Abstract
Legionella pneumophila is a ubiquitous, pathogenic, Gram-negative bacterium responsible for legionellosis. Like many other amoeba-resistant microorganisms, L. pneumophila resists host clearance and multiplies inside the cell. Through its Dot/Icm type IV secretion system, the bacterium injects more than three hundred effectors that modulate host cell physiology in order to promote its own intracellular replication. Here we report that L. pneumophila prevents proliferation of its natural host Acanthamoeba castellanii. Infected amoebae could not undergo DNA replication and no cell division was observed. The Dot/Icm secretion system was necessary for L. pneumophila to prevent the eukaryotic proliferation. The absence of proliferation was associated with altered amoebal morphology and with a decrease of mRNA transcript levels of CDC2b, a putative regulator of the A. castellanii cell cycle. Complementation of CDC28-deficient Saccharomyces cerevisiae by the CDC2b cDNA was sufficient to restore proliferation of CDC28-deficient S. cerevisiae and suggests for the first time that CDC2b from A. castellanii could be functional and a bona fide cyclin-dependent kinase. Hence, our results reveal that L. pneumophila impairs proliferation of A. castellanii and this effect could involve the cell cycle protein CDC2b.
Legionella pneumophila is a pathogenic Gram-negative bacterium found in natural and artificial aqueous environments1. It is responsible for legionellosis, a potentially lethal pneumonia that occurs after inhalation of aerosols containing the bacterium2. In water systems, L. pneumophila was found associated with free-living amoebae (FLA) which ensure its survival and replication3. The genus Acanthamoeba, among the most prevalent FLA found in the environment, has been characterized as a natural host of L. pneumophila3. The bacterium is phagocytized during Acanthamoeba grazing but it resists intracellular digestion3,4. L. pneumophila has evolved a number of mechanisms to modulate amoebal signal-transduction pathways to its advantage to support its replication. This successful strategy lies in the ability of L. pneumophila to perturb essential functions such as transcription, translation, cytoskeleton machineries, organelle function, vesicular trafficking, autophagy and host survival processes5. During infection of amoebae or alveolar macrophages, L. pneumophila injects more than three hundred effectors through a type IV secretion system (T4SS) called Dot/Icm (Defect in organelle trafficking; Intracellular multiplication) which is critical for resistance to host digestion, replication and exit of the bacterium from the cell6,7. The disturbance of eukaryotic functions by L. pneumophila is facilitated by bacterial proteins that contain a eukaryotic domain allowing the pathogen to mimic host cell functions8. To our knowledge, no study thus far has characterized the consequences of L. pneumophila infection on host-cell proliferation.
The cell cycle is a vital process that ensures growth and reproduction or proliferation of all living cells. It consists of duplication of the cell content (DNA and organelles) and repartition of the duplicated material into the daughter cells during mitosis and cytokinesis. The eukaryotic cell cycle is driven by a class of serine/threonine kinases named Cyclin-Dependent Kinases (CDKs) that act in concert with protein regulatory subunits called Cyclins9. Activities of some CDKs, such as the human protein CDK1 or the homologous protein in Saccharomyces cerevisiae CDC28, can be essential for cell proliferation10,11. Although CDK activity was detected in Acanthamoeba, the responsible protein had yet to be identified12. The sequencing of the genome of A. castellanii provided some clues as to the identity of the putative cell cycle regulator13.
Here, we show that L. pneumophila is able to prevent the proliferation of its natural host Acanthamoeba. This effect depends on the number of bacteria per amoebae and required the Dot/Icm secretion system. The arrest in Acanthamoeba multiplication provoked by L. pneumophila correlated with changes of the shape and motility of Acanthamoeba. Moreover, L. pneumophila induced a decrease in host mRNA levels of CDC2b, a putative CDK in Acanthamoeba. Furthermore, the genetic complementation of CDC2b cDNA from Acanthamoeba in CDC28-deficient S. cerevisiae demonstrates that CDC2b is a functional CDK and suggests that the cell cycle inhibition of Acanthamoeba upon infection could be related to down-regulation of CDC2b mRNA.
Results
L. pneumophila impairs proliferation of A. castellanii
In order to assess the impact of L. pneumophila on amoebal proliferation, A. castellanii 30010 were infected with L. pneumophila Paris or with Escherichia coli K12, which does not resist host digestion, at multiplicities of infection (MOIs) of 1, 5, 10 and 20. Proliferation of A. castellanii was evaluated for 48 h. In contrast to uninfected cells or A. castellanii co-cultured with E. coli, we did not observe any increase of the cell population when A. castellanii was infected with L. pneumophila (Fig. 1A). The difference between L. pneumophila infected and uninfected cells was significant as soon as 24 h post-infection (Fig. 1A). The inhibition of cell proliferation upon infection with L. pneumophila seemed dependent on the MOI used as this effect was more pronounced with higher MOIs (Fig. 1A). To address the possibility that the absence of proliferation of A. castellanii induced by L. pneumophila was bacterial or amoebal strain specific, we infected A. castellanii 30010 with the L. pneumophila Lens strain. Although compared to uninfected cells the difference was less pronounced at a MOI of 1, L. pneumophila Lens also impaired proliferation of A. castellanii (Fig. 1B). Similarly, another strain of A. castellanii (ATCC 30234) was co-cultured with L. pneumophila (Lens and Paris strains) and with E. coli K12 at a MOI of 20. Less proliferation over time was observed when infection was performed with L. pneumophila compared to uninfected cells or those infected with E. coli K12 (Fig. 1C). According to these results, L. pneumophila prevents proliferation of A. castellanii.
Figure 1. L. pneumophila prevents proliferation of A. castellanii.
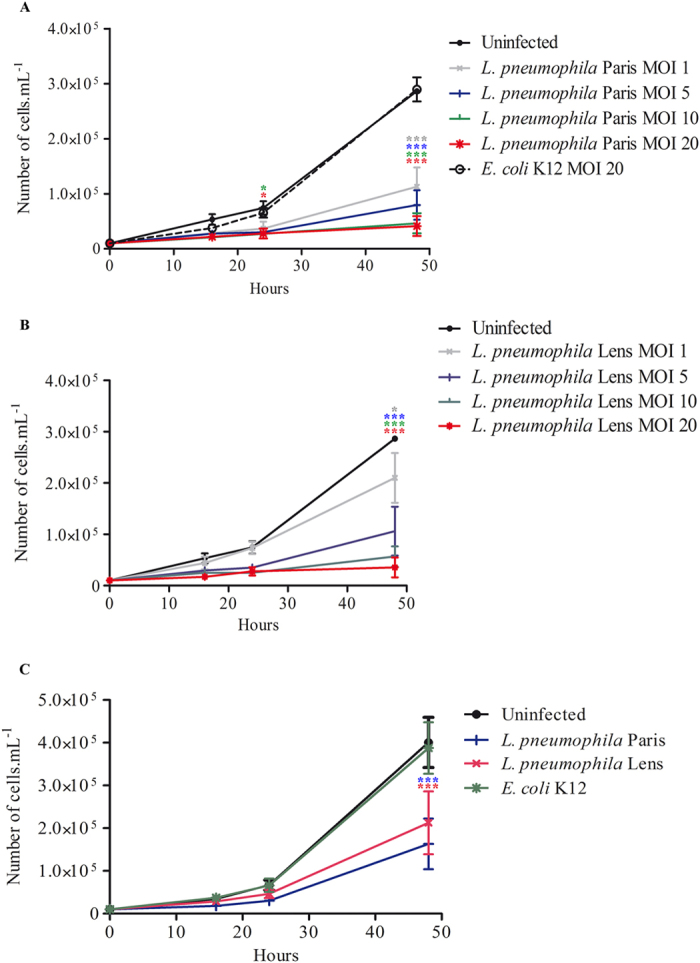
(A) A. castellanii ATCC 30010 were infected with L. pneumophila Paris and E. coli K12 or (B) with L. pneumophila Lens at a MOI of 1, 5, 10 and 20. Infections were carried out within the PAS solution for 2 h and cells were further incubated within the PYG medium containing gentamicin. Sixteen, twenty four and forty eight hours after infection, cells were harvested for counting. Results are average of three independent experiments and errors bars represent the standard error of the mean (±SEM). (C) A. castellanii ATCC 30234 were co-cultured with L. pneumophila Paris, L. pneumophila Lens and E. coli K12 at a MOI of 20. At different time points, cells were harvested for counting. Results are average of three independents experiments and errors bars represent the standard error of the mean (±SEM). The asterisks indicate conditions that are significantly different compared to uninfected cells (*p < 0.05; ***p < 0.001).
L. pneumophila inhibits proliferation of A. castellanii through the Dot/Icm secretion system
To address how L. pneumophila inhibited A. castellanii proliferation, amoebae were infected with live, heat-killed (65 °C) or ∆dotA mutant of L. pneumophila at a MOI of 20. The number of A. castellanii was assessed at different time-points to establish a kinetic of proliferation for each condition. In contrast to live L. pneumophila, infection with heat-killed or ∆dotA L. pneumophila did not impair proliferation of A. castellanii (Fig. 2). These results suggested that inhibition of A. castellanii proliferation requires live L. pneumophila with a functional Dot/Icm T4SS.
Figure 2. L. pneumophila prevents proliferation of A. castellanii through the Dot/Icm secretion system.
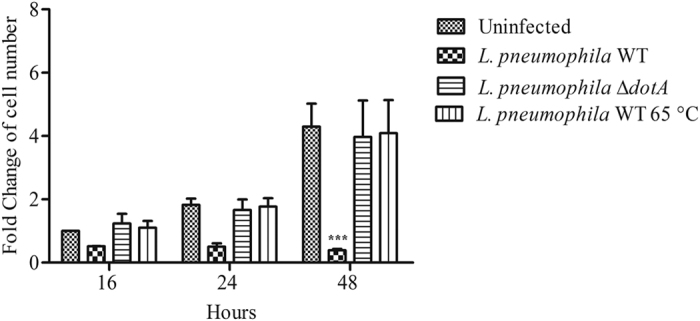
A. castellanii ATCC 30010 were infected with wild type (WT) L. pneumophila Paris, heat-killed (65 °C) and ∆dotA L. pneumophila Paris at a MOI of 20. Sixteen, twenty-four and forty-eight hours after infection, cells were harvested for counting. Results are average of three independents experiments and errors bars represent the standard error of the mean. Cell numbers are relative to uninfected cells at 16 h. The asterisks indicate conditions that are significantly different compared to uninfected cells (***p < 0.001).
L. pneumophila induces modifications in shape, motility and cell division of A. castellanii
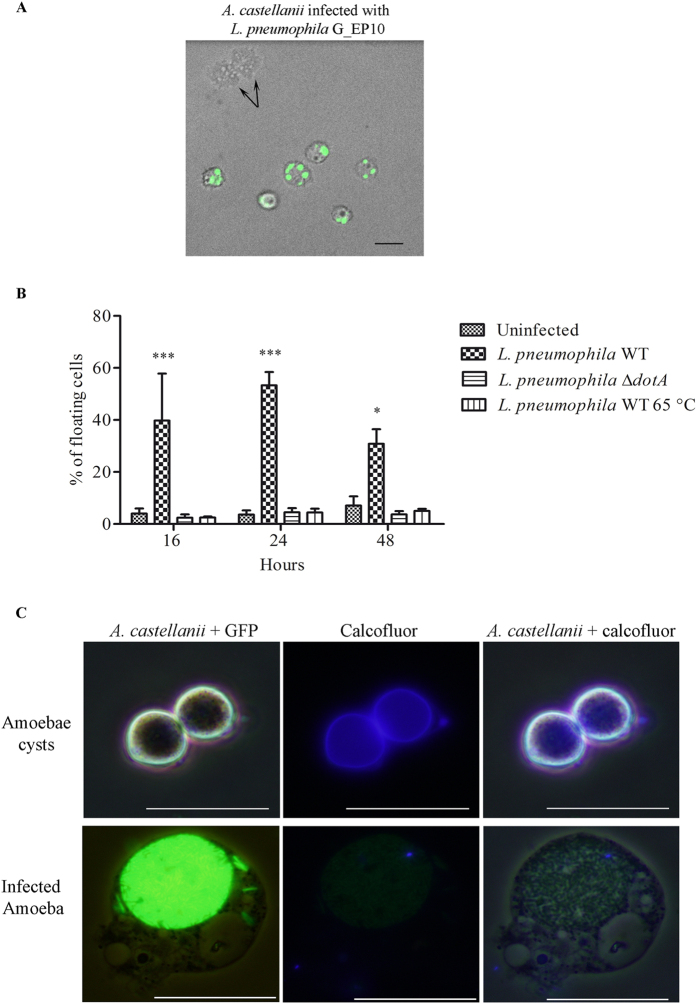
Time lapse microscopy was performed to visualize cell division of A. castellanii after infection with L. pneumophila Paris expressing the GFP protein (G_EP10). Videos were performed 16 h after infection and taken for approximately 17 h. As shown in the supplementary video 1, uninfected A. castellanii were highly motile and cell division was observed. Whereas in infected cells, we did not observe any cell division in GFP positive cells (supplementary video 2 and Fig. 3A). Infected cells seemed less motile with modifications of their shape as they became rounded (supplementary video 2 and Fig. 3A). In addition, cells positive for GFP seemed less adherent than the GFP negative cells (supplementary video 2).
Figure 3. L. pneumophila modifies the shape of A. castellanii.
(A) A. castellanii ATCC 30010 were infected with L. pneumophila Paris expressing GFP (G_EP10) at a MOI of 20. Green color represents the GFP signal. Images were captured 18 h post-infection. Black arrows show cell division of one mother cell into two daughter cells. The size bar represents 20 μm. (B) A. castellanii ATCC 30010 were infected with wild type (WT) L. pneumophila Paris, heat-killed and ∆dotA L. pneumophila Paris at a MOI of 20. At different time points, the percentage of floating cells was estimated. Results are average of three independents experiments and errors bars represent the standard error of the mean (±SEM). The asterisks indicate data that are significantly different compared to uninfected cells (*p < 0.05, ***p < 0.001). (C) A. castellanii ATCC 30010 were cultured in encystation medium or infected with L. pneumophila Paris G_EP10 at MOI of 20. Twenty-four hours after treatment or infection, cells were harvested and stained with calcofluor in order to reveal cyst forms. Transmitted light, green and blue channels represented respectively amoebae, GFP (L. pneumophila) and calcofluor (cellulose) signals. The size bar represents 20 μm.
In order to test cell adherence of A. castellanii following infection with L. pneumophila, floating cells were harvested after infection with live, heat-killed or ∆dotA mutant of L. pneumophila at a MOI of 20. We observed that in contrast to heat-killed and ∆dotA L. pneumophila, live wild-type L. pneumophila seemed to induce cell detachment (Fig. 3B). These data indicate that inhibition of cell division induced by L. pneumophila was associated with modifications of the shape and of the motility of A. castellanii. Moreover, L. pneumophila required a functional Dot/Icm secretion system to decrease adherence of A. castellanii from the surface.
A. castellanii has a life cycle that oscillates between a dormant and a replicative forms named cyst and trophozoite, respectively. Under unfavorable conditions, amoebae form cysts, which are resistant to environmental stress and metabolically inactive14. During encystation, A. castellanii retracts its pseudopodia and becomes rounded15,16. In order to verify that rounded-infected cells were not encysted, A. castellanii were stained with Calcofluor White to reveal cellulose in the cell wall of mature cysts17. We found that cells that were highly infected with L. pneumophila G_EP10 did not share phenotypic characteristics with A. castellanii cysts. Indeed, there was a striking difference in cell size and, in contrast to cysts, no cell wall containing cellulose was observed in infected amoebae with L. pneumophila G_EP10 (Fig. 3C). In our experimental conditions, round cells that seemed barely adherent upon infection with L. pneumophila were not cysts.
L. pneumophila impairs the ability of infected cells to replicate their DNA
Cell division and DNA replication represent the two essential cell cycle phases of any eukaryotic cell. Thus we assessed whether the absence of cell division upon L. pneumophila infection was associated to an inhibition of DNA synthesis. A. castellanii infected or not with L. pneumophila Paris expressing the DsRed protein (R_EP10) were incubated in a growth medium containing 5-ethynyl-2′-deoxyuridine (EdU) for 4 h. Incorporation of EdU, an analogue of thymidine, indicates DNA synthesis. Thus, as expected, uninfected cells were able to duplicate their DNA since around 20% of cells were positive for the EdU signal (Fig. 4A,C). However, in A. castellanii that were co-cultured with L. pneumophila R_EP10, the number of cells positive for EdU dropped to around 1% (Fig. 4B,C). In addition, we never detected cells positive both for EdU and DsRed signals (Fig. 4B). This result suggests that L. pneumophila prevented DNA replication in A. castellanii.
Figure 4. L. pneumophila prevents DNA synthesis in A. castellanii.
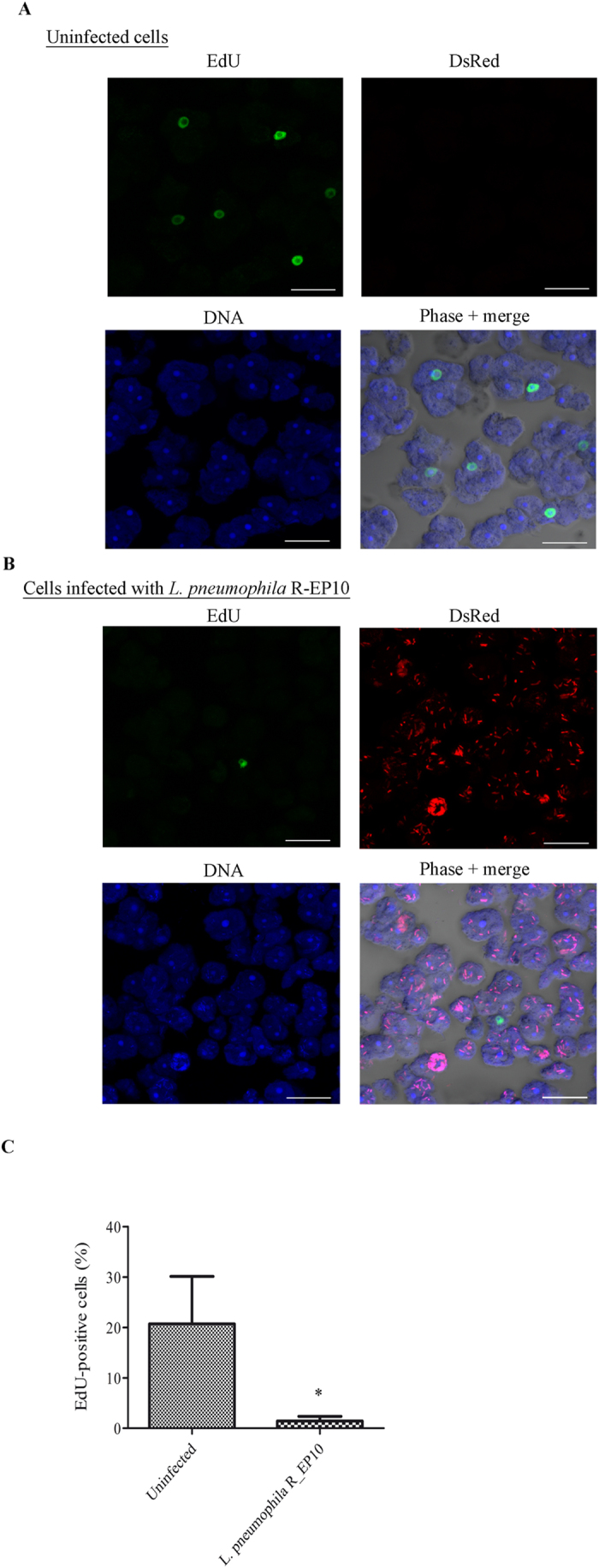
(A) Uninfected A. castellanii 30010 and (B) cells infected at a MOI of 20 with L. pneumophila Paris expressing DsRed (R_EP10) were incubated in PYG medium supplemented with gentamicin (20 μg/ml) and EdU (40 μM) for 4 h. For infected cells, the EdU treatment started just after infection. Cells were harvested for microscopic observation. EdU appears in green, DsRed signal in red, nuclei in blue (TO-PRO®-3 Iodide). The size bar represents 20 μm. (C) Quantification of EdU positive cells from 3 independent experiments, for a total of more than 350 A. castellanii counted. The graph shows the mean +/− SEM (*p ≤ 0.05).
The protein CDC2b from A. castellanii is a putative CDK
Eukaryotic cell cycle regulation is driven by cyclin dependent kinases (CDKs). Because Homo sapiens cells possess a considerable number of CDKs, we ran a blast search of protein sequences of human CDKs (supplementary Data 1)18 within the genome of A. castellanii (NCBI). Based on an E-value less than 1 and a percentage of identity of at least 35%, we obtained five candidates as putative CDKs of A. castellanii (Table 1). These five proteins were aligned with human CDKs in order to perform a phylogenetic analysis. Similarly to Cao et al.18, we used human (Hsa) cyclin-dependent kinase like-1 (CDKL1), glycogen synthase kinase-3 (GSK3) alpha and serine/threonine-protein kinase MAK isoform 1 as outgroups. Six of the human CDKs are directly related to the cell cycle (CDK1, CDK2, CDK3, CDK4, CDK6 and CDK7)19. As shown on the Fig. 5A, only the XP_004353710.1 CDC2b putative protein appeared phylogenetically similar to a cell cycle-related human CDK (CDK1).
Table 1. Putative A. castellanii CDK resulting from the blast of all human CDKs18.
| Accession number | Name |
|---|---|
| XP_004353710.1 | Cell division control protein 2b, putative (CDC2b) |
| XP_004336625.1 | Cdk10/11, putative |
| XP_004367653.1 | Protein kinase domain containing protein |
| XP_004335644.1 | Cyclindependent kinase |
| XP_004342572.1 | Protein kinase domain containing protein |
Figure 5. CDC2b from A. castellanii could be the regulator of the cell cycle.
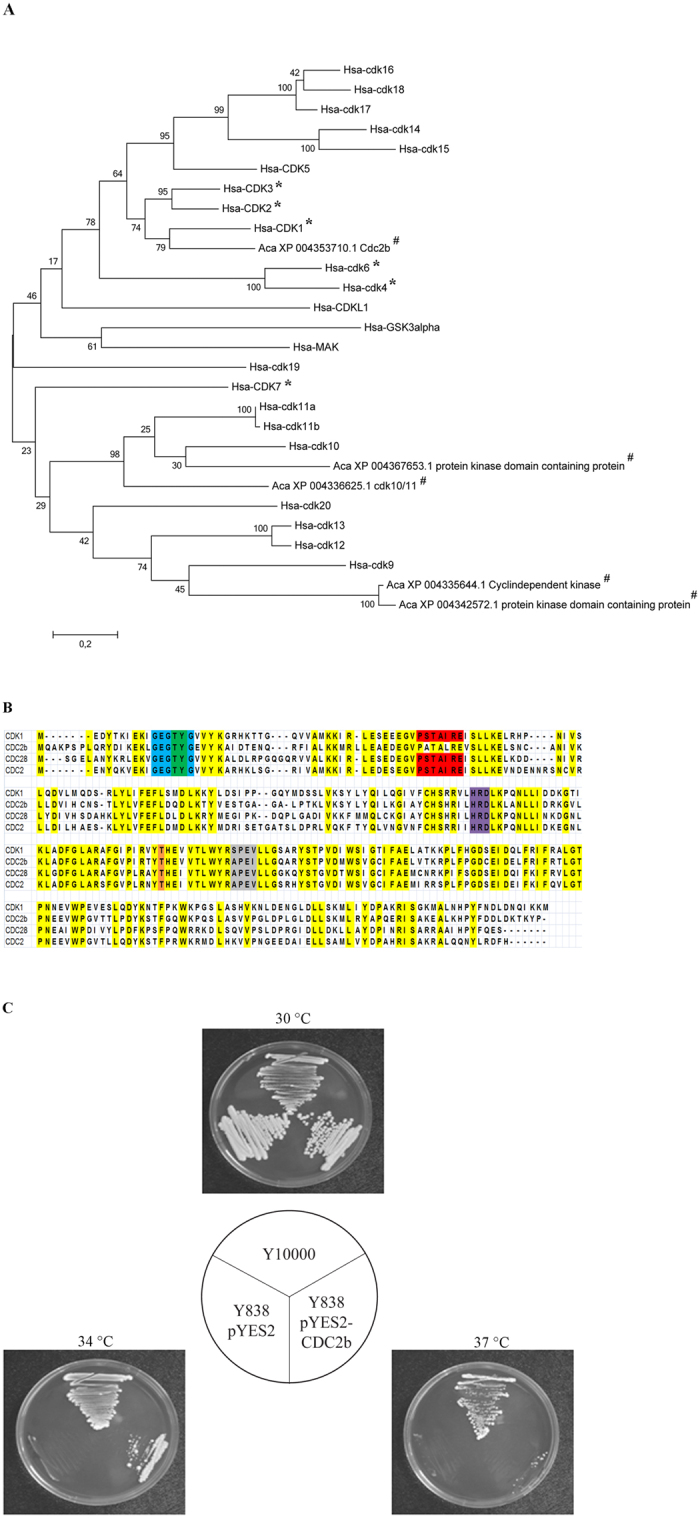
(A) CDK phylogeny inferred by Maximum Likelihood based on human (Hsa) CDKs18 and putative A. castellanii (Aca) CDKs. 1000 bootstrap replications were inferred and displayed at the nodes. The scale bar represents the number of amino acid substitution per site. All proteins were first labeled with their species name (Aca, A. castellanii; Hsa, Homo sapiens). *Indicates humans cell cycle-related proteins and # represents proteins from A. castellanii. (B) Sequence alignment between CDC2b (XP_004353710.1) from A. castellanii, CDK1 (NP_001777.1) from Homo sapiens, CDC28 (NP_009718.3) from Saccharomyces cerevisiae and CDC2 (CAC37513.1) from Schizosaccharomyces pombe using MUSCLE of the MEGA6 software. Yellow color highlight conserved residues. Motifs characteristics of CDK are displayed in blue (ATP-binding domain), green (inhibitory phosphorylation sites), red (cyclin-binding domain), purple (start of T-loop), orange (activating phosphorylation site) and grey (end of T-loop). (C) The S. cerevisiae strains Y10000 (wild type), Y838 (cdc28-4) transformed with the empty plasmid pYES2 or the recombinant plasmid pYES2-CDC2b were streaked on galactose-containing medium and cultivated at 30 °C, 34 °C or 37 °C for 4 days.
In order to further assess the similarity between CDC2b from A. castellanii and well described eukaryotic CDKs, we aligned the protein sequences with those of human (CDK1), and of Baker’s and fission yeast, Saccharomyces cerevisiae (CDC28) and Schizosaccharomyces pombe (CDC2), respectively. We found that the CDC2b protein sequence was highly conserved compared to the CDKs from the other organisms (Fig. 5B). Aside from the PSTAIRE cyclin-binding domain, all the CDKs we aligned displayed many highly conserved residues and domains that characterize CDKs19 such as an ATP-binding domain, an inhibitory phosphorylation site, an activating phosphorylation site and the start and end of a T-loop (Fig. 5B). Thus, the putative protein CDC2b from A. castellanii shares strong homology with other CDKs that are essential for the progression of the cell cycle.
Since S. cerevisiae can be used to test the function of proteins from A. castellanii20, we sought to determine whether heterologous expression of CDC2b could complement the loss of function of the CDC28 gene from yeast. To this end, the coding sequence of CDC2b was introduced into plasmid pYES2 under the control of an inducible GAL1 promoter. The yeast strain Y838, which harbors a temperature sensitive mutation cdc28-4, was transformed with the recombinant plasmid pYES2-CDC2b or the empty vector as a negative control. After propagation of the transformants at the permissive temperature, they were challenged for growth on YPGal medium at various temperatures. As shown in Fig. 5C, Y838 transformed with pYES-CDC2b was capable of robust growth at 30 °C or 34 °C but could not grow at 37 °C, whereas the same strain transformed with the empty plasmid was unable to grow above 30 °C. In contrast, the wild type strain Y10000 was able to grow regardless of the temperature of incubation (Fig. 5C). We conclude from this experiment that A. castellanii CDC2b can restore growth of the cdc28-4 mutant but not as well as a wild type yeast strain.
L. pneumophila provokes a decrease of the CDC2b gene expression
We next explored whether L. pneumophila modulates expression levels of CDC2b. We infected A. castellanii with L. pneumophila and monitored the level of CDC2b mRNA at different time-points. One found that L. pneumophila decreased CDC2b mRNA level as soon as 2 h after the beginning of the infection (0 h). The difference between uninfected and infected cells was significant at 0 and 2 h post-infection (Fig. 6). These results indicate that L. pneumophila infection leads to down-regulation of the mRNA levels of CDC2b, which correlates with a defect in amoebal proliferation during infection.
Figure 6. L. pneumophila induces a down-regulation of the A. castellanii cell cycle related protein CDC2b.
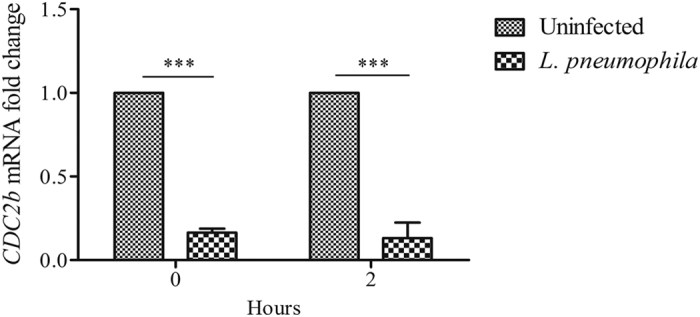
A. castellanii ATCC 30010 were infected with L. pneumophila Paris at a MOI of 20. RT-qPCR for CDC2b mRNA extracted was performed just after the end of infection (0 h) and 2 h latter (2 h). Expression levels were normalized to the 18S mRNA. CDC2b levels are presented as levels relative to the uninfected condition. Results are average of three independent experiments and errors bars represent the standard error of the mean (±SEM). The asterisks indicate a significant difference between uninfected and infected conditions (***p < 0.001).
Discussion
L. pneumophila is an intracellular bacterium that manipulates host functions to support its replication. However, the impact of Legionella infection on the host cell cycle has never been addressed. In this study, we demonstrate that L. pneumophila prevents multiplication of its natural host A. castellanii through the T4SS. Infection with L. pneumophila impairs cell division, DNA synthesis and motility of A. castellanii. The inability of infected cells to multiply correlates with a decrease of the mRNA levels of the putative amoebal cell cycle regulator CDC2b.
Some studies previously suggested a negative effect of L. pneumophila on the proliferation of Acanthamoeba21,22. In both studies, authors observed a decrease of the amoebal cell number upon infection, at least three days later, that was associated to an amoebal lysis21,22. Indeed, L. pneumophila provokes necrosis-mediated lysis of Acanthamoeba within 48 h after infection23. Here, we demonstrate that, even only several hours after infection with L. pneumophila, A. castellanii were unable to replicate their DNA, nor to undergo cytokinesis; two critical steps for cell proliferation. Thus, inhibition of proliferation, and not only Legionella-induced host lysis could account for this decrease in cell number. The type IV secretion system Dot/Icm was necessary for the inhibition of the host proliferation induced by L. pneumophila. Our ongoing work aims at identifying the L. pneumophila effectors involved in the regulation of proliferation of A. castellanii.
Our video microscopy experiments revealed that L. pneumophila induced a modification of the shape of amoebae which became rounded with some detachment from the substrate. The membrane of infected amoebae differed from mature Acanthamoeba cyst as no evidence for cellulose was observed by calcofluor stain. Modulation of encystment has been shown with other bacteria24,25. Regarding Legionella, controversially results could be found. Several papers have described cysts fully bound with L. pneumophila26,27 while some authors did not succeed to isolate infected cysts28. This suggests that some specific conditions might be required to obtain infected cysts with L. pneumophila. This question is of importance since cysts are thought to play a role in persistence and dissemination of L. pneumophila.
The alteration in amoebal shape and motility that we observed support previous results indicating that L. pneumophila modulates the cell host cytoskeletal molecules and migration29,30,31,32. Interestingly, treatment of the social amoeba Dictyostelium discoideum with the Legionella quorum sensing molecule LAI-1 induced down-regulation of genes involved in movement and in cell proliferation31. LAI-1 led to inactivation of the cytoskeletal-regulating protein CDC42 that was reported to induce cell cycle progression33,34. We aim to characterize the possible contribution of LAI-1 to the inhibition of A. castellanii proliferation in future studies.
Legionella pneumophila was shown to regulate transcription of genes implicated in proliferation of human monocyte-derived macrophages and of bone marrow-derived macrophages35,36. Furthermore, L. pneumophila modifies the host chromatin resulting in a repression of gene expression37,38. Since chromatin structure impinges on the cell cycle and vice-versa39, contribution of effectors implicated in chromatin-remodeling is a promising area to investigate.
To our knowledge, CDK proteins have never been characterized in A. castellanii. Thanks to genome sequencing of A. castellanii13, we found that the putative protein CDC2b shares a strong homology with other cell cycle-related CDKs. CDC2b could restore the ability of CDC28-deficient S. cerevisiae to grow although growth of transformants was limited at high temperature. Interestingly, the same conclusion was reported for the CDC2 gene of the distantly related amoeba Dictyostelium discoideum40. As CDK acts in concert with cyclin proteins, growth limitation of transformants carrying CDC2b could arise from the putative cyclin-binding domain of CDC2b that is different from the conserved PSTAIRE motif. This would be analogous to CDC2 from D. discoideum whose PSTAIRE is not completely conserved with an isoleucine substituted for leucine40.
From Amoebozoa to Vertebrata, several CDK proteins are present in one cell. The number of CDKs increases with eukaryotic evolution. While 20 CDKs are found in humans, only 6 and 8 CDKs are found in the yeast S. cerevisiae and in the amoeba D. discoideum, respectively18. As reported in yeast and mammalian systems, only one CDK is essential to drive the cell cycle from DNA replication to mitosis10,11. The ability of CDC2b to functionally complement CDC28-deficient S. cerevisiae suggests that CDC2b could represent the main cell cycle regulator in A. castellanii. Although as yet there is no method to generate mutants in A. castellanii, such experiments are needed to confirm our hypothesis.
Intracellular bacteria were reported to affect the growth rate of free-living amoebae41. Beyond, several bacteria are able to promote or to inhibit the eukaryotic cell cycle by using bacterial effectors called cyclomodulins42,43. Inhibitory cyclomodulins impair the function of cyclin/CDK complexes through activation of the DNA-damage response, tubulin protein sequestration, down-regulation of cyclins, stabilization of CDK inhibitors, interaction with proteins of the anaphase promoting complex or cleavage of the regulator of the endoplasmic reticulum (ER) function Bip44,45,46. Because cyclomodulins induce pleitropic effects on target cells, the next challenge will be to confirm that the decrease of CDC2b mRNA by L. pneumophila is responsible for the A. castellanii proliferation prevention and how this transcriptional/translational effect is important for L. pneumophila. Since down-regulation of CDK expression upon bacterial infection has never been reported, the down-regulation of the CDC2b mRNA level could be a novel strategy for a bacterium to regulate proliferation of its host.
In summary, this study shows for the first time that L. pneumophila prevents the proliferation of its host. Our work identifies a novel CDK of A. castellanii and shows that Legionella perturbs host proliferation, DNA replication and amoebal morphology during infection. Regulation of the host cell cycle contributes to the impressive list of the eukaryotic functions that are perturbed by L. pneumophila. We are currently attempting to decipher the signaling pathway responsible for A. castellanii cell cycle arrest and the bacterial effectors which mediate this effect during infection.
Methods
Strains, media and culture conditions
A. castellanii ATCC 30010 and ATCC 30234 were cultured in Peptone Yeast Glucose medium (2% proteose peptone, 0.1% yeast extract, 0.1 M glucose, 4 mM MgSO4, 0.4 mM CaCl2, 0.1% sodium citrate dehydrate, 0.05 mM Fe(NH4)2(SO4)2 6H2O, 2.5 mM NaH2PO3, 2.5 mM K2HPO3, pH 6.5) at 30 °C.
To induce encystation, A. castellanii were incubated in encystation buffer (0.1 M KCl, 8 mM MgSO4, 0.4 mM CaCl2, 20 mM Tris (2-amino-2hydroxymethyl-1,3-propanediol), 1 mM NaHCO3, pH 8.8) at 30 °C for 24 h47.
L. pneumophila strains were cultured on buffered charcoal yeast extract (BCYE) (1% ACES, 1% yeast extract, 0.2% charcoal, 1.5% agar, 0.025% Iron (III) pyrophosphate, 0.04% L-cysteine, pH 6.9) agar plate. For L. pneumophila Paris strains, 0.1% alpha-ketoglutarate was added to medium. Bacteria were cultured on BCYE at 37 °C for 3 days. Then, bacteria were inoculated at the optical density at 600 nm (OD600) of 0.1 in Buffered Yeast Extract (BYE) at 37 °C under agitation 160 rpm to reach OD600 above 2.3.
Different strains of L. pneumophila were used as L. pneumophila Paris CIP 107629T8 and L. pneumophila Lens CIP1082868. The ∆dotA mutant of L. pneumophila Paris (resistant to the kanamycin (15 μg/mL)) was a gift of Carmen Buchrieser from Institut Pasteur (Paris, France)48. E. coli K12 (ATCC 10798) was cultured on Lysogeny Broth (LB) medium at 37 °C under agitation at 160 rpm.
S. cerevisiae strains were cultured in minimal YNB medium (yeast nitrogen base with ammonium sulfate and 2% glucose) supplemented with the appropriate amino acids and bases, or complete YPGal medium (1% yeast extract, 1% peptone, 2% galactose) at 28 °C unless otherwise indicated. Yeast strains used in this study are Y10000 (BY4742; MATαura3Δ0leu2Δ0 his3Δ1 lys2Δ0) and Y838 (cdc28-4 ura3-52 lys2-801 leu2∆1 his3∆200).
Bacterial transformation
L. pneumophila Paris in exponential phase were washed with 10% cold glycerol solution and centrifuged (6000 g, 10 min at 4 °C). Pellets were washed in 20, 10 and 5 mL of 10% cold glycerol solution. Bacteria were re-suspended in 10% cold glycerol solution to reach an OD600 of 100. Electroporation was performed with the plasmid pSW00149 for bacteria producing the DsRed Fluorescent Protein and with pNT28 built from the plasmid pMMB207-Km14-GFPc50 for bacteria producing the Green Fluorescent Protein (GFP). One μg of plasmid and 100 μL of bacterial suspension were mixed. Electroporation was performed by using the EC2 programm (2.50 kV, 1 pulse) of the MicroPulser apparatus (Biorad). After the electroporation, 900 μL of liquid BYE were added to the bacteria followed by an incubation at 37 °C without agitation for two hours. Suspension was spread on BCYE supplemented with chloramphenicol (5 μg/mL) and the petri dishes were incubated at 37 °C during 3 or 4 days. The bacteria L. pneumophila Paris producing the DsRed and the GFP proteins were named L. pneumophila R_EP10 and L. pneumophila G_EP10 respectively.
Infection of A. castellanii
Three days old A. castellanii were seeded in 24-well plates at 1 × 104 cells per well in Page’s Amoeba Saline solution (PAS) (4 mM MgSO4, 0.4 M CaCl2, 0.1% sodium citrate dehydrate, 2.5 mM NaH2PO3, 2.5 mM K2HPO3, pH 6.5) for about 2 h at 30 °C. Once cells adhered on the plate, bacteria that reached an OD600 above 2.3 were added to the amoebae at different multiplicity of infection. The infection was synchronized by centrifugation (500 g, 10 min at room temperature). Co-cultures were incubated 2 h at 30 °C, rinsed with PAS and cultured within the PYG medium containing gentamicin (20 μg/mL). Plates were incubated at 30 °C.
To estimate the proliferation of A. castellanii, cells were harvested at different time-points and counted in triplicate for each condition using plastic counting slides FastRead 102 (Biosigma).
To obtain heat-killed L. pneumophila, bacteria were heated at 65 °C for 15 min.
Estimation of the cell detachment upon infection was performed by counting floating and adherent cells. The percentage of floating cells represents the number of floating cells under total cells, multiplied per 100.
Percentage of infected cells was assessed on epifluorescence microscope (Olympus BX41) using L. pneumophila Paris G_EP10. A. castellanii were seeded in 6-well plates at 1 × 106 cells/well and infection was performed with L. pneumophila Paris G_EP10 at MOI 20. After the infection, cells were incubated 30 min in PYG medium, and amoebae were harvested, centrifuged (500 g, 5 min at room temperature) and washed with Phosphate-Buffered Saline (PBS). Cells were fixed for 15 min with 4% paraformaldehyde at room temperature, washed twice with PBS and centrifuged. The pellet was suspended in 40 μL of CitiFluor® AF1 (Citifluor) and slide were analyzed under the microscope.
Cysts were assessed by a fluorescent microscopic detection using Calcofluor White Reagent Droppers (Becton Dickinson) according manufacturer’s instructions. 4.5 μL of A. castellanii and 0.5 μL of calcofluor were loaded on a glass slide. The mix was incubated 2 min at room temperature for staining followed by microscopic observations.
Video microscopy
A. castellanii 30010 (1.35 × 104 cells) were infected with L. pneumophila Paris G_EP10 at a MOI of 20 for 2 h in a μ-Slide 8 well IbiTreat microscopy chamber (Ibidi). Transmission images were acquired with confocal spinning disk from Andor technology mounted on an Olympus inverted IX81 microscope using Andor Ixon + 897 back illuminated EMCCD camera. Live imaging started 16 h after the end of the infection and run for around 17 h using the spinning disk confocal laser microscopy. Time lapse imaging was realized in 30 °C incubation chamber with x40 objective (512 × 512 pixels, 0.33 μm/pixel) and 1 image every 30 seconds.
DNA synthesis
The DNA synthesis was determined using the Click-iT® EdU Imaging Kits (Invitrogen™) following manufacturer’s recommendations with some modifications. Briefly, 1 × 106 A. castellanii cells were infected with L. pneumophila Paris R_EP10 at MOI 20 in 6-well plates. After infection period, EdU (40 μM) was added to the PYG medium supplemented with gentamicin (20 μg/mL) for 4 h. Amoebae were harvested and centrifuged (1000 g, 10 min at room temperature) before fixation for 15 min at room temperature with 3.7% paraformaldehyde. Amoebae were washed twice with 1 mL of washing solution (3% bovine serum albumin (BSA) (Sigma)) in PBS and lysed for 20 min at room temperature with 1% Triton® X-100 (Sigma). After two additional washes, 0.5 mL of Click-iT® reaction cocktail (according to the manufacturer’s instructions) was added to the pellet and incubated 1 h protected from the light. For DNA staining, amoebae were washed with 1 mL of PBS, treated with TO-PRO®-3 Iodide (dilution of 1:1000) and incubated for 1 h at room temperature in the dark. Amoebae were washed for the last time with PBS and suspended in washing buffer for analysis.
A. castellanii were examined with a laser scanning confocal microscope (FluoView-1000, Olympus) coupled to an inverted microscope IX-81(Olympus). Images were obtained with an Olympus UPLSAPO 60X W NA: 1.2 and zoom x2 (800 × 800 pixels, 0.13 μm/pixel). Samples were excited with 488/500–530 nm excitation/emission filters for EdU staining, 543/555–625 nm for bacteria producing DsRed and 633/LP650 nm for Topro-3 nuclear staining. Multiple fluorescence signals were acquired sequentially to avoid cross-talk between colour channels. For 3D acquisition, optical sectioning of the specimen (Z series) was driven by a Z-axis stepping motor and maximum intensity projection was further generated.
Extraction and purification of RNA
1 × 106 cells of A. castellanii were infected with L. pneumophila Paris at a MOI of 20 for 2 h in 6-well plates. After the infection (0 h), cells were harvested for RNA extraction at different time points 2 h, 16 h and 24 h. To increase the efficiency of the cell lysis, RNA extractions were preceded by a mechanical cell lysis using a FastPrep®-24 Instrument (MP) for 30 seconds at a speed of 5.0 (twice) with FastPrep Tubes containing glass breads 150–212 μm (Sigma). RNA extraction was performed using the High Pure RNA Isolation Kit (Roche).
An additional digestion of residual DNA was performed with Turbo DNA-free™ kit (Ambion®) according to manufacturer’s instructions.
Reverse transcription (RT) and quantitative PCR (qPCR)
Reverse transcription of RNA was performed using GoScript™ Reverse Transcription System (Promega) following manufacturer’s recommendations. Products of reverse transcription (RT) were purified with NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel). All cDNA were normalized (10 μg/mL) before proceeding to qRT-PCR reaction. All qPCR reactions were performed using the LightCycler® FastStart DNA MasterPlus SYBR Green I kit (Roche) on the LightCycler 1.5 instrument (Roche). Each tube contained 9 μL of reaction mix containing 0.5 μL of each primers CDC2b (XM_004353658.1) (5′ATGCAAGCCAAACCCAGTC 3′ and reverse: 5′ GAATCGCTGGTTCTCGGTATC 3′) or 18S primers51 at 10 μM, 2 μL of Master Mix 5X, 6 μL of water and 1 μL of cDNA or 1 μL of water for negative control. The qPCR program was: 10 min at 95 °C and 45 cycles of the amplification step (10 sec at 95 °C, 10 sec at 61 °C and 10 sec at 72 °C for extension time in a single acquisition mode), the melting curves step for 1 min at 65 °C for annealing and the cooling 30 seconds at 40 °C. The relative level of genes expression was calculated using the 2−∆∆Ct method52. Expression level were normalized to the 18S mRNA.
Phylogenetic analysis
To construct the phylogenetic tree, proteins were aligned using MUSCLE of the software MEGA6 with a Maximum likelihood and a bootstrap replications of 1000.
A. castellanii CDC2b cloning
The CDC2b (XM_004353658.1) cDNA from A. castellanii was amplified by PCR from total cDNA with flanking BamHI and XbaI restriction sites using primers ACA-cdc2b-5′ (TTTTTTTGGATCCTACACAATGCAAGCCAAACCCAGTCC) and ACA-cdc2b-3′-STOP (TTTTTTTTCTAGATTAGGGGTACTTGGTCTTGTCC) and inserted between BamHI and XbaI sites of the S. cerevisiae expression plasmid pYES2-CT (Invitrogen), yielding pWA-cdc2b-Ac. Plasmid DNA was prepared with a NucleoSpin Plasmid kit (Macherey-Nagel).
Yeast transformation of Y838 was done by the Li Acetate method as described by Gietz and Woods53. This strain is temperature sensitive for growth (ts−) due to a deficient CDC28 gene product. Transformants were selected at the permissive temperature (28 °C) on YNB medium supplemented with adequate amino acids and bases. For complementation assays of the cdc28-4ts− phenotype, transformants were streaked on YPGal medium and incubated at the indicated temperature (30 °C, 34 °C or 37 °C) for 4 days.
Statistical analyses
All experiments were performed three times. Results were analyzed by Two-way RM ANOVA with the Bonferroni post-test (GraphPad Prism5) or using a one-tailed Mann-Whitney test considering a statistical significance at p ≤ 0.05. All data are average of three independents experiments and error bars represent the standard error of the mean (±SEM).
Additional Information
How to cite this article: Mengue, L. et al. Legionella pneumophila prevents proliferation of its natural host Acanthamoeba castellanii. Sci. Rep. 6, 36448; doi: 10.1038/srep36448 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work has benefited from the facilities and expertise of ImageUP platform (Université de Poitiers). We thank Anne Cantereau (STIM-ERL7368 CNRS, Université de Poitiers) for her help on analysis in microscopy. We thank Dr. Carmen Buchrieser (Institut Pasteur de Paris, France) for the gift of ∆dotA L. pneumophila Paris and Dr. Hubert Hilbi (Max von Pettenkofer-Institute, Germany) for the gift of plasmids expressing GFP and DsRed. We thank as well Dr. Paule Seite (Université de Poitiers, France) for her technical assistance. We gratefully acknowledge Dr. Carl Mann (Université Paris-Saclay, France) for providing the cdc28-4 strain. We would like to thank Dr. Liliana Radoshevich (Institut Pasteur de Paris, France) for critical reading.
Footnotes
Author Contributions L.M., M.R., Y.H. and A.S.-L. conceived and designed the experiments; L.M., M.R., W.A. and E.P. performed the experiments; L.M., M.R., Y.H. and A.S.-L. analyzed data; L.M., M.R., W.A. and E.P. contributed reagents/materials/analysis tools; L.M., M.R. and A.S.-L. wrote the paper.
References
- Borella P. et al. Legionella infection risk from domestic hot water. Emerging infectious diseases 10, 457–464, doi: 10.3201/eid1003.020707 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields B. S., Benson R. F. & Besser R. E. Legionella and Legionnaires’ disease: 25 years of investigation. Clinical microbiology reviews 15, 506–526 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowbotham T. J. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. Journal of clinical pathology 33, 1179–1183 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greub G. & Raoult D. Microorganisms resistant to free-living amoebae. Clinical microbiology reviews 17, 413–433 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac D. T. & Isberg R. Master manipulators: an update on Legionella pneumophila Icm/Dot translocated substrates and their host targets. Future microbiology 9, 343–359, doi: 10.2217/fmb.13.162 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifshitz Z. et al. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proceedings of the National Academy of Sciences of the United States of America 110, E707–715, doi: 10.1073/pnas.1215278110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W. et al. Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PloS one 6, e17638, doi: 10.1371/journal.pone.0017638 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalet C. et al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet 36, 1165–1173, doi: 10.1038/ng1447 (2004). [DOI] [PubMed] [Google Scholar]
- Harashima H., Dissmeyer N. & Schnittger A. Cell cycle control across the eukaryotic kingdom. Trends in cell biology 23, 345–356, doi: 10.1016/j.tcb.2013.03.002 (2013). [DOI] [PubMed] [Google Scholar]
- Reed S. I. & Wittenberg C. Mitotic role for the Cdc28 protein kinase of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 87, 5697–5701 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria D. et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature 448, 811–815, doi: 10.1038/nature06046 (2007). [DOI] [PubMed] [Google Scholar]
- Jantzen H. & Schulze I. Growth condition-induced precocious activation of p34cdc2 kinase inhibits the expression of developmental competence. Developmental biology 166, 311–322, doi: 10.1006/dbio.1994.1317 (1994). [DOI] [PubMed] [Google Scholar]
- Clarke M. et al. Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome biology 14, R11, doi: 10.1186/gb-2013-14-2-r11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouque E. et al. Cellular, biochemical, and molecular changes during encystment of free-living amoebae. Eukaryotic cell 11, 382–387, doi: 10.1128/EC.05301-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers B. & Korn E. D. The fine structure of Acanthamoeba castellanii (Neff strain). II. Encystment. The Journal of cell biology 41, 786–805 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez-Munguia B. et al. Acanthamoeba castellanii cysts: new ultrastructural findings. Parasitology research 112, 1125–1130, doi: 10.1007/s00436-012-3261-7 (2013). [DOI] [PubMed] [Google Scholar]
- El-Sayed N. M. & Hikal W. M. Several staining techniques to enhance the visibility of Acanthamoeba cysts. Parasitol Res 114, 823–830, doi: 10.1007/s00436-014-4190-4 (2015). [DOI] [PubMed] [Google Scholar]
- Cao L. et al. Phylogenetic analysis of CDK and cyclin proteins in premetazoan lineages. BMC evolutionary biology 14, 10, doi: 10.1186/1471-2148-14-10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S. & Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development 140, 3079–3093, doi: 10.1242/dev.091744 (2013). [DOI] [PubMed] [Google Scholar]
- Régnacq M. et al. Identification of Atg8 from Acanthamoeba castellanii by genetic complementation in Saccharomyces cerevisiae. Molecular and biochemical parasitology. In press (2016). [DOI] [PubMed] [Google Scholar]
- Anacarso I. et al. Influence of Legionella pneumophila and other water bacteria on the survival and growth of Acanthamoeba polyphaga. Archives of microbiology 192, 877–882, doi: 10.1007/s00203-010-0618-0 (2010). [DOI] [PubMed] [Google Scholar]
- Dey R., Bodennec J., Mameri M. O. & Pernin P. Free-living freshwater amoebae differ in their susceptibility to the pathogenic bacterium Legionella pneumophila. FEMS microbiology letters 290, 10–17, doi: 10.1111/j.1574-6968.2008.01387.x (2009). [DOI] [PubMed] [Google Scholar]
- Gao L. Y. & Kwaik Y. A. The mechanism of killing and exiting the protozoan host Acanthamoeba polyphaga by Legionella pneumophila. Environ Microbiol 2, 79–90 (2000). [DOI] [PubMed] [Google Scholar]
- El-Etr S. H. et al. Francisella tularensis type A strains cause the rapid encystment of Acanthamoeba castellanii and survive in amoebal cysts for three weeks postinfection. Appl Environ Microbiol 75, 7488–7500, doi: 10.1128/AEM.01829-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitsch D. et al. Proteomic aspects of Parachlamydia acanthamoebae infection in Acanthamoeba spp. The ISME journal 4, 1366–1374, doi: 10.1038/ismej.2010.68 (2010). [DOI] [PubMed] [Google Scholar]
- Kilvington S. & Price J. Survival of Legionella pneumophila within cysts of Acanthamoeba polyphaga following chlorine exposure. The Journal of applied bacteriology 68, 519–525 (1990). [DOI] [PubMed] [Google Scholar]
- Skinner A. R., Anand C. M., Malic A. & Kurtz J. B. Acanthamoebae and environmental spread of Legionella pneumophila. Lancet 2, 289–290 (1983). [DOI] [PubMed] [Google Scholar]
- Bouyer S., Imbert C., Rodier M. H. & Hechard Y. Long-term survival of Legionella pneumophila associated with Acanthamoeba castellanii vesicles. Environ Microbiol 9, 1341–1344, doi: 10.1111/j.1462-2920.2006.01229.x (2007). [DOI] [PubMed] [Google Scholar]
- Chong A., Lima C. A., Allan D. S., Nasrallah G. K. & Garduno R. A. The purified and recombinant Legionella pneumophila chaperonin alters mitochondrial trafficking and microfilament organization. Infection and immunity 77, 4724–4739, doi: 10.1128/IAI.00150-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothmeier E. et al. Activation of Ran GTPase by a Legionella effector promotes microtubule polymerization, pathogen vacuole motility and infection. PLoS pathogens 9, e1003598, doi: 10.1371/journal.ppat.1003598 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S. et al. Inter-kingdom Signaling by the Legionella Quorum Sensing Molecule LAI-1 Modulates Cell Migration through an IQGAP1-Cdc42-ARHGEF9-Dependent Pathway. PLoS pathogens 11, e1005307, doi: 10.1371/journal.ppat.1005307 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S., Wagner M. A., Rothmeier E., Muller-Taubenberger A. & Hilbi H. Icm/Dot-dependent inhibition of phagocyte migration by Legionella is antagonized by a translocated Ran GTPase activator. Cellular microbiology 16, 977–992, doi: 10.1111/cmi.12258 (2014). [DOI] [PubMed] [Google Scholar]
- Lamarche N. et al. Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65PAK and the JNK/SAPK MAP kinase cascade. Cell 87, 519–529 (1996). [DOI] [PubMed] [Google Scholar]
- Olson M. F., Ashworth A. & Hall A. An essential role for Rho, Rac, and Cdc42 GTPases in cell cycle progression through G1. Science 269, 1270–1272 (1995). [DOI] [PubMed] [Google Scholar]
- Fortier A., Faucher S. P., Diallo K. & Gros P. Global cellular changes induced by Legionella pneumophila infection of bone marrow-derived macrophages. Immunobiology 216, 1274–1285, doi: 10.1016/j.imbio.2011.06.008 (2011). [DOI] [PubMed] [Google Scholar]
- Price C. T. & Abu Kwaik Y. The transcriptome of Legionella pneumophila-infected human monocyte-derived macrophages. PloS one 9, e114914, doi: 10.1371/journal.pone.0114914 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T. et al. SET-domain bacterial effectors target heterochromatin protein 1 to activate host rDNA transcription. EMBO reports 14, 733–740, doi: 10.1038/embor.2013.86 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolando M. et al. Legionella pneumophila effector RomA uniquely modifies host chromatin to repress gene expression and promote intracellular bacterial replication. Cell host & microbe 13, 395–405, doi: 10.1016/j.chom.2013.03.004 (2013). [DOI] [PubMed] [Google Scholar]
- Ma Y., Kanakousaki K. & Buttitta L. How the cell cycle impacts chromatin architecture and influences cell fate. Frontiers in genetics 6, 19, doi: 10.3389/fgene.2015.00019 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C. & Weeks G. Isolation and characterization of a cdc 2 cDNA from Dictyostelium discoideum. Biochimica et biophysica acta 1132, 35–42 (1992). [DOI] [PubMed] [Google Scholar]
- Collingro A. et al. Chlamydial endocytobionts of free-living amoebae differentially affect the growth rate of their hosts. Eur J Protistol 40, 57–60 (2004). [Google Scholar]
- Nougayrede J. P., Taieb F., De Rycke J. & Oswald E. Cyclomodulins: bacterial effectors that modulate the eukaryotic cell cycle. Trends in microbiology 13, 103–110, doi: 10.1016/j.tim.2005.01.002 (2005). [DOI] [PubMed] [Google Scholar]
- Oswald E., Nougayrede J. P., Taieb F. & Sugai M. Bacterial toxins that modulate host cell-cycle progression. Current opinion in microbiology 8, 83–91, doi: 10.1016/j.mib.2004.12.011 (2005). [DOI] [PubMed] [Google Scholar]
- Huang J., Lesser C. F. & Lory S. The essential role of the CopN protein in Chlamydia pneumoniae intracellular growth. Nature 456, 112–115, doi: 10.1038/nature07355 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai H. et al. A bacterial effector targets Mad2L2, an APC inhibitor, to modulate host cell cycling. Cell 130, 611–623, doi: 10.1016/j.cell.2007.06.043 (2007). [DOI] [PubMed] [Google Scholar]
- Samba-Louaka A., Taieb F., Nougayrede J. P. & Oswald E. Cif type III effector protein: a smart hijacker of the host cell cycle. Future microbiology 4, 867–877, doi: 10.2217/fmb.09.60 (2009). [DOI] [PubMed] [Google Scholar]
- Fouque E., Trouilhe M. C., Thomas V., Humeau P. & Hechard Y. Encystment of Vermamoeba (Hartmannella) vermiformis: Effects of environmental conditions and cell concentration. Experimental parasitology 145 Suppl, S62–68, doi: 10.1016/j.exppara.2014.03.029 (2014). [DOI] [PubMed] [Google Scholar]
- Gomez-Valero L. et al. Comparative analyses of Legionella species identifies genetic features of strains causing Legionnaires’ disease. Genome biology 15, 505, doi: 10.1186/PREACCEPT-1086350395137407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mampel J. et al. Planktonic replication is essential for biofilm formation by Legionella pneumophila in a complex medium under static and dynamic flow conditions. Applied and environmental microbiology 72, 2885–2895, doi: 10.1128/AEM.72.4.2885-2895.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiaden A. et al. The Legionella pneumophila response regulator LqsR promotes host cell interactions as an element of the virulence regulatory network controlled by RpoS and LetA. Cellular microbiology 9, 2903–2920, doi: 10.1111/j.1462-5822.2007.01005.x (2007). [DOI] [PubMed] [Google Scholar]
- Moon E. K., Chung D. I., Hong Y. C. & Kong H. H. Autophagy protein 8 mediating autophagosome in encysting Acanthamoeba. Molecular and biochemical parasitology 168, 43–48, doi: 10.1016/j.molbiopara.2009.06.005 (2009). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408, doi: 10.1006/meth.2001.1262 (2001). [DOI] [PubMed] [Google Scholar]
- Gietz R. D. & Woods R. A. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods in molecular biology 313, 107–120, doi: 10.1385/1-59259-958-3:107 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.