Abstract
DNA double-strand breaks (DSBs) are induced by SPO11 during meiosis to initiate recombination-mediated pairing and synapsis of homologous chromosomes. Germline genome integrity requires spatiotemporal control of DSB formation, which involves the proteinaceous chromosome axis along the core of each meiotic chromosome. In particular, a component of unsynapsed axes, HORMAD1, promotes DSB formation in unsynapsed regions where DSB formation must occur to ensure completion of synapsis. Despite its importance, the underlying mechanism has remained elusive. We identify CCDC36 as a direct interactor of HORMAD1 (IHO1) that is essential for DSB formation. Underpinning this function, IHO1 and conserved SPO11-auxiliary proteins MEI4 and REC114 assemble chromatin-bound recombinosomes that are predicted activators of DSB formation. HORMAD1 is needed for robust recruitment of IHO1 to unsynapsed axes and efficient formation and/or stabilization of these recombinosomes. Thus we propose that HORMAD1-IHO1 interaction provides a mechanism for the selective promotion of DSB formation along unsynapsed chromosome axes.
Introduction
Generation of haploid gametes requires segregation of homologous chromosomes (homologues) during the first meiotic division. In most taxa including mammals, each pair of homologues must engage and become physically linked via at least one crossover during the first prophase to achieve high fidelity segregation1,2. Inter-homologue crossovers are formed by homologous recombination, which initiates with SPO11 transesterase-mediated induction of multiple DSBs in early prophase3,4. Homology search initiated by multiple DSBs on each chromosome results in close juxtaposition of homologues along their lengths. The number of DSBs is tightly controlled: having too many may yield genotoxic effects, while too few would not ensure high fidelity homologue pairing. Feedback control from homologue pairing to DSB formation and repair may help meet these requirements5–8.
Spatiotemporal control of recombination relies on two meiosis-specific chromatin structures: the chromosome axis and the synaptonemal complex (SC). The axis is a rod-like proteinaceous structure that forms early in meiosis along the cohesin core of each sister chromatid pair. The SC is a structure that forms when homologue axes pair and become closely linked along their lengths in a zipper-like fashion by transverse filament proteins7,9. Meiosis-specific HORMA-domain proteins are axis components that mediate key functions in control of DSB formation and repair and/or in the quality control of recombination in diverse taxa7,9,10. In mammals, the HORMA-domain protein HORMAD1 preferentially associates with unsynapsed axes5,11 and is thought to have three main functions12–15. First, it ensures availability of sufficient DSBs for homology search by promoting DSB formation, and possibly by inhibiting premature DSB repair or inappropriate recombination between sister chromatids. Second, HORMAD1 supports SC formation. Third, HORMAD1 sets up checkpoints that prevent progression of meiocytes beyond prophase unless homologues are synapsed. SC formation is proposed to inhibit HORMAD1 functions and promote the depletion of HORMAD1 from axes5,12. This is one likely, but not exclusive, mechanism by which SC formation may also downregulate DSB formation and enable progression of meiocytes beyond prophase once homologues are successfully paired5,7,12,16. In this model, SC formation limits DSB numbers by restricting DSB formation to unsynapsed axes, precisely where DSBs are still needed to promote homologue engagement and SC formation5,7. Indeed, the SC appears to downregulate DSB formation both in budding yeast and mice6,8. A major goal is to identify the mechanisms that govern relationships between DSB formation and the chromosome axis.
The prevailing molecular model of meiotic DSB formation is based mainly on studies of yeast. Chromatin is arranged in loops emanating from the chromosome axis, and DSBs form preferentially in loop-forming DNA as opposed to axis-bound DNA17,18. However, it is thought that DSBs are introduced only after “loop DNAs” have been recruited to axes, because DSB-promoting protein complexes—recombinosomes—assemble only along axes17,18. In yeasts, complexes containing the conserved Mei4 and Rec114 proteins and a third coiled coil-containing protein (Mer2 and Rec15 in budding and fission yeasts, respectively) are thought to link Spo11 activity to axes9,18–22. MEI4 and REC114 are also present in mammals23. Mouse MEI4 is indispensable for DSB formation, and it interacts with REC114 and forms foci along unsynapsed chromosome axes23. These foci are thought to represent DSB-promoting recombinosomes because focus formation along axes correlates with DSB formation16,23. HORMAD1 that is associated with unsynapsed axes appears to be important for the function of these recombinosomes, as HORMAD1 is needed for efficient DSB formation12–14 and high MEI4 focus numbers16. However, pivotal questions remain unanswered. What are the composition and importance of axis-associated putative DSB-promoting recombinosomes, and what is the mechanism that targets their assembly and DSB formation to HORMAD1-rich unsynapsed axes?
Results
IHO1, a component of unsynapsed axes
To gain insights into axis functions, we screened for HORMAD1-interacting proteins by yeast two-hybrid assay and identified CCDC36, which we named Interactor of HORMAD1-1 (IHO1). Both mouse and human protein pairs interact. IHO1 is a coiled coil domain-containing protein without other recognizable domains or annotated functions and is conserved in vertebrates. Mouse ENCODE data24,25 and reverse transcription PCR (Supplementary Fig. S1a) showed testis enrichment of Iho1 transcripts, and anti-IHO1 antibodies preferentially stained spermatocytes in testis sections (Fig. 1a). HORMAD1 co-precipitated with IHO1 from testis extracts (Fig. 1b), and immuno-staining of spread meiocytes revealed that IHO1, like HORMAD1, associates with chromatin during prophase (Fig. 1c, d and Supplementary Fig. S1b–d). IHO1 and HORMAD1 foci appeared on chromatin during pre-leptotene, when pre-meiotic replication occurs and axes have not yet developed (Fig. 1d, and Supplementary Fig. S1b–d). In leptotene, IHO1 displayed increasing intensity along developing HORMAD1-rich axes. In zygotene, when axes elongate and synapsis begins, IHO1 remained associated with unsynapsed axes but disappeared from synapsed axes (Fig. 1c–d, Supplementary Fig. S1c–d). Although IHO1 and HORMAD1 largely colocalized, IHO1 staining was more discontinuous and more completely depleted from synapsed regions (Fig. 1c–d).
Figure 1.
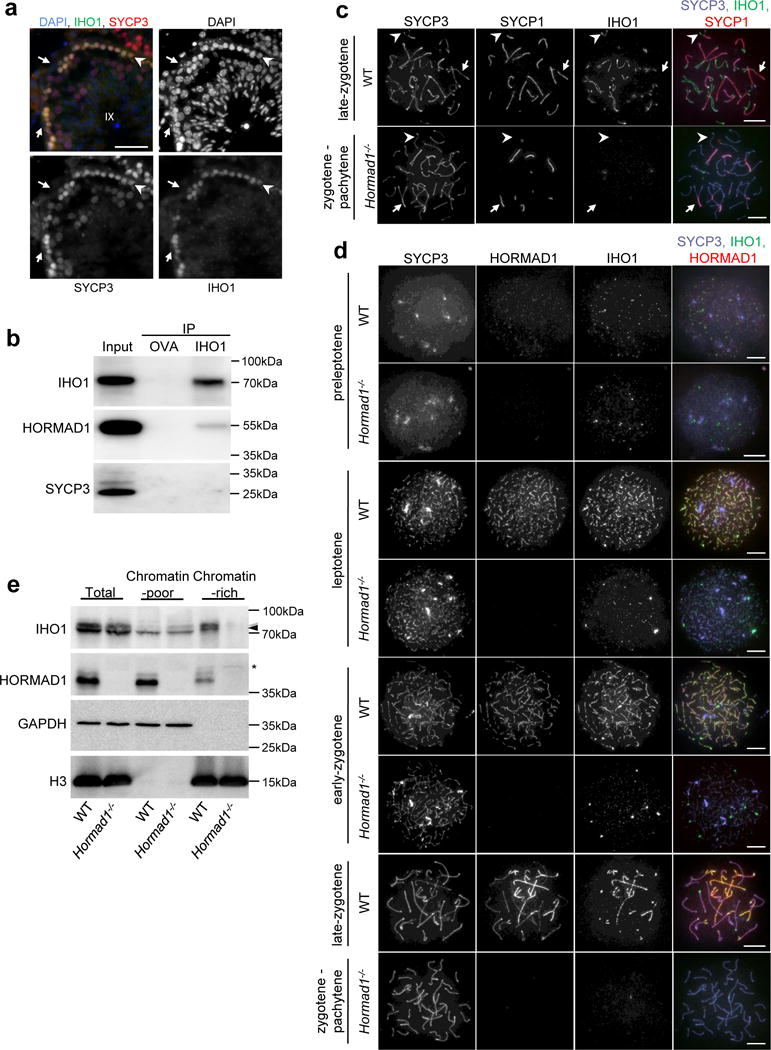
IHO1, interactor of HORMAD1, localizes to unsynapsed axes. (a) SYCP3 (meiotic axis marker) and IHO1 were detected by immunofluorescence, and DNA by DAPI, on wild-type (WT) testis cryosections. A stage IX tubule is shown. IHO1 signal is strong only in meiotic leptotene cells (arrowhead) and is absent from non-meiotic Sertoli cells (arrows). Scale bars, 50μm. (b) Solubilized chromatin fractions of wild-type testes were subjected to immunoprecipitation by anti-ovalbumin (OVA) or anti-IHO1 (IHO1) antibody. Input (equivalent to 5% of immunoprecipitation samples) and immunoprecipitated (IP) samples were analyzed by immunoblotting with anti-IHO1, anti-HORMAD1, and anti-SYCP3 antibodies. HORMAD1 but not SYCP3 co-precipitates with IHO1, indicating that IHO1-HORMAD1 co-precipitation is not mediated by the chromosome axis. Anti-OVA immunoprecipitation served as negative control. (c, d) IHO1, SYCP3, and either (c) SYCP1 (synaptonemal complex transverse filament) or (d) HORMAD1 were detected by immunofluorescence on nuclear surface spreads of wild-type and Hormad1−/− spermatocytes. Equal exposure images are shown. IHO1 appears on chromatin in preleptotene and preferentially accumulates on unsynapsed chromosome axes in wild-type (WT) cells at later stages. IHO1 levels are much lower on chromatin and unsynapsed axes in Hormad1−/− spermatocytes at corresponding stages. Stages corresponding to wild-type late-zygotene and early-pachytene cannot be distinguished in Hormad1−/− due to defective synaptonemal complex formation, hence we refer to this stage as “zygotene-pachytene”. (c) Arrows and arrowheads indicate synapsed and unsynapsed axes, respectively. Scale bars in c and d, 10μm. (e) Total, TritonX-100 soluble (Chromatin-poor) and insoluble (Chromatin-rich) extracts were prepared from testes of 13 days postpartum wild-type and Hormad1−/− littermates and analyzed by immunoblotting. IHO1 levels in the chromatin-rich TritonX-100 insoluble fraction are much lower in Hormad1−/− than in wild type. Black and grey arrowheads mark IHO1-specific bands that may have resulted from posttranslational modifications of IHO1. Asterisk marks a non-specific band. Unprocessed gel-scans of b and e are shown in Supplementary Fig. S8.
IHO1 localization diverged in the two sexes beyond zygotene. In spermatocytes, synapsis between the largely non-homologous X and Y chromosomes is mostly constrained to their short pseudoautosomal regions (PAR). At the zygotene-to-pachytene transition, high IHO1 and HORMAD1 levels remained on unsynapsed sex chromosome axes but also on PARs (Supplementary Fig. S2)5. However, by early pachytene IHO1 disappeared from both the synapsed and unsynapsed regions of sex chromosomes (Supplementary Fig. S2), contrasting persistent HORMAD1 association with unsynapsed axes in pachytene5,11. Curiously, IHO1 re-accumulated on unsynapsed axes of sex chromosomes in mid-late-pachytene and on desynapsing autosome axes in diplotene, but disappeared from chromatin concomitant with axis disassembly after diplotene (Supplementary Fig. S1c, S2). In oocytes, IHO1 disappeared from axes after all chromosomes synapsed and was undetectable on chromatin beyond zygotene (Supplementary Fig. S1d). Ovarian IHO1 levels also dramatically decreased as oocytes progressed to late prophase (Supplementary Fig. S1e).
The preferential enrichment of both IHO1 and HORMAD1 on unsynapsed axes (albeit with some differences in localization) supports the idea that they interact in vivo. Concordantly, HORMAD1 was required for robust IHO1 accumulation on chromatin and unsynapsed axes (Fig. 1d) and for high IHO1 levels in chromatin–rich fractions of testis extracts (Fig. 1e), suggesting that HORMAD1 recruits IHO1 to unsynapsed axes. To test the hypothesis that IHO1 may mediate some or all HORMAD1-dependent axis functions, we disrupted the Iho1 gene in mice (Supplementary Fig. S3).
Iho1−/− mice are defective in homologous synapsis
Iho1−/− mice were viable without obvious somatic defects, but both sexes were infertile (96 breeding-weeks, 2 males and 4 females). Oocytes were depleted in six-week-old Iho1−/− females, and spermatocytes in males underwent apoptosis at a stage equivalent to wild-type mid-pachytene (Supplementary Fig. S4), suggestive of meiotic recombination defects26–29. Cohesin cores, chromosome axes, and HORMAD1 recruitment to unsynapsed axes appeared normal, but axis alignment and SC formation were severely defective (Fig. 2b, Supplementary Fig. S5). Complete synapsis was never observed, but incomplete SCs were detected in meiocytes equivalent to late-zygotene and pachytene as judged by the presence of fully formed axes (n>500 cells, Fig. 2b). SC was observed between axes of unequal length and between multiple partners in most if not all meiocytes displaying extensive synapsis (n>100 cells, Fig. 2b, Supplementary Fig. S5h), thus SC often forms between non-homologous chromosomes. This contrasts Hormad1−/− meiocytes, where reduced DSB numbers yield incomplete SC formation that nonetheless occurs mostly between homologues12,13.
Figure 2.

Synaptonemal complex (SC) formation and alignment of chromosome axes is defective in Iho1−/− spermatocytes. (a–f) SYCP3 (axis marker) and SYCP1 (synaptonemal complex marker) were detected by immunofluorescence on nuclear surface spreads of wild-type (WT) pachytene and mutant zygotene-pachytene spermatocytes of indicated genotypes. Scale bars, 10μm. (b) Homologous chromosomes are not aligned in the presented image of an Iho1−/− spermatocyte, however SCs form between apparently non-homologous chromosomes, creating a meshwork of interconnected axes. Insets show enlarged view. Arrowheads mark sites where a chromosome axis engaged in synapsis with two different partners. This phenotype resembles those of Spo11−/− (c) and Iho1−/− Spo11−/− (d) spermatocytes, in which DNA strand invasion and homologue pairing are defective due to a lack of DSBs. SYCP1 stretches are much shorter (e, f) and less numerous in Hormad1−/− Spo11−/− (e) and Iho1−/− Hormad1−/− (f) spermatocytes than in Iho1−/− (b), Spo11−/− (c) or Iho1−/− Spo11−/− (d) spermatocytes. (g) The frequency distributions of SYCP1 stretch-numbers in zygotene-pachytene spermatocytes with fully condensed axes were plotted. n shows analyzed spermatocyte number from two (Hormad1−/− Spo11−/− and Iho1−/− Hormad1−/−) or three (Iho1−/−, Spo11−/− and Iho1−/−Spo11−/−) pooled experimental repeats. Numbers of SYCP1 stretches were not significantly different in Iho1−/−, Spo11−/− and Iho1−/− Spo11−/− (Iho1−/− versus Spo11−/− P= 0.3668, Iho1−/− versus Iho1−/− Spo11−/− P= 0.1596, Spo11−/− versus Iho1−/− Spo11−/− P= 0.5197, Mann-Whitney test), but SYCP1 stretch-numbers were higher in these three genotypes than in Iho1−/− Hormad1−/− and Hormad1−/− Spo11−/− spermatocytes (P< 2.20E-16 for all comparisons, Mann-Whitney test). SC formation was not significantly different in Iho1−/− Hormad1−/− and Hormad1−/− Spo11−/− spermatocytes (P= 0.9162, Mann-Whitney test). Primary source data of graphs are available in Supplementary Table 2.
Aberrant SC in Iho1−/− meiocytes may be explained by defective homologue pairing. Alternatively, IHO1 may function independent of pairing, similar to HORMAD1. The latter is required for non-homologous synapsis that occurs extensively in the DSB-deficient Spo11−/− background, where homologue pairing fails12,13. To distinguish between these possibilities, we compared effects of Iho1 and Hormad1 mutations on SC formation in the Spo11−/− background (Fig. 2). Iho1−/− Spo11−/− and Iho1−/− spermatocytes were equally proficient as Spo11−/− in forming non-homologous SC, whereas Hormad1 deficiency largely abolished SC in both the Spo11−/− and Iho1−/− backgrounds. Thus, whilst HORMAD1 has a function allowing homologue-pairing-independent SC in Spo11−/−, IHO1 does not.
IHO1 is required for DSB formation
The indistinguishable non-homologous SC formation phenotypes in Spo11−/− and Iho1−/− meiocytes could be explained if IHO1, like SPO11, is required for DSB formation. Consistent with this hypothesis, Iho1−/− spermatocytes formed few or no foci of recombination proteins that mark DSB ends, a number comparable to that in Spo11−/−, but much lower than in Hormad1−/− (Fig. 3a–f). Likewise, phospho-histone H2AX (γH2AX), a marker of the ATM/ATR-dependent DNA damage response, was virtually absent from chromatin in leptotene and early zygotene in both Spo11−/− and Iho1−/− spermatocytes, contrasting the reduced but still present staining in Hormad1−/− (Supplementary Fig. S6). Initial DSB processing generates SPO11-oligonucleotide complexes whose levels in testis extracts indicate DSB levels30. SPO11-oligonucleotide complexes were absent from both Spo11−/− and Iho1−/− testes (n=3 mice per genotype) (Fig. 3g–h), whereas they were detectable in Hormad1−/− testes at reduced levels compared with wild type (median 4.3-fold reduction, p=0.00027, n=7, one-sample t-test) or Dmc1−/− (median 2.4-fold reduction, p=0.0068, n=7, one-sample t-test based on current and published12 results). (Dmc1−/− mice provide DSB-formation-proficient controls for altered testis size and cellularity of Spo11−/−, Hormad1−/− and Iho1−/− mutants, because pachytene-equivalent spermatocytes are eliminated in all of these mutants at stage IV of the epithelial-cycle of seminiferous tubules12–14,27,31,32; Supplementary Fig. S4a.) These observations strongly suggest that programmed meiotic DSB formation is abolished in Iho1−/− mice.
Figure 3.

Recombination foci and SPO11-oligonucleotides are effectively absent from Iho1−/− spermatocytes and testes, respectively. (a–c) SYCP3 (axis marker) and DMC1 (a), RAD51 (b) or RPA (c) (markers of recombination/single-stranded DSB ends) were detected on nuclear surface spreads of wild-type (WT) and Iho1−/− zygotene spermatocytes. Scale bars, 10μm. (d–f) Numbers of DMC1 (d), RAD51 (e) and RPA (f) foci during leptotene (le) and early-zygotene (e-zy) in wild type, Spo11−/−, Hormad1−/− and Iho1−/−; late-zygotene (l-zy) and pachytene (pa) in wild type; and zygotene-pachytene (zy-pa) in Spo11−/−, Hormad1−/− and Iho1−/−. Median focus numbers are marked, n shows analyzed spermatocyte number from three (RAD51 and RPA) or four (DMC1) pooled experiments. P values were calculated by Mann-Whitney test. DMC1, RAD51 and RPA focus numbers in Hormad1−/− were on average 29, 22 and 39% of wild type, respectively, during the comparable early-zygotene stage. Virtually no recombination foci were detected in Iho1−/− (on average 2, 2 and 2% of wild-type DMC1, RAD51 and RPA, respectively) and Spo11−/− (on average 2, 2 and 2% of wild-type DMC1, RAD51, respectively). (g–h) SPO11-oligonucleotide complexes (g) and SPO11 protein (h) were detected in adult testis extracts. Dmc1−/− testes were used to control for altered cellularity in mutant testes; Dmc1−/−, Spo11−/−, Hormad1−/− and Iho1−/− testes are characterized by apoptosis of mid-pachytene-equivalent spermatocytes in tubules at stage IV of the seminiferous epithelial cycle. One representative of three experiments is shown. (g) SPO11-oligonucleotide complexes were immunoprecipitated and radioactively labelled. Bar marks SPO11-specific signals, asterisk indicates nonspecific labelling of contaminants, and arrowhead marks the immunoglobulin heavy-chain. Quantified levels of radioactive signals were background-corrected and normalized to wild type. (h) Blots from g were probed with anti-SPO11 antibody. Two alternative forms of SPO11 (α and β) protein were present in wild type30,47,48. Only SPO11β, the form that is expressed in early meiosis, was detected in Dmc1−/−, Hormad1−/− and Iho1−/− mutants. Unprocessed gel-scans are shown in Supplementary Fig. S8. Primary source data of recombination focus counts and SPO11-oligonucleotide quantifications are shown in Supplementary Table 2.
Similar to Iho1−/− single mutants, Iho1−/−Hormad1−/− double mutant spermatocytes had many fewer RAD51, DMC1, or RPA foci than Hormad1−/− spermatocytes (Fig. 4a–c). Thus, Iho1 is epistatic to Hormad1 in DSB formation, indicating that IHO1 is able to promote DSB formation even in the absence of HORMAD1. HORMAD1 may enhance this basal HORMAD1-independent IHO1 activity and IHO1 association with axes, thereby ensuring efficient DSB formation.
Figure 4.

IHO1 has HORMAD1-independent DSB formation-promoting activity. (a–c) SYCP3 and either DMC1 (a), RAD51 (b) or RPA (c) recombination markers were detected by immunofluorescence on nuclear surface spreads of Hormad1−/−, Iho1−/− and Iho1−/− Hormad1−/− zygotene spermatocytes. Whereas foci of all three marker proteins were readily detected in Hormad1−/−, they were essentially absent from Iho1−/− and Iho1−/− Hormad1−/− spermatocytes (n>200 cells examined for each staining). Scale bars, 10μm. (d) Model depicting the likely functional interplay between IHO1 and HORMAD1 in DSB formation. Blue arrows represent promotion of DSB formation by IHO1 proportionally to the thickness of the arrow. Black arrow represents HORMAD1-mediated recruitment of IHO1 to axes and promotion of IHO1 activity.
IHO1 is dispensable for HORMAD1-mediated meiotic prophase quality control
HORMAD1 and its paralogue HORMAD2 have crucial functions in quality control mechanisms that eliminate meiocytes in response to inappropriate asynapsis12,13,33,34. HORMAD1/2 promote recruitment of ATR activity to unsynapsed regions, resulting in H2AX phosphorylation and meiotic silencing of unsynapsed chromatin (MSUC)12,13,33–36. In pachytene spermatocytes, MSUC forms the sex body, the silenced chromatin that encompasses unsynapsed portions of the sex chromosomes and turns off genes whose expression is toxic at this stage26,35,37–39. Both extensive autosomal asynapsis and impaired HORMAD1 function cause spermatocyte elimination from failed sex chromosome silencing12–14,26,29. Autosomal asynapsis lowers ATR activity on sex chromosomes by redistributing ATR to unsynapsed autosomes29, whereas HORMAD1 deficiency impairs recruitment of ATR activity to all unsynapsed axes, including sex chromosomes12–14.
To test if IHO1 has a role similar to HORMAD1, we examined recruitment of ATR and the ATR activator BRCA1 as well as ATR activity (γH2AX accumulation) on unsynapsed chromatin. To control for the absence of DSBs in Iho1−/−, we compared it to Spo11−/− in addition to wild type. HORMAD1, ATR, and BRCA1 were all recruited to unsynapsed axes and/or chromatin similarly in Spo11−/− and Iho1−/− spermatocytes, and recruitment of ATR and BRCA1 required HORMAD1 in both backgrounds (Fig. 5a–d). Correspondingly, high fractions of Iho1−/− and Spo11−/− spermatocytes accumulated γH2AX in “pseudo-sex bodies,” which form around undefined subsets of unsynapsed axes due to spurious HORMAD1-dependent ATR signaling in DSB-defective mutants12,13,27,40. Importantly, efficient pseudo-sex body formation depended on HORMAD1 in both backgrounds (Fig. 5a, e). Thus, IHO1 is not involved in HORMAD1-mediated surveillance of asynapsis or ATR activation in unsynapsed regions.
Figure 5.

IHO1 is not required for ATR, BRCA1or γH2AX accumulation on unsynapsed chromatin. (a, b) SYCP3, γH2AX and either ATR (a) or BRCA1 (b) were detected by immunofluorescence on nuclear surface spreads of wild-type (WT) pachytene and mutant zygotene-pachytene spermatocytes. Scale bars, 10μm. (a) γH2AX accumulates on sex chromatin in wild type, and on distinct chromatin domains called pseudo-sex bodies in Iho1−/−, Spo11−/−, and Iho1−/− Spo11−/−. ATR accumulates both on axes and chromatin loops within these γH2AX-rich chromatin domains. In contrast, neither γH2AX nor ATR accumulates in a distinct chromatin domain in the shown Iho1−/− Hormad1−/− and Hormad1−/− Spo11−/− spermatocytes. (b) BRCA1 accumulates on axes in pseudo-sex bodies in Iho1−/− and Spo11−/− spermatocytes. (c–e) Quantification of the fraction of zygotene-pachytene spermatocytes where ATR (c), or BRCA1 (d) accumulated on unsynapsed chromatin or axis, respectively, or where a pseudo-sex-body(ies) (PSB) formed (e). Numbers of counted spermatocytes (n) are shown in the combined datasets from two experiments. In Iho1−/−, Spo11−/− and Iho1−/− Spo11−/−, ATR-rich chromatin domains (c) and pseudo-sex-bodies (e) are frequently observed, and BRCA1 (d) frequently associates with unsynapsed axes in these chromatin domains. In contrast, ATR (c) is absent from chromatin, and pseudo-sex-bodies (e) do not form in most Iho1−/− Hormad1−/− and Hormad1−/− Spo11−/− spermatocytes. BRCA1 localization to axes (d) was never observed in these two mutants. Primary source data of graphs in c–e are available in Supplementary Table 2.
This conclusion is supported by the near absence of oocytes in 6-week-old Iho1−/− mice (Supplementary Fig. S4b), which shows that IHO1, unlike HORMAD1, is dispensable for elimination of asynaptic oocytes. In females, persistent ATR signaling and/or inappropriate silencing of ad hoc sets of essential genes by MSUC (both dependent on HORMAD1) are thought to eliminate asynaptic oocytes during prophase41.
IHO1 is required for robust MEI4 focus formation on chromatin
How does IHO1 promote DSB formation? The budding yeast HORMA-domain protein Hop1 helps recruit Mei4, Rec114 and Mer2 to axes18 and mouse HORMAD1 is required for normal appearance of MEI4 foci that are presumptive DSB-promoting recombinosomes16. If IHO1 mediates HORMAD1 function in DSB formation, then IHO1 loss should also affect MEI4 complex assembly. MEI4 levels in testis extracts were comparable but MEI4 foci were 11-fold and 6-fold fewer in Iho1−/− leptotene spermatocytes than in wild type or Hormad1−/−, respectively (Fig. 6a–c). Iho1−/− and Iho1−/− Hormad1−/− spermatocytes were comparable (Fig. 6a), thus Iho1 is epistatic to Hormad1 in both DSB formation and MEI4 focus formation. MEI4 foci appear more stable in Spo11−/− than in wild type23, raising the possibility that SPO11 not only promotes DSB formation but also directly or indirectly destabilizes DSB-promoting protein complexes. IHO1 deficiency caused fewer MEI4 foci to form in both Spo11+/+ and Spo11−/− backgrounds (Fig. 6d–e), so this defect in MEI4 focus formation is not a consequence of increased sensitivity to a putative recombinosome destabilizing activity of SPO11.
Figure 6.

Robust MEI4 focus formation requires IHO1. (a, d) MEI4 and SYCP3 were detected in surface spread leptotene spermatocytes of indicated genotypes. Foci of MEI4 were preferentially detected on the forming axis both in wild type (WT) (a) and Spo11−/− (d). MEI4 foci were present in lower numbers or were nearly absent in spermatocytes that lacked HORMAD1 or IHO1, respectively, in both Spo11+/+ (a) and Spo11−/− (d) backgrounds. MEI4 foci were also nearly absent in Iho1−/− Hormad1−/−, indicating that IHO1 is needed for MEI4 focus formation in Hormad1−/−. Scale bars, 10μm. (b, e) Quantification of MEI4 focus numbers in spermatocytes. Stages: preleptotene (ple), leptotene (le) and early-zygotene (e-zy). Median focus numbers are marked, and n values indicate the number of analyzed spermatocytes. Pooled results from three (b) or two (e) experiments are shown. Because MEI4 foci are less robust in Spo11+/+ than in the Spo11−/− background, we detected MEI4 with both guinea pig and rabbit anti-MEI4 antibodies, and counted foci of MEI4 that were detected by both antibodies in Spo11+/+ background (b). This strategy increases the chance that the counted foci represent genuine chromatin-bound MEI4 protein complexes and may explain differences in MEI4 focus counts between the current and previous studies16,23 (e) Due to the higher signal-to-noise ratio of MEI4 staining in the Spo11−/− background, counts were based only on rabbit anti-MEI4 staining. MEI4 focus numbers were significantly lower in Hormad1−/− than in wild type, and were significantly lower in Iho1−/− than in wild type or Hormad1−/− in both Spo11+/+ (b) and Spo11−/− (e) backgrounds (Mann-Whitney test). (c) MEI4 was immunoprecipitated (IP) from wild-type, Hormad1−/− and Iho1−/− testis extracts from 13 days postpartum mice using anti-MEI4 antibodies crosslinked to magnetic beads. The same antibody was used to detect immunoprecipitated MEI4 by western blot (left image). Detection of histone H3 in the input served as a loading control (right image). MEI4 levels were not lower in Iho1−/− (or Hormad1−/−) than in wild type. Data represent one out of three experiments. See Supplementary Fig. S8 for unprocessed gel-scans. Primary source data of MEI4 focus counts (b, e) and MEI4 signal quantification (c) are shown in Supplementary Table 2.
We cannot exclude that HORMAD1 and IHO1 foster DSB formation through distinct mechanisms. It is also possible that HORMAD1 affects DSB formation indirectly, e.g., by altering timing of meiotic events via its checkpoint function, although available data argue against such a role for yeast Hop142. Nevertheless, our observations suggest that IHO1 and HORMAD1 collaborate in DSB formation. Specifically, IHO1 likely stimulates DSB formation through efficient formation of MEI4-containing recombinosomes, and HORMAD1 likely enhances this function. This hypothesis may explain why Iho1−/− spermatocytes do not form any programmed DSBs while Hormad1−/− spermatocytes do, albeit fewer than wild type.
Putative chromatin-bound DSB-promoting recombinosomes contain IHO1
We hypothesized that IHO1 might form complexes with MEI4 on chromatin. Supporting this, the majority of MEI4 foci colocalized with IHO1 in preleptotene wild-type cells, i.e., when IHO1 forms only foci and when elongated axis structures and DSBs have not yet formed (Fig. 7a, b). Note that only 53% (preleptotene, n=17 cells) and 65% (leptotene, n=43 cells) of foci detected by the anti-MEI4 guinea pig antibody overlapped with foci detected by the anti-MEI4 rabbit antibody (Supplementary Table 2, “gpMEI4-rbMEI4-coloc”), suggesting that half to two-thirds of foci detected by the guinea pig antibody represented genuine chromatin-associated MEI4 complexes. (For antibody validation, see Materials and Methods.) Given this observation, colocalizing MEI4-IHO1 foci (61% of MEI4 foci) may represent most, if not all, genuine MEI4-containing complexes. MEI4 foci were also preferentially detected along stretches of IHO1 staining that marked forming axes from leptotene onwards (Fig. 7a lower panel, b), indicating that MEI4 and IHO1 likely remain in a complex along axes in later stages. Importantly, the Hormad1−/− mutant displayed low-intensity IHO1 foci despite strongly reduced IHO1 levels and failure of IHO1 to spread along unsynapsed axes (Fig. 1d). A large fraction of these residual IHO1 foci (50%) and the majority of MEI4 foci (63%) colocalized in leptotene Hormad1−/− spermatocytes (Fig. 7c–e). Colocalization was similar in a Spo11−/− background where both MEI4 and IHO1 foci appeared more stable (Fig. 7f–h).
Figure 7.

MEI4 and IHO1 colocalize on unsynapsed chromosome axes. (a, c, f) MEI4, IHO1 and SYCP3 were detected by immunofluorescence on nuclear surface spreads of wild-type (WT, a), Hormad1−/− (c) and Hormad1−/− Spo11−/− (f) spermatocytes. (a, c, f right) Enlarged insets show colocalization of MEI4 with IHO1 in wild type, Hormad1−/− and Hormad1−/− Spo11−/−. Note that IHO1 has a “spread out” localization pattern along chromosome axes, whereas MEI4 forms only foci in wild-type leptotene cells. The red channel is shifted to the right by three pixels to allow better visualization of colocalizing green and red signals in the enlarged insets. Scale bars, 10μm. (b, d, g) Quantification of the fractions of MEI4 foci that colocalize with IHO1 foci or stretches (the latter only in wild-type leptotene cells) in wild-type preleptotene and leptotene (b), or Hormad1−/− (d) and Hormad1−/− Spo11−/− (g) leptotene spermatocytes. Quantification of the fractions of IHO1 foci that colocalize with MEI4 foci in Hormad1−/− (e) and Hormad1−/− Spo11−/− (h) leptotene spermatocytes (same cells were analysed as in d and g, respectively). To control for random overlap between anti-IHO1 and anti-MEI4 signals, the colocalization frequency was measured with and without rotating the IHO1 image 90° clockwise relative to the MEI4 image. The significantly lower (Mann-Whitney test) colocalization frequency after rotation indicates that MEI4 and IHO1 genuinely colocalize in spermatocytes. Pooled results from two repeats are shown for each experiment, median of colocalization frequency is marked, and n indicates number of analyzed nuclei in b, d, e, g, h. Primary source data of graphs are shown in Supplementary Table 2.
In yeast two-hybrid assays with known murine DSB-promoting proteins, HORMAD1 interacted only with IHO1 but IHO1 also interacted with REC114, which is a conserved binding partner of MEI4 (Fig. 8a, Supplementary Fig. S7a–d). REC114 is an assumed constituent of DSB-promoting recombinosomes in mammals based on functions of non-mammalian orthologues19–22,43. We also found that IHO1, REC114 and MEI4 self-interacted, which may help in forming cytologically detectable multiprotein-assemblies.
Figure 8.

IHO1 is a key component of putative DSB-promoting recombinosomes. (a) Schematic depicts yeast two-hybrid interactions between MEI4, REC114, IHO1 and HORMAD1. (b) SYCP3 and REC114 were detected in nuclear spreads of wild-type (WT) and Iho1−/− leptotene spermatocytes. (c) Quantification of REC114 foci showed significantly less REC114 foci in Iho1−/− than in wild type (Mann-Whitney test). (d) REC114 and histone H3 (loading control) were detected in testis extracts of wild-type and Iho1−/− 13dpp mice. REC114 levels were not lower in Iho1−/− than in wild type (Data represent one out of three experiments, see quantification in Supplementary Table 2). See Supplementary Fig. S8 for unprocessed gel-scans. (e) IHO1, REC114 and MEI4 were detected on spreads of wild-type preleptotene or zygotene spermatocytes. Green and red channels are shifted by three and six pixels to the right, respectively, to allow better visualization of colocalizing signals in the enlarged insets. REC114 and MEI4 foci colocalize with IHO1 foci or stretches in preleptotene and zygotene, respectively. Scale bars, 10μm. (f) Quantification of colocalization of IHO1, REC114 or MEI4 foci with co-foci of the other two proteins in wild-type preleptotene spermatocytes. Colocalization was measured with and without rotating IHO1, REC114 or MEI4 images by 90° clockwise to control for random colocalization. Strongly reduced colocalization after rotation (Mann-Whitney test) indicates genuine colocalization of MEI4, REC114 and IHO1. (c, f) Pools of two repeats are shown, medians are marked, see Supplementary Table 2 for source data. (g) Model for DSB formation in mammalian meiosis. Green arrows and red blocking arrow indicate promotion and inhibition, respectively. IHO1-REC114 and REC114-MEI4 interactions result in the formation of DSB-promoting recombinosomes that activate SPO11. Signalling from broken DNA ends is thought to prevent repeated DSB formation at nearby sites7,49, possibly by inhibiting or destabilizing DSB-promoting recombinosomes. MEI4-REC114-IHO1 recombinosomes connect to chromosome axes through IHO1-HORMAD1 interaction and as yet unidentified mechanisms (left, question mark). The IHO1-HORMAD1 interaction promotes recruitment of IHO1 and assembly or stabilization of DSB-promoting recombinosomes selectively along unsynapsed axes. This provides one likely, but not exclusive, mechanism for focusing DSB formation on unsynapsed regions, where additional DSBs are needed to promote completion of homologue engagement.
The two-hybrid interaction map suggests that REC114 connects MEI4 to IHO1 in DSB-promoting recombinosomes that interact with unsynapsed axis-bound HORMAD1 through HORMAD1-IHO1 linkages. We have thus far been unable to detect soluble complexes of recombinant REC114 and IHO1 in pull-down assays, or soluble complexes of MEI4, REC114 and IHO1 in anti-MEI4 and anti-IHO1 immunoprecipitates of testis extracts, so the putative MEI4-REC114-IHO1 complexes might be transient or their robust formation may require additional insoluble factors in a chromatin context. Therefore, to test if REC114 is part of MEI4-IHO1-containing complexes in vivo, we examined REC114 localization in spermatocytes. An anti-REC114 antibody detected foci on chromatin in preleptotene; these foci preferentially associated with unsynapsed axes from leptotene onwards and progressively disappeared as chromosomes synapsed and cells proceeded to pachytene (n=200 cells, Fig. 8b, Supplementary Fig. S7e–g). The majority of REC114 foci colocalized with MEI4 foci in leptotene spermatocytes of both wild-type (62%) and Hormad1−/− (53%) mice (Supplementary Fig. S7g–l). Robust anti-REC114 focus formation on chromatin, but not REC114 protein stability, required IHO1 (Fig. 8b–d). Triple immunofluorescence simultaneous staining of MEI4, REC114 and IHO1 displayed extensive colocalization in preleptotene cells (~50% of each protein’s foci; Fig. 8e–f). Co-foci of MEI4 and REC114 also localized along IHO1-rich unsynapsed axes at later stages (Fig. 8e lower panel). Considering technical limitations in identifying genuine chromatin-associated MEI4 foci, the measured colocalization appears highly significant. The combination of these observations supports the hypothesis that MEI4, REC114 and IHO1 are components of putative DSB-promoting recombinosomes that form on chromatin at the onset of meiosis.
Discussion
Here, we identified the previously uncharacterized IHO1 as a HORMAD1-interacting protein that is essential for DSB formation but not for HORMAD1 functions in pairing-independent SC formation, ATR recruitment to unsynapsed axes, MSUC, or meiotic surveillance. MEI4, REC114 and IHO1 form chromatin-bound complexes that appear crucial for DSB formation. These results are consistent with functions of the presumed Spo11-activating complex in yeast containing Mei4 and Rec114 plus a coiled-coil protein that lacks primary sequence conservation between fission (Rec15) and budding (Mer2) yeasts9,19–22. This complex (hereafter the MCD recombinosome: MEI4-containing DSB-promoting) interacts with Spo11 through intermediary proteins9,20–22. MCD recombinosomes may link Spo11 activity to the chromosome axis because chromatin immunoprecipitates of yeast MCD recombinosome components are enriched in DNA sequences that also associate with axis components18,21. Curiously, although there are no clear IHO1 orthologues beyond vertebrates, IHO1 is also a coiled-coil protein. Thus we hypothesise that IHO1 is a functional equivalent of Mer2/Rec15 and that SPO11 activity depends on a conserved MCD recombinosome in mice as in yeasts.
There is functional interplay between the axis and MCD recombinosomes for which HORMA-domain proteins are important, particularly in budding yeast18 and mice16, but to a lesser extent in fission yeast9,44,45. HORMA-domain proteins are needed for robust accumulation of MCD recombinosome components on chromatin and axes in mice (ref16 and this study), and on axis-binding DNA regions in budding yeast18. MCD recombinosome components are essential for DSB formation, whereas meiotic HORMA-domain proteins are only partially necessary9. Correspondingly, MCD recombinosomes were still observed in Hormad1−/− spermatocytes in mice, albeit at reduced numbers (ref16 and this study). HORMA-domain proteins may thus act by enhancing the essential function of MCD recombinosomes. Our epistasis experiments between Hormad1 and Iho1 tested this hypothesis, revealing that both DSB formation and residual MEI4 focus formation in Hormad1−/− spermatocytes depend on IHO1. Thus, IHO1 can promote DSB formation independently of HORMAD1, probably as part of the MCD recombinosome. Conversely, our observations support the idea that HORMAD1 promotes DSB formation by fostering assembly or stabilization of MCD recombinosomes. We hypothesise that HORMAD1 performs its DSB-promoting function through its interaction with IHO1, revealed by yeast two-hybrid, co-immunoprecipitation, and colocalization. HORMAD1 may induce conformational changes in IHO1 via direct binding, or HORMAD1-mediated recruitment of unidentified enzymatic activities may lead to activating modifications of MCD recombinosome components. HORMAD1 is required for “spread out” IHO1 localization along unsynapsed axes, but MEI4, REC114, and IHO1 form foci along unsynapsed axes even in the absence of HORMAD1, so IHO1-HORMAD1 interaction is not the sole link between MCD recombinosomes and axes (Fig. 8g). The nature of this HORMAD1-independent recruitment is unclear, but it may involve another known DSB-promoting protein, MEI146, because MEI4 foci do not associate robustly with axes in the absence of MEI1 even in a HORMAD1-proficient background16. This suggests that the primary function of IHO1-HORMAD1 interaction is not MCD-recombinosome recruitment to axes, but may instead be to support assembly or stabilization of MCD recombinosomes.
A model for the linkage of DSB formation to unsynapsed chromosome axes in mammals is presented in Fig. 8g. Pairwise interactions between IHO1-REC114 and REC114-MEI4 assemble chromatin-bound recombinosomes that promote DSB formation by activating SPO11 through an as yet unidentified mechanism. Robust assembly and/or stabilization of MCD recombinosomes require HORMAD1 starting from preleptotene, before distinguishable axis structures have formed. Parallel to DSB formation—presumably reliant on this initial set of MCD recombinosomes—axes form and HORMAD1 recruits IHO1 to elongating unsynapsed axes. This likely provides a platform for selective assembly and stabilization of additional MCD recombinosomes specifically along unsynapsed axes, where further DSB formation is desirable for successful homologue alignment. Following SC formation, however, HORMAD1 becomes depleted from axes. HORMAD1 depletion is a likely, but not sole, mechanism to protect against excessive DSB formation by destabilising MCD recombinosomes in regions where DSBs are no longer needed for homology search. IHO1 is removed more efficiently than HORMAD1 from synapsed axes, indicating that HORMAD1 displacement is not the only means by which SC formation displaces IHO1 and MCD recombinosomes. Additional mechanisms may include disruption of the IHO1-HORMAD1 interaction, localized degradation, and/or inactivating posttranslational modifications of MCD recombinosome components.
DSB formation is no longer needed in pachytene, and IHO1 disappears even from unsynapsed axes in spermatocytes through an unknown mechanism. IHO1 disappearance may also bring DSB formation to a halt on unsynapsed sex chromosome axes in pachytene. Curiously, IHO1 reappears in spermatocytes both on unsynapsed sex chromosomes and on desynapsing autosomes in late-pachytene and diplotene, respectively. Apoptosis of Iho1−/− spermatocytes at a mid-pachytene-equivalent stage precluded examining IHO1 functions at these late stages, where DSBs no longer form. Selective depletion after IHO1 performed its essential functions in DSB formation will be necessary to address possible late functions in spermatocytes.
Materials and Methods
Yeast two-hybrid, RNA-isolation and RT-PCR
To perform yeast two-hybrid screen, human Hormad1 full length gene (Source Bioscience, IMAGE ID: 5298160) was sub-cloned into a Saccharomyces cerevisiae yeast two-hybrid bait vector (pNGB, sequence available upon request). Full length HORMAD1 in fusion with C-terminal Gal4-DNA binding domain was used as a bait in combination with two prey libraries: a human testis cDNA library (Mate & Plate Library – Human, 630470, Clontech) and a human full length open reading frame (ORF) cDNA library (unpublished, DKFZ, Heidelberg). The prey libraries were supplied and the screens were performed by the Genome and Proteome Core Facility at the German Cancer Research Centre (DKFZ, Heidelberg) as described previously50. To test pairwise interactions between candidates by yeast two-hybrid (Supplementary Fig. S7a–d) we used published methods with minor modifications51. Interactions were tested in the Y2HGold Yeast strain (Cat.# 630498, Clontech). To transform Y2HGold with bait and prey vectors, yeasts were grown in 2xYPDA medium51 overnight at 30°C, 200rpm shaking. Afterwards yeast cells were diluted to 0.4 optical density (measured at 600nm) and incubated in 2xYPDA for 5h at 30°C, 200rpm shaking. Cells were harvested, washed with water and resuspended in 2ml of 100mM Lithium acetate (LiAc). 50μl of this cell suspension was used for each transformation. Transformation mix included 1μg of each vector (bait and prey), 60μl of Polyethylene glycol 50% (w/v in water), 9μl of 1.0M LiAc, 12,5μl of boiled single strand DNA from salmon sperm (AM9680, Ambion), and water up to 90μl in total. The transformation mix was incubated at 30°C for 30min, then at 42°C for 30min for the heat shock. The transformation mix was removed following centrifugation at 1000g for 10 minutes, then cells were resuspended in water, and plated first on –Leu –Trp plates to allow selective growth of transformants. After 2 days of growth transformants were plated both on –Leu –Trp and –Leu –Trp –Ade –His plates for three-to-seven days to test for interactions. For media and plate preparation we followed manufacturer’s instructions52 (See more at: http://www.sigmaaldrich.com/technical-documents/articles/biology/yeast-drop-out.html#sthash.zJzt2dEy.dpuf). To test Iho1 expression in testis and ovaries, RNA was isolated and RT-PCR was performed as described earlier5,53. The RNA of the somatic tissue mix in Supplementary Fig. S1a originated from 17 distinct tissues: liver, brain, thymus, heart, lung, spleen, kidney, mammary gland, pancreas, placenta, salivary gland, skeletal muscle, skin, small intestine, spinal cord, tongue and uterus.
Generation of knockouts and genotyping
The Ccdc36-targeting construct (clone HTGRS6006_A_G10, BACPAC Resources Center, Knockout Mouse Project KOMP repository) was based on a so called ‘knockout first’ multipurpose allele strategy54 (Supplementary Fig. S3). Targeting of R1 (agouti) ES cells was carried out as before12 and chimeras were generated by laser assisted C57BL/6 morula injections with ES cell clones heterozygote for the Iho1insertion allele (Supplementary Fig. S3). Progeny of the chimeric animals were crossed to the outbred wild-type CD-1® mouse line, and to pCAGGs-FLPo55 and PGK-Cre56 transgenic mice to generate first Iho1restored, and then Iho1deletion alleles from Iho1insertion allele (Supplementary Fig. S3). Mice were maintained on the outbred ICR (CD-1®) background. Mice were initially genotyped by Southern blotting (Supplementary Fig. S3), and by PCR in subsequent crosses using tail-tip genomic DNAs. Genotyping primers:
LacZfor 5′-CCCGTCGTTTTACAACGTC
LacZrev 5′-CATAACCACCACGCTCATC
Ccdc36RestFw 5′-CCACCAATACTCATTTACCCC
Ccdc36RestRv 5′-ACTATTTACTCTGCCTCTGAGC
Ccdc36LoxPRv1 5′-AACTTCAAAACACCTCGCC
5Ccdc36seq2 5′-TCTTCCTTTGCCAGTTGGGAAATC
CreFw 5′-GCCTGCATTACCGGTCGATGCAACGA
CreRv 5′-GTGGCAGATGGCGCGGCAACACCATT
FlpOFw 5′-GCTATCGAATTCCACCATGGCTCCTAAGAAGAA
FlpORv 5′-CAATGCGATGAATTCTCAGATCCGCCTGTTGATGTA
PCR product sizes: with LacZfor/LacZrev primers, Iho1insertion template-208 bp, other alleles-no specific product; with Ccdc36RestFw/Ccdc36RestRv primers, wild-type allele template-357 bp, Iho1insertion template-7423 bp, Iho1restored template-500 bp; with 5Ccdc36seq2/Ccdc36LoxPRv1 primers, wild-type allele template-404 bp, Iho1insertion and Iho1restored templates-366 bp, Iho1deletion template-no specific product; with Ccdc36RestFv/Ccdc36LoxPRv1 primers: wild-type allele-1096 bp, Iho1insertion template-no specific product, Iho1restored template-1201bp, Iho1deletion template-373 bp. FlpOFw/FlpORv were used to detect FlpO recombinase (1500 bp), CreFw/CreRv were used to detect Cre recombinase (750 bp).
Animal experiments
Mice carrying Hormad1-, Spo11-, Dmc1- or Mei4-null alleles were described earlier12,23,31,57. Western blotting reconfirmed the absence of full length IHO1 protein from testis extracts of both Iho1insertion/insertion and Iho1deletion/deletion mice (Supplementary Fig. S3c). Histology in testis, analysis of the synaptonemal complex and the axis, and focus counts of RAD51, DMC1 and RPA were carried out in both Iho1insertion and Iho1deletion strains. The rest of the experiments were carried out in the Iho1deletion lineage. Because no obvious differences were detected between the strains we refer to both of their genotypes as Iho1−/−. Iho1restored/restored mice were fertile and their spermatocytes were indistinguishable from wild-type spermatocytes. This supported the specificity of the observed phenotypes in the Iho1insertion and Iho1deletion strains. Whenever possible, experimental animals were compared with littermate controls or with age-matched non-littermate controls from the same colony. Adult mice between age 70-120 days were used for experiments unless indicated otherwise. No statistical method was used to predetermine sample size. The experiments were not randomized. The Investigators were not blinded to allocation during experiments and outcome assessment. All animals were used and maintained in accordance with the German Animal Welfare legislation (“Tierschutzgesetz”). All procedures pertaining to animal experiments were approved by the Governmental IACUC (“Landesdirektion Sachsen”) and overseen by the animal ethics committee of the Technische Universität Dresden. The license numbers concerned by the present experiments with mice are DD24-5131/287/1 and 24-9168.24-1/2006-13 (tissue collection without prior in vivo experimentation).
Cell lines
Only the R1 ES cell line was used in this study. This cell line was not found in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI Biosample. The cell line was not authenticated. The cell line was not tested for mycoplasma contamination.
Antibody generation
To produce anti-IHO1 antibody, full-length mouse Ccdc36 (Iho1) open reading frame and four non-overlapping fragments that together covered the full length Ccdc36 open reading frame were sub-cloned from plasmid DNA (Source Bioscience, IMAGE ID:5716219) into Escherichia coli expression vectors pDEST17 (Cat# 11803-012, Invitrogen) and petMM30 (gift from Dr. Drechsel, sequence available upon request), respectively. These expression vectors were used to produce N-terminal 6xHis-tagged full length IHO1, and 6xHis-1xGST-tagged IHO1 fragments in bacteria. All recombinant proteins were purified on Ni-Sepharose beads (Cat#17-5318-01, Amersham, GE Healthcare), and the tagged full-length IHO1 preparations were used for immunization of rabbits, guinea pigs and chickens. Polyclonal anti-IHO1 antibodies were affinity purified on antigen coupled NHS-Activated Sepharose 4 Fast Flow beads (Cat#17-0906-01, Amersham, GE Healthcare) according to standard methods58. Guinea pig and chicken anti-IHO1 serum was purified using IHO1-full length recombinant protein; rabbit anti-IHO1 serum was purified first using IHO1-full length and subsequently using recombinant IHO1-fragment peptides. Antibodies specific to IHO1 fragment-1 (144 amino-acids from 1M to V144) produced the best signal to noise ratio in immunofluorescence, hence we used this antibody in most IHO1 localization experiments where rabbit anti-IHO1 antibody was used. All antibodies were tested by immunofluorescence staining of spermatocyte nuclear spreads. Antibodies from all three species showed a similar staining pattern. The localization pattern of IHO1 was also reconfirmed by overexpression of GFP-tagged version of IHO1 following in vivo electroporation of testes59,60. In addition, all antibodies were tested by immunoblot analysis of testis extracts. Although all of the antibodies recognized several proteins of different molecular weights, only a protein that was approximately 65kDa based on its electrophoretic mobility was absent from Iho1−/− testis extracts (Supplementary Fig. S3c), and was detected by all the different antibodies in wild-type testis extracts. The size of this protein corresponds well to the predicted molecular mass of IHO1 (63.4 kDa).
To produce anti-MEI4 antibody, mouse full-length Mei4 open reading frame and two non-overlapping fragments corresponding to the N-terminal (130 amino acids: 1M to 130L) and C-terminal (129 amino acids: 261R to 389P) parts of MEI4 were sub-cloned from plasmid DNA23 into Escherichia coli expression vectors pDEST17, for full-length Mei4, or pETG41A (EMBL protein expression and purification facility), for both fragments. These expression vectors were used to produce N-terminal 6xHis-tagged full length MEI4 and 6xHis-1xMBP-tagged MEI4 fragments in bacteria. The recombinant full length MEI4 protein and MEI4 fragments were purified on Ni Sepharose beads (Cat#17-5318-01, Amersham, GE Healthcare) and 6xHis-tagged full-length MEI4 protein was used for immunization of guinea pigs. Polyclonal anti-MEI4 antibodies were affinity purified on antigen coupled NHS-activated Sepharose 4 fast flow beads (Cat#17-0906-01, Amersham, GE Healthcare). Guinea pig anti-MEI4 serum was purified first using MEI4-full length, and subsequently using recombinant MEI4-fragment peptides. Antibodies specific to MEI4 fragment-1 (130 amino-acids from 1M to L130) produced the best signal to noise ratio in immunofluorescence, hence we used this antibody in most MEI4 localization experiments. The specificity of the MEI4 fragment-1 antibody was tested by co-immunostaining wild-type and Mei4−/− spermatocyte nuclear spreads with the anti-MEI4 fragment-1 antibody and a published rabbit anti-MEI4 antibody23. The two antibodies showed similar staining patterns, and numbers of foci detected by both antibodies were dramatically reduced in Mei4−/− as compared to wild-type spermatocytes. Medians of focus numbers detected by the new guinea pig and the published23 rabbit antibodies were 5 and 47, respectively, and the median of co-focus numbers was 1 in Mei4−/− spermatocytes (n=36 leptotene spermatocytes; Supplementary Table 2, “gp&rb MEI4 WTvsMEI4ko”). Medians of focus numbers detected by the new guinea pig and the published rabbit antibodies were 304 and 361, respectively, and the median of co-focus numbers was 220 in wild-type spermatocytes (n=29 spermatocytes; Supplementary Table 2, “gp&rb MEI4 WTvsMEI4ko”). In addition, the guinea pig anti-MEI4 fragment-1 antibody was tested in immunoprecipitation and immunoblot analysis of testis extracts. This antibody recognizes a protein of approximately 50 kDa based on its electrophoretic mobility (Fig. 6c), which corresponds well to the predicted molecular mass of MEI4 (44.5 kDa).
Other Antibodies
In addition to antibodies that were previously described5,12 we used: rabbit anti-MEI423 (IF 1:50), guinea pig anti-MEI4 (IF 1:500, WB 1:2000), rabbit anti-REC114 (IF 1:500, WB 1:2000, unpublished antibody of S. Keeney and M. van Overbeek), guinea pig anti-IHO1 (IF 1:2000), rabbit anti-IHO1 fragment-1 (IF 1:2000, WB 1:3000), chicken anti-IHO1 (IF 1:3000), goat anti-Beta actin (WB 1:1000, ab8229 from Abcam), rabbit anti-SYCP3 (WB 1:6000, ab15093 from Abcam).
Immunofluorescence microscopy
Preparation and immunostaining of testis-ovary cryosections and nuclear surface spreads of meiocytes were adapted from methods described previously with minor modifications61. Briefly, testis cell suspensions were prepared in PBS pH 7.4, then mixed with hypotonic extraction buffer in 1:1 ratio and incubated for 8min at room temperature. After diluting the cell suspension five times in PBS pH 7.4, the cell suspension was centrifuged for 5min at 1000g, and cells were resuspended in the 1:2 mixture of PBS and 100 mM sucrose solution. Cell suspensions were added to seven times higher volume droplets of filtered (0.2 μm) 1% paraformaldehyde (PFA), 0.15% Triton X-100, 1 mM sodium borate pH 9.2 solution on diagnostic slides, and incubated for 60min at room temperature. Nuclei were then dried for at least 1 h under the hood. Finally, the slides were washed in 0.4% Photoflo (Kodak) and dried at room temperature. To compare oocyte numbers in adults, we sectioned ovaries of mice (8-μm-thick sections), identified oocytes by anti-NOBOX (oocyte marker62) immunostaining and counted oocytes in every tenth section. To immunostain spread nuclei and sections, slides were blocked with 2.5% w/v BSA in PBS for 30min, then slides were incubated with primary antibodies diluted in blocking solution either for 3 hours at room temperature or overnight at 4°C. Subsequently, slides were washed (3×10min) in PBS and incubated with secondary antibodies at room temperature for 90 minutes. Finally, slides were washed (3×10 minutes) in PBS and embedded in Slowfade gold antifade mounting media with or without DAPI (Invitrogen). For the quantification of the histone H1T, IHO1 and ɣH2AX signal in spread spermatocytes (Supplementary Fig. S2, S6), matched-exposure images were taken and signal intensities were measured with a similar strategy as described earlier5. To perform MEI4, REC114 or IHO1 focus quantifications and colocalization experiments, images were analyzed using Cell Profiler software63 and customized image analysis pipelines. Given that these proteins apparently formed foci along axes, thresholds were determined empirically to allow the automatic detection of most discernable axis-associated foci. To validate the specificity of colocalization we always compared colocalization in images where the channels of the tested proteins were either unturned or were turned 90 degrees clockwise relative to each other. We considered colocalization significant only if colocalization between foci was significantly reduced after turning one channel 90 degrees. To make sure that the shape of the cell did not influence the results we chose spreads where the nuclei were nearly circular and colocalization was measured only in the largest circular area that could be placed within these cells.
To measure colocalization of MEI4, REC114 or IHO1 foci in wild-type preleptotene spermatocytes, we stained nuclear spreads with guinea pig anti-MEI4, rabbit anti-REC114, and chicken anti-IHO1 antibodies. Preleptotene stage was identified based on the IHO1 staining pattern, which is characteristically punctate at this stage due to the lack of well developed elongated chromosome axis structures.
Due to the limitation of the nuclear surface spreading method, a variable amount of non-chromatin-bound nuclear content may remain fixed in the nuclear surface spreads. This can give rise to variable punctate background in immunostaining experiments. To reduce the effect of this technical variability and to acquire a more reliable estimate for the number of genuine chromatin-bound MEI4-containing protein assemblies, we co-stained nuclear surface spreads with guinea pig and rabbit anti-MEI4 antibodies. We based our focus counts only on foci that were detected by both antibodies in experiments where we addressed the effect of Iho1 or Hormad1 deficiency on MEI4 focus numbers. To validate this approach, we determined what fraction of axis-associated and non-axis-associated MEI4 foci were detected by both guinea pig and rabbit antibodies. “Background” foci that do not represent genuine chromatin-bound MEI4 complexes are expected to have a random distribution throughout the nucleus. In contrast, axis-associated foci are expected to represent genuine chromatin-bound MEI4 protein complexes with a higher likelihood. We found that a median of 63% or 34% of guinea pig antibody-detected foci colocalized with rabbit antibody-detected foci on axis or off axis, respectively, in zygotene stage wild-type spermatocytes (n=30; Supplementary Table 2, “wt-gpMEI4-rbMEI4vsAxis”). Similar numbers were obtained for the rabbit antibody stained foci (60% and 37%, respectively). Thus, axis-associated MEI4 foci are more likely to be detected by both antibodies than non-axis-associated foci. These observations are consistent with the idea that MEI4 focus counts that are based on co-staining with guinea pig and rabbit anti-MEI4 antibodies provide a more reliable estimate of the number of chromatin-bound MEI4 protein complexes than focus counts that are based on MEI4 detection with anti-MEI4 antibody from a single species.
Staging of meiotic prophase
Nuclear spreads were staged based on axis development, which was assessed through detection of SYCP3 on meiocyte spreads. Briefly, hazy/punctate SYCP3 staining corresponds to preleptotene, short stretches of axis corresponds to leptotene (most axis stretches shorter than 5× the widths of axes), relatively long and incomplete stretches of axes corresponds to early-zygotene and complete axes corresponds to late-zygotene and pachytene stages (see figure legend of Supplementary Fig. S5). Preleptotene cells are characterized by active DNA replication; therefore we also used EdU labelling (Supplementary Fig. S1b) in combination with SYCP3 stain to identify preleptotene spermatocytes and to reconfirm the axis morphology that characterizes preleptotene spermatocytes. To identify sub-stages in wild-type pachytene spermatocytes (Supplementary Fig. S2), we used the combination of SYCP3 and either histone H1T64 or γH2AX staining. Pachytene-zygotene transition: all autosomes appear synapsed, but the very ends of autosome axis pairs appear thicker, indicating that synaptonemal complex formation just finished; no H1T staining; γH2AX-marked chromatin of sex chromosomes is poorly condensed and is often irregular in shape, and γH2AX is detectable at a low level along synapsed autosomes. Early-pachytene: thickness of synapsed axis pairs is largely uniform along their entire length, synapsed PAR region is long (length is >5× the widths) or short (length is <5× but >3× longer than the widths) and no or very weak histone H1T signal. Histone γH2AX-marked chromatin of sex chromosomes is better condensed with a more regular shape than during zygotene-pachytene transition, γH2AX is still detectable at low levels along synapsed autosomes. Mid-pachytene: autosomal axis/synaptonemal complex morphology is the same as in early-pachytene. Synapsed PAR region is short or very short (length is <3× the widths), histone H1T signal is weak to intermediate strength. We first determined the signal intensity of histone H1T in cells that have very short PAR synapsis, hence were defined as mid-pachytene. The lowest histone H1T signal from these cells was chosen as a cut off to distinguish early- and mid-pachytene cells in which PAR synapsis was short. Late-pachytene: synapsed axes of autosomes are clearly thicker at ends than at interstitial sites. Synapsis at PAR is very short/dot like, or a gap appears between X and Y PAR regions. Histone H1T signal intensity is intermediate to strong. Histone γH2AX-marked chromatin of sex chromosomes is well condensed, γH2AX is not detectable along synapsed autosomal axes.
EdU incorporation
Testis cell suspensions were incubated with 10μM EdU in DMEM with 10% FCS at 37°C for 1 h for in vitro labeling. EdU incorporation was detected using Alexa Fluor 647 and the Click-iT EdU imaging kit according to the manufacturer’s instructions (Invitrogen).
Preparation of protein extracts
To prepare testis extract for IHO1 immunoprecipitation, adult mouse testes were detunicated and macerated by a scalpel in a petri dish, then resuspended in chromatin extraction buffer (8mM Tris-HCl pH 7.5, 1% Triton X-100), incubated for 5min at 4°C and centrifuged for 10min at 2000g. The pellet was resuspended in lysis buffer (50 mM Tris-HCl at pH 7.5, 500 mM NaCl, 1% Triton X-100, 1% NP40, 0.5% sodium deoxycholate, 10 mM MgCl2) for 60 min at 4°C. Benzonase (Merck Millipore) was used to digest DNA during lysis. Lysates were sonicated 5min 400w with Banson sonofier 450 and centrifuged at 20000g for 10min. Supernatants were diluted 1:4 with 50 mM Tris-HCl at pH 7.5 and used as input for the immunoprecipitation. To prepare testis extract for MEI4 immunoprecipitation, 12 days postpartum mouse testes were detunicated and macerated by a scalpel in a petri dish, then lysed in lysis buffer (50 mM Tris-HCl at pH 7.5, 500 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 10 mM MgCl2) for 60 min at 4°C. Lysates were processed further as described above for IHO1 immunoprecipitation. Preparation of TritonX-100-soluble and -insoluble testis extract fractions was performed as published previously34 (Fig. 1e). All extracts were prepared using protease and phosphatase inhibitors as described earlier34.
Antibody crosslinking, immunoprecipitation and western blotting
Either rabbit anti-IHO1 fragment 1 (3μg) antibodies or guinea pig anti-MEI4 (3μg) antibodies were cross-linked to 1.5mg of Dynabeads Protein A (Invitrogen) using 20 mM Dimethyl Suberimidate according to standard protocols58. After preclearing with empty protein A beads, protein extracts were incubated with affinity beads overnight at 4°C. Beads were then washed three times with washing buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.1% Tween20). Immunoprecipitated material was eluted from the beads by incubating the beads in 50ul Laemmli sample buffer for 10min at room temperature. The resulting elutions were analyzed with SDS-PAGE and immunoblotting using standard methods. Briefly, proteins from extracts were separated on SDS polyacrylamide gels and blotted onto either Nitrocellulose (Hybond ECL Cat#RPN2020D, GE Healthcare) (for IHO1 detection in total testis extracts) or PVDF membrane (Cat#P2938, Sigma). Membranes were then blocked using BSA 2.5%, 0.05% Tween, TBS blocking solution. To measure levels of SPO11-oligonucleotide complexes in testes, we carried out SPO11 immunoprecipitations and SPO11-oligonucleotide detection as published previously12,30. Both testes from one adult mouse were used for each experiment.
Statistics and Reproducibility
R “stats” or “exactRanktests” packages were used to calculate P values by one-sample t-test, paired t-test, Man Whitney/Wilcoxon rank sum test exact (non-paired samples) or Wilcoxon signed rank test exact (paired samples). Each conclusion in the manuscript was based on results that were reproduced in at least two independent experiments and at least two mice of each genotype.
Data availability
Supplementary Table 2 contains Statistics Source Data for Fig. 2, 3, 5, 6, 7, 8, Supplementary Fig. S2, S5, S6, S7 and for anti-MEI4 antibody validation and MEI4 foci axis association (sheets “gpMEI4-rbMEI4-coloc”, “gp&rb MEI4 WTvsMEI4ko” and “wt-gpMEI4-rbMEI4vsAxis”). All other data supporting the findings of this study are available from the corresponding author on request.
Supplementary Material
Acknowledgments
We thank R. Jessberger for sharing ideas, antibodies (anti-SYCP3 and anti-STAG3) and departmental support; M. Stevense for discussion, revising and proofreading the manuscript; G. Vader for sharing budding yeast protocols, E. Marcon for the anti-RPA antibody; Frank Schwarz and the protein interaction unit of Genomics and Proteomics Core Facility at the German Cancer Research Center – DKFZ (http://www.dkfz.de/gpcf/y2h.html) for yeast two-hybrid vectors and for performing yeast two hybrid screens; M.P. Thelen for the anti-SPO11 antibody, Handel MA for histone H1T antibody; David Drechsel and the EMBL protein expression and purification facility for protein expression vectors; K. Daniel for testis and somatic tissue cDNAs. We thank the DIGS-BB program for supporting M. Stanzione. The Deutsche Forschungsgemeinschaft (DFG; grants: TO421/3-2, SPP1384:TO421/4-2, TO421/5-1, TO421/6-1, TO421/8-1 and 8-2) supported M.S., M.B., F.P., I.D., A.R., D.T., and A.T.; The JSPS Research Fellowship supported H. S., Y.W.; BdM was funded by grants from the Centre National pour la Recherche Scientifique (CNRS) and the European Research Council Executive Agency under the European Community’s Seventh Framework Programme (FP7/2007-2013 Grant Agreement no. [322788]).; the NIH (R01 GM105421) supported J.L., M.J., and S.K., J.L. was supported in part by American Cancer Society fellowship PF-12-157-01-DMC. S.K. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Author contributions:
Most experiments were carried out by M.S. (Fig. 1a–c, 4, 6, 7, 8a, d, g, S1b–c, S2 b, d, S3, S5b–e, S6, S7a–d) and M.B. (Fig. 1e, 3a–f, S1a, d–e, S2a, c, S5a, f, h). Further experiments were performed by F.P. (Fig. 2, 5, S4, S5f, g), I.D. (Fig. 1d, 8b, c, e, f, S7e–l) and J.L. (Fig. 3g, h). Knockout ES cells were generated by D.T. with the supervision of A.R.; M.J. and S.K. contributed to SPO11-oligonucleotide measurements and S.K. supplied REC114 antibodies; B.d.M. supplied MEI4 antibodies, experimental material and critical unpublished information, H. S. and Y.W. contributed with critical supporting IHO1 localization data. A.T. and M.S. wrote the manuscript. All authors were involved in discussions and commented on the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Contributor Information
Marcello Stanzione, Email: marcello.stanzione@tu-dresden.de.
Marek Baumann, Email: marek.x.baumann@gsk.com.
Frantzeskos Papanikos, Email: frantzeskos.papanikos@mailbox.tu-dresden.de.
Ihsan Dereli, Email: ihsan.dereli@mailbox.tu-dresden.de.
Julian Lange, Email: langej@mskcc.org.
Angelique Ramlal, Email: aramlal_2000@yahoo.com.
Daniel Tränkner, Email: daniel.traenkner@tu-dresden.de.
Hiroki Shibuya, Email: hiroki.19851201@gmail.com.
Bernard de Massy, Email: bernard.de-massy@igh.cnrs.fr.
Yoshinori Watanabe, Email: ywatanab@iam.u-tokyo.ac.jp.
Maria Jasin, Email: m-jasin@ski.mskcc.org.
Scott Keeney, Email: s-keeney@ski.mskcc.org.
Attila Tóth, Email: attila.toth@mailbox.tu-dresden.de.
References
- 1.Baudat F, de Massy B. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res. 2007;15:565–577. doi: 10.1007/s10577-007-1140-3. [DOI] [PubMed] [Google Scholar]
- 2.Hunter N. Meiotic recombination in Molecular genetics of recombination. Springer-Verlag; Berlin Heidelberg: 2007. [Google Scholar]
- 3.Bergerat A, et al. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 4.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 5.Wojtasz L, et al. Mouse HORMAD1 and HORMAD2, Two Conserved Meiotic Chromosomal Proteins, Are Depleted from Synapsed Chromosome Axes with the Help of TRIP13 AAA-ATPase. PLoS genetics. 2009;5 doi: 10.1371/journal.pgen.1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thacker D, Mohibullah N, Zhu X, Keeney S. Homologue engagement controls meiotic DNA break number and distribution. Nature. 2014;510:241–246. doi: 10.1038/nature13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keeney S, Lange J, Mohibullah N. Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annual review of genetics. 2014;48:187–214. doi: 10.1146/annurev-genet-120213-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kauppi L, et al. Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes & development. 2013;27:873–886. doi: 10.1101/gad.213652.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lam I, Keeney S. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harbor perspectives in biology. 2015;7:a016634. doi: 10.1101/cshperspect.a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenberg SC, Corbett KD. The multifaceted roles of the HORMA domain in cellular signaling. J Cell Biol. 2015;211:745–755. doi: 10.1083/jcb.201509076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuda T, Daniel K, Wojtasz L, Toth A, Hoog C. A novel mammalian HORMA domain-containing protein, HORMAD1, preferentially associates with unsynapsed meiotic chromosomes. Experimental cell research. 2010;316:158–171. doi: 10.1016/j.yexcr.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Daniel K, et al. Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nature Cell Biology. 2011;13:599–U232. doi: 10.1038/ncb2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kogo H, et al. HORMAD1-dependent checkpoint/surveillance mechanism eliminates asynaptic oocytes. Genes to cells: devoted to molecular & cellular mechanisms. 2012;17:439–454. doi: 10.1111/j.1365-2443.2012.01600.x. [DOI] [PubMed] [Google Scholar]
- 14.Shin YH, et al. Hormad1 mutation disrupts synaptonemal complex formation, recombination, and chromosome segregation in mammalian meiosis. PLoS genetics. 2010;6:e1001190. doi: 10.1371/journal.pgen.1001190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin YH, McGuire MM, Rajkovic A. Mouse HORMAD1 is a meiosis i checkpoint protein that modulates DNA double-strand break repair during female meiosis. Biology of reproduction. 2013;89:29. doi: 10.1095/biolreprod.112.106773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar R, et al. MEI4: a central player in the regulation of meiotic DNA double strand break formation in the mouse. J Cell Sci. 2015 doi: 10.1242/jcs.165464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blat Y, Protacio RU, Hunter N, Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111:791–802. doi: 10.1016/s0092-8674(02)01167-4. [DOI] [PubMed] [Google Scholar]
- 18.Panizza S, et al. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell. 2011;146:372–383. doi: 10.1016/j.cell.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Hooker GW, Roeder GS. Saccharomyces cerevisiae Mer2, Mei4 and Rec114 form a complex required for meiotic double-strand break formation. Genetics. 2006;173:1969–1981. doi: 10.1534/genetics.106.058768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maleki S, Neale MJ, Arora C, Henderson KA, Keeney S. Interactions between Mei4, Rec114, and other proteins required for meiotic DNA double-strand break formation in Saccharomyces cerevisiae. Chromosoma. 2007;116:471–486. doi: 10.1007/s00412-007-0111-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyoshi T, et al. A central coupler for recombination initiation linking chromosome architecture to S phase checkpoint. Mol Cell. 2012;47:722–733. doi: 10.1016/j.molcel.2012.06.023. [DOI] [PubMed] [Google Scholar]
- 22.Arora C, Kee K, Maleki S, Keeney S. Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol Cell. 2004;13:549–559. doi: 10.1016/s1097-2765(04)00063-2. [DOI] [PubMed] [Google Scholar]
- 23.Kumar R, Bourbon HM, de Massy B. Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes & development. 2010;24:1266–1280. doi: 10.1101/gad.571710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin S, et al. Comparison of the transcriptional landscapes between human and mouse tissues. Proc Natl Acad Sci U S A. 2014;111:17224–17229. doi: 10.1073/pnas.1413624111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yue F, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burgoyne PS, Mahadevaiah SK, Turner JM. The consequences of asynapsis for mammalian meiosis. Nat Rev Genet. 2009;10:207–216. doi: 10.1038/nrg2505. [DOI] [PubMed] [Google Scholar]
- 27.Barchi M, et al. Surveillance of different recombination defects in mouse spermatocytes yields distinct responses despite elimination at an identical developmental stage. Molecular and cellular biology. 2005;25:7203–7215. doi: 10.1128/MCB.25.16.7203-7215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pacheco S, et al. The ATM signaling cascade promotes recombination-dependent pachytene arrest in mouse spermatocytes. PLoS genetics. 2015;11:e1005017. doi: 10.1371/journal.pgen.1005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahadevaiah SK, et al. Extensive meiotic asynapsis in mice antagonises meiotic silencing of unsynapsed chromatin and consequently disrupts meiotic sex chromosome inactivation. J Cell Biol. 2008;182:263–276. doi: 10.1083/jcb.200710195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pittman DL, et al. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell. 1998;1:697–705. doi: 10.1016/s1097-2765(00)80069-6. [DOI] [PubMed] [Google Scholar]
- 32.Yoshida K, et al. The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol Cell. 1998;1:707–718. doi: 10.1016/s1097-2765(00)80070-2. [DOI] [PubMed] [Google Scholar]
- 33.Kogo H, et al. HORMAD2 is essential for synapsis surveillance during meiotic prophase via the recruitment of ATR activity. Genes to cells: devoted to molecular & cellular mechanisms. 2012;17:897–912. doi: 10.1111/gtc.12005. [DOI] [PubMed] [Google Scholar]
- 34.Wojtasz L, et al. Meiotic DNA double-strand breaks and chromosome asynapsis in mice are monitored by distinct HORMAD2-independent and -dependent mechanisms. Genes & development. 2012;26:958–973. doi: 10.1101/gad.187559.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner JM, et al. BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr Biol. 2004;14:2135–2142. doi: 10.1016/j.cub.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 36.Royo H, et al. ATR acts stage specifically to regulate multiple aspects of mammalian meiotic silencing. Genes & development. 2013;27:1484–1494. doi: 10.1101/gad.219477.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turner JM, Mahadevaiah SK, Ellis PJ, Mitchell MJ, Burgoyne PS. Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Developmental cell. 2006;10:521–529. doi: 10.1016/j.devcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 38.Turner JM, et al. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet. 2005;37:41–47. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
- 39.Royo H, et al. Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr Biol. 2010;20:2117–2123. doi: 10.1016/j.cub.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 40.Bellani MA, Romanienko PJ, Cairatti DA, Camerini-Otero RD. SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm−/−spermatocytes. J Cell Sci. 2005;118:3233–3245. doi: 10.1242/jcs.02466. [DOI] [PubMed] [Google Scholar]
- 41.Cloutier JM, et al. Histone H2AFX Links Meiotic Chromosome Asynapsis to Prophase I Oocyte Loss in Mammals. PLoS genetics. 2015;11:e1005462. doi: 10.1371/journal.pgen.1005462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carballo JA, Johnson AL, Sedgwick SG, Cha RS. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell. 2008;132:758–770. doi: 10.1016/j.cell.2008.01.035. [DOI] [PubMed] [Google Scholar]
- 43.Sasanuma H, et al. Meiotic association between Spo11 regulated by Rec102, Rec104 and Rec114. Nucleic acids research. 2007;35:1119–1133. doi: 10.1093/nar/gkl1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Latypov V, et al. Roles of Hop1 and Mek1 in meiotic chromosome pairing and recombination partner choice in Schizosaccharomyces pombe. Molecular and cellular biology. 2010;30:1570–1581. doi: 10.1128/MCB.00919-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lorenz A, Estreicher A, Kohli J, Loidl J. Meiotic recombination proteins localize to linear elements in Schizosaccharomyces pombe. Chromosoma. 2006;115:330–340. doi: 10.1007/s00412-006-0053-9. [DOI] [PubMed] [Google Scholar]
- 46.Libby BJ, Reinholdt LG, Schimenti JC. Positional cloning and characterization of Mei1, a vertebrate-specific gene required for normal meiotic chromosome synapsis in mice. Proc Natl Acad Sci U S A. 2003;100:15706–15711. doi: 10.1073/pnas.2432067100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bellani MA, Boateng KA, McLeod D, Camerini-Otero RD. The expression profile of the major mouse SPO11 isoforms indicates that SPO11beta introduces double strand breaks and suggests that SPO11alpha has an additional role in prophase in both spermatocytes and oocytes. Molecular and cellular biology. 2010;30:4391–4403. doi: 10.1128/MCB.00002-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romanienko PJ, Camerini-Otero RD. Cloning, characterization, and localization of mouse and human SPO11. Genomics. 1999;61:156–169. doi: 10.1006/geno.1999.5955. [DOI] [PubMed] [Google Scholar]
- 49.Lange J, et al. ATM controls meiotic double-strand-break formation. Nature. 2011;479:237–240. doi: 10.1038/nature10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koegl M, Uetz P. Improving yeast two-hybrid screening systems. Briefings in functional genomics & proteomics. 2007;6:302–312. doi: 10.1093/bfgp/elm035. [DOI] [PubMed] [Google Scholar]
- 51.Gietz RD, Triggs-Raine B, Robbins A, Graham KC, Woods RA. Identification of proteins that interact with a protein of interest: applications of the yeast two-hybrid system. Molecular and cellular biochemistry. 1997;172:67–79. [PubMed] [Google Scholar]
- 52.Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics, a Cold Spring Harbor Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1994. [Google Scholar]
- 53.Wojtasz L, Daniel K, Toth A. Fluorescence Activated Cell Sorting of Live Female Germ Cells and Somatic Cells of the Mouse Fetal Gonad Based on Forward and Side Scattering. Cytometry Part A. 2009;75A:547–553. doi: 10.1002/cyto.a.20729. [DOI] [PubMed] [Google Scholar]
- 54.Testa G, et al. A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis. 2004;38:151–158. doi: 10.1002/gene.20012. [DOI] [PubMed] [Google Scholar]
- 55.Kranz A, et al. An improved Flp deleter mouse in C57Bl/6 based on Flpo recombinase. Genesis. 2010;48:512–520. doi: 10.1002/dvg.20641. [DOI] [PubMed] [Google Scholar]
- 56.Lallemand Y, Luria V, Haffner-Krausz R, Lonai P. Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site-specific recombinase. Transgenic Res. 1998;7:105–112. doi: 10.1023/a:1008868325009. [DOI] [PubMed] [Google Scholar]
- 57.Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell. 2000;6:989–998. doi: 10.1016/s1097-2765(00)00098-8. [DOI] [PubMed] [Google Scholar]
- 58.Halow E, Lane D. Using Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1999. [Google Scholar]
- 59.Shibuya H, Morimoto A, Watanabe Y. The dissection of meiotic chromosome movement in mice using an in vivo electroporation technique. PLoS genetics. 2014;10:e1004821. doi: 10.1371/journal.pgen.1004821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shoji M, Chuma S, Yoshida K, Morita T, Nakatsuji N. RNA interference during spermatogenesis in mice. Developmental biology. 2005;282:524–534. doi: 10.1016/j.ydbio.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 61.Peters AH, Plug AW, van Vugt MJ, de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 1997;5:66–68. doi: 10.1023/a:1018445520117. [DOI] [PubMed] [Google Scholar]
- 62.Suzumori N, Yan C, Matzuk MM, Rajkovic A. Nobox is a homeobox-encoding gene preferentially expressed in primordial and growing oocytes. Mechanisms of development. 2002;111:137–141. doi: 10.1016/s0925-4773(01)00620-7. [DOI] [PubMed] [Google Scholar]
- 63.Kamentsky L, et al. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics. 2011;27:1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Inselman A, Eaker S, Handel MA. Temporal expression of cell cycle-related proteins during spermatogenesis: establishing a timeline for onset of the meiotic divisions. Cytogenet Genome Res. 2003;103:277–284. doi: 10.1159/000076813. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary Table 2 contains Statistics Source Data for Fig. 2, 3, 5, 6, 7, 8, Supplementary Fig. S2, S5, S6, S7 and for anti-MEI4 antibody validation and MEI4 foci axis association (sheets “gpMEI4-rbMEI4-coloc”, “gp&rb MEI4 WTvsMEI4ko” and “wt-gpMEI4-rbMEI4vsAxis”). All other data supporting the findings of this study are available from the corresponding author on request.


