Abstract
The intermediate filament protein Nestin serves as a biomarker for stem cells and has been used to identify subsets of cancer stem-like cells. However, the mechanistic contributions of Nestin to cancer pathogenesis are not understood. Here we report that Nestin binds the hedgehog pathway transcription factor Gli3 to mediate the development of medulloblastomas of the hedgehog subtype. In a mouse model system, Nestin levels increased progressively during medulloblastoma formation resulting in enhanced tumor growth. Conversely, loss of Nestin dramatically inhibited proliferation and promoted differentiation. Mechanistic investigations revealed that the tumor-promoting effects of Nestin were mediated by binding to Gli3, a zinc finger transcription factor that negatively regulates hedgehog signaling. Nestin binding to Gli3 blocked Gli3 phosphorylation and its subsequent proteolytic processing, thereby abrogating its ability to negatively regulate the hedgehog pathway. Our findings show how Nestin drives hedgehog pathway-driven cancers and uncover in Gli3 a therapeutic target to treat these malignancies.
Keywords: Nestin, stem cells, hedgehog pathway, medulloblastoma, Gli3 processing
INTRODUCTION
The Hedgehog (Hh) signaling pathway controls cell growth, survival and fate in almost every tissue during development (1, 2). In the absence of Hh ligand, Patched1 (Ptch1), the antagonizing receptor of Hh, inhibits the activity of the seven-pass transmembrane protein Smoothened (Smo). Binding of Hh to Ptch1 releases Smo from inhibition, leading to the activation of downstream transcription factors including Gli1, Gli2 and Gli3. Gli1 is an early transcriptional target of Hh signaling and functions exclusively as a transcriptional activator. Gli2 and Gli3 exist in both full-length and repressor forms, although Gli2 is considered primarily a transcriptional activator. The majority of full-length Gli3 (Gli3FL) is proteolytically processed into a transcriptional repressor (Gli3R) that predominantly acts as a negative regulator of Hh signaling (Pan and Wang, 2007). Proteolytic processing of Gli3 requires phosphorylation, primarily mediated by protein kinase A (PKA) (3, 4). Phosphorylation targets Gli3FL for cleavage via the ubiquitin-proteasome pathway. The regulation of Gli3 processing is important to maintain an appropriate level of Hh pathway activity, which is essential for normal development (4, 5).
Aberrant activation of the Hh pathway is associated with several human malignancies, including medulloblastoma (MB), the most common malignant brain tumor in children (6, 7). Deletion of Ptch1 and consequent activation of the Hh pathway, in cerebellar granule neuron precursor cells (GNPs) leads to MB formation with 100% penetrance (8), suggesting that GNPs represent cells of origin for Hh pathway MB. Following Ptch1 deletion, the majority of GNPs ultimately differentiate, implying that loss of Ptch1 alone is insufficient to maintain constitutive Hh pathway activation in GNPs. However, a small number of Ptch1-deficient GNPs continue to proliferate and eventually develop into tumors (8). Importantly, how these GNPs preserve Hh pathway activity during MB progression has not been previously reported.
Nestin, a type VI intermediate filament protein, is commonly utilized as a marker for neural stem cell populations. However, it is expressed in a wide range of proliferating progenitor and stem cells during early stages of development (9, 10). In mature tissues, Nestin is expressed in situations that recapitulate developmental programs such as tissue regeneration, wound healing, and revascularization (11, 12). It is generally believed that Nestin participates in the assembly of intermediate filaments thus providing structural protection against mechanical stress (12). In addition, Nestin is present in various tumors and it is associated with poor prognosis in glioma and melanoma (13). However, the functional role of Nestin in tumorigenesis remains elusive.
Here, we show that Nestin levels increase progressively during MB formation, and that Nestin cooperates with Hh signaling to drive tumor growth. Conversely, suppression of Nestin dramatically inhibits tumor cell proliferation and promotes differentiation. The tumor-promoting effects of Nestin are a consequence of its interaction with Gli3, which blocks Gli3 phosphorylation and impairs its proteolytic processing. These data reveal a critical role for Nestin in the development of Hh subtype MB, and point to Nestin as a potential therapeutic target for Hh pathway-associated malignancies.
MATERIALS AND METHODS
Animals
Ptch1C/C mice and Nestin-CFP mice have been described previously (8, 10). Math1-Cre Mice, Actin-Ds-Red mice and R26R-SmoM2 mice were purchased from the Jackson Laboratory. CB17/SCID mice were bred in the Fox Chase Cancer Center Laboratory Animal Facility (LAF). All animals were maintained in the LAF at Fox Chase Cancer Center and all experiments were performed in accordance with procedures approved by the Fox Chase Cancer Center Animal Care and Use Committee.
Microdissection, flow cytometry and cell culture
Cerebella were harvested from Math1-Cre/Ptch1C/C/Nestin-CFP mice at P7. 600μm slices were prepared using a VT1000S vibratome (Leica). Under a fluorescent microscope, EGLs were carefully removed from the rest of the cerebellum using fine forceps. Dissected EGLs were dissociated as described previously (10). Briefly, EGLs were digested in a papain solution to obtain a single-cell suspension and then centrifuged through a 35% and 65% Percoll gradient. Cells from the 35% to 65% interface were suspended in Dulbecco’s PBS (DPBS) plus 0.5% BSA. CFP+ and CFP− were then purified using a FACSvantage SE (BD Bioscience).
GNPs and MB cells were suspended in NB-B27 (Neurobasal with 1 mM sodium pyruvate, 2 mM L-glutamine, B27 supplement and 1% Pen/Strep, all from Invitrogen) and plated on poly-D-lysine (PDL)-coated coverslips (BD Biosciences). For the neurosphere forming assay, cells were plated at 2×103 cells/ml in NSC proliferation medium (NeuroCult basal medium with proliferation supplemental, Stem Cell Technologies) plus 10 ng/ml basic fibroblast growth factor and 20 ng/ml epidermal growth factor (Pepro Tech). NIH3T3 cells were cultured in DMEM with 10% fetal bovine serum, 1% Pen/Strep and 2mM L-glutamine (Invitrogen).
NIH3T3 cells had been obtained from American Type Culture Collection (ATCC) prior to this study in 2010. Before being used in our studies, the FCCC in-house service was used for cell line authentication. This cell line was tested mycoplasma negative with the Venor GeM Mycoplasma Detection kit (Minerva Biolabs).
Intracranial transplantation
Nestin+ and Nestin− Ptch1-deficient GNPs or MB cells were injected into the cerebella of CB17/SCID mice using using a stereotaxic frame with a mouse adaptor (David Kopf Instruments), as described previously (8, 10). Before transplantation, MB cells were infected with lentivirus carrying RFP-tagged Nestin shRNA or scrambled shRNA for 24hrs, and infected MB cells (RFP+) were then purified by FACS. Survival was defined as the time from transplantation until symptom onset.
Immunostaining, immunoprecipitation and western blotting
Primary antibodies used in this study include: anti-Nestin (1:1000 for western blotting, 1:200 for immunofluorescent staining, Abcam), anti-GFP (1:500 for immunofluorescent staining, Invitrogen; 1:1000 for western blotting, Clontech), Zic1 (1:100, from Segal lab at Dana Farber Caner Institute), anti-Arl13b (1:500, Proteintech), anti-GAPDH (1:10000, Sigma), anti-DsRed (1:200, Santa Cruz), anti-Ki67 (1:500, Abcam), anti-Gli1 (1:1000, Abcam), anti-Gli2 (1:1000, R&D system), anti-Gli3 (1:1000, R&D system), anti-RFP (1:200, Life technology), anti-Flag (1:100 for immunoprecipitation, 1:1000 for western blotting, Sigma) and anti-HA (1:100 for immunoprecipitation, 1:1000 for western blotting, Covance).
Immunofluorescent staining of sections and cells was carried out according to standard methods. Briefly, sections or cells were blocked and permeabilized for 2 hrs with PBS containing 0.1% Triton X-100 and 10% normal goat serum, stained with primary antibodies overnight at 4°C, and incubated with secondary antibodies for 2 hrs at room temperature. Sections or cells were counterstained with DAPI and mounted with Fluoromount-G (Southern Biotech) before being visualized using a Nikon Eclipse Ti microscope.
For western blot analysis, cells were lysed in RIPA buffer (Thermo) supplemented with protease and phosphatase inhibitors (Thermo). Total lysate containing equal amount of protein were separated by SDS-PAGE gel and subsequently transferred onto PVDF membrane. Membranes were then subjected to probe with antibodies. Western blot signals were detected by using SuperSignal West Pico Chemiluminescent substrate (Thermo) and exposed on films.
For immunoprecipitation experiments, total cell lysate (250μg) were mixed with anti-HA or anti-Flag antibody and incubated at overnight at 4°C. Protein A-sepharose (GE Healthcare) (50μl) was added and incubated at 4°C for 2 hours. The beads were washed with lysis buffer, pelleted and resuspended in 80μl protein sample buffer and subjected to western blot analysis.
RT-PCR and microarray analysis
RNA was isolated using the RNAqueous kit (Ambion) and treated with DNA-free DNase (Ambion). cDNA was synthesized using oligo(dT) and Superscript II reverse transcriptase (Invitrogen). Quantitative PCR reactions were performed in triplicate using iQ SYBR Green Supermix (Bio-Rad) and the Bio-Rad iQ5 Multicolor Real-Time PCR Detection System. Primer sequences are available upon request.
RNAs isolated from Ptch1-deficient GNPs, MB cells and wild type GNPs were labeled and hybridized to Affymetrix Mouse Genome 430 2.0 arrays. Microarray data were preprocessed using robust multichip analysis (RMA). Principal component analysis (PCA) and identification of genes differentially expressed between Nestin+ and Nestin− Ptch1-deficient GNPs were preformed using Partek Genomics Suite 6.3 software. Microarray data of Nestin+ and Nestin− ptch1-deficient GNPs are publicly accessible at http://www.ncbi.nlm.nih.gov/projects/geo/, access code GSE84392.
Statistics
Unpaired t test was performed to determine the statistical significance of the difference. p<0.05 was considered statistically significant. Error bars represent the SEM. ANOVA was performed to examine the difference in cell cycle distribution. Overall survival in figure 2b and figure 5o was assessed using the Kaplan-Meier survival analysis and the Mantel-Cox log-rank test was used to assess the significance of difference between survival curves. Data handling and statistical processing was performed using Graphpad Prism Software.
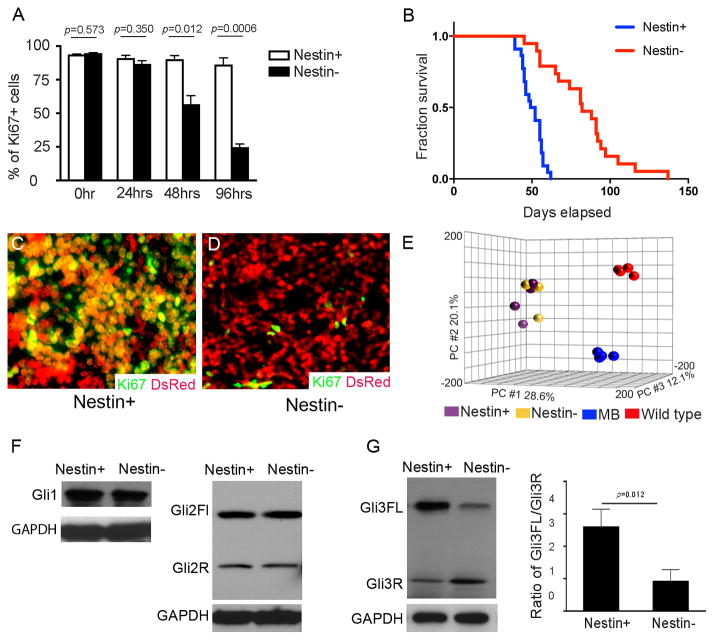
Figure 2. Nestin+ Ptch1-deficient GNPs exhibit increased proliferation and tumorigenicity.
A, Percentage of Ki67+ among Nestin+ and Nestin− GNPs immunostained for Ki67 at designated timepoints (0–96hrs). B, Survival curve of CB17/SCID mice after intracranial transplantation with Nestin+ and Nestin− Ptch1-deficient GNPs. n= 22 for transplantation with Nestin+ Ptch1-deficient GNPs, and n=19 for transplantation with Nestin− Ptch1-deficient GNPs. C–D, Nestin+ (C) and Nestin− (D) Ptch1-deficient GNPs from Nestin-CFP/Math1-Cre/Ptch1C/C/Actb-DsRed mice were immunostained for Ki67 and DsRed at 3 weeks after transplantation. E, Principal component analysis of Nestin+ and Nestin− GNPs from Nestin-CFP/Math1-Cre/Ptch1C/C mice at P7, GNPs from P7 wild type mice, and MB cells from Math1-Cre/Ptch1C/C mice at 8 weeks of age. F–G, Gli1 and Gli2 (F), Gli3 proteins (G) in Nestin+ and Nestin− Ptch1-deficient GNPs were examined by western blotting. The ratio of Gli3FL/Gli3R in Nestin+ and Nestin− cells was measured based on protein expression levels (G).
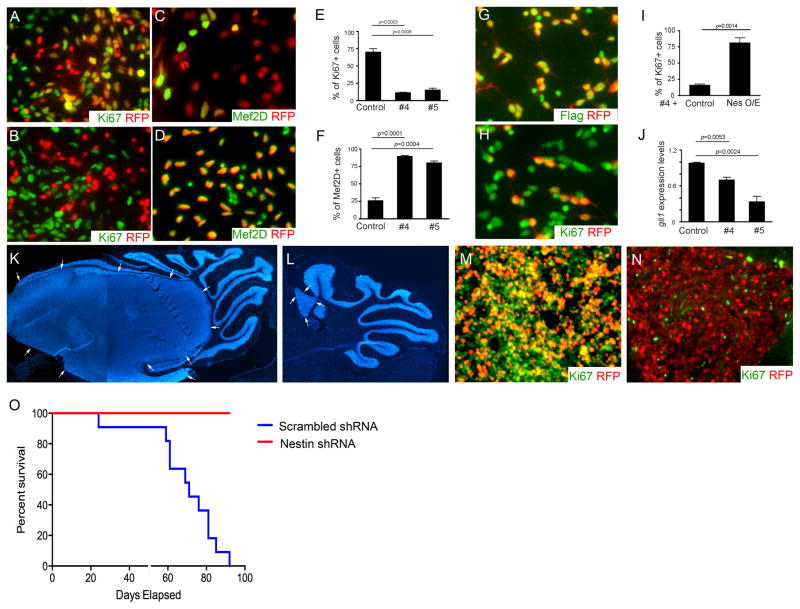
Figure 5. Nestin is critical for MB tumorigenesis.
A–D, MB cells infected with lentivirus carrying scrambled shRNA-RFP (A and C) and Nestin shRNA-RFP #4 (B and D), were immunostained for Ki67 and RFP (A and B) or Mef2D and RFP (C and D) at 48hrs following infection. E–F, The percentage of Ki67+ cells (E) and Mef2D+ (F) among infected MB cells (RFP+). G–H, MB cells were co-infected with lentivirus carrying RFP-tagged Nestin-shRNA #4 (targeting 3′-untranslational region of nestin gene) in combination with virus carrying Flag-tagged Nestin (containing only Nestin encoding sequence). 48hrs after the infection, MB cells were immunostained for Flag and RFP (G) or Ki67 and RFP (H). I, The percentage of Ki67+ cells among MB cells infected with Nestin shRNA #4 together with an empty vector (control) or a Nestin expressing construct (Nes O/E). J, gli1 mRNA expression in MB cells at 48hrs following the infection with Nestin shRNA #4, was examined by qRT-PCR. K–N, DAPI staining (K–L) and immunstaining for Ki67 and RFP (M–N) of cerebellar sections of CB17/SCID mice at 5 weeks after transplantation with MB cells infected with scrambled shRNA (K and M) and Nestin shRNA #4 (L and N). Tumor mass was surrounded by arrows in K, L and O. Survival curve of CB17/SCID mice after transplantation of infected MB cells. n=11 in each transplantation group.
RESULTS
MB cells express Nestin during tumor progression
Using Math1-Cre/Ptch1C/C mice, we demonstrated that deletion of Ptch1 in cerebellar GNPs resulted in cerebellar tumors resembling human MB (Fig. 1a) (8). Immunohistochemical analyses revealed that the majority of MB cells expressed Nestin and that it was located predominantly in the cytoplasm (Fig. 1b). Nestin expression was also detected in cells from tumors generated by overexpression of activated Smo as well as in MBs developed from Ptch1 heterozygous mice (Supplementary Fig. S1a, b) (14–16). However, Nestin was absent from conventional cerebellar GNPs (Fig. 1c, d), as previously reported (10). These data suggest that Nestin might be associated with tumorigenesis in Hh subtype MB.
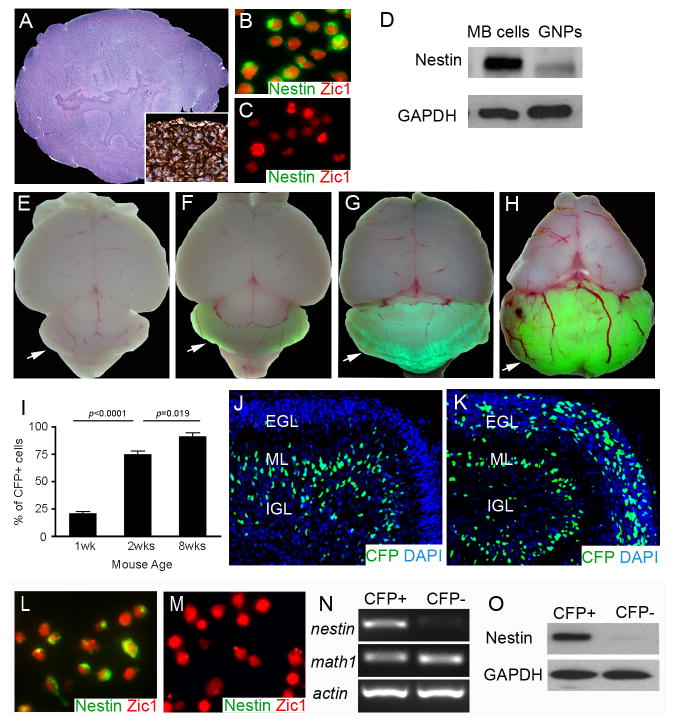
Figure 1. Cerebellar GNPs express Nestin after Ptch1 deletion.
A, Hematoxylin and eosin staining of MB from Math1-Cre/Ptch1C/C mouse at 8 weeks of age. Inset, immunostaining for Nestin in MB tissue. B–C, Immunostaining for Nestin and Zic1 on freshly isolated MB cells (B) and wildtype GNPs (C). D, Expression of Nestin and GAPDH proteins in GNPs and MB cells was examined by western blotting. e–h, Wholemount images of mouse brains from P7 Nestin-CFP mice (E), and Nestin-CFP/Math1-Cre/Ptch1C/C mice at P7 (F), P14 (G) and 8 weeks of age (H). Arrows point to mouse cerebella. I, Flow cytometry analysis of the percentage of CFP+ cells among GNPs isolated from Nestin-CFP/Math1-Cre/Ptch1C/C mice at designated stages. J–K, Immunostaining for CFP with a FITC-conjugated antibody on cerebellar sagittal sections from P7 Nestin-CFP mouse (J) and P7 Nestin-CFP/Math1-Cre/Ptch1C/C mouse (K). Cerebellar sections were counterstained with DAPI. L–M, CFP+ cells and CFP− cells were purified from cerebellar EGLs of Nestin-CFP/Math1-Cre/Ptch1C/C mice at P7 by microdissection followed by FACS. Expression of Nestin and Zic1 proteins in CFP+ cells (L) and CFP− cells (M) was examined by immunocytochemistry. N–O, mRNA expression of nestin, math1 and actin (N), and expression of Nestin and GAPDH proteins (O) in CFP+ and CFP− cells were examined by conventional RT-PCR and western blotting, respectively.
To examine Nestin expression during MB progression, we crossed Math1-Cre/Ptch1C/C mice with Nestin-CFP mice to generate tumor expressing a nuclear form of cyan fluorescent protein (CFP) in Nestin-containing cells (10, 17), hereafter these mice are referred to as Math1-Cre/Ptch1C/C/Nestin-CFP mice. No obvious CFP fluorescence was observed in whole-mount cerebella from Nestin-CFP mice in which Ptch1 was not deleted (Fig. 1e). A weak CFP signal was detected following Ptch1 deletion in Math1-Cre/Ptch1C/C/Nestin-CFP cerebella at postnatal day 7 (P7) (Fig. 1f) and the signal intensified as tumors developed (P14, Fig. 1g; 8 weeks of age, Fig. 1h). Next, we isolated GNPs from Math1-Cre/Ptch1C/C/Nestin-CFP cerebella and quantified the percentage of CFP+ cells by flow cytometry (Fig. 1i). We found that 21±4% of dissociated GNPs were positive for CFP in P7 Math1-Cre/Ptch1C/C/Nestin-CFP cerebella, with 70±5% of GNPs expressing CFP at 2 weeks of age. By 8 weeks of age, almost all (92±3%) MB cells were positive for CFP. These data indicate that Ptch1-deficient GNPs progressively increase expression of Nestin-CFP during MB formation.
Nestin+ Ptch1-deficient GNPs exhibit enhanced tumorigenicity
In the control cerebellum (from Nestin-CFP/Ptch1C/C mice) at P7 (Fig. 1j), CFP positive cells were rarely detected in the external germinal layer (EGL), where GNPs normally reside (18). As expected, Ptch1 deletion in Math1-Cre/Ptch1C/C/Nestin-CFP cerebellum resulted in a much thicker EGL compared with controls (Fig. 1k). Over 20% of cells in the mutant EGL were found to be CFP positive (Fig. 1k). To purify CFP+ Ptch1-deficient GNPs from Math1-Cre/Ptch1C/C/Nestin-CFP cerebella, we microdissected the mutant EGL to exclude CFP+ cells (namely Bergmann glia) located in the molecular layer and the internal granule cell layer (Supplementary Fig. S1c, d), as previously described (10, 19). Cells dissociated from the mutant EGL were then separated into CFP positive and negative populations by fluorescence-activated cell sorting (FACS). Nestin mRNA and protein expression were present in CFP positive cells, but absent from CFP negative cells (Fig. 1l–o) despite the same level of Ptch1 loss and comparable expression levels of the GNP specific markers, Math1 and Zic1 (20, 21), in both cell populations (Fig. 1l–n and Supplementary Fig. S1e). No difference was observed in the extent of Hh pathway activation (Gli1 and N-myc expression) between CFP+ Ptch1-deficient GNPs and their CFP negative counterparts (Supplementary Fig. S1f). These data suggest that following Ptch1 deletion, cerebellar GNPs segregate into two distinct populations, based on the presence or absence of Nestin, with equivalent levels of Hh pathway activity.
To determine the functional relevance of Nestin expression in tumorigenesis, we first compared the in vitro proliferation rates of CFP+ and CFP− cells purified from the EGL of P7 Math1-Cre/Ptch1C/C/Nestin-CFP cerebella. In the first 24hrs after plating, both cell populations were highly proliferative with more than 95% of cells positive for Ki67 (Fig. 2a), consistent with the comparable levels of Hh pathway activity (Supplementary Fig. S1f). After 48hrs in culture, Nestin-negative cells started to exit the cell cycle, and only 20±3% remained proliferative after 96 hrs in culture (Fig. 2a). In contrast, at that time point Nestin+ cells maintained high levels of proliferation in vitro (>95% of cells were Ki67+). To compare the tumorigenicity of Nestin+ and Nestin− Ptch1-deficient GNPs, we transplanted these two cell populations into the cerebellum of CB17/SCID mice as previously described (8, 10). As shown in Figure 2b, both cell populations generated tumors after transplantation. However, compared with Nestin− cells, Nestin+ Ptch1-deficient GNPs developed tumors much more rapidly (median survival: 50 days for Nestin+ cells vs 82 days for Nestin− cells, p<0.0001), demonstrating that Nestin+ Ptch1-deficient GNPs are more tumorigenic than the Nestin− GNPs.
To examine the basis for increased tumorigenicity in Ptch1-deficient GNPs after Nestin expression, Nestin positive (CFP+) and negative (CFP−) GNPs were purified from Math1-Cre/Ptch1C/C/Nestin-CFP/Actb-DsRed mice in which all cells express red fluorescent protein (22). Both cell populations were transplanted into the cerebella of CB17/SCID mice, and three weeks later, the recipient cerebella were sectioned to examine the proliferative state of transplanted cells (DsRed+) by immunostaining for Ki67. The majority of Nestin+ Ptch1-deficient GNPs (>90%) were proliferative (Ki67+ and DsRed+, Fig. 2c), whereas less than 2% of Nestin− Ptch1-deficient GNPs were proliferating (Ki67+ and DsRed+, Fig. 2d). These data indicate that the divergent growth rate of Nestin+ and Nestin− GNPs is a consequence of differential proliferative capacity. Importantly, tumor cells originating from either CFP+ or CFP− cells eventually became positive for CFP (Supplementary Fig. S2a, b), suggesting that Nestin expression may be necessary for MB progression.
Gli3 is stabilized in Nestin+ Ptch1-deficient GNPs
To determine the molecular basis for increased tumorigenicity of Nestin+ Ptch1-deficient GNPs, we performed microarray analysis on Nestin+ and Nestin− GNPs from P7 Math1-Cre/Ptch1C/C/Nestin-CFP cerebella, and compared the gene expression profiles with those of GNPs from P7 wild type mice, and MB cells isolated from Math1-Cre/Ptch1C/C mice at 8 weeks of age as controls. Principal component analysis revealed that despite being well separated from wildtype GNPs and MB cells, Nestin+ and Nestin− Ptch1-deficient GNPs co-clustered, indicating that they share similar gene expression profiles (Fig. 2e). No upregulation of stem cell-associated genes (except Nestin) was observed in Nestin+ Ptch1-deficient GNPs compared with Nestin− counterparts (Supplementary Table 1). Furthermore, Nestin+ Ptch1-deficient GNPs did not exhibit stem cell properties such as the capacity to form neurospheres or multipotency (23) (data not shown). These data indicate that Nestin does not alter the differentiation status or cell lineage of Ptch1-deficient GNPs.
Since no significant differences were detected between Nestin+ and Nestin− Ptch1-deficient GNPs at the level of gene expression, we evaluated differences between these two cell populations in proteins relevant to Hh signaling. The three Gli proteins (Gli1, Gli2 and Gli3) play important roles as Hh signaling effectors. Gli3 is unique among the three mammalian Gli proteins, in that it functions predominantly as a repressor of Hh signaling. Full-length Gli3 (Gli3FL) can be proteolytically processed into a truncated derivative (Gli3R) that potently inhibits Hh pathway activity. Comparable levels of Gli1 and Gli2 expression were detected in Nestin+ and Nestin− GNPs purified from Math1-Cre/Ptch1C/C/Nestin-CFP cerebellum at P7 (Fig. 2f). However, elevated levels of Gli3FL and decreased levels of Gli3R were observed in Nestin+ Ptch1-deficient GNPs compared to Nestin− GNPs, despite the presence of similar levels of gli3 mRNA in these two cell populations (Supplementary Fig. S3a). The protein ratio of Gli3FL to Gli3R (Gli3FL/Gli3R), indicative of Gli3 stability (4, 5), was dramatically increased in Nestin+ Ptch1-deficient GNPs (Fig. 2g). These data indicate that Nestin compromises Gli3 processing in Ptch1-deficient GNPs.
To investigate whether Nestin regulates Gli3 processing, we utilized NIH3T3 cells, which express low basal levels of Nestin (Fig. 3a). NIH3T3 cells were transfected with constructs encoding either Nestin or Nestin shRNAs (Fig. 3b), and cells were subsequently treated with recombinant Sonic Hedgehog (Shh) protein. Levels of Gli3 mRNA were comparable in NIH3T3 cells regardless of Nestin expression levels (Supplementary Fig. S3b, c). As shown in Figure 3c, when cells were cultured in control media, Gli3 levels were similar in NIH3T3 cells expressing empty vector, Nestin shRNA or Nestin cDNA. However, dramatic differences were evident when cells were treated with Shh for 48hrs (Fig. 3c). In Nestin shRNA expressing cells, the ratio of Gli3FL/Gli3R dropped significantly in Nestin-deficient NIH3T3 cells compared with wildtype cells (Fig. 3d), suggesting that Nestin suppression compromised Gli3 stability and potentiated Gli3 processing during Hh signal activation. In contrast, Nestin overexpression resulted in increased Gli3FL and diminished Gli3R in the presence of Shh (Fig. 3c); this was reflected by an elevated ratio of Gli3FL/Gli3R (Fig. 3d). No difference in cell cycle distribution was found among NIH3T3 cells or Ptch1-deficient GNPs in the presence or absence of Nestin expression (Supplementary Fig. S3d, e), indicating that differential Gli3 processing was not a result of altered mitotic rate. The above data indicate that Nestin inhibits proteolytic processing of Gli3.
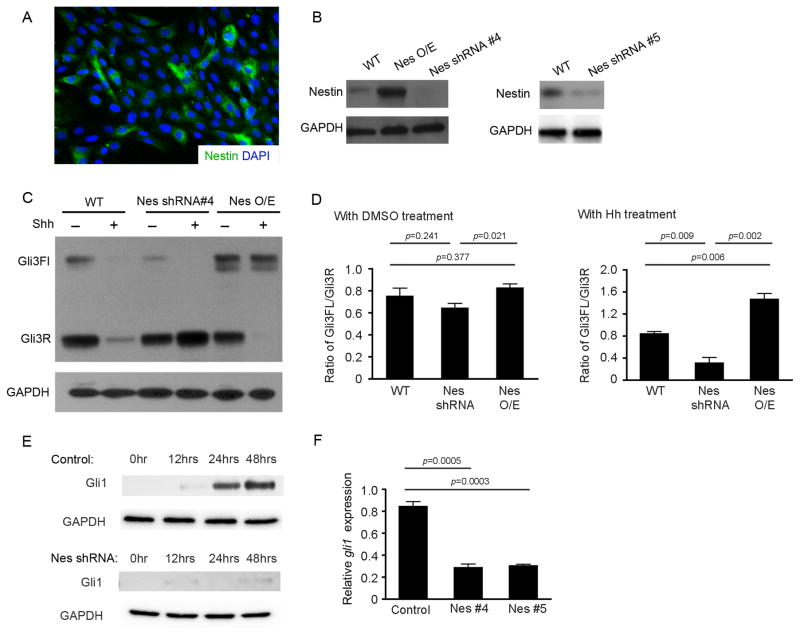
Figure 3. Nestin prevents Gli3 processing in GNPs and NIH3T3 cells.
A, NIH3T3 cells were immunostained for Nestin, and counterstained with DAPI. B, Nestin protein expression in NIH3T3 cells stably transfected with constructs carrying GFP (WT), Nestin (Nes O/E), or Nestin shRNAs (Nes shRNA #4 and #5). C, Western blotting analysis of Gli3 proteins in NIH3T3 cells transfected with GNP (WT), Nestin shRNA#4, or Nestin (NesO/E) after treatment with 3μg/ml Shh or DMSO for 48hrs. D, Gli3FL and Gli3R levels from c was quantified by densitometry and used to calculate the Gli3FL/Gli3R ratio. E–F, Gli1 protein expression (E) and gli1 mRNA expression (F) in control and Nestin-deficient NIH3T3 cells at designated timepoints (E) and 48hrs (F), after Shh treatment, were examined by western blotting and qRT-PCR, respectively. GAPDH in figure B, C and E was used as a loading control.
The level of Gli1 expression serves as a useful marker of Hh pathway activity (24). Following Shh treatment of NIH3T3 cells, the levels of Gli1 increased in a time-dependent manner (Fig. 3e). In Nestin-deficient NIH3T3 cells, Gli1 expression was detected at 12hrs in response to Shh treatment; however, upregulation of Gli1 protein and mRNA was not detected in these cells after 24hrs and 48hrs (Fig. 3e, f). Consistent with the role of Gli3 in negatively regulating Hh signaling, these data suggest that Nestin enhances and prolongs the output of Hh pathway activation by inhibiting Gli3 processing, thus abrogating a negative feedback mechanism.
Nestin inhibits Gli3 phosphorylation by physically interaction
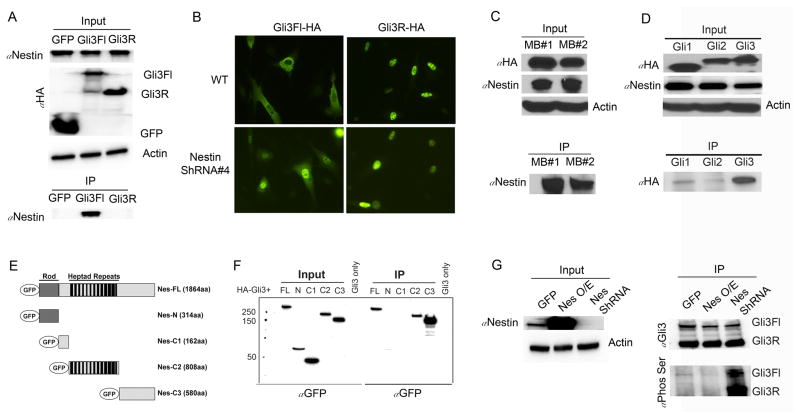
Previous studies have demonstrated that Gli proteins are capable of forming direct complexes with cytoskeletal proteins (25, 26), which prompted us to ask whether Nestin directly interacts with Gli3. Due to the lack of Gli3 or Nestin antibodies for immunoprecipitation, we transfected NIH3T3 cells with constructs encoding HA-tagged Gli3 or HA-tagged GFP vector, and used anti-HA antibodies to investigate a physical interaction between Gli3 and Nestin. As shown in Figure 4a, only Gli3FL, but not Gli3R, was co-immunoprecipitated with native Nestin, indicating that Nestin could form a complex with Gli3FL. Exogenous Gli3FL was predominantly located in the cytoplasm of wildtype NIH3T3 cells, while in the absence of Nestin, the majority of exogenous Gli3 protein migrated into the cell nucleus (Fig. 4b). However, Gli3R always resided in the nuclei of NIH3T3 cells, regardless of presence or absence of Nestin (Fig. 4b). These data suggest that Nestin can only interact with Gli3FL, not Gli3R. The physical association between Nestin and Gli3 was further confirmed by immunoprecipitation of native Nestin with exogenous Gli3 in MB cells (Fig. 4c). Moreover, it appears that Nestin preferentially binds to Gli3 compared with Gli1 and Gli2 (Fig. 4d).
Figure 4. Nestin prevents proteolytic processing of Gli3.
A, NIH3T3 cells were transfected with HA-tagged GFP, Gli3FL or Gli3R, and cell lysates were immunoprecipated with anti-Flag beads. Nestin, Actin and HA proteins in whole cell lysate, and precipitated Nestin were determined by western blotting. B, subcellular localization of exogenous Gli3FL or Gli3R in NIH3T3 cells transfected with HA-tagged Gli3FL or Gli3R, was examined by immunocytochemistry using anti-HA antibody. C, Cell lysates MB cells (isolated from 2 independent Math1-Cre/Ptch1C/C mice at 8 weeks of age) infected with a lentivirus carrying HA-tagged Gli3FL, were co-immunoprecipated with anti-HA beads. HA, Nestin and Actin proteins in the input and precipitated Nestin were examined by western blotting using designated antibodies. D, NIH3T3 cells transfected with HA-tagged Gli1, Gli2 or Gli3 in combination with Flag-tagged Nestin. Cell lysates were immunoprecipitated with anti-Flag beads. The presence of HA, Nestin and Actin proteins as well as precipitated Gli proteins were examined by western blotting using designated antibodies. E, Schematic representation of the GFP-tagged Nestin fragments. F, NIH3T3 cells were transfected with a HA-tagged Gli3 construct alone or together with GFP-tagged Nestin constructs. Nestin truncated proteins in the input and those precipitated with anti-HA beads were determined by western blotting using an anti-GFP antibody. G, NIH3T3 cells stably transfected with constructs carrying GFP (WT), Nestin (Nes O/E), or Nestin shRNA #4 (Nes null), were further transfected with HA-tagged Gli3FL. Whole cell lysates were immunoprecipitated with anti-HA beads, and phosphorylated Gli3 was examined by western blotting using an antibody recognizing phosphoserine.
Nestin is comprised of an N-terminal rod domain and 41 heptad repeats in an extensive C-terminal region (27). To identify the domain of Nestin responsible for interaction with Gli3, we constructed four Nestin fragments: one including the N-terminal domain (Nes-N), one containing containing all heptad repeats (Nes-C2), one containing the linker region between Nes-N1 and Nes-C2 (Nes-C1), and one including the C-terminal region (Nes-C3) (Fig. 4e). NIH3T3 cells were co-transfected with HA-tagged Gli3FL and GFP-tagged Nestin vectors to investigate the regions responsible for protein-protein interaction. Gli3 predominantly interacted with the C3-termimal region of Nestin (Fig. 4f). Gli3 also exhibited a weaker association with the Nestin-C2 region, but no Gli3-binding activity was observed with the C1 linker region of Nestin (Fig. 4f). These data suggest that Nestin regulates Gli3 processing through a physical interaction predominantly mediated by the C-terminal tail of Nestin.
Proteolytic processing of Gli3 requires phosphorylation, predominantly by the cyclic AMP-dependent protein kinase (PKA) (3, 4). Although there are multiple phosphorylation sites on Gli3, serine phosphorylation of the C-terminus by PKA has recently been reported to be critical for its proteolytic processing (3, 28). To investigate whether Nestin influences Gli3 phosphorylation, we introduced HA-tagged Gli3FL into wild type, Nestin-overexpressing, or Nestin-shRNA expressing NIH3T3 cells. We then immunoprecipitated Gli3 using an anti-HA antibody, and examined Gli3 phosphorylation using an antibody that recognizes phosphoserine (28). As shown in Figure 4g, Nestin depletion significantly enhanced Gli3 phosphorylation in NIH3T3 cells. Conversely, Nestin overexpression inhibited Gli3 phosphorylation. These data suggest that Nestin compromises Gli3 phosphorylation.
Nestin is critical for MB tumorigenesis
To determine if Nestin is essential for Ptch1-dependent tumorigenesis, we inhibited Nestin expression in MB cells by infection with lentiviruses encoding Nestin shRNAs. The efficiency of Nestin suppression in tumor cells after infection was confirmed by western blotting (Supplementary Fig. S4). At 48hrs following infection, approximately 70±9% of MB cells infected with scrambled shRNA were found to be Ki67+ (Fig. 5a), whereas less than 20% of MB cells were Ki67+ after Nestin suppression (Fig. 5b). The reduced proliferation among Nestin-deficient MB cells suggests that Nestin is required for MB cell proliferation (Fig. 5e). No obvious difference was observed in the levels of apoptosis (cleaved caspase 3+) in tumor cells after infection with either Nestin shRNA or scrambled shRNA (data not shown). Although only 25±5% of MB cells expressing scrambled shRNA were found to be differentiated, as determined by expression of the neuronal differentiation marker Mef2d (29) (Fig. 5c), the majority of Nestin-deficient MB cells were positive for Mef2d (Fig. 5d). These data indicate that Nestin deficiency promotes differentiation of MB cells (Fig. 5f). To confirm that inhibition of proliferation in MB cells infected with Nestin shRNA was indeed a consequence of Nestin deficiency, we performed a rescue experiment by infecting MB cells with a lentivirus expressing Nestin shRNA-RFP (targeting the 3′-untranslationed region of nestin gene) together with a lentivirus carrying Flag-tagged Nestin (containing only the protein encoding region of nestin) or empty vector as a control. As shown in figure 5g, Flag-tagged Nestin was present in virtually all MB cells (>95±2%) infected with Nestin shRNA. The percentage of proliferating Nestin deficient MB cells increased to 81±14% (Fig. 5h) after re-expression of Nestin, compared with overexpression of the control vector (16±3%, Fig. 5i), indicating that decreased proliferation of MB cells after Nestin knockdown is a consequence of Nestin deficiency. In addition, gli1 expression was significantly repressed after Nestin deletion in MB cells (Fig. 5j). Collectively, our results suggest that Nestin plays a critical role in maintaining MB cell proliferation.
We next asked whether Nestin is required for in vivo MB growth. MB cells were infected with lentivirus carrying Nestin shRNA-RFP or scrambled shRNA-RFP in vitro. At 24hrs following infection, before Nestin shRNA inhibits proliferation and Hh signaling in MB cells (Supplementary Fig. S5), 50,000 RFP positive cells purified by FACS were transplanted into the cerebella of CB17/SCID mice. 5 weeks after transplantation, MB cells expressing scrambled shRNA (Fig. 5k) gave rise to much larger tumor masses than MB cells expressing shRNA (Fig. 5l). The majority of MB cells infected with scrambled shRNA were highly proliferative (Ki67+, Fig. 5m), whereas less than 1% of Nestin-deficient MB cells were positive for Ki67 (Fig. 5n). No significant alterations in apoptosis were observed in MB with Nestin depletion (data not shown). Finally, all CB17/SCID mice developed tumors within 15 weeks of transplantation with MB cells infected with scrambled shRNA, whereas no tumors were generated using Nestin-deficient MB cells (Fig. 5o). These data indicate that Nestin is essential for the formation of Hh-dependent MB.
DISCUSSION
While research has illuminated many of the molecular events underlying tumor initiation, it has proven more difficult to define specific mechanisms responsible for tumor progression. Previously, we reported that despite loss of Ptch1, the majority of mouse cerebellar GNPs eventually differentiate following a period of prolonged proliferation, and only rare cells within the hyperplastic lesions actually develop into MB (8). The present results suggest an explanation for this observation: loss of Ptch1 alone may lead to a transient increase in Hh signaling, but in the absence of Nestin, pathway activity is quenched by Gli3R, and cells undergo differentiation. However, in the presence of high levels of Nestin, Gli3 is sequentered, and cells maintain high levels of Hh signaling and go on to form tumors. Our data demonstrate that Nestin expression is a necessary step in the enhancement and maintenance of Hh pathway activation leading to malignancy (Fig. 6). Studies have revealed the important role of negative-feedback loops that normally operate to attenuate various types of signaling during homeostatic regulation (2, 30). Our findings highlight the importance of disrupting these negative feedback loops in tumorigenesis. Beyond activation of proliferative signaling, cells need to develop mechanisms to override intrinsic negative controls to achieve full transformation.
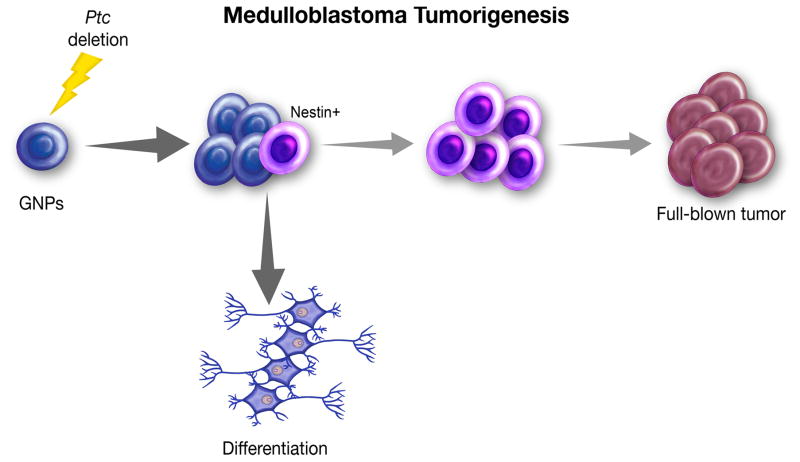
Figure 6. A model for MB tumorigenesis.
Cerebellar GNPs proliferate extensively after deletion of Ptch1; however, the majority of Ptch1-deficient GNPs finally exits the cell cycle and differentiates into mature neurons. During the initial proliferation, a proportion of Ptch1-deficient GNPs expresses Nestin, which further promotes the proliferation of GNPs and allows them to accumulate oncogenic alterations necessary for MB tumorigenesis.
Our studies reveal that in addition to being a putative marker for stem cells, Nestin acts to enhance Hh signaling. Although Nestin is not required for initiation of pathway activation, its presence significantly prolongs and elevates the level of Hh pathway activation. Nestin expression is widely observed during stem cell expansion and tissue regeneration where activation of Hh pathway is often involved. Indeed, deletion of Smo from neural progenitor cells using an inducible Cre recombinase under the control of the Nestin promoter markedly reduced the survival and proliferation of neural stem/progenitor cells in the mouse subventricular zone (31, 32). In addition, Nestin expression is frequently found in cancer stem cells, in which the Hh pathway is often active (33–36). Our studies suggest that Nestin may play an active role in modulating stem cell populations during normal development as well as in pathological processes.
Elucidating the mechanisms underlying the proteolytic processing of Gli3 is essential for understanding the role of the Hh pathway in normal development and disease. Our studies demonstrate that Nestin inhibits the proteolytic processing of Gli3 by direct binding, which sheds light on the role of intermediate filament proteins in regulating Gli3 stability. In mammalian cells, phosphorylation targets Gli3 for ubiquitination and processing into Gli3R (37, 38), which potently represses Hh signaling. Our studies demonstrate that the direct interaction with Nestin inhibits Gli3 phosphorylation thereby blocking Gli3 processing. It has been reported that in mammalian cells Gli3 processing relies on the primary cilium (39, 40). However, no difference in cilia formation or stability was observed in NIH3T3 cells with altered Nestin expression (Supplementary Fig. S6a). The percentage of cells with primary cilia among Nestin+ Ptch1-deficient GNPs was comparable to that among Nestin− Ptch1-deficient GNPs (Supplementary Fig. S6b, c). These data suggest that Nestin does not affect ciliogenesis in NIH3T3 cells and MB cells. However, we cannot rule out the possibility that interaction with Nestin may affect transport of Gli3 to the primary cilium.
Human MB comprises at least four subgroups: Wnt, Sonic Hedgehog (SHH), Group 3 and Group 4 (41–43). We examined Nestin expression by immunohistochemistry in 287 human MB samples on tissue microarrays and found that Nestin is present in a subset of tumors in each of the four subtypes (Supplementary Fig. S7). In our SHH subtype MB mouse model, Nestin expression accumulates progressively during tumor development, suggesting a stage-dependent modulation of Nestin expression during MB development. Thus, it is possible that Nestin expression increases during tumor development in human MB. In addition to regulating the Hh pathway by binding to Gli3 protein, Nestin may modulate other signaling pathways that are important for stem cells and tumorigenesis. Future studies are warranted to investigate the functions of Nestin in non-SHH type MB.
The observation of Hh pathway activation in tumors has made it as an attractive therapeutic target (44). All Hh pathway inhibitors currently being studied in clinical trials target Smo. Initial findings suggest that while tumors can exhibit initial dramatic responses, there is rapid emergence of drug-resistance (45),(46). In many cases, resistance arises as a consequence of mutations in Smo that prevent binding of antagonists, or genetic events that activate downstream components of the Hh pathway (47, 48). Since Nestin augments Hh signaling by regulating Gli3 downstream of Smo, it may represent a promising therapeutic target for treatment of drug-resistant MB. On the other hand, inhibition of Smo universally represses Hh signaling even in normal cells, which could potentially cause severe developmental side effects (49). Based on our studies, targeting Nestin expression in MB cells could restore the inhibitory functions of Gli3, and repress hyperactivation of the Hh pathway, in combination with Smo inhibitors or in Vismodegib-resistant tumors to improve patient outcomes. Potentially, this strategy could be expanded to non-Hh pathway tumors if Nestin functions in a similar way to enhance other tumorigenic signaling pathways.
Supplementary Material
Acknowledgments
Financial Support:
This research was supported by funds from the US National Cancer institute (CA178380 and CA185504, Z. Yang), Pennsylvania Department of Health (CURE 4100068716, Z. Yang), American Cancer Society (RSG1605301NEC, Z. Yang), the Children’s Brain Tumor Foundation (J.M.Y. Ng and T. Curran), the Brain Tumor Society (J.M.Y. Ng and T. Curran) and National Institutes of Health (CA 096832, J.M.Y. Ng and T. Curran).
We thank J. Oesterling for flow cytometric analysis; Y. Li for microarray analysis; Dr. A. Efimov for microscopy analysis; Dr. Q. Cai for histological analysis; Dr. J. Wu (University of Texas Southwestern Medical Center) for HA-tagged Gli constructs; Dr. R. Segal (Dana Farber Caner Institute) for anti-Zic1 antibody; Dr. B. Wang (Weill Cornell Medical College) for anti-Arl13b antibody and Dr. S. Scales (Genentech, Inc.) for anti-Gli3 antibodies.
Footnotes
No potential conflicts of interest were disclosed.
References
- 1.Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes & development. 2008;22:2454–72. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- 2.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes & development. 2001;15:3059–87. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 3.Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–34. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 4.Litingtung Y, Dahn RD, Li Y, Fallon JF, Chiang C. Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature. 2002;418:979–83. doi: 10.1038/nature01033. [DOI] [PubMed] [Google Scholar]
- 5.te Welscher P, Zuniga A, Kuijper S, Drenth T, Goedemans HJ, Meijlink F, et al. Progression of vertebrate limb development through SHH-mediated counteraction of GLI3. Science. 2002;298:827–30. doi: 10.1126/science.1075620. [DOI] [PubMed] [Google Scholar]
- 6.Gibson P, Tong Y, Robinson G, Thompson MC, Currle DS, Eden C, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468:1095–9. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romer JT, Kimura H, Magdaleno S, Sasai K, Fuller C, Baines H, et al. Suppression of the Shh pathway using a small molecule inhibitor eliminates medulloblastoma in Ptc1(+/−)p53(−/−) mice. Cancer Cell. 2004;6:229–40. doi: 10.1016/j.ccr.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 8.Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14:135–45. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lendahl U, Zimmerman LB, McKay RD. CNS stem cells express a new class of intermediate filament protein. Cell. 1990;60:585–95. doi: 10.1016/0092-8674(90)90662-x. [DOI] [PubMed] [Google Scholar]
- 10.Li P, Du F, Yuelling LW, Lin T, Muradimova RE, Tricarico R, et al. A population of Nestin-expressing progenitors in the cerebellum exhibits increased tumorigenicity. Nat Neurosci. 2013;16:1737–44. doi: 10.1038/nn.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.About I, Laurent-Maquin D, Lendahl U, Mitsiadis TA. Nestin expression in embryonic and adult human teeth under normal and pathological conditions. The American journal of pathology. 2000;157:287–95. doi: 10.1016/S0002-9440(10)64539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michalczyk K, Ziman M. Nestin structure and predicted function in cellular cytoskeletal organisation. Histol Histopathol. 2005;20:665–71. doi: 10.14670/HH-20.665. [DOI] [PubMed] [Google Scholar]
- 13.Krupkova O, Jr, Loja T, Zambo I, Veselska R. Nestin expression in human tumors and tumor cell lines. Neoplasma. 2010;57:291–8. doi: 10.4149/neo_2010_04_291. [DOI] [PubMed] [Google Scholar]
- 14.Jeong J, Mao J, Tenzen T, Kottmann AH, McMahon AP. Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes & development. 2004;18:937–51. doi: 10.1101/gad.1190304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14:123–34. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–13. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 17.Encinas JM, Michurina TV, Peunova N, Park JH, Tordo J, Peterson DA, et al. Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell stem cell. 2011;8:566–79. doi: 10.1016/j.stem.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldowitz D, Hamre K. The cells and molecules that make a cerebellum. Trends Neurosci. 1998;21:375–82. doi: 10.1016/s0166-2236(98)01313-7. [DOI] [PubMed] [Google Scholar]
- 19.Yuelling LW, Du F, Li P, Yang ZJ. Isolation of distinct cell populations from the developing cerebellum by microdissection. J Vis Exp. 2014:18. doi: 10.3791/52034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, et al. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–72. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- 21.Yokota N, Aruga J, Takai S, Yamada K, Hamazaki M, Iwase T, et al. Predominant expression of human zic in cerebellar granule cell lineage and medulloblastoma. Cancer research. 1996;56:377–83. [PubMed] [Google Scholar]
- 22.Vintersten K, Monetti C, Gertsenstein M, Zhang P, Laszlo L, Biechele S, et al. Mouse in red: red fluorescent protein expression in mouse ES cells, embryos, and adult animals. Genesis. 2004;40:241–6. doi: 10.1002/gene.20095. [DOI] [PubMed] [Google Scholar]
- 23.Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–10. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 24.Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:416–29. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 25.Dai P, Akimaru H, Tanaka Y, Maekawa T, Nakafuku M, Ishii S. Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. The Journal of biological chemistry. 1999;274:8143–52. doi: 10.1074/jbc.274.12.8143. [DOI] [PubMed] [Google Scholar]
- 26.Sisson JC, Ho KS, Suyama K, Scott MP. Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell. 1997;90:235–45. doi: 10.1016/s0092-8674(00)80332-3. [DOI] [PubMed] [Google Scholar]
- 27.Dahlstrand J, Zimmerman LB, McKay RD, Lendahl U. Characterization of the human nestin gene reveals a close evolutionary relationship to neurofilaments. J Cell Sci. 1992;103(Pt 2):589–97. doi: 10.1242/jcs.103.2.589. [DOI] [PubMed] [Google Scholar]
- 28.Niewiadomski P, Kong JH, Ahrends R, Ma Y, Humke EW, Khan S, et al. Gli protein activity is controlled by multisite phosphorylation in vertebrate Hedgehog signaling. Cell reports. 2014;6:168–81. doi: 10.1016/j.celrep.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam BY, Chawla S. MEF2D expression increases during neuronal differentiation of neural progenitor cells and correlates with neurite length. Neuroscience letters. 2007;427:153–8. doi: 10.1016/j.neulet.2007.09.030. [DOI] [PubMed] [Google Scholar]
- 30.Pan Y, Wang B. A novel protein-processing domain in Gli2 and Gli3 differentially blocks complete protein degradation by the proteasome. The Journal of biological chemistry. 2007;282:10846–52. doi: 10.1074/jbc.M608599200. [DOI] [PubMed] [Google Scholar]
- 31.Balordi F, Fishell G. Hedgehog signaling in the subventricular zone is required for both the maintenance of stem cells and the migration of newborn neurons. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2007;27:5936–47. doi: 10.1523/JNEUROSCI.1040-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balordi F, Fishell G. Mosaic removal of hedgehog signaling in the adult SVZ reveals that the residual wild-type stem cells have a limited capacity for self-renewal. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2007;27:14248–59. doi: 10.1523/JNEUROSCI.4531-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW, et al. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer research. 2006;66:6063–71. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peacock CD, Wang Q, Gesell GS, Corcoran-Schwartz IM, Jones E, Kim J, et al. Hedgehog signaling maintains a tumor stem cell compartment in multiple myeloma. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4048–53. doi: 10.1073/pnas.0611682104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zbinden M, Duquet A, Lorente-Trigos A, Ngwabyt SN, Borges I, Ruiz i Altaba A. NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. The EMBO journal. 2010;29:2659–74. doi: 10.1038/emboj.2010.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao C, Chen A, Jamieson CH, Fereshteh M, Abrahamsson A, Blum J, et al. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature. 2009;458:776–9. doi: 10.1038/nature07737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jia J, Tong C, Jiang J. Smoothened transduces Hedgehog signal by physically interacting with Costal2/Fused complex through its C-terminal tail. Genes & development. 2003;17:2709–20. doi: 10.1101/gad.1136603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruel L, Rodriguez R, Gallet A, Lavenant-Staccini L, Therond PP. Stability and association of Smoothened, Costal2 and Fused with Cubitus interruptus are regulated by Hedgehog. Nat Cell Biol. 2003;5:907–13. doi: 10.1038/ncb1052. [DOI] [PubMed] [Google Scholar]
- 39.Wen X, Lai CK, Evangelista M, Hongo JA, de Sauvage FJ, Scales SJ. Kinetics of hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Mol Cell Biol. 2010;30:1910–22. doi: 10.1128/MCB.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson SL, Wilson JP, Wang C, Wang B, McConnell SK. Primary cilia and Gli3 activity regulate cerebral cortical size. Developmental neurobiology. 2012;72:1196–212. doi: 10.1002/dneu.20985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng D, Stavrou T, Liu L, Taylor MD, Gold B, Dean M, et al. Retrospective family study of childhood medulloblastoma. Am J Med Genet A. 2005;134:399–403. doi: 10.1002/ajmg.a.30653. [DOI] [PubMed] [Google Scholar]
- 42.Northcott PA, Dubuc AM, Pfister S, Taylor MD. Molecular subgroups of medulloblastoma. Expert Rev Neurother. 2012;12:871–84. doi: 10.1586/ern.12.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Northcott PA, Korshunov A, Pfister SM, Taylor MD. The clinical implications of medulloblastoma subgroups. Nat Rev Neurol. 2012;8:340–51. doi: 10.1038/nrneurol.2012.78. [DOI] [PubMed] [Google Scholar]
- 44.Ng JM, Curran T. The Hedgehog’s tale: developing strategies for targeting cancer. Nature reviews Cancer. 2011;11:493–501. doi: 10.1038/nrc3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. The New England journal of medicine. 2009;361:1173–8. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gajjar A, Stewart CF, Ellison DW, Kaste S, Kun LE, Packer RJ, et al. Phase I study of vismodegib in children with recurrent or refractory medulloblastoma: a pediatric brain tumor consortium study. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:6305–12. doi: 10.1158/1078-0432.CCR-13-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Metcalfe C, de Sauvage FJ. Hedgehog fights back: mechanisms of acquired resistance against Smoothened antagonists. Cancer research. 2011;71:5057–61. doi: 10.1158/0008-5472.CAN-11-0923. [DOI] [PubMed] [Google Scholar]
- 48.Yauch RL, Dijkgraaf GJ, Alicke B, Januario T, Ahn CP, Holcomb T, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326:572–4. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kimura H, Ng JM, Curran T. Transient inhibition of the Hedgehog pathway in young mice causes permanent defects in bone structure. Cancer Cell. 2008;13:249–60. doi: 10.1016/j.ccr.2008.01.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.