Abstract
The aim of this study was to examine the changes in muscle proteome of the rainbow trout fed dietary β-glucan. The experimental diets contained 0 (control), 0.1% and 0.2% β-1,3/1,6 yeast glucan. First, feeding larvae were fed to apparent satiation nine times per day with their respective diets over two months. The percentage of body weight gain and feed efficiency of fish fed 0.2% diet was significantly higher than those of fish fed the control and 0.1% diets. Fish fed the control and 0.2% diets were subjected to proteomic analysis. Proteins of the muscle tissue were analyzed using two-dimensional electrophoresis and mass spectrometry. Spots that were found to differ significantly in abundance between control and β-glucan fed fish were selected for identification. Out of 8 protein spots showing differential expression, 7 spots were successfully identified. Two protein spots that were found to be increased in abundance in the β-glucan treated rainbow trout corresponded to tropomyosin alpha-1 chain (spot 1) and slow myotomal muscle tropomyosin (spot 2). The five spots that were down-regulated with dietary β-glucan supplementation were identified as different forms of myosin: myosin light polypeptide 3-2 (spot 3), myosin light chain 1 (spots 4 and 5), fast myosin light chain 2 (spot 6) and myosin heavy chain (spot 7). The altered expression of structural proteins in fish fed β-glucan may be related to higher growth rate in rainbow trout. These findings provide basic information to understand possible mechanisms of dietary β-glucan contribution to better growth in rainbow trout.
Key Words: Dietary β-glucan, Growth, Proteomics, Rainbow trout
Introduction
While aquaculture industry is growing at a higher rate than any other animal food-producing sector (Bostock et al., 2010 ▶), nutrition, welfare and health management have been thought to be the main limitations to an efficient production in aquaculture systems. One of the main goals of aquaculture industry is to produce fish with an optimal growth performance and health status (Rodrigues et al., 2012 ▶). Thus, food ingredients that benefit the fish by stimulating growth and improving the innate humoral and cellular defense mechanisms are of great importance in aquaculture production.
Prebiotics such as β-glucans are non-digestible food ingredients that benefit the host by stimulating the growth and/or the activity of selected bacteria that improve its health status. β-glucans are polysaccharides consisting of a backbone of repetitive D-glucose monomer units linked by β-(1, 3) glycosidic bonds with β-(1, 6) branching glucose side-chains. These carbo-hydrates are mostly found in algae, plants, fungi and in some bacteria where they represent a major component of the cell wall (Pionnier et al., 2013 ▶). It has been found that β-glucans incorporated in fish diet increase the growth performance and improve the humoral immune response of certain fish species (reviewed in Dalmo and Bøgwald (2008) ▶). Since β-glucans have the potential to exert positive effects on fish species, a major challenge is to reveal β-glucans related mechanisms that stimulate growth and humoral immune responses.
Proteomics has emerged as a powerful tool towards a deep understanding of aquatic organisms’ biology and provides data at a mechanistic level. Compared to genomics, it can also reveal changes in protein activity identified as post-translational modifications (PTMs) (Rodrigues et al., 2012 ▶).
Rainbow trout (Oncorhynchus mykiss) is one of the most commercially important species grown in Iran and its current annual production is more than 140,000 tonnes. The objective of this study was to look at dietary β-glucan related changes in muscle proteome of the rainbow trout fry, and identify possible relations to rainbow trout fry quality.
Materials and Methods
Diets
β-1,3/1,6 yeast glucan (MacroGard®, Biotec-Mackzymal, TromsØ, Norway) derived from cell wall of Saccharomyces cerevisiae, was included in feed before extrusion. A commercial rainbow trout diet lacking β-glucan (Beyza Feed Mill, Fars province, Iran) and containing 52% protein, 12.5% fat, 10% humidity and 4300 kcal kg-1 diet digestible energy was used. The basal diet was crushed, mixed with water and the supplements added to obtain diets containing 0.1% and 0.2% β-1,3/1,6 yeast glucan. Control feed was prepared in the same way without addition of the β-glucan. Extruded diets were pelleted using a mincer, air-dried at room temperature and stored at 4°C in sealed plastic bags until used.
Fish and experimental conditions
Fish rearing in the present study was done in a commercial rainbow trout farm in Sepidan, Iran. Rainbow trout eggs were obtained from 15 ovulated females of about 4 kg. The eggs were pooled, fertilized with milt from males from the same farm and were incubated in small trays. The mean water quality parameters were as follows: temperature 10.5°C, dissolved oxygen 7.9 mgl-1 and pH = 7.8. The hatched larvae were allocated to three different treatment groups: control fed, 0.1% β-glucan fed, and 0.2% β-glucan fed. First feeding larvae were fed to apparent satiation nine times per day with their respective diets over two months. All the treatments were carried out in triplicate.
Growth measurements
The fish in the different experimental groups were weighed at the end of 2-month feeding trial for estimation of growth. Based on records of the weight of fish, percentage of body weight gain (WG) and feed efficiency (FE) were calculated for each group as follows:
WG= 100 × (final body weight – initial body weight) / initial body weight
FE= (final body weight – initial body weight) / feed intake
In addition, survival rate was calculated at the end of the experiment:
Survival= (Nf/N0) × 100
where,
N0: The initial number of fish
Nf: The final number of fish
Fish sampling and protein extraction
Based on growth measurements (Table 1), fish fed diets with 0 and 0.2% β-glucan were selected for proteomic analysis. Fifteen fish from each group (five fish per tank) were sacrificed. The fish were anaesthetized with clove powder (150 ppm) prior to sampling. Muscle tissue samples were taken from behind the head and above the lateral line, snap frozen in liquid nitrogen and stored at -80°C until further analysis. In order to minimize the effects of individual variation, the muscle tissues of (each) five individuals were mixed before proteome extraction so that three pools were prepared for each experimental group. The tissue samples were washed in 40 mM Tris-HCl buffer (pH = 7), before being mechanically homogenized in cold lysis buffer containing 7 M urea, 2 M thiourea, 4% CHAPS, 50 mM DTT, 50 mM Tris (pH = 7), 0.2% carrier ampholyte, 1 mM PMSF, 0.25% RNase, and 1% DNase. Each sample was maintained for 1 h at room temperature for protein release. Cellular debris was removed by centrifugation at 12,000 × g for 10 min, and supernatants were collected (Keyvanshokooh and Tahmasebi-Kohyani, 2012 ▶). The protein concentration was determined according to the method of Bradford (Bradford, 1976 ▶) with BSA as standards.
Table 1.
Growth performance of rainbow trout fed different levels of dietary β-glucan for two months
| Diets (%) | Initial weight (mg) | Final weight (mg) | Weight gain (WG %) | Feed Efficiency (FE) | Survival (%) |
|---|---|---|---|---|---|
| 0 | 181.05 ± 3.5a | 884.2 ± 38.8a | 388.7 ± 28.6a | 1.15 ± 0.07a | 97.3 ± 0.7a |
| 0.1 | 184.7 ± 2.8a | 996.5 ± 56.1a | 438.3 ± 18.8ab | 1.32 ± 0.08ab | 98.7 ± 1.0a |
| 0.2 | 186.50 ± 3.5a | 1060.2 ± 84.9a | 493.3 ± 9.0b | 1.40 ± 0.05b | 98.4 ± 0.5a |
Values are mean ± SD of three replicate groups. Mean values with different superscripts are significantly different from each other. Significance level is defined as P<0.05
Two dimensional gel electrophoresis
Isoelectric focusing was performed using the IEF Cell (BioRad, USA) as described by (Keyvanshokooh and Tahmasebi-Kohyani, 2012 ▶). Each 17 cm immo-bilized pH gradient (IPG) strip with linear pH range of 3-10 (BioRad, USA) was passively rehydrated for at least 16 h with 2 mg protein in 300 µL of rehydration buffer containing 8 M urea, 4% CHAPS, 50 mM DTT, 0.2% ampholyte and bromophenol blue traces. Strips were focused at 20°C with the following program: 20 min with a linear ramp (0-250 V), 5 h with a linear ramp (250-10000 V) and 50000 Vh with a rapid ramp. After isoelectric focusing the strips were equilibrated in the first step with 6 M urea, 50 mM Tris-HCl pH = 8.8, 20% glycerol, 2% SDS and 2% DTT for 20 min; and the second step with the same buffer but 2.5% iodoacetamide instead of DTT for another 20 min.
Each IPG strip was then laid onto a 12% SDS-PAGE gel for second dimension electrophoresis. The running conditions were 16 mA/gel for 30 min and 24 mA/gel for approximately 5 h until the bromophenol blue front was 1 cm above the bottom of the gel. The gels were stained using colloidal coomassie brilliant blue G-250 procedure (Candiano et al., 2004 ▶). To confirm the reproducibility of the results, three 2-DE runs were performed for each experimental condition.
Analysis of 2-D gels
The gels were scanned at a resolution of 300 dpi using the Densitometer GS-800 scanner (BioRad, USA) and stored as TIF files. Spot detection, quantification, and matching were performed using Progenesis Samespots software (Nonlinear Dynamics, UK). A match set consisting of 6 images, 3 for control fed fish and 3 for 0.2% fed fish was created, and one image from control group was selected as the match set standard for spot matching. To correct for variability and reflect the quantitative variations of protein spot, the individual spot volumes were normalized by dividing their density (D) values by the total D values of all the spots present in the gel.
In-gel trypsin digestion, MALDI-TOF/TOF MS and database searching
Gel pieces were washed two times with 50% (v:v) aqueous acetonitrile containing 25 mM ammonium bicarbonate, then once with acetonitrile and dried in a vacuum concentrator for 20 min. Sequencing-grade, modified porcine trypsin (Promega) was dissolved in the 50 mM acetic acid supplied by the manufacturer, then diluted 5-fold with 25 mM ammonium bicarbonate to give a final trypsin concentration of 0.02 µg/µL. Gel pieces were rehydrated by adding 10 µL of trypsin solution, and after 10 min enough 25 mM ammonium bicarbonate solution was added to cover the gel pieces. Digests were incubated overnight at 37°C.
A 1 µL aliquot of each peptide mixture was applied to a ground steel MALDI target plate, followed immediately by an equal volume of a freshly-prepared 5 mg/ml solution of 4-hydroxy-α-cyano-cinnamic acid (Sigma) in 50% aqueous (v:v) acetonitrile containing 0.1%, trifluoroacetic acid (v:v).
Positive-ion MALDI mass spectra were obtained using a Bruker ultraflex III in reflectron mode, equipped with a Nd:YAG smart beam laser. MS spectra were acquired over a range of 800-5000 m/z. Final mass spectra were externally calibrated against an adjacent spot containing 6 peptides (des-Arg1-Bradykinin, 904.681; Angiotensin I, 1296.685; Glu1-Fibrinopeptide B, 1750.677; ACTH (1-17 clip), 2093.086; ACTH (18-39 clip), 2465.198; ACTH (7-38 clip), 3657.929). Monoisotopic masses were obtained using a SNAP averaging algorithm (C 4.9384, N 1.3577, O 1.4773, S 0.0417, H 7.7583) and a S/N threshold of 2.
For each spot the ten strongest precursors, with a S/N greater than 30, were selected for MS/MS fragmentation. Fragmentation was performed in LIFT mode without the introduction of a collision gas. The default calibration was used for MS/MS spectra, which were baseline-subtracted and smoothed (Savitsky-Golay, width 0.15 m/z, cycles 4); monoisotopic peak detection used a SNAP averaging algorithm (C 4.9384, N 1.3577, O 1.4773, S 0.0417, H 7.7583) with a minimum S/N of 6. Bruker FlexAnalysis software (version 3.3) was used to perform spectral processing and peak list generation.
Tandem mass spectral data were submitted to database searching using a locally-running copy of the Mascot program (Matrix Science Ltd., version 2.5.1), through the Bruker Protein Scape interface (version 2.1). Searches were performed against a subset of the 20150224 NCBInr database containing only Chordata sequences (61390244 sequences; 21889222377 residues). Search criteria specified: enzyme, trypsin; fixed modifications, carbamidomethyl (C); variable modifications, oxidation (M) and deamidated (NQ); peptide tolerance, 100 ppm; MS/MS tolerance, 0.5 Da; instrument, MALDI-TOF-TOF. Results were filtered to accept only peptides with an expected score of 0.05 or lower.
Statistical analysis
The data (means±SD) of growth performance were analyzed by one way analysis of variance (ANOVA) followed by Tukey’s test to compare the means between individual treatments with SPSS at P<0.05 level. The significance of the difference in protein expression between control fish and 0.2% group was estimated by Students’t-test, P<0.05 by using Progenesis Samespots software.
Results
Growth performance
There was no significant fish mortality during the 2-month feeding trial among the three treatment groups. At the end of feeding trial, WG and FE levels of fish fed 0.2% diet were significantly higher than those of fish fed the control and 0.1% diets (Table 1).
Proteomics
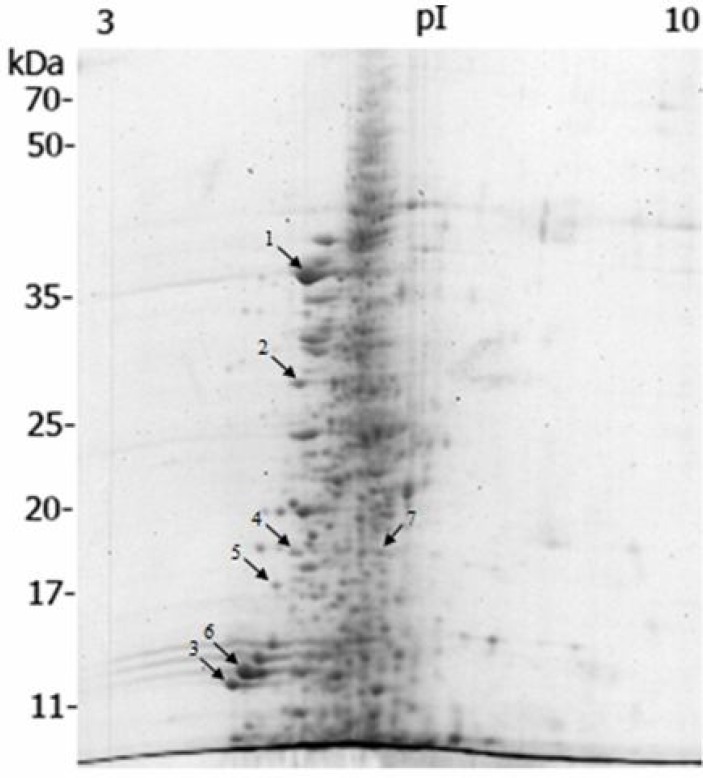
Coomassie stained 2-DE gel of rainbow trout muscle resolved over 580 spots having molecular weight (MW) values between approximately 5-70 kDa and pI values between 5-8 (Fig. 1).
Fig. 1.
2-DE map of muscle proteins of rainbow trout (Oncorhynchus mykiss), prepared by linear wide-range immobilized pH gradients (pH = 3-10, 17 cm; BioRad, USA) in the first dimension and on 12% SDS-PAGE for the second dimension. Proteins were stained with colloidal coomassie brilliant blue G-250. Labeled spots indicate identified proteins with significant altered expression profile after dietary β-glucan treatment (see Table 2
A total of 8 protein spots showed significant changes in the expression compared to the control group. Among these protein spots, 2 were upregulated and 6 down-regulated by dietary β-glucan supplementation. The differences in protein abundance between the two groups ranged from -1.9 to +1.3-fold.
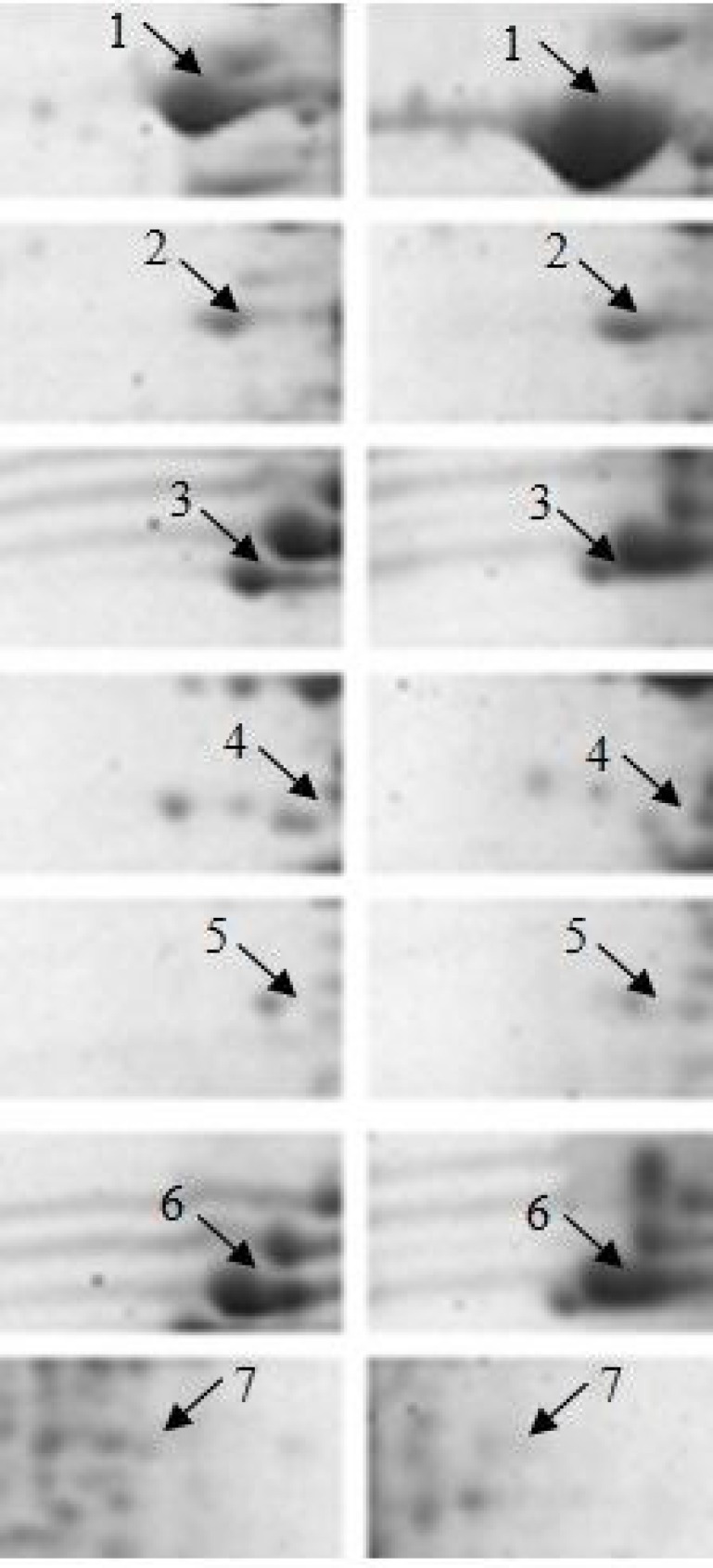
Based on the results from statistical analyses, spots that were found to differ significantly in abundance between control and 0.2% β-glucan fed fish were selected for identification. Out of 8 protein spots showing altered expression, 7 spots were successfully identified (Fig. 2). The protein identification success rate was 87.5% and among the 7 identified spots, 3 were identified by matching peptide data to Oncorhynchus mykiss protein sequences in the database, while the remaining spots were matched to proteins of Salmo salar, Salmo trutta and Cyprinus carpio. Table 2 indicates the accession number of identified proteins, fold change in abundance and the MASCOT database search results (score, percentage of sequence coverage and number of peptides matched).
Table 2.
Protein spots with significantly altered abundance between muscle from control and β-glucan fed rainbow trout
| Spot number | Accession numbera | Protein identification (species) | Fold changeb | Mascot score | SCc | PMd |
|---|---|---|---|---|---|---|
| Proteins increased in abundance in the β-glucan fed rainbow trout | ||||||
| 1 | gi|185132405 | Tropomyosin alpha-1 chain (Salmo salar) | +1.3 | 420 | 21% | 5 |
| 2 | gi|3063940 | Slow myotomal muscle tropomyosin (Salmo trutta) | +1.3 | 76 | 3% | 1 |
| Proteins decreased in abundance in the β-glucan fed rainbow trout | ||||||
| 3 | gi|197632465 | Myosin, light polypeptide 3-2 (Salmo salar) | -1.9 | 440 | 41% | 6 |
| 4 | gi|185134620 | Myosin light chain 1 (Oncorhynchus mykiss) | -1.6 | 149 | 11% | 2 |
| 5 | gi|185134620 | Myosin light chain 1 (Oncorhynchus mykiss) | -1.7 | 139 | 11% | 2 |
| 6 | gi|185134779 | Fast myosin light chain 2 (Oncorhynchus mykiss) | -1.5 | 384 | 24% | 4 |
| 7 | gi|806511 | Myosin heavy chain (Cyprinus carpio) | -1.4 | 263 | 3% | 3 |
NCBInr ID accession number,
Change in abundance in β-glucan fed rainbow trout relative to control,
Percentage of sequence coverage, and
Number of peptides matched
Two protein spots that were found to be increased in abundance in the β-glucan treated rainbow trout corresponded to tropomyosin alpha-1 chain (spot 1) and slow myotomal muscle tropomyosin (spot 2). The five spots that were down-regulated with dietary β-glucan supplementation were identified as different forms of myosin: myosin light polypeptide 3-2 (spot 3), myosin light chain 1 (spots 4 and 5), fast myosin light chain 2 (spot 6) and myosin heavy chain (spot 7).
Discussion
Oral administration of 0.2% β-glucan resulted in better growth performance in rainbow trout fry compared to fish fed the other diets. As a healthy and positive growth of fish that are in the early stages of development guarantees successful production in aquaculture industry, the size/age of fish used in our study was chosen. Similar to our results, several authors have reported that β-glucans such as yeast glucan obtained from Saccharomyces cerevisiae incorporated in fish feed increase the growth rate of certain species (Cook et al., 2003; Misra et al., 2006; Ai et al., 2007). However, in other reports no significant growth performance has been found after feeding fish with different β-glucans (Welker et al., 2007; Sealey et al., 2008). Growth enhancing effect of β-glucans may be dependent on the amount of β-glucan supplemented in the diet, duration of feeding, the size of fish and the species studied (Dalmo and Bøgwald, 2008 ▶).
As the characterization of the fish tissue proteomes is key to many aspects of fish aquaculture (Addis et al., 2010 ▶), the muscle proteome of the rainbow trout treated with dietary 0.2% β-glucan was analyzed in this study. The protein spots resolved on 2-DE map of rainbow trout muscle had pI values between 5 and 8. Similar pattern of protein resolution was also observed in a previous study on rainbow trout muscle proteome as an effect of dietary nucleotides (Keyvanshokooh and Tahmasebi-Kohyani, 2012 ▶). The protein identification rate in our study was 87.5% (7/8: number of identified/and number of differentially expressed protein spots), while using similar protein identification procedure, Keyvanshokooh and Tahmasebi-Kohyani (2012) ▶ reported higher protein discovery rate (100%; 8/8) in a rainbow trout muscle proteome experiment. Among the proteins identified, only 3 spots could be matched to rainbow trout proteins and the remaining spots were matched to the previously identified proteins of Salmo salar, Salmo trutta and Cyprinus carpio. Since the genome of rainbow trout is not completely sequenced, the protein identification success rate and peptide matching to rainbow trout proteins are expected to increase in future as more rainbow trout genomes are sequenced.
Fig. 2.
Comparison of 2-DE spots with significant altered expression level (P<0.05, Student’s t-test) between control and β-glucan-fed rainbow trout. The 2-D pattern of control is on the left and that of β-glucan treated fish is on the right
All of the identified proteins that changed in the muscle tissue of β-glucan treated rainbow trout were related to structural proteins including myosin and tropomyosin. The structural unit of muscular fibers is the sarcomere which is mainly composed of myosin, actin, tropomyosin, troponins and myosin light chains. Another major muscle protein fraction includes sarcoplasmic enzymes that are involved in glycolysis and energy metabolism (Addis et al., 2010 ▶). Myosin which is the most abundant protein in the muscle tissue is a protein of approximately 500 kDa and is composed of six subunits: two myosin heavy chains (MHCs) and four myosin light chains (MLCs) (Terova et al., 2011 ▶). In our study, four protein spots were identified as MLCs and one spot could be matched to MHC at low molecular weight (17 kDa), indicating that they are likely fragments of a protein with a high molecular weight. Regarding the lower abundance of myosin fragments in the muscle tissue of β-glucan fed fish, it can be concluded that dietary β-glucan improves the growth performance of rainbow trout by lessening the muscle atrophy caused by myosin degradation.
Tropomyosin is a dimer of α and β chains, which mediates interaction between troponin complex and actin that is needed for regulation of muscle contraction (Krasnov et al., 2003 ▶). In fish species, skeletal muscle growth is the result of both hyperplasia (an increase in myofiber number) and hypertrophy (an increase in myofiber size) (Greenlee et al., 1995 ▶). In contrast to mammals, in which the number of myocytes does not increase after birth, fish have a longer lasting capacity to recruit new myocytes (Goldspink et al., 2001 ▶). It is observed that small fish species like zebrafish grow mainly by hyperplasia, whereas species reaching a large adult body size grow mainly through hypertrophy (Rowlerson et al., 1997 ▶). Higher expression of tropomyosin and lower expression of myosin in our study may suggest that dietary β-glucan regulates the muscle growth of rainbow trout through an intermediate pattern of hyperplasia and hypertrophy. Consistent with these findings, growth enhancement of fish species through changes in muscle structural proteins has been reported in other feeding experiments (Alami-Durante et al., 2011 ▶; Keyvanshokooh and Tahmasebi-Kohyani, 2012 ▶).
In conclusion, we represented some changes in the muscle proteome of rainbow trout fed dietary β-glucan. The altered expression of structural proteins in fish fed β-glucan may be related to higher growth rate in rainbow trout. These findings provide basic information to understand possible mechanisms of dietary β-glucan contribution to better growth in fish.
Acknowledgements
This research was funded by Grant 93025104 from Iran National Science Foundation (Sandoogh-e Hemayat az Pajooheshgaran va Fannavaran-e Keshvar) and supported by Khorramshahr University of Marine Science and Technology. We thank Beyza Feed Mill (BFM), Beyza, Iran for supplying the experimental feed and also Sheshpir fish farm, Sepidan, Iran for supplying the experimental broodstocks and rearing facilities.
Conflict of interest
There are no known conflicts of interest associated with this publication.
References
- Addis, MF, Cappuccinelli, R, Tedde, V, Pagnozzi, D, Porcu, MC, Bonaglini, E, Roggio, T, Uzzau, S. Proteomic analysis of muscle tissue from gilthead sea bream (Sparus aurata, L) farmed in offshore floating cages. Aquaculture. 2010;309:245–252. [Google Scholar]
- Alami-Durante, H, Cluzeaud, M, Bazin, D, Mazurais, D, Zambonino-Infante, JL. Dietary cholecalci-ferol regulates the recruitment and growth of skeletal muscle fibers and the expressions of myogenic regulatory factors and the myosin heavy chain in European sea bass larvae. J. Nutr. 2011;141:2146–2151. doi: 10.3945/jn.111.146118. [DOI] [PubMed] [Google Scholar]
- Bostock, J, McAndrew, B, Richards, R, Jauncey, K, Telfer, T, Lorenzen, K, Little, D, Ross, L, Handisyde, N, Gatward, I, Corner, R. Aquaculture: global status and trends. Phil. Trans. R. Soc. B. 2010;365:2897–2912. doi: 10.1098/rstb.2010.0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Candiano, G, Bruschi, M, Musante, L, Santucci, L, Ghiggeri, GM, Carnemolla, B, Orecchia, P, Zardi, L, Righetti, PG. Blue silver: a very sensitive colloidal coomassie G-250 staining for proteome analysis. Electrophoresis. 2004;25:1327–1333. doi: 10.1002/elps.200305844. [DOI] [PubMed] [Google Scholar]
- Dalmo, RA, Bøgwald, J. β-glucans as conductors of immune symphonies. Fish shellfish Immunol. 2008;25:384–396. doi: 10.1016/j.fsi.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Ghaedi, G, Keyvanshokooh, S, Azarm, HM, Akhlaghi, M. Effects of dietary β-glucan on maternal immunity and fry quality of rainbow trout (Oncorhynchus mykiss) Aquaculture. 2015;441:78–83. [Google Scholar]
- Goldspink, G, Wilkes, D, Ennion, S. Myosin expression during ontogeny, post-hatching growth, and adaptation. Fish Physiol. 2001;18:43–72. [Google Scholar]
- Greenlee, AR, Dodson, MV, Yablonka-Reuveni, Z, Kersten, CA, Cloud, JG. In vitro differentia-tion of myoblasts from skeletal muscle of rainbow trout. J. Fish Biol. 1995;46:731–747. [Google Scholar]
- Keyvanshokooh, S, Tahmasebi-Kohyani, A. Proteome modifications of fingerling rainbow trout (Oncorhynchus mykiss) muscle as an effect of dietary nucleotides. Aquaculture. 2012;324:79–84. [Google Scholar]
- Krasnov, A, Teerijoki, H, Gorodilov, Y, Mölsä, H. Cloning of rainbow trout (Oncorhynchus mykiss) α-actin, myosin regulatory light chain genes and the 5′-flanking region of α-tropomyosin Functional assessment of promoters. J. Exp. Biol. 2003;206:601–608. doi: 10.1242/jeb.00116. [DOI] [PubMed] [Google Scholar]
- Pionnier, N, Falco, A, Miest, J, Frost, P, Irnazarow, I, Shrive, A, Hoole, D. Dietary β-glucan stimulate complement and C-reactive protein acute phase responses in common carp (Cyprinus carpio) during an Aeromonas salmonicida infection. Fish Shellfish Immunol. 2013;34:819–831. doi: 10.1016/j.fsi.2012.12.017. [DOI] [PubMed] [Google Scholar]
- Rodrigues, PM, Silva, TS, Dias, J, Jessen, F. Proteomics in aquaculture: applications and trends. J. Proteomics. 2012;75:4325–4345. doi: 10.1016/j.jprot.2012.03.042. [DOI] [PubMed] [Google Scholar]
- Rowlerson, A, Radaelli, G, Mascarello, F, Veggetti, A. Regeneration of skeletal muscle in two teleost fish: Sparus aurata and Brachydanio rerio. Cell Tissue Res. 1997;289:311–322. doi: 10.1007/s004410050878. [DOI] [PubMed] [Google Scholar]
- Terova, G, Addis, MF, Preziosa, E, Pisanu, S, Pagnozzi, D, Biosa, G, Gornati, R, Bernardini, G, Roggio, T, Saroglia, M. Effects of postmortem storage temperature on sea bass (Dicentrarchus labrax) muscle protein degradation: analysis by 2-D DIGE and MS. Proteomics. 2011;11:2901–2910. doi: 10.1002/pmic.201100073. [DOI] [PubMed] [Google Scholar]