Biological processes are inherently multi-scale, and supramolecular complexes at the nanoscale determine changes at the cellular scale and beyond. Single molecule localisation microscopy (SMLM)1–3 techniques have been established as important tools for studying cellular features with resolutions on the order of ~10 nm. However, in their current form these modalities are limited by a highly constrained field of view (FOV) and field-dependent image resolution. Here, we develop a low-cost microlens array (MLA)-based epi-illumination system called FIFI that can efficiently and homogeneously perform simultaneous imaging of multiple cells with nanoscale resolution. The optical principle of FIFI, which is an extension of the Köhler integrator, is further elucidated and modelled with a new, free simulation package. We demonstrate FIFI’s capabilities by imaging multiple Cos7 and bacteria cells in 100 × 100 µm2 SMLM images — more than quadrupling the size of a typical FOV, and producing near-gigapixel-sized images of uniformly high quality.
The development of super-resolution fluorescence techniques heralded a dramatic improvement over traditional wide-field and confocal microscopies by enabling resolutions surpassing the diffraction limit. When considering how excitation light is delivered to the sample in these techniques, two main strategies emerge. The first, which is applied in structured illumination microscopy (SIM)4,5, stimulated emission depletion (STED)6, and reversible saturable optical linear fluorescence transitions (RESOLFT)7,8, uses spatially patterned excitation light to control the photophysical states of fluorophores in a targeted way across a FOV. Alternatively, SMLM methods such as stochastic optical reconstruction microscopy (STORM) and (fluorescence) photoactivated localisation microscopy ((f)PALM)1–3 typically use a simpler strategy: wide-field laser epi-illumination. In this geometry, the sample is illuminated with a laser beam possessing a Gaussian-like irradiance profile a few tens of microns across. SMLM relies on the stochastic on-and-off switching of fluorophores, generating pointillist-like images of labelled structures with resolutions that depend on the density of the detected fluorophore positions—commonly referred to as localisations—and on the corresponding precision of the position estimates9. To achieve temporal separation of single-molecule emissions, such as from commonly used synthetic dyes, relatively large laser irradiances are delivered to the sample to cycle fluorophores from emitting to long-lived, non-emitting states10.
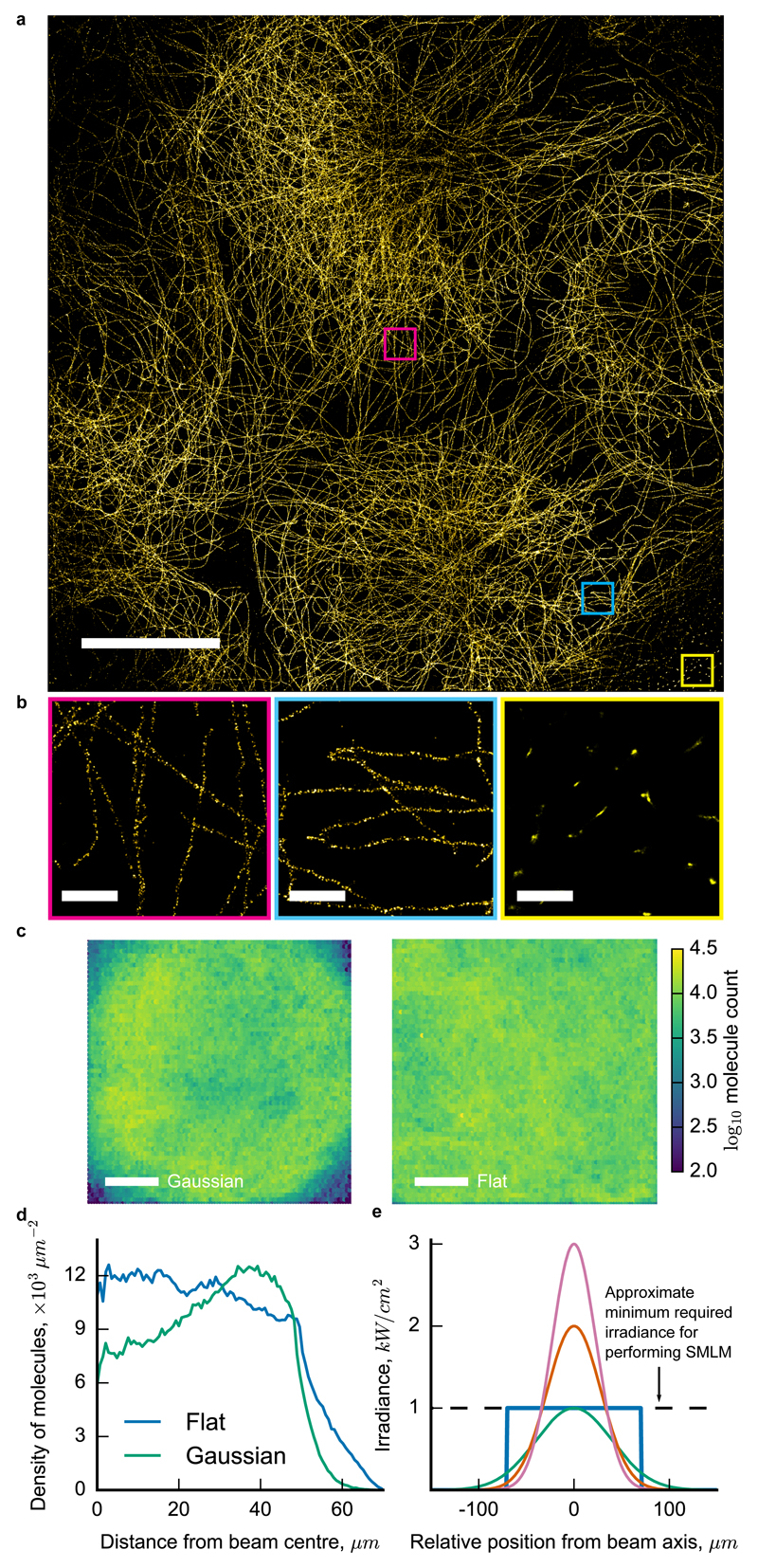
In practice, the size and quality of a SMLM image is severely limited by several factors related to the illumination. Wide-field epi-illumination is achieved by focussing the Gaussian excitation beam to the centre of the objective’s back focal plane (BFP) (Supp. Fig. 1). The resulting size of the illuminated spot on the sample is proportional to the numerical aperture (NA) of the input beam, which is restricted by apertures in the microscope’s mechanical housing. Additionally, SMLM image quality drops significantly and artefacts appear in the periphery of the illuminated region where the minimum irradiance requirement of 1 – 10 kW / cm2 is not satisfied (Fig. 1a, b)11–13. This is because the density of emitting molecules is too high to allow for precise and accurate localisation in these regions, therefore violating a basic assumption of SMLM reconstruction algorithms (Supp. Text, Section 4). On the other hand, in regions where the irradiance is too high, the acquisition time becomes longer than necessary because too few molecules are emitting at any given time. The result is a field-dependent density of detected localisations (Fig. 1b, c), localisation precisions and goodness of fits (Supp. Fig. 2, 3). The illumination is therefore critical in determining the quality and final resolution of a large FOV SMLM image (Supp. Text, Sections 4, 5).
Figure 1.
Wide-field epi-illumination is not optimised for SMLM imaging of large fields of view (FOV). a) Gaussian-illuminated SMLM image of microtubules in Cos7 cells with antibodies against α-tubulin (AlexaFluor 647) displays a non-uniform image quality. b) Highlighted regions of (a) show variation in the localisation density and artefacts in the periphery. c) Number of detected fluorescent molecules under Gaussian and flattened illumination patterns of the same power. Data are from ten combined FOVs, each of Cos7 microtubules spanning the full FOVs. d) Radially-averaged densities of detected molecules for the images in (c). e) Profiles of three different Gaussian illumination patterns (full widths at half maximum are 46 μm, 56 μm, and 80 μm) and the diagonal of a 100 × 100 μm2 flat-top pattern, all carrying 100 mW of power. The flat-top pattern produces the largest SMLM images without field-dependent fluorophore blinking. Scale bars: 20 μm (a) and (c), and 1 μm (b).
By contrast, a flat irradiance profile on the sample can produce uniform localisation density, precision, and single molecule fit quality across the FOV (Fig. 1c,d; Supp. Fig. 2). Conceptually, this occurs because optical power in the regions where the irradiance is too high in the Gaussian beam is redistributed to the periphery where it is too low. Additionally, the optimum irradiance required to perform SMLM imaging is ultimately set by the highest local density of fluorophores on the structures to be imaged12 (Supp. Text, Sections 4, 5). A flat profile that is adjusted to this irradiance will always span a larger area than a Gaussian beam of the same power (Fig. 1e), thereby producing larger images and increasing the throughput of the measurement.
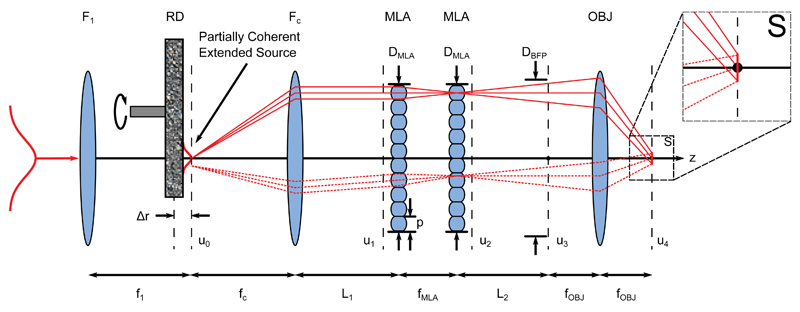
We developed FIFI, or Flat Illumination for Field Independent imaging, to efficiently achieve a large and uniform illumination pattern in laser fluorescence microscopes. FIFI is an analogue beam-shaping epi-illumination system and extends the Köhler integrator14–16 to SMLM (Fig. 2). In this system, a collimated beam is launched into a telescope whose lenses are spaced by the sum of their focal lengths. A high-efficiency diffuser with average grain size on the order of 10 × 10 µm2 and rotating at approximately 2000 revolutions per minute is placed at an offset distance from their common focal planes equal to ∆r. This offset determines the size of the beam on the diffuser and therefore the spatial coherence of the source. The beam is collected by the second telescope lens (focal length fc) before encountering the primary components of the system: two MLAs consisting of identical spherical lenslets. The first MLA splits the different field angles in the incoming partially coherent laser beam into independently propagating beamlets travelling parallel to one another through the system. The lenslets in the second MLA, which is located exactly one lenslet focal length after the first, cancel the quadratic phase curvature imparted by the first MLA17. The lenslets in the second MLA also image each lenslet in the first array to infinity. The beamlets are finally focussed through the infinity-corrected objective lens onto the sample plane where the images of the individual lenslets in the first array are overlapped. The ultimate effect is to illuminate the sample with a uniform irradiance pattern by averaging out heterogeneities in the input laser beam. Compared to other beam homogenizers, FIFI is easy to align, efficient, wavelength-insensitive, and features a continuously variable illumination size (Supp. Text, Section 3).
Figure 2.
Layout and design parameters for FIFI. An input Gaussian beam is launched into a telescope with focusing lens F1 and collimating lens FC. A rotating diffuser (RD) is offset from their shared focal plane, creating an extended source whose size depends on the offset Δr. Light from FC propagates a distance L1 to identical microlens arrays (MLAs) with periodicities p separated by one lenslet focal length fMLA. The MLAs create independent ray bundles that propagate another distance L2 and are redirected by the objective (OBJ) such that they overlap at the sample plane (S). Each illuminated point in the sample plane receives light from every point and from a range of solid angles in the source plane (solid vs dashed rays). Scalar fields u calculated in the simulations are marked with black dashed lines. f1, fc, and fOBJ: focal lengths of corresponding thin lenses; DMLA, DBFP: aperture sizes.
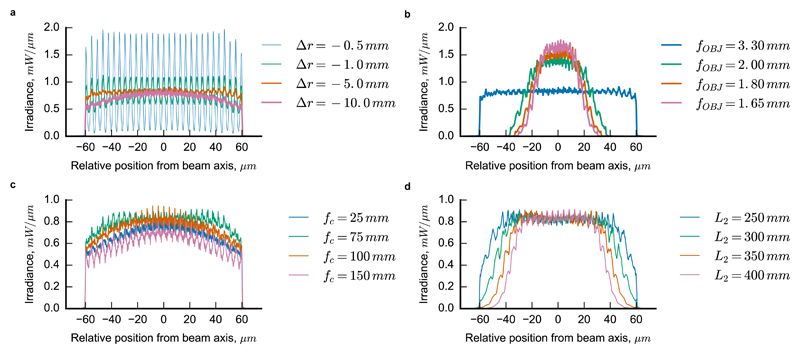
A geometrical optics design is sufficient to implement the system into the laser epi-illumination path of a low magnification system such as a flow cytometer18. However, the stringent requirements imposed by SMLM and the number of free parameters (Fig. 2) make the design of a FIFI system for super-resolution non-trivial. To overcome this difficulty, we developed a wave optics simulation platform that extends previous applications of scalar diffraction theory applied to MLAs15 and specifically accounts for the partial coherence and optical path through a high magnification fluorescence microscope. The simulations enabled us to identify the combination of design parameters that maximizes the two primary requirements of SMLM: the illumination uniformity and power throughput of the system (Fig. 3 and Supp. Fig. 4, 5). For example, the diffuser must be offset from the telescope focal plane to reduce non-uniformities from grating diffraction from the MLAs, but moving it too far from the focal plane results in overfilling of the objective back aperture and in a loss of uniformity (Fig. 3a) and power (Supp. Fig. 5a). The objective focal length and BFP diameter also have significant impacts on the sample space illumination area, uniformity and transmitted power (Fig. 3b, Supp. Fig. 5b). Additionally, the focal length of the telescope’s collimating lens fc has an optimal value (Fig. 3c, Supplementary Fig. 5c) and the distance between the second MLA and objective must be made as small as possible while still allowing for the insertion of dichroic filters (Fig. 3d, Supp. Fig. 5d). Our simulation platform is therefore a non-trivial and important tool for navigating this rich parameter space.
Figure 3.
Simulated 1D sample plane irradiance profiles through the centre of the illuminated region demonstrate the strong dependence of the illumination homogeneity on the design parameters. a) The offset of the rotating diffuser Δr has an optimum position that reduces the modulation in the grating pattern and prevents overfilling the microlens arrays (MLA). b) The objective lens focal length fOBJ largely determines the size of the sample plane irradiance. c) The focal length of the collimating lens fc for the partially coherent source controls both the field homogeneity and the spatial coherence of the source, as evidenced by the modulation depth of the grating fringes. d) The distance between the second MLA and the objective L2 must be small to prevent overfilling the objective.
We built a FIFI system into a STORM microscope with a sCMOS camera (Supp. Fig. 6 and Methods). The fluorescence profiles excited by the three lasers in a concentrated dye solution19 demonstrate the good uniformity, large size of the illuminated area, and accuracy of the simulations (Supp. Fig.’s 7-9). Curvature in the dichroic and/or introduction of a weak defocusing lens can help smoothen any residual non-uniformities, better enabling wide-field measurements requiring uniform illumination (Supp. Fig.’s 10-12). In practice, a few percent variation in illumination has no measurable effect on the STORM data when the diffuser is optimally placed (Fig. 1c, d, Fig.4). Furthermore, the illumination can be continuously resized simply by adjusting the MLA spacing (Supp. Fig. 13). The transmitted power at the objective’s BFP is about 70% for both lasers, with the diffuser and the MLAs each accounting for between 5% and 10% of the losses (Supp. Table 2). (The remaining losses are due to the dichroics, mirrors, and lenses.) The FOV, which corresponds to the area of homogeneous illumination onto the sample, is 100 × 100 μm2, which is approximately 5-10 times that of traditional wide-field SMLM. The throughput is further improved when combined with the high frame rates of sCMOS cameras20.
Figure 4.
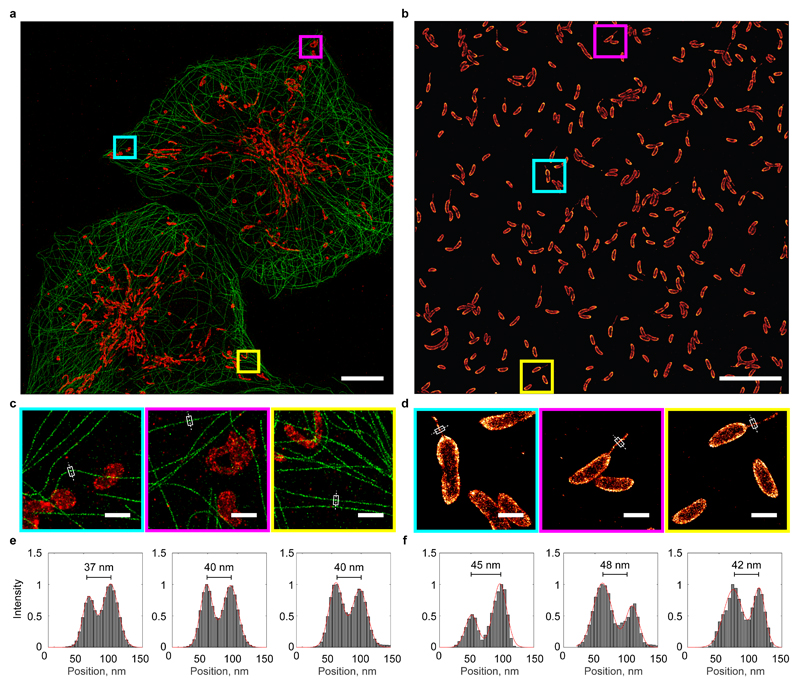
Uniform, large FOV STORM imaging of multiple eukaryotic cells or hundreds of bacterial cells in a single image. a) Dual colour STORM image of Cos7 cells labelled with antibodies against α-tubulin (AlexaFluor 647, green) and TOM20 (AlexaFluor 750, red). b) STORM image of Caulobacter crescentus labelled with wheat germ agglutinin (AlexaFluor 647). c) and d) Magnified views of marked regions in (a) and (b). e) and f) Pixel intensities through marked lines in (c) and (d) showing uniform resolution. Scale bars: 10 µm (a) and (b), and 1 µm (c) and (d).
The system is capable of performing STORM on multiple eukaryotic cells (1-3 Cos7 cells) or on a large number of bacteria (>400) in a single image (Fig. 4 and Supp. Fig.’s 14-17). The uniformly high quality is verified by line profiles of pixel intensities showing double-walled microtubules and bacterial stalk appendages, with sizes consistent with reported values21. For bacteria, the fast acquisition time and large FOV (3.3 minutes per FOV) mean that variability in cell shape or protein localisation can now be studied simultaneously at both nanometre and population scale.
We have demonstrated that an optimised laser epi-illumination system known as FIFI enables uniform, high-quality wide-field super-resolution fluorescence imaging. Importantly, we increased the throughput of SMLM by capturing multiple eukaryotic cells or hundreds of bacteria in each FOV, simultaneously resolving features that are smaller than the optical diffraction limit. The key design requirements are a uniform illumination and sufficient power delivered to the sample to induce fluorophore switching over a large area. Beyond imaging a larger FOV, FIFI improves the quality of the images by significantly reducing position-dependent photophysics which results in non-uniform resolution. FIFI is robust to misalignment and vibrations, and therefore serves as a simple, low-cost solution for fully exploiting the large sensor formats of new sCMOS cameras for SMLM13,22,23.
Methods
Sample Preparation
Cos7 cells were seeded in 6-well plates (TPP) on 25 mm glass slides (Menzel, #1.5) at a density of 1.5x105 cells per well. The cells were cultivated for 12-16 hours in DMEM (supplemented with 10% foetal calf serum) at 37°C and 5% CO2. The immunofluorescence staining procedure is previously described24 and briefly recapitulated here. The cells were washed twice in PBS and fixed for 10 minutes in pre-warmed fixation solution (4% paraformaldehyde (PFA), 0.1% glutaraldehyde (GA) in PBS). After washing in PBS, unreacted aldehyde groups were quenched with 0.1% sodium borohydride in PBS for 5 min. The sample was washed again in PBS followed by a permeabilisation and blocking incubation with 3% bovine serum albumin (BSA) and 0.1% Triton X-100 in PBS for 10 min. The cells were incubated with primary antibodies against tubulin (B512, Sigma, 1:100 dilution) and Tom20 (sc415, SantaCruz, 1:50 dilution) in blocking buffer (3% BSA, 0.1% Triton X-100) for 2 h. The sample was washed three times for 10 min in PBS supplemented with 0.2% BSA and 0.1% Triton X-100. Secondary antibodies against mouse (AlexaFluor 647-conjugate, A21237, Life Technologies) and rabbit (see antibody conjugation) were diluted in blocking buffer to a final concentration of 4-10 µg/ml. The sample was incubated with mixed secondary antibodies for 2 hours in the dark, then washed three times for 10 min in PBS (supplemented with 0.2% BSA and 0.1% Triton X-100) and post-fixed for 10 min in PBS with 4% PFA, 0.1% GA. Standard chemicals if not otherwise stated were purchased from Sigma Aldrich.
CB15N wild-type Caulobacter crescentus strain were grown in liquid M2G medium in mid-exponential phase for 12 to 16 hours. Bacteria were fixed with 2.5% PFA in PBS solution for 10 min then immediately resuspended in permeabilisation buffer (0.1% Triton X-100 in PBS) for 10 min. The bacteria were then resuspended in 100 µg/ml Wheat Germ Agglutinin (AlexaFluor 647-conjugate, W32466, Life Technologies) solution for 5 hours at room temperature then washed 3 times in PBS solution and post-fixed for 10 min in PBS with 2.5% PFA.
Preparation of Labelled Antibodies
Unconjugated goat anti-rabbit antibody (31239, ThermoFisher) was diluted to 0.6 mg/ml in 100 µl PBS (supplemented with 50 mM NaHCO3). AlexaFluor 750 NHS ester (A20011, Life Technologies) was added at a final concentration of (150 µM) and the solution was incubated for 30 min at room temperature. 100 µl PBS were added and the solution was applied to a NAP-5 size exclusion column (GE Healthcare) pre-equilibrated with PBS. 300 µl fractions were collected and analysed by UV-Vis spectroscopy (Nanodrop2000, ThermoFisher). Peak protein fractions were collected and the degree of labelling calculated. The labelled antibody fractions were stored at 4°C until further use.
Field-Dependent Blinking Statistics
The field dependence of SMLM imaging was assessed by performing direct STORM10 imaging of AlexaFluor 647-labeled α-tubulin in microtubules from Cos7 cells. Ten FOVs each were taken with microtubules filling the whole FOV for both flat-field illumination using FIFI and for standard wide-field illumination using an achromatic doublet as the field lens (AC254-250-B, Thorlabs). 25,000 frames at 10 ms exposure times and 1000 mW of laser light at a wavelength of 642 nm were recorded for each FOV, and care was taken to select FOVs where the density of microtubules was more or less uniform. The localisations from these images were combined across all ten FOVs, yielding approximately 108 localisations distributed across an area of 100 × 100 μm2 for each illumination. Localisations were computed using a previously described sCMOS-specific maximum-likelihood-based localisation algorithm13.
Field-dependent STORM simulations described in the Supplementary Material used SuReSim25 version 0.5.1 and ThunderSTORM26 version 1.3-2014-11-08.
Wave Optics Simulations
The epi-illuminator theory, developed in the Supplementary Text and based on previous work15, was simulated using standard numerical Fourier optics routines27 and a custom-written Python library called SimMLA freely available at https://github.com/kmdouglass/simmla. We did not use a ray optics simulation because ray optics cannot model the diffraction pattern and peak modulation. Scalar fields were propagated between the planes marked by subscripted u’s in Fig. 2 using fast Fourier transforms (FFT) for lens transformations and a FFT-based angular spectrum propagator for free-space propagations. FFT functionality was provided by Numpy (Version 1.10.4) and Scipy (Version 0.17.0) for Python (Version 3.5.1, 64-bits). To minimise aliasing artefacts, all fields were sampled on spatial grids with periodicities that were no more than approximately 12 nm. This necessitated large array sizes which restricted the field calculations to a single, one-dimensional (1D) line transverse to and intersecting the optical axis and parallel to one of the two symmetry lines of the square MLAs. Fig. 3 displays samples of the field along this line for the simulated conditions indicated in the figure.
The rotating diffuser was modelled as a random phase screen, as described previously28. The diffuser’s correlation length was set to 10 μm, which is the approximate size of a grain on its rough surface. The variance σr, which describes the roughness of the diffuser, was found by measuring the modulation depth of the fringes in the laser spot profile on a concentrated dye sample at a known diffuser offset distance from the telescope’s centre, and running the simulation with various values of σr. We found that a variance of 1.75 produced fringe modulation depths that matched the experimental ones and so used this number throughout all the simulations. All other simulation parameters are listed in Supplementary Table 1.
The MLA simulation code was verified by reproducing Fig. 7b and Fig. 7c in Ref.15. The partial coherence code was verified against Fig. 9.11 in Ref.27.
The simulated objective parameters were chosen from common commercial microscope objectives used in SMLM. From top to bottom for the objective parameters labelled in Fig. 3b, the objectives correspond to a 60x, NA 1.4 objective with a tube lens focal length fTL of 200 mm; a 100x, NA 1.45 objective with a tube lens focal length fTL of 200 mm; a 100x, NA 1.4 objective with a tube lens focal length of 180 mm; and a 100x, NA 1.46 objective with a tube lens focal length of 165 mm. The magnification and tube lens focal lengths determine the objective focal lengths and BFP diameters through the equations fOBJ = fTL/Magnification and DBFP = 2 × NA × fOBJ .
STORM Imaging
Two lasers with wavelengths of 642 nm (2RU-VFL-P-2000-642-B1R, MPB Communications) and 750 nm (2RU-VFL-P-500-750-B1R, MPB Communications) were used to switch off fluorophores on the sample, while a 405 nm laser (OBIS, Coherent) controlled the return rate of the fluorophores to the fluorescence-emitting state. These laser beams were expanded, combined using dichroic mirrors (ZT647rdc-UF2, Chroma and T425lpxr, Chroma), and then injected into the telescope (f1 = 100 mm, fc = 50 mm). The rotating diffuser (2.5º ± 0.25º FWHM at 650 nm, 24-00066, Süss MicroOptics SA) sat at an adjustable offset of -5 mm from the shared focal planes of the telescope lenses. A series of mirrors was then used to align the beam to both the MLA’s (500 μm pitch, 10 mm x 10 mm, f = 13.7 mm, square lenses, 18-00201, Süss MicroOptics SA) and the objective lens (CFI60 PlanApo Lambda 60x / NA 1.4, Nikon). A custom dichroic (ZT405/561/642/750/850rpc, Chroma) reflected the laser light and transmitted fluorescence emission. Emitted light from the sample was collected by the same objective, passed through the dichroic and was imaged by a tube lens (f = 200 mm, MXA20696, Nikon) onto the sCMOS camera (Zyla 4.2, Andor). The width of a square camera pixel corresponds to 106 ± 2 nm on the sample. One of two emission filters (ET700/75M, Chroma and ET810/90m, Chroma) were combined with a short-pass filter (FF01-842/SP, Semrock) and inserted into the emission path between the tube lens and camera depending on the fluorophore being imaged. A separate, 850 nm laser (0.9 mW CW circular beam, 85-238, Thorlabs) passed through a cleanup filter (LL01-852, Semrock) and was reflected from the coverslip by total internal reflection. The reflected beam was directed through an 850 nm band-pass filter (86-090, Edmund Optics) and onto a linear light sensor (TSL1401CL, AMS-TAOS USA, Inc.); changes in the beam position were read using the pgFocus open hardware autofocus module (http://big.umassmed.edu/wiki/index.php/PgFocus). This information was used to send a feedback signal to the piezo z-stage and lock the objective-coverslip distance to within a standard deviation of 10 nm. Microscope control was orchestrated with Micro-Manager (Version 1.4.22, nightly build 2015-07-27)29. Single molecule localisation analysis was performed using a sCMOS-specific maximum likelihood algorithm following a previously described calibration routine13.
MLAs and rotating diffuser were chosen based on the design specifications from the simulation package and the need to minimize losses in the transmitted laser power. Square lenses provide a complete fill factor and minimize scattering at the lenslet interfaces, while also projecting a square illumination pattern onto the sample. A diffuser with a small divergence angle specification helps to maintain a minimally-divergent laser beam profile through the system.
Imaging was performed using an optimized buffer as described previously30. Microtubules in Fig. 4a were imaged by acquiring 30,000 frames at 10 ms continuous exposure and approximately 1000 mW in the objective’s BFP at 642 nm. Mitochondria were imaged with the same parameters but a BFP laser power of 350 mW at 750 nm. The bacteria in Fig. 4b were imaged by acquiring 20,000 frames at 10 ms continuous exposure and 1400 mW of 642 nm laser light in the objective BFP.
Illumination Profile Measurement
The illumination profile in the sample plane just above the coverslip was measured using a highly concentrated fluorescent dye solution19. Briefly, 0.45 g/mL of acid blue #9, also known as erioglaucine disodium salt (3844-45-9, Sigma-Aldrich), was mixed with deionized water and vortexed for 5 minutes, followed by sonication for 1 hour until the dye was completely dissolved. A 4 μL drop of the solution was then pipetted onto a 25 mm diameter, #1.5 coverslip and covered with a 12 mm diameter coverslip to create a thin, uniform layer. During the profile measurements, the laser powers were set to be as low as possible while still obtaining a good signal-to-noise ratio from the fluorescence signal; doing so ensured that out-of-focus fluorescence did not erroneously smoothen out the measured profile and give a false sense of uniformity. The dye sample was used right away after preparation and discarded after use.
When measuring illumination profiles, only the emission filter was changed between profile measurements at different wavelengths.
Supplementary Material
Acknowledgements
The authors would like to thank Karl Bellvé for providing the pgFocus hardware, Fang Huang for his help in troubleshooting the sCMOS localisation software, David G. Voelz for providing code examples for modelling partially-coherent optical fields, Reinhard Völkel for discussions about the Köhler integrator, and Niklas Berliner, Daniel Ott, Matthew Weidman, and Pavan Ramdya for useful discussions. Research in SM’s laboratory is supported by the National Centre of Competence in Research Chemical Biology and by the European Research Council (ERC 243016-PALMassembly). KMD is supported by a SystemsX.ch Transition Postdoc Fellowship (2014/227).
Footnotes
Author Contributions
SM and KMD conceived the idea. KMD designed the epi-illumination system, built the microscope, and wrote the simulation code. CS and AL performed the cell preparation and imaging. CS and AA assisted KMD with building and testing the microscope. KMD, with contributions from all authors, wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–5. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–5. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 3.Hess ST, Girirajan TPK, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–72. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gustafsson MGL. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc. 2000;198:82–87. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- 5.Heintzmann R, Cremer CG. Laterally modulated excitation microscopy: improvement of resolution by using a diffraction grating. In: Bigio IJ, Schneckenburger H, Slavik J, Svanberg K, Viallet PM, editors. BiOS Europe ’98. International Society for Optics and Photonics; 1999. pp. 185–196. [DOI] [Google Scholar]
- 6.Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19:780. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 7.Schwentker MA, et al. Wide-field subdiffraction RESOLFT microscopy using fluorescent protein photoswitching. Microsc Res Tech. 2007;70:269–80. doi: 10.1002/jemt.20443. [DOI] [PubMed] [Google Scholar]
- 8.Chmyrov A, et al. Nanoscopy with more than 100,000 ‘doughnuts’. Nat Methods. 2013;10:737–40. doi: 10.1038/nmeth.2556. [DOI] [PubMed] [Google Scholar]
- 9.Nieuwenhuizen RPJ, et al. Measuring image resolution in optical nanoscopy. Nat Methods. 2013;10:557–62. doi: 10.1038/nmeth.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heilemann M, et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed Engl. 2008;47:6172–6. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- 11.Burgert A, Letschert S, Doose S, Sauer M. Artifacts in single-molecule localization microscopy. Histochem Cell Biol. 2015;144:123–31. doi: 10.1007/s00418-015-1340-4. [DOI] [PubMed] [Google Scholar]
- 12.van de Linde S, et al. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat Protoc. 2011;6:991–1009. doi: 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]
- 13.Huang F, et al. Video-rate nanoscopy using sCMOS camera-specific single-molecule localization algorithms. Nat Methods. 2013;10:653–8. doi: 10.1038/nmeth.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Völkel R, Weible KJ. Laser Beam Homogenizing: Limitations and Constraints. Proc SPIE. 2008;7102 71020J–71020J–12. [Google Scholar]
- 15.Büttner A, Zeitner UD. Wave optical analysis of light-emitting diode beam shaping using microlens arrays. Opt Eng. 2002;41:2393. [Google Scholar]
- 16.Brown DM, Dickey FM, Weichman LS. In: Laser Beam Shaping: Theory and Techniques. Dickey FM, Holswade SC, editors. CRC Press; 2000. [DOI] [Google Scholar]
- 17.Streibl N, Nölscher U, Jahns J, Walker S. Array generation with lenslet arrays. Appl Opt. 1991;30:2739–42. doi: 10.1364/AO.30.002739. [DOI] [PubMed] [Google Scholar]
- 18.Scholtens TM, et al. CellTracks TDI: An image cytometer for cell characterization. Cytom Part A. 2011;79A:203–213. doi: 10.1002/cyto.a.21024. [DOI] [PubMed] [Google Scholar]
- 19.Model MA, Blank JL. Concentrated dyes as a source of two-dimensional fluorescent field for characterization of a confocal microscope. J Microsc. 2008;229:12–6. doi: 10.1111/j.1365-2818.2007.01880.x. [DOI] [PubMed] [Google Scholar]
- 20.Lin Y, et al. Quantifying and optimizing single-molecule switching nanoscopy at high speeds. PLoS One. 2015;10:e0128135. doi: 10.1371/journal.pone.0128135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlimpert S, et al. General protein diffusion barriers create compartments within bacterial cells. Cell. 2012;151:1270–82. doi: 10.1016/j.cell.2012.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Almada P, Culley S, Henriques R. PALM and STORM: Into large fields and high-throughput microscopy with sCMOS detectors. Methods. 2015;88:109–21. doi: 10.1016/j.ymeth.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Huang Z-L, et al. Localization-based super-resolution microscopy with an sCMOS camera. Opt Express. 2011;19:19156–68. doi: 10.1364/OE.19.019156. [DOI] [PubMed] [Google Scholar]
- 24.Bates M, Dempsey GT, Chen KH, Zhuang X. Multicolor super-resolution fluorescence imaging via multi-parameter fluorophore detection. Chemphyschem. 2012;13:99–107. doi: 10.1002/cphc.201100735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venkataramani V, Herrmannsdörfer F, Heilemann M, Kuner T. SuReSim: simulating localization microscopy experiments from ground truth models. Nat Methods. 2016;13:319–321. doi: 10.1038/nmeth.3775. [DOI] [PubMed] [Google Scholar]
- 26.Ovesný M, Křížek P, Borkovec J, Svindrych Z, Hagen GM. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30:2389–90. doi: 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voelz DG. Computational Fourier Optics: A MATLAB Tutorial. SPIE Press; 2011. at < http://spie.org/Publications/Book/858456>. [Google Scholar]
- 28.Xiao X, Voelz D. Wave optics simulation approach for partial spatially coherent beams. Opt Express. 2006;14:6986. doi: 10.1364/oe.14.006986. [DOI] [PubMed] [Google Scholar]
- 29.Edelstein AD, et al. Advanced methods of microscope control using μManager software. J Biol methods. 2014;1:e10. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olivier N, Keller D, Gönczy P, Manley S. Resolution doubling in 3D-STORM imaging through improved buffers. PLoS One. 2013;8:e69004. doi: 10.1371/journal.pone.0069004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.