Abstract
Synthetic biology is a new discipline that combines science and engineering approaches to precisely control biological networks. These signaling networks are especially important in fields such as biomedicine and biochemical engineering. Additionally, biological networks can also be critical to the production of naturally occurring biological nanomaterials, and as a result, synthetic biology holds tremendous potential in creating new materials. This review introduces the field of synthetic biology, discusses how biological systems naturally produce materials, and then presents examples and strategies for incorporating synthetic biology approaches in the development of new materials. In particular, strategies for using synthetic biology to produce both organic and inorganic nanomaterials are discussed. Ultimately, synthetic biology holds the potential to dramatically impact biological materials science with significant potential applications in medical systems.
Keywords: Synthetic biology, Biomaterials, Gene circuits, Cellular engineering
Introduction: synthetic biology and its potential in materials science
Synthetic biology is revolutionizing approaches to cellular engineering and has already shown the potential to impact cell-based materials science. Whenever attempting to engineer materials, care should be taken to ensure robust design and synthesis processes, along with repeatable and precise structures. Biological systems naturally incorporate all of these requirements in their own synthesis processes. Reproducing these systems by imitating them through biomimicry is a widely used approach to capture the useful qualities of biological materials [1]. Yet, cells themselves hold potential as nanofactories for the production of biomaterials [2, 3]. Like all cellular functions, biomaterial synthesis processes are governed by underlying biological networks—programs encoded in their DNA—and synthetic biology aims to directly engineer these biological networks [4]. As a result, synthetic biology holds significant promise in materials science.
In order to engineer materials using these precisely controlled cellular processes, researchers must have access to the unique set of cellular programming tools provided by synthetic biology. Thirteen years ago, synthetic biology was launched with reports of two synthetic gene networks [5, 6], the ‘repressilator’ and the ‘toggle switch’. Rather than open-loop control of biological processes, these systems used feedback to provide a new level of complexity in engineered cellular control of gene expression. These types of engineered gene networks, also known as synthetic gene circuits, have rapidly advanced to include control structures such as counters, timers and logic gates [7–9]. The field has also expanded from circuits initially based on DNA–protein interactions, to include circuits based on protein–protein interactions [10, 11]. Furthermore, sophisticated algorithms and software tools have been developed to assist in engineering biological networks [12, 13]. These research thrusts have built a foundation for synthetic biology as a complete discipline. This review will explore this new field's increasing promise, particularly in the cellular production of materials.
Synthetic biology's potential in materials science and nanoengineering
Nanobiomaterials are synthesized across the different biological species ranging from bacteria to animals. For example, some species of bacteria can fix carbon dioxide in small nanoparticle structures called carboxysomes [14], allowing them to optimize their metabolism. Mechanisms to create metallic and magnetic particles are also present. For example, magnetotactic bacteria create ferromagnetic nanoparticles containing magnetite (Fe3O4) or greigite (Fe3S4) inside their bodies, allowing them to orient themselves using the geomagnetic field, thus optimizing their navigation toward food [15]. Similarly, clusters of magnetic particles have been found in several animals such as nanoscale particles in pigeons [16–18]. Clearly, biological mechanisms exist to create novel material particles. However, engineering these processes to create materials in a controlled fashion is critical for leveraging these processes in materials science.
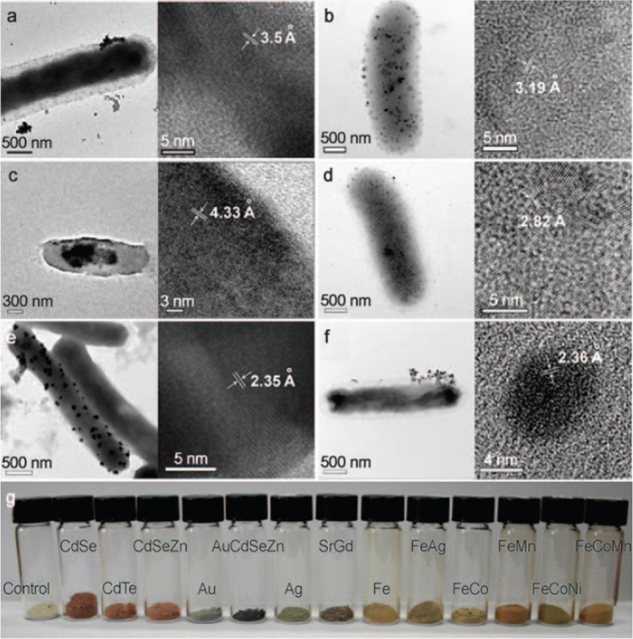
Along these lines, the group of Sang Yup Lee recently reported the intracellular formation of several types of metallic nanoparticles in Escherichia coli [19], including semiconducting, alkali earth and magnetic metal nanoparticles. They formed these metal nanoparticles by leveraging the metal binding properties of two proteins, phytochelatin (PC) and metallothionein (MT) in engineered E. coli. Examples of these particles are shown in figure 1. Furthermore, the researchers could control the size of these biogenic particles by varying metallic ion concentration in the cellular environment.
Figure 1.
TEM images of various nanoparticles synthesized by engineered E. coli in the group of Sang Yup Lee: (a) CdSeZn, (b) PrGd, (c) CdCs, (d) FeCo, (e) Au, (f) Ag. (g) Freeze dried E. coli cells containing diverse NPs each having crystalline nanostructures. Reprinted with permission from Wiley-VCH Verlag GmbH & Co: Angewandte Chemie [19], copyright 2010.
Engineering synthetic circuits
Synthetic gene circuits
For the first synthetic circuits to be engineered, several molecular biological control components were required, all of which are ultimately encoded in the cell's DNA. These DNA molecules inside cells contain the blueprint for cellular structure and function, as well as cellular control processes. This DNA blueprint can be read by several proteins with enzymatic activity. These enzymes are themselves encoded by the DNA blueprint. Other DNA-encoded enzymes are able to catalyze chemical reactions throughout the cell. Furthermore, other proteins contain regions that can bind metallic ions. Together, many structural and enzymatic proteins can form complexes that allow organic or inorganic atoms to be sequestered, resulting in the formation of nanomaterial particles.
These different molecular biological steps emerged in the last half of the 20th century as investigators began to understand what is now known as molecular biology's central dogma. Briefly, this paradigm refers to the fact that the order of DNA's sugar bases correspond directly to the order of amino acids in proteins monomers. More precisely, it refers to the fact that genetic information flows unidirectionally [20]. DNA is first transcribed by RNA polymerase into a messenger RNA (mRNA) molecule that mirrors the bases of DNA. This mRNA is then translated at intracellular ribosomes into protein monomers. These steps are critical to all biological functions, including the synthesis of materials, because proteins can serve as structural components or as catalysts (i.e. enzymes) in the reactions that produce biological materials (among many other functions).
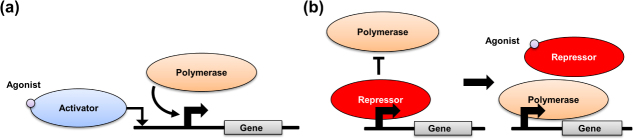
Although these steps are now well known, they are important to review, because synthetic biologists have built components to control each aspect of this process. For example, RNA polymerase must bind specifically to a promoter region of DNA to transcribe its downstream DNA bases into a complementary RNA molecule. As shown in figure 2(a), RNA polymerase can be helped by activator proteins that catalyze this binding event, or it can be blocked completely by repressor proteins, shown in figure 2(b), that bind directly to DNA. These polymerases and transcription factors (i.e. the activators and repressors) have known affinities for specific sequences of DNA. Similarly, the sequence transcribed in RNA affects its binding to ribosomes. By altering these sequences in a precise way, engineering control can be placed on gene expression events. In fact, extremely robust naturally occurring promoters were optimized as tools for controlling gene expression throughout the late 1990s. As an example, these tools include optimized versions of the PBAD promoter along with its activator AraC [21]. As another example, Lutz and Bujard [22] produced several engineered promoters, consisting of hybrids of viral and bacterial promoter regions, which displayed strong and reliable ON and OFF outputs in bacteria. These promoters responded well to the repressors lacI and tetR, and in one hybrid case, allowed both repression by lacI and activation by AraC.
Figure 2.
Regulation of gene expression by transcription factors: (a) upregulation by transcriptional activators and (b) downregulation by transcriptional repressors.
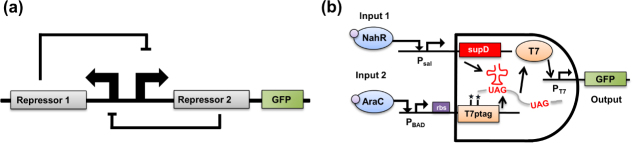
These new engineered promoters were critical in the design of networks like the aforementioned toggle switch developed in the laboratory of James Collins [5]. As shown in figure 3(a), the toggle consists of two mutually repressing genetic operons, and when the strength of each repression event is balanced, a bistable switch is formed. Over multiple generations of bacterial cell division, the toggle will remain in either an ON or OFF state, even without the presence of its corresponding inducer molecule, lactose and tetracycline. Yet, as noted, the potential for each operon to repress the other must be balanced to ensure bistability, and this potential repression is governed by multiple interactions, including the strength of the promoter (i.e. the binding strength of DNA and RNA polymerase), as well as the strength of ribosome binding site on the mRNA transcript (i.e. the binding strength of the initial few bases of a transcript and a ribosome). By strategically altering these bases, the toggle can be balanced. Furthermore, the toggle switch was based on strong fundamental engineering theory, as the Collins group first published an underlying theory showing that bistable memory could be achieved using the nonlinear dynamics of biological interactions [23]. Similar theoretical and experimental approaches have been harnessed to make other advances in synthetic biology.
Figure 1.
Synthetic gene circuits: (a) bistable ‘toggle’ switch [5] and (b) engineered AND logic gate [8].
For example, since these first circuits were reported, synthetic biology efforts have illustrated the potential for digital logic gate behavior in circuits [8, 24–26] using polymerases [8] as well as recombinases [27, 28]. In one case, Anderson et al engineered a synthetic AND gate in bacteria as shown in figure 3(b). This circuit consisted of an engineered viral RNA polymerase with a slight defect in the middle of its genetic code that blocked its complete translation [8]. Thus, when one input (such as arabinose) was provided, only a partial, useless portion of polymerase was expressed. When another input, such as salicylate, activated the transcription of a small RNA, this small molecule worked to reverse the defect, and the fully translated viral polymerase could then activate a viral promoter. In another study, the Voigt group showed that communities of bacteria could work together to produce NOR-gate behavior [26]. This was especially important as NOR-gates are Boolean complete and can be combined to form any other type of logic gate, thus suggesting that with enough discrete bacterial colonies, complexity in digital behavior could increase. Of course, applications may also require analog signals, and work in the lab of Timothy Lu at MIT has recently shown that synthetic techniques can be applied to create sophisticated analogue circuits in single bacteria cells [29].
Other synthetic control behaviors are also possible. For example, Ellis et al [9] described the development of synthetic timers in yeast, allowing flocculation to be precisely timed (thus potentially providing precise control to fermentation processes). In an especially interesting study, Stricker et al [30] used a minimal set of components to build an oscillator significantly more robust than the repressilator. In fact, multiple oscillators have now been developed in both bacterial and mammalian systems [6, 30–34]. Clearly, many synthetic circuit modules (bistable memory, oscillation, timing, counting, etc) can be further optimized.
Synthetic biology using proteins: components and circuits
As discussed above, engineering circuits that rely on DNA–protein interactions has been one major thrust in synthetic biology; yet, another key thrust has been the development of protein–protein signaling components. Just as genetic components have allowed the construction of eclectic synthetic circuits, protein components have allowed for new synthetic circuit designs as well. These engineered protein interactions have been important in providing new components to interface with the previously described genetic circuits, as well as allowing for the development of protein–protein interaction circuits. One key advantage of these protein-based circuits is the opportunity to take advantage of the speed at which proteins interact. In comparison to gene expression, protein-mediated (i.e. enzyme-mediated) single phosphorylation signals are evident on the order of seconds, while gene expression events frequently take on the order of minutes or longer to fully emerge in a cell's phenotype.
In the case of engineering new protein-based components to interface with synthetic gene circuits, several exciting components have been developed. Most notably, synthetic biology has significantly impacted the field of optogenetics. For example, in early work, a cyanobacterial photoreceptor was fused to an E. coli intracellular histidine kinase domain to control gene expression [10]. While researchers previously could measure gene circuit behavior by observing expression of GFP, light was not an accessible input to these engineered circuits. However, as shown in figure 4, it is now possible to use light directly as an input. This work has been expanded to include multiple different wavelengths and multiple applications in a significant expansion of the synthetic biologist's toolbox [35–38]. In yet another advance, June Medford's laboratory has developed a plant signaling receptor that allows plants to bind to trinitrotoluene (TNT) and activate internal reporters. Using this component, her laboratory engineered sensitive Arabidopsis plants to turn white in the presence of minute amounts of TNT explosives [39].
Figure 4.
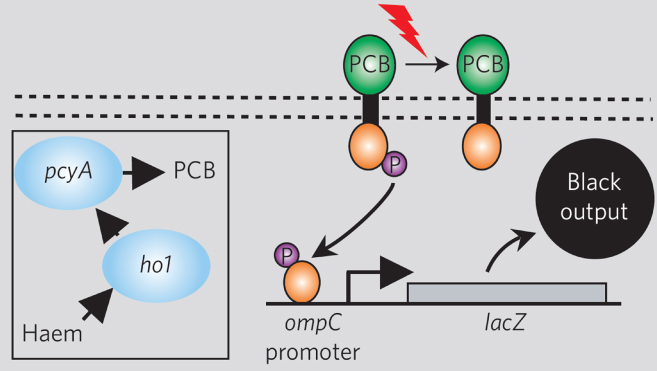
Synthetic light sensor based upon engineered proteins. Reprinted by permission from Nature Publishing Group: Nature [10], copyright 2005.
Perhaps the most interesting work in engineered protein circuits has come from the laboratory of Wendell Lim. For example, as shown in figure 5, Bashor et al [11] showed that positive and negative feedback loops could be engineered into living eukaryotes by anchoring synthetic protein signaling components directly to an engineered protein scaffold using leucine zippers.
Figure 5.
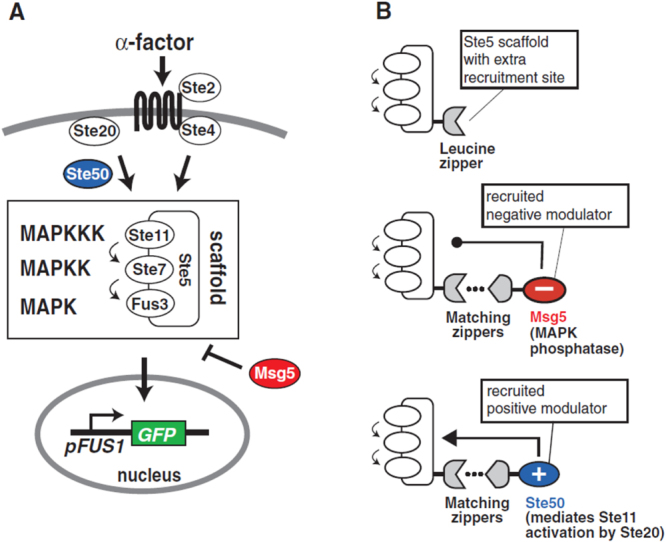
(a) Synthetic components and (b) circuits based upon engineered protein scaffold circuits. Reprinted by permission from The American Association for the Advancement of Science: Science [11], copyright 2008.
Synthetic biology: rapid prototyping
Several key challenges must be overcome to build and expand upon the systems described above. First, synthetic biology is plagued by a lack of components with which to develop circuits. Furthermore, engineering synthetic circuits can be extremely labor intensive. Finally, the behavior of designed circuits in vivo is often difficult to predict. Multiple approaches and technologies have been developed to address each of these challenges. In order to address the lack of synthetic components, several groups and consortia have focused on organizing repositories of biological components [41]. However, a key drawback to these repositories is often that the accompanying functional annotations (descriptions of how a component should behave) are often missing or unreliable. For example, synthetic components can behave quite differently in the various cell strains of a microbial or mammalian species. Thus, other approaches have focused on building sets of components that can be broadly extendable [42–46]. For example, Khalil et al [46] recently produced a framework for creating synthetic transcription factors in eukaryotes based on zinc-finger protein domains, which potentially can be extended to allow a broad range of designer-specified DNA sequences to function as promoters.
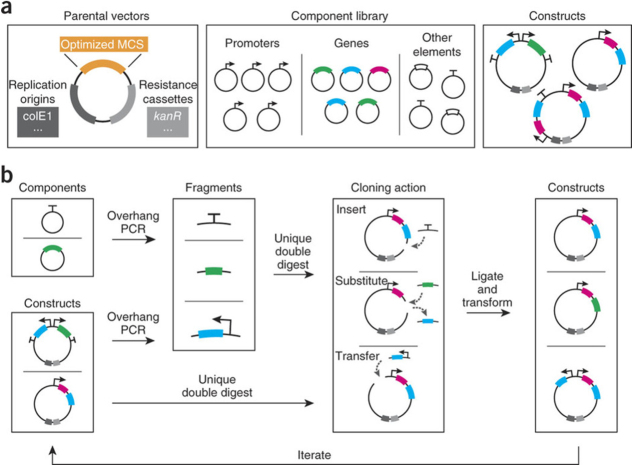
In order to address the challenge of quickly building synthetic systems, several exciting technologies have been recently developed. One of the biggest challenges in creating synthetic circuits is the laborious process of molecular cloning, which involves assembling multiple components over several days or weeks. Furthermore, as these components are cut and pasted together at specific DNA sequences (called restriction sites); the order of assembly is a challenge as many components contain internal restriction sites. One way around this challenge is to use techniques that allow long chains of DNA molecules to be assembled in a single, one-pot reaction, such as gene splicing by overlap extension [47, 48] or Gibson assembly [49]. However, just as electrical engineers can replace individual components to test circuit function, synthetic biologists would ideally be able to do the same in engineered biological circuits. Although cutting out individual components with restriction enzymes would be ideal, as mentioned, this could lead to unintended cuts in other components. Furthermore, the faster one-pot reactions typically avoid restriction sites altogether. In order to build systems rapidly while adding plug-and-play capabilities to molecular cloning, Litcofsky et al [40] recently reported a broad set of common synthetic biology components that had been reengineered to eliminate restriction sites. As a result, the authors could rapidly construct several unique gene circuits in just a few days, as opposed to weeks, while adding the ability to adjust single components. This approach is shown in figure 6.
Figure 6.
(a) Elements comprising the framework: parental cloning vectors harboring a custom multiple cloning site (MCS) of optimal restriction enzyme sites, a library of commonly used synthetic genetic components designed to exclude the restriction sites, and a repository of assembled constructs that includes synthetic modules, intermediates and circuits. (b) Generalized workflow for constructing and modifying synthetic gene networks, which prioritizes and streamlines the iterative process of arriving at functional networks and modules. Reprinted by permission from Macmillan Publishers Ltd: Nature Methods [40], copyright 2012.
One challenge of working with the molecular cloning techniques described above is that the inserted genetic components must provide an advantage to the host; otherwise, the genes will ‘fall out’ due to genetic drift or natural selection. As a result, most synthetic circuits must be inserted along with an advantageous component, such as a genetically encoded antibiotic resistance cassette. Cells must then be treated perpetually with antibiotics to ensure that the inserted components remain in the cell. A challenge is that most of these techniques are used to produce plasmids containing several genetic modifications, including the aforementioned antibiotic resistance cassette. Making several modifications directly to the host chromosome is onerous. To address these challenges, the group of George Church developed an automated method, multiplex automated genomic engineering [50], which enables direct genome modifications. Their approach initially allowed the optimization of up to 24 genes simultaneously, creating billions of genetic variants.
Moreover, key technologies have been developed to predict the behavior of synthetically designed components and circuits in vivo. As one of the best examples, Salis et al [51] reported the automated design of ribosome binding sites to control gene expression. Furthermore, the Anderson and Densmore laboratories have developed specific programming languages [52] and software [12] to enable the design of synthetic biology circuits using the repositories of genetic components mentioned above.
Biological material production
Engineering materials synthesis
In order to deploy synthetic biology in natural biological materials synthesis pathways, it is critical to understand how natural systems produce biomaterials. Extensive literature exists that discusses biological production of complex molecules and structures. Indeed, animal and plant anatomies are a testament to the level of complexity and specialization achievable by nature. For the purpose of understanding how synthetic biology can assist in producing engineered materials, it is important to consider how biological materials can be made, and how bioengineers have often approached this problem. For example, metabolic engineers have often tackled the problem of converting simple sugars and cellulosic biomass into useful substances. Over the past few decades [53], they have developed the catalytic capacity of organisms in the conversion of five-ring and six-ring carbon sources into useful organic molecules like drugs [54] and fuels [55]. By mapping the metabolic pathways of microbial organisms, they have been able to predictively model and experimentally confirm the conversion of these metabolites by cascades of intracellular reactions [56, 57]. Several of the key components now useful in synthetic biology are a direct result of these approaches and were specifically developed to control these processes [58–61]. While the line between these disciplines is blurry [62, 63], synthetic biology is certainly characterized by a drive toward the development of more complex pathways, cellular components and design-and-build approaches. In the case of materials synthesis, potential challenges loom in the production of complicated materials [64, 65] and patterns [66–68].
Natural and synthetic biological pattern formation
Biological patterning is critical in the material scaffold systems widely used in biomaterial design and synthesis. Just as the engineered protein scaffolds described above enabled the development of intracellular signaling circuits in yeast [11], patterned material scaffolds can coordinate extracellular binding events as well, and thus, extracellular material assembly. This paradigm has impacted broad fields ranging from tissue engineering [69] to molecular self-assembly for nanoelectronics (e.g. the engineered viral scaffolds [70–73] developed in the Belcher laboratory). As a result, strategies incorporating synthetic biology into biological patterning could be critical in leveraging cellular processes to build materials.
A robust example of biological patterning was reported by the Weiss group in work that coupled components of bacterial quorum sensing with engineered pigment changes. Some bacteria can naturally alter their behavior based upon their cellular density around them in a process known as quorum sensing [74, 75]. In these bacterial species, individual cells have the ability to secrete quorum sensing molecules. The bacteria then detect elevated levels of these secreted molecules and alter their behavior. Using this process, bacteria can regulate a range behaviors including bioluminescence [76], biosurfactant synthesis [77] and extracellular polymer production [78–80].
By transferring quorum sensing components to non-quorum sensing bacteria the Weiss group was able to engineer bacterial communication between cells that functioned as ‘sender’ and ‘receiver’ strains. As a result, when a colony of ‘sender’ cells was placed on a ‘receiver’ bacterial lawn, a bull's-eye pattern formed around the ‘sender’ colony. This pattern could be further altered by placing multiple ‘sender’ colonies on a lawn [67]. In another interesting study, the Voigt lab expanded upon their work with light-detection to develop a synthetic circuit that allowed a lawn of bacteria to distinguish the edges of projected silhouettes [68]. These first circuits, driving synthetic biological pattern formation, portend increasingly complicated engineered biological patterns. To this end, we can look at embryogenesis, the process by which a single fertilized egg grows into uniquely patterned tissues. In a now widely accepted model, Turing proposed that interactions between two molecular morphogens could give rise to the complicated patterns seen in development [81]. Rather that two molecules passively diffusing through a tissue and signaling cellular differentiation, in the proposed reaction–diffusion model, shown in figure 7, morphogens could also react with one another, and these reactions could give rise to increasingly complicated patterns [66]. These reaction networks are especially amenable to synthetic biology, and synthetic biological circuits that alter these networks could potentially control the patterning of molecular scaffolds through engineered cellular secretion machinery [82].
Figure 7.
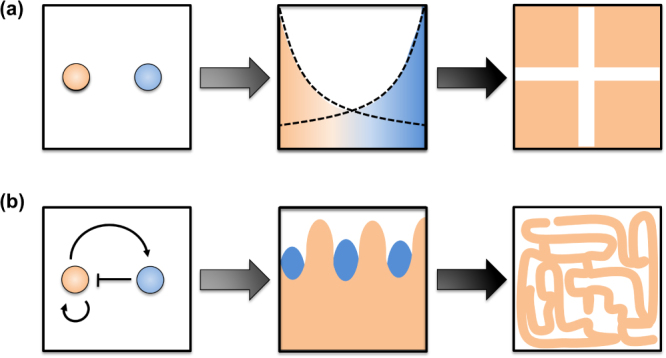
Pattern formation resulting from morphogens: (a) passively diffusing morphogens result in simple gradients capable yielding basic striped patterns and (b) interacting morphogens result in oscillations capable of yielding more complicated patterns [66].
Nanomaterial assembly in biological compartments
In addition to engineering biological systems to secrete patterned scaffolds for molecular assembly, control of biological systems producing more complex molecular assemblies is also possible. Although it has been widely understood that eukaryotic cells contain compartmental organelles, several important examples of primitive organelles have now been observed in bacteria as well. These microcompartments and nanocompartments can form either as a result of protein shells that allow the internal assembly of materials [84], or through complex processes that coordinate the assembly of internal material structures [83]. For example, in structures called carboxysomes [14], bacteria can synthesize protein shells that resemble virus capsids and contain carbon. These nanoscale inclusions have now been widely studied, and reengineered compartments can have been transferred between heterologous hosts [85]. These approaches may yield new intracellular nanoscale particles, as well as compartments housing cascades of designed reactions for the production of critical biomolecules [86], thus following an intracellular nanofactory paradigm [3].
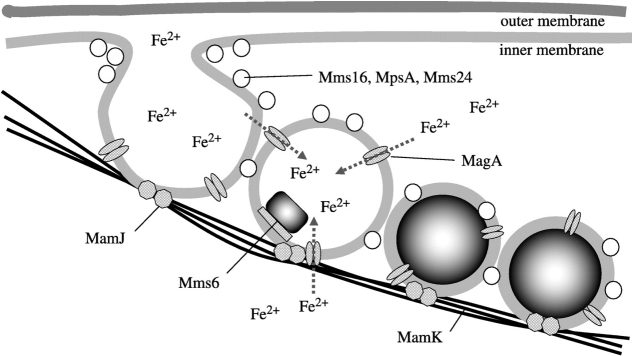
Beyond compartments formed by protein shells, some bacteria have the capacity to form even more complicated internal compartments. As an example, magnetotactic bacteria (MTB) synthesize linear chains of ferromagnetic particles that effectively serve as compass needles [87], allowing them to orient their movements with the geomagnetic field [15]. A well-studied genomic island contributes to the production of multiple proteins that coordinate the initial invagination of the MTB inner membrane, as well as the nucleation of Fe3O4 crystals [83, 88], as shown in figure 8. These crystals are spatially coordinated within the cell by structural scaffold proteins. In one example, when a scaffold gene was removed, crystals still formed but would no longer spatially arrange in a linear fashion [89]. Thus, synthetic circuits that control the expression of these key genes could potentially shape the assembly of these nanoparticles and their intracellular superstructure.
Figure 8.
Hypothesized mechanism of magnetosomes in MTB. Reprinted by permission from Royal Society Publishing: Journal of Royal Society Interface [83], copyright 2008.
Two efforts to synthetically engineer magnetic nanoparticles in living cells were recently published in separate studies by the groups of Martin Fussenegger and Pamela Silver, showing the production of magnetic nanoparticles within mammalian [90] and yeast cells [91], respectively. In the example from the Fussenegger group, shown in figure 9, both an iron transporter and a heavy-chain ferritin monomer were produced by synthetic networks in cells. In a process significantly less complicated than magnetosome formation, heavy-chain ferritin assembled into apoferritin, a protein shell capable of storing 4500 Fe3+ ions in its 8 nm cavity. While insufficient for ferromagnetic behavior, these particles produced paramagnetic behavior that was sufficient to allow cells to be separated from non-magnetic cells in complex cell mixtures.
Figure 9.
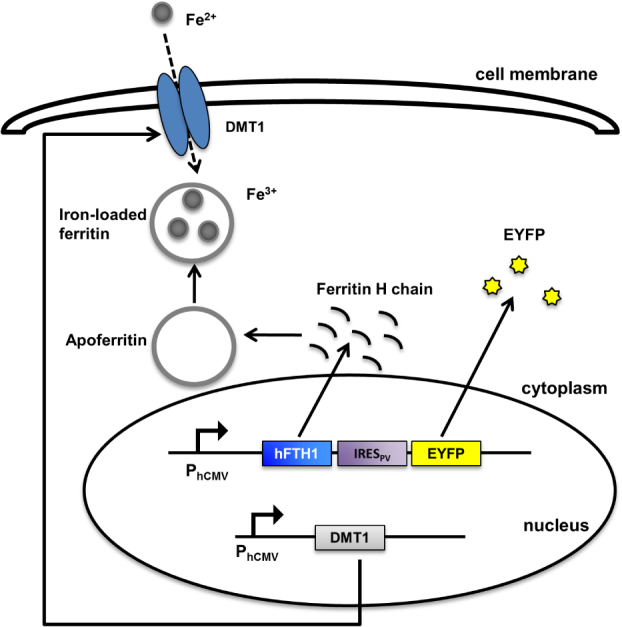
Superparamagnetic nanoparticles programmed by a synthetic network and synthesized in HEK 293 cells [90].
5. Conclusions and outlook
Synthetic biology has tremendous potential in creating cells that produce biological nanomaterials. Engineered biological circuits and control structures continue to improve, along with the computational and software tools necessary to optimize design with synthetic components. Concurrently, synthetic biologists are tackling problems in the development of new patterning approaches and the development of new microscale and nanoscale intracellular compartments. As these technologies evolve toward creating nanoscale biomaterials, it will be critical to integrate new synthetic patterning and materials synthesis components into collections of synthetic biology parts and tools. Ultimately, synthetic biology holds the potential to transform engineered biological cells beyond their role as metabolic catalysts in the production of simple organic molecules, allowing cells to serve as cellular foundries and nanofactories.
Acknowledgment
The authors gratefully acknowledge support from award FA9550-13-1-0108 from the Air Force Office of Scientific Research of the USA.
References
- Sanchez C, Arribart H and Guille M M. 2005. Biomimetism and bioinspiration as tools for the design of innovative materials and systems Nature Mater. 4 277–88 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ruder W C and LeDuc P R. 2008. Artificial cells: building bioinspired systems using small-scale biology Trends Biotechnol. 26 14–20 [DOI] [PubMed] [Google Scholar]
- Leduc P R. 2007. Towards an in vivo biologically inspired nanofactory Nature Nanotechnol. 2 3–7 [DOI] [PubMed] [Google Scholar]
- Ruder W C, Lu T and Collins J J. 2011. Synthetic biology moving into the clinic Science 333 1248–52 [DOI] [PubMed] [Google Scholar]
- Gardner T S, Cantor C R and Collins J J. 2000. Construction of a genetic toggle switch in Escherichia coli Nature 403 339–42 [DOI] [PubMed] [Google Scholar]
- Elowitz M B and Leibler S. 2000. A synthetic oscillatory network of transcriptional regulators Nature 403 335–8 [DOI] [PubMed] [Google Scholar]
- Friedland A E, Lu T K, Wang X, Shi D, Church G and Collins J J. 2009. Synthetic gene networks that count Science 324 1199–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J C, Voigt C A. and Arkin A P. Environmental signal integration by a modular AND gate. Mol. Syst. Biol. 2007;3:133. doi: 10.1038/msb4100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis T, Wang X and Collins J J. 2009. Diversity-based, model-guided construction of synthetic gene networks with predicted functions Nature Biotechnol. 27 465–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levskaya A. 2005. Synthetic biology: engineering Escherichia coli to see light Nature 438 441–2 [DOI] [PubMed] [Google Scholar]
- Bashor C J, Helman N C, Yan S and Lim W A. 2008. Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics Science 319 1539–43 [DOI] [PubMed] [Google Scholar]
- Xia B, Bhatia S, Bubenheim B, Dadgar M, Densmore D and Anderson J C. 2011. Developer's and user's guide to Clotho v2.0 a software platform for the creation of synthetic biological systems Methods Enzymol. 498 97–135 [DOI] [PubMed] [Google Scholar]
- Bilitchenko L. Eugene—a domain specific language for specifying and constraining synthetic biological parts, devices, and systems. PLoS ONE. 2011;6:e18882. doi: 10.1371/journal.pone.0018882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S. 2008. Atomic-level models of the bacterial carboxysome shell Science 319 1083–6 [DOI] [PubMed] [Google Scholar]
- Komeili A. 2012. Molecular mechanisms of compartmentalization and biomineralization in magnetotactic bacteria FEMS Microbiol. Rev. 36 232–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltschko R, Schiffner I, Fuhrmann P and Wiltschko W. 2010. The role of the magnetite-based receptors in the beak in pigeon homing Curr. Biol. 20 1534–8 [DOI] [PubMed] [Google Scholar]
- Treiber C D. 2012. Clusters of iron-rich cells in the upper beak of pigeons are macrophages not magnetosensitive neurons Nature 484 367–70 [DOI] [PubMed] [Google Scholar]
- Mora C V, Davison M, Wild J M and Walker M M. 2004. Magnetoreception and its trigeminal mediation in the homing pigeon Nature 432 508–11 [DOI] [PubMed] [Google Scholar]
- Park T J, Lee S Y, Heo N S and Seo T S. 2010. In vivo synthesis of diverse metal nanoparticles by recombinant Escherichia coli Angew. Chem. Int. Edn Engl. 49 7019–24 [DOI] [PubMed] [Google Scholar]
- Crick F. 1970. Central dogma of molecular biology Nature 227 561–3 [DOI] [PubMed] [Google Scholar]
- Guzman L M, Belin D, Carson M J and Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter J. Bacteriol. 177 4121–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz R and Bujard H. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1–I2 regulatory elements Nucleic Acids Res. 25 1203–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner T S, Dolnik M and Collins J J. 1998. A theory for controlling cell cycle dynamics using a reversibly binding inhibitor Proc. Natl Acad. Sci. USA 95 14190–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Wroblewska L, Prochazka L, Weiss R and Benenson Y. 2011. Multi-input RNAi-based logic circuit for identification of specific cancer cells Science 333 1307–11 [DOI] [PubMed] [Google Scholar]
- Rinaudo K, Bleris L, Maddamsetti R, Subramanian S, Weiss R and Benenson Y. 2007. A universal RNAi-based logic evaluator that operates in mammalian cells Nature Biotechnol. 25 795–801 [DOI] [PubMed] [Google Scholar]
- Tamsir A, Tabor J J and Voigt C A. 2011. Robust multicellular computing using genetically encoded NOR gates and chemical ‘wires’ Nature 469 212–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet J, Subsoontorn P and Endy D. 2012. Rewritable digital data storage in live cells via engineered control of recombination directionality Proc. Natl Acad. Sci. USA 109 8884–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siuti P, Yazbek J and Lu T K. 2013. Synthetic circuits integrating logic and memory in living cells Nature Biotechnol. 31 448–52 [DOI] [PubMed] [Google Scholar]
- Daniel R, Rubens J R, Sarpeshkar R and Lu T K. 2013. Synthetic analog computation in living cells Nature 497 619–23 [DOI] [PubMed] [Google Scholar]
- Stricker J, Cookson S, Bennett M R, Mather W H, Tsimring L S and Hasty J. 2008. A fast, robust and tunable synthetic gene oscillator Nature 456 516–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W W, Tsai T Y. and Liao J C. Single-cell zeroth-order protein degradation enhances the robustness of synthetic oscillator. Mol. Syst. Biol. 2007;3:130. doi: 10.1038/msb4100172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigges M, Marquez-Lago T T, Stelling J and Fussenegger M. 2009. A tunable synthetic mammalian oscillator Nature 457 309–12 [DOI] [PubMed] [Google Scholar]
- Tigges M, Denervaud N, Greber D, Stelling J and Fussenegger M. 2010. A synthetic low-frequency mammalian oscillator Nucleic Acids Res. 38 2702–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung E, Wong W W, Suen J K, Bulter T, Lee S G and Liao J C. 2005. A synthetic gene-metabolic oscillator Nature 435 118–22 [DOI] [PubMed] [Google Scholar]
- Auslander D and Fussenegger M. 2012. Optogenetic therapeutic cell implants Gastroenterology 143 301–6 [DOI] [PubMed] [Google Scholar]
- Bacchus W and Fussenegger M. 2012. The use of light for engineered control and reprogramming of cellular functions Curr. Opin. Biotechnol. 23 695–702 [DOI] [PubMed] [Google Scholar]
- Ye H, Daoud-El Baba M, Peng R W and Fussenegger M. 2011. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice Science 332 1565–8 [DOI] [PubMed] [Google Scholar]
- Tabor J J, Levskaya A and Voigt C A. 2011. Multichromatic control of gene expression in Escherichia coli J. Mol. Biol. 405 315–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes M S. Programmable ligand detection system in plants through a synthetic signal transduction pathway. PLoS ONE. 2011;6:e16292. doi: 10.1371/journal.pone.0016292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litcofsky K D, Afeyan R B, Krom R J, Khalil A S and Collins J J. 2012. Iterative plug-and-play methodology for constructing and modifying synthetic gene networks Nature Methods 9 1077–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl L J. and Endy D. A survey of enabling technologies in synthetic biology. J. Biol. Eng. 2013;7:13. doi: 10.1186/1754-1611-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacchus W, Weber W and Fussenegger M. 2013. Increasing the dynamic control space of mammalian transcription devices by combinatorial assembly of homologous regulatory elements from different bacterial species Metabolic Eng. 15 144–50 [DOI] [PubMed] [Google Scholar]
- Muller M, Auslander S, Auslander D, Kemmer C and Fussenegger M. 2012. A novel reporter system for bacterial and mammalian cells based on the non-ribosomal peptide indigoidine Metabolic Eng. 14 325–35 [DOI] [PubMed] [Google Scholar]
- Wieland M, Auslander D and Fussenegger M. 2012. Engineering of ribozyme-based riboswitches for mammalian cells. Methods 56 351–7 [DOI] [PubMed] [Google Scholar]
- Karlsson M, Weber W and Fussenegger M. 2012. Design and construction of synthetic gene networks in mammalian cells Methods Mol. Biol. 813 359–76 [DOI] [PubMed] [Google Scholar]
- Khalil A S. 2012. A synthetic biology framework for programming eukaryotic transcription functions Cell 150 647–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton R M, Ho S N, Pullen J K, Hunt H D, Cai Z and Pease L R. 1993. Gene splicing by overlap extension Methods Enzymol. 217 270–9 [DOI] [PubMed] [Google Scholar]
- Horton R M, Hunt H D, Ho S N, Pullen J K and Pease L R. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension Gene 77 61–8 [DOI] [PubMed] [Google Scholar]
- Gibson D G, Young L, Chuang R Y, Venter J C, Hutchison C A III and Smith H O. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases Nature Methods 6 343–5 [DOI] [PubMed] [Google Scholar]
- Wang H H. 2009. Programming cells by multiplex genome engineering and accelerated evolution Nature 460 894–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salis H M, Mirsky E A and Voigt C A. 2009. Automated design of synthetic ribosome binding sites to control protein expression Nature Biotechnol. 27 946–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilitchenko L, Liu A and Densmore D. 2011. The Eugene language for synthetic biology Methods Enzymol. 498 153–72 [DOI] [PubMed] [Google Scholar]
- Bailey J E. 1991. Toward a science of metabolic engineering Science 252 1668–75 [DOI] [PubMed] [Google Scholar]
- Paddon C J. 2013. High-level semi-synthetic production of the potent antimalarial artemisinin Nature 496 528–32 [DOI] [PubMed] [Google Scholar]
- Hanai T, Atsumi S and Liao J C. 2007. Engineered synthetic pathway for isopropanol production in Escherichia coli Appl. Environ. Microbiol. 73 7814–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J S. and Palsson B O. Metabolic flux balance analysis and the in silico analysis of Escherichia coli K-12 gene deletions. BMC Bioinformatics. 2000;1:1. doi: 10.1186/1471-2105-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J S, Ibarra R U and Palsson B O. 2001. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data Nature Biotechnol. 19 125–30 [DOI] [PubMed] [Google Scholar]
- Khlebnikov A, Skaug T and Keasling J D. 2002. Modulation of gene expression from the arabinose-inducible araBAD promoter J. Ind. Microbiol. Biotechnol. 29 34–7 [DOI] [PubMed] [Google Scholar]
- Khlebnikov A, Risa O, Skaug T, Carrier T A and Keasling J D. 2000. Regulatable arabinose-inducible gene expression system with consistent control in all cells of a culture J. Bacteriol. 182 7029–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khlebnikov A, Datsenko K A, Skaug T, Wanner B L and Keasling J D. 2001. Homogeneous expression of the P(BAD) promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter Microbiology 147 3241–7 [DOI] [PubMed] [Google Scholar]
- Fussenegger M, Moser S and Bailey J E. 1998. Regulated multicistronic expression technology for mammalian metabolic engineering Cytotechnology 28 111–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felnagle E A, Chaubey A, Noey E L, Houk K N and Liao J C. 2012. Engineering synthetic recursive pathways to generate non-natural small molecules Nature Chem. Biol. 8 518–26 [DOI] [PubMed] [Google Scholar]
- Boyle P M and Silver P A. 2012. Parts plus pipes: synthetic biology approaches to metabolic engineering Metabolic Eng. 14 223–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubeli R J, Burger K and Weber W. 2013. Synthetic biology for mammalian cell technology and materials sciences Biotechnol. Adv. 31 68–78 [DOI] [PubMed] [Google Scholar]
- Lee Y J. 2009. Fabricating genetically engineered high-power lithium-ion batteries using multiple virus genes Science 324 1051–5 [DOI] [PubMed] [Google Scholar]
- Kondo S and Miura T. 2010. Reaction–diffusion model as a framework for understanding biological pattern formation Science 329 1616–20 [DOI] [PubMed] [Google Scholar]
- Basu S, Gerchman Y, Collins C H, Arnold F H and Weiss R. 2005. A synthetic multicellular system for programmed pattern formation Nature 434 1130–4 [DOI] [PubMed] [Google Scholar]
- Tabor J J. 2009. A synthetic genetic edge detection program Cell 137 1272–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutos F T, Freed L E and Guilak F. 2007. A biomimetic three-dimensional woven composite scaffold for functional tissue engineering of cartilage Nature Mater. 6 162–7 [DOI] [PubMed] [Google Scholar]
- Nam Y S, Park H, Magyar A P, Yun D S Pollom T S Jr and Belcher A M. 2012. Virus-templated iridium oxide-gold hybrid nanowires for electrochromic application Nanoscale 4 3405–9 [DOI] [PubMed] [Google Scholar]
- Nam K T. 2006. Virus-enabled synthesis and assembly of nanowires for lithium ion battery electrodes Science 312 885–8 [DOI] [PubMed] [Google Scholar]
- Mao C. 2004. Virus-based toolkit for the directed synthesis of magnetic and semiconducting nanowires Science 303 213–7 [DOI] [PubMed] [Google Scholar]
- Mao C. 2003. Viral assembly of oriented quantum dot nanowires Proc. Natl Acad. Sci. USA 100 6946–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velicer G J. 2003. Social strife in the microbial world Trends Microbiol. 11 330–7 [DOI] [PubMed] [Google Scholar]
- West S A, Griffin A S, Gardner A and Diggle S P. 2006. Social evolution theory for microorganisms Nature Rev. Microbiol. 4 597–607 [DOI] [PubMed] [Google Scholar]
- Engebrecht J and Silverman M. 1984. Identification of genes and gene products necessary for bacterial bioluminescence Proc. Natl Acad. Sci. USA 81 4154–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M and Greenberg E P. 2006. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa Int. J. Med. Microbiol. 296 73–81 [DOI] [PubMed] [Google Scholar]
- Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W and Greenberg E P. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm Science 280 295–8 [DOI] [PubMed] [Google Scholar]
- Hammer B K and Bassler B L. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae Mol. Microbiol. 50 101–4 [DOI] [PubMed] [Google Scholar]
- Nadell C D, Xavier J B, Levin S A. and Foster K R. The evolution of quorum sensing in bacterial biofilms. PLoS Biol. 2008;6:e14. doi: 10.1371/journal.pbio.0060014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turing A M. The chemical basis of morphogenesis 1953. Bull. Math. Biol. 1990;52 doi: 10.1007/BF02459572. 153–97 (discussion 19–52) [DOI] [PubMed] [Google Scholar]
- Widmaier D M. Engineering the Salmonella type III secretion system to export spider silk monomers. Mol. Syst. Biol. 2009;5:309. doi: 10.1038/msb.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakaki A, Nakazawa H, Nemoto M, Mori T and Matsunaga T. 2008. Formation of magnetite by bacteria and its application J. R. Soc. Interface/ 5 977–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E Y and Tullman-Ercek D. 2012. Engineering nanoscale protein compartments for synthetic organelles Curr. Opin. Biotechnol. 24 627–32 [DOI] [PubMed] [Google Scholar]
- Bonacci W. 2012. Modularity of a carbon-fixing protein organelle Proc. Natl Acad Sci. USA 109 478–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary S, Quin M B, Sanders M A, Johnson E T. and Schmidt-Dannert C. Engineered protein nano-compartments for targeted enzyme localization. PLoS ONE. 2012;7:e33342. doi: 10.1371/journal.pone.0033342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakemore R. 1975. Magnetotactic bacteria Science 190 377–9 [DOI] [PubMed] [Google Scholar]
- Frankel R B, Blakemore R P and Wolfe R S. 1979. Magnetite in freshwater magnetotactic bacteria Science 203 1355–6 [DOI] [PubMed] [Google Scholar]
- Komeili A, Li Z, Newman D K and Jensen G J. 2006. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK Science 311 242–5 [DOI] [PubMed] [Google Scholar]
- Kim T, Moore D and Fussenegger M. 2012. Genetically programmed superparamagnetic behavior of mammalian cells J. Biotechnol. 162 237–45 [DOI] [PubMed] [Google Scholar]
- Nishida K. and Silver P A. Induction of biogenic magnetization and redox control by a component of the target of rapamycin complex 1 signaling pathway. PLoS Biol. 2012;10:e1001269. doi: 10.1371/journal.pbio.1001269. [DOI] [PMC free article] [PubMed] [Google Scholar]