ABSTRACT
Social bees collect carbohydrate-rich food to support their colonies, and yet, certain carbohydrates present in their diet or produced through the breakdown of pollen are toxic to bees. The gut microbiota of social bees is dominated by a few core bacterial species, including the Gram-negative species Gilliamella apicola. We isolated 42 strains of G. apicola from guts of honey bees and bumble bees and sequenced their genomes. All of the G. apicola strains share high 16S rRNA gene similarity, but they vary extensively in gene repertoires related to carbohydrate metabolism. Predicted abilities to utilize different sugars were verified experimentally. Some strains can utilize mannose, arabinose, xylose, or rhamnose (monosaccharides that can cause toxicity in bees) as their sole carbon and energy source. All of the G. apicola strains possess a manO-associated mannose family phosphotransferase system; phylogenetic analyses suggest that this was acquired from Firmicutes through horizontal gene transfer. The metabolism of mannose is specifically dependent on the presence of mannose-6-phosphate isomerase (MPI). Neither growth rates nor the utilization of glucose and fructose are affected in the presence of mannose when the gene encoding MPI is absent from the genome, suggesting that mannose is not taken up by G. apicola strains which harbor the phosphotransferase system but do not encode the MPI. Given their ability to simultaneously utilize glucose, fructose, and mannose, as well as the ability of many strains to break down other potentially toxic carbohydrates, G. apicola bacteria may have key roles in improving dietary tolerances and maintaining the health of their bee hosts.
IMPORTANCE
Bees are important pollinators of agricultural plants. Our study documents the ability of Gilliamella apicola, a dominant gut bacterium in honey bees and bumble bees, to utilize several sugars that are harmful to bee hosts. Using genome sequencing and growth assays, we found that the ability to metabolize certain toxic carbohydrates is directly correlated with the presence of their respective degradation pathways, indicating that metabolic potential can be accurately predicted from genomic data in these gut symbionts. Strains vary considerably in their range of utilizable carbohydrates, which likely reflects historical horizontal gene transfer and gene deletion events. Unlike their bee hosts, G. apicola bacteria are not detrimentally affected by growth on mannose-containing medium, even in strains that cannot metabolize this sugar. These results suggest that G. apicola may be an important player in modulating nutrition in the bee gut, with ultimate effects on host health.
INTRODUCTION
Gut bacteria possess a large repertoire of metabolic capabilities, and they play an important role in the fermentation of complex dietary carbohydrates in the intestines of insects, herbivorous vertebrates, and humans (1). Since animal hosts lack the enzymes to degrade most types of carbohydrates (2), these recalcitrant nutrients are often left to be digested by the intestinal microbiota, which, in turn, release fermentation products (i.e., short-chain fatty acids) that can be highly beneficial for host energy metabolism (3). For honey bees and bumble bees, foraging workers collect nectar for the carbohydrate needs of the bee colony, as well as pollen, which contains various carbohydrates, proteins, lipids, and other micronutrients that are critical for bee development and reproduction (4, 5). However, these food sources are not completely innocuous, as they may contain xenobiotics that are produced by plants or introduced during beekeeping practices (6).
The impact of carbohydrates on bee survival has been studied for nearly a century (7, 8), and it is well established that bees live longest on syrup containing sucrose, glucose, or fructose (9). Other sugars from natural nectar can exhibit strong toxicity, including the monosaccharides mannose (10), xylose, arabinose, and rhamnose, as well as some oligosaccharides; these can reduce the life span of adult bees at concentrations as low as 2% (11). Moreover, it has been shown that, during the process of pollen breakdown, hydrolysis of pectin produces a mixture of sugars that are toxic to bees (12).
Both honey bees (genus Apis) and bumble bees (genus Bombus) harbor characteristic gut microbial communities that are unique to social bees (13). The most predominant fermentative bacteria in the gut of bees are the Gram-negative species Gilliamella apicola (class Gammaproteobacteria, order Orbales) and the Gram-positive Lactobacillus species phylotypes Firm-4 and Firm-5 (14). It is believed that the gut microbiota play essential roles in honey bee and bumble bee health, with potential effects on pathogen defense and nutrient acquisition (15). G. apicola strains have complete glycolysis pathways in their genomes, as well as numerous genes encoding phosphotransferase systems (PTSs) (16), thus implicating their function as saccharolytic fermenters that participate in the digestion of the host’s carbohydrate-rich diet. In a metagenomic analysis, Engel et al. (17) identified genes encoding pectin-degrading enzymes in G. apicola that may help in the breakdown of the rigid polysaccharide walls of pollen grains and in the release of constituent monosaccharides. However, it remains unclear whether the gut bacteria can utilize these monosaccharides, some of which are toxic to the bee host.
In this study, we isolated 42 G. apicola strains from the guts of both honey bees and bumble bees and sequenced their genomes to assess their gene repertoires related to sugar utilization. The metabolism of specific carbohydrates was confirmed physiologically, and the ability to simultaneously utilize different sugars was determined. We analyzed genes for carbohydrate utilization in a phylogenetic framework and discovered that G. apicola has likely taken up relevant genes from species of Firmicutes. There was also substantial variation in sugar degradation capabilities among strains, suggesting niche differentiation or specialization to particular host species.
RESULTS
Whole-genome-based phylogeny.
We isolated 15 G. apicola strains from the gut homogenates of various honey bee species and 27 from bumble bee species, and the genomes of these isolates were then sequenced using the Illumina MiSeq platform (see Table S1 in the supplemental material). A complete genome of strain wkB7 was obtained using both the Illumina MiSeq and PacBio RS II platforms (see Materials and Methods). The G. apicola genomes range from 2.1 to 3.1 Mb in size, and the 16S rRNA gene sequences show high levels of similarity among all strains (overall mean similarity of 98.5%) (see Table S1). Nonetheless, the strains show considerable divergence at other loci, and the mean pairwise average nucleotide identity between orthologous genomic regions is only 83.0% (range, 77.9 to 99.9%).
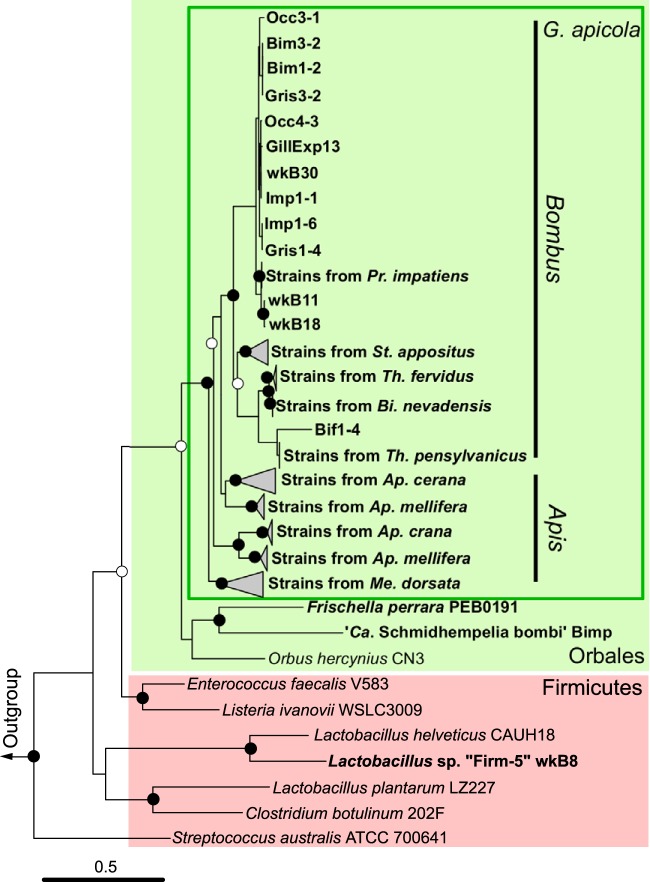
The genome sequences of these 42 G. apicola strains were then analyzed along with 6 previously published G. apicola genomes (16, 18). We identified a set of 225 orthologous genes present in all genomes. Phylogenetic analysis based on these shared orthologs revealed that the genomes from bumble bees clustered exclusively from those from honey bees (Fig. 1). The bumble bee-derived G. apicola strains appear to be monophyletic, while honey bee-derived strains were paraphyletic. Although isolates from the same honey bee species tend to cluster together, strains from Apis dorsata and Apis cerana are sisters to other G. apicola strains in the tree, supporting the occurrence of some host switching rather than strict host-microbe codiversification. Notably, the strains from the same subgenera of bumble bees are more closely related in the tree, and the strains from Pyrobombus, Cullumanobombus, and Bombus formed a separate cluster corresponding to a cluster within the phylogeny based on host DNA sequences (19).
FIG 1 .
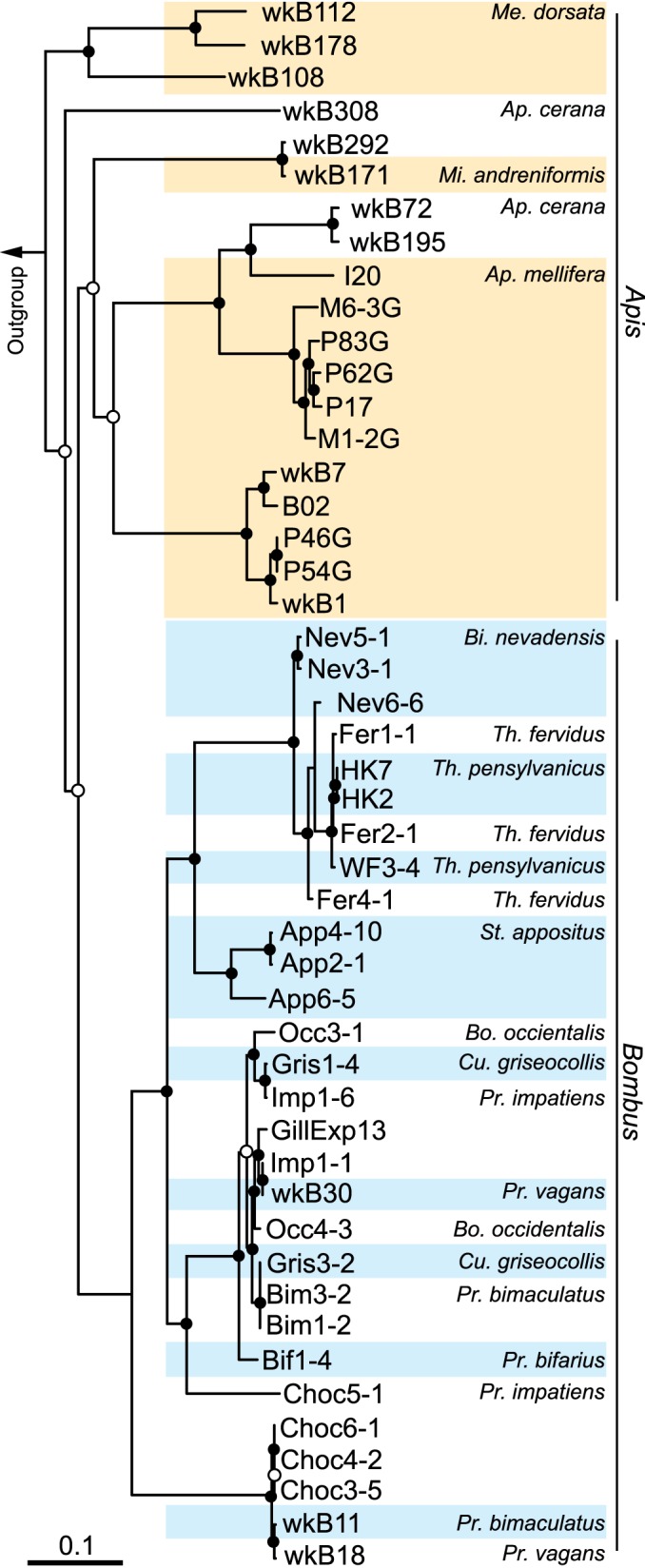
Phylogeny of 48 strains of Gilliamella apicola from guts of honey bees and bumble bees. Six previously published genomes (16, 18) and 42 newly sequenced genomes were included in the analysis. The tree was built using the maximum-likelihood algorithm based on the concatenated sequences of 225 single-copy genes (22,315 nucleotide positions) and rooted with sequences from Frischella perrara (51) and “Candidatus Schmidhempelia bombi” (52). Circles indicate node bootstrap support (○, >85%; ●, 100%, 1,000 replicates). Scale bar indicates 0.1 nucleotide substitutions per site. The subgenera of honey bee and bumble bee hosts are as follows: Me, Megapis; Mi, Micrapis; Ap, Apis; Bi, Bombias; Th, Thoracobombus; St, Subterraneobombus; Pr, Pyrobombus; Cu, Cullumanobombus; Bo, Bombus.
Evolution of mannose metabolism-related genes.
To investigate the ability of G. apicola to digest mannose, we identified the genes related to mannose metabolism in the genomes. In bacteria, the utilization of mannose typically requires two components: a PTS to take up extracellular mannose and mannose-6-phosphate (mannose-6-P) isomerase (MPI) to enable its catabolism via glycolysis pathways (20). The PTS is usually composed of enzyme I and HPr, which are general for all carbohydrates, and the substrate-specific enzyme II (EII) (21). EII complexes typically consist of three protein domains (IIA to -C), while the EII of the mannose PTS family is unique in having an additional IID domain (22).
We identified 11 EII complexes that have IID domains in the genomes of G. apicola strains (see Fig. S1 in the supplemental material); one of these is composed of a fused IIA/B domain and a downstream manO gene and is the only EII complex present in all G. apicola strains. The manO gene is a putative regulator of the mannose EII operon and, interestingly, has only been found in Firmicutes until now (23). Phylogenetic analysis of the manO-associated mannose EII operon revealed that it is also present in relatives of G. apicola within the order Orbales (Fig. 2). All other closely related sequences were from Firmicutes, suggesting that this PTS system, together with manO, was horizontally transferred from a Gram-positive bacterium to the common ancestor of the Gram-negative Orbales clade.
FIG 2 .
Maximum-likelihood phylogeny of the manO-associated mannose family PTS from G. apicola strains. The tree is based on the concatenated nucleotide sequences of three enzyme II genes (encoding EIIA/-B, -C, and -D). The tree was rooted with sequences from Escherichia coli and Klebsiella pneumoniae. Sequences from strains isolated from bee guts are shown in boldface. Sequences from the same host species are clustered in clades. Circles indicate node bootstrap support (○, >70%; ●, >95%). Scale bar indicates 0.5 nucleotide substitutions per site.
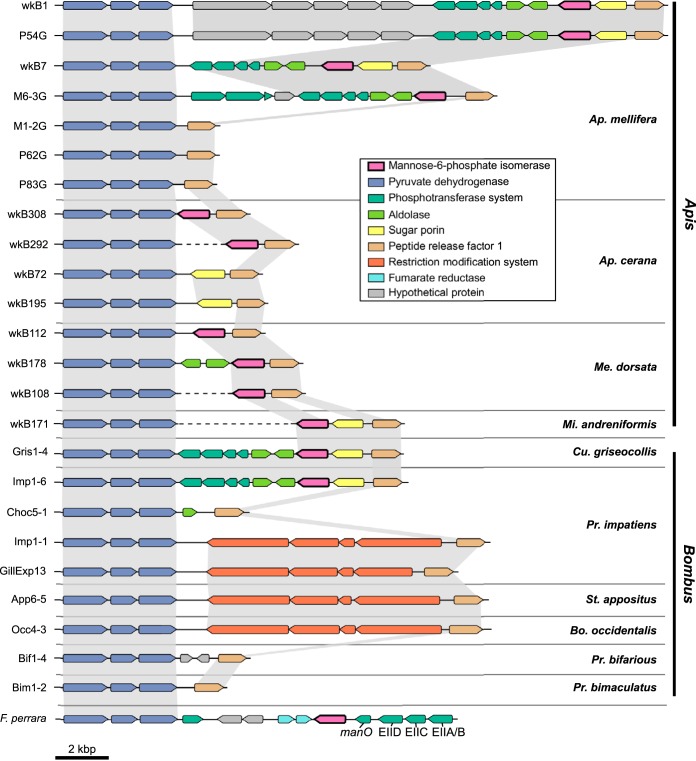
In contrast, the MPI gene (manA) was only present in 10 strains isolated from honey bees and two strains from bumble bees (Fig. 3). In the type strain wkB1, isolated from Apis mellifera (16), manA is flanked by genes encoding pyruvate dehydrogenase (PDH), PTS EIIA to -D, aldolase, a sugar porin, and peptide release factor 1 (RF1), together with several hypothetical genes (Fig. 3). In 23 of the newly sequenced genomes, the PDH and RF1 genes were detected in the same contig and seem to bracket a variable region that, in some genomes, contains manA. In four strains from bumble bees (strains Imp1-6, GillExp13, App6-5, and Occ4-3), restriction-modification system genes occupy this variable region instead of the typical genes. In the genomes of strains wkB292, wkB108, and wkB171, manA is also found next to the RF1 gene; however, the corresponding PDH genes are located on different contigs (due to incomplete genomic assembly). Intriguingly, the manA gene is also present in Frischella perrara, another core gut bacterium isolated from A. mellifera (24), where it appears to have become genetically linked with the manO-associated EII complex (Fig. 3).
FIG 3 .
The variable region containing the mannose-6-phosphate isomerase gene (manA) among G. apicola isolates from honey bees (Apis) and bumble bees (Bombus), and F. perrara. The variable region extends between the genes for pyruvate dehydrogenase and peptide release factor 1. Gray shading indicates orthologous regions. Dashed lines indicate that genes are present on different contigs. The subgenera of bee hosts are as listed in the legend to Fig. 1.
To investigate the origins of G. apicola MPI, the phylogenetic relationships of manA were reconstructed (Fig. 4). We found that sequences from G. apicola formed a single clade that clustered most closely with sequences from other Orbales (F. perrara and Orbus hercynius) and are more distantly related to sequences from other gammaproteobacteria (Fig. 4). The only two sequences obtained from bumble bee species (Bombus griseocollis and Bombus impatiens) form a monophyletic group that is nested within the larger clade representing strains from honey bees. Phylogenies of the neighboring genes for PDH and RF1 also show nesting of sequences from bumble bee-derived isolates within those of honey bee-derived isolates (see Fig. S2 in the supplemental material); the same phylogenetic relationship is evident from the whole-genome tree (Fig. 1). Altogether, these results indicate that the PDH, RF1, and MPI genes were present in the common ancestor of G. apicola strains and that the absence of manA in certain strains is due to gene loss during evolution.
FIG 4 .
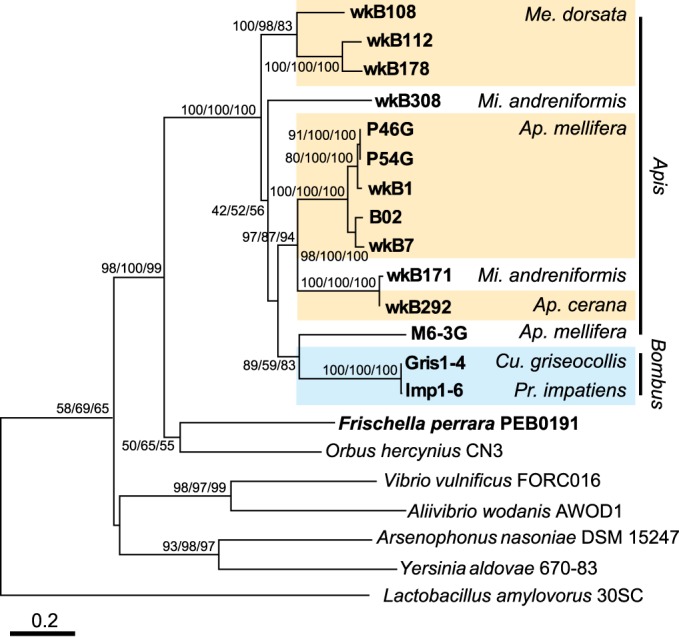
Phylogeny of the mannose-6-phosphate isomerase gene (manA). The tree is inferred from neighbor-joining/maximum-parsimony/maximum-likelihood methods based on nucleotide sequences and was rooted with the sequence from Lactobacillus amylovorus. Sequences from strains isolated from bee guts are shown in boldface. The numbers at nodes represent bootstrap confidence values (%) for each phylogeny-testing method. Scale bar indicates 0.2 nucleotide substitutions per site.
Growth ability on mannose, xylose, arabinose, and rhamnose.
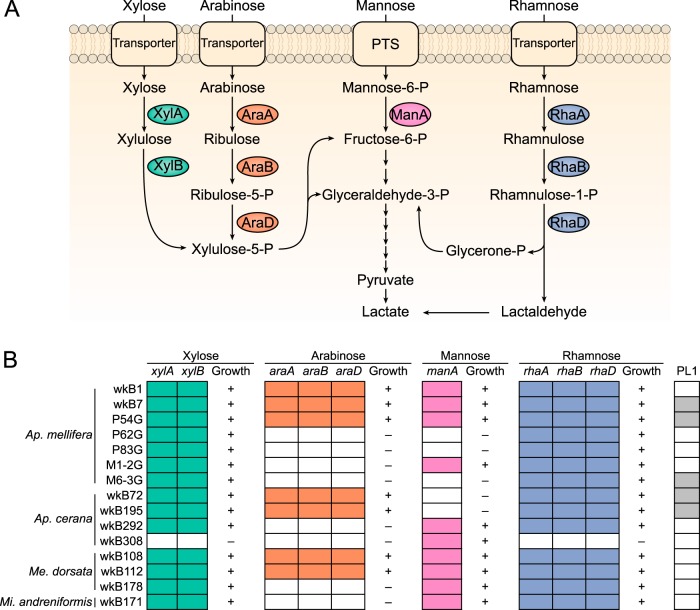
In addition to mannose, other polysaccharide hydrolysates can be toxic to bee hosts (9). We screened for the genes encoding enzymes related to the catabolism of xylose (xylA [xylose isomerase] and xylB [xylose kinase]), arabinose (araA [arabinose isomerase], araB [ribulokinase], and araD [l-ribulose-5-P 4-epimerase]), and rhamnose (rhaA [rhamnose isomerase], rhaB [rhamnulose kinase], and rhaD [rhamnulose-1-phosphate aldolase]) (Fig. 5A), and we tested the ability of the G. apicola isolates to grow on different sugars.
FIG 5 .
Variability in the presence of genes for sugar utilization among G. apicola strains. (A) Mannose, rhamnose, arabinose, and xylose metabolism pathways and related genes in G. apicola. (B) Presence of the genes related to the metabolism of the four sugars and the pectate lyase gene (PL1) in G. apicola strains isolated from honey bee guts. Colored boxes indicate gene presence, and white boxes indicate gene absence. The growth ability on the substrate is shown by plus (growth) and minus (no growth) signs.
G. apicola isolates were grown on a variety of monosaccharides (i.e., xylose, arabinose, mannose, and rhamnose) as the sole carbon and energy source. Since none of the G. apicola strains from bumble bee guts grew in the tested medium (see Materials and Methods) even with glucose and fructose, we only show the results for strains from honey bees (Fig. 5B). We found that growth abilities were consistent with the presence of the requisite genes for the catabolism of each sugar. Five strains grew on all four tested sugars. All of the strains from honey bees could grow on xylose and rhamnose, except strain wkB308 from A. cerana, which only grew on mannose. In contrast, the genes related to catabolizing all four sugars are underrepresented in the strains from bumble bees. Only araABD were detected from strains App2-1 and App2-10 from Bombus appositus (see Fig. S3 in the supplemental material), and only manA from strains Gris1-4 and Imp1-6 (Fig. 3; see also Fig. S3).
To determine their potential ability to degrade pectin and galacturonic acid, the main constituent of pectin, we also screened for the major pectate lyase (PL1) encoded in the honey bee gut metagenome (17) and genes related to galacturonic acid catabolism. The PL1 gene was detected in five genomes, and there is no clear correlation with monosaccharide degradation ability (Fig. 5B). All five genes in the pathway converting d-galacturonic acid to pyruvate and d-glyceraldehyde-3-phosphate, which have been described as active in Escherichia coli (25), are present in all G. apicola strains (data not shown), while the alternative oxidative pathway found in Pseudomonas (26) is totally absent from all G. apicola strains.
Simultaneous sugar utilization.
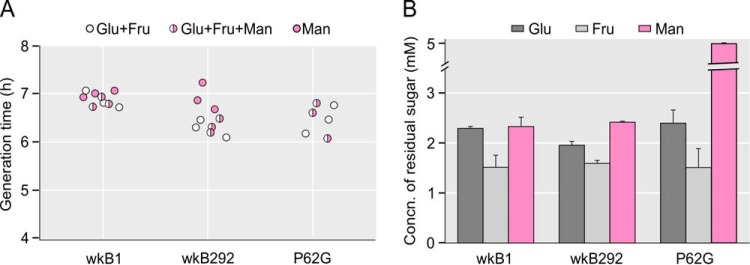
To determine if the presence of mannose affects the growth of G. apicola, strains wkB1 and P62G from A. mellifera and strain wkB292 from A. cerana were grown on (i) a 1:1 mixture of glucose and fructose, which approximates the natural ratio in honey, (ii) a 1:1:1 mixture of glucose, fructose, and mannose, and (iii) mannose alone.
On all substrates tested, all three strains typically grew to final cell densities of 0.5 × 108 to 1.5 × 108 cells/ml after 24 h at 35°C, with doubling times of 6.1 to 7.3 h (Fig. 6A). In the medium with all three sugars, strains wkB1 and wkB292 simultaneously utilized the individual sugars present (Fig. 6B). In all strains, fructose was consumed to the greatest extent. Although strain P62G cannot grow on mannose as it lacks MPI, the presence of equal amounts of mannose (5 mM) with glucose and fructose did not alter its growth; fructose and glucose were degraded to the same extent as in the other two strains, while all mannose added to the medium remained in the culture supernatant. This suggests that mannose was not transported into the cell through the PTS, despite the presence of a complete mannose PTS encoded in the genome. Altogether, these results indicate that G. apicola can utilize different sugars simultaneously and that the absence of MPI does not affect the growth rate, suggesting that mannose is not toxic to G. apicola, in contrast to what has been shown in its bee hosts.
FIG 6 .
Sugar utilization by G. apicola strains. (A) G. apicola strains wkB1, wkB292, and P62G grew at approximately the same rates on the different sugar mixtures tested. Each circle represents the value for a batch culture. (B) Simultaneous utilization of sugars during the growth of G. apicola strains in batch cultures. Glucose (Glu), fructose (Fru), and mannose (Man) were added to cultures at the time of inoculation to a concentration of 5 mM. The concentrations of each sugar remaining after 24 h are shown (n = 3). Error bars show standard deviations.
DISCUSSION
In this study, we document that one of the major bee gut symbionts, G. apicola, has the ability to metabolize mannose, xylose, arabinose, and rhamnose. These monosaccharides have been shown to reduce the life span of adult bees at low concentrations (11). Although the ability to metabolize these sugars varies among G. apicola strains, all strains tested from honey bee guts can utilize at least one of these sugars. Additionally, there is a strong correlation between the ability to grow on particular sugar substrates and the presence of the corresponding genes in catabolic pathways. Thus, we can predict with confidence the metabolic capabilities of strains from genomic sequence data.
The previous observations of sugar toxicity were based on bees that emerged naturally within hives and that presumably contained typical gut bacteria, so the contributions of gut symbionts to toxicity reduction were not assessed. G. apicola is acquired by all adult worker honey bees soon after their emergence within the colony, and this species comprises up to 39% of the bee gut microbiota (27). Our finding that many G. apicola strains can utilize certain sugars suggests that these gut symbionts may add to the energy budget of the host by helping to digest recalcitrant carbohydrates, as has been observed for bacteria in other insect guts (28) and in the human gut (1).
Although all G. apicola strains possess mannose family PTSs, including the manO-associated EII complex, the MPI enzyme is essential for mannose catabolism; in other bacteria, disruption of MPI can even repress growth on other carbon substrates (29). The mechanism of mannose toxicity in honey bees has been proposed to result from a deficiency of MPI. In bees, the ATP-driven phosphorylation of mannose to mannose-6-phosphate (mannose-6-P) is performed by hexokinase, and the imbalance of hexokinase and MPI expression leads to a competitive inhibition of glucose and fructose utilization when mannose is abundant (10). In a transcriptome data set from guts of A. cerana bees (30), the MPI transcript abundance was only ~6% of that of hexokinase transcripts. The result is an accumulation of intracellular mannose-6-P and ATP depletion due to the inefficiency of the bee host MPI in shunting mannose-6-P into glycolysis pathways (31).
We found that G. apicola lacking the gene encoding MPI, manA, is able to utilize glucose and fructose even when mannose is present in the medium, with no effect on growth. Thus, it seems that G. apicola does not experience the same mannose toxicity as its bee hosts. Our results suggest that this is due to mannose not being imported into G. apicola cells when manA is absent, despite the fact that all strains carry mannose family PTSs. How this is accomplished is unclear. A previous study of the manO-associated PTS EII complex in Streptococcus thermophilus also failed to observe cellular mannose uptake in vivo but detected mannose phosphorylation activity in cell-free experiments (32). Further gene expression studies will be required to unravel the regulatory mechanisms that allow G. apicola to suppress mannose import when mannose-6-P cannot be catabolized. It is likely that this metabolic flexibility permits the decoupling of mannose PTS and manA and the loss of manA in many G. apicola strains (Fig. 3).
Variability in carbohydrate utilization capabilities appears to be the norm among G. apicola strains. Carbohydrate metabolism genes constitute a large fraction of the accessory genome in six previously sequenced strains (16, 18). Overall, genes for sugar metabolism are highly represented in the honey bee gut community: a study of the bee gut metagenome documented that genes related to carbohydrate metabolism are specifically enriched compared to other gut-associated microbiomes (17), and genomic analyses of two abundant bee gut bacteria, G. apicola and Lactobacillus spp., uncovered a large proportion of genes in this functional group, particularly the mannose family PTSs (16, 33, 34), which are also shown to be preferentially transcribed in bee guts (35).
We showed that one mannose PTS with a manO regulator was acquired by the Orbales clade through horizontal gene transfer, likely from Firmicutes (Fig. 2). This PTS was shown to have undergone complex evolutionary events (23). This transfer is of particular interest here because it might allow the bee gut symbiont to digest toxic sugars. In Orbales, the manO-associated PTS was coupled to a gammaproteobacterial manA (Fig. 4), thus allowing mannose utilization; however, the manA gene was subsequently lost in many G. apicola strains, while the manO-associated PTS was retained. The comparison of gene arrangements in contigs containing manA reveals variation among different strains and supports the view that manA and other neighboring genes were present in the G. apicola ancestor and were gradually lost during strain diversification (Fig. 3). In four G. apicola strains from bumble bees, restriction-modification genes replace the missing genes. This is consistent with previous evidence that restriction-modification systems can behave as mobile genetic elements in the evolution of gut symbionts and can cause genome rearrangements (36).
Although almost identical in their 16S rRNA gene sequences, G. apicola strains exhibit extensive intraspecific variation in the presence of genes related to different sugar metabolism and growth abilities on different substrates (18). We found perfect correspondence between the predicted gene repertoires for sugar degradation in G. apicola genomes and the ability of strains to utilize the particular sugars in culture. Strains from the same host can differ in sugar metabolism ability, which is possibly due to the adaption to distinct metabolic niches. Notably, almost all G. apicola strains from bumble bees have lost the genes for the metabolism of the investigated sugars, which corroborates earlier suggestions that bumble bee G. apicola strains have fewer carbohydrate degradation capabilities than their honey bee counterparts (16). The diversification of G. apicola strains in different hosts is potentially shaped by the differing diets, longevities, and nest population sizes between honey bee and bumble bee species (37).
Bees have a carbohydrate-rich diet of nectar and pollen (38) and, probably, a high sugar concentration in their guts. Pectins and polysaccharides are present in the pollen wall, and some G. apicola strains possess pectate lyases and glycoside hydrolases, which can break down the pectin backbone and side chains (16, 17). We also found that some G. apicola strains have the PL1 gene; however, there is no apparent correlation between the ability to secrete enzymes for pectin degradation and the ability to utilize the constituent monosaccharides. Thus, pectin digestion and downstream sugar uptake and metabolism can be performed by different species or by different strains of the same species. Potentially, different hives have different profiles of G. apicola strains, with consequences for their ability to derive energy from pollen components and for their ability to resist the toxic effects of component sugars. Nevertheless, in vivo analyses of the ability of G. apicola strains to metabolize mannose in the bee gut will further characterize the effects of gut microbiota on bee health. High strain diversity within guts of individual bees or within hives could be beneficial by promoting nutrient availability, mitigating toxins, and improving immune response and microbiota stability, as has been suggested for human individuals with high gut microbiota richness (39, 40).
MATERIALS AND METHODS
Bee samples.
We collected honey bees (order Hymenoptera, family Apidae, genus Apis) comprising 4 species and bumble bees (genus Bombus) comprising 10 species belonging to different subgenera. The collection locations and dates of samples used in this study are shown in Table S1 in the supplemental material. The bees were identified based on their morphology. The dissected guts were either homogenized in 10 mM MgSO4 for immediate bacterial isolation or crushed in 19% glycerol and frozen directly after sampling.
Isolation and cultivation of G. apicola.
Pure cultures of G. apicola strains were isolated from bee guts as previously described (41). Briefly, fresh gut homogenates or glycerol stocks were plated on heart infusion agar (HIA) supplemented with defibrinated sheep’s blood (5% [vol/vol] final concentration; Hardy Diagnostics). After 2 days of incubation at 37°C under a CO2-enriched atmosphere (5%), visible colonies were identified by sequencing of the 16S rRNA gene.
The ability of G. apicola strains to grow on different substrates was tested by cultivation in BYZ medium containing a buffered salts solution (BSS) (42) composed of (per liter) 0.2 g KH2PO4, 0.25 g NH4Cl, 0.5 g KCl, 0.15 g CaCl2 ⋅ 2H2O, 1.0 g NaCl, 0.62 g MgCl2 ⋅ 6H2O, 2.84 g Na2SO4, and 10 mM MOPS (morpholinepropanesulfonic acid), as well as 0.05% (wt/vol) yeast extract. The medium was adjusted to a final pH of 7.0. Glucose, arabinose, xylose, or rhamnose (10 mM final concentration) was added as a carbon substrate. Cultures were established by inoculating the medium with single colonies growing on HIA plates and were incubated at 35°C under 5% CO2.
The ability of G. apicola strains to utilize different substrates simultaneously was tested by culturing the strains in medium containing BSS supplemented with 5 mM each of glucose, fructose, and/or mannose. Growth was determined spectrophotometrically by following the increase in optical density at 600 nm. After 24 h of incubation, 1-ml aliquots of culture medium were sampled and centrifuged at 12,000 × g for 5 min, and the supernatant was diluted 1:100 with water (high-performance liquid chromatography [HPLC] grade; Fisher Chemical). The concentration of residual glucose, fructose, or mannose in the diluted culture medium was determined by using the d-mannose/d-fructose/d-glucose assay kit (Megazyme, Inc.).
Genome sequencing and annotation.
Genomic DNA was extracted using phenol-chloroform and purified on DNeasy spin columns (Qiagen). Paired-end libraries of 2- to 4-kb insert sizes were constructed and sequenced on the Illumina MiSeq platform as described previously (16). The reads obtained were assembled with MaSuRCA version 3.1.0 (43). For strain wkB7, a complete genome was generated by sequencing on the PacBio RS II platform and assembly with the hierarchical genome assembly process (HGAP) method (Pacific Biosciences, Inc.); the assembly was closed and verified by mapping Illumina MiSeq reads onto the HGAP-generated contigs. All genomes were annotated with the RAST server (44). Annotated genomes were imported into Geneious version 9.1.4 (45) for visualization and further analysis. Pairwise average nucleotide identities were calculated using the pyani Python3 module (https://github.com/widdowquinn/pyani).
Phylogenetic analysis.
To generate the whole-genome tree, we used a concatenated alignment of 225 single-copy genes shared among all G. apicola strains. Orthologous genes were identified using best bidirectional hit with USEARCH (46) and clustered using the Markov cluster algorithm (47). The shared single-copy genes were aligned with MAFFT version 7 (48). The alignments of the nucleotide sequences were then concatenated, and maximum-likelihood trees were inferred using RAxML version 8 with the GTRGAMMA nucleotide substitution model (49).
For the phylogenetic analysis of manA and the mannose family EII complex, corresponding nucleotide sequences from G. apicola and close hits in the NCBI GenBank nr database were codon aligned using MAFFT version 7 (48). After manual refinement of the alignments, maximum-likelihood trees were constructed using MEGA7 (50) with 1,000 bootstraps and the GTR+G+I model, which was tested to best describe the substitution pattern using MEGA7.
Accession number(s).
All genome sequences of G. apicola strains isolated in this study have been deposited in the Whole Genome Shotgun projects at DDBJ/ENA/GenBank. The accession numbers are listed in Table S1.
SUPPLEMENTAL MATERIAL
Presence (+) of 11 mannose family phosphotransferase systems with enzyme IID domains in G. apicola strains. Download
Maximum-likelihood trees of the concatenated sequences of three pyruvate dehydrogenase (PDH) components and peptide release factor 1 (PRF) of G. apicola strains and F. perrara. Download
Presence of the genes related to the metabolism of the four sugars and the pectate lyase gene (PL1) in G. apicola strains isolated from bumble bee guts. Download
Genome features of the G. apicola strains isolated from the guts of honey bees and bumble bees.
ACKNOWLEDGMENTS
We thank Kim Hammond for beekeeping assistance. We also thank Louis-Marie Bobay and Drew Vander Wood for help in assembling genomes and in phylogenetic analyses.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Citation Zheng H, Nishida A, Kwong WK, Koch H, Engel P, Steele MI, Moran NA. 2016. Metabolism of toxic sugars by strains of the bee gut symbiont Gilliamella apicola. mBio 7(6):e01326-16. doi:10.1128/mBio.01326-16.
REFERENCES
- 1.Louis P, Hold GL, Flint HJ. 2014. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 2.White BA, Lamed R, Bayer EA, Flint HJ. 2014. Biomass utilization by gut microbiomes. Annu Rev Microbiol 68:279–296. doi: 10.1146/annurev-micro-092412-155618. [DOI] [PubMed] [Google Scholar]
- 3.Lee WJ, Hase K. 2014. Gut microbiota-generated metabolites in animal health and disease. Nat Chem Biol 10:416–424. doi: 10.1038/nchembio.1535. [DOI] [PubMed] [Google Scholar]
- 4.Frias BED, Barbosa CD, Lourenço AP. 2016. Pollen nutrition in honey bees (Apis mellifera): impact on adult health. Apidologie 47:15–25. doi: 10.1007/s13592-015-0373-y. [DOI] [Google Scholar]
- 5.Vaudo AD, Tooker JF, Grozinger CM, Patch HM. 2015. Bee nutrition and floral resource restoration. Curr Opin Insect Sci 10:133–141. doi: 10.1016/j.cois.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Johnson RM. 2015. Honey bee toxicology. Annu Rev Entomol 60:415–434. doi: 10.1146/annurev-ento-011613-162005. [DOI] [PubMed] [Google Scholar]
- 7.Bertholf LM. 1927. The utilization of carbohydrates as food by honeybee larvae. J Agric Res 35:429–452. [Google Scholar]
- 8.Phillips EF. 1927. The utilization of carbohydrates by honeybees. J Agric Res 35:385–428. [Google Scholar]
- 9.Barker RJ, Lehner Y. 1974. Acceptance and sustenance value of naturally occurring sugars fed to newly emerged adult workers of honey bees (Apis mellifera L.). J Exp Zool 187:277–285. doi: 10.1002/jez.1401870211. [DOI] [PubMed] [Google Scholar]
- 10.Sols A, Cadenas E, Alvarado F. 1960. Enzymatic basis of mannose toxicity in honey bees. Science 131:297–298. doi: 10.1126/science.131.3396.297. [DOI] [PubMed] [Google Scholar]
- 11.Barker RJ, Lehner Y. 1974. Influence of diet on sugars found by thin-layer chromatography in thoraces of honey bees, Apis mellifera L. J Exp Zool 188:157–164. doi: 10.1002/jez.1401880204. [DOI] [PubMed] [Google Scholar]
- 12.Barker RJ. 1977. Some carbohydrates found in pollen and pollen substitutes are toxic to honey bees. J Nutr 107:1859–1862. [DOI] [PubMed] [Google Scholar]
- 13.Martinson VG, Danforth BN, Minckley RL, Rueppell O, Tingek S, Moran NA. 2011. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol Ecol 20:619–628. doi: 10.1111/j.1365-294X.2010.04959.x. [DOI] [PubMed] [Google Scholar]
- 14.Kwong WK, Moran NA. 2016. Gut microbial communities of social bees. Nat Rev Microbiol 14:374–384. doi: 10.1038/nrmicro.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engel P, Kwong WK, McFrederick Q, Anderson KE, Barribeau SM, Chandler JA, Cornman RS, Dainat J, de Miranda JR, Doublet V, Emery O, Evans JD, Farinelli L, Flenniken ML, Granberg F, Grasis JA, Gauthier L, Hayer J, Koch H, Kocher S, Martinson VG, Moran N, Munoz-Torres M, Newton I, Paxton RJ, Powell E, Sadd BM, Schmid-Hempel P, Schmid-Hempel R, Song SJ, Schwarz RS, vanEngelsdorp D, Dainat B. 2016. The bee microbiome: impact on bee health and model for evolution and ecology of host-microbe interactions. mBio 7:e02164-15. doi: 10.1128/mBio.02164-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwong WK, Engel P, Koch H, Moran NA. 2014. Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc Natl Acad Sci U S A 111:11509–11514. doi: 10.1073/pnas.1405838111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engel P, Martinson VG, Moran NA. 2012. Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci U S A 109:11002–11007. doi: 10.1073/pnas.1202970109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engel P, Stepanauskas R, Moran NA. 2014. Hidden diversity in honey bee gut symbionts detected by single-cell genomics. PLoS Genet 10:e1004596. doi: 10.1371/journal.pgen.1004596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams PH, Cameron SA, Hines HM, Cederberg B, Rasmont P. 2008. A simplified subgeneric classification of the bumblebees (genus Bombus). Apidologie 39:46–74. doi: 10.1051/apido:2007052. [DOI] [Google Scholar]
- 20.Sharma V, Ichikawa M, Freeze HH. 2014. Mannose metabolism: more than meets the eye. Biochem Biophys Res Commun 453:220–228. doi: 10.1016/j.bbrc.2014.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deutscher J, Aké FM, Derkaoui M, Zébré AC, Cao TN, Bouraoui H, Kentache T, Mokhtari A, Milohanic E, Joyet P. 2014. The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol Mol Biol Rev 78:231–256. doi: 10.1128/MMBR.00001-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reizer J, Saier MH. 1997. Modular multidomain phosphoryl transfer proteins of bacteria. Curr Opin Struct Biol 7:407–415. doi: 10.1016/S0959-440X(97)80059-0. [DOI] [PubMed] [Google Scholar]
- 23.Zúñiga M, Comas I, Linaje R, Monedero V, Yebra MJ, Esteban CD, Deutscher J, Pérez-Martínez G, González-Candelas F. 2005. Horizontal gene transfer in the molecular evolution of mannose PTS transporters. Mol Biol Evol 22:1673–1685. doi: 10.1093/molbev/msi163. [DOI] [PubMed] [Google Scholar]
- 24.Engel P, Bartlett KD, Moran NA. 2015. The bacterium Frischella perrara causes scab formation in the gut of its honeybee host. mBio 6:e00193-15. doi: 10.1128/mBio.00193-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richard P, Hilditch S. 2009. d-Galacturonic acid catabolism in microorganisms and its biotechnological relevance. Appl Microbiol Biotechnol 82:597–604. doi: 10.1007/s00253-009-1870-6. [DOI] [PubMed] [Google Scholar]
- 26.Kilgore WW, Starr MP. 1959. Uronate oxidation by phytopathogenic pseudomonads. Nature 183:1412–1413. doi: 10.1038/1831412a0. [DOI] [PubMed] [Google Scholar]
- 27.Moran NA, Hansen AK, Powell JE, Sabree ZL. 2012. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS One 7:e36393. doi: 10.1371/journal.pone.0036393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engel P, Moran NA. 2013. The gut microbiota of insects—diversity in structure and function. FEMS Microbiol Rev 37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 29.Sasaki M, Teramoto H, Inui M, Yukawa H. 2011. Identification of mannose uptake and catabolism genes in Corynebacterium glutamicum and genetic engineering for simultaneous utilization of mannose and glucose. Appl Microbiol Biotechnol 89:1905–1916. doi: 10.1007/s00253-010-3002-8. [DOI] [PubMed] [Google Scholar]
- 30.Wang ZL, Liu TT, Huang ZY, Wu XB, Yan WY, Zeng ZJ. 2012. Transcriptome analysis of the Asian honey bee Apis cerana cerana. PLoS One 7:e47954. doi: 10.1371/journal.pone.0047954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De la Fuente M, Peñas PF, Sols A. 1986. Mechanism of mannose toxicity. Biochem Biophys Res Commun 140:51–55. doi: 10.1016/0006-291X(86)91056-9. [DOI] [PubMed] [Google Scholar]
- 32.Cochu A, Vadeboncoeur C, Moineau S, Frenette M. 2003. Genetic and biochemical characterization of the phosphoenolpyruvate:glucose/mannose phosphotransferase system of Streptococcus thermophilus. Appl Environ Microbiol 69:5423–5432. doi: 10.1128/AEM.69.9.5423-5432.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwong WK, Mancenido AL, Moran NA. 2014. Genome sequences of Lactobacillus sp. strains wkB8 and wkB10, members of the Firm-5 clade, from honey bee guts. Genome Announc 2:e01176-14. doi: 10.1128/genomeA.01176-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ellegaard KM, Tamarit D, Javelind E, Olofsson TC, Andersson SG, Vásquez A. 2015. Extensive intra-phylotype diversity in lactobacilli and bifidobacteria from the honeybee gut. BMC Genomics 16:284. doi: 10.1186/s12864-015-1476-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee FJ, Rusch DB, Stewart FJ, Mattila HR, Newton IL. 2015. Saccharide breakdown and fermentation by the honey bee gut microbiome. Environ Microbiol 17:796–815. doi: 10.1111/1462-2920.12526. [DOI] [PubMed] [Google Scholar]
- 36.Zheng H, Dietrich C, Hongoh Y, Brune A. 2016. Restriction-modification systems as mobile genetic elements in the evolution of an intracellular symbiont. Mol Biol Evol 33:721–725. doi: 10.1093/molbev/msv264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plowright RC, Laverty TM. 1984. The ecology and sociobiology of bumble bees. Annu Rev Entomol 29:175–199. doi: 10.1146/annurev.en.29.010184.001135. [DOI] [Google Scholar]
- 38.Brodschneider R, Crailsheim K. 2010. Nutrition and health in honey bees. Apidologie 41:278–294. doi: 10.1051/apido/2010012. [DOI] [Google Scholar]
- 39.Tap J, Furet JP, Bensaada M, Philippe C, Roth H, Rabot S, Lakhdari O, Lombard V, Henrissat B, Corthier G, Fontaine E, Doré J, Leclerc M. 2015. Gut microbiota richness promotes its stability upon increased dietary fibre intake in healthy adults. Environ Microbiol 17:4954–4964. doi: 10.1111/1462-2920.13006. [DOI] [PubMed] [Google Scholar]
- 40.Le Chatelier E, Nielsen T, Qin JJ, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, Leonard P, Li JH, Burgdorf K, Grarup N, Jørgensen T, Brandslund I, Nielsen HB, Juncker AS, Bertalan M, Levenez F, Pons N, Rasmussen S, Sunagawa S, Tap J, Tims S, Zoetendal EG, Brunak S, Clement K, Dore J, Kleerebezem M, Kristiansen K, Renault P, Sicheritz-Ponten T, de Vos WM, Zucker JD, Raes J, Hansen T, Bork P, Wang J, Ehrlich SD, Pedersen O, MetaHIT Consortium . 2013. Richness of human gut microbiome correlates with metabolic markers. Nature 500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 41.Kwong WK, Moran NA. 2013. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the Betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the order “Enterobacteriales” of the Gammaproteobacteria. Int J Syst Evol Microbiol 63:2008–2018. doi: 10.1099/ijs.0.044875-0. [DOI] [PubMed] [Google Scholar]
- 42.Wertz JT, Breznak JA. 2007. Stenoxybacter acetivorans gen. nov., sp. nov., an acetate-oxidizing obligate microaerophile among diverse O2-consuming bacteria from termite guts. Appl Environ Microbiol 73:6819–6828. doi: 10.1128/AEM.00786-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zimin AV, Marçais G, Puiu D, Roberts M, Salzberg SL, Yorke JA. 2013. The MaSuRCA genome assembler. Bioinformatics 29:2669–2677. doi: 10.1093/bioinformatics/btt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 47.Van Dongen S, Abreu-Goodger C. 2012. Using MCL to extract clusters from networks. Methods Mol Biol 804:281–295. doi: 10.1007/978-1-61779-361-5_15. [DOI] [PubMed] [Google Scholar]
- 48.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Engel P, Vizcaino MI, Crawford JM. 2015. Gut symbionts from distinct hosts exhibit genotoxic activity via divergent colibactin biosynthesis pathways. Appl Environ Microbiol 81:1502–1512. doi: 10.1128/AEM.03283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martinson VG, Magoc T, Koch H, Salzberg SL, Moran NA. 2014. Genomic features of a bumble bee symbiont reflect its host environment. Appl Environ Microbiol 80:3793–3803. doi: 10.1128/AEM.00322-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Presence (+) of 11 mannose family phosphotransferase systems with enzyme IID domains in G. apicola strains. Download
Maximum-likelihood trees of the concatenated sequences of three pyruvate dehydrogenase (PDH) components and peptide release factor 1 (PRF) of G. apicola strains and F. perrara. Download
Presence of the genes related to the metabolism of the four sugars and the pectate lyase gene (PL1) in G. apicola strains isolated from bumble bee guts. Download
Genome features of the G. apicola strains isolated from the guts of honey bees and bumble bees.