Abstract
Originally developed to study fundamental aspects of cellular biology, high‐content imaging (HCI) was rapidly adapted to study host–pathogen interactions at the cellular level and adopted as a technology of choice to unravel disease biology. HCI platforms allow for the visualization and quantification of discrete phenotypes that cannot be captured using classical screening approaches. A key advantage of high‐content screening technologies lies in the possibility to develop and interrogate physiologically significant, predictive ex vivo disease models that reproduce complex conditions relevant for infection. Here we review and discuss recent advances in HCI technologies and chemical biology approaches that are contributing to an increased understanding of the intricate host–pathogen interrelationship on the cellular level, and which will foster the development of novel therapeutic approaches for the treatment of human bacterial and protozoan infections. © 2016 The Authors. Cytometry Part A published by Wiley Periodicals, Inc. on behalf of ISAC
Keywords: drug discovery, antimicrobial, antiprotozoal, screening, hit identification, chemical biology
Introduction
The microscopic observation of single animalcules (later referred as bacteria and protozoans) by Anton van Leeuwenhoek provided an invaluable approach to observe and comprehend the multitude of microorganisms that surround our environment 1. In the 1870s, Ferdinand Cohn developed a bacterial classification solely based on microscopic observation of the shape of hundreds of bacteria, classification that is still in use today 2. More recently, the development of electron and fluorescence microscopy has revolutionized studies on microbes and host–pathogen interaction. The rapid development of multicolor probes and dyes has been instrumental in comprehending the intricate complexity of the interaction between microbes and a host cell at an unprecedented level. It has now become a routine experiment to use multicolor biological probes to decipher the infection process of infectious protozoans, bacteria, and viruses 3, 4, 5.
High‐content imaging coupled with high‐throughput screening robots and computer‐assisted image analysis, known as high‐content screening (HCS), has recently been adopted by pharmaceuticals and academic organizations as a technology of choice to discover and develop novel anti‐infective drugs. One of the main advantages of the technology is the possibility to screen for drug candidates in physiologically relevant contexts, allowing access to novel chemical entities targeting previously untapped metabolic pathways in both microbes and host cells.
In this review, we present and discuss recent promising HCS chemical biology approaches that are contributing to refuel the global drug pipeline for the treatment of human protozoan, bacterial, and viral infections.
HCI For The Development Of Antibacterial Drugs
The first report on HCI applied to the pathogenesis of intracellular bacteria was reported in 2007 6. A RNA interference screen (RNAi) of the human kinome in MCF7 cell revealed a series of kinases associated with the intracellular survival of Salmonella typhimurium. The screen was designed to monitor the rate of multiplication of a recombinant strain of GFP–S. typhimurium, as well as nuclear and membrane integrity. Genetics or pharmacological inhibition of PKB/akt1 protected infected cells and mice from a Salmonella challenge, validating host‐directed therapy for the treatment of Salmonellosis 6. Interestingly, PKB/akt1 inhibition was also shown to restrict the growth of intracellular mycobacteria, suggesting that host‐targeted therapy may have broad‐spectrum antibacterial activity 6. Other genome‐scale, microscopy‐based SiRNA screens for S. typhimurium have also enabled the identification of the role of novel host cell proteins and associated pathways, such as the COPI complex, in pathogen invasion 7.
The application of HCI to tuberculosis drug discovery opened new avenues to elicit novel host and bacteria targets involved in the intracellular survival of Mycobacterium tuberculosis in macrophages. While M. tuberculosis is a facultative intracellular pathogen, it is well recognized that intracellular mycobacteria are phenotypically resistant to a wide variety of antibiotics 8, 9, making them recalcitrant to drug treatment. Furthermore, intracellular mycobacteria depend on metabolic pathways required for virulence, making the macrophage model a more accurate model in contrast to the classical in vitro culture broth medium for studying the physiology of M. tuberculosis. The antecedent HCS assay to study the macrophage–mycobacteria interaction utilized a similar principle as the one used for S. typhimurium 10. Mouse macrophages were infected with GFP‐M. tuberculosis for 5–7 days, at which point the host nuclei were labelled with the SYTO 60 probe. GFP intensity and distribution as well as nuclei number was recorded on a HCS platform and quantified using proprietary software 6. The assay was applied to several large‐scale compound screenings and was instrumental to the discovery of novel lead series for the treatment of multi‐drug resistant tuberculosis (MDR‐TB) 10, 11. The most advanced compounds discovered on this platform are a series of imidazo‐pyridine amides that targets the mycobacterial bc1 complex 11. The lead drug candidate of the series is Q203 11, a drug candidate for the treatment of MDR‐TB that is currently in phase I clinical trial.
Subsequently, two alternate HCI platforms were further developed for small‐molecules or RNA interference screening in mycobacteria‐infected macrophages 12, 13. Using human macrophages and GFP‐Mycobacterium bovis BCG as a surrogate for M. tuberculosis, Sundaramurthy et al. identified several small‐molecules that trigger the killing of intracellular mycobacteria by modulating either autophagy or endosomal progression 13. Stanley et al. used mouse macrophages and virulent M. tuberculosis to uncover several novel targets that promote intracellular survival 12. Pharmacological inhibition of serotonin reuptake or of the Epidermal Growth Factor Receptor (EGFR) restricted intracellular growth of M. tuberculosis by promoting autophagy. Importantly, the specific EGFR inhibitor gefitinib demonstrated potency in both macrophages and in a mouse model of tuberculosis infection 12.
HCI technology has also started to gain traction as a method of choice to study the host–pathogen interaction of obligate intracellular pathogens such as Coxiella burnetii, the causative agent of Q fever. A HCS screen was performed to identify genes of C. burnetii required for survival inside nonphagocytic cells 14. A collection of 3,000 GFP‐tagged mutants constructed by transposon mutagenesis were used to perform a genetic screen in Vero cells. Designed to identify loci involved in specific steps of the infection process, the study revealed multiples genes essential for intracellular survival, including the first Coxiella invasion involved in host cell invasion 14.
Furthermore, HCI is not solely restricted to the study of intracellular bacteria, and has brought forth a technological solution for analyzing bacterial physiology in axenic culture. The team of John McKinney developed a platform based on microfluidic cultures and time‐lapse microscopy to study mycobacterial physiology at a single‐cell level, advancing HCI outputs beyond conventional average population measurements and accessing intrinsic bacterial stochastic variability in response to stress. In a stimulating application of the approach, the mycobacterial response to the prodrug isoniazid was shown to be influenced by stochastic pulse of expression of the catalase‐peroxidase katG, which activates isoniazid 15. Even more intriguingly, it was demonstrated that drug exposition resulting in a stable number of cells, usually referred as bacteriostatic effect, was in fact because of a dynamic balance of bacterial division and death 15. More recently, the same team demonstrated that single‐cell ATP tracking is a reliable marker of cell viability that can be used to evaluate antimicrobial mode of action 16. The adaptation of this platform to High‐Throughput Screening holds great promise for the development of new classes of antimicrobial agents.
A HCI platform has also been developed to study the architecture of bacteria biofilms and adapted to identify inhibitors of biofilm formation in Vibrio Cholera and Pseudomonas aeruginosa 17, 18, 19, 20. Numerous pathogenic bacteria form biofilm in infected tissues or on medical implant devices, causing clinical challenges because of their intrinsic resistance to antimicrobials. The assay relies on the detection of GFP‐tagged bacteria growing under conditions favoring biofilm formation. Several novel scaffolds that disrupt biofilm formation and increase their susceptibility to classical antibiotics were identified, further validating the therapeutic value of such approach. Of note, the cyclic depsipeptide skyllamycins act as biofilm inhibitors/dispensers in Pseudomonas aeruginosa and eliminate surface‐associated biofilms in combination with the antibiotic azithromycin 20.
The implementation of large‐scale HCS through join collaborations between academic groups, consortia and public–private partnerships represent a promising avenue for anti‐infective drug development. A noteworthy example is the Swiss initiative InfectX, made up of a consortium of eleven research groups with the aim to comprehensively identify components of the human infectome for important bacterial and viral pathogens. Utilizing genome‐wide RNAi and drug screens in combination with HCS to probe for host–pathogen interacting factors, this systems biology approach has enabled the identification of several host drug targets for anti‐infective treatments 21, 22, 23, 24.
HCI For The Development Of Antiprotozoal Drugs
Owing to their large size, HCI is particularly well‐adapted to study the interaction between pathogenic protozoans and their target cell. Multiple features such as number, size, mitochondria, and stage of development of intracellular protozoans can be recorded using commercial or custom‐made software 25. This technique was recently used to identify novel drug candidates for malaria 26. Malaria is caused by several Plasmodium species that are collectively responsible for >500,000 deaths, with several hundred million people at risk. Although falciparum malaria manifests as a blood‐disease, the parasite undergo an essential phase of maturation in the liver. Since the infection of hepatocytes by the sporozoite form is a very inefficient process ex vivo, an image‐based assay is pertinent for drug screening. The HCS assay was developed to visualize the infection process of exo‐erythrocytic Plasmodium yoelii (a surrogate for P. falciparum) in hepatome cells 26. Intracellular P. yoelii sporozoites were detected with αPyHSP70 antibodies, whereas the host nuclei were labeled with nucleic acid stain Hoechst 33342. A custom Acapella software was used for image analysis and processing. Strikingly, preliminary investigations revealed that promising drug candidates had a profound effect on the size of the parasites (but not necessarily on the infection ratio) 26, a feature that can only be captured by image analysis. The imidazolopiperazine drug candidate GNF179 displayed high activity against Plasmodium liver stage in hepatomes and in vivo, most likely by interfering with protein folding 26. A total of 275 hits active against liver‐stage parasites were identified by this approach. The open‐source release of these compounds will presumably fuel early‐stage drug discovery activities for malaria and mode of action studies.
In another sophisticated exemplification of this approach, a HCI assay was developed to decipher the multiple steps of the infection process of the parasites within red blood cells 25. The algorithm was designed to quantify multiple parameters including cell entry, number of intracellular parasites, lengths of the parasites, mitochondria, and stage of parasite development 25 and will therefore be useful to attribute a preliminary mode of action for these new antimalarial compound classes. The assay was recently used to identify and characterize a series of carbohybrid‐based 2‐aminopyrimidine analogues displaying antimalarial activity in red blood cells and in vivo 27. Mode of action studies and image analysis revealed that the series is a class of rapid‐acting antimalarial drug that inhibit schizont maturation and/or merozoite egress from infected cells 27. Notably, likely owing to a new mechanism of action, the series is active against drug resistant clinical isolates 27.
Image‐based assays can also be adapted to complex co‐culture systems to reproduce infectious steps relevant for human infections. Pregnant women and their unborn children form a disproportionately high burden of malaria disease 28 and placental malaria represent an unmet medical need. Recently, a HCI assay was developed to quantify the cytoadhesion of Plasmodium falciparum‐infected erythrocytes at the surface placental BeWo cells 29. The ability to replicate and quantify the initial cytoadhesion of Plasmodium falciparum‐infected erythrocytes in a test tube will likely accelerate the development of novel therapeutics for placental malaria.
Advances in screening approaches for chronic P. vivax malaria have stagnated in recent years, invoking the necessity for technological improvements in this area. P. vivax is less virulent than P. falciparum but can persist for an extensive period of time in the liver and cause recurrent episodes of malaria. Since a major barrier in targeting the liver stage has been the lack of robust, reliable, and reproducible in vitro liver‐stage cultures, attempts have been made to develop HCI assays that remain to be adapted to HCS requirements for the eventual identification of new drugs for the elimination of all P. vivax liver stages 30, 31, 32. These include a pilot study to reproducibly infect hepatoma cells with cryopreserved P. vivax sporozoites for subsequent quantification of the primary outcome variable of infected hepatoma cells 30, the recapitulation of the full liver stage of P. falciparum and the establishment of small forms in late liver stages of P. vivax in a microscale human liver platform 31, and the generation of transgenic fluorescent‐tagged P. cynomolgi for parasite identification in various liver stages, in particular permitting the isolation of hypnozoite‐forms 32.
HCI approaches are also reviving the drug pipeline for Trypanosome cruzii, the etiological agent of Chagas disease. >8 million people are infected globally, with the majority in Latin America in which the disease is endemic. HCI/HTS platforms were built for the screening of drug and siRNA 33, 34, 35, 36. Of particular interest is an assay developed in collaboration between the Diseases of the Developing World research center from GSK and the New York University School of Medicine 34. The HCI assay is based on the detection of cellular and kinetopastid DNA with the DRAQ5 DNA dye and subsequent detection and analysis of multiple parameters of infections using the Acapella High Content Imaging and Analysis Software 34. Since the methodology does not rely on a transgenic parasite, the assay can be adapted to any clinical isolates to screen up to 35,000 compounds per week. The assay was very recently used as a secondary screen to identify novel chemical starting point from a collection of 1.8 million compounds, resulting on the disclosure of 500 novels anti T. cruzi compounds provided as an open resource 37. This unique collection of compounds will most likely contribute to the development of novel therapies for Chagas disease in the coming years. Additionally, another large compound collection is currently being investigated by the Drug for neglected initiatives (DNDi) in the development of another type of HCI/HTS assay 35, 38.
Drug discovery for leishmaniasis, another deadly parasitic disease, has also made significant breakthroughs from progresses in HCI technologies. Leishmaniasis usually manifests as a cutaneous or visceral disease that is transmitted by the bite of a female sandflies. The cutaneous form is the most common, but visceral leishmaniasis is the most severe and causes the death of >20,000 patients every year. Recent HCI assay and innovative image‐analysis scripts were developed and adapted to medium‐throughput screening to quantify the infection process of the amastigote form of Leishmania donovani inside human macrophages 39, 40, 41. In one study, proprietary software was developed to record multiple parameters of infection and cytotoxicity, and validated against a collection of 26,500 small‐molecules 41. Remarkably, the data revealed that up to 50% of the positive hits were inactive against the extracellular promastigote form of the parasite 41, illustrating the relevance of a platform that recapitulate conditions relevant for infection in vivo. A similar conclusion was reached by independent investigators that also advocate for the use of an image‐based intramacrophage assay as a primary screen to identify novel drug candidates 39. Both assays revealed lead series that could represent opportunities for lead optimization programs 39, 42. An alternate HCI assay and associated software for the automated quantification of intracellular Leishmania parasites has also been established and subsequently released as open‐source software 43. The performance of this platform for screening remains to be determined.
Future Prospects
In closing, the technological advancements in HCI is revolutionizing anti‐infective drug discovery; forming an indispensable tool in accelerating drug development for infectious diseases. The various HCI/HCS platforms discussed in this review share common traits; typically enabling the rapid and simultaneous measurement of multiple output parameters of cell‐based assays to evaluate the therapeutic and cytotoxic characteristics of potential anti‐infectives (Fig. 1). Notably, this technique has been instrumental in the drug discovery process for intracellular pathogens by permitting insights into the microcellular environment in response to chemical identities on a large scale basis, which had conventionally been a laborious and impervious challenge to monitor. The ubiquitous applicability of HCS at various stages of the drug discovery process from target identification, small compound primary HTS, hit‐to‐lead progression, to mechanism of action studies have greatly fostered the development of drug candidates to combat the predicative notion of the return to a postantibiotic era 44.
Figure 1.
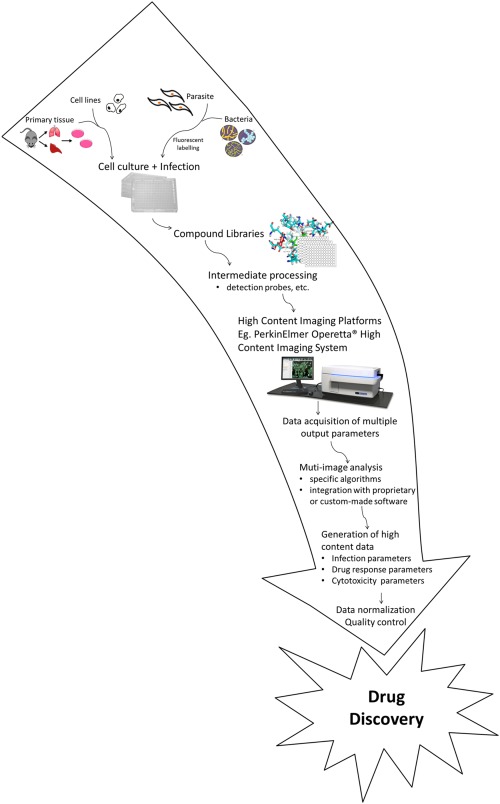
High‐content imaging for the development of anti‐infectives: A generalized workflow. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The recent development of three‐dimensional (3D) culture models and confocal screening technologies will likely be widely utilized in the near future. Transcending conventional two‐dimensional (2D) cell monolayer cultures, the adaptation of miniaturized 3D cell culture platforms and 3D organotypic culture models to HCI enable sophisticated analysis of host–pathogen interaction networks that will likely translate into the discovery of new levels of intervention for drug development. These advanced model systems better simulate the in vivo micro‐environments of human tissues, thus increasing the predictability of drug toxicity and in vivo efficacy 45. Several state‐of‐the‐art high‐throughput imaging systems with associated hardware and software compatible for this transition into 3D models have also been reported 46.
The implementation of HCS for infectious disease research has not just been limited to pharmaceutical companies, but has also become increasingly prevalent over the last five years in academic laboratories. This is due in part to the accessibility of high‐content microscopes and the development of HTS platforms 47. Furthermore, noncomplex multi‐image analyzing tools with built‐in algorithms such as Enhanced CellClassifier have been established to facilitate automated HCS for biologists and academic labs who lack programming skills 48. However, it is also critical to develop advanced data analysis methods in tandem for the analysis of large and multidimensional HCS data sets in order to optimally extract valuable phenotypic information 49. With the rapid advancements in technology and automation, it is anticipated that the next phase of development for HCS will ultimately entail in vivo profiling assays of higher complexity for the investigation of diverse subcellular heterogeneous phenotypes while integrating time dynamics. When utilized appropriately and precisely, the data generated from these platforms will immensely facilitate the assessment of the suitability of candidate compounds for augmentation into successful antibacterial drugs with minimal attrition rates.
Acknowledgments
This research is supported by the Singapore Ministry of Health's National Medical Research Council under its Cooperative Basic Research Grant (Project award no: NMRC/CBRG/0083/2015), and Lee Kong Chian School of Medicine, Nanyang Technological University Start‐Up Grant. The authors declare that they have no conflict of interest.
Literature Cited
- 1. Wollman AJ, Nudd R, Hedlund EG, Leake MC. From Animaculum to single molecules: 300 years of the light microscope. Open Biol 2015; 5: 150019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Drews G. The roots of microbiology and the influence of Ferdinand Cohn on microbiology of the 19th century. FEMS Microbiol Rev 2000; 24:225–249. [DOI] [PubMed] [Google Scholar]
- 3. Melican K, Richter‐Dahlfors A. Multiphoton imaging of host–pathogen interactions. Biotechnol J 2009; 4:804–811. [DOI] [PubMed] [Google Scholar]
- 4. Hoppe AD, Seveau S, Swanson JA. Live cell fluorescence microscopy to study microbial pathogenesis. Cell Microbiol 2009; 11:540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pegoraro G, Bavari S, Panchal RG. Shedding light on filovirus infection with high‐content imaging. Viruses 2012; 4:1354–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kuijl C, Savage ND, Marsman M, Tuin AW, Janssen L, Egan DA, Ketema M, van den Nieuwendijk R, van den Eeden SJ Geluk A, et al. Intracellular bacterial growth is controlled by a kinase network around PKB/AKT1. Nature 2007; 450:725–730. [DOI] [PubMed] [Google Scholar]
- 7. Misselwitz B, Dilling S, Vonaesch P, Sacher R, Snijder B, Schlumberger M, Rout S, Stark M, von Mering C Pelkmans L, et al. RNAi screen of Salmonella invasion shows role of COPI in membrane targeting of cholesterol and Cdc42. Mol Syst Biol 2011; 7:474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boshoff HI, Barry CE 3rd. Tuberculosis—metabolism and respiration in the absence of growth. Nat Rev Microbiol 2005; 3:70–80. [DOI] [PubMed] [Google Scholar]
- 9. Wayne LG, Sohaskey CD. Nonreplicating persistence of mycobacterium tuberculosis. Annu Rev Microbiol 2001; 55:139–163. [DOI] [PubMed] [Google Scholar]
- 10. Christophe T, Jackson M, Jeon HK, Fenistein D, Contreras‐Dominguez M, Kim J, Genovesio A, Carralot JP, Ewann F, Kim EH, et al. High content screening identifies decaprenyl‐phosphoribose 2' epimerase as a target for intracellular antimycobacterial inhibitors. PLoS Pathog 2009; 5:e1000645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pethe K, Bifani P, Jang J, Kang S, Park S, Ahn S, Jiricek J, Jung J, Jeon HK Cechetto J, et al. Discovery of Q203, a potent clinical candidate for the treatment of tuberculosis. Nat Med 2013; 19:1157–1160. [DOI] [PubMed] [Google Scholar]
- 12. Stanley SA, Barczak AK, Silvis MR, Luo SS, Sogi K, Vokes M, Bray MA, Carpenter AE, Moore CB Siddiqi N, et al. Identification of host‐targeted small molecules that restrict intracellular Mycobacterium tuberculosis growth. PLoS Pathog 2014; 10:e1003946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sundaramurthy V, Barsacchi R, Samusik N, Marsico G, Gilleron J, Kalaidzidis I, Meyenhofer F, Bickle M, Kalaidzidis Y, Zerial M. Integration of chemical and RNAi multiparametric profiles identifies triggers of intracellular mycobacterial killing. Cell Host Microbe 2013; 13:129–142. [DOI] [PubMed] [Google Scholar]
- 14. Martinez E, Cantet F, Fava L, Norville I, Bonazzi M. Identification of OmpA, a Coxiella burnetii protein involved in host cell invasion, by multi‐phenotypic high‐content screening. PLoS Pathog 2014; 10:e1004013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wakamoto Y, Dhar N, Chait R, Schneider K, Signorino‐Gelo F, Leibler S, McKinney JD. Dynamic persistence of antibiotic‐stressed mycobacteria. Science 2013; 339:91–95. [DOI] [PubMed] [Google Scholar]
- 16. Maglica Z, Ozdemir E, McKinney JD. Single‐cell tracking reveals antibiotic‐induced changes in mycobacterial energy metabolism. MBio 2015; 6:e02236–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peach KC, Bray WM, Shikuma NJ, Gassner NC, Lokey RS, Yildiz FH, Linington RG. An image‐based 384‐well high‐throughput screening method for the discovery of biofilm inhibitors in Vibrio cholerae. Mol Biosyst 2011; 7:1176–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peach KC, Cheng AT, Oliver AG, Yildiz FH, Linington RG. Discovery and biological characterization of the auromomycin chromophore as an inhibitor of biofilm formation in Vibrio cholerae. Chembiochem 2013; 14:2209–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peach KC, Bray WM, Winslow D, Linington PF, Linington RG. Mechanism of action‐based classification of antibiotics using high‐content bacterial image analysis. Mol Biosyst 2013; 9:1837–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Navarro G, Cheng AT, Peach KC, Bray WM, Bernan VS, Yildiz FH, Linington RG. Image‐based 384‐well high‐throughput screening method for the discovery of skyllamycins A to C as biofilm inhibitors and inducers of biofilm detachment in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2014; 58:1092–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Franceschini A, Meier R, Casanova A, Kreibich S, Daga N, Andritschke D, Dilling S, Rämö P, Emmenlauer M Kaufmann A, et al. Specific inhibition of diverse pathogens in human cells by synthetic microRNA‐like oligonucleotides inferred from RNAi screens. Proc Natl Acad Sci U S A 2014; 111:4548–4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meier R, Franceschini A, Horvath P, Tetard M, Mancini R, von Mering C, Helenius A, Lozach PY. Genome‐wide small interfering RNA screens reveal VAMP3 as a novel host factor required for Uukuniemi virus late penetration. J Virol 2014; 88:8565–8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kühbacher A, Emmenlauer M, Rämo P, Kafai N, Dehio C, Cossart P, Pizarro‐Cerdá J. Genome‐Wide siRNA Screen Identifies Complementary Signaling Pathways Involved in Listeria Infection and Reveals Different Actin Nucleation Mechanisms during Listeria Cell Invasion and Actin Comet Tail Formation. MBio 2015; 6:e00598‐15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mercer J, Snijder B, Sacher R, Burkard C, Bleck CK, Stahlberg H, Pelkmans L, Helenius A. RNAi screening reveals proteasome‐ and Cullin3‐dependent stages in vaccinia virus infection. Cell Rep 2012; 2:1036–1047. [DOI] [PubMed] [Google Scholar]
- 25. Moon S, Lee S, Kim H, Freitas‐Junior LH, Kang M, Ayong L, Hansen MA. An image analysis algorithm for malaria parasite stage classification and viability quantification. PLoS One 2013; 8:e61812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meister S, Plouffe DM, Kuhen KL, Bonamy GM, Wu T, Barnes SW, Bopp SE, Borboa R, Bright AT Che J, et al. Imaging of Plasmodium liver stages to drive next‐generation antimalarial drug discovery. Science 2011; 334:1372–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee S, Lim D, Lee E, Lee N, Lee HG, Cechetto J, Liuzzi M, Freitas‐Junior LH, Song JS Bae MA, et al. Discovery of carbohybrid‐based 2‐aminopyrimidine analogues as a new class of rapid‐acting antimalarial agents using image‐based cytological profiling assay. J Med Chem 2014; 57:7425–7434. [DOI] [PubMed] [Google Scholar]
- 28. Conroy AL, McDonald CR, Kain KC. Malaria in pregnancy: diagnosing infection and identifying fetal risk. Expert Rev anti Infect Ther 2012; 10:1331–1342. [DOI] [PubMed] [Google Scholar]
- 29. Ku MJ, Dossin Fde M, Hansen MA, Genovesio A, Ayong L, Freitas‐Junior LH. An image‐based drug susceptibility assay targeting the placental sequestration of Plasmodium falciparum‐infected erythrocytes. PLoS One 2012; 7:e41765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chattopadhyay R, Velmurugan S, Chakiath C, Andrews Donkor L, Milhous W, Barnwell JW, Collins WE, Hoffman SL. Establishment of an in vitro assay for assessing the effects of drugs on the liver stages of Plasmodium vivax malaria. PLoS One 2010; 5:e14275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. March S, Ng S, Velmurugan S, Galstian A, Shan J, Logan DJ, Carpenter AE, Thomas D, Sim BK Mota MM, et al. A microscale human liver platform that supports the hepatic stages of Plasmodium falciparum and vivax. Cell Host Microb 2013; 14:104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Voorberg‐van der Wel A, Zeeman AM, van Amsterdam SM, van den Berg A, Klooster EJ, Iwanaga S, Janse CJ, van Gemert GJ, Sauerwein R Beenhakker N, et al. Transgenic fluorescent Plasmodium cynomolgi liver stages enable live imaging and purification of Malaria hypnozoite‐forms. PLoS One 2013; 8:e54888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Genovesio A, Giardini MA, Kwon YJ, de Macedo Dossin F, Choi SY, Kim NY, Kim HC, Jung SY, Schenkman S Almeida IC, et al. Visual genome‐wide RNAi screening to identify human host factors required for Trypanosoma cruzi infection. PLoS One 2011; 6:e19733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Alonso‐Padilla J, Cotillo I, Presa JL, Cantizani J, Pena I, Bardera AI, Martin JJ, Rodriguez A. Automated high‐content assay for compounds selectively toxic to Trypanosoma cruzi in a myoblastic cell line. PLoS Negl Trop Dis 2015; 9:e0003493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moon S, Siqueira‐Neto JL, Moraes CB, Yang G, Kang M, Freitas‐Junior LH, Hansen MA. An image‐based algorithm for precise and accurate high throughput assessment of drug activity against the human parasite Trypanosoma cruzi. PLoS One 2014; 9:e87188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Engel JC, Ang KK, Chen S, Arkin MR, McKerrow JH, Doyle PS. Image‐based high‐throughput drug screening targeting the intracellular stage of Trypanosoma cruzi, the agent of Chagas' disease. Antimicrob Agents Chemother 2010; 54:3326–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pena I, Pilar Manzano M, Cantizani J, Kessler A, Alonso‐Padilla J, Bardera AI, Alvarez E, Colmenarejo G, Cotillo I Roquero I, et al. New compound sets identified from high throughput phenotypic screening against three kinetoplastid parasites: an open resource. Sci Rep 2015; 5:8771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Clayton J. Chagas disease: pushing through the pipeline. Nature 2010; 465:S12–S15. [DOI] [PubMed] [Google Scholar]
- 39. De Rycker M, Hallyburton I, Thomas J, Campbell L, Wyllie S, Joshi D, Cameron S, Gilbert IH, Wyatt PG Frearson JA, et al. Comparison of a high‐throughput high‐content intracellular Leishmania donovani assay with an axenic amastigote assay. Antimicrob Agents Chemother 2013; 57:2913–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aulner N, Danckaert A, Rouault‐Hardoin E, Desrivot J, Helynck O, Commere PH, Munier‐Lehmann H, Spath GF, Shorte SL Milon G, et al. High content analysis of primary macrophages hosting proliferating Leishmania amastigotes: application to anti‐leishmanial drug discovery. PLoS Negl Trop Dis 2013; 7:e2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Siqueira‐Neto JL, Moon S, Jang J, Yang G, Lee C, Moon HK, Chatelain E, Genovesio A, Cechetto J, Freitas‐Junior LH. An image‐based high‐content screening assay for compounds targeting intracellular Leishmania donovani amastigotes in human macrophages. PLoS Negl Trop Dis 2012; 6:e1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oh S, Kim S, Kong S, Yang G, Lee N, Han D, Goo J, Siqueira‐Neto JL, Freitas‐Junior LH, Song R. Synthesis and biological evaluation of 2,3‐dihydroimidazo[1,2‐a]benzimidazole derivatives against Leishmania donovani and Trypanosoma cruzi. Eur J Med Chem 2014; 84:395–403. [DOI] [PubMed] [Google Scholar]
- 43. Yazdanparast E, Dos Anjos A, Garcia D, Loeuillet C, Shahbazkia HR, Vergnes B. INsPECT, an open‐source and versatile software for automated quantification of (Leishmania) intracellular parasites. PLoS Negl Trop Dis 2014; 8:e2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Alanis AJ. Resistance to antibiotics: are we in the post‐antibiotic era? Arch Med Res 2005; 36:697–705. [DOI] [PubMed] [Google Scholar]
- 45. Joshi P, Lee MY. High Content Imaging (HCI) on Miniaturized Three‐Dimensional (3D) Cell Cultures. Biosensors (Basel) 2015; 5:768–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li L, Zhou Q, Voss TC, Quick KL, LaBarbera DV. High‐throughput imaging: Focusing in on drug discovery in 3D. Methods 2016; 96:97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mayr LM, Bojanic D. Novel trends in high‐throughput screening. Curr Opin Pharmacol 2009; 9:580–588. [DOI] [PubMed] [Google Scholar]
- 48. Misselwitz B, Strittmatter G, Periaswamy B, Schlumberger MC, Rout S, Horvath P, Kozak K, Hardt WD. Enhanced cell classifier: a multi‐class classification tool for microscopy images. BMC Bioinform 2010; 11:30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Singh S, Carpenter AE, Genovesio A. Increasing the content of high‐content screening: An overview. J Biomol Screen 2014; 19:640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]


