Abstract
Polymorphic major histocompatibility complex (MHC) molecules play a central role in the vertebrate adaptive immune system. By presenting short peptides derived from pathogen-derived proteins, these “classical” MHC molecules can alert the T cell branch of the immune system of infected cells and clear the pathogen. There exist other “non-classical” MHC molecules, which while similar in structure to classical MHC proteins, are contrasted by their limited polymorphism. While the functions of many class Ib MHC molecules have still to be elucidated, the nature and diversity of antigens (if any) that some of them might present to the immune system is expected to be more restricted and might function as another approach to distinguish self from non-self. The MHC-related 1 (MR1) molecule is a member of this family of non-classical MHC proteins. It was recently shown to present unique antigens in the form of vitamin metabolites found in certain microbes. MR1 is strongly conserved genetically, structurally, and functionally through mammalian evolution, indicating its necessity in ensuring an effective immune system for members of this class. Although MR1 will be celebrating 21 years this year since its discovery, most of our understanding of how this molecule functions has only been uncovered in the past decade. Herein, we discuss where MR1 is expressed, how it selectively is able to bind to its appropriate antigens and how it, then, is able to specifically activate a distinct population of T cells.
Keywords: MHC-Ib, MR1, MAIT, vitamin metabolites, TCR
Introduction
Vertebrate evolution over the past 500 million years has produced a form of immunity unique to this branch of life. This manifests itself partly with the existence of major histocompatibility complex (MHC) molecules, which serve as harbingers of a loss of homeostasis, usually due to a microbial infection. Classical MHC molecules capture peptides of various lengths and come in two flavors: MHC-I molecules, which bind to short peptides (8-9 residues long) sampled from the proteolytic products of intracellular proteins and MHC-II molecules, which bind to longer peptides (14-20 residues long) sampled from the extracellular milieu (Mohan and Unanue 2012; Neefjes et al. 2011). At steady-state, these molecules bind to peptides derived from host proteins exclusively. During an infection, however, these molecules bind to peptides derived from microbial proteins and present these protein fragments to T cells, which through the use of their diverse T cell antigen receptors (TCRs) can interact with the peptide-MHC (pMHC) ligands to which they are specific. Through these intricate interactions, infected cells alert and activate T cells, helping the host clear the infection. Peptides from the microbial proteome, though, are not the only antigens that hosts consider foreign. Microbes also produce lipids, modified lipids/peptides, and other small compounds not naturally present in vertebrates (Mori et al. 2016). Thus, vertebrates have taken a multi-pronged approach to tackle infections by employing different classes of molecules that bind to different antigens, including peptides and non-peptides.
Some of these molecules are the non-classical MHC-I proteins, or MHC-Ib as they are called, which are structurally similar to the classical MHC-I (MHC-Ia) proteins, but in contrast to the classical MHC-I proteins they display limited or no polymorphism, their expression tends to be more tissue-restricted and they might have functions other than antigen presentation to the immune system (Shawar et al. 1994). Although the existence of these molecules has been appreciated for quite some time (Stroynowski 1990), their individual functions have largely remained understudied.
Recently, though, one such molecule, the MHC-related 1 (MR1) protein, has received substantial attention due to the unusual ligands it presents to its cognate T-cell population, the mucosal associated invariant T (MAIT) cells. MAIT cells are αβ TCR-expressing T cells enriched in mucosal tissues, especially in the human liver where they comprise 20-40% of T cells (Howson et al. 2015). They originate in the thymus before migrating to mucosal sites for further maturation (Martin et al. 2009). Subsequently, they survey these sites for signs of infection and respond quickly upon encountering one. They primarily secrete TH1- and TH17-biased cytokines (such as IFNγ, TNFα, and IL-17) while also possessing the ability to lyse infected cells (Le Bourhis et al. 2013a; Tang et al. 2013). MAIT cells rely heavily on MR1 for both their development and activation. As such, understanding the role of this non-classical MHC-I molecule in the development, maintenance and activation of MAIT cells will be fundamental to our grasp of MAIT cell biology. Here, we review how MR1 functions as well as the antigens it presents.
The MR1 Gene and its Evolution
Classical MHC-I and MHC-II genes are clustered together on chromosome 6 and chromosome 17 in the human and murine genomes, respectively. Interestingly, many MHC-Ib genes are also interspersed within these loci, potentially resulting from gene duplication events (Amadou et al. 2003). For example, in mouse, over 20 MHC-Ib genes have been discovered in the telomeric region of this locus (Takada et al. 2003). As might be expected in gene duplication events, the duplicated genes either gain a novel function or lose gene function altogether due to redundancy with the original gene (Klein et al. 1998). To support this claim, there are several pseudogenes within this locus that have premature termination codons, frameshifts, or poor promoters limiting their expression (Geraghty et al. 1992). Many of the functional genes have also gained distinctive roles, as evidenced by their ability to present novel antigens or serve as stress ligands (Rodgers and Cook 2005). Despite this, they retain many features in common with their paralogous MHC-Ia genes. Genes in both families have similar exon-intron organization in the genome with each exon coding for a specific structural domain and together forming the heavy chain of the protein (Stroynowski and Lindahl 1994). Additionally, these heavy chains usually interact with the monomorphic β2-microglobulin (β2m) protein in order to be stably expressed on the surface. Of the three domains present in the heavy chains, the first two (α1 and α2) form the ligand-binding pocket, whereas the third (α3) tethers the protein to the cell surface and interacts with β2m (Stroynowski and Lindahl 1994). Therefore, it is the α1 and α2 domains that bestow specialized functions to these molecules by binding to antigens from different pools of potential ligands. It is also in these domains where the differences between MHC-Ia and MHC-Ib molecules are most apparent. Classical MHC-I proteins are considered the most polymorphic genes in the genome (Trowsdale and Knight 2013) and their polymorphism is primarily restricted to the α1 and α2 domains (Hughes et al. 1990). In this manner, different MHC-Ia alleles within a species can present various peptides to T cells and encompass a larger collection of antigens to promote stronger immunity on a population level. MHC-Ib proteins, on the other hand, display little polymorphism in these domains both within a species and across multiple species (Rodgers and Cook 2005). As a result, they might present limited sets of antigens, which might not overlap with the antigens presented by MHC-Ia proteins. Thus, over the course of evolution, MHC-Ib proteins have possibly undergone strong negative selection to guarantee binding of specific ligands (Hansen et al. 2007) while MHC-Ia proteins have not faced a similar pressure.
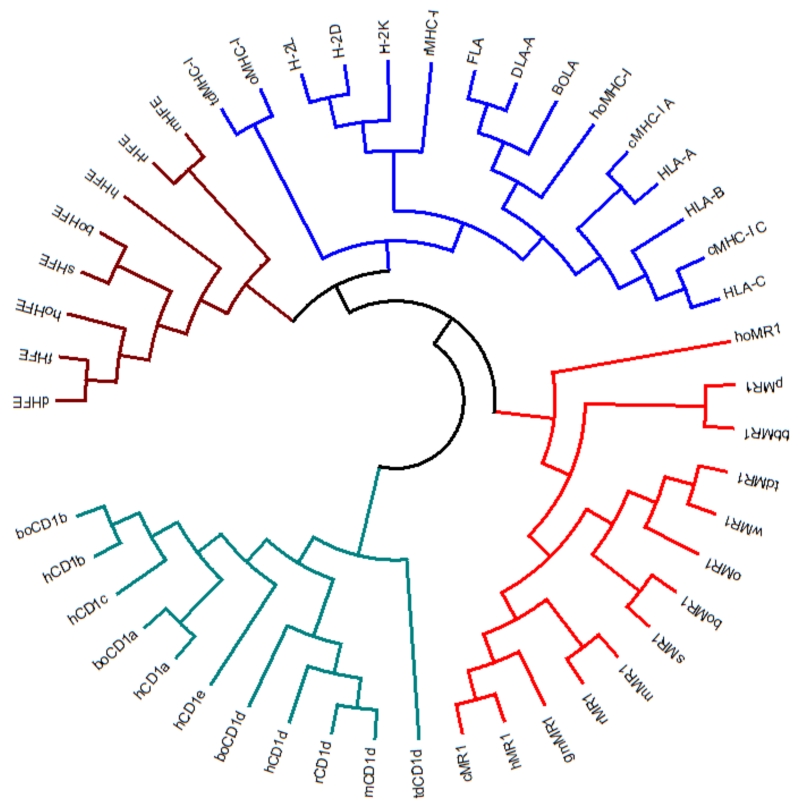
Although at least one MHC gene has been discovered in all jawed vertebrates studied thus far (Trowsdale 1995), some of the MHC-Ib genes, such as H2-M3 in mice and the group I CD1 genes in humans, are missing altogether in certain species and clades (Mori et al. 2016; Rodgers and Cook 2005), indicating that these specialized molecules are not necessary for some species occupying unique niches. Interestingly, a functional MR1 gene has been discovered in almost every mammalian species analyzed, except for monotremes (Goldfinch et al. 2010; Tsukamoto et al. 2013). Comparing the antigen-binding domains of the MR1 genes found in each of these species reveals a marked conservation, exemplified by the ~90% homology at the protein level between humans and mice (Yamaguchi et al. 1997) (Figure 1). Such conservation underscores the importance that this molecule might have in immune surveillance. In addition to sequence conservation, there is also a functional conservation since both murine and human MAIT cells have been observed to be xenoreactive to orthologs of the MR1 molecule (Huang et al. 2009). Other innate-like T cell populations such as iNKT cells also display similar xenoreactivity to MHC-Ib molecules like CD1d (Brossay et al. 1998; Kjer-Nielsen et al. 2006), suggesting that they MHC-Ib proteins tend to play similar roles in different hosts.
Figure 1. Phylogenetic Analysis by Maximum Likelihood Method of Different MHC-Ia and MHC-Ib Molecules.
The evolutionary history for various MHC-Ia and MHC-Ib molecules for different species was inferred by using the Maximum Likelihood method based on the JTT matrix-based model (Jones et al. 1992). The tree with the highest log likelihood (-7594.4549) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The analysis involved 47 amino acid sequences. All positions containing gaps and missing data were eliminated. A total of 167 positions were involved in the final analysis, all of which are located in the α1 and α2 domains so only the ligand-binding domains of the different proteins were analyzed. Only MHC-Ia, MR1, CD1, and HFE from various species were compared. Both MR1 and HFE are closer phylogenetically to MHC-Ia than each is to CD1. In lieu of using taxonomic classifications, each animal was assigned a 1-2 letter designation. Abbreviations used: bb – little brown bat; bo – bovine; c – chimpanzee; d – dog; f – feline; gm – green monkey; h – human; ho – horse; m – mouse; o – opossum; p – pig; r – rat; s – sheep; td – Tasmanian devil; w – wallaby. Common nomenclature was used for any proteins for which such naming already existed, such as HLA or H-2. Evolutionary analyses were conducted in MEGA7 (Kumar et al. 2016).
Curiously, MR1 shares greater homology with MHC-Ia genes than it does with other MHC-Ib genes (Hashimoto et al. 1995). Although the MR1 gene is considered part of the MHC-Ib family, it is not located within the MHC locus. It is instead located on chromosome 1 in both human and murine genomes. A detailed analysis of the genomic region surrounding MR1 has previously determined that it is in fact paralogous to the MHC locus on chromosome 6, providing evidence for a duplication event that occurred early in the evolution of vertebrates (Shiina et al. 2001). Given that such a whole genome duplication event transpired, why only placental and marsupial mammals possess this gene remains unclear, especially since many of the neighboring genes are conserved across other vertebrate species (Tsukamoto et al. 2013).
MR1 Protein Isoforms and Expression
Although MR1 is essentially a monomorphic molecule, various mRNA isoforms have been discovered in both humans and other species (Riegert et al. 1998). However, many of these isoforms have premature termination codons generating nonfunctional protein products (Riegert et al. 1998). One isoform, MR1B, has recently been characterized in greater depth. In humans, MR1B, which lacks the α3 domain, can be expressed at the cell surface in an overexpression system, although whether it is expressed as a monomer or a homodimer remains unresolved (Lion et al. 2013; Yamaguchi et al. 2014). Since the antigen-binding pocket is created by the α1 and α2 domains, this isoform can still present antigen to and stimulate MAIT cells. Additionally, due to the missing α3 domain, it cannot interact with β2m as the full-length isoform does (Lion et al. 2013; Yamaguchi et al. 2014). Many bacteria possess proteins promoting immune evasion and some especially target and interact with β2m (Bjorck et al. 1984; Sreejit et al. 2014). MR1B could, thus, potentially serve as a splice variant that circumvents the requirement for β2m for cell surface expression and antigen presentation during such a bacterial infection.
In contrast to other MHC-Ib genes, both MR1 and MR1B transcripts are expressed ubiquitously in various tissues and cell types, much like MHC-Ia transcripts (Hashimoto et al. 1995; Riegert et al. 1998). Such global expression might be advantageous to combat the expansive bacterial tropism and further highlights the crucial role MR1 plays in the immune system. Yet, observing MR1 protein expression on the cell surface has been notoriously difficult. B cells have been previously linked to MAIT cell activation and expansion (Treiner et al. 2003), implying that they might express MR1 and stimulate MAIT cells in the periphery. However, when the Hansen group stained several B cell lines to determine MR1 levels on the surface, they found little or no surface expression (Chua et al. 2011). Intriguingly, corroborating a previous study (Miley et al. 2003), the authors discovered that these cells have high intracellular expression of endogenous MR1. While the reasons for this remain unclear, one possible explanation could be that MR1 does not traffic to the cell surface efficiently due to low ligand availability. In agreement with this, infection of B cell lines with bacteria that carry ligands capable of binding to MR1 (discussed in further detail below) led to the expression of MR1 molecules on the cell surface (Salerno-Goncalves et al. 2014).
MAIT cell development in the thymus is also dependent upon MR1 expression (Treiner et al. 2003). Yet, the inability to detect MR1 on any cell surface obscured the cell type responsible for selecting MAIT cells. Work by two separate groups hinted that the thymic cells which expressed MR1 were the double positive (DP) thymocytes (Chua et al. 2011; Gold et al. 2013), in good agreement with their high MR1 mRNA expression level compared to other thymic cell populations (Seach et al. 2013). Furthermore, through the use of bone marrow chimera and thymic organ cultures, the Lantz lab demonstrated that DP cells are responsible for the positive selection of MAIT cells (Seach et al. 2013). Such a selection mechanism is reminiscent of iNKT cells that are also selected on CD1d-expressing DP thymocytes (Gapin 2016), and could directly affect the phenotype and/or functionality of the selected MAIT cells.
Antigens Presented by MR1
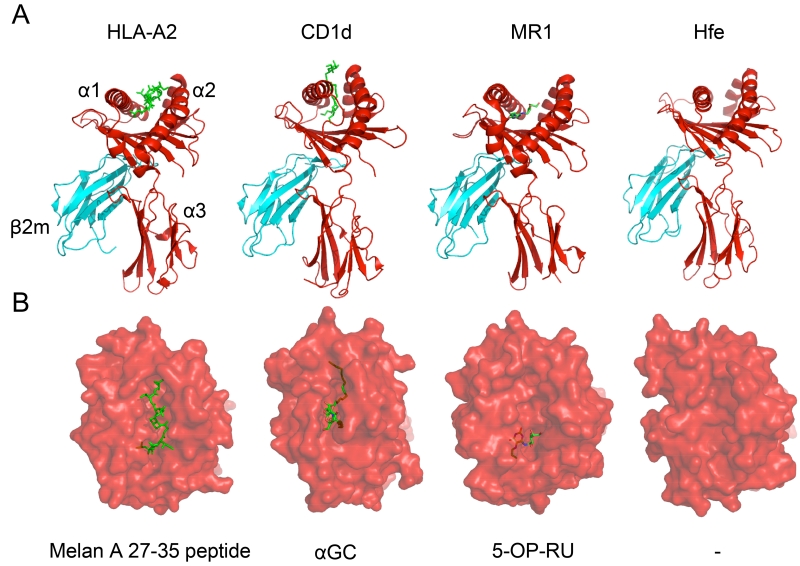
One of the distinctive features of classical MHC molecules is their ability to present peptides to certain T cells as a way to discriminate self from non-self. Antigen presentation is similarly important for MHC-Ib molecules, although they can present more than simply peptide fragments. Once MR1 was discovered and introduced as a member of the MHC-Ib family, investigators were uncertain as to what the antigens presented by this molecule were. The only known antigens presented by MHC or MHC-like molecules at the time were peptides and lipids but the MR1-bound antigen was protease-resistant and did not purify in the lipid fractions, perplexing investigators further (Young et al. 2013). However, there were several reasons to suggest that MR1 was indeed presenting a ligand and did not just serve to provide a danger signal. Firstly, unlike some MHC-Ib molecules such as Hfe (Lebron et al. 1998), MR1 does not have an antigen binding pocket that is flat and empty (Figure 2). Rather, its small ligand binding pocket is surrounded by aromatic and basic residues, indicating a small molecule could fit in the groove and stimulate MAIT cells (Kjer-Nielsen et al. 2012; Lopez-Sagaseta et al. 2013a). Secondly, when Hansen and colleagues used an epitope tagging strategy to distinguish between “open” (MR1 without a ligand) vs. “closed” (MR1 occupied by a ligand) conformations of MR1, they discovered that MR1 (similar to MHC-Ia molecules) undergoes ligand-induced folding (Huang et al. 2005; Miley et al. 2003). Finally, MAIT hybridoma cell stimulation was abrogated when many of the putative ligand binding residues of MR1 were mutated (Huang et al. 2005), hinting that the presumed ligand could no longer interact with MR1.
Figure 2. Structural Views of Ligands Bound to Different MHC-Ia and MHC-Ib Proteins.
In order to demonstrate the similarities and differences between MHC-Ia and MHC-Ib molecules, the HLA-A2 (PDB: 2GTW), hCD1d (PDB: 3SDX), hMR1 (PDB: 4PIC), and hHFE (PDB: 1A6Z) proteins are provided as examples. In panel A, the heavy chain is displayed in red with its three structural domains also denoted. The monomorphic β2m molecule, which associates with most MHC-Ib molecules, is portrayed in cyan as it interacts with the α3 domain of the MHC molecules. Finally, the antigen for each protein (except for hHFE) is illustrated in neon green. All proteins adopt a similar fold and use two α-helices to form the ligand-binding pocket while anti-parallel β-strands form the floor of the pocket. The pocket for hCD1d is much deeper than the ones present in HLA-A2 and hMR1, distinguishing the antigens each molecule presents. In panel B, a top-down illustration is used to represent the view of the antigen-MHC complex from the TCR’s perspective. HLA-A2 is shown presenting a melanoma-specific protein while hCD1d and hMR1 are presenting their prototypic stimulatory antigens α-galactosylceramide (α-GC) and 5-OP-RU, respectively. hHFE possesses too narrow and flat a groove to appropriately present an antigen. In this representation, it is easy to visualize that while HLA-A2 presents an antigen that spans the length of its groove, hMR1 presents an antigen that is sequestered exclusively in the A’ pocket.
Further work demonstrated that MAIT cells could be activated in a MR1-dependent manner by antigen presenting cells in the presence of some bacteria and yeasts but not viruses (Gold et al. 2010; Le Bourhis et al. 2010), yet the antigenic compounds unique to these microbes continued to be elusive. Eventually, in a seminal study by the McCluskey and Rossjohn groups, using various compounds to properly refold MR1, the authors discovered that vitamin metabolites, primarily derived from riboflavin (vitamin B2), were the antigens being presented by MR1 to MAIT cells (Kjer-Nielsen et al. 2012). Many bacteria harbor the genes required to synthesize vitamins and infection of antigen presenting cells with any of these microbes lead to the activation of MAIT cells. Thus, vitamins metabolites now provide another basis for self vs. non-self discrimination by the immune system since mammals do not naturally produce vitamins and the presence of these vitamin metabolites might serve as a sensor for infection (Birkinshaw et al. 2014).
The first antigen that was discovered to bind to MR1 was 6-formyl pterin (6-FP), which is a photo-degradation product of folic acid (vitamin B9). Intriguingly, in contrast to how other ligands interact with their respective MHC molecules, 6-FP forms a covalent Schiff base with one of the residues in the antigen-binding cleft for MR1 (K43), providing substantial stability for this complex (Kjer-Nielsen et al. 2012). When Kjer-Nielsen et al. solved the structure of 6-FP bound to MR1, it further clarified how the antigen bound to MR1 was discovered to be protease resistant. With bulky amino acids lining the antigen-binding pocket, the groove is too small to accommodate peptides (Figure 2). It similarly would not have been able to bind to hydrophobic lipids because of the charged residues surrounding the pocket. The authors, however, did encounter a roadblock in their analyses. While MR1 could fold properly with 6-FP loaded as the antigen, this metabolite was unable to stimulate the MAIT hybridoma clones against which they tested this antigen.
To successfully identify the stimulating ligand(s), the authors looked for other compounds with the pterin or analogous moieties by refolding MR1 in the presence of Salmonella typhimurium. This bacterium possesses a functional riboflavin synthesis pathway and MAIT cells are stimulated in its presence (Reantragoon et al. 2012). Thus, the authors reasoned that by using the bacterial supernatant to refold MR1 and subsequently performing mass spectrometry on the antigens bound to MR1, they could determine the identity of the stimulatory ligand. Eventually, they obtained reduced 6-hydroxymethyl-8-D-ribityllumazine (rRL-6-CH2OH), a derivative of riboflavin, as one of their products and it potently stimulated MAIT cells in a MR1-dependent manner. Thus, the ribityl group linked to this lumazine, a compound structurally similar to pterin, moiety, was essential for MAIT TCR recognition of the MR1-antigen complex (Kjer-Nielsen et al. 2012). Yet, what remained unresolved was how the stimulatory ligand was generated as an antigen since it had not been described as part of the riboflavin synthesis pathway. In a separate study, the same groups used a clever genetic approach and concluded that the antigen presented to MAIT cells during bacterial infections was in fact a riboflavin precursor that formed an adduct with intermediates from the host or microbial glycolytic pathway (Corbett et al. 2014). Interestingly, this chimeric antigen is an isomer of the stimulatory ligand previously described (Corbett et al. 2014). Additionally, a synthesized form of this antigen (5-OP-RU) has since been shown to bind to MR1 and stimulate MAIT cells when presented by MR1 expressed on APCs (Corbett et al. 2014; Gherardin et al. 2016; McWilliam et al. 2016). However, rRL-6-CH2OH has also previously been isolated from Escherichia coli supernatant and determined to be the primary stimulatory ligand bound to MR1 (Lopez-Sagaseta et al. 2013b). Thus, the origin of this antigen in E. coli remains unknown and requires further investigation.
Fluorescent MR1 tetramers loaded with 5-OP-RU label the vast majority of MAIT cells arguing that most, if not all, MAIT cells recognize this antigen in the context of MR1 (Gherardin et al. 2016). Nevertheless, whether this is the only antigen that MR1 can present and to which MAIT cells can respond remains unknown. Recently, the Rossjohn group stained human peripheral blood mononuclear cells (PBMCs) with various antigens loaded onto MR1 tetramers and consistently observed a small population of cells which stained with 6-FP/MR1 tetramers (Gherardin et al. 2016). Thus, in retrospect, the reason why the authors initially observed no stimulation with 6-FP as the antigen was due to incomplete sampling of MAIT cells since the 6-FP/MR1 tetramer+ cells only comprise ~0.1% of the T cells in PBMCs.
Antigens bound to MR1 discovered hitherto chiefly occupy the A’ pocket of the MR1 groove (Figure 2), opening up the possibility that larger ligands or ligands occupying the F’ pocket of the groove could be novel antigens (Eckle et al. 2015). Such antigens could promote the development of diverse subsets of MAIT cells with distinctive phenotypes, each of which might respond to in vivo bacterial infections to varying degrees. Already, the phenotypes of the 6-FP reactive MAIT cells are not identical to the phenotypes of the 5-OP-RU-specific cells (Gherardin et al. 2016). Further work needs to be performed in this area to characterize the full scope of ligands that could be captured and presented by MR1.
Finally, although evidence points to DP cells as the cells mediating the positive selection of MAIT cells in the thymus (Seach et al. 2013), the antigen(s) MR1 might present during selection is unknown. Many human commensal gut bacteria possess the riboflavin synthesis pathway (LeBlanc et al. 2013) and consequently, could potentially provide a steady supply of antigens that could be captured by MR1 to select MAIT cells. Yet, it is unclear how DP cells would be able to specifically take up the riboflavin precursors or the chimeric adducts in order to present these antigens for the selection of MAIT cells. Additionally, the development of MAIT cells has been observed in thymic organ cultures carried out in sterile conditions in vitro in the absence of any exogenously added ligands (Seach et al. 2013). These results suggest the possibility that endogenous ligand for MR1 might exist and be responsible for the selection of MAIT cells. Such hypothesis would be in agreement with the “autoreactivity” that some MAIT cell clones exhibited originally for cell lines engineered to overexpress MR1 molecules (Treiner et al. 2003). Whether these hypothetical “self” ligands will be structurally similar to the bacterial ligands is currently unknown, although one MAIT TCR has been shown to use different residues when responding to MR1-overexpressing cell lines in the presence or not of bacterial ligands, suggesting the possibility that they might be structurally different (Young et al. 2013).
MR1 Intracellular Trafficking and Antigen Loading
Nascent MHC-Ia molecules undergo proper folding and peptide loading in the ER. Soon thereafter, they are transported to the surface where they can present these peptides, whether self or foreign, to T cells (Neefjes et al. 2011). While this pathway has been studied for many years and has largely been characterized, the path MR1 takes from when it is translated to when it can travel to the surface has not yet been completely clarified. In contrast to MHC-Ia molecules, it appears that intracellular MR1 proteins are primarily sequestered in the ER. A high percentage of the protein is endoglycosidase H sensitive, indicating that intracellular MR1 is an ER-resident protein (Miley et al. 2003). In support of this, a recent study has demonstrated that MR1 is trapped in the ER in an incompletely folded conformation free of β2m until it can bind to its antigen. Upon binding to antigen, MR1 folds properly, associates with β2m and travels to the surface, furthering the claim that antigen availability truly does drive its surface expression (McWilliam et al. 2016).
Previously, MR1 has also been co-immunoprecipitated with many proteins involved in the peptide-loading complex (PLC) such as TAP, ERp57, and calnexin, all of which are ER-resident proteins (Miley et al. 2003). While this fits with the notion that MR1 is stuck in the ER intracellularly, this piece of evidence presents as an oddity since MR1 itself does not present peptides and can still be expressed on the surface of TAP-/- cells, which are deficient in MHC-Ia cell surface expression (Tilloy et al. 1999; Treiner et al. 2003). Perhaps one explanation, as noted when MR1 was first identified (Hashimoto et al. 1995), is that since MR1 has more sequence similarity to MHC-Ia molecules than to other MHC-Ib molecules (Figure 1), it bears the appropriate motifs to associate with the PLC. Therefore, while it does not require the PLC in order to be loaded with antigen, it might still interact with the complex merely due to sequence similarity.
Although MR1 has been demonstrated to traffic to the surface directly from the ER upon antigen binding, whether or not this reflects its natural trafficking pattern is unclear. Purified antigen was introduced exogenously in this system to promote MR1 folding and translocation to the surface (McWilliam et al. 2016). However, in the context of a pathogenic infection, how efficiently the MR1 ligand is transported to the ER currently remains unresolved. In addition, because at least one of the MAIT antigens is a chimeric product that requires the presence of host/microbial metabolic products (Corbett et al. 2014), the local concentration of the precursor molecules might determine the amount of 5-OP-RU generated. Thus, using synthesized antigen at high concentrations does not provide a complete picture for how antigen is efficiently captured and presented by MR1 in an infectious setting. Furthermore, given that many bacteria are internalized and lysed in the late endosomes/lysosomes (Stuart and Ezekowitz 2005), how the antigenic compounds might be transported to the ER remains unknown.
Strikingly, the Hansen group has previously observed that MR1 was also found in the late endosomal and lysosomal vesicles in association with invariant chain (Ii) (Huang et al. 2008). This protein has been historically coupled to assisting MHC-II molecules in trafficking to the endosome for antigen loading (Fortin et al. 2013). Notably, while MAIT cells continue to develop in Ii-/- mice (Treiner et al. 2003), MAIT cell stimulation was significantly augmented when Ii was overexpressed in the APCs expressing MR1, raising the question as to how this molecule was affecting MR1 trafficking and antigen presentation. Transmembrane proteins which are sorted into endosomal or lysosomal compartments contain specific sequences in their cytosolic regions called tyrosine signal-based motifs or dileucine-based motifs (Bonifacino and Traub 2003). CD1d is one such MHC-Ib protein that contains the tyrosine signal-based motif to target it to endosomes (Jayawardena-Wolf et al. 2001). MR1 lacks any such motif in its C-terminus and its physical association with Ii could help promote its effective trafficking to the late endosome (Huang et al. 2008). Thus, this mechanism might allow MR1 to directly have access to the locations where its cognate antigen concentration is highest. While MR1 has been shown to localize in endosomes upon internalization from the surface and undergo antigen exchange (McWilliam et al. 2016), direct transport to this compartment from the ER could serve as a pathway to efficiently present non-self ligands when they are limiting.
MAIT TCR Interaction with MR1-Ag
In order to interact with the myriad of potential antigens an individual might encounter, αβ TCRs have a theoretical diversity estimated to exceed 1015 unique sequences (Davis and Bjorkman 1988). Due to the nature of how these receptors are generated, the diversity is largely concentrated in the complementarity determining region 3 (CDR3) loops of both the TCRα and TCRβ chains (Schatz 2004), which are the loops that focus on the different antigens presented by MHC-Ia or MHC-Ib molecules. Through this mechanism, the most diverse parts of the TCR (the CDR3 loops) interact with the most diverse portions of the MHC molecule (antigen). MAIT cells, however, sample a restricted set of sequences from this enormous pool, likely because they interact with and recognize a limited set of ligands, which are precursors or byproducts of vitamin synthesis pathways. To add to the growing list of similarities between MAIT cells and iNKT cells, the TCRα chains for MAIT cells also primarily use a specific rearrangement between TRAV1-2 (TRAV1 in mice) and TRAJ33 (Rahimpour et al. 2015; Reantragoon et al. 2013). While the CDR3α sequence thus generated is not always exactly identical, the length is strictly conserved (Le Bourhis et al. 2013b). Interestingly, the TCRα chain utilized by MAIT cells has been previously shown to be similarly conserved in mammalian evolution through the use of orthologous gene rearrangements (Young et al. 2013). However, recent work has suggested that the TCRα rearrangements in MAIT cells are not thoroughly conserved in humans as several other TRAJ genes were found rearranged with the TRAV1-2 gene (Gold et al. 2014). Such diversity could provide increased coverage to different pathogens, which might each give rise to unique antigens bound to MR1. The TCRβ genes utilized by MAIT cells are predominantly TRBV6 and TRBV20 (TRBV13 and TRBV19 in mice) but these rearrangements are far more diverse in their CDR3 regions when compared to the TCRα chain with which they pair, although they still remain oligoclonal (Lepore et al. 2014). It is noteworthy that both iNKT cells and MAIT cells possess an invariant TCRα chain paired to diverse TCRβ chains. One possible explanation for this could be that because TCRβ chains are generated prior to TCRα chains during T cell development (Schatz and Ji 2011), an immature thymocyte with a functional TCRβ chain is not yet fated to develop into a specific subpopulation of T cells. Additionally, since immature thymocytes undergo a burst of proliferation after generating a functional TCRβ chain (Kreslavsky et al. 2012), many cells would exist with that specific TCRβ rearrangement, each of which can produce and pair with a completely unique TCRα chain. Upon TCRα rearrangement and expression, the complete TCR will then undergo selection on its appropriate ligand. Accordingly, the TCRα chain could provide specificity for a complex while the TCRβ chain could fine-tune the overall affinity. The MAIT TCRα chain, consequently, would be predicted to interact primarily with the antigen presented by MR1 to provide specificity.
In support of this, when the TCR-MR1-Ag complex structures were solved, one crucial residue in the CDR3α, Y95, contributed by TRAJ33 formed the major hydrogen bond with the ribityl moiety of the antigen (Lopez-Sagaseta et al. 2013a; Lopez-Sagaseta et al. 2013b; Patel et al. 2013). Surprisingly, when the structure was solved for the same TCR interacting with MR1 presenting the non-stimulatory ligand 6-FP, the Y95 could no longer form this hydrogen bond with the antigen, stressing the exquisite sensitivity of the TCR for the right antigen. The TCRβ chain was also observed to hover around the antigen and the CDR3β provided a key contact with the stimulatory ligand (Corbett et al. 2014). Thus, given that the MAIT TCRα chain can make its required contacts, the TCRβ chain can further strengthen this interaction and enhance the signal perceived by MAIT cells. Furthermore, this lends credence to the idea that individual MAIT cells might respond in distinct manners because of the individual TCRβ chains’ capabilities for ligand discrimination.
When MR1-tetramers loaded with an acetylated version of 6-FP were used to stain human PBMCs, a well-defined T cell population was revealed to be tetramer+ (Gherardin et al. 2016). Strangely, the majority of these cells were in fact TRAV1-2-. These T cells instead expressed a diverse array of TCRα chains, none of which possessed the Y95 residue in the CDR3α. Additionally, when these “atypical” MAIT cells were further phenotyped, they shared little similarity to typical MAIT cells and instead were phenotypically comparable to conventional T cells (Gherardin et al. 2016). One possible explanation could be that these cells were in fact not MAIT cells at all but rather conventional αβ T cells with TCRs that happened to interact with MR1 bound to antigen. Thus, they could have been selected on MHC-Ia molecules (since they were overwhelmingly CD8+) yet bore TCRs that could cross react with MR1 and its antigen. Though their role in immune responses is unclear, it is important not to conflate the ability of the TCR to interact with the MR1-Ag complex to the ability of the cell to respond to that stimulus. These cells could certainly follow the kinetics of conventional T cell activation before responding, even when stimulated by MR1, unlike typical MAIT cells, which secrete cytokines promptly upon stimulation (Kawachi et al. 2006).
In one of the few yet major differences between iNKT cell antigen recognition and MAIT cell antigen recognition, the MAIT TCR adopts a docking conformation that is orthogonal/diagonal to the axis created by the antigen-binding pocket (Gherardin et al. 2016; Patel et al. 2013). iNKT TCRs, on the other hand, dock in a parallel manner to CD1d presenting lipids, placing the CDR3α chain in a key location to dominate the interaction (Bhati et al. 2014). However, in the case of the MAIT TCR, while the CDR3α (or CDR1α for atypical MAIT TCRs) chain plays a larger role in antigen recognition, the CDR3β chain also participates, much like how both chains contribute to antigen recognition for TCRs from conventional T cells (Eckle et al. 2015). Even prior to the co-crystal of the TCR-MR1-Ag being solved, there was mutational evidence suggesting that the MAIT TCR interacted with MR1 in a manner similar to how CD8+ T-cell TCRs interact with MHC-Ia molecules (Huang et al. 2005). Using surface plasmon resonance (SPR), the affinity of MAIT TCRs for their cognate ligands was determined to be comparable to conventional TCR affinities for pMHC-Ia ligands (Patel et al. 2013) but lower than those observed for iNKT TCRs for lipid-CD1d (Wun et al. 2008). This is slightly puzzling since in a system where a MAIT TCR was reconstituted into a TCR- T-cell line, the cells were able to respond to stimulatory antigen with great sensitivity (Eckle et al. 2014). One explanation for this discrepancy could be that in situ TCR affinities for their cognate ligands have been previously shown to be substantially higher than their in vitro measured affinities (Huppa et al. 2010). Thus, even though the SPR measurements for MAIT TCRs complexed to antigen/MR1 are lower than those measured for iNKT TCRs in complex with their ligand, the MAIT cells themselves could respond with high sensitivity when stimulated by antigens presented by MR1. Various factors, such as the adhesion molecules maintaining the immune synapse, can increase the apparent affinities of TCRs for their ligands in situ. The co-receptors on T-cells, CD4 and CD8, for example, improve the affinities by interacting with MHC-II and MHC-I, respectively, and consequently, lengthening the dwell times of the TCRs on their ligands (Gao et al. 2002). In both humans and mice, many MAIT cells have been identified to express CD8 (Gherardin et al. 2016; Rahimpour et al. 2015). Since MR1 is part of the MHC-Ib family, CD8 could be hypothesized to stabilize the MAIT TCR/MR1 interaction by binding to MR1. Indeed, the region with acidic residues present in the α3 domain of MHC-Ia molecules that interacts with CD8 is also found in MR1 (Riegert et al. 1998; Walter and Gunther 1998). In addition, structurally, the CD8 co-receptor has a similar footprint on MR1 as it does on MHC-Ia molecules (Reantragoon et al. 2013). Furthermore, when CD8 was not allowed to engage MR1 due to the presence of a blocking antibody, Mtb-reactive CD8+ MAIT cell clones could not respond to Mtb-infected MR1+ APCs (Gold et al. 2013), thus demonstrating a critical role for CD8 in MAIT cell responses. However, many MAIT cells do not express any co-receptor on their surfaces (Gherardin et al. 2016). The TCRs expressed by these MAIT cells could have potentially higher affinities for MR1 and antigen to compensate for the absence of CD8. Future work will hopefully resolve how the CD8+ and CD8- MAIT cell TCR repertoires differ and whether any such differences amount to functional differences between the two populations as well.
Concluding Remarks
Innate-like T cell populations have long been considered attractive candidates for immune modulation (Cerundolo et al. 2009). Their conserved TCR usage, recognition of ligands with limited polymorphisms, and swift responses upon activation meant that these cells could be specifically targeted to quickly produce pro-inflammatory cytokines. Yet, the paucity of these specialized populations in addition to a lack of known markers to precisely identify these cells in humans has hampered the development of immunotherapies to exploit these cells. MAIT cells, however, do not conform to that stereotype as they are easily found in large numbers in human mucosal tissues and blood. Additionally, with the advent of antigen-loaded MR1 tetramers, they can now be specifically tracked and characterized. Their prevalence in humans has significantly contributed to their appeal, resulting in enormous interest in understanding these cells and how they respond to antigenic stimuli with a future hope to artificially direct their responses in predictable ways. From this perspective, gaining insight into the antigen presenting ligand, MR1, is fundamental to ultimately influence MAIT cell responses.
MR1 is not unlike its MHC-Ia counterparts from a broad structural point of view (Birkinshaw et al. 2014). Its monomorphism, though, sets it apart from conventional MHC-I proteins and has instead drawn parallels to pattern recognition receptors (Birkinshaw et al. 2014; Huang et al. 2009). By capturing and presenting vitamin metabolites to MAIT cells, MR1 can quickly activate its lymphocyte population similar to how TLRs binding their ligands can quickly activate innate immune cells. This comparison rings even truer since like TLRs, MR1 is specific for its antigen without being pathogen-specific.
But how can the power of MR1 and MAIT cells be harnessed? Frequently, when an immunodominant antigen for T cells is known, the host can be vaccinated with this antigen to generate a memory T cell population, which can then rapidly respond when the host encounters the actual pathogen possessing the antigen. However, MAIT cells already express markers associated with memory cells, such as CD44hi and CD62Llo (Martin et al. 2009; Rahimpour et al. 2015), which might make a vaccine strategy futile since the cells would respond with kinetics reminiscent of memory cells even prior to vaccination. Perhaps, though, since MAIT cells can bridge the adaptive and innate immune responses, MAIT cell antigens could be provided as adjuvants along with other peptidic antigens that are immunodominant epitopes in pathogens. By doing so, MAIT cells that respond would skew the conventional T cell response to the TH1/TH17 axes by preferentially secreting IFNγ and IL-17 (Tang et al. 2013). Employing this strategy could indeed generate a memory population of conventional T cells specific for a pathogen, especially ones that tend to infect mucosal tissues. Currently, whether the MAIT cell response can be modulated to exclusively the TH1 or the TH17 axis based on the stimulating antigen is unknown. Thus, it remains crucial to explore and identify any novel ligands that can be bound to MR1 that might specifically stimulate (certain) MAIT cells to secrete selective cytokines. In so doing, we will be able to better manipulate this arm of the immune system to influence immunity on a broader scale.
Acknowledgments
Research in the Gapin’s laboratory is supported by National Institute of Health grant (AI103736). We would like to acknowledge current and past members of the laboratory for discussions and Drs. Jennifer Matsuda and Manfred Brigl for critical reading of the manuscript. We apologize to colleagues whose works were not cited due to space constraints or omission.
References
- Amadou C, Younger RM, Sims S, Matthews LH, Rogers J, Kumanovics A, Ziegler A, Beck S, Lindahl KF. Co-duplication of olfactory receptor and MHC class I genes in the mouse major histocompatibility complex. Hum Mol Genet. 2003;12:3025–40. doi: 10.1093/hmg/ddg317. [DOI] [PubMed] [Google Scholar]
- Bhati M, Cole DK, McCluskey J, Sewell AK, Rossjohn J. The versatility of the alphabeta T-cell antigen receptor. Protein Sci. 2014;23:260–72. doi: 10.1002/pro.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkinshaw RW, Kjer-Nielsen L, Eckle SB, McCluskey J, Rossjohn J. MAITs, MR1 and vitamin B metabolites. Curr Opin Immunol. 2014;26:7–13. doi: 10.1016/j.coi.2013.09.007. [DOI] [PubMed] [Google Scholar]
- Bjorck L, Miorner H, Kuhnemund O, Kronvall G, Sundler R. On the interaction between beta 2-microglobulin and group A streptococci. Scand J Immunol. 1984;20:69–79. doi: 10.1111/j.1365-3083.1984.tb00979.x. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- Brossay L, Chioda M, Burdin N, Koezuka Y, Casorati G, Dellabona P, Kronenberg M. CD1d-mediated recognition of an alpha-galactosylceramide by natural killer T cells is highly conserved through mammalian evolution. J Exp Med. 1998;188:1521–8. doi: 10.1084/jem.188.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerundolo V, Silk JD, Masri SH, Salio M. Harnessing invariant NKT cells in vaccination strategies. Nat Rev Immunol. 2009;9:28–38. doi: 10.1038/nri2451. [DOI] [PubMed] [Google Scholar]
- Chua WJ, Kim S, Myers N, Huang S, Yu L, Fremont DH, Diamond MS, Hansen TH. Endogenous MHC-related protein 1 is transiently expressed on the plasma membrane in a conformation that activates mucosal-associated invariant T cells. J Immunol. 2011;186:4744–50. doi: 10.4049/jimmunol.1003254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett AJ, Eckle SB, Birkinshaw RW, Liu L, Patel O, Mahony J, Chen Z, Reantragoon R, Meehan B, Cao H, Williamson NA, Strugnell RA, Van Sinderen D, Mak JY, Fairlie DP, Kjer-Nielsen L, Rossjohn J, McCluskey J. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 2014;509:361–5. doi: 10.1038/nature13160. [DOI] [PubMed] [Google Scholar]
- Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- Eckle SB, Birkinshaw RW, Kostenko L, Corbett AJ, McWilliam HE, Reantragoon R, Chen Z, Gherardin NA, Beddoe T, Liu L, Patel O, Meehan B, Fairlie DP, Villadangos JA, Godfrey DI, Kjer-Nielsen L, McCluskey J, Rossjohn J. A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J Exp Med. 2014;211:1585–600. doi: 10.1084/jem.20140484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckle SB, Corbett AJ, Keller AN, Chen Z, Godfrey DI, Liu L, Mak JY, Fairlie DP, Rossjohn J, McCluskey J. Recognition of Vitamin B Precursors and Byproducts by Mucosal Associated Invariant T Cells. J Biol Chem. 2015;290:30204–11. doi: 10.1074/jbc.R115.685990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin JS, Cloutier M, Thibodeau J. Exposing the Specific Roles of the Invariant Chain Isoforms in Shaping the MHC Class II Peptidome. Front Immunol. 2013;4:443. doi: 10.3389/fimmu.2013.00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao GF, Rao Z, Bell JI. Molecular coordination of alphabeta T-cell receptors and coreceptors CD8 and CD4 in their recognition of peptide-MHC ligands. Trends Immunol. 2002;23:408–13. doi: 10.1016/s1471-4906(02)02282-2. [DOI] [PubMed] [Google Scholar]
- Gapin L. Development of invariant natural killer T cells. Curr Opin Immunol. 2016;39:68–74. doi: 10.1016/j.coi.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraghty DE, Koller BH, Pei J, Hansen JA. Examination of four HLA class I pseudogenes. Common events in the evolution of HLA genes and pseudogenes. J Immunol. 1992;149:1947–56. [PubMed] [Google Scholar]
- Gherardin NA, Keller AN, Woolley RE, Le Nours J, Ritchie DS, Neeson PJ, Birkinshaw RW, Eckle SB, Waddington JN, Liu L, Fairlie DP, Uldrich AP, Pellicci DG, McCluskey J, Godfrey DI, Rossjohn J. Diversity of T Cells Restricted by the MHC Class I-Related Molecule MR1 Facilitates Differential Antigen Recognition. Immunity. 2016;44:32–45. doi: 10.1016/j.immuni.2015.12.005. [DOI] [PubMed] [Google Scholar]
- Gold MC, Cerri S, Smyk-Pearson S, Cansler ME, Vogt TM, Delepine J, Winata E, Swarbrick GM, Chua WJ, Yu YY, Lantz O, Cook MS, Null MD, Jacoby DB, Harriff MJ, Lewinsohn DA, Hansen TH, Lewinsohn DM. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. 2010;8:e1000407. doi: 10.1371/journal.pbio.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold MC, Eid T, Smyk-Pearson S, Eberling Y, Swarbrick GM, Langley SM, Streeter PR, Lewinsohn DA, Lewinsohn DM. Human thymic MR1-restricted MAIT cells are innate pathogen-reactive effectors that adapt following thymic egress. Mucosal Immunol. 2013;6:35–44. doi: 10.1038/mi.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold MC, McLaren JE, Reistetter JA, Smyk-Pearson S, Ladell K, Swarbrick GM, Yu YY, Hansen TH, Lund O, Nielsen M, Gerritsen B, Kesmir C, Miles JJ, Lewinsohn DA, Price DA, Lewinsohn DM. MR1-restricted MAIT cells display ligand discrimination and pathogen selectivity through distinct T cell receptor usage. J Exp Med. 2014;211:1601–10. doi: 10.1084/jem.20140507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfinch N, Reinink P, Connelley T, Koets A, Morrison I, Van Rhijn I. Conservation of mucosal associated invariant T (MAIT) cells and the MR1 restriction element in ruminants, and abundance of MAIT cells in spleen. Vet Res. 2010;41:62. doi: 10.1051/vetres/2010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TH, Huang S, Arnold PL, Fremont DH. Patterns of nonclassical MHC antigen presentation. Nat Immunol. 2007;8:563–8. doi: 10.1038/ni1475. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Hirai M, Kurosawa Y. A gene outside the human MHC related to classical HLA class I genes. Science. 1995;269:693–5. doi: 10.1126/science.7624800. [DOI] [PubMed] [Google Scholar]
- Howson LJ, Salio M, Cerundolo V. MR1-Restricted Mucosal-Associated Invariant T Cells and Their Activation during Infectious Diseases. Front Immunol. 2015;6:303. doi: 10.3389/fimmu.2015.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Gilfillan S, Cella M, Miley MJ, Lantz O, Lybarger L, Fremont DH, Hansen TH. Evidence for MR1 antigen presentation to mucosal-associated invariant T cells. J Biol Chem. 2005;280:21183–93. doi: 10.1074/jbc.M501087200. [DOI] [PubMed] [Google Scholar]
- Huang S, Gilfillan S, Kim S, Thompson B, Wang X, Sant AJ, Fremont DH, Lantz O, Hansen TH. MR1 uses an endocytic pathway to activate mucosal-associated invariant T cells. J Exp Med. 2008;205:1201–11. doi: 10.1084/jem.20072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Martin E, Kim S, Yu L, Soudais C, Fremont DH, Lantz O, Hansen TH. MR1 antigen presentation to mucosal-associated invariant T cells was highly conserved in evolution. Proc Natl Acad Sci U S A. 2009;106:8290–5. doi: 10.1073/pnas.0903196106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Ota T, Nei M. Positive Darwinian selection promotes charge profile diversity in the antigen-binding cleft of class I major-histocompatibility-complex molecules. Mol Biol Evol. 1990;7:515–24. doi: 10.1093/oxfordjournals.molbev.a040626. [DOI] [PubMed] [Google Scholar]
- Huppa JB, Axmann M, Mortelmaier MA, Lillemeier BF, Newell EW, Brameshuber M, Klein LO, Schutz GJ, Davis MM. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature. 2010;463:963–7. doi: 10.1038/nature08746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayawardena-Wolf J, Benlagha K, Chiu YH, Mehr R, Bendelac A. CD1d endosomal trafficking is independently regulated by an intrinsic CD1d-encoded tyrosine motif and by the invariant chain. Immunity. 2001;15:897–908. doi: 10.1016/s1074-7613(01)00240-0. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–82. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kawachi I, Maldonado J, Strader C, Gilfillan S. MR1-restricted V alpha 19i mucosal-associated invariant T cells are innate T cells in the gut lamina propria that provide a rapid and diverse cytokine response. J Immunol. 2006;176:1618–27. doi: 10.4049/jimmunol.176.3.1618. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L, Borg NA, Pellicci DG, Beddoe T, Kostenko L, Clements CS, Williamson NA, Smyth MJ, Besra GS, Reid HH, Bharadwaj M, Godfrey DI, Rossjohn J, McCluskey J. A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition. J Exp Med. 2006;203:661–73. doi: 10.1084/jem.20051777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Bhati M, Chen Z, Kostenko L, Reantragoon R, Williamson NA, Purcell AW, Dudek NL, McConville MJ, O’Hair RA, Khairallah GN, Godfrey DI, Fairlie DP, Rossjohn J, McCluskey J. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012;491:717–23. doi: 10.1038/nature11605. [DOI] [PubMed] [Google Scholar]
- Klein J, Sato A, O’HUigin C. Evolution by gene duplication in the major histocompatibility complex. Cytogenet Cell Genet. 1998;80:123–7. doi: 10.1159/000014967. [DOI] [PubMed] [Google Scholar]
- Kreslavsky T, Gleimer M, Miyazaki M, Choi Y, Gagnon E, Murre C, Sicinski P, von Boehmer H. beta-Selection-induced proliferation is required for alphabeta T cell differentiation. Immunity. 2012;37:840–53. doi: 10.1016/j.immuni.2012.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016 doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bourhis L, Dusseaux M, Bohineust A, Bessoles S, Martin E, Premel V, Core M, Sleurs D, Serriari NE, Treiner E, Hivroz C, Sansonetti P, Gougeon ML, Soudais C, Lantz O. MAIT cells detect and efficiently lyse bacterially-infected epithelial cells. PLoS Pathog. 2013a;9:e1003681. doi: 10.1371/journal.ppat.1003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bourhis L, Martin E, Peguillet I, Guihot A, Froux N, Core M, Levy E, Dusseaux M, Meyssonnier V, Premel V, Ngo C, Riteau B, Duban L, Robert D, Huang S, Rottman M, Soudais C, Lantz O. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol. 2010;11:701–8. doi: 10.1038/ni.1890. [DOI] [PubMed] [Google Scholar]
- Le Bourhis L, Mburu YK, Lantz O. MAIT cells, surveyors of a new class of antigen: development and functions. Curr Opin Immunol. 2013b;25:174–80. doi: 10.1016/j.coi.2013.01.005. [DOI] [PubMed] [Google Scholar]
- LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol. 2013;24:160–8. doi: 10.1016/j.copbio.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Lebron JA, Bennett MJ, Vaughn DE, Chirino AJ, Snow PM, Mintier GA, Feder JN, Bjorkman PJ. Crystal structure of the hemochromatosis protein HFE and characterization of its interaction with transferrin receptor. Cell. 1998;93:111–23. doi: 10.1016/s0092-8674(00)81151-4. [DOI] [PubMed] [Google Scholar]
- Lepore M, Kalinichenko A, Colone A, Paleja B, Singhal A, Tschumi A, Lee B, Poidinger M, Zolezzi F, Quagliata L, Sander P, Newell E, Bertoletti A, Terracciano L, De Libero G, Mori L. Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRbeta repertoire. Nat Commun. 2014;5:3866. doi: 10.1038/ncomms4866. [DOI] [PubMed] [Google Scholar]
- Lion J, Debuysscher V, Wlodarczyk A, Hodroge A, Serriari NE, Choteau L, Ouled-haddou H, Plistat M, Lassoued K, Lantz O, Treiner E. MR1B, a natural spliced isoform of the MHC-related 1 protein, is expressed as homodimers at the cell surface and activates MAIT cells. Eur J Immunol. 2013;43:1363–73. doi: 10.1002/eji.201242461. [DOI] [PubMed] [Google Scholar]
- Lopez-Sagaseta J, Dulberger CL, Crooks JE, Parks CD, Luoma AM, McFedries A, Van Rhijn I, Saghatelian A, Adams EJ. The molecular basis for Mucosal-Associated Invariant T cell recognition of MR1 proteins. Proc Natl Acad Sci U S A. 2013a;110:E1771–8. doi: 10.1073/pnas.1222678110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Sagaseta J, Dulberger CL, McFedries A, Cushman M, Saghatelian A, Adams EJ. MAIT recognition of a stimulatory bacterial antigen bound to MR1. J Immunol. 2013b;191:5268–77. doi: 10.4049/jimmunol.1301958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E, Treiner E, Duban L, Guerri L, Laude H, Toly C, Premel V, Devys A, Moura IC, Tilloy F, Cherif S, Vera G, Latour S, Soudais C, Lantz O. Stepwise development of MAIT cells in mouse and human. PLoS Biol. 2009;7:e54. doi: 10.1371/journal.pbio.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam HE, Eckle SB, Theodossis A, Liu L, Chen Z, Wubben JM, Fairlie DP, Strugnell RA, Mintern JD, McCluskey J, Rossjohn J, Villadangos JA. The intracellular pathway for the presentation of vitamin B-related antigens by the antigen-presenting molecule MR1. Nat Immunol. 2016 doi: 10.1038/ni.3416. [DOI] [PubMed] [Google Scholar]
- Miley MJ, Truscott SM, Yu YYL, Gilfillan S, Fremont DH, Hansen TH, Lybarger L. Biochemical Features of the MHC-Related Protein 1 Consistent with an Immunological Function. The Journal of Immunology. 2003;170:6090–6098. doi: 10.4049/jimmunol.170.12.6090. [DOI] [PubMed] [Google Scholar]
- Mohan JF, Unanue ER. Unconventional recognition of peptides by T cells and the implications for autoimmunity. Nat Rev Immunol. 2012;12:721–8. doi: 10.1038/nri3294. [DOI] [PubMed] [Google Scholar]
- Mori L, Lepore M, De Libero G. The Immunology of CD1- and MR1-Restricted T Cells. Annu Rev Immunol. 2016 doi: 10.1146/annurev-immunol-032414-112008. [DOI] [PubMed] [Google Scholar]
- Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011;11:823–36. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- Patel O, Kjer-Nielsen L, Le Nours J, Eckle SB, Birkinshaw R, Beddoe T, Corbett AJ, Liu L, Miles JJ, Meehan B, Reantragoon R, Sandoval-Romero ML, Sullivan LC, Brooks AG, Chen Z, Fairlie DP, McCluskey J, Rossjohn J. Recognition of vitamin B metabolites by mucosal-associated invariant T cells. Nat Commun. 2013;4:2142. doi: 10.1038/ncomms3142. [DOI] [PubMed] [Google Scholar]
- Rahimpour A, Koay HF, Enders A, Clanchy R, Eckle SB, Meehan B, Chen Z, Whittle B, Liu L, Fairlie DP, Goodnow CC, McCluskey J, Rossjohn J, Uldrich AP, Pellicci DG, Godfrey DI. Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using MR1 tetramers. J Exp Med. 2015;212:1095–108. doi: 10.1084/jem.20142110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reantragoon R, Corbett AJ, Sakala IG, Gherardin NA, Furness JB, Chen Z, Eckle SB, Uldrich AP, Birkinshaw RW, Patel O, Kostenko L, Meehan B, Kedzierska K, Liu L, Fairlie DP, Hansen TH, Godfrey DI, Rossjohn J, McCluskey J, Kjer-Nielsen L. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J Exp Med. 2013;210:2305–20. doi: 10.1084/jem.20130958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reantragoon R, Kjer-Nielsen L, Patel O, Chen Z, Illing PT, Bhati M, Kostenko L, Bharadwaj M, Meehan B, Hansen TH, Godfrey DI, Rossjohn J, McCluskey J. Structural insight into MR1-mediated recognition of the mucosal associated invariant T cell receptor. J Exp Med. 2012;209:761–74. doi: 10.1084/jem.20112095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegert P, Wanner V, Bahram S. Genomics, isoforms, expression, and phylogeny of the MHC class I-related MR1 gene. J Immunol. 1998;161:4066–77. [PubMed] [Google Scholar]
- Rodgers JR, Cook RG. MHC class Ib molecules bridge innate and acquired immunity. Nat Rev Immunol. 2005;5:459–71. doi: 10.1038/nri1635. [DOI] [PubMed] [Google Scholar]
- Salerno-Goncalves R, Rezwan T, Sztein MB. B cells modulate mucosal associated invariant T cell immune responses. Front Immunol. 2014;4:511. doi: 10.3389/fimmu.2013.00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz DG. Antigen receptor genes and the evolution of a recombinase. Semin Immunol. 2004;16:245–56. doi: 10.1016/j.smim.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol. 2011;11:251–63. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- Seach N, Guerri L, Le Bourhis L, Mburu Y, Cui Y, Bessoles S, Soudais C, Lantz O. Double-positive thymocytes select mucosal-associated invariant T cells. J Immunol. 2013;191:6002–9. doi: 10.4049/jimmunol.1301212. [DOI] [PubMed] [Google Scholar]
- Shawar SM, Vyas JM, Rodgers JR, Rich RR. Antigen presentation by major histocompatibility complex class I-B molecules. Annu Rev Immunol. 1994;12:839–80. doi: 10.1146/annurev.iy.12.040194.004203. [DOI] [PubMed] [Google Scholar]
- Shiina T, Ando A, Suto Y, Kasai F, Shigenari A, Takishima N, Kikkawa E, Iwata K, Kuwano Y, Kitamura Y, Matsuzawa Y, Sano K, Nogami M, Kawata H, Li S, Fukuzumi Y, Yamazaki M, Tashiro H, Tamiya G, Kohda A, Okumura K, Ikemura T, Soeda E, Mizuki N, Kimura M, Bahram S, Inoko H. Genomic anatomy of a premier major histocompatibility complex paralogous region on chromosome 1q21-q22. Genome Res. 2001;11:789–802. doi: 10.1101/gr.175801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreejit G, Ahmed A, Parveen N, Jha V, Valluri VL, Ghosh S, Mukhopadhyay S. The ESAT-6 protein of Mycobacterium tuberculosis interacts with beta-2-microglobulin (beta2M) affecting antigen presentation function of macrophage. PLoS Pathog. 2014;10:e1004446. doi: 10.1371/journal.ppat.1004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroynowski I. Molecules related to class-I major histocompatibility complex antigens. Annu Rev Immunol. 1990;8:501–30. doi: 10.1146/annurev.iy.08.040190.002441. [DOI] [PubMed] [Google Scholar]
- Stroynowski I, Lindahl KF. Antigen presentation by non-classical class I molecules. Curr Opin Immunol. 1994;6:38–44. doi: 10.1016/0952-7915(94)90031-0. [DOI] [PubMed] [Google Scholar]
- Stuart LM, Ezekowitz RA. Phagocytosis: elegant complexity. Immunity. 2005;22:539–50. doi: 10.1016/j.immuni.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Takada T, Kumanovics A, Amadou C, Yoshino M, Jones EP, Athanasiou M, Evans GA, Fischer Lindahl K. Species-specific class I gene expansions formed the telomeric 1 mb of the mouse major histocompatibility complex. Genome Res. 2003;13:589–600. doi: 10.1101/gr.975303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang XZ, Jo J, Tan AT, Sandalova E, Chia A, Tan KC, Lee KH, Gehring AJ, De Libero G, Bertoletti A. IL-7 licenses activation of human liver intrasinusoidal mucosal-associated invariant T cells. J Immunol. 2013;190:3142–52. doi: 10.4049/jimmunol.1203218. [DOI] [PubMed] [Google Scholar]
- Tilloy F, Treiner E, Park SH, Garcia C, Lemonnier F, de la Salle H, Bendelac A, Bonneville M, Lantz O. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J Exp Med. 1999;189:1907–21. doi: 10.1084/jem.189.12.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, Affaticati P, Gilfillan S, Lantz O. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 2003;422:164–9. doi: 10.1038/nature01433. [DOI] [PubMed] [Google Scholar]
- Trowsdale J. “Both man & bird & beast”: comparative organization of MHC genes. Immunogenetics. 1995;41:1–17. doi: 10.1007/BF00188427. [DOI] [PubMed] [Google Scholar]
- Trowsdale J, Knight JC. Major histocompatibility complex genomics and human disease. Annu Rev Genomics Hum Genet. 2013;14:301–23. doi: 10.1146/annurev-genom-091212-153455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto K, Deakin JE, Graves JA, Hashimoto K. Exceptionally high conservation of the MHC class I-related gene, MR1, among mammals. Immunogenetics. 2013;65:115–24. doi: 10.1007/s00251-012-0666-5. [DOI] [PubMed] [Google Scholar]
- Walter L, Gunther E. Isolation and molecular characterization of the rat MR1 homologue, a non-MHC-linked class I-related gene. Immunogenetics. 1998;47:477–82. doi: 10.1007/s002510050385. [DOI] [PubMed] [Google Scholar]
- Wun KS, Borg NA, Kjer-Nielsen L, Beddoe T, Koh R, Richardson SK, Thakur M, Howell AR, Scott-Browne JP, Gapin L, Godfrey DI, McCluskey J, Rossjohn J. A minimal binding footprint on CD1d-glycolipid is a basis for selection of the unique human NKT TCR. J Exp Med. 2008;205:939–49. doi: 10.1084/jem.20072141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Hirai M, Kurosawa Y, Hashimoto K. A highly conserved major histocompatibility complex class I-related gene in mammals. Biochem Biophys Res Commun. 1997;238:697–702. doi: 10.1006/bbrc.1997.7379. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Tsukamoto K, Hashimoto K. Cell surface expression of MR1B, a splice variant of the MHC class I-related molecule MR1, revealed with antibodies. Biochem Biophys Res Commun. 2014;443:422–7. doi: 10.1016/j.bbrc.2013.11.096. [DOI] [PubMed] [Google Scholar]
- Young MH, U’Ren L, Huang S, Mallevaey T, Scott-Browne J, Crawford F, Lantz O, Hansen TH, Kappler J, Marrack P, Gapin L. MAIT cell recognition of MR1 on bacterially infected and uninfected cells. PLoS One. 2013;8:e53789. doi: 10.1371/journal.pone.0053789. [DOI] [PMC free article] [PubMed] [Google Scholar]