Abstract
Phytochromes constitute a major photoreceptor family found in plants, algae, fungi, and prokaryotes, including pathogens. Here, we report that Xanthomonas campestris pv. campestris (Xcc), the causal agent of black rot disease which affects cruciferous crops worldwide, codes for a functional bacteriophytochrome (XccBphP). XccBphP possesses an N‐terminal PAS2‐GAF‐PHY photosensory domain triad and a C‐terminal PAS9 domain as its output module. Our results show that illumination of Xcc, prior to plant infection, attenuates its virulence in an XccBphP‐dependent manner. Moreover, in response to light, XccBphP downregulates xanthan exopolysaccharide production and biofilm formation, two known Xcc virulence factors. Furthermore, the XccbphP null mutant shows enhanced virulence, similar to that of dark‐adapted Xcc cultures. Stomatal aperture regulation and callose deposition, both well‐established plant defense mechanisms against bacterial pathogens, are overridden by the XccbphP strain. Additionally, an RNA‐Seq analysis reveals that far‐red light or XccBphP overexpression produces genomewide transcriptional changes, including the inhibition of several Xcc virulence systems. Our findings indicate that Xcc senses light through XccBphP, eliciting bacterial virulence attenuation via downregulation of bacterial virulence factors. The capacity of XccBphP to respond to light both in vitro and in vivo was abolished by a mutation on the conserved Cys13 residue. These results provide evidence for a novel bacteriophytochrome function affecting an infectious process.
Keywords: bathy‐type phytochrome, infection, plant defenses, transcriptional regulation, virulence factors
Subject Categories: Microbiology, Virology & Host Pathogen Interaction; Plant Biology
Introduction
Biological photoreceptors are found in all kingdoms of life. They can detect the wavelength, duration, direction, and intensity of light and consequently transduce this information within cells to exert the corresponding biological outputs. Photoreceptors from photosynthetic autotrophs were the first to be identified and described, and were subsequently discovered in non‐photosynthetic organisms 1, 2. However, it was not until recently that they were reported to be functional in vivo in auxotrophic bacteria. In fact, several studies show that these proteins are key elements in bacterial physiology. Interestingly, it is the case of the blue‐light photoreceptors containing LOV (light, oxygen, voltage) domains that are known to regulate the infectious processes in some bacteria 3, 4, 5, among other activities.
Phytochrome photoreceptors can be found in plants, eukaryotic algae, fungi, and photosynthetic and non‐photosynthetic prokaryotes 6, 7. They are reversibly photoconverted, typically, between a red‐absorbing (Pr) and a far‐red‐absorbing (Pfr) state 8. These photoreceptors were originally discovered in plants where they are recognized to regulate many key processes 9, 10; subsequently, their bacterial homologues, bacteriophytochromes (BphP), were identified 11, 12. Interestingly, BphPs have been linked to some bacterial physiological responses 13, 14, yet most of the biological processes they regulate are still elusive.
Xanthomonas campestris pv. campestris (Xcc), a non‐photosynthetic phytopathogenic bacterium distributed worldwide, is responsible for the so‐called black rot disease in cruciferous plants, causing huge economic loses 15, 16. This pathogen has the ability to live epiphytically and also colonize the plant xylem by entering through the stomata or wounds 17. During the epiphytic stage, bacteria are exposed to nutrient limitation, fluctuating water availability, exposure to sunlight containing ultraviolet radiation, and subjected to the diurnal light cycle 18. The effect of light has been extensively studied in phototrophic bacteria and only recently examined in non‐phototrophic bacteria.
The Xcc genome codes for two identified photoreceptors, a LOV domain histidine kinase protein and a BphP. The purpose of this study was to determine the functionality of the Xanthomonas bacteriophytochrome photoreceptor, here designated XccBphP, and whether it plays a significant role in bacterial virulence.
Results
Xcc codes for a functional bacteriophytochrome
The XccbphP gene is located downstream from a heme oxygenase‐coding gene (XccbphO) forming a bicistronic operon (Fig 1A), as similarly found in other bacteria 19. The heme oxygenase enzyme is known to catalyze the conversion of heme into biliverdin‐IXα (BV), the open‐chain tetrapyrrole bilin chromophore bound in BphPs 8. In order to determine whether the XccbphO–XccbphP operon is functional in vivo, we performed RT–PCR from Xcc RNA purifications, confirming that it is actively transcribed and that there is a physical linkage between XccbphO and XccbphP mRNAs (Fig 1B). Moreover, XccBphP was detected in wild‐type extracts by SDS–PAGE and Western blotting displaying the expected molecular weight (Fig EV1B).
Figure 1. XccBphP genomic organization, domain topology, and UV–vis spectral properties.
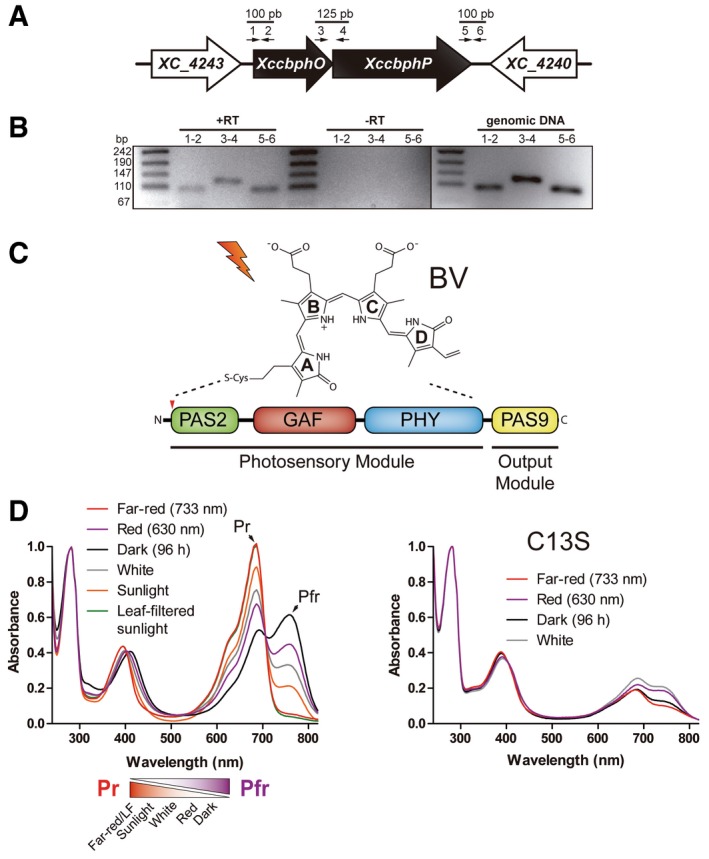
- The XccbphP gene (XC_4241) is located downstream the heme oxygenase gene (XccbphO) as a bicistronic operon. Specific primers (small numbered arrows) were designed to evaluate the physical linkage between XccbphO and XccbphP mRNAs.
- The confirmative RT–PCRs were resolved in a 2% agarose gel. Untreated reverse transcriptase RNAs (‐RT) and genomic DNA served as negative and positive controls, respectively.
- The XccBphP domain architecture. The photosensory module, composed by the PAS2‐GAF‐PHY domain triad, binds to the chromophore biliverdin‐IXα (BV) from a conserved cysteine residue indicated by a red arrow (Cys13). The output module comprises a single PAS9 domain.
- Left panel: Absorption spectra of dark‐adapted XccBphP after illumination with red, far‐red, fluorescent white lamp light, sunlight, or sunlight filtered through green leaves (LF). Pr/Pfr equilibrium scheme derived from absorption spectra is depicted in the bottom panel. Right panel: The XccBphP‐C13S mutant was purified in the presence of BV and its absorption spectra recorded upon illumination with red, far‐red, and white light or dark adaptation.
Figure EV1. Generation of the XccbphP mutant and detection of XccBphP.
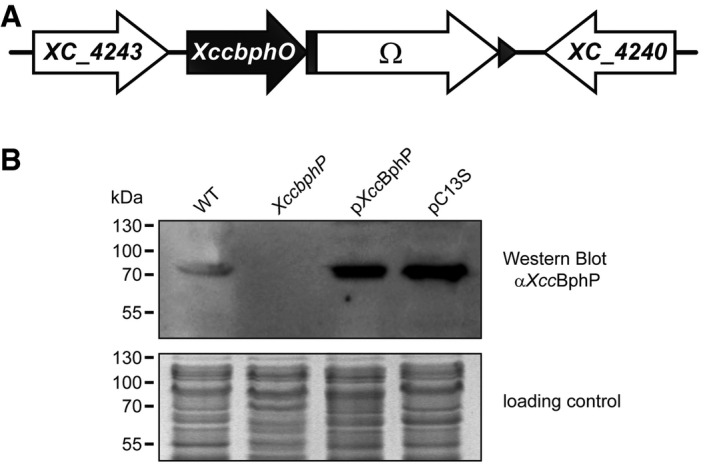
- XC_4241 ORF was partially deleted and replaced by a 2 kb Smr/Spcr cassette (Ω) through allelic exchange giving rise to the XccbphP null‐mutant strain.
- A Western blot was performed using mouse polyclonal antibodies against XccBphP in wild‐type, XccbphP, pXccBphP, and pC13S strains. Loading control SDS–PAGE stained with Coomassie blue is shown in the bottom panel.
The XccBphP sequence displays four conserved domains (Fig 1C): Per/Arndt/Sim (PAS2 family), cGMP phosphodiesterase/adenyl cyclase/FhlA (GAF), phytochrome‐associated (PHY), and output/effector module (PAS9 family), according to a Pfam database classification 20. The PAS2–GAF–PHY triad represents the photosensory module involved in BV binding, where the PAS2 domain bears a conserved cysteine at position 13 involved in the covalent chromophore linkage 21, 22. It is the BV chromophore buried in the protein structure that allows phytochromes to reversibly photoconvert between red‐absorbing (Pr) and far‐red‐absorbing (Pfr) states by photoisomerization. The PAS9 domain from XccBphP bears no predicted enzymatic activity; moreover, PAS domains are recognized to mediate interactions between proteins in signaling systems 23. The three‐domain photosensory module, typically coupled with an output module, is a conserved feature of bona fide phytochromes, and it can be found among phytochromes across different kingdoms of life comprising plant, fungi, algae, and bacteria, even while sharing low sequence identity. The composition of the output module varies among phytochromes and would govern the nature of the downstream signaling after light‐sensing events by the photosensor (Table EV1). To investigate the extent of BphP occurrence in the Xanthomonas genus, we identified all XccBphP homologous sequences bearing a PAS2–GAF–PHY photosensory module within the genus in UniProtKB database. A total of 75 different sequences were found from a wide variety of plant pathogenic Xanthomonas spp. exhibiting only five categories of output module domain sequences: (i) PAS4, (ii) PAS8, (iii) PAS9, (vi) histidine kinase (HisKA subfamily), and (v) absence of any conserved domain (Fig EV2). Single PAS domains correspond to almost 95% of the Xanthomonas BphP output modules, PAS4 being the most abundant (64%), followed by PAS9 (~21%) and PAS8 (~9%). Only two BphPs were found to bear a histidine kinase module, and two sequences did not show any recognizable output module.
Figure EV2. Phylogenetic tree of BphPs from the Xanthomonas genus.
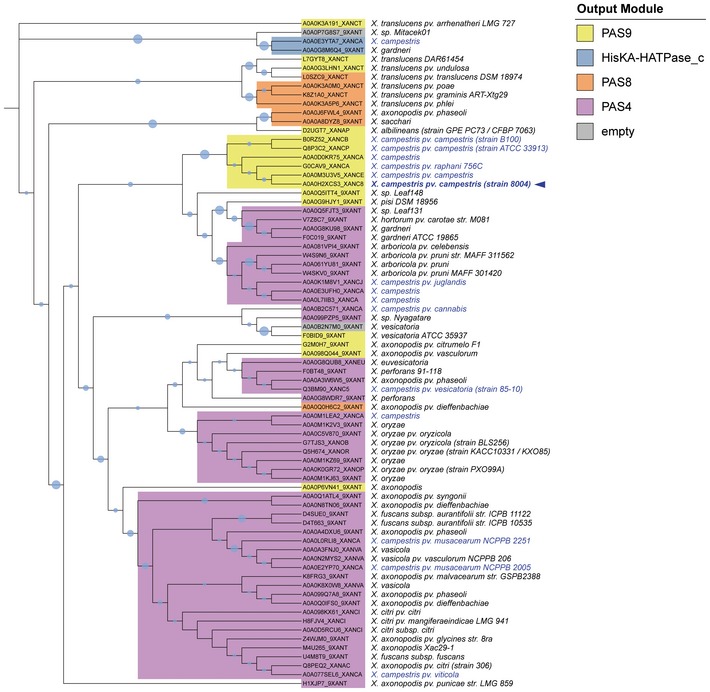
75 BphP sequences from UniProtKB database were identified in the Xanthomonas genus bearing a PAS2‐GAF‐PHY photosensory module. A molecular phylogenetic analysis by maximum‐likelihood method was performed using the respective PHY domain sequences. Species, strains, and UniProtKB accession numbers are indicated. Xanthomonas campestris species are indicated in blue letters, and the strain 8004 used in this work in bold letters and an arrow. Relative bootstrap values are represented by blue bubble sizes. The output module domains identified in Pfam database are depicted in colors that correspond to single PAS4, PAS8, PAS9 domains, and HisKA coupled with a HATPase_c domain. BphPs that showed no output module domains are indicated in gray (empty).
Recently, the XccBphP structure and basic photochemistry have been reported, demonstrating its capacity to function as a red/far‐red light photoreceptor in vitro 22. We sought to further characterize XccBphP spectroscopically. For that aim, recombinant full‐length XccBphP holoprotein was purified. BV bound to the photosensory module was evidenced by the presence of the Soret and Q bands (Abs400/Abs750) in the UV–vis absorption spectrum (Fig 1D). As expected, far‐red (733 nm) irradiation leads to a photoconversion of the holoprotein into a pure Pr species, indicated by a maximum absorption peak at 688 nm and the lack of the Pfr absorption band at 752 nm (Fig 1D). Irradiation with red (630 nm) and fluorescent white light lamp promotes the appearance of the Pfr form, corresponding to a Pfr:Pr ratio calculated of ~2 and ~1, respectively. In the dark, thermal conversion leads to the accumulation of the Pfr state exhibiting a final Pfr:Pr ratio calculated of ~6 at 21 h (Fig 1D). A Pr ground state and a dark conversion from Pfr to Pr are characteristic of prototypical phytochromes. Conversely, the bathy‐type phytochromes exhibit a Pfr ground state and a Pr to Pfr dark conversion 24, 25. Therefore, XccBphP fits into the bathy‐type phytochrome category, although an incomplete Pr‐to‐Pfr dark conversion and a Pr/Pfr equilibrium are observed. Interestingly, when XccBphP apoprotein is incubated with BV in the dark, the holoprotein is first constituted as pure Pr but then reaches the same Pfr:Pr ratio of ~6 22. To simulate natural conditions, XccBphP was exposed to direct sunlight or sunlight filtered through green leaves mimicking a canopy. Sunlight alone promotes a photoreceptor Pr enrichment with a Pr:Pfr ratio of ~3, contrary to the Pfr enrichment observed in dark equilibrium. Moreover, when XccBphP was subjected to leaf‐filtered sunlight a pure Pr spectrum could be observed (Fig 1D), similar to the one obtained after artificial far‐red LED irradiation. The functionality of the conserved cysteine residue at the PAS2 domain was addressed by constructing an XccBphP point mutant replacing Cys13 by a serine residue (XccBphP‐C13S) and evaluating its spectroscopic properties. Although XccBphP‐C13S still retains binding to BV, as evidenced by its absorption spectrum, it exhibits no photo‐inducible Pr‐Pfr changes (Fig 1E). Hence, the presence of Cys13 is necessary for the correct XccBphP photo‐activity.
XccBphP negatively regulates Xcc virulence in a light‐dependent manner
To test the role of XccBphP in Xcc virulence, we first generated a null mutant (XccbphP) by an allelic replacement of the XC_4241 ORF with an antibiotic resistance (Smr/Spcr cassette, Fig EV1A), confirming the absence of the protein in the mutant strain by Western blot (Fig EV1B). Once XccbphP growth curves were confirmed to be similar to the wild‐type strain (Fig EV3), we were prompted to evaluate the influence of XccBphP in bacterial virulence. For that purpose, 10‐day‐old Arabidopsis plant seedlings were inoculated with the bacterial strains cultures under normal laboratory conditions. After 1 or 3 days post‐infection (d.p.i.), colony forming units (CFU) per plant milligram were determined. Our results show that at 3 d.p.i. the XccbphP strain exhibits a significantly more virulent phenotype, meaning a higher capacity to replicate inside the host, compared to the wild type (Fig 2A). Interestingly, the complemented strain (pXccBphP), which displays XccBphP overexpression (Fig EV1B), behaves as an attenuated strain showing lower CFU counts than the wild type after 3 d.p.i. (Fig 2A). We then sought to examine whether these differences in bacterial infection are dependent on light. Consequently, before plant inoculation, all bacterial strains were either cultured in the dark or under continuous white illumination. Plants infected with wild‐type and pXccBphP‐complemented bacterial strains cultured in light displayed lower CFU counts at 2 d.p.i. compared to the cultures kept in the dark. XccbphP ability to infect plants remained comparable regardless of the light conditions, and always greater than the wild‐type strain. Similarly, when plants were infected with the XccbphP mutant complemented with the photo‐inactive XccBphP‐C13S version (pC13S), no statistical differences were detected between light and dark conditions (Fig 2B).
Figure EV3. Wild‐type, XccbphP, pXccbphP, and pC13S growth curves.
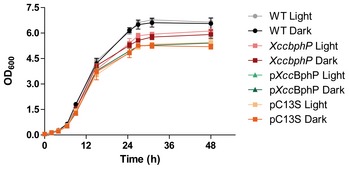
Wild‐type, XccbphP, pXccbphP and pC13S strains were cultured in PYM liquid medium under light or dark conditions (starting OD600 = 0.05) at 28°C and 250 rpm.
Figure 2. XccBphP functions as a negative regulator for Xanthomonas campestris infection affecting plant defenses.
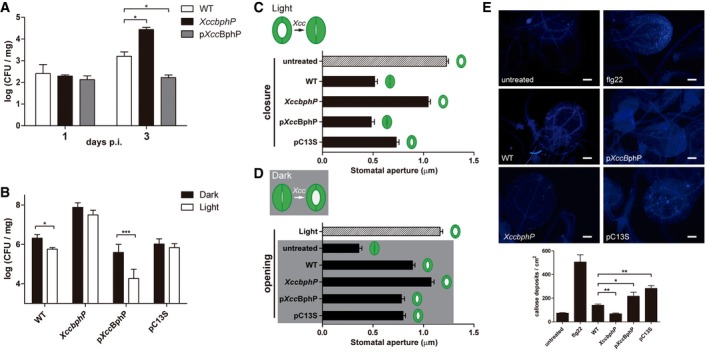
-
A, BArabidopsis thaliana plants were inoculated with wild‐type, XccbphP, pXccBphP, or pC13S bacterial strains cultured prior to inoculation under (A) normal laboratory illumination or (B) under light or dark conditions. After 1, 2, or 3 d.p.i., CFU per plant mg were determined (n = 3 replicates). Data presented here derive from more than four independent experiments.
-
C, D(C) Stomatal closure in the presence of bacterial strains or not (untreated) after 1 h under light conditions. (D) Stomatal opening after 3 h of incubation with bacterial strains or not (untreated) in the dark. A control treatment without bacteria and illuminated was included (Light). (C, D) Stomatal apertures were recorded (n = 80 replicates). Data are representative of two independent experiments.
-
EArabidopsis thaliana leaves were inoculated with wild‐type, XccbphP, pXccBphP, and pC13S strains, stained for callose deposits and observed by fluorescence microscopy. MgCl2 buffer (untreated) and flg22 peptide were used as negative and positive controls, respectively. Top panel: representative pictures of three independent experiments. Scale bar represents 200 μm. Bottom panel: the number of callose deposits per field of view (0.45 mm2) were determined (n = 8 replicates). Data are representative of two independent experiments.
The aerial part of terrestrial plants possesses microscopic pores called stomata, which allows the control of gas exchange (necessary for photosynthesis) and water loss. Plants regulate the opening and closing of stomata by different mechanisms 26. It has been reported that Xcc is able to reverse both pathogen‐ and abscisic acid‐induced stomatal closure in Arabidopsis through a virulence factor of a still unknown composition that is secreted to the extracellular medium 17. To start elucidating the mechanisms by which XccBphP modulates plant infection, we studied the pathogen‐induced stomatal closure as part of the plant innate immune response. In order to determine whether the increase in XccbphP virulence is related to its ability to penetrate into the host through stomata, we measured its capacity to promote stomatal closure in the light. Strikingly, stomata from epidermal peels incubated with XccbphP remained significantly opened at 1 h post‐incubation (h.p.i.). As expected, the wild‐type and complemented pXccBphP strains promoted normal stomatal closure. Stomata also remained opened when they were incubated with pC13S strain, although with lower amplitude compared to the XccbphP mutant (Fig 2C). Moreover, both wild‐type and XccbphP strains promote stomata opening at 3 h.p.i by producing the yet unidentified compound (Fig EV4) 17. Therefore, to evaluate a possible scenario in which the infection occurs in the dark (when stomata are closed) we assayed the capacity of Xcc strains to promote stomatal opening of dark‐adapted stomata. Results show that wild‐type Xcc is in fact able to open stomata at 3 h.p.i., while XccbphP shows an even better performance on this task (Fig 2D).
Figure EV4. Induced stomatal closure bioassay.
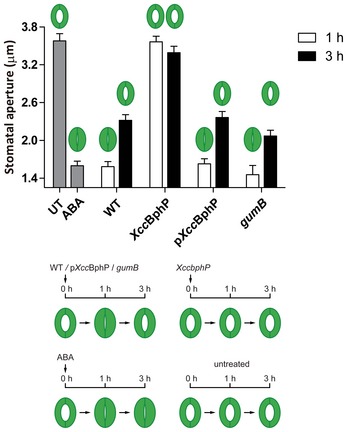
Promotion of stomatal closure induced by wild‐type, XccbphP, pXccBphP, and gumB bacterial strains. Stomatal apertures measurements recorded after 1 or 3 h.p.i.; UT: untreated (no bacterial treatment), ABA: abscisic acid. Bottom panel: Experimental design and results scheme: light‐irradiated stomata (opened) are treated for 1 or 3 h with the bacteria in light conditions. Values are expressed as mean ± s.e.m. (n = 80 replicates). Data are representative of two independent experiments.
Previous reports suggest that xanthan specifically suppresses local plant defense by the inhibition of callose (a β‐1, 3‐linear glucan that strengthens plant cell walls) deposition, which is required for disease resistance against Xcc 27, 28. Consequently, we measured callose biosynthesis during XccbphP infection. Leaves were challenged with wild‐type Xcc, XccbphP, pXccBphP, pC13S, and flg22 peptide, a 22‐amino acid sequence of the conserved flagellin N‐terminus that is known to activate the plant defense mechanisms 29. Twenty‐four hours after infection, the inoculated leaves were stained for callose with aniline blue and cytological observations were performed at the sites of infection with UV‐fluorescence microscopy. Callose depositions can be identified as bright‐blue points in leaves or veins (Fig 2E, top panel). The results indicate that XccbphP presents considerable reduced callose deposits compared to the wild type, the pXccBphP strain, and the flg22 peptide (Fig 2E, bottom panel). In turn, pC13S presents similar values to those of pXccBphP. These results are consistent with the results obtained in plant infection assays presented above.
XccBphP is involved in β‐1, 4‐endoglucanase production
We then focused on extracellular hydrolytic enzymes, which are virulence factors secreted by the type 2 secretion system (T2SS) that allow Xcc to degrade plant material and support infection 30. A direct influence on the plant–pathogen interaction was demonstrated for a secreted endoglucanase enzyme from Xcc 31. Endoglucanase activity from bacterial culture media can be estimated by measuring the degradation halo it produces in carboxymethyl cellulose (CMC) plates. Figure 3A shows that the overexpressing pXccBphP complemented strain displays significantly lower levels of endoglucanase activity compared to the wild type, although no light/dark regulation was detected. A quantitative colorimetric assay in solution was performed to measure β‐1, 4‐endoglucanase enzymatic units. Because we were not able to detect differences between the light/dark treatments before, we decided to perform the experiments under normal laboratory light conditions. CMC degradation plate assay results were corroborated in this system. Moreover, pXccBphP strain culture supernatants possess an almost 10‐fold reduction in endoglucanase units compared to the wild type (Fig 3B).
Figure 3. Xanthomonas campestris extracellular endoglucanase production is regulated by XccBphP.
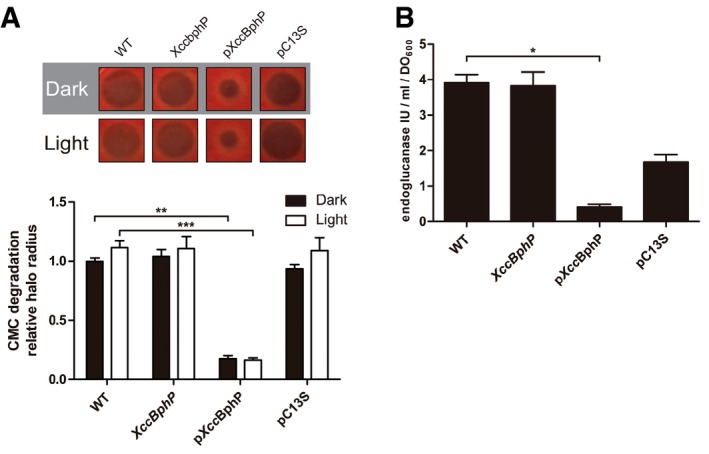
- Five microlitre supernatants from wild‐type, XccbphP, pXccBphP, and pC13 strains bacterial cultures (OD600 = 1) grown under light or dark conditions were plated onto PYM‐carboxymethyl cellulose (CMC) agar plates and revealed with Congo red staining (n = 2 replicates). The extracellular β‐1,4‐endoglucanase production levels correlated with CMC degradation halo radiuses (top panel). Halo measurements are presented in the bottom panel. Data derive from ten independent experiments.
- The extracellular β‐1,4‐endoglucanase activity from bacterial cultures supernatants (OD600 = 1) was determined by a colorimetric assay in solution (n = 2 replicates). Wild‐type, XccbphP, pXccBphP, and pC13 strains were assayed under normal laboratory conditions. Data derive from three independent experiments.
XccBphP regulates xanthan production and biofilm formation
Light regulates the plant endosymbiont R. leguminosarum infectivity levels through a blue‐light photoreceptor (LOV‐HK) 3. This regulation can be partially explained by the fact that exopolysaccharide (EPS) production and biofilm formation, intimately related to infection, are controlled by LOV‐HK in a light‐dependent manner 3. Similarly, the EPS xanthan produced by Xanthomonas is a virulence factor and is involved in biofilm development. Disruption of xanthan production impairs Xcc biofilm development in vitro and bacterial virulence 32, 33. Hence, we sought to associate the low Xcc infectivity levels induced by light treatments (Fig 2B) with low EPS production and poor biofilm development. To evaluate EPS production, bacteria were plated in rich media, cultured under light or dark conditions continuously for 48 h and EPS production quantified as a measure of colony diameter. As shown in Fig 4A, wild‐type Xcc produces significantly more EPS in the dark than in the light treatment. Interestingly, the XccbphP mutant is insensitive to light or dark and produces significantly more xanthan in both conditions compared to the wild‐type strain. In contrast, pXccBphP‐complemented strain colonies exhibited a severe decrement in xanthan content in both dark and light, although maintaining the tendency of light regulation. The XccbphP mutant was complemented with a gene coding for the photo‐inactive XccBphP‐C13S, which had no effect in restoring the wild‐type phenotype (Fig 4A) although the protein was overexpressed, similarly to pXccBphP (Fig EV1B). Xanthan content was also measured in liquid cultures confirming the results obtained in agar plates and evidencing that the only light‐responsive strains are the wild type and the complemented strain bearing a wild‐type copy of XccbphP (Fig 4B). Accordingly, when the sliding motility—a type of bacterial movement that depends on EPS, which reduces the friction between cells and substrate 34—was assayed, it was observed that sliding in XccbphP was strongly enhanced while distinctly impaired in pXccBphP compared to the wild type. As expected, the pC13S strain cannot restore the wild‐type phenotype and slides comparable to the XccbphP mutant (Fig EV5A).
Figure 4. Xanthomonas campestris virulence factors are inhibited by light through XccBphP.
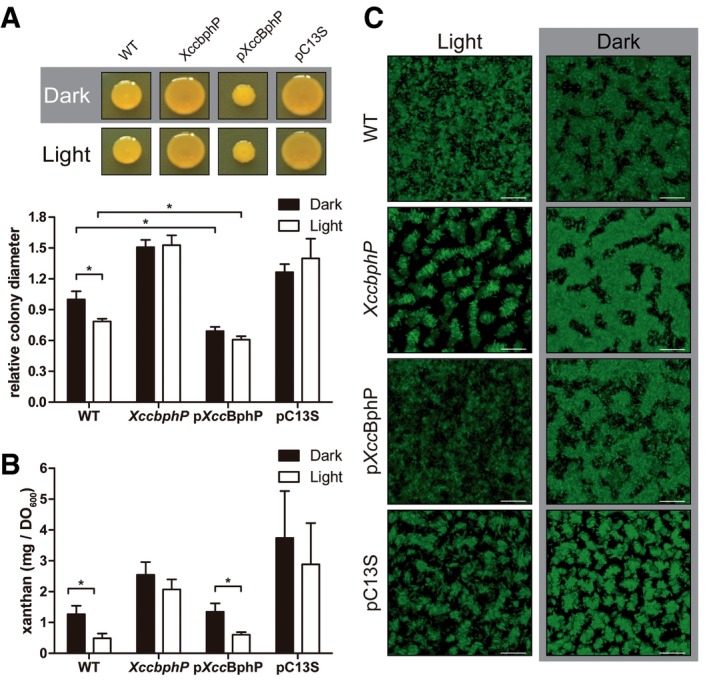
- Five microlitres from wild‐type, XccbphP, pXccBphP, and pC13S bacterial cultures (OD600 = 1) were plated onto PYM‐glucose agar plates under light or dark conditions. Xanthan production was determined measuring colony diameters (n = 4 replicates) for 20 independent experiments.
- Wild‐type, XccbphP, pXccBphP, and pC13S were grown in 20 ml PYM liquid medium under light or dark conditions. Extracellular xanthan was purified by KCl addition and ethanol precipitation, then dried, and weighed (n = 2 replicates) for four independent experiments.
- Representative confocal laser‐scanning microscopy pictures of biofilm development of bacteria cultured for 4 days in minimal medium in chambered cover slides under light or dark conditions. The scale bar represents 30 μm.
Figure EV5. Sliding and swimming motilities modulated by XccBphP.
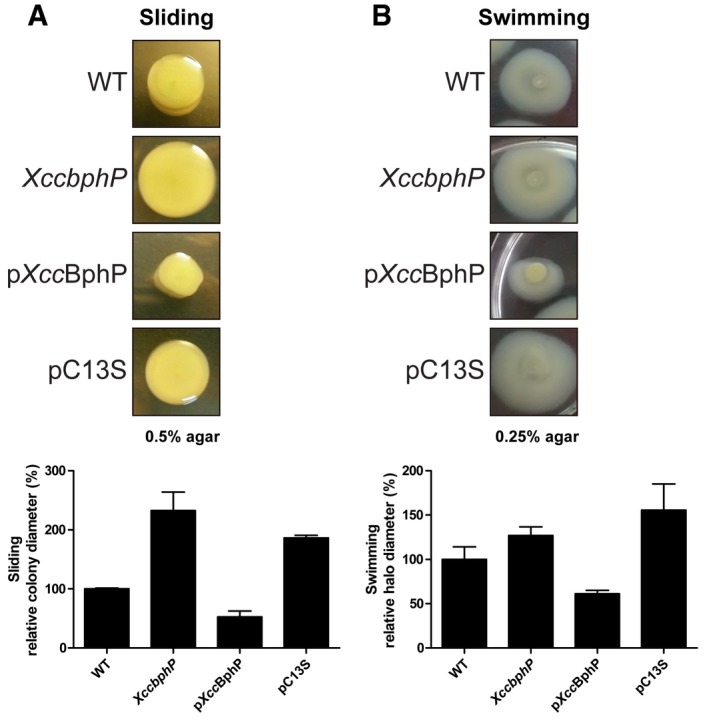
-
A, BThree microlitres from wild‐type, XccbphP, pXccBphP, and pC13S bacterial cultures (OD600 = 1) were plated onto PYM‐glucose 0.5% (A) or NYGB 0.25% (B) agar plates. (A) Sliding motility was assessed by measuring colony diameters, and (B) swimming motility was assessed by measuring external halo diameters. Bottom panels show quantifications as mean ± s.e.m. (N = 4).
The hypothesis that EPS regulation by light impacts on biofilm development was tested by confocal laser‐scanning microscopy (CLSM) studies of Xcc biofilms cultured in chambers in the dark or under continuous white light without agitation. After 4 days, a clear impairment in the wild‐type biofilm maturation in the light treatment was confirmed by the CLSM picture analysis while a completely mature biofilm is found to be formed in the dark condition (Fig 4C). In contrast, and reminiscent of the EPS experiments, the XccbphP and the pC13S strains were able to generate mature biofilms regardless of the light condition while the complemented strain exhibits its wild‐type phenotype restored (Fig 4C). Consistently, a biofilm COMSTAT analysis revealed that wild‐type and pXccBphP strains show increased values in biomass and average thickness in dark compared to the light treatment (Table EV2). Moreover, these variables are increased in XccbphP and pC13S in both light and dark conditions compared to the wild type. These results are in agreement with XccBphP sensing light and lowering EPS levels, which in turn produces an immature biofilm.
Far‐red light and XccBphP overexpression produce genomewide transcriptional changes
In order to find clues on the XccBphP signal transduction mechanism of the far‐red light pathway at the transcriptional level, we performed RNA‐Seq of the wild type cultured in the dark or under far‐red light, and the XccBphP overexpressing strain pXccBhpP in far‐red light. A principal component analysis (PCA) clearly shows that the biological replicates for each treatment cluster together but not the treatments themselves (Fig 5A). Hence, the transcriptomes from the treatments differ as if they were different “bacterial states”. To find differentially expressed (DE) genes with statistical significance (P < 0.05), pairwise comparisons between treatments were performed. Strikingly, 1,121 DE genes were found to be caused by illumination alone. This represents 25.6% of the Xcc genome which codes for 4,381 identified ORFs. In addition, when the dark wild type was compared with illuminated pXccBphP, 1196 DE genes arose. Furthermore, these two comparisons share 882 DE genes. In contrast, when pXccBphP and wild‐type far‐red datasets were compared, 196 DE genes appeared, representing 4.47% of the total genomic ORFs (Fig 5B). There are 81 out of these 196 DE genes which are common to all three comparisons making this group of particular interest. There is another group of genes that do not change in the wild type upon illumination but indeed change when XccBphP is overexpressed containing 272 DE genes. This group is composed of two subgroups: 256 DE genes exclusively found in the wild‐type dark/pXccBphP intersection and 16 DE genes that are exclusive in the intersection between the far‐red irradiated strains. These genes are dependent on XccBphP levels and do not show statistical differences due to illumination in the wild type (Fig 5B).
Figure 5. Genomewide differential expression RNA‐Seq analysis between treatments.
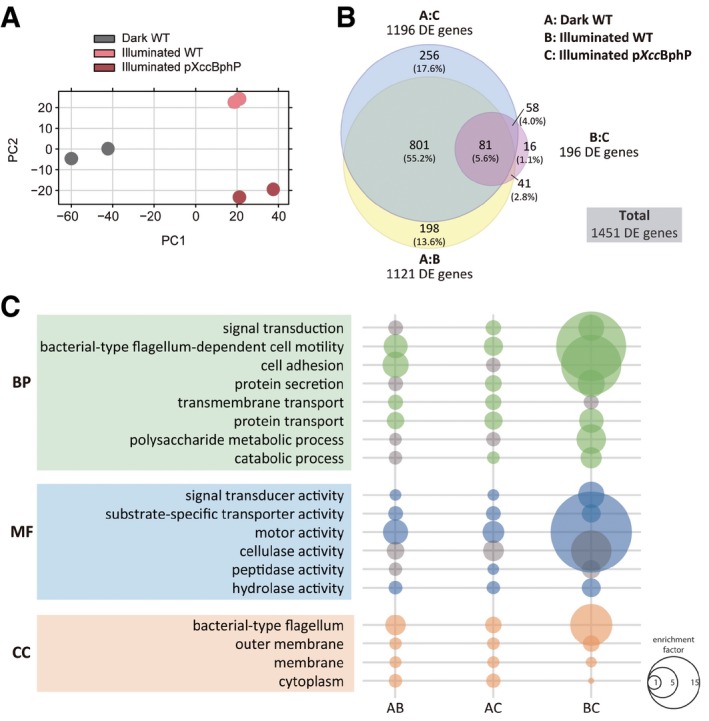
- Principal component analysis (PCA) plot of the first two components of the analyzed samples showing separation between the three treatments: far‐red illuminated wild‐type, far‐red illuminated pXccBphP (XccBphP overexpressing), and dark wild‐type Xcc treatments.
- Venn diagram showing overlap of differentially expressed (DE) genes between treatments (P‐values < 0.05), bubbles are drawn to scale. A total of 1,451 DE genes were found in comparisons between all treatments and relative percentages to this number are indicated.
- Gene Ontology (GO) enrichment analysis. GO enrichment was evaluated at three different levels: biological processes (BP), molecular function (MF), and cellular component (CC). Relevant categories showing enrichment of DE genes are depicted. Bubble size correlates with enrichment factor values; gray bubbles represent P‐values > 0.05.
A Gene Ontology (GO) analysis was performed to evaluate relative enrichment of GO terms in the three dataset comparisons. The three GO categories consist of biological process (BP), molecular function (MF), and cell compartment (CC). The datasets contained DE genes which correspond to GO terms with differential enrichment factors which can be inferred to be related to virulence and signal transduction in Xcc (Fig 5C). This analysis helped us visualized the GO terms of DE genes dependent on far‐red light and XccBphP involved in many of the experiments performed during this work, which implicate: (i) secretory systems of hydrolytic enzymes, including endoglucanases and proteases, (ii) adhesion, motility, and xanthan metabolism, necessary for proper biofilm development, and (iii) cell signaling systems that might include components of the red‐light‐XccBphP downstream signaling (Fig 5C). A comprehensive list of DE selected genes that fall into these categories was elaborated, comprising many virulence factors and pathogenic‐related systems (Dataset EV1).
XccBphP downregulates transcription of virulence systems
Finally, we focused on those genes coding for reported Xanthomonas virulence effectors and studied along the present work. Particularly, DE genes involved in extracellular endoglucanase activity, xanthan production, and motility were selected. The first group of genes that caught our attention were the endoglucanases and the T2SS (xps), involved in exporting degradative enzymes outside the cell 31, 35. Most endoglucanase and all xps T2SS DE genes are downregulated in pXccBphP compared to wild type cultured under any condition (Dataset EV1). This finding is consistent with the results shown in Fig 3. The second group of genes found to be XccBphP‐dependent is gathered in the gum operon, responsible for xanthan gum production 32. Again, we observed that pXccBphP strain shows statistically lower transcriptional levels of gum genes than the wild type (Dataset EV1), which may explain why this strain produces significant less xanthan in the plate assay than the wild type (Fig 4A). Flagellar genes (flg and fli) constitute the third group of selected DE genes, which are known to be necessary for Xanthomonas virulence 34. They exhibit a strong downregulation with XccBphP overexpression (Dataset EV1), suggesting an impairment in flagellar‐dependent motility for pXccBphP strain. Concordantly, our results clearly show that pXccBphP is indeed diminished in swimming motility (Fig EV5B).
Discussion
Bacteria inhabit all of our planet ecosystems, including the phyllosphere, the aerial parts of the plants, which is considered a hostile environment for the colonists. Bacteria are the most abundant members of the phyllosphere community and have been shown to colonize leaves at densities of up to 108 cells per cm2 36. During the day, bacteria are exposed to high visible light intensities and UV‐B radiation levels that can damage their metabolism and physiological processes 37, 38. Phytopathogenic bacteria living in the leaves take advantage of specific adaptations that allow them to survive and infect their hosts 18. However, there is still little information on how they respond to environmental light. Recent reports have shown that photoreceptors are involved in virulence and motility of different Pseudomonas syringae pathovars, suggesting that light is important for their pathogenicity 13, 39, 40, 41. Xcc is adapted to the phyllosphere environment, but so far there is a lack of knowledge on the role of light in its physiology, which stimulated us for the present study.
Our data demonstrate that XccBphP is a functional bathy‐type bacteriophytochrome in vitro as well as in vivo. We could establish in vitro that XccBphP can be photoconverted between the Pr and Pfr forms, the Pr/Pfr equilibrium ratio depends on the quality of sunlight, and that these light‐induced changes are dependent on the presence of the conserved Cys13 residue. Moreover, the absence of XccBphP in vivo boosts Xcc virulence likely by (i) an increment in many bacterial virulence factors and (ii) its ability to bypass the plant defense mechanism. In this sense, the excessive production of xanthan was discarded as a possible explanation for the XccBphP mutant strain not triggering stomatal closure as the gumB mutant, unable to produce xanthan, behaves similar to the wild‐type strain in stomatal closure experiments (Fig EV4). On the other hand, XccBphP overexpression attenuates virulence, likely through transcriptional downregulation of virulence factors as revealed by our RNA‐Seq results. These findings were gathered and summarized in Fig 6A.
Figure 6. XccBphP functions as a negative regulator for virulence through transcriptional regulation.
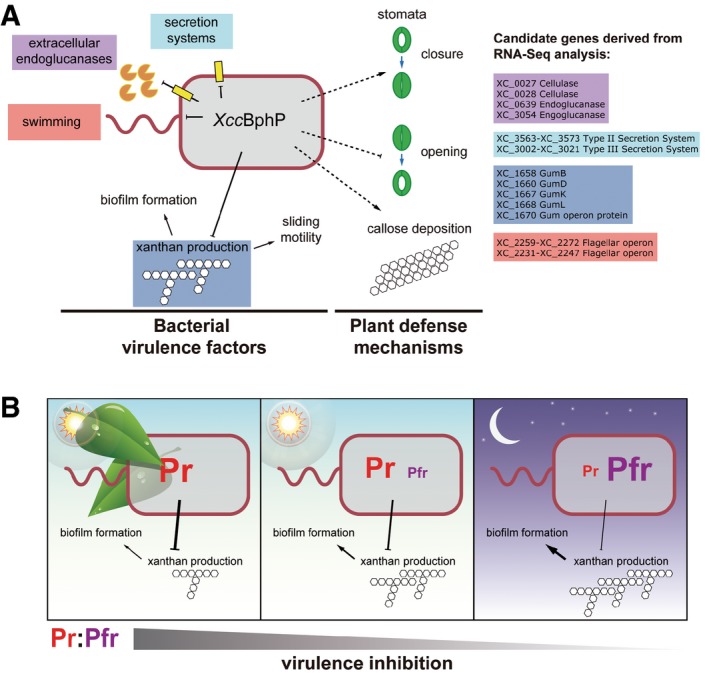
- Scheme derived from the results obtained throughout this work from the wild‐type, the XccbphP (null mutant), and the complemented pXccBphP (overexpressing) strains. Systems affected are indicated in different colors, and gene candidates involved in these systems found by RNA‐Seq analysis that are regulated by XccBphP are depicted in the same color code.
- Possible model for XccBphP light‐regulated EPS and biofilm results integrated with the spectroscopic data. XccBphP downregulates biofilm formation, xanthan production, and consequently Xcc virulence in a Pr:Pfr‐dependent manner. XccBphP exposed to sunlight filtered by a canopy (left), direct sunlight (middle), or darkness (right).
Moreover, Xcc is significantly less virulent in the light than in the dark and this is dependent on XccBphP. Taking together virulence, xanthan EPS, biofilm, and spectroscopy experiments, a simple hypothetical model can be envisaged for Xcc light/dark virulent behavior, which is consistent with light inhibition of xanthan production and biofilm maturation, probably triggered by a low Pfr:Pr ratio (Fig 6B). Hence, XccBphP may be described as a virulence negative regulator, which senses light and partially inhibits the Xcc virulence program.
Why a bacteriophytochrome photoreceptor downregulates virulence toward its host is an open question. It is possible that Xcc is constitutively primed for infection and as the light stimuli disappear, it can promptly respond by increasing its virulent behavior. Consistently, Xcc rapidly establishes infection in a period significantly shorter than a daily photoperiod. Another reason for virulence inhibition could be attributed to Xcc hemibiotrophic lifestyle 31, initially infecting live tissues and at later stages actively killing host cells during its life cycle, and an excess in its pathogenicity toward its host could result non‐adaptive. Therefore, XccBphP might have evolved to control and fine‐tune Xcc virulence by inhibiting it. It is worth mentioning that Xcc can also (i) survive in the soil independently from its host for several weeks and (ii) produce a seedborne disease 16. In this scenario, photoreception can also be playing a part in signaling (time or place) the bacteria for infection in its life cycle, similarly to Brucella or Rhizobium 3, 4. One possibility is that XccBphP serves as a time signaling system to distinguish between day and night. In line with this, it is known that light impacts positively on the plant immune system, probably making daytime the less favorable moment for infection. Plant defense mechanisms are modulated by light and circadian rhythm involving phytochrome photoreceptor signaling, among others 42, 43. In Arabidopsis, plant defense responses and disease resistance significantly depend on the time of day; morning or midday infections result in a more pronounced hypersensitive response than evening or night infections 44. Moreover, light is also associated with the production of ROS that in turn act in the plant–pathogen interaction 45. In this sense, it is reasonable to think that Xcc can synchronize infectivity to the plant dark‐susceptible period by means of its BphP. Synchronizing Xcc infectivity to an optimal moment of the host physiology and signaled by light/dark as environmental cues can be a feature shared by other infective bacteria that harbor photoreceptors. In an analogous fashion, the quorum‐sensing (QS) systems that are present in Xcc and in many other pathogenic bacteria, activates infection when the bacteria reach an appropriate situation (i.e. a certain population density) 46, 47. Another possibility is that XccBphP signals for a spatial cue. During the day, within a leaf canopy, most red light has been removed by the chlorophyll in shading leaves but almost none of the far‐red light 48. Hence, the light environment within the canopy may largely drive XccBphP from a Pfr to a Pr form, as observed in Fig 1D, decreasing Xcc virulence levels relative to direct sunlight exposure. Therefore, there should also be differential Xcc virulence during daytime in shaded and non‐shaded tissues. In this manner, Xcc might be optimizing its virulence levels (i.e. resources) according to the tissues susceptibilities as shaded leaves are more susceptible to infections that those ones that are exposed to direct light 42.
The XccBphP red‐light‐signaling pathway remains to be elucidated. It is plausible that the XccBphP first transducing events are transmitted to downstream elements via protein–protein interactions through the PAS9 output module 23. Our group has recently published a crystallographic structure of the full‐length XccBphP in the Pr state (PDB code 5AKP), revealing a parallel dimer arrangement with its complete PAS9 module, that may serve as starting material for studies on early signaling events for this photoreceptor 22. Moreover, the RNA‐Seq results show that there is a massive rearrangement of putative genes involved in this pathway, comprising (i) histidine kinases, (ii) two‐component systems, (iii) c‐di‐GMP‐modulating enzymes, and (iv) transcriptional regulators, all of which change due to far‐red illumination and/or XccBphP overexpression (Dataset EV1). To conclude, our findings strongly suggest that light is an important environmental cue sensed by XccBphP that regulates virulence‐associated mechanisms, which ultimately govern the plant–microbe interactions in Xcc. The understanding of Xanthomonas photobiology and its relationship with virulence may help in the task of finding novel ways to interfere with its infection cycle.
Materials and Methods
Bacterial strains and culture conditions
Xanthomonas campestris pv. campestris 8004 (Xcc) strains were cultured in PYM or in Y minimal medium (YMM) 49, 50 at 28°C, with agitation (250 r.p.m.). To examine biofilm development, bacteria were grown in YMM containing 1% (wt/vol) glucose as the carbon source 51. Escherichia coli strains were cultured at 37°C in Luria‐Bertani medium. When required, the antibiotics rifampicin (Rif), kanamycin (Km), spectinomycin (Spc), chloramphenicol (Cm), and ampicillin (Amp) were added in final concentrations of 30, 35, 100, 20, and 100 μg/ml, respectively.
A complete list of strains, plasmids, and primers used in this study is included in Table 1.
Table 1.
Strains, plasmids, and oligonucleotides used in this study
| Strains | Genotype/relevant characteristics | Source |
|---|---|---|
| Xcc | Xanthomonas campestris pv. campestris 8004, Rifr | Laboratory stock |
| XccbphP | Xcc, XC_4241::Ω, Rifr, Spcr | This work |
| pXccBphP | XccbphP + pBBR‐XccBphP, Rifr, Spcr, Kmr | This work |
| pC13S | XccbphP + pBBR‐C13S, Rifr, Kmr | This work |
| gumB | gumB::Tn5lac, EPS minus, Rifr, Kmr | 28 |
| DH5α | Escherichia coli, hsdR recA lacZYA Φ80 lacZDM15 | Gibco‐BRL |
| BL21(DE3)pLysE | Escherichia coli, high‐efficiency protein expression strain | Promega |
| pET‐XccBphP | BL21(DE3)pLysE, pET‐XccBphP, Cmr, Kmr | This work |
| pET‐C13S | BL21(DE3)pLysE, pET‐C13S, Cmr, Kmr | This work |
| Plasmids | Relevant characteristics | Source |
| pGEM‐T Easy | PCR cloning vector, lacZ, Ampr, | Promega |
| pG‐XccbphP::Ω | pGEM‐T Easy with Ω cassette (aadA gene) cloned between a 400‐bp and a 500‐bp fragments flanking XccbphP gene, Ampr, Spcr | This work |
| pKmobSacB | Suicide vector, sacB mob lacZ, Kmr | 53 |
| pK‐XccbphP::Ω | pKmobsacB with XccbphP::Ω construction, Kmr, Spcr | This work |
| pBBR1MCS2 | Broad‐host‐range cloning vector, Kmr | 54 |
| pBBR‐XccBphP | pBBR1MCS2 with a 2,708‐bp fragment containing XccbphO‐XccbphP operon and 5′ regulatory sequence cloned in HindIII site, Kmr | This work |
| pBBR‐C13S | Originated from pBBR‐XccBphP, containing mutation coding for XccBphP C13S, Kmr | This work |
| pET‐XccBphP | pET24a, Hisx6‐Full‐length XccBphP, Kmr | This work |
| pET‐C13S | pET24a, Hisx6‐Full‐length XccBphP‐C13S, Kmr | This work |
| Oligonucleotides | Sequence (5′ – 3′) | Source |
| ΔBphP_A | CTAAGCTTTACCTACGCGCAGGTGCTGCGCCGGCATC | This work |
| ΔBphP_B | ATGGATCCCGCGCGCAGACGTCCAGGTCCAACGGGTTG | This work |
| ΔBphP_C | TTGGATCCGCCGTTGCAAGTGAGCGACGGCGCACCGG | This work |
| ΔBphP_D | TCAAGCTTACGCTGCGCCTGTGCCGCAAGGCCGCTGC | This work |
| cBpbP_F | CTAAGCTTTGGCGCGCCGCTGCACCTGCCC | This work |
| cBpbP_R | TTCAAGCTTCTCTTATTCCGGATCGCGCAGCTGTAAC | This work |
| 5_Flank_F | GTGCCGCTGATGCAGGCGCTGGGGCAGG | This work |
| 3_Flank_R | GCAAGGCGATCAAGGTGATCGCACCGG | This work |
| C13S_F | TTGGACCTGGACGTCTCCGCGCGCGAACCCATC | This work |
| C13S_R | GATGGGTTCGCGCGCGGAGACGTCCAGGTCCAA | This work |
| BphP_F | ATCATATGCACCATCACCATCACCATAGCACTGCAACCAACCCGTTG | This work |
| BphP_R | TAGGATCCTTATTCCGGATCGCGCAGCTGTAAC | This work |
| RT_1 | CCCGGGTTGAGTCGGTGC | This work |
| RT_2 | GCCAGCAGCCGGTGATGC | This work |
| RT_3 | AGCAACGCCTGCAGAGCG | This work |
| RT_4 | CCAACGGGTTGGTTGCAGTG | This work |
| RT_5 | GCCAGTTCCACCTTGAACCTCT | This work |
| RT_6 | GCAGTCATTTTGATCGCTGCTC | This work |
Light, far‐red, and dark bacterial culture conditions
Light treatments were performed with continuous white light (2,700 K) from fluorescent tubes with a fluence of 15 μmol/m/s. Photon flux was measured using a Quantum Meter (Apogee Instruments model QMSW‐SS). Far‐red bacterial illumination was performed using 733 nm 0.7 W LEDs at a 15 cm distance during culture. Darkness was achieved covering cultures with two layers of aluminum foil.
Generation of the bacterial strains
The XccbphP mutant strain was obtained by allelic exchange. The XccbphP gene (XC_4241) was partially deleted and replaced by a 2 kb Smr/Spcr cassette (Ω) 52. Two fragments from the flanking regions of XccbphP gene were amplified using primers ΔBphP_A‐ΔBphP_B and ΔBphP_C‐ΔBphP_D, respectively. The fragments were BamHI‐digested, ligated to each other, amplified using the ΔBphP_A‐ΔBphP_D primers, and the product cloned into the pGEM‐T Easy vector (Promega). The resulting vector was BamHI‐digested, blunted with Klenow Fragment (New England Biolabs) and ligated to a SmaI‐digested Ω cassette, derived from the pHP45Ω plasmid, to yield pG‐XccbphP::Ω. The XccbphP::Ω construction was subcloned into pk18mobsacB 53 in the HindIII restriction site. The resulting pK‐XccbphP::Ω vector was used to transform Xcc. Sucrose‐resistant double recombinants were selected (Kms, Spcr). The mutation was confirmed by PCR sequencing, using specific primers (5_Flank_F‐3_Flank_R) flanking the mutation site and by Western blot. To construct the XccbphP complemented strain, a 2.7‐kb fragment containing the complete XC_4242‐XC_4241 operon and its regulatory sequences was amplified using specific primers (cBphP_F and cBphP_R), cloned into the HindIII restriction site of pBBR1MCS2 vector 54, to yield pBBR‐XccBphP, which was used to transform XccbphP to generate pXccBphP strain. The XccBphP‐C13S complemented strain was generated by site‐directed mutagenesis using Q5 High‐Fidelity DNA Polymerase (New England Biolabs), specific primers (C13S_F and C13_R) and the pBBR‐XccBphP vector as template to generate the pBBR‐C13S vector, which was used to transform XccbphP to generate pC13S strain.
Western blot
Bacterial extracts normalized by OD600 were loaded and separated in 12.5% SDS–PAGE electrophoresis. Similar amounts of protein were loaded into each lane of the gels, corroborated by Coomassie blue staining. Proteins were transferred to Immobilon‐P PVDF Membrane (Millipore). Membranes were blocked with non‐fat milk in PBS and incubated with an anti‐XccBphP polyclonal antibody (1:1,000) and then with anti‐mouse IgG (Fc specific)‐peroxidase antibody produced in goat (Sigma) (1:5,000). Detection was achieved using the Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare Life Sciences) on an ImageQuant LAS 400 apparatus (GE Healthcare Life Sciences).
RT–PCR of XccbphO‐XccbphP operon
Total Xcc bacterial RNA was isolated using the MasterPure™ RNA Purification Kit (Epicentre, Illumina). Reverse transcription was performed with a first‐strand SuperScript III cDNA kit (Invitrogen), using random decamer primers (Invitrogen) and RNasin ribonuclease inhibitor (Promega). The Primer‐BLAST program (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) was used to design primers for PCR products of 100–125 bp (Table 1). The cDNA samples were used as templates in PCR amplification, and products were resolved in a 2% agarose gel.
Bioinformatics
BphP homologs were retrieved from UniProtKB database (2016_01 release) by means of a BLAST search in the UniProt server (http://www.uniprot.org/) using default parameters and the photosensory module (PAS2‐GAF‐PHY) from XccBphP as query. All sequences were inspected using Pfam server (http://pfam.xfam.org/). A total of 75 sequences from Xanthomonas genus containing a complete PAS2‐GAF‐PHY domain triad were identified. PHY domain sequences inferred by Pfam from this dataset were used to build a multiple sequence alignment using MUSCLE program and conducted in MEGA7 software 55. Then, a molecular phylogenetic analysis by maximum‐likelihood method was performed to obtain a tree. The evolutionary history was inferred by using the maximum‐likelihood method based on the Whelan and Goldman model 56. The tree with the highest log‐likelihood (−1,872.6517) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor‐Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log‐likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites [five categories (+G, parameter = 3.3628)]. All positions containing gaps and missing data were eliminated. There were a total of 144 positions in the final dataset. The resulting tree was plotted using iTOL server (http://itol.embl.de/) 57.
Generation and purification of XccBphP and XccBphP C13S recombinant proteins
Full‐length XccBphP (residues 1–634) was cloned from the XC_4241 ORF into the NdeI/BamHI restriction sites of the pET24A vector (Novagen) to generate pET‐XccBphP as described elsewhere 58. The XccBphP‐C13S point mutant was generated by site‐directed mutagenesis using Q5 High‐Fidelity DNA Polymerase, specific primers (C13S_F and C13_R) and the pET‐XccBphP vector as template to generate the pET‐C13S vector. Escherichia coli BL21(DE3)pLysE strain cells were transformed with pET‐C13S. Cultures were induced with a final concentration of 0.4 mM IPTG overnight at 20°C with agitation (250 rpm), cells were harvested and ruptured. Next, the His‐tagged proteins were purified as previously described 58. The holoproteins were generated by incubating the apoproteins for 1 h at room temperature in the presence of biliverdin‐IXα (BV; Sigma‐Aldrich) before the size‐exclusion chromatography step. Protein concentration was estimated using the calculated molar extinction coefficient at λ = 280 nm provided by the ExPASy ProtParam tool (http://web.expasy.org/protparam/) based on the polypeptide sequence for the monomer (εXccBphP = ɛC13S = 74,370 M/cm); the biliverdin cofactor contribution was subtracted.
UV‐vis spectroscopy
Quartz cuvette containing holoprotein 1 mg/ml solutions of XccBphP or C13S in 50 mM Tris–HCl pH 7.7, 250 mM NaCl (buffer A) was irradiated for 10 min with a white (2,700 K) 15‐W fluorescent tube lamp, a red (630 nm) 1‐W LED, far‐red (733 nm) 0.7‐W LED, direct sunlight or sunlight filtered with two layers of fresh green Epipremnum aureum leaves. Absorption spectra were collected in an 8452A diode array spectrometer (Hewlett Packard). The dark states were determined after proteins were kept in the dark for 96 h.
Plant materials
Arabidopsis thaliana (L.) Heynh. ecotype Col‐0 seeds used for infection, callose staining, and stomatal aperture assays were surface sterilized with an ethanol:50%bleach:water (8:1:1) mixture for 5 min and rinsed three times with ethanol. Sterilized seeds were kept in water and in the dark for 3 days at 4°C. Seeds were germinated and cultivated in 60 × 15 cm Petri dishes containing Murashige and Skoog medium (MS), sucrose 0.5% and 0.5 g/l MES hydrate pH 5.7. Plants were grown at 22–23°C with a 12‐h photoperiod for 10 days.
Infection assays
Bacterial strains were grown overnight in PYM supplemented with the appropriate antibiotics and resuspended in water. Plants were infected adding the bacterial suspension to the plant growth medium to a final OD600 of 0.002, as previously described 28. Plants were kept at 22–23°C with a 12‐h photoperiod. Daily samples were taken for 3 days. Bacterial content was determined by plating a dilution series on PYM medium containing appropriate antibiotics of the homogenate leaves, and were expressed as CFU per gram of plant fresh weight.
Stomatal assays
Stomatal experiments were performed as previously described 17. Briefly, for stomatal closure assays, epidermal peels from 4‐week‐old leaves were floated in 10:10 buffer (10 mM KCl and 10 mM MES‐KOH pH 6.15) under light for 2 h, then 108 CFU/ml of each bacteria strain, flg22 peptide (5 μM, GL Biochem) or abscisic acid (20 μM, mixed isomers Sigma) were added to the medium and incubated for 1 or 3 h. Similarly, for stomatal opening assays, epidermal peels were incubated in buffer 10:10 for 2 h in the dark, then 108 CFU/ml of each bacterial strain were added to the medium and incubated for 3 h.
Apertures from 80 stomata for each experiment were measured in a Carl Zeiss microscope (4003) with the aid of an eyepiece micrometer.
Callose staining
Experiments were performed as described previously 29. Briefly, following treatment with bacteria (5 × 108 CFU/ml) or flg22 peptide (100 nM, GL Biochem), 10‐day‐old seedlings grown in 12‐well microtiter dishes were fixed in a 3:1 ethanol:acetic acid solution for several hours. The fixative was changed several times to ensure both thorough fixing and clearing of the tissues, which is essential for good callose detection. Seedlings were rehydrated in 70% (vol/vol) ethanol for 2 h, 50% ethanol for an additional 2 h, and water overnight. After three water washes, seedlings were treated with 10% (wt/vol) NaOH and placed at 37°C for 1–2 h to make the tissues transparent. After four water washes, seedlings were incubated in 150 mM K2HPO4 pH 9.5, and 0.01% (wt/vol) aniline blue (Sigma‐Aldrich) for several hours. Seedlings were mounted on slides, and callose was observed using a Nikon Eclipse E600 fluorescence microscope (excitation λ = 390 nm; emission λ = 460 nm).
Xanthan production: agar plate assay
Xanthan production was determined measuring the diameter of the colonies grown in PYM supplemented with 2% (wt/vol) glucose. Five microliters of bacterial cultures (OD600 = 1) for each strain was inoculated in the plates and cultured under light or dark conditions for 48 h. Colony diameters were determined analyzing plate pictures using the ImageJ 1.41 software.
Xanthan production: liquid culture assay
Xanthan quantification in liquid culture was performed as described before 59. Briefly, strains were cultured under light or dark conditions for 48 h at 28°C in 20 ml PYM liquid medium in 50‐ml flasks, using an orbital shaker rotating at 200 rpm. Then, OD600 were recorded, cells removed by centrifugation (25,000 g for 60 min), the supernatants supplemented with KCl at 1% (wt/vol) final concentration, and 40 ml of ethanol were added. The precipitated crude xanthan was collected, dried, and weighed.
In vitro analysis of biofilm formation by confocal laser‐scanning microscopy
Bacterial strains were cultured at 28°C in PYM medium supplemented with the proper antibiotics. Cultures were normalized by OD600 and diluted 1:2,000 in YMM and grown in coverglass slide chambers (no. 155411; Lab‐Tek, Nunc) for 4 days at 28°C as previously described 60, under light or dark conditions. Cells were stained with LIVE/DEAD cell viability assay (Thermo Fisher Scientific Inc.) before visualization. Biofilm formation was monitored with an Olympus Fluo View 1000 confocal laser‐scanning microscope (CLSM). Three‐dimensional images were generated with the ImageJ 1.41 CLSM software from the National Institutes of Health (http://rsbweb.nih.gov/ij/download.html). COMSTAT software was used for three‐dimensional biofilm structure quantifications 61.
CMC degradation halo assay
Production of extracellular β‐1,4‐endoglucanases was assessed inoculating 5 μl of supernatants onto PYM plates containing 0.125% (wt/vol) carboxymethyl cellulose (CMC, Sigma‐Aldrich). Supernatants were obtained by centrifugation of 1 ml of bacterial cultures (OD600 = 1) for 5 min at 12,400 g that were grown under light or dark conditions. The CMC degradation halo was developed after 24 h of incubation with an aqueous solution 0.1% (wt/vol) Congo red (Sigma‐Aldrich). Halo diameters were measured using the ImageJ 1.41 software. Data were normalized to the total area of the plate.
β‐1,4‐endoglucanase enzymatic activity: colorimetric assay in solution
The extracellular endoglucanase enzymatic activity measurements from liquid culture supernatants were performed by the colorimetric assay described before 62. Briefly, bacteria were culture for 24 h at 28°C in 5 ml PYM liquid medium. Then, bacterial cultures OD600 were recorded, 1 ml of culture was centrifuged for 10 min at 9,000 rpm, and supernatants were used for a following reaction. A total of 50 μl of a supernatant dilution and 50 μl 2% CMC (wt/vol) both prepared in 0.05 M citrate buffer pH 4.8 were transferred to a 96‐well plate, and the enzymatic reactions were incubated at 50°C for 60 min. The reducing sugar was measured by adding 100 μl of DNS (3,5‐dinitrosalicylic acid), followed by an incubation of the mix for 5 min at 95°C, and the absorbance was measured at 540 nm. The enzymatic units were calculated as CMC (units/ml) = 0.185 (units/ml)/(enzyme concentration) to release 0.5 mg glucose, where concentration = (vol. enzyme in dilution)/(total dilution volume). All the results were normalized by OD600.
Sliding motility assay
Bacteria were grown overnight in PYM medium; then, 3 μl of bacterial cultures (OD600 = 1) was inoculated in 0.5% (wt/vol) agar PYM plates as described before 34. After 72 h, motility was assessed measuring the circular halo formed by the growing bacterial cells. The assay was performed in triplicates.
Swimming motility assay
Swimming motility assays were carried out as previously described 51. Briefly, bacteria were grown overnight in PYM medium and 3 μl of bacterial cultures, normalized by OD600, was used to inoculate NYGB medium [0.5% (wt/vol) peptone extract, 0.3% (wt/vol) yeast extract, 2% (vol/vol) glycerol] 0.25% (wt/vol) agar plates. After 72 h, motility was assessed by measuring the outer colony halo. The assay was performed in triplicates.
RNA isolation and RNA‐Seq
Wild‐type Xcc or pXccBphP strains were cultured in red light or dark conditions up to logarithmic phase (0.7–0.8 OD600) at 28°C in PYM broth. Total bacterial RNA was isolated using the MasterPure™ RNA Purification Kit (Epicentre, Illumina). Samples corresponding to two biological replicates for each condition were submitted to Genome Québec for rRNA removal with Ribo‐Zero (Illumina) and TruSeq RNA‐seq library preparation. Fifty‐basepair single‐end sequencing of the libraries was performed using an Illumina HiSeq 2000 platform (Genome Québec). Removal of low‐quality reads and Illumina adapters, and assessment of the quality of the reads was performed using Trimmomatic 63 and FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), respectively. Reads were aligned to the Xcc genome obtained from GenBank (accession number: NC_007086.1) using SAMtools and the Burrows‐Wheeler Alignment software (BWA) 64, 65. Alignments were visualized using the software Integrated Genome Viewer (IGV) (http://broadinstitute.org/igv).
Gene annotation and RNA‐Seq differential expression analysis
Open reading frames (ORF) were primarily annotated using the GenBank NC_007086.1 “locus_tag” accession number. For subsequent functional analysis, each ORF entry was then mapped to the following fields: (i) from GenBank NC_007086.1: “old_locus_tag” and “product”, and (ii) from UniProtKB: “Protein names”, “EC number”, “Gene ontology (biological process)”, “Gene ontology (molecular function)”, “Gene ontology (cellular component)”, and “Gene ontology IDs”.
Read counts corresponding to annotated ORFs were quantified with the software FeatureCounts 66 using the strand‐specific mode. Differential expression analysis was performed using the software DESeq 67. Genes displaying adjusted P‐value < 0.05 and baseline read counts higher than the first quartile of baseMean were informed as differentially expressed genes.
Gene ontology analysis
Using GO.db Bioconductor annotation data package in R language, all Gene Ontology (GO) terms and ancestors were retrieved and annotated for all Xcc ORFs. For differentially expressed (DE) genes, an enrichment test was performed for the following categories: BP (biological process), MF (molecular function), and CC (cellular component). The enrichment factor (EF) was estimated as the ratio between the proportions of genes associated with a particular GO category present in the dataset under analysis, relative to the proportion of the number of genes in this category in the whole genome. P‐values were calculated using the Fisher's exact test.
Data availability
RNA‐Seq data are available in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under accession number E‐MTAB‐4958.
Author contributions
HRB and FM wrote the manuscript; HRB, FAG, AAV, and FM conceived and designed the experiments; HRB, LT, GS, and FM performed the experiments; HRB, LT, RS, AMT, GEG, FCL, and FM analyzed data.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Table EV2
Dataset EV1
Review Process File
Acknowledgements
This work was supported by the Argentinian Ministry of Science (MINCyT) and the Argentinian National Agency for Science and Technology (ANPCyT) grants 2011‐2672, 2012‐1545, 2014‐0710, and the Argentinian Research Council (CONICET) grant PIP‐2012 00677. All authors are supported by CONICET. We would like to thank Dr. Estefania Mancini for bioinformatics advices, and Dr. Roberto Bogomolni and Dr. Winslow Briggs for kindly revising the manuscript.
EMBO Reports (2016) 17: 1565–1577
Contributor Information
Hernán R Bonomi, Email: hbonomi@leloir.org.ar.
Adrián A Vojnov, Email: avojnov@fundacioncassara.org.ar.
Florencia Malamud, Email: fmalamud@iibintech.com.ar.
References
- 1. Auldridge ME, Forest KT (2011) Bacterial phytochromes: more than meets the light. Crit Rev Biochem Mol Biol 46: 67–88 [DOI] [PubMed] [Google Scholar]
- 2. Purcell EB, Crosson S (2008) Photoregulation in prokaryotes. Curr Opin Microbiol 11: 168–178 [DOI] [PubMed] [Google Scholar]
- 3. Bonomi HR, Posadas DM, Paris G, Carrica Mdel C, Frederickson M, Pietrasanta LI, Bogomolni RA, Zorreguieta A, Goldbaum FA (2012) Light regulates attachment, exopolysaccharide production, and nodulation in Rhizobium leguminosarum through a LOV‐histidine kinase photoreceptor. Proc Natl Acad Sci USA 109: 12135–12140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Swartz TE, Tseng TS, Frederickson MA, Paris G, Comerci DJ, Rajashekara G, Kim JG, Mudgett MB, Splitter GA, Ugalde RA et al (2007) Blue‐light‐activated histidine kinases: two‐component sensors in bacteria. Science 317: 1090–1093 [DOI] [PubMed] [Google Scholar]
- 5. Kraiselburd I, Alet AI, Tondo ML, Petrocelli S, Daurelio LD, Monzon J, Ruiz OA, Losi A, Orellano EG (2012) A LOV protein modulates the physiological attributes of Xanthomonas axonopodis pv. citri relevant for host plant colonization. PLoS ONE 7: e38226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rockwell NC, Lagarias JC (2010) A brief history of phytochromes. Chemphyschem 11: 1172–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rockwell NC, Duanmu D, Martin SS, Bachy C, Price DC, Bhattacharya D, Worden AZ, Lagarias JC (2014) Eukaryotic algal phytochromes span the visible spectrum. Proc Natl Acad Sci USA 111: 3871–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rockwell NC, Su YS, Lagarias JC (2006) Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol 57: 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Possart A, Fleck C, Hiltbrunner A (2014) Shedding (far‐red) light on phytochrome mechanisms and responses in land plants. Plant Sci 217–218: 36–46 [DOI] [PubMed] [Google Scholar]
- 10. Hughes J (2013) Phytochrome cytoplasmic signaling. Annu Rev Plant Biol 64: 377–402 [DOI] [PubMed] [Google Scholar]
- 11. Hughes J, Lamparter T, Mittmann F, Hartmann E, Gartner W, Wilde A, Borner T (1997) A prokaryotic phytochrome. Nature 386: 663 [DOI] [PubMed] [Google Scholar]
- 12. Yeh KC, Wu SH, Murphy JT, Lagarias JC (1997) A cyanobacterial phytochrome two‐component light sensory system. Science 277: 1505–1508 [DOI] [PubMed] [Google Scholar]
- 13. Wu L, McGrane RS, Beattie GA (2013) Light regulation of swarming motility in Pseudomonas syringae integrates signaling pathways mediated by a bacteriophytochrome and a LOV protein. mBio 4: e00334‐13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davis SJ, Vener AV, Vierstra RD (1999) Bacteriophytochromes: phytochrome‐like photoreceptors from nonphotosynthetic eubacteria. Science 286: 2517–2520 [DOI] [PubMed] [Google Scholar]
- 15. Williams PH (1980) Black rot: a continuing threat to world crucifers. Plant Dis 64: 736–742 [Google Scholar]
- 16. Vicente JG, Holub EB (2013) Xanthomonas campestris pv. campestris (cause of black rot of crucifers) in the genomic era is still a worldwide threat to brassica crops. Mol Plant Pathol 14: 2–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gudesblat GE, Torres PS, Vojnov AA (2009) Xanthomonas campestris overcomes Arabidopsis stomatal innate immunity through a DSF cell‐to‐cell signal‐regulated virulence factor. Plant Physiol 149: 1017–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vorholt JA (2012) Microbial life in the phyllosphere. Nat Rev Microbiol 10: 828–840 [DOI] [PubMed] [Google Scholar]
- 19. Bhoo SH, Davis SJ, Walker J, Karniol B, Vierstra RD (2001) Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature 414: 776–779 [DOI] [PubMed] [Google Scholar]
- 20. Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J et al (2014) Pfam: the protein families database. Nucleic Acids Res 42: D222–D230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lamparter T, Carrascal M, Michael N, Martinez E, Rottwinkel G, Abian J (2004) The biliverdin chromophore binds covalently to a conserved cysteine residue in the N‐terminus of Agrobacterium phytochrome Agp1. Biochemistry 43: 3659–3669 [DOI] [PubMed] [Google Scholar]
- 22. Otero LH, Klinke S, Rinaldi J, Velazquez‐Escobar F, Mroginski MA, Fernandez Lopez M, Malamud F, Vojnov AA, Hildebrandt P, Goldbaum FA et al (2016) Structure of the full‐length bacteriophytochrome from the plant pathogen Xanthomonas campestris provides clues to its long‐range signaling mechanism. J Mol Biol doi: 10.1016/j.jmb.2016.04.012 [DOI] [PubMed] [Google Scholar]
- 23. Moglich A, Ayers RA, Moffat K (2009) Structure and signaling mechanism of Per‐ARNT‐Sim domains. Structure 17: 1282–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rottwinkel G, Oberpichler I, Lamparter T (2010) Bathy phytochromes in rhizobial soil bacteria. J Bacteriol 192: 5124–5133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karniol B, Vierstra RD (2003) The pair of bacteriophytochromes from Agrobacterium tumefaciens are histidine kinases with opposing photobiological properties. Proc Natl Acad Sci USA 100: 2807–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Underwood W, Melotto M, He SY (2007) Role of plant stomata in bacterial invasion. Cell Microbiol 9: 1621–1629 [DOI] [PubMed] [Google Scholar]
- 27. Rigano LA, Payette C, Brouillard G, Marano MR, Abramowicz L, Torres PS, Yun M, Castagnaro AP, Oirdi ME, Dufour V et al (2007) Bacterial cyclic beta‐(1,2)‐glucan acts in systemic suppression of plant immune responses. Plant Cell 19: 2077–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yun MH, Torres PS, El Oirdi M, Rigano LA, Gonzalez‐Lamothe R, Marano MR, Castagnaro AP, Dankert MA, Bouarab K, Vojnov AA (2006) Xanthan induces plant susceptibility by suppressing callose deposition. Plant Physiol 141: 178–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Millet YA, Danna CH, Clay NK, Songnuan W, Simon MD, Werck‐Reichhart D, Ausubel FM (2010) Innate immune responses activated in Arabidopsis roots by microbe‐associated molecular patterns. Plant Cell 22: 973–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barber CE, Tang JL, Feng JX, Pan MQ, Wilson TJ, Slater H, Dow JM, Williams P, Daniels MJ (1997) A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol Microbiol 24: 555–566 [DOI] [PubMed] [Google Scholar]
- 31. Buttner D, Bonas U (2010) Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol Rev 34: 107–133 [DOI] [PubMed] [Google Scholar]
- 32. Torres PS, Malamud F, Rigano LA, Russo DM, Marano MR, Castagnaro AP, Zorreguieta A, Bouarab K, Dow JM, Vojnov AA (2007) Controlled synthesis of the DSF cell‐cell signal is required for biofilm formation and virulence in Xanthomonas campestris . Environ Microbiol 9: 2101–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rigano LA, Siciliano F, Enrique R, Sendin L, Filippone P, Torres PS, Questa J, Dow JM, Castagnaro AP, Vojnov AA et al (2007) Biofilm formation, epiphytic fitness, and canker development in Xanthomonas axonopodis pv. citri. Mol Plant Microbe Interact 20: 1222–1230 [DOI] [PubMed] [Google Scholar]
- 34. Malamud F, Torres PS, Roeschlin R, Rigano LA, Enrique R, Bonomi HR, Castagnaro AP, Marano MR, Vojnov AA (2011) The Xanthomonas axonopodis pv. citri flagellum is required for mature biofilm and canker development. Microbiology 157: 819–829 [DOI] [PubMed] [Google Scholar]
- 35. Baptista JC, Machado MA, Homem RA, Torres PS, Vojnov AA, do Amaral AM (2010) Mutation in the xpsD gene of Xanthomonas axonopodis pv. citri affects cellulose degradation and virulence. Genet Mol Biol 33: 146–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meyer KM, Leveau JH (2012) Microbiology of the phyllosphere: a playground for testing ecological concepts. Oecologia 168: 621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lindow SE, Brandl MT (2003) Microbiology of the phyllosphere. Appl Environ Microbiol 69: 1875–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gunasekera TS, Sundin GW (2006) Role of nucleotide excision repair and photoreactivation in the solar UVB radiation survival of Pseudomonas syringae pv. syringae B728a. J Appl Microbiol 100: 1073–1083 [DOI] [PubMed] [Google Scholar]
- 39. Rio‐Alvarez I, Rodriguez‐Herva JJ, Martinez PM, Gonzalez‐Melendi P, Garcia‐Casado G, Rodriguez‐Palenzuela P, Lopez‐Solanilla E (2013) Light regulates motility, attachment and virulence in the plant pathogen Pseudomonas syringae pv tomato DC3000. Environ Microbiol 16: 2072–2085 [DOI] [PubMed] [Google Scholar]
- 40. Ricci A, Dramis L, Shah R, Gartner W, Losi A (2015) Visualizing the relevance of bacterial blue‐ and red‐light receptors during plant‐pathogen interaction. Environ Microbiol Rep 7: 795–802 [DOI] [PubMed] [Google Scholar]
- 41. Moriconi V, Sellaro R, Ayub N, Soto G, Rugnone M, Shah R, Pathak GP, Gartner W, Casal JJ (2013) LOV‐domain photoreceptor, encoded in a genomic island, attenuates the virulence of Pseudomonas syringae in light‐exposed Arabidopsis leaves. Plant J 76: 322–331 [DOI] [PubMed] [Google Scholar]
- 42. de Wit M, Spoel SH, Sanchez‐Perez GF, Gommers CM, Pieterse CM, Voesenek LA, Pierik R (2013) Perception of low red: far‐red ratio compromises both salicylic acid‐ and jasmonic acid‐dependent pathogen defences in Arabidopsis . Plant J 75: 90–103 [DOI] [PubMed] [Google Scholar]
- 43. Hua J (2013) Modulation of plant immunity by light, circadian rhythm, and temperature. Curr Opin Plant Biol 16: 406–413 [DOI] [PubMed] [Google Scholar]
- 44. Griebel T, Zeier J (2008) Light regulation and daytime dependency of inducible plant defenses in Arabidopsis: phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiol 147: 790–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sorhagen K, Laxa M, Peterhansel C, Reumann S (2013) The emerging role of photorespiration and non‐photorespiratory peroxisomal metabolism in pathogen defence. Plant Biol 15: 723–736 [DOI] [PubMed] [Google Scholar]
- 46. He YW, Zhang LH (2008) Quorum sensing and virulence regulation in Xanthomonas campestris . FEMS Microbiol Rev 32: 842–857 [DOI] [PubMed] [Google Scholar]
- 47. Ryan RP, An SQ, Allan JH, McCarthy Y, Dow JM (2015) The DSF family of cell‐cell signals: an expanding class of bacterial virulence regulators. PLoS Pathog 11: e1004986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Smith H (2000) Phytochromes and light signal perception by plants–an emerging synthesis. Nature 407: 585–591 [DOI] [PubMed] [Google Scholar]
- 49. Cadmus MC, Rogovin SP, Burton KA, Pittsley JE, Knutson CA, Jeanes A (1976) Colonial variation in Xanthomonas campestris NRRL B‐1459 and characterization of the polysaccharide from a variant strain. Can J Microbiol 22: 942–948 [DOI] [PubMed] [Google Scholar]
- 50. Sherwood MT (1970) Improved synthetic medium for the growth of Rhizobium . J Appl Bacteriol 33: 708–713 [DOI] [PubMed] [Google Scholar]
- 51. Malamud F, Homem RA, Conforte VP, Yaryura PM, Castagnaro AP, Marano MR, do Amaral AM, Vojnov AA (2013) Identification and characterization of biofilm formation‐defective mutants of Xanthomonas citri subsp. citri. Microbiology 159: 1911–1919 [DOI] [PubMed] [Google Scholar]
- 52. Prentki P, Krisch HM (1984) In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29: 303–313 [DOI] [PubMed] [Google Scholar]
- 53. Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A (1994) Small mobilizable multi‐purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum . Gene 145: 69–73 [DOI] [PubMed] [Google Scholar]
- 54. Kovach ME, Phillips RW, Elzer PH, Roop RM II, Peterson KM (1994) pBBR1MCS: a broad‐host‐range cloning vector. Biotechniques 16: 800–802 [PubMed] [Google Scholar]
- 55. Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Whelan S, Goldman N (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum‐likelihood approach. Mol Biol Evol 18: 691–699 [DOI] [PubMed] [Google Scholar]
- 57. Letunic I, Bork P (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44: W242–W245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Klinke S, Otero LH, Rinaldi J, Sosa S, Guimaraes BG, Shepard WE, Goldbaum FA, Bonomi HR (2014) Crystallization and preliminary X‐ray characterization of the full‐length bacteriophytochrome from the plant pathogen Xanthomonas campestris pv. campestris . Acta Crystallogr F Struct Biol Commun 70: 1636–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vojnov AA, Zorreguieta A, Dow JM, Daniels MJ, Dankert MA (1998) Evidence for a role for the gumB and gumC gene products in the formation of xanthan from its pentasaccharide repeating unit by Xanthomonas campestris . Microbiology 144: 1487–1493 [DOI] [PubMed] [Google Scholar]
- 60. Malamud F, Conforte VP, Rigano LA, Castagnaro AP, Marano MR, Morais do Amaral A, Vojnov AA (2012) HrpM is involved in glucan biosynthesis, biofilm formation and pathogenicity in Xanthomonas citri ssp. citri. Mol Plant Pathol 13: 1010–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S (2000) Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146 (Pt 10): 2395–2407 [DOI] [PubMed] [Google Scholar]
- 62. Ortiz G, Blasco M, Albertó E (2016) An economical and high throughput alternative for endoglucanase activity determination. BioTechnology – An Indian Journal 12: 70–74 [Google Scholar]
- 63. Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Liao Y, Smyth GK, Shi W (2014) FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- 67. Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Table EV2
Dataset EV1
Review Process File
Data Availability Statement
RNA‐Seq data are available in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under accession number E‐MTAB‐4958.


