Abstract
Background
Ichthyoses are clinically characterized by scaling or hyperkeratosis of the skin or both. It can be an isolated condition limited to the skin or appear secondarily with involvement of other cutaneous or systemic abnormalities.
Methods
The present study investigated clinical and molecular characterization of three consanguineous families (A, B, C) segregating two different forms of autosomal recessive congenital ichthyosis (ARCI). Linkage in three consanguineous families (A, B, C) segregating two different forms of ARCI was searched by typing microsatellite and single nucleotide polymorphism marker analysis. Sequencing of the two genes TGM1 and ALOXE3 was performed by the dideoxy chain termination method.
Results
Genome-wide linkage analysis established linkage in family A to TGM1 gene on chromosome 14q11 and in families B and C to ALOXE3 gene on chromosome 17p13. Subsequently, sequencing of these genes using samples from affected family members led to the identification of three novel mutations: a missense variant p.Trp455Arg in TGM1 (family A); a nonsense variant p.Arg140* in ALOXE3 (family B); and a complex rearrangement in ALOXE3 (family C).
Conclusion
The present study further extends the spectrum of mutations in the two genes involved in causing ARCI. Characterizing the clinical spectrum resulting from mutations in the TGM1 and ALOXE3 genes will improve diagnosis and may direct clinical care of the family members.
Introduction
Autosomal recessive congenital ichthyosis (ARCI) is a rare, heterogeneous keratinization disorder of the skin. Classically, it is divided into lamellar ichthyosis (LI), congenital ichthyosiform erythroderma (CIE), and harlequin ichthyosis (HI).1 Patients with LI are often born encased in collodion membrane that later changes into large, dark-brown plate-like scales.2 Keratoderma is often also found on the palms and soles of patients affected with LI. Patients with CIE show variable erythroderma and generalized fine white scaling; additionally they may be born as collodion babies.3–5 Newborns with HI often show large, thick, plate-like scales with pronounced ectropion/eclabium, which is the most severe and lethal form among congenital ichthyoses.6 To date, mutations in nine genes have been implicated in ARCI; including five LI associated genes; TGM1 (MIM 242300), CYP4F22 (MIM 604777), NIPAL4 (MIM 612281), LIPN (MIM 613924), and PNPLA1 (MIM 612121); three CIE associated genes; ALOX12B (MIM 603741), ALOXE3 (MIM 607206), and CERS3 (MIM 615276); and a single HI associated gene; ABCA12 (MIM 601277).7–17 However, few studies also reported TGM1 mutations in patients with CIE.18,19
In the present study, we have investigated three unrelated consanguineous Pakistani families segregating LI and CIE phenotypes. Genotyping and DNA sequencing was used to identify sequence variants in the two genes (TGM1, ALOXE3) in these families.
Materials and methods
Subjects
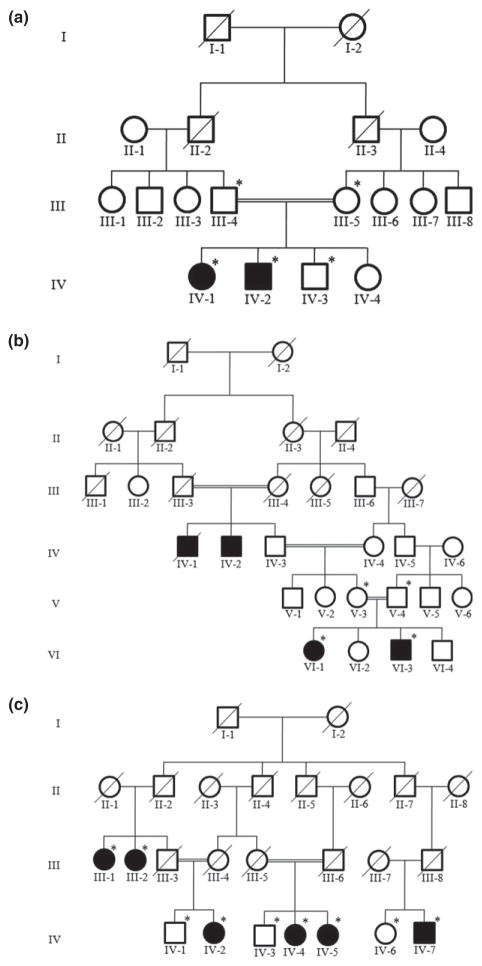
For the study presented here, three consanguineous families (A, B, C), segregating LI and CIE phenotypes, were located in remote regions of Pakistan (Fig. 1). Approval of the study was obtained from the Institutional Review Board of Quaid-i-Azam University, Islamabad, Pakistan, and Baylor College of Medicine and Affiliated Hospitals, Houston, TX, USA. Informed consent was obtained from all family members who participated in the study. Pedigree drawings of the families were based upon detailed question/answer sessions conducted with elders of the families.
Figure 1.
Pedigree drawings of three consanguineous Pakistani families segregating lamellar ichthyosis (a) and congenital ichthyosiform erythroderma (b,c). Filled symbols represent affected individuals. Symbols with crossed lines represent deceased individuals. Symbols with a star denote those individuals with an available DNA sample
Peripheral blood samples were collected from 18 pedigree members including two affected (IV-1, IV-2) and three unaffected (III-4, III-5, IV-3) individuals from family A two affected (VI-1, VI-3) and two unaffected (V-3, V-4) from family B, and six affected (III-1, III-2, IV-2, IV-4, IV-5, IV-7) and three unaffected (IV-1, IV-3, IV-6) members from family C (Fig. 1). Genomic DNA was extracted from peripheral blood samples using the GenElute™ blood genomic DNA kit (Sigma-Aldrich, St. Louis, MO, USA). DNA was quantified by Nanodrop-1000 spectrophotometer (Thermal Scientific, Wilmington, MA, USA) measuring its optical density at 260 nm and diluted to 40–50 ng/μL for amplification by polymerase chain reaction (PCR).
Genotyping and mutation analysis
Based upon the clinical features observed in the affected members and autosomal recessive mode of inheritance, homozygosity mapping in two families (A, B) was performed by genotyping several microsatellite markers flanking known ARCI genes. PCR amplification of microsatellite markers20 was performed by following standard procedures in a total volume of 25 μL. The amplified products were resolved on 8% non-denaturing polyacrylamide gel, stained with ethidium bromide, and genotypes were assigned by visual inspection.
DNA samples from available members of family C were submitted to the University of Washington Center for Mendelian Genomics to perform genome scan using Infinium® Human core exome chip (Illumina, San Diego, CA, USA), which consists of more than 500,000 single nucleotide polymorphism (SNP) markers. The genotype data were analyzed by homozygosity mapper to find common homozygous by descent regions,21 and linkage analysis was performed using Superlink.22 Analyses were performed by assuming a disease frequency 0.001 and recessive mode of inheritance. Allele frequencies for SNP markers were estimated using founders and reconstructed founders from additional Pakistani families genotyped along with family C. The genetic map positions were based on the Rutgers combined linkage-physical maps (build 37).20
For mutation identification, TGM1 (NM_000359) and ALOXE3 (NM_021628) genes were sequenced using Big Dye Terminator v3.1 Cycle Sequencing Kit on ABI Prism 310 Genetic Analyzer (Applera, Foster City, CA, USA). Bioedit sequence alignment tool (editor version 6.0.7; Ibis Biosciences Inc., Carlsbad, CA, USA) was used to align the sequence of each amplicon with reference sequence of both genes.
Deletion breakpoint mapping
In family C, ALOXE3 deletion breakpoint was identified using a PCR-based assay consisting of eight overlapping set of primers specifically designed to cover both sides of a putatively deleted genomic region. After several PCR reactions, a primer pair (5′-TGCTTGAACCCAGGAAGTG-3′; 5′-TCTTCCACACCCGTCACTTA-3′) was selected to amplify and sequence the deletion breakpoint. To determine the deletion coordinates, sequence data was mapped against reference human genome using BLAT tool from UCSC genome browser.23
Results
Clinical features
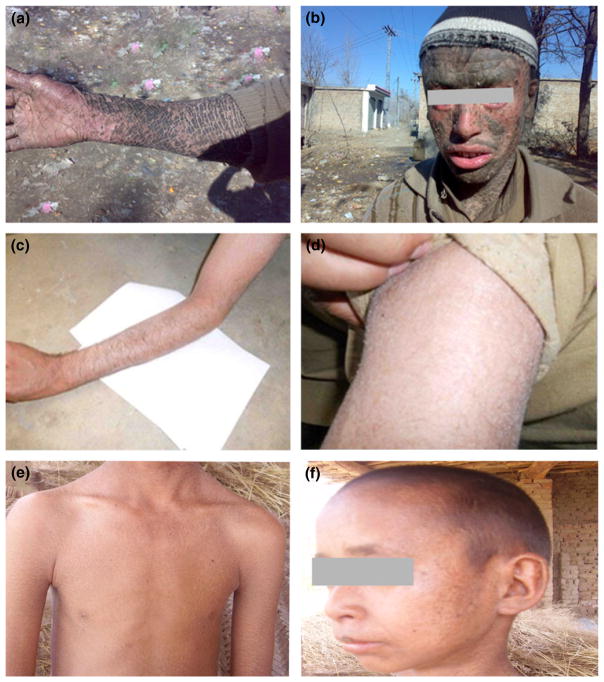
Affected individuals of the three families (A, B, C) were clinically investigated by dermatologists at the local government hospitals. In family A, the entire body surface of the affected members was covered with thick, large, dark-brown scales (Fig. 2a,b). Palms and soles showed severe keratoderma. Hairs were sparse and dry, and eyebrows were scanty. Ectropion, eclabium, and reduced sweating ability (hypohidrosis) were observed in the affected members. In family B, affected members displayed finer scales on arms, legs, and abdomen (Fig. 2c,d). In family C, affected individuals exhibited slightly thick dark-brown scales all over the body (Fig. 2e,f). No signs of ectropion, eclabium, or alopecia were observed in the affected individuals of family B and C. They had problems of minor sweating, severe heat intolerance, and bleeding from scaling skin, which occurs mostly in severe cold conditions. The clinical presentation of family A is compatible to LI, whereas milder phenotype of patients from family B and C is suggestive of CIE.
Figure 2.
Clinical presentation of ARCI in families A, B, and C. (a,b) Thick, large dark-brown scales on the arm and face in 21-year-old affected individual (IV-2) in family A. (c,d) Fine white ichthyotic scales on the arm of a 14-year-old affected individual (VI-3) in family B. (e,f) Slightly thick dark-brown scales on the neck, arms, chest, and face of a 16-year-old affected individual (IV-7) in family C
Affected members in all the three families were of normal height, growth, and mental health. Association of the phenotype with other ectodermal appendages such as nail and sebaceous glands was not observed in any of the affected members. Heterozygous carrier individuals had normal skin and were clinically indistinguishable from unaffected individuals of the respective families who are homozygous wild type.
Genotyping and mutation analysis
Homozygosity mapping in two families (A and B) was performed by using microsatellite markers flanking genes ABCA12 (2q34–q35), NIPAL4 (5q13), TGM1 (14q11), ALOX12B (17p13), ALOXE3 (17p13), and CYP4F22 (19p12–q12). Haplotype analysis showed mapping of family A to TGM1and family B toALOXE3 gene. In family C, data analysis with Homozygosity mapper21 indicated a 2.16 Mb homozygous region on chromosomes 17, which was delineated by markers rs14309 (6.91 Mb) and rs9906162 (9.08 Mb). The 2.16 Mb region of homozygosity was shared by all the affected individuals of family C and contains two known ARCI genes, ALOX12B and ALOXE3. Two-point linkage analyses yielded LOD score of 3.6 for several markers within the region of homozygosity. A maximum multipoint LOD score of 5.1 was obtained for the region containing the ALOXE3 gene.
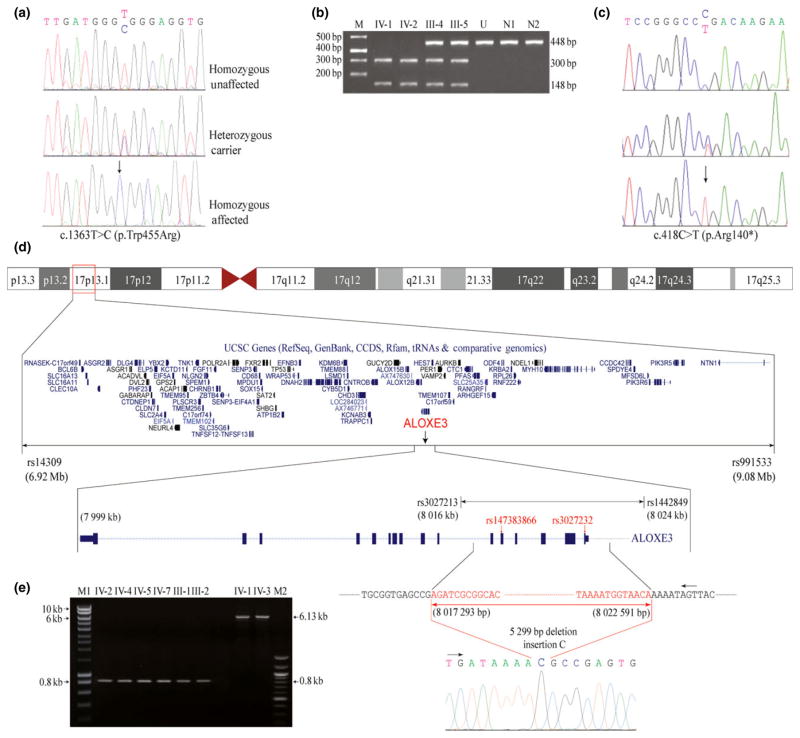
In family A, sequence analysis of exon 9 of the TGM1 gene revealed a novel homozygous missense mutation involving T to C transition at nucleotide position 1363 (c.1363T > C). This resulted in substitution of a codon for tryptophan at amino acid position 455 with arginine (p.Trp455Arg) (Fig. 3a). The missense mutation created a restriction site for the enzyme Ssi1 (Aci1) in exon 9 of the TGM1 gene. This was verified by Ssi1 (Aci1) restriction analysis of 448 bp PCR product encompassing exon 9 of TGM1 gene. Restriction enzyme analysis of PCR-amplified products produced two DNA fragments of 300 and 148 bp in affected members (IV-1, IV-2), three fragments of 448, 300, and 148 bp in carriers (III-4, III-5), and a single fragment of 448 bp in an unaffected (IV-3) member (Fig. 3b).
Figure 3.
Sequence analysis of TGM1 and ALOXE3 genes. a: a novel homozygous missense mutation (c.1363T > C; p.Trp455Arg) in TGM1 gene in family A. (b) Restriction enzyme Ssi1 (Aci1) analysis of 448 bp polymerase chain reaction (PCR) products in the family members IV-1 and IV-2 (affected), III-4 and III-5 (carrier), N1 and N2 (controls), and U (undigested PCR product of IV-1). (c) Sequence analysis of a novel nonsense mutation (c.418C > T; p.Arg140*) in ALOXE3 gene in family B. Arrows indicate position of mutations in the affected individuals. (d) Ideogram depicts a 2.1 Mb homozygous by descent region identified in family C. Deletion flanking single nucleotide polymorphism markers are indicated in red. (e) Amplified PCR product of ALOXE3 deletion flanking region of affected and normal individuals of family C. M1, 10,000 bp molecular marker and M2 represents 100 bp molecular marker
In family B, the sequence analysis of exon 4 of the ALOXE3 gene revealed a novel homozygous nonsense mutation involving a C to T transition at nucleotide position 418 (c.418C > T). This resulted in substitution of a codon for arginine at amino acid position 140 with a stop codon (p.Arg140*) (Fig. 3c).
In family C, a careful analysis of genotypes revealed missing data in all affected individuals for six consecutive SNP markers, from rs147383866 to rs3027232, within the mapped homozygous region. This corresponds to a large genomic region and is flanked by intact SNP markers rs3027213 (8.01 Mb) and rs1442849 (8.02 Mb) (Fig. 3d). To determine the exact breakpoint of the underlying deletion, long range DNA polymerase (Clontech Laboratories, Mountain View, CA, USA) was used to obtain 832 and 6130 bp products from DNA samples of affected and normal individuals, respectively (Fig. 3e). Sequencing analysis of the amplified PCR product revealed a complex rearrangement involving a 5299 bp deletion and a single nucleotide insertion (Fig. 3d). BLAT search of the sequence obtained from an affected individual indicates that the deletion occurs between chr17:8,017,292 and 8,022,592 (hg19), spanning a region of 5299 bp (hg19:g.chr17:8017293_8022591del). All affected individuals of this family were homozygous for cytosine insertion and 5299 bp deletion (hg19: g.chr17:8017293_8022591delinsC), which spans the first six exons of the ALOXE3 gene (Fig. 3d).
The novel sequence variants, identified in family A and B, were predicted as disease causing and damaging by mutation taster, Provean, SIFT, and polyphen-2 and segregate with disease in the respective families.24–26 The missense mutation (p.Trp455Arg), identified in family A, nonsense mutation (p.Arg140*), identified in family B, and gross deletion, identified in family C, were not found in panels of 300, 50, and 50 unaffected unrelated ethnically matched control individuals, respectively. These variants were also not found in the public variant database dbSNP27 and 1000 genomes,28 but p.Arg140* exists in the heterozygous state in the Exome Variant Server.29
Discussion
In the present investigation, we have described three consanguineous families segregating two different forms of ARCI. Most of the features associated with LI3,4 have been observed in affected members of family A. This included dark-brown plate-like scales on the trunk, face, scalp, and flexor areas, ectropion of eyelids, alopecia, reduced sweating ability (hypohidrosis), and erythema. However, photophobia,30 mixed dentition,31 and blepharitis32 reported in a few patients of LI were not found in our patients. Affected members in the other two families, B and C, exhibited erythroderma, hypohidrosis, and severe heat intolerance. However, ichthyotic scales were fine white on the skin of the affected members of family B, and slightly thick and dark brown on those of family C.
DNA sequence analysis led to the identification of a novel missense mutation (p.Trp455Arg) in the TGM1 gene in family A and two novel mutations (p.Arg140*, delEx1_6InsC) in the ALOXE3 gene in family B and C.
The TGM1 gene contains 15 exons, spanning 14.3 kb of genomic DNA on chromosome 14q11.2. The gene encodes calcium-dependent transglutaminase-1 (TGase-1) containing 817 amino acids. The TGase-1 contains N-terminal domain, a catalytic core domain and C-terminal domain, which is further divided into beta-barrel 1 and beta-barrel 2 (C-terminal end domain).33 To date several different mutations have been reported in the TGM1 gene, including missense, splicing, small and gross deletions, small insertions, and regulatory.34 The tryptophan at the 455th position is conserved in all vertebrates and is located in the catalytic core domain of TGase-1 enzyme. PolyPhen-2 analysis predicted that the p.Trp454Arg mutation is damaging with a probability score of 0.998. Transglutaminase-1 is a catalytic membrane-bound enzyme that functions in the formation of the cornified cell envelope. The cornified cell envelope is made up of different proteins, the cross-linking of which is facilitated by transglutaminase-1. The cornified cell envelope surrounds the skin cells and protects water loss and infection.32 Most of the ARCI causative TGM1 mutations are located in the catalytic core domain or its upstream part.17 All such mutations result in the defective intercellular lipid layers of the stratum corneum. This leads to the defective barrier function of stratum corneum resulting in ichthyotic skin phenotypes.35
In the present study the other two novel mutations (p.Arg140*, delEx1_6InsC) were identified in the ALOXE3 gene which contains 15 exons, spanning 22 kb of genomic DNA on chromosome 17p13.1. These two mutations are predicted to result in loss of function of the ALOXE3 protein either through nonsense-mediated mRNA decay (NMD) or transcription initiation failures. So far, only 13 mutations in the ALOXE3 gene, which cause CIE, have been reported. This included eight missense, three splicing, and two small deletions.34 The 843 amino acids lipoxygenase-3, encoded by ALOXE3 gene, contains the PLAT (Polycystin-1, Lipoxygenase, Alpha-Toxin) domain, the lipoxygenase homology 2 domain and a large lipoxygenase domain. The mutation (p.Arg140*) identified in family B is located in the lipoxygenase domain and could either lead to NMD or production of a truncated lipoxygenase-3 protein. This potentially shortened protein (p.Arg140*) should lack the major part of lipoxygenase domain that is required for lipid metabolism. The exome variant server29 has reported presence of the variant p.Arg140* in the heterozygous state only in a single African-American (AA = 0; AG = 1; GG = 6502). However, the absence of ARCI phenotype in individuals heterozygous for p.Arg140* variant, in our family, clearly ruled out its pathogenicity in the heterozygous state. It is highly likely that large deletion (delEx1_6InsC), identified in the family C, would result in loss of function mutation through NMD. However, in the case of the formation of a truncated protein PLAT, lipoxygenase homology 2 and a part of lipoxygenase domain would be missing.
Lipoxygenase-3 is non-heme iron-containing dioxygenase highly expressed in a suprabasal epidermis. This enzyme is involved in lipid metabolism of lamellar granule content or intercellular lipid layer by acting as hydroperoxide isomerase (epoxyalcohol synthase) using 12R-HPETE product of ALOX12B into a specific epoxy alcohol product 8R-hydroxy-11R, 12R-epoxyeicosa-5Z, 9E,14Z-trienoic acid.30,36 The pathomechanism involved behind the mutations in the ALOXE3 gene is the disruption of the normal permeability barrier of the skin, which might be due to the abnormal lipid metabolism in the keratinocytes.36
In summary, we have identified three novel sequence variants, one in TGM1 and two in ALOXE3, in three consanguineous families segregating LI and CIE types of ARCI. The mutations in ALOXE3 probably represent loss of function mutations, while the other mutation in the TGM1 may impair enzyme activity, supporting the crucial role played by these two genes during epidermal barrier formation.
Acknowledgments
We are grateful to all members of the three families for participating in the study. The work presented here was funded through research grants to W.A. and M.A. from the Higher Education Commission (Islamabad, Pakistan) and by National Institutes of Health grants U54 HG006493.
Footnotes
Conflicts of interest: None.
References
- 1.Oji V, Tadini G, Akiyama M, et al. Revised nomenclature and classification of inherited ichthyoses: results of the first ichthyosis consensus conference in Soreze 2009. J Am Acad Dermatol. 2010;63:607–641. doi: 10.1016/j.jaad.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 2.Richard G. Molecular genetics of the ichthyoses. Am J Med Genet. 2004;131:32–44. doi: 10.1002/ajmg.c.30032. [DOI] [PubMed] [Google Scholar]
- 3.Campbell JM, Banta-Wright SA. Neonatal skin disorders: a review of selected dermatologic abnormalities. J Perinat Neonat Nurs. 2000;14:63–83. doi: 10.1097/00005237-200006000-00006. [DOI] [PubMed] [Google Scholar]
- 4.Maurice L, Dorfman MB, Chaim Hershko MD, et al. Ichthyosiform dermatosis with systemic lipidosis. Arch Dermatol. 1974;110:261–266. [PubMed] [Google Scholar]
- 5.Fischer J. Autosomal recessive congenital ichthyosis. J Invest Dermatol. 2009;129:1319–1321. doi: 10.1038/jid.2009.57. [DOI] [PubMed] [Google Scholar]
- 6.Kelsell DP, Norgett EE, Unsworth H, et al. Mutations in ABCA12 underlie the severe congenital skin disease harlequin ichthyosis. Am J Hum Genet. 2005;76:794–803. doi: 10.1086/429844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akiyama M, Sugiyama-Nakagiri Y, Sakai K, et al. Mutations in ABCA12 in harlequin ichthyosis and functional rescue by corrective gene transfer. J Clin Invest. 2005;115:1777–1784. doi: 10.1172/JCI24834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grall A, Guaguere E, Planchais S, et al. PNPLA1 mutations cause autosomal recessive congenital ichthyosis in golden retriever dogs and humans. Nat Genet. 2012;44:140–147. doi: 10.1038/ng.1056. [DOI] [PubMed] [Google Scholar]
- 9.Huber M, Rettler I, Bernasconi K, et al. Mutations of keratinocyte transglutaminase in lamellar ichthyosis. Science. 1995;267:525–528. doi: 10.1126/science.7824952. [DOI] [PubMed] [Google Scholar]
- 10.Israeli S, Khamaysi Z, Fuchs-Telem D, et al. A mutation in LIPN, encoding epidermal lipase N, causes a late-onset form of autosomal-recessive congenital ichthyosis. Am J Hum Genet. 2011;88:482–487. doi: 10.1016/j.ajhg.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jobard F, Lefĕevre C, Karaduman A, et al. Lipoxygenase-3 (ALOXE3) and 12(R)-lipoxygenase (ALOX12B) are mutated in nonbullous congenital ichthyosiform erythroderma (NCIE) linked to chromosome 17p13.1. Hum Mol Genet. 2002;11:107–113. doi: 10.1093/hmg/11.1.107. [DOI] [PubMed] [Google Scholar]
- 12.Lefevre C, Audebert S, Jobard F, et al. Mutations in the transporter ABCA12 are associated with lamellar ichthyosis type 2. Hum Mol Genet. 2003;12:2369–2378. doi: 10.1093/hmg/ddg235. [DOI] [PubMed] [Google Scholar]
- 13.Lefevre C, Bouadjar B, Ferrand V, et al. Mutations in a new cytochrome P450 gene in lamellar ichthyosis type 3. Hum Mol Genet. 2006;15:767–776. doi: 10.1093/hmg/ddi491. [DOI] [PubMed] [Google Scholar]
- 14.Lefevre C, Bouadjar B, Karaduman A, et al. Mutations in ichthyin a new gene on chromosome 5q33 in a new form of autosomal recessive congenital ichthyosis. Hum Mol Genet. 2004;13:2473–2482. doi: 10.1093/hmg/ddh263. [DOI] [PubMed] [Google Scholar]
- 15.Natsuga K, Akiyama M, Kato N, et al. Novel ABCA12 mutations identified in two cases of non-bullous congenital ichthyosiform erythroderma associated with multiple skin malignant neoplasia. J Invest Dermatol. 2007;127:2669–2673. doi: 10.1038/sj.jid.5700885. [DOI] [PubMed] [Google Scholar]
- 16.Radner FP, Marrakchi S, Kirchmeier P, et al. Mutations in CERS3 cause autosomal recessive congenital ichthyosis in humans. PLoS Genet. 2013;9:e1003536. doi: 10.1371/journal.pgen.1003536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Russell LJ, DiGiovanna JJ, Rogers GR, et al. Mutations in the gene for transglutaminase-1 in autosomal recessive lamellar ichthyosis. Nat Genet. 1995;9:279–283. doi: 10.1038/ng0395-279. [DOI] [PubMed] [Google Scholar]
- 18.Hennies HC, Raghunath M, Wiebe V, et al. Genetic and immunohistochemical detection of mutations inactivating the keratinocyte transglutaminase in patients with lamellar ichthyosis. Hum Genet. 1998;102:314–318. doi: 10.1007/s004390050697. [DOI] [PubMed] [Google Scholar]
- 19.Huber M, Yee VC, Burri N, et al. Consequences of seven novel mutations on the expression and structure of keratinocyte transglutaminase. J Biol Chem. 1997;272:21018–21026. doi: 10.1074/jbc.272.34.21018. [DOI] [PubMed] [Google Scholar]
- 20.Matise TC, Chen F, Chen W, et al. A second-generation combined linkage physical map of the human genome. Genome Res. 2007;17:1783–1786. doi: 10.1101/gr.7156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seelow D, Schuelke M, Hildebrandt F, et al. HomozygosityMapper – an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37:593–599. doi: 10.1093/nar/gkp369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silberstein M, Tzemach A, Dovgolevsky N, et al. Online system for faster multipoint linkage analysis via parallel execution on thousands of personal computers. Am J Hum Genet. 2006;78:922–935. doi: 10.1086/504158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kent WJ. BLAT-the BLAST like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi Y, Sims GE, Murphy S, et al. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwarz JM, Rodelsperger C, Schuelke M, et al. Mutation Taster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 27.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The 1000 Genomes Project. [accessed 10 May 2014];A deep catalog of human genetic variation. Available via Dialog: http://www.1000genomes.org of subordinate document.
- 29.National Heart, Lung, and Blood Institute (NHLBI), Exome Sequencing Project (ESP) [accessed 10 May 2014];Exome Variant Server. Available via Dialog: http://evs.gs.washington.edu/EVS/ of subordinate document.
- 30.Wortsman X, Aranibar L, Morales C. Postnatal 2- and 3-dimensional sonography of the skin and nail in congenital autosomal recessive ichthyosis correlated with cutaneous histologic findings. J Ultrasound Med. 2011;30:1437–1443. doi: 10.7863/jum.2011.30.10.1437. [DOI] [PubMed] [Google Scholar]
- 31.Rathi NV, Rawlani SM, Hotwani KR. Oral manifestations of lamellar ichthyosis: a rare case report and review. JPAD. 2013;23:99–102. [Google Scholar]
- 32.Avrahami L, Maas S, Pasmanik-Chor M, et al. Autosomal recessive ichthyosis with hypotrichosis syndrome: further delineation of the phenotype. Clin Genet. 2008;74:47–53. doi: 10.1111/j.1399-0004.2008.01006.x. [DOI] [PubMed] [Google Scholar]
- 33.Terrinoni A, Serra V, Codispoti A, et al. Novel transglutaminase 1 mutations in patients affected by lamellar ichthyosis. Cell Death Dis. 2012;3:e416. doi: 10.1038/cddis.2012.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. [accessed 15 Jan 2014];The Human Gene Mutation Database. Available via Dialog: http://www.hgmd.org of subordinate document.
- 35.Elias PM, Schmuth M, Uchida Y, et al. Basis for the permeability barrier abnormality in lamellar ichthyosis. Exp Dermatol. 2002;11:248–256. doi: 10.1034/j.1600-0625.2001.110308.x. [DOI] [PubMed] [Google Scholar]
- 36.Yu Z, Schneider C, Boeglin WE, et al. The lipoxygenase gene ALOXE3 implicated in skin differentiation encodes a hydroperoxideisomerase. Proc Natl Acad Sci USA. 2003;100:9162–9167. doi: 10.1073/pnas.1633612100. [DOI] [PMC free article] [PubMed] [Google Scholar]