Abstract
The relationship between cyclin D1 (CCND1) rs9344 G>A polymorphism and colorectal cancer (CRC) risk is still ambiguous. To obtain a precise estimation of the relationship, we performed an extensive meta-analysis based on the eligible studies. Crude odds ratios with their 95% confidence intervals were harnessed to determine the strength of correlation between CCND1 rs9344 G>A polymorphism and CRC risk under the allele, the homozygote, the dominant, and the recessive genetic models, respectively (28 studies with 5,784 CRC cases and 7,858 controls). Our results indicated evidence of the association between CCND1 rs9344 G>A polymorphism and the increased risk of CRC in four genetic models: A vs G, AA vs GG, AA+GA vs GG, and AA vs GA+GG. In a stratified analysis by cancer type of CRC, there was an increased risk of sporadic CRC found in three genetic models: A vs G, AA vs GG, and AA+GA vs GG. In a stratified analysis by ethnicity, there was an increased CRC risk found among Asians in allele comparison genetic models, as well as Caucasians in two genetic models: AA+GA vs GG and A vs T. In summary, this meta-analysis demonstrates that CCND1 rs9344 G>A polymorphism may be a risk factor for CRC.
Keywords: polymorphism, CCND1, colorectal cancer, susceptibility, meta-analysis
Introduction
In 2012, colorectal cancer (CRC) is the third and second most commonly diagnosed malignancy in males and females, respectively, worldwide, with an estimated 1,360,600 new CRC cases and 693,900 CRC-related mortality occurring annually.1 This type of malignancy involves a more frequent sporadic CRC (sCRC) and a less frequent hereditary form. The increasing CRC incidence and mortality rate have been attributed to an increasingly “Westernized lifestyle,” including a decreased consumption of dietary fiber, drinking, smoking, overweight, and being physically inactive.2 However, the etiology of CRC is very complicated. A number of altered environmental and genetic factors have been considered as risk factors for CRC.3,4 Recently, a previous study showed that ~35% of CRC patients could be attributed to certain inherited genetic risk factors.5 Identification of these important genetic risk factors correlated with CRC may enrich our view of this complex disease.
The cyclin D1 (CCND1) gene located on chromosome 1q31–32. CCND1 is an important protein for the regulation of the G1–S phase transition of cell cycle. Overexpression or disordered regulation of the CCND1 gene will break the balance of cell cycle and might lead to abnormalities and consequently result in cellular transformation and malignancy. Recent studies showed that CCND1 was overexpressed in CRC, which was correlated with a poor clinical outcome and some clinicopathological characteristics.6,7
The human CCND1 gene is very polymorphic (http://www.ncbi.nlm.nih.gov/SNP). The CCND1 rs9344, a G to A polymorphism at nucleotide 870 in exon 4, increases the frequency of alternate splicing. Results of prior studies showed that the A allele of CCND1 rs9344 G>A resulted in an increasing level of mRNA (transcript-b) encoding CCND1 protein with an altered C-terminal domain.8,9 Results of some epidemiologic studies demonstrated that CCND1 rs9344 G>A polymorphism might confer CRC risk.10–18 Several meta-analyses showed that CCND1 rs9344 G>A polymorphism might be a risk factor for CRC, especially in the subgroups of sCRC and Caucasians.19–21 However, in these studies, as only a few case–control studies performed on the Asian populations, the power of these pooled analyses might be limited. Recently, more epidemiologic studies focusing on the relationship between CCND1 rs9344 G>A polymorphism and CRC risk were conducted among Asians. Considering the vital role of CCND1 rs9344 G>A polymorphism for CRC risk, an updated meta-analysis was needed to obtain a more precise assessment.
Materials and methods
Search strategy
PubMed and EMBASE online databases (updated to February 11, 2016) were searched using the corresponding keywords related to CCND1 rs9344 G>A polymorphism and CRC: cyclin D1 or CCND1; and polymorphism, variant, or single-nucleotide polymorphism; colorectal, rectal, or colon; and cancer, carcinoma, tumor, malignancy, or neoplasm. No language restriction was applied. We also searched the bibliography of reviews, meta-analyses, and all eligible articles to retrieve the potential publications.
Inclusion and exclusion criteria
The included studies were selected according to the major criteria as follows: 1) case–control studies; 2) the association of CCND1 rs9344 G>A polymorphism with CRC risk; 3) CRC cases diagnosed by histopathology; and 4) genotype frequencies to determine the pooled odds ratios (ORs) with their 95% confidence intervals (95% CIs). Accordingly, publications with insufficient data, reviews and meta-analyses, and comments were excluded.
Data extraction
For each included study, two authors (HQ and CC) extracted the data independently as follows: the first author’s surname; year of publication; country where the study was carried out; race (included Asians, Caucasians, and Mixed); the type of CRC (included hereditary non-polyposis colorectal cancer [HNPCC] and sCRC); the source of controls (included hospital-based study [HB], population-based study, and family-based study); genotyping method; sample size (numbers of cases/controls), genotypes; and the Hardy–Weinberg equilibrium (HWE) in the controls. If these two authors could not reach a consensus, the third author (YW) was consulted to resolve the dispute by discussion.
Statistical analysis
The distribution of genotypes in controls was calculated for departure from HWE by an online test (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl). The crude ORs with their 95% CIs were used to determine the strength of correlation between CCND1 rs9344 G>A polymorphism and CRC risk. Heterogeneity assumption was assessed by the chi-square-based Q-test and I2 test. I2>50% or P<0.10 indicates statistical heterogeneity among studies,22 so the pooled ORs and CIs were measured by the random-effects model (the DerSimoian and Laird method).23 Otherwise, the fixed-effects model (the Mantel–Haenszel method) was used.24 In order to check the ethnicity and the type of CRC effects, subgroup analyses were performed. Moreover, one-way sensitivity analysis was performed. Publication bias was tested by visual inspection of funnel plots and formally determined by Begg’s adjusted rank correlation test and Egger’ linear regression test.25 All statistical calculations were conducted with STATA version 12.0 (Stata Corporation, College Station, TX, USA). All P-values were two-sided, and P<0.05 was defined as statistically significant.
Results
Characteristics
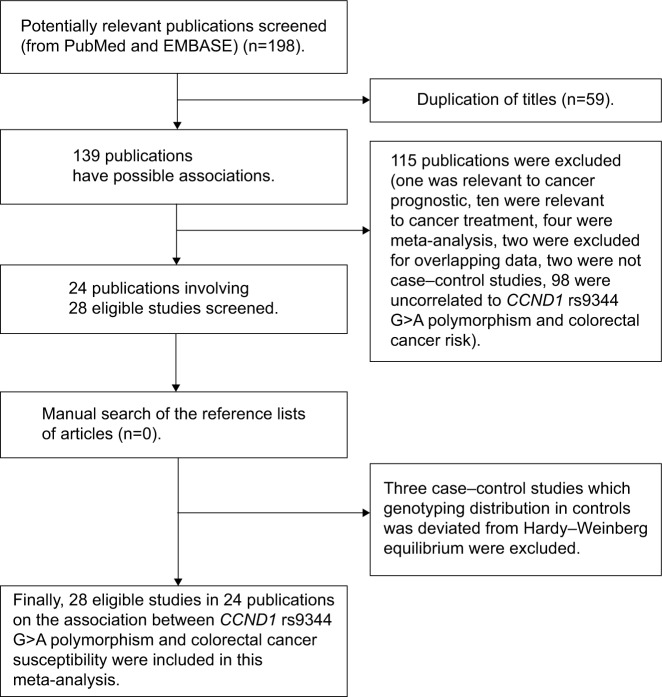
A total of 198 relevant publications were retrieved. There were several subgroups in certain publications,15,16,26 and we treated them separately. We listed the major screening process in Figure 1. Finally, there were 28 eligible studies included in the pooled analysis.12–18,26–42 There were 9 studies conducted in Asians,12,13,15,18,27,30,33,37 16 studies conducted in Caucasians,14–17,26,28,32,34–36,38–41 and 3 studies conducted in mixed populations.29,31,42 Of these articles, 22 investigated sCRC,12–18,26–38 and 6 investigated HNPCC.16,26,39–42 And the detailed characteristics of the included studies12–18,26–42 and the distribution of the CCND1 rs9344 G>A polymorphism as well as alleles are listed in Tables 1 and 2, respectively.
Figure 1.
Flow diagram of candidate studies selection process.
Abbreviation: CCND1, cyclin D1.
Table 1.
Characteristics of the candidate studies in the meta-analysis
| Study | Year | Country | Ethnicity | Type of CRC | Genotyping method | No of case/control | Source of controls |
|---|---|---|---|---|---|---|---|
| Govatati et al12 | 2014 | India | Asians | sCRC | DNA sequencing | 103/107 | HB |
| Sameer et al27 | 2013 | India | Asians | sCRC | PCR-RFLP | 130/160 | PB |
| Jelonek et al17 | 2010 | Poland | Caucasians | sCRC | PCR-RFLP | 50/153 | PB |
| Yaylim-Eraltan et al28 | 2010 | Turkey | Caucasians | sCRC | PCR-RFLP | 57/117 | HB |
| Kanaan et al29 | 2010 | USA | Mixed | sCRC | PCR-HLC | 75/93 | HB |
| Liu et al30 | 2010 | China | Asians | sCRC | PCR-RFLP | 373/838 | PB |
| Forones et al31 | 2008 | Brazil | Mixed | sCRC | PCR-RFLP | 123/120 | HB |
| Tan et al32 | 2008 | Germany | Caucasians | sCRC | PCR-RFLP | 498/600 | PB |
| Talseth et al39 | 2008 | Australia/Poland | Caucasians | HNPCC | TaqMan | 157/153 | HB |
| Grunhage et al26 | 2008 | Germany | Caucasians | HNPCC | PCR-RFLP | 98/218 | HB |
| Grunhage et al26 | 2008 | Germany | Caucasians | sCRC | PCR-RFLP | 96/218 | HB |
| Jing et al37 | 2008 | China | Asians | sCRC | TaqMan | 104/205 | HB |
| Josifovski et al38 | 2007 | Macedonia | Caucasians | sCRC | PCR-RFLP | 331/101 | HB |
| Kruger et al40 | 2006 | Germany | Caucasians | HNPCC | Multiplex-PCR | 315/245 | PB |
| Probst-Hensch et al33 | 2006 | Singapore | Asians | sCRC | TaqMan | 300/1,169 | PB |
| Schernhammer et al34 | 2006 | USA | Caucasians | sCRC | TaqMan | 610/1,237 | PB |
| Jiang et al13 | 2006 | India | Asians | sCRC | PCR-RFLP | 301/291 | HB |
| Hong et al18 | 2005 | Singapore | Asians | sCRC | PCR-RFLP | 254/101 | PB |
| Grieu et al35 | 2003 | Australia | Caucasians | sCRC | PCR-SSCP | 569/327 | HB |
| Le Marchand et al15 | 2003 | USA | Asians | sCRC | PCR-RFLP | 70/83 | PB |
| Le Marchand et al15 | 2003 | USA | Asians | sCRC | PCR-RFLP | 296/380 | PB |
| Le Marchand et al15 | 2003 | USA | Caucasians | sCRC | PCR-RFLP | 138/161 | PB |
| Porter et al16 | 2002 | UK | Caucasians | HNPCC | PCR-RFLP | 99/171 | PB |
| Porter et al16 | 2002 | UK | Caucasians | sCRC | PCR-RFLP | 235/171 | PB |
| Bala and Peltomaki41 | 2001 | Finland | Caucasians | HNPCC | PCR-SSCP | 146/186 | FB |
| Kong et al14 | 2001 | USA | Caucasians | sCRC | PCR-SSCP | 156/152 | PB |
| McKay et al36 | 2000 | UK | Caucasians | sCRC | PCR-RFLP | 100/101 | PB |
| Kong et al42 | 2000 | USA | Mixed | HNPCC | PCR-SSCP | 49/37 | FB |
Abbreviations: FB, family-based study; HB, hospital-based study; HNPCC, hereditary nonpolyposis colorectal cancer; PB, population-based; PCR-HLC, polymerase chain reaction high-performance liquid chromatography; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; PCR-SSCP, polymerase chain reaction single-stranded conformation polymorphism; sCRC, sporadic colorectal cancer.
Table 2.
Distribution of CCND1 rs9344 G>A polymorphism genotypes and alleles
| Study | Year | Case
|
Control
|
Case
|
Control
|
HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | GA | AA | GG | GA | AA | A | G | A | G | |||
| Govatati et al12 | 2014 | 54 | 39 | 10 | 71 | 33 | 3 | 59 | 147 | 39 | 175 | Yes |
| Sameer et al27 | 2013 | 19 | 70 | 41 | 41 | 76 | 43 | 152 | 108 | 162 | 158 | Yes |
| Jelonek et al17 | 2010 | 12 | 33 | 5 | 44 | 71 | 38 | 43 | 57 | 147 | 159 | Yes |
| Yaylim-Eraltan et al28 | 2010 | 9 | 28 | 20 | 29 | 60 | 28 | 68 | 46 | 116 | 118 | Yes |
| Kanaan et al29 | 2010 | 19 | 39 | 17 | 24 | 48 | 21 | 73 | 77 | 90 | 96 | Yes |
| Liu et al30 | 2010 | 66 | 187 | 120 | 160 | 429 | 249 | 427 | 319 | 927 | 749 | Yes |
| Forones et al31 | 2008 | 36 | 66 | 21 | 34 | 67 | 19 | 108 | 138 | 105 | 135 | Yes |
| Tan et al32 | 2008 | 120 | 263 | 115 | 147 | 310 | 143 | 493 | 503 | 596 | 604 | Yes |
| Talseth et al39 | 2008 | 34 | 78 | 45 | 42 | 80 | 31 | 168 | 146 | 142 | 164 | Yes |
| Grunhage et al26 | 2008 | 13 | 50 | 35 | 48 | 109 | 61 | 120 | 76 | 231 | 205 | Yes |
| Grunhage et al26 | 2008 | 24 | 43 | 29 | 48 | 109 | 61 | 101 | 91 | 231 | 205 | Yes |
| Jing et al37 | 2008 | 11 | 61 | 32 | 41 | 113 | 51 | 125 | 83 | 215 | 195 | Yes |
| Josifovski et al38 | 2007 | 77 | 153 | 100 | 25 | 51 | 25 | 353 | 307 | 101 | 101 | Yes |
| Kruger et al40 | 2006 | 110 | 144 | 61 | 73 | 121 | 51 | 266 | 364 | 223 | 267 | Yes |
| Probst-Hensch et al33 | 2006 | 56 | 132 | 112 | 207 | 548 | 414 | 356 | 244 | 1,376 | 962 | Yes |
| Schernhammer et al34 | 2006 | 125 | 311 | 174 | 264 | 593 | 380 | 659 | 561 | 1,353 | 1,121 | Yes |
| Jiang et al13 | 2006 | 46 | 130 | 125 | 56 | 145 | 90 | 380 | 222 | 325 | 257 | Yes |
| Hong et al18 | 2005 | 55 | 128 | 71 | 12 | 50 | 39 | 270 | 238 | 128 | 74 | Yes |
| Grieu et al35 | 2003 | 142 | 313 | 114 | 90 | 158 | 79 | 541 | 597 | 316 | 338 | Yes |
| Le Marchand et al15 | 2003 | 5 | 35 | 30 | 18 | 35 | 30 | 95 | 45 | 95 | 71 | Yes |
| Le Marchand et al15 | 2003 | 75 | 143 | 78 | 96 | 195 | 89 | 299 | 293 | 373 | 387 | Yes |
| Le Marchand et al15 | 2003 | 29 | 75 | 34 | 50 | 85 | 26 | 143 | 133 | 137 | 185 | Yes |
| Porter et al16 | 2002 | 30 | 47 | 22 | 60 | 81 | 30 | 91 | 107 | 141 | 201 | Yes |
| Porter et al16 | 2002 | 55 | 128 | 52 | 60 | 81 | 30 | 232 | 238 | 141 | 201 | Yes |
| Bala and Peltomaki41 | 2001 | 50 | 70 | 26 | 47 | 97 | 42 | 122 | 170 | 181 | 191 | Yes |
| Kong et al14 | 2001 | 36 | 71 | 49 | 45 | 84 | 23 | 169 | 143 | 130 | 174 | Yes |
| McKay et al36 | 2000 | 25 | 58 | 17 | 34 | 50 | 17 | 92 | 108 | 84 | 118 | Yes |
| Kong et al42 | 2000 | 9 | 36 | 4 | 10 | 21 | 6 | 44 | 54 | 33 | 41 | Yes |
Abbreviation: HWE, Hardy–Weinberg equilibrium.
Quantitative synthesis
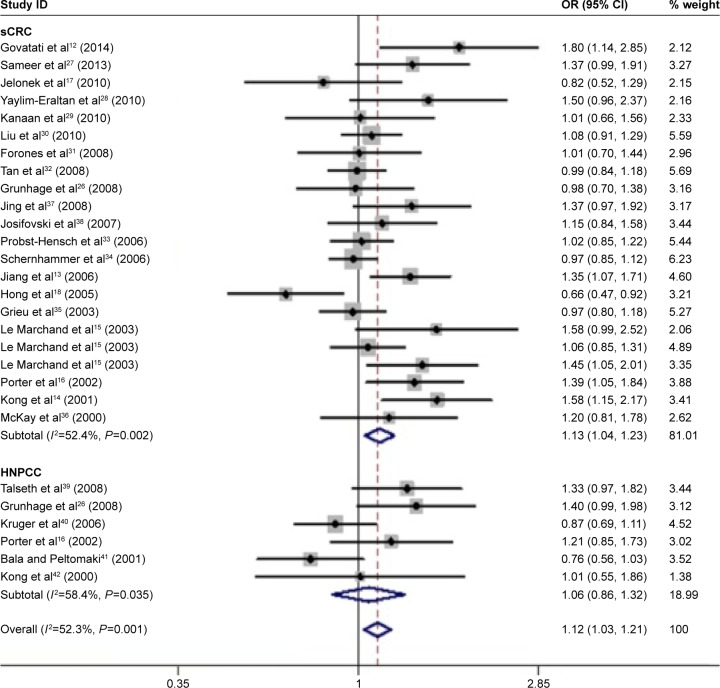
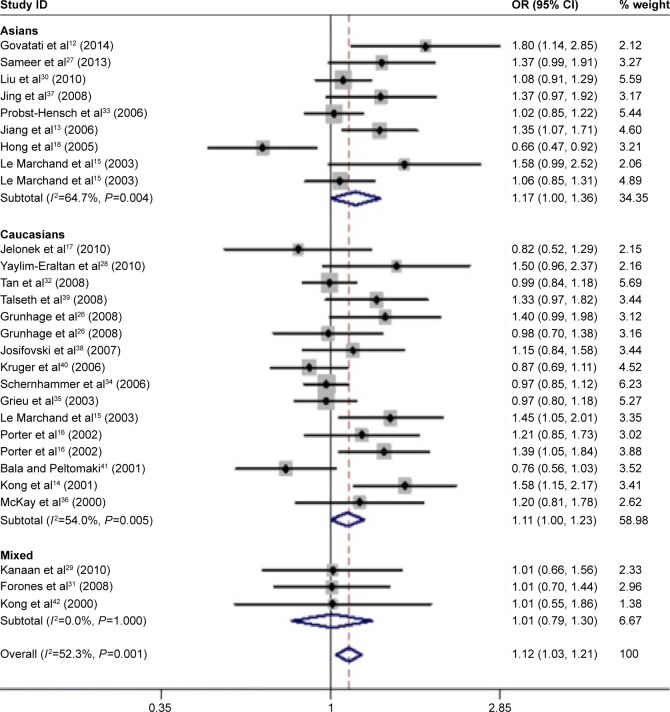
In total, 28 eligible studies12–18,26–42 with 5,784 CRC cases and 7,858 controls were included in our meta-analysis. Overall, the CCND1 rs9344 G>A polymorphism was associated with the overall CRC risk in four genetic models (A vs G: OR, 1.12; 95% CI: 1.03–1.21, P=0.005; AA vs GG: OR, 1.25; 95% CI: 1.06–1.48, P=0.008; AA+GA vs GG: OR, 1.18; 95% CI: 1.05–1.33, P=0.007; AA vs GA+GG: OR, 1.13; 95% CI: 1.05–1.28, P=0.042; Table 3 and Figure 2). In a subgroup analysis by CRC type, the CCND1 rs9344 G>A polymorphism was associated with an increased risk of sCRC in three genetic models (A vs G: OR, 1.13; 95% CI: 1.04–1.23, P=0.004; AA vs GG: OR, 1.28; 95% CI: 1.07–1.54, P=0.008; AA+GA vs GG: OR, 1.20; 95% CI: 1.06–1.36, P=0.004; Table 3 and Figure 2), but not of HNPCC. In a subgroup analysis by ethnicity, an increased CRC risk was found among Caucasians in two genetic models (A vs G: OR, 1.11; 95% CI: 1.00–1.23, P=0.049; AA+GA vs GG: OR, 1.16; 95% CI: 1.01–1.33, P=0.041; Table 3 and Figure 3), and among Asians in one genetic model (A vs G: OR, 1.17; 95% CI: 1.00–1.36, P=0.048; Table 3 and Figure 3), but not mixed populations.
Table 3.
Meta-analysis of the CCND1 rs9344 G>A polymorphism and CRC risk
| Group | No of study | A vs G
|
AA vs GG
|
AA+GA vs GG
|
AA vs GA+GG
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P-value |
P-value (Q-test) |
OR (95% CI) | P-value |
P-value (Q-test) |
OR (95% CI) | P-value |
P-value (Q-test) |
OR (95% CI) | P-value |
P-value (Q-test) |
||
| Total | 28 | 1.12 (1.03–1.21) | 0.005 | 0.001 | 1.25 (1.06–1.48) | 0.008 | <0.001 | 1.18 (1.05–1.33) | 0.007 | 0.013 | 1.13 (1.0–1.28) | 0.042 | 0.005 |
| Ethnicity | |||||||||||||
| Asians | 9 | 1.17 (1.00–1.36) | 0.048 | 0.004 | 1.38 (0.99–1.94) | 0.059 | 0.002 | 1.26 (0.96–1.65) | 0.092 | 0.005 | 1.18 (0.98–1.42) | 0.074 | 0.092 |
| Caucasians | 16 | 1.11 (1.00–1.23) | 0.049 | 0.005 | 1.23 (1.00–1.53) | 0.055 | 0.005 | 1.16 (1.01–1.33) | 0.041 | 0.090 | 1.13 (0.95–1.35) | 0.167 | 0.005 |
| Mixed | 3 | 1.01 (0.79–1.30) | 0.944 | 1.000 | 0.99 (0.58–1.71) | 0.978 | 0.925 | 1.06 (0.71–1.58) | 0.767 | 0.653 | 0.95 (0.60–1.51) | 0.830 | 0.519 |
| Type of CRC | |||||||||||||
| sCRC | 22 | 1.13 (1.04–1.23) | 0.004 | 0.002 | 1.28 (1.07–1.54) | 0.008 | 0.001 | 1.20 (1.06–1.36) | 0.004 | 0.045 | 1.14 (1.00–1.31) | 0.054 | 0.004 |
| HNPCC | 6 | 1.06 (0.86–1.32) | 0.578 | 0.035 | 1.13 (0.73–1.76) | 0.581 | 0.037 | 1.08 (0.78–1.51) | 0.630 | 0.051 | 1.10 (0.88–1.37) | 0.420 | 0.177 |
| Source of control | |||||||||||||
| HB | 11 | 1.19 (1.08–1.30) | <0.001 | 0.161 | 1.38 (1.14–1.68) | 0.001 | 0.140 | 1.27 (1.08–1.48) | 0.003 | 0.484 | 1.25 (1.07–1.45) | 0.004 | 0.134 |
| PB | 15 | 1.09 (0.99–1.21) | 0.085 | 0.003 | 1.21 (0.97–1.51) | 0.088 | 0.001 | 1.16 (0.99–1.37) | 0.065 | 0.012 | 1.09 (0.94–1.27) | 0.263 | 0.013 |
| FB | 2 | 0.80 (0.61–1.06) | 0.120 | 0.404 | 0.60 (0.34–1.08) | 0.089 | 0.778 | 0.77 (0.50–1.18) | 0.227 | 0.106 | 0.69 (0.42–1.15) | 0.157 | 0.516 |
Note: Statistically significant values are shown in bold.
Abbreviations: CRC, colorectal cancer; CI, confidence interval; FB, family-based study; HB, hospital-based study; HNPCC, hereditary nonpolyposis colorectal cancer; HWE, Hardy–Winberg equilibrium; OR, odds ratio; PB, population-based; sCRC, sporadic colorectal cancer.
Figure 2.
Meta-analysis with a random–effects model in the different type for the association between CCND1 rs9344 G>A polymorphism and CRC risk (A vs G genetic model).
Note: Weights are from random-effects analysis.
Abbreviations: CI, confidence interval; CRC, colorectal cancer; HNPCC, hereditary nonpolyposis colorectal cancer; OR, odds ratio; sCRC, sporadic colorectal cancer.
Figure 3.
Meta-analysis with a random–effects model in different races for the association between the CCND1 rs9344 G>A polymorphism and CRC risk (A vs G genetic model).
Note: Weights are from random-effects analysis.
Abbreviations: CI, confidence interval; CRC, colorectal cancer; OR, odds ratio.
Tests for publication bias, sensitivity analyses, and heterogeneity
Begg’s funnel plot and Egger’ linear regression test were harnessed to examine potential publication bias. As shown in Figure 4, no significant publication bias was detected in our study (Begg’s test P=0.514; Egger’s test P=0.259).
Figure 4.
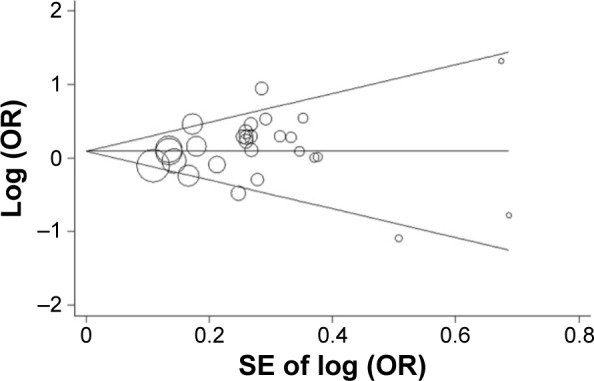
Begg’s funnel plot of meta-analysis of the relationship between the CCND1 rs9344 G>A polymorphism and CRC risk (AA vs GA+GG genetic model).
Abbreviations: CRC, colorectal cancer; OR, odds ratio; SE, standard error.
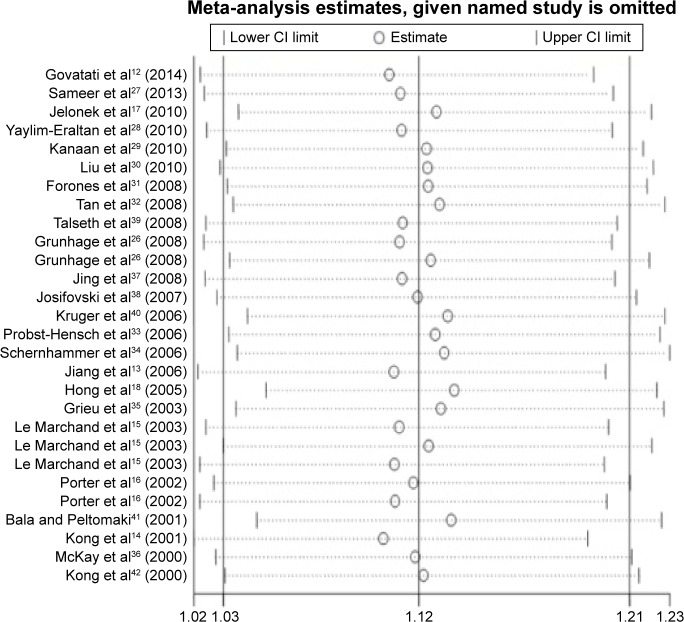
Influence of an individual study on the pooled ORs and CIs was also determined by omitting it in turn and repeating the meta-analysis.43 The results indicated that no individual study significantly altered the pooled ORs and CIs (Figure 5).
Figure 5.
Sensitivity analysis of the influence of A vs G genetic model in overall CRC meta-analysis (random–effects estimates).
Abbreviations: CI, confidence interval; CRC, colorectal cancer.
As shown in Table 3, there was significant heterogeneity in all genetic models. Because ethnicity, the type of CRC, and source of controls can affect the heterogeneity, subgroup analyses were conducted. Results showed that Asians, Caucasians, population-based study, HB study, and sCRC subgroups may contribute to the major source of heterogeneity.
Discussion
CCND1 may act as an important regulator in the evolution of malignancy by influencing cell proliferation, differentiation, and apoptosis. It has been reported that the G1–S transition of the cell cycle is controlled by sequential activation of cyclin/cyclin-dependent kinase (CDK) complexes.44 The CCND1, a vital cell cycle regulatory protein, regulates transition of G1–S phase during cell division. High activity of CCND1 leads to premature cell passage through the G1–S transition, resulting in proliferation of unrepaired DNA damage and genetic errors, thus leading to selective advantage for abnormal cell propagation.45 Previous studies indicated that CCND1 was overexpressed in a number of malignancies.6,46 Owing to these important roles in carcinogenesis, polymorphisms of CCND1 may be implicated in accelerating the development and/or progression of CRC.
Of late, numerous epidemiologic investigations focused on the relationship of the CCND1 polymorphism with CRC risk.12–18,26–42 The most prevalent CCND1 gene polymer phism, rs9344 G>A, has been most widely explored. High activity of CCND1 is common in a lot of human tumors.47,48 Several case–control studies have reported a positive signal of the CCND1 rs9344 G>A polymorphism with the risk of CRC;10–16 however, others have reported negative signal.17,18 Because of conflicting results and the insufficient sample size of individual studies, the final decision was far from certain. Because meta-analysis is a powerful way for pooling the results of all included studies with a more power, it can get more robust results than an individual study.49 Our findings showed that the presence of the CCND1 rs9344 A allele, which elevate CCND1 activity,8,9 might confer the susceptibility to CRC. In addition, subgroup analyses were performed regarding ethnicity and the type of CRC for this polymorphism. CCND1 rs9344 G>A polymorphism increased the risk of CRC among Asians, Caucasians, and sCRC. Results of the current meta-analysis indicated the influence of the CCND1 rs9344 G>A polymorphism and diversity on the type of CRC. However, our results should be interpreted with very caution. For HNPCC, only six studies with small sample sizes were included in this group, which may restrict the statistical power to obtain a final decision.16,26,39–42 When stratified by ethnicity, the CCND1 rs9344 G>A polymorphism was associated with CRC risk in both Asians and Caucasians. Additionally, in other genetic models, a borderline risk of CRC was also observed in these two ethnicities. Results of several previous meta-analyses showed that the CCND1 rs9344 G>A polymorphism might be a risk factor for CRC, especially in the subgroups of sCRC and Caucasians.19–21 Our results were very analogous to these pooled analyses. In addition, we also found that the CCND1 rs9344 G>A polymorphism might be a risk factor for CRC risk in Asians.
The CCND1 rs9344 G allele may provide an optimal splice donor site and produce a full transcript for CCND1 (transcript a), whereas the CCND1 rs9344 A allele results in a truncated transcript (transcript b).47,50,51 The well-described transcript (transcript a) interacts with and activates the downstream molecules, such as G1 CDK, CDK4, and CDK6. Then, the CCND1–CDK complex phosphorylates and inhibits the retinoblastoma tumor suppressor, which is necessary for the G1–S transition.52 However, a truncated transcript (transcript b) encodes the protein short of the point estimation by sequential testing (PEST) region in the C-terminal domain47 and decreases phosphorylation ability of retinoblastoma.53 On the other hand, the transcript b has a longer half-life than transcript a, which may result in an overexpression of CCND1. Subsequently, the CCND1 rs9344 G→A substitution could lead to facilitation of cell proliferation and increase the susceptibility of malignancy.50 The findings of our meta-analysis were consistent with the conclusion of previous functional studies mentioned earlier. The epidemiologic investigations provided evidence suggesting that CRC carcinogenesis may be multiple steps that involve both individual’s genetic and environmental factors. In the future, larger epidemiologic studies with a well-designed methodology are needed to confirm or refute these associations. Results of our pooled analysis may prompt further clinic investigation of diagnosis and prevention strategies.
There were some merits in this meta-analysis. First, the current meta-analysis was the most extensively study which explored the relationship of the CCND1 rs9344 G>A polymorphism with CRC susceptibility. Second, our results first confirmed that the CCND1 rs9344 G>A polymorphism was associated with CRC susceptibility among Asians.
Limitations
There were some limitations of our study. First, in some included studies, controls were selected from family member and non-cancer hospital patients, which might result in misclassification bias. Second, large heterogeneity was observed in our meta-analysis, which means our findings should be interpreted with caution. Finally, our findings were based on unadjusted ORs and CIs, while a more precise measurement should be adjusted by multiple risk factors, such as family history, smoking status, drinking, diabetes, body mass index, etc.
Conclusion
In summary, this meta-analysis suggests that the CCND1 rs9344 G>A polymorphism is correlated with increased risk of CRC. Moreover, these relationships were different across different cancer types of CRC, suggesting that large sample and well-designed epidemiologic studies are warranted to confirm or refute our findings.
Acknowledgments
The project was supported by the Natural Science Foundation of Universities and Colleges of Jiangsu Province (grant no 16KJB310002), the Natural Science Foundation of Fujian Province (grant no 2015J01435), the Medical Innovation Foundation of Fujian Province (grant no 2015-CX-9), and the National Clinical Key Specialty Construction Program.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Center MM, Jemal A, Smith RA, Ward E. Worldwide variations in colorectal cancer. CA Cancer J Clin. 2009;59(6):366–378. doi: 10.3322/caac.20038. [DOI] [PubMed] [Google Scholar]
- 3.Raskov H, Pommergaard HC, Burcharth J, Rosenberg J. Colorectal carcinogenesis – update and perspectives. World J Gastroenterol. 2014;20(48):18151–18164. doi: 10.3748/wjg.v20.i48.18151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinberg DS, Myers RE, Keenan E, et al. Genetic and environmental risk assessment and colorectal cancer screening in an average-risk population: a randomized trial. Ann Intern Med. 2014;161(8):537–545. doi: 10.7326/M14-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361(25):2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang MY, Chen HC, Yang IP, et al. Tumorigenesis and tumor progression related gene expression profiles in colorectal cancer. Cancer Biomark. 2013;13(4):269–279. doi: 10.3233/CBM-130350. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Wei J, Xu C, Zhao Z, You T. Prognostic significance of cyclin D1 expression in colorectal cancer: a meta-analysis of observational studies. PLoS One. 2014;9(4):e94508. doi: 10.1371/journal.pone.0094508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sobti RC, Kaur P, Kaur S, et al. Effects of cyclin D1 (CCND1) polymorphism on susceptibility to lung cancer in a North Indian population. Cancer Genet Cytogenet. 2006;170(2):108–114. doi: 10.1016/j.cancergencyto.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 9.Fan YZ, Fu JY, Zhao ZM, Chen CQ. Inhibitory effect of norcantharidin on the growth of human gallbladder carcinoma GBC-SD cells in vitro. Hepatobiliary Pancreat Dis Int. 2007;6(1):72–80. [PubMed] [Google Scholar]
- 10.Zahary MN, Ahmad Aizat AA, Kaur G, Yeong Yeh L, Mazuwin M, Ankathil R. Polymorphisms of cell cycle regulator genes G870A and C215G: association with colorectal cancer susceptibility risk in a Malaysian population. Oncol Lett. 2015;10(5):3216–3222. doi: 10.3892/ol.2015.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang CY, Tsai CW, Hsu CM, Chang WS, Shui HA, Bau DT. The significant association of CCND1 genotypes with colorectal cancer in Taiwan. Tumour Biol. 2015;36(8):6533–6540. doi: 10.1007/s13277-015-3347-9. [DOI] [PubMed] [Google Scholar]
- 12.Govatati S, Singamsetty GK, Nallabelli N, et al. Contribution of cyclin D1 (CCND1) and E-cadherin (CDH1) alterations to colorectal cancer susceptibility: a case-control study. Tumour Biol. 2014;35(12):12059–12067. doi: 10.1007/s13277-014-2505-9. [DOI] [PubMed] [Google Scholar]
- 13.Jiang J, Wang J, Suzuki S, et al. Elevated risk of colorectal cancer associated with the AA genotype of the cyclin D1 A870G polymorphism in an Indian population. J Cancer Res Clin Oncol. 2006;132(3):193–199. doi: 10.1007/s00432-005-0039-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong S, Wei Q, Amos CI, et al. Cyclin D1 polymorphism and increased risk of colorectal cancer at young age. J Natl Cancer Inst. 2001;93(14):1106–1108. doi: 10.1093/jnci/93.14.1106. [DOI] [PubMed] [Google Scholar]
- 15.Le Marchand L, Seifried A, Lum-Jones A, Donlon T, Wilkens LR. Association of the cyclin D1 A870G polymorphism with advanced colorectal cancer. JAMA. 2003;290(21):2843–2848. doi: 10.1001/jama.290.21.2843. [DOI] [PubMed] [Google Scholar]
- 16.Porter TR, Richards FM, Houlston RS, et al. Contribution of cyclin d1 (CCND1) and E-cadherin (CDH1) polymorphisms to familial and sporadic colorectal cancer. Oncogene. 2002;21(12):1928–1933. doi: 10.1038/sj.onc.1205245. [DOI] [PubMed] [Google Scholar]
- 17.Jelonek K, Gdowicz-Klosok A, Pietrowska M, et al. Association between single-nucleotide polymorphisms of selected genes involved in the response to DNA damage and risk of colon, head and neck, and breast cancers in a polish population. J Appl Genet. 2010;51(3):343–352. doi: 10.1007/BF03208865. [DOI] [PubMed] [Google Scholar]
- 18.Hong Y, Eu KW, Seow-Choen F, Fook-Chong S, Cheah PY. GG genotype of cyclin D1 G870A polymorphism is associated with increased risk and advanced colorectal cancer in patients in Singapore. Eur J Cancer. 2005;41(7):1037–1044. doi: 10.1016/j.ejca.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 19.Yang Y, Wang F, Shi C, Zou Y, Qin H, Ma Y. Cyclin D1 G870A polymorphism contributes to colorectal cancer susceptibility: evidence from a systematic review of 22 case-control studies. PLoS One. 2012;7(5):e36813. doi: 10.1371/journal.pone.0036813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J, Zhang G, Chen J. CCND1 G870A polymorphism is associated with increased risk of colorectal cancer, especially for sporadic colorectal cancer and in Caucasians: a meta-analysis. Clin Res Hepatol Gastroenterol. 2012;36(2):169–177. doi: 10.1016/j.clinre.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 21.Chen B, Cao L, Yang P, Zhou Y, Wu XT. Cyclin D1 (CCND1) G870A gene polymorphism is an ethnicity-dependent risk factor for digestive tract cancers: a meta-analysis comprising 20,271 subjects. Cancer Epidemiol. 2012;36(2):106–115. doi: 10.1016/j.canep.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 22.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 24.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 25.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grunhage F, Jungck M, Lamberti C, et al. Association of familial colorectal cancer with variants in the E-cadherin (CDH1) and cyclin D1 (CCND1) genes. Int J Colorectal Dis. 2008;23(2):147–154. doi: 10.1007/s00384-007-0388-6. [DOI] [PubMed] [Google Scholar]
- 27.Sameer AS, Parray FQ, Dar MA, et al. Cyclin D1 G870A polymorphism and risk of colorectal cancer: a case control study. Mol Med Rep. 2013;7(3):811–815. doi: 10.3892/mmr.2013.1287. [DOI] [PubMed] [Google Scholar]
- 28.Yaylim-Eraltan I, Arikan S, Yildiz Y, et al. The influence of cyclin D1 A870G polymorphism on colorectal cancer risk and prognosis in a Turkish population. Anticancer Res. 2010;30(7):2875–2880. [PubMed] [Google Scholar]
- 29.Kanaan Z, Eichenberger MR, Young M, et al. An alternative cyclin-D1 splice site is not linked to inflammatory bowel disease-associated neoplasia. Int J Biol Markers. 2010;25(1):27–31. doi: 10.1177/172460081002500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu B, Zhang Y, Jin M, et al. Association of selected polymorphisms of CCND1, p21, and caspase8 with colorectal cancer risk. Mol Carcinog. 2010;49(1):75–84. doi: 10.1002/mc.20579. [DOI] [PubMed] [Google Scholar]
- 31.Forones NM, de Lima JM, de Souza LG, da Silva ID. Cyclin D1 A870G polymorphism in Brazilian colorectal cancer patients. J Gastrointest Cancer. 2008;39(1–4):118–123. doi: 10.1007/s12029-009-9057-z. [DOI] [PubMed] [Google Scholar]
- 32.Tan XL, Nieters A, Kropp S, Hoffmeister M, Brenner H, Chang-Claude J. The association of cyclin D1 G870A and E-cadherin C-160A polymorphisms with the risk of colorectal cancer in a case control study and meta-analysis. Int J Cancer. 2008;122(11):2573–2580. doi: 10.1002/ijc.23363. [DOI] [PubMed] [Google Scholar]
- 33.Probst-Hensch NM, Sun CL, Van Den Berg D, Ceschi M, Koh WP, Yu MC. The effect of the cyclin D1 (CCND1) A870G polymorphism on colorectal cancer risk is modified by glutathione-S-transferase polymorphisms and isothiocyanate intake in the Singapore Chinese Health Study. Carcinogenesis. 2006;27(12):2475–2482. doi: 10.1093/carcin/bgl116. [DOI] [PubMed] [Google Scholar]
- 34.Schernhammer ES, Tranah GJ, Giovannucci E, et al. Cyclin D1 A870G polymorphism and the risk of colorectal cancer and adenoma. Br J Cancer. 2006;94(6):928–934. doi: 10.1038/sj.bjc.6603007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grieu F, Malaney S, Ward R, Joseph D, Iacopetta B. Lack of association between CCND1 G870A polymorphism and the risk of breast and colorectal cancers. Anticancer Res. 2003;23(5b):4257–4259. [PubMed] [Google Scholar]
- 36.McKay JA, Douglas JJ, Ross VG, et al. Cyclin D1 protein expression and gene polymorphism in colorectal cancer. Aberdeen Colorectal Initiative. Int J Cancer. 2000;88(1):77–81. doi: 10.1002/1097-0215(20001001)88:1<77::aid-ijc12>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 37.Jing JC XW, Jin H, Ji BN. Relationship between CCND1 gene A870G polymorphism and genetic susceptibility to colorectal cancer. World Chin J Digestol. 2008;16(22):4. [Google Scholar]
- 38.Josifovski T, Matevska N, Hiljadnikova-Bajro M, et al. Cyclin D1 G870A variant is associated with increased risk of microsatellite instability-positive colorectal cancer in young male patients. Balkan J Med Med Genetics. 2007;10(2):8. [Google Scholar]
- 39.Talseth BA, Ashton KA, Meldrum C, et al. Aurora-A and Cyclin D1 polymorphisms and the age of onset of colorectal cancer in hereditary nonpolyposis colorectal cancer. Int J Cancer. 2008;122(6):1273–1277. doi: 10.1002/ijc.23177. [DOI] [PubMed] [Google Scholar]
- 40.Kruger S, Engel C, Bier A, et al. Absence of association between cyclin D1 (CCND1) G870A polymorphism and age of onset in hereditary nonpolyposis colorectal cancer. Cancer Lett. 2006;236(2):191–197. doi: 10.1016/j.canlet.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 41.Bala S, Peltomaki P. CYCLIN D1 as a genetic modifier in hereditary nonpolyposis colorectal cancer. Cancer Res. 2001;61(16):6042–6045. [PubMed] [Google Scholar]
- 42.Kong S, Amos CI, Luthra R, Lynch PM, Levin B, Frazier ML. Effects of cyclin D1 polymorphism on age of onset of hereditary nonpolyposis colorectal cancer. Cancer Res. 2000;60(2):249–252. [PubMed] [Google Scholar]
- 43.Azzam A, Mathews CA. Meta-analysis of the association between the catecholamine-O-methyl-transferase gene and obsessive-compulsive disorder. Am J Med Genet B Neuropsychiatr Genet. 2003;123B(1):64–69. doi: 10.1002/ajmg.b.20013. [DOI] [PubMed] [Google Scholar]
- 44.Ekholm SV, Reed SI. Regulation of G(1) cyclin-dependent kinases in the mammalian cell cycle. Curr Opin Cell Biol. 2000;12(6):676–684. doi: 10.1016/s0955-0674(00)00151-4. [DOI] [PubMed] [Google Scholar]
- 45.Hall M, Peters G. Genetic alterations of cyclins, cyclin-dependent kinases, and Cdk inhibitors in human cancer. Adv Cancer Res. 1996;68:67–108. doi: 10.1016/s0065-230x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- 46.Quintayo MA, Munro AF, Thomas J, et al. GSK3beta and cyclin D1 expression predicts outcome in early breast cancer patients. Breast Cancer Res Treat. 2012;136(1):161–168. doi: 10.1007/s10549-012-2229-8. [DOI] [PubMed] [Google Scholar]
- 47.Knudsen KE, Diehl JA, Haiman CA, Knudsen ES. Cyclin D1: polymorphism, aberrant splicing and cancer risk. Oncogene. 2006;25(11):1620–1628. doi: 10.1038/sj.onc.1209371. [DOI] [PubMed] [Google Scholar]
- 48.Pabalan N, Bapat B, Sung L, Jarjanazi H, Francisco-Pabalan O, Ozcelik H. Cyclin D1 Pro241Pro (CCND1-G870A) polymorphism is associated with increased cancer risk in human populations: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2008;17(10):2773–2781. doi: 10.1158/1055-9965.EPI-08-0169. [DOI] [PubMed] [Google Scholar]
- 49.Munafo MR, Flint J. Meta-analysis of genetic association studies. Trends Genet. 2004;20(9):439–444. doi: 10.1016/j.tig.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 50.Betticher DC, Thatcher N, Altermatt HJ, Hoban P, Ryder WD, Heighway J. Alternate splicing produces a novel cyclin D1 transcript. Oncogene. 1995;11(5):1005–1011. [PubMed] [Google Scholar]
- 51.Solomon DA, Wang Y, Fox SR, et al. Cyclin D1 splice variants. Differential effects on localization, RB phosphorylation, and cellular transformation. J Biol Chem. 2003;278(32):30339–30347. doi: 10.1074/jbc.M303969200. [DOI] [PubMed] [Google Scholar]
- 52.Adams PD. Regulation of the retinoblastoma tumor suppressor protein by cyclin/cdks. Biochim Biophys Acta. 2001;1471(3):M123–M133. doi: 10.1016/s0304-419x(01)00019-1. [DOI] [PubMed] [Google Scholar]
- 53.Leveque C, Marsaud V, Renoir JM, Sola B. Alternative cyclin D1 forms a and b have different biological functions in the cell cycle of B lymphocytes. Exp Cell Res. 2007;313(12):2719–2729. doi: 10.1016/j.yexcr.2007.04.018. [DOI] [PubMed] [Google Scholar]