Figure 4D). Indeed, in RPE-1 cells CEP128 is localized to the base of the cilium (Figure 7E), and CEP128 knockdown (Figure S7B) led to abnormally high levels of ciliation in proliferating cells (Figure 7F). Expression of siRNA-resistant GFP-CEP128 abrogated this phenotype (Figure 7F). A CRISPR-generated RPE-1 CEP128 knockout cell line displayed elevated levels of ciliation in proliferating cells (Figures 7G, S7C), and CEP128 overexpression suppressed ciliation in serum-starved RPE-1 cells (Figure S7D (Kim et al., 2010; Spektor et al., 2007)). Finally, 57 CEP128 interacting proteins displayed a >two-fold change in peptide counts in response to ciliation, and knockdown of 25 (44%) of these polypeptides gave rise to ciliation phenotypes (Figure 7H; Table S5). Using BioID to map proximity interactions under both ciliating and non-ciliating conditions thus revealed CEP128 as a critical component of a ciliation modulator circuit. Additional analysis of other such dynamically-regulated nodes may be useful for better understanding ciliogenesis.
Discussion
Here we generated proximity profiles for 58 components of the centrosome-cilium interface and centriolar satellites, generating a network of >7000 interactions amongst >1700 unique proteins to reveal a sizeable, previously unexplored interaction space. Follow-up sub-diffraction imaging of 20 newly described centriole/satellite/cilium proteins allowed us to define or refine their localization, suggesting that further mining of this large dataset will yield additional valuable information regarding the higher-order organization of these structures. Our BioID map is also clearly enriched in functionally-relevant proteins: subjecting >30% of the new centrosome-cilium network constituents to a series of functional screens (centriole duplication, ciliation or satellite morphology) allowed us to ascribe putative functions in centriole/cilia regulation to 335 polypeptides.
Notably, 112 proteins yielded a phenotype in at least two of our functional assays, suggesting that the regulation of centriole biogenesis, ciliogenesis and centriolar satellites is intimately related. This idea is not without precedent, as satellite proteins have been shown to regulate ciliogenesis (this study; (Lee and Stearns, 2013; Lopes et al., 2011; Tang et al., 2013) and centriole assembly (Kodani et al., 2015; Tollenaere et al., 2015). Given the high degree of connectivity (Figure S3D), and their numerous specialized roles in centriole/cilia biogenesis, centriolar satellites are thus of considerable interest for future study.
Finally, analysis of the non-ciliated versus ciliated interactomes has allowed us to begin to understand higher order changes in protein proximity during ciliogenesis. Consistent with the earliest steps in ciliogenesis occurring at the distal end of the centriole (Lu et al., 2015; Tanos et al., 2013), the most dramatic changes in PxIs were observed for distal appendage bait proteins (e.g. CEP128 and others). Also of note, extensive interactions with TZ bait proteins, including Tz1-distal appendage interactions, were observed even in non-ciliated cells. Indeed, several TZ proteins are known to localize to the centrosome and centriolar satellites, or to associate with MTs, prior to ciliation (Hsiao et al., 2009), and IFT, Bardet–Biedl syndrome (BBS), and dynein-arm proteins and radial spoke complexes can preassemble for ciliogenesis (Nachury et al., 2007; Omran et al., 2008; Qin et al., 2004). This is consistent with our functional data; e.g. knockdown of several TZ proteins affected satellite morphology in non-ciliated cells (Figure 4F). This extensive functional interplay suggests that TZ assembly is likely to be “primed” in non-ciliated cells via coordination between satellites and the centriole distal end. The increase in interaction density between the Tz1, Tz2 and core groups, and the re-organization of TZ proteins in response to serum-starvation could then drive the ciliation process.
In summary, our results demonstrate that BioID coupled with directed functional screening can provide a basis for better understanding highly complex intracellular substructures such as the centrosome, and important biological processes such as ciliogenesis. Indeed, the combination of PxI profiling, localization studies and functional genomics could be applied to the study of many other cellular structures, organelles or processes.
Experimental Procedures
Detailed in Supplemental Experimental Procedures. Generation of cell lines, RT-PCR, co-IP, CRISPR, and affinity purification-MS followed standard procedures. Microscopy and centriole assays were as described (Comartin et al., 2013). BioID vector backbones and constructs (available from authors' labs) and siRNAs from this study are listed in Table S4. Unless otherwise stated, all p-values are from two-tailed unpaired Student t-tests: ***p<0.001, **p<0.01, *p<0.05.
Protein identification
Proteins identified with a Protein Prophet cut-off of 0.85 were analyzed with SAINT v. 3.3 (Choi et al., 2011; Teo et al., 2014) with the settings: total peptide >2, nburn 2,000, niter 5,000, lowMode 0, minFold 1, normalize 0. For BioID, twenty-two control runs (293 T-REx cells expressing FLAG-BirA* only, twelve from ciliating conditions, and ten from standard conditions) were collapsed to the highest 4 spectral counts for each hit. For FLAG IP-MS analysis, 20 control runs (four runs of FLAG-BirA* only, and four runs of each of four unrelated FLAG-BirA*-tagged proteins expressed in 293 T-REx cells) were collapsed to the highest 4 spectral counts for each protein ID. SAINT output was filtered with BFDR cut-off of 0.02. Common background proteins were removed manually (listed in Table S4).
Networks and clustering analysis
SAINT data were imported into Cytoscape 3.2.1 (www.cytoscape.org). Figures utilized the Dot Plot and Heatmap generator (http://prohitstools.mshri.on.ca/). All network files and parameters are available at http://prohits-web.lunenfeld.ca/. Protein SAINT score and peptide sum matrix were hierarchically clustered by Spearman rank correlation (both baits and interactors; centroid linkage; Cluster 3.0) and visualized with Java TreeView 1.1.6r4.
Functional screens
Cells were reverse transfected with Dharmacon ON-TARGETplus siRNA SMART pools or Invitrogen Stealth RNAi siRNAs (3,000 cells/well; 33.3-60nM) on 96-well coverslips, followed by respective downstream assays. All replicate data and Z-scores are listed in Table S5.
Supplementary Material
Highlights.
BioID conducted on 58 centriole, satellite and ciliary transition zone proteins
Centriole-cilium interface map comprises >1700 unique components, >7000 interactions
Microscopy and functional screens confirm new centriole-cilium regulatory modules
Dynamic modulation of interaction landscape observed during ciliogenesis program
Acknowledgments
We thank G. Pereira and O. Gruss for the RAB8A-GFP cell line and SSX2IP antibody, T. McKinnon for help with mouse cDNA synthesis, D. Holmyard for EM assistance, G. Bader for advice on network topology, F. Liu and J.-P. Zhang for expert bioinformatics advice and assistance, T. Srikumar for MS technical assistance, J.-P. Lambert for MED4 constructs, and W. Dunham, G. Hesketh and J.Y. Youn for sharing unpublished data. We thank D. Durocher, S. Angers, J. Wrana and L. Attisano for comments on the manuscript. J.G. and B.A.M. were funded by the Fundação para a Ciência e a Tecnologia (SFRH/BPD/75847/2011) and an Ontario Trillium studentship. A-C.G., B.R. and L.P. hold Canada Research Chairs in Functional Proteomics, Proteomics and Molecular Medicine, and Centrosome Biogenesis and Function, respectively. This work was funded by CIHR (OGB 137413), NSERC (RGPIN-2014-06434) and the Institute of Genetics to A-C.G., CIHR (MOP119289 and MOP130340) to B.R., and NSERC (RGPIN-355644-2008), CIHR (MOP-123468, MOP-130507), ORF-RE5 and the Krembil Foundation to L.P.
Footnotes
Author contributions: Creation of BioID cell lines for centriole and appendage components was carried out by B.A.M., S.W.T.C. and Q.W. BioID of TZ and satellite proteins was conceived and developed by J.G. E.C. and B.R. performed all BioID, MS and related analyses. Z.-Y.L. and S.W.T.C. performed FLAG IP-MS. Network and computational analysis was carried out by E.C. and G.D.G., with suggestions from J.G. Co-IP validation work was executed by J.G. and B.A.M. J.G. and M.B. conducted the cilia screen. J.G. conceived and executed WRAP73 experiments. Prey localization was carried out by J.G., L.G., B.A.M., M.H., G.D.G. and J.M. Centriole over-duplication screen was performed by G.D.G. and Q.W. Centriolar satellite screen and analysis were conducted by G.D.G. Work on ANK2 and CEP128 was performed by Y.L., D.C. and B.A.M. Analysis of RNAi screening data carried out by G.D.G. J.M.T. conducted all CRISPR experiments. A.-C.G. and J.K. assisted with MS data analysis, DotPlots and data porting to ProHits Web. M.S. and F.H. sequenced nephropathy patients. The paper was written by G.D.G., B.R. and L.P. with contributions from all authors. L.P and B.R. directed the project.
References
- Alves-Cruzeiro JM, Nogales-Cadenas R, Pascual-Montano AD. CentrosomeDB: a new generation of the centrosomal proteins database for Human and Drosophila melanogaster. Nucleic acids research. 2014;42:D430–436. doi: 10.1093/nar/gkt1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Balczon R, Bao L, Zimmer WE, Brown K, Zinkowski RP, Brinkley BR. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. The Journal of cell biology. 1995;130:105–115. doi: 10.1083/jcb.130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Larsen B, Lin ZY, Breitkreutz A, Mellacheruvu D, Fermin D, Qin ZS, Tyers M, Gingras AC, Nesvizhskii AI. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nature methods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comartin D, Gupta GD, Fussner E, Coyaud E, Hasegan M, Archinti M, Cheung SW, Pinchev D, Lawo S, Raught B, et al. CEP120 and SPICE1 cooperate with CPAP in centriole elongation. Curr Biol. 2013;23:1360–1366. doi: 10.1016/j.cub.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Edwards M, Zwolak A, Schafer DA, Sept D, Dominguez R, Cooper JA. Capping protein regulators fine-tune actin assembly dynamics. Nature reviews. 2014;15:677–689. doi: 10.1038/nrm3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat-Karalar EN, Rauniyar N, Yates JR, 3rd, Stearns T. Proximity interactions among centrosome components identify regulators of centriole duplication. Curr Biol. 2014;24:664–670. doi: 10.1016/j.cub.2014.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, Corbit KC, Sirerol-Piquer MS, Ramaswami G, Otto EA, Noriega TR, Seol AD, Robinson JF, Bennett CL, Josifova DJ, et al. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nature genetics. 2011;43:776–784. doi: 10.1038/ng.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho SA, Pellman D. Causes and consequences of centrosome abnormalities in cancer. Philos Trans R Soc Lond B Biol Sci. 2014;369 doi: 10.1098/rstb.2013.0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. Cep164, a novel centriole appendage protein required for primary cilium formation. The Journal of cell biology. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori A, Peddie CJ, Collinson LM, Toda T. Centriolar satellite- and hMsd1/SSX2IP-dependent microtubule anchoring is critical for centriole assembly. Mol Biol Cell. 2015;26:2005–2019. doi: 10.1091/mbc.E14-11-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao YC, Tong ZJ, Westfall JE, Ault JG, Page-McCaw PS, Ferland RJ. Ahi1, whose human ortholog is mutated in Joubert syndrome, is required for Rab8a localization, ciliogenesis and vesicle trafficking. Human molecular genetics. 2009;18:3926–3941. doi: 10.1093/hmg/ddp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen L, Vanselow K, Skogs M, Toyoda Y, Lundberg E, Poser I, Falkenby LG, Bennetzen M, Westendorf J, Nigg EA, et al. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J. 2011;30:1520–1535. doi: 10.1038/emboj.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Rogowski K, Wloga D, Regnard C, Kajava AV, Strub JM, Temurak N, van Dijk J, Boucher D, van Dorsselaer A, et al. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science (New York, NY. 2005;308:1758–1762. doi: 10.1126/science.1113010. [DOI] [PubMed] [Google Scholar]
- Keller LC, Romijn EP, Zamora I, Yates JR, 3rd, Marshall WF. Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr Biol. 2005;15:1090–1098. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Kim J, Lee JE, Heynen-Genel S, Suyama E, Ono K, Lee K, Ideker T, Aza-Blanc P, Gleeson JG. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 2010;464:1048–1051. doi: 10.1038/nature08895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger M, Wang W, Kuhns S, Barenz F, Drager-Meurer S, Pereira G, Gruss OJ. The novel centriolar satellite protein SSX2IP targets Cep290 to the ciliary transition zone. Mol Biol Cell. 2014;25:495–507. doi: 10.1091/mbc.E13-09-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodani A, Yu TW, Johnson JR, Jayaraman D, Johnson TL, Al-Gazali L, Sztriha L, Partlow JN, Kim H, Krup AL, et al. Centriolar satellites assemble centrosomal microcephaly proteins to recruit CDK2 and promote centriole duplication. eLife. 2015;4 doi: 10.7554/eLife.07519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Sasaki H, Yuba-Kubo A, Tsukita S, Shiina N. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. The Journal of cell biology. 1999;147:969–980. doi: 10.1083/jcb.147.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JP, Tucholska M, Go C, Knight JD, Gingras AC. Proximity biotinylation and affinity purification are complementary approaches for the interactome mapping of chromatin-associated protein complexes. J Proteomics. 2015;118:81–94. doi: 10.1016/j.jprot.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo S, Bashkurov M, Mullin M, Ferreria MG, Kittler R, Habermann B, Tagliaferro A, Poser I, Hutchins JR, Hegemann B, et al. HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr Biol. 2009;19:816–826. doi: 10.1016/j.cub.2009.04.033. [DOI] [PubMed] [Google Scholar]
- Lee JY, Stearns T. FOP is a centriolar satellite protein involved in ciliogenesis. PLoS One. 2013;8:e58589. doi: 10.1371/journal.pone.0058589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- Lin YC, Chang CW, Hsu WB, Tang CJ, Lin YN, Chou EJ, Wu CT, Tang TK. Human microcephaly protein CEP135 binds to hSAS-6 and CPAP, and is required for centriole assembly. EMBO J. 2013;32:1141–1154. doi: 10.1038/emboj.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes CA, Prosser SL, Romio L, Hirst RA, O'Callaghan C, Woolf AS, Fry AM. Centriolar satellites are assembly points for proteins implicated in human ciliopathies, including oral-facial-digital syndrome 1. Journal of cell science. 2011;124:600–612. doi: 10.1242/jcs.077156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Insinna C, Ott C, Stauffer J, Pintado PA, Rahajeng J, Baxa U, Walia V, Cuenca A, Hwang YS, et al. Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nature cell biology. 2015;17:531. doi: 10.1038/ncb3155. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peranen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- Nicholas AK, Khurshid M, Desir J, Carvalho OP, Cox JJ, Thornton G, Kausar R, Ansar M, Ahmad W, Verloes A, et al. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nature genetics. 2010;42:1010–1014. doi: 10.1038/ng.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omran H, Kobayashi D, Olbrich H, Tsukahara T, Loges NT, Hagiwara H, Zhang Q, Leblond G, O'Toole E, Hara C, et al. Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature. 2008;456:611–616. doi: 10.1038/nature07471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Diener DR, Geimer S, Cole DG, Rosenbaum JL. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. The Journal of cell biology. 2004;164:255–266. doi: 10.1083/jcb.200308132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, Blacque OE, Leroux MR. The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO reports. 2012;13:608–618. doi: 10.1038/embor.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nature reviews. 2013;14:7–12. doi: 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. The Journal of cell biology. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seixas C, Cruto T, Tavares A, Gaertig J, Soares H. CCTalpha and CCTdelta chaperonin subunits are essential and required for cilia assembly and maintenance in Tetrahymena. PLoS One. 2010;5:e10704. doi: 10.1371/journal.pone.0010704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir JH, Barr AR, Nicholas AK, Carvalho OP, Khurshid M, Sossick A, Reichelt S, D'Santos C, Woods CG, Gergely F. A primary microcephaly protein complex forms a ring around parental centrioles. Nature genetics. 2011;43:1147–1153. doi: 10.1038/ng.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulavie F, Piepenbrock D, Thomas J, Vieillard J, Duteyrat JL, Cortier E, Laurencon A, Gopfert MC, Durand B. hemingway is required for sperm flagella assembly and ciliary motility in Drosophila. Mol Biol Cell. 2014;25:1276–1286. doi: 10.1091/mbc.E13-10-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spektor A, Tsang WY, Khoo D, Dynlacht BD. Cep97 and CP110 suppress a cilia assembly program. Cell. 2007;130:678–690. doi: 10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- Staples CJ, Myers KN, Beveridge RD, Patil AA, Lee AJ, Swanton C, Howell M, Boulton SJ, Collis SJ. The centriolar satellite protein Cep131 is important for genome stability. Journal of cell science. 2012;125:4770–4779. doi: 10.1242/jcs.104059. [DOI] [PubMed] [Google Scholar]
- Stephan R, Goellner B, Moreno E, Frank CA, Hugenschmidt T, Genoud C, Aberle H, Pielage J. Hierarchical microtubule organization controls axon caliber and transport and determines synaptic structure and stability. Developmental cell. 2015;33:5–21. doi: 10.1016/j.devcel.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Lin MG, Stowe TR, Chen S, Zhu M, Stearns T, Franco B, Zhong Q. Autophagy promotes primary ciliogenesis by removing OFD1 from centriolar satellites. Nature. 2013;502:254–257. doi: 10.1038/nature12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos BE, Yang HJ, Soni R, Wang WJ, Macaluso FP, Asara JM, Tsou MF. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes & development. 2013;27:163–168. doi: 10.1101/gad.207043.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo G, Liu G, Zhang J, Nesvizhskii AI, Gingras AC, Choi H. SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. J Proteomics. 2014;100:37–43. doi: 10.1016/j.jprot.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollenaere MA, Mailand N, Bekker-Jensen S. Centriolar satellites: key mediators of centrosome functions. Cell Mol Life Sci. 2015;72:11–23. doi: 10.1007/s00018-014-1711-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam TJ, Wheway G, Slaats GG, Huynen MA, Giles RH Group, S.S. The SYSCILIA gold standard (SCGSv1) of known ciliary components and its applications within a systems biology consortium. Cilia. 2013;2:7. doi: 10.1186/2046-2530-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CL, Li C, Kida K, Inglis PN, Mohan S, Semenec L, Bialas NJ, Stupay RM, Chen N, Blacque OE, et al. MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. The Journal of cell biology. 2011;192:1023–1041. doi: 10.1083/jcb.201012116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.