Abstract
This study was conceived to detect skin mites in social mammals through real-time qPCR, and to estimate taxonomic Demodex and further Prostigmata mite relationships in different host species by comparing sequences from two genes: mitochondrial 16S rRNA and nuclear 18S rRNA. We determined the mite prevalence in the hair follicles of marmots (13%) and bats (17%). The high prevalence found in marmots and bats by sampling only one site on the body may indicate that mites are common inhabitants of their skin. Since we found three different mites (Neuchelacheles sp, Myobia sp and Penthaleus sp) in three bat species (Miotis yumanensis, Miotis californicus and Corynorhinus townsendii) and two different mites (both inferred to be members of the Prostigmata order) in one marmot species (Marmota flaviventris), we tentatively concluded that these skin mites 1) cannot be assigned to the same genus based only on a common host, and 2) seem to evolve according to the specific habitat and/or specific hair and sebaceous gland of the mammalian host. Moreover, two M. yumanensis bats harbored identical Neuchelacheles mites, indicating the possibility of interspecific cross-infection within a colony. However, some skin mites species are less restricted by host species than previously thought. Specifically, Demodex canis seems to be more transmissible across species than other skin mites. D. canis have been found mostly in dogs but also in cats and captive bats. In addition, we report the first case of D. canis infestation in a domestic ferret (Mustela putorius). All these mammalian hosts are related to human activities, and D. canis evolution may be a consequence of this relationship. The monophyletic Demodex clade showing closely related dog and human Demodex sequences also supports this likely hypothesis.
Introduction
Demodex mites are arthropods that belong to the class Arachnida and subclass Acari. Since Simon described the first Demodex in 1942, over 140 Demodex species have been identified in at least 11 orders of domestic and wild mammals [1–3]. Most mammals, including humans harbor Demodex mites on the skin without developing lesions or any other clinical signs [4–6]. However, changes in the host’s cutaneous environment and immune response can lead to mite overgrowth, lesions and other clinical signs [1,7]. Demodex proliferation in hair follicles and glands, such as sebaceous, Meibomian and/or ceruminous glands, can cause a severe and prevalent dermatitis in the host [1,2,8]. Canine and feline demodicosis is a well-known example of severe dermatitis caused by the proliferation of Demodex mites [9]. In humans, an overabundance of Demodex mites has been associated with rosacea, a facial skin disorder that affects well over 16 million Americans [10] (National Rosacea Society-Rosacea.org), although the role played by the mites in the pathogenesis of the disease remains unclear.
In addition to its medical importance, the study of Demodex parasites is of great interest for evolutionary biology. The widespread occurrence of skin mites throughout the mammalian class suggests that the parasitic relationship is very ancient and may have been established soon after the initial radiation of mammals 220 million years ago, when animals with hair follicles first appeared [11,12]. Approximately 100 million years ago, the clade Boreoeutheria split into two sister taxa: Euarchontoglires, including primates and rodents, and Laurasiatheria, constituted mainly by hoofed and carnivorous mammals, including cervids, mustelids, canids and felids but also chiroptera [13]. Demodex mites have been described (both morphologically and genetically) in both groups, reflecting the constancy of the hair follicle niche or the high transmissibility of Demodex mites between and within placental mammal species [5,14–20]. We have an extraordinary example of parasitic parallelism in the Demodex genus [6,12,21,22].
Historically, Demodex mite classification has been primarily based on sharing a common mammalian host and morphological features. For instance, species pairs D. folliculorum and D. brevis found on humans [23], D. canis and D. injai found on dogs [24–26] and D. cati, D. gatoi and D. felis found on cats [27–29] are assumed to be sister groups. More recently, genetic approaches have been used to establish phylogenetic relationships within the family Demodicidae. Specifically, randomly amplified polymorphic DNA (RAPD) has been used to analyze mite variability and relationships [30,31]. In addition, a 930 bp fragment of the mitochondrial Demodex genome was used to evaluate the genetic diversity and phylogeography of D. folliculorum [21]. However, one of the most effective genes known to detect, determine and classify Demodex mites is the mitochondrial 16S rRNA gene (16S rDNA). The taxonomic relationships of Demodex mites have also been estimated using the nuclear 18S rRNA gene [19,22]. While the 16S rRNA gene provides more discrimination at lower taxonomic levels [14,18,20,32,33], the 18S rRNA gene is considered more appropriate for classifications at the level of phyla and superphyla [34,35].
Acari form seven orders, four of which are parasitic: Metastigmata (ticks) and Mesostigmata, Astigmata and Prostigmata (mites). Prostigmata (= Trombidiformes) implies one or two pairs of stigmata in the region of the gnathosoma associated with the chelicerae (commonly referred to as “jaws”) [36]. Prostigmata is one of the most diverse orders within Acariformes. Several morphological and ecological differences reflect this diversity. The feeding ranges from phytophagous to fungivorous, algivorous and saprophagous, and the parasitic habitats affect both vertebrate and invertebrate hosts [36,37]. Prostigmata comprises two large clades, Eupodina (that includes Labidostommatina) and Anystina (that includes Eleutherengona) [38]. The cohort Eleutherengona comprises four superfamilies: Heterostigmata and Cheyletoidea and Tetranychoidea as sister groups, with Raphignathoidea as the sister group to the lineage giving rise to Cheyletoidea and Tetranychoidea [38]. Demodex mites belong to the superfamily Cheyletoidea and family Demodecidae. Finally, the Eupodina clade comprises six superfamilies: Bdelloida and Halacaroidea as sister groups with Labidostommatoidea as the sister group to the lineage and Eriophyoidea and Tydeoidea as sister groups with Eupodoidea [38].
The first aim of this investigation was to determine the prevalence of mites (using 16S rDNA primers that successfully amplified clinical Demodex samples [5,18,32,39]) in both captive and free-ranging wild mammals, including gray wolves (Canis lupus), yellow-bellied marmots (Marmota flaviventris), and four species of vespertillionid bats (Myotis californicus, M. lucifugus, M. yumanenses and Corynorhinus townsendii) distributed throughout North America. We chose these particular species because of their social behavior. Since physical contact among host individuals promotes parasite transmission, we expected high mite prevalence in colonial or social animals [40]. In the wild, wolves maintain a defined social hierarchy, which revolves around the reproductive pair [41]. A single wolf pack consists of a breeding pair and their offspring of the previous one to three years, while in large wolf packs multiple litters can coexist [42]. In contrast, wolves in captivity are often held in enclosures with both close-relatives and strangers. Yellow-bellied marmots are hibernating ground-dwelling squirrels that inhabit subalpine meadows, slopes and clearings [43]. Marmots are social mammals, with colony sizes ranging from one adult male, one adult female and their offspring to large colonies with several breeding females, sometimes including as many as thirty individuals [43,44]. Vespertillionid bats are widely distributed across North America, with summer maternity colony sizes ranging from a few dozen for M. californicus to upwards of several thousand individuals for M. yumanensis and hundreds of thousands for M. lucifugus [45,46]. Myotis spp. frequently forage near or over water (e.g. streams, ponds and lakes). Corynorhinus townsendii are distributed primarily in the western parts of US, Canada and Mexico. Summer maternity colony sizes for C. townsendii rarely reach above 100 individuals and colonies are typically in caves, abandoned mines, hollow trees and abandoned buildings [45]. Because group size has been shown to predict parasite risk in mammals, we expect skin mites to be prevalent within these populations [40].
The second aim was to determine the genetic variability of Demodex spp by comparing sequences from two genes: mitochondrial 16S rRNA and nuclear 18S rRNA. Demodex mites were additionally isolated from domestic dogs (Canis familiaris), cats (Felis catus) and one ferret (Mustela putorius) showing clinical signs and diagnosed with generalized demodicosis. Moreover, we isolated a Demodex folliculorum from a healthy human (Homo sapiens). The results were then used to establish the phylogenetic relationships within mites in social, wild, healthy mammals and unhealthy domestic mammals.
Material and Methods
Ethics Statement
Hair samples from captive gray wolves were collected from the California Wolf Center (CWC) (Julian, California, USA;”33.03236 Latidude, -116.54584 Longitude”), a 501(c)(3) non-profit center dedicated to the recovery of wolves in the wildlands they once roamed. Hair samples were collected in conjunction with annual health checks which CWC staff conducts on each wolf in January to provide vaccines, perform blood and fecal tests and assess overall physical condition. Hair samples from wild yellow-bellied marmots (Marmota flaviventris) were collected in the East River Valley around the Rocky Mountain Biological Laboratory (RMBL) in Gunnison County (Colorado, USA;”38.98831, -107.00979”). Marmots are not protected and they were trapped on publicly owned lands. Once hair samples were extracted, marmots were immediately released unharmed back into their environment. All procedures were approved under research protocol ARC 2001-191-01 as well as permits issued by the Colorado Division of Wildlife. The research protocol was approved by the University of California Los Angeles Animal Care Committee on 13 May 2002 and renewed annually. Hair samples were obtained from vespertillionid bats captured on the San Juan Islands (Washington, USA; “48.55137, -123.07811”), Douglas County (Washington, USA; “47.77906, -119.74746) and Whatcom County (Washington, USA; “48.87872, -121.97187”). Bats are not protected and they were trapped on publicly owned lands. Once hair samples were extracted, bats were immediately released unharmed back into their environment. All procedures were approved by the University of Washington and permits by the Washington department of Fish and Wildlife (IACUC number 4307–01). Hair samples from domestic animals were plucked from one of the co-authors and dogs, cats and a ferret affected by demodicosis. Owners gave their consent for participating in this study. Animals were not harmed in any way during the hair collection.
Sampling
Twenty-two hair samples from healthy captive gray wolves were collected from the CWC. The CWC keeps fourteen Mexican wolves (Canis lupus baileyi) in four different enclosures and eight Alaskan wolves (Canis lupus occidentalis) in a separate enclosure (no wolf remains alone).
Sixteen hair samples from wild yellow-bellied marmots (Marmota flaviventris) were plucked from the rumps of healthy marmots living in the East River Valley around the RMBL in Gunnison County (Colorado, USA). Six sites were sampled corresponding to six different colonies (Table 1). All sampled individuals appeared to be healthy at the time of capture (ranging from May to September 2014), there were no known diseases in the population at the time and no abnormalities were observed.
Table 1. Sample collection.
| Host | Sample | Area | Coll. Year | Collection Place | 16S qPCR Detection | GenBank sequence number 16S | GenBank sequence number 18S | Tree name |
|---|---|---|---|---|---|---|---|---|
| Bat Aa | B01 | Skin | 2014 | Site 6, WAi-US | Positive | KT259444 | KU253783 | mite_bat1UAB |
| Bat A | B02 | Skin | 2014 | Site 6, WA-US | Uncertain | x | x | |
| Bat A | B03 | Skin | 2014 | Site 6, WA-US | Negative | |||
| Bat A | B04 | Skin | 2014 | Site 6, WA-US | Negative | |||
| Bat A | B05 | Skin | 2014 | Site 6, WA-US | Negative | |||
| Bat A | B06 | Skin | 2014 | Site 6, WA-US | Negative | |||
| Bat A | B07 | Skin | 2014 | Site 6, WA-US | Negative | |||
| Bat A | B08 | Skin | 2014 | Site 6, WA-US | Negative | |||
| Bat A | B09 | Skin | 2014 | Site 6, WA-US | Negative | |||
| Bat A | B10 | Skin | 2014 | Site 8, WA-US | Negative | |||
| Bat B | B11 | Skin | 2014 | Site 8, WA-US | Positive | KU253782 | KU253785 | mite_bat3UAB |
| Bat A | B12 | Skin | 2014 | Site 8, WA-US | Positive | KT259444 | KU253783 | mite_bat1UAB |
| Bat A | B13 | Skin | 2014 | Site 8, WA-US | Negative | |||
| Bat B | B14 | Skin | 2014 | Site 7, WA-US | Negative | |||
| Bat A | B15 | Skin | 2014 | Site 6, WA-US | Negative | |||
| Bat A | B16 | Skin | 2014 | Site 6, WA-US | Positive | KT259444 | KU253783 | mite_bat1UAB |
| Bat A | B17 | Skin | 2014 | Site 6, WA-US | Negative | |||
| Bat A | B18 | Skin | 2014 | Site 6, WA-US | Negative | |||
| Bat Dd | B19 | Skin | 2014 | Site 12, WA-US | Positive | KT259445 | KU253784 | mite_bat2UAB |
| Bat D | B20 | Skin | 2014 | Site 12, WA-US | Negative | |||
| Bat A | B21 | Skin | 2014 | Site 11, WA-US | Negative | |||
| Bat Bb | B22 | Skin | 2014 | Site 10, WA- US | Negative | |||
| Bat A | B23 | Skin | 2014 | Site 10, WA- US | Negative | |||
| Bat A | B24 | Skin | 2014 | Site 10, WA- US | Negative | |||
| Bat A | B25 | Skin | 2014 | Site 9, WA-US | Negative | |||
| Bat Cc | B26 | Skin | 2014 | Site 9, WA-US | Negative | |||
| Bat A | B27 | Skin | 2014 | Site 6, WA-US | Negative | |||
| Bat A | B28 | Skin | 2014 | Site 6, WA-US | Negative | |||
| Marmote | M01 | Rump | 2014 | Col1j; CBk, COm-US | Negative | |||
| Marmot | M02 | Rump | 2014 | Col2; CB, CO-US | Positive | KT259446 | x | mite_marmot1UAB |
| Marmot | M03 | Rump | 2014 | Col3; CB, CO-US | Negative | |||
| Marmot | M04 | Rump | 2014 | Col3; CB, CO-US | Negative | |||
| Marmot | M05 | Rump | 2014 | Col1; CB, CO-US | Negative | |||
| Marmot | M06 | Rump | 2014 | Col4; CB, CO-US | Uncertain | x | x | |
| Marmot | M07 | Rump | 2014 | Col1; CB, CO-US | Positive | KT259447 | x | mite_marmot2UAB |
| Marmot | M08 | Rump | 2014 | Col5; CB, CO-US | Negative | |||
| Marmot | M09 | Rump | 2014 | Col1; CB, CO-US | Negative | |||
| Marmot | M10 | Rump | 2014 | Col4; CB, CO-US | Negative | |||
| Marmot | M11 | Rump | 2014 | Col4; CB, CO-US | Negative | |||
| Marmot | M12 | Rump | 2014 | Col4; CB, CO-US | Negative | |||
| Marmot | M13 | Rump | 2014 | Col1; CB, CO-US | Negative | |||
| Marmot | M14 | Rump | 2010 | Col1; CB, CO-US | Uncertain | x | x | |
| Marmot | M15 | Rump | 2010 | Col1; CB, CO-US | Negative | |||
| Marmot | M16 | Rump | 2014 | Col6; CB, CO-US | Negative | |||
| Dogf | D01 | Leg | 2015 | MAn-US | - | JX390978 | KU253790 | D. canisUAB |
| Dog | D02 | Leg | 2015 | MA-US | - | JF784000 | KU253790 | D. canis1/D. canisUAB |
| Dog | D03 | Face/Dorsum | 2015 | RIo-US | - | KT259448 | KU253790 | D. dog3/D. canisUAB |
| Dog | D04 | Dorsum | 2015 | MA-US | - | JX390978 | KU253790 | D. canisUAB |
| Dog | D05 | Face/neck | 2015 | MA-US | - | JX390978 | KU253790 | D. canisUAB |
| Dog | D07 | Thorax | 2015 | MA-US | - | JX390978 | KU253790 | D. canisUAB |
| Dog | D08 | Thorax | 2015 | Grenada, East Caribbean | - | JX390978 | KU253790 | D. canisUAB |
| Dog | DMX149 | Skin | 2012 | UABp-Spain | - | JX390978 | KU253790 | D. canisUAB |
| Dog | DMX169 | Leg | 2014 | Vienna, Austria | - | JX390978 | KU253790 | D. canisUAB |
| Dog | DMX154 | Skin | 2012 | UAB-Spain | - | JX390979 | KU253790 | D. corneiUAB/D. canisUAB |
| Dog | D09 | Dorsum | 2015 | MA-US | - | KT259449 | KU253789 | D. injaiUAB |
| Dog | DMX151 | Skin | 2012 | UAB-Spain | - | JX390980 | KU253789 | D. injaiUAB |
| Dog | DMX168 | Lumbar | 2014 | Vienna, Austria | - | JX390980 | KU253789 | D. injaiUAB |
| Catg | DMX155 | Neck | 2013 | Vienna, Austria | - | KF052996 | KU253786 | D. gatoiUAB |
| Cat | DMX156 | Skin | 2013 | UAB-Spain | - | KF052995 | KU253787 | D. felisUAB |
| Cat | DMX166 | Abdominal/Leg | 2013 | Vienna, Austria | - | JX193759 | KU253788 | D. catiUAB |
| Cat | DMX167 | Face | 2014 | Korneuburg, Austria | - | JX193759 | KU253788 | D. catiUAB |
| Ferreth | D11 | Leg | 2015 | MA-US | - | KU253781 | KU253791 | D. canisUAB |
| Human | DMX152 | Face | 2012 | UAB-Spain | - | JF783995 | KF745889 | D. folliculorumUAB |
aBat A (Myotis yumanensis)
bBat B (Myotis californicus)
cBat C (Myotis lucifugus)
dBat D (Corynorhinus townsendii)
eMarmot (Marmota flaviventris)
fDog (Canis familiaris)
gCat (Felis catus)
hFerret (Mustela putorius)
iWA: Washington
jCol: colony
kCB: Crested Butte
mCO: Colorado
nMA: Massachusetts
oRI: Rhode Island
pUAB: University Autonomous of Barcelona. In bold, new sequences.
Twenty-eight hair samples were obtained from healthy vespertillionid bats captured on the San Juan Islands (Washington, USA) and on the eastern side of Washington State from June to September 2014. Twenty-two samples were collected from M. yumanensis, three samples were from M. californicus, one sample from M. lucifugus and the remaining two samples were from Corynorhinus townsendii. All bats were caught using a mist net at seven different sites (Table 1); however, the number of colonies could not be determined since we did not know where the captured bats roost.
All hair samples were collected with gloved hands or surgical mosquito forceps. Hair was plucked in the direction of growth to include the hair bulb (root) for DNA extraction. Hair samples were obtained from one site on the body in marmots and bats and five sites in wolves. The specific sampling areas of the wolf skin were based on the best areas to detect Demodex mites in dog skin [6,32]: face, dorsum, lumbar, abdomen and feet. In three wolves, we also plucked hair from injury/scab areas. In total, we obtained hair samples from 22 wolves (113 samples), 28 bats and 16 marmots.
Demodex mites from domestic animals were isolated individually from skin scrapings of a healthy human and thirteen dogs, four cats and a ferret, affected by demodicosis, at the Cummings School of Veterinary Medicine at Tufts University (Massachusetts, USA), at the Veterinary School, Universitat Autònoma de Barcelona (UAB) (Spain) and at the Institute of Parasitology, University of Veterinary Medicine, Vienna (Austria) (Table 1 and S1 Table). The mites were examined microscopically and morphologically, and identified as: D. canis and D. canis variant cornei (ten samples; host: dog), D. injai (three samples; host: dog), D. cati (two samples; host: cat), D. gatoi (one sample; host: cat), D. felis (one sample; host: cat), D. folliculorum (one sample; host: human) and Demodex spp similar to D. canis (one sample, from the ferret). All samples were stored at –20°C until DNA extraction.
DNA extraction and real-time PCR (qPCR) amplification
DNA from hair bulb and skin scraping samples was extracted according to Ravera et al. [39]. Briefly, samples were centrifuged in a microcentrifuge at 16,000×g for 15 min. The pellet was re-suspended in 200 μl of digestion buffer (50 mM Tris–HCl, pH 8.5; 1 mM EDTA) and 10 μl of proteinase K solution (10 mg/ml, Roche Life Science). Samples were incubated at 56°C overnight. Then, the proteinase K was inactivated for 10 min at 95°C and the samples were centrifuged at 16,000×g for 15 min. Supernatant was diluted 1:5 for qPCR amplification.
Universal 16S primers, described by Ferreira et al. [5], were used to amplify an approximately 338-bp fragment of the mitochondrial 16S rRNA gene. Hair samples from wolves, marmots and bats were amplified using real-time qPCR. Positive Demodex qPCR controls were obtained from isolated mite DNA that was previously amplified and sequenced. Blank DNA extractions and negative (UltraPure™ Distilled Water, Invitrogen) qPCR controls were also used to detect exogenous DNA contamination. We amplified each sample in duplicate. Real-time qPCRs were carried out under a laminar flow hood in a final volume of 15 μL using FastStart Universal SYBR Green Master (Roche Life Science), 0.5 μM of each primer and 5 μL of 1:5 diluted DNA. Thermal cycling qPCR profiles were 95°C for 10 min followed by 45 cycles at 95°C for 15 s and 60°C for 1 min. Samples were placed in a LightCycler® 480 Multiwell Plate 384 (Roche), and real-time qPCRs were performed in a Roche LightCycler 480 Real-Time PCR System (Roche Life Science). qPCR specificity assessment was performed by adding a Melting Curve analysis at the end of the run. Universal 16S qPCR products showing melting curves between Tm = 72°C and 82°C and amplification cycles between Cp = 25 cycles and Cp = 39 cycles were sequenced.
Isolated Demodex mites from thirteen domestic dogs, four cats, one human and one domestic ferret were amplified using the same primers described above through conventional PCR. We amplified a 338-bp DNA fragment of the mitochondrial 16S rRNA gene. We used the PCR conditions described in Sastre et al. [18]. Briefly, PCR mixture was prepared in a 20μl reaction containing 5 μl of diluted DNA, PCR buffer (x1), 1.5 mmol⁄L MgCl2, 0.2 mmol⁄L of each dNTP, 0.5 μmol/L of each primer and 1 U AmpliTaq Gold DNA Polymerase (Life Technologies Corp.). The thermal cycling profile included 10 min at 94°C, followed by 35 cycles of 94°C (30 s), 57°C (30 s) and 72°C (30 s), and then completed with 10 min at 72°C.
For the 18S rRNA gene, we used conventional PCR to amplify a fragment of approximately 530 bp from the samples that had been sequenced successfully for the 16S rRNA gene. The primer pairs to amplify the fragment were as follows: 18S-F forward 5’-TCCAAGGAAGGCAGCAGGCA-3’ and 18S-R reverse 5’-CGCGGTAGTTCGTCTTGCGACG-3’. We used the same PCR conditions described above except for an increased annealing temperature of 60°C instead of 57°C.
All qPCR and PCR products were sequenced with BIG DyeTM Terminator Cycle Sequencing Ready Reaction Kit, version 3.1 (Life Technologies Corp.) following the manufacturer’s protocol. Sequences were purified using the Montage SEQ96 Sequencing Reaction Cleanup Kit (Millipore, MA, USA) and separated on an ABI PRISM 3730 automated sequencer (Life Technologies Corp.) according to the protocol provided by the manufacturer.
Genetic and phylogenetic analysis
The number of haplotypes (H), nucleotide diversity (π) and number of polymorphic sites (S) were estimated using DNASP 5.10 [47]. The nucleotide composition and pairwise genetic distances within and between populations were estimated with MEGA 6.06 [48]. Since p-distance may underestimate the true genetic distance because some of the nucleotide positions may have experienced multiple substitutions events, pairwise genetic distances were calculated using Kimura 2-parameter distances (K2P) [49]. All ambiguous positions were removed for each sequence pair.
For comparisons and phylogenetic analysis, we used the sequences of several Acari classes belonging to the orders Sarcoptiformes and Trombidiformes published in GenBank. All sequences were examined with Geneious v6.0.4 [50], aligned with Bioedit Sequence Alignment Editor [51] and compared with GenBank database (www.ncbi.nlm.nih.gov/BLAST). Phylogenetic analyses were performed using 25 sequences and 299 bp of the 16S rRNA gene, and 39 sequences and 483 bp of the 18S rRNA gene. For the combination data set (16S rDNA + 18S rDNA) we used 11 sequences and 764 bp. Two outgroups from the class Pycnogonida were used in order to root the trees: Achelia hispida (n: FJ862845) and Ammothea sp. (n: FJ862841) for the 16S rDNA tree, and Achelia echinata (n: AF005438) and Callipallene sp. (n: AF005439) for the 18S rDNA tree.
We used the Bayesian inference program MrBayes v3.2.5 [52] to estimate the phylogenetic trees. MrBayes uses Markov chain Monte Carlo (MCMC) methods to estimate the posterior distribution of model parameters. MODELTEST 3.7 [53,54] was used to select the best evolutionary model among 56 models of evolution by the Akaike Information Criterion. MCMC sampling was performed with 1,000,000 iterations using by default a burn-in of 25%. Bootstrap results are shown as a measures of tree repeatability and accuracy [55]. We then used the program FigTree to display the phylogenies (http://tree.bio.ed.ac.uk/software/figtree/).
Results
Mite prevalence in wolves, bats and marmots using qPCR for the 16S rRNA gene
We screened for mite DNA in hair samples from three social mammal populations using the 16S rRNA gene. We only considered qPCR samples to be positive when the melting curves (Tm) were close to the Demodex control value (Tm = 72°C± 2°C) and successfully sequenced. Samples were considered to be uncertain when the Tm was close to the Demodex control value but the sample was unseccusfully sequenced. The amplification cycle for Demodex qPCR control was on average Cp = 25 cycles. Three out of 22 wolf samples analyzed (14%) were uncertain. These three samples were from Mexican wolves and showed an average Tm = 72°C and Cp = 39 cycles. The most common skin location was the foot (N = 3), and one individual was also uncertain in the pre-auricular facial area. We could not use any mite sequence from wolves for further analyses since we obtained double PCR sequence products from each sample.
Five out of 28 bat samples (17%) were positive by qPCR and one was uncertain (Table 1). Three samples from M. yumanensis showed on average a Tm = 71°C and Cp = 36 cycles. A sample from C. townsendii showed a Tm = 72°C and Cp = 39 cycles. A sample from M. californicus dissociated at Tm = 71°C and Cp = 35 cycles. Finally, one M. yumanensis sample, unsuccessfully sequenced, dissociated at Tm = 73°C and Cp = 39 cycles.
Two out of 16 marmot samples (13%) were positive by qPCR and two were uncertain. Both positive samples showed a Tm = 73°C and Cp = 39 cycles. The other two uncertain samples showed a Tm = 72°C and 74°C and Cp = 39 cycles, respectively.
16S rDNA sequence variability
The mitochondrial 16S rDNA mite fragment from five bats, two marmots, thirteen dogs, four cats, one human and one ferret were successfully sequenced (Table 1 and S1 Table).
None of the mite sequences from bats and marmots were identical to any GenBank sequence previously submitted, including Demodex sequences (Table 1). The sequence called mite_bat1 was found in three different individuals and was submitted to GenBank (n: KT259444). Sequences named mite_bat2, mite_bat3, mite_marmot1 and mite_marmot2 were each found in one individual and submitted to GenBank (n: KT259445 and n: KU253782, n: KT259446 and KT259447, respectively). Mite sequences from seven dogs and the ferret were identical to the sequence of a D. canis obtained in dogs from Spain (Genbank accession number (n): JX390978). As no sequences from Demodex mites isolated from ferrets had been reported so far, we deposited the sequence (coverage and identity of 99%) in GenBank (n: KU253781). One sequence (named D. canis1) corresponded to D. canis found in dogs from China (n: JF784000) and one corresponded to D. canis variant cornei described in dogs from Spain (n: JX390979). Moreover, we found and submitted a new variant of D. canis (named dog3) in a dog sample from Rhode Island, US (n: KT259448), which had 98% identity with D. canis mites found in a Tibetan mastiff from China (n: JF784001). The last three mite sequences from dogs were identical to D. injai found in dogs from Italy and Spain (n: JX390980). Because one of our D. injai sequences was larger than the published sequences, we submitted it to GenBank (n: KT259449). The samples from the four cats corresponded to sequences published previously in GenBank: D. cati (2 samples), n: JX193759; D. gatoi, n: KF052996 and D. felis, n: KF052995. Finally, the D. folliculorum sequence was identical to the sequence of four Chinese D. folliculorum samples (n: JF783995, JF83996, FN42425 and FN42426).
In total, we aligned 25 16S rDNA fragments of 264 bp, including the 13 new sequences and 12 sequences—belonging to the Prostigmata (Trombidiformes) order—retrieved from GenBank (S1 Fig). Twenty-five different haplotypes were identified (14 Demodex mites and a further 11 Prostigmata mites) with nucleotide diversity π = 0.29, the number of polymorphic sites S = 138 and 261 total mutations. The nucleotide variability among D. canis (N = 5) was π = 0.02, D. folliculorum (N = 4) π = 0.02, and Demodex spp in cats (N = 3) π = 0.25. Table in S2 Table shows the overall percentage of adenine (A), cytosine (C), guanine (G) and thymine (T) nucleotides across all samples. The average nucleotide frequencies, excluding gaps, were 32.74% (A), 41.39% (T), 7.41% (C), and 18.46% (G). A bias toward A and T (74.13%; X2(24, N = 25) = 216, p<0.001) was consistent with the nucleotide composition of the mitochondrial DNA among Arthropoda phylum [12,56,57]. The distribution of the K2P distance for the 16S rRNA gene was wide, ranging from 0.004 to 0.828 (S2 Fig). The highest K2P distance was found between samples Diptilomiopus sp. (Eupodina suborder) and mite_marmot1 (Prostigmata order), where the number of nucleotide differences was 102 (up to 264 positions). The lowest K2P distance (one nucleotide substitution) was between D. canis variants, namely: D. canis (UAB) versus D. canis (1) and D. canis (cornei) versus D. canis (2).
18S rDNA sequence variability
The nuclear 18S rDNA mite fragment from five bats, thirteen dogs, four cats, one human and one ferret were successfully sequenced (Table 1 and S1 Table). Unfortunately, we could not obtain the 18S rRNA mite sequences from the two marmot samples.
We obtained ten different sequences (prefix UAB) and submitted nine to GenBank: sample mite_bat1 (N = 3), n: KU253783; mite_bat2, n: KU253784; mite_bat3, n: KU253785; D. canis (N = 10, including the two 16S rDNA-variants cornei and dog3), n: KU253790; D. canis (from the ferret sample) n: KU253791; D. injai (N = 3), n: KU253789; D. cati (N = 2), n: KU253788; D. gatoi, n: KU253786; D. felis, n: KU253787. The D. folliculorum sequence from one of the Spanish co-authors was identical to four obtained from American people and one obtained from a Chinese person (n: KF745889, KF745888, KF745886, KF745881, EU861211 and HQ728000).
In total, we aligned 39 18S rDNA fragments of 469 bp, including nine of our sequences (the sequence from the ferret and the sequences from the ten dogs were the same, named D. canis (UAB)) and 30 sequences belonging to the orders Prostigmata (Trombidiformes; 28) and Sarcoptiformes (two) were retrieved from GenBank (S3 Fig). We identified 39 different haplotypes (twenty-five were Demodex mites), with nucleotide diversity π = 0.1, the number of polymorphic sites S: 178 and 257 total mutations. The nucleotide diversity among D. canis (N = 5) was π = 0.04, D. folliculorum (N = 5) π = 0.02, D. brevis (N = 9) π = 0.01, and Demodex spp in cats (N = 3) π = 0.03.
The nucleotide frequencies were 29.02% (A), 29.61% (T), 17.59% (C), and 23.79% (G) (S3 Table). No bias toward A and T (58.63%; χ2(38, N = 39) = 29, p = 0.83) was observed among Acari sequences for this nuclear gene. The distribution of K2P distance for the 18S rRNA gene was narrower than for the 16S rRNA gene, ranging from 0.002 to 0.301 (S4 Fig). Interestingly, the highest K2P distances were found between Agistemus sp versus D. cati (115 nucleotide substitutions) and Syringophilidae sp (110 nucleotide substitutions), the three species belong of the Anystina suborder. The lowest K2P distances (one nucleotide substitution) were between species from the Demodex genus: D. folliculorum (127) versus D. folliculorum (UAB), D. canis (Dc2) versus D. canis (UAB) and D. brevis (141_3) versus D. brevis (127_5).
Phylogenetic relationships within Prostigmata mites
For the phylogenetic analyses, we used a 299 bp 16S rDNA fragment and a 483 bp 18S rDNA fragment, which include the regions of highest variability [58,59]. For the combination data set (16S rDNA + 18S rDNA), we used 11 sequences and 764 bp. All the trees were rooted using outgroups from the Pycnogonida class (Figs 1, 2 and 3). The transition (TIM+I+G) and the general-time-reversible (GTR+G) models were selected as the best-fitting models for 16S rRNA and 18S rRNA genes respectively, and for the combination data set, the Hasegawa-Kishino-Yano (HKY+G)(16S rRNA) and the Tamura-Nei (TrN+G)(18S rRNA) models were selected; (G: gamma distribution; I: proportion of invariable sites; for model details see Posada and Crandall 1998).
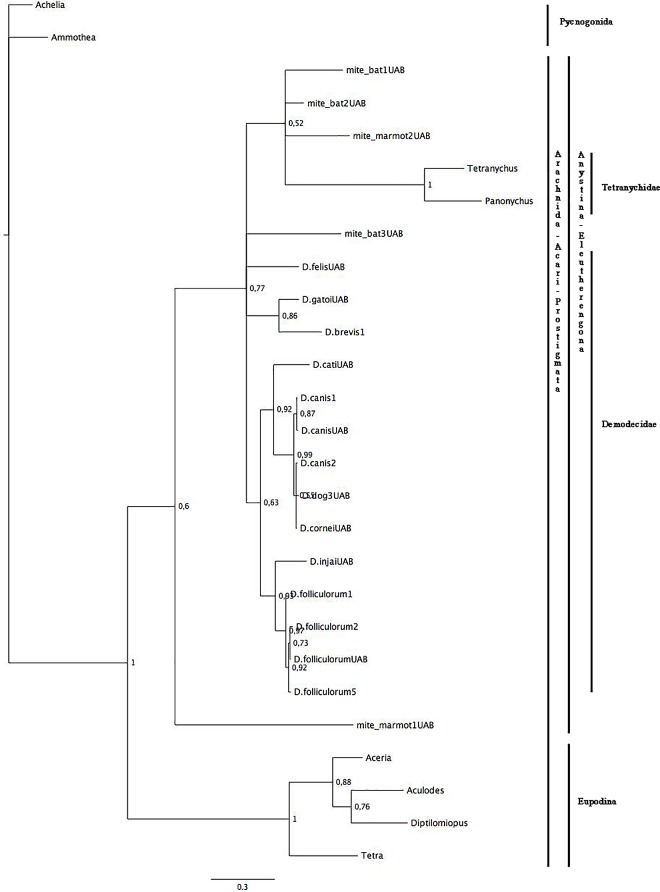
Fig 1. Phylogenetic analyses of Acari.
The tree was estimated using MrBayes based on aligned fragments of the 16S rDNAgene. Branch support is based on 10,000 boostrap replications. The scale at the bottom measures genetic distances in nucleotide substitutions per site.
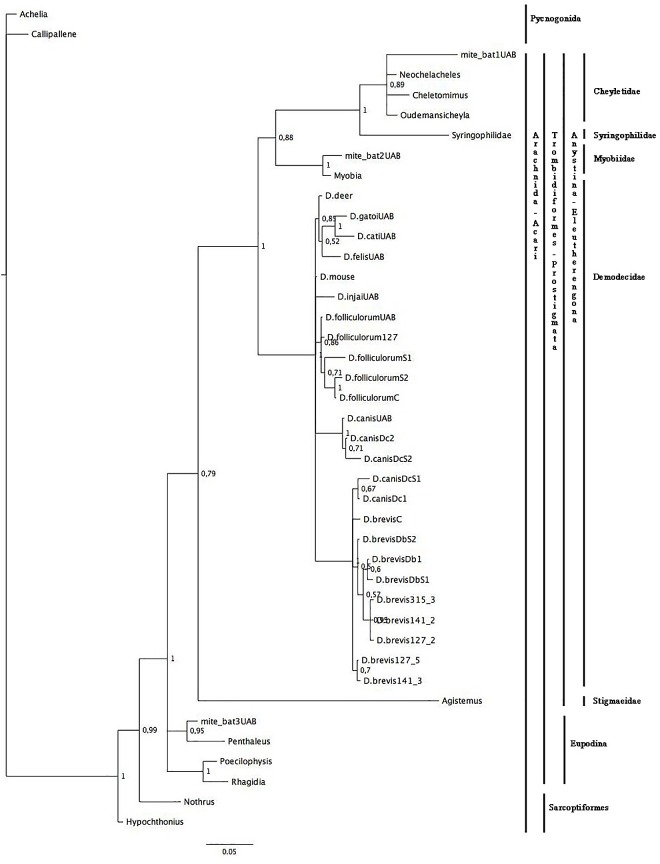
Fig 2. Phylogenetic analyses of Acari.
The three was estimated using MrBayes based on aligned fragments of the 18S rDNA gene. Branch support is based on 10,000 boostrap replications. The scale at the bottom measures genetic distances in nucleotide substitutions per site.
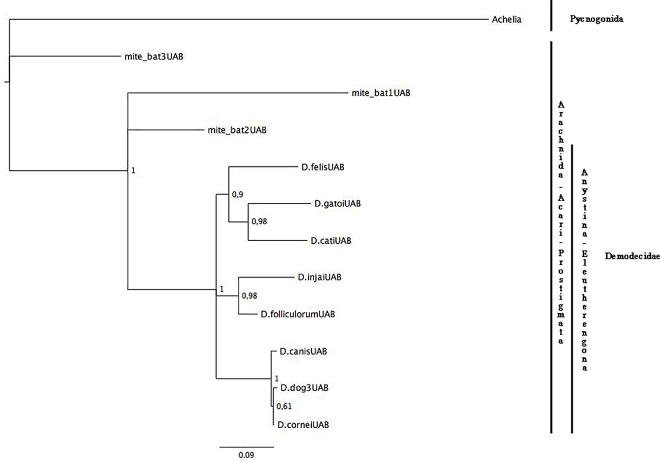
Fig 3. Phylogenetic analyses of Acari.
The tree was estimated using MrBayes based on aligned fragments of combinated 16S rDNA and 18S rDNA genes. Branch support is based on 10,000 boostrap replications. The scale at the bottom measures genetic distances in nucleotide substitutions per site.
The 16S-Bayesian tree (Fig 1) showed a major split between Eupodina and Anystina suborders with bootstrap values >70%, suggesting that all of our samples belong to the Anystina cohort. The second split separated the sample mite_marmot1 from the rest of the samples, suggesting that it does not belong to the Demodecidae or Tetranychidae families. However, due to the low bootstrap value of <70%, this split needs to be evaluated with caution. The third split showed a bootstrap value >70%, but the taxa did not group according to host, namely: 1) samples mite_bat1 and mite_bat2 clustered with sample mite_marmot2 and the Tetranychoidea clade, but not with sample mite_bat3; 2) D. gatoi (host: cat) and D. brevis (host: human) were sister taxa (boostrap value >70%); 3) D. cati (host: cat) and D. canis clade (host: dog) were sister taxa (boostrap value >70%); and finally 4) D. injai (host: dog) and D. folliculorum clade (host: human) were sister taxa (boostrap value >70%).
The 18S-Bayesian tree (Fig 2) showed a clear split (bootstrap value 99%) between the Sarcoptiformes and the Trombidiformes (Prostigmata) mites. Subsequently, Prostigmata split in two clusters: Eupodina and Anystina with bootstrap values >90%. The mite_bat3 sample fell within the Eupodina cluster and was sister to Penthaleus sp. On the other hand, Anystina clustered in five mite families with boostrap values >70%. The deepest split within the Anystina cohort was from the Stigmaeidae family. The mite_bat1 sample appeared to be a member of the family Cheyletidae with Syringophilidae as the sister group, while the mite_bat2 sample appeared to be a member of the Myobiidae family, since it was a sister group to Myobia sp. The monophyletic Demodecidae family included all the Demodex spp. Four large clusters constituted this family: 1) the D. canis cluster; 2) the D. folliculorum cluster; 3) the D. gatoi, D. cati and D. felis cluster (host: cat) (in contrast to the 16S-Bayesian tree), and 4) the D. brevis cluster (host: human) with two D. canis sequences (host: dog).
Due to the limited availability, we only used UAB samples and an outgroup (Achelia sp.) to estimate the 16S+18S Bayesian tree (Fig 3). As the 18S Bayesian tree showed, mite sequences from bats were not included in the Demodecidae family. The deepest split in the tree separated mite_bat3. Mite_bat1 and mite_bat2 were sisters to the monophyletic Demodex cluster. Paradoxically, mite_bat1 and mite_bat3 were collected in close proximity (San Juan County, WA), whereas mite_bat2 was collected over 400 km away (eastern WA). On the other hand, Demodex spp. from cats clustered together, all the three D. canis variants (canis, cornei, dog3) were assigned to the same clade and D. injai and D. folliculorum formed a sister group. Bootstrap values over 90% in most of the nodes show the robustness of the tree.
Sequence analysis: Divergence and interspecific genetic distances between and within Prostigmata mites
The phylogenetic trees did not cluster samples according to host species (Figs 1, 2 and 3). The K2P distances within groups were larger using the 16S rRNA gene (from 0.238 to 0.432) than the 18Sr RNA gene (from 0.025 to 0.068) (S2 and S4 Figs). For instance, within the family Demodecidae, 16S rDNA-K2P = 0.238 (including 14 sequences without sample mite_bat3) and 18S rDNA-K2P = 0.04 (including 25 sequences without sample mite_bat3). In the same way, the Eupodina cohort showed a 16S rDNA-K2P = 0.342 (four sequences) and an 18S rDNA-K2P = 0.069 (including three sequences and sample mite_bat3). The largest 16S rDNA-K2P distance (0.43) was observed within the Tetranychidae family, where samples from mite_bat1, mite_bat2 and mite_marmot2 were included. These results may suggest that these three samples belong to the Eleutherengona cohort, but not to the Tetranychidae family. In fact, according to the 18S rRNA gene (Fig 2), sample mite_bat1 belongs to the Cheyletidae family (N = 4; π = 0.07; K2P = 0.06), sample mite_bat2 belongs to the family Myobiidae (N = 2; π = 0.03; K2P = 0.03) and sample mite_bat3 belongs to the suborder Eupodina (N = 4; π3 = 0.05; K2P = 0.07). Unfortunately, we could not obtain mite sequences from marmots using the 18S rRNA gene. However, we may assume that the two marmot samples belong to the Anystina suborder since the greatest distance between groups was between the Eupodina cohort and sample mite_marmot1 (K2P = 0.74).
Discussion
Mite detection and prevalence in social mammals
We detected the presence of mite DNA in bats (17%) and marmots (12%) through real-time qPCR. The presence of mite DNA in wolves (14%) was uncertain since we obtained qPCR dissociation curves but not mite DNA sequences. Therefore, we recommend to carry out ethanol precipitation during DNA extraction to obtain higher mite DNA quality, and/or designed more specific PCR primers to obtain clear mite sequences from wolf samples. Identifying mites in wolves would help to elucidate whether mite evolution paralleled dog domestication about 15,000 years ago. In contrast, despite only sampling one site on the body, the high mite prevalence in bats and marmots may indicate that mites are common inhabitants of their skin.
16S rDNA and 18S rDNA sequence variability
This research was conceived to detect mites in social mammals by using specific primers that successfully amplified Demodex clinical samples. We found that bats and marmots do not harbor Demodex mites, at least at the limited sites that were sampled; instead, we detected mites from the Prostigmata order. The 16S rDNA-K2P distance between member of the Demodex genus (S2 Fig) ranged between 0.004 and 0.404 (average = 0.24). The 16S rDNA K2P-distance among the three bat samples and between the two marmot samples was on average 0.389 and 0.427, respectively, about two-fold greater than within the genus Demodex. Therefore, mites from bats and marmots cannot be assigned to the same genus based only on a common host. The lack of information in the GenBank database regarding Acari 16S rDNA sequences did not allow for the identification of these five mites. For that reason, we decided to sequence a fragment of the nuclear 18S rRNA gene, since more of these sequences are available in the GenBank database. Using this method, we found that 18S rDNA-K2P distances <7% between mite_bat1 and the family Cheyletidae, between mite_bat2 and the family Myobiidae, and between mite_bat3 and the suborder Eupodina. In general, the genetic distances among species (interspecific) are larger than intraspecific distances. Small genetic distances indicate that they are closely related and share a recent common ancestor. These results are consistent with previous research showing that intraspecific distances range from 0 to 0.05 and interspecific distances from 0.02 to 0.2 [18,60–62]. Therefore, we can conclude that samples from bats do not belong to the Demodex genus, but rather to the genera Neuchelacheles (mite_bat1), Myobia (mite_bat2) or Penthaleus (mite_bat3); these are all Prostigmata mites. We were unable to obtain 18S rDNA mite sequences in marmots; however, we can conclude that the mites found in marmots (as in bats) are not Demodex spp based on 16S rDNA sequences even though we used primers originally designed to detect Demodex mites.
Genetic variability values showed that the nuclear 18S rRNA gene is more conserved than the mitochondrial 16S rRNA gene. Nucleotide diversity, polymorphic sites and number of mutations were about two-fold greater for the 16S than the 18S rRNA gene. We recommend using the 18S rDNA fragment to identify mites for the classification of distantly related members of the Prostigmata order, and the 16S rDNA to discriminate at lower taxonomic levels, for instance among members of the Demodex genus [14,20,33–35].
Demodex mite variability and distribution
The nucleotide diversity using the 16S rRNA gene (π = 0.2) among members of the Demodex genus was five-fold greater than for the 18S rRNA gene (π = 0.04), showing again that it is better to use the 16S rRNA gene to discriminate at lower taxonomic levels. The nucleotide diversity (π) within D. canis, D. folliculorum and D. brevis was low and similar using both the 16S and 18S rRNA genes. However, among our samples (Table 1 and S1 Table), four variants of D. canis (UAB, 1, cornei and dog3) were differentiated using the 16S rRNA gene, but only one using the 18S rRNA gene (D. canis (UAB)). Our D. canis (UAB) sequences from dogs and a ferret were from Austria, Spain, the eastern Caribbean and the USA, and identical (except for one nucleotide) to a dog from Georgia (USA) [19]. Zhao et al. [20,63] found two and six D. canis variants using the 16S rRNA and 18S rRNA genes respectively, in dogs from China. Therefore, we can conclude that the distribution of D. canis (UAB) is wide, occurring in both the Old and the New Worlds. We found similar results for D. injai (host: dog) and D. folliculorum (host: human), which are distributed across Europe, Asia (for D. folliculorum) and the USA. Among Demodex mites in cats, we found significant π value differences between both genes. Nucleotide diversity was almost nine-fold greater using 16S rRNA than 18S rRNA. Paradoxically, despite having the highest π value, the three cats analyzed were exclusively from Europe. Additional cats should be analyzed throughout the world to better understand their mite variability and distribution. Demodex mites in cats were assigned differently according to the gene used. While the 18S rRNA gene and the combination data (16S rDNA + 18S rDNA) clustered D. gatoi, D. cati and D. felis according to their host (cat), the 16S rRNA gene clustered Demodex mites in cats with dogs and humans. Ferreira et al. [5] and Frank et al. [14] found similar results using 16S rDNA. There are no previously published results for cats using the 18S rRNA gene. D. brevis was also clustered differently according to 16S rRNA and 18S rRNA genes, and was closely related to D. gatoi (in agreement with Frank et al. [14]) and to D. canis (in agreement with Thoemmes et al. [22]). In the same way, D. injai was more closely related to D. folliculorum instead of D. canis. As mites in bats and marmots, some Demodex mites did not consistently cluster according to host species.
Demodex, Myobia, Neochelacheles and Penthaleus mites as potential parasites
The phylogenetic structure revealed by the Mr. Bayes analysis of 16S rDNA and 18S rDNA the new mite sequences (prefix UAB) all belong to members of the Prostigmata. Except for samples mite_bat3 and mite_marmot1, all our samples appeared to be members of the Anystina suborder, where most of the mites are known predators or parasites [36]. For instance, Demodex mite overgrowth causes severe and prevalent dermatitis in the host. Demodicosis has been extensively reported in domestic pets and even in humans [1,7,8,64]. Myobia sp is another mite species that infests marsupials, insectivores, bats and rodents [34,65], and Neochelacheles sp has been found in North America and the Philippines where it parasitizes several Tenebrionid beetles [66]. Sample mite_bat3 appeared to be a member of the Eupodina suborder, specifically the Penthaleus genus, considered a major agricultural pest in many temperate areas of the world, including the five continents [67]. Carrier wild hosts often go undiagnosed, allowing infestations to spread. Here, we have demonstrated an effective and easy molecular method to detect potential parasites in wild animals that, without control, can cause pest and disease.
Parasite-host specificity of skin Prostigmata mites?
Although mites are host-specific and usually do not cross-infest, transmission between individuals within the same colony may occur during allogrooming, breeding, fighting, aggregating, or from mother to offspring during birth or nursing [21,68–71]. Transmission does not necessarily result in an individual developing a disease (e.g. demodicosis), as this may occur only in the case of an underlying immune defect. In our study, there were no known diseases in the population at the time. Bats live in colonies ranging from a few dozen to hundreds of thousands of individuals [45,46], while marmot colonies can reach up to thirty individuals [43]. The high variability (one species carrying two different families of mites) in marmots may be partially explained by the fact that the two positive mite samples were taken from two different colonies that are behaviorally and geographically isolated from one another (Table 1). In fact, three positive samples (mite_marmot1 and the two unsuccessfully sequenced samples) were located on the east side of the East River and mite_marmot2 on the west side. More comprehensive surveys within this population could elucidate the patterns of social transmission and show whether mite speciation occurs according to colony rather than host species.
Regarding bats, we found three different mites in three different species of bats. Interestingly, the Myotis genus harbors two closely related mites (Neuchelacheles sp. and Myobia sp.) despite being collected over 400 km away from one another (Douglas County versus San Juan County, WA, USA) (Table 1). In contrast, Penthaleus mites from C. townsendii were collected at the same site and on the same day as Neuchelacheles mites (Douglas County). In this case, the geographic distance is negligible while the phylogenetic relationship is significant, since these mites belong to two different suborders (Anystina versus Eupodina). Therefore, it seems that each bat species harbors a mite that has evolved according to host species. Moreover, two M. yumanensis bats harbored identical Neuchelacheles mites, indicating feasible interspecific cross-infection at site six. Our results do not match with the results from other authors. Desch [72] described morphologically (but not genetically) a new species of hair follicle mite (Demodex nycticeii) in the evening bat (Nycticeius humeralis), and Lankton et al. [16] found a 99.6% homology to D. canis using the same 16S rRNA gene among four captive wild-caught big brown bats (Eptesicus fuscus) in eastern Tennessee (USA). Our bats were also captured in the USA and belong to the same family, Vespertilionidae, but not to the same genus. An additional difference was the captive status of Lankton’s bats.
Therefore, one question arises here: does captivity imply the transmission of Demodex in mammals, namely D. canis? Recently, Izdebska et al. [15] found Demodex mites morphologically similar to D. canis in otters (Lutra lutra). Interestingly, the sample from the ferret, a Mustelidae like the otter, was morphologically and genetically identical to D. canis. D. canis also have been found in cats [5,15,16]. All these mammals are directly related to human activities (e.g. domestication, captivity, hunting, furs). Furthermore, the monophyletic Demodex clade shows closely related dog and human Demodex sequences (Fig 2). Therefore, D. canis mite evolution may be a consequence of the relationship between “human-related” mammals and humans. Thoemmes et al. [22] concluded in their study that D. brevis may have colonized humans from wolves during their domestication. However, it could also be feasible that D. canis may have colonized dogs from humans after their domestication. Additional hair samples from wolves and mammals related to human activities and mite genes (e.g mtDNA COI or rDNA ITS2) should be analyzed to elucidate the evolution, adaptability and transmission of D. canis mites.
Supporting Information
(TIF)
Samples with the prefix UAB are those sequenced in the present study. Those in bold are new sequences. The rest are sequences retrieved from GenBank. Sequence distances were estimated using the Kimura 2-parameter model.
(TIF)
(TIF)
Samples with the prefix UAB are those sequenced from the present study. Those in bold are new sequences. The rest are sequences retrieved from GenBank. Analyses were conducted using the Kimura 2-parameter model.
(TIF)
(XLSX)
Samples with the prefix UAB are those sequenced from the present study.
(XLS)
Samples with the prefix UAB are those sequenced from the present study.
(XLS)
Acknowledgments
We are grateful to the personnel from the California Wolf Center, especially to Norm Switzer, for supplying wolf hair samples. We thank Ivan Ravera and Diana Ferreira from the Veterinary School, Universitat Autònoma de Barcelona for describing morphologically the Demodex samples and Melissa Nashat from the Rockefeller University, Memorial Sloan-Kettering Cancer Center for providing the 18S rDNA primers. Thanks are due also to Robert K. Wayne from the University of California, Los Angeles for supporting this research.
Data Availability
All relevant data are within the paper and its Supporting Information files. All new sequences are available from the GenBank database (accession numbers: KU253781, KU253782, KU253783, KU253784, KU253785, KU253786, KU253787, KU253788, KU253789, KU253790, KU253791, KT259444, KT259445, KT259446, KT259447, KT259448, KT259449).
Funding Statement
The authors received no specific funding for this work.
References
- 1.Lacey N, Raghallaigh S, Powell F. Demodex mites–commensals, parasites or mutualistic organisms? Dermatology. 2011;222: 128–130. 10.1159/000323009 [DOI] [PubMed] [Google Scholar]
- 2.Nutting WB. Hair follicle mites (Demodex spp.) of medical and veterinary concern. Cornell Vet. 1976;66: 214 [PubMed] [Google Scholar]
- 3.Nutting WB, Kettle PR, Tenquist JD, Whitten LK. Hair follicle mites (Demodex spp.) in New Zealand. N Z J Zool. 1975;2: 219–222. 10.1080/03014223.1975.9517871 [DOI] [Google Scholar]
- 4.Beale K. Feline demodicosis: a consideration in the itchy or overgrooming cat. J Feline Med Surg. 2012;14: 209–213. 10.1177/1098612X12439268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferreira D, Sastre N, Ravera I, Altet L, Francino O, Bardagí M, et al. Identification of a third feline Demodex species through partial sequencing of the 16S rDNA and frequency of Demodex species in 74 cats using a PCR assay. Vet Dermatol. 2015;26: 239–e53. 10.1111/vde.12206 [DOI] [PubMed] [Google Scholar]
- 6.Ravera I, Altet L, Francino O, Sánchez A, Roldán W, Villanueva S, et al. Small Demodex populations colonize most parts of the skin of healthy dogs. Vet Dermatol. 2013;24: 168–172.e37. 10.1111/j.1365-3164.2012.01099.x [DOI] [PubMed] [Google Scholar]
- 7.Kligman AM, Christensen MS. Demodex folliculorum: Requirements for Understanding Its Role in Human Skin Disease. J Invest Dermatol. 2011;131: 8–10. 10.1038/jid.2010.335 [DOI] [PubMed] [Google Scholar]
- 8.Scott D, Miller W, Griffin C. Parasitic diseases. Muller & Kirk’s small animal dermatology 6th ed. Philadelphia: WB Saunders; 2001. pp. 457–474. [Google Scholar]
- 9.Mueller RS. Treatment protocols for demodicosis: an evidence-based review. Vet Dermatol. 2004;15: 75–89. 10.1111/j.1365-3164.2004.00344.x [DOI] [PubMed] [Google Scholar]
- 10.Erbagˇci Z, Özgöztaşi O. The significance of Demodex folliculorum density in rosacea. Int J Dermatol. 1998;37: 421–425. 10.1046/j.1365-4362.1998.00218.x [DOI] [PubMed] [Google Scholar]
- 11.Zhou C-F, Wu S, Martin T, Luo Z-X. A Jurassic mammaliaform and the earliest mammalian evolutionary adaptations. Nature. 2013;500: 163–167. 10.1038/nature12429 [DOI] [PubMed] [Google Scholar]
- 12.Palopoli MF, Minot S, Pei D, Satterly A, Endrizzi J. Complete mitochondrial genomes of the human follicle mites Demodex brevis and D. folliculorum: novel gene arrangement, truncated tRNA genes, and ancient divergence between species. BMC Genomics. 2014;15 10.1186/1471-2164-15-1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy WJ, Eizirik E, O’Brien SJ, Madsen O, Scally M, Douady CJ, et al. Resolution of the Early Placental Mammal Radiation Using Bayesian Phylogenetics. Science. 2001;294: 2348–2351. 10.1126/science.1067179 [DOI] [PubMed] [Google Scholar]
- 14.Frank LA, Kania SA, Chung K, Brahmbhatt R. A molecular technique for the detection and differentiation of Demodex mites on cats. Vet Dermatol. 2013;24: 367–e83. 10.1111/vde.12030 [DOI] [PubMed] [Google Scholar]
- 15.Izdebska JN, Rolbiecki L. Demodex lutrae n. sp. (Acari) in European Otter Lutra lutra (Carnivora: Mustelidae) with Data from Other Demodecid Mites in Carnivores. J Parasitol. 2014;100: 784–789. 10.1645/14-532.1 [DOI] [PubMed] [Google Scholar]
- 16.Lankton JS, Chapman A, Ramsay EC, Kania SA, Newkirk KM. Preputial Demodex species in big brown bats (Eptesicus fuscus) in eastern Tennessee. J Zoo Wildl Med Off Publ Am Assoc Zoo Vet. 2013;44: 124–129. [DOI] [PubMed] [Google Scholar]
- 17.de Rojas M, Riazzo C, Callejón R, Guevara D, Cutillas C. Morphobiometrical and molecular study of two populations of Demodex folliculorum from humans. Parasitol Res. 2012;110: 227–233. 10.1007/s00436-011-2476-3 [DOI] [PubMed] [Google Scholar]
- 18.Sastre N, Ravera I, Villanueva S, Altet L, Bardagí M, Sánchez A, et al. Phylogenetic relationships in three species of canine Demodex mite based on partial sequences of mitochondrial 16S rDNA. Vet Dermatol. 2012;23: 509–e101. 10.1111/vde.12001 [DOI] [PubMed] [Google Scholar]
- 19.Yabsley MJ, Clay SE, Gibbs SEJ, Cunningham MW, Austel MG. Morphologic and Molecular Characterization of a Demodex (Acari: Demodicidae) Species from White-Tailed Deer (Odocoileus virginianus). ISRN Parasitol. 2013;2013: 1–7. 10.5402/2013/342918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y-E, Wu L-P. Phylogenetic relationships in Demodex mites (Acari: Demodicidae) based on mitochondrial 16S rDNA partial sequences. Parasitol Res. 2012;111: 1113–1121. 10.1007/s00436-012-2941-7 [DOI] [PubMed] [Google Scholar]
- 21.Palopoli MF, Fergus DJ, Minot S, Pei DT, Simison WB, Fernandez-Silva I, et al. Global divergence of the human follicle mite Demodex folliculorum: Persistent associations between host ancestry and mite lineages. Proc Natl Acad Sci. 2015; 201512609. 10.1073/pnas.1512609112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thoemmes MS, Fergus DJ, Urban J, Trautwein M, Dunn RR. Ubiquity and diversity of human-associated Demodex mites. PLoS ONE. 2014;9: e106265 10.1371/journal.pone.0106265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desch CE, Nutting WB. Demodex folliculorum (Simon) and D. brevis akbulatova of man: redescription and reevaluation. J Parasitol. 1972;58: 169–177. [PubMed] [Google Scholar]
- 24.Alvarez L, Medina OC, García M-E, García H. First report of an unclassified Demodex mite causing demodicosis in a Venezuelan dog. Ann Trop Med Parasitol. 2007;101: 529–532. 10.1179/136485907X193888 [DOI] [PubMed] [Google Scholar]
- 25.Desch CE, Hillier A. Demodex injai: a new species of hair follicle mite (Acari: Demodecidae) from the domestic dog (Canidae). J Med Entomol. 2003;40: 146–149. [DOI] [PubMed] [Google Scholar]
- 26.Saridomichelakis M, Koutinas A, Papadogiannakis E, Papazachariadou M, Liapi M, Trakas D. Adult-onset demodicosis in two dogs due to Demodex canis and a short-tailed demodectic mite. J Small Anim Pract. 1999;40: 529–532. [DOI] [PubMed] [Google Scholar]
- 27.Conroy JD, Healey MC, Bane AG. New Demodex spp. infesting a cat: A case report. J Am Anim Hosp Assoc. 1982;18: 405. [Google Scholar]
- 28.Desch CE, Nutting WB. Demodex cati Hirst 1919: a redescription. Cornell Vet. 1979;69: 280–285. [PubMed] [Google Scholar]
- 29.Löwenstein C, Beck W, Bessmann K, Mueller RS. Feline demodicosis caused by concurrent infestation with Demodex cati and an unnamed species of mite. Vet Rec. 2005;157: 290–292. 10.1136/vr.157.10.290 [DOI] [PubMed] [Google Scholar]
- 30.Toops E, Blagburn B, Lenaghan S, Kennis R, MacDonald J, Dykstra C. Extraction and characterization of DNA from Demodex canis. Int J Appl Res Vet Med. 2010;8: 31–43. [Google Scholar]
- 31.Zhao Y, Cheng H. RAPD analysis and sequence alignment of genomic DNA of hair follicle mites Demodex folliculorum and D. brevis (Acari: Demodicidae). Acta Entomol Sin. 2009;52: 1273–1279. [Google Scholar]
- 32.Sastre N, Ravera I, Ferreira D, Altet L, Sánchez A, Bardagí M, et al. Development of a PCR technique specific for Demodex injai in biological specimens. Parasitol Res. 2013;112: 3369–3372. 10.1007/s00436-013-3531-z [DOI] [PubMed] [Google Scholar]
- 33.Silbermayr K, Horvath-Ungerboeck C, Eigner B, Joachim A, Ferrer L. Phylogenetic relationships and new genetic tools for the detection and discrimination of the three feline Demodex mites. Parasitol Res. 2014;114: 747–752. 10.1007/s00436-014-4243-8 [DOI] [PubMed] [Google Scholar]
- 34.Feldman SH, Ntenda AM. Phylogenetic Analysis of Myobia musculi (Schranck, 1781) by Using the 18S Small Ribosomal Subunit Sequence. Comp Med. 2011;61: 484–491. [PMC free article] [PubMed] [Google Scholar]
- 35.Klompen H, Lekveishvili M, Black IV WC. Phylogeny of parasitiform mites (Acari) based on rRNA. Mol Phylogenet Evol. 2007;43: 936–951. 10.1016/j.ympev.2006.10.024 [DOI] [PubMed] [Google Scholar]
- 36.Woolley TA. Acarology: mites and human welfare. New York; 1988.
- 37.Kethley JB. Acariformes Synopsis and Classification of Living Organisms. New York: McGraw-Hill; 1982. pp. 117–145. [Google Scholar]
- 38.Lindquist EE. 1.5.2 Phylogenetic relationships In: Lindquist MWS E.E. and JB, editor. World Crop Pests. Elsevier; 1996. pp. 301–327. [Google Scholar]
- 39.Ravera I, Altet L, Francino O, Bardagí M, Sánchez A, Ferrer L. Development of a real-time PCR to detect Demodex canis DNA in different tissue samples. Parasitol Res. 2011;108: 305–308. 10.1007/s00436-010-2062-0 [DOI] [PubMed] [Google Scholar]
- 40.Altizer S, Nunn CL, Thrall PH, Gittleman JL, Antonovics J, Cunningham AA, et al. Social organization and parasite risk in mammals: integrating theory and empirical studies. Annu Rev Ecol Evol Syst. 2003; 517–547. [Google Scholar]
- 41.Mech LD. The wolf: the ecology and behavior of an endangerted species Natural History Press, Doubleday; New York; 1970. [Google Scholar]
- 42.Mech LD. Alpha status, dominance, and division of labor in wolf packs. Can J Zool. 1999;77: 1196–1203. [Google Scholar]
- 43.Frase BA, Hoffmann RS. Marmota flaviventris. Mamm Species. 1980;135: 1–8. [Google Scholar]
- 44.Armitage KB. Social and population dynamics of yellow-bellied marmots: results from long-term research. Annu Rev Ecol Syst. 1991; 379–407. [Google Scholar]
- 45.Arroyo-Cabrales J, Álvarez-Castañeda S. Myotis lucifugus. The IUCN Red List of Threatened Species. www.iucnredlist.org. 2008. p. Version 2015.2.
- 46.Brigham RM, Vonhof MJ, Barclay RMR, Gwilliam JC. Roosting Behavior and Roost-Site Preferences of Forest-Dwelling California Bats (Myotis californicus). J Mammal. 1997;78: 1231–1239. 10.2307/1383066 [DOI] [Google Scholar]
- 47.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25: 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 48.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16: 111–120. [DOI] [PubMed] [Google Scholar]
- 50.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28: 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41: 95–98. [Google Scholar]
- 52.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinforma Oxf Engl. 1998;14: 817–818. [DOI] [PubMed] [Google Scholar]
- 54.Swofford DL. Paup*: Phylogenetic analysis using parsimony (and other methods) 4.0. B5. 2001;
- 55.Hillis DM, Bull JJ. An Empirical Test of Bootstrapping as a Method for Assessing Confidence in Phylogenetic Analysis. Syst Biol. 1993;42: 182–192. 10.1093/sysbio/42.2.182 [DOI] [Google Scholar]
- 56.Mangold AJ, Bargues MD, Mas-Coma S. Mitochondrial 16S rDNA sequences and phylogenetic relationships of species of Rhipicephalus and other tick genera among Metastriata (Acari: Ixodidae). Parasitol Res. 1998;84: 478–484. [DOI] [PubMed] [Google Scholar]
- 57.Simon C, Frat F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, Weighting, and Phylogenetic Utility of Mitochondrial Gene Sequences and a Compilation of Conserved Polymerase Chain Reaction Primers. Ann Entomol Soc Am. 1994;87: 651–701. 10.1093/aesa/87.6.651 [DOI] [Google Scholar]
- 58.Klompen H, Lekveishvili M, Black WC. Phylogeny of parasitiform mites (Acari) based on rRNA. Mol Phylogenet Evol. 2007;43: 936–951. 10.1016/j.ympev.2006.10.024 [DOI] [PubMed] [Google Scholar]
- 59.Mangold AJ, Bargues MD, Mas-Coma S. 18S rRNA gene sequences and phylogenetic relationships of European hard-tick species (Acari: Ixodidae). Parasitol Res. 1998;84: 31–37. [DOI] [PubMed] [Google Scholar]
- 60.Hebert PD, Stoeckle MY, Zemlak TS, Francis CM. Identification of birds through DNA barcodes. PLoS Biol. 2004;2: 1657–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nei M. Molecular population genetics and evolution North-Holland Publishing Company; 1975. [PubMed] [Google Scholar]
- 62.Tsao W, Yeh W. DNA-Based Discrimination of Subspecies of Swallowtail Butterflies (Lepidoptera: Papilioninae) from Taiwan. Zool Stud. 2008;47: 633–643. [Google Scholar]
- 63.Zhao Y-E, Xu J-R, Hu L, Wu L-P, Wang Z-H. Complete sequence analysis of 18S rDNA based on genomic DNA extraction from individual Demodex mites (Acari: Demodicidae). Exp Parasitol. 2012;131: 45–51. 10.1016/j.exppara.2012.02.025 [DOI] [PubMed] [Google Scholar]
- 64.Nutting WB, Kettle PR, Tenquist JD, Whitten LK. Hair follicle mites (Demodex spp.) in New Zealand. N Z J Zool. 1975;2: 219–222. [Google Scholar]
- 65.Fain A. Adaptation, specificity and host-parasite coevolution in mites (Acari). Int J Parasitol. 1994;24: 1273–1283. [DOI] [PubMed] [Google Scholar]
- 66.Bochkov AV, OConnor BM. Phylogeny, taxonomy and biology of mites of the genera Chelacheles and Neochelacheles (Acari: Cheyletidae). Invertebr Syst. 2004;18: 547–592. [Google Scholar]
- 67.Umina PA, Hoffmann AA, Weeks AR. Biology, ecology and control of the Penthaleus species complex (Acari: Penthaleidae). Exp Appl Acarol. 2004;34: 211–237. 10.1007/s10493-004-1804-z [DOI] [PubMed] [Google Scholar]
- 68.Cao Y-S, You Q-X, Wang L, Lan H-B, Xu J, Zhang X-H, et al. Facial Demodex infection among college students in Tangshan. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2009;27: 271–273. [PubMed] [Google Scholar]
- 69.Fisher WF, Miller RW, Everett AL. Natural transmission of Demodex Bovis stiles to dairy calves. Vet Parasitol. 1980;7: 233–241. 10.1016/0304-4017(80)90027-8 [DOI] [Google Scholar]
- 70.Garven HSD. Demodex folliculorum in the human nipple. Lancet Lond Engl. 1946;2: 44–46. [DOI] [PubMed] [Google Scholar]
- 71.Greve J, Gaafar S. Natural transmission of Demodex canis in dogs. J Am Vet Med Assoc. 1966;148: 1043–5. [PubMed] [Google Scholar]
- 72.Desch CE. Demodex Nycticeii: a new species of hair follicle mite (Acari: Demodecidae) from the evening bat, Nycticeius humeralis (Chiroptera: Vespertilionidae). Int J Acarol. 1996;22: 187–191. 10.1080/01647959608684094 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
Samples with the prefix UAB are those sequenced in the present study. Those in bold are new sequences. The rest are sequences retrieved from GenBank. Sequence distances were estimated using the Kimura 2-parameter model.
(TIF)
(TIF)
Samples with the prefix UAB are those sequenced from the present study. Those in bold are new sequences. The rest are sequences retrieved from GenBank. Analyses were conducted using the Kimura 2-parameter model.
(TIF)
(XLSX)
Samples with the prefix UAB are those sequenced from the present study.
(XLS)
Samples with the prefix UAB are those sequenced from the present study.
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. All new sequences are available from the GenBank database (accession numbers: KU253781, KU253782, KU253783, KU253784, KU253785, KU253786, KU253787, KU253788, KU253789, KU253790, KU253791, KT259444, KT259445, KT259446, KT259447, KT259448, KT259449).