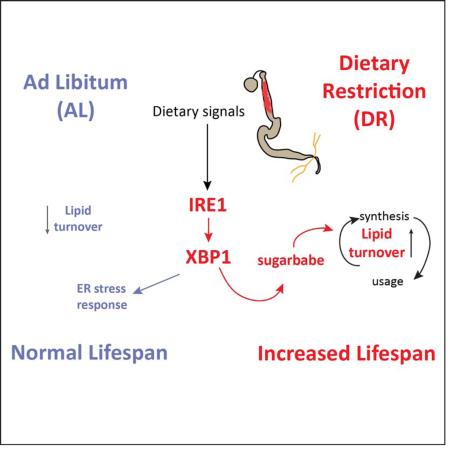
Abstract
Dietary restriction (DR) is one of the most robust lifespan-extending interventions in animals. The beneficial effects of DR involve a metabolic adaptation towards increased triglyceride usage. The regulatory mechanism and the tissue specificity of this metabolic switch remain unclear. Here we show that the IRE1/XBP1 ER stress signaling module mediates metabolic adaptation upon DR in flies by promoting triglyceride synthesis and accumulation in enterocytes (ECs) of the Drosophila midgut. Consistently, IRE1/XBP1 function in ECs is required for increased longevity upon DR. We further identify sugarbabe, a Gli-like zinc finger transcription factor, as a key mediator of the IRE1/XBP1 regulated induction of de novo lipogenesis in ECs. Overexpression of sugarbabe rescues metabolic and lifespan phenotypes of IRE1 loss of function conditions. Our study highlights the critical role of metabolic adaptation of the intestinal epithelium for DR-induced lifespan extension and explores the IRE1/XBP1 signaling pathway regulating this adaptation and influencing lifespan.
Graphical abstract
INTRODUCTION
Dietary Restriction (DR), defined as a regime of limited protein intake without malnutrition, leads to increased lifespan and healthspan in all tested model organisms (Fontana and Partridge, 2015; Kapahi et al., 2010). One of the conserved fundamental adaptations to DR, or to other low-nutrient conditions such as fasting, involves a metabolic shift towards increased triglyceride utilization. DR increases the conversion of dietary carbohydrates into lipids, elevates fat storage, and accelerates lipid turnover in flies, which appears to have a profound positive impact on longevity (Katewa et al., 2012; Longo and Mattson, 2014). Increased lipid turnover upon caloric restriction and increased adiposity upon protein restriction (which correlate with increased lifespan) has also been described in mice (Bruss et al., 2010).
Drosophila has emerged as an excellent model organism to explore the mechanisms driving diet and/or age-related changes in lipid metabolism. Importantly, Drosophila provides critical technical advantages that allow characterizing tissue-tissue coordination during metabolic adaptation (Chatterjee et al., 2014; Katewa et al., 2012; Leopold and Perrimon, 2007). While lipids are stored in the fatbody and transferred to oenocytes for mobilization (Chatterjee et al., 2014; Gutierrez et al., 2007), the Drosophila intestine also contributes to lipid synthesis and cholesterol homeostasis (Sieber and Thummel, 2012; Song et al., 2014). The Drosophila intestine plays a key role in modulating healthspan by modulation of immune responses, metabolic homeostasis and stress signaling (Biteau et al., 2011; Wang et al., 2014a).
The adult intestine is regenerated by intestinal stem cells (ISCs), which divide to replace functional enterocytes (ECs) and enteroendocrine cells when needed (Biteau et al., 2011). The intestine is also central to longevity in Drosophila, as gut function rapidly declines in aging flies (Biteau et al., 2011; Guo et al., 2014). Furthermore, in old flies, the ability of the intestine to generate and store lipids is severely compromised (Karpac et al., 2013), and restoring the adequate metabolic function of this tissue increases healthspan (Biteau et al., 2010; Karpac et al., 2013). The age-related decline in intestinal function in flies is a consequence of complex inflammatory conditions that are associated with increased protein misfolding (Wang et al., 2014b, 2015). How ER stress and ER stress response pathways influence diet and/or age-related metabolic function of the intestinal epithelium remains unclear.
The ER stress transducer IRE1 triggers one of the three signaling pathways engaged by ER stress. Interestingly, IRE1 also influences lipid homeostasis (Nikawa and Yamashita, 1992; Volmer and Ron, 2015). IRE1 is also required for S6K and HIF-1 mediated lifespan extension under DR in C elegans, though the mechanisms mediating this effect remain unclear (Chen et al., 2009). During ER stress, IRE1 dimerizes and splices the mRNA of XBP1, leading to translation of a functional transcription factor that induces genes involved in ER biogenesis, protein folding and degradation, to restore ER homeostasis (Ron and Walter, 2007). IRE1 splices and degrades several other mRNAs in a process dubbed RIDD (Regulated IRE1-Dependent Decay), alleviating the protein load into the ER (Hollien and Weissman, 2006). IRE1alpha further regulates a shift to a ketogenic metabolism under starvation conditions in the mouse liver (Shao et al., 2014), and mice deficient for XBP1 develop insulin resistance and are prone to diabetes (Ozcan et al., 2004). To regulate lipogenesis, XBP1 controls target genes that are functionally distinct from its canonical ER stress-related targets, and are regulated by distinct sequence motifs (Acosta-Alvear et al., 2007; Sha et al., 2009).
The role of IRE1/XBP1 in the regulation of lipid homeostasis has not been explored in the context of a DR intervention or during conditions of obligatory lipid recruitment, such as prolonged fasting/starvation. Here we identify IRE1 as a player in DR-induced lifespan extension in flies. Our data suggest that IRE1 is required for the metabolic shift towards elevated triglyceride turnover occurring during DR, and that absence of IRE1 is detrimental under this dietary intervention. Moreover, we identify the transcription factor Sugarbabe as a downstream target of the IRE1/XBP1 module that is required for increased lipid turnover under DR. Our results provide insights into physiological mechanisms that link tissue-specific metabolic adaptation to lifespan extension under DR conditions.
RESULTS
IRE1 is required for DR-mediated increases in lifespan, starvation resistance and triglyceride synthesis
The increase in fat stores and triglyceride turnover during DR results in increased starvation tolerance in flies. Whole body depletion of IRE1 via ubiquitous RNAi knockdown (using the RU486-inducible gene-switch GAL4 drivers daughterless-gs (da-gs) or tubulin-gs (tub-gs)), led to a striking decrease in the starvation resistance of flies previously fed on DR food for 10days (Fig S1A and S1B). We assessed the tissue-specificity of this effect by inhibiting IRE1 specifically in the brain (ElavGS-GAL4), the muscle (MHC-GAL4), oenocytes (pE(800)GS-GAL4) and differentiated cells of the gut (5966GS-GAL4) (Fig S1C-E). These experiments revealed that IRE1 expression in ECs (5966GS-GAL4) in the midgut is required for starvation tolerance (Fig 1A and Fig S1F). We confirmed efficient IRE1 down-regulation in dissected guts by qPCR using two EC drivers 5966GS-GAL4 and NP1-GAL4 (Fig S1G, H). To determine if IRE1 depletion in ECs affects overall gut integrity, we monitored intestinal stem cell (ISC) activity, which increases with age and in response to gut damage, as well as overall EC morphology. IRE1-deficiency in ECs did not result in significant changes in mitotic activity in the gut, and staining against the beta-catenin homolog Armadillo (which outlines cell boundaries in the intestinal epithelium) did not reveal any morphological changes (Fig S1I and S1J).
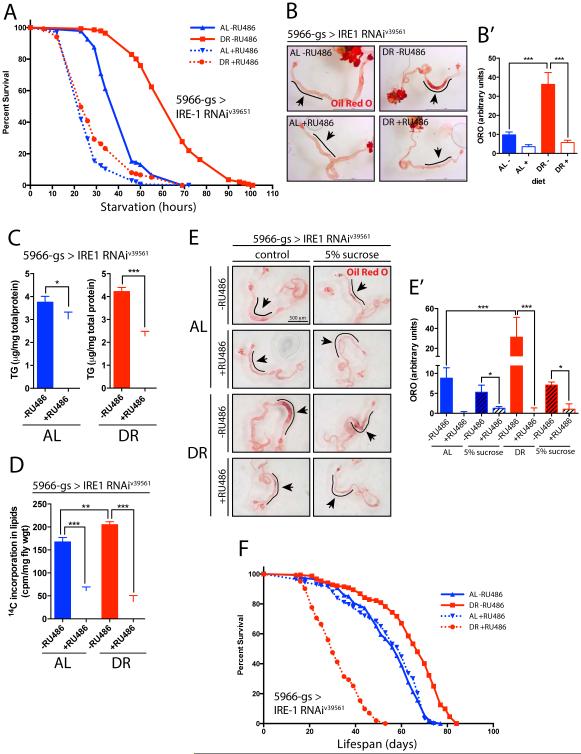
Figure 1. Metabolic effects of depleting IRE1 in the midgut enterocytes of Drosophila.
A. Starvation resistance upon midgut-specific IRE1 knockdown (v39561 line was used throughout, unless otherwise specified) via the tissue-specific driver 5966GS-GAL4. Females were fed on either Ad Libitum (AL) or Dietary Restriction (DR) food, either with (+) or without (−) RU486 to induce GAL4 expression. n=140-150 flies/condition. B. Oil Red O lipid staining of midguts from 10-12 days flies upon IRE1 depletion via 5966GS-GAL4 (+RU486); scale bar = 1mm. B’. Quantification of the intensity of ORO stain from (B) in the anterior midgut; n=13-15 flies/condition/replicate, 2 biological replicates. C. Total triglycerides (TAG) levels of whole bodies (heads removed). Flies were 10-12 days at collection; n=48 flies/condition. D. Quantification of lipid synthesis, via incorporation of 14C labeled glucose in diet for 36h; 14C radioactivity quantified in total lipid fraction. E. 5966GS-GAL4>IRE1RNAi flies were reared on AL or DR (−/+RU486) food for 10 days. Half of the flies on each condition were then placed on 5% sucrose-only containing vials while the other half was kept on the same AL/DR (−/+RU486) diet. 36h later guts were dissected to measure local neutral lipid content via ORO staining. E’. Quantification of ORO stain from (E) in anterior midgut; n=8-12 flies/condition. F. Lifespan curves of 5966GS-GAL4>IRE1RNAi females flies. Adults flipped into fresh vials and survival scored every 2-3 days. See Sup Data for statistics and replicates. n=140-150 flies/condition.
In all panels, error bars show SEM; Statistical significance and interaction between Diet and IRE1RNAi, was determined by two-way ANOVA with Bonferonni’s post hoc test. Details in Sup Information. See also related Figure S1.
It has been reported that depleting IRE1alpha in mouse intestinal cells induces high levels of apoptosis leading to colitis (Zhang et al., 2015). To examine apoptotic cells, we employed the APOLINER apoptosis-reporter (Fig S1K) and measured transcript levels of Reaper and Grim. We observed no significant increase in apoptosis in the midguts of flies as old as 22 days after IRE1 depletion (Fig S1L and S1M). Since 5966GS-GAL4>IRE1RNAi flies on DR do not show evident accumulation of damage in the intestine of young animals, we reasoned that another mechanism must be mediating the decrease in starvation resistance.
Flies under DR undergo a metabolic shift from a glycolytic system towards fatty acid oxidation, using triglycerides as a primary source of energy. Although the carbohydrate amount in the experimental food is the same in both diets (5% sucrose and 8% cornmeal), the lower protein content in our model of DR (0.5% DR vs 5% AL) leads to enhanced triglyceride synthesis (Katewa et al., 2012). Lipid homeostasis and synthesis are known to decrease with age in the midgut (Karpac et al., 2013). Under DR, this loss in lipid levels in ECs with age is delayed, when compared to AL (Fig S2A and S2A’). Depletion of IRE1 in the ECs resulted in a dramatic decrease in lipid levels under DR in the anterior midgut (knockdown via 5966GS-GAL4 in Fig 1B and 1B’, or via NP1-GAL4 in Fig S2B and S2B’). We confirmed that RU486 (100 μM), used to induce the geneswitch system does not affect lipid levels in the intestine (Fig S2C and S2C’) nor lifespan (Fig S2D) on either diet. We also observed a 50% decrease in overall triglyceride (TG) levels, as measured in whole-fly TG content (Fig 1C), or by sudan black (ideal to stain abdominal fat, Fig S1E). While we measured this lower amount of fat stores on flies fed on both diets, the decrease is more dramatic on DR (Fig 1B, interaction p-value = 0.0002 and Fig 1C, interaction p-value = 0.0141; determined by two-way-ANOVA). These diminished fat stores are consistent with the lower starvation resistance upon IRE1 depletion.
The lower fat content in flies in which IRE1 is knocked down in ECs could be a consequence of decreased TG synthesis or increased consumption. To assess these possibilities we examined the dynamic changes in TGs after feeding radiolabeled glucose for a 36h period as described previously (Katewa et al., 2012). As expected, control flies on DR accumulate higher levels of TG compared to flies on AL food (Katewa et al., 2012). Upon loss of IRE1, however, much lower incorporation rates are observed on both diets (Fig 1D), suggesting a defect in de novo lipid synthesis. To further characterize the role of IRE1 in lipid synthesis and its requirement for survival we assessed the ability of flies to regain starvation tolerance upon short-term sugar-only feeding. We reasoned that feeding only sugar after a starvation period would allow flies to replenish their lipid stores via de novo conversion of dietary carbohydrates to fatty acids and enhance starvation resistance. We first tested directly the capacity to convert dietary sucrose into lipids by feeding control flies (−RU486) or 5966GS-GAL4>IRE1RNAi flies (+RU496) a sucrose-only diet for 36h, after being previously reared on either AL or DR (Fig 1E and E’). Upon sucrose-only feeding we noticed that overall lipid levels were decreased when compared to their fully fed controls on both diets, suggesting that a minimum protein content is required for normal metabolic function and lipid accumulation (compare “control” with “5% sucrose” for -RU486 on both diets). In line with the lower conversion of glucose to fat (Fig 1D) we observed that depletion of IRE1 led to lower lipid content in the intestine on both diets. We then tested whether this lack of conversion of sucrose into lipids correlates with a lower resistance to food deprivation. Indeed, after control flies, already under starvation for 10h and 30h, are placed for a short period (8-10h) on sucrose-only agar food (represented by vertical green dashed lines on Fig S1F) they can increase their starvation resistance and survive longer than their constantly starved siblings (solid green line versus solid red line). However, flies in which IRE1 had been depleted in ECs did not exhibit this increase (dotted red and green curves), suggesting that absence of IRE1 impairs the capacity to enhance starvation resistance upon refeeding dietary carbohydrates.
We conclude that IRE1 expression in the intestinal epithelium is required for lipid synthesis, a function that is essential for survival upon DR. Consequently, upon depleting IRE1 in the midgut, overall longevity was drastically reduced specifically upon DR (Fig1F red curves, S2G and S2H). When fed a rich diet (AL), IRE1 depletion in the midgut did not significantly influence lifespan (Fig 1F blue curves, S2G and S2H; Cox proportional hazards interaction p-value <2×10−10 for both repeats). These results indicate that IRE1 has an unexpected role in DR-induced longevity, promoting metabolic adaptation to nutrient restricted conditions in the midgut.
XBP1 acts downstream of IRE1 to regulate lipid levels under DR
Consistent with a role of XBP1 as a downstream target of IRE1, we found that depletion of XBP1 using 5966GS-GAL4 (Fig S3A) reduces the lipid accumulation in the anterior midgut normally observed upon DR (Fig 2A and 2A’). Similar to loss of IRE1, this decrease in lipid content is associated with a lower capacity of survival upon nutrient depletion (Fig 2B and S3B). To confirm that IRE1 acts via XBP1 to regulate lipid levels under DR, we tested whether expression of a spliced form of XBP1 mRNA (UAS-XBP1s construct) in ECs is sufficient to rescue the IRE1RNAi phenotypes. Adding a second UAS-regulated transcript to recombinant flies (5966GS-GAL4; IRE1RNAi) did not affect the IRE1 knockdown-induced lower lipid stores (Fig S3C). While on its own, XBP1s did not further increase starvation resistance of animals fed on DR nor AL (Fig S3D) and did not generally increase intestinal lipid accumulation on either diet (Fig S3E and S3E’), both local lipid accumulation in the anterior midgut and starvation resistance were recovered in animals co-expressing IRE1RNAi and XBP1s (Fig 2C-D).
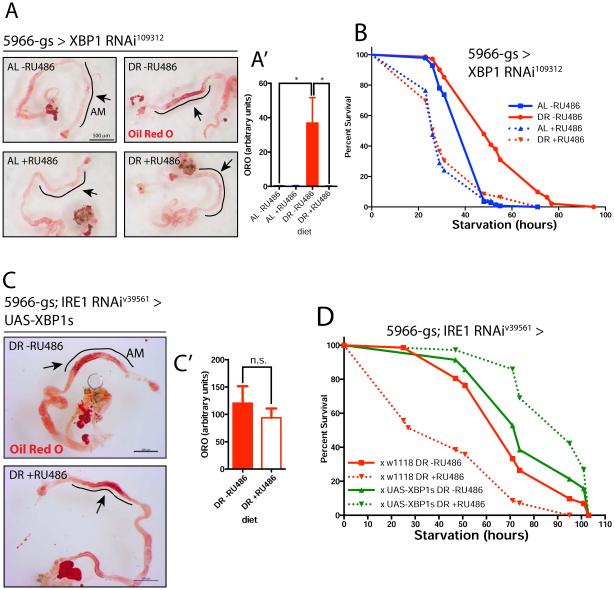
Figure 2. The IRE1 target, XBP1, is required for lipid accumulation and starvation survival in the midgut and is sufficient to rescue IRE1 depletion.
A. Oil Red O staining of midguts. Flies fed for 10-12 days on AL or DR (−/+RU486 for XBP1 depletion; the black line denotes the anterior midgut area where lipids accumulate when fed a DR diet. A’. Quantification of ORO signal from (A) in the anterior midgut. n=10 flies/condition. B. Starvation resistance upon XBP1 depletion in midgut enterocytes, after 10-12 days of feeding in differentiated food. n=100 flies/condition, from 4 independent crosses. C. Oil Red O lipid staining of midguts from flies fed on DR shows rescue of lipid stores after over-expression of spliced XBP1 (UAS-XBP1s) in an IRE1 depletion background. Scale bar 200 µm, n=10-12 flies/condition. C’. Quantification of ORO signal from (C), in the anterior midgut. n=9-10 flies/condition. D. Spliced XBP1 (UAS-XBP1s) was over-expressed in a 5966GS-GAL4;IRE1RNAi background and is sufficient to rescue the starvation sensitivity upon IRE1 depletion.
In all panels, error bars show SEM; statistical significance and interaction between Diet and RNAi, was determined by two-way ANOVA with Bonferonni’s post hoc test. See also related Figure S2.
To characterize the role of canonical ER stress signaling in the DR-induced metabolic switch, we determined if flies under DR have altered levels of ER stress. We used an XBP1 reporter line (Ryoo et al., 2013) to assess tissue-specific activation of the ER stress response mediated by IRE1. Surprisingly, the XBP1-DsRed signal was much higher in the guts of flies fed on AL and this activation was dependent on IRE1 (Fig S3F and S3G). This result contrasts with the fact that both IRE1 and XBP1 are required for metabolic adaptation to DR, and may be explained by different constitutive and inducible roles of the ER stress pathway, where genetic programs downstream of XBP1 are selectively engaged dependent on whether acute ER stress is activating IRE1/XBP1 signaling, or whether changes in nutrient conditions induce chronic metabolic adaptation. Such selective programs engaged by IRE1 signaling have been proposed in other contexts as well (Shen et al., 2005; Volmer and Ron, 2015; Volmer et al., 2013). Supporting a complex role for IRE1/XBP1 signaling in intestinal homeostasis, overexpression of either IRE1 or spliced XBP1 in ECs is not sufficient to increase lifespan, but slightly reduces lifespan (Fig S3H and S3I). Overexpression of IRE1 was also not sufficient to prevent the age-dependent loss of homeostasis that results in increased ISC activation (Fig S3J).
The dietary-sensor Sugarbabe is required for DR and mediates IRE1-regulated lipid homeostasis
To further dissect the mechanism by which the IRE1/XBP1 module regulates dietary carbohydrate-derived lipid synthesis, we analyzed the total transcriptome of Drosophila midgut ECs during DR using ribosome tagging and RNAseq. Flag-tagged RPL13A was used to pull down ribosomes expressed exclusively in midgut ECs, via the NP1-GAL4 driver, and used to isolate mRNAs directly bound to polysomes (Thomas et al., 2012). Using this approach, we profiled changes induced in EC gene expression after a 10 day DR intervention (Sup Table S1). A large number of up-regulated genes relate to digestive activity, including peptidases and proteases, as well as amino acid sensing and transport (Fig 3A and Sup Table S2). DR also induced a large array of G-protein coupled receptors, as well as genes encoding proteins acting in ion transport and the mitochondrial electron transport chain (ETC) (Fig 3A).
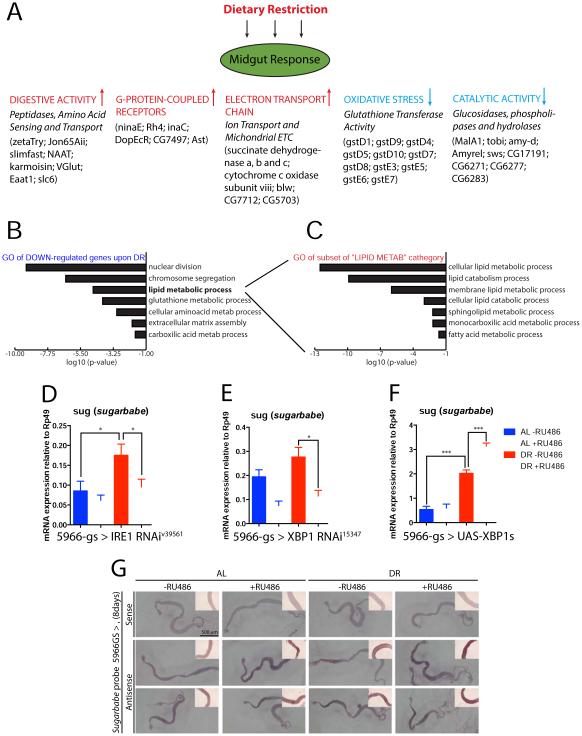
Figure 3. IRE1 is required in the midgut for DR-mediated regulation of the sugar-sensor Sugarbabe.
A. Enterocyte-specific genome-wide mRNA translation profiling from flies fed AL or DR for 10 days, by mass sequencing of polysome-bound mRNAs. Up- and Down-regulated genes (>2 fold and p-value <0.05) were analyzed for significant Gene Ontology (GO) classifiers (Flymine). A list of some regulated genes (complete list in tables S1 and S2) are shown as an illustration of the midgut response to DR. B. The most significantly represented GO terms for genes that are DOWN-regulated by DR. See also Table S2. C. List of the 31 genes classified under the general “lipid metabolic process” emphasized in (B) was further analyzed, revealing most are involved in lipid catabolism. D-F. sugarbabe (sug) mRNA levels (from dissected guts) measured by RT-qPCR in flies fed under AL or DR for 10 days: (D) midgut depletion of IRE1, (E) midgut depletion of XBP1, (F) midgut over-expression of spliced XBP1. (G) in situ hybridization of sugarbabe mRNA shows its levels are induced by DR and XBP1 in a region-specific manner. Flies were fed 8 days on indicated food prior to staining. Middle panel shows UAS-sug overexpression via the same driver used for UAS-XBP1s (bottom panel), illustrating induction of GAL4 over the entire midgut; red lines mark the anterior midgut.
Statistical significance was determined by two-way ANOVA with Bonferonni’s post hoc test; See also related Figure S3 and Tables S1-5.
Interestingly, there is a strong suppression of glutathione oxidative response genes, such as GstD9, GstD6, GstD5 (Fig 3A and Sup Table S2). Gene ontology analysis further shows downregulation of genes related to processes that are repressed under DR (Fig 3B and Sup Table S2). We also observe that a large set of genes related to lipid regulation is altered. The “lipid metabolic processes” category is comprised of 31 target genes (Sup Table S3) and further GO analysis of this subset list - comprising triacylglycerol lipases (Yp2, Yp3), alpha and beta-hydrolases (Lip3, CG10383, CG8093) and phospholipases (swiss cheese, CG17191, CG15533) - reveals that they are related mostly to lipid breakdown and catabolic processes (Fig 3C). This suggests that in the midgut, DR inhibits lipid breakdown. It has previously been suggested that such a repression is an initial step before conversion of dietary sugars into lipids (Zinke et al., 2002). Thus, we further compared changes induced by DR with the ones observed in response to high sugar-feeding from two studies (Zinke et al. 2002 and Matilla et al. 2015). We observe a partial overlap in repressed genes (of 15.5% with the first and 11.6% with the second). Commonly down regulated targets related to lipid catabolism include the lipases Lip3, bmm, CG6277, CG6283 and CG8093, among others, which should result in halting lipid breakdown, shifting the balance to their synthesis and accumulation, for subsequent transport to other tissues (Sup Table S4). We confirmed the regulation of some of these genes in independent samples via qPCR and found that the DR-mediated repression of these lipases is dependent on IRE1 (Fig S4A-D).
Under DR conditions, we also observe up-regulation of genes involved in lipid synthesis, like FASN2, Akhr, and, importantly, a strong IRE1-dependent up-regulation of sugarbabe, a homologue of mammalian C2H2 Gli-like zinc finger transcription factors (Fig. 3D), and likely a requirement for XBP1 in this induction (Fig. 3E). sugarbabe is induced in the midgut, fat body and Malphigian tubules upon sugar feeding, and coordinates the repression of a network of lipases and induction of key drivers of lipogenesis (such as ACC and FAS), setting the stage for lipid accumulation (Mattila et al., 2015; Zinke et al., 2002). Its expression is strongly repressed in melt mutants, which have almost 50% less fat stores than normal (Teleman et al., 2005). Overexpression of spliced XBP1 (UAS-XBP1s) further increased the induction of sugarbabe under DR, but did not affect its expression under AL (Fig 3F), suggesting that XBP1 interacts with other factors only present under DR or a “sugar response” condition, to induce sugarbabe expression. In accordance with these data, in situ hybridization shows that in response to DR or XBP1s over-expression, sugarbabe levels are specifically induced in the anterior midgut region, where lipid accumulation is also observed (Fig 3G).
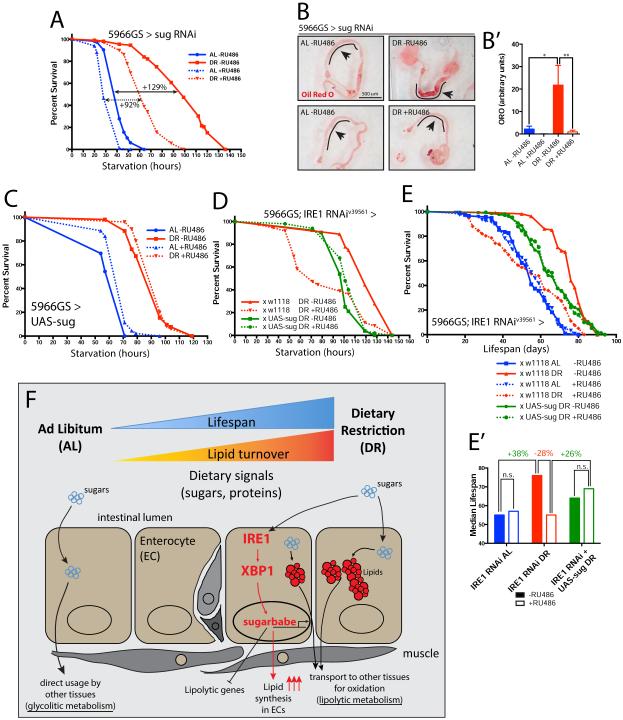
Depletion of sugarbabe in ECs resulted in lower starvation resistance (Fig 4A) and reduced lipid levels in anterior midgut ECs (Fig 4B and 4B’). To test whether Sugarbabe acts by repressing lipid breakdown, and thus facilitating the metabolic shift towards lipid usage under DR downstream of IRE1, we examined if restoring sugarbabe expression levels (using 5966GS-GAL4 and UAS-sug) would be sufficient to rescue IRE1/XBP1 loss of function phenotypes in midgut ECs. In wild-type backgrounds, this perturbation did not alter the starvation resistance of flies raised either on AL or DR nor their lifespan (Fig 4C and S4E). However, induction of sugarbabe in 5966GS-GAL4>IRE1RNAi animals was sufficient to rescue the increased starvation sensitivity, placing sugarbabe downstream of IRE1 in the metabolic shift towards lipid usage under DR (Fig 4D). Supporting this interpretation, sugarbabe over-expression also rescued the shorter lifespan of IRE1-depleted flies on DR (Fig 4E and 4E’).
Figure 4. Sugarbabe is required for lipid accumulation and is sufficient to rescue the IRE1-depletion starvation and lifespan phenotypes on DR.
A. Starvation resistance upon RNAi-mediated sugarbabe depletion in midgut enterocytes. Flies were fed for 10-12days in AL or DR food (−/+ RU486) before starvation. n=108-118 flies/condition, from 5 independent crosses. B. Oil Red O staining of dissected midguts, from 5966GS-GAL4>sugRNAi flies, fed for 10-12 days; the black line denotes the anterior midgut area where lipids accumulate. B’. Quantification of ORO intensity from (B) in the anterior midgut. n=10-12 flies/condition. C. Sugarbabe was over-expressed (UAS-sug) via 5966GS-GAL4. Flies were reared on AL or DR −/+RU486 food for 10 days prior testing starvation resistance. Increased sugarbabe does not lead to any clear difference over control flies on either diet. D. sugarbabe was ectopically expressed in the midguts of 5966GS-GAL4>IRE1RNAi flies fed on DR for 10 days and starvation resistance was then tested. Increased sugarbabe in a IRE1RNAi background was sufficient to restore control levels. E. The shorter lifespan of flies depleted of IRE1 in the midgut under DR (red curves) is rescued by restoring sugarbabe expression in the same cells (green curves). Flies were set up as in (D). E’. Median lifespans of cohorts plotted in (E). F. Model of the IRE1/XBP1/sugarbabe signaling module regulating increased lipid stores in the intestine under DR. Triglycerides are necessary for oxidative phosphorylation under DR due to a shift towards a lypolitic system. See also supplementary statistical analysis and related Figure S4.
DISCUSSION
Our results identify an IRE1/XBP1/Sugarbabe signaling module that is required to establish a permissive environment for de novo lipogenesis and lipid accumulation under DR (Fig 4F). While our data indicate transcriptional regulation of lipases as a mechanism by which IRE1/XBP1/Sug influences lipid synthesis, it remains possible that this module also influences the absorptive capacity of ECs during digestion. RNA-Seq data indicate strong induction of genes involved in nutrient transport upon DR, yet additional studies are needed to test this possibility. Also, the less dramatic effect of sugarbabe knockdown on starvation resistance might be a reflection of being just one of several downstream mechanisms by which IRE1/XBP1 influence metabolic adaptation. Several studies have linked IRE1 and XBP1, proteins predominantly associated with the ER stress pathway, with increased lipogenesis and lipid usage (Acosta-Alvear et al., 2007; Lee et al., 2008; Shao et al., 2014). Our study highlights sugarbabe as a downstream mediator of this response in the Drosophila intestine. We propose that under the low protein and high sugar diet conditions of DR, IRE1/XBP1 signaling in ECs is required for the induction of a lipid anabolic response that establishes a beneficial metabolic state of the animal and extends lifespan. As part of this function, XBP1 may also contribute to the previously described rapid induction of sugarbabe by dietary sugars (Mattila et al., 2015; Zinke et al., 2002). In addition to the regulation of XBP1 and Sugarbabe, IRE1 may also influence lipogenesis by regulating the transcripts of several lipid metabolism genes via a mechanism called RIDD (Regulated IRE1-Dependent Decay) (Coelho et al., 2013; Hollien and Weissman, 2006). However, a role for RIDD in mediating a metabolic shift under DR remains to be explored.
ER stress has been extensively linked to lipid homeostasis as well as diabetes and metabolic syndrome (Ozcan et al., 2004; Wang and Kaufman, 2012). Interestingly, we find that XBP1 levels are not increased, but in fact downregulated in DR when compared to AL. This observation contrasts with the DR-specific requirement for IRE1 for survival, and may be explained by the fact that under AL, the high dietary protein drives increased protein production (increasing ER stress) when compared to DR (Tatar et al., 2014). We speculate distinct roles for IRE1 under the two dietary conditions akin to the proposed ‘constitutive’ and ‘induced’ roles of ER stress response pathways (Shen et al., 2005). We speculate that loss of IRE1 under AL condition is not sufficient to disrupt the UPR in ECs and lead to premature death, likely as other UPR effectors cope with increased protein folding requirement in the ER. However, it is enough to lower lipid production and storage, critical for survival under DR. We propose that this specific requirement is a direct consequence of a UPR-independent function of IRE1/XBP1 signaling in lipogenesis. A UPR-independent function for both IRE1 and XBP1 has previously been proposed in the context of development (Shen et al., 2005). The activation of IRE1/XBP1 in the absence of acute ER stress is likely mediated by changes in cellular lipid content, known to allow IRE1 dimerization independently of the UPR (Volmer and Ron, 2015; Volmer et al., 2013).
A recent study of the role of IRE1alpha in the intestine of mice showed that depletion of IRE1 in EC generated mice with impaired intestinal epithelial barrier function and that were short lived (Zhang et al., 2015). While the authors explain their colitis-like phenotype with increased apoptosis in the gut, it will be interesting to test whether a loss of lipid homeostasis contributes to these phenotypes. Since DR-fed flies depleted of IRE1 die at a very young age, we cannot exclude that IRE1 deficiency causes gut damage later in life, with subsequent ISC activation and dysplasia. Our data suggest that such deleterious effects of IRE1 deficiency on epithelial homeostasis are preceded by changes in the homeostatic metabolism of ECs.
Based on the evolutionary conservation of intestinal morphology and function, as well as of the signaling molecules involved, it can be anticipated that the IRE1/XBP1/Sugarbabe-mediated metabolic adaptation of the intestinal epithelium is conserved in vertebrates and may be part of the metabolic adaptation required for DR-induced lifespan extension.
EXPERIMENTAL PROCEDURES
Fly husbandry and stocks
Fly strains and fly media recipes are in Sup Materials. Dietary Restriction (DR) and Ad Libitum (AL) differ by their concentration of yeast of 0.5% and 5%, respectively. To induce gene switch drivers, 100uM RU486 was diluted in the (+) food and the same amount of ethanol in the (−) control. All fly lines used in this study have been backcrossed at least 6 generations to our lab w1118 stock.
Survival Analysis
All longevity survival assays were carried out on AL or DR media as described previously (Katewa et al., 2012). Details in Sup Materials.
Starvation Assays
Flies were maintained as for a longevity survival assay. After 10-12 days feeding on either DR or AL food, they were transferred to vials containing 1% agar and flipped into fresh agar-only every 24 hrs. 25 mated females were used per vial, from 3-6 independent mating bottles per condition.
Lipid analysis
Triglyceride and free fatty acid content (TG) were measured via a Stanbio labs kit (Boerne, TX) and normalized to total protein. 12 replicates of 4-5 flies each, from 3 independent matting bottles per condition were used. Heads were removed to avoid interference of the red eye pigment with the signal produced.
Oil Red O (ORO) staining: guts were dissected in PBS, fixed in 4% PFA/PBS for 20 min and incubated in fresh Oil Red O for 30 min. ORO signal was quantified in the anterior half of the midguts using ImageJ.
Lipid synthesis assay: After 14 days feeding on regular AL and DR media, 150 flies were transferred to AL or DR media with 14C labeled glucose for 36 hrs. The de novo synthesis of lipids is expressed as the amount of 14C radioactivity incorporated in lipids extracted from 1 mg of fly wt. (details in Sup Materials).
In situ Hybridization
Protocol was adapted from (Zinke et al., 2002) using Digoxigenin (DIG) labeled RNA probes detected by NBT/BCIP assay (DIG nucleic acid detection kit, Roche). Primers used to generate RNA probes for sugarbabe: 5´-ccc acc aca gac aac aaa c-3´ and 5´-gtt cag att cga tgg atg c-3´
Statistical Analysis
Survival assays were analyzed with GraphPad Prism (v.5.0). Significance of the interaction between two variables in survival outcomes was tested by Cox Proportional Hazards. Interaction between genetic manipulations with different diets was evaluated by two-way analysis of variance (ANOVA) with Bonferroni’s post hoc test. Other data were analyzed by the student’s Ttest (analysis in Sup Materials). p-values shown: *<0.05, **<0.01 and ***<0.001.
Supplementary Material
ACKNOWLEDGEMENTS
This work was funded by grants from the American Federation of Aging Research (SDK, PK and HJ), NIH R01AG038688 & AG045835 (PK) and R01AG028127 (HJ), Larry L. Hillblom Foundation postdoctoral fellowship (NML) and start-up grant (SDK), and Ellison Medical Foundation/AFAR postdoctoral fellowship (LW). We thank Drs. M. Pankratz, H. Don Ryoo, P. Domingos, the Bloomington and the Vienna Drosophila stock centers for flies. We thank Christopher Nelson (Buck Institute) and Sabrina Carpentier (CIML, Marseille) for assistance with survival data analysis. We thank Matt Laye, Pedro Sousa-Victor and members of the Kapahi and Jasper labs for helpful discussions. N.M.L. thanks Cedric Maurange for providing reagents, resources and helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization, N.M.L., J.K., H.J. and P.K.; Methodology, N.M.L., J.K., H.J., and P.K.; Investigation, N.M.L., L.W., M.O., H.D., S.D.K. and P.W.L.; Writing – Original Draft, N.M.L. and H.J.; Writing – Review & Editing, N.M.L., J.K., H.J. and P.K.; Project Administration, N.M.L.; Supervision, H.J. and P.K.; Funding Acquisition, H.J. and P.K.
ACESSIONS NUMBERS
The RNA-Seq data is accessible through GEO Series accession number GSE76396.
REFERENCES
- Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. XBP1 Controls Diverse Cell Type- and Condition-Specific Transcriptional Regulatory Networks. Mol. Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Biteau B, Karpac J, Supoyo S, DeGennaro M, Lehmann R, Jasper H. Lifespan extension by preserving proliferative homeostasis in Drosophila. PLoS Genet. 2010;6:1–15. doi: 10.1371/journal.pgen.1001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Hochmuth CE, Jasper H. Maintaining Tissue Homeostasis: Dynamic Control of Somatic Stem Cell Activity. Cell Stem Cell. 2011;9:402–411. doi: 10.1016/j.stem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruss MD, Khambatta CF, Ruby MA, Aggarwal I, Hellerstein MK. Calorie restriction increases fatty acid synthesis and whole body fat oxidation rates. Am. J. Physiol. Endocrinol. Metab. 2010;298:E108–E116. doi: 10.1152/ajpendo.00524.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee D, Katewa SD, Qi Y, Jackson S. a., Kapahi P, Jasper H. Control of metabolic adaptation to fasting by dILP6-induced insulin signaling in Drosophila oenocytes. Proc. Natl. Acad. Sci. 2014;111:17959–17964. doi: 10.1073/pnas.1409241111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Thomas EL, Kapahi P. HIF-1 modulates dietary restriction-mediated lifespan extension via IRE-1 in Caenorhabditis elegans. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho DS, Cairrão F, Zeng X, Pires E, Coelho AV, Ron D, Ryoo HD, Domingos PM. Xbp1-Independent Ire1 Signaling Is Required for Photoreceptor Differentiation and Rhabdomere Morphogenesis in Drosophila. Cell Rep. 2013;5:791–801. doi: 10.1016/j.celrep.2013.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Partridge L. Promoting Health and Longevity through Diet: From Model Organisms to Humans. Cell. 2015;161:106–118. doi: 10.1016/j.cell.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Karpac J, Tran SL, Jasper H. PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell. 2014;156:109–122. doi: 10.1016/j.cell.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E, Wiggins D, Fielding B, Gould AP. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature. 2007;445:275–280. doi: 10.1038/nature05382. [DOI] [PubMed] [Google Scholar]
- Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Chen D, Rogers AN, Katewa SD, Li PWL, Thomas EL, Kockel L. With TOR, less is more: A key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010;11:453–465. doi: 10.1016/j.cmet.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpac J, Biteau B, Jasper H. Misregulation of an Adaptive Metabolic Response Contributes to the Age-Related Disruption of Lipid Homeostasis in Drosophila. Cell Rep. 2013;4:1250–1261. doi: 10.1016/j.celrep.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katewa SD, Demontis F, Kolipinski M, Hubbard A, Gill MS, Perrimon N, Melov S, Kapahi P. Intramyocellular fatty-acid metabolism plays a critical role in mediating responses to dietary restriction in drosophila melanogaster. Cell Metab. 2012;16:97–103. doi: 10.1016/j.cmet.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A-H, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopold P, Perrimon N. Drosophila and the genetics of the internal milieu. Nature. 2007;450:186–188. doi: 10.1038/nature06286. [DOI] [PubMed] [Google Scholar]
- Longo VD, Mattson MP. Fasting: molecular mechanisms and clinical applications. Cell Metab. 2014;19:181–192. doi: 10.1016/j.cmet.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila J, Havula E, Suominen E, Teesalu M, Surakka I, Hynynen R, Kilpinen H, Väänänen J, Hovatta I, Käkelä R, et al. Mondo-Mlx Mediates Organismal Sugar Sensing through the Gli-Similar Transcription Factor Sugarbabe. Cell Rep. 2015:1–15. doi: 10.1016/j.celrep.2015.08.081. [DOI] [PubMed] [Google Scholar]
- Nikawa J, Yamashita S. IRE1 encodes a putative protein kinase containing a membrane-spanning domain and is required for inositol phototrophy in Saccharomyces cerevisiae. Mol. Microbiol. 1992;6:1441–1446. doi: 10.1111/j.1365-2958.1992.tb00864.x. [DOI] [PubMed] [Google Scholar]
- Ozcan U, Cao Q, Yilmaz E, Lee A-H, Iwakoshi NN, Ozdelen E, Tuncman G, Görgün C, Glimcher LH, Hotamisligil GS. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Li J, Kang M-J. Drosophila XBP1 expression reporter marks cells under endoplasmic reticulum stress and with high protein secretory load. PLoS One. 2013;8:e75774. doi: 10.1371/journal.pone.0075774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha H, He Y, Chen H, Wang C, Zenno A, Shi H, Yang X, Zhang X, Qi L. The IRE1alpha-XBP1 Pathway of the Unfolded Protein Response Is Required for Adipogenesis. Cell Metab. 2009;9:556–564. doi: 10.1016/j.cmet.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao M, Shan B, Liu Y, Deng Y, Yan C, Wu Y, Mao T, Qiu Y, Zhou Y, Jiang S, et al. Hepatic IRE1α regulates fasting-induced metabolic adaptive programs through the XBP1s-PPARα axis signalling. Nat. Commun. 2014;5:3528. doi: 10.1038/ncomms4528. [DOI] [PubMed] [Google Scholar]
- Shen X, Ellis RE, Sakaki K, Kaufman RJ. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genet. 2005;1:e37. doi: 10.1371/journal.pgen.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber MH, Thummel CS. Coordination of triacylglycerol and cholesterol homeostasis by DHR96 and the drosophila lipa homolog magro. Cell Metab. 2012;15:122–127. doi: 10.1016/j.cmet.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Veenstra JA, Perrimon N. Control of Lipid Metabolism by Tachykinin in Drosophila. Cell Rep. 2014;9:40–47. doi: 10.1016/j.celrep.2014.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M, Post S, Yu K. Nutrient control of Drosophila longevity. Trends Endocrinol. Metab. 2014;25:509–517. doi: 10.1016/j.tem.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman AA, Chen Y-W, Cohen SM. Drosophila Melted modulates FOXO and TOR activity. Dev. Cell. 2005;9:271–281. doi: 10.1016/j.devcel.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Thomas A, Lee P-J, Dalton JE, Nomie KJ, Stoica L, Costa-Mattioli M, Chang P, Nuzhdin S, Arbeitman MN, Dierick HA. A versatile method for cell-specific profiling of translated mRNAs in Drosophila. PLoS One. 2012;7:e40276. doi: 10.1371/journal.pone.0040276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volmer R, Ron D. Lipid-dependent regulation of the unfolded protein response. Curr. Opin. Cell Biol. 2015;33:67–73. doi: 10.1016/j.ceb.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volmer R, van der Ploeg K, Ron D. Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc. Natl. Acad. Sci. U. S. A. 2013;110:4628–4633. doi: 10.1073/pnas.1217611110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Kaufman RJ. The impact of the unfolded protein response on human disease. J. Cell Biol. 2012;197:857–867. doi: 10.1083/jcb.201110131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Karpac J, Jasper H. Promoting longevity by maintaining metabolic and proliferative homeostasis. J. Exp. Biol. 2014a;217:109–118. doi: 10.1242/jeb.089920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zeng X, Ryoo HD, Jasper H. Integration of UPRER and oxidative stress signaling in the control of intestinal stem cell proliferation. PLoS Genet. 2014b;10:e1004568. doi: 10.1371/journal.pgen.1004568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Ryoo HD, Qi Y, Jasper H. PERK Limits Drosophila Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress. PLoS Genet. 2015;11:e1005220. doi: 10.1371/journal.pgen.1005220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H-S, Chen Y, Fan L, Xi Q-L, Wu G-H, Li X-X, Yuan T-L, He S-Q, Yu Y, Shao M-L, et al. The Endoplasmic Reticulum Stress Sensor IRE1α in Intestinal Epithelial Cells Is Essential for Protecting against Colitis. J. Biol. Chem. 2015;290:15327–15336. doi: 10.1074/jbc.M114.633560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinke I, Schütz CS, Katzenberger JD, Bauer M, Pankratz MJ. Nutrient control of gene expression in Drosophila: Microarray analysis of starvation and sugar-dependent response. EMBO J. 2002;21:6162–6173. doi: 10.1093/emboj/cdf600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.