Abstract
Responses to pathogens, including host transcriptional reprogramming, require partially antagonistic signalling pathways dependent on the phytohormones salicylic (SA) and jasmonic (JA) acids. However, upstream factors modulating the interplay of these pathways are not well characterized. Here, we identify the transcription factor ANAC032 from Arabidopsis thaliana as one such regulator in response to the bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (Pst). ANAC032 directly represses MYC2 activation upon Pst attack, resulting in blockage of coronatine‐mediated stomatal reopening which restricts entry of bacteria into plant tissue. Furthermore, ANAC032 activates SA signalling by repressing NIMIN1, a key negative regulator of SA‐dependent defence. Finally, ANAC032 reduces expression of JA‐responsive genes, including PDF1.2A. Thus, ANAC032 enhances resistance to Pst by generating an orchestrated transcriptional output towards key SA‐ and JA‐signalling genes coordinated through direct binding of ANAC032 to the MYC2, NIMIN1 and PDF1.2A promoters.
Keywords: Arabidopsis, jasmonic acid, pathogens, salicylic acid, transcription factor
Subject Categories: Microbiology, Virology & Host Pathogen Interaction; Plant Biology; Transcription
Introduction
During evolution, both plants and pathogens have evolved complex strategies to overcome virulence or defence mechanisms. The final outcome of a plant–pathogen interaction depends on the competence of the host to recognize the pathogen and trigger an appropriate, rapid defence response, as well as on the ability of the pathogen to mask its attack and activate suppression of the plant's innate immune system. Fine‐tuning of hormone signalling plays an essential role for governing the defence response of plants against pathogen challenge. The phytohormone salicylic acid (SA) is crucial for the defence against (hemi)biotrophic pathogens such as Pseudomonas syringae, while jasmonic acid (JA) and/or ethylene (ET) function as key signalling molecules in response to necrotrophic pathogens or insects 1, 2, 3. Prioritizing one hormonal signalling pathway over the other in dependence of the pathogen encountered is crucial for attaining specificity and success of the overall plant's defence response 2.
Interestingly, as a counter strategy, pathogens manipulate phytohormone homeostasis or produce toxins that interfere with plant defence mechanisms 4, 5, 6, 7. For example, Pseudomonas syringae, a foliar bacterial pathogen, produces the phytotoxin coronatine (COR), which is structurally similar to bioactive jasmonate, that is JA‐isoleucine (JA‐Ile) 8, 9. COR functionally mimics JA and promotes susceptibility of the host by manipulating its defence hormone signalling 4, 8, 9, 10, 11, 12, 13, 14. The role of COR in combating the pathogen‐associated molecular pattern (PAMP)‐induced stomatal defence received much attention in recent years 15, 16, 17, 18, 19, 20, 21 and led to the identification of a COR‐induced signalling cascade that involves the SCFCOI1 (Skp1‐Cdc53‐F‐box protein, CORONATINE1) ubiquitin‐ligase complex, jasmonate ZIM domain (JAZ) repressor proteins, MYC2 (a bHLH domain‐containing transcription factor) and NAC (NAM, ATAF and CUC) transcription factors (TFs) that jointly represent essential components in the suppression of the plant immune response by Pst 22. Interestingly, the myc2 mutant exhibits enhanced resistance to Pst with increased expression of PATHOGENESIS‐RELATED (PR) genes and SA accumulation when compared to wild‐type plants giving MYC2 an important position in the JA‐SA antagonistic interplay 23, 24. Several reports clearly support the mutually antagonistic relation between the SA and JA pathways either mounted by plants as a defence response or manipulated by microbes for their own survival 25, 26.
Activation of the plant's defence response involves complex transcriptional reprogramming. The central modulator of systemic acquired resistance (SAR), NPR1 (NONEXPRESSOR of PR GENE1), was identified as a key signalling node in the regulation of the SA‐JA interplay 27, 28. By interacting with bZIP family TGA TFs, NPR1 induces the expression of PR genes required for SAR establishment 29, 30, 31, 32, 33. Moreover, interaction with TGA TFs was shown to be necessary for suppression of the JA‐responsive gene PDF1.2 (PLANT DEFENSIN1.2) by GRX480 (GLUTAREDOXIN480), an SA‐responsive glutaredoxin gene whose SA inducibility itself is NPR1‐dependent 34. Additionally, NIMIN (NIM1 INTERACTING) proteins were identified to differentially interact with NPR1 and affect PR1 gene expression at different stages of SAR, thus representing negative regulators of SAR 35, 36. Recently, a JA‐responsive GCC‐box motif was found to be enriched in the promoters of JA‐responsive genes that are suppressed by SA 37. Furthermore, COR‐mediated MYC2‐dependent activation of three NAC family TFs, namely ANAC019, ANAC055 and ANAC072, and their tomato homologs JA2 (JASMONIC ACID2) and JA2L (JASMONIC ACID2 LIKE), were reported to promote bacterial propagation by inhibiting SA accumulation 22, 38. Despite the identification of several components involved in plant defence responses, the molecular mechanisms involved in fine‐tuning the hormonal pathways still remain largely unexplored. Here, we identified a novel TF of the NAC family, ANAC032, as a positive regulator of the defence response against Pseudomonas syringae pv. tomato DC3000 (Pst). ANAC032 promotes pathogen‐induced defence responses by activating SA, but repressing JA signalling including COR‐mediated stomatal reopening. ANAC032 acts upstream of the MYC2‐ANAC019/55/72 transcriptional cascade and also regulates the expression of the key pathogen‐response genes NIMIN1 and PDF1.2A by binding to their promoters. Our study provides new insights into the molecular mechanisms that underlie the antagonistic interaction between SA and JA signalling in response to Pst.
Results
Expression of ANAC032 is induced by various defence signals
Pathogen‐induced global transcriptome analysis and functional genomic studies have identified several members of NAC TF family as key components in transcriptional regulation of gene expression during pathogen attack 39. We identified ANAC032 (AT1G77450), an uncharacterized member of the ATAF clade of the Arabidopsis NAC family, as a plausible candidate involved in the regulation of the plant's response to pathogens. Members of this clade exhibit strong expression responses to various types of stresses including those induced by pathogen attack. ANAC032 expression is induced upon infection with the bacterial pathogens P. syringae pv. tomato DC3000 (Pst), Pst avrRPM1, Pst avrRpt2, P. syringae pv. Maculicola ES4326 (Psm ES4326) and with the leaf fungus Alternaria brassicicola (Genevestigator) which suggests an active role of the ANAC032 transcription factor in plant immune responses.
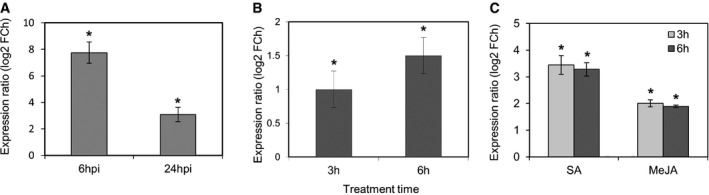
To confirm the induction of ANAC032 by Pst, we monitored its expression in Arabidopsis Col‐0 (wild type; WT) challenged by surface inoculation with Pst (1 × 108 c.f.u. ml−1). ANAC032 expression was highly induced 6 h post‐inoculation (hpi) and showed a moderate increase at 24 hpi, suggesting that this NAC TF preferentially functions during the early pathogen‐response phase (Fig 1A). As Pst can simultaneously trigger synthesis of both SA and JA 27, we next examined the effect of the two hormones on ANAC032 expression. Treatment with methyl jasmonate (MeJA) and SA rapidly (3 h) induced ANAC032 expression (Fig 1B). Ethylene has been shown to act synergistically with JA in response to pathogens and that it induces the expression of ANAC032 in cooperation with JA 40. Finally, we tested the effect of coronatine (COR) on ANAC032 expression. COR, a phytotoxin produced by various strains of Pseudomonas syringae 8, is structurally similar to JA‐isoleucine (JA‐Ile) and can activate JA signalling 4, 8, 9, 10, 11, 12. ANAC032 expression was induced in WT seedlings within 3 h and 6 h upon treatment with COR (Fig 1C). Taken together, these observations support a role for ANAC032 in response of Arabidopsis to pathogen (Pst) attack.
Figure 1. Enhanced expression of ANAC032 by Pst DC3000‐derived signals.
-
AANAC032 expression in WT plants sprayed with Pst DC3000, 6 and 24 hpi, compared to control (sprayed with 10 mM MgCl2 (mock)). Bars represent means ± SD (n = 3 independently performed experiments, each including the rosette leaves of at least three plants grown in individual pots).
-
B, CANAC032 expression in WT treated with (B) SA or MeJA or (C) COR for 3 and 6 h compared to non‐treated controls. Means ± SD are given (n = 3 independently performed experiments, each including at least 20 seedlings).
ANAC032 promotes plant disease resistance against P. syringae pv. tomato DC3000
To elucidate the possible involvement of ANAC032 in plant defence, we tested transgenic Arabidopsis plants with modified ANAC032 expression levels for their response to Pst infection. This we first tested by pressure‐infiltrating leaves of ANAC032 overexpressors (hereafter, 35S:ANAC032) and anac032‐1 (a T‐DNA insertion knockout mutant, SALK_012253; Fig EV1) with Pst (1 × 106 c.f.u. ml−1). While anac032‐1 plants showed increased susceptibility towards Pst compared to WT, 35S:ANAC032 plants exhibited a strongly enhanced disease resistance (Fig 2A). We also observed greater Pst proliferation 3 days postinoculation (dpi) in anac032‐1 compared to WT (Fig 2B), while in 35S:ANAC032 plants, the bacterial titre was less compared to WT, suggesting that ANAC032 restricts bacterial growth. Since pressure infiltration bypasses the normal mode of pathogen entry through natural openings (stomata), we next sprayed plants with inocula containing Pst (1 × 108 c.f.u. ml−1). While severe chlorotic lesions occurred in anac032‐1 (Fig 2C), 35S:ANAC032 plants showed only minimal disease symptoms 4–5 dpi (Fig 2C and D).
Figure EV1. Identification of anac032‐1 knockout mutant and ANAC032 expression analysis in anac032‐1 knockout, 35S:ANAC032, ANAC032 complementation (comp) and ANAC032 prom‐ANAC032‐GFP plants.
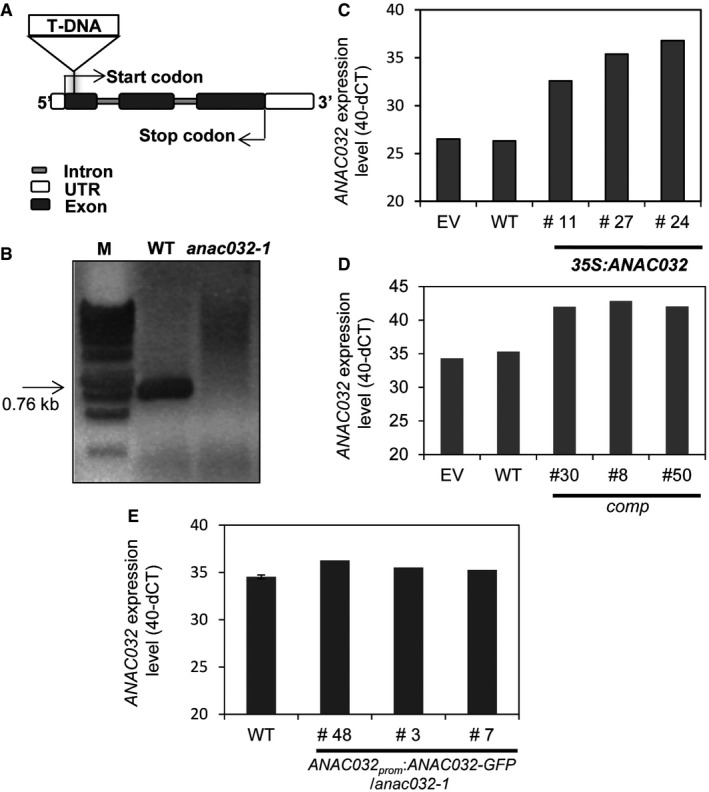
- Schematic representation of the ANAC032 gene showing the position of the T‐DNA insertion in the anac032‐1 mutant.
- Analysis of anac032‐1 and wild‐type (WT) plants for the presence of full‐length ANAC032 transcript using end point PCR. M, DNA size marker.
- Different 35S:ANAC032 plant lines (individual T0 plants) showing increased expression of ANAC032 compared to empty‐vector (EV) and wild‐type (WT) plants. Expression was analysed by qRT–PCR. Numbers on the y‐axis indicate 40 − ΔC T.
- Different complemented (35S:ANAC032 in anac032‐1) plant lines (individual T0 plants) showing increased expression of ANAC032 compared to empty‐vector (EV) and wild‐type (WT) plants. Expression was analysed by qRT–PCR. Numbers on the y‐axis indicate 40 − ΔC T.
- Different ANAC032prom‐ANAC032‐GFP plant lines (individual T0 plants) showing increased expression of ANAC032 compared to wild‐type (WT) upon treatment with Pst for 6 h. Expression was analysed by qRT–PCR. Numbers on the y‐axis indicate 40 − ΔC T.
Figure 2. Role of ANAC032 in resistance against Pst DC3000.
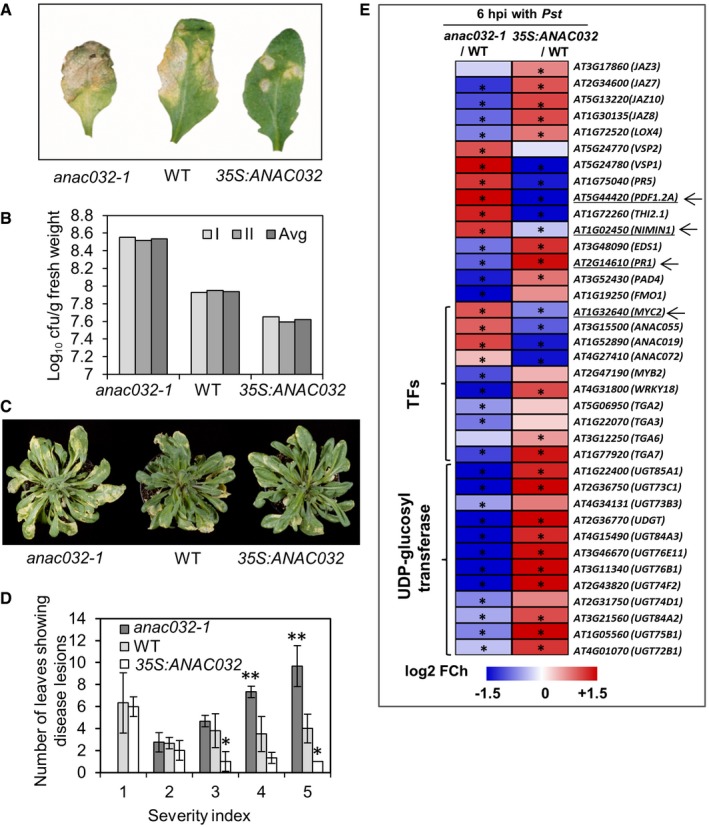
- Plants after pressure infiltration with Pst DC3000, at 4 dpi. The experiment was repeated three times (each time 9–12 plants) with similar results.
- Pst DC3000 growth in ANAC032 transgenics and WT plants 3 dpi after pressure infiltration. Two independent experiments were performed with three replications per experiment, each replicate consisting of three plants grown in individual pots (six measurements in total). The graph shows data points of the two individual experiments (I and II) along with their mean (Avg).
- Disease status of plants sprayed with Pst DC3000, 5 dpi. The experiment was repeated four times with similar results.
- Disease severity index (1, small chlorotic lesions; 5, large lesions) scored 5 dpi after spraying with Pst DC3000. Data are from three independent experiments with at least six plants per genotype in each. Means ± SD are shown. Asterisks indicate significant (*P < 0.05 and **P < 0.005) differences between transgenic and WT plants in chi‐square test analysis.
- Heat map showing the fold change (log2 basis) in the expression ratio of defence‐/stress‐related differentially expressed genes in anac032‐1 and 35S:ANAC032 compared to WT after spraying with Pst DC3000 (6 hpi) normalized to their respective controls. Blue, downregulated; red, upregulated. Data represent means of three independent experiments, each including the rosettes leaves of at least three plants grown in individual pots. Asterisks indicate significant differences from WT plants (Student's t‐test, P ≤ 0.05).
To verify that the observed phenotypes of anac032‐1 plants resulted from a loss of ANAC032 function, we generated complementation lines of the mutant (hereafter comp) (Fig EV1D). To this end, the ANAC032 coding sequence was placed under the control of the cauliflower mosaic virus (CaMV) 35S promoter and introduced into anac032‐1 plants. Upon challenge with Pst (both by pressure infiltration and by surface inoculation), comp plants clearly displayed a recovery from the phenotype of anac032‐1 plants with reduced progression of disease symptoms and bacterial load compared to WT (Fig EV2), proving that the lack of ANAC032 function is responsible for the phenotypes observed. Taken together, our data clearly indicate that ANAC032 functions as a positive regulator of the plant defence response against Pst.
Figure EV2. Loss of ANAC032 increases disease susceptibility to Pst DC3000.
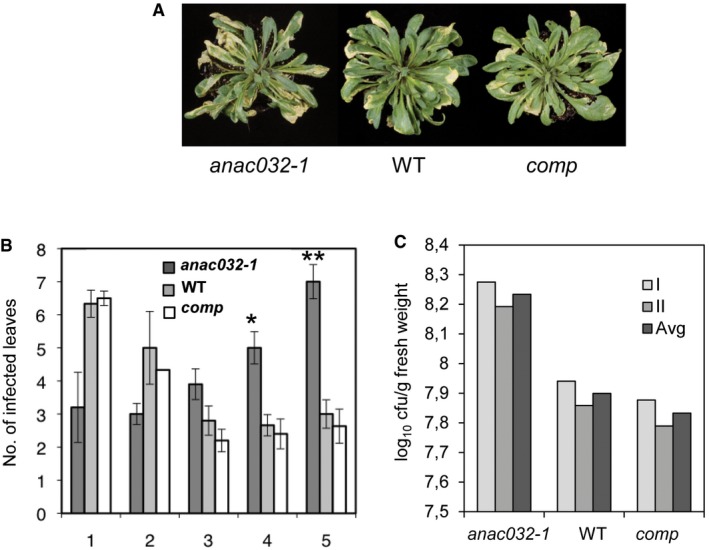
- Disease symptoms of plants sprayed with Pst DC3000, 5 dpi. The experiment was repeated four times with similar results.
- Disease severity index (1, small chlorotic lesions; 5, large lesions) scored 5 dpi after spraying with Pst DC3000. Data are from three independent experiments with at least six plants per genotype in each. Means ± SD are shown. Asterisks indicate significant (*P < 0.05 and **P < 0.005) differences between transgenic and WT plants in chi‐square test analysis.
- Bacterial growth in ANAC032 transgenic and WT plants 3 dpi after pressure infiltration. Two independent experiments were performed with three replications per experiment, each replicate consisting of three plants grown in individual pots. The graph shows data points of the two individual experiments (I and II) along with their mean (Avg). “Comp” indicates the anac032‐1 mutant transformed with the 35S:ANAC032 construct.
ANAC032 activates the SA‐mediated defence response, but antagonizes JA‐responsive gene expression
To elucidate the molecular mechanisms underlying ANAC032‐mediated defence, we tested the expression of 123 defence‐/stress‐related genes and 15 genes encoding UDP‐glucosyl transferases (UGTs) in 6‐week‐old ANAC032 transgenic and WT plants after Pst infection (at 6 hpi) by qRT–PCR (quantitative real‐time polymerase chain reaction). Genes included in the platform (Table EV1) were extracted from reports on previous studies on plant defence responses against invading pathogens, in particular Pseudomonas syringae 25, 26, 37, 41, 42. All genes chosen are induced by Pst infection. Our data revealed that 37 genes, including JA and SA defence responsive genes, biotic stress‐induced TFs, and several UGTs, were differentially expressed after Pst treatment in ANAC032 transgenic plants compared to the WT (Fig 2E). Upon Pst treatment, expression of 12 UGTs (UGT85A1, UGT73C1, UGT73B3, UGT84A3, UGT76E11, UGT76B1, UGT74F2, UGT74D1, UGT84A2, UGT75B1, UGT72B1 and At2g36770) was significantly reduced in anac032‐1 plants compared to WT, whereas their transcript levels were enhanced in 35S:ANAC032 plants. Among the JA‐related genes, expression of JAZ3, JAZ7, JAZ8 and JAZ10, which encode JAZ proteins, was downregulated in anac032‐1, but upregulated in 35S:ANAC032 upon Pst treatment (compared to WT). In contrast, expression of MYC2, a key transcription factor in JA signalling 43, 44, 45, was upregulated in anac032‐1, but downregulated in 35S:ANAC032 plants upon Pst treatment (compared to WT). Expression of other JA‐responsive genes including PDF1.2A, THI2.1, VSP1 and VSP2 was also upregulated in anac032‐1, but downregulated in 35S:ANAC032 plants 6 h after Pst treatment (Fig 2E). JAZ proteins act as repressors of JA signalling under basal conditions, while the accumulation of JA leads to the release of their repressive effect, for example on JA activator TFs such as MYC2, which results in induced JA responses 44, 45, 46, 47, 48. In addition to the negative regulation of JA signalling, JAZ10 was reported to attenuate the development of disease symptoms upon Pst infection 49. The elevated expression of MYC2 and other JA‐responsive genes in anac032‐1 suggests a role for ANAC032 as a negative regulator of JA signalling in response to Pst attack.
Among the SA‐related genes, significant downregulation in the anac032‐1 mutant was observed for PAD4 (PHYTOALEXIN DEFICIENT4) and EDS1 (ENHANCED DISEASE SUSCEPTIBILITY1), both of which are involved in SA accumulation 50, 51, 52. Transcript levels of these genes were increased in 35S:ANAC032 plants compared to WT upon Pst treatment. In contrast, the transcript level of NIMIN1, a central but negative regulator of SA‐dependent pathogen defence 35, 36, was significantly increased in anac032‐1 but reduced in 35S:ANAC032 plants compared to WT. Consistent with this observation, the transcript level of PR1, the SA‐dependent defence marker gene negatively affected by NIMIN1 36, was significantly reduced in anac032‐1 and increased in 35S:ANAC032 compared to WT upon Pst treatment (Fig 2E). Activation of SA‐responsive defence genes and the concomitant suppression of JA‐responsive genes suggest that ANAC032 modulates the interplay between SA‐ and JA‐dependent defence signalling in response to Pst.
ANAC032 directly regulates MYC2, PDF1.2A and NIMIN1 expression in vivo
To further elucidate the regulatory function of ANAC032 in the SA/JA interplay and defence against Pst, we aimed to identify genes directly regulated by ANAC032. To this end, we first identified cis‐elements recognized by ANAC032 (ANAC032 binding sites), using an in vitro binding site selection experiment (BSSE) employing the CELD‐fusion method 53. In vitro binding site selection revealed two consensus sequences bound by ANAC032, that is RgWannCAAnnnnnnYACGnMWCY (24 bp) and RgWKnCGTRnnnnnYACGtMWcY (23 bp). The nucleotides underlined are fully conserved in all 18 and 8, respectively, sequenced clones suggesting that ANAC032 binds to a bipartite cis‐regulatory site with two core motifs separated by a flexible linker region (Table EV2). Mutation analysis identified several nucleotides essential for high binding affinity (Table EV3).
We then analysed the promoters of genes expressed differentially between ANAC032 transgenic and WT plants upon Pst treatment and identified NIMIN1, MYC2 and PDF1.2A harbouring ANAC032 binding sites in their 500‐bp upstream promoter regions as putative direct targets of ANAC032. All three genes showed reduced and increased expression, respectively, in 35S:ANAC032 and anac032‐1 plants (Fig 3A), suggesting that they are directly and negatively controlled by ANAC032.
Figure 3. ANAC032 directly regulates MYC2, PDF1.2A and NIMIN1 .
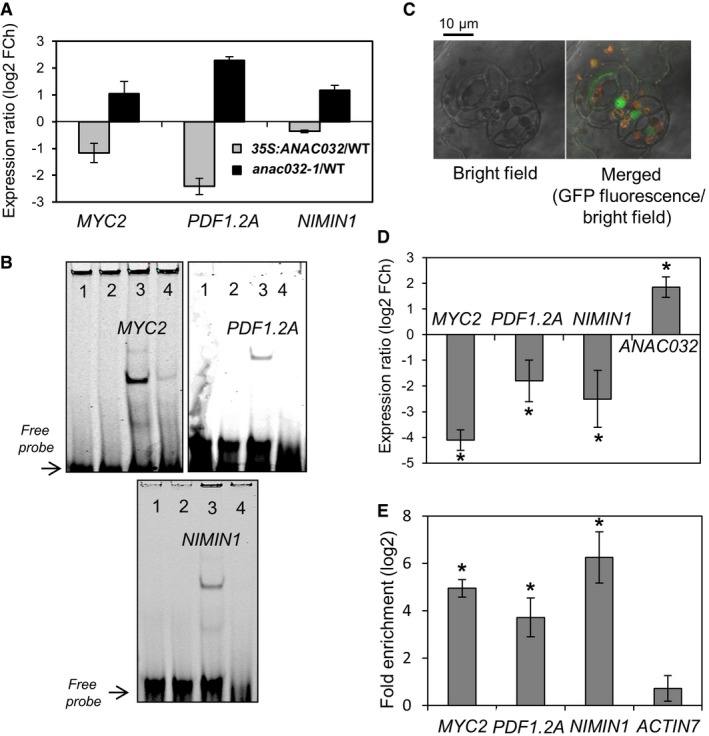
- MYC2, PDF1.2A and NIMIN1 expressions in ANAC032 transgenics compared to WT after spraying with Pst DC3000 (24 hpi) normalized to their respective controls. FCh, fold change. Means ± SD (n = 3 independently performed experiments, each including the rosette leaves of at least three plants grown in individual pots).
- EMSA showing binding of ANAC032 to MYC2, PDF1.2A and NIMIN1 promoter regions (in 5′‐DY682‐labelled double‐stranded oligonucleotides) harbouring ANAC032 binding sites. 1, labelled probe only; 2, labelled probe plus GST protein; 3, labelled probe plus ANAC032‐GST protein; 4, labelled probe, ANAC032‐GST protein and 100× competitor (unlabelled oligonucleotide containing ANAC032 binding site).
- Confocal microscope image showing nuclear localization of ANAC032‐GFP fusion protein expressed from the ANAC032 promoter in ANAC032 prom :ANAC032‐GFP/anac032‐1 seedlings treated with Pst at 6 hpi. Left, bright field; right, chlorophyll auto‐fluorescence (red) and GFP fluorescence (green) under bright field.
- Expression of MYC2, PDF1.2A, NIMIN1 and ANAC032 in 5‐week‐old ANAC032 prom :ANAC032‐GFP/anac032‐1 plants compared to WT at 6 hpi with Pst, normalized to their respective controls. FCh, fold change. Means ± SD (n = 3 independently performed experiments, each including the rosette leaves of at least three plants grown in individual pots). Asterisks indicate a significant difference from WT, normalized to their respective controls (*P < 0.05; Student's t‐test).
- ChIP‐qPCR shows enrichment of MYC2, PDF1.2A and NIMIN1 promoter regions containing ANAC032 binding site compared to a promoter region lacking the ANAC032 binding site (AT5G09810; ACTIN7). Means ± SD (n = 3 independently performed experiments, each including the rosette leaves of at least three plants grown in individual pots). Asterisks indicate a significant difference from negative control (*P < 0.01; Student's t‐test).
We next employed electrophoretic mobility shift assays (EMSAs) to test for physical interaction of ANAC032 with the respective promoter sequences of MYC2, PDF1.2A and NIMIN1. ANAC032 protein fused to glutathione S‐transferase (GST) was incubated with 5′‐DY‐682‐labelled 40‐bp double‐stranded DNA fragments containing the ANAC032 binding sites (Table EV1). As evident from the retarded bands seen in Fig 3B, ANAC032 interacts with the promoter sequences of all three genes. This interaction is significantly reduced when unlabelled promoter fragments (competitor) are added in excess, indicating the specificity of the interaction.
To test whether ANAC032 also interacts in vivo with the MYC2, PDF1.2A and NIMIN1 promoters, we performed chromatin immunoprecipitation coupled with qRT–PCR (ChIP‐qPCR) using a transgenic line that expresses an ANAC032‐GFP fusion protein from the native ANAC032 promoter in anac032‐1 plants (Figs EV1E and 3C). ANAC032 is a nuclear localized protein (Fig 3C) and ANAC032 prom:ANAC032‐GFP/anac032‐1 plants showed reduced expression of MYC2, PDF1.2A and NIMIN1 upon Pst treatment (Fig 3D), demonstrating that fusion to GFP did not impair the biological function of ANAC032. Significant ChIP enrichment was observed for the three promoter regions harbouring ANAC032 binding sites compared to a promoter lacking the ANAC032 binding site (negative control) (Fig 3E). Thus, the ChIP‐qPCR results confirmed direct transcriptional regulation of MYC2, PDF1.2A and NIMIN1 by ANAC032 in vivo.
SA‐induced NIMIN1 expression is suppressed by ANAC032
NIMIN1 is a key regulator of SA‐dependent systemic acquired resistance (SAR) 36. At the protein level, it targets the NPR1‐TGA complex and negatively regulates expression of NPR1‐dependent SAR genes such as PR1. NIMIN1 expression has been shown to be transiently enhanced soon after SA application, before induction of PR1, thereby preventing premature activation of the latter 36. To investigate the SA‐ANAC032‐NIMIN1‐PR1 regulatory network further, we analysed NIMIN1 and PR1 transcript abundance in ANAC032 transgenic and WT plants upon SA treatment compared to control condition. As shown in Fig 4A, NIMIN1 expression increased markedly (~fourfold) within 3 h of SA treatment in WT plants, while its expression was attenuated at 6 h (up ~twofold compared to the initial level), consistent with previous reports showing early and transient induction of NIMIN1 upon SA treatment 36. PR1 transcript level was not increased 3 h after SA treatment (possibly due to a high level of the PR1 suppressor, NIMIN1, at this time point), whereas it increased significantly after 6 h (Fig 4B). Induction of NIMIN1 by SA was further enhanced at both time points in anac032‐1, while it was greatly diminished in 35S:ANAC032 plants, suggesting that ANAC032 is a negative regulator of NIMIN1 expression in response to SA. In accordance with this model, PR1 induction by SA is completely suppressed in anac032‐1. Furthermore, PR1 expression is induced at 3 h after SA treatment in 35S:ANAC032 plants, possibly due to reduced NIMIN1 expression (Fig 4B). Collectively, our data convincingly implicate ANAC032 as a regulator of SA‐dependent defence gene expression. It acts upstream of NIMIN1 and modulates time‐specific SA‐dependent PR1 regulation by NIMIN1.
Figure 4. ANAC032 facilitates the SA‐mediated transcriptional repression of NIMIN1 and PDF1.2A .
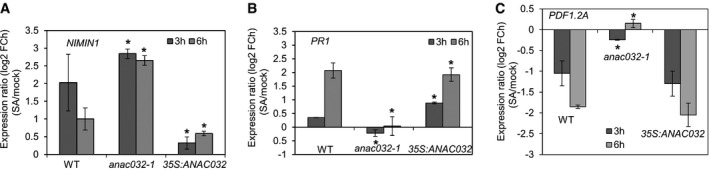
-
A, BTranscript levels of (A) NIMIN1 and (B) PR1 in WT, anac032‐1 and 35S:ANAC032 plants after 3 and 6 h of treatment with SA compared to their respective controls.
-
CExpression of PDF1.2A in WT and anac032‐1 plants after 3 and 6 h of treatment with SA compared to their respective controls.
ANAC032 is required for SA‐mediated suppression of PDF1.2A
SA strongly antagonizes JA signalling, resulting in the downregulation of JA‐responsive genes 25, 34, 37. Our study shows that ANAC032 negatively regulates the transcript levels of various JA‐responsive genes (such as PDF1.2A) upon Pst infection (Figs 2 and 3). To test whether ANAC032 is required for SA‐mediated suppression of JA signalling, we treated 2‐week‐old WT and anac032‐1 and 35S:ANAC032 seedlings with SA for 3 h and 6 h and analysed (compared to untreated control) expression of PDF1.2A, the direct target of ANAC032, by qRT–PCR. In WT, SA treatment reduced PDF1.2A transcript level at both time points (~twofold and 3.5‐fold after 3 and 6 h SA treatment, respectively), consistent with previous reports 27, 37, 54. PDF1.2 expression upon SA treatment was slightly more reduced at both time points (~2.6‐ and fourfold after 3 and 6 h SA treatment, respectively), in 35S:ANAC032 seedlings compared with WT. However, repression of PDF1.2A by SA was completely abolished in anac032‐1 (Fig 4C), clearly indicating ANAC032 as a regulator of SA‐mediated PDF1.2A suppression.
ANAC032 inhibits Pst‐induced stomatal reopening
Stomata play an active role in restricting bacterial invasion 16, 17, 18; within ~1 h after Pst DC3000 infection, stomata close in Arabidopsis Col‐0, a process dependent on ABA (abscisic acid)‐mediated signalling and PAMPs such as the flg22 (Flagellin 22) or lipopolysaccharide 16. However, stomata reopen at ~4 hpi, a process which requires COR signalling 16. ANAC032 is induced by COR (Fig 1C), ABA 55 and flg22 56, and it enhances resistance towards Pst as shown here, which provoked us to check whether ANAC032 is involved in regulating stomata movements during bacterial infection.
We found that stomatal closing upon Pst treatment was normal in all genotypes analysed (Figs 5A and C, and EV3A), showing that ANAC032 is not needed for ABA/PAMP‐triggered stomatal closure. However, at 4 hpi, stomata of WT, anac032‐1 and ANAC032‐complemented plants reopened, whereas stomata of 35S:ANAC032 plants remained closed, suggesting an inhibitory role for ANAC032 in COR‐dependent stomatal reopening or signalling (Figs 5B and C, and EV3B). To test this hypothesis further, we treated all genotypes for 1 h with ABA or COR. While ABA closed stomata in all genotypes, COR alone did not affect stomatal closure (Figs 5D and EV3C). However, when COR was applied together with ABA, stomata remained open in WT, ANAC032‐complemented and anac032‐1 plants, but they closed in 35S:ANAC032 plants (Figs 5D and EV3C), suggesting that ANAC032 suppresses the inhibitory effect of COR on ABA‐mediated stomatal closure.
Figure 5. ANAC032 inhibits Pseudomonas‐induced reopening of stomata.
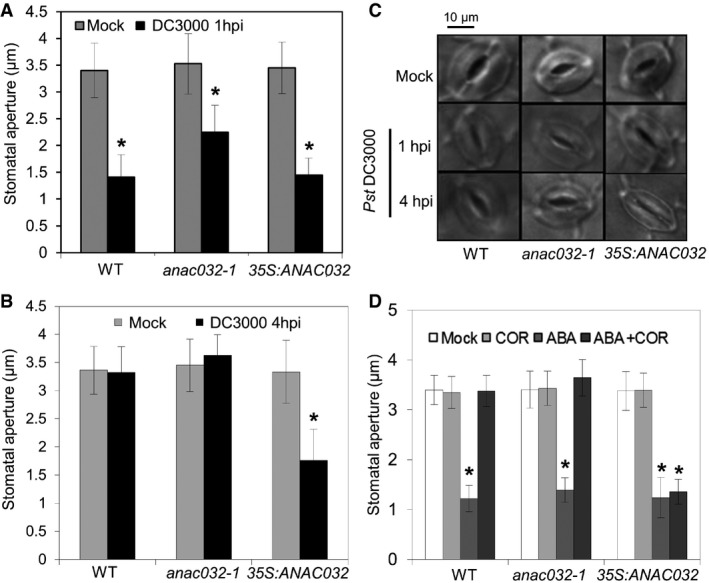
-
A, BStomatal aperture 1 h (A) or 4 h (B) after spraying with Pst DC3000 or mock treatment.
-
CMicroscope images of stomata at 1 and 4 hpi with Pst DC3000 (mock at 4 h).
-
DStomatal aperture 1 h after treatment with COR, ABA or COR plus ABA, compared to mock.
Figure EV3. Pseudomonas‐induced reopening of stomata in ANAC032 complementation plants.
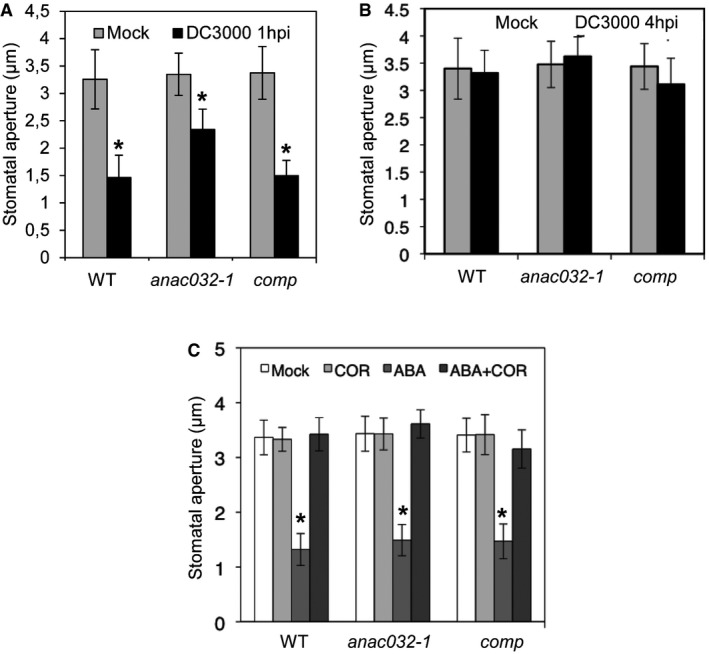
-
A, BStomatal aperture 1 h (A) or 4 h (B) after spraying with Pst DC3000 or mock treatment.
-
CStomatal aperture 1 h after treatment with COR, ABA, or COR plus ABA, compared to mock.
Finally, we surface inoculated anac032‐1 and WT plants with a COR‐deficient (COR−) Pst DC3000 strain and used Pst DC3000 in control experiments. Compared to WT, the anac032‐1 mutant exhibited increased susceptibility to both strains of DC3000; however, the severity of the disease symptoms upon infection with DC3000 COR− was more strongly diminished in anac032‐1 than in WT (Fig EV4), suggesting that ANAC032 regulation of disease response may, at least in part, be mediated through inhibition of COR‐induced stomatal reopening.
Figure EV4. Differential susceptibility of anac032‐1 and WT plants to Pst DC3000 and DC3000 COR− strains.
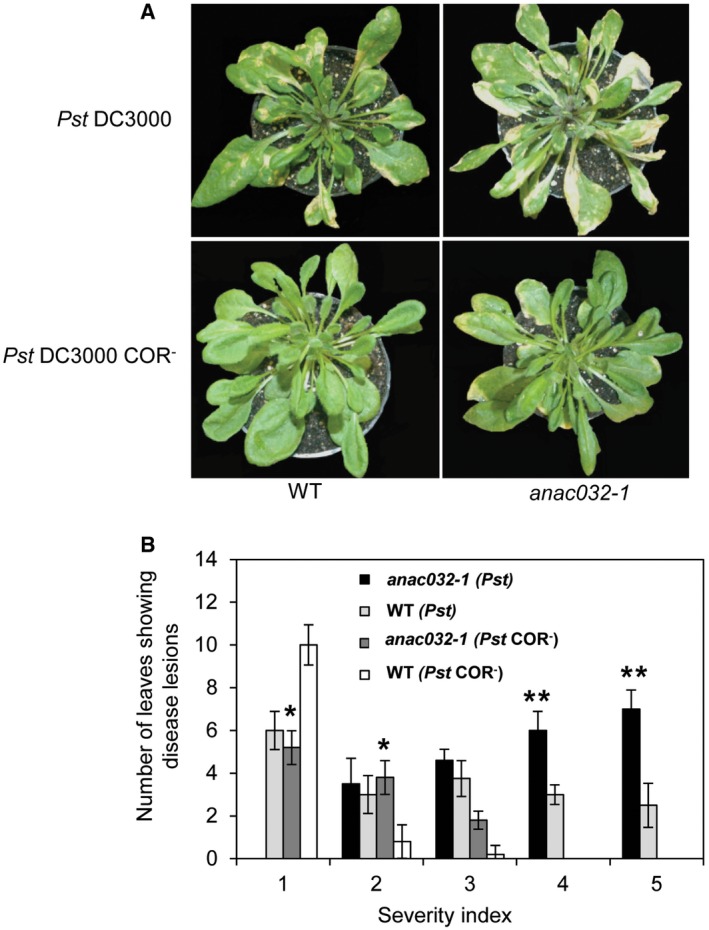
- Disease symptoms of WT and anac032‐1 plants surface inoculated with either Pst DC3000 or DC3000 COR− at 5 dpi.
- Disease severity index (1, small chlorotic lesions; 5, large lesions) scored 5 dpi after spraying with Pst DC3000 or DC3000 COR−. The experiment was repeated three times (n = 3) with similar results. Asterisks indicate significant (*P < 0.05 and **P < 0.005) differences between transgenic and respective controls (WT plants) in chi‐square test analysis.
ANAC032 directly suppresses MYC2 expression (Fig 3), and MYC2 positively regulates expression of the three closely related NAC genes ANAC019, ANAC055 and ANAC072, which mediate COR‐induced stomatal reopening after Pst infection 22. Interestingly, transcript abundance of all three NACs was enhanced in anac032‐1, but repressed in ANAC032 overexpressors upon Pst infection (Fig 2E), further supporting the role of ANAC032 as a negative regulator of COR signalling.
Discussion
Interplay between SA‐ and JA‐dependent pathways is essential for generating an appropriate physiological output upon pathogen attack 2, 26. Here, we identified transcription factor ANAC032 from Arabidopsis thaliana as a central regulator of the plant′s defence response against Pseudomonas syringae pv. tomato DC3000 (Pst); while overexpression of ANAC032 resulted in enhanced resistance to Pst, the ANAC032 loss‐of‐function mutant (anac032‐1) exhibited severe disease symptoms (Figs 2A–D and EV2). ANAC032 directly controls the expression of key elements of SA signalling (NIMIN1) and JA signalling (MYC2 and PDF1.2A) (Fig 3), suggesting that it exerts its physiological function by simultaneously enhancing SA, but decreasing JA signalling thereby shifting the plant′s response activity towards biotrophic over necrotrophic invaders.
NIMIN1 is one of the central components involved in the differential regulation of the transcriptional outputs of PR gene expression at different stages of SAR 35, 36. At an early stage of SAR, NIMIN1 interacts with NPR1 (resulting in the formation of the NIMIN1‐NPR1‐TGA complex) and negatively affects expression of the late SAR gene, PR1. At a later stage of SAR, accumulation of SA results in the removal of repressing NIMIN1 from the NPR1 complex, which is a prerequisite for induction of PR1 gene expression 36. Although tightly controlled, transcriptional control of NIMIN1 by upstream regulators is not well understood at present. A previous study demonstrated that a TGA2‐binding motif located in the NIMIN1 promoter is necessary for SA inducibility of NIMIN1 expression 57, 58. Here, we identified ANAC032 as a direct negative transcriptional regulator of NIMIN1 (Figs 2, 3, 4), which to our knowledge is the first TF reported to directly control NIMIN1 expression in planta.
Suppression of NIMIN1 transcription by ANAC032 may be considered as one of the mechanisms to relieve PR gene expression in the course of SAR. Additionally, analysis of ANAC032 transgenics and WT plants at different time points upon SA treatment revealed differential expression levels of NIMIN1 and PR1. In WT plants, the transcript levels of both genes increased upon SA treatment. However, while NIMIN1 expression was more pronounced at the earlier time point (3 h), PR1 was higher at the later time point (6 h), which is in agreement with previous reports 36. Interestingly, SA‐induced expression of NIMIN1 was further enhanced in anac032‐1 plants compared to WT, at the earlier time point and remained high even at the later time point, resulting in the suppression of PR1 expression at both time points tested. In contrast, NIMIN1 induction by SA was significantly suppressed in transgenic plants overexpressing ANAC032, which correlated with increased PR1 expression (Fig 4A and B). These data provide strong evidence that ANAC032 regulates PR1 gene expression through its negative effect on SA‐mediated NIMIN1 induction and therefore suggest ANAC032 as a key component regulating the SAR response.
MYC2 is a major regulator of JA signalling and functions as a negative regulator of SA biosynthesis and metabolism 8, 22, 23, 45. myc2/jin1 mutants exhibit reduced sensitivity to Pst DC3000 as well as to COR treatment due to higher accumulation of SA and enhanced expression of PR1 23, 24. MYC2 mediates COR signalling by directly activating three NAC factors involved SA biosynthesis and metabolism (ANAC019, ANAC055 and ANAC072). The three NACs repress expression of the SA synthesis gene ICS1 and activate expression of the SA metabolism gene BSMT1 22. Expression of the three NACs was enhanced in anac032‐1 but lowered in ANAC032 overexpressors upon Pst infection, further supporting the role of ANAC032 as a negative regulator of COR signalling and COR‐mediated stomatal reopening (Fig 2E), in accordance with our observation that ANAC032 overexpressors are impaired in stomatal reopening 4 h after surface inoculation with Pst and are less sensitive to COR (Figs 5 and EV3). Furthermore, anac032‐1 mutants exhibit an increased susceptibility to both Pst strains (DC3000 and DC3000 COR−) compared to WT, although disease symptoms induced by COR‐deficient DC3000 were more reduced in anac032‐1 than in WT plants indicating that ANAC032 contributes to the disease response by restricting COR‐induced stomatal reopening (Fig EV4). Stomatal closure establishes a physical barrier to restrict pathogen entry and is regulated by both, SA and ABA signalling 16, 17, 18. To counteract the plant′s defence response, Pseudomonas syringae produces the polyketide toxin COR to reopen stomata 16, 21, 22. In addition to overcoming stomatal defence, COR also aids to apoplastic bacterial propagation and promotes systemic susceptibility by inducing cell death leading to the formation of disease‐associated chlorosis and necrosis 4, 8, 13, 14. Interestingly, COR has been shown to more actively trigger the COI1‐JAZ interaction and the consecutive JAZ degradation than the plant hormone JA‐Ile 9, 11. It has recently been demonstrated that COR exerts its virulence by executing MYC2 signalling to suppress SA‐mediated defence response 22.
MYC2 has also been identified to act as a key defence response regulator against herbivores 59. The repressive effect of ANAC032 on MYC2 transcription suggests that it negatively regulates the response to herbivory. However, as ANAC032 expression is induced by MeJA, this observation indicates the presence of a negative feedback loop in herbivore defence/JA signalling, an interesting aspect to study in the future.
We furthermore observed that expression of PAD4, EDS1, PBS3, several TGA TFs and FMO1 is upregulated in 35S:ANAC032, but downregulated in anac032‐1 plants upon pathogen infection (Fig 2E). The eds1 and pad4 mutants in Arabidopsis are defective in SA production and exhibit enhanced disease susceptibility 50, 51, 52, and FMO1 has been shown to be critical for the establishment of SAR against necrotizing bacteria; in addition, it is required for the establishment of the EDS1/PAD4‐induced defence response 60.
Loss of ANAC032 also leads to transcriptional repression of JAZ genes (JAZ3, JAZ7, JAZ8 and JAZ10) while it enhances expression of MYC2 and several JA‐responsive genes including PDF1.2A, THI2.1, VSP1 and VSP2 (Fig 2E). JAZ proteins are major repressors of JA signalling under basal conditions 48. These gene expression profiles support the conclusion that ANAC032 acts as a negative regulator of JA responses upon infection by Pst.
Although previous studies had demonstrated the involvement of other TFs from diverse families including WRKYs (e.g. WRKY70, WRKY50, WRKY51), NACs (e.g. ANAC055, ANAC019, ANAC072) and bZIPs (e.g. TGA2, TGA5, TGA6) in the regulation of the SA/JA interplay 2, 22, 25, 26, 34, 61, 62, information about their direct target genes is currently limited.
Model for ANAC032 action
Collectively, our study identifies ANAC032 as a negative and positive regulator of JA and SA signalling, respectively, thereby channelling defence towards the SA pathway for a more efficient immune response against Pst. Our data suggest the following working model for the action of ANAC032 in mediating the interplay between SA‐ and JA‐dependent defence signalling (Fig 6). Expression of ANAC032 is induced by a number of pathogen‐derived signals and ANAC032 regulates the plant′s response to Pst infection through a network that involves genes associated with multiple layers of defence: (i) ANAC032 negatively regulates JA signalling through the suppression of JA‐responsive defence genes (MYC2, PDF1.2A, THI2.1, VSP1 and VSP2). (ii) ANAC032 positively affects SA signalling and expression of an SAR marker gene (PR1) by direct repression of SA‐induced NIMIN1 expression. (iii) Inhibition of pathogen‐triggered stomatal reopening by ANAC032 may be achieved by direct repression of the MYC2‐ANAC019/55/72 transcriptional cascade in the JA/COR signalling pathway, thereby restricting pathogen invasion.
Figure 6. Working model for the role of ANAC032 in mediating the interplay between SA‐ and JA‐dependent defence signalling in response to Pst DC3000.

ANAC032 expression is induced by pathogen‐derived signals. ANAC032 represses MYC2 and PDF1.2A, but stimulates JAZ repressors thereby reducing JA/COR signalling. COR‐mediated stomatal reopening and thus bacterial re‐entry into host cells is negatively affected by ANAC032 possibly through the MYC2‐ANAC019/55/72 transcriptional cascade. Simultaneously, ANAC032 activates PR1 gene expression and SA signalling by direct transcriptional repression of NIMIN1. Lines ending in arrows and bars indicate positive and negative interactions, respectively.
In summary, ANAC032—and likely its orthologs in crops (http://bioinformatics.psb.ugent.be/plaza/) —plays a decisive role in regulating the plant′s immune response against Pst DC3000 and possibly other biotrophic pathogens. This involves orchestrating the expression of multiple genes that are key to the SA/JA interplay, leading to enhanced SA, but reduced JA signalling. The upstream elements that regulate ANAC032 transcription in response to Pst infection remain to be identified.
Materials and Methods
General
Oligonucleotides (Table EV1) were obtained from Eurofins MWG Operon (Ebersberg, Germany). Tools provided by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), the Arabidopsis Information Resource (http://www.arabidopsis.org/), the Plant Transcription Factor Database (http://plntfdb.bio.uni-potsdam.de/v3.0/) and Genevestigator (http://genevestigator.com) were used for computational analyses.
Growth conditions
Arabidopsis thaliana (L.) Heynh. (Col‐0) was used. For experiments with seedlings, surface sterilized seeds were sown on 0.5× Murashige and Skoog (MS) agar medium containing 1% (w/v) sucrose. Plants were grown at 22°C under a 16‐h day (140 μmol m−2 s−1)/8‐h night regime. For growth under short‐day conditions, seedlings were grown in soil (Einheitserde GS90; Gebrüder Patzer) in a climate‐controlled chamber with an 8‐h day length at 120 μmol m−2 s−1 and a day/night temperature regime of 20/16°C, relative humidity 60/75%.
The T‐DNA insertion line SALK_012253 (anac032‐1) was obtained from the European Arabidopsis Stock Centre (http://arabidopsis.info/). Homozygous plants were identified by PCR using a T‐DNA left border primer and the gene‐specific primers LP and RP.
Constructs
Constructs were generated by PCR and restriction enzyme‐mediated or directional cloning. PCR‐generated amplicons were checked by DNA sequence analysis (MWG or Seqlab). Constructs were transformed into Arabidopsis using Agrobacterium tumefaciens.
For 35S:ANAC032, the ANAC032 open‐reading frame was amplified by PCR from Arabidopsis leaf cDNA, inserted into pUni/V5‐His‐TOPO (Invitrogen), and then cloned, via added PmeI‐PacI sites, into a modified pGreen0229‐35S plant transformation vector. For 35S:ANAC032‐GFP and ANAC032‐GST, the ANAC032 open‐reading frame was PCR‐amplified without the stop codon and cloned into the pENTR/D‐TOPO vector using the pENTR Directional TOPO Cloning kit (Invitrogen). The verified entry clones were then transferred to the pK7FWG2 vector (Ghent University) and the Gateway pDEST24 vector (Invitrogen), respectively, by LR recombination. For ANAC032 prom ‐ANAC032‐GFP, the 35S promoter in the 35S:ANAC032‐GFP construct was replaced with the ANAC032 upstream promoter (1 kb) using SLiCE (Seamless Ligation Cloning Extract) 63. ANAC032 prom ‐ANAC032‐GFP/anac032‐1 plants were produced by transforming the ANAC032 prom ‐ANAC032‐GFP construct into anac032‐1 plants. For anac032‐1 complementation, the above‐described 35S:ANAC032 construct was transformed into anac032‐1 plants. For ANAC032‐CELD, the ANAC032 CDS was inserted into pCR2.1‐TOPO (Invitrogen) and then cloned into the plasmid pTacLCELD6XHis 53 to create an in‐frame fusion construct, pTacANAC032CELD6XHis.
Treatments
For hormone treatments, 2‐week‐old seedlings grown on solid MS medium were transferred to flasks containing liquid MS medium (1% sucrose) and 1 mM SA or 100 μM MeJA or 1 ng/ml COR (with 0.015% [v/v] Silwet 77); flasks containing the seedlings were kept on a rotary shaker at 75–80 rpm. Medium with only 0.015% [v/v] Silwet 77 served as controls. Samples (whole seedlings) were harvested by flash‐freezing them in liquid nitrogen, and expression analysis was performed using quantitative real‐time PCR (qRT–PCR). Three independent experiments were performed for hormone treatments and gene expression analyses. Each replicate consisted of at least 20 seedlings.
Pseudomonas syringae pv. tomato DC3000 (Pst) or Pst DC3000 COR− (DB29, kindly provided by Dr. Barbara Kunkel, USA) was grown on King's B medium plates with appropriate antibiotics at 28°C for 2 days 13. Pseudomonas infection was performed as described 64. Briefly, bacterial cells were collected by centrifugation (2,500 g) and resuspended in 10 mM MgCl2. Pressure infiltration of Pst (c.f.u. 1 × 106 ml−1; OD600 nm = 0.002) was carried out using a needleless syringe. For inoculation by spraying, a Pst suspension with 1 × 108 c.f.u. ml−1 (OD600 nm = 0.2) or 10 mM MgCl2 mock inoculum solution containing 0.02% [v/v] Silwet L‐77 was used; the boxes containing the plants were covered with plastic lids. Whole rosette leaves were harvested at the indicated time points after bacterial challenge, frozen immediately in liquid nitrogen and used for ChIP and gene expression analysis. Three independent experiments were performed for gene expression analysis and ChIP. Each replicate consisted of rosettes leaves of at least three plants grown in individual pots.
Bacterial growth count and disease severity index
Bacterial growth was quantified by assessing the Pst population [c.f.u. (g fresh weight)−1] 3 days postbacterial challenge (by pressure infiltration) as described 64. The disease severity index was scored as described 64.
Stomatal assay
Pst DC3000 infection was performed by evenly spraying bacterial suspension (1 × 108 c.f.u. ml−1, containing 0.02% [v/v] Silwet L‐77) onto leaves of 6‐week‐old, soil‐grown plants. Leaves were fixed using formaldehyde fixative solution 16, 22 at the indicated time points and used for microscopic measurements 16, 22. Plants treated with 10 mM MgCl2 were used as mock infection controls. For stomatal studies after ABA, COR, or ABA + COR treatment, detached leaves were transferred to solutions containing either 15 μM ABA, 1 ng/μl COR, or 15 μM ABA + 1 ng/μl COR and placed on a rotary shaker (75–80 rpm) under constant light for 1 hour. Leaves were fixed using formaldehyde fixative solution 65; an epi‐fluorescence microscope was used to take images at random of at least 30 stomata for each sample and time point. Measurement of stomatal apertures was performed using Leica Cell software (http://www.leica-microsystems.com/).
Expression profiling by qRT–PCR
Total RNA extraction, synthesis of cDNA and qRT–PCR were performed as described 66. ACTIN2 served as reference gene. PCRs were run on an ABI PRISM 7900HT sequence detection system (Applied Biosystems Applera), and amplification products were visualized using SYBR Green (Life Technologies).
In vitro binding site selection assay
In vitro binding site selection was performed using the CELD method with the pTacANAC032CELD6XHis protein, employing biotin‐labelled double‐stranded oligonucleotides (Bio‐RS‐Oligo 3), which contained 30‐nucleotide random sequences 53. ANAC032‐selected oligonucleotides were cloned and sequenced. The DNA binding activity of ANAC032‐CELD was measured using methylumbelliferyl‐beta‐D‐cellobioside as substrate 53. DNA binding assays with a biotin‐labelled single‐stranded oligonucleotide or a biotin‐labelled double‐stranded oligonucleotide without a target binding site were used as controls.
EMSA
ANAC032‐GST fusion protein was purified from Escherichia coli expression strain BL21 Star (DE3) pRARE, which was generated by transforming the pRARE plasmid isolated from Rosetta (DE3) pRARE cells (Merck) into E. coli BL21 Star (DE3) (Invitrogen). Protein was purified using a GSTrap HP column (GE Healthcare) coupled to the ÄKTA purifier FPLC system (GE Healthcare). EMSA was performed as described 67 using an Odyssey Infrared EMSA kit (LI‐COR). 5′‐DY682‐labelled DNA fragments (Table EV1) were purchased from Eurofins MWG Operon.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was carried out on chromatin extracted from 5‐week‐old Arabidopsis plants expressing the ANAC032‐GFP fusion protein in anac032‐1 background (ANAC032prom‐ANAC032‐GFP/anac032‐1), 6 hpi with Pst DC3000. Anti‐GFP antibody was employed to immunoprecipitate protein–DNA complexes 68. Wild‐type (Col‐0) plants treated with Pst DC3000 for 6 h served as negative control. Three independent experiments of the ChIP experiment were run. The qPCR primers (Table EV1) for the target promoters were designed to flank the ANAC032 binding sites. As a negative control, we used primers annealing to a promoter region of an Arabidopsis gene (ACTIN7; AT5G09810) lacking an ANAC032 binding site. ChIP‐qPCR data were analysed as described 68.
Author contributions
SB conceived the study and wrote the manuscript. ADA performed the experiments. YB contributed to the Pst infection experiments and performed the chi‐square test analysis. G‐PX identified the ANAC032 binding sites.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Review Process File
Acknowledgements
SB thanks the Deutsche Forschungsgemeinschaft (BA 4769/2‐1) for funding. We thank Barbara N. Kunkel (Department of Biology, Washington University) for providing Pst DC3000 strains, Karin Koehl and colleagues for plant care, Eugenia Maximova for help with microscopy work and Ke Xu for help with Pst infection. Support by the University of Potsdam and the MPI of Molecular Plant Physiology is gratefully acknowledged.
EMBO Reports (2016) 17: 1578–1589
References
- 1. Koornneef A, Pieterse CMJ (2008) Cross talk in defence signalling. Plant Physiol 146: 839–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pieterse CMJ, van der Does D, Zamioudis C, Leon‐Reyes A, van Wees SCM (2012) Hormonal modulation of plant immunity. Ann Rev Cell Dev Biol 28: 489–521 [DOI] [PubMed] [Google Scholar]
- 3. Glazebrook J (2005) Contrasting mechanisms of defence against biotrophic and necrotrophic pathogens. Ann Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- 4. Mittal S, Davis KR (1995) Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato . Mol Plant Microbe Interact 8: 165–171 [DOI] [PubMed] [Google Scholar]
- 5. Weingart H, Ullrich H, Geider K, Volksch B (2001) The role of ethylene production in virulence of Pseudomonas syringae pv. glycinea and phaseolicola . Phytopathology 91: 511–518 [DOI] [PubMed] [Google Scholar]
- 6. Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, Deboy RT, Durkin AS, Kolonay JF et al (2003) The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA 100: 10181–10186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Siewers V, Kokkelink L, Smedsgaard J, Tudzynski P (2006) Identification of an abscisic acid gene cluster in the grey mold Botrytis cinerea . Appl Environ Microbiol 72: 4619–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brooks DM, Hernández‐Guzmán G, Kloek AP, Alarcón‐Chaidez F, Sreedharan A, Rangaswamy V, Peñaloza‐Vázquez A, Bender CL, Kunkel BN (2004) Identification and characterization of a well‐defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact 17: 162–174 [DOI] [PubMed] [Google Scholar]
- 9. Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, Miersch O, Wasternack C, Solano R (2009b) (+)‐7‐iso‐Jasmonoyl‐L‐isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol 5: 344–350 [DOI] [PubMed] [Google Scholar]
- 10. Brooks DM, Bender CL, Kunkel BN (2005) The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid‐dependent defences in Arabidopsis thaliana . Mol Plant Pathol 6: 629–639 [DOI] [PubMed] [Google Scholar]
- 11. Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao Y, Thilmony R, Bender CL, Schaller A, He SY, Howe GA (2003) Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J 36: 485–499 [DOI] [PubMed] [Google Scholar]
- 13. Cui J, Bahrami AK, Pringle EG, Hernandez‐Guzman G, Bender CL, Pierce NE, Ausubel FM (2005) Pseudomonas syringae manipulates systemic plant defences against pathogens and herbivores. Proc Natl Acad Sci USA 102: 1791–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Geng X, Cheng J, Gangadharan A, Mackey D (2012) The coronatine toxin of Pseudomonas syringae is a multifunctional suppressor of Arabidopsis defense. Plant Cell 24: 4763–4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Uppalapati SR, Ishiga Y, Wangdi T, Urbanczyk‐Wochniak E, Ishiga T, Mysore KS, Bender CL (2008) Pathogenicity of Pseudomonas syringae pv. tomato on tomato seedlings: phenotypic and gene expression analyses of the virulence function of coronatine. Mol Plant Microbe Interact 21: 383–395 [DOI] [PubMed] [Google Scholar]
- 16. Melotto M, Underwood W, Koczan J, Nomura K, He SY (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- 17. Melotto M, Underwood W, He SY (2008) Role of stomata in plant innate immunity and foliar bacterial diseases. Ann Rev of Phytopathol 46: 101–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Underwood W, Melotto M, He SY (2007) Role of plant stomata in bacterial invasion. Cell Microbiol 9: 1621–1629 [DOI] [PubMed] [Google Scholar]
- 19. Lee S, Ishiga Y, Clermont K, Mysore KS (2013) Coronatine inhibits stomatal closure and delays hypersensitive response cell death induced by nonhost bacterial pathogens. Peer J 1: e34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zeng W, He SY (2010) A prominent role of the flagellin receptor FLAGELLIN‐SENSING2 in mediating stomatal response to Pseudomonas syringae pv. tomato DC3000 in Arabidopsis. Plant Physiol 153: 1188–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zeng W, Brutus A, Kremer JM, Withers JC, Gao X, Jones AD, He SY (2011) A genetic screen reveals Arabidopsis stomatal and/or apoplastic defenses against Pseudomonas syringae pv. tomato DC3000. PLoS Pathog 7: e1002291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zheng XY, Spivey NW, Zeng W, Liu PP, Fu ZQ, Klessig DF, He SY, Dong X (2012) Coronatine promotes Pseudomonas syringae virulence in plants by activating a signalling cascade that inhibits salicylic acid accumulation. Cell Host Mic 11: 587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laurie‐Berry N, Joardar V, Street IH, Kunkel BN (2006) The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid‐dependent defense during infection by Pseudomonas syringae . Mol Plant Microbe Interact 19: 789–800 [DOI] [PubMed] [Google Scholar]
- 24. Nickstadt A, Thomma BP, Feussner I, Kangasjärvi J, Zeier J, Loeffler C, Scheel D, Berger S (2004) The jasmonate‐insensitive mutant jin1 shows increased resistance to biotrophic as well as necrotrophic pathogens. Mol Plant Pathol 5: 425–434 [DOI] [PubMed] [Google Scholar]
- 25. Gimenez‐Ibanez S, Solano R (2013) Nuclear jasmonate and salicylate signaling and crosstalk in defense against pathogens. Front Plant Sci 4: 72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robert‐Seilaniantz A, Grant M, Jones JD (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate‐salicylate antagonism. Annu Rev Phytopathol 49: 317–343 [DOI] [PubMed] [Google Scholar]
- 27. Spoel SH, Koornneef A, Claessens SM, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Métraux JP, Brown R, Kazan K et al (2003) NPR1 modulates cross‐talk between salicylate‐ and jasmonate‐dependent defense pathways through a novel function in the cytosol. Plant Cell 15: 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leon‐Reyes A, Spoel SH, De Lange ES, Abe H, Kobayashi M, Tsuda S, Millenaar FF, Welschen RA, Ritsema T, Pieterse CM (2009) Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS‐RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiol 149: 1797–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Y, Fan W, Kinkema M, Li X, Dong X (1999) Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR‐1 gene. Proc Natl Acad Sci USA 96: 6523–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Després C, DeLong C, Glaze S, Liu E, Fobert PR (2000) The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12: 279–290 [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou JM, Trifa Y, Silva H, Pontier D, Lam E, Shah J, Klessig DF (2000) NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR‐1 gene required for induction by salicylic acid. Mol Plant Microbe Interact 13: 191–202 [DOI] [PubMed] [Google Scholar]
- 32. Fan W, Dong X (2002) In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid‐mediated gene activation in Arabidopsis. Plant Cell 14: 1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dong X (2004) NPR1, all things considered. Curr Opin Plant Biol 7: 547–552 [DOI] [PubMed] [Google Scholar]
- 34. Ndamukong I, Abdallat AA, Thurow C, Fode B, Zander M, Weigel R, Gatz C (2007) SA‐ inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA‐responsive PDF1.2 transcription. Plant J 50: 128–139 [DOI] [PubMed] [Google Scholar]
- 35. Weigel RR, Pfitzner UM, Gatz C (2005) Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell 17: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hermann M, Maier F, Masroor A, Hirth S, Pfitzner AJ, Pfitzner UM (2013) The Arabidopsis NIMIN proteins affect NPR1 differentially. Front Plant Sci 4: 88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Van der Does D, Leon‐Reyes A, Koornneef A, Van Verk MC, Rodenburg N, Pauwels L, Goossens A, Körbes AP, Memelink J, Ritsema T et al (2013) Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1‐JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25: 744–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Du M, Zhaia Q, Denga L, Lia S, Lia H, Yana L, Huang Z, Wang B, Jiang H, Huang T et al (2014) Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell 26: 3167–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nuruzzaman M, Sharoni AM, Kikuchi S (2013) Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front Microbiol 4: 248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakano T, Suzuki K, Ohtsuki N, Tsujimoto Y, Fujimura T, Shinshi H (2006) Identification of genes of the plant‐specific transcription factor families co‐operatively regulated by ethylene and jasmonate in Arabidopsis thaliana . J Plant Res 119: 407–413 [DOI] [PubMed] [Google Scholar]
- 41. Beckers GJM, Spoel SH (2006) Fine‐Tuning Plant Defence Signalling: Salicylate versus Jasmonate. Plant Biol 8: 1–10 [DOI] [PubMed] [Google Scholar]
- 42. Sendon PM, Seo HS, Song JT (2011) Salicylic Acid Signaling: Biosynthesis, Metabolism, and Crosstalk with Jasmonic Acid. J Korean Soc Appl Biol Chem 54: 501–506 [Google Scholar]
- 43. Boter M, Ruiz‐Rivero O, Abdeen A, Prat S (2004) Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev 18: 1577–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM et al (2007) MYC2 differentially modulates diverse jasmonate‐dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lorenzo O, Chico JM, Sanchez‐Serrano JJ, Solano R (2004) JASMONATE‐INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate‐regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, Garcia‐Casado G, Lopez‐Vidriero I, Lozano FM, Ponce MR et al (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- 47. Fonseca S, Chico JM, Solano R (2009) The jasmonate pathway: the ligand, the receptor and the core signalling module. Curr Opin Plant Biol 12: 539–547 [DOI] [PubMed] [Google Scholar]
- 48. Fernandez‐Calvo P, Chini A, Fernández‐Barbero G, Chico JM, GimenezIbanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco‐Zorrilla JM et al (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Demianski AG, Chung KM, Kunkel BN (2012) Analysis of Arabidopsis JAZ gene expression during Pseudomonas syringae pathogenesis. Mol Plant Pathol 13: 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Falk A, Feys BJ, Frost LN, Jones JDG, Daniels MJ, Parker JE (1999) EDS1, an essential component of R gene‐mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc Natl Acad Sci 96: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J (1998) PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10: 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J (1999) Arabidopsis thaliana PAD4 encodes a lipase‐like gene that is important for salicylic acid signaling. Proc Natl Acad Sci USA 96: 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xue GP (2005) A CELD‐fusion method for rapid determination of the DNA‐binding sequence specificity of novel plant DNA‐binding proteins. Plant J 41: 638–649 [DOI] [PubMed] [Google Scholar]
- 54. Van Wees SC, Luijendijk M, Smoorenburg I, van Loon LC, Pieterse CM (1999) Rhizobacteria‐mediated induced systemic resistance (ISR) in Arabidopsis is not associated with a direct effect on expression of known defense‐related genes but stimulates the expression of the jasmonate‐inducible gene Atvsp upon challenge. Plant Mol Biol 41: 537–549 [DOI] [PubMed] [Google Scholar]
- 55. Goda H, Sasaki E, Akiyama K, Maruyama‐Nakashita A, Nakabayashi K, Li W, Ogawa M, Yamauchi Y, Preston J, Aoki K et al (2008) The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J 55: 526–542 [DOI] [PubMed] [Google Scholar]
- 56. Denoux C, Galletti R, Mammarella N, Gopalan S, Werck D, De Lorenzo G, Ferrari S, Ausubel FM, Dewdney J (2008) Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol Plant 1: 423–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fonseca JP, Menossi M, Thibaud‐Nissen F, Town CD (2010) Functional analysis of a TGA factor‐binding site located in the promoter region controlling salicylic acid‐induced NIMIN‐1 expression in Arabidopsis. Genet Mol Res 2: 167–175 [DOI] [PubMed] [Google Scholar]
- 58. Fode B, Siemsen T, Thurow C, Weigel R, Gatz C (2008) The Arabidopsis GRAS protein SCL14 interacts with class II TGA transcription factors and is essential for the activation of stress‐inducible promoters. Plant Cell 20: 3122–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Woldemariam MG, Baldwin IT, Galis I (2011) Transcriptional regulation of plant inducible defenses against herbivores, a mini‐review. J Plant Interact 6: 113–119 [Google Scholar]
- 60. Mishina TE, Zeier J (2006) The Arabidopsis flavin‐dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol 141: 1666–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li J, Brader G, Palva ET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate‐mediated and salicylate‐mediated signals in plant defense. Plant Cell 16: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gao Q‐M, Venugopal S, Navarre D, Kachroo A (2011) Low oleic acid‐derived repression of jasmonic acid‐inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol 155: 464–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang Y, Werling U, Edelmann W (2012) SLiCE: a novel bacterial cell extract‐based DNA cloning method. Nuc Acid Res 40: e55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kover PX, Wolf JB, Kunkel BN, Cheverud JM (2005) Genetic architecture of Arabidopsis thaliana response to infection by Pseudomonas syringae . Heredity 94: 507–517 [DOI] [PubMed] [Google Scholar]
- 65. Ruzin SE (1999) Plant microtechniques and microscopy. Oxford, UK: Oxford University Press; [Google Scholar]
- 66. Balazadeh S, Riano‐Pachon DM, Mueller‐Roeber B (2008) Transcription factors regulating leaf senescence in Arabidopsis thaliana . Plant Biol (Stuttg) 10: 63–75 [DOI] [PubMed] [Google Scholar]
- 67. Wu A, Allu AD, Garapati P, Siddiqui H, Dortay H, Zanor MI, Asensi‐Fabado MA, Munné‐Bosch S, Antonio C, Tohge T et al (2012) JUNGBRUNNEN1, a reactive oxygen species‐responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24: 482–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kaufmann K, Muiño JM, Østerås M, Farinelli L, Krajewski P, Angenent GC (2010) Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP‐SEQ) or hybridization to whole genome arrays (ChIP‐CHIP). Nature Protoc 5: 457–472 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Review Process File


