Abstract
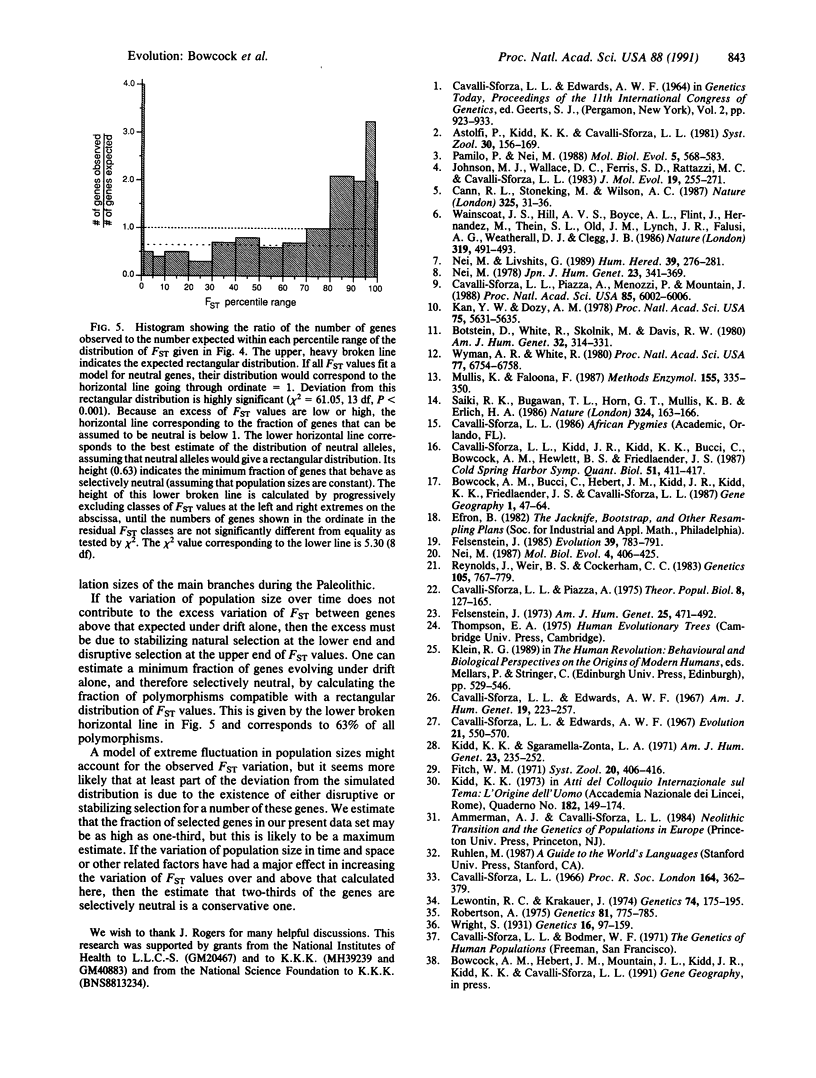
Accuracy of evolutionary analysis of populations within a species requires the testing of a large number of genetic polymorphisms belonging to many loci. We report here a reconstruction of human differentiation based on 100 DNA polymorphisms tested in five populations from four continents. The results agree with earlier conclusions based on other classes of genetic markers but reveal that Europeans do not fit a simple model of independently evolving populations with equal evolutionary rates. Evolutionary models involving early admixture are compatible with the data. Taking one such model into account, we examined through simulation whether random genetic drift alone might explain the variation among gene frequencies across populations and genes. A measure of variation among populations was calculated for each polymorphism, and its distribution for the 100 polymorphisms was compared with that expected for a drift-only hypothesis. At least two-thirds of the polymorphisms appear to be selectively neutral, but there are significant deviations at the two ends of the observed distribution of the measure of variation: a slight excess of polymorphisms with low variation and a greater excess with high variation. This indicates that a few DNA polymorphisms are affected by natural selection, rarely heterotic, and more often disruptive, while most are selectively neutral.
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Botstein D., White R. L., Skolnick M., Davis R. W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980 May;32(3):314–331. [PMC free article] [PubMed] [Google Scholar]
- Bowcock A. M., Bucci C., Hebert J. M., Kidd J. R., Kidd K. K., Friedlaender J. S., Cavalli-Sforza L. L. Study of 47 DNA markers in five populations from four continents. Gene Geogr. 1987 Apr;1(1):47–64. [PubMed] [Google Scholar]
- Cann R. L., Stoneking M., Wilson A. C. Mitochondrial DNA and human evolution. Nature. 1987 Jan 1;325(6099):31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza L. L., Edwards A. W. Phylogenetic analysis. Models and estimation procedures. Am J Hum Genet. 1967 May;19(3 Pt 1):233–257. [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza L. L., Kidd J. R., Kidd K. K., Bucci C., Bowcock A. M., Hewlett B. S., Freidlaender J. S. DNA markers and genetic variation in the human species. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):411–417. doi: 10.1101/sqb.1986.051.01.049. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza L. L., Piazza A. Analysis of evolution: evolutionary rates, independence and treeness. Theor Popul Biol. 1975 Oct;8(2):127–165. doi: 10.1016/0040-5809(75)90029-5. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza L. L., Piazza A., Menozzi P., Mountain J. Reconstruction of human evolution: bringing together genetic, archaeological, and linguistic data. Proc Natl Acad Sci U S A. 1988 Aug;85(16):6002–6006. doi: 10.1073/pnas.85.16.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza L. L. Population structure and human evolution. Proc R Soc Lond B Biol Sci. 1966 Mar 22;164(995):362–379. doi: 10.1098/rspb.1966.0038. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Maximum-likelihood estimation of evolutionary trees from continuous characters. Am J Hum Genet. 1973 Sep;25(5):471–492. [PMC free article] [PubMed] [Google Scholar]
- Johnson M. J., Wallace D. C., Ferris S. D., Rattazzi M. C., Cavalli-Sforza L. L. Radiation of human mitochondria DNA types analyzed by restriction endonuclease cleavage patterns. J Mol Evol. 1983;19(3-4):255–271. doi: 10.1007/BF02099973. [DOI] [PubMed] [Google Scholar]
- Kan Y. W., Dozy A. M. Polymorphism of DNA sequence adjacent to human beta-globin structural gene: relationship to sickle mutation. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5631–5635. doi: 10.1073/pnas.75.11.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd K. K., Sgaramella-Zonta L. A. Phylogenetic analysis: concepts and methods. Am J Hum Genet. 1971 May;23(3):235–252. [PMC free article] [PubMed] [Google Scholar]
- Lewontin R. C., Krakauer J. Distribution of gene frequency as a test of the theory of the selective neutrality of polymorphisms. Genetics. 1973 May;74(1):175–195. doi: 10.1093/genetics/74.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullis K. B., Faloona F. A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- Nei M., Livshits G. Genetic relationships of Europeans, Asians and Africans and the origin of modern Homo sapiens. Hum Hered. 1989;39(5):276–281. doi: 10.1159/000153872. [DOI] [PubMed] [Google Scholar]
- Nei M. The theory of genetic distance and evolution of human races. Jinrui Idengaku Zasshi. 1978 Dec;23(4):341–369. doi: 10.1007/BF01908190. [DOI] [PubMed] [Google Scholar]
- Pamilo P., Nei M. Relationships between gene trees and species trees. Mol Biol Evol. 1988 Sep;5(5):568–583. doi: 10.1093/oxfordjournals.molbev.a040517. [DOI] [PubMed] [Google Scholar]
- Reynolds J., Weir B. S., Cockerham C. C. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics. 1983 Nov;105(3):767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson A. Gene frequency distributions as a test of selective neutrality. Genetics. 1975 Dec;81(4):775–785. doi: 10.1093/genetics/81.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Bugawan T. L., Horn G. T., Mullis K. B., Erlich H. A. Analysis of enzymatically amplified beta-globin and HLA-DQ alpha DNA with allele-specific oligonucleotide probes. Nature. 1986 Nov 13;324(6093):163–166. doi: 10.1038/324163a0. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Wainscoat J. S., Hill A. V., Boyce A. L., Flint J., Hernandez M., Thein S. L., Old J. M., Lynch J. R., Falusi A. G., Weatherall D. J. Evolutionary relationships of human populations from an analysis of nuclear DNA polymorphisms. Nature. 1986 Feb 6;319(6053):491–493. doi: 10.1038/319491a0. [DOI] [PubMed] [Google Scholar]
- Wright S. Evolution in Mendelian Populations. Genetics. 1931 Mar;16(2):97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman A. R., White R. A highly polymorphic locus in human DNA. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6754–6758. doi: 10.1073/pnas.77.11.6754. [DOI] [PMC free article] [PubMed] [Google Scholar]