Abstract
Pathogenicity of most Gram-negative plant-pathogenic bacteria depends on the type III secretion (T3S) system, which translocates bacterial effector proteins into plant cells. Type III effectors modulate plant cellular pathways to the benefit of the pathogen and promote bacterial multiplication. One major virulence function of type III effectors is the suppression of plant innate immunity, which is triggered upon recognition of pathogen-derived molecular patterns by plant receptor proteins. Type III effectors also interfere with additional plant cellular processes including proteasome-dependent protein degradation, phytohormone signaling, the formation of the cytoskeleton, vesicle transport and gene expression. This review summarizes our current knowledge on the molecular functions of type III effector proteins with known plant target molecules. Furthermore, plant defense strategies for the detection of effector protein activities or effector-triggered alterations in plant targets are discussed.
Keywords: type III effector, plant immunity, MAPK signaling, proteasome, cytoskeleton, phytohormones
Translocated type III effector proteins from Gram-negative plant-pathogenic bacteria promote bacterial virulence by interfering with defense responses, signaling pathways, protein degradation, gene expression or the formation of the cytoskeleton.
INTRODUCTION
Gram-negative plant-pathogenic bacteria cause a variety of diseases in economically important crop plants and thus often lead to major yield losses. The bacterial ability to infect plants and to multiply inside the plant tissue depends on secreted proteins such as adhesins, toxins and degradative enzymes. Furthermore, bacterial pathogens often inject effector proteins directly into plant cells. Bacterial virulence factors are delivered by specialized protein secretion systems, which are grouped into at least six different classes, designated type I to type VI (Hayes, Aoki and Low 2010; Costa et al. 2015). Virulence-associated proteins can also be secreted via outer membrane vesicles, which allow the transport of large quantities of proteins from the bacterial periplasm to the extracellular milieu (Ellis and Kuehn 2010; Bonnington and Kuehn 2013).
Most Gram-negative plant-pathogenic bacteria depend on a type III secretion (T3S) system to conquer their host plants and to multiply inside the plant tissue. T3S systems translocate bacterial effector proteins directly into eukaryotic cells and thus allow the manipulation of host cellular pathways to the benefit of the pathogen (He, Nomura and Whittam 2004; Dean 2011; Lee et al.2013). T3S systems are present in plant- and animal-pathogenic bacteria as well as in several non-pathogenic bacteria and species (spp.) of the symbiotic bacterium Rhizobium, suggesting that T3S is not exclusively linked to pathogenicity (Troisfontaines and Cornelis 2005; Tampakaki 2014). According to phylogenetic analyses, T3S systems from plant-pathogenic bacteria have been grouped into different families including Hrp1 (hypersensitive response and pathogenicity 1; in spp. of Pseudomonas syringae and Erwinia) and Hrp2 T3S systems (in Ralstonia solanacearum, spp. of Xanthomonas, Acidovorax and Burkholderia) (Alfano and Collmer 1997; Troisfontaines and Cornelis 2005). In addition to the Hrp1 T3S system, some P. syringae strains also contain a rhizobial-like T3S system, designated Hrp3 (Gazi et al.2012). Interestingly, gene clusters with homologies to T3S gene clusters from plant-pathogenic bacteria are also present in several animal-pathogenic bacteria including Vibrio parahaemolyticus strains isolated from patients, a clinical strain of Pantoea, and Burkholderia pseudomallei (Troisfontaines and Cornelis 2005; Kirzinger, Butz and Stavrinides 2015). Pantoea and Burkholderia spp. are cross-kingdom pathogens, which infect humans and plants (Kirzinger, Nadarasah and Stavrinides 2011). Several plant-pathogenic bacteria including Xanthomonas albilineans and X. axonopodis pv. phaseoli also contain a SPI-1 (Salmonella pathogenicity island 1) T3S gene cluster, which is usually present in animal-pathogenic bacteria (Alavi et al.2008; Pieretti et al.2015).
T3S systems are complex membrane-spanning nanomachines and contain an extracellular pilus-like appendage, which provides a transport channel for secreted proteins to the plant–pathogen interface (Jin and He 2001; Li et al.2002; Blocker et al.2008; Büttner 2012). The translocation of effector proteins into the plant cell is mediated by a bacterial T3S translocon, which presumably inserts as a pore-forming complex into the plant plasma membrane (Mattei et al.2011; Guignot and Tran Van Nhieu 2016). According to the final destination of T3S substrates, protein transport into the extracellular milieu is hereafter referred to as ‘secretion’, and transport into the eukaryotic cell cytosol as ‘translocation’. Secreted substrates of the T3S system mostly include extracellular components of the secretion apparatus such as T3S pilus and translocon proteins, whereas effectors are directly translocated into eukaryotic cells. Type III-dependent secretion and translocation depends on a specific export signal, which is often located in the N-terminal region of T3S substrates. T3S signals are not conserved on the amino acid level but are often associated with specific amino acid patterns or compositions and are structurally disordered. The lack of tertiary structures in the T3S signal might facilitate the binding of interaction partners such as components of the T3S system, which are involved in the recognition of secreted proteins (Arnold et al.2009; Löwer and Schneider 2009; Samudrala, Heffron, McDermott 2009; Buchko et al. 2010; Schechter et al. 2012; Wang et al.2013). In addition to the T3S signal, the efficient targeting of many secreted proteins to the T3S system also depends on cytoplasmic T3S chaperones, which bind to and often stabilize T3S substrates and presumably promote the recognition of secreted proteins by the secretion apparatus (e.g. Menard et al. 1994; Frithz-Lindsten et al. 1995; Tucker and Galan 2000; Gaudriault, Paulin and Barny 2002; Feldman and Cornelis 2003).
Given the essential contribution of pilus and translocon proteins to effector protein translocation, T3S is presumably a hierarchical process, suggesting that the T3S substrate specificity switches from the secretion of extracellular components of the secretion apparatus to effector proteins (e.g. Magdalena et al.2002; Edqvist et al. 2003; Journet et al. 2003; Lara-Tejero et al. 2011). Furthermore, it is assumed that different effector proteins are secreted at different timepoints after the assembly of the T3S system. While experimental evidence for a secretion hierarchy of effector proteins in plant-pathogenic bacteria is still missing, differences in the timing of translocation were reported for effector proteins with antagonistic activities from animal pathogens (Enninga et al. 2005; Schlumberger et al. 2005; Mills et al. 2008; Van Engelenburg and Palmer 2008; Winnen et al.2008). A hierarchy in effector protein translocation might help to avoid possible functional interferences between different effectors and could also prevent the clogging of the T3S channel by the simultaneous transport of multiple effector molecules.
Plant-pathogenic bacteria usually possess a large pool of different effector proteins. Genome sequence analyses of P. syringae strains revealed a meta-repertoire of 94 effector families with variable numbers of nine up to 39 effectors in individual strains (Baltrus et al. 2011; Lindeberg, Cunnac and Collmer 2012). R. solanacearum strains contain 60 to 75 effectors, which belong to 57 families including 32 core effectors, which are present in most of the strains (Peeters et al.2013; Deslandes and Genin 2014). In Xanthomonas spp., the core effector set is limited to 3 out of 32 known effectors as was recently revealed by comparative genome sequence analysis (Roux et al.2015). In several bacteria, deletion of single effector genes often has little influence on virulence, suggesting that effectors share redundant functions. The generation of multi-effector mutant strains in P. syringae revealed that the deletion of 18 effector genes from six genomic clusters is required to impair the in planta bacterial growth (Kvitko et al.2009). A minimal set of eight effectors promotes bacterial virulence and suppresses plant defense responses (Cunnac et al. 2011; Wei et al.2015).
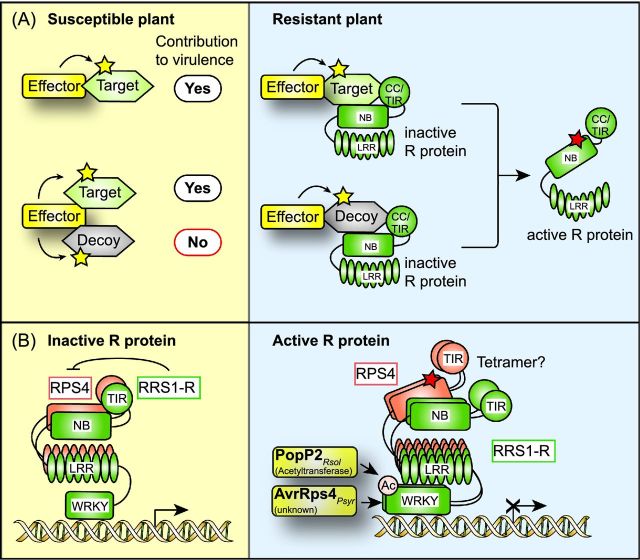
Plants usually defend themselves against microbial attacks by two levels of defense responses, which are referred to as PAMP (pathogen-associated molecular pattern)-triggered immunity (PTI) and effector-triggered immunity (ETI). PTI is one of the first defense responses and is activated upon recognition of PAMPs by plant pattern recognition receptors (PRRs) (Zipfel 2014). PAMPs are conserved microbial molecules such as flagellin, bacterial elongation factor (EF)-Tu, peptidoglycan, chitin or cell-wall-derived molecules, which are essential for pathogen survival or fitness. PTI responses can be overcome by the actions of translocated type III effector proteins, which interfere with PTI responses and thus promote bacterial virulence. This so-called effector-triggered susceptibility is counteracted by a second line of plant defense responses, designated ETI, which is activated by the products of plant resistance (R) genes upon detection of individual effector proteins (Wu et al.2014; Cui, Tsuda and Parker 2015) (Fig. 1A). Plant R genes often encode NB (nucleotide binding, also termed NB ARC [nucleotide-binding adaptor shared by Apaf1])-LRR (leucine-rich repeat) receptors (NLRs; see below) (Wu et al.2014; Cui, Tsuda and Parker 2015). Plant PTI and ETI responses are discussed in more detail below.
Figure 1.
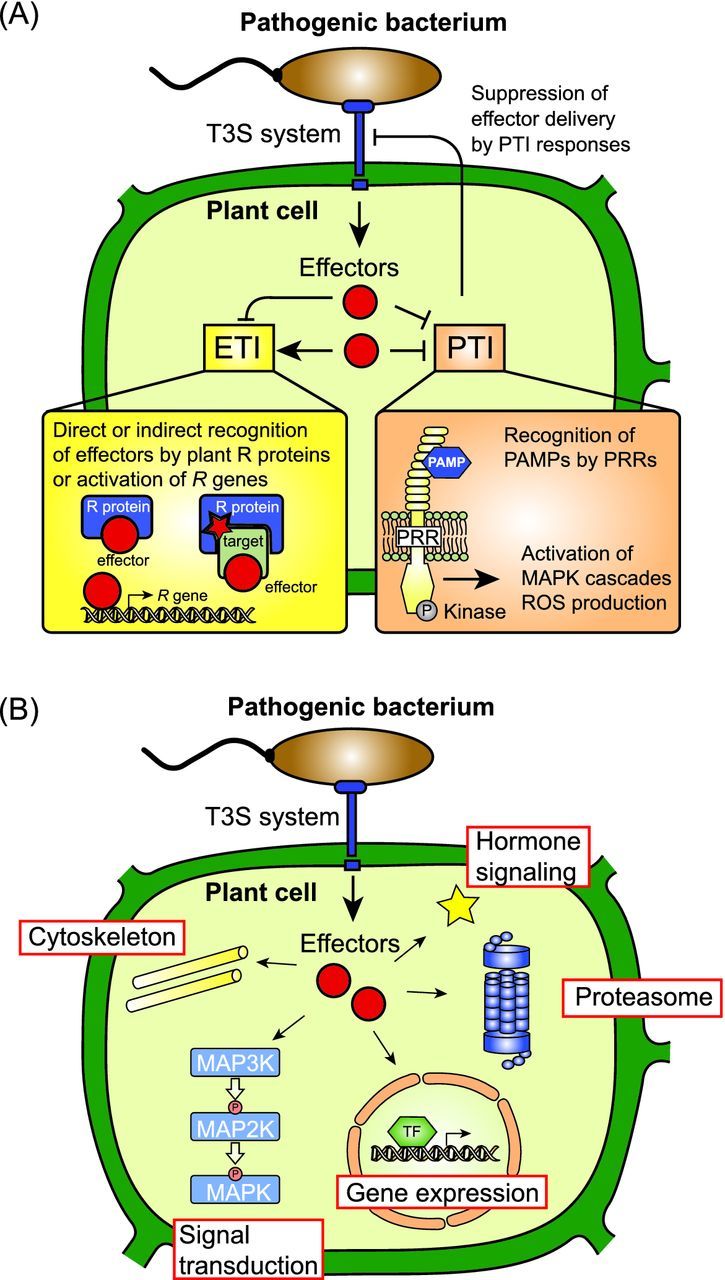
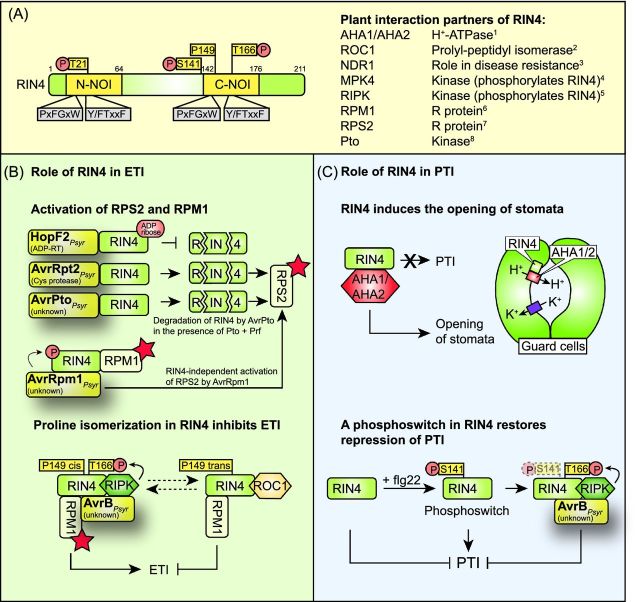
Interference of type III effector proteins with plant cellular pathways. (A) Type III effectors elicit and suppress plant defense reponses. Plant-pathogenic bacteria translocate effector proteins via the T3S system into plant cells. In resistant plants, individual effectors are directly or indirectly recognized by corresponding plant R proteins or activate plant R genes, and elicit defense responses, which are designated ETI. The indirect recognition of effector proteins by plant R proteins depends on effector-mediated modifications of plant target proteins, which are sensed by matching R proteins. Several type III effectors suppress ETI and/or PTI responses and thus promote bacterial virulence. PTI responses are triggered upon recognition of bacterial PAMPs by plant PRRs (see the text for details), and do not only interfere with pathogen survival but can also affect T3S-mediated delivery of effector proteins. (B) Overview on known plant targets of type III effectors. Translocated effector proteins interfere with the assembly of the cytoskeleton, MAPK cascades, gene expression, proteasome-dependent protein degradation or hormone signaling pathways (see the text for details).
In the past years, significant progress has been made in the functional characterization of effector proteins from plant-pathogenic bacteria. The results of numerous studies revealed that type III effectors interfere with multiple plant cellular pathways (Fig. 1B). The aim of this review is to summarize the known molecular functions of type III effectors from plant-pathogenic bacteria and their influence on plant target molecules. The interference of type III effectors with PTI responses, signal transduction pathways, proteasome-dependent protein degradation, phytohormone signaling, plant gene expression and the plant cytoskeleton is discussed in six separate sections. The last section describes known mechanisms underlying effector-mediated activation of plant defense responses. All sections are focused on interactions between type III effectors and plant targets, which have been confirmed by several independent experimental approaches. Predicted functions of type III effectors and known interaction partners are also summarized in Table 1. All abbreviations are listed in Table S1 (Supporting Information). For the description of general virulence functions of type III effectors, the reader is referred to excellent recent review articles (Dean 2011; Canonne and Rivas 2012; Deslandes and Rivas 2012; Feng and Zhou 2012; Lindeberg, Cunnac and Collmer 2012; Lee et al. 2013; Kazan and Lyons 2014; Macho and Zipfel 2015).
Table 1.
Targets and biochemical functions of type III effector proteins.
| Effectora | Known or predicted biochemical function | Plant targets (organism)b | Effector-triggered modifications and/or functions of plant targetsc | References |
|---|---|---|---|---|
| Pseudomonas syringae | ||||
| AvrB | n.d.d | RAR1 (Arabidopsis) | RAR1 mediates the interaction of AvrB with Hsp90 | Cui et al. (2010) |
| RIN4 (tomato, Arabidopsis) | Phosphorylation of RIN4 in NOI domains and activation of the R protein RPM1; induction of AHA1-mediated stomatal opening; increased interaction between COI1 and JAZ9 | Mackey et al. (2002); Liu et al. (2009); Cui et al. (2010); Zhou et al. (2015) | ||
| MPK4 (Arabidopsis) | Phosphorylation and activation of MPK4; the interaction with AvrB is increased in the presence of RAR1 | Cui et al. (2010) | ||
| RIPK (Arabidopsis) | Disruption of the RIPK-RIN4 complex; phosphorylation of RIN4; phosphorylation of AvrB by RIPK | Liu et al. (2011) | ||
| AvrPphB (HopAR1) | Cysteine protease | PBS1, PBS1-like kinases, RIPK (Arabidopsis) | Cleavage of PBS1 and PBS1-like proteins including RIPK; activation of the R protein RPS5, which guards PBS1 | Shao et al. (2003); Zhang et al. (2010); Russell, Ashfield and Innes (2015) |
| BIK1 (Arabidopsis) | Cleavage of BIK1; interference with PTI | Zhang et al. (2010) | ||
| AvrPto | n.d. | Pto (tomato) | Phosphorylation of Pto; activation of Prf-Pto-mediated ETI responses | Scofield et al. (1996); Tang et al. (1996); Kim, Lin and Martin (2002); Ntoukakis et al. (2013) |
| FLS2 kinase domain (Arabidopsis, tomato) | Inhibition of autophosphorylation of FLS2; suppression of BIK1 phosphorylation | Xiang et al. (2008, 2011) | ||
| EFR kinase domain (Arabidopsis) | Inhibition of autophosphorylation of EFR | Xiang et al. (2008) | ||
| BAK1 kinase domain (Arabidopsis) | Disruption of FLS2-BAK1 complexe | Shan et al. (2008); Xiang et al. (2008); Zhou et al. (2014) | ||
| RIN4 (tomato) | Degradation of RIN4 (dependent on Pto, Prf and the proteasome) | Luo et al. (2009) | ||
| Kinases At2g23200, BSK1, BSK3, CDG1 (Arabidopsis) | n.d. | Xiang et al. (2008, 2011) | ||
| AvrPtoB (HopAB1) | E3 ubiquitin ligase | EFR (Arabidopsis) | Ubiquitination of EFR | Göhre et al. (2008) |
| Kinase Bti9 (tomato) | Suppression of Bti9 kinase activity | Zeng et al. (2012) | ||
| CERK1 (Arabidopsis) | Degradation of CERK1 | Gimenez-Ibanez et al. (2009) | ||
| Pto (tomato) | Phosphorylation of Pto and AvrPtoB; activation of Prf-Pto-mediated ETI responses | Kim, Lin and Martin (2002); Ntoukakis et al. (2009) | ||
| Kinase Fen (tomato) | Degradation of Fen; suppression of Fen-triggered ETI | Rosebrock et al. (2007) | ||
| FLS2 (Arabidopsis) | Dissociation of FLS2-BAK1 complex; degradation of FLS2 | Göhre et al. (2008); Shan et al. (2008) | ||
| BAK1 (Arabidopsis) | Dissociation of FLS2-BAK1 complex; inhibition of BAK1 kinase activity | Göhre et al. (2008); Shan et al. (2008); Cheng et al. (2011) | ||
| RIN4 (tomato) | Degradation of RIN4 (dependent on Pto and Prf) | Luo et al. (2009) | ||
| AvrRpm1 | n.d. | RIN4 (Arabidopsis) | Phosphorylation of RIN4 in NOI domains; activation of RPM1 | Mackey et al. (2002) |
| AvrRps4 | n.d. | Lipase/esterase-like protein EDS1 (Arabidopsis) | Disruption of the complex between EDS1 and the R protein RPS4f | Bhattacharjee et al. (2011) |
| WRKY domains of RRS1-R and RRS1-S, WRKY41, WRKY70, WRKY33, WRKY60 | Activation of RPS4-dependent ETI | Sarris et al. (2015) | ||
| AvrRpt2 | Cysteine protease | RIN4 (Arabidopsis) | Cleavage of RIN4 and activation of RPS2 | Axtell et al. (2003b); Mackey et al. (2003); Kim et al. (2005a) |
| ROC1 (Arabidopsis) | Activation of AvrRpt2 protease activity | Coaker, Falick and Staskawicz (2005); Coaker et al. (2006) | ||
| Unknown | Degradation of the Aux/IAA repressor protein AXR2 (depends on protease activity of AvrRpt2 and proteasome; no direct cleavage) | Cui et al. (2013) | ||
| HopA1 (HopPsyA) | n.d. | EDS1 (Arabidopsis) | Disruption of the complex between EDS1 and RPS4f | Bhattacharjee et al. (2011) |
| HopAF1 | n.d. | Methylthioadenosine nucleosidase proteins MTN1 and MTN2 (Arabidopsis) | MTN1 and MTN2 are involved in the methionine recycling pathway and required for PAMP-induced ethylene production; HopAF1 inhibits MTN1 activity in vitro and displays structural homology to deamidases | Washington et al. (2016) |
| HopAI1 (HolPtoAI) | Phosphothreonine lyase | MPK3 (Arabidopsis) | Dephosphorylation and inactivation of MPK3 | Zhang et al. (2007) |
| MPK4 (Arabidopsis) | Reduced kinase activity of MPK4 | Zhang et al. (2012) | ||
| MPK6 (Arabidopsis, tomato) | Dephosphorylation and inactivation of MPK6 | Singh et al. (2014); Zhang et al. (2007) | ||
| Kinase BSK7 (tomato) | n.d. | Singh et al. (2014) | ||
| HopAO1 (HopPtoD2) | Tyrosine phosphatase | FLS2 kinase domain (Arabidopsis) | Suppression of FLS2-mediated PTI responses | Macho et al. (2014) |
| EFR kinase domain (Arabidopsis) | Reduced phosphorylation of EFR | Macho et al. (2014) | ||
| HopD1 (AvrPphD) | n.d. | Transcription factor NTL9 (Arabidopsis) | Suppression of NTL9-regulated gene expression during ETI | Block et al. (2014) |
| HopE1 | n.d. | Calmodulin (Arabidopsis) | Dissociation of MAP65 from microtubules in the presence of calmodulin | Guo et al. (2016) |
| MAP65 (Arabidopsis) | ||||
| HopF2 | ADP-RT | MKK5, MAP2Ks (Arabidopsis) | ADP-ribosylation of MKK5 at R313, which presumably leads to the inactivation of MKK5 | Wang et al. (2010); Singh et al. (2014) |
| MPK6 (tomato) | n.d. | Singh et al. (2014) | ||
| RIN4 (Arabidopsis) | ADP-ribosylation of RIN4; inhibition of RIN4 degradation by AvrRpt2 | Wilton et al. (2010); Hurley et al. (2014) | ||
| BAK1 (Arabidopsis) | Interference with BIK1 phosphorylation and MAPK signaling | Wang et al. (2010); Wu et al. (2011); Zhou et al. (2014) | ||
| Kinase BSK7 (tomato) | n.d. | Singh et al. (2014) | ||
| HopG1 | n.d. | Mitochondrial-localized kinesin motor protein (Arabidopsis) | HopG1 associates with actin presumably via its interaction with kinesin and induces actin filament bundling | Shimono et al. (2016) |
| HopI1 (HopPmaI) | n.d. | Hsp70 (Arabidopsis) | Increased ATPase activity of Hsp70 in vitro; recruitment of cytosolic Hsp70 to chloroplasts | Jelenska, van Hal and Greenberg (2010) |
| HopM1 | n.d. | ARF-GEF MIN7 (Arabidopsis) | Degradation of MIN7; inhibition of vesicle trafficking | Nomura et al. (2006, 2011) |
| 14-3-3 protein GRF8/MIN10 (Arabidopsis) | GRF8/MIN10 retains TF BZR1 in the cytoplasm; HopM1 presumably alters the activity of GRF8/MIN10 and leads to increased accumulation of TF BZR1 in the nucleus and downregulation of BZR1 target genes (the ortholog of MIN10 is TFT1 which is targeted by XopN) | Nomura et al. (2006); Lozano-Duran et al. (2014) | ||
| HopN1 | Cysteine protease | Photosystem II component PsbQ (tomato) | Destabilization of PsbQ; reduced activity of PSII; suppression of ROS production | Rodriguez-Herva et al. (2012) |
| HopQ1 | n.d. | 14-3-3 proteins (N. benthamiana, tomato) | n.d. | Giska et al. (2013); Li et al. (2013b); Hann et al. (2014) |
| HopU1 (HopPtoS2) | ADP-RT | RNA-binding proteins including GRP7 and GRP8 (Arabidopsis) | ADP-ribosylation of GRP7; reduced binding of GRP7 to PRR transcripts; reduced FLS2 protein levels; no influence on FLS2 transcript levels | Fu et al. (2007); Jeong et al. (2011); Nicaise et al. (2013) |
| HopW1 | n.d. | Actin (Arabidopsis) | Disruption of actin cytoskeleton; inhibition of protein trafficking and interference with endocytosis | Kang et al. (2014) |
| Acetylornithine transaminase WIN1, protein phosphatase WIN2, firefly luciferase superfamily protein WIN3 (Arabidopsis) | Overexpression of WIN1 in Arabidopsis increases growth of P. syringae pv. tomato DC3000 delivering HopW1 | Lee, Jelenska and Greenberg (2008) | ||
| HopX1 (AvrPphE) | Cysteine protease | JAZ proteins (Arabidopsis) | Proteaseome-dependent degradation of JAZ proteins | Gimenez-Ibanez et al. (2014) |
| HopZ1a | Acetyltransferase | JAZ proteins (Soybean, Arabidopsis) | Acetylation and degradation of JAZ proteins; induction of JA-responsive genes; Hopz1a activity depends on phytate | Jiang et al. (2013) |
| Tubulin (Arabidopsis) | Acetylation of tubulin and destruction of MT; interference with vesicle trafficking | Lee et al. (2012) | ||
| GmHID1 (Soybean) | Degradation of GmHID1 | Zhou et al. (2011) | ||
| Pseudokinase ZED1 (Arabidopsis) | Acetylation of ZED1 and activation of the R protein ZAR1 | Lewis et al. (2013) | ||
| HopZ4 | Protease/ acetyltransferase | ATPase of proteasome RPT6 (Arabidopsis) | Inhibition of proteasome activity | Üstün et al. (2014) |
| Xanthomonas spp. | ||||
| AvrAC (XopACXcc) | Uridylyltransferase | BIK1 (Arabidopsis) | Uridylylation of BIK1; inhibition of BIK1 phosphorylation | Feng et al. (2012) |
| RIPK (Arabidopsis) | Uridylylation of RIPK | Feng et al. (2012); Guy et al. (2013) | ||
| RLCKs of family VII including PBL2 (Arabidopsis) | Uridylylation of RLCKs; activation of R protein ZAR1, which guards PBL2 interaction partner RKS1 | Guy et al. (2013); Wang et al. (2015) | ||
| AvrBsTXcv | Acetyltransferase | Tubulin-binding protein ACIP1 (Arabidopsis) | Acetylation of ACIP1; formation of ACIP1 aggregates | Cheong et al. (2014) |
| Arginine decarboxylase CaADC1 (pepper) | n.d. | Kim, Kim and Hwang (2013) | ||
| SGT1 (pepper) | Inhibition of SGT1 phosphorylation | Kim et al. (2014) | ||
| Hsp70 (pepper) | n.d. | Kim and Hwang (2015b) | ||
| SNF1-related kinase SnRK1 (pepper) | n.d. | Szczesny et al. (2010) | ||
| 19S subunit of proteasome RPN8 (pepper) | n.d. | Szczesny et al. (2010) | ||
| Aldehyde dehydrogenase ALDH1 (pepper) | n.d. | Kim and Hwang (2015a) | ||
| AvrGf2Xfa (XopAG family) | n.d. | Cyclophilin (grapefruit) | The interaction depends on the predicted cyclophilin binding domain of AvrGf2 | Gochez et al. (2016) |
| AvrXccBXccB186 | Cysteine protease/ acetyltransferase (YopJ family) | S-Adenosyl-L-methionine-dependent methyltransferases (SAM-MT1 and SAM-MT2) (Arabidopsis) | The interaction with SAM-MT2 was only observed in vivo but not detectable by in vitro approaches. | Liu et al. (2016) |
| XopDXcv | SUMO protease | Transcription factor MYB30 (Arabidopsis) | Reduced MYB30 activity; relocalization of MYB30 to subnuclear foci | Canonne et al. (2011) |
| Transcription factor ERF4 (tomato) | DeSUMOylation of ERF4; destabilization of ERF4 and reduced ethylene levels | Kim, Stork and Mudgett (2013) | ||
| XopDXccB100 | SUMO protease | Transcription factor MYB30 (Arabidopsis) | Reduced MYB30 activity; relocalization of MYB30 from nucleus to subnuclear foci; reduced expression of MYB30 target genes | Canonne et al. (2011) |
| XopDXcc8004 | SUMO protease | DELLA proteins (Arabidopsis) | Delay of the GA-induced degradation of the DELLA protein RGA; no effect on transcription of GA-responsive genes | Tan et al. (2014) |
| Transcription factor HFR1 (Arabidopsis) | DeSUMOylation of HFR1 | Tan et al. (2015) | ||
| XopJXcv | Cysteine protease/ acetyltransferase | ATPase RPT6 of the proteasome (Arabidopsis, tobacco) | Degradation of RPT6; inhibition of proteasome activity | Üstün, Bartetzko and Börnke (2013); Üstün and Börnke (2015) |
| XopLXcv | E3 ubiquitin ligase | n.d. | Ubiquitination of N. benthamiana proteins | Singer et al. (2013) |
| XopNXcv | n.d. | Tomato atypical receptor-like kinase (TARK) 1 (tomato) | Stabilization of a TARK1/TFT1 complex; suppression of PTI responses | Kim et al. (2009a); Taylor et al. (2012) |
| 14-3-3 proteins TFT1, TFT3, TFT5, TFT6 (tomato) | Interaction was observed in yeast | Kim et al. (2009a) | ||
| XopNXoo | n.d. | Putative zinc finger protein OsVOZ2 (rice) | n.d. | Cheong et al. (2013) |
| Putative thiamine synthase OsXNP (rice) | n.d. | Cheong et al. (2013) | ||
| XopPXoo | n.d. | U box domain of E3 ubiquitin ligase OsPUB44 (rice) | Inhibition of the E3 ubiquitin ligase activity of OsPUB44 | Ishikawa et al. (2014). |
| XopQXcv | n.d. | 14-3-3 protein TFT4 (tomato, pepper, N. benthamiana) | n.d. | Teper et al. (2014) |
| 14-3-3 proteins (tomato) | n.d. | Teper et al. (2014) | ||
| XopQXoo | n.d. | n.d. | XopQXoo is present in complex with adenosine diphosphate ribose | Yu, Hwang and Rhee (2014) |
| Xoo2875 | n.d. | BAK1 (rice) | n.d. | Yamaguchi et al. (2013a) |
| Xoo1488 (XopY) | n.d. | Kinase RLCK55 (rice) | n.d. | Yamaguchi et al. (2013b) |
| Kinase RLCK185 (rice) | Suppression of CERK1-mediated phosphorylation of RLCK185 | Yamaguchi et al. (2013b) | ||
| TAL effectors g | ||||
| AvrBs3Xcv | Importin alpha (pepper) | Szurek et al. (2001) | ||
| PthA4X.citri | DNA- and RNA-binding proteins (Citrus sinensis) | de Souza et al. (2012) | ||
| Putative chromatin-associated protein HMG (Citrus sinensis) | PthA4 binds to poly(U) RNA and forms higher molecular weight complexes with poly(U) RNA in the presence of CsHMG | de Souza et al. (2012) | ||
| RNA polymerase III repressor MAF1 (Citrus sinensis) | PthA4 presumably counteracts MAF1 activity to increase transcription of host genes | Soprano et al. (2013) | ||
| PthA1, PthA2, PthA3, PthA4Xac306 | Importin alpha, cyclophilin CsCyp, thioredoxin, ubiquitin-conjugating enzyme complex (Citrus sinensis) | Inhibition of the peptidyl-prolyl cis–trans isomerase activity of cyclophilin | Domingues et al. (2010, 2012) | |
| C-terminal domain of RNA polymerase II (Citrus sinensis) | Domingues et al. (2012) | |||
| Ralstonia solanacearum | ||||
| GALA proteins (RipG family) | F box proteins | SKP1-like proteins (Arabidopsis) | n.d. | Remigi et al. (2011) |
| PopP2 (RipP2) | Acetyltransferase | WRKY domains of RRS1-R and RRS1-S (Arabidopsis) | Stabilization of RRS1-R and RRS1-S; acetylation of WRKY domains and reduced DNA binding of RRS1-R and RRS1-S; elicitation of RRS1-R-dependent ETI responses | Deslandes et al. (2003); Tasset et al. (2010); Le Roux et al. (2015); Sarris et al. (2015) |
| WRKY41, WRKY70, WRKY60, WRKY33 | Acetylation of WRKY41, WRKY70 and WRKY33 | Sarris et al. (2015) | ||
| Protease RD19 (Arabidopsis) | Relocalization of RD19 from vacuole-associated cellular compartments to the nucleus; RD19 is not acetylated by PopP2 but contributes to the RRS1-R-triggered ETI | Bernoux et al. (2008); Tasset et al. (2010) | ||
| RipTps | Trehalose phosphate synthase | n.d. | Synthesis of trehalose-6-phosphate | Poueymiro et al. (2014) |
| RipAY | γ-Glutamyl cyclo-transferase | Thioredoxins (Arabidopsis) | RipAY presumably reduces glutathione levels in eggplant; activity of RipAY is stimulated by thioredoxin AtTrx-h5 from Arabidopsis | Fujiwara et al. (2016)h |
| Degradation of glutathione; RipAY is activated by the cytosolic thioredoxins AtTrx-h1, -h2, -h4 and -h5, and the mitochondrial thioredoxin AtTrx-o1 from Arabidopsis | Mukaihara et al. (2016)i | |||
Alternative names of effectors are given in brackets. Xac, Xanthomonas axonopodis pv. citri; Xcc, X. campestris pv. campestris; Xcv, X. campestris pv. vesicatoria; Xfa, Xanthomonas fuscans subsp. aurantifolii; Xoo, X. oryzae pv. oryzae.
The plants, in which the effector targets were identified and/or studied, are given in brackets.
For details, see also the text.
n.d., not determined.
This result could not be reproduced in an independent study (Xiang et al.2011) (see the text for details).
EDS1 (enhanced disease susceptibility 1) contributes to both ETI and PTI responses and was shown to interact with the NLRs RPS4 and RPS6 (Bhattacharjee et al. 2011; Heidrich et al.2011). The interaction of EDS1 with RPS4 and RPS6 was reduced in the presence of the effector proteins AvrRps4 and HopA1 from P. syringae, which were detected in a complex with EDS1 (Bhattacharjee et al. 2011; Heidrich et al.2011). EDS1 was therefore proposed to be a guarded effector target. Notably, however, the in planta interaction of EDS1 with AvrRps4 could not be reproduced in an independent study (Sohn et al.2012).
TAL effectors induce the expression of plant target genes, which are summarized in Table S2.
First published on 1 February 2016.
Published on 12 April 2016.
INTERFERENCE OF TYPE III EFFECTORS WITH BASAL PLANT DEFENSE RESPONSES
PTI responses include the production of reactive oxygen species (ROS), callose deposition in the plant cell wall, stomatal closure and the activation of defense-related genes, and interfere with the survival and multiplication of non-adapted microbial invaders (Wu, Shan and He 2014b; Bigeard, Colcombet and Hirt 2015). PTI can also lead to a reduction in type III-dependent effector protein translocation, suggesting that plants actively interfere with the expression of T3S genes and/or T3S-dependent protein delivery (Crabill et al. 2010; Oh, Park and Collmer 2010; Anderson et al. 2014).
PAMP perception depends on PRRs, which often contain an extracellular domain with the PAMP-binding site, a transmembrane domain and an intracellular kinase domain (Dodds and Rathjen 2010; Schwessinger and Ronald 2012; Böhm et al. 2014; Zipfel 2014). Upon PAMP binding, PRRs activate downstream signaling components such as receptor-like cytoplasmic kinases (RLCKs) and mitogen-activated protein kinase (MAPK) cascades (Fig. 2A). Among the well-studied plant PRRs are EFR (EF-Tu receptor), the chitin receptor CERK1 (chitin elicitor receptor kinase 1) and the flagellin receptor FLS2 (flagellin sensitive 2) (Gómez-Gómez and Boller 2000; Schwessinger and Ronald 2012; Wu, Shan and He 2014). PTI elicitors, which are frequently used for the activation of FLS2 and EFR, are peptides of flagellin (flg22) or EF-Tu (elf18). FLS2 belongs to the group of non-RD receptor-like kinases (RLK), which lack a conserved arginine residue (R) next to the catalytic aspartate (D) in the activation loop. FLS2 is presumably autophosphorylated at Ser-938 and associates with the RLCK BIK1 (Botrytis-induced kinase 1) prior to ligand perception (Gómez-Gómez, Bauer and Boller 2001; Lu et al.2010; Cao et al. 2013). Upon binding of flg22, FLS2 forms a complex with the membrane-associated RLK BAK1 (BRI1 [brassinosteroid-insensitive 1]-associated receptor kinase 1), which also interacts with BIK1 (Chinchilla et al.2007). BAK1 phosphorylates BIK1, which subsequently phosphorylates FLS2 and BAK1, dissociates from the FLS2-BAK1 complex and activates downstream signaling pathways including MAPK cascades (Lu et al.2010; Kim, Kim and Nam 2013; Lin et al. 2014) (Fig. 2A). BIK1 also phosphorylates the NADPH oxidase RBOHD (respiratory burst oxidase homolog D), which contributes to ROS production (Kadota et al. 2014; Li et al.2014a) (Fig. 2A). In addition to BIK1, FLS2-mediated signaling events involve heterotrimeric G proteins. The Gα subunit XLG2, the Gβ subunit AGB1 and the Gγ subunit AGG1/2 from Arabidopsis were recently shown to suppress proteasome-dependent degradation of BIK1 (Liang et al.2016). BIK1 interacts with and phosphorylates XLG2, which dissociates from BIK1 after flg22 perception and interacts with RBOHD, suggesting that it is involved in the regulation of the ROS burst (Liang et al.2016).
Figure 2.
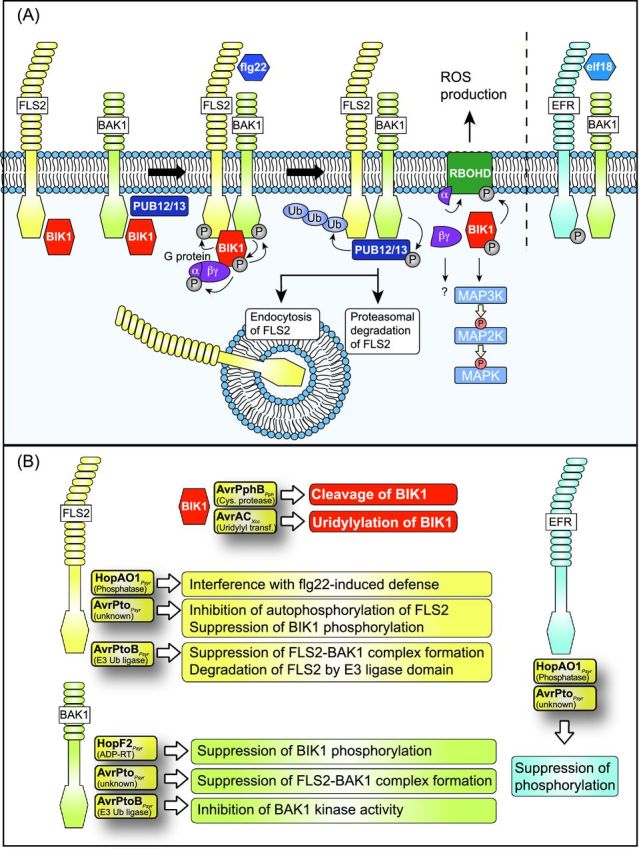
Interference of type III effectors with FLS2- and EFR-dependent signaling pathways during PTI. (A) Schematic model of signaling pathways triggered by the flagellin receptor FLS2 and the EF-Tu receptor EFR. FLS2 and EFR consist of a cytoplasmic kinase, a transmembrane and an extracellular LRR domain and insert into the plant plasma membrane. Signaling by FLS2 and EFR involves the transmembrane kinase BAK1, which associates with FLS2 and EFR upon binding of their corresponding ligands. FLS2 and BAK1 are associated with the kinase BIK1, which is phosphorylated by BAK1 upon flg22 perception by FLS2 and subsequently phosphorylates FLS2 and BAK1. BIK1 also phosphorylates an associated heterotrimeric G protein, which stabilizes BIK1 and dissociates after flg22 perception. The Gα subunit interacts with the NADPH oxidase RBOHD, which is involved in ROS production. flg22 perception also leads to the dissociation of BIK1 from the FLS2/BAK1 complex. BIK1 subsequently phosphorylates RBOHD and activates MAPK signaling pathways. FLS2-associated BAK1 phosphorylates the E3 ubiquitin ligases PUB12 and PUB13, which interact with and ubiquitinate FLS2. The proteasomal degradation of FLS2 and the endocytosis of FLS2 after flg22 perception presumably prevent the constitutive activation of FLS2-mediated PTI responses. flg22, peptide of flagellin; elf18, peptide of elongation factor EF-Tu. (B) As indicated, several type III effectors target FLS2, BAK1, BIK1 and EFR. Known effector-triggered alterations in PRRs, BAK1 and BIK1 are listed in boxes (see the text for details). Contradictory data were published about the interaction of AvrPto with BAK1 (see the text for details).
A possible constitutive activation of FLS2-mediated PTI responses is counteracted by the endocytosis of FLS2 after ligand perception (Robatzek, Chinchilla and Boller 2006; Ben Khaled, Postma and Robatzek 2015) (Fig. 2A). Furthermore, FLS2-mediated signaling is suppressed by the proteasomal degradation of FLS2, which is controlled by the E3-ubiquitin ligases PUB12 and PUB13. Both E3 ligases interact with BAK1 and are recruited to FLS2 upon ligand perception (Li, Lu and Shan 2014). Recently, it was shown that FLS2-mediated PTI responses are also suppressed by the MAP kinase kinase kinase MKKK7, which interacts with FLS2 and is phosphorylated upon flg22 perception (Mithoe et al.2016).
Given the inhibitory effect of PTI on bacterial pathogeni-city, successful pathogens have developed skills to counteract basal defense responses to establish themselves in the plant tissue (Figs 1A and 2B). Several effectors were shown to target PRRs and associated proteins. Known examples include AvrPto, AvrPtoB and HopAO1 (Hrp-dependent outer protein AO1) from Pseudomonas syringae as is outlined below. AvrPto presumably inhibits the kinase activities of FLS2 and EFR, whereas the E3 ubiquitin ligase AvrPtoB degrades PRRs including FLS2 and CERK1. The tyrosine phosphatase HopAO1 was shown to interfere with the phosphorylation of the PRR EFR (see below). Additional effectors from P. syringae and Xanthomonas campestris pv. campestris including the mono-ADP-ribosyltransferase (mADP-RT) HopF2, the cysteine protease AvrPphB and the uridylyl transferase AvrAC target the PRR-associated proteins BAK1 and BIK1 (see below). Several effectors also modulate PTI responses by interfering with PTI-associated downstream MAPK signaling cascades. These effectors and their specific mode of action will be detailed in the section ‘Modulation of MAPK cascades by type III effectors’ below.
AvrPto from P. syringae targets the PRRs FLS2 and EFR and presumably interacts with BAK1
AvrPto from P. syringae interacts with the kinase domains of the PRRs FLS2 and EFR, and leads to the suppression of PTI responses including MAPK signaling pathways (Xiang et al.2008) (Fig. 2B). Given that AvrPto inhibits the autophosphorylation of FLS2 and EFR, it likely acts as a kinase inhibitor. In agreement with this hypothesis, a point mutation in AvrPto (Y89D), which abolishes the interaction of AvrPto with FLS2 and EFR, interferes with the AvrPto-mediated suppression of MAPK activity (Xiang et al.2008). The interaction between AvrPto and FLS2 presumably suppresses BIK1 phosphorylation (Xiang et al.2011).
AvrPto was also reported to bind to the RLK BAK1 and to prevent the formation of the FLS2-BAK1 complex (Shan et al. 2008; Zhou et al.2014). Experimental evidence for the interaction between AvrPto and BAK1 was provided by the results of coimmunprecipitation studies in protoplasts and transgenic Arabidopsis seedlings, bimolecular fluorescence complementation (BiFC) studies and in vitro pull-down assays (Shan et al. 2008; Zhou et al.2014). No interaction with BAK1 was observed for an AvrPtoS46P point mutant derivative, which does not suppress PAMP-triggered activation of MAPK signaling cascades (He et al.2006). This suggests that the observed interaction with BAK1 was specific for AvrPto. In an independent study, however, similar interaction experiments did not reveal an interaction between AvrPto and BAK1 whereas the interaction between AvrPto and FLS2 was detected (Xiang et al.2011). It is possible that the observed lack of interaction between AvrPto and BAK1 was caused by differences in the experimental conditions. Notably, Xiang et al. (2011) also did not detect the postulated AvrPto-induced dissociation of the FLS2-BAK1 complex in the presence of an AvrPto-nYFP (N-terminal region of yellow fluorescent protein) fusion protein. However, it cannot be excluded that the presence of the nYFP fusion partner interfered with the ability of AvrPto to dissociate the FLS2-BAK1 complex.
The E3 ubiquitin ligase AvrPtoB from P. syringae degrades the PRRs FLS2 and CERK1 and inhibits the kinase activity of BAK1
In addition to AvrPto, the distantly related effector AvrPtoB suppresses PTI responses (Fig. 2B). AvrPtoB is presumably activated in planta by phosphorylation of the serine residue at position 258, suggesting that it mimics a substrate of a plant kinase (Xiao, Giavalisco and Martin 2007). Given that the exchange of S258 to alanine leads to a loss of the virulence activity of AvrPtoB, phosphorylation of AvrPtoB is presumably required for protein function (Xiao, Giavalisco and Martin 2007).
AvrPtoB contains a C-terminal E3 ubiquitin-ligase domain, which leads to the proteasomal degradation of most of its plant targets (Abramovitch et al. 2006; Janjusevic et al.2006; Göhre et al. 2008; Gimenez-Ibanez et al. 2009). Interaction partners of AvrPtoB include FLS2, BAK1 and additional receptor kinases such as the chitin receptor CERK1 (Göhre et al. 2008; Shan et al.2008; Gimenez-Ibanez et al. 2009) (Table 1). AvrPtoB degrades FLS2 and CERK1 and inhibits the kinase activity of BAK1, thus suppressing PTI responses (Göhre et al.2008; Gimenez-Ibanez et al. 2009; Cheng et al. 2011). The E3 ubiquitin ligase activity of AvrPtoB is described in more detail in the section ‘Interference of type III effectors with the 26S proteasome’.
The tyrosine phosphatase HopAO1 interferes with EFR phosphorylation and FLS2-mediated signaling pathways
The tyrosine phosphatase HopAO1 (formerly known as HopPtoD2) from P. syringae interacts with the kinase domain of the PRR EFR (Macho et al.2014). HopAO1 leads to reduced tyrosine phosphorylation of EFR after ligand binding, suggesting that the interaction of HopAO1 with EFR suppresses EFR autophosphorylation (Macho et al.2014). Reduced phosphorylation of EFR presumably interferes with downstream signaling pathways which involve BAK1. The targeted mutagenesis of tyrosine residues in the cytoplasmic domain of EFR revealed that tyrosine residue Y836 is essential for EFR-mediated downstream signaling but dispensable for the kinase activity of EFR (Macho et al.2014). It remains to be investigated whether HopAO1 suppresses EFR-mediated signaling by interfering with the phosphorylation of Y836 of EFR. The suppression of EFR phosphorylation by HopAO1 presumably does not solely depend on its tyrosine phosphatase activity because a catalytically inactive HopAO1 derivative led to 20% reduction in EFR phosphorylation (Macho et al.2014). Notably, in addition to EFR, HopAO1 also interacts with the kinase and cytoplasmic domain of FLS2 and interferes with FLS2-mediated defense signaling by a yet unknown mechanism (Macho et al.2014) (Fig. 2B).
The mADP-RT HopF2 from P. syringae interacts with BAK1 and interferes with BIK1 phosphorylation
An additional effector from P. syringae pv. tomato DC3000, which suppresses PTI responses, is the mADP-RT HopF2 (Wu et al. 2011; Zhou et al.2014; Lo et al. 2016). ADP-RTs hydrolyze NAD+ to transfer ADP ribose to their cognate substrate molecules (Deng and Barbieri 2008). HopF2 interacts with BAK1 (Zhou et al.2014) and interferes with flg22-induced BIK1 phosphorylation (Wu et al.2011) (Fig. 2B). It is still unknown whether BAK1 is a substrate of the HopF2 ADP-RT activity. Given that HopF2 interacts with and ADP-ribosylates the MAP kinase kinase (MAP2K) MKK5 as well as the immune regulator RIN4 (RPM1-interacting protein 4, see below), BAK1 is not the only target of HopF2 (Wang et al. 2010; Wilton et al.2010). HopF2 was also found in a complex with the autoinhibited plasma membrane H(+) ATPase AHA2, which associates with RIN4 and mediates stomatal closure (Liu et al.2009; Hurley et al. 2014) (see below). HopF2 presumably interferes with stomatal immunity independently of its ADP-RT activity because transgenic Arabidopsis plants expressing HopF2 or the catalytically inactive HopF2D175A derivative were impaired in stomatal closure upon treatment with P. syringae pv. tomato DC3000 (Hurley et al.2014).
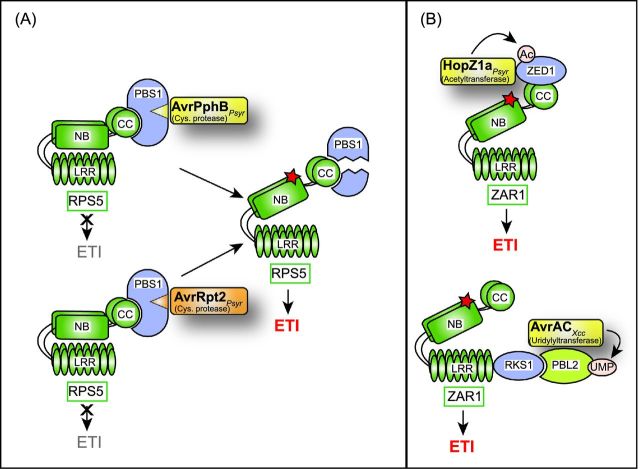
The cysteine protease AvrPphB and the uridylyl transferase AvrAC target BIK1
AvrPphB from P. syringae pv. phaseolicola and AvrAC (also known as Xop [Xanthomonas outer protein] AC) from X. campestris pv. campestris both target BIK1. AvrPphB is a cysteine protease, which is secreted by the T3S system as a preprotease and is autocatalytically processed after the tripeptide Gly-Asp-Lys, which is the recognition site of AvrPphB (Puri et al. 1997; Shao et al.2002). The mature AvrPphB is myristoylated inside the plant cell and associates with the plant plasma membrane (Dowen et al.2009). AvrPphB interacts with and cleaves BIK1 and thus presumably interferes with PTI (Zhang et al.2010) (Fig. 2B). Additional substrates of AvrPphB include the membrane-associated serine/threonine kinase PBS1 and PBS1-like (PBL) proteins, which all contain the Gly-Asp-Lys recognition site (Shao et al. 2003; Zhang et al.2010) (Table 1). Cleavage of PBS1 by AvrPphB leads to the activation of ETI in the presence of the R protein RPS5 (Shao et al.2003) (see below). AvrPphB also cleaves the kinase RIPK (RPM1-induced protein kinase), which is required for the phosphorylation of the immune regulator RIN4 (Russell, Ashfield and Innes 2015) (see below).
AvrAC from X. campestris pv. campestris contains an N-terminal LRR domain and a C-terminal Fic (filamentation-induced by c-AMP) domain and acts as uridylyl transferase, i.e. it transfers uridine 5′-monophosphate (UMP) to plant target proteins (Feng et al.2012). The results of in vitro and in vivo assays suggest that AvrAC interacts with and uridylylates BIK1 (Feng et al.2012) (Fig. 2B). UMP modification of BIK1 depends on amino acid residues S236 and T237 of BIK1, which are conserved phosphorylation sites in the activation loop (Lu et al. 2010; Zhang et al.2010; Laluk et al. 2011). It was, therefore, suggested that the AvrAC-mediated transfer of UMP inhibits BIK1 activity (Feng et al.2012). In addition to BIK1, AvrAC interacts with and uridylates other RLCKs of the family VII, which is the largest family of RLCKs and includes several RLCKs involved in plant immune responses. Among the AvrAC targets are RIPK and PBL2 (Feng et al. 2012; Guy et al. 2013; Wang et al.2015). Uridylylation of PBL2 by AvrAC triggers ETI responses, which depend on the pseudokinase RKS1 and the R protein ZAR1 (see below).
MODULATION OF MAPK CASCADES BY TYPE III EFFECTORS
PRR-mediated immune responses often involve the activation of MAPK cascades. These signaling pathways are attractive targets for type III effectors because they contribute to various cellular pathways. MAPK signaling is triggered by MAP kinase kinase kinases (also termed MAP3K or MEKK), which are directly or indirectly activated by receptor proteins including PRRs (Rodriguez, Petersen and Mundy 2010; Rasmussen et al. 2012). MAP3Ks are serine or threonine kinases and activate MAP2Ks (also designated MEK) via phosphorylation (Rodriguez, Petersen and Mundy 2010). MAP2Ks subsequently phosphorylate threonine or tyrosine residues of MAPKs, leading to their activation (Rodriguez, Petersen and Mundy 2010; Rasmussen et al. 2012). Among the well-studied MAPKs from plants are MPK3, MPK4 and MPK6, which are involved in defense responses. MPK3 and MPK6 are part of a signaling cascade, which is activated by the MAP3K MEKK1 and the two MAP2Ks MKK4 and MKK5 (Meng and Zhang 2013) (Fig. 3A). A second signaling cascade involves the MAP3K MEKK1, the two MAP2Ks MKK1 and MKK2 as well as MPK4 (Meng and Zhang 2013). Known substrates of MPK4 are the MPK4 interaction partner RIN4 (Cui et al.2010), the MAP3K MEKK2 (also designated SUMM1 [suppressor of mkk1 mkk2 1]) (Kong et al.2012) and MKS1 (MPK4 substrate 1), which forms a complex with MPK4 and the transcription factor WRKY33 in the nucleus (Rasmussen et al.2012). Phosphorylation of MKS1 by MPK4 leads to the release of WRKY33, which in turn induces the expression of its target genes and initiates PTI responses (Rasmussen et al.2012) (Fig. 3A).
Figure 3.
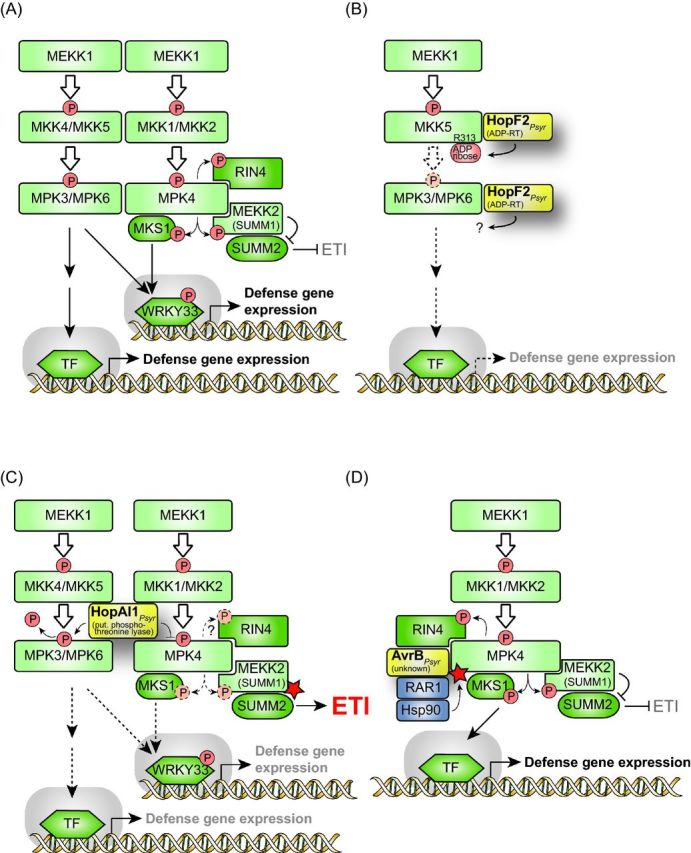
Influence of type III effectors on MAPK signaling pathways. (A) Schematic overview on MAPK signaling pathways involved in plant defense responses. During plant defense responses, two MAPK signaling pathways are activated which involve (i) MPK3/MPK6 and MKK4/MKK5 and (ii) MPK4 and MKK1/MKK2, respectively. The MP2Ks of both pathways are activated by the MP3K MEKK1, however, MPK3 and MPK6 can also be activated independently of MEKK1 (Suarez-Rodriguez et al.2007). The activation of MAPKs directly or indirectly leads to the release of transcription factors (TFs), which trigger the expression of defense genes. Known substrates of MPK4 are MKS1, RIN4 and the MP3K MEKK2 (also designated SUMM1). Phosphorylation of MKS1 by MPK4 leads to the release of the MKS1-bound TF WRKY33, which subsequently activates gene expression. MPK4 also phosphorylates the MAP3K MEKK2 and presumably results in its inactivation. MPK4-mediated inactivation of MEKK2 leads to the suppression of ETI responses triggered by the CC-NB-LRR R protein SUMM2, which likely guards MEKK2 (see the text for details). (B) HopF2 from P. syringae interferes with MPK3/MPK6 signaling pathway. The mono-ADP-RT HopF2 ADP-ribosylates and thus inactivates MKK5 and suppresses the MPK3/MPK6-mediated signaling pathway (indicated by dashed arrows). HopF2 also interacts with MPK6, yet, the outcome of this interaction is unknown. (C) The putative phosphothreonine lyase HopAI1 from P. syringae inhibits the activity of MAPKs. HopAI1 dephosphorylates MPK3 and MPK6 and thus interferes with the MPK3/MPK6 signaling pathway. Furthermore, HopAI1 suppresses the kinase activity of MPK4 and thus the phosphorylation of the MPK4 substrates MKS1 and MEKK2. It has not yet been shown whether HopAI1 also interferes with the MPK4-mediated phosphorylation of RIN4 (indicated by a dashed arrow and a questionmark). The loss of MPK4 activity leads to the activation of MEKK2 (indicated by a red asterisk), which in turn triggers SUMM2-mediated ETI (see the text for details). (D) AvrB from P. syringae activates MPK4. AvrB interacts with MPK4 and leads to its phosphorylation and thus activation. The efficient interaction between AvrB and MPK4 depends on RAR1, which presumably acts as a linker between AvrB and Hsp90. Hsp90 promotes the activity of MPK4 as is indicated by a red asterisk (see the text for details).
Notably, MPK4 was initially identified as a negative regulator of plant immunity because mutations in MPK4 lead to the activation of defense responses (Meng and Zhang 2013). Genetic screens for suppressor mutations in mkk1mkk2 plants led to the identification of SUMM1 and the R protein SUMM2. Mutations in SUMM1 or SUMM2 abolish the constitutive activation of plant defense responses in mkk1mkk2 plants, suggesting that SUMM1 and SUMM2 are both involved in the activation of defense responses in the absence of MPK4 (Kong et al. 2012; Zhang et al.2012) (Fig. 3A). The R protein SUMM2 has probably evolved to sense changes in the MEKK1-MKK1/MKK2-MPK4 signaling cascade and likely guards SUMM1, which interacts with and is phosphorylated by MPK4 (Kong et al. 2012; Zhang et al.2012). Given the finding that overexpression of SUMM1 activates SUMM2-dependent defense responses, it was proposed that SUMM1 is negatively regulated by MPK4. Thus, activation of SUMM1 in the absence of MEKK1-MKK1/MKK2-MPK4 signaling triggers SUMM2-dependent defense responses (Kong et al.2012).
Effectors, which interfere with MAPK signaling pathways and SUMM2-mediated defense, include HopF2, HopAI1 and AvrB from P. syringae. As is detailed below, the ADP-RT HopF2 presumably inactivates the MP2K MKK5, whereas HopAI1 suppresses the activities of MAPKs. AvrB, however, activates the MAPK MPK4, suggesting that effector proteins from P. syringae have opposing functions with regard to the interference with MAPK signaling cascades. The functions of HopF2, HopAI1 and AvrB are also summarized in Fig. 3B-D.
HopF2 from P. syringae ADP-ribosylates MKK5
The ADP-RT HopF2 from P. syringae does not only interfere with BIK1 phosphorylation (see above) but also with MAPK signaling. HopF2 interacts with MPK6 (Singh et al.2014) as well as with several MAP2Ks including MKK5. HopF2 ADP-ribosylates the arginine residue at position 313 (R313) of MKK5, which is important for MKK5 function (Wang et al.2010). It was, therefore, proposed that HopF2 inactivates MKK5 and thus interferes with the MKK5-dependent signaling cascade (Wang et al.2010) (Fig. 3B). This hypothesis is supported by the finding that the phosphorylation of MPK6 by a constitutively active MKK5DD derivative is suppressed by HopF2 in vitro (Wang et al.2010).
HopAI1 from Pseudomonas syringae suppresses MAPK activities
An additional effector, which suppresses MAPK activities, is HopAI1 from P. syringae pv. tomato (Zhang et al. 2007, 2012). HopAI1 interacts with several MAPKs including MPK3, MPK4 and MPK6 (Zhang et al. 2007, 2012; Singh et al. 2014) (Fig. 3C). The analysis of hopAI1-transgenic Arabidopsis plants revealed that HopAI1 suppresses the kinase activities of MPK3, MPK4 and MPK6 and thus interferes with plant defense responses, e.g. flg22-induced gene expression, ROS production and callose deposition (Zhang et al. 2007, 2012). In agreement with the HopAI1-mediated suppression of MPK4 activity, the in planta expression of hopAI1 leads to the induction of SUMM2-dependent ETI (Zhang et al.2012) (see above; Fig. 3C). HopAI1 presumably directly targets MAPKs because the HopAI1-mediated suppression of MPK3 and MPK6 kinase activities was also observed in the presence of the constitutively active MAP2K derivative MKK5DD (Zhang et al.2007). It was shown that HopAI1 inactivates MPK3 and MPK6 via dephosphorylation of phosphothreonine residues in vitro, suggesting that HopAI1 acts as a phosphothreonine lyase (Zhang et al.2007).
AvrB from Pseudomonas syringae activates MPK4
In contrast to the HopAI1-mediated dephosphorylation and inactivation of MPK4, AvrB from P. syringae leads to the phosphorylation and activation of MPK4 (Cui et al.2010) (Fig. 3D). Thus, HopAI1 and AvrB have opposing activities with regard to the phosphorylation of MPK4. Coimmunprecipitation studies revealed that the interaction of AvrB with MPK4 is increased in the presence of RAR1, which is a cochaperone of the heat shock protein 90 (HSP90) and interacts with AvrB (Cui et al.2010). It was, therefore, suggested that the interaction between AvrB and RAR1 promotes the association of AvrB with MPK4 (Cui et al.2010). Similarly to AvrB, HSP90 positively regulates the activity of MPK4 (Cui et al.2010). RAR1 and MPK4 are presumably not the only virulence targets of AvrB because AvrB also interacts with RIN4 and triggers its phosphorylation (Mackey et al.2002; Cui et al. 2010) (see below). As RIN4 was found in a complex with MPK4 and AvrB and presumably acts downstream of MPK4, it was speculated that the AvrB-mediated phosphorylation of RIN4 involves MPK4 (Cui et al.2010) (Fig. 3D).
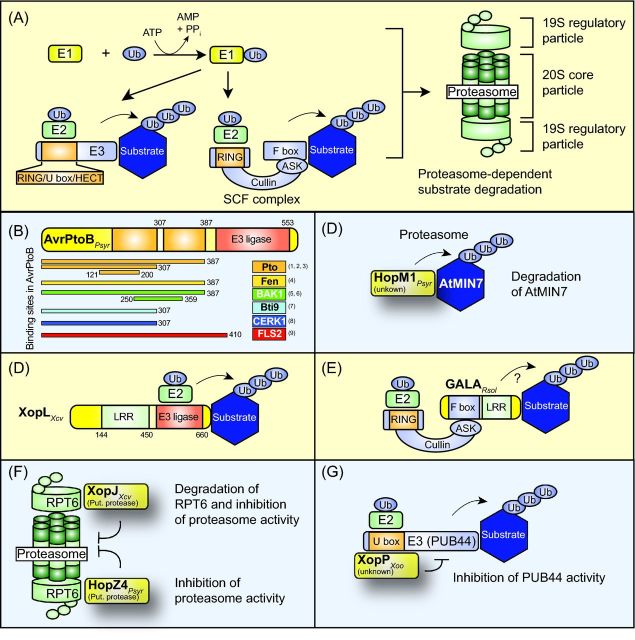
INTERFERENCE OF TYPE III EFFECTORS WITH THE 26S PROTEASOME
The eukaryotic 26S proteasome plays a central role in many cellular processes including hormone signaling and defense responses and is a virulence target of several type III effectors (Price and Kwaik 2010; Dudler 2014; Duplan and Rivas 2014; Banfield 2015). The proteasome is composed of a 20S core particle and two 19S regulatory subunits, which recognize ubiquitinated proteins. Proteins destined for degradation are covalently attached via the ɛ-amino group of a lysine residue to at least four ubiquitin molecules. Ubiquitin is a highly conserved 76-amino acid polypeptide and is often linked to other ubiquitin molecules via the lysine residue at position 48 (Pickart 2001). Ubiquitin is adenylated by the ubiquitin-activating enzyme (E1), transferred to a conjugating E2 enzyme and subsequently bound to a lysine residue of the target protein by an E3 ubiquitin ligase (Sadanandom et al.2012) (Fig. 4A). Plants possess only one or two E1 proteins but a significantly higher number of E2 and E3 proteins (e.g. 1500 E3 ligases in Arabidopsis) (Chen and Hellmann 2013). E3 ubiquitin ligases are single or multisubunit proteins, which interact with E2 enzymes via a HECT (homologous to E6-associated protein C-terminus), a RING (really interesting new gene) or a U-box domain (Chen and Hellmann 2013). Very well studied are SCF (SKP1 [S-phase kinase-associated protein 1]-like-cullin 1-F-box) complexes, which are multimeric RING-finger E3 ligases and play a central role in phytohormone signaling (Shabek and Zheng 2014). SCF complexes consist of a cullin protein as a central scaffold, which associates via its C-terminal region with the RING protein RBX1 (RING box 1). The N-terminal part of the cullin protein is connected via a SKP1-like protein with a member of the F-box protein family, which provides the binding sites for the substrates of the SCF complex (Vierstra 2009; Chen and Hellmann 2013) (Fig. 4A). F-box proteins contain the F-box motif, i.e. a short motif of approximately 50 amino acids, which mediates the interaction of F-box proteins with SKP1-like proteins of the SCF complex (Schulman et al.2000).
Figure 4.
Contribution of type III effectors to proteasome-dependent protein degradation. (A) Model of the proteasome-dependent protein degradation pathway. Ubiquitin (Ub) is activated by the ubiquitin-activating enzyme (E1) and transferred to the ubiquitin-conjugating enzyme (E2), which interacts with the ubiquitin ligase (E3). E3 ubiquitin ligases are divided into several classes according to the presence of a HECT, RING or U-box domain. RING domain-containing E3 ubiquitin ligases can be part of a multimeric protein complex such as the SCF complex, which consists of a RING-box protein, the molecular scaffold protein cullin, an Arabidopsis SKP1-like protein (ASK1) and an F-box protein, which binds the substrate of the E3 ligase. E3 enzymes mediate the transfer of ubiquitin molecules to the substrate, thus leading to the formation of poly-ubiquitin chains, which allow the targeting of proteins to the proteasome. The proteasome consists of two 19S regulatory and a 20S core particle and catalyzes the unfolding and degradation of polyubiquitinated proteins. (B) Domain structure of the effector protein AvrPtoB from P. syringae. The regions of AvrPtoB, which provide binding sites for the AvrPtoB interaction partners Pto, Bti9, Fen, FLS2 and BAK1, are indicated. References: (1) Dong et al. 2009; (2) Xiao et al. 2007; (3) Mathieu, Schwizer and Martin 2014; (4) Rosebrock et al. 2007; (5) Shan et al. 2008; (6) Cheng et al. 2011; (7) Zeng et al. 2012; (8) Gimenez-Ibanez et al. 2009; (9) Göhre et al.2008. Experimental evidence for the presence of two Pto-binding sites in AvrPtoB (indicated as orange boxes) was reported by Mathieu, Schwizer and Martin (2014). Numbers refer to amino acid positions in AvrPtoB. (C) HopM1 from P. syringae induces the degradation of its interaction partner AtMIN7. The HopM1-mediated degradation of AtMIN7 depends on the activity of the proteasome. (D) The effector protein XopL from X. campestris pv. vesicatoria triggers the ubiquitination of plant proteins. XopL contains an N-terminal LRR and a C-terminal E3 ubiquitin ligase domain. The plant targets of XopL are unknown. Numbers refer to amino acid positions in XopL. (E) GALA proteins from R. solancearum contain an F-box domain and were proposed to associate with components of the SCF complex. In agreement with this hypothesis, an interaction between GALA proteins and ASK proteins has been shown. A contribution of GALA proteins to the ubiquitination of substrates of the SCF complex remains to be demonstrated. (F) XopJ from Xanthomonas spp. and HopZ4 from P. syringae interact with the proteasome subunit RPT6 and suppress the activity of the proteasome. XopJ leads to the degradation of RPT6. (G) XopP from X. oryzae pv. oryzae interacts with the U box E3 ubiquitin ligase PUB44 from rice and inhibits its activity.
Some effectors bind to E3 ligases or act themselves as E3 ligases and exploit the proteasome for the degradation of specific plant proteins. Examples are AvrPtoB and HopM1 from P. syringae, XopL from X. campestris pv. vesicatoria and GALA proteins from R. solanacearum, which are discussed below. Other effectors such as XopD from Xanthomonas spp. can indirectly destabilize plant proteins by cleaving small ubiquitin-related modifier (SUMO) from SUMOylated proteins. SUMO is structurally related to ubiquitin and can reversibly modify proteins, thus leading to alterations in protein localization, stability and/or activity (Gill 2004; Park and Yun 2013). The functions of XopD family members are discussed in the section ‘Modulation of phytohormone signaling by type III effectors’.
In contrast to effectors that stimulate protein degradation, type III effectors of the YopJ (Yersinia outer protein J) family of predicted cysteine proteases and/or acetyltransferases such as XopJ from X. campestris pv. vesicatoria and HopZ4 from P. syringae suppress proteasome activity (Hotson and Mudgett 2004; Lewis et al.2011). A similar effect is achieved by the bacterial tripeptide derivative syringolin, which is produced by several strains of P. syringae (Groll et al.2008; Dudler 2014). Furthermore, the effector XopP from Xanthomonas oryzae pv. oryzae was shown to interfere with the activity of an E3 ubiquitin ligase as is discussed below. The apparent contradictory activities of bacterial virulence factors, which suppress or promote the activity of the proteasome, might be caused by different spatial distributions of effectors in the plant cell or temporal differences in their synthesis or translocation. The interference of single effector proteins with the proteasome and with proteasome-dependent protein degradation is summarized below and in Fig. 4B-G.
AvrPtoB from P. syringae contains an E3 ubiquitin ligase domain
As mentioned above, AvrPtoB from P. syringae contains a C-terminal E3 ubiquitin ligase domain and degrades interaction partners including CERK1, FLS2 and RIN4 (Fig. 4B; Table 1). An additional substrate of the E3 ubiquitin ligase activity of AvrPtoB is the kinase Fen, which is a member of the Pto (resistance to P. syringae pv. tomato) kinase family and binds to the N-terminal region of AvrPtoB (Martin et al. 1994; Rosebrock et al.2007). Fen is ubiquitinated and degraded by AvrPtoB (Rosebrock et al.2007). In the presence of AvrPtoB1-387, however, which lacks the E3 ubiquitin ligase domain, Fen triggers defense responses in tomato. It was, therefore, suggested that AvrPtoB evades its own Fen-mediated recognition by acquiring E3 ubiquitin ligase activity (Rosebrock et al.2007). Fen-dependent defense responses elicited by E3 ubiquitin ligase-inactive AvrPtoB derivatives were previously referred to as Rsb (resistance suppressed by AvrPtoB C terminus) (Abramovitch et al.2003) and depend on the R protein Prf (Rosebrock et al.2007). As is described below, Prf also detects full-length AvrPtoB and AvrPto and elicits ETI responses, which depend on the kinase Pto (Oh and Martin 2011).
In addition to its role in suppression of Fen-mediated defense responses, the C-terminal E3 ubiquitin ligase domain of AvrPtoB might also contribute to other virulence activities of AvrPtoB. Thus, AvrPtoB derivatives without E3 ubiquitin ligase activity did not suppress ROS production after chitin but after flg22 perception (Gimenez-Ibanez et al.2009). Furthermore, AvrPtoB1-387 failed to restore in planta growth of a P. syringae avrPtoB deletion mutant, suggesting that the E3 ubiquitin ligase domain contributes to the virulence function of AvrPtoB (Göhre et al.2008). Complementation of the avrPtoB mutant phenotype by full-length AvrPtoB, however, was not analyzed (Göhre et al.2008). Notably, a virulence function of the E3 ligase domain of AvrPtoB was not observed in additional studies, in which AvrPtoB1-387 and catalytically inactive AvrPtoB derivatives were shown to promote virulence of P. syringae avrPtoB mutants on susceptible tomato lines and Nicotiana benthamiana plants (Xiao, Giavalisco and Martin 2007; Xiao et al.2007; Zeng et al.2012; Wei et al. 2015).
HopM1 leads to the proteasome-dependent degradation of its interaction partner AtMIN7
The effector HopM1 from P. syringae pv. tomato triggers the degradation of its plant interaction partners including AtMIN7 (Arabidopsis HopM interactor 7), which is an ADP ribosylation factor-guanine nucleotide exchange factor (ARF-GEF) involved in vesicle trafficking (Nomura et al.2006) (Fig. 4C). AtMIN7 colocalizes with HopM1 to the trans-Golgi network/early endosome (Nomura et al.2011). This is in agreement with the role of ARF-GEF proteins in vesicle trafficking. The analysis of Arabidopsis AtMIN7 mutants revealed that AtMIN7 contributes to PTI and ETI responses but is dispensable for ROS production and stomatal closure upon flg22 treatment (Nomura et al.2011; Lozano-Duran et al. 2014). In agreement with its role in plant defense, AtMIN7 levels increase upon activation of PTI and are stabilized during ETI (Nomura et al.2011). HopM1 leads to the destabilization of AtMIN7 when hopM1 and AtMIN7 are coexpressed in leaves of N. benthamiana or when HopM1 is delivered by the T3S system of P. syringae pv. tomato DC3000 into leaves of Arabidopsis (Nomura et al.2006). The HopM1-mediated destabilization of AtMIN7 in N. benthamiana was reduced in the presence of proteasome inhibitor, suggesting that HopM1 exploits the proteasome for the degradation of AtMIN7 (Nomura et al.2006) (Fig. 4C). Given the contribution of AtMIN7 to plant defense, HopM1 was suggested to suppress plant defense responses via degradation of AtMIN7. HopM1 failed to trigger the degradation of AtMIN7 when delivered as heterologous protein by P. syringae pv. phaseolicola, which lacks a native hopM1 gene (Gangadharan et al.2013). However, delivery of HopM1 by P. syringae pv. phaseolicola led to the suppression of PTI responses, suggesting that AtMIN7 is not the only virulence target of HopM1 (Gangadharan et al.2013). This hypothesis was supported by the finding that HopM1 suppresses PTI in Arabidopsis atmin7 mutant plants (Gangadharan et al. 2013; Lozano-Duran et al.2014). One additional interaction partner, which is degraded by HopM1, is the 14-3-3 protein GRF8 (general regulatory factor 8) (Nomura et al.2006). GRF8 interacts with and controls the transcriptional repressor BZR1 (brassinazole resistant 1), which is involved in brassinosteroid signaling (Nomura et al. 2006; Gampala et al. 2007; Ryu et al.2007). As is discussed below, HopM1 presumably interferes with the function of GRF8 and leads to the nuclear accumulation of BZR1 (Lozano-Duran et al.2014).
XopL from X. campestris pv. vesicatoria acts as an E3 ubiquitin ligase
The type III effector XopL from X. campestris pv. vesicatoria contains a C-terminal E3 ubiquitin ligase domain with a novel fold and likely acts as a RING/U-box E3 ligase (Singer et al.2013) (Fig. 4D). XopL leads to the ubiquitination of plant proteins, yet the plant substrates of the XopL E3 ubiquitin ligase activity remain to be identified (Singer et al.2013). The analysis of different XopL protein regions revealed that the C-terminal E3 ubiquitin ligase domain is required for the XopL-mediated elicitation of plant cell death but dispensable for the suppression of PTI, which was observed in the presence of XopL. It was, therefore, speculated that the virulence function of XopL does not solely depend on its E3 ubiquitin ligase activity but also on its N-terminal LRR domain (Singer et al.2013).
GALA proteins from R. solanacearum contain an F box motif
GALA proteins (also designated Rip [Ralstonia protein injected into plant cells] G family) from R. solanacearum are effector proteins, which contain a conserved GAxALA amino acid motif in the C-terminal LRR region and an F-box motif in the N-terminal region (Cunnac et al.2004) (Fig. 4E). At least four GALA proteins from R. solanacearum including GALA6 were shown to interact with SKP1-like proteins from Arabidopsis (Angot et al.2006). However, it is yet unknown whether GALA proteins contribute to protein degradation. Notably, effector proteins with F-box motifs have also been identified as substrates of the type IV secretion systems from Agrobacterium tumefaciens and Legionella spp. (Price and Kwaik 2010).
XopJ from X. campestris pv. vesicatoria and HopZ4 from P. syringae interact with RPT6 and interfere with proteasome activity
Several members of the YopJ family of putative proteases and acetyltransferases including XopJ from X. campestris pv. vesicatoria strain 85-10 and HopZ4 from P. syringae interfere with the activity of the proteasome. The analysis of fluorogenic peptide substrates revealed that the proteasome activity in N. benthamiana was reduced in the presence of XopJ but not of a derivative thereof with a mutation in the catalytic cysteine residue (Üstün, Bartetzko and Börnke 2013). This suggests that the enzymatic activity of XopJ is required for the suppression of the proteasome. In agreement with the observed influence of XopJ on the proteasome, infection of pepper leaves with an X. campestris pv. vesicatoria xopJ deletion mutant led to increased proteasome activity when compared with leaves infected with the wild-type strain (Üstün, Bartetzko and Börnke 2013). XopJ interacts with and degrades the ATPase RPT6 (regulatory particle ATPase 6) of the 19S regulatory particle of the 26S proteasome at the plant plasma membrane (Üstün, Bartetzko and Börnke 2013; Üstün and Börnke 2015) (Fig. 4F). It was, therefore, proposed that XopJ acts as a protease and interferes with the activity of the proteasome by targeting RPT6. Similarly to XopJ, the homologous HopZ4 protein from P. syringae pv. lacrymans binds to RPT6 and interferes with proteasome activity (Üstün et al.2014) (Fig. 4F). Furthermore, the related effector protein AvrBsT from X. campestris pv. vesicatoria interacts with the 19S subunit RPN8 of the proteasome in yeast (Szczesny et al.2010), suggesting that the proteasome is a virulence target of several YopJ family members.
XopP from X. oryzae pv. oryzae inhibits the activity of the E3 ubiquitin ligase OsPUB44 from rice
The effector protein XopP from X. oryzae pv. oryzae interacts with the E3 ubiquitin ligase OsPUB44 from rice (Ishikawa et al.2014) (Fig. 4G). OsPUB44 presumably contributes to basal plant defense responses because rice OsPUB44 RNAi lines promote growth of X. oryzae pv. oryzae wild-type and non-pathogenic mutant strains (Ishikawa et al.2014). The results of interaction studies and in vitro activity assays revealed that XopP binds to the U-box domain of OsPUB44 and inhibits its E3 ubiquitin ligase activity (Ishikawa et al.2014). XopP was, therefore, suggested to suppress plant defense by interfering with the activity of OsPUB44.
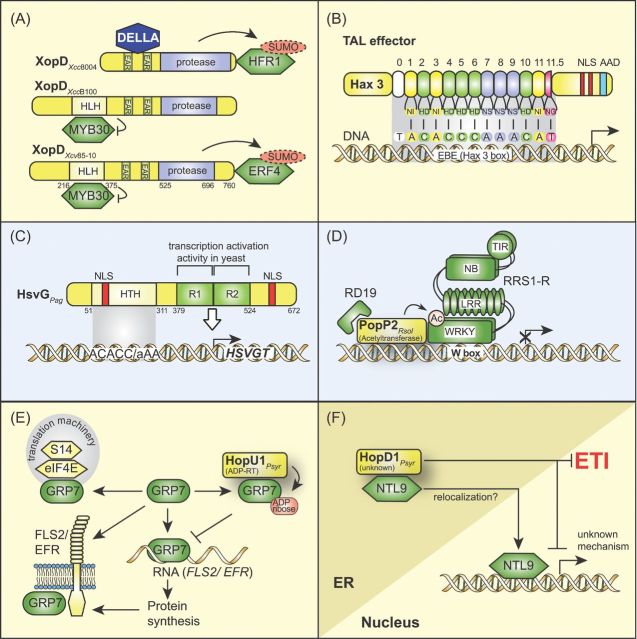
MODULATION OF PHYTOHORMONE SIGNALING BY TYPE III EFFECTORS
Phytohormones are chemical messengers, which initiate signaling responses during various cellular processes such as plant growth, development, reproduction and responses to biotic and abiotic stress. Phytohormones usually do not function independently of each other but are often controlled by a regulatory network, which links different hormone responses (Robert-Seilaniantz, Grant and Jones 2011; Gimenez-Ibanez and Solano 2013). Signaling by several hormones such as auxin, jasmonic acid (JA), ethylene (ET) and salicylic acid (SA) involves the proteasomal degradation of transcriptional repressors and the release or activation of transcription factors, which lead to hormone-induced gene expression. Research in the model plant Arabidopsis revealed that JA, SA and ET are key players in plant defense against microbial pathogens. While SA is usually involved in resistance against biotrophic and hemibiotrophic pathogens, JA and ET can act antagonistically to SA and promote resistance against necrotrophic pathogens (Glazebrook 2005) (Fig. 5). Due to the antagonistic interplay between JA and SA, the activation of JA-dependent defense responses often represses SA-induced signaling pathways, which are usually mounted upon infection by biotrophic pathogens (Gimenez-Ibanez and Solano 2013; Kazan and Lyons 2014). In addition to JA, SA and ET, recent studies revealed a role of other phytohormones including auxin, cytokinins, brassinosteroids, abscisic acid and gibberellins in plant–pathogen interactions (Gimenez-Ibanez and Solano 2013; Kazan and Lyons 2014).
Figure 5.
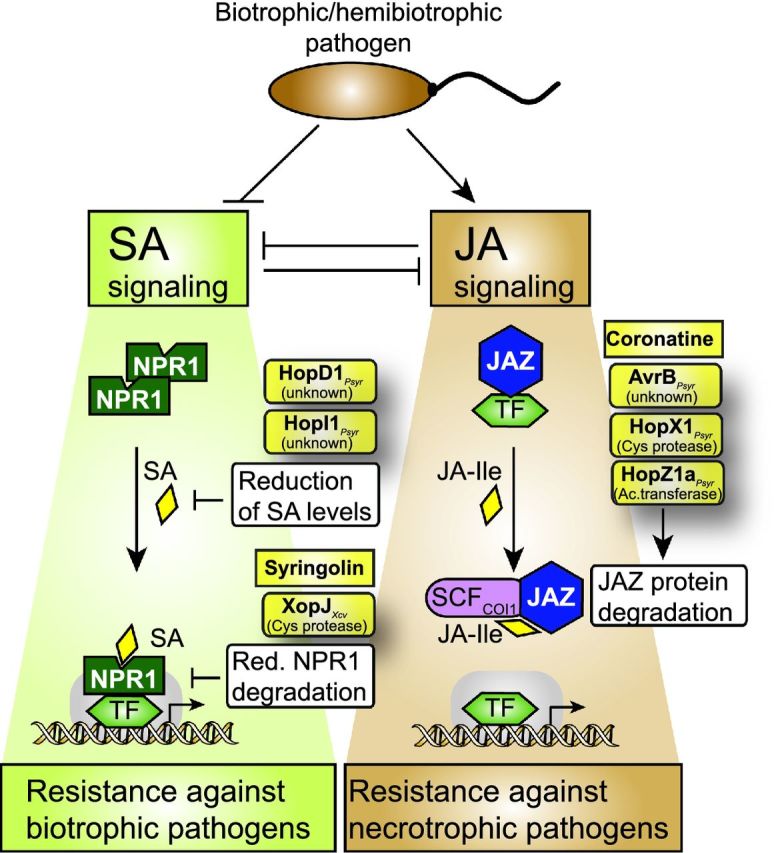
Interference of type III effectors with SA and JA signaling pathways. SA-dependent defense responses are required for plant resistance against biotrophic pathogens whereas JA-dependent defense is mounted against necrotrophic pathogens. SA and JA pathways thus act antagonistically and can suppress each other. Type III effectors from biotrophic or hemibiotrophic pathogens activate JA signaling pathways and suppress SA-mediated defense by the actions of translocated type III effectors. SA-dependent defense responses depend on NPR1 (non-expressor of PR genes), which is present in an oligomeric inactive state in the absence of SA. Upon binding of SA, monomeric NPR1 binds to and activates transcription factors (TF) and thus induces the expression of SA-dependent genes (Gimenez-Ibanez and Solano 2013). The effectors HopD1 and HopI1 from P. syringae lead to reduced SA levels whereas the bacterial toxin syringolin and the effector XopJ from X. campestris pv. vesicatoria interfere with the degradation of NPR1. The turnover of NPR1 is required for the expression of SA-responsive genes. Stabilization of NPR1, therefore, suppresses SA signaling (Robert-Seilaniantz, Grant and Jones 2011). JA signaling pathways involve JAZ proteins and the SCF complex. The bacterial toxin coronatine and the effector proteins AvrB, HopX1 and HopZ1 from P. syringae lead to the degradation of JAZ proteins and thus activate the expression of JA-responsive genes (see the text for details).
Plant-pathogenic bacteria produce phytohormone mimics to interfere with hormone signaling pathways. One prominent example is the phytotoxin coronatine, which is synthesized by a few pathovars of P. syringae and mimicks the action of bioactive JA-isoleucine (JA-Ile) (Katsir et al.2008). In addition to phytohormone mimics, plant-pathogenic bacteria deliver type III effector proteins to interfere with hormone signaling pathways. Effectors, which interfere with JA signaling pathways, include the cysteine protease HopX1, the acetyltransferase HopZ1a and AvrB from P. syringae (see below). Furthermore, the cysteine protease AvrRpt2 from P. syringae was shown to interfere with auxin signaling, whereas XopD from X. campestris pv. campestris modulates gibberellic acid (GA) and ET pathways. The functions of HopX1, HopZ1a, AvrB, AvrRpt2 and XopD are desribed below and summarized in Figs 5 and 6.
Figure 6.
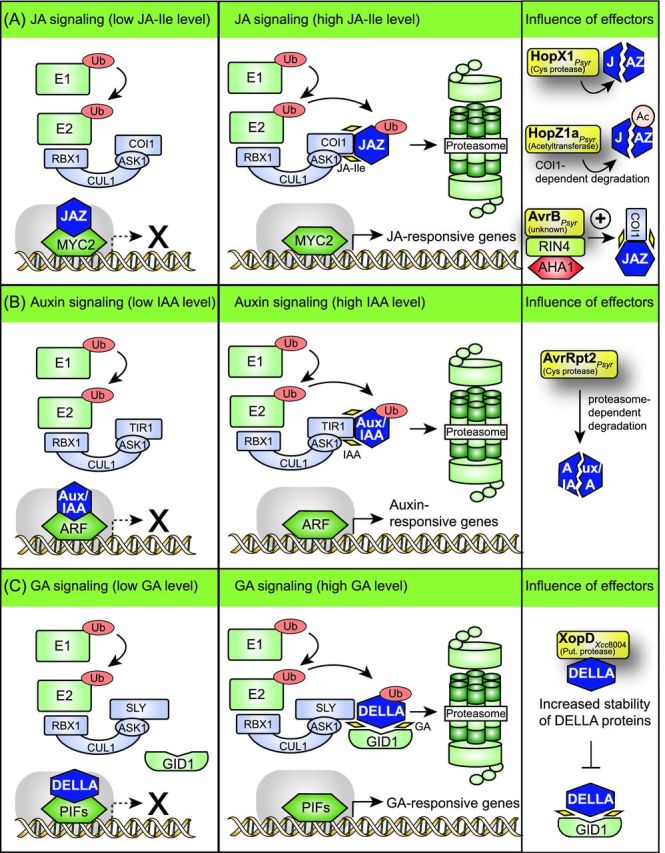
Modulation of JA, auxin and GA signaling pathways by type III effectors. (A) HopX1, HopZ1a and AvrB from P. syringae interfere with JA signaling pathways. Bioactive JA-Ile promotes the interaction between JAZ proteins and the F-box protein COI1, which is a component of the SCF complex. The subsequent degradation of JAZ proteins leads to the release of JAZ-interacting transcription factors (e.g. MYC2), which activate the expression of JA-responsive genes. The cysteine protease HopX1 directly or indirectly degrades several JAZ proteins independently of the JA receptor COI1 and thus activates the expression of JA-responsive genes. The acetyltransferase HopZ1a acetylates JAZ proteins and leads to their proteasome-dependent degradation. The effector protein AvrB from P. syrinage interacts with RIN4, which is a negative regulator of PTI and associates with the H+ ATPase AHA1. The interaction of AvrB with the RIN4-AHA1 complex promotes the interaction between JAZ proteins and COI1 and leads to the activation of JA-responsive genes. (B) Auxin signaling pathways are targeted by the cysteine protease AvrRpt2 from P. syringae. IAA promotes the interaction between Aux/IAA proteins and the F-box protein TIR1. This leads to the proteasome-dependent degradation of Aux/IAA proteins and to the release and activation of ARFs. ARFs subsequently activate the expression of auxin-responsive genes. The cysteine protease AvrRpt2 directly or indirectly induces the degradation of Aux/IAA proteins by the proteasome and thus activates the expression of auxin-responsive genes. (C) XopD from X. campestris pv. campestris strain 8004 interferes with the stability of DELLA proteins, which are negative regulators of GA-responsive genes. GA-dependent signaling is controlled by DELLA proteins, which inactivate PIF (phytochrome interacting factors) transcription factors. Binding of GA to its receptor GID1 leads to a conformational change in GID1, which subsequently binds to DELLA proteins. The formation of a GID1-DELLA complex promotes the interaction between DELLA proteins and the F-box protein SLY and thus the proteasome-dependent degradation of DELLA proteins. This leads to the release of PIF transcription factors, which activate the expression of GA-responsive genes. XopDXcc8004 presumably interferes with the binding of GID1 to DELLA proteins and delays the GA-induced degradation of the DELLA protein RGA. Notably, however, an influence of XopDXcc8004 on the transcription of GA-responsive genes has not yet been detected.
The P. syringae effectors HopX1 and HopZ1a promote the degradation of JAZ proteins
HopX1 and HopZ1a from P. syringae target JAZ (jasmonate ZIM-domain) proteins, which are involved in JA signaling. JAZ proteins act as transcriptional repressors and interact with and inhibit transcription factors. JAZ proteins are degraded by the proteasome in the presence of JA-Ile, which is perceived by the JA receptor and F-box protein COI1 (coronatine insensitive 1). COI1 is a component of the SCF complex (Shabek and Zheng 2014) and associates upon binding of JA-Ile with JAZ proteins to promote their proteasome-dependent degradation (Chini et al. 2007; Thines et al.2007; Sheard et al. 2010). This leads to the release of JAZ-interacting transcription factors and thus to the activation of JA-responsive gene expression (Robert-Seilaniantz, Grant and Jones 2011) (Fig. 6A).
The effector protein HopX1 is a cysteine protease and is delivered by P. syringae pv. tabaci strain 11528, which does not produce coronatine (Gimenez-Ibanez et al.2014). Transient coexpression studies in N. benthamiana revealed that HopX1 directly or indirectly degrades at least eight out of 12 JAZ protein family members (Gimenez-Ibanez et al.2014). The HopX1-mediated degradation of JAZ proteins occurs independently of the JA receptor COI1 and leads to the activation of JA-responsive genes as well as to the repression of SA-induced signaling pathways. It was, therefore, assumed that HopX1 directly or indirectly degrades JAZ proteins independently of the SCF complex (Gimenez-Ibanez et al.2014) (Fig. 6A).
JAZ proteins are also targeted by HopZ1a from P. syringae. Yeast two-hybrid, pull-down and BiFC studies revealed that HopZ1a interacts with JAZ proteins from soybean and Arabidopsis at the plasma membrane and in the nucleus (Jiang et al.2013). HopZ1a leads to the degradation of JAZ proteins and the induction of JA-responsive genes in Arabidopsis when delivered by a coronatine-deficient mutant derivative of P. syringae pv. tomato DC3000 (Jiang et al.2013) (Fig. 6A). No effect of HopZ1a on JAZ proteins was observed in Arabidopsis coi1 mutants, suggesting that HopZ1a exploits the proteasome to induce the degradation of JAZ proteins (Jiang et al.2013). A transferase assay with 14C-labeled acetyl-CoA revealed that HopZ1a acetylates JAZ proteins as well as additional plant targets including tubulin (see below) and the pseudokinase ZED1 (hopZ1-ETI deficient) (Lee et al. 2012; Jiang et al. 2013; Lewis et al.2013). The activity of HopZ1a depends on the presence of phytic acid (also known as phytate or IP6), which is a cofactor of YopJ family members, and induces a conformational change in HopZ1a (Lee et al. 2012; Ma et al.2015). It remains to be investigated whether the acetylation of JAZ proteins by HopZ1a facilitates their COI1-dependent degradation.
AvrB promotes the interaction between JAZ proteins and COI1 through RIN4 and AHA1
The type III effector AvrB from P. syringae can substitute coronatine for the induction of JA-responsive genes and stomatal opening (He et al.2004; Zhou et al.2015). The regulation of stomatal opening involves the autoinhibited plasma membrane H(+) ATPases AHA1 and AHA2, which pump protons from the cytosol into the apoplast. This leads to the establishment of a proton electrochemical gradient, which is utilized by channel and carrier proteins to mediate the uptake of charged solutes into cells (Sondergaard, Schulz and Palmgren 2004; Liu et al.2009; Elmore and Coaker 2011). Increased concentrations of charged solutes in the guard cells result in a water uptake and elevated turgor, thus leading to stomatal opening. The induction of stomatal opening by AvrB depends on the F-box protein COI1 and the immune regulator RIN4, which interacts with AvrB (Mackey et al. 2002; He et al.2004; Cui et al. 2010; Zhou et al.2015) (see above). RIN4 also interacts with AHA1 and promotes its activity (Liu, Elmore and Coaker 2009) (see below). Transient expression studies in N. benthamiana revealed that AHA1, RIN4 and AvrB trigger the degradation of JAZ proteins, suggesting a link between JA signaling, the RIN4-AvrB interaction and stomatal opening (Zhou et al.2015). Furthermore, coimmunprecipitation experiments showed that AvrB and AHA1 promote the interaction between COI1 and the JAZ protein JAZ9 (Zhou et al.2015). Given that AvrB induces the phosphorylation of RIN4 and that RIN4 positively regulates AHA proteins (Liu et al.2009), it was suggested that AvrB promotes the COI1-JAZ interaction through RIN4 and AHA1 and thus leads to the degradation of JAZ proteins (Zhou et al.2015) (Fig. 6A). The biochemical mechanism underlying the AvrB-RIN4-AHA1-mediated induction of COI1-JAZ interactions is yet unknown.
The cysteine protease AvrRpt2 promotes the degradation of Aux/IAA proteins and leads to the activation of auxin-responsive genes
The cysteine protease AvrRpt2 from P. syringae interferes with auxin signaling and was shown to induce the expression of auxin-responsive genes (Chen et al.2007). Expression studies in Arabidopsis protoplasts and transgenic plants revealed that AvrRpt2 enhances the degradation of the Aux/IAA (auxin/indole acetic acid) transcription repressor protein AXR2 and promotes the effect of auxin (Cui et al.2013). Members of the Aux/IAA family of transcription suppressors bind to and inactivate auxin response factors (ARFs) and thus suppress the expression of auxin-responsive genes (Fu and Wang 2011). In the presence of auxin, Aux/IAA proteins interact with the auxin receptor and F-box protein TIR1 (transport inhibitor response 1), which is a component of the SCF complex (Fig. 6B). This promotes the degradation of Aux/IAA proteins by the proteasome and leads to the release of ARFs, which activate the expression of auxin-responsive genes (Fu and Wang 2011).
The AvrRpt2-induced degradation of the Aux/IAA protein AXR2 was abolished in the presence of proteasome inhibitor or upon mutation of catalytic residues in AvrRpt2. This suggests that the proteasome and the cysteine protease activity of AvrRpt2 are required to induce the degradation of Aux/IAA proteins (Cui et al.2013). Given that a direct cleavage of AXR2 by AvrRpt2 was not observed in vitro, the AvrRpt2-triggered degradation of Aux/IAA proteins likely involves additional plant proteins (Cui et al.2013). In addition to the interference with auxin-regulated gene expression, AvrRpt2 was recently shown to promote bacterial growth in a COI1-dependent manner (Geng et al.2016). It was, therefore, proposed that AvrRpt2 also interferes with JA signaling pathways and thus suppresses SA-mediated defense (Geng et al.2016).
XopD proteins from Xanthomonas modify the stability of an ET-responsive transcription factor (ERF) and DELLA proteins
Members of the XopD family of nuclear-localized effector proteins from Xanthomonas spp. contain a C-terminal cysteine protease domain and ERF-associated amphiphilic repression (EAR) motifs, which were previously described for plant transcriptional regulators (Kazan 2006). Sequence comparisons revealed variations in the domain organization of XopD family members. While XopD from X. campestris pv. vesicatoria (XopDXcv) and X. campestris pv. campestris strain B100 (XopDXccB100) contains N-terminal extensions and a central putative DNA-binding helix-loop-helix (HLH) domain, these regions are absent in XopD from X. campestris pv. campestris strain 8004 (XopDXcc8004) (Canonne et al. 2010; Kim, Taylor and Mudgett 2011) (Fig. 7A). The XopD cysteine protease domain shares structural similarity with the yeast ubiquitin-like protease ULP1 and was shown to cleave tomato SUMO from SUMOylated proteins (Hotson et al.2003; Chosed et al. 2007). XopDXcv deSUMOylates and thus destabilizes SlERF4 from Solanum lycopersicum (Kim, Stork and Mudgett 2013). As the presence of proteasome inhibitor interferes with the XopD-induced destabilization of SlERF4, it was suggested that XopD facilitates the degradation of SlERF4 by the proteasome (Kim, Stork and Mudgett 2013). SlERF4 is presumably involved in the regulation of ET biosynthesis and colocalizes with XopD to subnuclear foci (Kim, Stork and Mudgett 2013). In agreement with the observed XopD-mediated destabilization of SlERF4, XopDXcv leads to reduced ET levels in infected plant tissue and suppresses the expression of genes involved in ET production (Kim, Stork and Mudgett 2013). Given that ET production is required for plant immunity, XopDXcv likely deSUMOylates SlERF4 to suppress plant defense responses (Kim, Stork and Mudgett 2013). Notably, however, XopDXcv also suppresses SA- and JA-induced gene transcription (Kim et al.2008a).
Figure 7.
Interference of type III effector proteins with plant gene expression. (A) Domain organization of XopD proteins from Xanthomonas spp. XopD family members consist of a C-terminal cysteine protease domain and N-terminal EAR motifs. Additionally, XopD from X. campestris pv. vesicatoria strain 85-10 (XopDXcv85-10) and X. campestris pv. campestris strain B100 (XopDXccB100) contain N-terminal extensions and a central putative DNA-binding HLH domain. XopDXcc8004 was shown to interact with and stabilize DELLA proteins via the EAR motif-containing region. Furthermore, XopDXcc8004 interacts with and deSUMOylates the transcription factor HFR1. XopDXccB100 and XopDXcv85-10 bind to the transcription factor MYB30 via the HLH domain and suppress its transcriptional activity. XopDXcv deSUMOylates and thus destabilizes the transcription factor ERF4. Numbers refer to amino acid positions in XopDXcv85-10. (B) Domain organization and DNA-binding specificity of the TAL effector Hax (homolog of AvrBs3 in Xanthomonas) 3 from X. campestris pv. armoraciae. TAL effectors contain a C-terminal acidic activation domain (AAD), two NLSs and a central protein region with repeats. The RVDs of Hax3 and the matching bases in the EBE in the promoter regions of Hax3-induced genes are indicated. (C) Domain organization of HsvG from P. agglomerans (Pag). HsvG contains N- and C-terminal NLSs, an N-terminal HTH region and two repeats of 71 and 74 amino acids (R1 and R2), which confer transcription activation activity in yeast. The repeats determine the specificity of plant gene activation (indicated by a white arrow) but are dispensable for DNA binding of HsvG, which depends on the N-terminal region. Numbers refer to amino acid positions in HsvG. (D) Modification of RRS1-R by the effector protein PopP2 from R. solanacearum. The TIR-NB-LRR protein RRS1-R forms a dimer and binds via its WRKY domain to a DNA motif (W box) present in promoters of target genes of WRKY transcription factors. PopP2 interacts with and acetylates the WRKY domain of RRS1-R and thus interferes with its DNA binding. The additional PopP2 interaction partner RD19, which is a predicted protease, is presumably not acetylated by PopP2. RRS1-R also interacts with the R protein RPS4, which is required for the induction of ETI and is not shown in this figure (see the text for details). (E) The mono-ADP-RT HopU1 from P. syringae pv. tomato DC3000 ADP-ribosylates the RNA-binding protein GRP7, which interacts with components of the translational machinery including the cap-binding protein eIF4E and the ribosomal subunit S14. GRP7 also interacts with the PRRs FLS2 and EFR and with FLS2 and EFR transcripts, and was, therefore, assumed to promote PRR translation. ADP-ribosylation of GRP7 by HopU1 reduces the ability of GRP7 to bind to RNA and might suppress FLS2 and EFR protein synthesis. (F) HopD1 from P. syringae pv. tomato DC3000 interacts with the transcription factor NTL9 at the ER and leads to reduced expression of NTL9-induced genes during ETI. Furthermore, HopD1 suppresses ETI responses. The mechanisms underlying the HopD1-mediated inhibition of NTL9-dependent gene expression are unknown.
Interference with hormone signaling was also shown for the XopD family member XopDXcc8004, which interacts with DELLA proteins (Tan et al.2014) (Figs 6C and 7A). DELLA proteins are negative regulators of GA response activators and colocalize with XopDXcc8004 to the plant nucleus. The degradation of DELLA proteins by the proteasome is stimulated in the presence of GA, which binds to its receptor GID1 (gibberellin insensitive dwarf 1) (Hauvermale, Ariizumi and Steber 2012). GA promotes the interaction between GID1 and DELLA proteins, which are subsequently targeted to the F-box protein SLY and degraded by the proteasome. The degradation of DELLA proteins leads to the activation of GA response activators, which induce the expression of GA-responsive genes (Hauvermale, Ariizumi and Steber 2012) (Fig. 6C). XopDXcc8004 delays the GA-induced degradation of the DELLA protein RGA (Tan et al.2014). The N-terminal EAR domain of XopDXcc8004 interacts with the DELLA domain of RGA, which contains the conserved DELLA motif and is required for the GA-induced degradation of DELLA proteins (Sun and Gubler 2004; Tan et al.2014). It is speculated that the N-terminal EAR domain of XopDXcc8004 interferes with the binding of GID1 to DELLA proteins (Tan et al.2014). Notably, however, despite the observed effect of XopDXcc8004 on DELLA protein stability, no influence on the transcription of GA-responsive genes was detected (Tan et al.2014).
MODULATION OF PLANT GENE EXPRESSION BY TYPE III EFFECTORS
One effective strategy employed by type III effectors to interfere with plant cellular processes is the manipulation of gene expression on the transcriptional or posttranscriptional level. Effector proteins, which are directly imported into the nucleus and either bind to DNA or to components of the plant transcription machinery, are transcription activator-like (TAL) effectors from Xanthomonas spp. and the effector protein HsvG from Pantoea agglomerans (see below). Type III effectors, which target plant transcription factors and RNA-binding proteins, include XopD proteins from Xanthomonas spp., PopP2 from R. solanacearum as well as HopU1, HopD1 and HopM1 from P. syringae. Known mechanisms underlying type III effector-mediated modulation of plant gene expression are summarized below and in Fig. 7.
TAL effectors from Xanthomonas spp. bind to sequence-specific promoter elements of plant target genes
Members of the TAL effector family were mainly isolated from Xanthomonas spp. However, related proteins are also present in R. solanacearum and Burkholderia rhizoxinica (de Lange et al. 2013, 2014; Li et al.2013a; Juillerat et al. 2014). Characteristic features of TAL effectors include a C-terminal acidic activation domain and nuclear localization signals (NLSs), which are required for the import of TAL effectors into the plant nucleus (Boch and Bonas 2010). DNA binding is mediated by the central region of TAL effectors, which consists of 1.5 to 33.5 repeats with an average of approximately 17 repeats (Boch and Bonas 2010) (Fig. 7B). A minimum of 6.5 repeats is required to induce target gene expression (Boch et al.2009). The repeats are nearly amino acid sequence identical and usually 33 to 35 amino acids long, but longer and shorter repeats have also been described (Boch and Bonas 2010).
Sequence-specific binding to DNA bases depends on the polymorphic amino acids at positions 12 and 13 of each repeat of the TAL effector. These amino acids are referred to as RVDs (repeat variable diresidues) and are exposed on a short loop located between two nearly identical alpha helices (Deng et al. 2012; Mak et al.2012). Direct DNA contact is mediated by the RVD at position 13 (also referred to as base-specifying residue), whereas the RVD at position 12 stabilizes the loop (Deng et al. 2012; Mak et al.2012). The RVDs determine the binding specificity of TAL effectors to DNA. Common RVDs are HD, NG, NN and NI (letters refer to amino acid residues), which bind to cytosine (HD), thymine (NG), guanine or adenine (NN), and adenine (NI), respectively (Boch et al. 2009; Moscou and Bogdanove 2009).
In the past years, numerous studies have focused on the analysis of the binding specificity of TAL effectors to the effector-binding elements (EBEs) in the promoter regions of plant target genes. Repeat number and RVDs determine the number and nature of DNA bases, which are bound by the TAL effectors. Furthermore, different RVDs differ in DNA-binding affinity and thus contribute differently to TAL effector activity (Streubel et al.2012; Meckler et al. 2013). Replacement of the natural RVDs of specific repeats by all possible 400 RVD combinations revealed that not all artificial RVDs are functional. Optimal reporter gene activation was observed with TAL effectors containing naturally occurring RVD combinations (Yang et al.2014). In addition to the RVD composition, the length of the repeats affects the binding to DNA bases. Experimental evidence suggests that shorter or longer repeats bind to matching nucleotides of the EBE or are excluded from binding (Richter et al.2014). The looping-out of repeats allows a shift of the following repeats by one nucleotide position in the EBE. This provides flexibility in the DNA-binding activities of TAL effectors and enables TAL effectors to bind to promoter elements in which the EBEs have been modified by insertion/deletion mutations (Richter et al.2014). Notably, however, DNA binding of TAL effectors to EBEs is not only determined by the RVDs but also by N-terminal non-canonical repeats as well as C-terminal protein regions and can be affected by epigenetic modifications of the DNA (Boch, Bonas and Lahaye 2014; Schreiber et al.2015).
The groundbreaking discovery of the TAL-DNA-binding code has marked the beginning of a new era in genome engineering because it has led to the design of various genome editing tools (e.g. TAL effector nucleases), which allow sequence-specific binding of DNA-modifying enzymes by the use of TAL effector repeats as fusion partners (Scharenberg, Duchateau and Smith 2013; Mahfouz, Piatek and Stewart 2014). The mechanisms leading to transcriptional activation of plant genes by TAL effectors are not yet understood. The results of interaction studies suggest that TAL effectors interact with RNA polymerase II (Domingues et al.2012), and with negative regulators of RNA polymerase II and III, respectively (de Souza et al. 2012; Soprano et al.2013) (Table 1). It is, therefore, assumed that TAL effectors do not only bind to DNA but also associate with components of the plant transcription machinery to activate gene expression.
Among the plant genes targeted by TAL effectors are those encoding transcription factors and proteins involved in senescence, development, stress response and sugar transport (Table S2, Supporting Information). Examples are the SWEET genes from rice, which are involved in sucrose or fructose transport and are induced by TAL effectors from the systemic rice pathogen X. oryzae pv. oryzae (Boch, Bonas and Lahaye 2014; Chen 2014) (Table S2). TAL target genes, which contribute to virulence, are also referred to as plant susceptibility genes. Notably, however, TAL effectors can also induce the expression of plant resistance (R) genes and thus trigger ETI responses (Boch, Bonas and Lahaye 2014). TAL effector-responsive R genes have been categorized into different groups including recessive and dominant R genes. Recessive R genes have evolved from S genes and contain mutations in the EBE, which abolish TAL effector binding (Hutin et al.2015). In this case, resistance is the result of the loss of induction of an S gene. Dominant R genes such as Bs4 from tomato, which elicits ETI in response to the TAL effector AvrBs4 (Schornack et al.2004), confer non-transcriptional-based resistance upon recognition of corresponding TAL effectors. The third group of TAL effector-responsive R genes are executor R genes, which contain EBEs in their promoter regions and are specifically activated by matching TAL effectors (Zhang, Yin and White 2015). Examples are Bs3 from pepper, and Xa10, Xa23 and Xa27 from rice (Table S2). The engineering of executor R gene promoters allows gene induction by various TAL effectors and might help to improve strategies for plant resistance and disease control (Boch, Bonas and Lahaye 2014).
The effector protein HsvG from P. agglomerans binds to DNA and activates plant gene expression
DNA binding has also been described for the effector protein HsvG, which is an important pathogenicity factor of the gall-forming plant-pathogenic bacterium P. agglomerans pv. gypsophilae (Valinsky et al.1998; Nissan et al. 2006). HsvG is homologous to the type III effector HsvB, which is required for pathogenicity of Pa. agglomerans pv. betae on beet (Nissan et al.2006). Both effectors localize to the plant cell nucleus and harbor N- and C-terminal NLSs as well as an N-terminal helix-turn-helix (HTH) domain (Nissan et al. 2006; Weinthal et al.2011) (Fig. 7C). HsvG and HsvB mainly differ in the C-terminal region, which contains two tandem direct repeats of 71 and 74 amino acids in HsvG whereas one repeat is missing in HsvB (Nissan et al.2006). The analysis of deletion derivatives of HsvG revealed that the presence of two repeats is required for the contribution of HsvG to bacterial pathogenicity on gypsophila (Nissan et al.2006).
The results of yeast one-hybrid assays suggest that HsvG acts as a transcriptional activator. One repeat of HsvG is sufficient for the transcriptional activation of reporter genes in yeast (Nissan et al.2006). Notably, in contrast to TAL effectors, DNA binding of HsvG does not depend on the repeats but on an N-terminal protein region consisting of 14 predicted alpha helices and one beta strand (Nissan et al.2012). A random binding-site selection procedure revealed that HsvG binds to DNA sequences with the consensus motif ACACC/aAA (Nissan et al.2006), which is present in the promoter of the HsvG target gene HSVGT from gypsophila (Nissan et al.2012). HSVGT encodes a protein with homology to the chaperone DnaJ and contains predicted NLSs as well as DNA-binding motifs, suggesting that it acts as a transcription factor (Nissan et al.2012). Expression of HSVGT is specifically induced by HsvG as well as by a modified HsvB derivative containing one additional repeat from HsvG (Nissan et al.2012). These findings suggest that the repeats and not the DNA-binding region of HsvG and HsvB determine the specificity in target gene activation.
XopD proteins from Xanthomonas spp. target plant transcription factors
Among the nuclear localized effectors, which presumably interfere with plant gene expression, are members of the XopD family from Xanthomonas spp. As described above, XopD family members interfere with hormone signaling and cleave SUMO from SUMOylated proteins. Furthermore, XopDXcc8004 binds to and deSUMOylates the basic HLH transcription factor HFR1 in subnuclear foci (Tan et al.2015) (Fig. 7A), suggesting that XopD family members target plant transcription regulators to promote bacterial virulence. This hypothesis is supported by the finding that XopDXcv and XopDXccB100 interact with the transcription factor MYB30, which acts as a positive regulator of hypersensitive cell death responses and resistance to avirulent pathogens in Arabidopsis (Canonne et al. 2011; Raffaele and Rivas 2013) (Fig. 7A). No interaction with AtMYB30 was observed for XopDXcc8004, which lacks the HLH domain (Canonne et al.2011). Domain swapping experiments with XopDXcv and XopDXcc8004suggest that the central HLH domain of XopDXcv is required for the interaction with AtMYB30 (Canonne et al.2011).
XopDXcv and XopDXccB100 induce the relocalization of MYB30 from the nucleus to subnuclear foci and suppress its transcriptional activity (Canonne et al.2011). In agreement with this finding, XopDXccB100reduces the expression of AtMYB30 target genes in Arabidopsis during the natural infection (Canonne et al.2011). It was, therefore, suggested that XopD family members suppress plant defense responses by targeting MYB30.
PopP2 from R. solanacearum acetylates RRS1-R and WRKY transcription factors and interferes with the transcription of plant genes
Another effector protein, which modulates plant gene expression, is the YopJ family member and acetyltransferase PopP2 from R. solanacearum. PopP2 localizes to the plant cell nucleus and acetylates lysine residues of WRKY transcription factors (Deslandes et al. 2003; Le Roux et al. 2015; Sarris et al.2015). WRKY transcriptional regulators contain an N-terminal WRKY amino acid motif and a zinc finger structure, and bind to a conserved sequence motif (W box; TTGACC/T) in the promoter of target genes (Rushton et al.2010; Bakshi and Oelmuller 2014; Llorca, Potschin and Zentgraf 2014). Acetylation by PopP2 suppresses the DNA binding of WRKY transcription factors and the transcriptional activation of their target genes (Le Roux et al. 2015; Sarris et al.2015). In addition to WRKY transcription factors, PopP2 binds to and acetylates lysine residues in the C-terminal WRKY domain of the R protein RRS1-R from Arabidopsis (Deslandes et al. 2003; Le Roux et al. 2015; Sarris et al.2015) (Fig. 7D). Acetylation of RRS1-R by PopP2 interferes with the binding of RRS1-R to W box-containing DNA sequences and thus with the RRS1-R-mediated activation of gene expression (Le Roux et al. 2015; Sarris et al.2015) (see below). Notably, PopP2 does not only acetylate but also stabilizes RRS1-R. Stabilization of RRS1-R by PopP2 is independent of the acetyltransferase activity of PopP2 (Tasset et al.2010).
The mADP-RT HopU1 from P. syringae targets RNA-binding proteins and reduces transcript levels of PRR genes during PTI
Type III effectors from plant-pathogenic bacteria do not only bind to DNA or transcription factors but can also interact with RNA-binding proteins. One example is the mADP-RT HopU1 from P. syringae pv. tomato DC3000, which targets several plant RNA-binding proteins (Fu et al.2007). Among the HopU1 targets are the glycine-rich protein (GRP) 7 and GRP8, which colocalize with HopU1 to the cytoplasm and possibly to the nucleus (Fu et al. 2007; Jeong et al.2011). GRP7 is abundantly expressed in guard cells, affects stomatal opening and closing, and contributes to PTI responses in Arabidopsis (Fu et al. 2007; Kim et al.2008b; Jeong et al. 2011). In line with its contribution to PTI, GRP7 interacts with the PRRs FLS2 and EFR at the plant plasma membrane (Nicaise et al.2013). Furthermore, GRP7 binds to RNAs including its own transcript as well as FLS2 and EFR transcripts (Nicaise et al.2013). Given that GRP7 was also found in a complex containing components of the translational machinery such as the cap-binding protein eIF4E and the ribosomal subunit S14, it was assumed that GRP7 promotes PRR translation (Nicaise et al.2013) (Fig. 7E).
The results of coimmunprecipitation experiments suggest that HopU1 interferes with the association of GRP7 with PRR transcripts (Nicaise et al.2013). A site-directed mutagenesis approach revealed that the arginine residue at position 49 of GRP7 is mono-ADP-ribosylated by HopU1 (Jeong et al.2011). R49 is located in the conserved ribonucleoprotein consensus sequence 1 (RNP-1) motif of the RNA recognition motif of GRP7 and is essential for the ability of GRP7 to bind to RNA and to contribute to plant immunity (Fu et al. 2007; Jeong et al.2011). ADP-ribosylation of R49 reduces the ability of GRP7 to bind to RNA in vitro and therefore likely blocks complex formation between GRP7 and the 3′-untranslated region of FLS2 (Jeong et al.2011) (Fig. 7E). Taken together, these findings suggest that HopU1 suppresses the GRP7-induced accumulation of FLS2 by ADP-ribosylation of GRP7.
HopD1 from P. syringae targets the transcription factor NTL9 at the ER and inhibits ETI responses
HopD1 from P. syringae pv. tomato DC3000 is an effector protein with unknown enzymatic function, which interacts with the transcription factor NTL9 from Arabidopsis (Block et al.2014). NTL9 is a member of the NTLM1 (NAC with transmembrane motif 1) family of transcription factors. This family is one of the largest families of plant transcription factors and is involved in various processes including developmental and stress-related signaling (Nuruzzaman, Sharoni and Kikuchi 2013). At least 13 out of more than 110 NAC transcription family members contain C-terminal transmembrane domains, suggesting that they do not exclusively localize to the nucleus (Kim et al.2007). NTL9 contains an N-terminal DNA-binding domain and a C-terminal transmembrane motif and colocalizes with HopD1 to the endoplasmic reticulum (ER) (Block et al.2014) (Fig. 7F).
NTL9 is preferentially expressed in guard cells (Yoon et al.2008) and activates the isochorismate synthase 1 gene ICS1, which encodes a key enzyme in the biosynthesis of SA (Wildermuth et al. 2001; Zheng et al.2015). SA is involved in various cellular processes including plant stomatal immunity, i.e. the closure of stomata upon pathogen attack. Stomatal immunity is abolished in Arabidopsis ntl9 mutant plants and this phenotype is suppressed upon application of SA. It was, therefore, proposed that the contribution of NTL9 to SA synthesis is required for stomatal immunity (Zheng et al.2015). NTL9 was also shown to contribute to ETI responses in Arabidopsis (Block et al.2014). A search for genes with a putative DNA-binding site for NTL9 led to the identification of genes, which are induced by NTL9 during ETI (Block et al.2014). Expression of NTL9-induced genes during ETI is reduced in the presence of HopD1. Several NTL9-regulated genes were also induced upon treatment of plants with flg22; however, no effect of HopD1 on NTL9-regulated gene expression was observed, suggesting that HopD1 specifically interferes with NTL9-regulated gene expression during ETI (Block et al.2014). This is in agreement with the finding that HopD1 specifically suppresses ETI responses (Jamir et al.2004; Guo et al. 2009). It was therefore suggested that HopD1 targets NTL9 at the ER to inhibit NTL9-mediated gene expression during ETI (Block et al.2014) (Fig. 7F). Given that HopD1 did not alter the localization or activity of NTL9 (Block et al.2014), the mechanisms underlying the HopD1-mediated inhibition of NTL9-dependent gene expression are still unknown.
HopM1 indirectly targets the transcription factor BZR1
As mentioned above, HopM1 from P. syringae triggers the degradation of its interaction partners including the ARF/GEF AtMIN7 and the 14-3-3 protein GRF8/AtMIN10 (Nomura et al. 2006, 2011). GRF8/AtMIN10 interacts with BZR1, a major transcription factor in brassinosteroid signaling, and retains BZR1 in the cytoplasm (Gampala et al. 2007; Ryu et al.2007). HopM1 leads to an increased accumulation of BZR1 in the nucleus and thus likely interferes with the function of GRF8/AtMIN10 (Lozano-Duran et al.2014). A similar phenomenon was observed upon treatment of plants with AICAR (5-aminoimidazole-4-carboxamide-1-beta-D-ribofuranoside), which inhibits the interaction of 14-3-3 proteins with their targets (Lozano-Duran et al.2014). The HopM1-induced nuclear accumulation of BZR1 results in the downregulation of BZR1 target genes (Lozano-Duran et al.2014). Taken together, these results suggest that HopM1 targets a 14-3-3 protein to interfere with the activity of a transcriptional repressor.
INTERFERENCE OF TYPE III EFFECTORS WITH THE PLANT CYTOSKELETON
The plant cytoskeleton mainly consists of actin filaments and microtubules and contributes to many processes including cell division and growth, organelle movement, vesicle trafficking, endocytosis, opening of stomata and plant defense responses (Day et al. 2011; Henty-Ridilla et al.2013a). The analysis of actin filaments using a GFP (green fluorescent protein)-fABD2 (filamentous actin-binding domain 2) reporter fusion in Arabidopsis epidermis cells revealed a transient increase in actin filaments upon infection with Pseudomonas syringae pv. tomato DC3000 wild-type or T3S mutant strains (Henty-Ridilla et al.2013b) (Fig. 8A). A similar formation of actin filaments was observed upon treatment of plants with PAMPs and was shown to depend on FLS2, BAK1 and BIK1. It was, therefore, suggested that the increase in actin filaments is part of the PTI response (Henty-Ridilla et al.2013b). Twenty-four hours after infection with the P. syringae wild-type strain, a reduced number of actin filments and an increase in bundled actin structures were observed (Fig. 8A). No changes were induced by P. syringae strains lacking a functional T3S system or multiple effectors, suggesting that the delivery of type III effectors leads to a decrease in actin filaments (Henty-Ridilla et al.2013b; Shimono et al.2016). Infiltration of latrunculin B, which inhibits actin polymerization, promotes susceptibility of Arabidopsis leaves to bacterial infections and leads to an increased growth of P. syringae pv. tomato DC3000 wild-type or T3S mutant strains (Henty-Ridilla et al.2013b). This is in agreement with the predicted contribution of actin filaments to PTI and the observed effector-mediated decrease in actin filaments (Henty-Ridilla et al.2013b; Shimono et al.2016). The actin filament network might also be involved in ETI responses because the actin-depolymerizing factor 4 (ADF4), which is required to sever and disassemble F-actin, contributes to the expression of the R gene RPS5 (Porter et al.2012; Henty-Ridilla et al. 2014).
Figure 8.
Influence of type III effectors on actin filaments and microtubules. (A) Infection of Arabidopsis cells with P. syringae pv. tomato DC3000 leads to alterations in the actin cytoskeleton. The infection with wild-type or T3S mutant strains leads to an increase in actin filaments 6 hours post infection. Twenty-four hours post infection, the wild-type strain induces the formation of actin bundles and leads to a reduced number of actin filaments. Actin filaments and bundles are indicated as yellow dashes. The plant cell wall is represented in green. The following cell organelles are shown: chloroplasts (green), mitochondria (beige), vacuole (blue), nucleus (beige), ER (light brown) and Golgi apparatus (red). (B) HopW1 leads to the disruption of actin filaments. The effector protein HopW1 binds to filamentous actin and leads to the disruption of actin filaments. (C) HopG1 induces the formation of actin bundles. HopG1 binds to a mitochondrial-localized kinesin motor protein, which associates with microtubules and presumably links microtubules to actin filaments. HopG1 induces the formation of actin bundles, presumably via its interaction with kinesin. (D) HopE1 leads to the dissociation of MAP65 from microtubules. HopE1 interacts with calmodulin (CaM) and the microtubule-associated protein MAP65 and leads to the dissociation of MAP65 from microtubules. No effect of HopE1 on the microtubule network was observed. (E) HopZ1a from P. syringae dissociates microtubules. The acetyltransferase HopZ1a binds to and acetylates tubulin and disrupts microtubules.
In addition to actin filaments, plant defense against bacterial infections also involves microtubules. Treatment of Arabidopsis plants with oryzalin, which disrupts microtubules, enhances the growth of P. syringae pv. tomato DC3000 but not of a T3S-deficient strain (Lee et al.2012). It was, therefore, suggested that effector proteins from P. syringae interfere with microtubule formation to promote bacterial virulence (Lee et al.2012). Given that oryzalin did not enhance the growth of a T3S mutant strain, the disruption of microtubules is presumably not sufficient to counteract PTI responses (Lee et al.2012). The influence of type III effector proteins including HopW1, HopG1 and HopE1 from P. syringae and members of the YopJ family on the plant cytoskeleton is described below and summarized in Fig. 8.
HopW1 from P. syringae disrupts the actin cytoskeleton
HopW1 from P. syringae pv. maculicola was found in a complex with actin after transient expression in N. benthamiana (Kang et al.2014). Confocal microscopy using the reporter protein Lifeact-GFP, which binds to filamentous actin, showed that the delivery of HopW1 by P. syringae pv. tomato DC3000 leads to the disruption of the actin cytoskeleton (Kang et al.2014) (Fig. 8B). A disruption of actin filaments was also observed when hopW1 was transiently expressed in N. benthamiana or Arabidopsis protoplasts (Kang et al.2014). Furthermore, recombinant HopW1 disassembles filamentous actin in vitro (Kang et al.2014). In agreement with the HopW1-mediated destabilization of actin filaments and the contribution of the actin cytoskeleton to protein trafficking, HopW1 inhibits protein trafficking to the ER or the vacuole (Kang et al.2014). As expected, a similar effect on protein trafficking was observed for latrunculin B (Kang et al.2014). HopW1 also interferes with endocytosis as was shown in Arabidopsis cotyledons, which were stained with the lipophilic dye FM4-64 (Kang et al.2014). Thus, the delivery of HopW1 by P. syringae pv. tomato DC3000 leads to a reduction in the number of endosomes during the early stages of infection (Kang et al.2014). The mechanisms by which HopW1 destabilizes actin remain to be elucidated.
HopG1 from P. syringae induces actin filament bundling
HopG1 from P. syringae pv. tomato DC3000 is targeted to the mitochondria of infected plant cells and was shown to interact with a mitochondrial-localized kinesin motor protein (Mukhtar et al. 2011; Shimono et al.2016). Kinesin motor proteins are known as microtubule-associated proteins but can also interact with actin filaments and might be involved in the crosstalk and crosslinking between microtubules and actin filaments (Schneider and Persson 2015). The analysis of a hopG1 deletion mutant revealed that HopG1 induces actin bundling and promotes disease symptom development (Shimono et al.2016). The results of coimmunprecipitation experiments suggest that HopG1 and kinesin associate with actin. No interaction between actin and HopG1 was observed, suggesting that the interaction between HopG1 and actin is indirect and requires the HopG1-interacting kinesin motor protein (Shimono et al.2016). Given the predicted role of kinesin motor proteins in cytoskeleton assembly, it was proposed that HopG1 targets kinesin to induce the formation of actin bundles and thus to promote disease (Shimono et al.2016) (Fig. 8C).
HopE1 from P. syringae interacts with calmodulin and the microtubule-associated protein MAP65
The effector HopE1 from P. syringae pv. tomato DC3000 contributes to virulence and suppresses PTI responses when transiently expressed in Arabidopsis (Guo et al.2016). Interaction studies revealed that HopE1 interacts with calmodulin and the microtubule-associated protein MAP65, which is required for the crosslinking of anti-parallel microtubule bundles (Guo et al.2016). The results of pull-down experiments and BiFC assays suggest that HopE1, calmodulin and MAP65 are present in the same complex and that calmodulin is required for the interaction between HopE1 and MAP65 (Guo et al.2016). HopE1 induces the dissociation of a MAP65-GFP fusion protein from microtubules and inhibits secretion of secGFP, which is a fusion protein between GFP and the secretion signal of the Arabidopsis PR-3 protein (Guo et al.2016). No effects were observed for HopE1 derivatives with a mutated calmodulin-binding site, which is located between amino acids 171 and 190 of HopE1 and required for the contribution of HopE1 to in planta growth of P. syringae pv. tomato DC3000 (Guo et al.2016). It was, therefore, proposed that HopE1 in association with calmodulin binds to MAP65 and dissociates MAP65 from microtubules to inhibit protein secretion (Guo et al.2016) (Fig. 8C). Arabidopsis map65 mutants are impaired in PTI responses and more susceptible to infections with P. syringae pv. tomato DC3000. Reduced growth of hopE1 mutants was observed in wild-type but not in map65 mutant plants, suggesting that MAP65 is an important virulence target of HopE1 (Guo et al.2016). Notably, however, the microtubule network was not significantly altered in the presence of HopE1 (Guo et al.2016).
Members of the YopJ effector family target tubulin or tubulin-binding proteins
The YopJ effector family members HopZ1a from P. syringae and AvrBsT from Xanthomonas spp. were shown to interfere with microtubule formation. As mentioned above, HopZ1a acts as an acetyltransferase and acetylates plant targets including JAZ proteins and tubulin in the presence of phytic acid (Lee et al.2012; Jiang et al. 2013). Fluorescence microscopy of Arabidopsis seedlings containing GFP-labeled microtubule markers revealed that delivery of HopZ1a by P. syringae pv. tomato DC3000 leads to the destruction of microtubules 16 hours post infection (Fig. 8D). In contrast, no effect of HopZ1a on the actin cytoskeleton was observed (Lee et al.2012). Twenty-two hours post infection, microtubules were destroyed even in the absence of HopZ1a, suggesting that microtubules are also targeted by additional effectors from P. syringae pv. tomato DC3000 (Lee et al.2012). In agreement with the HopZ1a-mediated destruction of microtubules and the role of microtubules in vesicle trafficking, the transient expression of HopZ1a in N. benthamiana interferes with the secretion of the secGFP reporter protein to the apoplast (Boutte, Vernhettes, Satiat-Jeunemaitre 2007; Lee et al.2012). An interference with secretion of secGFP was also reported for the YopJ homolog XopJ from X. campestris pv. vesicatoria (Bartetzko et al.2009).
AvrBsT from X. campestris pv. vesicatoria is an additional member of the YopJ effector family, which might indirectly interfere with microtubule formation. AvrBsT interacts with and acetylates the putative tubulin-binding protein ACIP1 (acetylated interacting protein 1), which is presumably involved in plant immunity (Cheong et al.2014). Infection experiments revealed that the in planta growth of virulent and avirulent strains of P. syringae pv. tomato DC3000 was increased in ACIP1-silenced Arabidopsis Pi-0 plants, suggesting that ACIP1 and ACIP family members contribute to PTI and ETI responses (Cheong et al.2014). The analysis of a GFP-ACIP1 fusion revealed that ACIP1 colocalizes with microtubules. In the presence of AvrBsT, GFP-ACIP1 forms aggregates, suggesting that AvrBsT alters the localization of ACIP1 (Cheong et al.2014). It remains to be investigated whether AvrBsT acetylates ACIP1 to interfere with plant defense responses and to alter microtubule formation. Notably, ACIP1 is presumably not the only plant target of AvrBsT because an arginine decarboxylase, the defense-related protein SGT1, Hsp70, the aldehyde dehydrogenase ALDH1 and the SNF1-related kinase 1 (see Table 1) were also reported as AvrBsT interaction partners. The relevance of most of these interactions for the virulence function of AvrBsT has not yet been adressed and should be in the focus of future studies.
EFFECTOR PROTEINS AND PLANT DEFENSE RESPONSES
Type III effectors do not always act as virulence factors but can also trigger defense responses in plants, which possess corresponding R genes and can thus recognize individual effector proteins. R genes often encode NLR proteins and are divided into TIR (toll/interleukin 1 receptor-like)-NB-LRR or CC (coiled-coil)-NB-LRR proteins according to their N-terminal domains (Takken and Goverse 2012; Wu et al.2014; Cui, Tsuda and Parker 2015). NLR protein activity is tightly regulated to avoid unnecessary and harmful activation of defense responses. On the posttranscriptional level, intramolecular interactions between the LRR and the NB domain of the R protein might prevent NB-mediated nucleotide exchange and thus NLR activation (Bonardi and Dangl 2012; Takken and Goverse 2012). Activation of R proteins upon direct or indirect recognition of effectors is triggered by molecular rearrangements, which allow nucleotide exchanges and/or the interaction of the NLR protein with other signaling molecules (Fig. 9A).
Figure 9.
Activation of R protein-mediated defense responses by type III effectors. (A) Detection of effector protein-triggered modifications in plant target proteins by R proteins. Type III effectors interact with and modify plant target proteins to promote bacterial virulence. According to the guard model, plant R proteins in resistant plants detect effector-triggered modifications (indicated by yellow asterisks) in plant target molecules. Many R proteins consist of CC/TIR, NB and LRR domains. The activity of the NB domain is often regulated by intramolecular interactions with the LRR domain. The detection of effector-triggered modifications in plant targets leads to intramolecular rearrangements in the R proteins and thus to the activation of the NB domain (indicated by a red asterisk). According to an alternative model, plants have evolved decoy molecules, which resemble virulence targets of effectors but do not contribute to bacterial virulence. Effector-mediated modifications in plant decoys can also lead to the activation of R protein-mediated resistance. (B) Model of the activation of RRS1-R/RPS4-triggered plant defense responses. The TIR-NB-LRR proteins RPS4 and RRS1-R form a heterodimer in the plant nucleus, and RRS1-R negatively regulates RPS4. The WRKY domain of RRS1-R binds to DNA and presumably triggers the RRS1-R-dependent expression of plant genes. The effector proteins PopP2 from R. solanacearum and AvrRps4 from P. syringae interact with the WRKY domain of RRS1-R, which is acetylated by PopP2. Interactions with effectors and/or acetylation by PopP2 lead to molecular rearrangements in the RPS4/RRS1-R immune receptor complex and thus to the activation of RPS4 (indicated by a red asterisk). It has been suggested that the WRKY domain of RRS1-R acts as an integrated decoy, which allows the elicitation of AvrRps4- and PopP2-triggered ETI responses. The activated complex of RRS1-R and RPS4 was proposed to be a tetramer.
Direct recognition of a bacterial type III effector was reported for the R protein RRS1-R from Arabidopsis which interacts with the type III effector PopP2 from R. solanacearum as is described below. Most other known NLR proteins recognize their cognate effectors presumably upon detection of effector-triggered changes in plant target molecules. According to the so-called guard model, a small repertoire of R proteins can detect a wide variety of effector proteins by guarding common effector targets (Van der Biezen and Jones 1998) (Fig. 9A). Alternatively, plant R proteins were proposed to detect effector-mediated changes in a non-functional effector target mimic, which acts as a decoy to trap the effector (Van der Hoorn and Kamoun 2008) (Fig. 9A). Decoys might have evolved from effector targets by gene duplication and do not contribute to the pathogen's fitness. In an evolutionary point of view, the presence of decoys guarantees the persistence of NLR-mediated effector recognition, even upon changes in the virulence targets of effector proteins. Known examples of guarded effector targets are detailed below and include (i) the Pto kinase, which interacts with AvrPto and AvrPtoB; (ii) RIN4, which is a negative regulator of plant immunity; (iii) the RLCK PBS1, which is cleaved by AvrPphB; and (iv) the pseudokinase ZED1, which mediates the recognition of HopZ1. Effector-R protein interactions and guarded plant targets or decoys of effector proteins are also summarized in Figs 9–12.
Figure 12.
Detection of effector-triggered modifications in plant target molecules by the R proteins RPS5 and ZAR1. (A) RPS5 activates ETI upon cleavage of PBS1. The CC-NB-LRR protein RPS5 interacts via the CC domain with the kinase PBS1. Cleavage of PBS1 by the effector protein and cysteine protease AvrPphB leads to the activation of RPS5. Exchange of the AvrPphB cleavage site (indicated by a yellow triangle) against the recognition site of the cysteine protease AvrRpt2 (indicated by an orange triangle) leads to cleavage of PBS1 in the presence of AvrRpt2 and thus to the activation of RPS5. (B) The R protein ZAR1 detects modifications in ZED1 and PBL2. The CC-NB-LRR protein ZAR1 interacts via the CC domain with the pseudokinase ZED1. Acetylation of ZED1 by the effector protein HopZ1a leads to the activation of ZAR1 (indicated by a red asterisk) and thus to ETI responses. ZAR1 also interacts via the LRR domain with the ZED1-related pseudokinase RKS1. The ZED1-RKS1 complex detects modifications in PBL2, which is a target of the uridylyltransferase AvrAC from X. campestris pv. campestris (see the text for details). Uridylylation of PBL2 by AvrAC leads to the activation of ZAR1-dependent ETI responses.
PopP2 from R. solanacearum is recognized by the R proteins RRS1-R and RPS4
One example for a putative plant decoy, which mimics an effector target, is the WRKY domain of the R protein RRS1-R, which is bound and acetylated by PopP2 from R. solancearum (Figs 7D and 9B). Acetylation of RRS1-R interferes with its DNA binding and thus with the RRS1-R-mediated activation of gene expression (Le Roux et al. 2015; Sarris et al.2015). Given that PopP2 acetylates multiple WRKY transcription factors, it was suggested that the WRKY domain of RRS1-R acts as an integrated decoy, which allows the detection of PopP2-mediated modifications of WRKY domains (Le Roux et al. 2015; Sarris et al.2015). In agreement with this model, the exchange of the lysine residue at position 1221 of RRS1-R by glutamine, which mimics the acetylation of K1221, interferes with DNA binding of RRS1-R and leads to the activation of plant defense responses (Le Roux et al.2015).
The recognition of PopP2 in Arabidopsis does not only depend on RRS1-R but also on the TIR-NB-LRR R protein RPS4 (Gassmann, Hinsch and Staskawicz 1999; Sohn et al. 2014; Williams et al.2014). RRS1-R and RPS4 form a functional immune receptor complex via their TIR domains and trigger defense responses upon recognition of PopP2 (Narusaka et al. 2009; Williams et al.2014; Sarris et al. 2015) (Fig. 9B). Transient expression studies in tobacco revealed that the TIR domain of RPS4 triggers an effector-independent cell death, which is suppressed upon coexpression of the TIR domain of RRS1-R (Williams et al.2014). It was, therefore, suggested that RRS1-R negatively regulates RPS4 via the TIR domain (Cesari et al. 2014; Williams et al.2014). The RRS1-R/RPS4 immune receptor pair also detects AvrRps4 from P. syringae. Similarly to PopP2, AvrRps4 interacts with the WRKY domain of RRS1-R and activates defense responses in the presence of RPS4 (Sarris et al.2015) (Fig. 9B). Mutations in the P loop (phosphate-binding loop) of RPS4 but not of RRS1-R abolish AvrRps4- and PopP2-triggered ETI, suggesting that the function of RRS1-R is independent of NB (Williams et al.2014). It was proposed that RRS1-R acts as a sensor R protein, which binds to the effector proteins PopP2 and AvrRps4 and triggers plant defense via the executor R protein RPS4 (Delga, Le Roux and Deslandes 2015).
In addition to RRS1-R, AvrRps4 is also recognized by RRS1-S, which is a derivative of RRS1-R and lacks the C-terminal 96 amino acids of RRS1-R (Deslandes et al. 1998, 2002; Sarris et al.2015). The RRS1-S-mediated recognition of AvrRps4 depends on the WRKY domain, which provides the binding site for AvrRps4 and is also acetylated by PopP2 (Sarris et al.2015). PopP2-mediated acetylation of the WRKY domain of RRS1-S does not trigger ETI but leads to reduced binding of RRS1-S to DNA and interferes with the RRS1-S-mediated recognition of AvrRps4 (Sarris et al.2015). Notably, the WRKY domains of RRS1-R and RRS1-S are not the only targets of PopP2 and AvrRps4 because coimmunprecipitation experiments revealed that both effectors associate with additional WRKY proteins (Le Roux et al. 2015; Sarris et al.2015). The contribution of these interactions to the virulence functions of PopP2 and AvrRps4 remains to be investigated.
The Pto kinase is guarded by the CC-NB-LRR protein Prf
The guard model was initially proposed for the recognition of the effector proteins AvrPto and AvrPtoB from P. syringae. AvrPto and AvrPtoB are two unrelated effectors, which differ significantly in their tertiary structures (Xing et al.2007; Dong et al. 2009). Both effectors interact with the serine/threonine kinase Pto from tomato, which elicits ETI responses upon recognition of AvrPto or AvrPtoB in concert with its interaction partner Prf (Oh and Martin 2011). Prf is a CC-NB-LRR protein, which is encoded in the PTO locus (Martin et al. 1993; Salmeron et al.1996; Pedley and Martin 2003). Prf contains a large N-terminal region, which is required for dimerization and interacts with Pto as well as with the kinase Fen (Mucyn et al.2006; Gutierrez et al. 2010) (Fig. 10A). Fen shares 80% amino acid identity with Pto but does not confer resistance to AvrPto or AvrPtoB (Martin et al.1994). As mentioned above, Fen is degraded by AvrPtoB and only elicits a cell death reaction in the presence of a C-terminally truncated AvrPtoB derivative, which lacks the C-terminal E3 ubiquitin ligase domain (Rosebrock et al.2007).
Figure 10.
Model of Prf/Pto-triggered activation of plant defense responses. (A) AvrPtoB triggers Prf/Pto-dependent ETI responses. The CC-NB-LRR protein Prf contains an SD (Solanaceae domain) domain with weak homology to other solanaceous R proteins and an N-terminal domain (N), which interacts with the Pto kinase and mediates the Pto-independent dimerization of Prf (Gutierrez et al.2010). The Prf/Pto complex contains at least two molecules of each protein. Pto is autophosphorylated at S198 and required to maintain Prf in an inactive conformation. The sensor Pto molecule interacts with and phosphorylates the E3 ubiquitin ligase AvrPtoB from P. syringae and evades AvrPtoB-mediated degradation. Effector binding to the P+1 loop of the Pto sensor presumably triggers a conformational change in this loop, which activates the Pto helper protein. The Pto helper molecule transphosphorylates the sensor at amino acid residue T199. Transphosphorylation of the sensor Pto leads to the activation of Prf (indicated by a red asterisk) and thus to Prf/Pto-triggered ETI responses. (B) A kinase-inactive Pto helper molecule (PtoD164N) does not transphosphorylate the kinase-active Pto sensor and therefore does not activate Prf/Pto-dependent ETI responses.
Pto is probably not the main virulence target of AvrPto and AvrPtoB because both effectors also interact with other kinase domains of plant proteins including kinase domains of PRRs (see above; Table 1). Given the assumed roles of AvrPto and AvrPtoB in the suppression of PTI, Pto might have evolved as a decoy to detect the activities of AvrPto and AvrPtoB inside the plant cell. Binding of AvrPto or AvrPtoB to Pto likely disturbs the Pto-mediated negative regulation of the R protein Prf and thus leads to ETI. Both effectors interact with the catalytic region of Pto including the P+1 loop and can act as kinase inhibitors (Xing et al.2007; Dong et al. 2009; Cheng et al. 2011; Ntoukakis et al. 2014). The P+1 loop of kinases is involved in substrate binding and is located adjacent to the activation loop (Huse and Kuriyan 2002). The P+1 loop and the activation loop of kinases are part of the activation segment, which controls kinase activity (Nolen, Taylor and Ghosh 2004). In many kinases, phosphorylation events within the activation segment lead to conformational changes and thus alterations in the kinase activity (Nolen, Taylor and Ghosh 2004). In agreement with this model, mutations within the P+1 loop of Pto interfere with the kinase activity of Pto, suggesting that the P+1 loop is involved in the regulation of the Pto kinase activity (Xing et al.2007). Furthermore, the P+1 loop also interferes with the Pto-mediated regulation of Prf-dependent ETI because mutations within this region lead to constitutive gain-of-function Pto mutant derivatives, which trigger a ligand-independent HR in Nicotiana benthamiana (Rathjen et al. 1999; Wu et al. 2004; Mucyn et al. 2006; Xing et al.2007; Dong et al. 2009) (see Table 2). Similar findings were observed for a kinase-inactive Pto derivative with a mutation in the P+1 loop (Wu et al.2004). This suggests that the conformation of the P+1 loop rather than the kinase activity of Pto is important for the regulation of Prf activity by Pto. Some of the results of the numerous coexpression studies with Prf, Pto and avrPto/avrPtoB in N. benthamiana are summarized in Table 2. According to a current model, effector binding to the P+1 loop of Pto presumably triggers a conformational change in this loop, which derepresses Prf activity and thus leads to the activation of ETI (Ntoukakis et al.2014).
Table 2.
Results of selected transient expression assays with Pto, Prf and avrPto(B) in N. benthamiana.
| Prf (tomato)a | Pto (tomato)a | Effectora | Plant reactionb | References | Commentsc |
|---|---|---|---|---|---|
| Prf and Pto trigger a ligand-independent HR | |||||
| Prf35S | Pto35S | – | HR | Mucyn et al. (2006) | Overexpression of Pto and Prf under control of the 35S promoter leads to a ligand-independent HR, which occurs in the absence of AvrPto or AvrPtoB. Overexpression of Prf under control of the DEX-inducible promoter induces a Pto-independent HR, suggesting that Prf alone can activate downstream signaling. The PrfDEX-induced HR is presumably independent of Pto because it is not abolished upon silencing of the Pto homolog Pth1 from N. benthamiana with a tobacco rattle virus (TRV) silencing vector (Mucyn et al.2006). |
| Prf35S | PtoPnat | – | HR | Mucyn et al. (2006) | |
| Pto35S | – | – | Mucyn et al. (2006) | ||
| Prf35S | – | – | – | Mucyn et al. (2006) | |
| PrfDEX | – | – | HR | Mucyn et al. (2006) | |
| PrfDEX | Pto35S | – | HR | Mucyn et al. (2006) | |
| PrfDEX | TRV:Pth1 | – | HR | Mucyn et al. (2006) | |
| The Prf/Pto-triggered ligand-independent HR depends on the kinase activity of Pto | |||||
| Prf35S | PtoD164N/35S | – | – | Mucyn et al. (2006) | The ligand-independent HR induced by Prf and Pto depends on the kinase activity of Pto. The PrfDEX-induced HR is suppressed by the kinase-inactive PtoD164N derivative, but not by PtoG50S, which has residual kinase activity. |
| Prf35S | PtoG50S/35S | – | – | Mucyn et al. (2006) | |
| PrfDEX | PtoD164N/35S | – | – | Mucyn et al. (2006) | |
| PrfDEX | PtoG50S/35S | – | HR | Mucyn et al. (2006) | |
| Pto and AvrPto trigger an HR in N. benthamiana | |||||
| – | – | AvrPto35S | – | Rathjen et al. (1999) | Expression of Pto and avrPto in N. benthamiana induces the HR. Silencing of the Prf homolog from N. benthamiana abolishes the HR, suggesting that the elicitation of ETI by Pto and AvrPto depends on Prf. |
| – | Pto (transgene) | AvrPto35S | HR | Scofield et al. (1996) | |
| – | Pto35S | AvrPto35S | HR | Rathjen et al. (1999) | |
| TRV:Prf | Pto35S | AvrPto35S | – | Wu et al. (2004) | |
| Recognition of AvrPtoB depends on both Pto and Prf | |||||
| – | Pto35S (transgene) | AvrPtoB35S | – | Kim, Lin and Martin (2002); Mucyn et al. (2006) | Coexpression of Pto and avrPtoB does not trigger the HR, suggesting that the intrinisic Prf gene from N. benthamiana is not sufficient for the recognition of AvrPtoB. Coexpression of Prf, Pto and AvrPtoB triggers the HR, which appears earlier than the ligand-independent HR induced by Prf and Pto (see above). |
| Prf35S | – | AvrPtoB35S | – | Mucyn et al. (2006) | |
| Prf35S | Pto35S (transgene) | AvrPtoB35S | HR | Mucyn et al. (2006) | |
| AvrPtoB deleted in the E3 ubiquitin ligase domain triggers Prf-dependent defense reactions | |||||
| – | Pto35S | AvrPtoB35S | – | Abramovitch et al. (2003) | AvrPtoB1-387 lacks the C-terminal E3 ubiquitin ligase domain and triggers a Prf-dependent defense reaction, designated Rsb (resistance suppressed by AvrPtoB C terminus) (Abramovitch et al.2003). The C-terminal E3 ubiquitin ligase domain of AvrPtoB presumably suppresses the Rsb phenotype and also the recognition of AvrPtoB by Pto (Abramovitch et al.2003). Note that AvrPtoB1-308 is recognized by Pto but is not sufficient to elicit the Rsb phenotype (Abramovitch et al.2003). |
| – | Pto35S | AvrPtoB1-387/35S | HR | Abramovitch et al. (2003) | |
| – | – | AvrPtoB1-387/35S | HR | Abramovitch et al. (2003); Rosebrock et al. (2007) | |
| TRV:Prf | – | AvrPtoB1-387/35S | – | Rosebrock et al. (2007) | |
| – | Pto35S | AvrPtoB1-308/35S | HR | Abramovitch et al. (2003) | |
| – | – | AvrPtoB1-308/35S | – | Abramovitch et al. (2003) | |
| The kinase activity of Pto is required for ETI | |||||
| – | PtoG50S/35S | AvrPto35S | HR | Mucyn et al. (2006) | PtoG50S, which has residual kinase activity (Mucyn et al.2006), induces the HR in the presence of AvrPto as well as of Prf and AvrPtoB. It was, therefore, proposed that the kinase activity of Pto is dispensable for the activation of ETI and for the suppression of the AvrPtoB-mediated degradation of Pto (Mathieu, Schwizer and Martin 2014). This is in contrast to the finding that the kinase-inactive PtoD164N derivative does not trigger the HR in the presence of Prf and AvrPto or AvrPtoB. In contrast, PtoD164N triggers the HR in the presence of the E3 ligase mutant derivative AvrPtoBF479A, suggesting that the Pto kinase activity is required to evade AvrPtoB-mediated degradation. |
| – | PtoD164N/35S | AvrPto35S | – | Rathjen et al. (1999); Wu et al. (2004) | |
| Prf35S | PtoD164N/35S | AvrPtoDEX (transgene) | – | Mucyn et al. (2009) | |
| Prf35S | Pto35S | AvrPtoB35S | HR | Mathieu, Schwizer and Martin (2014) | |
| Prf35S | PtoG50S/35S | AvrPtoB35S | HR | Mathieu, Schwizer and Martin (2014) | |
| Prfpnat (transgene) | PtoD164N/Pro(Pto)d | AvrPtoB35S | – | Ntoukakis et al. (2009) | |
| Prfpnat (transgene) | PtoD164N/Pro(Pto) | AvrPtoBF479A/35S | HR | Ntoukakis et al. (2009) | |
| Degradation of Pto by AvrPtoB might depend on the Pto-binding site in AvrPtoB | |||||
| Prf35S | Pto35S | AvrPtoBF173A/35S | – | Mathieu, Schwizer and Martin (2014) | The N-terminal Pto-binding site is mutated in AvrPtoBF173A/35S. Based on the results of yeast two-hybrid studies, it was suggested that Pto binds adjacent to the E3 ligase domain of AvrPtoBF173A and is therefore degraded. In contrast, Pto is not degraded by AvrPtoBF173A/E3-LOF/35S, which contains an inactive E3 ubiquitin ligase domain. |
| Prf35S | Pto35S | AvrPtoBF173A/E3-LOF/35S | HR | Mathieu, Schwizer and Martin (2014) | |
| The kinase activity of Pto is dispensable for ETI if Pto has been made active by mutation | |||||
| – | PtoL205D/35S | – | HR | Wu et al. (2004); de Vries et al. (2006) | The Pto derivative PtoL205D with a mutation in the P+1 loop induces the HR in the absence of AvrPto. Similar results were observed with the kinase-inactive PtoL205D/D164N, suggesting that the kinase activity of Pto is dispensable for the HR induction by PtoL205D. The PtoL205D-triggered HR depends on the Prf gene from N. benthamiana because no HR induction was observed in Prf-silenced plants (TRV:Prf). |
| – | PtoL205D/D164N/35S | – | HR | Wu et al. (2004) | |
| TRV:Prf | PtoL205D/35S | – | – | Wu et al. (2004) | |
| Double phosphorylation of Pto is required for signaling | |||||
| PrfPnat (transgene) | Pto35S | AvrPtoBF479A/35S | HR | Ntoukakis et al. (2013) | The Pto derivative PtoS198A/T199A with mutations in both phosphorylation sites is an active kinase but does not trigger the HR in the presence of the E3 ubiquitin ligase-deficient AvrPtoBF479A. This suggests that both phosphorylation sites of Pto rather than the kinase activity per se are required for the induction of ETI. The phosphomimic PtoS198D/T199D derivative induces cell death in the presence of AvrPto or AvrPtoB. AvrPto also induces the HR in the presence of the kinase-inactive PtoS198D/T199D/D164N, suggesting that the kinase activity of Pto is dispensable after phosphorylation of S198 and T199. AvrPtoB is not recognized by the kinase-inactive PtoS198D/T199D/D164N derivative, which is presumably degraded by AvrPtoB. The L205D mutation in the P+1 loop of Pto leads to the induction of an AvrPto/B-independent cell death (see above), which is abolished in the presence of S198A/T199A mutations in Pto. This suggests that the phosphorylation of Pto at S198 and T199 is required for the ETI induction by the gain-of-function derivative PtoL205D. |
| PrfPnat (transgene) | PtoS198A/T199A/35S | AvrPtoBF479A/35S | – | Ntoukakis et al. (2013) | |
| PrfPnat (transgene) | PtoS198A/35S | AvrPto/B35S | Cell death | Ntoukakis et al. (2013) | |
| PrfPnat (transgene) | PtoT199A/35S | AvrPto/B35S | Cell death | Ntoukakis et al. (2013) | |
| PrfPnat (transgene) | PtoS198A/T199A/35S | AvrPto/B35S | – | Ntoukakis et al. (2013) | |
| PrfPnat (transgene) | PtoS198D/T199D/35S | AvrPto/B35S | Cell death | Ntoukakis et al. (2013) | |
| PrfPnat (transgene) | PtoS198D/T199D/D164N/35S | AvrPto35S | Cell death | Ntoukakis et al. (2013) | |
| PrfPnat (transgene) | PtoS198D/T199D/D164N/35S | AvrPtoB35S | – | Ntoukakis et al. (2013) | |
| PrfPnat (transgene) | PtoL205D/S198A/T199A/35S | – | – | Ntoukakis et al. (2013) | |
| Pto is transphosphorylated after effector binding | |||||
| PrfPnat (transgene) | Pto35S + PtoD164N/35S | AvrPto35S | – | Ntoukakis et al. (2013) | AvrPto binds to autophosphorylated Pto, which cannot be transphosphorylated by PtoD164N and does not trigger ETI. |
| PrfPnat (transgene) | PtoD164N/35S + PtoS198D/T199D/35S | AvrPto35S | Cell death | Ntoukakis et al. (2013) | AvrPto triggers ETI in the presence of the kinase-inactive PtoD164N upon coexpression of the phosphomimic Pto derivative PtoS198D/T199D. |
| PrfPnat (transgene) | PtoS198D/T199D/35S | – | – | Ntoukakis et al. (2013) | The phosphomimic PtoS198D/T199D derivative does not induce the ligand-independent HR, suggesting that phosphorylation of Pto is not sufficient and that the induction of Pto-dependent defense responses depends on the disruption of the P+1 loop of Pto. |
| PrfPnat (transgene) | PtoL205D/35S | – | Cell death | Ntoukakis et al. (2013) | PtoL205D induces a cell death because of the disruption of the P+1 loop. The authors speculate that PtoL205D is transphosphorylated by a Pto homolog from N. benthamiana. The presence of PtoD164N prevents transphopshorylation of PtoL205D and, therefore, suppresses the induction of ETI. |
| PrfPnat (transgene) | PtoL205D/35S + PtoD164N/35S | – | – | Ntoukakis et al. (2013) | |
| PrfPnat (transgene) | PtoL205D/35S + PtoS198D/T199D/35S | – | Cell death | Ntoukakis et al. (2013) | PtoL205D is transphosphorylated by the kinase-active derivative PtoS198D/T199D or PtoS198A/T199A and induces ETI. |
| PrfPnat (transgene) | PtoL205D/35S + PtoS198A/T199A/35S | – | Cell death | Ntoukakis et al. (2013) | |
The promoters, which were used for transient expression or the expression of transgenes, are indicated in all cases for which this information was provided in the publications. Pnat, native promoter of Prf; 35S, 35S promoter; DEX, DEX-inducible promoter; transgene, integration of the gene into the genome of N. benthamiana.
HR, hypersensitive response; -, no visible plant reactions.
For the better understanding of some of the results of the selected transient expression studies, conclusions provided by the authors of the indicated publications are shortly summarized. See also the text for details.
The role of the Pto kinase activity in the elicitation of ETI has been intensively studied by several research groups. With regard to the recognition of AvrPtoB, the Pto kinase activity was proposed to be required for the phosphorylation of AvrPtoB and thus for the inhibition of its E3 ligase activity (Ntoukakis et al.2009). In agreement with this model, the transient expression of the kinase-inactive Pto derivative PtoD164N did not trigger the HR in the presence of AvrPtoB but of an E3 ubiquitin ligase mutant derivative thereof (Mucyn et al. 2009; Ntoukakis et al.2009) (Table 2). According to an alternative hypothesis, however, Pto evades the degradation by AvrPtoB by binding to two distinct binding sites in the N-terminal and central region of AvrPtoB (Mathieu, Schwizer and Martin 2014). It was suggested that binding of Pto to the central region of AvrPtoB adjacent to the E3 ubiquitin ligase domain leads to the degradation of Pto. In contrast, Pto bound to the N-terminal-binding site in AvrPtoB was reported to be stable (Mathieu, Schwizer and Martin 2014). This model was mainly based on the results of yeast two-hybrid studies, which revealed that Pto does not interact with an AvrPtoB derivative containing the N-terminal Pto-binding site in fusion with the E3 ubiquitin ligase domain (Mathieu, Schwizer and Martin 2014). The authors concluded from this observation that the binding of Pto adjacent to the E3 ubiquitin ligase domain of AvrPtoB leads to its AvrPtoB-dependent degradation. Biochemical evidence for this hypothesis is still missing. Mathieu, Schwizer and Martin (2014) also observed that a Pto derivative with a G50S mutation and significantly reduced kinase activity triggers ETI in the presence of AvrPtoB and Prf (Table 2). Based on this finding, it was proposed that the Pto kinase activity is dispensable for the recognition of AvrPtoB and the elicitation of ETI. This hypothesis is in contrast to the finding that the kinase-inactive Pto derivative PtoD164N fails to trigger the HR in the presence of AvrPtoB (see above; Table 2).
The activation of Prf-mediated defense signaling correlates with a phosphorylation of serine and threonine residues at positions 198 and 199 in Pto (Ntoukakis et al.2013) (Fig. 10A). Mutation of both S198 and T199 did not interfere with the kinase activity of Pto but with the elicitation of ETI in the presence of AvrPto or an E3 ligase mutant derivative of AvrPtoB (Ntoukakis et al.2013) (Table 2). However, a phosphomimetic derivative of Pto (PtoS198D/T199D) did not trigger ligand-independent defense, suggesting that the phosphorylation of Pto per se is not sufficient for the signaling (Ntoukakis et al.2013). Thus, the activation of Prf by Pto presumably depends on the additional disruption of the P+1 loop after binding of AvrPto or AvrPtoB (Ntoukakis et al.2013). This hypothesis is supported by the finding that the coexpression of AvrPto or AvrPtoB with PtoS198D/T199D and Prf triggers a cell death (Table 2) (Ntoukakis et al.2013). An additional mutation leading to the kinase-inactive PtoS198D/T199D/D164N abolished cell death induction by AvrPtoB but not by AvrPto (Table 2) (Ntoukakis et al.2013). This is in agreement with the model that the kinase activity of Pto is dispensable for signaling per se but required for Pto to evade degradation by AvrPtoB (see above).
The double phosphorylation of Pto is presumably the result of a transphosphorylation event which might be stimulated upon disruption of the P+1 loop of one Pto molecule (the sensor) upon binding of AvrPto or AvrPtoB (Ntoukakis et al.2013). It was proposed that the sensor Pto molecule is initially autophosphorylated at S198 and subsequently transphosphorylated by a second Pto molecule (the helper) upon binding of AvrPto or AvrPtoB (Ntoukakis et al.2014). This model is supported by the finding that the constitutively active PtoL205D (with a mutation in the P+1 loop) does not trigger cell death upon coexpression of the kinase-inactive PtoD164N because PtoL205D likely needs to be transphosphorylated by a kinase-active Pto derivative (Table 2; Fig. 10B) (Ntoukakis et al.2013). Taken together, binding of AvrPto or AvrPtoB is assumed to trigger a conformational change in the P+1 loop of an autophosphorylated sensor Pto molecule and leads to the transphosphorylation by a second helper Pto molecule. The double phosphorylation of Pto finally activates Prf, which triggers plant defense responses.
The effector target RIN4 is guarded by the R proteins RPS2 and RPM1
RIN4 was identified as a regulator of both PTI and ETI, and as interaction partner of at least two R proteins, RPS2 and RPM1 (Mackey et al.2002; Axtell and Staskawicz 2003). RIN4 contains N- and C-terminal NOI (NO3-induced) domains, a C-terminal cysteine-rich membrane anchoring motif and was shown to be acylated and anchored in the plasma membrane (Mackey et al.2002; Kim et al.2005b; Afzal, Kim and Mackey 2013) (Fig. 11A). Intrinsically disordered protein regions in RIN4 presumably allow the specific binding of RIN4 to various interaction partners (Sun et al.2014; Lee et al. 2015) including at least four different effector proteins from P. syringae (see below). RIN4 also interacts with and activates the plant plasma membrane H(+) ATPases AHA1 and AHA2 and thus presumably promotes stomatal opening (Liu et al.2009). Plant interaction partners, effector-triggered modifications of RIN4 and their effect on plant defense responses are summarized below and in Fig. 11.
Figure 11.
Effector-triggered modifications of RIN4 and their contributions to PTI and ETI responses. (A) Domain organization of RIN4 and list of known plant interaction partners of RIN4. RIN4 contains N- and C-terminal NOI domains with conserved PxFGxW and Y/FTxxF amino acid motifs. The PxFGxW motif is the cleavage site of the effector protein AvrRpt2 from P. syringae. The Y/FTxxF motif contains a conserved threonine residue, which is phosphorylated by the effector proteins AvrB and AvrRpm1 from P. syringae. Additional important amino acids are indicated (see the text for details). Known plant interaction partners of RIN4 and their predicted functions are listed. References: 1, Liu et al. (2009); 2, Li et al. (2014b); 3, Day, Dahlbeck and Staskawicz (2006); 4, Cui et al. (2010); 5, Liu et al. (2011); 6, Mackey et al. (2002); 7, Axtell and Staskawicz (2003); 8, Luo et al. (2009). (B) Contribution of RIN4 to RPS2- and RPM1-triggered ETI responses. The ADP-RT HopF2 from P. syringae ADP-ribosylates RIN4 and suppresses the degradation of RIN4 by the cysteine protease AvrRpt2 from P. syringae. RIN4 is also degraded by AvrPto from P. syringae in the presence of Pto and Prf. The cleavage products of RIN4 are detected by the R protein RPS2, which triggers ETI. The effector proteins AvrRpm1 and AvrB from P. syringae lead to the phosphorylation of RIN4 and thus to the activation of the R protein RPM1 (indicated by a red asterisk). Effector-triggered phosphorylation of RIN4 presumably depends on the kinase RIPK, which interacts with RIN4 and AvrB and phosphorylates RIN4 at several amino acid residues including T166. The phosphorylation of T166 of RIN4 interferes with the interaction of RIN4 with the cyclophilin ROC1, which catalyzes the cis/trans isomerization of the proline residue at position 149 of RIN4. The cis/trans isomerization of P149 presumably leads to a specific conformational change in RIN4 and thus inhibits the activation of RPM1 and RPS2. Phosphorylation of RIN4 in the presence of AvrB likely induces conformational changes, which interfere with ROC1-mediated isomerization of RIN4 and lead to the activation of RPM1 (see the text for details). (C) Model of the role of RIN4 during PTI. RIN4 is a suppressor of PTI responses and interacts with and activates the H+ ATPases AHA1 and AHA2, which pump protons from the cytosol of guard cells into the apoplast. The activity of AHA1/2 leads to the establishment of a proton electrochemical gradient, which is used by channel and carrier proteins to mediate the uptake of ions into guard cells, thus leading to stomatal opening. Upon detection of flg22 during PTI, RIN4 is phosphorylated at amino acid S141, which leads to the derepression (= activation) of PTI. The phosphorylation of RIN4 at amino acid T166 by AvrB and AvrRpm1 from P. syringae presumably counteracts the effect of the S141 phosphorylation and restores the repression of PTI (see the text for details).
Effector-triggered cleavage of RIN4 and activation of the R protein RPS2
RIN4 is cleaved by the effector protein AvrRpt2 from P. syringae, which is a cysteine protease and is self-processed inside the plant cell (Axtell and Staskawicz 2003; Axtell et al. 2003; Mackey et al.2003). The self-cleavage of AvrRpt2 requires the plant cyclophilin ROC1, which catalyzes the isomerization between cis and trans isoforms of X-prolyl peptide bonds (Coaker, Falick and Staskawicz 2005; Coaker et al. 2006; Aumüller et al. 2010; Kumari et al.2013). AvrRpt2 cleaves RIN4 within the NOI domains at cleavage sites (consensus sequence: PXFGXW), which are similar to the AvrRpt2 processing site (Chisholm et al. 2005; Kim et al.2005a) (Fig. 11A). At first glance, the cleavage of a PTI suppressor such as RIN4 by AvrRpt2 does not appear to be favorable for the pathogen. However, RIN4 cleavage products were shown to be even more active PTI suppressors than the uncleaved protein (Afzal, da Cunha and Mackey 2011). RIN4 is presumably not the only target of AvrRpt2 because predicted AvrRpt2 cleavage sites are also present in other plant proteins. Furthermore, RIN4 is not essential for the virulence function of AvrRpt2 as was shown by infection studies with Arabidopsis rin4 rps2 plants (Belkhadir et al. 2004; Lim and Kunkel 2004; Chisholm et al. 2005).
The cleavage of RIN4 by AvrRpt2 is detected by the R protein RPS2, which interacts with the C-terminal nine amino acids of RIN4 and is negatively regulated by RIN4 (Mackey et al.2003; Day et al. 2005) (Fig. 11B). The cleavage of RIN4 abolishes the interaction between RIN4 and RPS2 and results in the activation of the RPS2-dependent ETI (Mackey et al.2003; Day et al. 2005). The AvrRpt2-mediated ETI is compromised by the ADP-RT HopF2 (see above), which interacts with and ADP-ribosylates RIN4 and thus attenuates the AvrRpt2-induced cleavage of RIN4 (Wilton et al.2010). Interestingly, RPS2 can also be activated by the effector protein AvrRpm1 from P. syringae, which interacts with and phosphorylates RIN4 (Mackey et al.2002; Chung et al. 2011) (Fig. 11B). As described below, the AvrRpm1-mediated phosphorylation of RIN4 is detected by the R protein RPM1. Notably, however, AvrRpm1 also triggers RPS2-dependent defense, which is independent of RIN4, suggesting that RPS2 guards additional plant proteins (Kim et al.2009b; Cherkis et al. 2012).
In tomato, RIN4 is also cleaved in the presence of AvrPto, which interacts with RIN4, Pto and Prf (Luo et al.2009). Furthermore, RIN4 degradation is induced in the absence of AvrPto by the Pto derivative PtoL205D, which contains a mutation in the P+1 loop and triggers a ligand-independent HR (see above, Table 2). This suggests that AvrPto-triggered RIN4 degradation depends on Pto and on a plant-specific protease (Luo et al.2009). In the absence of Pto, AvrPto interacts with RIN4 without triggering RIN4 degradation (Luo et al.2009). Experimental evidence suggests that RIN4 is also degraded in the presence of additional effectors including HopQ1 and HopAM1 (Luo et al.2009).
Phosphorylation of RIN4 and activation of the R protein RPM1
The type III effectors AvrRpm1 and AvrB from P. syringae phosphorylate the threonine residue of the conserved Y/FTXXF motif within the NOI domains of RIN4 (Chung et al. 2011; Liu et al.2011). The AvrB/AvrRpm1-mediated phosphorylation of RIN4 leads to the activation of the R protein RPM1, which guards RIN4 (Mackey et al.2002) (Fig. 11B). Phosphomimic mutations in RIN4 cause an effector-independent activation of RPM1 (Chung et al. 2011; Liu et al.2011) as well as increased disease susceptibility and stomatal opening in the absence of RPM1 (Chung et al. 2014; Lee et al.2015). The latter finding suggests that AvrB and AvrRpm1 induce the phosphorylation of RIN4 to suppress PTI responses. The RPM1-mediated recognition of AvrB and AvrRpm1 is suppressed by AvrRpt2, which cleaves RIN4 (Mackey et al.2002) (see above).
Phosphorylation of RIN4 by AvrB and AvrRpm1 presumably depends on the plant kinase RIPK, which interacts with AvrB and RIN4, and phosphorylates RIN4 at the amino acid residues T21, S160 and T166 (Chung et al. 2011; Liu et al.2011). RIPK is also targeted by the cysteine protease AvrPphB from P. syringae, which cleaves RIPK (Russell, Ashfield and Innes 2015). Notably, AvrPphB suppresses the AvrB- but not the AvrRpm1-mediated activation of RPM1, suggesting that RIPK is dispensable for the RPM1-dependent recognition of AvrRpm1 (Liu et al. 2011; Russell, Ashfield and Innes 2015). AvrB does not only target RIPK but also interacts with MPK4, which is an additional RIN4 interaction partner (see above). As MPK4 can phosphorylate RIN4, AvrB might interact with different plant kinases to induce the phosphorylation of RIN4 (Cui et al.2010).
The biochemical mechanisms leading to the AvrRpm1- and AvrB-induced phosphorylation of RIN4 and their impact on ETI and PTI are not yet completely understood. Experimental evidence suggests that the phosphorylation of T166 of RIN4 by AvrRpm1 interferes with the interaction of RIN4 with the cyclophilin ROC1, which was identified as RIN4 interaction partner (Li et al.2014b). ROC1 catalyzes the isomerization between cis and trans isoforms of X-prolyl peptide bonds and promotes the cleavage of RIN4 (Coaker et al. 2006; Li et al.2014b). Thus, a gain-of-function derivative of ROC1 (ROC1S58F) leads to enhanced RIN4 cleavage (Ma et al.2013; Li et al.2014b). Notably, however, the increased RIN4 cleavage in the presence of ROC1S58F does not promote the RPS2-specific HR. On the contrary, ROC1S58F interferes with RPM1 and RPS2 activation and this inhibitory effect on ETI responses is dependent on the catalytic activity of ROC1 (Li et al.2014b).
Experimental evidence suggests that ROC1 interacts with the C-terminal region of RIN4 and catalyzes the cis/trans isomerization of P149. This presumably induces a conformational change in RIN4, which interferes with the AvrB-induced phosphorylation of RIN4 as well as with RPM1 and RPS2 activation (Li et al.2014b) (Fig. 11B). A targeted mutagenesis approach led to the identification of P149 as essential residue for RPM1 activation. Thus, the mutant derivative RIN4P149V did not trigger the RPM1-dependent HR (Li et al.2014b). Furthermore, the introduction of the P149V mutation into the phosphomimic RIN4 derivative RIN4T166D abolished the RPM1-dependent HR induced by RIN4T166D (Li et al.2014b). It was, therefore, suggested that the activation of RPM1 does not only depend on the phosphorylation of RIN4 at T166 but also on P149.
Deletion of P149 leads to a constitutive phosphorylation of RIN4 and thus to the activation of RPM1, suggesting that the phosphorylation of RIN4 is regulated by protein conformation. The HR triggered by RIN4ΔP149 was not suppressed by the constitutively active ROC1S58F and was not affected upon introduction of an additional T166A mutation into RIN4ΔP149 (Li et al.2014b). It was, therefore, concluded that RIN4ΔP149 is recognized by RPM1 even in the absence of T166 phosphorylation and that RPM1 senses a specific conformation of RIN4, which is adopted by RIN4ΔP149 (Li et al.2014b). Notably, phosphorylation of RIN4 at T166 leads to a reduced interaction of RIN4 with ROC1 and could thus interfere with the ROC1-mediated isomerization of P149. The authors of this study proposed that ROC1 induces a specific conformational change in RIN4, which interferes with the activation of RPM1 and RPS2. Phosphorylation of RIN4 in the presence of AvrB likely leads to conformational changes that suppress the ROC1-mediated isomerization of RIN4. Thus, the AvrB-mediated phosphorylation of T166 of RIN4 presumably leads to a conformation similar to that of RIN4ΔP149 and is sensed by RPM1 (Li et al.2014b) (Fig. 11B).
Notably, the AvrB- and AvrRpm1-dependent phosphorylation of RIN4 also affects its role in PTI. RIN4 is phosphorylated at the conserved serine residue at position 141 upon perception of flg22 (Chung et al.2014). Phosphorylation of S141 requires BIK1 and/or the BIK1 paralog PBL1 and leads to the derepression (i.e. activation) of PTI (Chung et al.2014). Phosphorylation of S141 is reduced in the phosphomimic RIN4 derivative RIN4T166D, suggesting that the T166 phosphorylation is epistatic to S141 phosphorylation (Fig. 11C). Thus, the AvrB/AvrRpm1-induced phosphorylation of T166 of RIN4 presumably counteracts the effect of the S141 phosphorylation and restores the repression of PTI (Chung et al.2014).
The RLCK PBS1 acts as a decoy for AvrPphB and is guarded by the R protein RPS5
The cysteine protease AvrPphB from P. syringae was shown to cleave several plant kinases including BIK1 and RIPK as well as the membrane-associated serine/threonine kinase PBS1 and PBL proteins (Puri et al. 1997; Shao et al. 2003; Zhang et al.2010; Russell, Ashfield and Innes 2015) (see above). The cleavage of PBS1 by AvrPphB leads to the activation of the CC-NB-LRR R protein RPS5 from Arabidopsis, which interacts with PBS1 via the N-terminal CC domain (Swiderski and Innes 2001; Shao et al. 2003; Ade et al. 2007). RPS5 also interacts with kinase-inactive PBS1 derivatives, suggesting that the kinase activity of PBS1 is not required for the interaction with RPS5 (DeYoung et al.2012). In planta expression and interaction studies revealed that both cleavage products of PBS1 interact with RPS5 and are required for HR induction (DeYoung et al.2012). It is assumed that RPS5 detects a conformational change in PBS1 and thus triggers ETI (Fig. 12A). This model is based on the observation that the insertion of five amino acids at the cleavage site of PBS1 interferes with the cleavage of PBS1 and leads to the activation of RPS5 in the absence of AvrPphB (DeYoung et al.2012). RPS5 presumably senses the movement of a loop-exposed five-amino acid motif in the kinase domain of PBS1 (Qi et al.2014). As this motif is polymorphic in different PBL proteins, the AvrPphB-induced cleavage of PBL proteins other than PBS1 does not trigger the RPS5-dependent ETI (Qi et al.2014).
In the absence of PBS1 cleavage, the activity of RPS5 is presumably suppressed by intramolecular interactions between the C-terminal LRR domain and the NB domain (Ade et al.2007). Cleavage of PBS1 by AvrPphB presumably leads to conformational changes in RPS5, which relieves the negative regulation of the NB by the LRR domain and thus leads to the activation of RPS5. Notably, PBS1 is not the main virulence target of AvrPphB because pbs1 mutants are only marginally affected in PTI defense (Zhang et al.2010). It was, therefore, suggested that PBS1 acts as a decoy, which is guarded by RPS5 and mimicks true virulence targets of AvrPphB such as BIK1 and RIPK (Zhang et al.2010) (see above). A recent study showed that PBS1 can be engineered to mediate recognition of bacterial effectors with protease activity other than AvrPphB. Thus, the exchange of the AvrPphB cleavage site against the cleavage site of AvrRpt2 led to a PBS1 derivative, which is cleaved in the presence of AvrRpt2 and subsequently triggers the RPS5-dependent HR (Kim et al.2016) (Fig. 12A).
The pseudokinase ZED1 acts as a decoy and mediates recognition of HopZ1a by the R protein ZAR1
Another plant decoy is the pseudokinase ZED1 from Arabidopsis, which interacts with and is acetylated by the YopJ family member and acetyltransferase HopZ1a from P. syringae. Acetylation of ZED1 is presumably recognized by the CC-NB-LRR protein ZAR1, which guards ZED1, and leads to ETI (Lewis et al. 2010, 2013) (Fig. 12B). Given that ZED1 is dispensable for the virulence function of HopZ1a, it has probably evolved as a decoy to trigger ZAR1-dependent defense in response to the acetylation by HopZ1a (Lewis et al.2013). As mentioned above, HopZ1a likely targets additional plant proteins including kinases, JAZ proteins and GmHID1, which is involved in isoflavone biosynthesis (Zhou et al.2011; Jiang et al. 2013) (Table 1).
In addition to ZED1, ZAR1 interacts with the ZED1-related pseudokinase RKS1. ZAR1 and RKS1 mediate the recognition of the effector protein and uridylyltransferase AvrAC from Xanthomonas (see above) (Feng et al. 2012; Wang et al.2015). AvrAC uridylylates several RLCKs including BIK1 and RIPK as well as the BIK1 paralog PBL2, which interacts with AvrAC (Feng et al. 2012; Guy et al. 2013; Wang et al.2015). Uridylylated PBL2 is recruited to the ZAR1-RKS1 complex and triggers ZAR1-dependent ETI (Fig. 12B). As uridylylation of PBL2 is required for AvrAC recognition but dispensable for the virulence activity of AvrAC, PBL2 presumably acts as a decoy for the detection of the enzymatic activity of AvrAC (Wang et al.2015).
CONCLUDING REMARKS
In the past years, significant progress has been made in the identification and molecular characterization of type III effector proteins from plant-pathogenic bacteria. The increasing number of available genome sequences paved the way for comparative studies of type III effectomes and led to the identification of core sets of effectors. The analysis of single- and multi-effector mutants revealed that individual effector proteins do not always significantly contribute to bacterial virulence, presumably because they share functional redundancies. Key for the understanding of the molecular functions of type III effectors in plant cells is the identification of plant targets and the characterization of effector-triggered modifications in plant cellular processes. To date, effectors from plant-pathogenic bacteria were shown to interfere with PTI and ETI responses, MAPK signaling, proteasome-dependent protein degradation, phytohormone signaling, photosynthesis, plant gene expression and the formation of the cytoskeleton. The results of various experimental approaches revealed that effectors from different pathogens employ common as well as pathogen-specific strategies to subvert plant immunity and to promote pathogen survival. It is interesting to note that several effectors appear to have multiple plant targets and thus presumably interfere with different cellular pathways. Examples are HopM1, which interferes with vesicle trafficking and gene expression, and HopZ1a, which targets JAZ proteins, tubulin and the isoflavone biosynthesis enzyme GmHID1. Given the multiple functions of some effectors and the large size of the effectome in different bacterial species, the analysis of effector-triggered alterations in plant cellular pathways and their contribution to bacterial virulence remains a major challenge for ongoing and future research projects.
Several effectors do not only target different plant proteins but also have antagonistic activities as was for instance reported for effectors that suppress or activate the proteasome or MAPK signaling pathways. It is yet largely unknown how effector activities inside the plant cell are regulated. Are all effectors simultaneously translocated after the assembly of the T3S system or is there a hierarchy in type III-dependent effector protein delivery as was reported for animal-pathogenic bacteria? It also remains to be investigated whether effector delivery is not only regulated by the bacterium but also by translocated effectors, which possibly interfere with the insertion of the translocon of the T3S system into the plant plasma membrane. Notably, translocated effectors from animal-pathogenic bacteria were shown to control effector protein translocation from inside the host cell (Dewoody et al. 2011; Gaus et al.2011; Berger et al. 2012). The efficient translocation of effector proteins into animal cells depends on host actin filaments, which are one of the targets of type III effectors (Viboud and Bliska 2001; Mejia, Bliska and Viboud 2008; Verove et al. 2012). Similar findings have not yet been described for effectors from plant-pathogenic bacteria. Intriguingly, however, it was reported that the recognition of AvrBs2 from X. campestris pv. vesicatoria in plants possessing the R gene Bs2 suppresses the translocation of additional type III effectors into the plant cell (Zhao et al.2011). It remains to be investigated whether the Bs2-triggered ETI affects the insertion of the translocon or the expression of T3S genes.
In conclusion, we are just beginning to understand the highly complex interplay between type III effectors and plant cellular processes. The characterization of effector activities in plant cells and the analysis of mechanisms underlying the control of effector translocation will, therefore, remain major projects for future research.
Supplementary Material
Acknowledgments
I am grateful to U. Bonas for critical reading of the manuscript and express my special thanks to the reviewers of this article for their numerous helpful comments.
FUNDING
Work in my group is supported by funds from the (grant numbers , ) and the Collaborative Research Centre 648 (Molecular mechanisms of information processing in plants, project A8).
Conflict of interest. None declared.
REFERENCES
- Abramovitch RB, Janjusevic R, Stebbins CE, et al. Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. P Natl Acad Sci USA. 2006;103:2851–6. doi: 10.1073/pnas.0507892103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramovitch RB, Kim YJ, Chen S, et al. Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J. 2003;22:60–9. doi: 10.1093/emboj/cdg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ade J, DeYoung BJ, Golstein C, et al. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. P Natl Acad Sci USA. 2007;104:2531–6. doi: 10.1073/pnas.0608779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afzal AJ, da Cunha L, Mackey D. Separable fragments and membrane tethering of Arabidopsis RIN4 regulate its suppression of PAMP-triggered immunity. Plant Cell. 2011;23:3798–811. doi: 10.1105/tpc.111.088708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afzal AJ, Kim JH, Mackey D. The role of NOI-domain containing proteins in plant immune signaling. BMC Genomics. 2013;14:327. doi: 10.1186/1471-2164-14-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alavi SM, Sanjari S, Durand F, et al. Assessment of the genetic diversity of Xanthomonas axonopodis pv. phaseoli and Xanthomonas fuscans subsp. fuscans as a basis to identify putative pathogenicity genes and a type III secretion system of the SPI-1 family by multiple suppression subtractive hybridizations. Appl Environ Microb. 2008;74:3295–301. doi: 10.1128/AEM.02507-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano JR, Collmer A. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J Bacteriol. 1997;179:5655–62. doi: 10.1128/jb.179.18.5655-5662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JC, Wan Y, Kim YM, et al. Decreased abundance of type III secretion system-inducing signals in Arabidopsis mkp1 enhances resistance against Pseudomonas syringae. P Natl Acad Sci USA. 2014;111:6846–51. doi: 10.1073/pnas.1403248111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angot A, Peeters N, Lechner E, et al. Ralstonia solanacearum requires F-box-like domain-containing type III effectors to promote disease on several host plants. P Natl Acad Sci USA. 2006;103:14620–5. doi: 10.1073/pnas.0509393103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold R, Brandmaier S, Kleine F, et al. Sequence-based prediction of type III secreted proteins. PLoS Pathog. 2009;5:e1000376. doi: 10.1371/journal.ppat.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumüller T, Jahreis G, Fischer G, et al. Role of prolyl cis/trans isomers in cyclophilin-assisted Pseudomonas syringae AvrRpt2 protease activation. Biochemistry. 2010;49:1042–52. doi: 10.1021/bi901813e. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Chisholm ST, Dahlbeck D, et al. Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol Microbiol. 2003;49:1537–46. doi: 10.1046/j.1365-2958.2003.03666.x. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–77. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- Bakshi M, Oelmuller R. WRKY transcription factors: Jack of many trades in plants. Plant Signal Behav. 2014;9:e27700. doi: 10.4161/psb.27700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltrus DA, Nishimura MT, Romanchuk A, et al. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog. 2011;7:e1002132. doi: 10.1371/journal.ppat.1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfield MJ. Perturbation of host ubiquitin systems by plant pathogen/pest effector proteins. Cell Microbiol. 2015;17:18–25. doi: 10.1111/cmi.12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartetzko V, Sonnewald S, Vogel F, et al. The Xanthomonas campestris pv. vesicatoria type III effector protein XopJ inhibits protein secretion: evidence for interference with cell wall-associated defense responses. Mol Plant Microbe In. 2009;22:655–64. doi: 10.1094/MPMI-22-6-0655. [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Nimchuk Z, Hubert DA, et al. Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell. 2004;16:2822–35. doi: 10.1105/tpc.104.024117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Khaled S, Postma J, Robatzek S. A moving view: subcellular trafficking processes in pattern recognition receptor-triggered plant immunity. Annu Rev Phytopathol. 2015;53:379–402. doi: 10.1146/annurev-phyto-080614-120347. [DOI] [PubMed] [Google Scholar]
- Berger CN, Crepin VF, Baruch K, et al. EspZ of enteropathogenic and enterohemorrhagic Escherichia coli regulates type III secretion system protein translocation. MBio. 2012;3:e00317–12. doi: 10.1128/mBio.00317-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M, Timmers T, Jauneau A, et al. RD19, an Arabidopsis cysteine protease required for RRS1-R-mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell. 2008;20:2252–64. doi: 10.1105/tpc.108.058685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S, Halane MK, Kim SH, et al. Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science. 2011;334:1405–8. doi: 10.1126/science.1211592. [DOI] [PubMed] [Google Scholar]
- Bigeard J, Colcombet J, Hirt H. Signaling mechanisms in pattern-triggered immunity (PTI) Mol Plant. 2015;8:521–39. doi: 10.1016/j.molp.2014.12.022. [DOI] [PubMed] [Google Scholar]
- Block A, Toruno TY, Elowsky CG, et al. The Pseudomonas syringae type III effector HopD1 suppresses effector-triggered immunity, localizes to the endoplasmic reticulum, and targets the Arabidopsis transcription factor NTL9. New Phytol. 2014;201:1358–70. doi: 10.1111/nph.12626. [DOI] [PubMed] [Google Scholar]
- Blocker AJ, Deane JE, Veenendaal AK, et al. What's the point of the type III secretion system needle? P Natl Acad Sci USA. 2008;105:6507–13. doi: 10.1073/pnas.0708344105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol. 2010;48:419–36. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- Boch J, Bonas U, Lahaye T. TAL effectors - pathogen strategies and plant resistance engineering. New Phytol. 2014;204:823–32. doi: 10.1111/nph.13015. [DOI] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, et al. Breaking the code of DNA-binding specificity of TAL-type III effectors. Science. 2009;326:1509–12. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Böhm H, Albert I, Fan L, et al. Immune receptor complexes at the plant cell surface. Curr Opin Plant Biol. 2014;20:47–54. doi: 10.1016/j.pbi.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Bonardi V, Dangl JL. How complex are intracellular immune receptor signaling complexes? Front Plant Sci. 2012;3:237. doi: 10.3389/fpls.2012.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnington KE, Kuehn MJ. Protein selection and export via outer membrane vesicles. Biochim Biophys Acta. 2013;1843:1612–9. doi: 10.1016/j.bbamcr.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutte Y, Vernhettes S, Satiat-Jeunemaitre B. Involvement of the cytoskeleton in the secretory pathway and plasma membrane organisation of higher plant cells. Cell Biol Int. 2007;31:649–54. doi: 10.1016/j.cellbi.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Buchko GW, Niemann G, Baker ES, et al. A multi-pronged search for a common structural motif in the secretion signal of Salmonella enterica serovar Typhimurium type III effector proteins. Mol Biosyst. 2010;6:2448–58. doi: 10.1039/c0mb00097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner D. Protein export according to schedule – architecture, assembly and regulation of type III secretion systems from plant and animal pathogenic bacteria. Microbiol Mol Biol R. 2012;76:262–310. doi: 10.1128/MMBR.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canonne J, Marino D, Jauneau A, et al. The Xanthomonas type III effector XopD targets the Arabidopsis transcription factor MYB30 to suppress plant defense. Plant Cell. 2011;23:3498–511. doi: 10.1105/tpc.111.088815. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Canonne J, Marino D, Noël LD, et al. Detection and functional characterization of a 215 amino acid N-terminal extension in the Xanthomonas type III effector XopD. PLoS One. 2010;5:e15773. doi: 10.1371/journal.pone.0015773. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Canonne J, Rivas S. Bacterial effectors target the plant cell nucleus to subvert host transcription. Plant Signal Behav. 2012;7:217–21. doi: 10.4161/psb.18885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Aceti DJ, Sabat G, et al. Mutations in FLS2 Ser-938 dissect signaling activation in FLS2-mediated Arabidopsis immunity. PLoS Pathog. 2013;9:e1003313. doi: 10.1371/journal.ppat.1003313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari S, Bernoux M, Moncuquet P, et al. A novel conserved mechanism for plant NLR protein pairs: the “integrated decoy” hypothesis. Front Plant Sci. 2014;5:606. doi: 10.3389/fpls.2014.00606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Hellmann H. Plant E3 ligases: flexible enzymes in a sessile world. Mol Plant. 2013;6:1388–404. doi: 10.1093/mp/sst005. [DOI] [PubMed] [Google Scholar]
- Chen LQ. SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol. 2014;201:1150–5. doi: 10.1111/nph.12445. [DOI] [PubMed] [Google Scholar]
- Chen Z, Agnew JL, Cohen JD, et al. Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. P Natl Acad Sci USA. 2007;104:20131–6. doi: 10.1073/pnas.0704901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Munkvold KR, Gao H, et al. Structural analysis of Pseudomonas syringae AvrPtoB bound to host BAK1 reveals two similar kinase-interacting domains in a type III effector. Cell Host Microbe. 2011;10:616–26. doi: 10.1016/j.chom.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H, Kim CY, Jeon JS, et al. Xanthomonas oryzae pv. oryzae type III effector XopN targets OsVOZ2 and a putative thiamine synthase as a virulence factor in rice. PLoS One. 2013;8:e73346. doi: 10.1371/journal.pone.0073346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong MS, Kirik A, Kim JG, et al. AvrBsT acetylates Arabidopsis ACIP1, a protein that associates with microtubules and is required for immunity. PLoS Pathog. 2014;10:e1003952. doi: 10.1371/journal.ppat.1003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkis KA, Temple BR, Chung EH, et al. AvrRpm1 missense mutations weakly activate RPS2-mediated immune response in Arabidopsis thaliana. PLoS One. 2012;7:e42633. doi: 10.1371/journal.pone.0042633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernandez G, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–71. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Dahlbeck D, Krishnamurthy N, et al. Molecular characterization of proteolytic cleavage sites of the Pseudomonas syringae effector AvrRpt2. P Natl Acad Sci USA. 2005;102:2087–92. doi: 10.1073/pnas.0409468102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chosed R, Tomchick DR, Brautigam CA, et al. Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ubiquitin-like protein proteases. J Biol Chem. 2007;282:6773–82. doi: 10.1074/jbc.M608730200. [DOI] [PubMed] [Google Scholar]
- Chung EH, da Cunha L, Wu AJ, et al. Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe. 2011;9:125–36. doi: 10.1016/j.chom.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung EH, El-Kasmi F, He Y, et al. A plant phosphoswitch platform repeatedly targeted by type III effector proteins regulates the output of both tiers of plant immune receptors. Cell Host Microbe. 2014;16:484–94. doi: 10.1016/j.chom.2014.09.004. [DOI] [PubMed] [Google Scholar]
- Coaker G, Falick A, Staskawicz B. Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science. 2005;308:548–50. doi: 10.1126/science.1108633. [DOI] [PubMed] [Google Scholar]
- Coaker G, Zhu G, Ding Z, et al. Eukaryotic cyclophilin as a molecular switch for effector activation. Mol Microbiol. 2006;61:1485–96. doi: 10.1111/j.1365-2958.2006.05335.x. [DOI] [PubMed] [Google Scholar]
- Costa TR, Felisberto-Rodrigues C, Meir A, et al. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol. 2015;13:343–59. doi: 10.1038/nrmicro3456. [DOI] [PubMed] [Google Scholar]
- Crabill E, Joe A, Block A, et al. Plant immunity directly or indirectly restricts the injection of type III effectors by the Pseudomonas syringae type III secretion system. Plant Physiol. 2010;154:233–44. doi: 10.1104/pp.110.159723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui F, Wu S, Sun W, et al. The Pseudomonas syringae type III effector AvrRpt2 promotes pathogen virulence via stimulating Arabidopsis auxin/indole acetic acid protein turnover. Plant Physiol. 2013;162:1018–29. doi: 10.1104/pp.113.219659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE. Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol. 2015;66:487–511. doi: 10.1146/annurev-arplant-050213-040012. [DOI] [PubMed] [Google Scholar]
- Cui H, Wang Y, Xue L, et al. A Pseudomonas syringae effector protein perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe. 2010;7:164–75. doi: 10.1016/j.chom.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnac S, Chakravarthy S, Kvitko BH, et al. Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. P Natl Acad Sci USA. 2011;108:2975–80. doi: 10.1073/pnas.1013031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnac S, Occhialini A, Barberis P, et al. Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol Microbiol. 2004;53:115–28. doi: 10.1111/j.1365-2958.2004.04118.x. [DOI] [PubMed] [Google Scholar]
- Day B, Dahlbeck D, Huang J, et al. Molecular basis for the RIN4 negative regulation of RPS2 disease resistance. Plant Cell. 2005;17:1292–305. doi: 10.1105/tpc.104.030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day B, Dahlbeck D, Staskawicz BJ. NDR1 interaction with RIN4 mediates the differential activation of multiple disease resistance pathways in Arabidopsis. Plant Cell. 2006;18:2782–91. doi: 10.1105/tpc.106.044693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day B, Henty JL, Porter KJ, et al. The pathogen-actin connection: a platform for defense signaling in plants. Annu Rev Phytopathol. 2011;49:483–506. doi: 10.1146/annurev-phyto-072910-095426. [DOI] [PubMed] [Google Scholar]
- de Lange O, Schreiber T, Schandry N, et al. Breaking the DNA-binding code of Ralstonia solanacearum TAL effectors provides new possibilities to generate plant resistance genes against bacterial wilt disease. New Phytol. 2013;199:773–86. doi: 10.1111/nph.12324. [DOI] [PubMed] [Google Scholar]
- de Lange O, Wolf C, Dietze J, et al. Programmable DNA-binding proteins from Burkholderia provide a fresh perspective on the TALE-like repeat domain. Nucleic Acids Res. 2014;42:7436–49. doi: 10.1093/nar/gku329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza TA, Soprano AS, de Lira NP, et al. The TAL effector PthA4 interacts with nuclear factors involved in RNA-dependent processes including a HMG protein that selectively binds poly(U) RNA. PLoS One. 2012;7:e32305. doi: 10.1371/journal.pone.0032305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries JS, Andriotis VM, Wu AJ, et al. Tomato Pto encodes a functional N-myristoylation motif that is required for signal transduction in Nicotiana benthamiana. Plant J. 2006;45:31–45. doi: 10.1111/j.1365-313X.2005.02590.x. [DOI] [PubMed] [Google Scholar]
- Dean P. Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiol Rev. 2011;35:1100–25. doi: 10.1111/j.1574-6976.2011.00271.x. [DOI] [PubMed] [Google Scholar]
- Delga A, Le Roux C, Deslandes L. Plant immune receptor decoy: pathogens in their own trap. Oncotarget. 2015;6:15748–9. doi: 10.18632/oncotarget.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng D, Yan C, Pan X, et al. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science. 2012;335:720–3. doi: 10.1126/science.1215670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Barbieri JT. Molecular mechanisms of the cytotoxicity of ADP-ribosylating toxins. Annu Rev Microbiol. 2008;62:271–88. doi: 10.1146/annurev.micro.62.081307.162848. [DOI] [PubMed] [Google Scholar]
- Deslandes L, Genin S. Opening the Ralstonia solanacearum type III effector tool box: insights into host cell subversion mechanisms. Curr Opin Plant Biol. 2014;20:110–7. doi: 10.1016/j.pbi.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Peeters N, et al. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. P Natl Acad Sci USA. 2003;100:8024–9. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Theulieres F, et al. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. P Natl Acad Sci USA. 2002;99:2404–9. doi: 10.1073/pnas.032485099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Pileur F, Liaubet L, et al. Genetic characterization of RRS1, a recessive locus in Arabidopsis thaliana that confers resistance to the bacterial soilborne pathogen Ralstonia solanacearum. Mol Plant Microbe In. 1998;11:659–67. doi: 10.1094/MPMI.1998.11.7.659. [DOI] [PubMed] [Google Scholar]
- Deslandes L, Rivas S. Catch me if you can: bacterial effectors and plant targets. Trends Plant Sci. 2012;17:644–55. doi: 10.1016/j.tplants.2012.06.011. [DOI] [PubMed] [Google Scholar]
- Dewoody R, Merritt PM, Houppert AS, et al. YopK regulates the Yersinia pestis type III secretion system from within host cells. Mol Microbiol. 2011;79:1445–61. doi: 10.1111/j.1365-2958.2011.07534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung BJ, Qi D, Kim SH, et al. Activation of a plant nucleotide binding-leucine rich repeat disease resistance protein by a modified self protein. Cell Microbiol. 2012;14:1071–84. doi: 10.1111/j.1462-5822.2012.01779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11:539–48. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- Domingues MN, Campos BM, de Oliveira ML, et al. TAL effectors target the C-terminal domain of RNA polymerase II (CTD) by inhibiting the prolyl-isomerase activity of a CTD-associated cyclophilin. PLoS One. 2012;7:e41553. doi: 10.1371/journal.pone.0041553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues MN, De Souza TA, Cernadas RA, et al. The Xanthomonas citri effector protein PthA interacts with citrus proteins involved in nuclear transport, protein folding and ubiquitination associated with DNA repair. Mol Plant Pathol. 2010;11:663–75. doi: 10.1111/j.1364-3703.2010.00636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Xiao F, Fan F, et al. Crystal structure of the complex between Pseudomonas effector AvrPtoB and the tomato Pto kinase reveals both a shared and a unique interface compared with AvrPto-Pto. Plant Cell. 2009;21:1846–59. doi: 10.1105/tpc.109.066878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen RH, Engel JL, Shao F, et al. A family of bacterial cysteine protease type III effectors utilizes acylation-dependent and -independent strategies to localize to plasma membranes. J Biol Chem. 2009;284:15867–79. doi: 10.1074/jbc.M900519200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudler R. The role of bacterial phytotoxins in inhibiting the eukaryotic proteasome. Trends Microbiol. 2014;22:28–35. doi: 10.1016/j.tim.2013.10.010. [DOI] [PubMed] [Google Scholar]
- Duplan V, Rivas S. E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front Plant Sci. 2014;5:42. doi: 10.3389/fpls.2014.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edqvist PJ, Olsson J, Lavander M, et al. YscP and YscU regulate substrate specificity of the Yersinia type III secretion system. J Bacteriol. 2003;185:2259–66. doi: 10.1128/JB.185.7.2259-2266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol R. 2010;74:81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore JM, Coaker G. The role of the plasma membrane H+-ATPase in plant-microbe interactions. Mol Plant. 2011;4:416–27. doi: 10.1093/mp/ssq083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enninga J, Mounier J, Sansonetti P, et al. Secretion of type III effectors into host cells in real time. Nat Methods. 2005;2:959–65. doi: 10.1038/nmeth804. [DOI] [PubMed] [Google Scholar]
- Feldman MF, Cornelis GR. The multitalented type III chaperones: all you can do with 15 kDa. FEMS Microbiol Lett. 2003;219:151–8. doi: 10.1016/S0378-1097(03)00042-9. [DOI] [PubMed] [Google Scholar]
- Feng F, Yang F, Rong W, et al. A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature. 2012;485:114–8. doi: 10.1038/nature10962. [DOI] [PubMed] [Google Scholar]
- Feng F, Zhou JM. Plant-bacterial pathogen interactions mediated by type III effectors. Curr Opin Plant Biol. 2012;15:469–76. doi: 10.1016/j.pbi.2012.03.004. [DOI] [PubMed] [Google Scholar]
- Frithz-Lindsten E, Rosqvist R, Johansson L, et al. The chaperone-like protein YerA of Yersinia pseudotuberculosis stabilizes YopE in the cytoplasm but is dispensible for targeting to the secretion loci. Mol Microbiol. 1995;16:635–47. doi: 10.1111/j.1365-2958.1995.tb02426.x. [DOI] [PubMed] [Google Scholar]
- Fu J, Wang S. Insights into auxin signaling in plant-pathogen interactions. Front Plant Sci. 2011;2:74. doi: 10.3389/fpls.2011.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Guo M, Jeong BR, et al. A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature. 2007;447:284–8. doi: 10.1038/nature05737. [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Kawazoe T, Ohnishi K, et al. RipAY, a plant pathogen effector protein exhibits robust gamma-glutamyl cyclotransferase activity when stimulated by eukaryotic thioredoxins. J Biol Chem. 2016;291:6813–30. doi: 10.1074/jbc.M115.678953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala SS, Kim TW, He JX, et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell. 2007;13:177–89. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangadharan A, Sreerekha MV, Whitehill J, et al. The Pseudomonas syringae pv. tomato type III effector HopM1 suppresses Arabidopsis defenses independent of suppressing salicylic acid signaling and of targeting AtMIN7. PLoS One. 2013;8:e82032. doi: 10.1371/journal.pone.0082032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann W, Hinsch ME, Staskawicz BJ. The ArabidopsisRPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 1999;20:265–77. doi: 10.1046/j.1365-313x.1999.t01-1-00600.x. [DOI] [PubMed] [Google Scholar]
- Gaudriault S, Paulin JP, Barny MA. The DspB/F protein of Erwinia amylovora is a type III secretion chaperone ensuring efficient secretion of the DspA/E essential pathogenicity factor. Mol Plant Pathol. 2002;3:313–20. doi: 10.1046/j.1364-3703.2002.00124.x. [DOI] [PubMed] [Google Scholar]
- Gaus K, Hentschke M, Czymmeck N, et al. Destabilization of YopE by the ubiquitin-proteasome pathway fine-tunes Yop delivery into host cells and facilitates systemic spread of Yersinia enterocolitica in host lymphoid tissue. Infect Immun. 2011;79:1166–75. doi: 10.1128/IAI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazi AD, Sarris PF, Fadouloglou VE, et al. Phylogenetic analysis of a gene cluster encoding an additional, rhizobial-like type III secretion system that is narrowly distributed among Pseudomonas syringae strains. BMC Microbiol. 2012;12:188. doi: 10.1186/1471-2180-12-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X, Shen M, Kim JH, et al. The Pseudomonas syringae type III effectors AvrRpm1 and AvrRpt2 promote virulence dependent on the F-box protein COI1. Plant Cell Rep. 2016;35:921–31. doi: 10.1007/s00299-016-1932-z. [DOI] [PubMed] [Google Scholar]
- Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Gene Dev 2004182046–59. [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Boter M, Fernandez-Barbero G, et al. The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS Biol. 2014;12:e1001792. doi: 10.1371/journal.pbio.1001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Hann DR, Ntoukakis V, et al. AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol. 2009;19:423–9. doi: 10.1016/j.cub.2009.01.054. [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Solano R. Nuclear jasmonate and salicylate signaling and crosstalk in defense against pathogens. Front Plant Sci. 2013;4:72. doi: 10.3389/fpls.2013.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giska F, Lichocka M, Piechocki M, et al. Phosphorylation of HopQ1, a type III effector from Pseudomonas syringae, creates a binding site for host 14-3-3 proteins. Plant Physiol. 2013;161:2049–61. doi: 10.1104/pp.112.209023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–27. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- Gochez AM, Shantharaj D, Potnis N, et al. Molecular characterization of XopAG effector-AvrGf2 from Xanthomonas fuscans subsp. aurantifolii in grapefruit. Mol Plant Pathol. 2016 doi: 10.1111/mpp.12408. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göhre V, Spallek T, Haweker H, et al. Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr Biol. 2008;18:1824–32. doi: 10.1016/j.cub.2008.10.063. [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Bauer Z, Boller T. Both the extracellular leucine-rich repeat domain and the kinase activity of FSL2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell. 2001;13:1155–63. [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T. FLS2: An LRR Receptor–like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–11. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- Groll M, Schellenberg B, Bachmann AS, et al. A plant pathogen virulence factor inhibits the eukaryotic proteasome by a novel mechanism. Nature. 2008;452:755–8. doi: 10.1038/nature06782. [DOI] [PubMed] [Google Scholar]
- Guignot J, Tran Van Nhieu G. Bacterial control of pores induced by the type III secretion system: mind the gap. Front Immunol. 2016;7:84. doi: 10.3389/fimmu.2016.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Kim P, Li G, et al. A bacterial effector co-opts calmodulin to target the plant microtubule network. Cell Host Microbe. 2016;19:67–78. doi: 10.1016/j.chom.2015.12.007. [DOI] [PubMed] [Google Scholar]
- Guo M, Tian F, Wamboldt Y, et al. The majority of the type III effector inventory of Pseudomonas syringae pv. tomato DC3000 can suppress plant immunity. Mol Plant Microbe In. 2009;22:1069–80. doi: 10.1094/MPMI-22-9-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez JR, Balmuth AL, Ntoukakis V, et al. Prf immune complexes of tomato are oligomeric and contain multiple Pto-like kinases that diversify effector recognition. Plant J. 2010;61:507–18. doi: 10.1111/j.1365-313X.2009.04078.x. [DOI] [PubMed] [Google Scholar]
- Guy E, Lautier M, Chabannes M, et al. xopAC-triggered immunity against Xanthomonas depends on Arabidopsis receptor-like cytoplasmic kinase genes PBL2 and RIPK. PLoS One. 2013;8:e73469. doi: 10.1371/journal.pone.0073469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann DR, Dominguez-Ferreras A, Motyka V, et al. The Pseudomonas type III effector HopQ1 activates cytokinin signaling and interferes with plant innate immunity. New Phytol. 2014;201:585–98. doi: 10.1111/nph.12544. [DOI] [PubMed] [Google Scholar]
- Hauvermale AL, Ariizumi T, Steber CM. Gibberellin signaling: a theme and variations on DELLA repression. Plant Physiol. 2012;160:83–92. doi: 10.1104/pp.112.200956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes CS, Aoki SK, Low DA. Bacterial contact-dependent delivery systems. Annu Rev Genet. 2010;44:71–90. doi: 10.1146/annurev.genet.42.110807.091449. [DOI] [PubMed] [Google Scholar]
- He P, Chintamanani S, Chen Z, et al. Activation of a COI1-dependent pathway in Arabidopsis by Pseudomonas syringae type III effectors and coronatine. Plant J. 2004;37:589–602. doi: 10.1111/j.1365-313x.2003.01986.x. [DOI] [PubMed] [Google Scholar]
- He SY, Nomura K, Whittam TS. Type III protein secretion mechanism in mammalian and plant pathogens. Biochim Biophys Acta. 2004;1694:181–206. doi: 10.1016/j.bbamcr.2004.03.011. [DOI] [PubMed] [Google Scholar]
- He P, Shan L, Lin NC, et al. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell. 2006;125:563–75. doi: 10.1016/j.cell.2006.02.047. [DOI] [PubMed] [Google Scholar]
- Heidrich K, Wirthmueller L, Tasset C, et al. Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science. 2011;334:1401–4. doi: 10.1126/science.1211641. [DOI] [PubMed] [Google Scholar]
- Henty-Ridilla JL, Li J, Blanchoin L, et al. Actin dynamics in the cortical array of plant cells. Curr Opin Plant Biol. 2013a;16:678–87. doi: 10.1016/j.pbi.2013.10.012. [DOI] [PubMed] [Google Scholar]
- Henty-Ridilla JL, Li J, Day B, et al. Actin depolymerizing factor4 regulates actin dynamics during innate immune signaling in Arabidopsis. Plant Cell. 2014;26:340–52. doi: 10.1105/tpc.113.122499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henty-Ridilla JL, Shimono M, Li J, et al. The plant actin cytoskeleton responds to signals from microbe-associated molecular patterns. PLoS Pathog. 2013b;9:e1003290. doi: 10.1371/journal.ppat.1003290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotson A, Chosed R, Shu H, et al. Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol Microbiol. 2003;50:377–89. doi: 10.1046/j.1365-2958.2003.03730.x. [DOI] [PubMed] [Google Scholar]
- Hotson A, Mudgett MB. Cysteine proteases in phytopathogenic bacteria: identification of plant targets and activation of innate immunity. Curr Opin Plant Biol. 2004;7:384–90. doi: 10.1016/j.pbi.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Hurley B, Lee D, Mott A, et al. The Pseudomonas syringae type III effector HopF2 suppresses Arabidopsis stomatal immunity. PLoS One. 2014;9:e114921. doi: 10.1371/journal.pone.0114921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–82. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- Hutin M, Perez-Quintero AL, Lopez C, et al. MorTAL Kombat: the story of defense against TAL effectors through loss-of-susceptibility. Front Plant Sci. 2015;6:535. doi: 10.3389/fpls.2015.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K, Yamaguchi K, Sakamoto K, et al. Bacterial effector modulation of host E3 ligase activity suppresses PAMP-triggered immunity in rice. Nat Commun. 2014;5:5430. doi: 10.1038/ncomms6430. [DOI] [PubMed] [Google Scholar]
- Jamir Y, Guo M, Oh HS, et al. Identification of Pseudomonas syringae type III effectors that can suppress programmed cell death in plants and yeast. Plant J. 2004;37:554–65. doi: 10.1046/j.1365-313x.2003.01982.x. [DOI] [PubMed] [Google Scholar]
- Janjusevic R, Abramovitch RB, Martin GB, et al. A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science. 2006;311:222–6. doi: 10.1126/science.1120131. [DOI] [PubMed] [Google Scholar]
- Jelenska J, van Hal JA, Greenberg JT. Pseudomonas syringae hijacks plant stress chaperone machinery for virulence. P Natl Acad Sci USA. 2010;107:13177–82. doi: 10.1073/pnas.0910943107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong BR, Lin Y, Joe A, et al. Structure function analysis of an ADP-ribosyltransferase type III effector and its RNA-binding target in plant immunity. J Biol Chem. 2011;286:43272–81. doi: 10.1074/jbc.M111.290122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Yao J, Ma KW, et al. Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS Pathog. 2013;9:e1003715. doi: 10.1371/journal.ppat.1003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q, He SY. Role of the Hrp Pilus in type III protein secretion in Pseudomonas syringae. Science. 2001;294:2556–8. doi: 10.1126/science.1066397. [DOI] [PubMed] [Google Scholar]
- Journet L, Agrain C, Broz P, et al. The needle length of bacterial injectisomes is determined by a molecular ruler. Science. 2003;302:1757–60. doi: 10.1126/science.1091422. [DOI] [PubMed] [Google Scholar]
- Juillerat A, Bertonati C, Dubois G, et al. BurrH: a new modular DNA binding protein for genome engineering. Sci Rep. 2014;4:3831. doi: 10.1038/srep03831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y, Sklenar J, Derbyshire P, et al. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell. 2014;54:43–55. doi: 10.1016/j.molcel.2014.02.021. [DOI] [PubMed] [Google Scholar]
- Kang Y, Jelenska J, Cecchini NM, et al. HopW1 from Pseudomonas syringae disrupts the actin cytoskeleton to promote virulence in Arabidopsis. PLoS Pathog. 2014;10:e1004232. doi: 10.1371/journal.ppat.1004232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, et al. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. P Natl Acad Sci USA. 2008;105:7100–5. doi: 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K. Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends Plant Sci. 2006;11:109–12. doi: 10.1016/j.tplants.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Kazan K, Lyons R. Intervention of phytohormone pathways by pathogen effectors. Plant Cell. 2014;26:2285–309. doi: 10.1105/tpc.114.125419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BH, Kim SY, Nam KH. Assessing the diverse functions of BAK1 and its homologs in Arabidopsis, beyond BR signaling and PTI responses. Mol Cells. 2013;35:7–16. doi: 10.1007/s10059-013-2255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Desveaux D, Singer AU, et al. The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. P Natl Acad Sci USA. 2005a;102:6496–501. doi: 10.1073/pnas.0500792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JG, Li X, Roden JA, et al. Xanthomonas T3S effector XopN suppresses PAMP-triggered immunity and interacts with a tomato atypical receptor-like kinase and TFT1. Plant Cell. 2009a;21:1305–23. doi: 10.1105/tpc.108.063123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JG, Stork W, Mudgett MB. Xanthomonas type III effector XopD desumoylates tomato transcription factor SlERF4 to suppress ethylene responses and promote pathogen growth. Cell Host Microbe. 2013;13:143–54. doi: 10.1016/j.chom.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JG, Taylor KW, Hotson A, et al. XopD SUMO protease affects host transcription, promotes pathogen growth, and delays symptom development in Xanthomonas-infected tomato leaves. Plant Cell. 2008a;20:1915–29. doi: 10.1105/tpc.108.058529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JG, Taylor KW, Mudgett MB. Comparative analysis of the XopD type III secretion (T3S) effector family in plant pathogenic bacteria. Mol Plant Pathol. 2011;12:715–30. doi: 10.1111/j.1364-3703.2011.00706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Jung HJ, Lee HJ, et al. Glycine-rich RNA-binding protein 7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J. 2008b;55:455–66. doi: 10.1111/j.1365-313X.2008.03518.x. [DOI] [PubMed] [Google Scholar]
- Kim MG, da Cunha L, McFall AJ, et al. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell. 2005b;121:749–59. doi: 10.1016/j.cell.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Kim MG, Geng X, Lee SY, et al. The Pseudomonas syringae type III effector AvrRpm1 induces significant defenses by activating the Arabidopsis nucleotide-binding leucine-rich repeat protein RPS2. Plant J. 2009b;57:645–53. doi: 10.1111/j.1365-313X.2008.03716.x. [DOI] [PubMed] [Google Scholar]
- Kim NH, Hwang BK. Pepper aldehyde dehydrogenase CaALDH1 interacts with Xanthomonas effector AvrBsT and promotes effector-triggered cell death and defence responses. J Exp Bot. 2015a;66:3367–80. doi: 10.1093/jxb/erv147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NH, Hwang BK. Pepper heat shock protein 70a interacts with the type III effector AvrBsT and triggers plant cell death and immunity. Plant Physiol. 2015b;167:307–22. doi: 10.1104/pp.114.253898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NH, Kim BS, Hwang BK. Pepper arginine decarboxylase is required for polyamine and gamma-aminobutyric acid signaling in cell death and defense response. Plant Physiol. 2013;162:2067–83. doi: 10.1104/pp.113.217372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NH, Kim DS, Chung EH, et al. Pepper suppressor of the G2 allele of skp1 interacts with the receptor-like cytoplasmic kinase1 and type III effector AvrBsT and promotes the hypersensitive cell death response in a phosphorylation-dependent manner. Plant Physiol. 2014;165:76–91. doi: 10.1104/pp.114.238840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Qi D, Ashfield T, et al. Using decoys to expand the recognition specificity of a plant disease resistance protein. Science. 2016;351:684–7. doi: 10.1126/science.aad3436. [DOI] [PubMed] [Google Scholar]
- Kim SY, Kim SG, Kim YS, et al. Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucleic Acids Res. 2007;35:203–13. doi: 10.1093/nar/gkl1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Lin NC, Martin GB. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell. 2002;109:589–98. doi: 10.1016/s0092-8674(02)00743-2. [DOI] [PubMed] [Google Scholar]
- Kirzinger MW, Butz CJ, Stavrinides J. Inheritance of Pantoea type III secretion systems through both vertical and horizontal transfer. Mol Genet Genomics. 2015;290:2075–88. doi: 10.1007/s00438-015-1062-2. [DOI] [PubMed] [Google Scholar]
- Kirzinger MW, Nadarasah G, Stavrinides J. Insights into cross-kingdom plant pathogenic bacteria. Genes. 2011;2:980–97. doi: 10.3390/genes2040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Qu N, Gao M, et al. The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell. 2012;24:2225–36. doi: 10.1105/tpc.112.097253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S, Roy S, Singh P, et al. Cyclophilins: proteins in search of function. Plant Signal Behav. 2013;8:e22734. doi: 10.4161/psb.22734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitko BH, Park DH, Velasquez AC, et al. Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 2009;5:e1000388. doi: 10.1371/journal.ppat.1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laluk K, Luo H, Chai M, et al. Biochemical and genetic requirements for function of the immune response regulator botrytis-induced kinase1 in plant growth, ethylene signaling, and PAMP-triggered immunity in Arabidopsis. Plant Cell. 2011;23:2831–49. doi: 10.1105/tpc.111.087122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Tejero M, Kato J, Wagner S, et al. A sorting platform determines the order of protein secretion in bacterial type III systems. Science. 2011;331:1188–91. doi: 10.1126/science.1201476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux C, Huet G, Jauneau A, et al. A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell. 2015;161:1074–88. doi: 10.1016/j.cell.2015.04.025. [DOI] [PubMed] [Google Scholar]
- Lee AH, Hurley B, Felsensteiner C, et al. A bacterial acetyltransferase destroys plant microtubule networks and blocks secretion. PLoS Pathog. 2012;8:e1002523. doi: 10.1371/journal.ppat.1002523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH-Y, Middleton MA, Guttman DS, et al. Phytopathogen type III effectors as probes of biological systems. Microbiol Biotechnol. 2013;6:230–40. doi: 10.1111/1751-7915.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Bourdais G, Yu G, et al. Phosphorylation of the plant immune regulator RPM1-INTERACTING PROTEIN4 enhances plant plasma membrane H+-ATPase activity and inhibits flagellin-triggered immune responses in Arabidopsis. Plant Cell. 2015;27:2042–56. doi: 10.1105/tpc.114.132308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MW, Jelenska J, Greenberg JT. Arabidopsis proteins important for modulating defense responses to Pseudomonas syringae that secrete HopW1-1. Plant J. 2008;54:452–65. doi: 10.1111/j.1365-313X.2008.03439.x. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Lee A, Ma W, et al. The YopJ superfamily in plant-associated bacteria. Mol Plant Pathol. 2011;12:928–37. doi: 10.1111/j.1364-3703.2011.00719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Lee AH, Hassan JA, et al. The Arabidopsis ZED1 pseudokinase is required for ZAR1-mediated immunity induced by the Pseudomonas syringae type III effector HopZ1a. P Natl Acad Sci USA. 2013;110:18722–7. doi: 10.1073/pnas.1315520110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Wu R, Guttman DS, et al. Allele-specific virulence attenuation of the Pseudomonas syringae HopZ1a type III effector via the Arabidopsis ZAR1 resistance protein. PLoS Genet. 2010;6:e1000894. doi: 10.1371/journal.pgen.1000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Lu D, Shan L. Ubiquitination of pattern recognition receptors in plant innate immunity. Mol Plant Pathol. 2014;15:737–46. doi: 10.1111/mpp.12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CM, Brown I, Mansfield J, et al. The Hrp pilus of Pseudomonas syringae elongates from its tip and acts as a conduit for translocation of the effector protein HrpZ. EMBO J. 2002;21:1909–15. doi: 10.1093/emboj/21.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Atef A, Piatek A, et al. Characterization and DNA-binding specificities of Ralstonia TAL-like effectors. Mol Plant. 2013a;6:1318–30. doi: 10.1093/mp/sst006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li M, Yu L, et al. The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RBOHD to control plant immunity. Cell Host Microbe. 2014a;15:329–38. doi: 10.1016/j.chom.2014.02.009. [DOI] [PubMed] [Google Scholar]
- Li M, Ma X, Chiang YH, et al. Proline isomerization of the immune receptor-interacting protein RIN4 by a cyclophilin inhibits effector-triggered immunity in Arabidopsis. Cell Host Microbe. 2014b;16:473–83. doi: 10.1016/j.chom.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Yadeta KA, Elmore JM, et al. The Pseudomonas syringae effector HopQ1 promotes bacterial virulence and interacts with tomato 14-3-3 proteins in a phosphorylation-dependent manner. Plant Physiol. 2013b;161:2062–74. doi: 10.1104/pp.112.211748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Ding P, Lian K, et al. Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. Elife. 2016;5:e13568. doi: 10.7554/eLife.13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim MT, Kunkel BN. The Pseudomonas syringae type III effector AvrRpt2 promotes virulence independently of RIN4, a predicted virulence target in Arabidopsis thaliana. Plant J. 2004;40:790–8. doi: 10.1111/j.1365-313X.2004.02251.x. [DOI] [PubMed] [Google Scholar]
- Lin W, Li B, Lu D, et al. Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. P Natl Acad Sci USA. 2014;111:3632–7. doi: 10.1073/pnas.1318817111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg M, Cunnac S, Collmer A. Pseudomonas syringae type III effector repertoires: last words in endless arguments. Trends Microbiol. 2012;20:199–208. doi: 10.1016/j.tim.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Coaker G. Investigating the functions of the RIN4 protein complex during plant innate immune responses. Plant Signal Behav. 2009;4:1107–10. doi: 10.4161/psb.4.12.9944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Fuglsang AT, et al. RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol. 2009;7:e1000139. doi: 10.1371/journal.pbio.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Lin ZJ, et al. A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe. 2011;9:137–46. doi: 10.1016/j.chom.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Wang Y, Cui F, et al. The type III effector AvrXccB in Xanthomonascampestris pv. campestris targets putative methyltransferases and suppresses innate immunity in Arabidopsis. Mol Plant Pathol. 2016 doi: 10.1111/mpp.12435. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca CM, Potschin M, Zentgraf U. bZIPs and WRKYs: two large transcription factor families executing two different functional strategies. Front Plant Sci. 2014;5:169. doi: 10.3389/fpls.2014.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo T, Koulena N, Seto D, et al. The HopF family of Pseudomonas syringae type III secreted effectors. Mol Plant Pathol. 2016 doi: 10.1111/mpp.12412. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwer M, Schneider G. Prediction of type III secretion signals in genomes of gram-negative bacteria. PLoS One. 2009;4:e5917. doi: 10.1371/journal.pone.0005917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Duran R, Bourdais G, He SY, et al. The bacterial effector HopM1 suppresses PAMP-triggered oxidative burst and stomatal immunity. New Phytol. 2014;202:259–69. doi: 10.1111/nph.12651. [DOI] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, et al. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. P Natl Acad Sci USA. 2010;107:496–501. doi: 10.1073/pnas.0909705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Caldwell KS, Wroblewski T, et al. Proteolysis of a negative regulator of innate immunity is dependent on resistance genes in tomato and Nicotiana benthamiana and induced by multiple bacterial effectors. Plant Cell. 2009;21:2458–72. doi: 10.1105/tpc.107.056044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma KW, Jiang S, Hawara E, et al. Two serine residues in Pseudomonas syringae effector HopZ1a are required for acetyltransferase activity and association with the host co-factor. New Phytol. 2015;208:1157–68. doi: 10.1111/nph.13528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Song L, Yang Y, et al. A gain-of-function mutation in the ROC1 gene alters plant architecture in Arabidopsis. New Phytol. 2013;197:751–62. doi: 10.1111/nph.12056. [DOI] [PubMed] [Google Scholar]
- Macho AP, Schwessinger B, Ntoukakis V, et al. A bacterial tyrosine phosphatase inhibits plant pattern recognition receptor activation. Science. 2014;343:1509–12. doi: 10.1126/science.1248849. [DOI] [PubMed] [Google Scholar]
- Macho AP, Zipfel C. Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr Opin Microbiol. 2015;23:14–22. doi: 10.1016/j.mib.2014.10.009. [DOI] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, et al. Arabidopsis RIN4 Is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–89. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- Mackey D, Holt BF, Wiig A, et al. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–54. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- Magdalena J, Hachani A, Chamekh M, et al. Spa32 regulates a switch in substrate specificity of the type III secreton of Shigella flexneri from needle components to Ipa proteins. J Bacteriol. 2002;184:3433–41. doi: 10.1128/JB.184.13.3433-3441.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz MM, Piatek A, Stewart CN., Jr Genome engineering via TALENs and CRISPR/Cas9 systems: challenges and perspectives. Plant Biotechnol J. 2014;12:1006–14. doi: 10.1111/pbi.12256. [DOI] [PubMed] [Google Scholar]
- Mak AN, Bradley P, Cernadas RA, et al. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. 2012;335:716–9. doi: 10.1126/science.1216211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GB, Brommonschenkel SH, Chunwongse J, et al. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 1993;262:1432–6. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- Martin GB, Frary A, Wu T, et al. A member of the tomato Pto gene family confers sensitivity to fenthion resulting in rapid cell death. Plant Cell. 1994;6:1543–52. doi: 10.1105/tpc.6.11.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Schwizer S, Martin GB. Pto kinase binds two domains of AvrPtoB and its proximity to the effector E3 ligase determines if it evades degradation and activates plant immunity. PLoS Pathog. 2014;10:e1004227. doi: 10.1371/journal.ppat.1004227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattei PJ, Faudry E, Job V, et al. Membrane targeting and pore formation by the type III secretion system translocon. FEBS J. 2011;278:414–26. doi: 10.1111/j.1742-4658.2010.07974.x. [DOI] [PubMed] [Google Scholar]
- Meckler JF, Bhakta MS, Kim MS, et al. Quantitative analysis of TALE-DNA interactions suggests polarity effects. Nucleic Acids Res. 2013;41:4118–28. doi: 10.1093/nar/gkt085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejia E, Bliska JB, Viboud GI. Yersinia controls type III effector delivery into host cells by modulating Rho activity. PLoS Pathog. 2008;4:e3. doi: 10.1371/journal.ppat.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard R, Sansonetti P, Parsot C, et al. Extracellular association and cytoplasmic partitioning of the IpaB and IpaC invasins of S. flexneri. Cell. 1994;7984:515–25. doi: 10.1016/0092-8674(94)90260-7. [DOI] [PubMed] [Google Scholar]
- Meng X, Zhang S. MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol. 2013;51:245–66. doi: 10.1146/annurev-phyto-082712-102314. [DOI] [PubMed] [Google Scholar]
- Mills E, Baruch K, Charpentier X, et al. Real-time analysis of effector translocation by the type III secretion system of enteropathogenic Escherichia coli. Cell Host Microbe. 2008;3:104–13. doi: 10.1016/j.chom.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Mithoe SC, Ludwig C, Pel MJ, et al. Attenuation of pattern recognition receptor signaling is mediated by a MAP kinase kinase kinase. EMBO Rep. 2016;17:441–54. doi: 10.15252/embr.201540806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Mucyn TS, Clemente A, Andriotis VM, et al. The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell. 2006;18:2792–806. doi: 10.1105/tpc.106.044016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucyn TS, Wu AJ, Balmuth AL, et al. Regulation of tomato Prf by Pto-like protein kinases. Mol Plant Microbe In. 2009;22:391–401. doi: 10.1094/MPMI-22-4-0391. [DOI] [PubMed] [Google Scholar]
- Mukaihara T, Hatanaka T, Nakano M, et al. Ralstonia solanacearum type III effector RipAY is a glutathione-degrading enzyme that is activated by plant cytosolic thioredoxins and suppresses plant immunity. MBio. 2016;7 doi: 10.1128/mBio.00359-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar MS, Carvunis AR, Dreze M, et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 2011;333:596–601. doi: 10.1126/science.1203659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka M, Shirasu K, Noutoshi Y, et al. RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 2009;60:218–26. doi: 10.1111/j.1365-313X.2009.03949.x. [DOI] [PubMed] [Google Scholar]
- Nicaise V, Joe A, Jeong BR, et al. Pseudomonas HopU1 modulates plant immune receptor levels by blocking the interaction of their mRNAs with GRP7. EMBO J. 2013;32:701–12. doi: 10.1038/emboj.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan G, Manulis-Sasson S, Chalupowicz L, et al. The type III effector HsvG of the gall-forming Pantoeaagglomerans mediates expression of the host gene HSVGT. Mol Plant Microbe In. 2012;25:231–40. doi: 10.1094/MPMI-06-11-0173. [DOI] [PubMed] [Google Scholar]
- Nissan G, Manulis-Sasson S, Weinthal DM, et al. The type III effectors HsvG and HsvB of gall-forming Pantoea agglomerans determine host specificity and function as transcriptional activators. Mol Microbiol. 2006;61:1118–31. doi: 10.1111/j.1365-2958.2006.05301.x. [DOI] [PubMed] [Google Scholar]
- Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15:661–75. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- Nomura K, Debroy S, Lee YH, et al. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science. 2006;313:220–3. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- Nomura K, Mecey C, Lee YN, et al. Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. P Natl Acad Sci USA. 2011;108:10774–9. doi: 10.1073/pnas.1103338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntoukakis V, Balmuth AL, Mucyn TS, et al. The tomato Prf complex is a molecular trap for bacterial effectors based on Pto transphosphorylation. PLoS Pathog. 2013;9:e1003123. doi: 10.1371/journal.ppat.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntoukakis V, Mucyn TS, Gimenez-Ibanez S, et al. Host inhibition of a bacterial virulence effector triggers immunity to infection. Science. 2009;324:784–7. doi: 10.1126/science.1169430. [DOI] [PubMed] [Google Scholar]
- Ntoukakis V, Saur IM, Conlan B, et al. The changing of the guard: the Pto/Prf receptor complex of tomato and pathogen recognition. Curr Opin Plant Biol. 2014;20:69–74. doi: 10.1016/j.pbi.2014.04.002. [DOI] [PubMed] [Google Scholar]
- Nuruzzaman M, Sharoni AM, Kikuchi S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front Microbiol. 2013;4:248. doi: 10.3389/fmicb.2013.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh CS, Martin GB. Effector-triggered immunity mediated by the Pto kinase. Trends Plant Sci. 2011;16:132–40. doi: 10.1016/j.tplants.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Oh HS, Park DH, Collmer A. Components of the Pseudomonas syringae type III secretion system can suppress and may elicit plant innate immunity. Mol Plant Microbe In. 2010;23:727–39. doi: 10.1094/MPMI-23-6-0727. [DOI] [PubMed] [Google Scholar]
- Park HJ, Yun DJ. New insights into the role of the small ubiquitin-like modifier (SUMO) in plants. Int Rev Cel Mol Bio. 2013;300:161–209. doi: 10.1016/B978-0-12-405210-9.00005-9. [DOI] [PubMed] [Google Scholar]
- Pedley KF, Martin GB. Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu Rev Phytopathol. 2003;41:215–43. doi: 10.1146/annurev.phyto.41.121602.143032. [DOI] [PubMed] [Google Scholar]
- Peeters N, Carrere S, Anisimova M, et al. Repertoire, unified nomenclature and evolution of the Type III effector gene set in the Ralstonia solanacearum species complex. BMC Genomics. 2013;14:859. doi: 10.1186/1471-2164-14-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–33. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Pieretti I, Pesic A, Petras D, et al. What makes Xanthomonas albilineans unique amongst xanthomonads? Front Plant Sci. 2015;6:289. doi: 10.3389/fpls.2015.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter K, Shimono M, Tian M, et al. Arabidopsis Actin-Depolymerizing Factor-4 links pathogen perception, defense activation and transcription to cytoskeletal dynamics. PLoS Pathog. 2012;8:e1003006. doi: 10.1371/journal.ppat.1003006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poueymiro M, Cazale AC, Francois JM, et al. A Ralstonia solanacearum type III effector directs the production of the plant signal metabolite trehalose-6-phosphate. MBio. 2014;5:e02064–14. doi: 10.1128/mBio.02065-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CT, Kwaik YA. Exploitation of host polyubiquitination machinery through molecular mimicry by eukaryotic-like bacterial F-box effectors. Front Microbiol. 2010;1:122. doi: 10.3389/fmicb.2010.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri N, Jenner C, Bennett M, et al. Expression of avrPphB, an avirulence gene from Pseudomonas syringae pv. phaseolicola, and the delivery of signals causing the hypersensitive reaction in bean. Mol Plant Microbe In. 1997;10:247–56. doi: 10.1094/MPMI.1997.10.2.247. [DOI] [PubMed] [Google Scholar]
- Qi D, Dubiella U, Kim SH, et al. Recognition of the protein kinase AvrPphB susceptible1 by the disease resistance protein resistance to pseudomonas syringae5 is dependent on s-acylation and an exposed loop in AvrPphB susceptible1. Plant Physiol. 2014;164:340–51. doi: 10.1104/pp.113.227686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S, Rivas S. Regulate and be regulated: integration of defense and other signals by the AtMYB30 transcription factor. Front Plant Sci. 2013;4:98. doi: 10.3389/fpls.2013.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen MW, Roux M, Petersen M, et al. MAP Kinase cascades in Arabidopsis innate immunity. Front Plant Sci. 2012;3:169. doi: 10.3389/fpls.2012.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathjen JP, Chang JH, Staskawicz BJ, et al. Constitutively active Pto induces a Prf-dependent hypersensitive response in the absence of avrPto. EMBO J. 1999;18:3232–40. doi: 10.1093/emboj/18.12.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remigi P, Anisimova M, Guidot A, et al. Functional diversification of the GALA type III effector family contributes to Ralstonia solanacearum adaptation on different plant hosts. New Phytol. 2011;192:976–87. doi: 10.1111/j.1469-8137.2011.03854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter A, Streubel J, Blücher C, et al. A TAL effector repeat architecture for frameshift binding. Nat Commun. 2014;5:3447. doi: 10.1038/ncomms4447. [DOI] [PubMed] [Google Scholar]
- Robatzek S, Chinchilla D, Boller T. Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Gene Dev. 2006;20:537–42. doi: 10.1101/gad.366506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol. 2011;49:317–43. doi: 10.1146/annurev-phyto-073009-114447. [DOI] [PubMed] [Google Scholar]
- Rodriguez MC, Petersen M, Mundy J. Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol. 2010;61:621–49. doi: 10.1146/annurev-arplant-042809-112252. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Herva JJ, Gonzalez-Melendi P, Cuartas-Lanza R, et al. A bacterial cysteine protease effector protein interferes with photosynthesis to suppress plant innate immune responses. Cell Microbiol. 2012;14:669–81. doi: 10.1111/j.1462-5822.2012.01749.x. [DOI] [PubMed] [Google Scholar]
- Rosebrock TR, Zeng L, Brady JJ, et al. A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature. 2007;448:370–4. doi: 10.1038/nature05966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B, Bolot S, Guy E, et al. Genomics and transcriptomics of Xanthomonascampestrisspecies challenge the concept of core type III effectome. BMC Genomics. 2015;16:975. doi: 10.1186/s12864-015-2190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, et al. WRKY transcription factors. Trends Plant Sci. 2010;15:247–58. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Russell AR, Ashfield T, Innes RW. Pseudomonas syringae effector AvrPphB suppresses AvrB-induced activation of RPM1 but not AvrRpm1-induced activation. Mol Plant Microbe In. 2015;28:727–35. doi: 10.1094/MPMI-08-14-0248-R. [DOI] [PubMed] [Google Scholar]
- Ryu H, Kim K, Cho H, et al. Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell. 2007;19:2749–62. doi: 10.1105/tpc.107.053728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadanandom A, Bailey M, Ewan R, et al. The ubiquitin-proteasome system: central modifier of plant signalling. New Phytol. 2012;196:13–28. doi: 10.1111/j.1469-8137.2012.04266.x. [DOI] [PubMed] [Google Scholar]
- Salmeron JM, Oldroyd GED, Rommens CMT, et al. Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell. 1996;86:123–33. doi: 10.1016/s0092-8674(00)80083-5. [DOI] [PubMed] [Google Scholar]
- Samudrala R, Heffron F, McDermott JE. Accurate prediction of secreted substrates and identification of a conserved putative secretion signal for type III secretion systems. PLoS Pathog. 2009;5:e1000375. doi: 10.1371/journal.ppat.1000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarris PF, Duxbury Z, Huh SU, et al. A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell. 2015;161:1089–100. doi: 10.1016/j.cell.2015.04.024. [DOI] [PubMed] [Google Scholar]
- Scharenberg AM, Duchateau P, Smith J. Genome engineering with TAL-effector nucleases and alternative modular nuclease technologies. Curr Gene Ther. 2013;13:291–303. doi: 10.2174/15665232113139990026. [DOI] [PubMed] [Google Scholar]
- Schechter LM, Valenta JC, Schneider DJ, et al. Functional and computational analysis of amino acid patterns predictive of type III secretion system substrates in Pseudomonas syringae. PLoS One. 2012;7:e36038. doi: 10.1371/journal.pone.0036038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlumberger MC, Muller AJ, Ehrbar K, et al. Real-time imaging of type III secretion: Salmonella SipA injection into host cells. P Natl Acad Sci USA. 2005;102:12548–53. doi: 10.1073/pnas.0503407102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Persson S. Connecting two arrays: the emerging role of actin-microtubule cross-linking motor proteins. Front Plant Sci. 2015;6:415. doi: 10.3389/fpls.2015.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornack S, Ballvora A, Gürlebeck D, et al. The tomato resistance protein Bs4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severely truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J. 2004;37:46–60. doi: 10.1046/j.1365-313x.2003.01937.x. [DOI] [PubMed] [Google Scholar]
- Schreiber T, Sorgatz A, List F, et al. Refined requirements for protein regions important for activity of the TALE AvrBs3. PLoS One. 2015;10:e0120214. doi: 10.1371/journal.pone.0120214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman BA, Carrano AC, Jeffrey PD, et al. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature. 2000;408:381–6. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- Schwessinger B, Ronald PC. Plant innate immunity: perception of conserved microbial signatures. Annu Rev Plant Biol. 2012;63:451–82. doi: 10.1146/annurev-arplant-042811-105518. [DOI] [PubMed] [Google Scholar]
- Scofield SR, Tobias CM, Rathjen JP, et al. Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science. 1996;274:2063–5. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- Shabek N, Zheng N. Plant ubiquitin ligases as signaling hubs. Nat Struct Mol Biol. 2014;21:293–6. doi: 10.1038/nsmb.2804. [DOI] [PubMed] [Google Scholar]
- Shan L, He P, Li J, et al. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe. 2008;4:17–27. doi: 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, et al. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science. 2003;301:1230–3. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- Shao F, Merritt PM, Bao Z, et al. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell. 2002;109:575–88. doi: 10.1016/s0092-8674(02)00766-3. [DOI] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468:400–5. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono M, Lu YJ, Porter K, et al. The Pseudomonas syringae type-III effector HopG1 induces actin remodeling to promote symptom development and susceptibility during infection. Plant Physiol. 2016;171:2239–55. doi: 10.1104/pp.16.01593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer AU, Schulze S, Skarina T, et al. A pathogen type III effector with a novel E3 ubiquitin ligase architecture. PLoS Pathog. 2013;9:e1003121. doi: 10.1371/journal.ppat.1003121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DK, Calvino M, Brauer EK, et al. The tomato kinome and the tomato kinase library ORFeome: novel resources for the study of kinases and signal transduction in tomato and solanaceae species. Mol Plant Microbe In. 2014;27:7–17. doi: 10.1094/MPMI-08-13-0218-TA. [DOI] [PubMed] [Google Scholar]
- Sohn KH, Hughes RK, Piquerez SJ, et al. Distinct regions of the Pseudomonas syringae coiled-coil effector AvrRps4 are required for activation of immunity. P Natl Acad Sci USA. 2012;109:16371–6. doi: 10.1073/pnas.1212332109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn KH, Segonzac C, Rallapalli G, et al. The nuclear immune receptor RPS4 is required for RRS1SLH1-dependent constitutive defense activation in Arabidopsis thaliana. PLoS Genet. 2014;10:e1004655. doi: 10.1371/journal.pgen.1004655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondergaard TE, Schulz A, Palmgren MG. Energization of transport processes in plants. roles of the plasma membrane H+-ATPase. Plant Physiol. 2004;136:2475–82. doi: 10.1104/pp.104.048231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soprano AS, Abe VY, Smetana JH, et al. Citrus MAF1, a repressor of RNA polymerase III, binds the Xanthomonas citri canker elicitor PthA4 and suppresses citrus canker development. Plant Physiol. 2013;163:232–42. doi: 10.1104/pp.113.224642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streubel J, Blucher C, Landgraf A, et al. TAL effector RVD specificities and efficiencies. Nat Biotechnol. 2012;30:593–5. doi: 10.1038/nbt.2304. [DOI] [PubMed] [Google Scholar]
- Suarez-Rodriguez MC, Adams-Phillips L, Liu Y, et al. MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 2007;143:661–9. doi: 10.1104/pp.106.091389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Gubler F. Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol. 2004;55:197–223. doi: 10.1146/annurev.arplant.55.031903.141753. [DOI] [PubMed] [Google Scholar]
- Sun X, Greenwood DR, Templeton MD, et al. The intrinsically disordered structural platform of the plant defence hub protein RPM1-interacting protein 4 provides insights into its mode of action in the host-pathogen interface and evolution of the nitrate-induced domain protein family. FEBS J. 2014;281:3955–79. doi: 10.1111/febs.12937. [DOI] [PubMed] [Google Scholar]
- Swiderski MR, Innes RW. The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J. 2001;26:101–12. doi: 10.1046/j.1365-313x.2001.01014.x. [DOI] [PubMed] [Google Scholar]
- Szczesny R, Büttner D, Escolar L, et al. Suppression of the AvrBs1-specific hypersensitive response by the YopJ effector homolog AvrBsT from Xanthomonas depends on a SNF1-related kinase. New Phytol. 2010;187:1058–74. doi: 10.1111/j.1469-8137.2010.03346.x. [DOI] [PubMed] [Google Scholar]
- Szurek B, Marois E, Bonas U, et al. Eukaryotic features of the Xanthomonas type III effector AvrBs3: protein domains involved in transcriptional activation and the interaction with nuclear import receptors from pepper. Plant J. 2001;26:523–34. doi: 10.1046/j.0960-7412.2001.01046.x. [DOI] [PubMed] [Google Scholar]
- Takken FL, Goverse A. How to build a pathogen detector: structural basis of NB-LRR function. Curr Opin Plant Biol. 2012;15:375–84. doi: 10.1016/j.pbi.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Tampakaki AP. Commonalities and differences of T3SSs in rhizobia and plant pathogenic bacteria. Front Plant Sci. 2014;5:114. doi: 10.3389/fpls.2014.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CM, Li MY, Yang PY, et al. Arabidopsis HFR1 is a potential nuclear substrate regulated by the Xanthomonas type III effector XopD(Xcc8004) PLoS One. 2015;10:e0117067. doi: 10.1371/journal.pone.0117067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Rong W, Luo H, et al. The Xanthomonas campestris effector protein XopDXcc8004 triggers plant disease tolerance by targeting DELLA proteins. New Phytol. 2014;204:595–608. doi: 10.1111/nph.12918. [DOI] [PubMed] [Google Scholar]
- Tang X, Frederick RD, Zhou J, et al. Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science. 1996;274:2060–3. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- Tasset C, Bernoux M, Jauneau A, et al. Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1-R-mediated immunity in Arabidopsis. PLoS Pathog. 2010;6:e1001202. doi: 10.1371/journal.ppat.1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KW, Kim JG, Su XB, et al. Tomato TFT1 is required for PAMP-triggered immunity and mutations that prevent T3S effector XopN from binding to TFT1 attenuate Xanthomonas virulence. PLoS Pathog. 2012;8:e1002768. doi: 10.1371/journal.ppat.1002768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teper D, Salomon D, Sunitha S, et al. Xanthomonas euvesicatoria type III effector XopQ interacts with tomato and pepper 14-3-3 isoforms to suppress effector-triggered immunity. Plant J. 2014;77:297–309. doi: 10.1111/tpj.12391. [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–5. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- Troisfontaines P, Cornelis GR. Type III secretion: more systems than you think. Physiol Plantarum. 2005;20:326–39. doi: 10.1152/physiol.00011.2005. [DOI] [PubMed] [Google Scholar]
- Tucker SC, Galan JE. Complex function for SicA, a Salmonella enterica serovar typhimurium type III secretion-associated chaperone. J Bacteriol. 2000;182:2262–8. doi: 10.1128/jb.182.8.2262-2268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Üstün S, Bartetzko V, Börnke F. The Xanthomonas campestris type III effector XopJ targets the host cell proteasome to suppress salicylic-acid mediated plant defence. PLoS Pathog. 2013;9:e1003427. doi: 10.1371/journal.ppat.1003427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Üstün S, Börnke F. The Xanthomonas campestris type III effector XopJ proteolytically degrades proteasome subunit RPT6. Plant Physiol. 2015;168:107–19. doi: 10.1104/pp.15.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Üstün S, König P, Guttman DS, et al. HopZ4 from Pseudomonas syringae, a member of the HopZ type III effector family from the YopJ superfamily, inhibits the proteasome in plants. Mol Plant Microbe In. 2014;27:611–23. doi: 10.1094/MPMI-12-13-0363-R. [DOI] [PubMed] [Google Scholar]
- Valinsky L, Manulis S, Nizan R, et al. A pathogenicity gene isolated from the pPATH plasmid of Erwiniaherbicola pv. gypsophilae determines host specificity. Mol Plant Microbe In. 1998;11:753–62. doi: 10.1094/MPMI.1998.11.8.753. [DOI] [PubMed] [Google Scholar]
- Van der Biezen EA, Jones JDG. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci. 1998;23:454–6. doi: 10.1016/s0968-0004(98)01311-5. [DOI] [PubMed] [Google Scholar]
- Van der Hoorn RAL, Kamoun S. From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell. 2008;20:2009–17. doi: 10.1105/tpc.108.060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Engelenburg SB, Palmer AE. Quantification of real-time Salmonella effector type III secretion kinetics reveals differential secretion rates for SopE2 and SptP. Chem Biol. 2008;15:619–28. doi: 10.1016/j.chembiol.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verove J, Bernarde C, Bohn YS, et al. Injection of Pseudomonas aeruginosa Exo toxins into host cells can be modulated by host factors at the level of translocon assembly and/or activity. PLoS One. 2012;7:e30488. doi: 10.1371/journal.pone.0030488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viboud GI, Bliska JB. A bacterial type III secretion system inhibits actin polymerization to prevent pore formation in host cell membranes. EMBO J. 2001;20:5373–82. doi: 10.1093/emboj/20.19.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Bio. 2009;10:385–97. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- Wang G, Roux B, Feng F, et al. The decoy substrate of a pathogen effector and a pseudokinase specify pathogen-induced modified-self recognition and immunity in plants. Cell Host Microbe. 2015;18:285–95. doi: 10.1016/j.chom.2015.08.004. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li J, Hou S, et al. A Pseudomonas syringae ADP-ribosyltransferase inhibits Arabidopsis mitogen-activated protein kinase kinases. Plant Cell. 2010;22:2033–44. doi: 10.1105/tpc.110.075697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sun M, Bao H, et al. T3_MM: a Markov model effectively classifies bacterial type III secretion signals. PLoS One. 2013;8:e58173. doi: 10.1371/journal.pone.0058173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington EJ, Mukhtar MS, Finkel OM, et al. Pseudomonas syringae type III effector HopAF1 suppresses plant immunity by targeting methionine recycling to block ethylene induction. P Natl Acad Sci USA. 2016;113:E3577-86. doi: 10.1073/pnas.1606322113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei HL, Chakravarthy S, Mathieu J, et al. Pseudomonas syringae pv. tomato DC3000 type III secretion effector polymutants reveal an interplay between HopAD1 and AvrPtoB. Cell Host Microbe. 2015;17:752–62. doi: 10.1016/j.chom.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinthal DM, Barash I, Tzfira T, et al. Characterization of nuclear localization signals in the type III effectors HsvG and HsvB of the gall-forming bacterium Pantoea agglomerans. Microbiology. 2011;157:1500–8. doi: 10.1099/mic.0.047118-0. [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, et al. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414:562–5. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- Williams SJ, Sohn KH, Wan L, et al. Structural basis for assembly and function of a heterodimeric plant immune receptor. Science. 2014;344:299–303. doi: 10.1126/science.1247357. [DOI] [PubMed] [Google Scholar]
- Wilton M, Subramaniam R, Elmore J, et al. The type III effector HopF2Pto targets Arabidopsis RIN4 protein to promote Pseudomonas syringae virulence. P Natl Acad Sci USA. 2010;107:2349–54. doi: 10.1073/pnas.0904739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnen B, Schlumberger MC, Sturm A, et al. Hierarchical effector protein transport by the SalmonellaTyphimurium SPI-1 type III secretion system. PLoS One. 2008;3:e2178. doi: 10.1371/journal.pone.0002178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AJ, Andriotis VM, Durrant MC, et al. A patch of surface-exposed residues mediates negative regulation of immune signaling by tomato Pto kinase. Plant Cell. 2004;16:2809–21. doi: 10.1105/tpc.104.024141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Chen H, Curtis C, et al. Go in for the kill. How plants deploy effector-triggered immunity to combat pathogens. Virulence. 2014;5:710–21. doi: 10.4161/viru.29755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Lu D, Kabbage M, et al. Bacterial effector HopF2 suppresses Arabidopsis innate immunity at the plasma membrane. Mol Plant Microbe In. 2011;24:585–93. doi: 10.1094/MPMI-07-10-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Shan L, He P. Microbial signature-triggered plant defense responses and early signaling mechanisms. Plant Sci. 2014;228:118–26. doi: 10.1016/j.plantsci.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T, Zong N, Zhang J, et al. BAK1 is not a target of the Pseudomonas syringae effector AvrPto. Mol Plant Microbe In. 2011;24:100–7. doi: 10.1094/MPMI-04-10-0096. [DOI] [PubMed] [Google Scholar]
- Xiang T, Zong N, Zou Y, et al. Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol. 2008;18:74–80. doi: 10.1016/j.cub.2007.12.020. [DOI] [PubMed] [Google Scholar]
- Xiao F, Giavalisco P, Martin GB. Pseudomonas syringae type III effector AvrPtoB is phosphorylated in plant cells on serine 258, promoting its virulence activity. J Biol Chem. 2007;282:30737–44. doi: 10.1074/jbc.M705565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, He P, Abramovitch RB, et al. The N-terminal region of Pseudomonas type III effector AvrPtoB elicits Pto-dependent immunity and has two distinct virulence determinants. Plant Journal. 2007;52:595–614. doi: 10.1111/j.1365-313X.2007.03259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing W, Zou Y, Liu Q, et al. The structural basis for activation of plant immunity by bacterial effector protein AvrPto. Nature. 2007;449:243–7. doi: 10.1038/nature06109. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Nakamura Y, Ishikawa K, et al. Suppression of rice immunity by Xanthomonas oryzae type III effector Xoo2875. Biosci Biotech Bioch. 2013a;77:796–801. doi: 10.1271/bbb.120929. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Yamada K, Ishikawa K, et al. A receptor-like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe. 2013b;13:347–57. doi: 10.1016/j.chom.2013.02.007. [DOI] [PubMed] [Google Scholar]
- Yang J, Zhang Y, Yuan P, et al. Complete decoding of TAL effectors for DNA recognition. Cell Res. 2014;24:628–31. doi: 10.1038/cr.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HK, Kim SG, Kim SY, et al. Regulation of leaf senescence by NTL9-mediated osmotic stress signaling in Arabidopsis. Mol Cells. 2008;25:438–45. [PubMed] [Google Scholar]
- Yu S, Hwang I, Rhee S. The crystal structure of type III effector protein XopQ from Xanthomonas oryzae complexed with adenosine diphosphate ribose. Proteins. 2014;82:2910–4. doi: 10.1002/prot.24656. [DOI] [PubMed] [Google Scholar]
- Zeng L, Velasquez AC, Munkvold KR, et al. A tomato LysM receptor-like kinase promotes immunity and its kinase activity is inhibited by AvrPtoB. Plant J. 2012;69:92–103. doi: 10.1111/j.1365-313X.2011.04773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 2010;7:290–301. doi: 10.1016/j.chom.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Zhang J, Shao F, Li Y, et al. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe. 2007;1:175–85. doi: 10.1016/j.chom.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Zhang J, Yin Z, White F. TAL effectors and the executor R genes. Front Plant Sci. 2015;6:641. doi: 10.3389/fpls.2015.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wu Y, Gao M, et al. Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe. 2012;11:253–63. doi: 10.1016/j.chom.2012.01.015. [DOI] [PubMed] [Google Scholar]
- Zhao B, Dahlbeck D, Krasileva KV, et al. Computational and biochemical analysis of the Xanthomonas effector AvrBs2 and its role in the modulation of Xanthomonas type three effector delivery. PLoS Pathog. 2011;7:e1002408. doi: 10.1371/journal.ppat.1002408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XY, Zhou M, Yoo H, et al. Spatial and temporal regulation of biosynthesis of the plant immune signal salicylic acid. P Natl Acad Sci USA. 2015;112:9166–73. doi: 10.1073/pnas.1511182112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Lin J, Johnson A, et al. Pseudomonas syringae type III effector HopZ1 targets a host enzyme to suppress isoflavone biosynthesis and promote infection in soybean. Cell Host Microbe. 2011;9:177–86. doi: 10.1016/j.chom.2011.02.007. [DOI] [PubMed] [Google Scholar]
- Zhou J, Wu S, Chen X, et al. The Pseudomonas syringae effector HopF2 suppresses Arabidopsis immunity by targeting BAK1. Plant J. 2014;77:235–45. doi: 10.1111/tpj.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Wu Y, Yang Y, et al. An Arabidopsis plasma membrane proton ATPase modulates JA signaling and is exploited by the Pseudomonas syringae effector protein AvrB for stomatal invasion. Plant Cell. 2015;27:2032–41. doi: 10.1105/tpc.15.00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C. Plant pattern-recognition receptors. Trends Immunol. 2014;35:345–51. doi: 10.1016/j.it.2014.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.