ABSTRACT
Export of macromolecules via extracellular membrane-derived vesicles (MVs) plays an important role in the biology of Gram-negative bacteria. Gram-positive bacteria have also recently been reported to produce MVs; however, the composition and mechanisms governing vesiculogenesis in Gram-positive bacteria remain undefined. Here, we describe MV production in the Gram-positive human pathogen group A streptococcus (GAS), the etiological agent of necrotizing fasciitis and streptococcal toxic shock syndrome. M1 serotype GAS isolates in culture exhibit MV structures both on the cell wall surface and in the near vicinity of bacterial cells. A comprehensive analysis of MV proteins identified both virulence-associated protein substrates of the general secretory pathway in addition to “anchorless surface proteins.” Characteristic differences in the contents, distributions, and fatty acid compositions of specific lipids between MVs and GAS cell membrane were also observed. Furthermore, deep RNA sequencing of vesicular RNAs revealed that GAS MVs contained differentially abundant RNA species relative to bacterial cellular RNA. MV production by GAS strains varied in a manner dependent on an intact two-component system, CovRS, with MV production negatively regulated by the system. Modulation of MV production through CovRS was found to be independent of both GAS cysteine protease SpeB and capsule biosynthesis. Our data provide an explanation for GAS secretion of macromolecules, including RNAs, lipids, and proteins, and illustrate a regulatory mechanism coordinating this secretory response.
IMPORTANCE
Group A streptococcus (GAS) is a Gram-positive bacterial pathogen responsible for more than 500,000 deaths annually. Establishment of GAS infection is dependent on a suite of proteins exported via the general secretory pathway. Here, we show that GAS naturally produces extracellular vesicles with a unique lipid composition that are laden with proteins and RNAs. Interestingly, both virulence-associated proteins and RNA species were found to be differentially abundant in vesicles relative to the bacteria. Furthermore, we show that genetic disruption of the virulence-associated two-component regulator CovRS leads to an increase in vesicle production. This study comprehensively describes the protein, RNA, and lipid composition of GAS-secreted MVs and alludes to a regulatory system impacting this process.
INTRODUCTION
Infections by the Gram-positive human pathogen Streptococcus pyogenes, also known as group A streptococcus (GAS), are responsible for global mortality and high societal costs in both developed and developing countries. Conservative estimates of the global mortality burden of GAS disease broach 500,000 deaths per year (1). Noninvasive GAS disease also represents a significant financial burden, with pediatric cases of pharyngitis in the United States alone inflicting a societal cost of $539 million per year (2). Despite concerted effort, no vaccines are currently licensed for commercial use against GAS infections (3).
Both mild and severe forms of GAS diseases are mediated by a suite of secreted and membrane-associated virulence factors that interact with the human host (3, 4). Trafficking and secretion of GAS proteins have long been assumed to be solely dependent on the canonical general secretory (Sec) translocation pathway. In GAS, the Sec pathway is associated with the ExPortal, a cell membrane microdomain enriched for proteins of the pathway (5). Interestingly, previous studies illustrate the presence of multiple factors present in the GAS secretome that lack the N-terminal secretion signal peptide or C-terminal cell wall anchor motif required for Sec-mediated translocation (6). These factors, dubbed “anchorless surface proteins,” comprise many important GAS virulence determinants and vaccine candidates; however, the mechanisms responsible for transport of these factors across the cell membrane remain unexplained (7).
In Gram-negative bacteria, outer membrane vesicles (OMVs) are actively formed through the budding and release of the outer membrane and contain numerous virulence-associated and immunomodulatory factors (8). OMVs have been implicated in many aspects of Gram-negative bacterial pathogenesis, including delivery of protein toxins, triggering host inflammatory responses and transfer of nucleic acids and providing protection against phages and antimicrobial peptides (9). Recent seminal works describe production of extracellular membrane-derived vesicles (MVs) by several clinically relevant species of Gram-positive bacteria, such as Staphylococcus aureus, Bacillus anthracis, and Streptococcus pneumoniae (10–12). Two recent reports indicate that GAS also releases MVs and that this process increases following sublethal penicillin or LL-37 treatment (13, 14). Mycobacterium tuberculosis and other mycobacterial species also actively produce MVs, which are able to modulate host inflammatory responses (15). These studies hint at a more broadly conserved export strategy utilized by both Gram-negative and Gram-positive bacteria (16). In the present work, we provide a comprehensive analysis of naturally produced GAS MVs and assess the involvement of genetic factors influencing MV production in GAS.
RESULTS
GAS exhibits active MV production during in vitro growth.
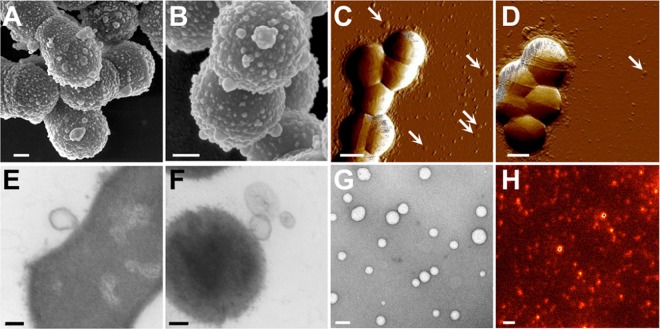
We first undertook morphological examination of GAS clinical isolate ISS3348 and reference strain SF370 (both M1 serotype) axenic cultures using several high-resolution microscopy approaches. Both strains displayed multiple spherical structures protruding from their cell membranes, with diameters ranging between 10 and 272 nm (Fig. 1A and B). These vesicle-like structures appeared liable to be released from the membrane, as similarly sized vesicular structures could also be discerned both on the cell surface and in the vicinity of the streptococci (Fig. 1C to F). To determine whether these structures were indeed freely released from the membrane, GAS culture supernatants were filtered twice through 0.22-µm-pore membranes prior to ultracentrifugation. Pellets obtained from ISS3348 cultures were examined by negative-staining transmission electron microscopy (TEM) and exhibited a heterogeneously sized population of circular structures suggestive of extracellular membrane-derived vesicles (MVs) (Fig. 1G; see Fig. S1A in the supplemental material). To determine whether MV production is an active process, MVs from late-logarithmic-phase GAS culture supernatants and the same cultures incubated in fresh media following heat inactivation were quantified by flow cytometry (see Fig. S1B). MV production of both cultures following heat inactivation was reduced to the level of flow cytometric events recorded with buffer alone; interestingly, ISS3348 demonstrated significantly higher MV production than SF370 (see Fig. S1B). These data support the assumption that production of MVs is an active process requiring live, growing bacteria. MVs were also isolated from GAS treated with a sublethal concentration of lysozyme, leading to an altered MV protein profile in lysozyme-treated bacteria (see Fig. S1C). An altered MV protein profile in lysozyme-treated bacteria indicates that weakening of the cell wall via lysozyme treatment releases membrane fragments distinct from natively isolated MVs as has been previously noted (17), refuting the possibility that MVs are formed through nonspecific assembly of shed membrane fragments.
FIG 1 .
GAS cells exhibit MV protrusions during in vitro growth. Cellular morphologies of ISS3348 (A, C, and E) and SF370 (B, D, and F) were examined by SEM (A and B), AFM (C and D), and TEM (E and F). Isolated MVs from ISS3348 were visualized using negative-stain TEM (G) and staining for M1 protein and STED microscopy (H). White arrowheads indicate vesicle-like structures. Scale bars are drawn to 200 nm (A and B and E to G) or 500 nm (C, D, and H).
As a complementary approach, ISS3348 MVs were visualized by immunofluorescent staining of M1 protein using stimulated emission depletion (STED) microscopy (Fig. 1H). STED microscopic analysis permitted the accurate resolution and measurement of MV particles (Fig. 1H; see Fig. S1D in the supplemental material), with these data supporting previous TEM image analysis (see Fig. S1A). No fluorescent labeling in STED images was noted in the absence of MV samples, and as such, particle measurements were unlikely to result from freely reactive dye molecules (see Fig. S1E to G). Size distribution measurements were further confirmed by dynamic laser light scattering (DLS), with isolated MVs from ISS3348 and SF370 exhibiting single, broadly dispersed populations (see Fig. S1H). Taken together, these data strongly support the notion that GAS actively produces MVs during normal cell growth. As the clinical isolate ISS3348 produced more MVs relative to other strains, we focused primarily on this isolate for further characterization of MV composition.
Proteomic characterization of GAS MVs.
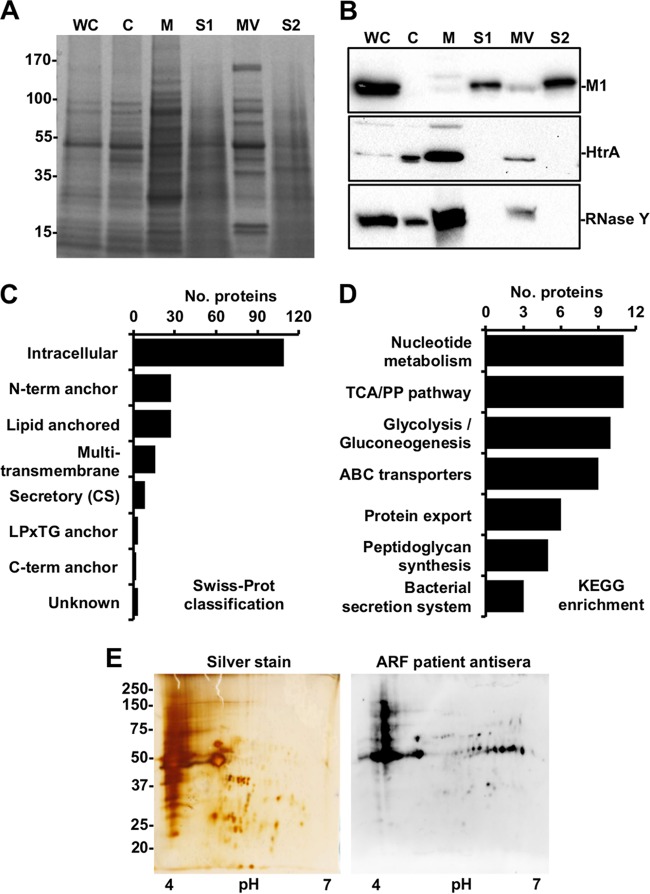
Previous studies of diverse Gram-negative bacteria have indicated that outer membrane proteins are differentially enriched in OMVs relative to the outer membrane (18–20). Whether Gram-positive bacteria, which lack an outer membrane, differentially enrich proteins in MVs relative to the bacterial membrane is unknown. We isolated ISS3348 subcellular fractions and compared total protein profiles by one-dimensional (1D) SDS-PAGE (Fig. 2A). In comparison to the secreted and membrane fractions, MV protein profiles exhibited similarities to and differences from both compartments (Fig. 2A), a phenomenon also observed in protein fractions from SF370 and other M1 GAS strains (see Fig. S2A in the supplemental material). Further complementary proteomic analysis by 2D difference gel electrophoresis (DIGE) and high-resolution nano-liquid chromatography-tandem mass spectrometry (nano-LC-MS/MS) analyses revealed a high degree of complexity and overlap between GAS MVs and membranes (see Fig. S2B). We consistently identified 905 proteins in MVs and 1,027 proteins in membranes, of which 23 proteins were more prominently found in MVs compared to membranes according to our filter criteria and 3 proteins were entirely unique to MVs (see Fig. S2C and Data Set S1 in the supplemental material). Label-free quantification revealed 169 proteins that were >2-fold enriched in MVs relative to membranes (see Table S1 in the supplemental material), resulting in a consensus MV proteome of 195 proteins. The subcellular distribution of a subset of proteins identified in the MV proteome, included M1 protein, serine protease HtrA, and RNase Y, the presence of which was confirmed by immunoblotting (Fig. 2B). Grouping of MV proteins by putative location indicated that more than half were cytoplasmic, with the next largest group being putatively membrane associated (Fig. 2C). Furthermore, we tested for enrichment of MV proteins involved in discrete biological processes, revealing that a number of proteins in seven KEGG categories were statistically overrepresented (P < 0.0001; false discovery rate [FDR], <5%), including the nucleotide metabolism, glycolysis/gluconeogenesis, tricarboxylic acid/pentose phosphate (TCA/PP) pathway, ABC transports, protein export, peptidoglycan synthesis, and bacterial secretion (Fig. 2D). A complement of virulence-associated proteins, including M protein, C5a peptidase (ScpA), and streptolysin O were identified in both MVs and the membrane fraction (see Fig. S2D).
FIG 2 .
GAS MVs carry classically secreted and anchorless virulence proteins. (A) Protein profiles of ISS3348 whole-cell extract (WC), cytoplasm (C), membrane (M), secreted fraction prior to ultracentrifugation (S1), MV, and secreted fraction after centrifugation (S2). (B) Western blot of virulence-associated proteins identified in MVs. The samples in each lane are the same as in panel A. (C) Swiss-Prot classification of nano-LC-MS/MS-identified MV proteins. (D) KEGG categories enriched in MV proteins. (E) Silver-stained 2D protein profile and respective 2D Western blot of ISS3348 MV proteins probed with ARF patient antisera and anti-human IgG-Fc-horseradish peroxidase (HRP).
In comparison with previous studies, we identified MV proteins arising from both the GAS secretome and the cell surface (13, 21) (see Fig. S2E and F and Data Set S1 in the supplemental material). A number of the identified MV proteins have recently been proposed as candidates of a non-M protein-based prophylactic vaccine (22) (see Fig. S2G). To examine the immunogenicity and pathological relevance of MV proteins, we immunoblotted vesicle samples after 2D SDS-PAGE and probed with pooled antisera from patients suffering from acute rheumatic fever (ARF) (Fig. 2E). The presence of multiple immunoreactive IgG epitopes in MV preparations suggests that protein factors present in MVs are expressed during GAS infection in humans.
Lipid components of GAS MVs.
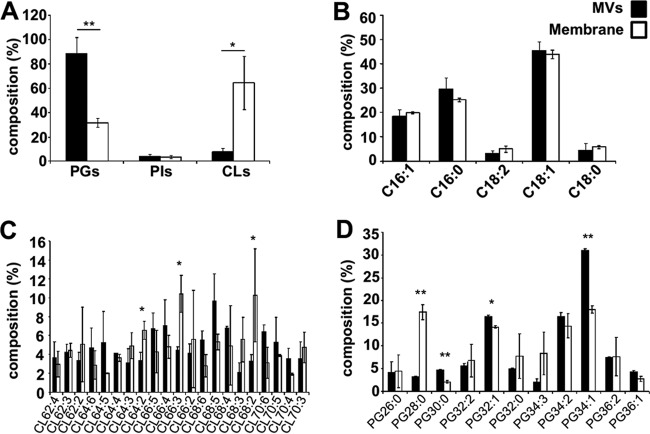
To further characterize GAS MV composition, the lipid components of ISS3348 cell membranes and MVs were analyzed using a previously described matrix-assisted laser desorption ionization (MALDI)-based mass spectrometry approach (see Fig. S3A to C in the supplemental material) (23). More than 85 individual lipid species corresponding to glycoglycerolipids and both anionic and cationic phospholipids (PLs) could be identified. These lipid classes primarily contained 16:1 (palmitoleic), 16:0 (palmitic), 18:1 (oleic/vaccenic), 18:2 (linoleic), and 18:0 (stearic) fatty acids (FAs) (see Table S2 in the supplemental material). Thin-layer chromatography (TLC)-based quantification of membrane and MV lipid species identified anionic phosphatidylglycerol (PG) as being significantly enriched in GAS MVs (Fig. 3A; see Fig. S3D). In contrast, cardiolipin (CL) comprised only a minor fraction of MV lipids relative to the GAS membrane (Fig. 3A). Other lipid classes, including glycoglycerolipids and cationic PLs, were found to be equally distributed between MVs and the GAS membrane (see Fig. S3E and F). Although the overall saturation levels of medium-chain (C16 and C18) FAs were not different between MVs and membranes (Fig. 3B), characteristic differences in the FA distributions of individual CL and PG species (Fig. 3C and D) and other less abundant lipids were observed (see Fig. S3G and H). Differential enrichment of MV lipid species relative to the membrane provides further evidence for the existence of an ordered mechanism contributing to MV biogenesis, as opposed to nonspecific membrane shedding, which would generate MVs with an identical lipid composition.
FIG 3 .
PG is the dominant anionic phospholipid in GAS MVs. (A) Characterization of anionic phospholipids phosphatidylglycerol (PG), phosphatidylinositol (PI), and cardiolipin (CL) in ISS3348 MVs and membranes. (B) General distribution of GAS medium-chain fatty acids (FAs) in MVs and membranes. (C and D) Comparison of GAS MVs and membrane FA saturation level and arrangement in anionic lipids CL and PG. The results presented are means ± standard deviations (SD) from 3 experiments. Asterisks indicate statistical significance by one-way analysis of variance (ANOVA) with Tukey’s post hoc test: *, P < 0.05; **, P < 0.01.
GAS MVs contain differentially abundant RNA species.
Packaging of DNA into OMVs has been noted historically as a mechanism facilitating horizontal transfer of antibiotic resistance genes between Gram-negative bacteria (24–26). Recent studies also report that RNA molecules are present in OMVs in marine ecosystems and in pathogenic Vibrio cholerae OMVs (27–29). We initially examined ISS3348 MVs for binding of fluorescent nucleic acid dyes following rigorous nuclease treatment (see Fig. S4A in the supplemental material). MVs were negative for the membrane-impermeable DNA/RNA dye propidium iodide; however, they were positive for a membrane-permeable RNA-specific dye (see Fig. S4A), indicating that GAS may secrete RNA species protected in the lumen of MVs.
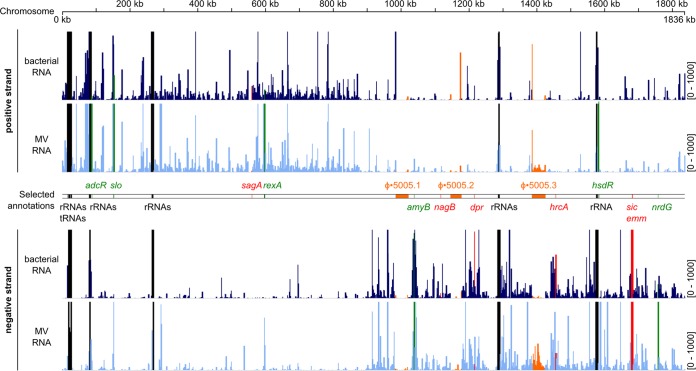
We characterized RNA species present in MVs using deep RNA sequencing (RNA-seq), comparing the abundance of vesicular RNAs relative to bacterial cellular RNAs (Fig. 4). Both bacterial and MV-associated mapped reads were well distributed over the reference genome (Fig. 4). The majority of vesicular and bacterial sequences obtained corresponded to rRNAs and tRNAs, with approximately 14% of MV RNAs sequenced mapping uniquely to loci in the reference genome, many of which were differentially abundant between MVs and bacteria. At a conservative cutoff (≥log2 fold change, 1.5; adjusted P value, <0.01), a total of 207 RNA species were found to be differentially abundant in MVs relative to bacteria (see Table S3 in the supplemental material). Of these, 120 RNAs were more abundant in MVs, with 47 of the 50 most highly differentially abundant RNAs being more enriched in the MV fraction. Highly abundant MV RNAs mapped within the zinc-responsive adhesive competence repressor adcR, anaerobic ribonucleoside triphosphate reductase nrdG, exonuclease-helicase subunit A rexA, and type 1 restriction endonuclease hsdR genes (see Fig. S4B in the supplemental material). Conversely, 87 RNAs were less abundant in the MV fraction, including streptolysin S gene sagA and glucosamine-6-phosphate isomerase gene nagB (see Fig. S4B).
FIG 4 .
RNA species are differentially abundant between GAS cells and MVs. Coverage of reads from MVs and bacterial cellular RNAs mapped to the reference M5005 genome. Chromosomal coordinates are indicated at the top in kilobases. The scale for mapped reads is indicated to the right of the coverage map. tRNA and rRNA loci are indicated in black. The positions of prophage regions are indicated in orange. Selected RNA species more abundant in MVs are highlighted in green, and RNA species less abundant in MVs are highlighted in red. The vesicular and bacterial coverages shown are from a representative sample from biological triplicates.
We further analyzed a number of differentially abundant vesicular RNAs by quantitative PCR (qPCR) (see Fig. S4C in the supplemental material). Seven more abundant (adcR, rexA, nrdG, amyB, salR, artP, and slo) and three less abundant (sagA, nagB, and pfl) vesicular RNA species were quantified by qPCR from triplicate samples isolated independently from those used for transcriptome sequencing. qPCR data obtained demonstrated a strong correlation with the RNA-seq fold change values (Pearson correlation, R = 0.977; P < 0.0001). Both RNA-seq and qPCR analyses strongly indicate that GAS MVs contain RNA species that are differentially abundant from the originating bacteria. Previous studies have shown that Escherichia coli OMVs contain abundant tRNAs and small, noncoding RNA species (29). To our knowledge, this is the first report describing Gram-positive bacterial secretion of differentially enriched intragenic RNAs via extracellular membrane-derived vesicles.
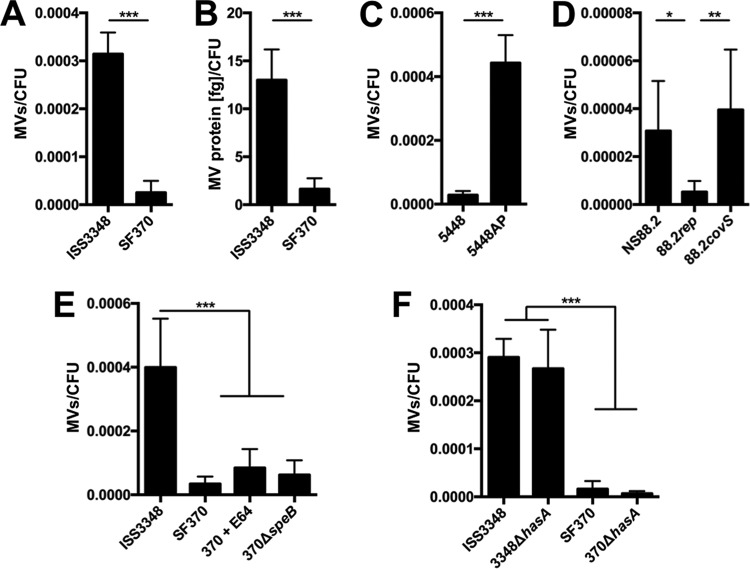
GAS vesiculogenesis is affected by activity of the two-component regulator CovRS.
During our characterization of GAS MV production and composition, we noted a consistently higher quantity of recoverable MVs from ISS3348 culture than from SF370 culture. This difference was significant and reproducible using two different methodologies: (i) flow cytometric counting of MV particles and (ii) total MV protein abundance (Fig. 5A and B) (P < 0.001). Of note, both methodologies demonstrated a high correlation in the quantification of MV production (see Fig. S4D in the supplemental material). We hypothesized that the genomic backgrounds of these strains influenced their level of MV production. ISS3348 has been described to bear inactivating mutations in the two-component “control of virulence regulator-sensor” operon (covRS) (30). This finding was verified by direct sequencing of the ISS3348 covRS operon, which exhibited a large deletion in the covS (sensor) catalytic domain (covSΔ1262–1306 of SF370 sequence). Inactivating mutations in covRS are frequently acquired by M1 strains and result in broad regulatory alterations (31). Accordingly, we found several virulence-associated, covRS-regulated genes (hasA, slo, speB, and grab) differentially expressed in ISS3348 relative to SF370 (see Fig. S4E), suggesting that this mutation disrupts CovS function and may be responsible for the increased virulence of this strain in animal infection models (30).
FIG 5 .
CovRS signaling impacts MV production. Shown is quantification of MV production by GAS strains on the basis of FM1-43 dye staining expressed as MVs per CFU (A and C to F) or total vesicular protein abundance estimated by Bradford protein determination expressed as femtograms of MV protein per CFU (B). Asterisks indicate statistical significance by Student’s unpaired t test (A to C) or one-way ANOVA with Tukey’s post hoc test (D to F): *, P < 0.05; **, P < 0.01; and, ***, P < 0.001. The results presented are means ± SD from 3 to 5 independent experiments, each of which was conducted with 2 technical replicates.
Recent evolutionary tracing of the M1 serotype GAS genomic lineage illustrates that the laboratory reference clinical isolate SF370 differs significantly in both mobile and core genomic compositions from more contemporary M1 isolates, including ISS3348 (32). To distinguish whether the differences in the genomic contents of ISS3348 and SF370 or disruption of covRS signaling influences MV formation, MVs were isolated from M1 GAS strains 5448 and 5448AP. These strains carry identical genomes, except for a single base pair insertion in covS leading to expression of truncated, inactive CovS protein in the 5448AP strain (33). We show that the 5448AP strain produces a significantly higher number of MVs relative to 5448 (Fig. 5C). This finding suggests that genes under control of the CovRS regulator may affect GAS MV production. To examine whether this phenomenon applied exclusively to M1 GAS, we analyzed the virulent covS-deficient M98.1 GAS strain NS88.2, covS-intact derivative 88.2 rep, and complemented strain 88.2 covS (34). Consistent with a model of covRS-mediated regulation of MV production, covS-defective strains NS88.2 and 88.2 covS exhibited significantly higher MV production, albeit with a smaller degree of difference in comparison to M1 covRS mutant strains (Fig. 5D). Altogether, the increased MV production by strains harboring defective covRS loci supports the conclusion that the CovRS two-component system impacts MV-mediated release of proteins, nucleic acids, and lipids.
GAS MV production is independent of SpeB activity and capsule synthesis.
Previous studies demonstrate that extracellular cysteine protease SpeB drastically remodels the GAS secretome in response to covRS mutations (35). Consistent with the strong downregulation of speB by covRS-deficient strain ISS3348 relative to SF370 (see Fig. S4E in the supplemental material), this strain exhibited extracellular cysteine protease activity equivalent to that of an SF370 SpeB deletion strain (370 ΔspeB) and SF370 culture supernatant incubated with cysteine protease inhibitor E64 (see Fig. S4F). To determine whether an absence of extracellular SpeB activity resulted in a larger quantity of recoverable MVs, MVs were isolated from 370 ΔspeB and SF370 grown in the presence of E64 (Fig. 5E). No significant differences in MV production between SF370 and SpeB activity-deficient conditions were observed (Fig. 5E), suggesting that GAS MV production is insensitive to SpeB-mediated degradation of the soluble GAS secretome.
Capsule has previously been suggested to hinder S. pneumoniae MV release (12). In concordance with previous studies (31), mutation of covRS in ISS3348 resulted in upregulation of expression of the hyaluronic acid capsule biosynthesis gene hasA (see Fig. S4E). To assess whether capsule expression impacted GAS MV release, MVs were isolated from capsule-deficient ISS3348 (3348 ΔhasA) and SF370 (370 ΔhasA) mutants (Fig. 5F). The 3348 ΔhasA and 370 ΔhasA strains showed no significant differences in MV production relative to the parental strains (Fig. 5F). Thus, although MV production appears directly or indirectly regulated by CovRS activity, two phenotypically important genes under direct control of covRS, hasA and speB, do not impact GAS vesiculogenesis.
DISCUSSION
Vesicular packaging of biomolecules enables cellular compartmentalization and export of proteins, nucleic acids, and lipids in eukaryotes. This mechanism is also utilized by Gram-negative bacteria and has been the subject of intense study (9). Until recently, MV production by Gram-positive bacteria has been disregarded, due to precluding differences in cell wall structure (16). Here, we present a comprehensive analysis of the protein, nucleic acid, and lipid composition of GAS MVs and provide evidence for a two-component system regulating MV release.
To characterize MV protein cargo and examine potential specific enrichment of vesicular versus membrane proteins, we undertook both 2D electrophoretic and mass spectrometric approaches. Both analyses demonstrated a high overlap in protein content and enrichment of MV proteins relative to the membrane. Similar findings have been reported for outer membrane proteins in Gram-negative bacteria (18–20); however, a global comparative analysis of protein enrichment in Gram-positive MVs has been lacking. Multiple putatively soluble (intracellular and secreted) proteins were identified in the membrane fraction, as previously noted (6, 7). A limitation of this study is that despite identification of putatively soluble proteins in membrane, the true localization of anchorless surface proteins on a global scale is difficult to predict. We calculated enrichment of putatively soluble MV-associated factors relative to the membrane compartment; however, the intracellular concentrations of these factors may lead to differences in enrichment values relative to whole-cell concentrations. Of the enriched MV factors, we identified multiple virulence-associated and metabolic proteins. Mechanistic explanation of how this enrichment occurs should be the focus of further investigation.
GAS protein secretion has been linked to a distinct lipid microdomain, the ExPortal, which is enriched in anionic phospholipids PG and CL and serves as a cue for localization and accumulation of Sec pathway translocons (5). We therefore performed lipidomic analysis to link cellular lipid asymmetry to GAS vesiculogenesis and Sec-independent secretion via ExPortal-like microdomains. Our analysis demonstrates a selective release of PG over CL in MVs compared to whole bacterial membranes. Detailed lipidomic analysis revealed characteristic differences in the compositions and distributions of individual lipid species and differences in the chain lengths/saturation levels of FAs between MVs and the GAS membrane. Based on these results, we hypothesize that enrichment of PG relative to CL, a factor known to dictate membrane curvature (36), and increased monounsaturated FA content in MV lipid classes could determine GAS MV formation and export of cargo.
Why and how MVs containing asymmetrical RNA species relative to the bacteria arise are unclear. It is feasible that bacterial subpopulations with distinct transcriptomic profiles are more prone to produce MVs and contribute to the differential enrichment of MV RNAs relative to the total bacterial population. This hypothesis is supported by recent evidence of a subpopulation of Pseudomonas aeruginosa strains that contribute to MV formation via explosive cell lysis (37). In comparison to previous studies, we found a high degree of intragenic RNA species, as opposed to intergenic or tRNA species, differentially enriched in MVs relative to bacterial cells (29). Transcription factor adcR mRNA was highly enriched in GAS MV fractions and recently was shown to play a central role in GAS adaptive responses to zinc availability (38). Thus, secretion of selected RNAs may allow rapid intercellular communication analogous to secretion of mammalian exosomal shuttle RNA (39).
Most bacterial species express alternative transcription factors (sigma factors [σ]) that direct transcription to distinct gene subsets, enabling adaptation to changing conditions. Multiple reports indicate OMV production is increased in response to defects in the σE pathway, whereby OMV formation enables cells to eject toxic accumulations of misfolded periplasmic proteins (40). This mechanism may also apply in the case of general stress-responsive σB present in Gram-positive bacteria (41). The CovRS system in GAS acts as a global regulator of approximately 15% of the GAS genome, including many known GAS virulence determinants, in response to general stress conditions (42, 43). GAS does not encode recognizable stress response sigma factors; however, CovRS has been hypothesized to act as a functional analogue of these transcription factors (43). The observation that disruption of E. coli EnvZ/OmpR response regulator (to which CovRS is homologous) similarly bestows a hypervesiculation phenotype supports our finding that CovRS signaling impacts MV production in GAS (44). This finding is further emphasized by the recent observation of vesicle-like structures on the GAS cell surface following incubation with CovS ligands LL-37 and RI-10 (14). Collectively these data could indicate a general trend in regulation of MV synthesis through two-component regulators, particularly in the absence of alternative sigma factors.
Our finding that GAS MV production is unaffected by SpeB protease activity is noteworthy, given that this factor effects broad proteolytic degradation of the GAS secretome and is directly regulated by covRS (35). It is also feasible that SpeB-expressing GAS have an altered MV protein composition. Transcriptomic profiles of covRS-defective M1 GAS have previously been examined and implicate a multitude of other regulators and virulence factors as potential contributors to MV production (31). Whether one factor solely contributes or multiple covRS-regulated factors act in concert to influence MV biogenesis is unknown. In an analogous study of Bacillus subtilis extracellular membrane-derived vesicles, it was determined that differences in recoverable MV quantities between an environmental strain and laboratory strain were linked to the secretion of lipopeptide antibiotic surfactin, which destabilized MV integrity (45). It is possible that a covRS-regulated factor acts in a similar manner to either disrupt MV integrity or inhibit MV release. This notion is supported by the morphological evidence of MV formation on the SF370 strain cell surface, despite lower quantifiable MV yield.
During the preparation of the manuscript for this article, Biagini et al. (13) demonstrated that GAS releases high-molecular-weight vesicular structures principally composed of lipoproteins. In agreement with their proteomic analysis, we identified all except two (M5005_Spy0249 and M5005_Spy_1732) abundant vesicular lipoproteins found in reference 13 in our 60 highest-label-free quantification (LFQ)-value MV proteome, as well as 80 out of the 83 vesicular non-lipoproteins. (M5005_Spy0012, M5005_Spy1434, and M5005_Spy0107 were not found in our study.) We furthermore consistently identified 50 of 55 proteins detected in the ISS3348 chemically defined media secretome from reference 13 in our MV preparations (see Fig. S2E in the supplemental material). (M5005_Spy0249, M5005_Spy0996, M5005_Spy0469, M5005_Spy2009, and M5005_Spy1436 were not found in our study.) The MV lipidomic analyses of both our study and the study by Biagini et al. (13) indicate an asymmetrical relationship between the vesicular and overall membrane lipid composition, corroborating our identification of the MV-associated ExPortal microdomain proteins and lipid components. It should be noted that despite weakening the GAS cell wall using sublethal penicillin treatment, Biagini et al. (13) noted no significant changes in either the MV proteome or membrane lipidome.
Collectively, we propose GAS exports differentially enriched RNA species and proteins lacking traditional secretion signal peptides through production of MVs of a characteristic lipid composition. These findings should stimulate further studies of MV biogenesis mechanisms and elucidation of the downstream effects of MV-borne factors on both neighboring bacteria and the human host during GAS infection.
MATERIALS AND METHODS
For detailed methodologies, see the Text S1 in the supplemental material.
Bacterial strains.
GAS strains used in this study (see Table S4 in the supplemental material) were routinely cultured at 37°C in 5% CO2 on tryptone soy agar (TSA) sheep blood agar or in Todd-Hewitt broth (THB) without agitation. Construction of the 370 ΔspeB and 3348 ΔhasA strains is described in detail in Text S1 in the supplemental material.
MV isolation and quantification.
MVs were routinely isolated from late-logarithmic- or early-stationary-phase GAS cultures in THB. Bacteria were pelleted at 2,500 × g, and the supernatant was decanted and filtered twice through 0.22-µm-pore polyethylsulfone membranes prior to ultracentrifugation (175,000 × g, 4 h, 4°C). Pellets obtained after ultracentrifugation were directly quantified or were washed once in phosphate-buffered saline (PBS) and further processed for other analyses (described below). MVs were quantified by Bradford protein determination assay (Sigma) or by incubation with 0.5 µg/ml of lipophilic membrane dye FM1-43 (Life Technologies) for 15 min, prior to flow cytometric counting using Count Bright counting particles (Life Technologies). MV production was normalized to the CFU of each strain. For detection of MV RNA species, MVs and GAS were incubated with Benzonase (see the protocol for vesicular RNA isolation below) and stained with both 0.5 µg/ml of FM4-64 and 1 µM Syto RNAselect dyes (Life Technologies) or with propidium iodide (BioLegend) for 15 min prior to detection by flow cytometry.
AFM and STED microscopic analysis.
Atomic force microscopy (AFM) of bacteria was performed as previously described (46). STED microscopy was conducted by blocking MVs using naive rabbit serum, followed by labeling with mouse anti-M1 antiserum, washing once in PBS, and secondary labeling with goat anti-mouse Alexa Fluor 488 IgG (Dako). Measurements of labeled MVs were taken using a home-built STED microscope setup previously described (47). Data acquisition and analysis was performed using Imspector software (Imspector Image Acquisition and Analysis software v0.10; http://www.imspector.de).
Proteomics.
Delipidized protein samples with disulfide bonds reduced and thiol groups alkylated were trypsinized and further processed for nano-LC-MS/MS using the Q Exactive Orbitrap mass spectrometer (Thermo Fisher). Raw data were analyzed using MaxQuant, with protein localization and function predicted using LocateP and DAVID.
Lipidomic analysis.
Lipids extracted with methanol were profiled using FlexiMass-DS sample plates (Shimadzu). Mass spectra were recorded using an AXIMA-CFRplus (Shimadzu) curved-field reflectron time-of-flight mass spectrometer in positive or negative mode using delayed ion extraction for unit mass resolution.
Vesicular RNA isolation and RNA-seq.
Following MV isolation from early-stationary-phase GAS cultures, samples were treated with Benzonase nuclease (Sigma) for 30 min to degrade all forms of extracellular nucleic acids prior to being washed once with PBS. Vesicular RNA was isolated from biological triplicates using TRI reagent (Sigma), followed by treatment with DNA-free Turbo DNase (Ambion) to remove any contaminating genomic DNA (gDNA). The RNA concentration was assessed using a Bioanalyzer (Agilent) prior to library construction. cDNA libraries were generated without rRNA depletion using the ScriptSeq v2 library preparation kit (Epicentre) and sequenced on an Illumina HiSeq 2500 platform. Short or low-quality reads were filtered and trimmed prior to mapping of trimmed reads to the most closely related reference genome (MGAS5005; GenBank accession no. NC_007297) using the STAR alignment tool (48). Visualization and manual inspection of read coverage were conducted using the Integrative Genomics Viewer (IGV) (49). Differentially abundant RNAs in MV samples and bacteria were computed using the DESeq2 package (50). Prophage-associated genes (φ5005.1, M5005_0995 to -M5005_1054, φ5005.2, M5005_1168 to M5005_1222; and φ5005.3, M5005_1414 to -M5005_1467) were removed from differential expression results.
Accession number(s).
RNA sequencing data have been deposited at NCBI under accession no. SRP089791.
Mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD005167.
SUPPLEMENTAL MATERIAL
Supplemental materials and methods. Download
Detailed proteomic analysis of GAS MVs and membranes by nano-LC-MS/MS. Shown is a comprehensive list of annotated proteins identified by at least 2 peptides in GAS ISS3348 MV or respective membranes in any of the biological triplicates. Download
Biophysical analyses of GAS MVs. (A) Size distribution of isolated ISS3348 MVs as determined by ImageJ-based vesicle diameter measurements taken from negative-stain TEM micrographs (n = 147). Cumulative size frequency is given on the right y axis. (B) Quantification of MV production by ISS3348 and SF370 on the basis of FM1-43 dye staining pre- and post-heat inactivation (HI). MVs were harvested from late-logarithmic-growth-phase culture supernatants, purified, and quantified as described in Materials and Methods in the main text. GAS cultures were subsequently washed twice in PBS and heat inactivated at 80°C for 1 h, prior to inoculation into fresh media for 4 h, with MVs purified and quantified from HI GAS cultures as described in Materials and Methods in the main text. The results presented are pooled data ± SD from two independent experiments. Asterisks indicate statistical significance by one-way ANOVA with Tukey’s post hoc test: ***, P < 0.0001. (C) Treatment of mid-logarithmic-growth-phase ISS3348 and 5448AP cultures with or without 1 mg/ml lysozyme for 2 h prior to MV isolation. Cultures were lysozyme treated prior to MV isolation and SDS-PAGE analysis. (D) Size distribution of FM1-43-labeled ISS3348 MVs as determined by the full-width half-maximum (FWHM) profile of pixel intensities of individual anti-M1-stained MVs using stimulated emission depletion (STED) microscopy. (E and F) STED analysis of isolated ISS3348 MVs stained with mouse anti-M1 and anti-mouse Alexa Fluor 488 (E) or Alexa Fluor 488 alone (no MVs) (F). (G) Confocal microscopy analysis of isolated ISS3348 MVs stained with anti-M1 and anti-mouse Alexa Fluor 488. (H) Size distribution of ISS3348 and SF370 MVs assessed by dynamic laser light scattering analysis. Download
GAS MV and membrane protein profiles. (A) Protein profiles of cytoplasmic (C), membrane (M), and MV (MV) samples from strains 5448, 5448AP, and SF370. (B) Representative 2D DIGE images from Cy5-labeled ISS3348 MVs and respective Cy3-labeled membranes. Individual channels are shown in black and white, and the merged image was created in ImageJ. (C) Venn diagram showing the overlap of proteins identified in ISS3348 MVs and membranes by at least 2 peptides in each biological triplicate using nano-LC-MS/MS. (D) Venn diagram showing the overlap between virulence-associated proteins (http://www.iedb.org and http://www.mgc.ac.cn/VFs), proteins identified in ISS3348 MVs (unique and >2-fold-enriched compared to membranes [MV+M]; see Data Set S1 in the supplemental material), and all identified proteins in membrane and MV analysis (ALL [see Data Set S1]). (E) Venn diagram showing the overlap of recently published (13) secreted and ISS3348 MV proteins with proteins identified in this study. (F) Venn diagram showing the overlap of surface-associated proteins (21) with proteins identified in this study. (G) Venn diagram showing the overlap of immunogenic proteins described by Fritzer et al. (22) and proteins identified in this study. (H) Representative Coomassie-stained ISS3348 MV protein profile and immunoreactive MV protein profile after incubation with ARF patient antiserum. Download
Supporting GAS MV and membrane lipidomic data. (A to C) Representative annotated ionization/mass-spectrometry spectra of different lipid classes showing mono- and diglycoglycerolipids (MG and DG, respectively [A]), anionic phospholipids (PG, CL, and PI [B]), and cationic phospholipids (PC [C]). (D) Representative TLC of lipid extracts from ISS3348 GAS MVs and corresponding bacterial membrane preparations (M). Sample loading was normalized to MV protein abundance from Coomassie staining. (E) Quantitative distribution of mono- and diglycosyldiacylglycerols (MGDG and DGDG, respectively) in GAS MVs and membranes. (F) Quantitative distribution of sphingomyelin (SM) and phosphatidylcholine (PC) in GAS MVs and membranes. (G and H) Acyl chain length and saturation level in MGDG (G) and PC (H). Download
Fluorescence-activated cell sorter (FACS) and qPCR analysis of GAS MV RNA. (A) Analysis of ISS3348 MV nucleic acid content by flow cytometry. MVs were isolated as described for RNA-seq analysis and incubated with propidium iodide (PI), singly with 0.5 µg/ml of the lipophilic dye FM4-64 (Life Technologies) or with FM4-64 together with 1 µM of Syto RNAselect (Life Technologies) in the dark for 15 min, prior to being washed once in PBS and analysis of 30,000 events by flow cytometry. Negative controls consisting of PBS plus nucleic acid dyes alone had minimal fluorescence (not shown). Fluorescence of stained MVs or GAS (blue) is shown relative to that of unstained samples (top and bottom rows) or samples stained singly with FM4-64 (red) (middle row). The data shown are representative of two experiments with similar results. (B) Reads per kilobase of transcript per million mapped reads (RPKM) values for selected differentially abundant RNA species. RNA species more abundant in MVs are shown in the right panel and RNA species more abundant in bacteria in the left panel. The results presented are means ± SD. (C) Correlation of differentially abundant RNA species by RNA-seq and qPCR analyses. The Pearson’s correlation coefficient R, P value, and R2 of the linear regression line are indicated. Differences in RNA abundance determined by the DESeq2 algorithm and qPCR were log transformed and are expressed as fold change in MV RNA relative to bacterial RNA. The results presented are pooled means from independently triplicated experiments. (D) Correlation of FM1-43-based flow cytometric counting and total vesicular protein abundance quantification using Bradford protein determination. MV abundance is expressed as number of FM1-43-positive events per CFU or femtograms of MV protein per CFU. The Pearson’s correlation coefficient R and P value are indicated. (E) Expression of ISS3348 covRS-regulated genes relative to SF370 during the late logarithmic growth phase. The results presented are pooled means ± SD from two independent experiments conducted in triplicate. (F) Cysteine protease activity of GAS supernatants as assessed by azocasein degradation. The results presented are pooled means ± SD from three independent experiments. Download
Proteins exclusively present or enriched in GAS MVs.
Detailed MALDI-MS lipidomic analysis of ISS3348 MVs and membranes.
DESeq2 analysis of GAS MV and bacterial cellular RNAs.
Strains and primers used in this study.
ACKNOWLEDGMENTS
We thank M. Wuczkowski for performing the TLC experiments and O. Belgacem (Kratos Analytical Ltd./Shimadzu Corp.) for providing the MALDI-MS instrumentation used in this study. We are grateful to M. Zellner (Medical University of Vienna) for providing 2D DIGE instrumentation, M. Persson (MIMS) for atomic force microscopy, and I. Schleicher (HZI) for assistance with electron microscopic preparations. We thank Mikael Huss (SciLifeLab) for assistance with RNA sequencing analysis. We thank W. Smith (Newcastle University), G. Teti (University of Messina), M. Walker (University of Queensland), and M. Sanderson-Smith (University of Wollongong) for providing GAS strains. RNA sequencing computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under project no. b20133265.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Statement
Funding was also provided to Emmanuelle Charpentier by the Max Planck Society and by the Louis Jeantet Prize for Medicine given by the Louis-Jeantet Foundation and the Göran Gustafsson Prize given by the Göran Gustafsson Foundation.
Footnotes
Citation Resch U, Tsatsaronis JA, Le Rhun A, Stübiger G, Rohde M, Kasvandik S, Holzmeister S, Tinnefeld P, Nyunt Wai S, Charpentier E. 2016. A two-component regulatory system impacts extracellular membrane-derived vesicle production in group A Streptococcus. mBio 7(6):e00207-16 doi:10.1128/mBio.00207-16.
REFERENCES
- 1.Carapetis JR, Steer AC, Mulholland EK, Weber M. 2005. The global burden of group A streptococcal diseases. Lancet Infect Dis 5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 2.Pfoh E, Wessels MR, Goldmann D, Lee GM. 2008. Burden and economic cost of group A streptococcal pharyngitis. Pediatrics 121:229–234. doi: 10.1542/peds.2007-0484. [DOI] [PubMed] [Google Scholar]
- 3.Walker MJ, Barnett TC, McArthur JD, Cole JN, Gillen CM, Henningham A, Sriprakash KS, Sanderson-Smith ML, Nizet V. 2014. Disease manifestations and pathogenic mechanisms of group A Streptococcus. Clin Microbiol Rev 27:264–301. doi: 10.1128/CMR.00101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsatsaronis JA, Walker MJ, Sanderson-Smith ML. 2014. Host responses to group A Streptococcus: cell death and inflammation. PLoS Pathog 10:e1004266. doi: 10.1371/journal.ppat.1004266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosch J, Caparon M. 2004. A microdomain for protein secretion in Gram-positive bacteria. Science 304:1513–1515. doi: 10.1126/science.1097404. [DOI] [PubMed] [Google Scholar]
- 6.Lei B, Mackie S, Lukomski S, Musser JM. 2000. Identification and immunogenicity of group A Streptococcus culture supernatant proteins. Infect Immun 68:6807–6818. doi: 10.1128/IAI.68.12.6807-6818.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chhatwal GS. 2002. Anchorless adhesins and invasins of Gram-positive bacteria: a new class of virulence factors. Trends Microbiol 10:205–208. doi: 10.1016/S0966-842X(02)02351-X. [DOI] [PubMed] [Google Scholar]
- 8.Kulp A, Kuehn MJ. 2010. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol 64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis TN, Kuehn MJ. 2010. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev 74:81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee E-Y, Choi D-Y, Kim D-K, Kim J-W, Park JO, Kim S, Kim S-H, Desiderio DM, Kim Y-K, Kim K-P, Gho YS. 2009. Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics 9:5425–5436. doi: 10.1002/pmic.200900338. [DOI] [PubMed] [Google Scholar]
- 11.Rivera J, Cordero RJ, Nakouzi AS, Frases S, Nicola A, Casadevall A. 2010. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc Natl Acad Sci U S A 107:19002–19007. doi: 10.1073/pnas.1008843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olaya-Abril A, Prados-Rosales R, McConnell MJ, Martín-Peña R, González-Reyes JA, Jiménez-Munguía I, Gómez-Gascón L, Fernández J, Luque-García JL, García-Lidón C, Estévez H, Pachón J, Obando I, Casadevall A, Pirofski L-A, Rodríguez-Ortega MJ. 2014. Characterization of protective extracellular membrane-derived vesicles produced by Streptococcus pneumoniae. J Proteomics 106:46–60. doi: 10.1016/j.jprot.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 13.Biagini M, Garibaldi M, Aprea S, Pezzicoli A, Doro F, Becherelli M, Taddei AR, Tani C, Tavarini S, Mora M, Teti G, D’Oro U, Nuti S, Soriani M, Margarit I, Rappuoli R, Grandi G, Norais N. 2015. The human pathogen Streptococcus pyogenes releases lipoproteins as lipoprotein-rich membrane vesicles. Mol Cell Proteomics 14:2138–2149. doi: 10.1074/mcp.M114.045880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uhlmann J, Rohde M, Siemens N, Kreikemeyer B, Bergman P, Johansson L, Norrby-Teglund A. 2016. LL-37 triggers formation of Streptococcus pyogenes extracellular vesicle-like structures with immune stimulatory properties. J Innate Immun 8:243–257. doi: 10.1159/000441896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prados-Rosales R, Baena A, Martinez LR, Luque-Garcia J, Kalscheuer R, Veeraraghavan U, Camara C, Nosanchuk JD, Besra GS, Chen B, Jimenez J, Glatman-Freedman A, Jacobs WR, Porcelli SA, Casadevall A. 2011. Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J Clin Invest 121:1471–1483. doi: 10.1172/JCI44261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown L, Wolf JM, Prados-Rosales R, Casadevall A. 2015. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat Rev Microbiol 13:620–630. doi: 10.1038/nrmicro3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metruccio MM, Evans DJ, Gabriel MM, Kadurugamuwa JL, Fleiszig SM. 2016. Pseudomonas aeruginosa outer membrane vesicles triggered by human mucosal fluid and lysozyme can prime host tissue surfaces for bacterial adhesion. Front Microbiol 7:871. doi: 10.3389/fmicb.2016.00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lappann M, Otto A, Becher D, Vogel U. 2013. Comparative proteome analysis of spontaneous outer membrane vesicles and purified outer membranes of Neisseria meningitidis. J Bacteriol 195:4425–4435. doi: 10.1128/JB.00625-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reales-Calderón JA, Corona F, Monteoliva L, Gil C, Martínez JL. 2015. Quantitative proteomics unravels that the post-transcriptional regulator Crc modulates the generation of vesicles and secreted virulence determinants of Pseudomonas aeruginosa. J Proteomics 127:352–364. doi: 10.1016/j.jprot.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 20.Veith PD, Chen Y-Y, Gorasia DG, Chen D, Glew MD, O’Brien-Simpson NM, Cecil JD, Holden JA, Reynolds EC. 2014. Porphyromonas gingivalis outer membrane vesicles exclusively contain outer membrane and periplasmic proteins and carry a cargo enriched with virulence factors. J Proteome Res 13:2420–2432. doi: 10.1021/pr401227e. [DOI] [PubMed] [Google Scholar]
- 21.Severin A, Nickbarg E, Wooters J, Quazi SA, Matsuka YV, Murphy E, Moutsatsos IK, Zagursky RJ, Olmsted SB. 2007. Proteomic analysis and identification of Streptococcus pyogenes surface-associated proteins. J Bacteriol 189:1514–1522. doi: 10.1128/JB.01132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fritzer A, Senn BM, Minh DB, Hanner M, Gelbmann D, Noiges B, Henics T, Schulze K, Guzman CA, Goodacre J, von Gabain A, Nagy E, Meinke AL. 2010. Novel conserved group A streptococcal proteins identified by the antigenome technology as vaccine candidates for a non-M protein-based vaccine. Infect Immun 78:4051–4067. doi: 10.1128/IAI.00295-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stübiger G, Aldover-Macasaet E, Bicker W, Sobal G, Willfort-Ehringer A, Pock K, Bochkov V, Widhalm K, Belgacem O. 2012. Targeted profiling of atherogenic phospholipids in human plasma and lipoproteins of hyperlipidemic patients using MALDI-QIT-TOF-MS/MS. Atherosclerosis 224:177–186. doi: 10.1016/j.atherosclerosis.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 24.Dorward DW, Garon CF, Judd RC. 1989. Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae. J Bacteriol 171:2499–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fulsundar S, Harms K, Flaten GE, Johnsen PJ, Chopade BA, Nielsen KM. 2014. Gene transfer potential of outer membrane vesicles of Acinetobacter baylyi and effects of stress on vesiculation. Appl Environ Microbiol 80:3469–3483. doi: 10.1128/AEM.04248-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rumbo C, Fernández-Moreira E, Merino M, Poza M, Mendez JA, Soares NC, Mosquera A, Chaves F, Bou G. 2011. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob Agents Chemother 55:3084–3090. doi: 10.1128/AAC.00929-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biller SJ, Schubotz F, Roggensack SE, Thompson AW, Summons RE, Chisholm SW. 2014. Bacterial vesicles in marine ecosystems. Science 343:183–186. doi: 10.1126/science.1243457. [DOI] [PubMed] [Google Scholar]
- 28.Sjöström AE, Sandblad L, Uhlin BE, Wai SN. 2015. Membrane vesicle-mediated release of bacterial RNA. Sci Rep 5:15329. doi: 10.1038/srep15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghosal A, Upadhyaya BB, Fritz JV, Heintz-Buschart A, Desai MS, Yusuf D, Huang D, Baumuratov A, Wang K, Galas D, Wilmes P. 2015. The extracellular RNA complement of Escherichia coli. Microbiol Open 4:252–266. doi: 10.1002/mbo3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiappini N, Seubert A, Telford JL, Grandi G, Serruto D, Margarit I, Janulczyk R. 2012. Streptococcus pyogenes SpyCEP influences host-pathogen interactions during infection in a murine air pouch model. PLoS One 7:e40411. doi: 10.1371/journal.pone.0040411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. 2006. Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog 2:e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nasser W, Beres SB, Olsen RJ, Dean MA, Rice KA, Long SW, Kristinsson KG, Gottfredsson M, Vuopio J, Raisanen K, Caugant DA, Steinbakk M, Low DE, McGeer A, Darenberg J, Henriques-Normark B, Van Beneden CA, Hoffmann S, Musser JM. 2014. Evolutionary pathway to increased virulence and epidemic group A Streptococcus disease derived from 3,615 genome sequences. Proc Natl Acad Sci U S A 111:E1768–E1776. doi: 10.1073/pnas.1403138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker MJ, Hollands A, Sanderson-Smith ML, Cole JN, Kirk JK, Henningham A, McArthur JD, Dinkla K, Aziz RK, Kansal RG, Simpson AJ, Buchanan JT, Chhatwal GS, Kotb M, Nizet V. 2007. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat Med 13:981–985. doi: 10.1038/nm1612. [DOI] [PubMed] [Google Scholar]
- 34.Maamary PG, Sanderson-Smith ML, Aziz RK, Hollands A, Cole JN, McKay FC, McArthur JD, Kirk JK, Cork AJ, Keefe RJ, Kansal RG, Sun H, Taylor WL, Chhatwal GS, Ginsburg D, Nizet V, Kotb M, Walker MJ. 2010. Parameters governing invasive disease propensity of non-M1 serotype group A streptococci. J Innate Immun 2:596–606. doi: 10.1159/000317640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aziz RK, Pabst MJ, Jeng A, Kansal R, Low DE, Nizet V, Kotb M. 2004. Invasive M1T1 group A Streptococcus undergoes a phase-shift in vivo to prevent proteolytic degradation of multiple virulence factors by SpeB. Mol Microbiol 51:123–134. doi: 10.1046/j.1365-2958.2003.03797.x. [DOI] [PubMed] [Google Scholar]
- 36.Barák I, Muchová K. 2013. The role of lipid domains in bacterial cell processes. Int J Mol Sci 14:4050–4065. doi: 10.3390/ijms14024050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turnbull L, Toyofuku M, Hynen AL, Kurosawa M, Pessi G, Petty NK, Osvath SR, Carcamo-Oyarce G, Gloag ES, Shimoni R, Omasits U, Ito S, Yap X, Monahan LG, Cavaliere R, Ahrens CH, Charles IG, Nomura N, Eberl L, Whitchurch CB. 2016. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat Commun 7:11220. doi: 10.1038/ncomms11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanson M, Makthal N, Flores AR, Olsen RJ, Musser JM, Kumaraswami M. 2015. Adhesin competence repressor (AdcR) from Streptococcus pyogenes controls adaptive responses to zinc limitation and contributes to virulence. Nucleic Acids Res 43:418–432. doi: 10.1093/nar/gku1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. 2007. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 40.McBroom AJ, Kuehn MJ. 2007. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol 63:545–558. doi: 10.1111/j.1365-2958.2006.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JH, Choi C-W, Lee T, Kim SI, Lee J-C, Shin J-H. 2013. Transcription factor σB plays an important role in the production of extracellular membrane-derived vesicles in Listeria monocytogenes. PLoS One 8:e73196. doi: 10.1371/journal.pone.0073196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graham MR, Smoot LM, Migliaccio CA, Virtaneva K, Sturdevant DE, Porcella SF, Federle MJ, Adams GJ, Scott JR, Musser JM. 2002. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc Natl Acad Sci U S A 99:13855–13860. doi: 10.1073/pnas.202353699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dalton TL, Scott JR. 2004. CovS inactivates CovR and is required for growth under conditions of general stress in Streptococcus pyogenes. J Bacteriol 186:3928–3937. doi: 10.1128/JB.186.12.3928-3937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McBroom AJ, Johnson AP, Vemulapalli S, Kuehn MJ. 2006. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J Bacteriol 188:5385–5392. doi: 10.1128/JB.00498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown L, Kessler A, Cabezas-Sanchez P, Luque-Garcia JL, Casadevall A. 2014. Extracellular vesicles produced by the Gram-positive bacterium Bacillus subtilis are disrupted by the lipopeptide surfactin. Mol Microbiol 93:183–198. doi: 10.1111/mmi.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kouokam JC, Wai SN, Fällman M, Dobrindt U, Hacker J, Uhlin BE. 2006. Active cytotoxic necrotizing factor 1 associated with outer membrane vesicles from uropathogenic Escherichia coli. Infect Immun 74:2022–2030. doi: 10.1128/IAI.74.4.2022-2030.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beater S, Holzmeister P, Pibiri E, Lalkens B, Tinnefeld P. 2014. Choosing dyes for cw-STED nanoscopy using self-assembled nanorulers. Phys Chem Chem Phys 16:6990–6996. doi: 10.1039/c4cp00127c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods. Download
Detailed proteomic analysis of GAS MVs and membranes by nano-LC-MS/MS. Shown is a comprehensive list of annotated proteins identified by at least 2 peptides in GAS ISS3348 MV or respective membranes in any of the biological triplicates. Download
Biophysical analyses of GAS MVs. (A) Size distribution of isolated ISS3348 MVs as determined by ImageJ-based vesicle diameter measurements taken from negative-stain TEM micrographs (n = 147). Cumulative size frequency is given on the right y axis. (B) Quantification of MV production by ISS3348 and SF370 on the basis of FM1-43 dye staining pre- and post-heat inactivation (HI). MVs were harvested from late-logarithmic-growth-phase culture supernatants, purified, and quantified as described in Materials and Methods in the main text. GAS cultures were subsequently washed twice in PBS and heat inactivated at 80°C for 1 h, prior to inoculation into fresh media for 4 h, with MVs purified and quantified from HI GAS cultures as described in Materials and Methods in the main text. The results presented are pooled data ± SD from two independent experiments. Asterisks indicate statistical significance by one-way ANOVA with Tukey’s post hoc test: ***, P < 0.0001. (C) Treatment of mid-logarithmic-growth-phase ISS3348 and 5448AP cultures with or without 1 mg/ml lysozyme for 2 h prior to MV isolation. Cultures were lysozyme treated prior to MV isolation and SDS-PAGE analysis. (D) Size distribution of FM1-43-labeled ISS3348 MVs as determined by the full-width half-maximum (FWHM) profile of pixel intensities of individual anti-M1-stained MVs using stimulated emission depletion (STED) microscopy. (E and F) STED analysis of isolated ISS3348 MVs stained with mouse anti-M1 and anti-mouse Alexa Fluor 488 (E) or Alexa Fluor 488 alone (no MVs) (F). (G) Confocal microscopy analysis of isolated ISS3348 MVs stained with anti-M1 and anti-mouse Alexa Fluor 488. (H) Size distribution of ISS3348 and SF370 MVs assessed by dynamic laser light scattering analysis. Download
GAS MV and membrane protein profiles. (A) Protein profiles of cytoplasmic (C), membrane (M), and MV (MV) samples from strains 5448, 5448AP, and SF370. (B) Representative 2D DIGE images from Cy5-labeled ISS3348 MVs and respective Cy3-labeled membranes. Individual channels are shown in black and white, and the merged image was created in ImageJ. (C) Venn diagram showing the overlap of proteins identified in ISS3348 MVs and membranes by at least 2 peptides in each biological triplicate using nano-LC-MS/MS. (D) Venn diagram showing the overlap between virulence-associated proteins (http://www.iedb.org and http://www.mgc.ac.cn/VFs), proteins identified in ISS3348 MVs (unique and >2-fold-enriched compared to membranes [MV+M]; see Data Set S1 in the supplemental material), and all identified proteins in membrane and MV analysis (ALL [see Data Set S1]). (E) Venn diagram showing the overlap of recently published (13) secreted and ISS3348 MV proteins with proteins identified in this study. (F) Venn diagram showing the overlap of surface-associated proteins (21) with proteins identified in this study. (G) Venn diagram showing the overlap of immunogenic proteins described by Fritzer et al. (22) and proteins identified in this study. (H) Representative Coomassie-stained ISS3348 MV protein profile and immunoreactive MV protein profile after incubation with ARF patient antiserum. Download
Supporting GAS MV and membrane lipidomic data. (A to C) Representative annotated ionization/mass-spectrometry spectra of different lipid classes showing mono- and diglycoglycerolipids (MG and DG, respectively [A]), anionic phospholipids (PG, CL, and PI [B]), and cationic phospholipids (PC [C]). (D) Representative TLC of lipid extracts from ISS3348 GAS MVs and corresponding bacterial membrane preparations (M). Sample loading was normalized to MV protein abundance from Coomassie staining. (E) Quantitative distribution of mono- and diglycosyldiacylglycerols (MGDG and DGDG, respectively) in GAS MVs and membranes. (F) Quantitative distribution of sphingomyelin (SM) and phosphatidylcholine (PC) in GAS MVs and membranes. (G and H) Acyl chain length and saturation level in MGDG (G) and PC (H). Download
Fluorescence-activated cell sorter (FACS) and qPCR analysis of GAS MV RNA. (A) Analysis of ISS3348 MV nucleic acid content by flow cytometry. MVs were isolated as described for RNA-seq analysis and incubated with propidium iodide (PI), singly with 0.5 µg/ml of the lipophilic dye FM4-64 (Life Technologies) or with FM4-64 together with 1 µM of Syto RNAselect (Life Technologies) in the dark for 15 min, prior to being washed once in PBS and analysis of 30,000 events by flow cytometry. Negative controls consisting of PBS plus nucleic acid dyes alone had minimal fluorescence (not shown). Fluorescence of stained MVs or GAS (blue) is shown relative to that of unstained samples (top and bottom rows) or samples stained singly with FM4-64 (red) (middle row). The data shown are representative of two experiments with similar results. (B) Reads per kilobase of transcript per million mapped reads (RPKM) values for selected differentially abundant RNA species. RNA species more abundant in MVs are shown in the right panel and RNA species more abundant in bacteria in the left panel. The results presented are means ± SD. (C) Correlation of differentially abundant RNA species by RNA-seq and qPCR analyses. The Pearson’s correlation coefficient R, P value, and R2 of the linear regression line are indicated. Differences in RNA abundance determined by the DESeq2 algorithm and qPCR were log transformed and are expressed as fold change in MV RNA relative to bacterial RNA. The results presented are pooled means from independently triplicated experiments. (D) Correlation of FM1-43-based flow cytometric counting and total vesicular protein abundance quantification using Bradford protein determination. MV abundance is expressed as number of FM1-43-positive events per CFU or femtograms of MV protein per CFU. The Pearson’s correlation coefficient R and P value are indicated. (E) Expression of ISS3348 covRS-regulated genes relative to SF370 during the late logarithmic growth phase. The results presented are pooled means ± SD from two independent experiments conducted in triplicate. (F) Cysteine protease activity of GAS supernatants as assessed by azocasein degradation. The results presented are pooled means ± SD from three independent experiments. Download
Proteins exclusively present or enriched in GAS MVs.
Detailed MALDI-MS lipidomic analysis of ISS3348 MVs and membranes.
DESeq2 analysis of GAS MV and bacterial cellular RNAs.
Strains and primers used in this study.