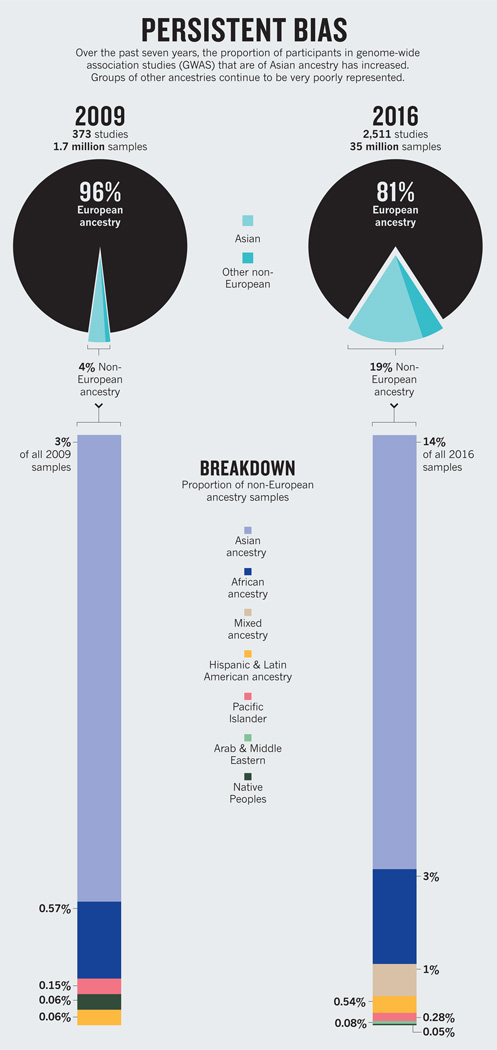
A 2009 analysis revealed that 96% of participants in genome-wide association studies (GWAS) were of European descent1. Such studies scan the genomes of thousands of people to find variants associated with disease traits. The finding prompted warnings that a much broader range of populations should be investigated2 to avoid genomic medicine being of benefit merely to “a privileged few”.
Seven years on, we’ve updated that analysis. Our findings indicate that the proportion of individuals included in GWAS who are not of European descent has increased to nearly 20%. Much of this rise, however, is a result of more studies being done in Asia on populations of Asian ancestry. The degree to which people of African and Latin American ancestry, Hispanic people and indigenous peoples are represented in GWAS has barely shifted.
Thus, more than 20 years after the US National Institutes of Health (NIH) mandated the inclusion of diverse participants in the biomedical research it funds, GWAS funded by the NIH and other sources are continuing to miss a vast portion of the world’s genetic variation.
Over the past decade, GWAS have been the preferred tool for discovering the genetic factors involved in common diseases. Tens of thousands of significant associations between genetic variants and biological traits have now been found, and many of these associations have helped geneticists to uncover biological mechanisms underpinning conditions from diabetes to schizophrenia.
The most comprehensive, publicly accessible summary of human genetic association research is the GWAS Catalog (www.ebi.ac.uk/gwas) produced by the US National Human Genome Research Institute in partnership with the European Bioinformatics Institute. Every week, the curators of the catalogue receive automatic alerts of any new English-language GWAS reported in PubMed. These studies are then put through two rounds of data extraction and validation before being added to the catalogue. Among the data extracted from each study are the race, ethnicity or ancestry (as described by the authors of the study) of the subjects whose samples were analysed.
DATA GATHERING
To determine ancestry, we analysed the sample descriptions included in the GWAS Catalog with an approach similar to that used in 2009 (see Supplementary Information; go.nature.com/2dv2faf).
As of August, 2,511 studies involving nearly 35 million samples were included in the GWAS Catalog. This is a more than 2,000% increase in sample number from the 2009 analysis (which looked at roughly 1.7 million samples across 373 independent studies1.)
We found considerable heterogeneity in descriptions. For example, 26 terms, including ‘black cases’ and ‘sub-Saharan African’, were used to describe people of African ancestry. The most geographically specific and informative descriptions were those used for samples of European origin, as previous studies have shown3.
During the past seven years, the proportion of samples used in catalogued GWAS from participants who are not of European descent has increased fivefold (see ‘Persistent bias’). Yet nearly 78% of this growth is due to an increase in the number of samples from Japan, China, Korea, India and other populations from east Asia, south Asia and southeast Asia.
Together, individuals of African and Latin American ancestry, Hispanic people (individuals descended from Spanish-speaking cultures in central or South America living in the United States) and native or indigenous peoples represent less than 4% of all samples analysed. Collectively, these are the most vulnerable and traditionally underserved populations in many of the world’s richest nations.
The proportion of samples from individuals of African ancestry has increased by 2.5%, and the proportion of people of Hispanic or of Latin American ancestry by around 0.5%. In the case of indigenous peoples (including Native Americans, Australian Aboriginals and Pacific Islanders), representation has decreased slightly since 2009.
By looking up GWAS involving only Asian participants in PubMed (349 studies), we found the institution of the first author of each study. Around 93% of these studies were conducted in Asian countries. That the number of GWAS involving local populations has risen so much in Asia is heartening. But with such a large increase overall in the number of GWAS performed in the past seven years, the lack of growth in representation from other populations is remarkable and deeply disconcerting.
Of course, our analysis does not account for the resampling of data sets across independent studies. Information from some cohorts in publicly available databases has been used multiple times for different GWAS (see Supplementary Information). So the numerous samples of European ancestry used in GWAS could come from a smaller number of actual individuals. Yet if European-ancestry data sets are resampled more often than others, this in itself reflects population-specific differences in research effort.
WHY THE BIAS?
The continuing European bias in GWAS is likely to be the result of logistical, systemic and historical factors.
The more populations that are included in a study, the more variables there are to control for. In trying to keep things as simple as possible, geneticists probably favour the use of existing cohorts, such as that of the Framingham Heart Study, or other large data sets generated by well-established medical centres.
Such organizations collect samples and information from people in the same geographic location, who are presumed to be exposed to shared environmental factors, using uniform collection practices. But for various reasons, some populations are easily bypassed. People may have limited access to certain medical centres, for example, or, for cultural or historical reasons, elect not to contribute their samples to research.
Genotype and phenotype information from diverse populations is available. Researchers using NIH funding are required to submit any such information they have collected to dbGaP, a public database of genotypes and phenotypes. Analogous recommendations are made by other major biomedical funders outside the United States. In Europe, geneticists are encouraged to share similar data through the European Genome-phenome Archive (EGA). Yet for various reasons (such as the difficulties of getting certain kinds of studies funded, a preference for larger sample sizes, a perception that the analysis will be simplified by using data from one ancestry group or a lack of awareness of the diversity of data sets available) geneticists seem to be preferentially using cohorts of European ancestry.
Repeated sampling is almost certainly exacerbating the problem. Indeed, to some degree, the over-representation of people of European ancestry in GWAS may be a legacy of earlier biases.
WHAT’S MISSED
Irrespective of what’s driving it, the continued under-representation of populations of mixed ancestry or of people whose ancestry is not European is a problem.
Until they are able to conduct amply powered GWAS on each major ancestral population across the world, geneticists will continue to miss important information about disease biology. They won’t know how many of the thousands of associations between variants and diseases, and between variants and responses to drugs, observed in populations of European ancestry replicate in other groups. And opportunities will be missed to discover new associations with disease traits in other populations.
For example, for 25% of the variants in European Americans that GWAS have identified as being associated with body mass index, type 2 diabetes and lipid levels, the strength of the association differs in at least one out of five populations of non-European ancestry4. This means that a variant that is associated with diabetes may confer a different risk of disease in someone of European ancestry than in, say, an individual of African ancestry.
Likewise, population-specific differences in the frequencies of variants associated with drug metabolism may mean that certain drugs will be safer and more effective in some populations than in others. The CYP2D6 gene, for instance, is involved in the metabolism of many commonly prescribed drugs, including tamoxifen, which is used to treat breast cancer. More than 100 different variants of this gene (alleles) — many of which affect an individual’s ability to safely digest and use a drug5 — occur at different frequencies in different populations.
Several associations between drug responses and clinically relevant genetic variants have already been identified with GWAS. In some cases in which the effect sizes are large, significant results have been found with as few as 51 cases and 282 controls6. (In this case, patients had different reactions to the lipid-lowering drug simvastatin.) Although physicians must weigh the costs and benefits of using pharmacogenetic testing to guide prescription and dosage decisions for individual patients, these findings suggest that the small samples that have already been collected from under-represented populations could yield leads that have not been identified in populations of European ancestry.
Conducting analyses in other populations is also crucial for assessing the accuracy and broader relevance of a finding. It is possible, for example, that associations between certain disease traits and variants found in European populations that cannot be replicated in other populations are actually false positives. In fact, the analysis of a broader representation of populations can reveal insights that would have otherwise been missed.
A genome-wide scan in a Greenlandic Inuit population, for example, found last year that a single-nucleotide polymorphism (SNP) in a fatty-acid enzyme affects height in both this population and Europeans7. The authors suggest that previous GWAS may have missed this variant because of its low frequency in Europeans (0.017 compared to 0.98 in the Greenlandic Inuit population) — even though it has a much greater effect on height than others previously identified through GWAS.
NEW DIRECTIONS
Increasingly, the sequencing of whole genomes and whole exomes (that is, the complete set of protein-coding genes) are beginning to be used more widely for discovery as costs fall. These may prove more fruitful than GWAS for individual-level diagnosis and treatment. Certainly, they are better suited to revealing rare variations that are clinically informative. (GWAS identify known genetic markers associated with a trait, but not necessarily the mutations that cause the disease.)
Studies that use these new approaches have been slightly more successful than GWAS at recruiting a greater diversity of populations. For example, the international Exome Aggregation Consortium hosts data on genetic variants from more than 60,000 samples, of which 8.6% are from people of African ancestry, 9.5% are from people of Latin American ancestry, and 60.4% are from people of European ancestry8 (see page 154). The remaining samples (21.5%) are from south Asia, east Asia and the Middle East. Similarly, the Trans-Omics for Precision Medicine whole-genome sequencing project of the US National Heart, Lung and Blood Institute is growing and currently holds 62,000 samples, of which 50% are from European Americans, 30% are from African Americans, 10% are from Hispanics or Latin Americans, and 8% are from Asians.
Often, large sample sizes are needed to uncover rare genetic variants associated with disease traits. In fact, this realization — from the first generation of exome discovery studies — is driving new interest in ultracheap genotyping arrays (collections of targeted fragments of DNA). Using such arrays, geneticists can speed up the sequencing process and analyse many targeted samples in one go. Exome sequencing combined with the use of genotyping arrays is likely to be the favoured approach over the next decade. Nonetheless, GWAS remains a useful precursor to such studies, as well as to those involving whole-genome sequencing.
And emerging data indicate that inequalities in health care are being exacerbated by findings from whole-exome and -genome sequencing, despite their greater sample diversity compared with GWAS. Patients of African and Asian ancestry are currently more likely than those of European ancestry to receive ambiguous genetic test results after exome sequencing, or be told that they have variants of unknown significance9. Furthermore, patients of African ancestry are more likely than those of European ancestry to be wrongly told that a mutation they carry increases their risk of developing a life-threatening heart condition known as hypertrophic cardiomyopathy10. Had more ethnically diverse controls been included in the candidate-gene studies that identified these associations, population-specific differences in the frequency of presumed disease-causing variants would have revealed a false positive at the outset.
WHAT NOW?
The message being broadcast by the scientific and medical genomics community to the rest of the world is currently a harmful and misleading one: the genomes of European descendants matter the most.
Certain efforts, combined with newer data-gathering initiatives, can help to move the needle in the right direction. Some investigators in genomics focus exclusively on diverse populations. For instance, landmark trans-ethnic studies have identified genes associated with traits such as diabetes, levels of lipids and other metabolites, prostate cancer and gene expression11. Also, various ventures aim to boost genomics studies in under-represented populations worldwide. The Human Heredity and Health in Africa Consortium, for example, was established by the NIH and the Wellcome Trust in London in 2012 to help build infrastructure and genomics expertise across Africa.
In our view, more fundamental changes are needed — both top-down and bottom-up. Funding agencies should develop financial incentives for the creation of diverse cohorts of study participants. One way for them to do this would be to prioritize grant requests that propose investigations in populations of non-European (and especially of African) ancestry. Given limited budgets, this may need to happen hand in hand with a reduction in the funding of research on existing cohorts of European ancestry for traits and diseases that have been relatively well characterized. (Around 850 genetic associations with height have now been reported by roughly 30 independent GWAS — the vast majority of which have been conducted using individuals of European ancestry.)
Further, all genomics researchers need to recognize the importance of studying under-represented populations to ensure that the benefits of research are distributed fairly and to maximize the potential for discovery. On a practical level, training programmes and new infrastructure, such as good health-care clinics that provide genetic testing in predominantly black or Hispanic neighbourhoods, could enhance trust and allow people to engage in projects as stakeholders rather than as study participants.
A culture shift is required at every level. Efforts to recruit participants for biomedical research in under-represented communities have been most successful when conducted by investigators of concordant racial or ethnic background, and in partnership with institutions trusted by those communities12 — such as historically black colleges and universities in the United States.
Indeed, to a large extent, the persistent bias in sampling in genomics mirrors the employment trends evident in biomedical institutions worldwide. In the United States in 2012, less than 4% of the tenured and tenure-track faculty members in research-intensive biomedical departments were African American, Hispanic or Native American13.
A complex web of historical, cultural, scientific and logistical factors is sustaining an embarrassing bias in genomics. Before precision medicine takes hold in clinical practice, we must correct its course.
Certain drugs may be less effective, or even unsafe, in some populations because of genetic differences.
Terms for ethnicity are those used in the GWAS Catalog. Some have changed between 2009 and 2016 as sampling has increased. Samples of European origin have the most specific descriptions of population ancestry.
A study of Greenlandic Inuits revealed a previously missed genetic variant associated with height.
Contributor Information
Alice B. Popejoy, Email: popejoy@uw.edu, Institute for Public Health Genetics (IPHG) at the University of Washington, Seattle, USA.
Stephanie M. Fullerton, Email: smfllrtn@uw.edu, Bioethics and humanities at the University of Washington, Seattle, USA.
References
- 1.Need AC, Goldstein DB. Trends Genet. 2009;25:489–494. doi: 10.1016/j.tig.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 2.Bustamante CD, De La Vega FM, Burchard EG. Nature. 2011;475:163–165. doi: 10.1038/475163a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fullerton SM, Yu J-H, Crouch J, Fryer-Edwards K, Burke W. Hum. Genet. 2010;127:563–572. doi: 10.1007/s00439-010-0800-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson CS, et al. PLoS Biol. 2013;11:e1001661. doi: 10.1371/journal.pbio.1001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desta Z, Ward BA, Soukhova NV, Flockhart DA. J. Pharmacol. Exp. Ther. 2004;310:1062–1075. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- 6.Daly AK. Nature Rev. Genet. 2010;11:241–246. doi: 10.1038/nrg2751. [DOI] [PubMed] [Google Scholar]
- 7.Fumagalli M, et al. Science. 2015;349:1343–1347. doi: 10.1126/science.aab2319. [DOI] [PubMed] [Google Scholar]
- 8.Lek M, et al. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petrovski S, Goldstein DB. Genome Biol. 2016;17:157. doi: 10.1186/s13059-016-1016-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manrai AK, et al. N. Engl. J. Med. 2016;375:655–665. doi: 10.1056/NEJMsa1507092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li YR, Keating BJ. Genome Med. 2014;6:91. doi: 10.1186/s13073-014-0091-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yancey AK, Ortega AN, Kumanyika SK. Annu. Rev. Public Health. 2006;27:1–28. doi: 10.1146/annurev.publhealth.27.021405.102113. [DOI] [PubMed] [Google Scholar]
- 13.Leboy PS, Madden JF. DNA Cell Biol. 2012;31:1365–1371. doi: 10.1089/dna.2012.1756. [DOI] [PubMed] [Google Scholar]