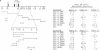Abstract
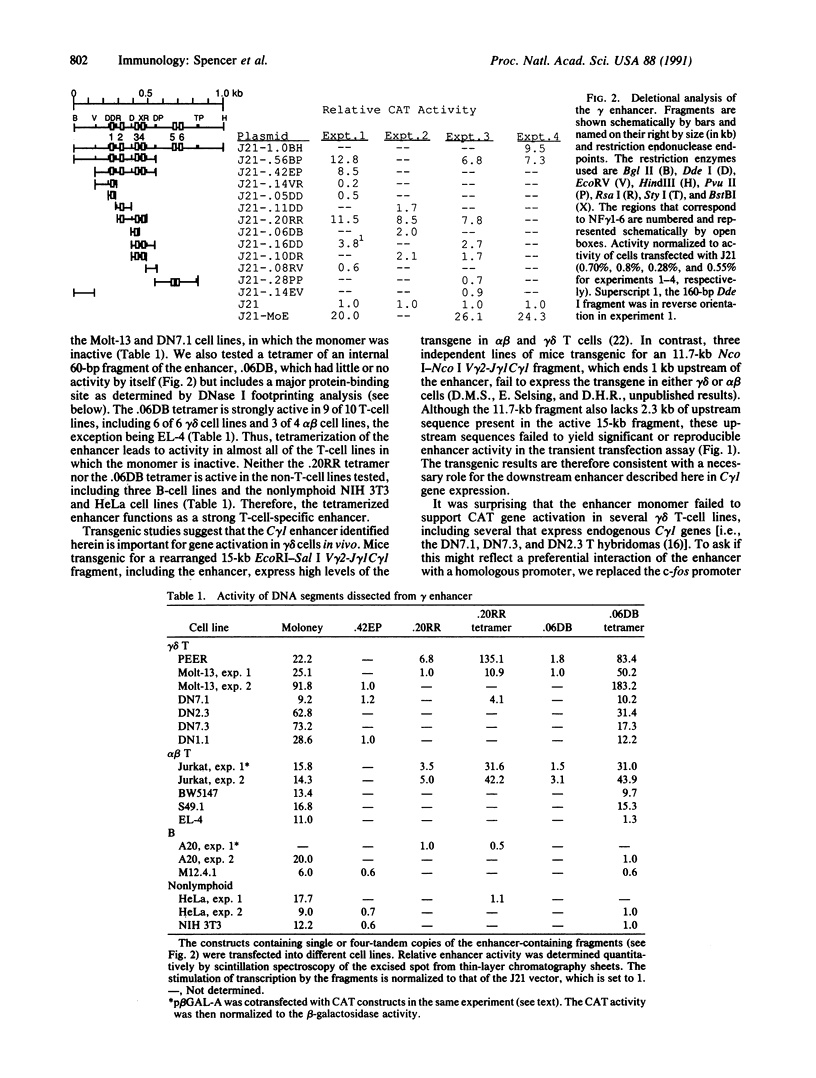
A transcriptional enhancer element has been localized 3 kilobases 3' of the murine T-cell receptor C gamma 1 locus using a chloramphenicol acetyltransferase reporter gene construct. As a monomer the enhancer functions only in PEER gamma delta cells and Jurkat alpha beta cells of the T-cell lines tested. However, a tetramer of the enhancer functions in virtually all T-cell lines tested, including alpha beta T-cell lines, but not in other cell types. These results suggest that elements other than the enhancer are responsible for the failure of rearranged C gamma 1 genes to be expressed in alpha beta T cells. The enhancer has been localized to a 200-base-pair Rsa I restriction fragment, which contains sequence motifs similar to those found in the other T-cell receptor enhancers but not in the immunoglobulin enhancers.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison J. P., Lanier L. L. Structure, function, and serology of the T-cell antigen receptor complex. Annu Rev Immunol. 1987;5:503–540. doi: 10.1146/annurev.iy.05.040187.002443. [DOI] [PubMed] [Google Scholar]
- Asarnow D. M., Kuziel W. A., Bonyhadi M., Tigelaar R. E., Tucker P. W., Allison J. P. Limited diversity of gamma delta antigen receptor genes of Thy-1+ dendritic epidermal cells. Cell. 1988 Dec 2;55(5):837–847. doi: 10.1016/0092-8674(88)90139-0. [DOI] [PubMed] [Google Scholar]
- Bonneville M., Ishida I., Mombaerts P., Katsuki M., Verbeek S., Berns A., Tonegawa S. Blockage of alpha beta T-cell development by TCR gamma delta transgenes. Nature. 1989 Dec 21;342(6252):931–934. doi: 10.1038/342931a0. [DOI] [PubMed] [Google Scholar]
- Bonneville M., Janeway C. A., Jr, Ito K., Haser W., Ishida I., Nakanishi N., Tonegawa S. Intestinal intraepithelial lymphocytes are a distinct set of gamma delta T cells. Nature. 1988 Dec 1;336(6198):479–481. doi: 10.1038/336479a0. [DOI] [PubMed] [Google Scholar]
- Brenner M. B., McLean J., Dialynas D. P., Strominger J. L., Smith J. A., Owen F. L., Seidman J. G., Ip S., Rosen F., Krangel M. S. Identification of a putative second T-cell receptor. Nature. 1986 Jul 10;322(6075):145–149. doi: 10.1038/322145a0. [DOI] [PubMed] [Google Scholar]
- Chien Y. H., Iwashima M., Wettstein D. A., Kaplan K. B., Elliott J. F., Born W., Davis M. M. T-cell receptor delta gene rearrangements in early thymocytes. Nature. 1987 Dec 24;330(6150):722–727. doi: 10.1038/330722a0. [DOI] [PubMed] [Google Scholar]
- Clevers H., Lonberg N., Dunlap S., Lacy E., Terhorst C. An enhancer located in a CpG-island 3' to the TCR/CD3-epsilon gene confers T lymphocyte-specificity to its promoter. EMBO J. 1989 Sep;8(9):2527–2535. doi: 10.1002/j.1460-2075.1989.tb08390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986 Jan 31;44(2):273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- Elliott J. F., Rock E. P., Patten P. A., Davis M. M., Chien Y. H. The adult T-cell receptor delta-chain is diverse and distinct from that of fetal thymocytes. Nature. 1988 Feb 18;331(6157):627–631. doi: 10.1038/331627a0. [DOI] [PubMed] [Google Scholar]
- Garman R. D., Doherty P. J., Raulet D. H. Diversity, rearrangement, and expression of murine T cell gamma genes. Cell. 1986 Jun 6;45(5):733–742. doi: 10.1016/0092-8674(86)90787-7. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K., van den Elsen P., Bier E., Maxam A., Terhorst C. A T cell-specific enhancer is located in a DNase I-hypersensitive area at the 3' end of the CD3-delta gene. EMBO J. 1988 Aug;7(8):2401–2407. doi: 10.1002/j.1460-2075.1988.tb03085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman T., Lefrançois L. Expression of the gamma-delta T-cell receptor on intestinal CD8+ intraepithelial lymphocytes. Nature. 1988 Jun 30;333(6176):855–858. doi: 10.1038/333855a0. [DOI] [PubMed] [Google Scholar]
- Havran W. L., Allison J. P. Developmentally ordered appearance of thymocytes expressing different T-cell antigen receptors. Nature. 1988 Sep 29;335(6189):443–445. doi: 10.1038/335443a0. [DOI] [PubMed] [Google Scholar]
- Havran W. L., Allison J. P. Origin of Thy-1+ dendritic epidermal cells of adult mice from fetal thymic precursors. Nature. 1990 Mar 1;344(6261):68–70. doi: 10.1038/344068a0. [DOI] [PubMed] [Google Scholar]
- Herr W., Clarke J. The SV40 enhancer is composed of multiple functional elements that can compensate for one another. Cell. 1986 May 9;45(3):461–470. doi: 10.1016/0092-8674(86)90332-6. [DOI] [PubMed] [Google Scholar]
- Ho I. C., Yang L. H., Morle G., Leiden J. M. A T-cell-specific transcriptional enhancer element 3' of C alpha in the human T-cell receptor alpha locus. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6714–6718. doi: 10.1073/pnas.86.17.6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida I., Verbeek S., Bonneville M., Itohara S., Berns A., Tonegawa S. T-cell receptor gamma delta and gamma transgenic mice suggest a role of a gamma gene silencer in the generation of alpha beta T cells. Proc Natl Acad Sci U S A. 1990 Apr;87(8):3067–3071. doi: 10.1073/pnas.87.8.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. A., Yamamoto K. R., Tjian R. Two distinct transcription factors bind to the HSV thymidine kinase promoter in vitro. Cell. 1985 Sep;42(2):559–572. doi: 10.1016/0092-8674(85)90113-8. [DOI] [PubMed] [Google Scholar]
- Korman A. J., Marusic-Galesic S., Spencer D., Kruisbeek A. M., Raulet D. H. Predominant variable region gene usage by gamma/delta T cell receptor-bearing cells in the adult thymus. J Exp Med. 1988 Sep 1;168(3):1021–1040. doi: 10.1084/jem.168.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimpenfort P., de Jong R., Uematsu Y., Dembic Z., Ryser S., von Boehmer H., Steinmetz M., Berns A. Transcription of T cell receptor beta-chain genes is controlled by a downstream regulatory element. EMBO J. 1988 Mar;7(3):745–750. doi: 10.1002/j.1460-2075.1988.tb02871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder P., Battey J., Lenoir G., Moulding C., Murphy W., Potter H., Stewart T., Taub R. Translocations among antibody genes in human cancer. Science. 1983 Nov 18;222(4625):765–771. doi: 10.1126/science.6356357. [DOI] [PubMed] [Google Scholar]
- Lenardo M., Pierce J. W., Baltimore D. Protein-binding sites in Ig gene enhancers determine transcriptional activity and inducibility. Science. 1987 Jun 19;236(4808):1573–1577. doi: 10.1126/science.3109035. [DOI] [PubMed] [Google Scholar]
- Luria S., Gross G., Horowitz M., Givol D. Promoter and enhancer elements in the rearranged alpha chain gene of the human T cell receptor. EMBO J. 1987 Nov;6(11):3307–3312. doi: 10.1002/j.1460-2075.1987.tb02650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrack P., Kappler J. The T cell receptor. Science. 1987 Nov 20;238(4830):1073–1079. doi: 10.1126/science.3317824. [DOI] [PubMed] [Google Scholar]
- Mercola M., Wang X. F., Olsen J., Calame K. Transcriptional enhancer elements in the mouse immunoglobulin heavy chain locus. Science. 1983 Aug 12;221(4611):663–665. doi: 10.1126/science.6306772. [DOI] [PubMed] [Google Scholar]
- Meyer K. B., Neuberger M. S. The immunoglobulin kappa locus contains a second, stronger B-cell-specific enhancer which is located downstream of the constant region. EMBO J. 1989 Jul;8(7):1959–1964. doi: 10.1002/j.1460-2075.1989.tb03601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Wang C., Tjian R. Positive and negative regulation of transcription in vitro: enhancer-binding protein AP-2 is inhibited by SV40 T antigen. Cell. 1987 Sep 11;50(6):847–861. doi: 10.1016/0092-8674(87)90512-5. [DOI] [PubMed] [Google Scholar]
- Perry R. P. What controls the transcription of immunoglobulin genes? Nature. 1984 Jul 5;310(5972):14–15. doi: 10.1038/310014a0. [DOI] [PubMed] [Google Scholar]
- Raulet D. H., Garman R. D., Saito H., Tonegawa S. Developmental regulation of T-cell receptor gene expression. Nature. 1985 Mar 7;314(6006):103–107. doi: 10.1038/314103a0. [DOI] [PubMed] [Google Scholar]
- Raulet D. H. The structure, function, and molecular genetics of the gamma/delta T cell receptor. Annu Rev Immunol. 1989;7:175–207. doi: 10.1146/annurev.iy.07.040189.001135. [DOI] [PubMed] [Google Scholar]
- Redondo J. M., Hata S., Brocklehurst C., Krangel M. S. A T cell-specific transcriptional enhancer within the human T cell receptor delta locus. Science. 1990 Mar 9;247(4947):1225–1229. doi: 10.1126/science.2156339. [DOI] [PubMed] [Google Scholar]
- Saito H., Kranz D. M., Takagaki Y., Hayday A. C., Eisen H. N., Tonegawa S. Complete primary structure of a heterodimeric T-cell receptor deduced from cDNA sequences. 1984 Jun 28-Jul 4Nature. 309(5971):757–762. doi: 10.1038/309757a0. [DOI] [PubMed] [Google Scholar]
- Shaw J. P., Utz P. J., Durand D. B., Toole J. J., Emmel E. A., Crabtree G. R. Identification of a putative regulator of early T cell activation genes. Science. 1988 Jul 8;241(4862):202–205. doi: 10.1126/science.3260404. [DOI] [PubMed] [Google Scholar]
- Snodgrass H. R., Kisielow P., Kiefer M., Steinmetz M., von Boehmer H. Ontogeny of the T-cell antigen receptor within the thymus. Nature. 1985 Feb 14;313(6003):592–595. doi: 10.1038/313592a0. [DOI] [PubMed] [Google Scholar]
- Speck N. A., Renjifo B., Golemis E., Fredrickson T. N., Hartley J. W., Hopkins N. Mutation of the core or adjacent LVb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 1990 Feb;4(2):233–242. doi: 10.1101/gad.4.2.233. [DOI] [PubMed] [Google Scholar]
- Veldman G. M., Lupton S., Kamen R. Polyomavirus enhancer contains multiple redundant sequence elements that activate both DNA replication and gene expression. Mol Cell Biol. 1985 Apr;5(4):649–658. doi: 10.1128/mcb.5.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- Weiss A., Newton M., Crommie D. Expression of T3 in association with a molecule distinct from the T-cell antigen receptor heterodimer. Proc Natl Acad Sci U S A. 1986 Sep;83(18):6998–7002. doi: 10.1073/pnas.83.18.6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winoto A., Baltimore D. A novel, inducible and T cell-specific enhancer located at the 3' end of the T cell receptor alpha locus. EMBO J. 1989 Mar;8(3):729–733. doi: 10.1002/j.1460-2075.1989.tb03432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winoto A., Baltimore D. Alpha beta lineage-specific expression of the alpha T cell receptor gene by nearby silencers. Cell. 1989 Nov 17;59(4):649–655. doi: 10.1016/0092-8674(89)90010-x. [DOI] [PubMed] [Google Scholar]
- Winoto A., Baltimore D. Developmental regulation of the TCR alpha delta locus. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):87–92. doi: 10.1101/sqb.1989.054.01.011. [DOI] [PubMed] [Google Scholar]
- Yancopoulos G. D., Alt F. W. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985 Feb;40(2):271–281. doi: 10.1016/0092-8674(85)90141-2. [DOI] [PubMed] [Google Scholar]
- Yancopoulos G. D., Alt F. W. Regulation of the assembly and expression of variable-region genes. Annu Rev Immunol. 1986;4:339–368. doi: 10.1146/annurev.iy.04.040186.002011. [DOI] [PubMed] [Google Scholar]