Abstract
Strains of the species Komagataella phaffii are the most frequently used “Pichia pastoris” strains employed for recombinant protein production as well as studies on peroxisome biogenesis, autophagy and secretory pathway analyses. Genome sequencing of several different P. pastoris strains has provided the foundation for understanding these cellular functions in recent genomics, transcriptomics and proteomics experiments. This experimentation has identified mistakes, gaps and incorrectly annotated open reading frames in the previously published draft genome sequences. Here, a refined reference genome is presented, generated with genome and transcriptome sequencing data from multiple P. pastoris strains. Twelve major sequence gaps from 20 to 6000 base pairs were closed and 5111 out of 5256 putative open reading frames were manually curated and confirmed by RNA-seq and published LC-MS/MS data, including the addition of new open reading frames (ORFs) and a reduction in the number of spliced genes from 797 to 571. One chromosomal fragment of 76 kbp between two previous gaps on chromosome 1 and another 134 kbp fragment at the end of chromosome 4, as well as several shorter fragments needed re-orientation. In total more than 500 positions in the genome have been corrected. This reference genome is presented with new chromosomal numbering, positioning ribosomal repeats at the distal ends of the four chromosomes, and includes predicted chromosomal centromeres as well as the sequence of two linear cytoplasmic plasmids of 13.1 and 9.5 kbp found in some strains of P. pastoris.
Keywords: P. pastoris, genome, splicing, killer plasmid, centromere, RNA-seq
1. Introduction
Methanol utilizing yeast isolates from the Yosemite region of California were used to establish the species Pichia pastoris, and selected by Phillips Petroleum for the large-scale production of single cell protein. Subsequently, P. pastoris clones capable of high cell density growth on simple defined medium in 100,000 liter fermenters were deposited into the yeast culture collections by Phillips Petroleum for patent protection; and work was initiated in collaboration with scientists at the Salk Institute/Biotechnology Associate, SIBIA, to use these P. pastoris strains for the expression of recombinant proteins (Cregg et al., 2009). In 2009, a reclassification dictated that the P. pastoris strains most commonly used around the world for protein production now belong to the species Komagataella phaffii, and include the strains: NRRL Y-11430 from the Agriculture Research Service culture collection (Peoria IL, USA), and NRRL Y-48124 (X-33, Invitrogen expression kit strains, Carlsbad CA, USA) (Kurtzman, 2009, 2005). The same strain deposited in Peoria, IL, as NRRL Y-11430 was also deposited in Utrecht, The Netherlands as CBS7435.
Although the genome sequence of the K. phaffii type strain NRRL Y-7556 (=CBS2612) is not yet known, the first draft P. pastoris genome (De Schutter et al., 2009), and subsequent CBS7435 genomic data (Küberl et al., 2011) have accelerated P. pastoris research. Both sequenced P. pastoris strains, like most gene expression studies, build on the NRRL Y-11430/CBS7435 strain or strains directly derived from those. For example, P. pastoris GS115 was derived from the NRLL Y11430 strain by chemical mutagenesis and selection for histidine auxotrophy (US Patent 4,879,231 A) and became one of the most frequently used P. pastoris strains. More recently we reported the construction of an alcohol oxidase (AOX1) gene knock out (mutS) variant of the CBS7435 strain by homologous recombination and marker recycling employing an FRT/flipper recombinase based strategy (Näätsaari et al., 2012). However, initial sequence data from this CBS7435 strain (aox1-) did not match the published draft genome sequence as expected (i.e. outside of the AOX1 deletion). Although most data of RNA-seq experiments (Liang et al., 2012) and LC-MS/MS based proteomics (Renuse et al., 2014) mapped to predicted open reading frames (ORFs) of the published draft genomes, many were miscalled and additional new ORFs and alternative splice sites were recently identified. Here, state-of-the-art sequencing technologies including long read sequencing is used in resequencing the genome of P. pastoris CBS7435 mutS.
With the advent of next generation sequencing technologies, Sanger-based shotgun sequencing was replaced by massively parallel, short read sequencing methods such as ABI’s SOLiD or Illumina’s Solexa platforms. While this development allowed for higher base coverage and cheaper per base sequencing costs, the assembly of short reads to generate full length genome sequences remained challenging. Current draft genomes (English et al., 2012) including the genomes of P. pastoris CBS7435 (Küberl et al., 2011) and P. pastoris GS115 (Schutter et al., 2009) reflect these previous limitations which can be observed in gaps, insertions, deletions and rearrangements. Typically, repetitive genome features, skewed GC distributions and other genomic complexities limit the methods used for genome sequencing and assembly, giving rise to such errors (Quail et al., 2012a; Roberts et al., 2013).
Currently Pacific Biosciences (PacBio) single molecule, real-time sequencing technology, SMRT, provides an alternative. SMRT sequencing is a sequencing-by-synthesis approach based on the real-time imaging of fluorescently tagged nucleotides which are incorporated by a polymerase affixed at the bottom of a zero-mode waveguide (ZMW) well (Mccarthy, 2010). The advantages offered by this sequencing technology are two-fold. Generally, the average read length of the PacBio RS platform is 8 kb – 15 kb. The availability of such long reads acting as anchoring sequences substantially improves eukaryotic genome assemblies and therefore the generation of high quality full length genome sequences. Compared to other techniques, PacBio is limited by modest per base throughput and a high error rate of approximately 13% observed in raw reads (Quail et al., 2012b); these errors are however corrected for by the increased sequencing depth offered by shorter reads present in the sequencing reaction.
Here new genomic sequence data from multiple closely related P. pastoris strains were combined to provide a new reference genome for this powerful eukaryotic expression system. For the first time, de novo sequencing of P. pastoris strains has been performed employing the Pacific Biosciences RSII platform (PacBio). In relation to the genome sequence published in 2011 (Küberl et al., 2011) deletions, insertions, repeats and larger inversions have been identified. By integrating the PacBio derived de novo genome sequence with new Illumina HiSeq data, as well as more traditional Sanger sequencing data, a first complete P. pastoris reference genome sequence has been generated.
2. Materials and methods
2.1 Strains
Strains used in this study are listed in table 1.
Table 1.
P. pastoris strains used in this study.
| strain | Description | reference |
|---|---|---|
| P. pastoris CBS7435 | wildtype strain received from CBS | (Küberl et al., 2011) |
| P. pastoris CBS7435 mutS | AOX1 knockout derived from P. pastoris CBS7435 | (Näätsaari et al., 2012) |
| P. pastoris CBS7435 Δdas1Δdas2 | das1/das2double knockout derived from P. pastoris CBS7435 | (Geier et al., 2015a) |
| P. pastoris CBS7435 crtEBIY | β-carotene producing strain derived from P. pastoris CBS7435 | (Geier et al., 2014) |
| P. pastoris PPY12 | his4 arg4 auxotrophic strain | (Gouldi et al., 1992) |
| P. pastoris BG08 | BioGrammatics Inc. | |
| P. pastoris BG10 | BioGrammatics Inc. | Cat. No. PS001-01 |
2.2 Strain Cultivation and DNA extraction
P. pastoris CBS7435 mutS (Näätsaari et al., 2012) and related strains overexpressing the s-carotene biosynthesis pathway from Pantoea ananatis (Geier et al. 2015) were grown overnight in 50 ml YPD medium (20 g/L peptone, 10 g/L yeast extract, 20 g/L dextrose) at 28°C and 120 rpm. 1.5x109 cells were removed from the supernatant by centrifugation and washed once with TE Buffer before resuspension in 1 ml yeast lysis buffer (1 M sorbitol, 100 mM EDTA, 14 mM β-mercaptoethanol). Spheroplasts were generated by addition of 100 μL of a zymolyase stock solution (1000 U/ml) and incubation of the suspension at 30°C for 30 min. Spheroplasts, pelleted by centrifugation at 3220xg, 4°C for 10 min were resuspended in 2 ml digestion buffer (800 mM guanidine HCl, 30 mM Tris-HCl, pH 8, 30 mM EDTA, 5% Tween-20 and 0.5% Triton X-100) supplemented with RNase A. 45 μL of a Proteinase K stock solution (21.4 mg/ml) were then added to the suspension followed by incubation at 50°C for 30 min. Cellular debris were removed by centrifugation at 3220xg (4°C for 10 min) and genomic DNA was then isolated from the obtained supernatant using Genomic-tips 20/G (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The concentration and quality of the isolated gDNA was determined spectrophotometrically and via agarose gel electrophoresis.
P. pastoris strain BG08 (BioGrammatics Inc., Carlsbad; CA, USA) is a single colony isolate from the Phillips Petroleum strain NRRL Y-11430 obtained from the Agriculture Research Service culture collection. For genomic DNA isolation, zymolyase, Proteinase K and RNase A were used. P. pastoris BG10 (BioGrammatics Inc, Carlsbad, CA, USA) was derived from BG08 using Hoechst dye selection to remove cytoplasmic killer plasmids.
2.3 Strain Cultivation and RNA extraction
The wildtype P. pastoris strain CBS7435, as well as related strains with deletions of the dihydroxyacetone synthase (Δdas1, Δdas2, Δdas1/das2) (Geier et al., 2015a), were cultivated for 24 h on BMD2% (200 mM KPi, pH 6.0, 20 g/L dextrose, 13.4 g/L yeast nitrogen base and 0.4 mg/L biotin) at 28°C and 100 rpm. Cells were harvested by centrifugation (1000xg, 5 min) and used to inoculate 200 ml of BMM (as BMD2% but supplemented with 0.5% methanol instead of dextrose) for growth to an OD600 of 8 at 28°C and 100 rpm. Samples for RNA-seq analysis (150–300 mg wet cell weight, wcw) were drawn after 24 h growth on glucose and 5 h growth on methanol; all cell samples were immediately frozen in liquid nitrogen after removing the supernatant. Total RNA samples were prepared in duplicate from 8 samples using a FastRNA™ Yeast SPIN kit (MP Biomedicals, Santa Ana, CA, USA). Briefly, cell disruption was performed with 3×2 minute bursts, using a BioSpec Products Mini–Beadbeater–96 (Bartlesville, OK, USA) and purified RNA samples were flash frozen in liquid nitrogen before storage at –80°C. cDNA libraries were constructed using Illumina TruSeq stranded mRNA library preparation kits and sequenced on an Illumina HiSeq 2500 platform. TruSeq libraries of two additional P. pastoris strains expressing the β-carotene biosynthetic pathway, either regulated by the constitutive GAP promoter or the inducible AOX1 promoter, were also analyzed by RNA-seq; these samples were similarly drawn after 48 h growth on glucose and after 24 h of methanol induction, respectively. Additional libraries were prepared from BG10 strains expressing a variety of heterologous proteins, both intracellular and secreted. In total, 57 RNA-seq libraries were created and sequenced.
2.4 Sequencing and genome assembly
PacBio sequencing was performed on 10 μg of DNA by GATC Biotech (Konstanz, Germany). Preparation of a large insert library and subsequent sequencing was performed on a PacBio RS II instrument. No manual filtering of sequence reads was attempted and the assembly could be done by using the HGAP 3 based de-novo assembly protocol with standard settings, except for p_assembleunitig.genomeSize = 9000000 and p_assembleunitig.xCoverage = 15. The software was provided by GATC in the SMRT Portal Version 2.2.0. For P. pastoris BG08 and BG10 paired-end genomic DNA sequencing was performed by GeneWiz Inc. (New Jersey, USA) on an Illumina HiSeq 2500 platform. The de novo assembled reference genome sequence was evaluated by comparison with previously generated wildtype sequence data (Küberl et al., 2011). In order to obtain the full genome sequence, the deleted AOX1 ORF of the sequenced mutS strain was adjusted by in silico complementation into the PacBio genome assembly employing CLC Genomics Workbench software (Qiagen, Hilden, Germany).
2.5 Sequencing and transcriptome mapping
Library preparation was performed at the University of California, San Diego, Institute for Genomic Medicine. RNA samples were analyzed on an Agilent 2200 TapeStation to visualize intact ribosomal RNA prior to library preparation. Libraries were prepared using an Illumina TruSeq mRNA preparation kit and libraries were barcoded for multiplexing during sequencing. Subsequently, the libraries were size selected for >200 bp, with modes of approximately 300 bp and 50 cycles of single-end sequencing was performed on an Illumina HiSeq 2500 machine. Data was provided in standard FASTQ (Cock et al., 2009) format and TopHat2 (Trapnell et al., 2009) was used to map reads to the PacBio genome assembly.
2.6 Gene prediction and functional sequence annotation
A de novo transcriptome assembly was generated from the single-end strand-specific RNA-seq library using Trinity (Trinity version: trinityrnaseq_r20140717) (Manfred G. Grabherr et al., 2013). Open reading frames (ORFs) were identified in the assembled transcripts using the Perl script, transcripts_to_best_scoring_ORFs.pl (Trinity version: trinityrnaseq_r2012-01-25p1). Consensus gene models were flagged in assembled transcripts using a combination of the predicted ORFs and homology-based annotation as evidence. Blast matches were made to the non-redundant database at NCBI, and Hidden Markov Models (HMM) matches were made using hmmer3 (Finn et al., 2011; Sonnhammer ELL, 1998) to models from PFAM (Finn et al., 2015) and TIGRfam (Haft et al., 2003). These consensus gene models, based on the de novo transcriptome assembly were in turn mapped to the P. pastoris genome assembly using gmap. These genome-mapped transcriptome-derived gene models were then used to improve the SGI/ArchetypeR eukaryotic gene prediction pipeline. The SGI/ArchetypeR eukaryotic gene prediction pipeline is divided into two primary components – the first trains a HMM for gene prediction and the second uses the trained HMM and any supporting gene evidence (e.g., genome-mapped transcriptome-derived gene models) to predict the final set of genes. A high-quality training set of P. pastoris gene models was obtained from the following series of steps: 1) blastx search of the input genomic sequences against eukaryotic protein sequences in UniProt (Consortium, 2015), 2) GeneWise (Birney Clamp, M., Durbin, R, 2004) generation of more precise genomic alignments, with splice sites as needed, from the blastx matches, and 3) filtering of the GeneWise output to ensure that predicted coding sequences each have a valid start codon, a valid stop codon, no inner stop codons, overlap with a transcriptome-derived gene model. The resulting filtered gene models were then used to train an HMM for the AUGUSTUS gene prediction algorithm (Grabherr et al., 2011; Stanke et al., 2008). AUGUSTUS was then used to predict genes using the trained HMM, the original input genomic sequences, and evidence provided in the form of transcriptome-derived gene models, protein alignments, and negative evidence in the form of internal data (which provides regions that are likely RNA non-coding and are therefore not good candidates for gene coding regions). Finally, any full-length transcriptome-derived gene models mapped to regions in the genomic sequence without any AUGUSTUS-predicted genes were added to the final catalog of predicted genes. Having generated the ORFs they were annotated through the ArchetypeR annotation pipeline (Robson et al., 2015).
2.7 Identification of P. pastoris killer plasmids
In addition to the assemblies described above, a Velvet de novo assembly (Zerbino and Birney, 2008) of paired-end Illumina data from the P. pastoris BG08 strain and PacBio sequence data of the P. pastoris CBS7435 mutS strain resulted in the discovery of two high coverage contigs with homology to K. lactis killer plasmid sequences (Schickel et al., 1996). Subsequently the ORFs on both plasmids were identified by manually checking for ORFs and gene by gene blastp. Both assembled contigs were flanked by inverted repeats.
2.8 Analysis of intron splicing
The TopHat 2.1.0 software tool was used to map individual RNA-seq data sets to the genome sequence presented here with intron length limited to 3000 bases. In this analysis, typically >98% of reads aligned to the genome sequence. A custom BioRuby (Goto et al., 2010) script was then employed to combine all junction files and identify introns with GT—AG ends. All TopHat alignments were then rerun, this time forcing the use of only GT—AG introns with the “--no-novel-juncs” option. In all cases, forcing alignments to predetermined splicing sites resulted in an ~0.1% increase in mapping. The resulting BAM files were filtered using SAMtools (Li et al., 2009) to regions spanning the predetermined splicing sites. The filtered BAM files for all experiments were combined and a mpileup output was generated. Subsequently a custom BioRuby script was used to analyze the mpileup output and to determine the splicing density at each predicted spliced nucleotide.
2.9 Identification of centromere regions
The P. pastoris strain used to visualize centromeres was a derivative of PPY12 (his4 arg4) (Gould et al., 1992). To label the endoplasmic reticulum, this strain was transformed with a HIS4 integrating vector encoding DsRed.T1-HDEL as previously described (Bevis et al., 2002). To label Cse4, PPY12 genomic DNA was used as a PCR template to amplify the CSE4 gene, including 589 bp of upstream sequence and 294 bp of downstream sequence, and this fragment was inserted into the polylinker of a pUC19 derivative containing the Saccharomyces cerevisiae ARG4 gene (Rossanese et al., 1999). In-Fusion cloning was then used to generate a chimeric gene encoding msGFP (Fitzgerald and Glick, 2014) fused to the C-terminus of Cse4 with an intervening GSSGSSGSSGSS linker. This construct was linearized with SpeI for integration at the CSE4 locus, resulting in a tandem duplication of CSE4 in which one copy of the gene was fused to msGFP. Fluorescence microscopy was performed as previously described (Papanikou et al., 2015). In brief, cells grown to logarithmic phase in a non-fluorescent minimal medium were compressed beneath a coverslip, and a Z-stack of images in red, green, and transmitted light channels was collected using a Leica SP5 confocal microscope. This Z-stack was then deconvolved and average projected. Planning and simulation of cloning procedures, visualization of chromosome organization, and identification of inverted repeats were performed using SnapGene or SnapGene Viewer software (GSL Biotech, Chicago, IL).
3. Results and Discussion
3.1 Sequencing and assembly of the P. pastoris CBS7435 genome
Pacific Bioscience’s single, molecule real-time sequencing platform (SMRT) enabled the de novo sequencing and assembly of the methylotrophic yeast strain P. pastoris CBS7435 mutS genome. Sequencing of a PacBio RS II library with an insert size of 8–12 kbp in a 1 movie run mode resulted in 185,064 sequence reads with 948,348,000 sequenced bases. The de novo assembly of PacBio sequence reads resulted in the identification of 31 unitigs with four large unitigs of size 2.9 Mbp, 2.4 Mbp, 2.3 Mbp and 1.8 Mbp (table 2). Focus is on these four large unitgs since most of the remaining unitigs correspond to fragmented mitochondrial and killer plasmid DNA generated during the fragmentation and size selection of PacBio libraries. By including the mitochondrial DNA sequence published in 2011, as well as, the two killer plasmid sequences, and remapping the PacBio raw reads onto this combinatorial data, 99.96% of all reads are correctly aligned. Two unitigs of 13.1 kbp and 9.5 kbp bearing homology to Kluveromyces lactis killer plasmid sequences were also identified (Schickel et al., 1996).
Table 2. Summary of unitigs identified in the assembly of P. pastoris genomic DNA.
The length, mean coverage and all protein coding sequences of the CBS7435 mutS strain are presented here. Assembly metrics can be found in supplementary figure S1.
| unitig | designation | length [bp] | mean coverage | protein-coding sequences |
|---|---|---|---|---|
| 1 | chromosome 1 | 2894792 | 66.42 | 1601 |
| 2 | chromosome 2 | 2396129 | 64.72 | 1355 |
| 3 | chromosome 3 | 2263199 | 66.21 | 1264 |
| 4 | chromosome 4 | 1825687 | 66.70 | 1036 |
Based on a comparison with published genome data of different P. pastoris wildtype strains (Küberl et al., 2011; Schutter et al., 2009), and by using blast algorithm to align all four unitigs to the P. pastoris CBS7435 genome, the four large unitigs correspond to the four chromosomes of P. pastoris. The entire length of the P. pastoris genome sequence presented here is 9.38 Mbp.
For the first time, a reference genome with four un-gapped chromosomes, telomere sequences on each chromosome and ribosomal repeats is created. The chromosomes of different yeast species, including P. pastoris, show regions of rDNA tandem repeats, variable in number and located at the end of chromosomes (Küberl et al., 2011; Rustchenko et al., 1993; Schutter et al., 2009). Here, the new genome sequence data orient the chromosomes so that these ribosomal repeats are at the distal “end” of each chromosome. This is done to stabilize the more proximal genome and annotation numberings employed for cataloging the genetic information. This results in “flipping” chromosome 1 relative to the first published draft genome (figure 1, (Küberl et al., 2011)). Moreover, chromosomal rearrangements ranging from as few as 1 kbp to 134 kbp were found. One chromosomal fragment of 76 kbp between two previous gaps on chromosome 1, 134.2 kbp fragment at the end of chromosome 4, as well as, several shorter fragments of 2–3 kbp were reoriented (De Schutter et al., 2009; Küberl et al., 2011).
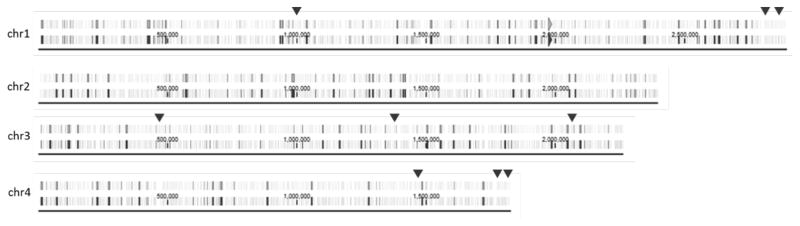
Figure 1. Open reading frames (ORFs) identified in the closed gaps of the P. pastoris chromosomes.
Six small gaps of 60–200 bp as well as six larger gaps of 2–6 kbp present in the CBS7435 genome sequence could be closed. The ORFs found in these regions are marked with dark triangles. In addition, the orientation of the four chromosomes was standardized to show the ribosomal clusters at the 3′ ends. The vertical lines marked on all four chromosomes represent the annotation of ORFs.
The genome sequence of P. pastoris CBS7435 published in 2011 (Küberl et al., 2011) shows 12 gaps ranging from 20 bp to 6 kbp in size. Genome assembly with short sequence reads cannot assemble longer, highly repetitive sequence elements. Here, the longer PacBio reads of up to 15 kbp allow these repetitive structure elements to be assembled and previously existing gaps to be closed. Within these regions nine ORFs with altered annotations relative to the previous genome sequences were identified and summarized in table 3. An increase in size is also caused by additional bases within the open reading frame at the 3′ end, and the correction of splicing events occurring in these genes as exemplified in figure 2. Interestingly, blastp database searches for protein homologues identified proteins involved in cell flocculation (agglutination) and cell surface recognition.
Table 3. Putative ORFs identified in the gaps of the P. pastoris CBS7435 genome sequence of 2011.
Putative ORFs differing in length were found on each of the four chromosomes. All 9 genes identified in these regions are characterized by the presence of highly repetitive sequence motifs. Amongst these, one previously undiscovered and new putative open reading frame (location 2016 genome sequence: 1752938..1759474) was identified. n.p. not present.
| gene location | chr | length [bp] | spliced | description | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| 2011 | 2016 | 2011 | 2016 | 2011 | 2016 | ||
|
|
|
||||||
| 2804402..2805474 | 2798500..2802711 | 1 | 1020 | 4149 | Yes | yes | plaque matrix protein-like |
| 475309..477983 | 475237..478581 | 2 | 2394 | 3345 | Yes | no | cell surface glycoprotein |
| 1319551..1320953 | 1319750..1321156 | 3 | 1257 | 1407 | Yes | no | zinc metalloprotease zmpB |
| 2208981..2209753 | 2214003..2221310 | 3 | 1503 | 7308 | Yes | no | zonadhesin |
| 2217232..2219859 | 2224962..2228516 | 3 | 2628 | 3555 | No | no | flocculation protein flo9 |
| 2241445..2242652 | 2250102..2253884 | 3 | 1131 | 4407 | Yes | yes | agglutinin-like protein 3 |
| 1666..2583 | 1491065..1492294 | 4 | 918 | 1230 | No | no | cell surface glycoprotein |
| n.p. | 1752938..1759474 | 4 | - | 6537 | - | no | cell agglutination protein |
| 1805300..1806330 | 1811058..1819145 | 4 | 981 | 8088 | Yes | no | zonadhesin |
Figure 2. Multiple sequence alignment of P. pastoris CBS7435 genome sequence (2011) and the P. pastoris CBS7435 reference genome sequence presented here.
All alignments were performed using CLC Bio’s proprietary alignment algorithm. The dark areas correspond to perfect nucleotide matches whereas white areas denote mismatches or missing bases. N denotes bases missing in the 2011 genome sequence. A. The genes at location 475309..477983 (chr2) of the 2011 sequence (top) and 475237..478581 (chr2) of the 2016 sequence (bottom) were aligned against each other. B. The genes at location 2208981..2209753 (chr3) of the 2011 sequence (top) and 2214003..2221310 (chr3) of the 2016 sequence (bottom) were aligned against each other.
In total we found more than 500 sequence differences relative to the 2011 published genome (Küberl et al., 2011). PCR amplification and Sanger sequencing of insertions and deletions observed in putative coding regions confirmed the validity of the new reference sequence. In 34 out of 35 regions tested, the de novo assembled Pacific Biosciences sequence was confirmed (data not shown).
Additionally, the improvement with this new reference genome is confirmed by mapping the Roche 454 GS FLX Titanium reads to both the new reference genome and the 2011 genome (Küberl et al., 2011). More of the reads map to a combinatorial data set (de novo genome, killer plasmids and mitochondrial DNA sequence) of the new reference genome than the 2011 sequence (99.96% vs 99.63%, respectively).
Furthermore, paired-end Illumina HiSeq reads from the strains BG08 and BG10 (BioGrammatics Inc., Carlsbad, USA) were aligned to the new reference genome sequence to determine the differences between closely related P. pastoris strains. 2 base differences are evident between BG08 and BG10 outside of the killer plasmids, and 24 differences were found between the BioGrammatics strains and the CBS7435 sequence presented here (supplementary table S3). Except for one triple nucleotide insertion, all of the changes affected single nucleotide deletions and insertions; no inversions or larger rearrangements are found. Only small clonal variations occur between these closely related P. pastoris strains, even after storage at different sites, for many years indicating defined molecular manipulation can be precise with relatively little clonal drift.
In this study, 5256 potential ORFs are identified, of which 5111 can be verified on the basis of either RNA-seq data or published peptide sequences (Renuse et al., 2014). In this manner more than 50 new reading frames were identified (supplementary table S4) relative to the 2011 draft genome sequence (Küberl et al., 2011). Under the growth conditions described in this manuscript, no considerable transcript levels were found for 145 previously annotated open reading frames. Neither evidence by RNA-seq experiments nor data from published proteomics experiments (Renuse et al., 2014) find transcripts in these regions (supplementary table S5). Additionally, 304 regions were identified to which transcript sequence reads map without the presence of either an ORF >50 aa or ORFs showing significant similarity to NCBInr protein database deposited entries (supplementary table S6). These regions might contain non-coding RNA regulatory elements, such as described in S. cerevisiae (Thompson and Parker, 2007; Wu et al., 2014) and Schizosaccharomyces pombe (Volpe et al., 2003; Wilhelm et al., 2008). One can speculate that similar cryptic unstable transcripts (CUTs), such as those involved in the regulation of meiosis (Lardenois et al., 2011), histone methylation (van Dijk et al., 2011) and telomere length (Luke et al., 2008) in S. cerevisiae, might exist Pichia species as well. The RNA-seq data also contains reads for overlapping ORFS, e.g. transcripts of two genes with opposite transcriptional orientation showing elevated read coverage. These overlaps of sense-antisense gene pairs might have a regulatory function in gene expression and silencing such as described in S. cerevisiae (David et al., 2006; Drinnenberg et al., 2009; Nagalakshmi et al., 2008).
In order to create a reference sequence for the wildtype genome, the deleted AOX1 gene of the sequenced mutS strain was complemented in silico to generate the new full reference sequence, which can be accessed from the NCBI web interface. Due to the large number of changes in this genome compared to the previously published genomes, we propose to use this new and completed whole genome sequence as a reference sequence for P. pastoris, which should facilitate future omics and systems biology studies, as well as, precise genome engineering approaches.
3.2 Alternative splicing and RNA-seq data mapping
To further refine the automated annotation performed on the genome sequence of P. pastoris CBS7435, we performed an RNA-seq analysis using Illumina HiSeq sequencing data. An Illumina HiSeq mRNA library was run with an average read length of 50 bp reads from several different RNA samples of wildtype strains and strains harboring heterologous expression cassettes. These data were then mapped against the genome sequence which resulted in more than 98% of reads correctly aligned.
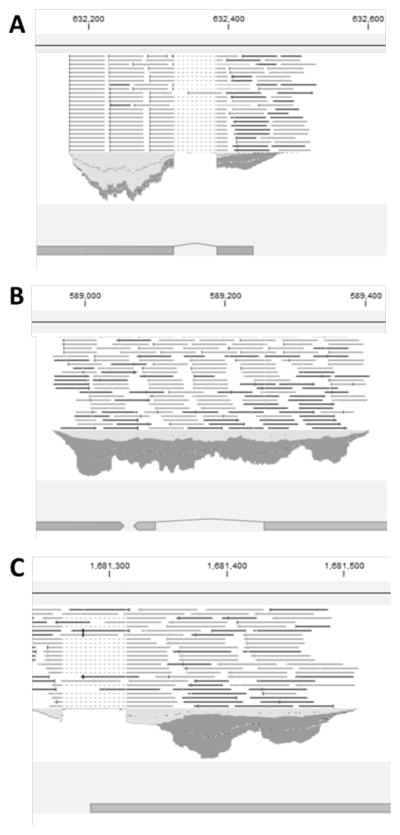
Figure 3 shows an example of three different events occurring in the analysis. In panel A, the RNA-seq data confirm the automated annotation as successful mapping depends on a gap opening in the reads. Alternatively, panel B and C depict two different examples in which the RNA-seq reads do not confirm previous predicted splicing events. Previous intron mis-calls in the 2011 draft genome sequence demonstrate: 1) in panel B, the mapped reads do not substantiate the presence of an intron in this position, and 2) in panel C, the RNA-seq mapping is forced to open a gap to correctly align to the genome sequence and therefore indicated the presence of an intron at this position. Based on these strategies, all four P. pastoris chromosomes and their corresponding open reading frames were manually corrected. The analysis resulted in the identification of 571 experimentally confirmed spliced genes in the genome sequence presented here compared with 797 reported in 2011 (based on computational predictions).
Figure 3. Exemplary intron splicing site prediction as identified by mapping RNA-seq reads to the P. pastoris reference sequence.
A Gene at location 2262918..2263848 (chr1). Mapped reads verify the automated annotation. B Gene at location 586755..589073 (chr4). Due to the presence of mapped reads in the intron sequence the automated annotation had to be corrected. C Gene at location 579086..582205 (chr3). The intron identified in the RNA-seq data was incorporated into the automated annotation. The bottom part of each figure shows the gene as present in the genome sequence of P. pastoris CBS7435 published in 2011. The middle part corresponds to the RNA-seq reads. CLC Genomics Workbench version 7 was used for visualizations and manual corrections.
In order to accurately map RNA-seq data onto the reference genome, it is important to allow mapping software to consider not only the introns annotated to create the protein encoding ORFs, but also splicing events that occur either outside these ORFs or as variants of the major splicing events. Forty-six variant splice acceptors and 33 variant splice donors were identified in the bed file outputs from TopHat. In addition, 11 exon “jumping” events were found where genes containing multiple introns were spliced from the donor site of one intron to the branch / acceptor site of a second intron. In general, alternative splicing occurs at low frequency and is the result of faulty selection of the proper donor or acceptor. For most cases that use an alternative splice acceptor, the same splice branch site is used and either the next upstream or downstream AG relative to the proper splice acceptor is used. In addition to alternative splicing, a low-level of cryptic splicing events is observed in some highly transcribed mRNAs. Additionally, a number of mRNAs have introns in their 5′ UTRs, which were previously not annotated; annotations for these probable locations have been included to help with the future identification of promoter-gene regions. A very small number of genes appear to have 3′ UTR introns. TopHat analysis of the complete set of RNA-seq data revealed 828 splicing events where the intron was flanked by GT—AG and there were at least 10 reads distributed across at least 4 experiments to support the existence of an intron. These 828 loci were hand curated to remove all locations that fell on the wrong strand of a highly transcribed mRNA or appeared to be the result of mis-mapping in genes with repetitive sequences. A final set of 771 junctions was used to generate mapping data for all RNA-seq experiments. A “.juncs” file (for use with TopHat –no-novel-juncs –raw-juncs options) containing the 771 locations is provided in the supplementary data. 771 possible splice sites can be confirmed by the current RNA-seq data. Wherever possible, the RNA-seq data set and proteomics data was used to confirm the predicted ORFs, to evaluate discrepancies in respect to translational starts and splice sites, and to re-evaluate or confirm sequence differences from the transcriptome comparisons (Renuse et al., 2014). Not surprisingly, for many ORFs the number of identified peptides was too low to clearly confirm deep sequencing RNA-seq data from the currently available proteomics data sets.
3.3 Identification of putative P. pastoris centromeres
Budding yeast centromere sequences were originally identified in S. cerevisiae, where they are as short as 120 bp (Biggins, 2013; Clarke and Carbon, 1985). Those centromeres can be incorporated into plasmids to confer replicative stability (Clarke and Carbon, 1985), and can be used to generate yeast artificial chromosomes (Noskov et al., 2010). It would therefore be of value to identify centromere sequences for P. pastoris. However, centromere sequences diverge rapidly during evolution and are quite variable among yeast species (Malik and Henikoff, 2009; Roy and Sanyal, 2011), so homology searches of the P. pastoris genome were unlikely to identify putative centromeres.
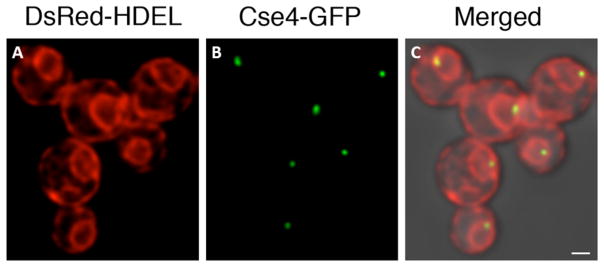
As a first step toward identifying P. pastoris centromeres, we asked whether P. pastoris resembles other yeasts in possessing the centromere-specific histone H3 variant Cse4 (Biggins, 2013). Indeed, a clear CSE4 homolog on chromosome two (2011 genome location 1026080..1026598 or 2016 genome location 1026960..1027478) was present on Chr2. P. pastoris Cse4 was tagged with the monomeric superfolder GFP variant msGFP (Fitzgerald and Glick, 2014). For visualizing nuclei, the cells also expressed ER-targeted DsRed-HDEL, which fills the nuclear envelope with red fluorescence (Bevis et al., 2002). Figure 6 shows that each cell contained a single green fluorescent spot in close proximity to the nuclear envelope. This pattern is typical for budding yeasts, in which the centromeres are clustered near the spindle pole bodies (Pearson et al., 2001). Therefore, it is inferred that P. pastoris likely has centromeres that are functionally similar to those found in other budding yeasts.
Figure 6. Visualization of clustered centromeres in P. pastoris by confocal microscopy.
This strain expressed DsRed-HDEL to label the endoplasmic reticulum in red. The ring visible in each cell is the nuclear envelope. In addition, the strain expressed Cse4-GFP to label centromeres in green. The merged image shows the two fluorescence signals overlaid on a transmitted light image of the cells. A cluster of centromeres is visible at the nuclear periphery in each cell. Scale bar, 2 μm.
A clue to the possible locations of P. pastoris centromeres came from visual inspection of the chromosome annotation patterns. Figure 4 shows a portion of P. pastoris Chr1 with the predicted ORFs annotated in dark gray. The chromosome is densely packed with protein-coding genes, but a single gap of 9 kbp was observed. Upon closer inspection, this region was found to contain a perfect inverted repeat of 1991 bp. A similar inspection of the other three chromosomes revealed that each of them also has a single region of 9–11 kbp that is largely devoid of predicted ORFs, and that contains perfect or near perfect inverted repeats of 1991–2699 bp (table 4). Because centromeres tend to have few transcribed genes and sometimes contain inverted repeats (McFarlane and Humphrey, 2010), the ORF-free regions with inverted repeats are putative centromeres for P. pastoris.
Figure 4. P. pastoris chr1 with the predicted ORFs annotated.
The annotated ORFs are marked in dark grey. The putative centromere unique region is marked in bright gray.
Table 4. Locations of Hi-C centromere calls and RNA-seq mapping based prediction for P. pastoris CBS7435 centromeres.
Varoquaux et al. used the chromatin conformation capture assay, Hi-C, to predict the centromere regions of P. pastoris GS115 to within a 20 kbp region. Based on RNA-seq data mapping to the reference sequence presented here we were able observe a drastic drop in signal strength in the regions below, indicating a low transcriptional status in those regions. In those regions we identified near perfect inverted repeats. The reorientation of chr1, chr3 and chr4 described above resulted in a differing value when compared to GS115. The slightly differing value between GS115 and CBS7435 on chromosome 4 arises from the shorter length of GS115 chr4.
| GS115 2009 | CBS7435 2016 predicted centromeres | ORF-free space [bp] | Inverted Repeats [bp] | |||||
|---|---|---|---|---|---|---|---|---|
| chr | Hi-C | predicted | before reorientation | after reorientation | individual repeats | total sequence spanned | Identity [%] | |
| 1 | 1408908 ± 20000 | 1400423..1409375 | 1487825..1493796 | 1401559..1407530 | 8,955 | 1991 | 5354 | 99 |
| 2 | 1556231 ± 20000 | 1542915..1551466 | 1545323..1551977 | 844482..851136 | 10,413 | 2699 | 6655 | 99 |
| 3 | 2226823 ± 20000 | 2202870..2211602 | 2222793..2228973 | 34486..40666 | 8,734 | 2649 | 6183 | 99 |
| 4 | 1719280 ± 20000 | 1701016..1712046 | 1762920..1769148 | 58794..65022 | 9976 | 2559 | 6229 | 99 |
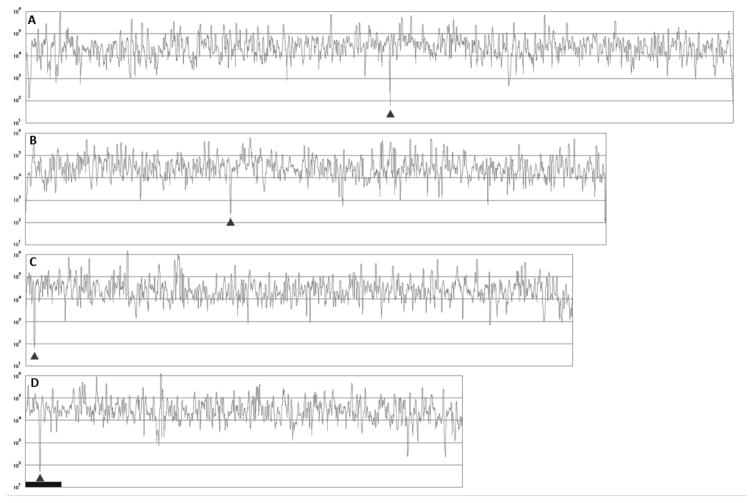
To confirm that the putative centromeres are largely devoid of transcribed genes, we examined RNA-seq data. As shown in figure 5, each of the putative centromere regions corresponds to a sharp and pronounced drop in the RNA-seq signal strength. When transcriptional profiles were generated for the full chromosomes using 4 kbp windows at 100 bp intervals, the predicted centromeres corresponded to the minimum values in the plots after telomere sequences were excluded (figure 5).
Figure 5. Putative location of P. pastoris centromeres indicated by RNA-seq reads mapped to this reference sequence.
A-D corresponds to chromosomes 1–4. The putative centromere regions are largely devoid of transcribed genes as can be seen by the marked drop in the RNA-seq signal strength. The dark triangles correspond to the location of the putative centromere on each chromosome. The 138 kbp mating type chromosomal inversion region is indicated by the dark bar on chr4. The log scale plot shows the transcriptome density with 4 kbp windows at 100 bp intervals normalized to the maximum density window of 900,000 for chr1.
Independent evidence that these regions are P. pastoris centromeres came from a recent analysis using a chromatin conformation capture assay called Hi-C (Varoquaux et al., 2015). That study took advantage of centromere clustering, of the type shown in figure 6, to map predicted centromere locations within approximately 20 kbp. The predicted centromere locations closely match the regions identified here (table 4). Interestingly, the MATa1 and a2 locus on chr4 is found in between the P. pastoris centromere inverted repeats. Hanson et al. have determined that the flanking mating type inverted repeats are found in all four orientations (Hanson et al., 2014). While the centromere repeats are smaller, it is still possible that these can also undergo flipping and result in eight combinations at the end of chr4. If the centromere function is defined solely by the inverted repeats and their spacing, the resulting four possible arrangements of the centromeric core relative to immediate flanking regions and the chromosome as a whole would be functionally identical. The core and its relative orientation might potentially play a role during meiotic recombination and chromosome segregation during sporulation of a diploid cell.
The combined data give high confidence that we have identified the four centromeres in P. pastoris. However, a rigorous demonstration will require further evidence, such as crosslinking of nucleosome-associated Cse4 to the putative centromeres (Meluh et al., 1998) or confirmation that the putative centromeres confer replicative stability to plasmids (Clarke and Carbon, 1985).
3.4 P. pastoris killer plasmids
The dairy yeast K. lactis was one of the first yeast species proven to harbor a set of two different linear DNA fragments, which enabled the cells to kill other yeasts by secreting an exotoxin (Gunge et al., 1981). These plasmids have been termed yeast killer plasmids and have so far been identified in several yeast genera such as Botryascus, Pichia, Debaryomyces and Wingea (Cong et al., 1994; Hayman and Bolen, 1991; Wickner and Leibowitz, 1976; Wickner, 1979; Worsham and Bolen, 1990). The genetic composition of these linear plasmids seems to be quite conserved among those species. The K. lactis pGKL1/pGKL2 system has been extensively studied and resulted in the elucidation of plasmid encoded gene functions (Butler et al., 1991; Gunge and Kitada, 1988; Gunge, 1986; Kikuchi et al., 1984; Sor et al., 1983; Stam et al., 1986; Tokunaga et al., 1987). Banerjee and colleagues were the first to describe the presence of RNase resistant double stranded DNA molecules sensitive towards DNase I digestion in the methylotrophic yeast P. pastoris (Banerjee et al., 1998).
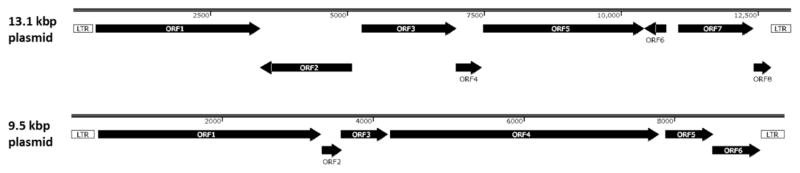
Using paired-end Illumina data from a P. pastoris wildtype strain (BG08) and a P. pastoris CBS7435 crtEBIY strain expressing the β-carotene synthesis pathway (Geier et al., 2015b) as well as PacBio sequence data of the P. pastoris CBS7435 mutS strain, two so far unpublished sequences of differently sized linear plasmids with intact LTR sequences were identified. These linear plasmids are 13.1 and 9.5 kbp in size and show homologues of several annotated ORFs frequently found on killer plasmids from other yeast species such as Kluveromyces lactis, Pichia accaciae and S. cerevisiae. Among the coding sequences found on the two plasmids are DNA polymerases, an RNA polymerase, a helicase, an mRNA capping enzyme and several homologues to K. lactis killer plasmid proteins. Due to the DNA size selection performed during PacBio library construction, the smaller killer plasmid was lost entirely and the larger killer plasmid was significantly underrepresented in the PacBio library. In Illumina data from a library prepared with ~ 500 bp inserts, the copy number of the two killer plasmids was estimated at 80–100 in BG08 and P. pastoris CBS7435 crtEBIY. In sequencing other wild type P. pastoris strains, approximately 25% of sequencing data maps to killer plasmid sequences (supplementary figure S2), indicating about 3 Mbp of total killer plasmid DNA relative to 9.38 Mbp of genomic DNA.
The 13.1 and 9.5 kbp large plasmids identified here show 8 and 6 ORFs, respectively (figure 7). These ORFs have high sequence identity to already known K. lactis killer plasmid proteins as seen in table 5. Both P. pastoris killer plasmids showed very similar spatial organization compared to K. lactis and S. cerevisiae plasmids (Schickel et al., 1996) with regard to the order of the ORFs. Also the coding density is similarly high, with 90.9% and 91.8% of all nucleotides coding for proteins in the 13.1 kbp and 9.5 kbp P. pastoris killer plasmids, respectively. ORF1 on the 13.1 kbp large plasmid shows high sequence identity to a plasmid specific DNA polymerase. Together with ORF5 and ORF6, annotated as RNA polymerase and RNA polymerase subunits, respectively, these putative genes most likely allow for the replication and transcription of plasmid encoded genes (Jung et al., 1987; Wilson and Meacock, 1988). ORF2, a potential mRNA capping enzyme and ORF3, a helicase, could potentially stabilize the linear plasmid in the cytosolic space (Larsen et al., 1998). On the basis of K. lactis killer plasmid protein functions we could also identify potential DNA binding proteins (ORF4) and a terminal recognition factor (ORF8) which is potentially responsible for the protection of linear DNA present in the cytosol by binding to the LTR 12 sequences found at the outer boundaries of the plasmids (Schaffrath and Meacock, 2001). As described, in S. cerevisiae the smaller 9.5 kbp plasmid contains a DNA polymerase (ORF1) and several different ORFs with sequence identities to killer plasmid toxins. The elements responsible for conferring toxicity are encoded on the smaller plasmid. In K. lactis and other yeast species the heterotrimeric killer toxin is made up of three different subunits, termed alpha, beta and gamma while a fourth protein coded on the same plasmid functions as an anti-toxin (Stark and Boyd, 1986). Based on sequence homology and localization on the plasmid, ORF4 might contain the alpha and beta subunit (Larsen et al., 1998). Neither ORF2, ORF3, ORF5 nor ORF6 could be identified as the killer toxin gamma subunit or anti-toxin protein. Although Banerjee and colleagues were able to isolate a DNase I digestible double stranded DNA fragment that was not susceptible to RNase degradation in P. pastoris (Banerjee et al., 1998), evaluation of 14 different P. pastoris strains for their killer activity showed no killer phenotype (Banerjee and Verma, 2000).
Figure 7. Genetic Organization of the two linear plasmids identified in P. pastoris.
Based on the plasmid sequences we identified 8 open reading frames on the 13.1 kbp killer plasmid and 6 open reading frames on the 9.5 kbp killer plasmid. Both plasmids are flanked by long terminal repeat sequences (LTR).
Table 5. Putative open reading frames identified on the 13.1 kbp and 9.5 kbp plasmids of P. pastoris.
All protein sequences were analyzed using blastp (protein-protein blast) against the non-redundant protein database. The protein entries showing the highest sequence identity to the query are summarized. No homologues of the gamma toxin subunit or the antitoxin gene were identified on the two P. pastoris killer plasmids.
| 3.1 kbp plasmid | Description | Organism and sequence identity |
|---|---|---|
| ORF1 | DNA-polymerase | Debaryomyces hansenii 67% |
| ORF2 | mRNA capping enzyme | Pichia etchellsii 49% |
| ORF3 | Helicase | K. lactis 59% |
| ORF4 | DNA binding protein | Millerozyma acacia 54% |
| ORF5 | RNA-polymerase | K. lactis 57% |
| ORF6 | RNA-polymerase subunit | M. acacia 41% |
| ORF7 | Killer toxin protein | K. lactis 50% |
| ORF8 | terminal recognition factor | M. acacia 58% |
|
| ||
| 9.5 kbp plasmid | ||
|
| ||
| ORF1 | DNA-polymerase | D. hansenii 55% |
| ORF2 | Hypothetical protein | no similarity |
| ORF3 | Hypothetical protein | no similarity |
| ORF4 | Killer toxin protein | K. lactis 46% |
| ORF5 | Hypothetical protein | no similarity |
| ORF6 | Hypothetical protein | no similarity |
In the sequencing data presented here major components of the killer plasmid system, such as the presence of DNA and RNA polymerases and putative open reading frames responsible for plasmid integrity, have been found. However, none of the remaining ORFs could be identified as homologues of the K. lactis toxin gamma subunit or the anti-toxin protein. The lack of these proteins could potentially cause a loss of the killer phenotype and therefore provide support for the finding of Banerjee and colleagues (Banerjee et al., 1998; nath Banerjee and Verma, 2000).
Supplementary Material
Acknowledgments
The research leading to these results has received funding from the Innovative Medicines Initiative Joint Undertaking project CHEM21 under grant agreement n°115360, resources of which are composed of financial contribution from the European Union’s Seventh Framework Program (FP7/2007–2013) and EFPIA companies’ in kind contribution. In addition, this work has been supported by the Federal Ministry of Science, Research and Economy (BMWFW), the Federal Ministry of Traffic, Innovation and Technology (bmvit), the Styrian Business Promotion Agency SFG, the Standortagentur Tirol, the Government of Lower Austria and ZIT -Technology Agency of the City of Vienna through the COMET-Funding Program managed by the Austrian Research Promotion Agency FFG. BSG was supported by NIH grant R01 GM104010, and KJD was supported by NIH training grant T32 GM007183. AG gratefully acknowledges the support and fruitful discussion of the presented work with Prof. Suresh Subramani and group members during a sabbatical at UCSD.
Abbreviations
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- ORF
open reading frame
- blast
basic local alignment search tool
- blastx
nucleotide 6-frame translation-protein basic local alignment search tool
- blastp
protein-protein basic local alignment search tool
- NCBI
National Center for Biotechnology Information
- HMM
Hidden Markov Model
- SGI
Synthetic Genomics Inc
- LTR
long terminal repeat
- PCR
polymerase chain reaction
- CBS
Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands
Footnotes
Author’s Contributions
LS, TC, MG, IT, JC, KM and AG planned and started the project collaboration. Wet laboratory work was carried out by LS, TC, MG, AS, KD and KM. In silico analysis was performed by LS, TC, MG, FK, UV, ST, TR and LSo. Experimental work and analysis of centromeres was done by LS, TC, KD and BG. Peptide mapping and analysis of proteome data was performed by TC, BD and RBG. LS, TC, BG, TR, IT, JC, KM, and AG wrote the manuscript.
References
- Banerjee H, Kopvak C, Curley D. Identification of Linear DNA Plasmids of the Yeast Pichia pastoris. Plasmid. 1998;40:58–60. doi: 10.1006/plas.1998.1341. [DOI] [PubMed] [Google Scholar]
- Banerjee H, Verma M. Search for a novel killer toxin in yeast Pichia pastoris. Plasmid. 2000;43:181–3. doi: 10.1006/plas.1999.1452. [DOI] [PubMed] [Google Scholar]
- Bevis BJ, Hammond AT, Reinke CA, Glick BS. De novo formation of transitional ER sites and Golgi structures in Pichia pastoris. Nat Cell Biol. 2002;4:750–756. doi: 10.1038/ncb852. [DOI] [PubMed] [Google Scholar]
- Biggins S. The composition, functions, and regulation of the budding yeast kinetochore. Genetics. 2013;194:817–846. doi: 10.1534/genetics.112.145276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney Clamp M, Durbin RE. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler AR, O’Donnell RW, Martin VJ, Gooday GW, Stark MJ. Kluyveromyces lactis toxin has an essential chitinase activity. Eur J Biochem. 1991;199:483–488. doi: 10.1111/j.1432-1033.1991.tb16147.x. [DOI] [PubMed] [Google Scholar]
- Clarke L, Carbon J. The structure and function of yeast centromeres. Ann Rev Genet. 1985;19:29–56. doi: 10.1146/annurev.ge.19.120185.000333. [DOI] [PubMed] [Google Scholar]
- Cock PJA, Fields CJ, Goto N, Heuer ML, Rice PM. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2009;38:1767–1771. doi: 10.1093/nar/gkp1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Yarrow D, Li Y, Fukuhara H. Debaryomyces hansenii and Wingea robertsiae. Yeast. 1994:1327–1335. doi: 10.1099/00221287-140-6-1327. [DOI] [PubMed] [Google Scholar]
- Consortium TU. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregg JM, Tolstorukov I, Kusari A, Sunga J, Madden K, Chappell T. Chapter 13 Expression in the Yeast Pichia pastoris. In: RRB, MPDBT-M, editors. Enzymology. 2. Academic Press; 2009. pp. 169–189. Guide to Protein Purification. [DOI] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci U S A. 2006;103:5320–5. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schutter K, Lin YC, Tiels P, Van Hecke A, Glinka S, Weber-Lehmann J, Rouzé P, Van de Peer Y, Callewaert N. Genome sequence of the recombinant protein production host Pichia pastoris. Nat Biotechnol. 2009;27:561–566. doi: 10.1038/nbt.1544. [DOI] [PubMed] [Google Scholar]
- Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP. RNAi in budding yeast. Science (80) 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AC, Richards S, Han Y, Wang M, Vee V, Qu J, Qin X, Muzny DM, Reid JG, Worley KC, Gibbs RA. Mind the Gap: Upgrading Genomes with Pacific Biosciences RS Long-Read Sequencing Technology. PLoS One. 2012;7:1–12. doi: 10.1371/journal.pone.0047768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011;39:1–9. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2015:gkv1344. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald I, Glick BS. Secretion of a foreign protein from budding yeasts is enhanced by cotranslational translocation and by suppression of vacuolar targeting. Microb Cell Fact. 2014;13:125. doi: 10.1186/s12934-014-0125-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geier M, Brandner C, Strohmeier Ga, Hall M, Hartner FS, Glieder A. Engineering Pichia pastoris for improved NADH regeneration: A novel chassis strain for whole-cell catalysis. Beilstein J Org Chem. 2015a;11:1741–1748. doi: 10.3762/bjoc.11.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geier M, Fauland P, Vogl T, Glieder A. Compact multi-enzyme pathways in P. pastoris. Chem Commun. 2015b;51:1643–1646. doi: 10.1039/c4cc08502g. [DOI] [PubMed] [Google Scholar]
- Geier M, Fauland PC, Vogl T, Glieder A. Compact multi enzyme pathways in Pichia pastoris. Chem Commun. 2014 doi: 10.1039/c4cc08502g. [DOI] [PubMed] [Google Scholar]
- Goto N, Prins P, Nakao M, Bonnal R, Aerts J, Katayama T. BioRuby: Bioinformatics software for the Ruby programming language. Bioinformatics. 2010;26:2617–2619. doi: 10.1093/bioinformatics/btq475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouldi SJ, Mccollum D, Spong AP, Heyman JA. Development of the Yeast Pichia pastoris as a Model Organism for a Genetic and Molecular Analysis of Peroxisome Assembly. 1992:8. doi: 10.1002/yea.320080805. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotech. 2011;29:644– 652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunge N. Linear DNA killer plasmids from the yeast Kluyveromyces. Yeast. 1986;2:153–62. doi: 10.1002/yea.320020303. [DOI] [PubMed] [Google Scholar]
- Gunge N, Kitada K. Replication and maintenance of the Kluyveromyces linear pGKL plasmids. Eur J Epidemiol. 1988;4:409–414. doi: 10.1007/BF00146390. [DOI] [PubMed] [Google Scholar]
- Gunge N, Tamaru A, Ozawa F, Sakaguchi K. Isolation and Characterization of Linear Deoxyribonucleic Acid Plasmids from Kluyveromyces lactis and the Plasmid-Associated Killer Character. J Bacteriol. 1981;145:382–390. doi: 10.1128/jb.145.1.382-390.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SJ, Byrne KP, Wolfe KH. Mating-type switching by chromosomal inversion in methylotrophic yeasts suggests an origin for the three-locus Saccharomyces cerevisiae system. Proc Natl Acad Sci U S A. 2014:1–8. doi: 10.1073/pnas.1416014111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman GT, Bolen PL. Linear DNA plasmids of Pichia inositovora are associated with a novel killer toxin activity. Curr Genet. 1991;19:389–393. doi: 10.1007/BF00309600. [DOI] [PubMed] [Google Scholar]
- Jung GH, Leavitt MC, Ito J. Yeast killer plasmid pGKL1 encodes a DNA polymerase belonging to the family B DNA polymerases. Nucleic Acids Res. 1987;15:9088. doi: 10.1093/nar/15.21.9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Hirai K, Hishinuma F. The yeast linear DNA killer plasmids, pGKL1 and pGKL2, possess terminally attached proteins. Nucleic Acids Res. 1984;12:5685–5692. doi: 10.1093/nar/12.14.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küberl A, Schneider J, Thallinger GG, Anderl I, Wibberg D, Hajek T, Jaenicke S, Brinkrolf K, Goesmann A, Szczepanowski R, Pühler A, Schwab H, Glieder A, Pichler H. High-quality genome sequence of Pichia pastoris. CBS7435. 2011;154:312–320. doi: 10.1016/j.jbiotec.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP. Biotechnological strains of Komagataella (Pichia) pastoris are Komagataella phaffii as determined from multigene sequence analysis. J Ind Microbiol Biotechnol. 2009;36:1435–1438. doi: 10.1007/s10295-009-0638-4. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP. Description of Komagataella phaffii sp nov and the transfer of Pichia pseudopastoris to the methylotrophic yeast genus Komagataella. Int J Syst Evol Microbiol. 2005;55:973–976. doi: 10.1099/ijs.0.63491-0. [DOI] [PubMed] [Google Scholar]
- Lardenois A, Liu Y, Walther T, Chalmel F, Evrard B, Granovskaia M, Chu A, Davis RW, Steinmetz LM, Primig M. Execution of the meiotic noncoding RNA expression program and the onset of gametogenesis in yeast require the conserved exosome subunit Rrp6. Proc Natl 15 Acad Sci U S A. 2011;108:1058–1063. doi: 10.1073/pnas.1016459108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M, Gunge N, Meinhardt F. Kluyveromyces lactis killer plasmid pGKL2: evidence for a viral-like capping enzyme encoded by ORF3. Plasmid. 1998;40:243–246. doi: 10.1006/plas.1998.1367. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Wang B, Pan L, Ye Y, He M, Han S, Zheng S, Wang X, Lin Y. Comprehensive structural annotation of Pichia pastoris transcriptome and the response to various carbon sources using deep paired-end RNA sequencing. BMC Genomics. 2012;13:1. doi: 10.1186/1471-2164-13-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J. The Rat1p 5′ to 3′ Exonuclease Degrades Telomeric Repeat-Containing RNA and Promotes Telomere Elongation in Saccharomyces cerevisiae. Mol Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Malik HS, Henikoff S. Major Evolutionary Transitions in Centromere Complexity. Cell. 2009;138:1067–1082. doi: 10.1016/j.cell.2009.08.036. [DOI] [PubMed] [Google Scholar]
- Grabherr Manfred G, Haas Brian J, Yassour Moran, Levin Joshua Z, Thompson Dawn A, Amit Ido, Adiconis Xian, Fan Lin, Raychowdhury Raktima, Zeng Qiandong, Chen Zehua, Mauceli Evan, Hacohen Nir, Gnirke Andreas, Rhind Nicholas, di Palma Federica, Bruce WN, Friedman AR. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol. 2013;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccarthy A. Third generation DNA sequencing: Pacific biosciences’ single molecule real time technology. Chem Biol. 2010;17:675–676. doi: 10.1016/j.chembiol.2010.07.004. [DOI] [PubMed] [Google Scholar]
- McFarlane RJ, Humphrey TC. A role for recombination in centromere function. Trends Genet. 2010;26:209–213. doi: 10.1016/j.tig.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- Näätsaari L, Mistlberger B, Ruth C, Hajek T, Hartner FS, Glieder A. Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology. PLoS One. 2012;7:e39720. doi: 10.1371/journal.pone.0039720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The Transcriptional Landscape of the Yeast Genome Defined by RNA Sequencing. Science (80) 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- nath Banerjee H, Verma M. Search for a Novel Killer Toxin in Yeast Pichia pastoris. Plasmid. 2000;43:181–183. doi: 10.1006/plas.1999.1452. [DOI] [PubMed] [Google Scholar]
- Noskov VN, Chuang RY, Gibson DG, Leem SH, Larionov V, Kouprina N. Isolation of circular yeast artificial chromosomes for synthetic biology and functional genomics studies. Nat Protoc. 2010;6:89–96. doi: 10.1038/nprot.2010.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Maddox PS, Salmon EDD, Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J Cell Biol. 2001;152:1255–66. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail M, Smith ME, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y. A tale of three next generation sequencing platforms: comparison of Ion torrent, pacific biosciences and illumina MiSeq sequencers. BMC Genomics. 2012a;13:1. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renuse S, Madugundu AK, Kumar P, Nair BG, Gowda H, Prasad TSK, Pandey A. Proteomic analysis and genome annotation of Pichia pastoris, a recombinant protein expression host. PROTEOMICS. 2014 doi: 10.1002/pmic.201400267. [DOI] [PubMed] [Google Scholar]
- Roberts RJ, Carneiro MO, Schatz MC. The advantages of SMRT sequencing. Genome Biol. 2013;14:405. doi: 10.1186/gb-2013-14-7-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson RL, Jones R, Robson RM, Schwartz A, Richardson TH. Azotobacter Genomes: The Genome of Azotobacter chroococcum NCIMB 8003 (ATCC 4412) PLoS One. 2015;10:e0127997. doi: 10.1371/journal.pone.0127997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Sanyal K. Diversity in requirement of genetic and epigenetic factors for centromere function in fungi. Eukaryot Cell. 2011;10:1384–1395. doi: 10.1128/EC.05165-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustchenko EP, Curran TM, Sherman F. Variations in the number of ribosomal DNA units in morphological mutants and normal strains of Candida albicans and in normal strains of Saccharomyces cerevisiae. J Bacteriol. 1993;175:7189–7199. doi: 10.1128/jb.175.22.7189-7199.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffrath R, Meacock PA. An SSB encoded by and operating on linear killer plasmids from Kluyveromyces lactis. Yeast. 2001;18:1239–1247. doi: 10.1002/yea.773. [DOI] [PubMed] [Google Scholar]
- Schickel J, Helmig C, Meinhardt F. Kluyveromyces lactis killer system: Analysis of cytoplasmic promoters of the linear plasmids. Nucleic Acids Res. 1996;24:1879–1886. doi: 10.1093/nar/24.10.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutter K, De Lin Y, Tiels P, Hecke A, Van Glinka S, Peer Y, Van De Callewaert N, Weber-lehmann J, Rouze P. Genome sequence of the recombinant protein production host. Pichia pastoris. 2009 doi: 10.1038/nbt.1544. [DOI] [PubMed] [Google Scholar]
- Sonnhammer ELL, BEES Multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 1998;26:320–2. doi: 10.1093/nar/26.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sor F, Wèsolowski M, Fukuhara H. Inverted terminal repetitions of the two linear DNA associated with the killer character of the yeast Kluyveromyces lactis. Nucleic Acids Res. 1983;11:5037–5044. doi: 10.1093/nar/11.15.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam JC, Kwakman J, Meijer M, Stuitje AR. Efficient isolation of the linear DNA killer plasmid of Kluyveromyces lactis: evidence for location and expression in the cytoplasm and characterization of their terminally bound proteins. Nucleic Acids Res. 1986;14:6871–6884. doi: 10.1093/nar/14.17.6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- Stark MJ, Boyd A. The killer toxin of Kluyveromyces lactis: characterization of the toxin subunits and identification of the genes which encode them. EMBO J. 1986;5:1995–2002. doi: 10.1002/j.1460-2075.1986.tb04455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Parker R. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:92–101. doi: 10.1128/MCB.01023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga M, Wada N, Hishinuma F. Expression and identification of immunity determinants on linear DNA killer plasmids pGKL1 and pGKL2 in Kluyveromyces lactis. Nucleic Acids Res. 1987;15:1031–1046. doi: 10.1093/nar/15.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq 17. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk EL, Chen CL, d’Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Né P, Loeillet S, Nicolas A, Thermes C, Morillon A. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. TL -475. Nature. 2011;475 VN:114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- Varoquaux N, Liachko I, Ay F, Burton JN, Shendure J, Dunham MJ, Vert JP, Noble WS. Accurate identification of centromere locations in yeast genomes using Hi-C. Nucleic Acids Res. 2015;43:5331–5339. doi: 10.1093/nar/gkv424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe T, Schramke V, Hamilton GL, White SA, Teng G, Martienssen RA, Allshire RC. RNA interference is required for normal centromere function in fission yeast. Chromosom Res. 2003;11:137–146. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- Wickner RB. The killer double-stranded RNA plasmids of yeast. Plasmid. 1979;2:303–322. doi: 10.1016/0147-619x(79)90015-5. [DOI] [PubMed] [Google Scholar]
- Wickner RB, Leibowitz MJ. Chromosomal genes essential for replication of a double-stranded RNA plasmid of Saccharomyces cerevisiae: The killer character of yeast. J Mol Biol. 1976;105:427–443. doi: 10.1016/0022-2836(76)90102-9. [DOI] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bähler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- Wilson DW, Meacock PA. Extranuclear gene expression in yeast: evidence for a plasmid-encoded RNA polymerase of unique structure. Nucleic Acids Res. 1988;16:8097–8112. [PMC free article] [PubMed] [Google Scholar]
- Worsham PL, Bolen PL. Killer toxin production in Pichia acaciae is associated with linear DNA plasmids. Curr Genet. 1990;18:77–80. doi: 10.1007/BF00321119. [DOI] [PubMed] [Google Scholar]
- Wu J, Delneri D, Keefe RTO. Europe PMC Funders Group Non-coding RNAs in Saccharomyces cerevisiae : What is the function? 2014;40:907–911. doi: 10.1042/BST20120042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.