Abstract
Previous genome-wide association studies using P-values to select single nucleotide polymorphisms (SNPs) have suffered from high false-positive and false-negative results. This case-control study recruited 713 late-onset Alzheimer’s disease (LOAD) cases and controls aged ≥65 from three teaching hospitals in northern Taiwan from 2007 to 2010. Performance metrics were used to select SNPs in stage 1, which were then genotyped to another dataset (stage 2). Four SNPs (CPXM2 rs2362967, APOC1 rs4420638, ZNF521 rs7230380, and rs12965520) were identified for LOAD by both traditional P-values (without correcting for multiple tests) and performance metrics. After correction for multiple tests, no SNPs were identified by traditional P-values. Simultaneous testing of APOE e4 and APOC1 rs4420638 (the SNP with the best performance in the performance metrics) significantly improved the low sensitivity of APOE e4 from 0.50 to 0.78. A point-based genetic model including these 2 SNPs and important covariates was constructed. Compared with elders with low-risks score (0–6), elders belonging to moderate-risk (score = 7–11) and high-risk (score = 12–18) groups showed a significantly increased risk of LOAD (adjusted odds ratio = 7.80 and 46.93, respectively; Ptrend < 0.0001). Performance metrics allow for identification of markers with moderate effect and are useful for creating genetic tests with clinical and public health implications.
Dementia is an important health issue in the elderly (aged ≥65 years). Alzheimer’s disease (AD) is the most common subtype of dementia and cannot currently be cured, prevented, or even slowed1. In 2010, the prevalence of AD among the elderly in the United States was 11%, and the disease was the fifth leading cause of death in this age group2. Taiwan has a very high aging rate3, and a large survey conducted between 2011 and 2012 showed that the prevalence of dementia was 4.84% in the elderly4. Several risk factors, including genetic factors, old age, female gender, vascular risk factors, life style, social factors, and environmental exposure, have been related to AD5. Among various genetic factors, single-nucleotide polymorphisms (SNPs), which are germline polymorphisms that remain stable throughout life, are particularly suitable for predicting disease risk at an early stage.
Apolipoprotein E (APOE) e4 has been associated with both familial and sporadic AD since 19936 and is the only genetic marker strongly associated with late-onset AD (LOAD). A meta-analysis showed that one APOE e4 allele (e4/e2 or e4/e3) was associated with a 2.7-fold (Caucasian) to 5.6-fold (Japanese) increased risk of AD; this risk increased further in people carrying two APOE e4 alleles (11.8-fold for Caucasians, 33.1-fold for the Japanese)7. Despite the strong association between APOE e4 status and the disease, the sensitivity of APOE e4 for predicting AD risk is only approximately 0.48, which indicates it has limited clinical implications.
In the past decade, genome-wide association studies (GWASs) have been widely conducted to identify genetic markers for AD9,10,11. GWASs in east-Asian populations are limited; only one Japanese GWAS12 has been conducted. Although several studies in China have validated SNPs identified from GWASs in the western countries13,14,15, no GWAS has been conducted in a Chinese population using microarray chips customized for Chinese populations. For LOAD, some genes have been consistently identified across GWASs, and these genes mainly belong to three biological pathways, i.e., the metabolic, trafficking, and signaling pathways16,17,18. These genes include APOE e4, CLU, CR1, PICALM, B1N1, ABCA7, the MS4A gene cluster, CD2AP, CD33, EPHA1, and TREM219,20,21,22,23,24. However, studies applying traditional P-values to identify genetic markers for various research outcomes have suffered from high false-positive and false-negative results25,26. Hence, these studies are unable to identify SNPs with moderate effects. In addition, these studies have mainly focused on Caucasians and may not be representative of other ethnic groups. Therefore, a recent trend of genetic analysis for resolving the issue of SNP selection includes approaches using high-dimensional multivariable modeling (e.g., penalized logistic regression and Bayesian analysis), clinical validity [i.e., sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV)], etc.27,28,29.
Clinical validity (i.e., sensitivity, specificity) has been proposed to be a more practical measure for selecting SNPs in GWASs30 and is important for evaluating and translating genetic tests for public health and clinical implications (Centers for Disease Control and Prevention, USA) (http://www.cdc.gov/genomics/gtesting/ACCE/). However, to the best of our knowledge, no GWAS has yet used clinical validity and other performance metrics [e.g., sensitivity, specificity, Youden index, PPV, NPV, diagnostic odds ratio (DOR), accuracy, net sensitivity, net specificity, and area under the receiver operating characteristic curve (AUC)] to identify SNPs for health outcomes. In this study, a two-stage design was adopted, and performance metrics were used to identify SNPs for predicting LOAD. Although APOE e4, which is determined by rs429358 and rs7412, is the best-known and strongest genetic factor for LOAD, it suffers from low sensitivity (0.4) for discriminating LOAD8. Simultaneous testing of APOE e4 and the identified SNPs was applied here in an attempt to improve sensitivity. In addition, a point-based genetic model including these two SNPs and important covariates was constructed to differentiate elders with low, moderate, and high risk of LOAD.
Results
Characteristics of the study population
After restriction and matching between the LOAD cases and controls in stage 1 (training set, n = 94), the distributions of age, years of education, and the APOE e4 status were significantly different between LOAD and the control groups (Table 1). In stage 2 (validation set, n = 619), age, years of education, body mass index (BMI, kg/m2), sex, APOE e4 status, alcohol consumption, smoking status, history of stroke, diabetes mellitus (DM), and hypercholesterolemia significantly differed between the LOAD and control groups.
Table 1. Demographics of the study population.
| Variables | Stage 1: Training set (N = 94) |
Stage 2: Validation set (N = 619) |
||||
|---|---|---|---|---|---|---|
| LOAD | Control | P-value | LOAD | Control | P-value | |
| N = 55 | N = 39 | N = 196 | N = 423 | |||
| Mean ± SD | Mean ± SD | |||||
| Age (years) | 78.3 ± 5.6 | 75.4 ± 4.8 | 0.01 | 79.9 ± 6.5 | 73.1 ± 5.9 | <0.0001 |
| Years of education | 8.3 ± 5.5 | 11.4 ± 4.4 | 0.004 | 7.9 ± 5.0 | 12.7 ± 4.1 | <0.0001 |
| BMI (kg/m2) | 23.3 ± 1.7 | 23.4 ± 1.9 | 0.75 | 22.8 ± 3.6 | 24.2 ± 3.1 | <0.0001 |
| N (%) | N (%) | |||||
| Female | 38 (69) | 24 (62) | 0.45 | 121 (62) | 210 (50) | 0.01 |
| APOE e4 carriers | 27 (50) | 4 (11) | <0.0001 | 72 (37) | 59 (15) | <0.0001 |
| Alcohol drinking | 4 (7) | 1 (3) | 0.32 | 27 (14) | 50 (12) | 0.50 |
| Ever smoking | — | — | — | 56 (29) | 75 (18) | <0.002 |
| Cardiovascular disease | 11 (20) | 7 (18) | 0.85 | 50 (26) | 137 (33) | 0.08 |
| Stroke | 2 (4) | 1 (3) | 0.77 | 7 (4) | 5 (1) | 0.05 |
| Hypertension | 24 (44) | 18 (46) | 0.81 | 74 (38) | 236 (56) | <0.0001 |
| DM | — | — | — | 48 (24) | 67 (16) | 0.01 |
| Hypercholesterolemia | — | — | — | 46 (24) | 141 (33) | 0.02 |
Abbreviations: LOAD, late-onset Alzheimer’s disease; SD, sample standard deviation; BMI, body mass index; APOE, apolipoprotein E; DM, diabetes mellitus.
The P-value was obtained by comparing the distribution of LOAD with the controls.
Numbers in bold indicate significant differences between cases and controls.
Because of restriction and matching in the training set (stage 1), some variables showed empty cells in the table.
Identification of SNPs in stage 1 (Training set)
In stage 1, 858 SNPs deviated from Hardy-Weinberg Equilibrium (HWE) tests and were excluded from further analysis. After quality control, the quantile-quantile (QQ) plot for the observed chi-square P-values of 500,941 SNPs revealed no deviation from the expected values, i.e., there was no significant association with the AD outcome as demonstrated by the diagonal line (Figure S1). Five SNPs (CCDC81 rs10501617, CPXM2 rs2362967, APOC1 rs4420638, ZNF521 rs7230380, and rs12965520) with the best performance (the largest number of performance metrics with the highest value) among the performance metrics were selected for genotyping in stage 2 (Table 2). The performance of these five SNPs in stage 1 was as follows: sensitivity (0.56–0.89), specificity (0.49–0.90), Youden index (0.38–0.46), PPV (0.71–0.89), NPV (0.60–0.78), DOR (6.4–11.6), accuracy (0.71–0.74), net sensitivity (0.78–0.95), net specificity (0.44–0.81), and AUC (0.860–0.888, Table 2).
Table 2. Comparison between two APOE SNPs and the top SNPs selected by performance metrics or traditional measures in stage 1 and stage 2.
| Gene | SNP | Performance metrics |
Traditional measures |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Youden | PPV | NPV | DOR | Accuracy | Net sensitivitya | Net specificitya | AUC (95% CI) | P-valueb | AOR (95% CI) | ||
| Stage 1 (Training set, N = 94) | |||||||||||||
| APOE e4 | rs429358 & rs7412c | 0.50 | 0.89 | 0.39 | 0.87 | 0.56 | 8.5 | 0.66 | — | — | 0.835 (0.749–0.922) | 3.0 × 10−4 | 11.68 (3.13–43.53) |
| CCDC81 | rs10501617 | 0.71 | 0.73 | 0.43 | 0.78 | 0.64 | 6.4 | 0.72 | 0.85 | 0.65 | 0.888 (0.822–0.955) | 5.8 × 10−4 | 3.3 (1.6–6.5) |
| CPXM2 | rs2362967 | 0.89 | 0.49 | 0.38 | 0.71 | 0.76 | 7.8 | 0.72 | 0.95 | 0.44 | 0.873 (0.801–0.944) | 3.4 × 10−5 | 3.6 (1.9–6.6) |
| APOC1 | rs4420638 | 0.56 | 0.90 | 0.46 | 0.89 | 0.60 | 11.6 | 0.71 | 0.78 | 0.81 | 0.873 (0.799–0.947) | 6.2 × 10−6 | 8.9 (3.0–26.2) |
| ZNF521 | rs7230380d | 0.89 | 0.53 | 0.42 | 0.72 | 0.78 | 9.0 | 0.74 | 0.95 | 0.47 | 0.879 (0.812–0.947) | 1.3 × 10−4 | 3.3 (1.8–6.1) |
| ZNF521 | rs12965520d | 0.89 | 0.53 | 0.42 | 0.72 | 0.78 | 9.0 | 0.74 | 0.95 | 0.47 | (0.812–0.947) | 1.3 × 10−4 | 3.3 (1.8–6.1) |
| BHLHB2 | rs2137946 | 0.38 | 0.18 | −0.44 | 0.39 | 0.17 | 0.13 | 0.29 | 0.43 | 0.15 | 0.860 (0.781–0.938) | 4.8 × 10−5 | 0.4 (0.2–0.7) |
| BHLHB2 | rs2137947 | 0.38 | 0.18 | −0.44 | 0.39 | 0.17 | 0.13 | 0.29 | 0.43 | 0.15 | 0.862 (0.785–0.939) | 4.8 × 10−5 | 0.4 (0.2–0.7) |
| Stage 2 (Validation set, N = 619) | |||||||||||||
| APOE e4 | rs429358 & rs7412c | 0.37 | 0.85 | 0.22 | 0.55 | 0.73 | 3.26 | 0.69 | — | — | 0.879 (0.850–0.909) | <0.0001 | 3.99 (2.36–6.75) |
| CCDC81 | rs10501617 | 0.44 | 0.54 | −0.02 | 0.31 | 0.67 | 0.91 | 0.51 | 0.64 | 0.46 | 0.879 (0.849–0.909) | 6.3 × 10−1 | 0.92 (0.64–1.31) |
| CPXM2 | rs2362967 | 0.73 | 0.30 | 0.03 | 0.33 | 0.71 | 1.16 | 0.44 | 0.83 | 0.25 | 0.879 (0.850–0.909) | 6.4 × 10−1 | 0.93 (0.68–1.28) |
| APOC1 | rs4420638 | 0.42 | 0.79 | 0.21 | 0.48 | 0.75 | 2.73 | 0.67 | 0.63 | 0.67 | 0.881 (0.852–0.911) | 9.0 × 10−2 | 1.85 (0.90–3.79) |
| ZNF521 | rs7230380 | 0.71 | 0.25 | −0.04 | 0.30 | 0.65 | 0.82 | 0.40 | 0.82 | 0.21 | 0.879 (0.850–0.909) | 2.4 × 10−1 | 3.07 (0.47–20.1) |
| ZNF521 | rs12965520 | 0.71 | 0.25 | −0.03 | 0.31 | 0.66 | 0.85 | 0.40 | 0.82 | 0.22 | 0.879 (0.850–0.909) | 2.8 × 10−1 | 0.36 (0.06–2.32) |
Abbreviations: SNP, single nucleotide polymorphism; PPV, positive predictive value; NPV, negative predictive value; DOR, diagnostic odds ratio; AOR, adjusted odds ratio; AUC, area under the receiver operating characteristic curve; CI, confidence interval.
aNet sensitivity and net specificity were obtained for each selected SNP and APOE e4 for simultaneous screening purposes.
bNone of the SNPs reached statistical significance (P < 10−7) after correction for multiple tests using the traditionally measured P-value.
cAPOE e4 status was determined by rs429358 & rs7412; the performance metrics estimation was based on APOE e4 carriers versus non-carriers.
dBecause of ties (P = 1.3 × 10−4 for ZNF521 rs7230380 and rs12965520), 6 SNPs were selected using the traditional P-value.
Except for APOE e4, the performance metrics or traditional measures for the remaining SNPs were estimated based on dominant genetic models.
For SNPs selected based on performance metrics or P-value, numbers in bold indicate the highest value of the corresponding indices or the lowest for P-value.
Italicised numbers indicate the top 5 SNPs selected by performance metrics and the top 6 SNPs (because of ties) selected by traditional P-values (regardless of statistical significance after correction for multiple tests), respectively.
BHLHB2 rs2137946 and rs2137947 were selected using traditional P-value regardless of statistical significance. Because these two SNPs were not identified by performance metrics, they were not genotyped in stage 2.
Numbers in bold indicate the largest value of each performance matrix or traditional measure.
For comparison with traditional GWASs, a Manhattan plot (Figure S2) was produced to assess the distribution of P-values by chromosome. Without correction for multiple tests, the top six SNPs (because of ties) identified by traditional P-value were CPXM2 rs2362967, APOC1 rs4420638, ZNF521 rs7230380 and rs12965520 and BHLHB2 rs2137946 and rs2137947. No single SNP was significantly associated with LOAD after correction for multiple tests because the P-values of these top six SNPs ranged from 1.3 × 10−4 to 6.2 × 10−6 (Table 2), and these values were larger than the Bonferroni-corrected P-values (α/number of SNPs = 10−7).
Construction of the genetic prediction model in stage 2 (Validation set)
The top five SNPs identified by performance metrics in stage 1 were genotyped in stage 2 (validation set). These SNPs showed no significant deviation from HWE among the controls (n = 423) after correction for multiple tests, and their minor allele frequencies (MAFs, 11–50%) were similar to those in the Han Chinese in Beijing from the HapMap dataset (12–50%, Table 3). Because of the strong effect of APOE e4 on LOAD risk observed in this study [e3/e4 vs. e3/e3: adjusted odds ratio (AOR) = 3.5; e4/e4 vs. e3/e3: AOR = 17.7] and a previous meta-analysis of Japanese subjects [e3/e4 vs. e3/e3: odds ratios (OR) = 3.9; e4/e4 vs. e3/e3: OR = 21.8]7, APOE e4 status was forced into the genetic model for AD. In stage 2, among the top five SNPs, APOC1 rs4420638 showed the best overall performance in performance metrics, especially in terms of specificity, Youden index, PPV, NPV, DOR, accuracy, net specificity, and AUC. However, AUC performance was very similar for the top 5 SNPs (AUC = 0.879–0.881); therefore, the other SNPs would likely be informative as well. This SNP was therefore selected for entry into the final model with APOE e4.
Table 3. Characteristics of 2 APOE SNPs and the top 5 SNPs selected by performance metrics in stage 2 (N = 619).
| Gene | SNP | Nucleotide change | Chromosome/Location | HapMap or dbSNP MAF |
Controls (n = 423) |
Cases (n = 196) |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| CEU | CHB | JPT | MAF | HWEP-value | MAF | HWE P-value | ||||
| APOE | rs429358 (APOE112) | T → C | 19/exon | 0.15* | 0.00 | 0.01 | 0.08 | 0.68 | 0.23 | 0.27 |
| APOE | rs7412 (APOE158) | C → T | 19/exon | 0.08* | 0.11 | 0.05 | 0.08 | 0.22 | 0.06 | 0.97 |
| APOC1 | rs4420638 | G → A | 19/3′UTR | 0.18 | 0.12 | 0.07 | 0.11 | 0.93 | 0.23 | 0.75 |
| CCDC81 | rs10501617 | T → C | 11/intronic | 0.40 | 0.20 | 0.16 | 0.27 | 0.37 | 0.26 | 0.64 |
| ZNF521 | rs7230380 | A → C | 18/intronic | 0.47 | 0.49 | 0.50 | 0.50 | 0.90 | 0.48 | 0.36 |
| ZNF521 | rs12965520 | T → C | 18/intronic | 0.50 | 0.45 | 0.50 | 0.50 | 0.58 | 0.48 | 0.33 |
| CPXM2 | rs2362967 | C → T | 10/intronic | 0.26 | 0.50 | 0.58 | 0.48 | 0.03 | 0.48 | 0.78 |
Abbreviations: SNP, single nucleotide polymorphism; UTR, untranslated region; CEU, Utah residents with Northern and Western European ancestry from the CEPH collection; JPT, Japanese in Tokyo, Japan; CHB, Han Chinese in Beijing; MAF, minor allele frequency; HWE, Hardy-Weinberg equilibrium test.
*MAF data were obtained from dbSNP because the data were not available from the HapMap Project.
Five factors that were identified as the most predictive of LOAD risk in the final (point-based) model, along with the coefficient values and numbers of points for each predictor, are shown in Table 4. In the point-based model, total risk scores ranged from 0 to 18, with a mean of 6.2 (standard deviation = 5.1). Key binary predictors included age (7 points if age >75), sex (1 point for women), years of education (6 points for ≤6 years), APOE e4 status (2 point for carriers), and APOC1 rs4420638 (2 point for carriers). The total point score showed ideal prediction ability for LOAD risk [AUC = 0.87; 95% confidence interval (CI) = 0.83–0.90]. Validation of the final model using leave-one-out cross-validation (LOOCV) techniques estimated optimism as 0.004 (AUC corrected for optimism = 0.866). The mean age of the study participants was 75 years, and the predictive ability of the model was ideal with slight differences in participants aged <75 years (AUC = 0.87) and ≥75 years (0.81).
Table 4. Final genetic model for predicting late-onset Alzheimer’s disease.
| Variables | Coefficient | Point |
|---|---|---|
| Age groups | ||
| 65–75 | 0 | 0 |
| >75 | 2.42 | 7 |
| Sex | ||
| Men | 0 | 0 |
| Women | 0.37 | 1 |
| Education | ||
| >6 years | 0 | 0 |
| ≤6 years | 2.11 | 6 |
| APOE e4 status | ||
| Non-carriers | 0 | 0 |
| Carriers | 0.91 | 2 |
| rs4420638 (APOC1) | ||
| Non-carriers | 0 | 0 |
| Carriers | 0.64 | 2 |
| Total range | 0–18 | |
| Mean score ± SD | 6.2 ± 5.1 | |
| AUC (95% CI) | 0.87 (0.83–0.90) | |
| Corrected for optimism | 0.866 | |
Abbreviations: SD, sample standard deviation; AUC, area under the receiver operating characteristic curve; CI, confidence interval.
The correction for optimism was performed by leave-one-out cross-validation (LOOCV) in stage 2 (validation set).
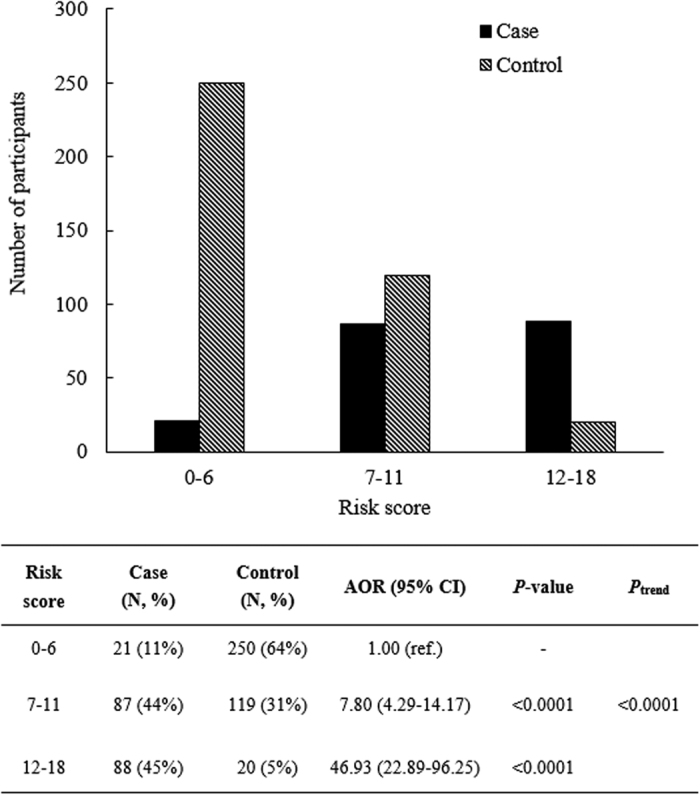
When participants were grouped based on their risk scores, 11% of the cases showed low risk (0–6 points, n = 21), 44% of the cases showed moderate risk (7–11 points, n = 87), and 45% of cases showed high risk (12–18 points, n = 88, Fig. 1). In addition, compared with the low-risk group, the moderate- and high-risk groups showed significantly increased risks of LOAD (AOR = 7.80 and 46.93, respectively; Ptrend < 0.0001, Fig. 1).
Figure 1. Distribution of late-onset Alzheimer’s disease cases and controls with low, moderate, and high risk.
Discussion
Five SNPs for predicting LOAD risk were identified in stage 1 (training set) by using performance metrics. These SNPs were located at four genes, including apolipoprotein C-I (APOC1) for rs4420638, carboxypeptidase-2 (CPXM2) for rs2362967, coiled-coil domain containing 81 (CCDC81) for rs10501617, and zinc finger protein521 (ZNF521) for rs7230380 and rs12965520. If SNPs were sorted by P-value regardless of statistical significance, four of the top SNPs identified by P-value (CPMX2 rs2362967, APOC1 rs4420638 and ZNF521 rs7230380 and rs12965520) were identical to the top five SNPs identified by performance metrics (Table 2). However, in both stages 1 and 2, none of the 500,941 SNPs reached statistical significance (based on P-value < 10−7) after correction for multiple tests.
APOE e4 is a strong risk factor of LOAD6; however, previous studies have indicated that its sensitivity was low (approximately 0.40)8,31, and this finding was confirmed in the current study (0.50 in stage 1 and 0.37 in stage 2). In an attempt to resolve this issue, simultaneous testing using APOE e4 and SNPs selected from the training set was applied in the present study. Among the top five SNPs, APOC1 rs4420638 showed the best overall performance across different indices (criteria were defined in the Materials and Methods section). In the Caucasian population, APOC1 rs4420638 and APOE rs429358 were in strong LD (D’ = 0.96, R2 = 0.72) based on the genotype data from the 1000 Genome Project (http://analysistools.nci.nih.gov/LDlink/?tab=home). However, these two SNPs were not in strong LD (D’ = 0.6, R2 = 0.012) in our population, and APOC1 rs4420638 remained significant conditioning on APOE e4 status (adjusted odds ratio = 8.9, 95% CI = 3.0–26.2). The discrepancy may result from different ethnic groups. Therefore, the inclusion of APOE e4 status and APOC1 rs4420638 does not indicate the same signal in this population. Point-based genetic model showed that elders belonging to moderate-risk (risk score = 7–11) and high-risk (score = 12–18) groups presented significantly increased risk of LOAD (AOR = 7.80 and 46.93, respectively; Ptrend < 0.0001, Fig. 1) compared with elders with low risk scores (0–6). Although addition of APOC1 rs4420638 did not significantly improve the prediction ability of the model, simultaneous testing with APOE e4 significantly increased the sensitivity of APOE e4 from 0.50 to 0.78, which indicates simultaneous testing could have useful clinical and public health implications.
Although the top five SNPs selected by performance metrics did not reach statistical significance based on traditional P-values, they have been directly or indirectly related to LOAD as discussed below. A GWAS in Caucasians found that APOC1 rs4420638 was associated with LOAD risk32, which is consistent with our findings. APOC1 is produced by astrocytes33 and regulates lipoprotein metabolism via its interaction with APOE34 by masking or altering the conformation of APOE on lipoprotein particles35. In addition, an animal study showed that apoC-1 may affect cognitive functions by lowering the expression of apoE or offsetting the effects of apoE on lipid distribution in the brains of mice36.
The SNP rs236967 is located on the CPXM2 gene, which plays an important role in synaptic integrity and remodeling, cell adhesion37,38, and upregulation of clusterin (CLU), a gene previously linked to LOAD39. CPXM2 is also related to AD40, Parkinson’s disease, and schizophrenia38,41. However, the only epidemiologic study to investigate this gene did not identify a significant association with LOAD42, probably because of the moderate effects of this SNP.
CCDC81 has been associated with colorectal cancer43, but direct evidence to link this gene with LOAD is not available. Recent studies found that AD risk was inversely associated with cancer risk because of upregulation of oxidative phosphorylation in AD and glycolysis in cancer44,45,46. Therefore, the observed association between CCDC81 rs10501617 and LOAD risk in this study (AOR = 3.3, Table 2) may be a result of this indirect relationship.
Both mice and human studies have shown that ZNF521 plays a role in sustaining neural differentiation, regulating neural differentiation in stem cells and brain development47,48. These findings may explain the associations between ZNF521 rs7230380 and rs12965520 and LOAD in the present study (AOR = 3.3), which have not been previously reported. Because of the complexity of AD, the mechanism underlying the modulation of disease progression by multiple genes remains to be elucidated. BHLHB2 rs2137946 and rs2137947 were among the top SNPs selected by traditional P-values if no correction of multiple tests was applied. However, because they were not identified by the performance metrics and did not reach statistical significance after Bonferroni correction, their mechanisms are not described here.
The strengths of this study are as follows. First, this study compared SNPs selected by traditional P-values, which suffer from high false-positive and false-negative results49, with those selected by performance metrics. The application of performance metrics appears to allow for greater inclusion of factors with moderate effects and helps balance the risks, benefits, and costs of genetic markers for application in public health or clinical units30. Second, this study used extensive matching and restriction in stage 1, thereby allowing greater statistical efficiency, reducing the sample size, and subsequently lowering the cost of microarray assays. Third, simultaneous testing using APOE e4 and APOC1 rs4420638, the SNP identified by performance metrics, significantly enhanced the low sensitivity of APOE e4 from 0.50 to 0.78 and thus made the genetic model more applicable.
This study presents a number of limitations. This work is a case-control study and may suffer from recall bias. However, a high concordance rate was observed between self-reports and medical record-confirmed vascular disease based on a random sample of 5% of all participants in the study. In addition, previous studies showed that participants’ awareness of major health issues diagnosed by physicians tended to be correct50,51,52. Finally, the matching ratio for cases to controls is sub-optimal (<1) because of the multiple matching criteria applied, which allowed us to control for several covariates at a time but also limited the number of controls who met the criteria.
To the best of our knowledge, this study is the first to compare SNPs for LOAD prediction selected by performance metrics with those selected by traditional P-values. Simultaneous testing using APOC1 rs4420638 and APOE e4 to predict LOAD risk significantly improved the low sensitivity of APOE e4. A point-based genetic model based on these two SNPs and important covariates successfully differentiated elders with a low, moderate, and high risk of LOAD. Our findings revealed that performance metrics are an excellent alternative for identifying SNPs for disease prediction and are highly applicable for creating genetic tests with public health and clinical implications.
Materials and Methods
Study population
The present work was a two-stage case-control study including 294 mild to moderate LOAD cases and 503 controls recruited from the neurology clinics of three teaching hospitals in northern Taiwan from 2007 to 2010. All participants were aged 65 years or older. Each participant provided blood samples for genotyping and microarray analysis. Participants were excluded (n = 84) if blood samples were not obtained or if they had any of the following conditions or diseases: depression, Parkinson’s disease, hemorrhagic stroke, cerebral infarction, and/or organic brain tumors. After exclusion, a total of 251 LOAD cases and 462 controls were included for statistical analysis.
In stage 1 (training set), LOAD cases and controls were matched in terms of age (±5 years), gender, birthplace of parents/grandparents, and comorbidities (hypercholesterolemia, hypertension, DM, and head injury). Participants were further selected based on the following restriction criteria: 18.5 kg/m2 < BMI < 27 kg/m2 and no history of cigarette smoking. In total, 55 matched pairs (55 LOAD cases and 39 controls; one control may have matched with more than one LOAD case) were selected for genome-wide microarray scans. For comparison, five SNPs with the best performance in the performance metrics (SNPs with the largest number of indices with the highest performance metrics) and the smallest P-values (traditional approach) were selected. These SNPs were genotyped in another dataset (stage 2: 196 LOAD cases and 423 controls) for validation.
Performance metrics
Performance metrics estimated in this study included clinical validity [sensitivity = TP/(TP + FN) and specificity = TN/(FP + TN)], Youden index (sensitivity + specificity − 1), positive predictive value [PPV = TP/(TP + FP)], negative predictive value [NPV = TN/(FN + TN)], diagnostic odds ratio [DOR = (TP/FP)/(FN/TN)], accuracy [(TP + TN)/(TP + TN + FP + FN)], net sensitivity [(sensitivity of APOE e4 + sensitivity of selected SNP) − (sensitivity of APOE e4 × sensitivity of selected SNP)], net specificity (specificity of APOE e4 × specificity of selected SNP) and AUC. Here TP denotes true positive, FN denotes false negative, FP denotes false positive, and TN denotes true negative.
Ethics statement
The study protocol was approved by the Institutional Review Boards of National Taiwan University Hospital, En Chu Kong Hospital, and Cardinal Tien Hospital. Written informed consent was obtained from each study participant. Consent from the legal guardian/next of kin was obtained when patients had serious cognitive impairment. This study complies with the World Medical Association Declaration of Helsinki.
Covariates
A self-reporting questionnaire (Table S1) was administered to collect information on demography, lifestyle (e.g., cigarette smoking and alcohol consumption), and disease comorbidity (e.g., hypertension, DM, cardiovascular disease, and hypercholesterolemia).
Assessment of Alzheimer’s disease
A neurologist at each hospital diagnosed potential dementia cases. The Mini-Mental State Examination53 and Clinical Dementia Rating54 were used to evaluate cognitive function. The diagnosis of probable dementia was evaluated using the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition55. Head magnetic resonance imaging scans and computed tomography were taken to exclude participants with organic lesions. LOAD diagnosis was based on the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s Disease and Related Disorders Association Alzheimer’s Criteria56. Controls with complete independence in activity of daily living and instrumental activity of daily living were assessed using a Short Portable Mental Status Questionnaire57, and those with possible cognitive impairment and other mental disorders were excluded from this study.
Collection and pretreatment of biospecimens
Blood samples were collected from each participant in tubes containing sodium EDTA for genotyping. After centrifugation, genomic DNA was extracted from the buffy coat using a QuickGene-Mini 80 system (Fujifilm, Tokyo, Japan) and then stored at −80 °C. To ensure the quality of the microarray genotyping assay, all DNA samples were required to meet the following criteria: OD260/OD280 (DNA quality) = 1.8–2.0, OD260/OD230 (remaining organic solvent) >1.5 and zero fragments of genomic DNA.
Genotyping assays
For stage 1, the Axiom™ Genome-Wide CHB 1 Array Plate (Affymetrix Inc., Santa Clara, California), which includes 563,746 SNPs, was used for genotyping. This array chip is optimized for the best coverage of common variants in the Chinese population. For the microarray data, SNPs were excluded based on the following quality control criteria: (1) genotyping success rate <90%, (2) call rate <98%, (3) MAF <0.05, or (4) P-value of HWE tests in controls <0.0001. In total, 511,718 SNPs were included after frequency and genotyping pruning. Because frequencies differed between SNPs on sex and autosomal chromosomes, SNPs located on sex chromosomes were also excluded from analysis. After further exclusion of 10,877 SNPs, 500,941 SNPs remained for further analysis.
APOE e4 genotypes were determined by rs429358 and rs741258. Because these two SNPs were not included in the array chip, they were genotyped together with the top five SNPs selected by performance metrics at stage 1 for all participants (stages 1 and 2) by employing TaqMan Genomic Assays using an ABI 7900HT Fast Real-time PCR system (Applied Biosystems Inc., CA, USA). We selected the top 5 SNPs because these SNPs showed moderate association with the outcome and the inclusion of more SNPs may not improve AD prediction ability. In addition, based on the principle of parsimony for model building and better generalizability, we avoided including too many predictors. The genotyping call rate was greater than 95% for SNPs determined by the TaqMan assays. The internal quality control obtained from 5% of the samples in duplicate had a concordance rate of 100%.
Statistical analyses
To compare the distribution between cases and controls, Student’s t-tests and Mann-Whitney U tests were used for normally and non-normally distributed continuous variables, respectively. Normality of continuous variables were checked by visual inspection if there was strong deviation from the diagonal line on QQ plots. Chi-square tests were used for categorical variables.
The PLINK program (http://pngu.mgh.harvard.edu/~purcell/plink/) was used to estimate genotype frequency, P-value of the HWE test, and MAF for each SNP. Instead of using an additive model, a dominant genetic model was used to build a contingency table. Performance metrics, e.g., sensitivity, specificity, PPV and NPV and others, were estimated by classifying the genotype data into positive (carriers of a variant allele) and negative (wildtype) disease state predictions. Five SNPs with the best performance in the performance metrics were selected for genotyping in stage 2 (validation set). Traditional P-values and AORs were calculated for comparison. Simultaneous testing of APOE e4 and each of the five SNPs selected in stage 1 was performed to estimate the net sensitivity and net specificity. The SNP with the best overall performance in the performance metrics was used to construct the genetic model for predicting LOAD. APOE e4 was forced into the genetic model because it is a well-known genetic risk factor for LOAD.
Multivariable logistic regression models were used to estimate AORs and 95% CIs for LOAD cases in the dominant genetic model. For stage 1 (training set), the regression model was conditioned on the matched set. For stage 2 (validation set), the study participants were conditioned on an age interval of 5 years to control for the confounding effect of age, i.e., cases and controls were compared within each stratum in the multivariable analysis. Continuous age variable was also adjusted in the model to control for the residual confounding within each 5-year age stratum. In the final genetic model, age (65–75 and >75 years old), sex, years of education (>6 and ≤6 years), APOE e4 status (carriers vs. non-carriers), and SNPs identified by performance metrics were adjusted. Each variable was then assigned a point value by dividing its model coefficient value with the coefficient for sex, i.e., the smallest coefficient for a dichotomous variable in the final model, rounding up to the nearest integer. This point-based approach has previously been successfully used to develop clinical prediction tools59,60. In addition, the AUC statistic was also used for model discrimination, where AUC ≥ 0.7 indicates acceptable discriminative ability. LOOCV was used to evaluate the internal validity of the final model.
For comparison with traditional GWAS results, a Manhattan plot and a QQ plot were produced using PLINK and R (http://www.r-project.org/), respectively, to assess whether the P-values deviated from the expected distribution (i.e., H0: no association between SNPs and LOAD). SAS version 9.4 (SAS Institute, Cary, NC) was used to conduct all statistical analyses, and all statistical tests were two-sided.
Additional Information
How to cite this article: Chen, Y.-C. et al. Performance Metrics for Selecting Single Nucleotide Polymorphisms in Late-onset Alzheimer’s Disease. Sci. Rep. 6, 36155; doi: 10.1038/srep36155 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
Funding for this study was provided by grants from the Ministry of Science and Technology in Taiwan (96-2314-B-002-197, 97-2314-B-002-168-MY3, 104-0210-01-09-02, and 105-0210-01-13-01) and grants from Academia Sinica. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions Y.-C.C., J.-H.C., C.-J.C. and H.-I.Y. conceived and designed the experiments. C.-J.H. and H.-H.H. performed the experiments. Y.-C.C., C.-J.H. and C.-C.J. analyzed the data. Y.-C.C., W.-C.L., J.-M.C., C.-J.C. and H.-I.Y. contributed experimental and analysis tools. Y.-C.C. and C.-J.H. contributed to the writing of the manuscript. J.-H.C., T.-F.C., Y.S., L.-L.W., P.-K.Y. and Y.-M.C. were responsible for sample pretreatment and participant recruitment. All authors reviewed the manuscript.
References
- Alzheimer’s Association. Alzheimer’s disease facts and figures. Alzheimer’s Association 10, e47–e92 (Alzheimer’s Association 2014). [DOI] [PubMed] [Google Scholar]
- Murphy S., Xu J. & Kochanek K. D. Deaths: Final Data for 2010. (Center for Disease Control and Prevention, U.S. Department of Health and Human Services, Hyattsville, MD, 2013). [PubMed] [Google Scholar]
- Council for Economic Planning and Development. Situation and trend of global population aging, Taiwan (2013).
- Sun Y. et al. A nationwide survey of mild cognitive impairment and dementia, including very mild dementia, in Taiwan. PLOS ONE 9, e100303 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. H., Lin K. P. & Chen Y. C. Risk factors for dementia. J. Formos. Med. Assoc. 108, 754–764 (2009). [DOI] [PubMed] [Google Scholar]
- Corder E. H. et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261, 921–923 (1993). [DOI] [PubMed] [Google Scholar]
- Bertram L. et al. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat. Genet. 39, 17–23 (2007). [DOI] [PubMed] [Google Scholar]
- Wang M. & Jia J. The interleukin-6 gene −572C/G promoter polymorphism modifies Alzheimer’s risk in APOE ɛ4 carriers. Neurosci. Lett. 482, 260–263 (2010). [DOI] [PubMed] [Google Scholar]
- Bertram L. & Tanzi R. E. Genome-wide association studies in Alzheimer’s disease. Hum. Mol. Genet. 18, R137–R145 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuyvers E. & Sleegers K. Genetic variations underlying Alzheimer’s disease: evidence from genome-wide association studies and beyond. Lancet Neurol. 15, 857–868 (2016). [DOI] [PubMed] [Google Scholar]
- Kamboh M. et al. Genome-wide association study of Alzheimer’s disease. Transl. Psychiatr. 2, e117 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano A. et al. A genome-wide association study of late-onset Alzheimer’s disease in a Japanese population. Psychiatr. Genet. 25, 139–146 (2015). [DOI] [PubMed] [Google Scholar]
- Tan L. et al. Association of GWAS-linked loci with late-onset Alzheimer’s disease in a northern Han Chinese population. Alzheimers Dement. 9, 546–553 (2013). [DOI] [PubMed] [Google Scholar]
- Wang H. Z. et al. Validating GWAS-identified risk loci for Alzheimer’s disease in Han Chinese populations. Mol. Neurobiol. 53, 379–390 (2016). [DOI] [PubMed] [Google Scholar]
- Zhu X. C. et al. Association of Parkinson’s disease GWAS-linked loci with Alzheimer’s disease in Han Chinese. Mol. Neurobiol., 1–11 (2016). [DOI] [PubMed] [Google Scholar]
- Yao P. J. Synaptic frailty and clathrin-mediated synaptic vesicle trafficking in Alzheimer’s disease. Trends Neurosci. 27, 24–29 (2004). [DOI] [PubMed] [Google Scholar]
- Mattson M. P. & Chan S. L. Neuronal and glial calcium signaling in Alzheimer’s disease. Cell Calcium 34, 385–397 (2003). [DOI] [PubMed] [Google Scholar]
- Blass J. P. Brain metabolism and brain disease: is metabolic deficiency the proximate cause of Alzheimer dementia? J. Neurosci. Res. 66, 851–856 (2001). [DOI] [PubMed] [Google Scholar]
- Naj A. C. et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat. Genet. 43, 436–441 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingworth P. et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat. Genet. 43, 429–435 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri S. et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. J. Am. Med. Assoc. 303, 1832–1840 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J. C. et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat. Genet. 41, 1094–1099 (2009). [DOI] [PubMed] [Google Scholar]
- Harold D. et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet. 41, 1088–1093 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro R. et al. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 368, 117–127 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter D. J. & Kraft P. Drinking from the fire hose–statistical issues in genomewide association studies. N. Engl. J. Med. 357, 436–439 (2007). [DOI] [PubMed] [Google Scholar]
- Johnson R. C. et al. Accounting for multiple comparisons in a genome-wide association study (GWAS). BMC genomics 11, 724 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft P. & Haiman C. A. GWAS identifies a common breast cancer risk allele among BRCA1 carriers. Nat. Genet. 42, 819–820 (2010). [DOI] [PubMed] [Google Scholar]
- Sambo F. et al. Bag of Naïve Bayes: biomarker selection and classification from genome-wide SNP data. BMC bioinformatics 13, S2 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber V., Silva A. P. D. & Strimmer K. A novel algorithm for simultaneous SNP selection in high-dimensional genome-wide association studies. BMC bioinformatics 13, 284 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft P. et al. Beyond odds ratios-communicating disease risk based on genetic profiles. Nat. Rev. Genet. 10, 264–269 (2009). [DOI] [PubMed] [Google Scholar]
- Chen Y. C. et al. Sequence variants of toll like receptor 4 and late-onset Alzheimer’s disease. PLOS ONE 7, e50771 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L. et al. Genome-wide association analysis reveals putative Alzheimer’s disease susceptibility loci in addition to APOE. Am. J. Hum. Genet. 83, 623–632 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit-Turcotte C. et al. Apolipoprotein C-I expression in the brain in Alzheimer’s disease. Neurobiol. Dis. 8, 953–963 (2001). [DOI] [PubMed] [Google Scholar]
- Carter C. J. Convergence of genes implicated in Alzheimer’s disease on the cerebral cholesterol shuttle: APP, cholesterol, lipoproteins, and atherosclerosis. Neurochem. Int. 50, 12–38 (2007). [DOI] [PubMed] [Google Scholar]
- Jong M. C., Hofker M. H. & Havekes L. M. Role of ApoCs in lipoprotein metabolism: functional differences between ApoC1, ApoC2, and ApoC3. Arterioscler. Thromb. Vasc. Biol. 19, 472–484 (1999). [DOI] [PubMed] [Google Scholar]
- Abildayeva K. et al. Human apolipoprotein C-I expression in mice impairs learning and memory functions. J. Lipid. Res. 49, 856–869 (2008). [DOI] [PubMed] [Google Scholar]
- Ihara M. et al. Association of the cytoskeletal GTP-binding protein Sept4/H5 with cytoplasmic inclusions found in Parkinson’s disease and other synucleinopathies. J. Biol. Chem. 278, 24095–24102 (2003). [DOI] [PubMed] [Google Scholar]
- Hoepken H. H. et al. Parkinson patient fibroblasts show increased alpha-synuclein expression. Exp. Neurol. 212, 307–313 (2008). [DOI] [PubMed] [Google Scholar]
- Lin Y. L. et al. Genetic polymorphisms of clusterin gene are associated with a decreased risk of Alzheimer’s disease. Eur. J. Epidemiol. 27, 73–75 (2012). [DOI] [PubMed] [Google Scholar]
- Li Y. H. et al. IT in Medicine & Education, 2009. IEEE International Symposium on. 1, 38–49 (2009).
- Hashimoto R. et al. Genome-wide association study of cognitive decline in schizophrenia. Am. J. Psychiatry. 170, 683–684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grupe A. et al. A scan of chromosome 10 identifies a novel locus showing strong association with late-onset Alzheimer disease. Am. J. Hum. Genet. 78, 78–88 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonso P. et al. Proteome analysis of membrane fractions in colorectal carcinomas by using 2D-DIGE saturation labeling. J. Proteome. Res. 7, 4247–4255 (2008). [DOI] [PubMed] [Google Scholar]
- Behrens M. I., Lendon C. & Roe C. M. A common biological mechanism in cancer and Alzheimer’s disease? Curr. Alzheimer. Res. 6, 196–204 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thinnes F. P. Nonmelanoma skin cancer is associated with reduced Alzheimer disease risk. Neurology 81, 2056 (2013). [DOI] [PubMed] [Google Scholar]
- Musicco M. et al. Inverse occurrence of cancer and Alzheimer disease: a population-based incidence study. Neurology 81, 322–328 (2013). [DOI] [PubMed] [Google Scholar]
- Kamiya D. et al. Intrinsic transition of embryonic stem-cell differentiation into neural progenitors. Nature 470, 503–509 (2011). [DOI] [PubMed] [Google Scholar]
- Shen S., Pu J., Lang B. & McCaig C. D. A zinc finger protein Zfp521 directs neural differentiation and beyond. Stem Cell Res. Ther. 2, 20 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens A. C. & van Duijn C. M. Genome-based prediction of common diseases: advances and prospects. Hum. Mol. Genet. 17, R166–R173 (2008). [DOI] [PubMed] [Google Scholar]
- El Fakiri F., Bruijnzeels M. A. & Hoes A. W. No evidence for marked ethnic differences in accuracy of self-reported diabetes, hypertension, and hypercholesterolemia. J. Clin. Epidemiol. 60, 1271–1279 (2007). [DOI] [PubMed] [Google Scholar]
- St Sauver J. L. et al. Agreement between patient reports of cardiovascular disease and patient medical records. Mayo. Clin. Proc. 80, 203–210 (2005). [DOI] [PubMed] [Google Scholar]
- Okura Y. et al. Agreement between self-report questionnaires and medical record data was substantial for diabetes, hypertension, myocardial infarction and stroke but not for heart failure. J. Clin. Epidemiol. 57, 1096–1103 (2004). [DOI] [PubMed] [Google Scholar]
- Folstein M. F., Folstein S. E. & McHugh P. R. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res. 12, 189–198 (1975). [DOI] [PubMed] [Google Scholar]
- Morris J. C. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology 43, 2412–2414 (1993). [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders, Fourth Edition (2000). [Google Scholar]
- McKhann G. et al. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA work group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s disease. Neurology 34, 939–944 (1984). [DOI] [PubMed] [Google Scholar]
- Pfeiffer E. A short portable mental status questionnaire for the assessment of organic brain deficit in elderly patients. J. Am. Geriatr. Soc. 23, 433–441 (1975). [DOI] [PubMed] [Google Scholar]
- Ghebranious N., Ivacic L., Mallum J. & Dokken C. Detection of ApoE E2, E3 and E4 alleles using MALDI-TOF mass spectrometry and the homogeneous mass-extend technology. Nucleic. Acids. Res. 33, e149 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes D. E. et al. A point-based tool to predict conversion from mild cognitive impairment to probable Alzheimer’s disease. Alzheimers Dement. 10, 646–655 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. J., Lindquist K., Segal M. R. & Covinsky K. E. Development and validation of a prognostic index for 4-year mortality in older adults. J. Am. Med. Assoc. 295, 801–808 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


