Abstract
The microRNA (miR)‐29 family is closely associated with fibrotic processes by virtue of its low expression in many tissues during organ fibrosis. The present study investigated whether miR‐29b overexpression suppressed hepatic stellate cell (HSC) activation and its interactions with transforming growth factor (TGF)‐β1/mothers against decapentaplegic homolog 3 (Smad3), a classical signal transduction pathway contributing to the activation of HSCs. The results showed that transfection of LX‐2 (human HSC) cells with miR‐29b mimic or pSUPER‐Smad3 silencing (si)RNA resulted in significantly increased expression of miR‐29b and decreased expression of Smad3. miR‐29b overexpression inhibited proliferation of LX‐2 cells 24 h after transfection. Both miR‐29b overexpression and Smad3 silencing antagonized the effects of TGF‐β1 on the expression of α‐smooth muscle actin (α‐SMA) and collagen type I (col‐1). Furthermore, infection with miR‐29b mimics suppressed Smad3 and TGF‐β1 expression, suggesting that miR‐29b inhibited LX‐2 activation mediated by both Smad3 and TGF‐β1. Nevertheless, primary miR‐29a/b1, miR‐29b2/c and mature miR‐29b were downregulated by TGF‐β1 and stimulated by Smad3 silencing, suggesting that TGF‐β1/Smad3 signalling pathway regulate not just mature miR‐29b but also its transcription. In summary, our results show overwhelming evidence corroborating the suppressive effect of miR‐29b on TGF‐β1‐induced LX‐2 cell activation. The results also revealed the existence of crosstalk between miR‐29b and TGF‐β1/Smad3 during LX‐2 activation, suggesting a feedback loop between miR‐29b and TGF‐β1/Smad3 signalling that promotes liver fibrosis. Copyright © 2016 The Authors. Cell Biochemistry and Function published by John Wiley & Sons, Ltd.
Keywords: hepatic stellate cell, miR‐29b, TGF‐β1/Smad3, gene transfection, crosstalk
Introduction
Liver fibrosis is characterized by increased deposition of extracellular matrix (ECM), which is an inescapable path from hepatitis to hepatocirrhosis. Since first discovered and described by Karl Wilhelm von Kupffer in 1876, hepatic stellate cells (HSCs) have been established as the main effector cells and the primary source of ECM in liver fibrosis.1, 2 After liver injury, HSCs become activated and trans‐differentiate into myofibroblast‐like cells characterized by loss of stored vitamin A droplets, expression of α‐smooth muscle actin (SMA) and production of ECM, increasing proliferation and other phenotypic changes.3
MicroRNAs (miRNAs) are RNAs 20–25 nucleotides in length involved in translational regulation of gene expression.4 More than 2000 human miRNAs have been described since the first miRNA, Caenorhabditis elegans lin‐4, was cloned by Rosalind Lee, Rhonda Feinbaum and Victor Ambros in 1993.5, 6 miRNAs play important roles in cell differentiation, proliferation and apoptosis.5, 7, 8
The miR‐29 family, composed of miR‐29a, b and c, represents a small noncoding RNA family crucially involved in the modulation of cellular processes leading to fibrosis of multiple tissues and organs including liver fibrosis.9 miR‐29b is considered as a delegate for co‐expressing with both miR‐29a and miR‐29c. miR‐29s (miR‐29a/b/c) have been shown to have dozens of direct target genes. The majority of them are ECM, migration‐related proteins and some signal transduction pathway‐related proteins.
Transforming growth factor (TGF)‐β1/mothers against decapentaplegic homolog 3 (Smad3) is one of the most prominent signal transduction pathways contributing to activation of HSCs 10, 11 and other mesenchymal cells intimately involved with fibrosis, including renal mesangial cells,12 pulmonary macrophages and fibroblasts.13 Recent studies have shown that Smad3, but not Smad2, plays the main role in TGF‐β1‐mediated fibrosis.14, 15, 16
miR‐29b has been demonstrated to suppress type I collagen (col‐1) production in cultured human stellate cells by its direct interaction with the col1A1 3′ untranslated region and transcription factor SP1.17 Additionally, the inhibitory effect of miR‐29b on TGF‐β1 has been described in trabecular meshwork cells.18 Moreover, recent studies have shown that TGF‐β1 modulates expression of miR‐29b by modulating Smad3.19, 20 This research thus suggests a close cooperative relationship between miR‐29b and TGF‐β1/Smad3 during liver fibrosis. However, our knowledge about the interactions between miR‐29 and TGF‐β1/Smad3 is still incomplete. Accordingly, we investigated the inhibitory effect of miR‐29b on HSC activation and its crosstalk with TGF‐β1/Smad3.
Materials and Methods
Cell culture and treatments
Human hepatic stellate cells (LX‐2) were purchased from XiangYa School of Medicine (Changsha, China). Cells were maintained in high‐glucose Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and incubated at 37 °C in a humidified chamber supplemented with 5% CO2.
Seven separate experimental groups were studied: blank group, negative control (NC) group, NC transfection + TGF‐β1 group, miR‐29b transfection group, miR‐29b transfection + TGF‐β1 group, Smad3 transfection group and Smad3 transfection + TGF‐β1 group. For TGF‐β1 treatment groups, the cells were exposed to 10 ng/ml TGF‐β1 for 24 h after transfection. TGF‐β1 was obtained from PeproTech (Rocky Hill, NJ, USA).
Transfection
Cells were transfected at 80% confluency with X‐tremeGENE siRNA transfection reagent (F. Hoffmann‐La Roche, Ltd., Basel, Switzerland) plus siRNA or miRNA (Shanghai GenePharma, Ltd., Shanghai, China), 10 µl and 2 µg/well, respectively, for 6‐well plates and 0.8 µl and 0.15 µg/well, respectively, for 96‐well plates, 24 h after seeding. Medium was refreshed after 6 h, and transfection efficiency was evaluated after 24 h. siRNA sequences are listed in Table 1. Two different sequences of Smad3 siRNA‐labelled Smad3 and Smad3‐1309 were designed and synthesized for transfection, and the one with a stronger inhibitory effect was used in the following experiments.
Table 1.
siRNA and miRNA sequences
| Gene | Strand | Sequence |
|---|---|---|
| miR‐29b mimics | Sense | 5′‐UAGCACCAUUUGAAAUCAGUGUU‐3′ |
| Antisense | 5′‐CACUGAUUUCAAAUGGUGCUAUU‐3′ | |
| Smad3‐siRNA | Sense | 5′‐CGUCAACACCAAGUGCAUCTT‐3′ |
| Antisense | 5′‐GAUGCACUUGGUGUUGACGTT‐3′ | |
| Smad3‐siRNA‐1309 | Sense | 5′‐GCAACCUGAAGAUCUUCAATT‐3′ |
| Antisense | 5′‐UUGAAGAUCUUCAGGUUGCTT‐3′ | |
| NC‐siRNA | Sense | 5′‐UUCUCCGAACGUGUCACGUTT‐3′ |
| Antisense | 5′‐ACGUGACACGUUCGGAGAATT‐3′ |
Cell Counting Kit‐8
LX‐2 cells were re‐seeded at a density of 7 × 103 cells per well in 96‐well plates 24 h prior to transfection and divided into three groups: blank group, NC group and miR‐29b transfection group. Cells from each group were seeded in triplicate. Medium was refreshed 24 h after transfection with 10 µl/well Cell Counting Kit‐8 (CCK‐8) reagent (Dojindo Laboratories, Kumamoto, Japan). Cells were incubated in 10% CCK‐8 diluted in normal culture media at 37 °C until visual colour conversion appeared. Proliferation rates were determined at 24, 48 and 72 h post‐transfection, and quantification was performed on a microplate reader set according to the manufacturer's protocol. The proliferation inhibition rate was calculated from the following model: proliferation inhibition rate = [1 − (A450 nmExperimental group − A450nmblank group) / (A450 nmControl group − A450 nmblank group)] × 100%.
Real‐time RT‐PCR
Total RNA was extracted from LX‐2 cells using an RNA extraction kit (Shanghai Fastagen Biotech, Shanghai, China). Different reaction systems and conditions from Takara Bio were used for the reverse transcription of miR‐29b and other RNAs. Small nuclear RNA U6 was used as an endogenous control for miR‐29b expression. Primer sequences (Sangon Biotech, Shanghai, China) are listed in Table 2. The PCR amplification was performed at 0.5 min at 94 °C followed by 40 cycles of 10 s at 94 °C and 30 s at 60 °C using TaKaRa SYBR Premix TaqTM II (TliRNaseN Plus) kit (TaKaRa Biotechnology Co., Ltd., Dalian, Liaoning, China). Gene expression profiles were calculated using ∆∆Ct (2−∆∆Ct) levels.
Table 2.
Primer sequences
| Gene | Primer | Sequence |
|---|---|---|
| TGF‐β1 | F | 5′‐GTGACAGCAGGGATAACACACTG‐3′ |
| R | 5′‐CATGAATGGTGGCCAGGTC‐3′ | |
| Smad3 | F | 5′‐TCCTGGCTACCTGAGTGAAGA‐3′ |
| R | 5′‐GTTGGGAGACTGGACGAAAA‐3′ | |
| α‐SMA | F | 5′‐CATCATGCGTCTGGATCTGG‐3′ |
| R | 5′‐GGACAATCTCACGCTCAGCA‐3′ | |
| Col‐1 | F | 5′‐CCCGGGTTTCAGAGACAACTTC‐3′ |
| R | 5′‐TCCACATGCTTTATTCCAGCAATC‐3′ | |
| β‐Actin | F | 5′‐TGAAGTACCCCATCGAGCACG‐3′ |
| R | 5′‐CAAACATGATCTGGGTCATCTTCT‐3′ | |
| miR‐29b | RT primer | 5′‐GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAACACTGATT‐3′ |
| F | 5′‐GCGCGCTAGCACCATTTG‐3′ | |
| R | 5′‐CAGTGCAGGGTCCGAGGT‐3′ | |
| Primary miR‐29a/b1 exon 1 | F | 5′‐TACTGAACTGTCACGGCAGA‐3′ |
| R | 5′‐TGTAGTTAGCGACCTCTGCT‐3′ | |
| Primary miR‐29a/b1 exon 4 | F | 5′‐TTGCACCCTCACGACATGCT‐3′ |
| R | 5′‐TGACTCTCAGCAGGCCTCA‐3′ | |
| Primary miR‐29b2/c exon 1 | F | 5′‐ACTTCTTTAGGGGTGTGCGTA‐3′ |
| R | 5′‐ACCCATCTCCCTAGCATTCT‐3′ | |
| Primary miR‐29b2/c exon 6 | F | 5′‐TCAGACTTGCCACCTGGACT‐3′ |
| R | 5′‐AGTTGGCATGAGGCTTCGA‐3′ | |
| U6 | RT primer | 5′‐AACGCTTCACGAATTTGCGT‐3′ |
| U6‐F | 5′‐CTCGCTTCGGCAGCACA‐3′ | |
| U6‐R | 5′‐AACGCTTCACGAATTTGCGT‐3′ |
Western blot
Total cellular protein was extracted in radioimmunoprecipitation assay lysis buffer (1% Nonidet P‐40, 0.1% SDS, 1 mmol/l phenylmethylsulfonyl fluoride, 0.5% sodium deoxycholate, 1 mmol/l sodium orthovanadate and 1 mmol/l sodium fluoride in phosphate‐buffered saline), and the protein concentration was measured using a bicinchoninic acid protein assay kit (Pierce Biotechnology, Rockford, IL, USA). From each sample, 50 µg of protein were subjected to sodium dodecyl sulfate‐polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes (Millipore, Boston, MA, USA). Membranes were incubated with primary antibodies (rabbit monoclonal antibodies against human α‐SMA, phospho‐Smad3 and TGF‐β1, all obtained from Abcam, Cambridge, UK) at 4 °C overnight and secondary antibodies (goat anti‐rabbit IgG, Abcam) for 1 h at room temperature. The specific protein was detected using an enhanced chemiluminescence system (Amersham, Arlington Heights, IL, USA). β‐Actin was used as the loading control in all cases.
Enzyme‐linked immunosorbent assay
Cellular supernatant was collected for col‐1 protein analysis (Wuhan Xinqidi Biological Tech Co., Wuhan, China) after centrifugation of protein extracts at 14 000×g for 30 min. According to the manufacturer's instructions, the sample solution was diluted four times and optical density values were detected at a wavelength of 450 nm using a microplate reader. A standard curve was established to calculate col‐1 concentrations.
Statistical analysis
Results are presented as means ± SD ( ) or percentage (%). Statistical analysis was performed using Student's t‐test, and P < 0.05 was considered statistically significant.
Results
Transfection efficiency
Because transfection efficiency will have a great impact on the interpretation of our results, we first determined transfection efficiency by transfecting cells with 50 nM fluorescein (FAM)‐labelled NC‐siRNA. Observation of increased fluorescence intensity (Figure 1) suggested satisfactory transfection efficiency. We then examined gene expression by real‐time RT‐PCR after transfecting LX‐2 cells with miR‐29b mimics or pSUPER‐Smad3 siRNA (Figure 2). The results showed that Smad3 RNA could be scarcely detected after transfection (Figure 2A), while miR‐29b levels showed a remarkable enhancement after transfection (Figure 2B). p‐SUPER‐Smad3 siRNA was used in the following experiments because it exhibited a stronger inhibitory effect compared with pSUPER‐Smad3‐1309.
Figure 1.
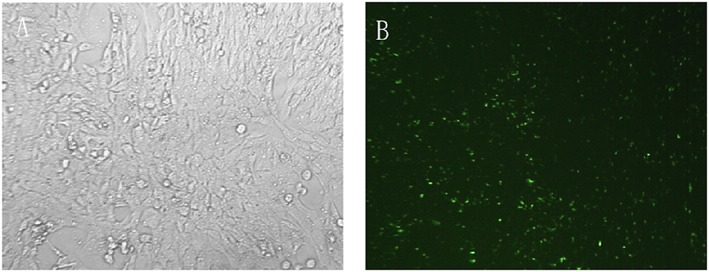
Transfection efficiency. Cultured LX‐2 cells transfected with FAM‐NC siRNA for 12–24 h using X‐tremeGENE siRNA transfection reagent (A). Substantial numbers of green fluorescent granules can be identified inside LX‐2 cells by fluorescence microscopy (B)
Figure 2.
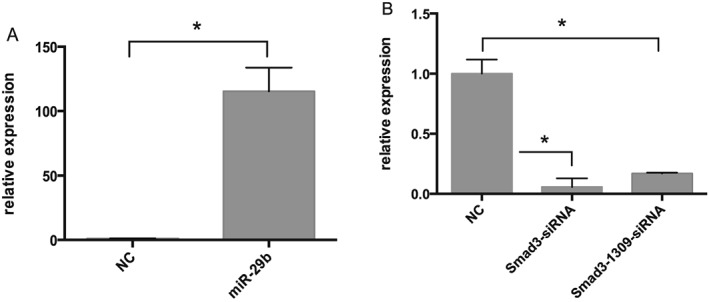
Quantitative measurement of transfection efficiency. Real‐time RT‐PCR revealed that miR‐29b and Smad3 expression were significantly changed 24 h after transfection with miR‐29b mimics, pSUPER‐Smad3 or pSUPER‐Smad3‐1309. Compared with untreated controls, miR‐29b expression was significantly upregulated (A; *P < 0.05), while Smad3 was significantly downregulated (B; *P < 0.05) by both pSUPER‐Smad3 and pSUPER‐Smad3‐1309 transfection
miR‐29b reduces LX‐2 cell proliferation and suppresses col‐1 and α‐SMA expression driven by TGF‐β1
The miRanda software system predicted that the miR‐29 family had almost 7000 target genes,21 some of which are involved in cell proliferation. We therefore utilized the CCK‐8 assay to detect the effects of miR‐29b overexpression on LX‐2 cell proliferation 24, 48 and 72 h after transfection with miR‐29b mimics. Cells transfected with miR‐29b had reduced relative absorbance compared with normal controls (Figure 3A; P < 0.05), with a proliferation inhibition rate of 6.95 ± 1.43% at 24 h (Figure 3B). This effect weakened at 48 and 72 h, showing a limited influence of miR‐29b overexpression on LX‐2 cell proliferation.
Figure 3.
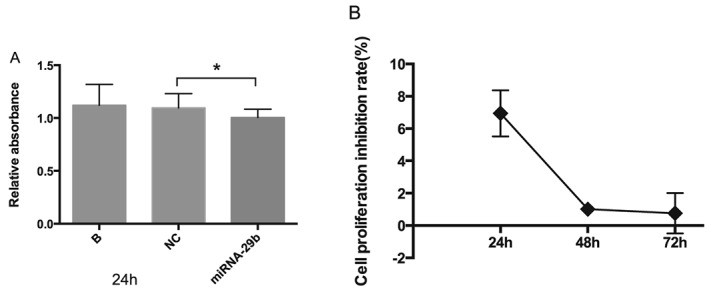
Effects of miR‐29b on LX‐2 cell proliferation. CCK‐8 assays indicated a statistically significant proliferation inhibitory effect of miR‐29b on LX‐2 cells 24 h after transfection (A; *P < 0.05). This effect weakened at 48 and 72 h (B). These data suggest repressive effect of miR‐29b on LX‐2 cell proliferation
The miR‐29 family has been demonstrated to be an important downstream mediator of TGF‐β1‐mediated fibrogenesis in renal tubular epithelium‐like cells and podocytes.22 In order to evaluate the effects of miR‐29b on LX‐2 activation stimulated by TGF‐β1, we transfected LX‐2 cells with miR‐29b mimics, incubated them with TGF‐β1 for 24 h, then examined col‐1 and α‐SMA gene expression by real‐time RT‐PCR. α‐SMA protein levels were detected by western blot, and col‐1 protein levels in cellular supernatants were monitored by ELISA. The influence of Smad3 silencing on col‐1 and α‐SMA expression was also determined. The results showed that both miR‐29b overexpression and Smad3 silencing significantly reduced the upregulation of col‐1 and α‐SMA mRNA by TGF‐β1 (Figure 4A and B; P < 0.05), with comparable effects on col‐1 and α‐SMA protein (Figure 4B, D and E, respectively), suggesting a notable inhibitory effect of miR‐29b on TGF‐β1‐mediated LX‐2 activation and a pivotal role for Smad3 in TGF‐β1‐mediated fibrosis.
Figure 4.
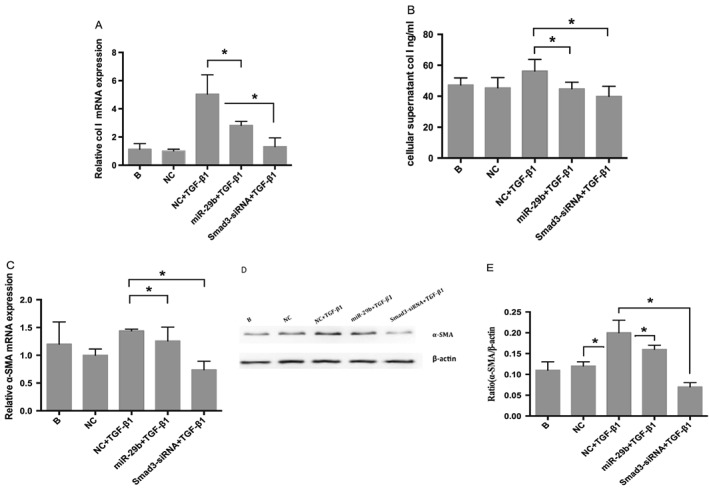
Effects of miR‐29b on expression of col‐1 and α‐SMA. Real‐time RT‐PCR (A, C), ELISA (B) and western blot (D, E) showed that col‐1/α‐SMA were upregulated by TGF‐β1, then significantly downregulated by miR‐29b overexpression or Smad3 silencing (*P < 0.05). Col‐1/α‐SMA levels in the Smad3 silencing group were even lower than the miR‐29b transfection group. The results suggest that miR‐29b has a notable inhibitory effect on TGF‐β1‐mediated LX‐2 activation and that Smad3 plays a key role in TGF‐β1‐mediated fibrosis
miR‐29b inhibits TGF‐β1/Smad3 signalling pathway
In an effort to identify the interaction between miR‐29b and TGF‐β1/Smad3, we simultaneously detected TGF‐β1 and Smad3/p‐Smad3 levels in LX‐2 cells transfected with miR‐29b mimics. The result showed that transfection with miR‐29b resulted in a significant (P < 0.05) decrease in TGF‐β1 (Figure 5A, B and C), Smad3 (Figure 5D) and p‐Smad3 (Figure 5E and F) levels compared with untreated control cells. These results indicate that miR‐29b inhibits both Smad3 and TGF‐β1 expression. To further confirm the contribution of miR‐29 family to TGF‐β1/Smad3 signalling pathway, we also determined the effect of miR‐29a, another miR‐29 family member, on Smad3 and TGF‐β1 expression. The results showed that transfection with miR‐29a also resulted in a significant (P < 0.05) decrease in TGF‐β1 (Figure 6A, B and C), Smad3 (Figure 6D) and p‐Smad3 (Figure 6E and F) levels compared with untreated control cells. These results indicate that miR‐29a inhibits Smad3 and TGF‐β1 expression. Taken together, these results suggest that miR‐29b as a delegate of miR‐29 family inhibits TGF‐β1/Smad3 signalling pathway.
Figure 5.
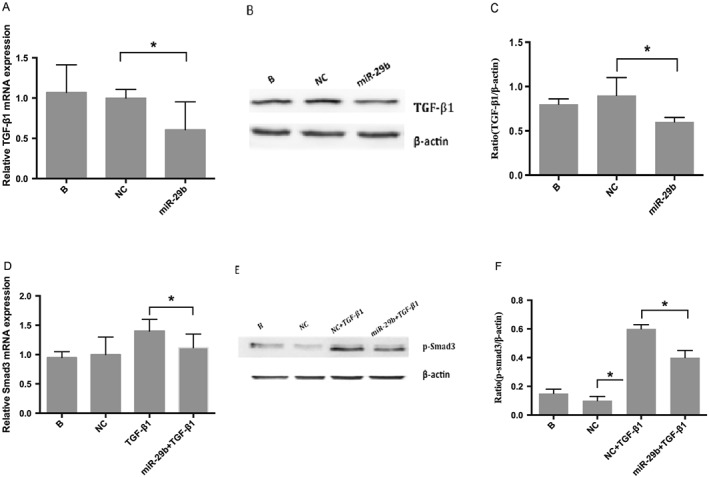
Effects of miR‐29b on TGF‐β1 and Smad3. Real‐time RT‐PCR (A) and western blot (B, C) indicated that TGF‐β1 levels decreased significantly (P < 0.05) after transfection with miR‐29b mimics. Panel D depicts a decrease in Smad3 RNA after transfection with miR‐29b mimics detected by real‐time RT‐PCR, whereas panels E and F reveal a decrease in p‐Smad3 (active form of Smad3) protein examined by western blot
Figure 6.
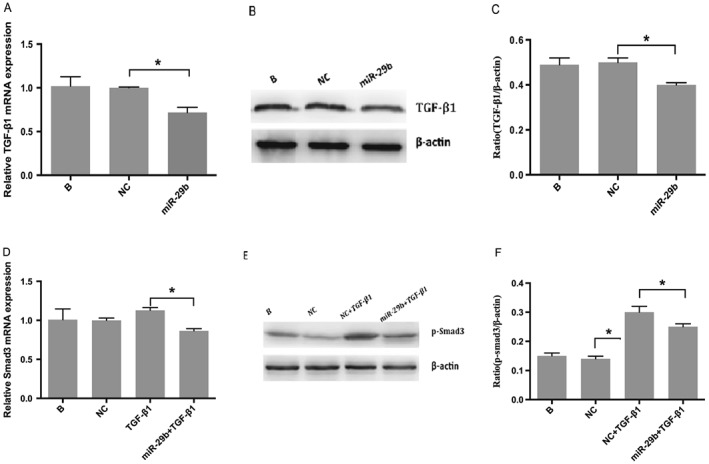
Effects of miR‐29a on TGF‐β1 and Smad3. Real‐time RT‐PCR (A) and western blot (B, C) indicated that TGF‐β1 levels decreased significantly (P < 0.05) after transfection with miR‐29a mimics. Panel D depicts a decrease in Smad3 RNA after transfection with miR‐29b mimics detected by real‐time RT‐PCR, whereas panels E and F reveal a decrease in p‐Smad3 (active form of Smad3) protein examined by western blot
TGF‐β1/Smad3 signalling pathway regulates miR‐29b
Several studies have shown that TGF‐β1 downregulated miR‐29b in vivo and in vitro,19, 22 suggesting that miR‐29b mediated TGF‐β1‐induced LX‐2 activation. In the current work, we not only checked the expression of miR‐29b in TGF‐β1‐stimulated LX‐2 cells but also examined the impact of Smad3 silencing on miR‐29b expression. Mature miR‐29b, primary miR‐29a/b1 and miR‐29b2/c levels were detected respectively by real‐time RT‐PCR. The results showed that the expression of primary miR‐29a/b1 (Figure 7A and B; P < 0.05), miR‐29b2/c (Figure 7C and D; P < 0.05) and mature miR‐29b (Figure 7E; P < 0.05) was significantly downregulated by TGF‐β1 and stimulated by Smad3 silencing. This suggests that TGF‐β1/Smad3 signalling pathway regulate not just mature miR‐29b but also its transcription.
Figure 7.
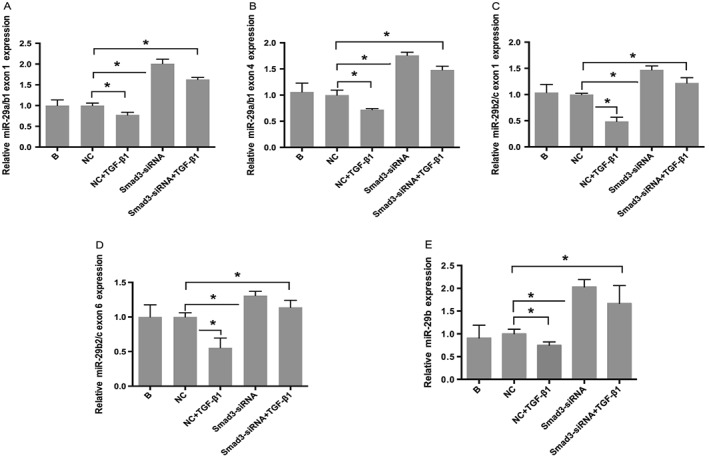
Effects of TGF‐β1/Smad3 on miR‐29b. Real‐time RT‐PCR showed that TGF‐β1 downregulated expression of primary miR‐29a/b1 exon 1 (A), exon 4 (B), primary miR‐29b2/c exon 1 (C), exon 6 (D) and mature miR‐29b (E), which was then significantly upregulated by Smad3 silencing
Discussion
MicroRNAs are noncoding single‐stranded RNA molecules about 22 nucleotides in length, ubiquitous in eukaryotic cells. Previously established knowledge shows that microRNAs modulate approximately one‐third of human genes.23 miR‐29a was first described in 2001 by Lagos‐Quintana and colleagues,24 followed by descriptions of miR‐29b and miR‐29c in the ensuing 2 years.25, 26 Further research demonstrated that the miR‐29 family is closely connected with the fibrotic process because it is downregulated in cardiac,27 pulmonary,28 renal,20 hepatic19 and other29 tissue or organ fibrosis.
It has been previously suggested that microRNAs are highly organ‐ and cell‐type specific.30 Roderburg and colleagues found that the miR‐29b expression level in primary HSCs was more than 100 times that of hepatocytes, Kupffer's cells and endothelial cell,19 suggesting a very intimate connection between miR‐29b and liver fibrosis. Another important study was carried out by Ogawa and colleagues in 2010, in which miR‐29b was found to suppress collagen 1 RNA and protein production in cultured HSCs by directly binding to the 3′ untranslated region of the col‐1 gene.17 In the current study, we detected a suppressive effect of miR‐29b on LX‐2 activation, primarily presented as significant downregulation of not only col‐1 but also α‐SMA expression stimulated by TGF‐β1. In addition, we also examined the effect of miR‐29b on LX‐2 proliferation. miR‐29b inhibited cell proliferation 24 h after transfection of miR‐29b mimics showing a repressive effect on cell proliferation. These data confirm the negative regulation of miR‐29b on HSC, suggesting that miR‐29b may be a downstream inhibitor of TGF‐β1‐induced LX‐2 activation.
TGF‐β/Smad is one of the most important signalling pathways involved in liver fibrosis. TGF‐β1 has been shown to be a key mediator of liver fibrosis by activating its downstream mediators, Smad2 and Smad3, which interact with the kinase‐deficient TGF‐β type I receptor after phosphorylation by TGF‐β type II kinase.31, 32 In recent years, the research community has formed the opinion that it is Smad3 and not Smad2 that plays the key role in TGF‐β1‐mediated fibrosis.33, 34, 35 In our study, we have provided evidence that silencing of Smad3 suppresses col‐1 and α‐SMA expression stimulated by TGF‐β1. This inhibitory effect was far more substantial than the effect of miR‐29b overexpression. The data thus support the theory that Smad3 plays a key role in TGF‐β1‐mediated fibrosis.
Despite multiple research studies, the interactions between miR‐29b and TGF‐β1/Smad3 still remain unclear. Smad3 is predicted to be one of the target genes of miR‐29b by miRanda, PICTAR5 and Targetscan and was also detected by Vincenza Leone in PC C1 3 rat thyroid epithelial cells.36 A target coding sequence for miR‐29b in exon 3 of the TGF‐β1 gene was detected in cardiac fibroblasts in 2014.37 In the current study, we not only detected a suppressive effect of miR‐29b on Smad3 RNA and p‐Smad3 (active form of Smad3) protein but also found decreased TGF‐β1 in the miR‐29b overexpression group. To further confirm the contribution of miR‐29 family to TGF‐β1/Smad3 signalling pathway, we also determined the effect of miR‐29a, another miR‐29 family member, on Smad3 and TGF‐β1 expression. The data showed a similar effect of miR‐29a with miR‐29b on TGF‐β1 and Smad3. The results indicate that miR‐29b inhibits LX‐2 activation by directly suppressing the TGF‐β1/Smad3 signalling pathway in LX‐2 cells.
Despite our results, Roderburg and colleagues found in 2011 that TGF‐β1 downregulated expression of miR‐29b in GRX‐HSC myofibroblasts and primary HSCs of C57BL/6 mice.19 Furthermore, a Smad3 binding site conserved in rat, mouse and in the human miR‐29b2 promoter was detected and confirmed by chromatin immunoprecipitation assay. In contrast, Coralia and colleagues detected an interaction between the miR‐29 and TGF‐β families in trabecular meshwork cells and failed to find any regulatory effect of TGF‐β1 on miR‐29b.18 In the current study, our results showed that expression of primary miR‐29a/b1, miR‐29b2/c and mature miR‐29b was significantly downregulated in LX‐2 cells after incubation with TGF‐β1 for 24 h and was stimulated by Smad3 silencing, which suggests that TGF‐β1/Smad3 signalling pathway regulate not just mature miR‐29b but also its transcription. As a transcription factor, Smad3 likely mediates the inhibitory effect of TGF‐β1 on miR‐29.
In summary, the results of our study demonstrate the suppressive effect of miR‐29b on LX‐2 activation and the essential role of Smad3 in TGF‐β1‐mediated liver fibrosis. The results also reveal that crosstalk between miR‐29b and the TGF‐β1/Smad3 signal transduction pathway is conserved during LX‐2 activation. Smad3 plays an important role in the interaction between miR‐29b and TGF‐β1, promoting a vicious cycle between miR‐29b and TGF‐β1/Smad3 that ultimately stimulates liver fibrosis.
Conflict of Interest
The authors have declared that there is no conflict of interest.
Acknowledgements
This study was supported by research grants from the Science and Technology Commission of Jinshan District Shanghai Municipality (grant number: JSKJ‐KTQN‐2013‐09) and key clinical disciplines construction of Jinshan District(no. JSZK2015A06).
Liang, C. , Bu, S. , and Fan, X. (2016) Suppressive effect of microRNA‐29b on hepatic stellate cell activation and its crosstalk with TGF‐β1/Smad3. Cell Biochem Funct, 34: 326–333. doi: 10.1002/cbf.3193.
References
- 1. Friedman SL. Seminars in medicine of the Beth Israel Hospital, Boston. The cellular basis of hepatic fibrosis. Mechanisms and treatment strategies. N Engl J Med 1993; 328: 1828–1835. [DOI] [PubMed] [Google Scholar]
- 2. Friedman SL. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J Biol Chem 2000; 275(4): 2247–2250. [DOI] [PubMed] [Google Scholar]
- 3. Friedman SL. Cytokines and fibrogenesis. Semin Liver Dis 1999; 19(2): 129–140. [DOI] [PubMed] [Google Scholar]
- 4. Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol 2007; 8(1): 23–36. [DOI] [PubMed] [Google Scholar]
- 5. Lee RC, Feinbaum RL, Ambros V. The C.elegans heterochronic gene lin‐4 encodes small RNAs with antisense complementarity to lin‐14. Cell 1993; 75(5): 843–854. [DOI] [PubMed] [Google Scholar]
- 6. McKiernan PJ, Greene CM. MicroRNA dysregulation in cystic fibrosis. Mediators Inflamm 2015; 2015: 529642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leaman D, Chen PY, Fak J, et al. Antisense‐mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell 2005; 121: 1097–1108. [DOI] [PubMed] [Google Scholar]
- 8. Brennecke J, Hipfner DR, Stark A, et al. Bantam encodes a developmentaaly regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hide in Drosophila. Cell 2003; 113: 25–36. [DOI] [PubMed] [Google Scholar]
- 9. Patel V, Noureddine L. MicroRNAs and fibrosis. Curr Opin Nephrol Hypertens 2012; 21: 410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Breitkopf K, Godoy P, Ciuclan L, et al. TGF‐beta/Smad signaling in the injured liver. Z Gastroenterol 2006; 44: 57–66. [DOI] [PubMed] [Google Scholar]
- 11. Gressner AM, Weiskirchen R. Modern pathogenetic concepts of liver fibrosis suggest stellate cells and TGF‐beta as major players and therapeutic targets. J Cell Mol Med 2006; 10: 76–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schnaper HW, Hayashida T, Hubchak SC, et al. TGF‐beta signal transduction and mesangial cell fibrogenesis. Am J Physiol Renal Physiol 2003; 284(2): 243–252. [DOI] [PubMed] [Google Scholar]
- 13. Usuki J, Matsuda K, Azuma A, et al. Sequential analysis of myofibroblast differentiation and transforming growth factor‐β1/Smad pathway activation in murine pulmonary fibrosis. Nippon Med Sch 2012; 79(1): 46–59. [DOI] [PubMed] [Google Scholar]
- 14. Roberts AB, Tian F, Byfield SD, et al. Smad3 is key to TGF‐beta‐mediated epithelial‐to‐mesenchymal transition, fibrosis, tumor suppression and metastasis. Cytokine Growth Factor Rev 2006; 17: 19–27. [DOI] [PubMed] [Google Scholar]
- 15. Chung AC, Zhang H, Kong YZ, et al. Advanced glycation end‐products induce tubular CTGF via TGF‐beta‐independent Smad3 signaling. J Am Soc Nephrol 2010; 21: 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meng XM, Huang XR, Chung AC, et al. Smad2 protects against TGF‐beta/Smad3‐mediated renal fibrosis. J Am Soc Nephrol 2010; 21: 1477–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ogawa T, Lizuka M, Sekiya Y, et al. Suppression of Type|collagen production by microRNA‐29b in cultured human stellate cells[J]. Biochem Biophys Res Commun 2010; 391(1): 316–321. [DOI] [PubMed] [Google Scholar]
- 18. Luna C, Li G, Qiu J, et al. Cross‐talk between miR‐29 and transforming growth factor‐betas in trabecular meshwork cells(J). Invest Ophthalmol Vis Sci 2011; 52(6): 3567–3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roderburg C, Urban GW, Bettermann K, et al. MicroRNA profiling reveals a role for miR‐29 in human and murine liver fibrosis. Hepatology 2011; 53: 208–218. [DOI] [PubMed] [Google Scholar]
- 20. Qin W, Chung AC, Huang XR, et al. TGF‐β/Smad3 signaling promotes renal fibrosis by inhibiting miR‐29. J Am Soc Nephrol 2011; 22(8): 1462–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. John B, Enright AJ, Aravin A, et al. Human microRNA targets[J]. Cell 2009; 136(2): 215–233.19167326 [Google Scholar]
- 22. Wang B, Komers R, Carew R, et al. Suppression of microRNA‐29 expression by TGF‐β1 promotes collagen expression and renal fibrosis. J Am Soc Nephrol 2012; 23(2): 252–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120: 15–20. [DOI] [PubMed] [Google Scholar]
- 24. Lagos‐Quintana M, Rauhut R, Lendeckel W, et al. Identification of novel genes coding for small expressed RNAs(J). Science 2001; 294(5543): 853–858. [DOI] [PubMed] [Google Scholar]
- 25. Lagos‐Quintana M, Rauhut R, Yalcin A, et al. Idenfication of tissue‐specific microRNAs from mouse(J). Curr Biol 2002; 12(9): 735–739. [DOI] [PubMed] [Google Scholar]
- 26. Dostie J, Mourelatos Z, Yang M, et al. Numerous mircoRNPs in neuronal cells containing novel microRNAs(J). RNA 2003; 9(2): 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Rooij E, Sutherland LB, Thatcher JE, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR‐29 in cardiac fibrosis. Proc Natl Acad Sci U S A 2008; 105(35): 13027–13032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cushing L, Kuang PP, Qian J, et al. MIR‐29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol 2011; 45(2): 287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maurer B, Stanczyk J, Junqel A, et al. MicroRNA‐29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum 2010; 62(6): 1733–1743. [DOI] [PubMed] [Google Scholar]
- 30. Wheeler G, Valoczi A, Havelda Z, et al. In situ detection of animal and plant microRNAs. DNA Cell Biol 2007; 26: 251–255. [DOI] [PubMed] [Google Scholar]
- 31. Heldin CH, Miyazono K, ten Dijke P. TGF‐beta signaling from cell membrane to nucleus through SMAD proteins. Nature 1997; 390(6659): 465–471. [DOI] [PubMed] [Google Scholar]
- 32. Nakao A, Imamura T, Souchelnytskyi S, et al. TGF‐beta receptor‐mediated signaling through Smad2, Smad3 and Smad4. EMBO J 1997; 16(17): 5353–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roberts AB, Tian F, Byfield SD, et al. Smad3 is key to TGF‐beta‐mediated epithelial‐to‐mesenchymal transition, fibrosis, tumor suppression and metastasis. Cytokine Growth Factor Rev 2006; 17: 19–27. [DOI] [PubMed] [Google Scholar]
- 34. Chung AC, Zhang H, Kong YZ, et al. Advanced glycation end‐products induce tubular CTGF via TGF‐beta‐independent Smad3 signaling. J Am Soc Nephrol 2010; 21: 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meng XM, Huang XR, Chung AC, et al. Smad2 protects against TGF‐beta/Smad3‐mediated renal fibrosis. J Am Soc Nephrol 2010; 21: 1477–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Leone V, D'Angelo D, Pallante P, et al. Thyrotropin regulates thyroid cell proliferation by up‐regulating miR‐23b and miR‐29b that target SMAD3. J Clin Endocrinol Metab 2013; 97: 3292–3301. [DOI] [PubMed] [Google Scholar]
- 37. Zhang Y, Wei LH, Huang XR, et al. miR‐29b as a therapeutic agent for angiotensin II‐induced cardiac fibrosis by blocking TGF‐b/Smad3 pathway. Mol Ther 2014; 22: 974–985. [DOI] [PMC free article] [PubMed] [Google Scholar]


