Abstract
The interferon‐induced transmembrane (IFITM) proteins protect host cells from diverse virus infections. IFITM proteins also incorporate into HIV‐1 virions and inhibit virus fusion and cell‐to‐cell spread, with IFITM3 showing the greatest potency. Here, we report that amino‐terminal mutants of IFITM3 preventing ubiquitination and endocytosis are more abundantly incorporated into virions and exhibit enhanced inhibition of HIV‐1 fusion. An analysis of primate genomes revealed that IFITM3 is the most ancient antiviral family member of the IFITM locus and has undergone a repeated duplication in independent host lineages. Some IFITM3 genes in nonhuman primates, including those that arose following gene duplication, carry amino‐terminal mutations that modify protein localization and function. This suggests that “runaway” IFITM3 variants could be selected for altered antiviral activity. Furthermore, we show that adaptations in IFITM3 result in a trade‐off in antiviral specificity, as variants exhibiting enhanced activity against HIV‐1 poorly restrict influenza A virus. Overall, we provide the first experimental evidence that diversification of IFITM3 genes may boost the antiviral coverage of host cells and provide selective functional advantages.
Keywords: evolution, HIV, IFITM, innate immunity, virus
Subject Categories: Microbiology, Virology & Host Pathogen Interaction; Immunology; Evolution
Introduction
Viruses depend on numerous cellular factors and pathways to complete the viral life cycle and spread to new target cells. Known as virus cofactors, these are host molecules that are commandeered by the virus in order to promote efficient infection 1. To prevent virus takeover, cells express a battery of antiviral effectors that repress viral replication at multiple stages. These proteins make up the cell‐intrinsic innate immune response and are also known as host restriction factors. The establishment of a hostile intracellular environment sets the stage for ongoing host–virus coevolution, a phenomenon that is particularly well characterized between primates and lentiviruses 2. As their name implies, host restriction factors influence the host tropism of virus infections in vitro and in vivo because they are rapidly evolving and thus divergent between species.
The immune‐related IFITM proteins (IFITM1, IFITM2, and IFITM3 in humans) may represent the earliest acting restriction factors yet identified. The IFITM family also includes the IFITM5 and IFITM10 genes with no known immune function 3. IFITM genes are present among a wide range of vertebrate animal species, and they may have originated in early unicellular eukaryotes via horizontal gene transmission from a bacterium 4. Since then, expansions within genomes have given rise to unique IFITM gene repertoires that vary at the level of sequence and copy number. To date, all immune‐related IFITM proteins identified in animals (metazoans) display antiviral function 5, 6, 7. Even IFITM‐like genes in Mycobacteria inhibit virus infection when expressed in human cells 8.
As residents of membranes at the interior and exterior of the cell, they block the entry step of diverse viruses 9, 10 by inhibiting virus–cell fusion. The mechanisms behind this protection involve altering the biophysical properties 11, 12, 13 or cholesterol content 14 of the cellular membranes in which they are found. These proteins inhibit many enveloped viruses, including influenza A virus (IAV), West Nile virus, dengue virus, severe acute respiratory syndrome coronavirus, hepatitis C virus, and Ebola virus 15, as well as lentiviruses including HIV‐1 and SIV 16, 17, 18. While the majority of studies have relied on in vitro infection systems, it is well established that IFITM3 restricts virus infection in vivo. Ifitm3‐deficient mice fail to clear infection by otherwise mild strains of IAV 19, 20.
Using a cellular coculture system, we and others identified new antiviral functions of IFITM proteins during HIV‐1 infection 21, 22, 23. We showed that IFITM proteins, in particular IFITM3, block the spread of HIV‐1 from infected to uninfected T cells. Surprisingly, this function was primarily the result of interactions occurring between IFITM3 and virus in the infected cells. We found that IFITM3 in the virus‐producing cell decreases virion infectivity at the level of virus–cell fusion and that IFITM3 becomes incorporated into nascent virions. This mechanism of antiviral function may apply to other lentiviruses as well as diverse families of enveloped viruses. Therefore, IFITM proteins are restriction factors of increasingly broad scope.
While human IFITM3 is described as a resident of endosomal and lysosomal membranes 24, we observed that a portion of IFITM3 is detectable at the surface of transfected 293T cells and in primary CD4+ T cells 21. In fact, IFITM3 can be observed at many different subcellular compartments and its localization is regulated by various post‐translational modifications 25. The fate of IFITM3 is governed by phosphorylation 16, 26, ubiquitination 27, palmitoylation 27, 28, and methylation 29. These regulatory processes impact the localization and turnover of IFITM3 in the cell, and as a result, they are critical to its antiviral function. During biosynthesis, IFITM3 protein is trafficked to the plasma membrane and endocytosed. This latter step is driven by an internalization motif (YxxΦ, YEML in human IFITM3) that is recognized by the μB unit of the AP‐2 complex, resulting in the sorting of IFITM3 into late endosomes, multivesicular bodies, and lysosomes 26, 30. Furthermore, an overlapping motif (PPxY, PPNY in human IFITM3) recruits the E3 ubiquitin ligase NEDD4 to promote IFITM3 ubiquitination and turnover via lysosomes 31. Phosphorylation of tyrosine residue 20 (Y20) common to the two motifs inhibits both endocytosis and ubiquitination, causing an accumulation of IFITM3 and redistribution to the plasma membrane 26. Mutation or deletion of Y20 produces a similar effect 30. Interestingly, a single nucleotide polymorphism (SNP) (rs12252‐C) in human IFITM3 is predicted to produce an alternatively spliced transcript that encodes a protein lacking these two regulatory motifs. This putative truncated form of IFITM3 (Δ1–21) is largely confined to the plasma membrane when expressed in cell lines 16. Intriguingly, rs12252‐C is associated with severe outcomes following IAV infection 20, 32, 33, 34. These epidemiologic studies coincide with in vitro experiments showing that IFITM3 Δ1–21 fails to accumulate in the endosomal compartment, where IAV undergoes pH‐dependent fusion to access the cytoplasm. However, endogenous expression of the putative truncated form of human IFITM3 has not been demonstrated to date. While these previous reports reveal how synthetic mutations in IFITM3 affect the restriction of RNA viruses such as IAV and vesicular stomatitis virus (VSV), it remains unclear how they may regulate the activity against other viruses with different cell entry strategies. Furthermore, very little is known about natural, genome‐encoded mutations that may affect IFITM function in diverse animal species.
HIV‐1 is believed to carry out the initial (fusion) and terminal (budding) steps of its life cycle at the plasma membrane of T lymphocytes 35. Given that the amount of IFITM3 at the cell surface most likely dictates its ability to restrict HIV‐1 entry and the infectivity of nascent budding virions, we hypothesized that post‐translational modifications affecting cell surface association will impact the activity of IFITM3 against HIV‐1. Here, we report that endocytosis and ubiquitination are negative regulators of IFITM3 anti‐HIV activity. Furthermore, we show that post‐translational regulation of IFITM3 may result in a functional trade‐off: alterations that improve the function against one virus can inhibit its activity against other viruses. As a testament to their functional importance, periodic diversification of residues within the two regulatory motifs of IFITM3 is observed during primate evolution, resulting in variants with different potency and specificity. Finally, our multifaceted analysis has allowed us to retrace the origins of IFITM in primates and provides an explanation for the recurrent gene duplication observed in this antiviral gene family.
Results
Mutation in IFITM3 alters subcellular localization and anti‐HIV activity
IFITM3 contains two hydrophobic domains conserved in other IFITM family members, but is divergent in its amino‐ and carboxy‐termini. The amino‐terminal sequence motifs recognized by NEDD4 and the AP‐2 complex are adjacent and overlap at the tyrosine at residue 20, found just upstream of an internal methionine (which serves as the translation start site in IFITM1 and the Δ1–21 construct of IFITM3) 25 (Fig 1A).
Figure 1. Mutation in IFITM3 alters protein subcellular localization and anti‐HIV activity.

- A schematic of the IFITM3 protein is shown, with gray boxes indicating transmembrane domains and colored bars indicating residues targeted for post‐translational modification: ubiquitinated lysines (blue), palmitoylated cysteines (purple), and a phosphorylated tyrosine (red). Residues 17–24 are shown in detail to highlight two adjacent, overlapping motifs that recruit NEDD4 (PPNY) and the AP‐2 complex (YEML). The IFITM3 Δ1–21 variant initiates translation at an internal methionine residue (green) and thus lacks these motifs.
- Confocal fluorescence microscopy of 293T cells transfected with 0.5 μg pQCXIP encoding IFITM3 variants containing N‐terminal FLAG (pQCXIP‐FLAG‐IFITM3) and immunostained with anti‐FLAG M2 antibody. Scale bar, 5 μm.
- Flow cytometric analysis of transfected cells in (B) immunostained with anti‐FLAG M2 antibody. Corresponding mean fluorescence intensity values are shown.
- SDS–PAGE of cell lysates derived from 293T cells transfected in (B) followed by immunoblotting with anti‐FLAG M2 and anti‐actin antibodies.
- SDS–PAGE of OptiPrep‐purified supernatants (25 ng p24 equivalent) derived from 293T cells transfected with pQCXIP‐FLAG‐IFITM3 variants, HIV‐1 pNL4‐3, and Vpr‐Blam plasmid followed by immunoblotting with anti‐FLAG M2 and anti‐p24 Gag (183‐H12‐5C).
- OptiPrep‐purified supernatants from (E) were incubated with SupT1 target cells for 4 h and virus–cell fusion was scored by flow cytometry. Mean + SD of 5–10 experiments (each performed with virus produced from an independent transfection) is shown.
- Viruses used in (E) were used to infect SupT1 cells and productive infection was scored at 48–60 h by immunostaining with the KC57 antibody and flow cytometry. Expression of pCMV‐MxA served as a negative control. The mean + SD of 5–10 experiments is shown.
- Confocal fluorescence microscopy of 293T cells transfected with 0.5 μg pQCXIP‐FLAG‐IFITM3 variants and immunostained with anti‐FLAG M2 antibody.
- SDS–PAGE of cell lysates derived from 293T cells transfected in (H) followed by immunoblotting with anti‐FLAG M2 and anti‐actin antibodies. Scale bar, 5 μm.
- SDS–PAGE of OptiPrep‐purified supernatants (25 ng p24 equivalent) derived from 293T cells transfected with pQCXIP‐FLAG‐IFITM3 constructs, HIV‐1 pNL4‐3, and Vpr‐Blam plasmid followed by immunoblotting with anti‐FLAG M2 and anti‐p24 Gag.
- OptiPrep‐purified supernatants from (J) were incubated with SupT1 target cells for 4 h and virus–cell fusion was scored by flow cytometry. The mean + SD of three experiments is shown.
293T cells were selected as a suitable system to study the impact of post‐translational modifications on IFITM3 function because they express very low levels of constitutive IFITM3. Transfection of FLAG‐tagged IFITM3 into 293T results in expression levels comparable to, but less uniform than, those induced by type‐I interferon (Fig EV1A) and localization to disperse intracellular vesicles and the plasma membrane (Fig 1B). As previously described 16, the subcellular localization of the Δ1–21 variant is mostly restricted to the plasma membrane (Fig 1B). A mutant deficient in cysteine residues (referred to as Δpalm because it prevents palmitoylation of IFITM3 27) results in a predominantly perinuclear localization with additional diffuse staining throughout the cytoplasm. Western blot and flow cytometric analysis reveal quantitative differences in protein expression. Levels of the Δ1–21 variant are elevated relative to full‐length protein, while Δpalm is expressed at lower levels (Fig 1C and D).
Figure EV1. Dissection of two negative regulatory motifs in IFITM3 reveals additive roles in anti‐HIV activity.

- Flow cytometric analysis of IFITM3 protein levels in 293T, HeLa, Tet‐ON SupT1, and primary CD4+ T cells. Staining was performed with the mouse monoclonal anti‐IFITM3 antibody (Proteintech, 66081‐1‐Ig) with or without stimulation by type‐I IFN (1,000 U/ml) or doxycycline for 24 h.
- Confocal fluorescence microscopy of 293T cells transfected with 1 μg pNL4‐3 and 0.5 μg pQCXIP‐FLAG‐IFITM3 or pQCXIP‐FLAG‐IFITM3 Δ1–21. Immunostaining was performed with anti‐FLAG M2 and anti‐Gag p17 (ARP342). Scale bars, 5 μm.
- Confocal fluorescence microscopy of 293T cells transfected with 0.5 μg pQCXIP‐FLAG‐IFITM3 ΔY20 or pCMV‐myc‐IFITM3 L23Q and immunostained with anti‐FLAG M2 or anti‐IFITM3 (Proteintech, 66081‐1‐Ig), respectively.
- SDS–PAGE of OptiPrep‐purified supernatants (25 ng p24 equivalent) derived from 293T cells transfected with pQCXIP‐FLAG‐IFITM3 or pQCXIP‐FLAG‐IFITM3 ΔY20, HIV‐1 pNL4‐3, and Vpr‐Blam plasmid followed by immunoblotting with anti‐FLAG M2 and anti‐p24 Gag.
- Virus supernatants from (D) were incubated with SupT1 target cells for 4 h and virus–cell fusion was scored by flow cytometry. Mean + SD of three experiments is shown.
- Flow cytometric analysis of 293T cells transfected with 0.5 μg of pQCXIP‐FLAG‐IFITM3 constructs and immunostained with anti‐FLAG M2 antibody. Corresponding mean fluorescence intensity values are shown.
- Virus produced in Fig 1K was used to infect SupT1 cells and productive infection was scored at 48–60 h by immunostaining with the KC57 antibody and flow cytometry. Mean + SD of three experiments is shown.
- OptiPrep‐purified supernatants (25 ng p24 equivalent) derived from 293T cells transfected with pCMV‐myc‐IFITM3 or pCMV‐myc‐IFITM3 L23Q or pCMV‐MxA constructs, HIV‐1 pNL4‐3, and Vpr‐Blam plasmid were incubated with SupT1 target cells for 4 h and virus–cell fusion was scored by flow cytometry.
- Flow cytometric analysis of 293T cells transfected with 0.5 μg of pCMV constructs and immunostained with anti‐IFITM3 or anti‐MxA antibodies. For these experiments, IFITM3 and the L23Q mutant were expressed from a different plasmid background (pCMV) and protein levels and antiviral activity were generally increased. Of note, the L23Q mutant did not achieve higher relative protein expression, suggesting that its enhancement at the cell surface is responsible for its superior anti‐HIV activity.
To address how mutation in IFITM3 affects its activity against HIV‐1, virus was produced in 293T cells via co‐transfection with the HIV‐1 molecular clone pNL4‐3, pVpr‐Blam (which allows the detection of virion access into the cytoplasm), and plasmids expressing IFITM3 mutants (as performed in 21). Consistent with its enhanced localization to the cell surface, which also serves as the site for virus assembly and budding, the Δ1–21 variant exhibits increased levels of virion incorporation relative to full‐length IFITM3 (Fig 1E). Importantly, viruses produced in the presence of the Δ1–21 variant are further reduced in virion infectivity when incubated with fresh SupT1 T‐cell targets, as evidenced by reduced virus–cell fusion at 4 h (Fig 1F) and reduced productive viral infection at 48 h (Fig 1G). Virus produced in the presence of MxA was used as an additional negative control. Confocal immunofluorescence microscopy confirms that surface‐associated IFITM3 can be localized to sites of HIV‐1 budding tagged by the presence of mature Gag protein (Fig EV1B). Results with an IFITM3 mutant encoding a single residue deletion at position 20 (ΔY20) reproduced those of Δ1–21, strongly suggesting that the tyrosine residue and the two overlapping regulatory motifs to which it belongs are crucial to the enhancement of anti‐HIV activity observed (Fig EV1C–E). Thus, the extent of inhibition of HIV‐1 infectivity is associated with cell surface localization and the increased detection of IFITM3 in virions. We previously demonstrated that IFITM1, which naturally lacks the amino‐terminus present in IFITM3, only modestly restricts HIV‐1 infectivity 21. Thus, the absence of an amino‐terminus per se is not sufficient to enhance anti‐HIV‐1 activity. Rather, IFITM3 contains other intrinsic determinants that enable a potent HIV‐1 restriction that can be amplified by the removal of its amino‐terminus.
To assess the involvement of distinct regulatory activities in the regulation of IFITM3 activity, we took advantage of additional IFITM3 mutants in which the NEDD4‐binding site alone (Δ17–18) or both the NEDD4‐ and the AP‐2‐binding sites (Δ17–20) are disrupted. Immunofluorescence microscopy confirmed previously published results 16 showing that Δ17–20 is enriched at the cell surface (Fig 1H). Similar to Δ1–21, the Δ17–18 mutant exhibits slightly elevated protein levels relative to full‐length IFITM3. Importantly, the Δ17–20 variant does not share this feature, and it is even expressed at lower levels relative to full‐length IFITM3 (Figs 1I and EV1F). Both the Δ17–18 and Δ17–20 variants display an increased propensity for virion incorporation, with the mutant in which endocytosis is disrupted (Δ17–20) showing the greatest phenotype (Fig 1J). Consistently, the Δ17–20 variant demonstrates the most profound inhibition of HIV‐1 particle fusogenicity (Fig 1K) and infectivity (Fig EV1G). Therefore, the enhancement of IFITM3 antiviral activity is associated with its subcellular localization and is not solely dependent on protein levels. When we employed a L23Q mutant to inhibit protein endocytosis 26 without disturbing the NEDD4‐binding motif, we found that this single amino acid change also results in more pronounced restriction of HIV‐1 infectivity relative to wild‐type IFITM3 (Fig EV1H and I). These results collectively suggest that motifs interacting with endogenous NEDD4 and AP‐2 act in an additive fashion to negatively regulate the anti‐HIV activity of IFITM3.
The E3 ubiquitin ligase NEDD4 regulates the activity of IFITM3 against HIV‐1
We directly tested the regulatory effect of NEDD4 in virus‐producing cells using a system in which NEDD4 is co‐expressed by transfection with IFITM3 or the Δ1–21 variant in 293T cells. The co‐expression of NEDD4 resulted in lower levels of IFITM3 in cell lysates, an effect that was not observed for the catalytically inactive NEDD4 mutant C867A (Fig EV2A). These results confirm that NEDD4‐mediated ubiquitination accelerates the turnover of IFITM3 31. Moreover, NEDD4 co‐expression diminished the levels of IFITM3 incorporated into HIV‐1 virions (Fig EV2A). In contrast to its typical expression pattern in the cytoplasm and on the cell surface, confocal immunofluorescence microscopy revealed that IFITM3 becomes mostly intracellular in cells co‐expressing NEDD4, but not in those with inactive NEDD4 C867A (Fig EV2B and C). NEDD4 induced the accumulation of IFITM3 in large vesicular compartments that partially colocalize with LAMP1 (Fig EV2D). In contrast, protein levels of the Δ1–21 variant in cell lysates or in virions were unaffected by NEDD4, and the localization remained predominantly cell surface oriented (Fig EV3D). Collectively, these findings suggest that NEDD4 regulates IFITM3 levels in cells producing HIV‐1 and thus may regulate its antiviral effect. They also highlight how variants of IFITM3 missing key elements of the amino‐terminus may confer escape from NEDD4 and lead to enhanced antiviral function in the cell.
Figure EV2. The E3 ubiquitin ligase NEDD4 regulates IFITM3 in HIV‐1‐infected cells.

- SDS–PAGE of cell lysates (upper) and OptiPrep‐purified virus (lower) derived from 293T cells transfected with 0.2 μg of pQCXIP‐FLAG‐IFITM3 or pQCXIP‐FLAG‐IFITM3 Δ1–21, 1.0 μg of HIV‐1 pNL4‐3, and 0.5 μg pCI‐HA‐NEDD4 WT or pCI‐HA‐NEDD4 C867A plasmids. Immunoblotting was performed with anti‐NEDD4, anti‐FLAG M2, and anti‐actin on whole‐cell lysates, while anti‐FLAG M2 and anti‐p24 Gag were used on 15 ng p24 equivalents of purified virus.
- Confocal fluorescence microscopy of 293T cells transfected with pQCXIP‐FLAG‐IFITM3 and pCI‐HA‐NEDD4 or pCI‐HA‐NEDD4 C867A or pCI‐Empty, followed by immunostaining with anti‐FLAG M2 and anti‐HA. A single medial Z slice is shown.
- A second example of cells from (B) in which pQCXIP‐FLAG‐IFITM3 and pCI‐HA‐NEDD4 are co‐transfected. A single medial Z slice (upper) and a 3D reconstruction of Z‐stacks (lower) are shown.
- 293T cells were transfected with pQCXIP‐FLAG‐IFITM3 or pQCXIP‐FLAG‐IFITM3 Δ1–21 and pCI‐HA‐NEDD4, and immunostaining was performed with anti‐FLAG M2 and anti‐LAMP1.
Figure EV3. Assessment of virion composition by supernatant fractionation.

Virus‐containing supernatants produced from Tet‐ON IFITM3+ or Δ1–21+ cells were fractionated using a 6–18% OptiPrep gradient velocity gradient. Fractions were analyzed by SDS–PAGE and immunoblotted with anti‐p24 Gag and anti‐FLAG M2 antibodies. Western blot images are representative of two independent experiments.
The amino‐terminus of IFITM3 regulates activity against HIV‐1 in T cells
To study the regulation of IFITM3 protein in the natural target cells of HIV‐1, we took advantage of T‐lymphocyte cell lines (SupT1) engineered to allow for Dox‐inducible expression of IFITM3 protein variants 17, 21. Following Dox treatment of these cells, expression levels are uniform and full‐length IFITM3 localizes mostly to the cell interior, while the Δ1–21 variant is predominantly found at the surface (Fig 2A), mirroring the results in 293T cells. Importantly, we observed the induction of higher levels of full‐length IFITM3 compared to the Δ1–21 variant in its respective cell line (Fig 2B). Using this T cell‐based system, we tested the relative antiviral potential of IFITM3 in both donor and target cells. We produced virus in the presence or absence of IFITM3 variants and subsequently infected target T cells in which IFITM3 variants were induced or not. As previously reported 21, IFITM3 restricts HIV‐1 infection within donor and target cells in an additive fashion (Fig 2C). However, the Δ1–21 variant exhibits superior antiviral activity at both stages, such that infection is almost completely blocked when it is present in both donor and target cells (Fig 2C). These data demonstrate that removal of the amino‐terminus enhances both antiviral functions fulfilled by IFITM3 during HIV‐1 infection. Despite the relatively lower level of protein induction in this system, and in agreement with its localization at the cell surface, the Δ1–21 variant is enriched in virions (Fig 2D). Importantly, we did not detect IFITM3 in supernatants from noninfected cells, strongly suggesting that the majority of signal detected originates from within virus particles 21 (Fig 2D). Nonetheless, because IFITM proteins can also be found in exocytic vesicles 36, we performed density gradient purification of virions produced in the presence of full‐length IFITM3 and the Δ1–21 variant. As previously reported 21, full‐length IFITM3 co‐sediments with HIV‐1 Gag protein and is nearly absent from fractions containing lower‐density microvesicles, and the pattern was similar for Δ1–21 (Fig EV3).
Figure 2. Mutation of N‐terminal regulatory motifs of IFITM3 in T cells enhances the restriction of HIV‐1.

- Confocal fluorescence microscopy of SupT1 T cells transduced with Tet‐inducible pQCXIP‐FLAG‐IFITM3 or pQCXIP‐FLAG‐IFITM3 Δ1–21 (Tet‐ON) immunostained with anti‐FLAG M2 antibody following overnight treatment with 500 ng/ml doxycycline. Scale bar, 10 μm.
- Tet‐ON SupT1 cell lines were treated or not with 500 ng/ml doxycycline overnight and induction of IFITM3 protein was assessed by anti‐FLAG M2 immunostaining and flow cytometry.
- Tet‐ON SupT1 cell lines were productively infected with NL4‐3 VSV‐G and then treated with 500 ng/ml doxycycline overnight to produce virus from IFITM3− and IFITM3+ cells; 25 ng p24 equivalents of purified supernatants was used to infect fresh Tet‐ON SupT1 cells, which were previously treated with 500 ng/ml doxycycline or not. Infection was scored at 72 h postinfection by immunostaining with KC57 and flow cytometry. Mean + SD of three experiments is shown.
- About 25 ng p24 equivalents of virus used in (C) was subjected to SDS–PAGE. Immunoblotting was performed using anti‐FLAG M2, anti‐p24 Gag, and anti‐gp120 Env (NIH #288) antibodies.
- 293T cells were transfected with the indicated amount of pQCXIP‐FLAG‐IFITM3 or pQCXIP‐FLAG‐Δ1–21 or pCMV‐MxA and 1.0 μg of HIV‐1 pNL4‐3. SDS–PAGE was performed on whole‐cell lysates followed by immunoblotting with anti‐FLAG M2, anti‐p24 Gag, anti‐Env gp120 (NIH #288), and anti‐tubulin antibodies.
A recent publication reported that IFITM3 antagonizes HIV‐1 envelope protein, altering both the maturation of gp120 and inhibiting its incorporation into virions produced in 293T cells 23. However, and consistent with our previous publication 21, the induction of IFITM3 or the Δ1–21 variant did not affect gp120 levels in virions produced in T‐cell lines (Fig 2D). To explore further the effect of IFITM3 on HIV‐1 Env in virus‐producing cells, we co‐transfected 293T cells with HIV‐1 pNL4‐3 and titrating amounts of plasmid encoding IFITM3, the Δ1–21 variant, or MxA. We observed that gp120 levels were slightly reduced only at the highest level of IFITM3 overexpression (Fig 2E). The Δ1–21 variant reduced gp120 levels somewhat further in a manner consistent with its elevated expression, while MxA overexpression had no effect. Interestingly, we also found that reduction in gp120 was accompanied by a concomitant decrease in HIV‐1 p24 levels, possibly suggestive of a global effect on virus translation (Fig 2E). Therefore, while IFITM3‐mediated restriction of HIV‐1 infectivity may involve Env antagonism, this effect is cell type dependent and expression level dependent. We propose that the mechanistic basis for IFITM3 antiviral function against HIV‐1 lies primarily with its cell surface localization, which is associated with its incorporation into virions budding from the same compartment.
Recurrent duplication and divergence of primate IFITM3 over evolutionary time
Given our finding that synthetic mutants in the amino‐terminus of IFITM3 exhibit enhanced anti‐HIV‐1 activity, we chose to examine the functional consequences of standing variation in IFITM3 genes from nonhuman primates. Dozens of species harbor simian immunodeficiency virus (SIV) strains in the wild, infections that have likely persisted for millions of years in coevolution with their hosts 2. Experimental approaches combining in silico genomic analysis and in vitro or in vivo infection assays have revealed that primate immunity genes, such as APOBEC3G, TRIM5, tetherin, and SAMHD1, are undergoing adaptive evolution in ways that impact lentiviral infections 37. Our results prompted us to ask whether the antiviral function and specificity of IFITM3 may also be dynamic and subject to natural selection on evolutionary timescales.
Some previous reports focusing on the evolution of the IFITM gene family in vertebrates have shown that, compared to the five IFITM genes present on human chromosome 11, the number of IFITM genes per species may vary as a result of gene duplication events 4, 38. To build a comprehensive understanding of IFITM evolution in primates, we used comparative genomics to identify IFITM genes from full reference genomes. Aided by the Genomicus toolkit 39 and the Ensembl gene database (www.ensembl.org), we characterized the IFITM locus in simian and prosimian primate species. Simians consist of haplorhine primates, including hominoids (humans, chimpanzee, gorilla, orangutan, and gibbon), Old World Monkeys [rhesus macaque, olive baboon, and vervet monkey (African Green Monkey)], and New World Monkeys (common marmoset), while prosimians consist of strepsirrhine primates (mouse lemur and bushbaby) as well as a Tarsiiforme relict species (tarsier). This analysis yielded several novel insights into IFITM evolution. First, the “canonical” IFITM gene locus found in humans is atypical with regard to gene number and gene composition. For example, the IFITM locus of the prosimian bushbaby species is syntenic with that of humans, but it does not contain IFITM1 or IFITM2. Instead, it contains two adjacent copies of IFITM3 resulting from gene duplication (a third copy of IFITM3, also the product of gene duplication, is located on a separate chromosome) (Fig 3A). In fact, while the last common primate ancestor (LCPA) is predicted to contain only three IFITM genes, primate evolution is punctuated by IFITM gene gain and gene loss events (Fig 3B). Many of the modern primate genomes analyzed show evidence of IFITM3 gene duplication, while the tarsier contains only a single IFITM gene, and it is IFITM3 (Fig 3B). The number of IFITM3 genes listed for each species is a minimal estimate, as we only considered IFITM3 sequences predicted to be protein coding by Ensembl (sequences containing an intact open reading frame and lacking premature stop codons). These data suggest that IFITM3 is the oldest of the IFITM family members with known antiviral activities. Furthermore, the genomes of hominoids (apes) revealed that IFITM2 is a relatively recent genetic innovation specific to humans, chimpanzee, and gorilla (Fig 3B). Overall, the evolutionary trajectory of IFITM in primates suggests a recurrent need for functional diversification.
Figure 3. Retracing the origins of IFITM in primates.

- A comparison of the IFITM locus on chromosome 11 of the human reference genome (GRCh38.p5) and the corresponding locus in the bushbaby reference genome (OtoGar3) visualized using Genomicus v84.01. Synteny is observed between human IFITM and bushbaby IFITM, as evidenced by the conserved flanking genes B4GALNT4 and ATHL1. The bushbaby locus lacks IFITM2 and IFITM1 genes. In addition, it contains two adjacent IFITM3 genes, as well as a third IFITM3 gene located in a different genomic context, indicative of gene duplication events.
- A gene gain/loss tree summarizing the phylogenetic history of the IFITM gene family in primates, using annotated reference genomes in Ensembl. The number at a branch tip refers to the number of IFITM genes present in a given extant species, while the number at each node refers to the number present in an ancestral species. Differences in gene number at tips and nodes represent significant gene gain events (expansions) or gene loss events (contractions) based on a P‐value calculation of < 0.01 determined by CAFE (Computational Analysis of gene Family Evolution) 55. The makeup of the IFITM repertoire for each species, as determined by comparative genomics and the IFITM gene family tree in Ensembl, is detailed to the right. When multiple copies of IFITM genes were identified, the total number is listed inside the box. The gene numbers estimated for the mouse lemur and bushbaby in Ensembl were adjusted following phylogenetic analysis of IFITM homologs in Genomicus as well as BLAST searches of the NCBI GenBank database. Blue and red arrows indicate the emergence of IFITM1 and IFITM2 genes, respectively.
We used BLAST analysis to confirm the identity of sequences in Ensembl and also to supplement our IFITM3 sequence list with additional species (such as an additional strepsirrhine species (the sifaka) and an Old World Monkey of the Colobinae subfamily, the colobus monkey). From these combined analyses, we compiled an alignment of IFITM3 coding sequences and performed phylogenetic reconstructions (Fig 4A and B, and Appendix Fig S1). Notably, our analysis uncovered recurrent mutation in the amino‐terminus of primate IFITM3, including many examples found in the motifs controlling ubiquitination and endocytosis (selected examples shown in Fig 4A). IFITM3 sequences from prosimians demonstrate that these motifs were likely intact in the common ancestor of all primate species (Fig 4A). However, each one of the strepsirrhine species contains two IFITM3 genes in their respective IFITM locus, and one copy of each pair contains mutations that disrupt motifs for ubiquitination and/or endocytosis. This finding suggests that a tandem duo of IFITM3 genes with potentially divergent functional properties has evolved independently on multiple occasions. Meanwhile, similar observations were made in simian species. It was previously recognized that the marmoset monkey genome contains dozens of IFITM3 sequences in its genome 38. Interestingly, the array of 25 IFITM3 variants present in this species carry many iterations of mutations or deletions that disrupt the motifs necessary for interaction with NEDD4 and/or AP‐2 (Fig 4A and Appendix Fig S1).
Figure 4. Recurrent duplication and divergence of primate IFITM3 over evolutionary time.

- A partial protein alignment of select IFITM3 genes from various primate species for which a reference genome is available and accessible in Genomicus. Sequences from the mouse lemur and colobus monkey were identified by BLAST analysis of reference genomes in NCBI GenBank. Gene location is indicated, when available, by chromosome number, and an asterisk indicates that the given gene sequence falls into a canonical IFITM locus with flanking B4GALNT4 and ATHL1 genes. The suborder and parvorder (from left to right) indicate taxonomical relationships between species. Residues bolded in red highlight nonsynonymous substitutions that disrupt canonical motifs for the recruitment of NEDD4 and AP‐2. A complete protein alignment of all IFITM sequences analyzed in this study is available in Appendix.
- A phylogenetic gene tree (cladogram) indicates the relationship among IFITM3 sequences analyzed in this study. The final consensus tree was made by merging results from neighbor joining and maximum‐likelihood phylogenetic reconstructions using both nucleotide and amino acid sequences in Ensembl (see Fig EV4 for robustness measurement by Bayesian inference). Branch lengths are normalized and arbitrary. The size of triangles at the tree tips roughly corresponds to the number of sequences contained therein. Note that the IFITM2 gene is the most recent member of the IFITM locus, appearing only in humans, chimpanzee, and gorilla genomes.
- A graphical representation of naturally occurring truncated forms of IFITM3 identified in the vervet and marmoset reference genomes, as compared to the putative Δ1–21 variant in humans.
These cases of IFITM3 duplication and divergence were not isolated to far‐removed primate species, since we also found similar patterns in the catarrhine parvorder to which humans belong. We identified a single IFITM3 sequence in the colobus monkey that contains a deletion (Δ22–24) eliminating the motif necessary for AP‐2‐mediated endocytosis as well as a neighboring lysine residue (K24) targeted for ubiquitination 27 (Fig 4A). Furthermore, codons 17 and 18 of IFITM3, corresponding to the critical proline residues necessary for its interaction with and ubiquitination by NEDD4 31, have diversified in IFITM3 genes in the vervet, gorilla, and orangutan. This latter species contains an IFITM3 that is syntenic with human IFITM3 on chromosome 11 and contains a P17L mutation. Phylogenetic analysis of primate IFITM3 showed branching patterns that reproduce major taxonomic groups, with a few exceptions (Figs 4B and EV4A). While IFITM3 from prosimians clearly represents the basal clade, some IFITM3 sequences found in gorilla, vervet, and orangutan genomes form two outlier branches (in gray). These sequences appear to predate other IFITM3 sequences found in the same species, suggesting that they most likely represent duplication events that took place in simian ancestors. Importantly, the maintenance of these sequences in multiple species until present day may indicate that they retain functional importance (Figs 4B and EV4A).
Figure EV4. A Bayesian phylogenetic reconstruction of primate IFITM3 and IFITM2 .

- A phylogenetic tree based on select IFITM3 and IFITM2 genes aligned pairwise using T‐Coffee. Ambiguous regions, including gaps, were resolved with Gblocks. The tree was reconstructed using the Bayesian inference method implemented in the MrBayes program (v3.2.3). The number of substitution types was fixed to 6. The standard (4 × 4) model of nucleotide substitutions was used, with rates variation across sites fixed to “invariable + gamma”. Four Markov chain Monte Carlo (MCMC) chains were run for 100,000 generations, with sampling every 10 generations and the first 500 sampled trees discarded as “burn‐in”. A 50% majority rule was used for the placement of ancestral nodes. Branch annotations are Bayesian posterior probabilities used as a measure of tree robustness. Sequences in bold are those discussed in Fig 4 and functionally characterized in Fig 5.
- The full proteins of putative human IFITM3 variant Δ1–21, marmoset IFITM3 ENSCJAG00000037392, and vervet IFITM3 ENSCSAG00000019450 were aligned via Clustal Omega.
Since we found that many naturally occurring mutations identified in primate IFITM3 effectively mimic the synthetic mutants tested in this study, we predicted that they might similarly confer escape from the cellular processes of ubiquitination and/or endocytosis. A further observation that is especially pertinent to the data shown in this study is that certain IFITM3 sequences identified in the vervet and marmoset genome closely resemble the human Δ1–21 construct, with each lacking the amino‐terminus and initiating translation at an internal methionine (Fig 4C). In addition, we observed a few IFITM3 sequences in which the initial methionine is absent or mutated (Appendix Fig S1), which could also give rise to truncated IFITM3 proteins similar to the Δ1–21 construct.
Naturally occurring mutations in IFITM3 disrupt regulatory motifs and toggle antiviral activity
We next characterized the functional consequences of select nonhuman IFITM3 mutations by introducing them into the human IFITM3 background. Transfection into 293T cells and confocal immunofluorescence microscopy showed that Δ22–24 resulted in a shift to the cell surface (Fig 5A). Predominant plasma membrane staining was also observed for marmoset‐specific IFITM3 mutations, reiterating an effect on protein endocytosis through ablation of the AP‐2‐recruiting motif (Fig 5A). As expected, primate mutations in the NEDD4‐binding motif (P17L found in orangutan and P18H found in vervet) did not grossly affect the subcellular localization of IFITM3. To assess the extent to which each variant could be ubiquitinated in cells, we immunoprecipitated IFITM3 from cells co‐expressing NEDD4 or catalytically inactive NEDD4 C867A. In agreement with a previous report 31, ubiquitinated forms of human full‐length IFITM3 can be detected following the overexpression of NEDD4, while the Δ1–21 variant (which lacks the NEDD4‐binding motif) was less ubiquitinated (Fig 5B). The P17L and P18H mutations also led to lower levels of ubiquitination, especially pools modified with two or three ubiquitin moieties (as indicated by ** and ***, respectively). The IFITM3‐specific antibody used here recognized all but the Δ22–24 variant. We confirmed the presence of ubiquitin using an antibody capable of recognizing mono‐ and polyubiquitinated proteins (Fig 5B, colored image). The direct labeling of ubiquitin showed that the Δ22–24 variant is modified by NEDD4 to an intermediate extent, since it contains an intact NEDD4‐binding site yet lacks one nearby lysine residue. Ubiquitination was also detected in the presence of catalytically inactive NEDD4 C867A, albeit at lower levels (Fig 5B). These results reveal that the natural P17L, P18H, and Δ22–24 mutations prevent normal protein ubiquitination not only by ectopic NEDD4, but also by endogenous E3 ligases in 293T cells. Examination of whole‐cell lysate fractions confirmed the expression of all IFITM3 variants (Fig 5C). In accordance with the degree of ubiquitination observed in the immunoprecipitated fractions, certain NEDD4‐sensitive variants of IFITM3 were detected at lower levels (relative to the inactive NEDD4 C867A control). Notably, these naturally occurring mutations that disrupt subcellular localization (Fig 5A) and/or diminish NEDD4‐mediated ubiquitination (Fig 5B) also exhibit enhanced restriction of HIV‐1 virion infectivity relative to full‐length human IFITM3 (Fig 5D).
Figure 5. Natural mutations in IFITM3 disrupt regulatory motifs and toggle antiviral activity.

- Confocal fluorescence microscopy of 293T cells transfected with 0.5 μg of pQCXIP‐FLAG‐IFITM3 mutated at the indicated residues and immunostained with anti‐FLAG M2 antibody. Scale bars, 5 μm. The P17L, P18H, Δ22–24, P18L/L23P, and P18H/Δ19–23 mutations were introduced into the human IFITM3 background.
- SDS–PAGE of FLAG‐immunoprecipitated fractions from 293T cells following transfection of 0.5 μg of pQCXIP‐FLAG‐IFITM3 variants and 0.5 μg of pCI‐HA‐NEDD4 or pCI‐HA‐NEDD4 C867A. Immunoblotting was performed with anti‐FLAG M2, anti‐IFITM3 (Abcam, EPR5242), and an anti‐ubiquitin antibody (Enzo Life Sciences, recognizing K29‐, K48‐, and K63‐linked mono‐ and polyubiquitinylated proteins). “L” indicates the presence of light chain antibody derived from the FLAG antibody‐coupled beads used for immunoprecipitation. *, **, and *** indicate mono‐, di‐, and triubiquitinated forms of IFITM3.
- SDS–PAGE of whole‐cell lysate input fractions from 293T cells used in (B). Immunoblotting was performed with anti‐FLAG M2, anti‐NEDD4, and anti‐actin.
- 293T cells were transfected with 1.0 μg of pNL4‐3 and 0.5 μg of pQCXIP‐FLAG‐IFITM3 variants. Virus‐containing supernatants were harvested and 10 ng p24 equivalents were used to reinfect fresh SupT1 T cells. Productive infection was scored by immunostaining with the KC57 antibody and flow cytometry at 48 h postinfection. Mean + SD of three experiments is shown.
- About 2.0 × 105 cells were seeded and challenged with 10 ng of HIV‐1 NL4‐3 in the presence of 2 μg/ml DEAE‐dextran. Productive infection was scored by immunostaining with KC57 and flow cytometry at 48 h postinfection. The “Colobus native” sequence represents the full IFITM3 sequence identified in the colobus monkey.
- As in (E), except that cells were challenged with influenza A virus (H1N1 PR/8/34, Charles River Laboratories), at a dose that resulted in ˜50% infection of control (Empty) cells (equivalent to 103 TCID50/0.2 ml). Infection was scored by immunostaining with an anti‐IAV NP antibody and flow cytometry at 18 h postinfection. Results are presented in a logarithmic scale. Mean + SD of 3–5 experiments is shown.
We previously showed that IFITM3 performs two antiviral roles in the setting of HIV‐1 infection: (i) inhibition of virus particle infectivity within infected cells (donor cell effect) and (ii) protection of uninfected cells from incoming cell‐free virus particles (target cell effect) 21. To measure the functional impact of IFITM3 variation in both of its antiviral roles, we tested the ability of primate IFITM3 variants to inhibit HIV‐1 entry when expressed in uninfected target cells. To this end, we achieved the stable expression of IFITM3 variants in 293T cells transduced with CD4 and CXCR4 (293T 4 × 4 cells) 40 and tested their relative resistance to infection by HIV‐1. Importantly, we achieved similar levels of protein expression across all conditions (Fig EV5A), and the differential subcellular localization displayed between IFITM3 variants was maintained (Fig EV5B). When these cells were challenged with HIV‐1, synthetic and natural IFITM3 variants containing amino‐terminus mutations conferred elevated resistance to incoming HIV‐1 relative to human IFITM3 (Fig 5E). The greatest degree of virus restriction was achieved by mutants with the enhanced surface localization (ΔY20, Δ1–21, and Δ22–24).
Figure EV5. Further characterization of 293T 4 × 4 cells stably expressing IFITM proteins.
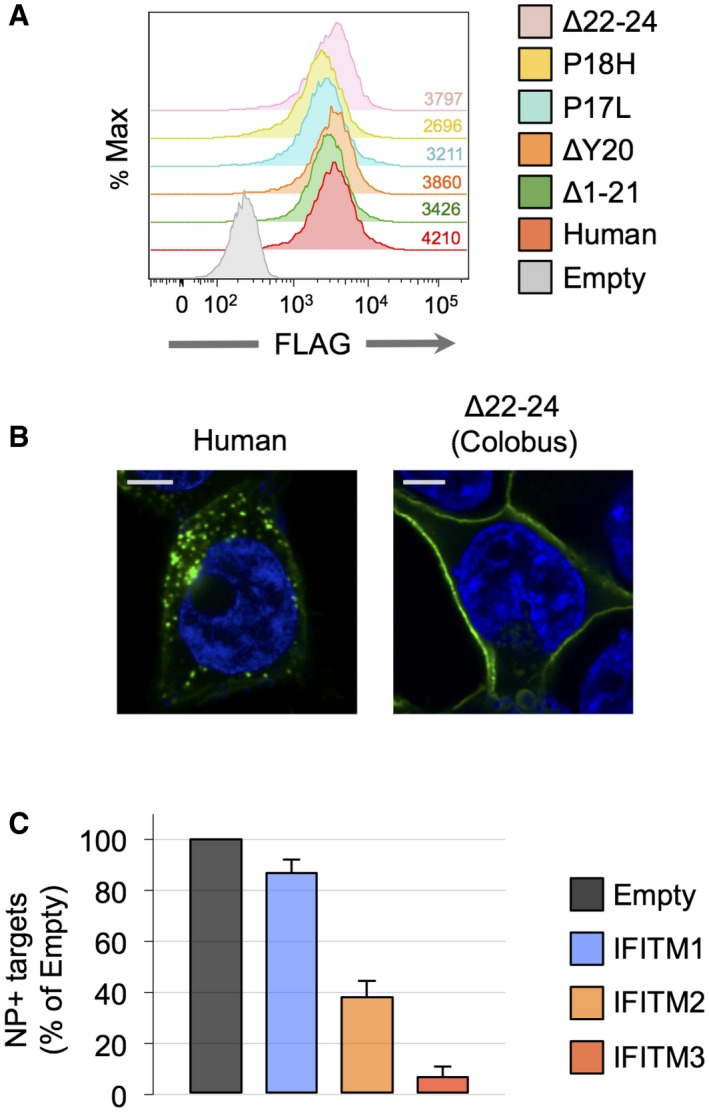
- 293T cells transduced to express CD4 and CXCR4 were transfected with 0.5 μg of pQCXIP‐FLAG‐IFITM3 variants, and stable expression was selected using puromycin. Corresponding mean fluorescence intensity values are shown.
- The subcellular localization of IFITM3 variants expressed in 293T following stable transfection was assessed by confocal immunofluorescence microscopy. Representative images of two variants, human IFITM3 and human IFITM3 encoding the Δ22–24 deletion, are shown.
- 293T cells were transfected with 0.5 μg of pQCXIP‐FLAG‐IFITM1, 2, or 3 and stable expression was selected using puromycin; 105 cells were seeded and challenged with influenza A virus (H1N1 PR/8/34, Charles River Laboratories), at a dose that resulted in ˜50% infection of control (Empty) cells (equivalent to 103 TCID50/0.2 ml). Infection was scored by immunostaining with an anti‐IAV NP antibody and flow cytometry at 18 h postinfection. Mean + SD of three experiments is shown.
Since synthetic mutations in IFITM3 that affect regulation by NEDD4 and the AP‐2 complex have previously been shown to alter sensitivity to RNA viruses such as IAV and VSV, we also examined the extent of IAV entry restriction in the same 293T 4 × 4 cell lines. In line with previous findings 9, we confirmed that IFITM3 is a more potent inhibitor of IAV compared to IFITM1 and IFITM2 (Fig EV5C). While human IFITM3 restricts IAV infection by nearly 50‐fold, ΔY20 and the Δ1–21 variant are markedly less active (Fig 5F), consistent with previous studies 20. Furthermore, while IFITM3 carrying the P17L and P18H mutations retained the potent restriction of IAV, the Δ22–24 deletion specific to colobus monkey IFITM3 reduces anti‐IAV activity by approximately 20‐fold (Fig 5G). We also tested the antiviral activity of a full‐length “native” colobus IFITM3 protein, which encodes all amino acid substitutions that differ from human IFITM3 (Appendix Fig S1), and the results were similar (Fig 5G). These findings suggest that the functionally informative mutations that govern antiviral potency and specificity are primarily those found among the NEDD4‐ and AP2‐binding motifs.
These results demonstrate that select IFITM3 sequences derived from nonhuman primates exhibit elevated activity against HIV‐1 when expressed in either donor or target cells, suggesting that naturally occurring mutation has led to IFITM3 variants with enhanced anti‐lentiviral potential. However, we show that mutations arising in IFITM3 have opposing effects against HIV‐1 and IAV, suggesting a protective trade‐off for host antiviral immunity.
Discussion
Despite the conserved antiviral activities displayed by immune‐related IFITM proteins, regulatory mechanisms are in place to control their abundance and localization in the cell, allowing for a fine‐tuning of the antiviral response. This study identifies the determinants that regulate the potency and specificity with which IFITM3 proteins inhibit HIV‐1 infection. We show that post‐translational modifications targeting the amino‐terminus of IFITM3 impact antiviral activity, and furthermore, the sequence motifs found therein may be subject to natural selection over evolutionary time. Together, these data provide a functional rationale for the recurrent duplication of IFITM genes observed in vertebrate genomes. The duplication and subsequent divergence of IFITM genes may allow them to be conferred with novel cellular or antiviral functions, a process known as neo‐functionalization. Following gene duplication, diversification of IFITM3 coding sequence via alternative splicing (in the case of the putative Δ1–21 form in humans) or by germ line‐encoded codon substitution mutations (in the case of nonhuman primates) may allow the cell to expand its antiviral arsenal and to protect different compartments from viral invaders.
Specifically, our findings suggest that synthetic and naturally occurring mutation of single amino acid residues allows IFITM3 to escape regulation by the E3 ubiquitin ligase NEDD4 and the endocytosis adaptor complex AP‐2. However, our description of natural mutations in primate IFITM3 sequences offers only a first glimpse at the potential variation that exists. It was previously reported that the IFITM3 locus is duplicated or polymorphic in the rhesus macaque and the African Green Monkey 18, 41. Due to a lack of complete genome sequences, we did not assess IFITM3 variation in additional members of the primate family Cercopithicinae, which comprises most of the monkey species that naturally harbor SIV (and other virus) infections in the wild. Previous reports have demonstrated that these species carry adaptive mutations in restriction factor genes APOBEC3G 42, 43, SAMHD1 44, and TRIM5α 45, 46, which impact lentivirus infection in vitro and in vivo. Thus, cloning of further IFITM3 genes in diverse species is warranted and may reveal other instances of functional diversification. Future experiments focused on the NEDD4–IFITM3 interaction need to be performed in cells from different tissue types and from different species. For example, while we show that certain primate genomes contain IFITM3 genes in which the NEDD4‐binding site (PPxY) is altered, experiments were performed in human cells (and thus with human NEDD4). It is unknown whether NEDD4 from different species targets the same structural motif as human NEDD4. The prospect that NEDD4 E3 ligases in different species exhibit different substrate specificities is improbable, however, because the NEDD4 homolog in yeast, Rsp5, also targets substrates bearing the same PPxY signature 47. A departure from this expectation would be exciting, however, since species‐specific NEDD4 activities may be indicative of coevolution between NEDD4 and its cellular substrates in a novel form of genetic conflict 37. Similar questions regarding the cell‐ or species‐specific function of the endocytic adaptor complex, AP‐2, must also be addressed.
Previous publications examining the antiviral activity of IFITM3 mutants demonstrated that the hypervariable amino‐terminus governs its subcellular localization and, as a result, antiviral activity against IAV. As such, variants of IFITM3 carrying Δ1–21 or ΔY20 were deemed “defective” or “nonfunctional” 20. On the contrary, we report here that these and other mutations to the amino‐terminus of IFITM3 retain and even enhance antiviral function during HIV infection. Furthermore, the use of these hyperactive mutants allowed us to assess the involvement of two distinct, but not mutually exclusive, mechanisms underlying this restriction activity: (i) IFITM3 incorporation into viral membranes and (ii) inhibition of HIV‐1 Env maturation and virion incorporation. While our results in 293T cells show that IFITM3 overexpression can lead to slightly decreased gp120 levels in virus‐producing cells, as proposed by Yu et al 23, we do not observe this phenomenon when IFITM3 is induced in T cells. Previously published studies illustrating an effect of IFITM proteins on the synthesis and maturation of many lentiviral proteins 48, including Gag 17, and even other cellular proteins 49, raise the possibility that IFITM proteins, when overexpressed, promote wholesale changes that may affect protein trafficking in general. Furthermore, an HIV‐1 Env‐specific effect on restriction activity cannot explain the finding that VSV‐G pseudotyping of virions produced in the presence of IFITM proteins does not rescue their infectivity 22. Overall, while changes to Env may be partially responsible for restriction in 293T cells, our results suggest that the situation may be different in lymphocytes.
The roles played by human IFITM3 in susceptibility to infection have been actively researched, yet many questions remain. Numerous studies have drawn an association between the human SNP rs12252‐C in IFITM3 and a severe outcome to IAV infection, yet the protein variant (Δ1–21) predicted to derive from this SNP has not been directly detected in human cells. Paradoxically, a study has recently reported an association between rs12252‐C and rapid progression to AIDS in HIV‐infected individuals in China 50. Since we show here that the Δ1–21 variant exhibits an elevated anti‐HIV‐1 activity in both of its antiviral roles when ectopically expressed in human cells, efforts are needed to establish a direct functional link between rs12252‐C and the truncated protein variant. Importantly, other polymorphisms exist in human IFITM3, such as one in the promoter that is associated with susceptibility to tuberculosis 51, further experimental advances will be necessary to directly assess the relationship between genotype, IFITM3 protein, and disease susceptibility. There is evidence for recent positive selection in human IFITM3 20, so a scenario whereby “hitchhiking” (linkage disequilibrium) occurs between neighboring SNPs must also be considered.
While mutations to the NEDD4‐binding motif (PPxY) did not interfere with anti‐IAV function, our description of single amino acid changes and deletions disrupting the endocytosis motif (YxxL) demonstrates that some naturally occurring mutations carry a cost in the form of a protective trade‐off. The recurrent appearance of similar mutations in different primate lineages may reflect the presence of a common selective pressure in the past and may even reveal the nature of the selective pressure itself. Our results suggest that certain nonhuman primate IFITM3 may exhibit greater activity against HIV‐1, suggesting that evolution may have fine‐tuned their function against lentiviruses or other viruses entering and exiting from the cellular plasma membrane. Future studies examining the activities of primate IFITM3 proteins against their autologous SIV strains will be important for confirming their anti‐lentiviral potential and may reveal viral countermeasures that allow antagonism or evasion of IFITM proteins. Regardless, this enhancement of function against lentiviruses comes with a loss of activity against other viruses entering at endosomes, like IAV and VSV. Considering that IFITM3 is critical to the antiviral response to IAV in mice and humans 19, 20, the appearance of mutations in nonhuman IFITM3 that suppress this activity is surprising. Thus, our observation that many primate genomes contain multiple IFITM3 genes may reflect an evolutionary mechanism to provide balanced antiviral protection to the cell. Since some species covered in this study, such as the orangutan, the gorilla, and the common marmoset, have been studied with regard to susceptibility to human virus infections 52, 53 it will be of great interest to address how IFITM3 genotype and gene copy number impact in vivo virus replication and transmission.
Materials and Methods
Cells, viruses, and reagents
HEK293T cells (293T, ATCC) and derivatives were cultured in DMEM supplemented with 10% FBS and 1% penicillin–streptomycin. Primary CD4+ T cells and HeLa cells were treated with type‐I interferon as previously described 21. 293T 4 × 4 cells were generated by lentiviral transduction with constructs encoding CD4 and CXCR4, as previously described 40. pQCXIP plasmids encoding IFITM3 variants with amino‐terminal FLAG (FLAG‐IFITM3) were previously described 16, 17, and additional mutants were generated by site‐directed mutagenesis. 293T cells were transfected with Metafectene (Biontex) or Lipofectamine 2000 (Invitrogen) reagent. Tet‐ON SupT1 cells were treated with doxycycline (500 ng/ml) overnight to induce FLAG‐IFITM3 variants as previously described 16, 17, 21. Stocks of HIV‐1 were produced via CaPO4 transfection of 293T cells with pNL4‐3. HIV‐1 incorporating Vpr‐B‐lactamase (Vpr‐Blam) fusion protein was produced via co‐transfection of pNL4‐3, pVpr‐Blam, and pQCXIP encoding IFITM3 variants or an empty control or pCMV‐MxA, at a respective plasmid ratio of 3:1.5:1. HIV‐1 produced from Tet‐ON SupT1 cell lines was produced by first infecting the cells with VSV‐G‐pseudotyped NL4‐3 in the presence of DEAE‐dextran (2 μg/ml) followed by overnight doxycycline treatment to induce IFITM3 protein expression.
Western blot
Cells were lysed in 1% Triton‐X/PBS in the presence of protease inhibitor cocktail (cOmplete, Roche) and clarified by centrifugation. Virions were purified from supernatants by overlaying on a 6% OptiPrep cushion and pelleted by ultracentrifugation (50,000 × g, 4°C) for 1 h. An aliquot was removed for p24 Gag ELISA, and normalized amounts were loaded on a Bis–Tris 12% acrylamide SDS–PAGE gel. The following antibodies were used: mouse anti‐FLAG‐M2 (Sigma), mouse anti‐IFITM3 (Proteintech, 66081‐1‐Ig), rabbit anti‐IFITM3 (Abcam, EPR5242), mouse anti‐Gag p24 (183‐H12‐5C, NIH #1513), sheep anti‐Env gp120 (NIH #288), mouse anti‐HA (Covance, HA.11 clone 16B12), rabbit anti‐actin (Santa Cruz Biotechnology), mouse anti‐tubulin (Santa Cruz Biotechnology) and mouse anti‐ubiquitin antibody (FK2, Enzo Life Sciences, recognizing K29‐, K48‐, and K63‐linked mono‐ and polyubiquitinated proteins). DyLight‐coupled secondary antibodies (Thermo Fisher) were used for protein detection on a Li‐Cor Odyssey imaging system.
Flow cytometry
Cells were fixed in 4% PFA and permeabilized with 0.5% saponin. The same antibodies used for Western blot were also used for flow cytometry. In addition, the mouse anti‐Gag KC57 (Beckman Coulter) and mouse anti‐influenza A group (NP) (Argene) antibodies were used to score HIV‐1 and IAV infections, respectively. FLAG M2‐PE (Abcam) was used to measure the expression of FLAG‐IFITM3 from pQCXIP. Acquisition was performed on a FACS Canto II (Becton Dickinson).
Confocal immunofluorescence microscopy
Transfected 293T cells were cultured on 12‐mm glass coverslips, fixed in 4% PFA, and permeabilized with 0.5% saponin. The same antibodies used for Western blot were also used for confocal microscopy. In addition, the mouse anti‐p17 Gag (ARP342, AIDS Reagents from Programme EVA Centre) and the mouse anti‐LAMP1 (clone H4A3, Santa Cruz Biotechnology) antibodies were used. Analysis was carried out on a Zeiss LSM700 using a 63× objective. Images were analyzed using FIJI software and assembled with the Magic Montage plugin.
Vpr‐Blam virus fusion assay
A total of 25 ng p24 equivalent of Vpr‐Blam‐containing virions was incubated with 1 × 105 SupT1 cells. After 2 h, the cells were washed and incubated with the CCF2‐AM substrate (CCF2‐AM β‐lactamase loading kit, Invitrogen) for an additional 2 h. Following fixation in 4% PFA, cleavage of CCF2‐AM was measured by flow cytometry.
IFITM3 co‐immunoprecipitation and ubiquitination assays
About 5 × 105 293T cells were transfected with 0.5 μg pCl‐HA‐NEDD4 or pCI‐HA‐NEDD4 C867A and 0.2 μg pQCXIP encoding FLAG‐tagged IFITM3 variants. The cells were harvested at 24 h post‐transfection and lysed for 30 min on ice in a lysis buffer consisting of 50 mM Tris, 300 mM NaCl, 1 mM EDTA, 1% NP‐40, H2O, and a protease inhibitor cocktail. Cell lysates were incubated with prewashed FLAG‐M2 agarose beads (Sigma) for 2 h at 4°C. The beads were washed and mixed directly with SDS–PAGE loading buffer (2× Laemmli plus reducing agent). Samples were then denatured by heating at 95°C for 5 min and stored at −20°C until ready for SDS–PAGE loading.
Purification of HIV‐1 particles by velocity gradients
About 7 × 106 Tet‐ON SupT1 cells (encoding inducible FLAG‐IFITM3 or FLAG‐IFITM3 Δ1–21) were infected with HIV‐1 NL4‐3 to achieve approximately 50% Gag+ cells. IFITM3 variants were induced by doxycycline addition (500 ng/ml). After two additional days, supernatants from HIV‐1‐infected, IFITM3+ cells were filtered on a 0.22‐μm filter. Clarified supernatants were ultracentrifuged at 70,000 × g for 1 h. Pellets containing both cellular microvesicles and viral particles were then suspended in PBS and subjected to second ultracentrifugation at 200,000 × g for 1.5 h through a discontinuous 6–18% OptiPrep (iodixanol) gradient, stratified by increments of 1.2%. Ten to eleven fractions were collected and further ultracentrifuged at 435,000 × g for 30 min to concentrate the samples. Pellets were resuspended in 30 μl of 1% Triton‐X lysis buffer supplemented with protease inhibitors and directly loaded on a 4–12% acrylamide Bis–Tris SDS–PAGE gel.
Primate IFITM3 sequence retrieval, molecular analysis, and cloning
IFITM sequences were assembled from reference genome sequences for the following species: mouse lemur (Microcebus murinus; micMur1), bushbaby (Otolemur garnettii; OtoGar3), tarsier (Tarsius syrichta; tarSyr1), common marmoset (Callithrix jacchus; C_jacchus3.2.1), rhesus macaque (Macaca mulatta; MMUL 1.0), olive baboon (Papio anubis; PapAnu2.0), vervet (African Green Monkey‐sabeus) (Chlorocebus sabaeus; ChlSab1.1), orangutan (Pongo abelii; PPYG2), gorilla (Gorilla gorilla gorilla; gorGor3.1), chimpanzee (Pan troglodytes; CHIMP2.1.4), humans (Homo sapiens; GRCh38.p5), and gibbon (Nomascus leucogenys; Nleu1.0) using Genomicus v84.01 and Ensembl (www.ensembl.org), with additional sequences provided by BLASTN or BLASTP query in NCBI GenBank. Nucleotide and protein sequence alignments were performed using T‐Coffee or Clustal Omega and were adjusted manually in CLC Sequence Viewer. Phylogenetic analysis was performed using the IFITM gene tree function in Ensembl, and the tree visualized consisted of a final consensus tree constructed using distance and maximum‐likelihood methods of nucleotide and amino acid sequences. Further phylogenetic analysis was performed using the Bayesian inference method implemented in the MrBayes program (v3.2.3) 54. Mutations were introduced into the human IFITM3 sequence by site‐directed mutagenesis, while select primate IFITM3 sequences were directly synthesized by Eurofins Genomics and cloned into the BamHI/EcoRI sites of pQCXIP.
Author contributions
AAC, CL, JSY, and OS conceived the study. AAC, NR, FP, AB, and NC performed the experiments and data analysis. AAC and OS wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Acknowledgements
We thank Matthew Daugherty and members of the Virus & Immunity Unit for critical reading of the manuscript and Lluis Quintana‐Murci, Helene Quach, Guillaume Laval, and Jean‐François Zagury for helpful discussions. Work was supported by grants from the Agence Nationale de Recherches sur la Sida et les Hépatites Virales (ANRS), Sidaction, the LabEx IBEID program, the Investissements d'Avenir program, and the Pasteur Foundation.
EMBO Reports (2016) 17: 1657–1671
Contributor Information
Alex A Compton, Email: alex.compton@pasteur.fr.
Olivier Schwartz, Email: schwartz@pasteur.fr.
References
- 1. Bushman FD, Malani N, Fernandes J, D'Orso I, Cagney G, Diamond TL, Zhou H, Hazuda DJ, Espeseth AS, Konig R et al (2009) Host cell factors in HIV replication: meta‐analysis of genome‐wide studies. PLoS Pathog 5: e1000437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Compton AA, Malik HS, Emerman M (2013) Host gene evolution traces the evolutionary history of ancient primate lentiviruses. Philos Trans R Soc B Biol Sci 368: 20120496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chesarino NM, McMichael TM, Hach JC, Yount JS (2014) Phosphorylation of the antiviral protein IFITM3 dually regulates its endocytosis and ubiquitination. J Biol Chem 289: 11986–11992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sällman Almén M, Bringeland N, Fredriksson R, Schiöth HB (2012) The dispanins: a novel gene family of ancient origin that contains 14 human members. PLoS One 7: e31961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blyth GAD, Chan W‐F, Webster RG, Magor KE (2015) Duck IFITM3 mediates restriction of influenza viruses. J Virol 90: 103–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith SE, Gibson MS, Wash RS, Ferrara F, Wright E, Temperton N, Kellam P, Fife M (2013) Chicken interferon‐inducible transmembrane protein 3 restricts influenza viruses and lyssaviruses in vitro . J Virol 87: 12957–12966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhu R, Wang J, Lei X‐Y, Gui J‐F, Zhang Q‐Y (2013) Evidence for Paralichthys olivaceus IFITM1 antiviral effect by impeding viral entry into target cells. Fish Shellfish Immunol 35: 918–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Melvin W, McMichael T, Chesarino N, Hach J, Yount J (2015) IFITMs from mycobacteria confer resistance to influenza virus when expressed in human cells. Viruses 7: 3035–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E et al (2009) The IFITM proteins mediate cellular resistance to influenza a H1N1 virus, West Nile virus, and dengue virus. Cell 139: 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang IC, Bailey CC, Weyer JL, Radoshitzky SR, Becker MM, Chiang JJ, Brass AL, Ahmed AA, Chi X, Dong L et al (2011) Distinct patterns of IFITM‐mediated restriction of filoviruses, SARS coronavirus, and influenza a virus. PLoS Pathog 7: e1001258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Desai TM, Marin M, Chin CR, Savidis G, Brass AL, Melikyan GB (2014) IFITM3 restricts influenza a virus entry by blocking the formation of fusion pores following virus‐endosome hemifusion. PLoS Pathog 10: e1004048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li K, Markosyan RM, Zheng YM, Golfetto O, Bungart B, Li M, Ding S, He Y, Liang C, Lee JC et al (2013) IFITM proteins restrict viral membrane hemifusion. PLoS Pathog 9: e1003124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin TY, Chin CR, Everitt AR, Clare S, Perreira JM, Savidis G, Aker AM, John SP, Sarlah D, Carreira EM et al (2013) Amphotericin B increases influenza a virus infection by preventing IFITM3‐mediated restriction. Cell Rep 5: 895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Amini‐Bavil‐Olyaee S, Choi YJ, Lee JH, Shi M, Huang IC, Farzan M, Jung JU (2013) The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe 13: 452–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Perreira JM, Chin CR, Feeley EM, Brass AL (2013) IFITMs restrict the replication of multiple pathogenic viruses. J Mol Biol 425: 4937–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jia R, Pan Q, Ding S, Rong L, Liu SL, Geng Y, Qiao W, Liang C (2012) The N‐terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. J Virol 86: 13697–13707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lu J, Pan Q, Rong L, Liu SL, Liang C (2011) The IFITM proteins inhibit HIV‐1 infection. J Virol 85: 2126–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qian J, Le Duff Y, Wang Y, Pan Q, Ding S, Zheng YM, Liu SL, Liang C (2015) Primate lentiviruses are differentially inhibited by interferon‐induced transmembrane proteins. Virology 474: 10–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bailey CC, Huang IC, Kam C, Farzan M (2012) Ifitm3 limits the severity of acute influenza in mice. PLoS Pathog 8: e1002909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, Chin CR, Feeley EM, Sims JS, Adams DJ et al (2012) IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484: 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Compton AA, Bruel T, Porrot F, Mallet A, Sachse M, Euvrard M, Liang C, Casartelli N, Schwartz O (2014) IFITM proteins incorporated into HIV‐1 virions impair viral fusion and spread. Cell Host Microbe 16: 736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tartour K, Appourchaux R, Gaillard J, Nguyen XN, Durand S, Turpin J, Beaumont E, Roch E, Berger G, Mahieux R et al (2014) IFITM proteins are incorporated onto HIV‐1 virion particles and negatively imprint their infectivity. Retrovirology 11: 103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu J, Li M, Wilkins J, Ding S, Swartz TH, Esposito AM, Zheng YM, Freed EO, Liang C, Chen BK et al (2015) IFITM proteins restrict HIV‐1 infection by antagonizing the envelope glycoprotein. Cell Rep 13: 145–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smith SE, Weston S, Kellam P, Marsh M (2014) IFITM proteins—cellular inhibitors of viral entry. Curr Opin Virol 4: 71–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chesarino NM, McMichael TM, Yount JS (2014) Regulation of the trafficking and antiviral activity of IFITM3 by post‐translational modifications. Future Microbiol 9: 1151–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chesarino NM, McMichael TM, Hach JC, Yount JS (2014) Phosphorylation of the antiviral protein interferon‐inducible transmembrane protein 3 (IFITM3) dually regulates its endocytosis and ubiquitination. J Biol Chem 289: 11986–11992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yount JS, Karssemeijer RA, Hang HC (2012) S‐palmitoylation and ubiquitination differentially regulate interferon‐induced transmembrane protein 3 (IFITM3)‐mediated resistance to influenza virus. J Biol Chem 287: 19631–19641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yount JS, Moltedo B, Yang YY, Charron G, Moran TM, López CB, Hang HC (2010) Palmitoylome profiling reveals S‐palmitoylation‐dependent antiviral activity of IFITM3. Nat Chem Biol 6: 610–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shan Z, Han Q, Nie J, Cao X, Chen Z, Yin S, Gao Y, Lin F, Zhou X, Xu K et al (2013) Negative regulation of interferon‐induced transmembrane protein 3 by SET7‐mediated lysine monomethylation. J Biol Chem 288: 35093–35103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jia R, Xu F, Qian J, Yao Y, Miao C, Zheng YM, Liu SL, Guo F, Geng Y, Qiao W et al (2014) Identification of an endocytic signal essential for the antiviral action of IFITM3. Cell Microbiol 16: 1080–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chesarino NM, McMichael TM, Yount JS (2015) E3 ubiquitin ligase NEDD4 promotes influenza virus infection by decreasing levels of the antiviral protein IFITM3. PLoS Pathog 11: e1005095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang YH, Zhao Y, Li N, Peng YC, Giannoulatou E, Jin RH, Yan HP, Wu H, Liu JH, Liu N et al (2013) Interferon‐induced transmembrane protein‐3 genetic variant rs12252‐C is associated with severe influenza in Chinese individuals. Nat Commun 4: 1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xuan Y, Wang LN, Li W, Zi HR, Guo Y, Yan WJ, Chen XB, Wei PM (2015) IFITM3 rs12252 T>C polymorphism is associated with the risk of severe influenza: a meta‐analysis. Epidemiol Infect 143: 2975–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Z, Zhang A, Wan Y, Liu X, Qiu C, Xi X, Ren Y, Wang J, Dong Y, Bao M et al (2014) Early hypercytokinemia is associated with interferon‐induced transmembrane protein‐3 dysfunction and predictive of fatal H7N9 infection. Proc Natl Acad Sci USA 111: 769–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Herold N, Anders‐Osswein M, Glass B, Eckhardt M, Muller B, Krausslich HG (2014) HIV‐1 entry in SupT1‐R5, CEM‐ss, and primary CD4+ T cells occurs at the plasma membrane and does not require endocytosis. J Virol 88: 13956–13970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhu X, He Z, Yuan J, Wen W, Huang X, Hu Y, Lin C, Pan J, Li R, Deng H et al (2014) IFITM3‐containing exosome as a novel mediator for anti‐viral response in dengue virus infection. Cell Microbiol 17: 105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Duggal NK, Emerman M (2012) Evolutionary conflicts between viruses and restriction factors shape immunity. Nat Rev Immunol 12: 687–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Z, Liu J, Li M, Yang H, Zhang C (2012) Evolutionary dynamics of the interferon‐induced transmembrane gene family in vertebrates. PLoS One 7: e49265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Louis A, Muffato M, Roest Crollius H (2013) Genomicus: five genome browsers for comparative genomics in eukaryota. Nucleic Acids Res 41 (Database issue): D700–D705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lepelley A, Louis S, Sourisseau M, Law H, Pothlichet J, Schilte C, Chaperot L, Plumas J, Randall RE, Si‐Tahar M et al (2011) Innate sensing of HIV‐infected cells. PLoS Pathog 7: e1001284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wrensch F, Karsten CB, Gnirss K, Hoffmann M, Lu K, Takada A, Winkler M, Simmons G, Pohlmann S (2015) Interferon‐induced transmembrane protein‐mediated inhibition of host cell entry of ebolaviruses. J Infect Dis 212(Suppl. 2): S210–S218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Compton AA, Emerman M (2013) Convergence and divergence in the evolution of the APOBEC3G‐Vif interaction reveal ancient origins of simian immunodeficiency viruses. PLoS Pathog 9: e1003135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Krupp A, McCarthy KR, Ooms M, Letko M, Morgan JS, Simon V, Johnson WE (2013) APOBEC3G polymorphism as a selective barrier to cross‐species transmission and emergence of pathogenic SIV and AIDS in a primate host. PLoS Pathog 9: e1003641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Spragg CJ, Emerman M (2013) Antagonism of SAMHD1 is actively maintained in natural infections of simian immunodeficiency virus. Proc Natl Acad Sci 110: 21136–21141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McCarthy KR, Kirmaier A, Autissier P, Johnson WE (2015) Evolutionary and functional analysis of old world primate TRIM5 reveals the ancient emergence of primate lentiviruses and convergent evolution targeting a conserved capsid interface. PLoS Pathog 11: e1005085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kirmaier A, Wu F, Newman RM, Hall LR, Morgan JS, O'Connor S, Marx PA, Meythaler M, Goldstein S, Buckler‐White A et al (2010) TRIM5 suppresses cross‐species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol 8: e1000462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gupta R, Kus B, Fladd C, Wasmuth J, Tonikian R, Sidhu S, Krogan NJ, Parkinson J, Rotin D (2007) Ubiquitination screen using protein microarrays for comprehensive identification of Rsp5 substrates in yeast. Mol Syst Biol 3: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chutiwitoonchai N, Hiyoshi M, Hiyoshi‐Yoshidomi Y, Hashimoto M, Tokunaga K, Suzu S (2013) Characteristics of IFITM, the newly identified IFN‐inducible anti‐HIV‐1 family proteins. Microbes Infect 15: 280–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wee YS, Roundy KM, Weis JJ, Weis JH (2012) Interferon‐inducible transmembrane proteins of the innate immune response act as membrane organizers by influencing clathrin and v‐ATPase localization and function. Innate Immun 18: 834–845 [DOI] [PubMed] [Google Scholar]
- 50. Zhang Y, Makvandi‐Nejad S, Qin L, Zhao Y, Zhang T, Wang L, Repapi E, Taylor S, McMichael A, Li N et al (2015) Interferon‐induced transmembrane protein‐3 rs12252‐C is associated with rapid progression of acute HIV‐1 infection in Chinese MSM cohort. AIDS 29: 889–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shen C, Wu XR, Jiao WW, Sun L, Feng WX, Xiao J, Miao Q, Liu F, Yin QQ, Zhang CG et al (2013) A functional promoter polymorphism of IFITM3 is associated with susceptibility to pediatric tuberculosis in Han Chinese population. PLoS One 8: e67816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Buitendijk H, Fagrouch Z, Niphuis H, Bogers W, Warren K, Verschoor E (2014) Retrospective serology study of respiratory virus infections in captive great apes. Viruses 6: 1442–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Moncla LH, Ross TM, Dinis JM, Weinfurter JT, Mortimer TD, Schultz‐Darken N, Brunner K, Capuano SV III, Boettcher C, Post J et al (2013) A novel nonhuman primate model for influenza transmission. PLoS One 8: e78750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- 55. De Bie T, Cristianini N, Demuth JP, Hahn MW (2006) CAFE: a computational tool for the study of gene family evolution. Bioinformatics 22: 1269–1271 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File


