Abstract
Pattern recognition receptors are critical for the detection of invading microorganisms. They activate multiple pathways that lead to the induction of proinflammatory responses and pathogen clearance. The intensity and duration of this immune reaction must be tightly controlled spatially and temporally in every tissue by different negative regulators. We hypothesized that monocyte chemoattractant protein-1-induced protein-1 (MCPIP-1) might play a role in maintaining immune homeostasis in the epithelium both under physiological conditions and upon bacterial infection. To this end, we examined the distribution of the MCPIP-1 transcript and protein in various tissues. The MCPIP-1 protein level was higher in epithelial cells than in myeloid cells. MCPIP-1 exerted RNase activity towards the interleukin (IL)-8 transcript and the lifespan of IL-8 was determined by the presence of the stem-loops/hairpin structures at the 3′UTR region of IL-8 mRNA. Moreover, using fully active, purified recombinant MCPIP-1 protein, we elucidated the mechanism by which MCPIP-1 controls the IL-8 mRNA level. In conclusion, we uncovered a novel IL-8-dependent mechanism via which MCPIP-1 maintains epithelial homeostasis. This study reveals for the first time that MCPIP-1 plays a crucial anti-inflammatory role not only in myeloid cells but also in epithelial cells.
Key Words: MCPIP-1, Epithelium, Interleukin-8, Innate immunity, Infection
Introduction
A layer of epithelial cells is an essential component of the physiological barrier and the first line of defense against microbial pathogens. An important innate immune function of the epithelium is to recruit immune cells and maintain homeostasis. Moreover, epithelial cells constantly exposed to microbial ligands recognize pathogen-associated molecular patterns (PAMPs) and may initiate innate immune inflammatory responses needed for pathogen clearance during infection [1, 2, 3]. The crucial cytokine produced by epithelium in response to bacterial invasion is CXCL8/interleukin (IL)-8, which determines the recruitment of neutrophils to the site of infection [4, 5]. The uncontrolled secretion of this proinflammatory cytokine can lead to excessive activation and chemotaxis of neutrophils [6]. Therefore, inflammation in the host epithelium must be regulated to prevent tissue damage and maintain epithelial homeostasis after microbial infection. Thus, the crucial task of the epithelial tissue is to control responses to PAMPs, not only by limiting TLR receptor expression, but also by expressing cytoplasmic negative regulators of cell signaling. These regulators can be constitutively expressed or upregulated during inflammation, thereby preventing excessive destruction of inflamed tissue [7, 8, 9, 10].
Monocyte chemoattractant protein-1-induced protein-1 (MCPIP-1), also known as Regnase-1, was recently described as a multifunctional regulator of innate immunity [11]. Originally, MCPIP-1 was identified as a protein encoded by the ZC3H12A gene, a member of the CCCH-type zinc finger gene family [12]. Increased ZC3H12A expression has been observed in myeloid cells upon stimulation with various molecules, such as MCP-1, IL-1β, PMA, and TLRs agonists, and in response to bacterial infections [13, 14]. Moreover, recently published data indicate that MCPIP-1 plays an important role in the suppression of miRNA activity and biogenesis [15], angiogenesis [16], adipogenesis [17], and osteoclastogenesis [18]. Furthermore, the protein was initially characterized as a transcription factor for apoptotic gene families, since its structure possesses a putative nuclear localization signal sequence and two potential proline-rich activation domains [12]. Other studies have focused on the role of MCPIP-1 in inflammation exerted via its ubiquitin-associated domain, which provides the deubiquitinase activity that results in the negative regulation of the NF-κB signaling pathway [19, 20]. The PilT N-terminus domain of MCPIP-1 possesses RNase activity that targets the mRNA of proinflammatory cytokines such as IL-6, IL-1β, and IL-12p40 [13, 21, 22]. Moreover, in vivo studies have revealed that MCPIP-1-deficient mice develop hyperresponsiveness upon exposure to some PAMPs [20, 21]. However, despite the general consensus that MCPIP-1 is a potent negative regulator of inflammation, the mechanism via which it exerts this function remains poorly understood.
Although, MCPIP-1 has already been described as a potent inhibitor of proinflammatory cytokine secretion in myeloid cells [23, 24], its role in the pathophysiology and homeostasis of the epithelium remains unexplored. This study investigated the regulatory function of MCPIP-1 and the underlying mechanism by which it affects the IL-8 level in epithelial cells under physiological conditions as well as upon infection.
Materials and Methods
Reagents
Fetal bovine serum (FBS), high-glucose DMEM, RPMI-1640, Dulbecco's PBS without Ca2+/Mg2+, penicillin/streptomycin, and Opti-MEM medium were from Gibco. SYBR Green JumpStart Taq ReadyMix, PMSF (phenylmethanesulfonyl fluoride), and puromycin were obtained from Sigma.
Cell Cultures
HeLa human cervix epithelial carcinoma, Caco-2 human colon epithelial cancer, HEK293T human embryonic kidney, Hep-G2 human hepatocellular liver carcinoma, KATO III human stomach carcinoma, and KB human cervix epithelial carcinoma cell lines were obtained from the American Type Culture Collection (ATCC). Human skin keratinocytes, kindly provided by Dr. Krystyna Staliska (Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University) were maintained in high-glucose DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. THP-1 human monocytic cells and U937 human leukemic monocyte lymphoma cells, obtained from ATCC, were cultured in RPMI-1640 medium supplemented with 10% FBS. TIGKs (telomerase-immortalized gingival keratinocytes), kindly provided by Prof. Richard Lamond (University of Louisville School of Dentistry), were routinely cultured in KBM-Gold™ keratinocyte basal medium supplemented with Single Quotes™ (Lonza). For experiments, cells were cultured under standard conditions (37°C and 5% CO2) and seeded on 12-well plates (TPP). Human monocyte-derived macrophages (hMDMs) differentiated from peripheral blood mononuclear cells (PBMCs) as described previously were seeded on 24-well plates (Sarstedt) and cultured under standard conditions in RPMI-1640 medium supplemented with 10% FBS. The purity of monocyte-derived macrophages was verified by the analysis of macrophage surface markers by flow cytometry. Briefly, the hMDM phenotype was evaluated by immunofluorescence staining for CD14 (DakoCytomation), CD16 (DakoCytomation), CD11b (Becton Dickinson), and CD209 (Becton Dickinson). The routine procedure used in our laboratory yields at least 90% cells positive for the first three antigens, with fewer than 1% of cells staining with anti-CD209 antibodies [25]. Primary monocytes (Mo) were isolated from the PBMC fraction using the Human Monocyte Enrichment Set-DM (BD IMag). Further supplementation of Mo culture media (RPMI with 10% FBS and 1% penicillin/streptomycin) with IL-4 (500 U/ml) and GM-CSF (10 ng/ml) (R&D Systems) led to differentiation of the primary monocytes into monocyte-derived dendritic cells (moDCs). Neutrophils were isolated from granulocyte-enriched fractions by mixing with a 1% solution of polyvinyl alcohol in PBS. After 30 min at room temperature, neutrophils were separated from red blood cells by harvesting the upper phase; the remaining erythrocytes were subsequently removed by hypotonic lysis. Blood was obtained from the Red Cross, Krakow, Poland, which deidentifies blood materials as appropriate for human subject confidentiality assurances. Thus, this manuscript adheres to appropriate exclusions from human subject approval.
Bacteria Preparation, Cell Infection, and Cell Stimulation
Escherichia coli strain (ATCC 25922) was inoculated from preculture stocks into 10 ml of LB Broth (Bio Shop) media and grown to a stationary phase at 37°C overnight under constant rotation (180 rpm). Prior to infection, bacteria were collected by centrifugation (5,000 rpm for 5 min), washed with PBS, and resuspended in PBS at the desired OD600. Infection of HeLa and Caco-2 cells with E. coli was carried out at an MOI (multiplicity of infection) of 1:5, in DMEM without antibiotics at 37°C. Three hours postinfection, the cells were rinsed four times with ice-cold PBS. After the final wash, medium supplemented with antibiotics (1% penicillin/streptomycin) was added, and the cells were further cultured for the desired time.
For colony-forming unit (CFU) determination, after the washing step, cells were cultured in media containing antibiotics for an additional hour, followed by the plating of cell lysates onto LB agar for the counting of intracellular bacteria.
Calcium Mobilization
Neutrophils resuspended in HBSS (Hanks Balanced Salt Solution; Lonza) were loaded with calcium indicator Fura-2 AM (Molecular Probes) at 37°C for 30 min. Cells were washed twice with HBSS, and Ca2+ mobilization was measured in response to conditioned media (CM) from MCPIP-1-silenced cells infected or not infected with E. coli. After excitation at 340 and 380 nm, the fluorescence-related [Ca2+] changes were monitored at 505 nm, and the results were presented as the fluorescence ratio (340 nm/380 nm).
Cell Transduction
Silencing of the ZC3H12A gene in HeLa cells was achieved using a lentiviral system. Packaging of plasmids psPAX2 (Addgene plasmid cat. No. 12,260) and envelope plasmid pMD2.G (Addgene plasmid cat. No. 12,259; kindly provided by Didier Trono) into lentiviruses was carried out using a second-generation packaging system. For silencing, a plasmid expressing an shRNA sequence targeting the ZC3H12A gene (Sigma, cat. No. TRCN0000130710) or a nonspecific scrambled control sequence (Sigma, cat. No. SHC002) was used. Overexpression of ZC3H12A in HeLa cells was achieved using a pMX retrovirus system with expression plasmids encoding wild-type (WT) human MCPIP-1 (pMX-MCPIP-1) or empty pMX-puro. Transduction was carried out in suspension, in growth medium mixed 1:2 with infection medium and 6 µg/ml of Polybrene. After 24 h, medium was exchanged for normal growth medium; after 3 days puromycin (6 µg/ml) was added to the medium to select transduced cells. Overexpression was verified by Western blot analysis after 7 days.
Cell Transfection
Silencing of MCPIP-1 was achieved by transfection of HeLa and Caco-2 cells with the specific siRNAs listed in table 1 or STEALTH siRNA negative control (NC) duplexes (Invitrogen). Transfections were performed with cells at 40% confluence. Briefly, 50 pmol of siRNA was combined with 2 µl of Lipofectamine 2000 (Invitrogen) in Opti-MEM medium, and cells were transfected for 6 h, followed by 48 h of normal cell growth in DMEM medium alone prior to assays. The efficiency of silencing was verified by Western blot analysis.
Table 1.
List of silencing sequences used in the study
| siRNA | Strand | Sequence |
|---|---|---|
| l | Sense | 5′-UCCACCUCGAGAGUACUGGUCUGAA-3′ |
| Antisense | 5′-UUCAGACCAGUACUCUCGAGGUGGA-3′ | |
| 2 | Sense | 5′-AGCUGGGCUAUUCAUCCACGGAGAU-3′ |
| Antisense | 5′-AUCUCCGUGGAUGAAUAGCCCAGCU-3′ | |
| 3 | Sense | 5′-CCCUGUUGAUACACAUUGUtt-3′ |
| Antisense | 5′-ACAAUGUGUAUCAACAGGGtg-3′ | |
| 4 | Sense | 5′-GGAAACCCACAAACCAAAGtt-3′ |
| Antisense | 5′-CUUUGGUUUGUGGGUUUCCtt-3′ | |
| 5 | Sense | 5′-GAAGGAAACCCACAAACCAAAGAUA-3′ |
| Antisense | 5′-UAUCUUUGGUUUGUGGGUUUCCUUC-3′ | |
| 6 | Sense | 5′-GAUACAAUGUGUAUCAACAGGGUGA-3′ |
| Antisense | 5′-UCACCCUGUUGAUACACAUUGUAUC-3′ |
Cytokine Assays
The levels of IL-8 and IL-6 in HeLa and Caco-2 supernatants were determined using commercially available ELISA kits (BD Bioscience). Estimation of MCP-1 concentration was performed using the BD™ Cytometric Bead Array Human Chemokine Kit (BD Biosciences).
Quantitative Reverse-Transcription PCR
Total cellular RNA was extracted from HeLa cells using TRIzol (Ambion). Reverse transcription was performed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Five hundred nanograms of RNA from each sample were used for cDNA synthesis with random primers in a total volume of 20 µl. Quantitative PCR (qPCR) was performed by the SYBR Green method in a reaction volume of 15 µl containing 1 µl of cDNA sample, 10 µM of each primer, and 1× SYBR Green JumpStart Taq ReadyMix. qPCRs were performed using forward and reverse primers for MCPIP-1 (ZC3H12A), IL-8, IL-6, iNOS, and EF2 (housekeeping gene, used for normalization). After 5 min of initial denaturation at 95°C, reactions were carried out for 40 cycles, followed by a final elongation step at 72°C for 10 min. Primer sequences and conditions for denaturation, annealing, and extension for each pair of primers are provided in table 2. All reactions were performed in duplicate. Mean threshold cycle (Ct) values were calculated and analyzed using the ΔΔCt method [26]. To evaluate the quality of qPCRs, samples were routinely resolved on nondenaturing 1.5% agarose gels and visualized by staining with ethidium bromide.
Table 2.
Oligonucleotides used in the study
| Oligonucleotide | Sequence | Temperature program1 |
|---|---|---|
| EF2_R | 5′-TTCAGCACACTGGCATAGAGGC-3′ | 1. 95° C, 30 s |
| EF2_F | 5′-GACATCACCAAGGGTGTGCAG-3′ | 2. 62° C, 30 s |
| MCPIP-1_R | 5′-TCCAGGCTGCACTGCTCACTC-3′ | 3. 72° C, 30 s |
| MCPIP-1_F | 5′-GGAAGCAGCCGTGTCCCTATG-3′ | |
| IL-6_R | 5′-AGTGCCTCTTTGCTGCTTTCACAC-3′ | 1. 95° C, 30 s |
| IL-6_F | 5′-AAATTCGGTACATCCTCGACGGCA-3′ | 2. 58° C, 30 s |
| MCP-1_R | 5′-CCAGGTGGTCCATGGAATCCTG-3′ | 3. 72° C, 45 s |
| MCP-1_F | 5′-CTTCTGTGCCTGCTGCTCATAGC-3′ | |
| IL-8_R | 5′-TCTCAGCCCTCTTCAAAAACTTCT-3′ | 1. 95° C, 30 s |
| IL-8_F | 5′-ATGACTTCCAAGCTGGCCGTGGCT-3′ | 2. 60° C, 1 min |
| iNOS_R | 5′-GGTTGGGGGTGTGGTGATGT-3′ | 3. 72° C, 45 s |
| iNOS_F | 5′-CCGAGGCAAACAGCACATTC-3′ | |
Denaturation (1); annealing (2); extension (3).
Protein Isolation and Immunoblotting
Whole cellular extracts from control, modified, and stimulated cells were prepared using 100 µl of RIPA-lysis buffer (0.25% Na-deoxycholate, 0.5% Nonidet P-40, 0.05% SDS, protease inhibitor cocktail, 2.5 mM EDTA in PBS). The protein concentration was determined using the BCA protein assay (ThermoFisher Scientific). Equal amounts of protein (20 μg/well) were separated on 12% SDS-PAGE gels and electrotransferred onto PVDF membranes (Millipore) in buffer consisting of 25 mM Tris, 0.2 M glycine, and 20% methanol (100 V, 90 min). Nonspecific binding sites were blocked with 5% skimmed milk in TBST buffer (20 mM Tris, pH 7.5, 0.5 M NaCl, 0.05% Tween 20) for 2 h, followed by overnight incubation with the relevant primary antibody: rabbit anti-MCPIP-1 (1:2,000, GeneTex) or mouse anti-β-actin (1:10,000, BD Bioscience). Membranes were washed extensively in TBST buffer, and then incubated for 2 h in TBST buffer containing 5% skimmed milk and horseradish peroxidase-conjugated secondary antibodies: goat anti-rabbit IgG (1:3,000, Cell Signaling) and goat anti-mouse (1:10,000, BD Bioscience). All incubations were performed at 4°C. Membranes were washed (5 × 5 min) in TBST buffer, and blots were developed using Luminata Crescendo Substrate (Millipore). Membranes were exposed to Kodak medical X-ray film and developed.
Densitometric Analysis
Densitometric analyses of Western blots were performed using Kodak digital software. The results are presented as mean values in arbitrary densitometric units, corrected for background intensity, or as increases over the corresponding levels in nonstimulated cells.
Protein Expression and Purification
The WT human MCPIP-1 nucleotide sequence (NM_025079.2) was optimized for efficient expression in E. coli strains and synthetized by GeneScript (Piscataway, N.J., USA). A synthetic MCPIP-1 gene was cloned into vector pET-21a (Novagen). Mutant MCPIP-1 (D141N) was generated by site-directed mutagenesis. WT MCPIP-1 and MCPIP-1 (D141N) proteins were overexpressed in BL21 RIL E. coli strain (Novagen) in LB broth at 37°C, and protein overexpression was induced by the addition of 0.5 mM of IPTG when the culture reached an OD600 of 0.5. MCPIP-1 was purified from inclusion bodies by several washing steps combined with sonication. Final inclusion body solubilization was performed at room temperature in buffer containing 25 mM Tris, 7 M urea, 1 mM DTT, pH 9. To obtain a pure fraction of MCPIP-1 protein, TMAE resin (Merck) ion-exchange chromatography was performed under denaturing conditions. For elution, the buffer was enriched to 1 M NaCl. The first step of the refolding procedure was performed by dilution of MCPIP-1 to 1 mg/ml in buffer containing 25 mM Tris, 500 mM NaCl, 1.8 M urea, 5 mM DTT, and 1 mM EDTA, pH 8.3 at 4°C. The diluted sample was incubated at 4°C for 6 h. Subsequently, overnight dialysis at 4°C was performed against native buffer: 25 mM Tris, 300 mM NaCl, 1 mM DTT, 1 mM EDTA, pH 8.3. Refolded fractions were concentrated to 10 mg/ml by ultracentrifugation using Amicon Ultra 30K centrifugal filters (Merck), and then purified by size-exclusion chromatography using Superdex 200 prep grade HR 30/10 column (GE Healthcare). All recombinant MCPIP-1 activity assays were performed on protein that had never been frozen.
RNA Synthesis
The cDNA sequence of IL-8 (CDS + 3′UTR) [NM_000584.3 from 133 to 1705 nt.] was amplified from HeLa cDNA with primers 5′-ACCATCTCACTGTGTGTAAACA-3′ and 5′-TTACTTTGACAACAAATTATATTTTAAATGT-3′. The PCR product was phosphorylated and cloned into pcDNA3.0 vector (Invitrogen) at blunted HindIII and EcoRI sites. Prior to mRNA synthesis, the template was linearized at the EcoRV site. mRNA was synthesized in vitro using the TranscriptAid T7 High Yield Transcription Kit (ThermoFisher), followed by purification by phenol-chloroform extraction and ethanol precipitation [27].
In vitro RNA Cleavage Assay
Endonuclease activity tests of WT MCPIP-1 were performed on IL-8 transcript (CDS + 3′UTR). Samples were mixed at a molar ratio of 1:5 (1 pmol of RNA to 5 pmol of WT MCPIP-1) and incubated at 37°C in reaction buffer (50 mM Tris, 150 mM NaCl, 2.5 mM MgCl2, 2.5 mM DTT, 0.5 mM EDTA, 0.025 mM ZnCl2, pH 8.3). Final sample volumes were 20 µl. Samples in each time point were frozen to stop the reaction. After cleavage, samples were denatured for 15 min at 70°C and analyzed by 4% denaturing PAGE in TBE buffer. Gel was stained in 100 ml of TBE buffer with the addition of 10 µl of SimpleSafe™ dye (EuRX) and visualized using a ChemiDoc system (Bio-Rad).
EMSA Assay
MCPIP-1 (D141N) was incubated with synthesized IL-8 (CDS + 3′UTR) mRNA in reaction buffer containing 25 mM Tris, 150 mM NaCl, 2.5 mM MgCl2, 2.5 mM DTT, 0.5 mM EDTA, 0.025 mM ZnCl2, 5% w/v glycerol, pH 8.3. Samples were mixed with 2 pmol of RNA (RNA:protein molar ratio as 1:0, 1:25, or 1:50) and incubated for 20 min on ice. Subsequently, complexes were separated by electrophoresis through 0.8% agarose gel that was prepared using 1× TAE buffer and SimpleSafe™ dye.
Computational Analysis
Calculations of ΔG of RNA stem-loop structures were performed using mFOLD software [28].
RNA-Protein Immunoprecipitation
HeLa cells were infected with E. coli for 8 h. Subsequently, cells were crosslinked with 1% formaldehyde for 10 min, and then washed with PBS containing 1 mM PMSF and 125 mM glycine. Cells were scraped from the plate with a rubber policeman, centrifuged at 280 g for 8 min at 4°C, and then incubated on ice for 30 min in lysis buffer (100 mM KCl, 25 mM EDTA, 2 mM DTT, 10 mM HEPES, 0.5% NP-40, 1× complete protease inhibitor cocktail). Lysates were centrifuged at 14,000 g for 5 min at 4°C, and the resultant supernatants were diluted with IP buffer (50 mM NaCl, 1 mM EDTA, 2 mM DTT, 50 mM HEPES, pH 7.5, 10% glycerol, 0.5% Triton X-100, complete protease inhibitor cocktail). Protein A agarose (Roche) was prepared for precleaning of the lysate by washing with IP buffer and blocking with 1% BSA, followed by the addition of anti-MCPIP-1 antibodies. Beads were incubated overnight at 4°C with constant mixing, and lysates were added the next day. After incubation for an additional 10 h, the immunoprecipitates were centrifuged at 400 g for 1 min at room temperature and washed five times with IP buffer. RNA-protein complexes were eluted from beads with RIP buffer (100 mM NaCl, 5 mM EDTA, 10 mM DTT, 50 mM HEPES, pH 7.5, 10% glycerol, 0.5% Triton X-100, 1% SDS, 1× complete protease inhibitor cocktail) with RNase inhibitor (Invitrogen). After elution, crosslinks were reversed by incubation at 70°C for 1 h, and the samples were centrifuged at 400 g for 1 min at room temperature. From the resultant supernatant, RNA was isolated using the TRIzol reagent, reverse transcribed, and subjected to qPCR analysis.
Statistical Analysis
All experiments were performed at least in triplicate, and results were analyzed for statistical significance using parametric and nonparametric Student's t tests. All values are expressed as means ± SD, and differences were considered significant at p < 0.05.
Results
High Constitutive Expression of MCPIP-1 in Epithelial Cells
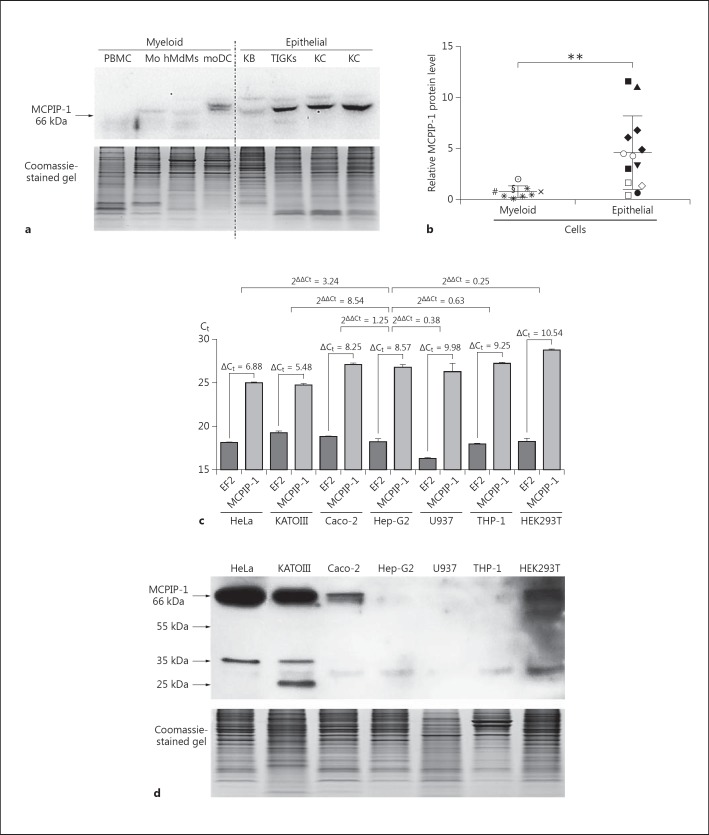
In contrast to the known function of MCPIP-1 in myeloid cells, its physiological role in the epithelium is poorly understood [14, 21, 29]. Moreover, unlike its transcript distribution, the levels of MCPIP-1 protein in different human tissues have not yet been assessed [13]. Thus, we evaluated the distribution of MCPIP-1 protein in several epithelial and myeloid cell lines as well as primary cell cultures. Surprisingly, this analysis revealed that the MCPIP-1 protein was constitutively produced in epithelial cells. Robust MCPIP-1 protein expression was higher in skin and gingival keratinocytes, hepatocytes, and cervix, kidney, stomach, and colon epithelium than in myeloid cells such as PBMCs, monocytes, macrophages, and moDCs (fig. 1a, b, d). This observation suggested that MCPIP-1 is functionally important in the epithelium. Moreover, we observed weak correlations between transcript and protein levels, suggesting that MCPIP-1 is regulated by diverse mechanisms in different cell types (fig. 1c, d). Because MCPIP-1 mRNA and protein were both abundantly expressed in HeLa cells, we chose this cell line as a model for studying the role of this regulator in the epithelium.
Fig. 1.
Comparison of mRNA and protein MCPIP-1 expression in various epithelial and myeloid cells. TotalRNA and protein was isolated from 1 million cells of each type. a Representative Western blots of protein lysates isolated from PBMCs, Mo, hMdMs, moDCs, KB, TIGKs, and primary skin keratinocytes (KC) (upper panel). Coomassie staining shows equal protein loading in each well (lower panel). b Comparison of MCPIP-1 protein expression between different myeloid [U937 (×), THP-1(#), hMDMs (*) obtained from 5 different donors, monocytes (§), MoDCs (☉)] and epithelial cells [primary skin keratinocytes obtained from 3 different donors (◆), TIGKs (⚪), HEK 293T (◇), HeLa (◼), KB (◻), Caco-2 (▼), KATO III (▲), Hep-G2 (⚫)]. The graph represents the MCPIP-1 protein level, estimated by densitometric analysis of Western blots. c MCPIP-1 mRNA and protein levels in seven different cell lines. Transcript levels were determined by qRT-PCR. The graph represents the cycle number at which the fluorescence signal crossed the threshold (Ct) for EF2 (reference gene) and MCPIP-1. For differences in cycle number between the gene of interest and the reference gene (ΔCt) and the changes in gene expression across samples (2ΔΔCt), representative results from three independent experiments are presented. d Representative results of Western blotting with specific anti-MCPIP-1 antibodies (upper panel). Coomassie staining confirms equal protein loading in each well (lower panel). Bars represent mean relative expression levels ± SD; * p < 0.05, indicating a significant difference; ** p < 0.01.
The Role of MCPIP-1 in the Regulation of IL-8 Expression in Epithelial Cells
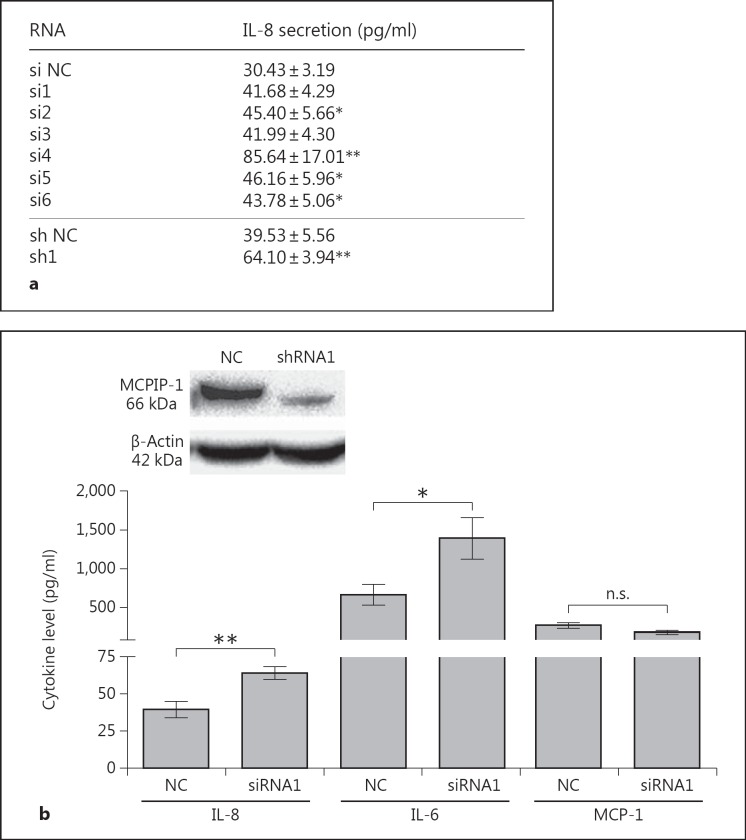
Epithelia represent dynamic barriers between the external environment and the organism's interior. Accordingly, their integrity is of central importance, and they play crucial roles in response to injury. Moreover, epithelia are a source of vitamin D and keratins, which are involved in intracellular signaling pathways with regulatory functions such as stress-response and wound healing [30, 31]. In innate immunity, one critical role of epithelia is the detection of pathogens and mobilization of neutrophil recruitment to the site of infection [5]. Therefore, we decided to investigate the potential role of MCPIP-1 in the regulation of IL-8 expression, the main chemokine involved in neutrophil attraction to an infection site [4]. Briefly, siRNA and shRNA were used to silence the MCPIP-1 gene. Treatment of cells with MCPIP-1-specific siRNA and shRNA, but not a nonspecific control siRNA/shRNA (NC), resulted in the upregulation of IL-8 secretion up to 2.4-fold (for the si4 sequence) relative to that in control cells (fig. 2a). In further experiments, we found that an shRNA1 that lowered the constitutive MCPIP-1 protein expression by 77% (4.35 ± 0.12-fold decrease; fig. 2b) resulted in higher IL-8 secretion than in cells treated with the control shRNA (fig. 2b). Simultaneously, we tested the effect of the same shRNA (shRNA1) on the expression levels of IL-6 [21], which is modulated by the RNase activity of MCPIP-1, and MCP-1 [32] (fig. 2b), which is not. The results revealed that a reduction in the intracellular MCPIP-1 level caused a significant increase in IL-8 and IL-6 expression, but not in MCP-1 expression (fig. 2a, b). The same correlation was observed in primary keratinocytes, in which downregulation of MCPIP-1 led to elevated secretion of IL-8 (data not shown). Thus, MCPIP-1 negatively regulates IL-8 expression in epithelial cells.
Fig. 2.
Influence of MCPIP-1 on the constitutive expression of IL-8 in HeLa cells. a MCPIP-1 was silenced in HeLa cells using specific siRNA or shRNA sequences; scrambled nonselective sequences were used as NCs. The level of secreted IL-8 was determined by ELISA. b Levels of the secreted cytokines IL-8 and IL-6 in shRNA1 MCPIP-1-silenced HeLa cells were estimated by ELISA, and the MCP-1 level was detected using a cytometric bead array. Data represent mean secreted cytokine concentrations from three independent experiments ± SD. * p < 0.05; ** p < 0.01. n.s. = Not significant. The efficiency of silencing was verified by Western blot (above).
The Role of MCPIP-1 in the Postinfectious Regulation of IL-8 in the Epithelium
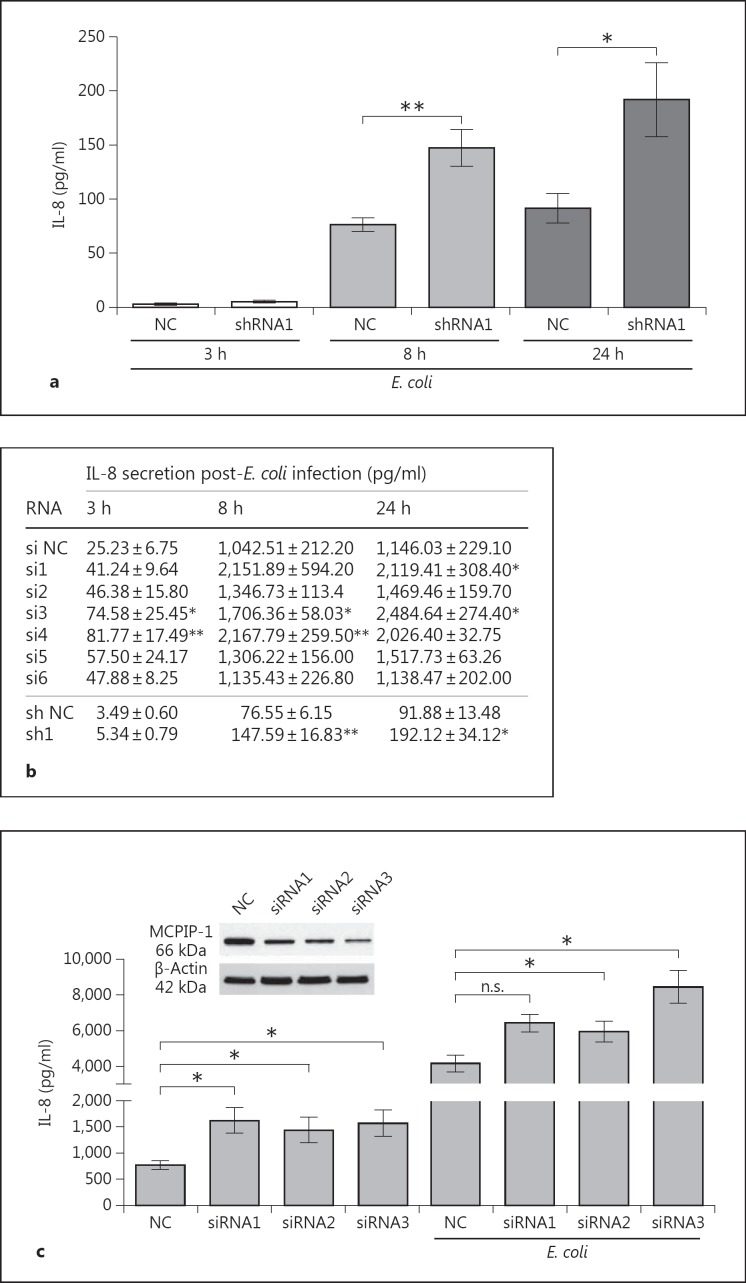
IL-8 expression is upregulated under multiple pathological conditions, including infections [33]. We confirmed increased IL-8 synthesis in epithelial cells upon infection with Gram-negative bacteria (fig. 3a) and examined the impact of MCPIP-1 on the expression of proinflammatory mediators during the time course of infection. Briefly, using the same silencing sequence (sh1), we evaluated MCPIP-1-dependent regulation of IL-8 secretion after E. coli infection. The results revealed that silencing of MCPIP-1 expression significantly increased the IL-8 concentration in CM as early as 8 h postinfection and that the effect lasted up to 24 h (fig. 3a). Using other silencing sequences, we not only confirmed the suppression of IL-8 expression by MCPIP-1, but also showed that this effect was rapid and started as early as 3 h postinfection (fig. 3b).
Fig. 3.
Influence of MCPIP-1 on E. coli-induced expression of IL-8 in HeLa and Caco-2 cell lines. a, b MCPIP-1 was silenced in HeLa cells using specific siRNA or shRNA sequences; scrambled nonselective sequences were used as NCs. The cells were subsequently infected with E. coli. After the indicated times, culture media were collected and the level of IL-8 was estimated by ELISA. c MCPIP-1 was silenced in Caco-2 cells using three specific siRNAs; scrambled nonselective sequences were used as NCs. The level of MCPIP-1 protein after transfection was determined by Western blotting (above). The IL-8 level was estimated in culture media of noninfected and E. coli-infected cells (24 h postinfection). Levels of secreted IL-8 from three independent experiments are presented as absolute values (pg/ml) ± SD. * p < 0.05; ** p < 0.01. n.s. = Not significant.
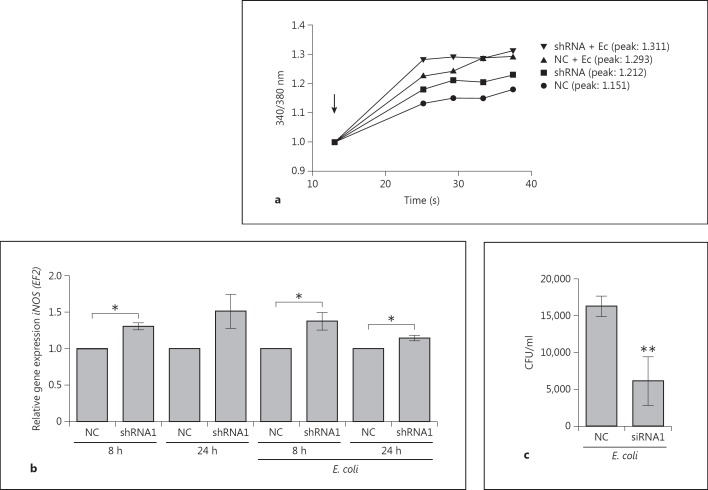
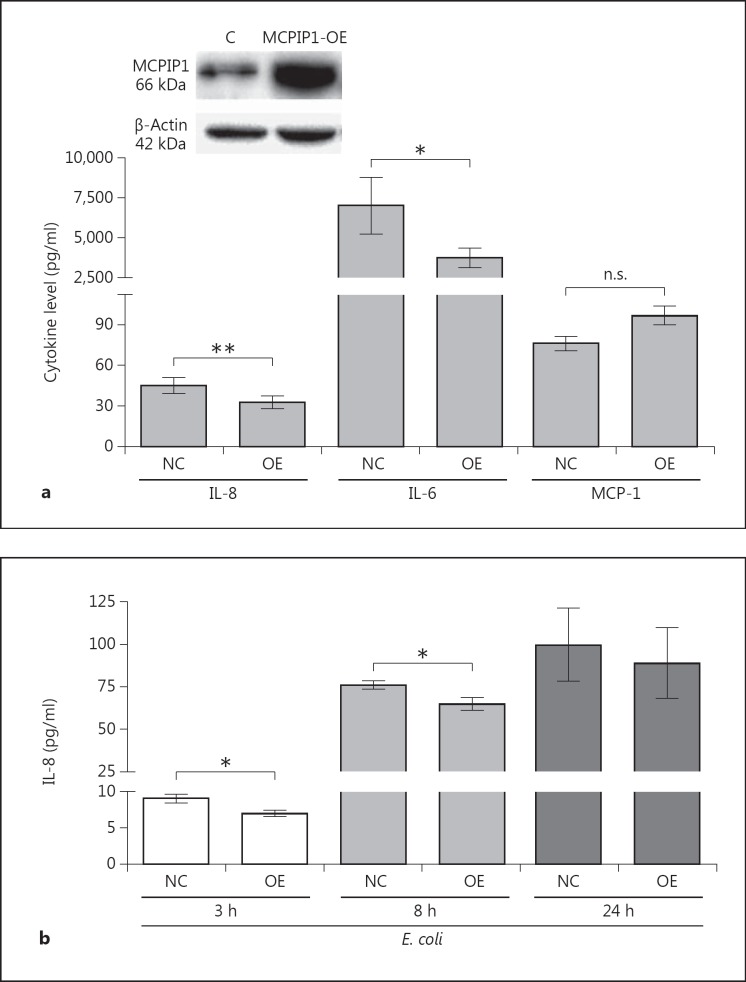
We investigated the biological effects of IL-8 regulation by MCPIP-1 in primary granulocytes (PMNs), which represent a population of cells that are highly responsive to this chemokine. In PMNs treated with CM from MCPIP-1-silenced HeLa cells, elevated production of IL-8 increased mobilization of calcium ions (fig. 4a). This effect was augmented when cells were exposed to CM collected from E. coli-infected cells (fig. 4a). Additionally, to confirm the paracrine influence of IL-8 secretion on transcriptional activation in HeLa cells, we measured expression of iNOS, which encodes a potent proinflammatory factor and is strongly upregulated by IL-8 [34]. The results revealed that iNOS expression was elevated in response to higher IL-8 secretion in MCPIP-1-silenced HeLa cells in comparison to NC cells, irrespective of whether they were infected with E. coli (fig. 4b). Finally, to investigate the role of MCPIP-1 in the progress of E. coli infection in epithelial cells, we monitored the rate of bacterial eradication. MCPIP-1-silenced cells cleared infection significantly more efficiently than control cells (fig. 4c). Taken together, these data indicate that MCPIP-1 plays an important role in the regulation of inflammation during bacterial infection of HeLa cells. To verify if the same mechanism operates in other epithelial cells, we used Caco-2 cells silenced with three of the tested siRNA sequences. Because silencing in Caco-2 cells was not as efficient as in HeLa cells (fig. 3c), the high endogenous expression of IL-8 flattened the final readout. Nonetheless, the results once again confirmed that both constitutive and infection-induced levels of IL-8 were higher in MCPIP-1-deficient cells (fig. 3c), indicating that the suppression of IL-8 expression by MCPIP-1 is a common feature of epithelial cells. Additionally, we investigated whether high constitutive MCPIP-1 expression in epithelial cells contributes to the preservation of immune homeostasis by assessing the effect of MCPIP-1 overexpression (from a transfected plasmid) in HeLa cells. Overexpression of MCPIP-1 led to a 5.6-fold increase in its protein level (fig. 5a), which correlated significantly with lower secretion of IL-8 by both noninfected and infected cells (fig. 5a, b). A similar effect was noted for the IL-6 transcript specifically targeted by MCPIP-1, whereas the level of the control transcript (MCP-1) was unaffected (fig. 5a). Taken together, the results show that MCPIP-1 regulates both the physiological and postinfectious expression of IL-8 in the epithelial cells.
Fig. 4.
Biological consequences of MCPIP-1 downregulation. a Mobilization of intracellular free calcium levels in Fura-2-loaded cells was monitored by spectrofluorometry after the treatment of neutrophils with culture media collected from noninfected and E. coli-infected HeLa cells (shNC - NC, shRNA MCPIP-1 silenced); the arrow indicates the time of media addition. Representative ratios of fluorescence intensities (340/380 nm) from three independent experiments are shown. b MCPIP-1 was silenced in HeLa cells using specific siRNA or shRNA sequences; scrambled nonselective sequences were used as NCs. Forty-eight hours later, cells were infected with E. coli for 8 and 24 h. The level of iNOS mRNA, determined by RT-PCR, was calculated across the samples; the value in the NC sample was defined as 1. c Four hours after E. coli infection of MCPIP-1-silenced HeLa cells (MOI = 5), cell lysates were collected and plated onto LB agar for the counting of intracellular bacteria (CFU). Data represent mean values from three independent experiments ± SD. * p < 0.05; ** p < 0.01.
Fig. 5.
Effect of MCPIP-1 overexpression in HeLa cells on constitutive and E. coli-induced IL-8 levels. a Levels of MCPIP-1 in HeLa cells after transduction with empty vector or expression plasmid-harboring WT MCPIP-1 was verified by Western blotting (above). Using conditioned media, IL-8 and IL-6 protein levels were determined by ELISA, and the MCP-1 level by cytometric bead array. Bars represent the mean concentrations of secreted cytokines ± SD. b Control and MCPIP-1-overexpressing cells were infected with E. coli. At the indicated times postinfection, media were harvested and levels of secreted IL-8 were estimated by ELISA. Data represent mean values from three or four independent experiments ± SD. * p < 0.05; ** p < 0.01. n.s. = Not significant.
The Mechanism of IL-8 Regulation by MCPIP-1
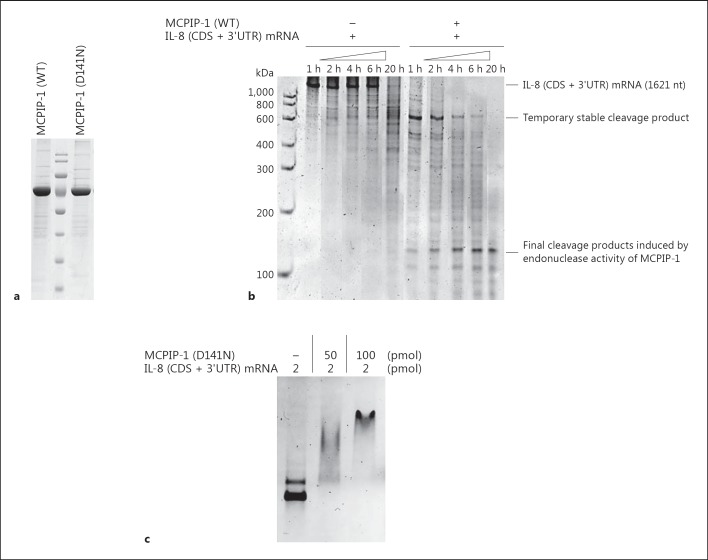
It was previously shown that the regulation of the expression of several proinflammatory cytokines by MCPIP-1 depends on its deubiquitinase or RNase activity [13, 22]. Therefore, to determine the mechanism of action and its impact on IL-8 transcript degradation, we expressed and purified recombinant MCPIP-1 (rMCPIP-1) with and without the D141N mutation, which inactivates its nuclease activity [13, 19, 21] (fig. 6a). First, we performed the RNA cleavage assay. Incubation of IL-8 (CDS + 3′UTR) mRNA with WT rMCPIP, but not with the D141N mutant, resulted in time-dependent degradation of IL-8 mRNA, confirming its endonuclease activity (fig. 6b). The full-length IL-8 transcript (1,621 nt) was degraded in 1 h to a ∼600-nt transiently stable product, which then underwent further gradual cleavage into several ∼100 nt products after 20 h incubation. This degradation pattern indicates that some elements in the IL-8 (CDS + 3′UTR) transcript were preferentially recognized and processed by MCPIP-1. Thereafter, we examined recognition of IL-8 mRNA by MCPIP-1 using the D141N mutated rMCPIP-1 protein, which is devoid of nuclease activity. The IL-8 transcript showed retarded mobility, indicating complex formation between MCPIP-1 and IL-8 (CDS + 3′UTR) mRNA (fig. 6c).
Fig. 6.
MCPIP-1 directly regulates the stability of IL-8 mRNA. a Purity of recombinant MCPIP-1 protein (WT and D141N) used in the experiments, documented by SDS PAGE. b Degradation of IL-8 transcript by recombinant MCPIP-1 protein (WT), estimated by in vitro RNA cleavage assay. c Interaction of recombinant MCPIP-1 lacking RNase activity (D141N) with the IL-8 transcript (CDS + 3′UTR), estimated by EMSA. Results are representative of three independent experiments.
The above findings motivated us to search for stem-loop elements that might be important for the regulation of IL-8 mRNA stability and its recognition and cleavage by MCPIP-1. Briefly, the alignment of stem-loop sequences of IL-8 mRNA in different species revealed a high structural similarity among transcripts (online suppl. fig. S1; for all online suppl. material, see www.karger.com/doi/10.1159/000448038). Additionally, preliminary analysis of the 3′UTR region of the IL-8 mRNA sequence revealed that is has the potential to form conserved stem-loop structures (online suppl. fig. S1). Moreover, the low ΔG values (ΔG1 in the range from −11.2 to −11.3) of these particular IL-8 structures indicate their stability and suggest that they are important for IL-8 recognition by MCPIP-1 RNase (online suppl. fig. S1). These results are consistent with the fact that other stem-loop motifs are recognized by MCPIP-1, such as the conserved loop-structures in the IL-6 transcript [22]. Nevertheless, we confirmed the importance of the stem-loop motifs by performing an immunoprecipitation assay.
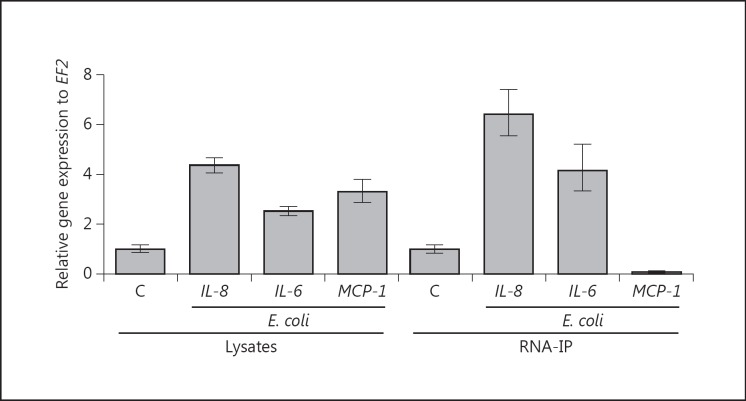
Lysates from noninfected HeLa cells and cells obtained 8 h after infection with E. coli were incubated with specific MCPIP-1 antibodies. The pull-down fraction was enriched in cytokine mRNAs that are substrates for MCPIP-1 RNase (fig. 7, right panel). The left panel shows increased expression of the indicated cytokines in unprocessed whole-cell lysates infected and noninfected with E. coli. The right panel shows the level of transcripts in the pull-down fraction obtained by immunoprecipitation with the MCPIP-1 antibody, in which the IL-6 and MCP-1 transcript served as positive and negative controls, respectively. These results showed that, despite the increase in the levels of different cytokine mRNAs in the lysates of infected cells, only IL-8 and IL-6 were detected in the immunoprecipitates, confirming that IL-8 mRNA was explicitly recognized by the MCPIP-1 protein. Collectively, the results indicate that MCPIP-1 regulates epithelial homeostasis in a specific manner via recognition and degradation of particular cytokine transcripts.
Fig. 7.
Specificity of transcript recognition by MCPIP-1. Expression levels of proinflammatory cytokines (IL-6, IL-8, and MCP-1) in E. coli-infected HeLa cells and mock-infected cells (C) were compared by qPCR. The left panel (lysates) shows transcript levels in unprocessed whole-cell lysates. The right panel (RNA-IP) shows transcript levels in the pull-down fraction obtained by immunoprecipitation of cell lysates with anti-MCPIP-1 antibodies. For each cytokine, the transcript level in mock-infected cells was defined as 1. Therefore, in both panels, the C bar represents the transcript level for each cytokine in mock-infected cells. Results are representative of three independent experiments.
Discussion
Pattern recognition receptors are critical players in innate and adaptive immune systems for sensing invading microorganisms and activating multiple pathways that lead to the induction of proinflammatory responses essential for pathogen clearance. The intensity and duration of defense against invading pathogens must be tightly controlled in every tissue at various time points. These features are maintained by an array of positive and negative regulators. We hypothesized that MCPIP-1, an important negative regulator of proinflammatory responses in myeloid cells, might also play an important role in maintaining homeostasis in the epithelium, both under physiological conditions as well as upon bacterial infection.
Previous reports on the tissue distribution of MCPIP-1 focused mainly on its transcript level [13]. However, the transcript level of a gene does not always provide information about the true level of protein due to posttranscriptional RNA processing. Therefore, we determined the protein level of MCPIP-1 in different cell types. The results showed ubiquitous expression of MCPIP-1 in various epithelial cells and surprisingly higher levels MCPIP-1 protein in epithelial cells than in myeloid cells, which contradicts the results of previous analyses based on transcriptome analysis. The divergent expression levels could be related to differences in the proliferation rate or cell culture conditions; nonetheless, significant correlations were observed (online suppl. table S1). It is also reasonable to postulate the presence of distinct, cell-specific regulatory mechanisms that affect MCPIP-1 expression and transcript processing. The elevated level of MCPIP-1 protein in epithelial cells may indicate that the MCPIP-1 transcript is more stable in epithelial cells. Alternatively, reduced ubiquitin modification or proteolytic truncation exerted by MALT-1 may be responsible for the higher MCPIP-1 protein level. Indeed, a number of studies provide evidence that MALT-1 is expressed and functions in NF-κB activation in nonimmune cells, including epithelial cells [35]. However, the extent to which MALT-1 affects MCPIP-1 function or helps determine its mechanism of action in immune and nonimmune cells remains unclear and represents a challenge for future investigations [36]. In summary, our observations suggest that MCPIP-1 has an important role not only in the myeloid lineage, but also in epithelial cells. In tissues that are continually exposed to microorganisms, such as skin, oral cavity, or colon, increased expression of negative regulators of the innate immune responses, such as MCPIP-1, may constitute an important mechanism for dampening immune responses. This could open up new opportunities for the development of novel treatments for inflammatory diseases involving mucosal as well as epidermal linings.
One of the major inflammatory mediators secreted by epithelial cells in response to invaders is chemokine IL-8. Abnormalities in epithelial IL-8 secretion are associated with various inflammatory diseases, such as inflammatory bowel disease, asthma, and psoriasis, indicating that its strict control is pivotal for the maintenance of epithelial hyporesponsiveness [37, 38, 39]. Our results clearly showed that MCPIP-1 plays an important role in this process by exerting RNase activity towards the IL-8 transcript. We found that the lifespan of IL-8 is determined by the presence of stem-loops/hairpin structures in the 3′UTR region of IL-8 mRNA, which are conserved between species as shown by the results of computational analysis. These conserved structures are targeted by the MCPIP-1 PIN domain and share many similarities with those that regulate the stability of the IL-6 transcript [22]. Our results showed that the recombinant MCPIP-1 mutant protein lacking RNase activity recognizes IL-8 mRNA but does not affect its integrity. However, using fully active, purified rMCPIP-1, we demonstrated rapid degradation of the IL-8 transcript only 1 h after the coincubation of the IL-8 transcript with rMCPIP-1 RNase. Our results complement and expand the results published by Mino et al. [40], who showed that IL-8 mRNA associated with MCPIP-1 exclusively in noninfected HeLa cells. In line with these findings, we observed higher levels of IL-8, both at the mRNA level and protein level, in MCPIP-1 knockdown cells than in native cells. Although this effect was specific for IL-8, and the expression of other chemokines, such as CXCL-10 and CCL-5, was not significantly affected by MCPIP-1 (data not shown), it would be useful to investigate the MCPIP-1-dependent posttranscriptional regulation of other relevant chemokines. Taken together, these results suggest that a high level of MCPIP-1 in the epithelium can downregulate the constitutive expression of IL-8. The loss of MCPIP-1-mediated control following depletion leads to a wide range of physiological effects, including granulocyte activation (calcium mobilization) and upregulation of inflammatory mediators such as iNOS, which is induced in an autocrine fashion by IL-8 in the epithelium. Finally, using the E. coli infection model in HeLa cells, we investigated the role of MCPIP-1 in epithelial cells upon exposure to pathogens. The results suggest that MCPIP-1 expression in epithelial tissue decreases the level of IL-8 transcript, as well as translated IL-8, in the early stage of infection, as in noninfected cells. At this stage of infection, the level of IL-8 expression is regulated not only by de novo synthesis mediated by NF-κB activation [33], but also posttranscriptionally by MCPIP-1. However, because the modulation of MCPIP-1 by pathogens cannot be excluded, our proposed mechanism for IL-8 regulation may be oversimplified. Recent work showed that the MCPIP-1 mRNA is controlled by the activation of TLRs; however, the posttranslational modification of this protein in response to infection has not been analyzed. The importance of high MCPIP-1 expression in the epithelium was confirmed by overexpression of MCPIP-1, which resulted in significant but nonetheless minor downregulation of IL-8 secretion. The role of highly abundant MCPIP-1 was also highlighted by the more efficient eradication of E. coli, concomitant with elevated expression of iNOS, in MCPIP-1-silenced cells. Although the involvement of ROS/RNS in the response to infection in epithelial cells remains poorly documented, some studies suggest that this pathway plays an important role in eliminating bacterial infection [41]. Accordingly, it is reasonable to speculate that bacterial invasion initiates the process of MCPIP-1 depletion, leading to increased susceptibility to pathogens. Therefore, it would be worthwhile to investigate the regulation of MCPIP-1 expression and its posttranslational modification during infection.
Pattern recognition receptor pathway activation in the epithelium is controlled by low expression of PAMP receptors but also by increases in the levels of its negative regulators [8, 10, 42]. Some of these regulators are expressed constitutively, whereas others are upregulated during inflammation. The latter category is represented by A20, the concentration of which increases rapidly in epithelial cells following stimulation with TNF-α or TLR ligands [7]. In this respect, MCPIP-1 cannot be classified as an ‘acute-phase’ negative regulator in epithelial cells, as we did not observe its upregulation during infection (data not shown). Interestingly, SIGIRR, another negative IL-1R/TLR signaling regulator, shows a similar expression pattern to that of MCPIP-1 and is highly expressed in many epithelial cell lines but not in primary macrophages, fibroblasts, or endothelial cells [8].
In conclusion, we uncovered a novel IL-8-dependent mechanism by which MCPIP-1 maintains immune homeostasis, which is of significance for epithelial linings. This study reveals for the first time that MCPIP-1 plays an anti-inflammatory role not only in myeloid cells, but also in epithelial cells. On the one hand, inhibition of the host's immune response and restriction of IL-8-dependent neutrophil infiltration may help to quickly restore the immunological balance but, on the other hand, may contribute to the onset of infection. Additional investigations are necessary to elucidate its precise role in orchestrating the host's defense mechanisms in diverse epithelial tissues. Elucidation of these mechanisms in detail might contribute to the development of immunomodulatory therapeutic agents as well as help to identify individuals predisposed to prolonged activation of inflammatory responses. Finally, we showed that MCPIP-1 maintains cytokine balance in normal as well as bacteria-stimulated cells, indicating that it contributes to homeostasis in bacteria-exposed epithelial surfaces.
Supplementary Material
Supplementary data
Supplementary data
Acknowledgements
This work was supported by National Science Center (Poland) grants 2011/03/B/NZ6/00053 (to J.K.), 2013/11/B/NZ2/00125 (to J.J.), and 2013/08/W/NZ1/00696 to J.P., who is also partially supported by the National Institute of Health (NIDCR/R21DE023207). The Faculty of Biochemistry, Biophysics and Biotechnology of the Jagiellonian University is a part of the Leading National Research Center (KNOW) program supported by the Ministry of Science and Higher Education in Poland.
References
- 1.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaykhiev R, Bals R. Interactions between epithelial cells and leukocytes in immunity and tissue homeostasis. J Leukoc Biol. 2007;82:1–15. doi: 10.1189/jlb.0207096. [DOI] [PubMed] [Google Scholar]
- 4.Hammond ME, Lapointe GR, Feucht PH, Hilt S, Gallegos CA, Gordon CA, Giedlin MA, Mullenbach G, Tekamp-Olson P. IL-8 induces neutrophil chemotaxis predominantly via type I IL-8 receptors. J Immunol. 1995;155:1428–1433. [PubMed] [Google Scholar]
- 5.Kucharzik T, Hudson JT, 3rd, Lugering A, Abbas JA, Bettini M, Lake JG, Evans ME, Ziegler TR, Merlin D, Madara JL, Williams IR. Acute induction of human IL-8 production by intestinal epithelium triggers neutrophil infiltration without mucosal injury. Gut. 2005;54:1565–1572. doi: 10.1136/gut.2004.061168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kruger P, Saffarzadeh M, Weber AN, Rieber N, Radsak M, von Bernuth H, Benarafa C, Roos D, Skokowa J, Hartl D. Neutrophils: between host defence, immune modulation, and tissue injury. PLoS Pathog. 2015;11:e1004651. doi: 10.1371/journal.ppat.1004651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oshima N, Ishihara S, Rumi MA, Aziz MM, Mishima Y, Kadota C, Moriyama I, Ishimura N, Amano Y, Kinoshita Y. A20 is an early responding negative regulator of Toll-like receptor 5 signalling in intestinal epithelial cells during inflammation. Clin Exp Immunol. 2010;159:185–198. doi: 10.1111/j.1365-2249.2009.04048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wald D, Qin J, Zhao Z, Qian Y, Naramura M, Tian L, Towne J, Sims JE, Stark GR, Li X. SIGIRR, a negative regulator of Toll-like receptor-interleukin 1 receptor signaling. Nat Immunol. 2003;4:920–927. doi: 10.1038/ni968. [DOI] [PubMed] [Google Scholar]
- 9.Kinjyo I, Hanada T, Inagaki-Ohara K, Mori H, Aki D, Ohishi M, Yoshida H, Kubo M, Yoshimura A. SOCS1/JAB is a negative regulator of LPS-induced macrophage activation. Immunity. 2002;17:583–591. doi: 10.1016/s1074-7613(02)00446-6. [DOI] [PubMed] [Google Scholar]
- 10.Wang Z, Fayngerts S, Wang P, Sun H, Johnson DS, Ruan Q, Guo W, Chen YH. TIPE2 protein serves as a negative regulator of phagocytosis and oxidative burst during infection. Proc Natl Acad Sci USA. 2012;109:15413–15418. doi: 10.1073/pnas.1204525109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jura J, Skalniak L, Koj A. Monocyte chemotactic protein-1-induced protein-1 (MCPIP1) is a novel multifunctional modulator of inflammatory reactions. Biochim Biophys Acta. 2012;1823:1905–1913. doi: 10.1016/j.bbamcr.2012.06.029. [DOI] [PubMed] [Google Scholar]
- 12.Zhou L, Azfer A, Niu J, Graham S, Choudhury M, Adamski FM, Younce C, Binkley PF, Kolattukudy PE. Monocyte chemoattractant protein-1 induces a novel transcription factor that causes cardiac myocyte apoptosis and ventricular dysfunction. Circ Res. 2006;98:1177–1185. doi: 10.1161/01.RES.0000220106.64661.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizgalska D, Wegrzyn P, Murzyn K, Kasza A, Koj A, Jura J, Jarzab B, Jura J. Interleukin-1-inducible MCPIP protein has structural and functional properties of RNase and participates in degradation of IL-1β mRNA. FEBS J. 2009;276:7386–7399. doi: 10.1111/j.1742-4658.2009.07452.x. [DOI] [PubMed] [Google Scholar]
- 14.Blazusiak E, Florczyk D, Jura J, Potempa J, Koziel J. Differential regulation by Toll-like receptor agonists reveals that MCPIP1 is the potent regulator of innate immunity in bacterial and viral infections. J Innate Immun. 2013;5:15–23. doi: 10.1159/000339826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki HI, Arase M, Matsuyama H, Choi YL, Ueno T, Mano H, Sugimoto K, Miyazono K. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol Cell. 2011;44:424–436. doi: 10.1016/j.molcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 16.Roy A, Kolattukudy PE. Monocyte chemotactic protein-induced protein (MCPIP) promotes inflammatory angiogenesis via sequential induction of oxidative stress, endoplasmic reticulum stress and autophagy. Cell Signal. 2012;24:2123–2131. doi: 10.1016/j.cellsig.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Lipert B, Wegrzyn P, Sell H, Eckel J, Winiarski M, Budzynski A, Matlok M, Kotlinowski J, Ramage L, Malecki M, Wilk W, Mitus J, Jura J. Monocyte chemoattractant protein-induced protein 1 impairs adipogenesis in 3T3-L1 cells. Biochim Biophys Acta. 2014;1843:780–788. doi: 10.1016/j.bbamcr.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 18.Wang K, Niu J, Kim H, Kolattukudy PE. Osteoclast precursor differentiation by MCPIP via oxidative stress, endoplasmic reticulum stress, and autophagy. J Mol Cell Biol. 2011;3:360–368. doi: 10.1093/jmcb/mjr021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skalniak L, Mizgalska D, Zarebski A, Wyrzykowska P, Koj A, Jura J. Regulatory feedback loop between NF-κB and MCP-1-induced protein 1 RNase. FEBS J. 2009;276:5892–5905. doi: 10.1111/j.1742-4658.2009.07273.x. [DOI] [PubMed] [Google Scholar]
- 20.Liang J, Saad Y, Lei T, Wang J, Qi D, Yang Q, Kolattukudy PE, Fu M. MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-κB signaling. J Exp Med. 2010;207:2959–2973. doi: 10.1084/jem.20092641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T, Satoh T, Kato H, Tsujimura T, Nakamura H, Akira S. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458:1185–1190. doi: 10.1038/nature07924. [DOI] [PubMed] [Google Scholar]
- 22.Iwasaki H, Takeuchi O, Teraguchi S, Matsushita K, Uehata T, Kuniyoshi K, Satoh T, Saitoh T, Matsushita M, Standley DM, Akira S. The IκB kinase complex regulates the stability of cytokine-encoding mRNA induced by TLR-IL-1R by controlling degradation of regnase-1. Nat Immunol. 2011;12:1167–1175. doi: 10.1038/ni.2137. [DOI] [PubMed] [Google Scholar]
- 23.Yu F, Du F, Wang Y, Huang S, Miao R, Major AS, Murphy EA, Fu M, Fan D. Bone marrow deficiency of MCPIP1 results in severe multi-organ inflammation but diminishes atherogenesis in hyperlipidemic mice. PLoS One. 2013;8:e80089. doi: 10.1371/journal.pone.0080089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miao R, Huang S, Zhou Z, Quinn T, Van Treeck B, Nayyar T, Dim D, Jiang Z, Papasian CJ, Eugene Chen Y, Liu G, Fu M. Targeted disruption of MCPIP1/Zc3h12a results in fatal inflammatory disease. Immunol Cell Biol. 2013;91:368–376. doi: 10.1038/icb.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koziel J, Maciag-Gudowska A, Mikolajczyk T, Bzowska M, Sturdevant DE, Whitney AR, Shaw LN, DeLeo FR, Potempa J. Phagocytosis of staphylococcus aureus by macrophages exerts cytoprotective effects manifested by the upregulation of antiapoptotic factors. PLoS One. 2009;4:e5210. doi: 10.1371/journal.pone.0005210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 28.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang J, Wang J, Azfer A, Song W, Tromp G, Kolattukudy PE, Fu M. A novel CCCH-zinc finger protein family regulates proinflammatory activation of macrophages. J Biol Chem. 2008;283:6337–6346. doi: 10.1074/jbc.M707861200. [DOI] [PubMed] [Google Scholar]
- 30.Sun J. Vitamin D and mucosal immune function. Curr Opin Gastroenterol. 2010;26:591–595. doi: 10.1097/MOG.0b013e32833d4b9f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bragulla HH, Homberger DG. Structure and functions of keratin proteins in simple, stratified, keratinized and cornified epithelia. J Anat. 2009;214:516–559. doi: 10.1111/j.1469-7580.2009.01066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koziel J, Bryzek D, Sroka A, Maresz K, Glowczyk I, Bielecka E, Kantyka T, Pyrc K, Svoboda P, Pohl J, Potempa J. Citrullination alters immunomodulatory function of LL-37 essential for prevention of endotoxin-induced sepsis. J Immunol. 2014;192:5363–5372. doi: 10.4049/jimmunol.1303062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmann E, Dittrich-Breiholz O, Holtmann H, Kracht M. Multiple control of interleukin-8 gene expression. J Leukoc Biol. 2002;72:847–855. [PubMed] [Google Scholar]
- 34.Bruch-Gerharz D, Fehsel K, Suschek C, Michel G, Ruzicka T, Kolb-Bachofen V. A proinflammatory activity of interleukin 8 in human skin: expression of the inducible nitric oxide synthase in psoriatic lesions and cultured keratinocytes. J Exp Med. 1996;184:2007–2012. doi: 10.1084/jem.184.5.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McAllister-Lucas LM, Ruland J, Siu K, Jin X, Gu S, Kim DS, Kuffa P, Kohrt D, Mak TW, Nunez G, Lucas PC. CARMA3/Bcl10/MALT1-dependent NF-κB activation mediates angiotensin II-responsive inflammatory signaling in nonimmune cells. Proc Natl Acad Sci USA. 2007;104:139–144. doi: 10.1073/pnas.0601947103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uehata T, Iwasaki H, Vandenbon A, Matsushita K, Hernandez-Cuellar E, Kuniyoshi K, Satoh T, Mino T, Suzuki Y, Standley DM, Tsujimura T, Rakugi H, Isaka Y, Takeuchi O, Akira S. Malt1-induced cleavage of regnase-1 in CD4+ helper T cells regulates immune activation. Cell. 2013;153:1036–1049. doi: 10.1016/j.cell.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 37.Shute JK, Vrugt B, Lindley IJ, Holgate ST, Bron A, Aalbers R, Djukanovic R. Free and complexed interleukin-8 in blood and bronchial mucosa in asthma. Am J Respir Crit Care Med. 1997;155:1877–1883. doi: 10.1164/ajrccm.155.6.9196089. [DOI] [PubMed] [Google Scholar]
- 38.Lemster BH, Carroll PB, Rilo HR, Johnson N, Nikaein A, Thomson AW. IL-8/IL-8 receptor expression in psoriasis and the response to systemic tacrolimus (FK506) therapy. Clin Exp Immunol. 1995;99:148–154. doi: 10.1111/j.1365-2249.1995.tb05525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanian S, Rhodes JM, Hart CA, Tam B, Roberts CL, Smith SL, Corkill JE, Winstanley C, Virji M, Campbell BJ. Characterization of epithelial IL-8 response to inflammatory bowel disease mucosal E. coli and its inhibition by mesalamine. Inflamm Bowel Dis. 2008;14:162–175. doi: 10.1002/ibd.20296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mino T, Murakawa Y, Fukao A, Vandenbon A, Wessels HH, Ori D, Uehata T, Tartey S, Akira S, Suzuki Y, Vinuesa CG, Ohler U, Standley DM, Landthaler M, Fujiwara T, Takeuchi O. Regnase-1 and Roquin regulate a common element in inflammatory mRNAs by spatiotemporally distinct mechanisms. Cell. 2015;161:1058–1073. doi: 10.1016/j.cell.2015.04.029. [DOI] [PubMed] [Google Scholar]
- 41.Hedl M, Abraham C. NLRP1 and NLRP3 inflammasomes are essential for distinct outcomes of decreased cytokines but enhanced bacterial killing upon chronic Nod2 stimulation. Am J Physiol Gastrointest Liver Physiol. 2013;304:G583–G596. doi: 10.1152/ajpgi.00297.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gourbeyre P, Berri M, Lippi Y, Meurens F, Vincent-Naulleau S, Laffitte J, Rogel-Gaillard C, Pinton P, Oswald IP. Pattern recognition receptors in the gut: analysis of their expression along the intestinal tract and the crypt/villus axis. Physiol Rep. 2015;3:e12225. doi: 10.14814/phy2.12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data