Abstract
Myotonic Dystrophy type 1 (DM1) originates from alleles of the DMPK gene with hundreds of extra CTG repeats in the 3′ untranslated region (3′ UTR). CUG repeat RNAs accumulate in foci that sequester Muscleblind-like (MBNL) proteins away from their functional target transcripts. Endogenous upregulation of MBNL proteins is, thus, a potential therapeutic approach to DM1. Here we identify two miRNAs, dme-miR-277 and dme-miR-304, that differentially regulate muscleblind RNA isoforms in miRNA sensor constructs. We also show that their sequestration by sponge constructs derepresses endogenous muscleblind not only in a wild type background but also in a DM1 Drosophila model expressing non-coding CUG trinucleotide repeats throughout the musculature. Enhanced muscleblind expression resulted in significant rescue of pathological phenotypes, including reversal of several mis-splicing events and reduced muscle atrophy in DM1 adult flies. Rescued flies had improved muscle function in climbing and flight assays, and had longer lifespan compared to disease controls. These studies provide proof of concept for a similar potentially therapeutic approach to DM1 in humans.
Myotonic dystrophy type 1 (DM1) is an incurable neuromuscular disorder that is caused by an expanded CTG*CAG repeat in the 3′-untranslated region (3′ UTR) of the dystrophia myotonica-protein kinase (DMPK) gene (for a recent review, see ref. 1). The normal human DMPK gene harbors 5–37 copies of the trinucleotide motif, but a dynamic mutation may increase this number to over 5000 repeat copies. Clinically, DM1 is a multisystemic disorder, which mainly affects skeletal muscle, the heart and the nervous system. Severity of disease correlates with the expansion size and typical disease features are myotonia, muscle weakness and atrophy, smooth and cardiac muscle involvement, CNS dysfunction, somnolence, endocrine disorders and reduced life span2,3.
Expression of expanded alleles in DM1 results in the nuclear retention of mutant DMPK mRNA and reduced DMPK protein levels4. Mutant transcripts sequester Muscleblind-like (MBNL) splicing factors, leading to the abnormal alternative splicing of a multitude of other transcripts and the expression of fetal forms of their protein products in DM1 adults5,6,7. Spliceopathy is therefore thought to be the major factor underlying the pathogenesis of DM1. However, alternative mechanisms such as additional changes in gene expression, antisense transcripts, translation efficiency, misregulated alternative polyadenylation and miRNA deregulation may also contribute to the pathogenesis of DM18,9,10,11,12,13,14,15.
Several therapeutic approaches have been tested in DM1 animal models. Among them, the most exciting results derived from blocking the interaction between MBNL and toxic RNA using small molecules, peptides, morpholinos or antisense oligonucleotides, and gapmers to degrade the mutant transcripts16,17,18,19,20,21. A less explored alternative in DM1 is the therapeutic modulation of MBNL gene expression. Although the expression of CUG expansions triggers different molecular alterations, current evidence points to MBNL depletion as the main cause of disease symptoms. A Mbnl1 knock-out (KO) mouse model displays myotonia, missplicing of muscular transcripts and cataracts, which are all characteristic symptoms of DM1 disease22. More recently, relevant cardiac dysfunction features have been described in 2 month-old Mbnl1 mutant mice (hypertrophy, interstitial fibrosis and splicing alterations), which suggests a role for Mbnl1 reduction in the cardiac problems in DM123. However, Mbnl1 KO mice do not display the whole set of symptoms of DM1. Therefore, it has been hypothesized that Mbnl2 could compensate for Mbnl1 loss of function in these mice. In fact, Mbnl1 KO mice with reduced expression of Mbnl2 (Mbnl1−/−; Mbnl2+/−), are viable but develop most of the cardinal defects of the disease, including reduced lifespan, cardiac blockage, severe myotonia, atrophic fibers and progressive weakness of skeletal muscles. In support of the compensation hypothesis, levels of Mbnl2 are increased in Mbnl1−/− KO mice and Mbnl2 can regulate the splicing of exons which are normally regulated by Mbnl124. Furthermore, genetic polymorphisms in the human MBNL1 gene promoter have been associated with the severity of the disease25.
Several observations suggest that MBNL1 overexpression has potential for treating DM1 pathology. Firstly, administration of recombinant Mbnl1 protein to a HSALR mouse model of DM1, rescues myotonia and the splicing alterations characteristic of DM126. Secondly, we showed that the overexpression of a muscleblind isoform partially rescues muscle atrophy in a Drosophila DM1 model27. Finally, MBNL1 overexpression is well tolerated in skeletal muscle in transgenic mice where it causes only relatively minor splicing changes but no effect on longevity28.
In this proof of concept study, we use the Drosophila DM1 model to explore the therapeutic potential of silencing specific microRNAs (miRNAs) and thus boost muscleblind (mbl) expression. The fundamental roles of miRNAs in the regulation of gene expression have been well-established. These endogenous ∼22 nucleotide-long non-coding RNAs act post-transcriptionally and exert their regulatory effects mainly by binding to the 3′ UTR of target mRNAs, which results in mRNA deadenylation and decay, translational suppression or, rarely, mRNA cleavage29,30,31,32,33. Starting from a set of miRNAs predicted as muscleblind regulators, we confirmed that specific silencing of two of them, using sponge constructs, which sequester the miRNAs, upregulated muscleblind mRNA and protein. Similar effects were observed in flies co-expressing 480 CTG interrupted repeats (i(CTG)480) and either of the two sponge constructs in muscle. Muscleblind upregulation was sufficient to rescue characteristic DM1 model phenotypes such as missplicing events, reduced lifespan, and muscle atrophy. Importantly, the rescue of muscle atrophy resulted in improved climbing and flight ability in DM1 model flies. These data provide proof-of-principle for the therapeutic potential of Muscleblind upregulation by specific miRNA inhibitors in DM1 patients.
Results
Silencing of dme-miR-277 or dme-miR-304 derepresses muscleblind in Drosophila muscle
Muscleblind sequestration in RNA foci and subsequent loss of function of the protein is a main triggering factor in DM1 molecular pathology. In order to identify miRNAs that repress muscleblind we selected candidate miRNAs and blocked their activity using specific miRNA sponges. We selected dme-miR-92a, dme-miR-100 and dme-miR-124 based on data generated in our laboratory and their orthology with human miRNAs. To widen the search of miRNA set of candidates, we used TargetScan34 to analyze miRNA recognition sites in the muscleblind 3′UTR and identified sites for two additional miRNAs: dme-miR-277 and dme-miR-304 (Table 1). Importantly, profiling of Drosophila microRNA expression in dissected thoracic muscles, had previously demonstrated miR-124, miR-100, miR-277 and miR-304 expression in these muscles35.
Table 1. Number of miRNA recognition sites predicted in muscleblind 3′ UTR according to different algorithms.
| miRNA | Miranda | TargetScan | mbl isoform |
|---|---|---|---|
| miR-92a | — | — | mblA |
| — | — | mblB | |
| — | — | mblC | |
| — | — | mblD | |
| miR-100 | — | — | mblA |
| — | — | mblB | |
| — | — | mblC | |
| — | — | mblD | |
| miR-124 | 1 site | — | mblA |
| — | — | mblB | |
| — | — | mblC | |
| 1 site | — | mblD | |
| miR-277 | 1 site | — | mblA |
| 2 sites | — | mblB | |
| — | — | mblC | |
| 2 sites | 1 site | mblD | |
| miR-304 | — | — | mblA |
| — | — | mblB | |
| 1 site | — | mblC | |
| — | 1 site | mblD |
-: no predictions.
To validate that these miRNAs regulate Muscleblind, we targeted the expression of miRNA sponge constructs35, UAS-miR-XSP, to the Drosophila muscles using the Myosin heavy chain (Mhc)-Gal4 driver line and analyzed muscleblind transcript levels by qRT-PCR. We used specific primers to amplify a region in muscleblind exon 2, which is shared by all known transcript isoforms36,37. As a control, we used a scramble miRNA sponge line (UAS-scramble-SP). No significant increase in muscleblind expression level was detected in flies expressing miR-92aSP, miR-100SP or miR-124SP under the control of Mhc-Gal4. In contrast, muscleblind transcripts were significantly increased in flies that expressed miR-277SP or miR-304SP in muscle compared with scramble-SP controls (Fig. 1a). Muscleblind levels were 14-fold higher when dme-miR-277 was inhibited while dme-miR-304 silencing resulted into a 6-fold increase. Consistently, the quantification of the expression of the mCherry reporter contained in the SP constructs showed that miR-277SP and miR-304SP were the two SPs with the highest expression in the flies (Fig. S1). Thus, we cannot disprove that miR-92aSP, miR-100SP or miR-124SP regulate muscleblind since their SP constructs had comparatively lower expression levels. As we were not interested in a complete description of muscleblind regulation by microRNAs but in providing proof of concept of their usefulness as therapeutic targets in DM1, we continued our studies with the two confirmed muscleblind regulators; miR-277 and miR-304. To assess the efficiency of miRNA downregulation by driving sponge constructs with the Mhc-Gal4 driver, we performed qRT-PCRs to detect the levels of the corresponding RNAs and confirmed that flies expressing miR-277SP or miR-304SP had reduced levels of the corresponding microRNA (Fig. 1b,c). dme-miR-277 was silenced ~60% while a robust reduction of ~80% was detected for dme-miR-304. Therefore, these results demonstrate that silencing of dme-miR-277 or dme-miR-304 derepresses muscleblind.
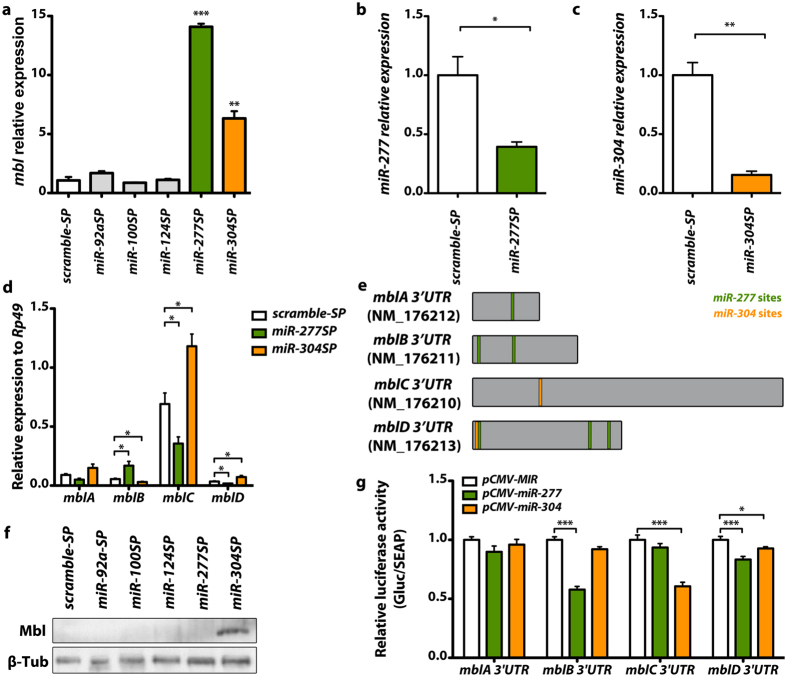
Figure 1. Tissue-specific silencing of dme-miR-277 and dme-miR-304 upregulates muscleblind mRNA and protein in Drosophila muscle.
(a) qRT-PCR amplification of muscleblind from flies expressing miRNA sponge constructs for dme-miR-92a, dme-miR-100, dme-miR-124, dme-miR-277 and dme-miR-304 in muscle. muscleblind expression levels were strongly upregulated in miR-277SP and miR-304SP flies. (b) Analysis of the levels of muscleblind isoforms by qRT-PCR. dme-miR-277 silencing in muscle caused an upregulation of the mblB isoform whilst expression levels of the mblC and mblD isoforms were reduced in miR-277SP flies. Conversely, mblC and mblD levels were increased and the mblB isoform was reduced in miR-304SP flies. (c) Detection of Muscleblind protein by Western blot. An increase of Muscleblind protein was only detected in miR-304SP flies. All the indicated transgenes were driven in muscle using Mhc-Gal4. Histogram showing dme-miR-277 (d) and dme-miR-304 (e) relative expression levels according to qRT-PCR data. Both miRNAs were significantly silenced in flies expressing the corresponding sponge constructs under the control of Mhc-Gal4 compared to flies that expressed scramble-SP (control). (f) Scheme of the predicted binding sites for dme-miR-277 and dme-miR-304 in muscleblind 3′ UTRs (mblA to mblD). Reference sequence accessions and size (in nt) are also included. Representation is to scale. (g) Quantification of Gaussian luciferase activity relative to alkaline phosphatase (Gluc/SEAP) of HeLa cells cotransfected with the indicated mbl 3′ UTR sensor constructs and plasmids expressing dme-miR-277 or dme-miR-304. Significantly reduced relative luminescence compared to empty vector (pCMV-MIR, control) reveals direct binding of dme-miR-277 to mblB and mblD 3′UTRs and of dme-miR-304 to mblC and mblD 3′ UTRs. The graphs show means±s.e.m. *p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t-test).
dme-miR-277 and dme-miR-304 regulate different Muscleblind isoforms
Drosophila muscleblind is a large gene, spanning more than 110 kb, which gives rise to different 3′ UTRs through the use of alternative 3′ exons36,37. Experimental evidence suggests that muscleblind isoforms are not functionally redundant38. To determine which muscleblind isoforms are regulated by dme-miR-277 or dme-miR-304, we used the Miranda algorithm39 to identify dme-miR-277 and dme-miR-304 recognition sites in Muscleblind isoform 3′ UTRs (Table 1, Fig. 1e). Importantly, note that Miranda database allowed the search in mblA, mblB, mblC and mblD transcripts named according to36 but did not include the recently identified isoforms mblH, mblH′, mblJ and mblK37. We found one potential recognition site for dme-miR-277 in the mblA isoform and two in mblB and mblD. qRT-PCR analyses revealed that the level of mblB significantly increased when dme-miR-277 was inhibited. mblD expression levels were reduced in Mhc-Gal4 miR-277SP flies and no significant differences were detected for mblA compared with scramble-SP control flies (Fig. 1d). Intriguingly, expression level of mblC, an isoform with no predicted recognition sites for dme-miR-277, were significantly reduced in Mhc-Gal4 miR-277SP flies. For dme-miR-304, we found one recognition site in the mblC and mblD 3′ UTR and observed a significant upregulation of both isoforms in Mhc-Gal4 miR-304SP flies (Fig. 1d). Notably, silencing of dme-miR-304 in muscle triggered a strong increase in the level of mblC, which is the most expressed isoform in adult flies38. The fact that dme-miR-277 and dme-miR-304 silencing cause isoform-specific changes in muscleblind expression levels suggest that these miRNAs directly regulate the mbl transcripts. To confirm direct binding of these miRNAs to the corresponding 3′ UTRs of mbl transcripts, we performed luciferase reporter gene assays in HeLa cells. In these studies, the 3′ UTR of the different mbl transcripts were cloned downstream of Gaussia luciferase and the interaction of the microRNAs to their targets in these regions, was detected as a decrease in the luminescence measurements. These experiments confirmed direct binding of dme-miR-277 to the 3′ UTR of the mblB and D isoforms and direct binding of dme-miR-304 to mbl isoforms C and D (Fig. 1e,g).
Given that miRNAs can act either by reducing target transcript levels or blocking their translation, we decided to analyze Muscleblind protein levels to validate the regulatory miRNA candidates. With this aim, we used an anti-Mbl antibody that has previously been optimized to detect overexpression of MblA, MblB and MblC protein, but not their endogenous expression40,41. Western blotting analyses revealed an increase in Muscleblind protein levels only in Mhc-Gal4 miR-304SP flies (Fig. 1f). Consistently with the qRT-PCR determinations, we only detected one band in the Western blot corresponding to MblC protein. To further analyze the effect of dme-miR-277 or dme-miR-304 silencing we stained Muscleblind distribution in longitudinal sections of indirect flight muscles (IFMs). We had previously shown that endogenous Muscleblind protein is localized mainly in sarcomeric Z and H bands of muscle42. Consistently, we detected Muscleblind proteins in the bands of muscle sarcomeres in control flies that express the scramble-SP construct (Fig. 2a–c). Interestingly, dme-miR-277 and dme-miR-304 exhaustion, had different effects on Mbl protein distribution. dme-miR-277 silencing increased cytoplasmic Mbl, (Fig. 2d–f), while a strong nuclear localization was detected in Mhc-Gal4 miR-304SP flies (Fig. 2g–i). Taken together, these results demonstrate that endogenous Muscleblind isoforms can be upregulated by blocking dme-miR-277 and dme-miR-304 inhibitory activity.
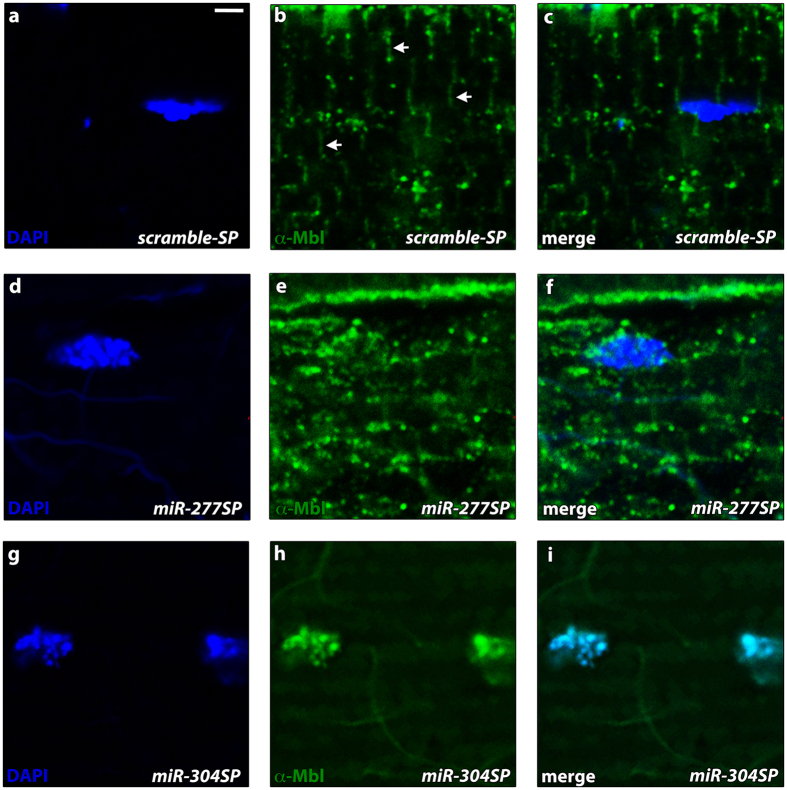
Figure 2. dme-miR-277 and dme-miR-304 silencing upregulates Muscleblind proteins with different subcellular localization.
Representative confocal images of longitudinal sections of IFMs showing anti-Mbl staining (green). Nuclei were counterstained with DAPI (blue) (a–c) Endogenous Muscleblind expression was preferentially detected in sarcomeric bands while a low signal was detected in some cell nuclei. Arrow-heads in (b) point to the sarcomeric bands. (d–f) An increase in the Muscleblind cytoplasmic signal was detected in miR-277SP flies. In contrast, silencing of dme-miR-304 in IFMs boosted the Muscleblind nuclear signal (g–i). Scale bar = 2 μm.
dme-miR-277 or dme-miR-304 silencing upregulates Muscleblind expression in a Drosophila DM1 model
Previous Drosophila models of DM1 displayed ribonuclear foci in muscle cells containing Muscleblind proteins43,44,45. To test the effect of specific silencing of the miRNA repressors of muscleblind in a Drosophila DM1 model, we studied Muscleblind expression in flies expressing 480 interrupted CTG repeats under the control of the muscle-specific driver Myosin heavy chain with simultaneous expression of sponge constructs (Mhc-Gal4 UAS-i(CTG)480 UAS-miR-XSP). Analyses of muscleblind transcript levels by qRT-PCR showed that silencing of dme-miR-277 or dme-miR-304 upregulated muscleblind in DM1 flies (Fig. 3a). muscleblind transcript levels were 19-fold higher in flies expressing both i(CTG)480 and miR-277SP and 7-fold higher in those expressing i(CTG)480 and miR-304SP compared to controls. Moreover, in agreement with protein analyses in miR-SP (Fig. 1c), silencing of dme-miR-304 triggered an increase of MblC protein levels in DM1 flies (Fig. 3b).
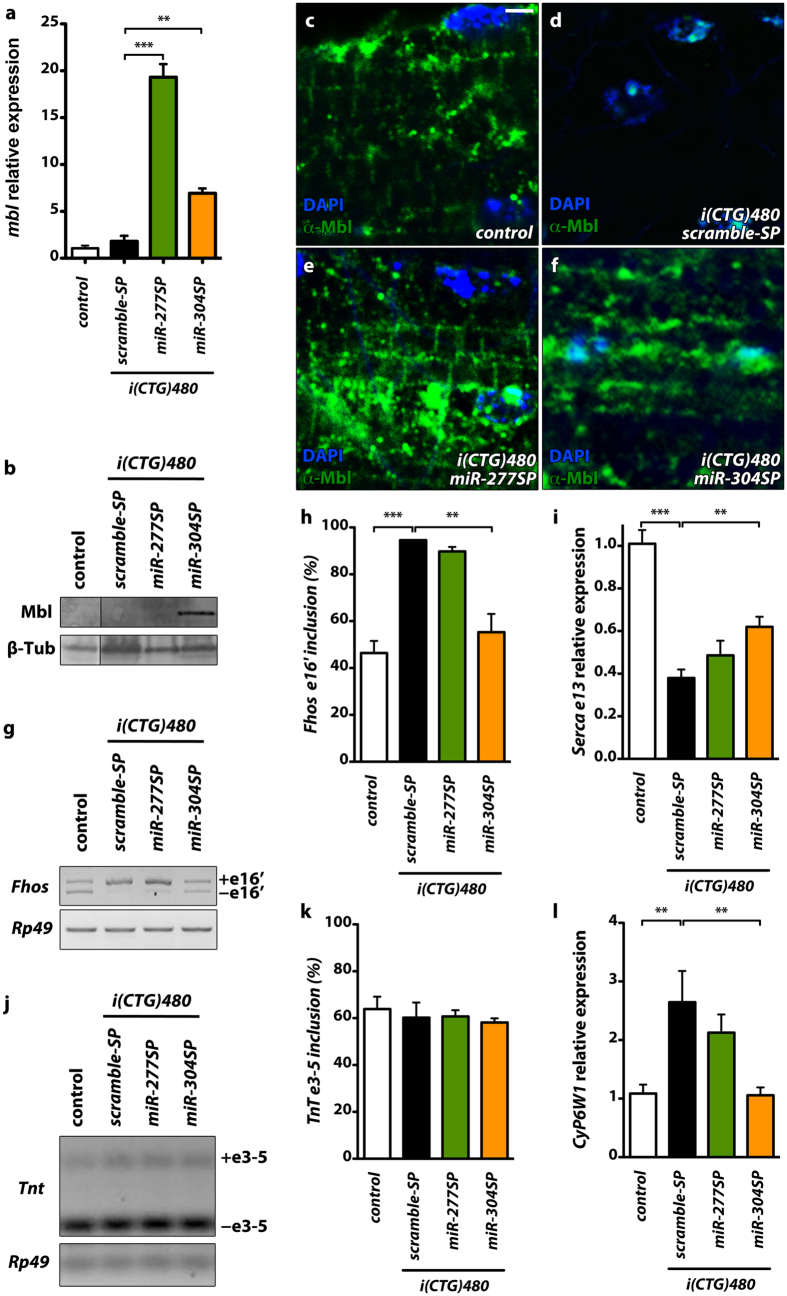
Figure 3. dme-miR-277 or dme-miR-304 silencing enhance muscleblind expression and rescues missplicing events in a DM1 background.
(a) Bar graph showing muscleblind expression levels qRT-PCR data. muscleblind mRNA was significantly upregulated in model flies expressing miR-277SP and miR-304SP in comparison to flies that did not express the expansions (control, Mhc-Gal4/+) or model flies expressing scramble-SP. (b) Western blot analysis showed additional Muscleblind C only in model flies expressing miR-304SP. (c–f) Confocal images of longitudinal sections of IFMs reveal Muscleblind (green) distribution to sarcomeric bands in control flies (c). In contrast, Muscleblind was found in nuclear aggregates in IFMs expressing CTG expansions (d). Expression of miR-277SP in model flies released Muscleblind from aggregates and restored its distribution to sarcomeric bands (e). miR-304SP expression achieved a dispersed overexpression of Muscleblind in both nuclei and cytoplasm (f). Nuclei were counterstained with DAPI (blue). (g) RT-PCR to assess inclusion of Fhos exon 16′ in flies with different genotypes. Rp49 transcripts were detected as endogenous control. (h) Quantification of percentage of exon inclusion (according to g) confirmed an improvement of Fhos missplicing in model flies expressing miR-304SP. (i,l) qRT-PCR results of Serca exon 13 and CyP6W1 expression relative to Rp49, confirmed a significant rescue of both events in model flies expressing miR-304SP. (j) RT-PCR showing inclusion of TnT exon 3–5, which did not differ in the studied genotypes. (k) Quantification of exon percentage inclusion according to (j). All the indicated genotypes were driven to muscle using Mhc-Gal4. Scale bar = 2 μm. *p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t-test).
To study the effect of dme-miR-277 or dme-miR-304 silencing on Muscleblind subcellular localization in DM1 flies, we examined Muscleblind protein distribution by inmunodetection in IFMs. Expression of either miR-277SP or miR-304SP in DM1 model flies released Muscleblind from ribonuclear foci and increased the level of protein, both in nuclei and in cytoplasm (Fig. 3c–f). In the case of model flies expressing miR-277SP, Muscleblind distribution in sarcomeric bands of muscle, which is characteristic of control flies not expressing the repeats, was significantly rescued. Similarly, expression of miR-304SP led to a detectable increase of Muscleblind dispersed in nuclei and cytoplasm. Of note, in flies that did not express the CTG repeats, dme-miR-304 silencing results in increase of Muscleblind only in nuclei (compare Figs 2i and 3f). The overall greater derepression of mbl in a CTG background, compared to wild type, may stem from the higher transcription or stability of muscleblind transcripts, perhaps as a compensatory mechanism, in DM1 flies (1.8 fold; Fig. 3a). Hence, when exposed to the same levels of miR-304SP sponge, more muscleblind transcripts may be available to translation in a DM1 background. Therefore, silencing of dme-miR-277 or dme-miR-304 upregulates Muscleblind levels and rescues its subcellular distribution in DM1 fly muscles.
dme-miR-304 silencing rescues molecular defects in a Drosophila DM1 model
RNA metabolism alterations are the major biochemical hallmark in DM1. Specifically, spliceopathy is the only molecular alteration that has been directly linked with DM1 symptoms. To test whether Muscleblind increase, triggered by dme-miR-277 or dme-miR-304 silencing, was enough to rescue missplicing in the DM1 model flies we studied two altered splicing events (Fig. S2g) and the alteration in the expression level of a specific transcript. First, we identified Fhos exon 16′ missplicing in DM1 flies and demonstrated that this splicing event and Serca exon 13 inclusion, previously identified as altered in the Drosophila DM1 model, both are regulated by Muscleblind C (Fig. S2a,b)45. Second, we confirmed that the expression level of the CyP6W1 gene was also dependent on mblC expression (Fig. S2c,f). In DM1 model flies, we confirmed a 2-fold increase of Fhos exon 16′ inclusion, a 2.4-fold reduction of Serca transcripts with exon 13 and a 3-fold increase of CyP6W1 expression in comparison to control flies not expressing the repeats. Expression of miR-304SP in these flies achieved a complete rescue of Fhos splicing and CyP6W1 expression and a significant 20% increase of Serca transcripts including exon 13 (Fig. 3g–i,l). Notably, dme-miR-304 silencing in muscle caused a strong increase in the level of mblC (Fig. 3b), which is an isoform previously shown to act as splicing regulator38. Conversely, expression of miR-277SP, which rescued Muscleblind expression in cytoplasm, and reduced mblC expression levels, did not modify these splice events. As a control, we confirmed that the splicing pattern of Tnt exons 3–5, which is not altered in DM1 adult flies44, was neither modified by the expression of the sponge constructs nor by mbl expression alterations (Fig. 3j,k; Fig. S2d,e). These results show that the level of muscleblind derepression achieved with miRNA sponges is enough to trigger significant molecular rescues.
dme-miR-277 or dme-miR-304 silencing rescues muscle atrophy and motor function in a Drosophila model of DM1
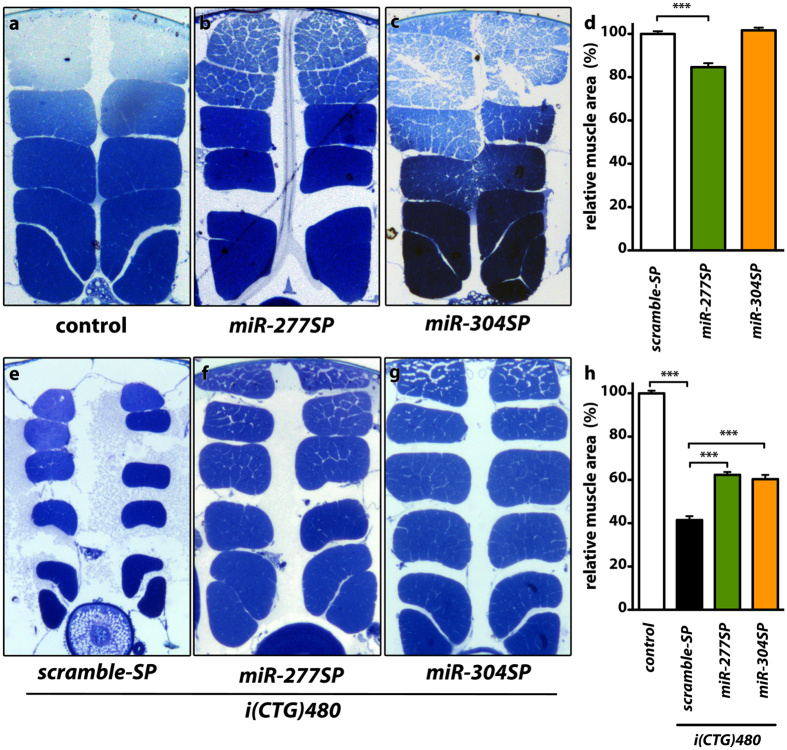
To assess the functional relevance of Muscleblind increase achieved by the expression of specific sponge constructs, we studied the effect of dme-miR-277 or dme-miR-304 silencing on muscle atrophy, which is a characteristic alteration in DM1 individuals. To study muscle atrophy we first measured muscle area in dorsoventral sections of IFMs in control flies expressing either miR-277SP or miR-304SP in muscle (Fig. 4a–d). dme-miR-277 inhibition induced a reduction of 15% in IFM area, in comparison to flies expressing scramble-SP as control. Importantly, miR-304SP expression had no effect on this parameter. We have previously reported muscle atrophy in i(CTG)480 flies27,44. In these DM1 model flies, we found that tissue-specific silencing of dme-miR-277 or dme-miR-304 was enough to rescue muscle area percentage significantly (Fig. 4e–h). In comparison to control flies that did not express the CUG repeats, the mean area of IFMs in model flies expressing the scramble-SP was significantly reduced to 40%. Concomitant expression of CUG repeats and either miR-277SP or miR-304SP resulted in a 20% increase of muscle area in these flies. These data confirm that derepression of muscleblind by miRNA silencing was sufficient to rescue muscle atrophy in Drosophila.
Figure 4. dme-miR-277 or dme-miR-304 silencing rescue muscle atrophy in model flies.
(a–c,e–g) Representative dorsoventral sections of resin-embedded thoraces of flies with the indicated relevant genotypes. Compared to control flies (a) muscle-specific expression of miR-277SP resulted into a significant reduction of indirect flight muscle (IFM) area (b) whereas miR-304SP expression had no effect on this phenotype (c) In DM1 model flies the IFM muscle area was reduced to 40% of normal (e) In model flies co-expressing either miR-277SP or miR-304SP and i(CTG)480 the muscle area increased to 60% of normal (f,g). (d,h) Quantification of the mean percentage of muscle area per genotype. The graphs show means ± s.e.m. All the indicated genotypes were driven to muscle using Mhc-Gal4. *p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t-test). In all images the dorsal side is on top.
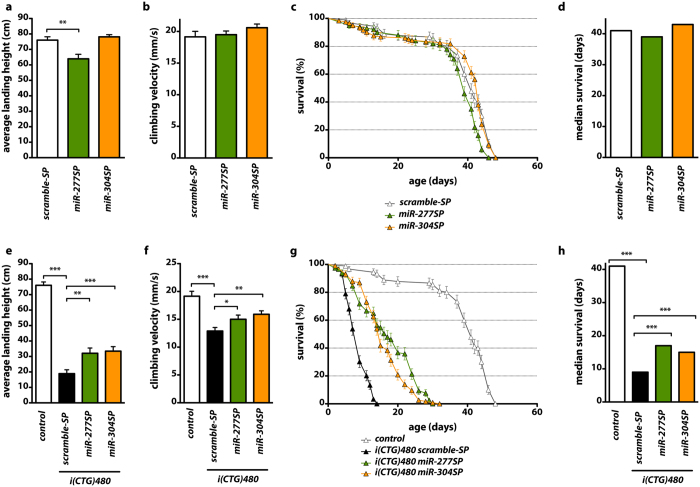
To assess the correlation between muscle area and locomotor activity we analyzed the flight and climbing ability in flies of different genotypes. Expression of miR-277SP in otherwise wild type muscle resulted in a reduction of the average landing height of around 10% in comparison to control flies expressing the scramble-SP, which indicates that the reduction of muscle area observed in these flies has a functional correlation (Fig. 5a). However, the muscle atrophy was apparently specific to IFMs since climbing velocity was unchanged in these flies (Fig. 5b). In contrast, silencing of dme-miR-304 in muscle did not affect locomotor activity of flies (Fig. 5a,b). In DM1 model flies, in comparison to controls not expressing the repeats, concomitant expression of CUG repeats and the scramble-SP construct resulted in a drastic reduction of average landing height and climbing velocity (Fig. 5e,f). However, expression of either miR-277SP or miR-304SP in model flies resulted in a significant partial rescue of both of these parameters to similar levels (Fig. 5e,f). Thus, these results demonstrate that specific silencing of miRNAs regulating muscleblind can rescue the muscle atrophy and functional phenotypes characteristic of DM1.
Figure 5. Inhibition of dme-miR-277 or dme-miR-304 improved locomotion and survival of DM1 model flies.
(a,e) Average landing height for flies with the indicated relevant genotypes. In control individuals (a) dme-miR-277 silencing decreased landing height while dme-miR-304 silencing did not affect flight. In DM1 model flies (e) expression of miR-277SP or miR-304SP rescued the reduced flight ability observed. (b,f) Histograms showing the climbing velocity as mean speed ± SEM in mm/s. In control flies (b) silencing of either dme-miR-277 or dme-miR-304 had no effect on climbing velocity. However, in DM1 flies (f) which have very reduced climbing velocity, expression of miR-277SP or miR-304SP significantly rescued this phenotype. (c,g) Survival curves and (d,h) median survival, showing that expression of miR-277SP or miR-304SP had no effect on control but improved survival of DM1 model flies. Between 140 and 160 individuals from each genotype were analyzed. All the indicated transgenes were driven in muscle with Mhc-Gal4. *p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t-test).
Functional depletion of dme-miR-277 or dme-miR-304 extends lifespan of DM1 flies
Muscle wasting, particularly in the respiratory system, is the leading cause of death in DM1. We have previously reported that flies expressing i(CTG)480 in the musculature had a reduced lifespan and median survival compared with control flies44. To study whether dme-miR-277 or dme-miR-304 silencing rescues lifespan of DM1 flies, we performed survival curves analyses in flies of different genotypes. Importantly, survival curves for flies expressing miR-277SP or miR-304SP in otherwise wild type muscle were not different to scramble-SP control indicating that dme-miR-277 or dme-miR-304 silencing did not alter lifespan (Fig. 5c,d). Lifespan of DM1 model flies expressing the scramble-SP was significantly reduced compared with control flies that did not express the CTG repeats (Fig. 5g,h). Expression of either miR-277SP or miR-304SP in model flies increased the median survival. dme-miR-277 silencing increased median survival by eight days while an increase of six days was detected for DM1 flies expressing miR-304SP. Thus, muscleblind upregulation triggered by dme-miR-277 or dme-miR-304 silencing, improved survival of DM1 model flies. Taken together, our results demonstrate that silencing of specific miRNAs in Drosophila triggers an increase of muscleblind levels that is sufficient to rescue several molecular and physiological DM1-like features thus supporting miRNA-based derepression of Muscleblind as a potential strategy to treat human DM1.
Discussion
DM1 presents a considerable disease burden as it is the most common adult-onset muscle dystrophy, and includes cognitive dysfunction, malignant heart arrhythmia, and respiratory failure, ultimately leading to shortened life expectancy. Inhibition of MBNL activity due to sequestration by microsatellite expansion RNAs is a major pathogenic event in DM1. Using a Drosophila model of this disease, we confirmed that upregulation of endogenous muscleblind by specific microRNA silencing, can rescue DM1-like phenotypes. Endogenous gene modulation to alleviate pathology has been successful in breast cancer where estrogen receptor antagonists are regularly used in clinical practice46. Furthermore, pharmacological enhancement of utrophin expression, a gene exclusively expressed in fetus but that can compensate Dystrophin loss of function in Duchenne Muscular Dystrophy is currently under investigation47. One of the most promising therapeutic strategies for endogenous gene regulation is based on miRNA derepression. These strategies have proven to be beneficial in animal models, to be highly efficient in specific target silencing, and to have appropriate pharmacokinetic parameters to be developed as drugs. In this study we aimed to upregulate endogenous muscleblind expression by silencing defined miRNAs, which regulate it in muscle. miRNAs have been extensively associated with several neuromuscular disorders in valuable in-vivo systems, which highlights the importance of studying miRNA-based regulation of dystrophy-associated genes as potential therapeutic strategy48,49,50,51,52. Specifically, we used miRNA sponge constructs, which are transgenes, containing multiple, tandem binding sites of a microRNA of interest, expressed from strong promoters. From our previous studies and TargetScan predictions, we selected a group of microRNAs as potential regulators of Drosophila muscleblind and confirmed that sponge constructs for dme-miR-277 and dme-miR-304, which had the highest expression among the sponge constructs tested, reduced the abundance of their respective target miRNAs and achieved upregulation of muscleblind at the RNA and protein levels. Isoform-specific quantitative analysis confirmed that each of these sponge construct upregulated different muscleblind isoforms and luciferase reporter assays revealed that this regulation was mediated by direct binding of the miRNAs to the 3′ UTR of the different muscleblind isoforms. We confirmed direct binding of dme-miR-277 to the 3′ UTR of the mbl B and D isoforms and direct binding of dme-miR-304 to mbl isoforms C and D. Accordingly, the mblB and mblC isoforms were increased by miR-277SP and miR-304SP, which reduced the levels of these microRNAs. In the case of mblD, we have confirmed direct binding of both microRNAs to its 3′ UTR. Consistently, miR-304SP produced an increase of the transcript. However, we have observed a slight but significant decrease of mblD transcript as a result of miR-277SP, which might be explained by an inter-isoform regulation as dme-miR-277 also produces a strong upregulation of the mblB isoform, which might have a negative effect on mblD. Of note, binding sites of dme-miR-277 and dme-miR-304 overlap in the mblD 3′ UTR, which could explain the difference observed between in vivo and luciferase assays for mblD regulation by dme-miR-277. Interestingly, miR-304SP and miR-277SP were able to downregulate the expression of mblB and mblC, respectively, instead of increasing expression, which could also be a consequence of inter-isoform regulation as it has been previously shown for MBNL proteins53,54.
Since different subcellular localizations, indicative of specific functions, have been reported for different Muscleblind isoforms38,55, we immunodetected the protein in fly muscle tissue expressing either miR-277SP or miR-304SP. In both cases we observed Muscleblind overexpression but in different subcellular locations. Whereas miR-277SP preferentially increased Muscleblind in sarcomeric bands, miR-304SP enhanced Muscleblind expression in nuclei. Consistently, when we analyzed the effects of the sponge constructs on DM1 model flies expressing pathogenic expansions, miR-277SP rescued Muscleblind localization in sarcomeric bands and miR-304SP increased Muscleblind in nuclei and cytoplasm. Of note, in both cases Muscleblind retention in ribonuclear foci was no longer detectable and protein levels in DM1 flies seemed higher than in normal individuals, suggesting that, in addition to an upregulation of muscleblind expression, there might be a release of the protein from foci. Finally, we have shown that MblC localizes to nuclei55 and, as we show in this study, is preferentially upregulated by miR-304SP expression. Supporting a role for MblC in nuclei, we also confirmed that miR-304SP expression rescued a number of Muscleblind-dependent molecular events. Taken together, these data confirm that the upregulation of muscleblind achieved by silencing specific regulatory miRNAs is sufficient to rescue critical molecular features that are altered in DM1 model flies.
We also checked the effect of miR-277SP and miR-304SP expression on muscle atrophy, which is a characteristic DM1 phenotype. We have previously reported that muscle atrophy stems, at least in part, from hyperactivation of the autophagy process and that this had a Muscleblind component. Specifically, we showed that MblC overexpression partially rescued muscle atrophy in the DM1 model flies27 and, consistent with these previous observations, we confirmed that miR-304SP expression in model flies also rescued muscle atrophy. However, processes other than splicing must be involved in triggering this phenotype as miR-277SP expression, which preferentially upregulates Muscleblind in sarcomeric bands, was also able to rescue muscle area in the DM1 model flies. Importantly, the increase in IFM muscle area achieved by expression of sponge constructs had a functional correlation, as survival, climbing and flight abilities also improved.
By expressing the sponge constructs with the Mhc-Gal4 driver we also tested the effects of long-term Muscleblind overexpression. In control flies, we observed that miR-304SP expression, caused a 6-fold increase in muscleblind relative expression and had no effect on muscle area, survival or locomotor function. However, miR-277SP expression, which produced a 15-fold upregulation of muscleblind caused a significant reduction in muscle area, which correlated with decreased landing height. In a CTG expressing background, however, expression of either of the sponge constructs brought about beneficial effects suggesting that limited overexpression of additional natural miRNA target transcripts are negligible compared to the positive effects of boosting muscleblind. Previous studies have confirmed that long-term overexpression of MBNL1 in mouse models is well tolerated when limited to skeletal muscle. MBNL1 overexpression, in a range of 10 to 17 fold, caused no detectable histopathology or functional abnormalities28. Deletereous effects of miR-277SP could originate from overexpression of several targets in addition to musclebind, as dme-miR-277 is one of the miRNAs with highest expression in muscle35. Importantly, we show for the first time functional locomotor defects in DM1 model flies expressing CUG repeats in skeletal muscle and, according to our data, flight assays seem more sensitive than climbing assays as small differences in muscle area translate into detectable differences in flight ability.
Conceptually similar to sponge constructs, several drugs targeting specific miRNAs are currently being developed for the treatment of human diseases. Miravirsen, a drug targeting hsa-miR-122, the hepatocyte-specific microRNA that Hepatitis C virus hijacks and uses to self-replicate56, is one the most advanced. Our study with the Drosophila model of DM1 sets the stage for the evaluation of miRNA blockers to de-repress muscleblind expression as a valid and powerful therapeutic target for treatment of DM1.
Materials and Methods
Drosophila stocks
Mhc-Gal4 flies were described in ref. 57. miRNA sponge lines (UAS-miR-SP) for dme-miR-92a, dme-miR-100, dme-miR-124, dme-miR-277, dme-miR-304 and scramble-SP (control) were obtained from Dr. T. Fulga35. Briefly, miR-SP constructs were designed with a silencing cassette of 20 repetitive miRNA complementary sequences separated by variable four-nucleotide linker sequences. The recombinant line Mhc-Gal4 UAS-i(CTG)480 was generated in ref. 42. UAS-mblC flies58 and UAS-IR-mbl flies42 were previously reported. All crosses were carried out at 25 °C with standard fly food. Transgene doses were the same for control and experimental conditions throughout this work.
RNA extraction, RT-PCR and qRT-PCR
For each biological replicate, total RNA from 10 adult males was extracted using Trizol (Sigma). One microgram of RNA was digested with DNaseI (Invitrogen) and reverse-transcribed with SuperScript II (Invitrogen) using random hexanucleotides. 20 ng of cDNA were used in a standard PCR reaction with GoTaq polymerase (Promega) and specific primers were used to analyze Fhos exon 16′ and Tnt exon 3–5 splicing (Table S2). Rp49 was used as endogenous control using 0.2 ng of cDNA. qRT-PCR was carried out on 2 ng of cDNA template with SYBR Green PCR Master Mix (Applied Biosystems) and specific primers (Table S1). For reference gene, Rp49, qRT-PCR was carried out on 0.2 ng of cDNA. Thermal cycling was performed with Step One Plus Real Time PCR System (Applied Biosystems). Three biological replicates and three technical replicates per biological sample were carried out. Relative expression to endogenous gene and the control group was obtained by the 2−∆∆Ct method. Pairs of samples were compared using two-tailed t-test (α = 0.05), applying Welch’s correction when necessary.
Luciferase reporter assay
A luciferase assay using the pEZX-MT05 vector was performed to validate the binding of dme-miR-277 and dme-miR-304 to mblA, mblB, mblC, and/or mblD 3′ UTR regions. pEZX-MT05 vector contains a secreted Gaussia luciferase (GLuc) ORF driven by SV40 promoter as a reporter of the 3′ UTR expression and a secreted Alkaline Phosphatase (SEAP) reporter driven by a CMV promoter as an internal control. Firstly, the 3′ UTR mbl isoforms were generated by amplifying genomic Drosophila DNA with specific primers using Pfu DNA polymerase (Biotools) (Table S1). The PCR products were cloned into the EcoRV site of pBluescript II KS vector. The plasmids containing the 3′ UTR mbl isoforms were cut out and subcloned into the pEZX-MT05 vector at the XhoI and SfaAI sites. The constructs were verified by sequencing the plasmids from both ends. Secondly HeLa cells were used for the 3′ UTR luciferase assay. Cells were maintained in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin at 37 °C and 5% CO2. HeLa cells were seeded (5 × 104/well) in 24-well plates. A total of 500 ng/well of pEZX-MT05 vector containing mblA, mblB, mblC or mblD were cotrasfected with 500 ng/well of pCMVMIR vector (Blue Heron) containing dme-miR-277 or dme-miR-304, using X-tremeGENE TM HP DNA Transfection Reagent (Sigma-Aldrich). 48 h and 72 h after transfection, Gaussia luciferase (GLuc) and alkaline phosphatase (SEAP) activities were measured by luminescence in conditioned medium using the secreted-pair dual luminescence kit (GeneCopoeia). Gaussia luciferase activity was normalized to alkaline phosphatase activity (GLuc/SEAP). The statistical differences were analyzed using the Student’s t test (p < 0.05) on normalized data.
MicroRNA quantification
UAS-miR-277SP, UAS-miR-304SP and UAS-Scramble-SP were expressed in flies under the control of a muscle specific Mhc-Gal4 driver. Total RNA from thoraces was isolated according to the miRNeasy miRNA kit protocol without enrichment for miRNAs (Qiagen). The expression analysis of dme-miR-277 and dme-miR-304 was performed by real-time PCRs with specific miRCURY LNA microRNA PCR primers (Exiqon) according to the manufacturer’s instructions. As reference genes we used dme-miR-7 and dme-miR-8. Expression level determinations were performed using an Applied Biosystems Step One Plus Real Time PCR System and the values were calculated using the 2−∆∆Ct.
Western blotting
For total protein extraction 20 female thoraces were homogenized in RIPA buffer (150 mM NaCl, 1.0% IGEPAL, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl pH 8.0) plus protease and phosphatase inhibitor cocktails (Roche Applied Science). Total proteins were quantified with BCA protein assay kit (Pierce) using bovine serum albumin as standard. 20 μg of samples were denatured for 5 min at 100 °C, resolved in 12% SDS-PAGE gels and transferred onto polyvinylidene difluoride (PVDF) membranes. The membranes were blocked with 5% nonfat dried milk in PBS-T (8 mM Na2HPO4, 150 mM NaCl, 2 mM KH2PO4, 3 mM KCl, 0.05% Tween 20, pH 7.4) and immunodetected following standard procedures. For Mbl protein detection anti-Mbl antibody40 was pre-absorbed against early stage wild type embryos (0–6 h after egg laying) to eliminate non-specific binding of antibody. Membranes were incubated with pre-absorbed primary (overnight, 1:1000) followed by horseradish peroxidase (HRP)-conjugated anti-sheep-IgG secondary antibody (1 h, 1:5000, Sigma-Aldrich). Loading control was anti-Tubulin (overnight, 1:5000, Sigma-Aldrich) followed by incubation with HRP-conjugated anti-mouse-IgG secondary antibody (1 h, 1:3000, Sigma-Aldrich). Bands were detected using ECL Western Blotting Substrate (Pierce). Images were acquired with an ImageQuant LAS 4000 (GE Healthcare).
Histological analysis
Inmunofluorescence detection of Muscleblind in fly muscle and analysis of the IFM area in Drosophila thoraces were performed as previously described42. Briefly, six adult female thoraces were embedded in Epon following standard procedures. After drying the resin, semi-thin sections of 1.5 μm were obtained using an ultramicrotome (Ultracut E, Reichert-Jung and Leica). Images were taken at 100× magnification with a Leica DM2500 microscope. To quantify muscle area, sections were counterstained with toluidine blue and five images containing IFMs per fly were converted into binary images. Considering the complete image as 100% of the area, we used NIH ImageJ software to calculate the percentage occupied by pixels corresponding to IFMs. P-values were obtained using a two-tailed, non-paired t-test (α = 0.05), applying Welch’s correction when necessary.
Drosophila lifespan analyses
A total of 120 newly hatched flies with the appropriate genotypes were collected and kept at 29 °C. Flies were transferred to new fresh nutritive media every second day and the decline in number was scored on a daily basis. Survival curves were obtained using the Kaplan-Meier method and statistical analysis was performed with a log-rank (Mantel-Cox) test (α = 0.05) using the GraphPad Prism5 software.
Functional assays
Flight assays were performed at day 5 according to ref. 59 using 100 male flies per group. Landing distance was compared between groups using two-tailed t-test (α = 0.05). To assess climbing velocity groups of ten 5-day-old males were transferred into disposable pipettes (1.5 cm in diameter and 25 cm height) after a period of 24 h without anesthesia. The height reached from the bottom of the vial by each fly in a period of 10 s was recorded with a camera. For each genotype, two groups of 30 flies were tested. Two-tailed t-test (α = 0.05) was used for comparisons of pairs of samples applying Welch’s correction whenever necessary.
Additional Information
How to cite this article: Cerro-Herreros, E. et al. Derepressing muscleblind expression by miRNA sponges ameliorates myotonic dystrophy-like phenotypes in Drosophila. Sci. Rep. 6, 36230; doi: 10.1038/srep36230 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Dr. Tudor Fulga for kindly providing miRNA sponge flies. This project was carried out with research grants SAF2012-36854 and SAF2015-64500-R, which include European Regional Development Funds, awarded to R.A. by the Ministerio de Economia y Competitividad. E.C.H. was supported by a predoctoral fellowship (BES-2013-064522) from the Ministerio de Economia y Competitividad.
Footnotes
Author Contributions R.A. provided the conceptual framework for the study. All the authors conceived and designed the experiments. E.C.H., J.M.F.C., M.S.A. and B.L.L. performed the experiments and analyzed data. J.M.F.C. and B.L.L. prepared the manuscript with input by R.A.
References
- Thornton C. A. Myotonic dystrophy. Neurol Clin 32, 705–719, viii, doi: 10.1016/j.ncl.2014.04.011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper P. Myotonic dystrophy. (Saunders, 2001). [Google Scholar]
- Gagnon C. et al. Towards an integrative approach to the management of myotonic dystrophy type 1. Journal of Neurology, Neurosurgery & Psychiatry 78, 800–806 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B. M., McCurrach M. E., Taneja K. L., Singer R. H. & Housman D. E. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc Natl Acad Sci USA 94, 7388–7393 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X. et al. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet 15, 2087–2097, doi: 10.1093/hmg/ddl132 (2006). [DOI] [PubMed] [Google Scholar]
- Mankodi A. et al. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell 10, 35–44 (2002). [DOI] [PubMed] [Google Scholar]
- Du H. et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol 17, 187–193, doi: 10.1038/nsmb.1720 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra R. et al. Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol Cell 56, 311–322, doi: 10.1016/j.molcel.2014.08.027 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin M. et al. MBNL Sequestration by Toxic RNAs and RNA Misprocessing in the Myotonic Dystrophy Brain. Cell Rep 12, 1159–1168, doi: 10.1016/j.celrep.2015.07.029 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A. et al. The Mef2 transcription network is disrupted in myotonic dystrophy heart tissue, dramatically altering miRNA and mRNA expression. Cell Rep 6, 336–345, doi: 10.1016/j.celrep.2013.12.025 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Costa J. M. et al. Expanded CTG repeats trigger miRNA alterations in Drosophila that are conserved in myotonic dystrophy type 1 patients. Hum Mol Genet 22, 704–716, doi: 10.1093/hmg/dds478 (2013). [DOI] [PubMed] [Google Scholar]
- Rau F. et al. Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat Struct Mol Biol 18, 840–845, doi: 10.1038/nsmb.2067 (2011). [DOI] [PubMed] [Google Scholar]
- Moseley M. L. et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet 38, 758–769, doi: 10.1038/ng1827 (2006). [DOI] [PubMed] [Google Scholar]
- Cho D. H. et al. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell 20, 483–489, doi: 10.1016/j.molcel.2005.09.002 (2005). [DOI] [PubMed] [Google Scholar]
- Timchenko N. A. et al. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J Biol Chem 279, 13129–13139, doi: 10.1074/jbc.M312923200 (2004). [DOI] [PubMed] [Google Scholar]
- Mulders S. A. et al. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc Natl Acad Sci USA 106, 13915–13920, doi: 10.1073/pnas.0905780106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Lopez A., Llamusi B., Orzaez M., Perez-Paya E. & Artero R. D. In vivo discovery of a peptide that prevents CUG-RNA hairpin formation and reverses RNA toxicity in myotonic dystrophy models. Proc Natl Acad Sci USA 108, 11866–11871, doi: 10.1073/pnas.1018213108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler T. M. et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature 488, 111–115, doi: 10.1038/nature11362 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf M. B., Nakamori M., Matthys C. M., Thornton C. A. & Berglund J. A. Pentamidine reverses the splicing defects associated with myotonic dystrophy. Proc Natl Acad Sci USA 106, 18551–18556, doi: 10.1073/pnas.0903234106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkesh R. et al. Design of a bioactive small molecule that targets the myotonic dystrophy type 1 RNA via an RNA motif-ligand database and chemical similarity searching. J Am Chem Soc 134, 4731–4742, doi: 10.1021/ja210088v (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. E., Bennett C. F. & Cooper T. A. RNase H-mediated degradation of toxic RNA in myotonic dystrophy type 1. Proc Natl Acad Sci USA 109, 4221–4226, doi: 10.1073/pnas.1117019109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia R. N. et al. A muscleblind knockout model for myotonic dystrophy. Science 302, 1978–1980, doi: 10.1126/science.1088583 (2003). [DOI] [PubMed] [Google Scholar]
- Dixon D. M. et al. Loss of muscleblind-like 1 results in cardiac pathology and persistence of embryonic splice isoforms. Sci Rep 5, 9042, doi: 10.1038/srep09042 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. Y. et al. Compound loss of muscleblind-like function in myotonic dystrophy. EMBO Mol Med 5, 1887–1900, doi: 10.1002/emmm.201303275 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huin V. et al. MBNL1 gene variants as modifiers of disease severity in myotonic dystrophy type 1. J Neurol 260, 998–1003, doi: 10.1007/s00415-012-6740-y (2013). [DOI] [PubMed] [Google Scholar]
- Kanadia R. N. et al. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc Natl Acad Sci USA 103, 11748–11753, doi: 10.1073/pnas.0604970103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargiela A. et al. Increased autophagy and apoptosis contribute to muscle atrophy in a myotonic dystrophy type 1 Drosophila model. Dis Model Mech 8, 679–690, doi: 10.1242/dmm.018127 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain C. M. & Ranum L. P. Mouse model of muscleblind-like 1 overexpression: skeletal muscle effects and therapeutic promise. Hum Mol Genet 21, 4645–4654, doi: 10.1093/hmg/dds306 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada T. P. et al. MicroRNA silencing through RISC recruitment of eIF6. Nature 447, 823–828, doi: 10.1038/nature05841 (2007). [DOI] [PubMed] [Google Scholar]
- Lytle J. R., Yario T. A. & Steitz J. A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA 104, 9667–9672, doi: 10.1073/pnas.0703820104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. & Xie X. Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression. Genome Biol 7, R85, doi: 10.1186/gb-2006-7-9-r85 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez A. J. et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75–79, doi: 10.1126/science.1122689 (2006). [DOI] [PubMed] [Google Scholar]
- Guo H., Ingolia N. T., Weissman J. S. & Bartel D. P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840, doi: 10.1038/nature09267 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheradpour P., Stark A., Roy S. & Kellis M. Reliable prediction of regulator targets using 12 Drosophila genomes. Genome Res 17, 1919–1931, doi: 10.1101/gr.7090407 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulga T. A. et al. A transgenic resource for conditional competitive inhibition of conserved Drosophila microRNAs. Nat Commun 6, 7279, doi: 10.1038/ncomms8279 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann G. et al. muscleblind, a gene required for photoreceptor differentiation in Drosophila, encodes novel nuclear Cys3His-type zinc-finger-containing proteins. Development 124, 4321–4331 (1997). [DOI] [PubMed] [Google Scholar]
- Irion U. Drosophila muscleblind codes for proteins with one and two tandem zinc finger motifs. PLoS One 7, e34248, doi: 10.1371/journal.pone.0034248 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente M. et al. Muscleblind isoforms are functionally distinct and regulate alpha-actinin splicing. Differentiation 75, 427–440, doi: 10.1111/j.1432-0436.2006.00156.x (2007). [DOI] [PubMed] [Google Scholar]
- Enright A. J. et al. MicroRNA targets in Drosophila. Genome Biol 5, R1, doi: 10.1186/gb-2003-5-1-r1 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J. M. et al. Myotonic dystrophy associated expanded CUG repeat muscleblind positive ribonuclear foci are not toxic to Drosophila. Hum Mol Genet 14, 873–883, doi: 10.1093/hmg/ddi080 (2005). [DOI] [PubMed] [Google Scholar]
- Vicente-Crespo M. et al. Drosophila muscleblind is involved in troponin T alternative splicing and apoptosis. PLoS One 3, e1613, doi: 10.1371/journal.pone.0001613 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamusi B. et al. Muscleblind, BSF and TBPH are mislocalized in the muscle sarcomere of a Drosophila myotonic dystrophy model. Dis Model Mech 6, 184–196, doi: 10.1242/dmm.009563 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haro M. et al. MBNL1 and CUGBP1 modify expanded CUG-induced toxicity in a Drosophila model of myotonic dystrophy type 1. Hum Mol Genet 15, 2138–2145, doi: 10.1093/hmg/ddl137 (2006). [DOI] [PubMed] [Google Scholar]
- Garcia-Lopez A. et al. Genetic and chemical modifiers of a CUG toxicity model in Drosophila. PLoS One 3, e1595, doi: 10.1371/journal.pone.0001595 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picchio L., Plantie E., Renaud Y., Poovthumkadavil P. & Jagla K. Novel Drosophila model of myotonic dystrophy type 1: phenotypic characterization and genome-wide view of altered gene expression. Hum Mol Genet 22, 2795–2810, doi: 10.1093/hmg/ddt127 (2013). [DOI] [PubMed] [Google Scholar]
- Shiau A. K. et al. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95, 927–937 (1998). [DOI] [PubMed] [Google Scholar]
- Tinsley J., Robinson N. & Davies K. E. Safety, tolerability, and pharmacokinetics of SMT C1100, a 2-arylbenzoxazole utrophin modulator, following single- and multiple-dose administration to healthy male adult volunteers. J Clin Pharmacol 55, 698–707, doi: 10.1002/jcph.468 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone A. K., Edeleva E. V., Kucherenko M. M., Hsiao N. H. & Shcherbata H. R. Dg-Dys-Syn1 signaling in Drosophila regulates the microRNA profile. BMC Cell Biol 13, 26, doi: 10.1186/1471-2121-13-26 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsenko A. S. & Shcherbata H. R. Drosophila miR-9a targets the ECM receptor Dystroglycan to canalize myotendinous junction formation. Dev Cell 28, 335–348, doi: 10.1016/j.devcel.2014.01.004 (2014). [DOI] [PubMed] [Google Scholar]
- Yatsenko A. S., Marrone A. K. & Shcherbata H. R. miRNA-based buffering of the cobblestone-lissencephaly-associated extracellular matrix receptor dystroglycan via its alternative 3′-UTR. Nat Commun 5, 4906, doi: 10.1038/ncomms5906 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorillo A. A. et al. TNF-alpha-Induced microRNAs Control Dystrophin Expression in Becker Muscular Dystrophy. Cell Rep 12, 1678–1690, doi: 10.1016/j.celrep.2015.07.066 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N. et al. microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J Clin Invest 122, 2054–2065, doi: 10.1172/JCI62656 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino Y. et al. Nuclear localization of MBNL1: splicing-mediated autoregulation and repression of repeat-derived aberrant proteins. Hum Mol Genet 24, 740–756, doi: 10.1093/hmg/ddu492 (2015). [DOI] [PubMed] [Google Scholar]
- Terenzi F. & Ladd A. N. Conserved developmental alternative splicing of muscleblind-like (MBNL) transcripts regulates MBNL localization and activity. RNA Biol 7, 43–55 (2010). [DOI] [PubMed] [Google Scholar]
- Fernandez-Costa J. M. & Artero R. A conserved motif controls nuclear localization of Drosophila Muscleblind. Mol Cells 30, 65–70, doi: 10.1007/s10059-010-0089-9 (2010). [DOI] [PubMed] [Google Scholar]
- Ottosen S. et al. In vitro antiviral activity and preclinical and clinical resistance profile of miravirsen, a novel anti-hepatitis C virus therapeutic targeting the human factor miR-122. Antimicrob Agents Chemother 59, 599–608, doi: 10.1128/AAC.04220-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek K. W. et al. A genetic analysis of synaptic development: pre- and postsynaptic dCBP control transmitter release at the Drosophila NMJ. Neuron 25, 537–547 (2000). [DOI] [PubMed] [Google Scholar]
- Garcia-Casado M. Z., Artero R. D., Paricio N., Terol J. & Perez-Alonso M. Generation of GAL4-responsive muscleblind constructs. Genesis 34, 111–114, doi: 10.1002/gene.10147 (2002). [DOI] [PubMed] [Google Scholar]
- Babcock D. T. & Ganetzky B. An improved method for accurate and rapid measurement of flight performance in Drosophila. J Vis Exp, e51223, doi: 10.3791/51223 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.