Abstract
Seasonal environmental heterogeneity is cyclic, persistent and geographically widespread. In species that reproduce multiple times annually, environmental changes across seasonal time may create different selection regimes that may shape the population ecology and life history adaptation in these species. Here, we investigate how two closely related species of Drosophila in a temperate orchard respond to environmental changes across seasonal time. Natural populations of Drosophila melanogaster and Drosophila simulans were sampled at four timepoints from June through November to assess seasonal change in fundamental aspects of population dynamics as well as life history traits. D. melanogaster exhibit pronounced change across seasonal time: early in the season, the population is inferred to be uniformly young and potentially represents the early generation following overwintering survivorship. D. melanogaster isofemale lines derived from the early population and reared in a common garden are characterized by high tolerance to a variety of stressors as well as a fast rate of development in the laboratory environment that declines across seasonal time. In contrast, wild D. simulans populations were inferred to be consistently heterogeneous in age distribution across seasonal collections; only starvation tolerance changed predictably over seasonal time in a parallel manner as in D. melanogaster. These results suggest fundamental differences in population and evolutionary dynamics between these two taxa associated with seasonal heterogeneity in environmental parameters and associated selection pressures.
Keywords: demography, Drosophila, life history, seasonal change
Introduction
Understanding how populations adapt to environmental variability is a fundamental interest in evolutionary biology. Environmental heterogeneity is commonly partitioned into two basic axes: variation in space and in time. Although parallels between spatial and temporal environmental parameters may exist, greater emphasis has been placed on evaluating spatial variation in evolutionary dynamics of natural populations (Endler, 1977, 1986; Slatkin, 1987; Kingsolver et al., 2001). Inferences regarding spatial variation in selection pressures can be evaluated using population samples collected at a single point in time, whereas determining the significance of temporal variation often requires longitudinal studies over various timescales (Hendry & Kinnison, 1999; Grant & Grant, 2002; Carroll et al., 2007; Siepielski et al., 2009). Temporal variation can be evaluated over short timescales using environmental parameters that change predictably as a function of time across seasons within a year. This includes change in abiotic factors, such as temperature and photoperiod, as well as biotic variation such as ecological interactions within and among taxa. The traits and selection pressures associated with high fitness may be quite distinct between seasons that are favourable for reproduction and population expansion (e.g. summer) and those that are not and must be endured (e.g. winter). Such alternating selection pressures across seasons may be integral in the maintenance of genetic variation in natural populations (Levene, 1953; Dempster, 1955; Haldane & Jayakar, 1963; Gillespie, 1973; Ewing, 1979; MacKay, 1980; Turelli, 1981; Ellner & Hairston, 1994; Hedrick, 1995, 2002). However, there is limited empirical work on how populations respond ecologically and evolutionary to seasonal changes in environmental parameters, and therefore, there is a need for longitudinal studies in natural populations across seasonal time.
For organisms that have multiple generations each year (multivoltine), there are several predicted outcomes in response to the seasonal environmental differences experienced by subsequent generations; the null hypothesis is that populations either do not respond or exhibit only stochastic differences across seasonal time. Alternatively, changes in traits over generational time may occur at an individual level as direct result of the different environments experienced (e.g. phenotypic plasticity) or they may reflect differences in the genetic composition of the population due to differential fitness over generational time. Phenotypic plasticity is a commonly predicted response to short-term environmental changes over seasonal timescales (Brakefield & Reitsma, 1991; Bradford & Roff, 1993) as seen in the change in body size throughout the summer in the dung fly Sepsis cynipsea (Blanckenhorn et al., 1999) and seasonal shifts in frequency of colour morphs of the ladybird beetle Adalia bipunctata (Brakefield, 1985). By comparison, seasonal change in the genetic composition of the population due to differential fitness across the changing environments has been less well studied. Although seasonal changes have been documented at the genetic level in annual cycling of Drosophila pseudoobscura chromosomal arrangements (Dobzhansky, 1943, 1948) and Drosophila melanogaster allele frequencies (Cogni et al., 2013, 2014; Bergland et al., 2014; Paaby et al., 2014), the phenotypic basis for seasonal cycling in allele frequencies remains largely unknown. Reproductive diapause incidence in D. melanogaster is one example of change over seasonal time for both phenotype (Schmidt & Conde, 2006) and underlying allele frequencies (Cogni et al., 2013). Here, we use the numerous seasonal changes in allele frequencies in D. melanogaster as a point of departure to investigate phenotypic change over seasonal time for a subset of traits that have been implicated in the adaptive response of D. melanogaster to spatially variable selection.
Drosophila melanogaster has long been used as a model system to evaluate the role of environmental heterogeneity and spatially variable selection on evolutionary pattern and process. The species is native to sub-Saharan Africa and, as a human commensal, has colonized temperate habitats on multiple continents (David & Capy, 1988; Andolfatto, 2005) on which they exhibit latitudinal clines for a variety of traits (Capy et al., 1993; James & Partridge, 1995; James et al., 1997; Azevedo et al., 1998; Karan et al., 1998; Robinson et al., 2000; Mitrovski & Hoffmann, 2001; Hoffmann et al., 2002; De Jong & Bochdanovits, 2003; Schmidt et al., 2005; Trotta et al., 2006; Schmidt & Paaby, 2008) as well as allele frequencies at specific loci (Berry & Kreitman, 1993; Verrelli & Eanes, 2000; Bettencourt et al., 2002; Frydenberg et al., 2003; Sezgin, 2004; Tauber et al., 2007; McKechnie et al., 2010; Paaby et al. 2010; Cogni et al., 2013; Paaby et al., 2014). Although such patterns of spatial variation may reflect aspects of demography and colonization history (Roesti et al., 2014), the latitudinal clines in D. melanogaster are commonly interpreted as an adaptive result of spatially variable selection (Verrelli & Eanes, 2000; Mitrovski & Hoffmann, 2001; Bettencourt et al., 2002; Sezgin, 2004; Schmidt & Paaby, 2008). As many of the climatic factors that change over latitudinal gradients also vary seasonally in temperate environments (e.g. temperature, photoperiod, humidity), D. melanogaster populations may also respond adaptively to environmental heterogeneity over short seasonal timescales. Thus, we can use these parallels with latitudinal clines to make concrete predictions about how traits will change over seasonal time: as temperature and associated parameters increase from spring to summer, we predict that traits will change in the same pattern as from high to low latitudes.
We compare two closely related species to identify generality in seasonal response and to dissect particular aspects unique to each species. In temperate North American orchards, the closely related species D. melanogaster and Drosophila simulans co-occur both over seasonal time and with geography. The species share common ecologies and exhibit at least some degree of parallel response with respect to phenotypic (McKenzie & Parsons, 1974; Watada et al., 1986; Gibert et al., 2004; Arthur et al., 2008; Van Heerwaarden et al., 2012), allozyme (Anderson & Oakeshott, 1984) and transcriptional clines (Zhao et al., 2015), although there is at least one instance of an opposing phenotypic cline between these species (Van Heerwaarden et al., 2012). However, phenotypic and allele frequency clines in D. simulans are less abundant and shallower than those observed in D. melanogaster (McKenzie & Parsons 1974; Watada et al., 1986; Capy et al., 1993; Arthur et al., 2008) and D. simulans has less physiological tolerance to cold and starvation stresses when compared to D. melanogaster (Hoffmann & Harshman, 1999); this suggests distinct aspects of demography, physiology or selective response to environmental variance associated with the latitudinal extremes. Additionally, these sibling species exhibit different patterns of relative abundance over seasonal time. In temperate North American orchards, D. melanogaster are first evident in the late spring (Schmidt & Conde, 2006) and the population appears persistent over time. In contrast, D. simulans populations appear in midsummer and expand throughout the agricultural growing season to outnumber D. melanogaster by autumn, but are not present the following spring. The ecological parallels lead to the prediction that the sister taxa will have similar response to seasonal change, but the less robust clinal patterns in D. simulans and different frequencies over seasonal time suggest that their seasonal response may similarly be of reduced magnitude compared to D. melanogaster.
Natural orchard populations of both species were sampled from spring through autumn to assess seasonal changes in ecological and evolutionary population dynamics and life history traits. Seasonal changes in ecological parameters are documented using wild-caught individuals and their offspring to measure a subset of ecologically relevant traits: age distribution, reproductive output and development time. Population age structure is a fundamental component in population dynamics in the wild (Cole, 1957) and has been shown to change across seasonal time in other insect species (Carey et al., 2008). It is predicted that adults overwintering in a temperate location will emerge from dormancy fairly synchronously in response to environmental cues, which will result in a single young cohort in the spring that becomes more heterogeneous in age as nonoverlapping generations reproduce throughout the summer (Tauber et al., 1986). In temperate North America, D. melanogaster populations appear persistent over time (Ives, 1970; Bergland et al., 2014), potentially due to the expression of an adult reproductive diapause that is associated with overwintering (Saunders et al., 1989; Izquierdo, 1991; Mitrovski & Hoffmann, 2001); therefore, it is predicted that the post-dormancy populations will be young and age heterogeneity will increase across seasonal time.
We examine seasonal genetic change for a subset of traits previously shown to vary with latitude in D. melanogaster using wild-derived isofemale lines reared in a common laboratory environment for several generations. We predict traits favoured for survival at high latitudes will also be favoured during the winter because of similarities in their environments; likewise, parallels are predicted in low latitude and summer traits. In North America, adaptation to northern environments is associated with increased investment in stress resistance (Capy et al., 1993; Hoffmann et al., 2001; De Jong & Bochdanovits, 2003; Schmidt et al., 2005; Schmidt & Paaby, 2008). Therefore, we predict that winter environments also select for increased stress tolerance and that early-season generations in the spring will be characterized by elevated stress resistance. As the environment becomes more conducive to population growth throughout the summer, we predict generalized stress tolerance to decline, due to correlations and trade-offs with other aspects of fitness (Roff, 1992).
Materials and methods
Samples
Drosophila melanogaster and D. simulans were collected from Linvilla Orchards in Media, PA (39.884179°N, −75.411227°E], using baited traps and aspiration at four timepoints spaced approximately every 8 weeks: 1–4 June, 31 July, 26 September and 9 November 2011. Under light carbon dioxide anaesthetic, flies were sorted to species subgroup and allowed to recover on standard cornmeal molasses food. Isofemale lines were established by placing gravid females into individual vials of standard medium, and the species were identified through examination of the posterior lobe of male offspring.
Characteristics of the natural populations were measured on wild-caught flies: demography, reproductive output and F1 development time. Isofemale lines were maintained in a common laboratory environment (25 °C, 12:12 L:D, standard cornmeal molasses food) for four generations to remove environmental effects so that any difference in traits among the collections represented evolutionary change in the genetics of the population. After the generations in the laboratory, heat knockdown, chill recovery, starvation resistance and development time were measured under standard laboratory conditions.
Age distribution
The age distribution of the sampled populations was estimated at each collection timepoint to assess whether demography changes across seasonal time as it does in other taxa (Carey et al., 2008). Age structure was estimated utilizing the deconvolution model (Müller et al., 2007; Carey et al., 2008) that compares the post-capture survivorship of wild individuals to the full lifespan of their offspring to back-calculate the age distribution of the wild population. This model relies on the assumption that the age of an individual caught in the wild is reflected in post-capture survival, with young individuals surviving proportionally longer than old individuals in the laboratory. By extension, a relative or absolute change in estimated age distribution of the population between collection timepoints indicates a shift in population age structure. All flies were reared in individual vials of standard cornmeal medium that were changed every day for the first ten days in captivity and every three to five days thereafter. Mortality was recorded daily. The deconvolution model was implemented using MatLab (Math Works, Natick, MA, USA). Kaplan–Meier survivorship curves for the post-capture survivorship of wild individuals and the full lifespan of the F1 offspring was graphed using the ‘survival’ package (Therneau, 2012) in the R statistical analysis software (R Core Team 2012).
Fecundity and development time
Reproductive output was measured through daily transfers and egg counts of wild females during the first ten days post-capture. We analysed fecundity in two ways: the mean fecundity is a function of the population and is affected by the age distribution (e.g. Tatar et al., 1996; Novoseltsev et al., 2003), and the maximum fecundity is a function of the individual.
Vials containing eggs laid during the first 24 h of captivity were used to measure development time in the F1 post-capture generation. At three timepoints per day, the number of puparia and eclosed adults was recorded to determine the time to pupation and time to eclosion. After four to five generations in standard laboratory culture, development time to eclosion was again measured in the same way. Thus, the full development time from egg to eclosion was estimated at two time-points for each line: in the F1 generation and after several generations in common-garden laboratory culture. The measurements conducted on F1 generation reflected a combination of genetic, environmental and associated effects, whereas the measurements conducted in the common laboratory environment primarily reflected genetic variance.
Stress tolerance
Tolerance to a variety of stressors was examined for each isofemale line in the common laboratory environment after four to five generations of culture. For starvation resistance, groups of 12 individuals per sex for each line were sorted under light carbon dioxide anaesthesia and recovered on food for 24 h before transfer to vials that contained a cotton ball and 2 mL of deionized water. The number of live and dead flies was observed at three standardized timepoints every 24 h until all experimental flies died.
Thermal stress assays measuring response to high and low temperatures used DAM2 activity monitors (TriKinetics, Waltham, MA, USA) to record locomotor activity every 10 s. Eight flies per sex per line were placed into individual glass tubes and given an hour to recover from the carbon dioxide anaesthetic. To examine response to low temperature, groups of flies were buried in ice, placed in a 4 °C incubator for 2 h and then transferred to 25 °C; chill coma recovery time was estimated at the time required for each fly to resume an upright stance and locomotor activity. To evaluate response to high temperature, collections were placed at 25 °C in a Percival I36VL incubator programmed to increase temperature by 1 °C per min to 37 °C. The temperature remained constant at 37 °C, and time to thermal knockdown was recorded as the time at which locomotor activity ceased.
Statistical analysis of life history traits
Mixed-model anovas were used to assess seasonal change in all phenotypic traits with month and sex as fixed effects and line[month] as a random effect. Species were analysed separately to address the species-specific question of changes over seasonal time and because the absence of D. simulans in June made the data and models nonorthogonal. When both species were present, a direct comparison was made between the two species using t-tests corrected for multiple comparisons using Tukey's honestly significant difference tests. All statistical analyses were conducted in JMP v10.0.0 (SAS Institute, Cary, NC, USA).
Genetic variance/covariance estimates
For each species, genetic correlations among stress tolerance traits for all seasonal collections were calculated using Pearson's product-moment coefficients. Genetic correlations were estimated using isofemale lines to generate line means (Via, 1984; Roff, 1997). Sample sizes were as follows: D. melanogaster, n = 66, 83, 47 and 43 isofemale lines per timepoint, in chronological order; D. simulans, n = 0, 50, 87 and 58 isofemale lines, respectively. Significance probabilities indicate whether a particular genetic correlation differed from zero; P-values were obtained by treating the test statistic as coming from a t distribution. To test whether line mean genetic variance/covariance (G) matrices were statistically different among species by season combinations, manova was used on jack-knifed genetic variance/covariance values (Roff, 2002). Comparisons of variance/covariance matrices with this manova method have been shown to produce the same statistical results as the Flury method (e.g. Phillips & Arnold, 1999) for comparing G matrices, but environmental effects are easier to incorporate into the manova approach. For every trait pair, genetic variance/covariance pseudovalues were created by jack-knifing. Each set of variance/covariance pseudovalues was coded for species and season of collection. manova was then used to test whether sets of variance/covariance values significantly differed between seasons for each taxon (Roff, 2002); for example, chill recovery variance, heat knockdown variance and chill/heat covariance pseudovalues were treated as multiple response variables in a manova that included seasonal timing (early vs. late) as a predictor variable. F-tests and associated P-values were calculated from Wilks’ lambda values.
Results
Over seasonal time, the relative abundance of D. melanogaster and D. simulans changed dramatically. In the spring, the flies we sampled were exclusively D. melanogaster, but by the end of the autumn, D. simulans outnumbered D. melanogaster five-fold (Table 1). This suggests fundamental differences in population dynamics between the two taxa.
Table 1.
Post-capture survival of wild-caught Drosophila and full lifespan of offspring measured in days.
|
Drosophila melanogaster
|
Drosophila simulans
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Month | Sex | n | Mean | SE | Med. | Max. | n | Mean | SE | Med. | Max. |
| Wild fly post-capture survival | |||||||||||
| June | F | 59 | 45.92 | 2.59 | 44 | 82 | – | – | – | – | – |
| M | 96 | 43.9 | 1.74 | 40 | 75 | – | – | – | – | – | |
| July | F | 114 | 24.62 | 1.48 | 19 | 68 | 78 | 30.94 | 1.75 | 37 | 57 |
| M | 110 | 24.67 | 1.39 | 22 | 61 | 92 | 34.09 | 1.43 | 34.5 | 61 | |
| September | F | 34 | 29.53 | 3.07 | 30 | 70 | 126 | 25.93 | 1.4 | 30 | 60 |
| M | 136 | 20.57 | 1.49 | 16 | 75 | 114 | 29.69 | 1.62 | 32.5 | 60 | |
| November | F | 56 | 26.39 | 1.75 | 24 | 62 | 239 | 26.76 | 0.77 | 27 | 52 |
| M | 94 | 21.89 | 1.51 | 21 | 58 | 122 | 31.36 | 1.18 | 30 | 56 | |
| F1 full lifespan | |||||||||||
| June | F | 68 | 44.44 | 1.82 | 46 | 73 | – | – | – | – | – |
| M | 63 | 57.54 | 1.61 | 62 | 74 | – | – | – | – | – | |
| July | F | 96 | 47.15 | 1.30 | 48.5 | 67 | 66 | 47.79 | 1.46 | 46.5 | 67 |
| M | 81 | 46.99 | 1.71 | 52 | 66 | 68 | 46.37 | 1.66 | 52 | 65 | |
| September | F | 45 | 52.78 | 2.56 | 56 | 81 | 151 | 44.75 | 0.98 | 44 | 73 |
| M | 49 | 53.73 | 2.10 | 56 | 83 | 130 | 56.65 | 1.18 | 58 | 84 | |
| November | F | 47 | 46.06 | 2.04 | 43 | 73 | 194 | 43.88 | 0.78 | 44 | 77 |
| M | 45 | 52.16 | 2.35 | 54 | 75 | 193 | 54.22 | 1.03 | 58 | 78 | |
Age structure
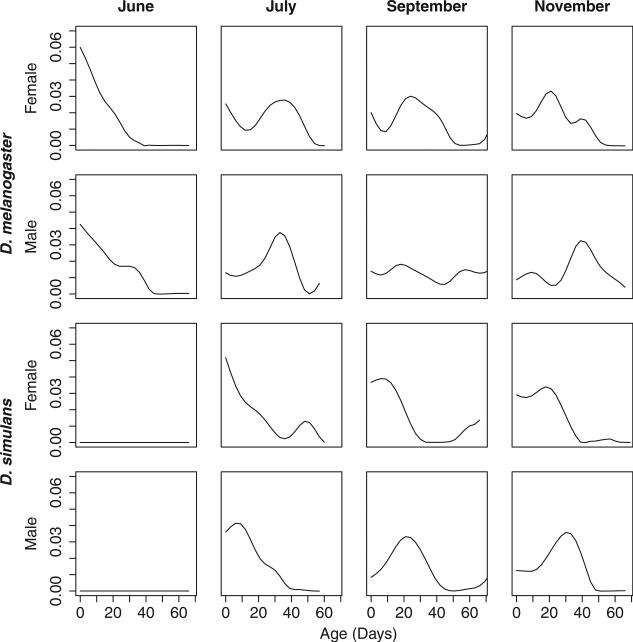
Multiple measurements of population age distribution suggest that the early D. melanogaster collection was unimodal and young; in contrast, the collections throughout the rest of the year contained more individuals inferred to come from older age classes. This is seen in the population age structure distributions estimated using the deconvolution model (Fig. 1). The Kaplan–Meier survivorship curves also demonstrated the same pattern: the wild-caught flies in June were inferred to be young because their post-capture survivorship curve was so similar to the full-lifespan survivorship curve of the corresponding F1 flies (Fig. S1). The other collections are inferred to be older: when caught, their post-capture survival was truncated compared to the full F1 lifespan. Consistent with these patterns, the median post-capture survival time of wild flies was shorter than the mean in the earliest collection and shifted towards the mean in the later months as the age heterogeneity increased (Table 1). Together, these data provide a consistent picture of a population that appears uniformly young early in the spring, but increases in age heterogeneity as time progresses.
Fig. 1.
Seasonal changes in age distribution of wild Drosophila estimated using the deconvolution model. Each graph plots the density of the estimated age distribution of the population in nature vs. age in days. Using this method, the June population of Drosophila melanogaster is inferred to be relatively young and is distinct from the subsequent collections that are more heterogeneous and contain older flies. Not collected in June, Drosophila simulans consistently contained flies of old age classes when the species was present in the orchard.
In contrast, the same demographic measurements yield distinct patterns for the D. simulans populations. Based on the inferences using the deconvolution model, D. simulans populations contained flies of older age classes when the species first appeared in July and there was much less change in age structure across the rest of the year relative to D. melanogaster (Fig. 1). The Kaplan–Meier survivorship curves for D. simulans exhibited minimal change across seasonal time, suggesting that the age distribution remained consistent across the measured timepoints (Fig. S1). The age distribution of wild flies shifted slightly younger as seasonal time progressed; the median survival time was initially skewed left of the mean and subsequently shifted towards the mean age throughout the autumn (Table 1). These data together depict a population that is consistently age-structured and does not have pronounced shifts in age distribution across seasonal time.
Seasonal change in natural population
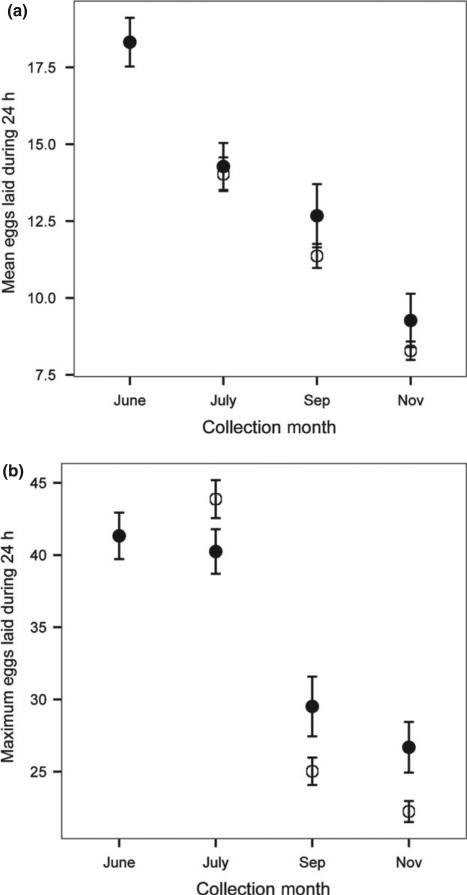
The measurements on wild females and their F1 offspring include multiple components, including maternal effects influenced by habitat quality and the environment. In both Drosophila species, reproductive output of wild-caught females declined over seasonal time (Fig. 2). The mean number of eggs laid declined, whereas the maximum showed a bimodal pattern with more eggs laid by flies collected in the first half of the study (June and July) than later (September and November). Although there was no difference between species in the early collections, the autumn D. melanogaster laid more eggs than D. simulans (Table 2).
Fig. 2.
Wild flies of both species have a seasonal decline in mean (a) and maximum (b) number of eggs (±SE) laid per day during the first 10 days of captivity. Drosophila melanogaster are indicated in filled circles and Drosophila simulans in empty circles. There is no difference between species in the mean reproductive output; however, D. simulans has a higher maximum fecundity compared to D. melanogaster in July but lower during the rest of the season.
Table 2.
Mixed-model anovas for environmental and common-garden traits measured across seasons for each species with month and sex as fixed effects and line[month] as a random effect.
|
Drosophila melanogaster
|
Drosophila simulans
|
|||||
|---|---|---|---|---|---|---|
| DF | SS | F ratio | DF | SS | F ratio | |
| Environment | ||||||
| Mean eggs per day | ||||||
| Month | 3 | 26 222 | 20.479*** | 2 | 18 315.9 | 49.623*** |
| Line[month] | 236 | 100 785 | 3.3052*** | 389 | 71 992.5 | 2.5087*** |
| Maximum eggs per day | ||||||
| Month | 3 | 9870.932 | 18.749*** | 2 | 23 027.047 | 105.639*** |
| F1 development time | ||||||
| Month | 3 | 264 618 | 81.784*** | 2 | 439 181 | 98.709*** |
| Sex | 1 | 14 097.8 | 87.686*** | 1 | 30 101.3 | 166.695*** |
| Month × sex | 3 | 1074.77 | 2.283 | 2 | 606.947 | 1.6806 |
| Line[month] | 281 | 696 134 | 15.409*** | 430 | 1 244 342 | 16.025*** |
| Common garden | ||||||
| Development time | ||||||
| Month | 3 | 1 317 083 | 172.41*** | 2 | 1 362 253 | 286.562*** |
| Sex | 1 | 1863 | 4.481* | 1 | 4521.36 | 11.796** |
| Month × sex | 3 | 58 446 | 46.859*** | 2 | 41 179.3 | 53.717*** |
| Line[month] | 227 | 1 058 577 | 11.216*** | 397 | 1 992 859 | 13.096*** |
| Heat knockdown | ||||||
| Month | 3 | 7.23E+09 | 27.061*** | 2 | 6.66E+08 | 14.409*** |
| Sex | 1 | 1 812 750 | 0.771 | 1 | 93 538.8 | 0.055 |
| Month × sex | 3 | 8 405 978 | 1.192 | 2 | 3.42 + 06 | 0.999 |
| Line[month] | 228 | 2.20E+10 | 40.636*** | 208 | 6.51E+09 | 18.327*** |
| Chill recovery | ||||||
| Month | 3 | 2.51E+07 | 3.773* | 2 | 6.74E+07 | 11.517*** |
| Sex | 1 | 2 015 167 | 2.593 | 1 | 4 525 210 | 4.726* |
| Month × sex | 3 | 1 452 574 | 0.623 | 2 | 2 497 510 | 1.304 |
| Line[month] | 237 | 5.36E+08 | 2.908*** | 430 | 6.01E+08 | 3.171*** |
| Starvation resistance | ||||||
| Month | 3 | 1 375 073 | 138.684*** | 2 | 226 248 | 67.8033*** |
| Sex | 1 | 2 474 462 | 7062.638*** | 1 | 1 070 297 | 3779.517*** |
| Month × sex | 3 | 85 167.5 | 81.0287*** | 2 | 33 365.4 | 58.9112*** |
| Line[month] | 227 | 1 919 109 | 10.0876*** | 397 | 465 053 | 6.0823*** |
P < 0.05
P < 0.01
P < 0.0001.
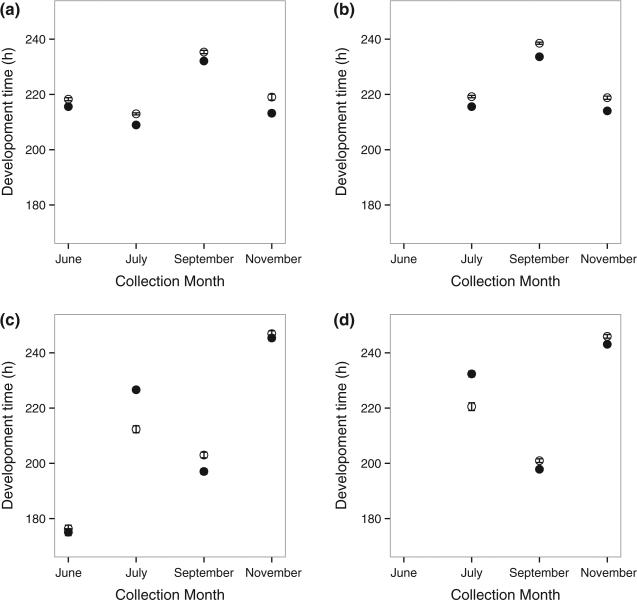
The development time from egg to adult for F1 offspring differed by month for both species (Table 3), but there was no directionality in these changes: the development time oscillated around 220 h across the collection timepoints (Fig. 3). Early-season D. melanogaster developed faster than D. simulans, but a difference between taxa was not evident in the autumn collections (Table 3). Larval development time (egg to pupation) mirrored that of the total development time (egg to eclosion) although the time in the puparium did not change by species or season (data not shown).
Table 3.
Mean and standard error by species for traits measured in the common garden and T-test comparing the trait performance when both species were present.
|
D. melanogaster
|
D. simulans
|
Species comparison |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Assay | Collection month | Sex | Mean | SE | Mean | SE | t-ratio (S-M) | DF | p-value |
| Chill recovery (minutes) | June | F | 52.292 | 2.819 | – | – | – | – | – |
| M | 58.658 | 3.173 | – | – | – | – | – | ||
| July | F | 57.741 | 2.987 | 75.602 | 4.77 | 17.861 | 592 | ** | |
| M | 61.919 | 3.396 | 66.37 | 4.542 | 4.452 | 673 | |||
| September | F | 62.683 | 4.555 | 66.076 | 3.114 | 3.393 | 669 | ||
| M | 72.779 | 5.261 | 65.181 | 3.097 | 7.598 | 574 | |||
| November | F | 85.327 | 6.18 | 78.507 | 5.016 | 6.820 | 553 | ||
| M | 84.848 | 6.439 | 68.161 | 4.089 | 16.687 | 491 | * | ||
| Heat knockdown (minutes) | June | F | 93.586 | 4.454 | – | – | – | – | – |
| M | 83.755 | 4.103 | – | – | – | – | – | ||
| July | F | 40.663 | 1.056 | 37.159 | 1.623 | 2.961 | 518 | ||
| M | 41.859 | 1.168 | 38.692 | 2.143 | 3.361 | 549 | |||
| September | F | 57.419 | 1.533 | 60.405 | 1.479 | 3.133 | 756 | ||
| M | 60.167 | 1.536 | 58.984 | 1.417 | 1.186 | 737 | |||
| November | F | 49.067 | 1.367 | 49.945 | 1.107 | 0.489 | 801 | ||
| M | 50.322 | 1.194 | 49.175 | 1.027 | 1.441 | 823 | |||
| Starvation survival (hours) | June | F | 119.664 | 1.042 | – | – | – | – | – |
| M | 75.152 | 0.777 | – | – | – | – | – | ||
| July | F | 112.456 | 0.917 | 95.734 | 0.974 | 18.963 | 1714 | *** | |
| M | 74.328 | 0.655 | 58.236 | 0.631 | 10.784 | 1871 | *** | ||
| September | F | 113.886 | 1.384 | 84.95 | 0.779 | 28.209 | 686 | *** | |
| M | 66.187 | 0.929 | 47.407 | 0.397 | 20.264 | 610 | *** | ||
| November | F | 72.216 | 0.886 | 68.515 | 0.695 | 3.701 | 683 | ** | |
| M | 47.144 | 0.374 | 44.558 | 0.211 | 2.586 | 548 | *** | ||
| Development time (hours) | June | F | 214.362 | 0.494 | – | – | – | – | – |
| M | 216.560 | 0.455 | – | – | – | – | – | ||
| July | F | 211.043 | 0.371 | 218.704 | 0.475 | 1.098 | 1517 | ||
| M | 214.955 | 0.379 | 221.856 | 0.492 | 0.368 | 818 | |||
| September | F | 234.133 | 0.605 | 236.069 | 0.442 | 0.397 | 983 | ||
| M | 237.707 | 0.595 | 241.151 | 0.443 | 2.801 | 972 | * | ||
| November | F | 214.839 | 1.059 | 215.697 | 0.720 | 6.841 | 1170 | *** | |
| M | 221.812 | 1.1502 | 220.063 | 0.787 | 2.766 | 1057 | * | ||
p<0.05
p<0.01
p<0.0001.
Fig. 3.
Mean (±SE) development time in hours for F1 individuals (a and b) and flies that had been in a common-garden laboratory environment (c and d) for Drosophila melanogaster (a and c) and Drosophila simulans (b and d). Females are indicated in filled circles and males in empty circles. The F1 development time includes maternal effects that may reflect environmental quality and oscillates around the same duration for both species across seasonal time. The common-garden development time removes such environmental effects and increases drastically for D. melanogaster; however, it does not have a directional change for D. simulans.
Seasonal change in genetic composition
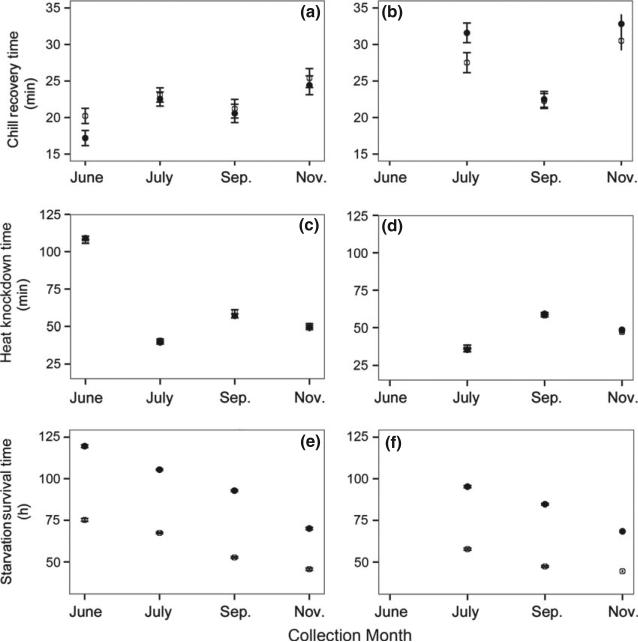
Drosophila melanogaster demonstrated a consistent and strong pattern of directional decline in performance over seasonal time for the phenotypes assayed in the common garden: tolerance to heat, cold and starvation stressors was highest in the early-season collection and declined predictably over time (Fig. 4, Table 3). Similarly, development time in the common garden increased linearly in D. melanogaster such that the last collection took nearly three full days longer to eclose than the earliest, with mid-season collections being intermediate (Fig. 3).
Fig. 4.
Mean (±SE) recovery time from chill (a and b), knockdown time from heat (c and d) and survival time without food (e and f) for flies that had been in a common-garden laboratory environment. Drosophila melanogaster (a, c, e) shows a seasonal decline in quality for all of these traits, whereas Drosophila simulans (b, d, f) demonstrates no clear pattern for thermal traits and a seasonal decline in starvation resistance. Females are indicated in filled circles and males in empty circles.
In contrast to patterns observed for D. melanogaster, D. simulans did not demonstrate a consistent change in thermal tolerance over seasonal time, although the linear decline in starvation tolerance paralleled that observed for D. melanogaster (Fig. 4). Compared to D. melanogaster, D. simulans were more susceptible in the chill and starvation assays, taking longer to recover from the chill and surviving a shorter time without food; however, there was no difference between the species in heat tolerance (Table 3).
Genetic variance/covariance
The estimated variance–covariance matrices significantly differed between species and between the early and late endpoints of the collections; furthermore, the change in the G matrix over seasonal time was distinct between D. simulans and D. melanogaster (Table 4, Table S1). For either species, only a single variance/covariance estimate (between starvation tolerance and development time in D. melanogaster) did not significantly vary between the early and late collections. In D. melanogaster, knockdown time under heat stress and recovery time from chill coma demonstrated a signifi-cant positive correlation both in the early and in the late season collections, although the correlation was significantly stronger in the former. The positive correlation indicated a negative functional association, as increasing values for high-temperature knockdown were indicative of increased stress tolerance, whereas decreasing values were indicative of the same for tolerance to cold. This suggests the potential for a pronounced trade-off between performance under high and low temperatures. In contrast, the correlation between development time and high-temperature knockdown did not indicate a functional trade-off, as increased resistance to heat stress was associated with a faster rate of development; this was evident in the early-season collection only and was distinct from the observed pattern in the late season collection.
Table 4.
Correlated traits throughout the season with early season (June) above the diagonal and late (November) below the diagonal. Bolded correlations are significantly different than zero at P = 0.05 with Pearson's product-moment correlation tests; italicized and bolded correlations are marginally different than zero at P = 0.10. Italicized correlations indicate trait variance/covariances that did not significantly differ between months at P = 0.05; all other variance/covariances differed between months for a particular trait combination at P = 0.001 with manova.
| Chill | Heat | Starvation | Development | |
|---|---|---|---|---|
| Drosophila melanogaster | ||||
| Chill | 1 | 0.48429018 | −0.0880632 | 0.01198028 |
| Heat | 0.39263658 | 1 | 0.00142993 | −0.2642816 |
| Starvation | −0.124261 | 0.00840717 | 1 | −0.0276306 |
| Development | −0.0668889 | −0.0847142 | 0.02486902 | 1 |
| Drosophila simulans | ||||
| Chill | 1 | −0.2277738 | −0.3282139 | 0.10125605 |
| Heat | −0.0513712 | 1 | −0.3038521 | 0.12133322 |
| Starvation | −0.0636857 | −0.186429 | 1 | 0.10700678 |
| Development | −0.0845394 | −0.1804966 | 0.15166336 | 1 |
Although all variance/covariance matrices were significantly distinct between early and late collections for D. simulans, the only individual correlation that approached significance was that between starvation tolerance and chill coma recovery time (Table 4). This correlation was negative: that is, this demonstrated a positive functional association where lines with an increased starvation resistance also recovered more quickly from exposure to cold.
Discussion
Mode of seasonal change
There are several ways in which multivoltine species may respond to changing selection pressures caused by seasonal environmental heterogeneity. Populations may not respond to the seasonal change in environmental parameters, with phenotypes that are either fixed over time or fluctuate in a stochastic, nondirectional manner. Although phenotypic plasticity may be elicited in response to cyclic environmental heterogeneity (e.g. Brakefield & Reitsma, 1991; Bradford & Roff, 1993), in this study the isofemale lines were reared in a common-garden laboratory environment to remove environmental effects that may have reflected phenotypic plasticity. Alternatively, natural selection for traits associated with high fitness in a specific environment may result in a rapid adaptive response if the population contains standing genetic variation for those traits; for example, in D. melanogaster seasonal shifts in diapause incidence (Schmidt & Conde, 2006) may result in seasonal change in allele frequencies at the gene couch potato (Cogni et al., 2013).
The significant differences in traits across collection timepoints in both species were indicative of some degree of seasonal response. Changes were seen in measurements of wild flies (i.e. fecundity) that reflect environmental variation as well as in the traits measured in the common garden (i.e. stress traits) that indicate genetic change in the population. The predictable pattern of decline for some traits suggests that the changes across seasonal time were not due to random chance. Additionally, the traits measured in the common garden showed rapid changes between subsequent collections that were unlikely to be explained by genetic drift. Therefore, it seemed unlikely that the seasonal patterns described were due to random stochasticity, but instead represented deterministic ecological and evolutionary processes.
The change across seasonal time in stress resistance traits measured in the common laboratory environment demonstrated seasonal change in the genetic composition of the population. In D. melanogaster, the decline in stress resistance from spring through summer was consistent with the operation of natural selection following the prediction of selection for high stress resistance during the winter and relaxed selection on stress resistance throughout the summer. However, the observed data may have reflected migration because D. melanogaster populations at lower latitudes are characterized by reduced tolerance to at least some stressors (Hoffmann et al., 2001; Paaby & Schmidt, 2008) and an influx of migrants from lower-latitude locales throughout the summer would be predicted to result in a decrease in stress tolerance over seasonal time. Such patterns of migration should have affected allele frequency profiles as well as phenotypes. Pooled sequencing of this Pennsylvania orchard population over three successive years, including the collections analysed here, has demonstrated that migration alone is insufficient to explain observed seasonal changes in allele frequencies genomewide (Bergland et al., 2014). By extension, migration from southern regions on the east coast of the United States cannot explain the rapid and pronounced change in phenotypic profiles that we observed and describe here.
The demographic and phenotypic patterns across seasonal time were different for D. simulans compared to D. melanogaster. Drosophila simulans was absent in the earliest collection, and when it appeared in late July, the population was composed of a diversity of age classes that remained consistent throughout the autumn. The absence of D. simulans during the first half of the calendar year may be because they are generally considered to be a more tropical taxon than D. melanogaster (Capy et al., 1993; Hoffmann & Harshman, 1999) with no overwintering mechanism identified, and therefore, they may be less able to maintain a resident population in temperate climates (Schmidt & Conde, 2006; Schmidt, 2011). The stability of age heterogeneity across seasonal time was consistent with the hypothesis of either annual recolonization or a longer residence time in refugia. Migration from a southern refuge could have caused the delay in appearance and explained the age heterogeneity, as Drosophila of all ages are thought to be transported passively by wind or humans over long distances (Dobzhansky, 1973). Alternatively, the delayed appearance could have been due to a longer residence in local refugia that support continuous populations, such that upon return to the orchard, the flies were a mixture of ages. Both scenarios would result in the patterns collected here; D. simulans reappeared in the orchard when environmental conditions were suitable and exhibited less directional phenotypic change in comparison with D. melanogaster. Only starvation resistance declined from spring to autumn in a parallel way between the two species. Distinguishing between the recolonization and local refuge hypotheses requires a targeted study that involves direct field measurements over time (e.g. mark–release–recapture) or inference from longitudinal sampling and sequencing. However, both hypotheses are consistent with the inference that, relative to D. melanogaster, D. simulans was less temporally persistent in temperate orchards and may exhibit a relatively weaker adaptive response to seasonal change in environmental parameters.
Seasonal population dynamics
The initial young composition of D. melanogaster was consistent with the hypothesis that after overwintering, adults were cued by environmental stimuli to emerge synchronously from dormancy to produce an initial cohort of uniform age composition (Tauber et al., 1986). The June collection analysed here was likely among the first post-dormancy cohorts; based on slower development time at cool spring temperatures (Trotta et al., 2006), it was estimated that the eggs were laid in April, the time at which D. melanogaster were first collected in appreciable numbers in Pennsylvania (Schmidt & Conde, 2006). After the initial uniformly young sample, the age composition followed the predicted increase in heterogeneity as the population grew and reproduced throughout the summer (Tauber et al., 1986); such seasonal changes in population age composition have also been documented in the medfly Ceratitis capitata (Carey et al., 2008). Together, the seasonal changes in demography of C. capitata and D. melanogaster suggest that the age composition of many multivoltine species may be dynamic across seasonal time. However, the different seasonal demographic dynamics in D. melanogaster and D. simulans indicate that seasonal changes in age composition is one component of a suite of traits that may respond to environmental heterogeneity.
Whereas D. melanogaster and D. simulans are commonly considered to have a short lifespan (~1–6 days) in nature (Rosewell & Shorrocks, 1987), the data presented here suggest that individuals of both species reached old ages of one to 2 months in the wild; as with other short-lived insects (Bonduriansky & Brassil, 2002), senescence may be pronounced in natural populations of Drosophila. As many life history traits have age-specific properties (e.g. Minois & Le Bourg, 1999; Nghiem et al., 2000; Zerofsky et al., 2005), any seasonal change over time in the demographic composition of the population may also have significant effects on selection dynamics in the field. Early in the season, the unimodal young D. melanogaster population would be expected to have a uniform age-specific response to the environment. In contrast, the increase in age heterogeneity throughout the summer and autumn would lead to a predicted wider range of responses to the same stress.
The change in population age structure across the season in D. melanogaster suggests that antagonistic pleiotropy associated with age-specific fitness parameters may add an additional layer of complexity to the population dynamics (Williams, 1957). Antagonistic pleiotropy can maintain additive genetic variation for fitness components and may allow for protected polymorphism in the absence of over dominance in populations with overlapping and nonoverlapping generations (Rose, 1982, 1983, 1985). In this way, it is possible that antagonistic pleiotropy may contribute to adaptive seasonal polymorphism in these populations (Bergland et al., 2014).
Implications of seasonal selection
In D. melanogaster populations, traits associated with fitness change in a nonrandom manner along with the environment over seasonal time; this may be due to environmentally mediated selection over short time-scales that previously may have been considered evolutionarily static. Whole-genome resequencing of the same population over three consecutive years has demonstrated that hundreds of SNPs consistently oscillate in allele frequency between spring and autumn (Bergland et al., 2014). Taken together, these results suggest that selection in D. melanogaster can act in a rapid fashion and that temporal variation in fitness may result in seasonal oscillations at both the phenotypic and genomic levels. However, the rapidity of environmental change may result in maladaptation because of a delay between the traits being selected in the parental environmental conditions that may not have the highest fitness in the subsequent generation.
The rapid response to environmental variables that characterize seasons may result in cyclic selection that maintains diversity in the population. Based on the observed change in traits from spring to autumn, we predict that the distinct selection regimes associated with summer population expansion and winter collapse will produce annual cycles in these traits as seen in reproductive diapause frequency in D. melanogaster (Schmidt & Conde, 2006). This alternating selection for winter and summer phenotypes is a special case of microevolution for an intermediate optimum known as fluctuating–stabilizing selection (Wright, 1968; Istock, 1981); this selection for bet hedging can be applied hierarchically to a broad range of evolutionary scales (Simons, 2002). It is expected to maintain phenotypic and genetic variation within a population and, in doing so, seasonal environmental selection may limit or slow evolutionary processes including population divergence (Levins, 1968; Sasaki & Ellner, 1997) and local adaptation (Kawecki & Ebert, 2004).
The phenotypic change we observe here from spring through autumn should be considered in the larger context of annual cycles of seasonal selection across seasons. Such cycling selection may yield underestimates in the strength or direction of selection by averaging trait values across seasonal time; this may contribute to low estimates of the strength of selection over an entire breeding season (Kingsolver et al., 2001). Additionally, seasonal changes in the variance–covariance matrices demonstrate that strong selection on this timescale effects genetic correlations; this alters the basic assumption of stable covariance over time when making phenotypic evolutionary predictions.
The magnitude and rapidity of the phenotypic change observed over seasonal time leads to the hypothesis that seasonal dynamics may also contribute to the formation and persistence of latitudinal clines. Differential length of seasons could generate latitudinal clines if a favoured phenotype reaches high frequency in the winter but selection against it during the summer decreases its frequency in proportion to the length of the growing season (Rhomberg & Singh, 1986). Our data demonstrate that the range and variance associated with temporal sampling of Drosophila in a temperate orchard is equivalent to that previously observed in collections of natural populations spanning 20° latitude in the eastern United States (Schmidt & Paaby, 2008). Given the extent of phenotypic change throughout the climatic period favourable for Drosophila population growth and reproduction, temporal variation in selection pressures could be at least partially responsible for the generation of latitudinal clines that appear so pervasive in D. melanogaster (e.g. Capy et al., 1993; James & Partridge, 1995; Mitrovski & Hoffmann, 2001; Schmidt et al., 2005; Trotta et al., 2006). Such systematic changes in season length along a latitudinal gradient can generate either simple or ‘saw-tooth’ clines (Roff, 1980). However, the seasonal phase cline hypothesis and the connection between temporal and spatial evolutionary dynamics of life histories remain to be comprehensively tested in nature.
Supplementary Material
Acknowledgments
We thank Annalise Paaby, Thomas Flatt, Wolf Blanckenhorn and two anonymous reviewers for providing useful feedback on this manuscript. This work was supported by NSF GRF DGE-0822 (ELB) and by NSF DEB 0921307 and NIH RO1GM100366 (PSS).
Footnotes
Supporting information
Additional Supporting Information may be found in the online version of this article:
Figure S1 Kaplain–Meier survivorship curves for wild (solid) and F1 (dashed) Drosophila across seasonal time.
Table S1 Correlation matrices between traits by species across time.
References
- Anderson PR, Oakeshott JG. Parallel geographical patterns of allozyme variation in two sibling Drosophila species. Nature. 1984;308:729–731. [Google Scholar]
- Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- Arthur AL, Weeks AR, Sgró CM. Investigating latitudinal clines for life history and stress resistance traits in Drosophila simulans from eastern Australia. J. Evol. Biol. 2008;21:1470–1479. doi: 10.1111/j.1420-9101.2008.01617.x. [DOI] [PubMed] [Google Scholar]
- Azevedo RB, James AC, McCabe J, Partridge L. Latitudinal variation of wing: thorax size ratio and wing-aspect ratio in Drosophila melanogaster. Evolution. 1998;52:1353–1362. doi: 10.1111/j.1558-5646.1998.tb02017.x. [DOI] [PubMed] [Google Scholar]
- Bergland AO, Behrman EL, O'Brien KR, Schmidt PS, Petrov DA. Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. PLoS Genet. 2014;10:e1004775. doi: 10.1371/journal.pgen.1004775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry AJ, Kreitman M. Molecular analysis of an allozyme cline: alcohol dehydrogenase in Drosophila melanogaster on the east coast of North America. Genetics. 1993;134:869–893. doi: 10.1093/genetics/134.3.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt BR, Kim I, Hoffmann AA, Feder ME. Response to natural and laboratory selection at the Drosophila Hsp70 genes. Evolution. 2002;56:1796–1801. doi: 10.1111/j.0014-3820.2002.tb00193.x. [DOI] [PubMed] [Google Scholar]
- Blanckenhorn WU, Morf C, Muhlhauser C, Reusch T. Spatiotemporal variation in selection on body size in the dung fly Sepsis cynipsea. J. Evol. Biol. 1999;12:563–576. [Google Scholar]
- Bonduriansky R, Brassil CE. Rapid and costly ageing in wild male flies. Nature. 2002;420:377–377. doi: 10.1038/420377a. [DOI] [PubMed] [Google Scholar]
- Bradford MJ, Roff DA. Bet hedging and the dia-pause strategies of the cricket Allonemobius fasciatus. Ecology. 1993;74:1129–1135. [Google Scholar]
- Brakefield PM. Differential winter mortality and seasonal selection in the polymorphic ladybird Adalia bipunctata (L) in the Netherlands. Biol. J. Linn. Soc. 1985;24:189–206. [Google Scholar]
- Brakefield PM, Reitsma N. Phenotypic plasticity, seasonal climate and the population biology of Bicyclus butter-flies (Satyridae) in Malawi. Ecol. Entomol. 1991;16:291–303. [Google Scholar]
- Capy P, Pla E, David J. Phenotypic and genetic variability of morphometrical traits in natural populations of Drosophila melanogaster and D simulans. I. Geographic variations. Genet. Sel. Evol. 1993;25:517–536. [Google Scholar]
- Carey JR, Papadopoulos NT, Müller HG, Katsoyannos BI, Kouloussis NA, Wang JL, et al. Age structure changes and extraordinary lifespan in wild medfly populations. Aging Cell. 2008;7:426–437. doi: 10.1111/j.1474-9726.2008.00390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SP, Hendry AP, Reznick DN, Fox CW. Evolution on ecological time-scales. Funct. Ecol. 2007;21:387–393. [Google Scholar]
- Cogni R, Kuczynski C, Koury S, Lavington E, Behrman E, O'Brien K, et al. The intensity of selection acting on the couch potato gene –spatial-temporal variation in a diapause cline. Evolution. 2013;68:538–548. doi: 10.1111/evo.12291. [DOI] [PubMed] [Google Scholar]
- Cogni R, Kuczynski C, Lavington E, Koury S, Behrman E, O'Brien K, et al. Variation in Drosophila melanogaster central metabolic genes appears driven by natural selection both within and between populations. Proc. R. Soc. B. 2014;282:20142688. doi: 10.1098/rspb.2014.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole LC. Sketches of general and comparative demography. Cold Spring Harb. Symp. Quant. Biol. 1957;22:1–15. doi: 10.1101/sqb.1957.022.01.004. [DOI] [PubMed] [Google Scholar]
- David JR, Capy P. Genetic variation of Drosophila melanogaster natural populations. Trends Genet. 1988;4:106–111. doi: 10.1016/0168-9525(88)90098-4. [DOI] [PubMed] [Google Scholar]
- De Jong G, Bochdanovits Z. Latitudinal clines in Drosophila melanogaster: body size, allozyme frequencies, inversion frequencies, and the insulin-signalling pathway. J. Genet. 2003;82:207–223. doi: 10.1007/BF02715819. [DOI] [PubMed] [Google Scholar]
- Dempster ER. Maintenance of genetic heterogeneity. Cold Spring Harbor Symp. Quant. Biol. 1955;20:25–32. doi: 10.1101/sqb.1955.020.01.005. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics of natural populations IX. Temporal changes in the composition of populations of Drosophila Pseudoobscura. Genetics. 1943;28:162. doi: 10.1093/genetics/28.2.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics of natural populations. Xvi. Altitudinal and seasonal changes produced by natural selection in certain populations of Drosophila Pseudoobscura and Drosophila Persimilis. Genetics. 1948;33:158. doi: 10.1093/genetics/33.2.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Active dispersal and passive transport in Drosophila. Evolution. 1973;27:565–575. doi: 10.1111/j.1558-5646.1973.tb00706.x. [DOI] [PubMed] [Google Scholar]
- Ellner S, Hairston NG. Role of overlapping generations in maintaining genetic variation in a fluctuation environment. Am. Nat. 1994;14:403–417. [Google Scholar]
- Endler JA. Geographic Variation, Speciation, and Clines. Princeton University Press; Princeton, NJ.: 1977. [PubMed] [Google Scholar]
- Endler JA. Natural Selection in the Wild. Princeton University Press; Princeton, NJ.: 1986. [Google Scholar]
- Ewing EP. Genetic variation in a heterogeneous environment VII. Temporal and spatial heterogeneity in infinite populations. Am. Nat. 1979;114:197–212. [Google Scholar]
- Frydenberg J, Hoffmann A, Loeschcke V. DNA sequence variation and latitude associations in hsp23, hsp26 and hsp27 from populations of Drosophila melanogaster. Mol. Ecol. 2003;12:2025–2032. doi: 10.1046/j.1365-294x.2002.01882.x. [DOI] [PubMed] [Google Scholar]
- Gibert P, Capy P, Imasheva A, Moreteau B, Morin JP, P etavy G, et al. Comparative analysis of morphological traits among Drosophila melanogaster and D. simulans: genetic variability, clines and phenotypic plasticity. Genetica. 2004;120:165–179. doi: 10.1023/b:gene.0000017639.62427.8b. [DOI] [PubMed] [Google Scholar]
- Gillespie JH. Natural selection with varying selection coefficients –a haploid model. Genet. Res. 1973;21:115–120. [Google Scholar]
- Grant PR, Grant BR. Unpredictable evolution in a 30-year study of Darwin's finches. Science. 2002;296:707–711. doi: 10.1126/science.1070315. [DOI] [PubMed] [Google Scholar]
- Haldane JBS, Jayakar SD. Polymorphism due to selection of varying direction. J. Genet. 1963;58:237–242. [Google Scholar]
- Hedrick PW. Genetic polymorphism in a temporally varying environment: effects of delayed germination or dia-pause. Heredity. 1995;75:164–170. [Google Scholar]
- Hedrick PW. Pathogen resistance and genetic variation at MHC loci. Evolution. 2002;56:1902–1908. doi: 10.1111/j.0014-3820.2002.tb00116.x. [DOI] [PubMed] [Google Scholar]
- Hendry AP, Kinnison M. The pace of modern life: measuring rates of contemporary microevolution. Evolution. 1999;53:1637–1653. doi: 10.1111/j.1558-5646.1999.tb04550.x. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Harshman LG. Desiccation and starvation resistance in Drosophila: patterns of variation at the species, population and intrapopulation levels. Heredity. 1999;83:637–643. doi: 10.1046/j.1365-2540.1999.00649.x. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Hallas R, Sinclair C, Mitrovski P. Levels of variation in stress resistance in Drosophila among strains, local populations, and geographic regions: patterns for desiccation, starvation, cold resistance, and associated traits. Evolution. 2001;55:1621–1630. doi: 10.1111/j.0014-3820.2001.tb00681.x. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Anderson A, Hallas R. Opposing clines for high and low temperature resistance in Drosophila melanogaster. Ecol. Lett. 2002;5:614–618. [Google Scholar]
- Istock CA. The extent and consequences of heritable variation for fitness characters. In: King CR, Dawson PS, editors. Population biology. Retrospect and Prospect. Oregon State University Colloquium: Biology. Columbia Univ. Press; New York: 1981. pp. 61–96. [Google Scholar]
- Ives PT. Further genetic studies of the south Amherst population of Drosophila melanogaster. Evolution. 1970;24:507–518. doi: 10.1111/j.1558-5646.1970.tb01785.x. [DOI] [PubMed] [Google Scholar]
- Izquierdo JI. How does Drosophila melanogaster overwinter? Entomol. Exp. Appl. 1991;59:51–58. [Google Scholar]
- James A, Partridge L. Thermal evolution of rate of larval development in Drosophila melanogaster in laboratory and field populations. J. Evol. Biol. 1995;8:315–330. [Google Scholar]
- James AC, Azevedo RBR, Partridge L. Genetic and environmental responses to temperature of Drosophila melanogaster from a latitudinal cline. Genetics. 1997;146:881–890. doi: 10.1093/genetics/146.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karan E, Dahiya N, Munjal AK, Gilbert P, Moreteau B, Parkash R, et al. Desiccation and starvation tolerance of adult Drosophila: opposing latitudinal clines in natural populations of three different species. Evolution. 1998;53:825–831. doi: 10.1111/j.1558-5646.1998.tb03706.x. [DOI] [PubMed] [Google Scholar]
- Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecol. Lett. 2004;7:1225–1241. [Google Scholar]
- Kingsolver JG, Hoekstra HE, Hoekstra JM, Berrigan D, Vignieri SN, Hill CE, et al. The strength of phenotypic selection in natural populations. Am. Nat. 2001;157:245–261. doi: 10.1086/319193. [DOI] [PubMed] [Google Scholar]
- Levene H. Genetic equilibrium when more than one ecological niche is available. Am. Nat. 1953;87:331–333. [Google Scholar]
- Levins R. Evolution in Changing Environments. Princeton University Press; Princeton, NJ.: 1968. [Google Scholar]
- MacKay T. Genetic variance, fitness, and homeostasis in varying environments: an experimental check of the theory. Evolution. 1980;34:1219–1222. doi: 10.1111/j.1558-5646.1980.tb04070.x. [DOI] [PubMed] [Google Scholar]
- McKechnie SW, Blacket MJ, Song SV, Rako L, Carroll X, Johnson TK, et al. A clinally varying promoter polymorphism associated with adaptive variation in wing size in Drosophila. Mol. Ecol. 2010;19:775–784. doi: 10.1111/j.1365-294X.2009.04509.x. [DOI] [PubMed] [Google Scholar]
- McKenzie A, Parsons PA. The genetic architecture of resistance to desiccation in populations of Drosophila melanogaster and D. simulans. Aust. J. Biol. Sci. 1974;27:441–456. doi: 10.1071/bi9740441. [DOI] [PubMed] [Google Scholar]
- Minois N, Le Bourg E. Resistance to stress as a function of age in Drosophila melanogaster living in hypergravity. Mech. Ageing Dev. 1999;109:53–64. doi: 10.1016/s0047-6374(99)00025-1. [DOI] [PubMed] [Google Scholar]
- Mitrovski P, Hoffmann AA. Postponed reproduction as an adaptation to winter conditions in Drosophila melanogaster: evidence for clinal variation under semi-natural conditions. Proc. Biol. Sci. 2001;268:2163–2168. doi: 10.1098/rspb.2001.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller HG, Wang JL, Yu W, Delaigle A, Carey JR. Survival and aging in the wild via residual demography. Theor. Popul. Biol. 2007;72:513–522. doi: 10.1016/j.tpb.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghiem D, Gibbs AG, Rose MR, Bradley TJ. Postponed aging and desiccation resistance in Drosophila melanogaster. Exp. Gerontol. 2000;35:957–969. doi: 10.1016/s0531-5565(00)00163-7. [DOI] [PubMed] [Google Scholar]
- Novoseltsev VN, Novoseltseva JA, Boyko SI, Yashin AI. What fecundity patterns indicate about aging and longevity: insights from Drosophila studies. J. Gerontol. A Biol. Sci. Med. Sci. 2003;58:484–494. doi: 10.1093/gerona/58.6.b484. [DOI] [PubMed] [Google Scholar]
- Paaby A, Schmidt P. Functional significance of allelic variation in methuselah, an aging gene in Drosophila. PLoS One. 2008;3:e1987. doi: 10.1371/journal.pone.0001987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby AB, Blacket MJ, Hoffmann AA, Schmidt PS. Identification of a candidate adaptive polymorphism for Drosophila life history by parallel independent clines on two continents. Mol. Ecol. 2010;19:760–774. doi: 10.1111/j.1365-294X.2009.04508.x. [DOI] [PubMed] [Google Scholar]
- Paaby AB, Bergland AO, Behrman EL, Schmidt PS. An amino acid polymorphism in the Drosophila insulin receptor demonstrates pleiotropic and adaptive function in life history traits. Evolution. 2014;68:3395–3409. doi: 10.1111/evo.12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips PC, Arnold SJ. Hierarchical comparison of genetic variance-covariance matrices. I. Using the Flury hierarchy. Evolution. 1999;53:1506–1515. doi: 10.1111/j.1558-5646.1999.tb05414.x. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2012. ISBN 3-900051-07-0, http://www.R-project.org/ [Google Scholar]
- Rhomberg LR, Singh RS. Evidence for a link between local and seasonal cycles in gene frequencies and latitudinal gene clines in a cyclic parthenogen. Genetica. 1986;78:73–79. doi: 10.1007/BF00058677. [DOI] [PubMed] [Google Scholar]
- Robinson S, Zwaan B, Partridge L. Starvation resistance and adult body composition in a latitudinal cline of Drosophila melanogaster. Evolution. 2000;54:1819–1824. doi: 10.1111/j.0014-3820.2000.tb00726.x. [DOI] [PubMed] [Google Scholar]
- Roesti M, Gavrilets S, Hendry A, Salzburger W, Berner D. The genomic signature of parallel adaptation from shared genetic variation. Mol. Ecol. 2014;23:3944–3956. doi: 10.1111/mec.12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roff D. Optimizing development time in a seasonal environment: the ‘ups and downs' of clinal variation. Oecologia. 1980;45:202–208. doi: 10.1007/BF00346461. [DOI] [PubMed] [Google Scholar]
- Roff D. The Evolution of Life Histories: Theory and Analysis. Chapman and Hall, New York: 1992. [Google Scholar]
- Roff D. Evolutionary Quantitative Genetics. Chapman and Hall; New York: 1997. [Google Scholar]
- Roff D. Comparing G-matrices: a MANOVA approach. Evo lution. 2002;56:1286–1291. [PubMed] [Google Scholar]
- Rose MR. Antagonistic pleiotropy, dominance, and genetic variation. Heredity. 1982;48:63–78. [Google Scholar]
- Rose MR. Further models of selection with antagonistic pleiotropy. In: Freedman HI, Stro-beck C, editors. Population Biology. Spring-Verlag; Berlin: 1983. pp. 47–53. [Google Scholar]
- Rose MR. Life history evolution with antagonistic pleiotropy and overlapping generations. Theor. Popul. Biol. 1985;28:342–358. [Google Scholar]
- Rosewell J, Shorrocks B. The implication of survival rates in natural populations of Drosophila: capture-recapture experiments on domestic species. Biol. J. Linn. Soc. 1987;32:373–384. [Google Scholar]
- Sasaki A, Ellner S. Quantitative genetic variance maintained by fluctuating selection with overlapping generations: variance components and covariances. Evolution. 1997;51:682–696. doi: 10.1111/j.1558-5646.1997.tb03652.x. [DOI] [PubMed] [Google Scholar]
- Saunders DS, Henrich VC, Gilbert LI. Induction of diapause in Drosophila melanogaster: photoperiodic regulation and the impact of arrhythmic clock mutations on time measurement. Proc. Natl. Acad. Sci. USA. 1989;86:3748–3752. doi: 10.1073/pnas.86.10.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt PS. Evolution and Mechanisms of Inset Reproductive Diapause: A Plastic and Pleiotropic Life History Syndrome. Oxford Univ Press; New York: 2011. [Google Scholar]
- Schmidt PS, Conde DR. Environmental heterogeneity and the maintenance of genetic variation for reproductive dia-pause in Drosophila melanogaster. Evolution. 2006;60:1602–1611. [PubMed] [Google Scholar]
- Schmidt PS, Paaby AB. Reproductive Diapause and life-history clines in North American Populations of Drosophila melanogaster. Evolution. 2008;62:1204–1215. doi: 10.1111/j.1558-5646.2008.00351.x. [DOI] [PubMed] [Google Scholar]
- Schmidt PS, Paaby AB, Heschel MS. Genetic variance for diapause expression and associated life histories in Drosophila melanogaster. Evolution. 2005;59:2616–2625. [PubMed] [Google Scholar]
- Sezgin E. Single-locus latitudinal clines and their relationship to temperate adaptation in metabolic genes and derived alleles in Drosophila melanogaster. Genetics. 2004;168:923–931. doi: 10.1534/genetics.104.027649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepielski A, DiBattista J, Carlson S. It's about time: the temporal dynamics of phenotypic selection in the wild. Ecol. Lett. 2009;12:1261–1276. doi: 10.1111/j.1461-0248.2009.01381.x. [DOI] [PubMed] [Google Scholar]
- Simons AM. The continuity of microevolution and macroevolution. J. Evol. Biol. 2002;15:688–701. [Google Scholar]
- Slatkin M. Gene flow and the geographic structure of populations. Science. 1987;236:787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- Tatar M, Promislow DL, Khazaeli AA, Curtsinger JW. Age-specific patterns of genetic variance in Drosophila melanogaster. II Fecundity and its genetic covariances with age-specific mortality. Genetics. 1996;143:849–858. doi: 10.1093/genetics/143.2.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber M, Tauber C, Masaki S. Seasonal Adaptations of Insects. Oxford University Press; New York: 1986. [Google Scholar]
- Tauber E, Zordan M, Sandrelli F, Pegoraro M, Osterwalder N, Breda C, et al. Natural selection favors a newly derived timeless allele in Drosophila melanogaster. Science. 2007;316:1895–1898. doi: 10.1126/science.1138412. [DOI] [PubMed] [Google Scholar]
- Therneau T. A Package for Survival Analysis in S. R package version 2. 2012:36–14. http://cran.r-project.org/package=survival.
- Trotta V, Calboli FC, Ziosi M, Guerra D, Pezzoli MC, David JR, et al. Thermal plasticity in Drosophila melanogaster: a comparison of geographic populations. BMC Evol. Biol. 2006;6:67. doi: 10.1186/1471-2148-6-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M. Temporally varying selection on multiple alleles. A diffusion analysis. J. Math. Biol. 1981;13:115–129. [Google Scholar]
- Van Heerwaarden B, Lee RFH, Begener B, Weeks AR, Sgro CM. Complex patterns of local adaptation in heat tolerance in Drosophila simulans from eastern Australia. J. Evol. Biol. 2012;25:1765–1778. doi: 10.1111/j.1420-9101.2012.02564.x. [DOI] [PubMed] [Google Scholar]
- Verrelli BC, Eanes WF. Clinal variation for amino acid polymorphisms at the Pgm locus in Drosophila melanogaster. Genetics. 2000;157:1649–1663. doi: 10.1093/genetics/157.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via S. The quantitative genetics of polyphagy in an insect herbivore. II. Genetic correlations in larval performance within and among host plants. Evolution. 1984;38:896–905. doi: 10.1111/j.1558-5646.1984.tb00360.x. [DOI] [PubMed] [Google Scholar]
- Watada M, Ohba S, Tobari Y. Genetic differentiation in Japanese populations of Drosophila simulans and D. melanogaster. II Morphological variation. Jpn. J. Genet. 1986;61:469–480. [Google Scholar]
- Williams GC. Pleiotropy, natural selection and the evolution of senescence. Evolution. 1957;4:398–411. [Google Scholar]
- Wright S. Evolution and the Genetics of Populations. Vol. 1. Genetic and Biometric Foundations. University of Chicago Press; Chicago, IL.: 1968. [Google Scholar]
- Zerofsky M, Harel E, Silverman N, Tatar M. Aging of the innate immune response in Drosophila melanogaster. Aging Cell. 2005;4:103–108. doi: 10.1111/j.1474-9728.2005.00147.x. [DOI] [PubMed] [Google Scholar]
- Zhao L, Wit J, Svetec N, Begun D. Parallel gene expression differences between low and high latitude populations of Drosophila melanogaster and D. simulans. PLoS Genet. 2015;11:e1005184. doi: 10.1371/journal.pgen.1005184. doi:10.1371/journal.pgen.1005184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.