Abstract
Meningitis infection is one of the major threats during Hajj season in Mecca. Meningitis vaccines are available, but their uses are limited in some countries due to religious reasons. Furthermore, they only give protection to certain serogroups, not to all types of meningitis-inducing bacteria. Recently, research on epitope-based vaccines has been developed intensively. Such vaccines have potential advantages over conventional vaccines in that they are safer to use and well responded to the antibody. In this study, we developed epitope-based vaccine candidates against various meningitis-inducing bacteria, including Streptococcus pneumoniae, Neisseria meningitidis, and Haemophilus influenzae type b. The epitopes were selected from their protein of polysaccharide capsule. B-cell epitopes were predicted by using BCPred, while T-cell epitope for major histocompatibility complex (MHC) class I was predicted using PAProC, TAPPred, and Immune Epitope Database. Immune Epitope Database was also used to predict T-cell epitope for MHC class II. Population coverage and molecular docking simulation were predicted against previously generated epitope vaccine candidates. The best candidates for MHC class I- and class II-restricted T-cell epitopes were MQYGDKTTF, MKEQNTLEI, ECTEGEPDY, DLSIVVPIY, YPMAMMWRNASNRAI, TLQMTLLGIVPNLNK, ETSLHHIPGISNYFI, and SLLYILEKNAEMEFD, which showed 80% population coverage. The complexes of class I T-cell epitopes–HLA-C*03:03 and class II T-cell epitopes–HLA-DRB1*11:01 showed better affinity than standards as evaluated from their ΔGbinding value and the binding interaction between epitopes and HLA molecules. These peptide constructs may further be undergone in vitro and in vivo testings for the development of targeted vaccine against meningitis infection.
Keywords: meningitis, immunoinformatics, epitope-based vaccine, epitope prediction, molecular docking
Introduction
Meningitis infection is one of the serious threats during Hajj due to its tendency to cause outbreaks and epidemics.1 According to WHO, this disease affects more than 400 million people who live in the area of “African meningitis belt” (from Senegal to Ethiopia). More than 800,000 people in this area were infected, with a case fatality rate of 10%.2 In Saudi Arabia, the epidemics of meningitis usually occur during or after Hajj and Umrah seasons, due to massive gathering of people in certain areas. Pattern evolution confirmed that 48% of meningitis cases were reported at the two holy cities, namely, Mecca and Medina.2 In 2000, Indonesian pilgrims were infected by Neisseria meningitidis serogroup A and W135. Among 253 identified cases from Saudi Arabia, 93 cases were caused by N. meningitidis serogroup W135, while 60 cases were caused by N. meningitidis serogroup A. Statistically, there were nine cases caused by serogroup W135 and six cases caused by serogroup A per 100,000 population.3
Generally, meningitis can be induced by certain species of virus and bacteria. Three bacterial species, namely, Haemophilus influenzae (45%), Streptococcus pneumoniae (18%), and N. meningitidis (14%), are known to cause the majority of cases, where the case fatality rates vary according to the type of bacteria. The highest fatality rate is caused by S. pneumoniae (19%), while the case fatality rates caused by N. meningitidis and H. influenzae are 13% and 3%, respectively.4
The Indonesian government requests all Hajj pilgrims to be vaccinated against meningitis bacteria before departing to Mecca. Currently, the government allows the use of Meningitis vaccine only from Novartis, Menveo®. This vaccine contains polysaccharides from each of serogroups A, C, W, and Y conjugated to a mutant of diphtheria toxin, CRM197, that differs from the wild type by the substitution of one amino acid.5,6 There have been some efforts to develop alternative vaccine candidates that could incorporate broader types of meningitis-inducing bacteria.7–10
Recently, novel approaches have been directed toward the rational design of B- and T-cell epitope-based vaccine, on account of the advancement of recombinant DNA technology, cell culture technique, immunoinformatics, big data projects, and rational design of antigens.11,12 The epitope-based vaccine has several advantages over conventional vaccines, and some are moving forward to the clinical trial pipeline.12 This next-generation vaccine has high specificity in evoking immune response, high capacity of production, and effective cost of production. Moreover, peptides consisting of epitopes are easily synthesized, purified, stored, and handled. Generally, epitope-based vaccines are also considered safer than traditional vaccines.12
Immunoinformatics approach has been used to develop subunit vaccine candidates against meningitis-inducing bacteria. Some previous studies generated several vaccine candidates: epitope FMILPIFNV against human leukocyte antigen (HLA) class II from ABC transporter protein of S. pneumoniae and epitope KGLVDDADI against HLA class I from outer membrane protein of N. meningitidis.13,14 The epitopes such as FMILPIFNV and KGLVDDADI were used as standards in this study.
This study designed epitope-based meningitis vaccine in silico by using polysaccharide capsule protein of S. pneumoniae, N. meningitidis serogroup A, N. meningitidis serogroup W, and H. influenzae type b and analyzed the complex stability between predicted epitopes and HLA molecules using molecular docking approach. The designed epitopes may serve as promising candidates for the development of epitope-based vaccine against the meningitis-inducing bacteria.
Research Methodology
Tools and materials
This study was conducted in silico. The pipeline used in this study was adjusted and extended from the existing ones.15–19 Polysaccharide proteins of S. pneumoniae, N. meningitidis, and H. influenzae type b were obtained from National Center Biotechnology information (NCBI). The 3D structure of HLA was obtained from Research Collaboratory for Structural Bioinformatics (RCSB). Online and offline software including the latest version of BCPred,20 VaxiJen v2.0,21 PAPRoc I,22 TAPPRed,23,24 Immune Epitope Database (IEDB) 3.0,25 PEP-FOLD,26 MOE 2009,27 and Chimera 1.928 were used in this study.
Procedure
Retrieving protein sequences from database
The sequences of polysaccharide protein of S. pneumoniae, N. meningitidis, and H. influenzae type b were searched in GenBank of NCBI (http://www.ncbi.nlm.nih.gov/). After that, antigenicity of these sequences was predicted using VaxiJen v2.0, which can be obtained at http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html.21
B-cell epitope prediction
The B-cell epitope was predicted using BCPred, which can be accessed at http://ailab.cs.iastate.edu/bcpreds/.20 Antigenicity was also predicted against B-cell epitope afterward using VaxiJen.
HLA class I and class II T-cell epitope prediction from the conserved sequences
Epitopes from S. pneumoniae, N. meningitidis, and H. influenzae type b against HLA class I were predicted using several online softwares including PAProC,22 TAPPred,23 and IEDB.25 Proteasome cleavage site of consensus protein sequence was predicted using PAProC (http://www.paproc2.de/paproc1/paproc1.html). PAProC then generated peptide sequences and score of estimated strength.29 The peptide binding to the transporter associated with antigen processing (TAP) binding was predicted using TAPPred, http://www.imtech.res.in/raghava/tappred/, which generated protein sequences, their position, and score of predicted binding affinity to each peptide sequence.23 The antigenicity of peptide sequence generated from TAPPred prediction was analyzed using VaxiJen,21 and the antigen-bearing sequences were used to predict epitopes that bind to HLA class I and HLA class II using IEDB analysis resource.25 The binding character of epitopes to HLA was taken into consideration for the selection of the best epitopes. Epitopes with more number of bonds with HLA were considered as better than those with fewer bonds.16
The 3D structure of epitopes for HLA class I and class II
The 3D structures of the best epitopes for each HLA class I and class II were predicted using PEP-FOLD (http://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD/)26 and saved in.pdb format, while the 3D structures of HLA class I and class II were obtained from RCSB (www.rcsb.org).30
The prediction of coverage population of the selected epitopes
The human population coverage is one aspect that has to be taken into consideration in selecting the best epitopes, besides their ability to bind to HLA molecules.16 Human coverage population for previously selected epitopes was predicted using IEDB analysis resource for population coverage calculation (http://tools.immuneepitope.org/tools/population/iedb_input).31
Molecular docking study of HLA–epitope interaction
The interaction between the best predicted epitopes and HLA alleles was analyzed by means of molecular docking using MOE 2009. The 3D structure of HLA, as a target molecule, and the epitopes must be prepared before docking. Preparation and docking steps were performed according to the established pipeline from previous research.32,33
Results
Protein sequence searching
A total of 4 polysaccharide protein sequences of H. influenzae type b, 3069 sequences of S. pneumoniae, 19 sequences of N. meningitidis serogroup A, and 19 sequences of N. meningitidis serogroup W were retrieved from NCBI. These sequences were subjected to antigenicity prediction in order to estimate the presence of antigen in their sequences. The peptide sequences having the VaxiJen value above the threshold level ($0.4) were used for further analysis.
B-cell epitope prediction
B-cell epitopes from H. influenzae type b, S. pneumoniae, and N. meningitidis were predicted using BCPred where criteria were set to have 75% specificity, including only nonoverlapping epitopes. BCPred prediction generated peptide sequences along with their scores (Table 1). Peptides having higher scores mean that they are easily recognized by B-cell, thus having a higher probability as epitopes.34
Table 1.
B-cell epitope prediction.
| EPITOPE | BCpred | ANTIGENICITY |
|---|---|---|
| Haemophilus influenzae type b | ||
| GDKTTFKQS | 0.863 | Antigen |
| NFSKGVEPQ | 0.715 | Antigen |
| LGLIICAIA | 0.704 | Non |
| GKIWGTLSF | 0.696 | Non |
| WRNASNRAI | 0.693 | Antigen |
| Streptococcus pneumoniae | ||
| DRVPEEASR | 0.99 | Antigen |
| QDVLEEVVS | 0.99 | Non |
| PATSPSSPN | 0.99 | Non |
| SDVTTLEEA | 0.94 | Antigen |
| TLQMTLLGI | 0.92 | Antigen |
| VVNRDQGEK | 0.88 | Antigen |
| LKLDLTPKD | 0.80 | Antigen |
| Neisseria meningitidis serogroup W | ||
| PNTRYRTPN | 0.99 | Antigen |
| ATTFSYLDG | 0.97 | Non |
| PILSNENVE | 0.97 | Non |
| DGSKDGSED | 0.97 | Antigen |
| RNTGIKNSN | 0.96 | Antigen |
| EKEVYAEDI | 0.94 | Antigen |
| PGSACNKII | 0.92 | Antigen |
| YYRQGRKDS | 0.91 | Antigen |
| VPIYNVESY | 0.87 | Non |
| LEKNAEMEF | 0.86 | Antigen |
| KYDKGSVSH | 0.83 | Non |
| IDSDDFINC | 0.81 | Non |
| YIYQDNQGT | 0.75 | Antigen |
| Neisseria meningitidis serogroup A | ||
| WQELYKKYK | 0.99 | Non |
| NANTLLEKE | 0.99 | Non |
| NSDATSTSR | 0.99 | Antigen |
| YFSAKKFAK | 0.98 | Non |
| EGEPDYLNG | 0.94 | Antigen |
| ILNNRKWRK | 0.93 | Antigen |
| EMEKKYPEE | 0.93 | Antigen |
| EISSLPYEE | 0.92 | Non |
| LNEEWNVQV | 0.91 | Antigen |
| LCILESHKE | 0.91 | Non |
| KLNNVVTLT | 0.89 | Non |
| TNISKAQSN | 0.88 | Antigen |
| QLFKEGIRN | 0.86 | Non |
| LEFCKEDKD | 0.86 | Non |
| FTWVNSEDK | 0.86 | Antigen |
| NCAPPAWLD | 0.85 | Non |
| ETSLHHIPG | 0.85 | Antigen |
| LSRDELKFA | 0.80 | Antigen |
| SLDDIAVTG | 0.79 | Antigen |
| HEEIMPQSA | 0.78 | Non |
| AWGNVNGEC | 0.77 | Antigen |
| YSNDDFLLT | 0.77 | Non |
| FFNFEYIVK | 0.73 | Antigen |
| FPLPSSFEK | 0.71 | Non |
| DPSAFFRDS | 0.69 | Non |
| NPKSVNEIW | 0.68 | Non |
Besides being recognized by B-cells, the peptide sequences must also possess antigen as evaluated by VaxiJen. The peptides with antigenic properties are necessary to raise the immune responses.18 Not all peptides from each bacteria passed these criteria, as listed in Table 1. H. influenzae protein, S. pneumoniae protein, N. meningitidis serogroup A, and N. meningitidis serogroup W protein generated 3, 5, 12, and 8 probable epitopes, respectively, with antigen attribute. The predicted B-cell epitopes from each bacteria protein, which produced the best BCPred score, were GDKTTFKQS, DRVPEEASR, PNTRYRTPN, and WQELYKKYK.
Identification of T-cell epitopes
T-cell epitopes must be recognized by T-cell receptor (TCR) in order to induce immune response. The epitopes that TCRs recognized are presented by major histocompatibility complex (MHC) molecules on the surface of cells. There are two major types of MHC protein molecules, namely, class I and class II. Cytotoxic T-lymphocyte TCRs recognize endogenous antigen presented on MHC class I, while helper T-cell and inflammatory T-cell TCRs recognize exogenous antigen presented on MHC class II.35
Identification of MHC class I epitope
A peptide having a proteasomal recognition site is not favorable as an epitope vaccine candidate because it will be degraded during antigen processing. The determination of proteasomal cleavage site was conducted using PAProC.22 Fragments having ≥9 amino acid residues were used for further analysis of TAP binding preference (Table 2). TAP binding preference has significant influence in selecting T-cell epitopes because antigenic peptides must first be transported by TAP from cytosol to endoplasmic reticulum to be presented on MHC class I.24,36 The higher the score, the higher the affinity between them and the higher the possibility for the epitope to be transported by TAP. In our result, we found that despite having high affinity toward TAP, several peptide sequences failed to have antigenicity, as calculated using VaxiJen.
Table 2.
The result of TAP prediction.
| PEPTIDE RANK | START POSITION | SEQUENCE | SCORE | PREDICTED AFFINITY | ANTIGENICITY |
|---|---|---|---|---|---|
| Haemophilus influenzae type b | |||||
| 34 | 1 | MQYGDKTTF | 6.05 | High | Antigen |
| 70 | 244 | DLALLLLGL | 4.71 | Intermediate | Antigen |
| 91 | 167 | IICAIAQQF | 4.18 | Intermediate | Non |
| 167 | 245 | LALLLLGLV | 2.37 | Low or undetectable | Non |
| 193 | 189 | LLPISGAFF | 1.73 | Low or undetectable | Non |
| 204 | 166 | LIICAIAQQ | 1.39 | Low or undetectable | Non |
| 245 | 243 | SDLALLLLG | −0.32 | Low or undetectable | Non |
| Streptococcus pneumoniae | |||||
| 82 | 1 | MKEQNTLEI | 3.13 | Intermediate | Antigen |
| 99 | 202 | DTRVKRPKD | 2.42 | Low or undetectable | Non |
| 149 | 222 | IVPNLNKLK | 1.42 | Low or undetectable | Non |
| 161 | 201 | LDTRVKRPK | 1.07 | Low or undetectable | Antigen |
| Neisseria meningitidis serogroup A | |||||
| 23 | 66 | RKQDMLIPI | 6.86 | High | Non |
| 39 | 65 | NRKQDMLIP | 6.31 | High | Non |
| 76 | 245 | SEDKNWQEL | 5.11 | Intermediate | Non |
| 137 | 110 | LESHKEDFL | 4.13 | Intermediate | Non |
| 252 | 68 | QDMLIPINF | 2.76 | Low or undetectable | Antigen |
| 313 | 67 | KQDMLIPIN | 2.15 | Low or undetectable | Non |
| 432 | 387 | ECTEGEPDY | 0.69 | Low or undetectable | Antigen |
| 486 | 386 | GECTEGEPD | −0.50 | Low or undetectable | Antigen |
| 520 | 385 | NGECTEGEP | −1.52 | Low or undetectable | Antigen |
| Neisseria meningitidis serogroup W | |||||
| 96 | 4 | SIVVPIYNV | 3.89 | Intermediate | Antigen |
| 123 | 21 | SSIEPILSN | 3.22 | Intermediate | Non |
| 160 | 2 | DLSIVVPIY | 2.58 | Low or undetectable | Antigen |
| 226 | 3 | LSIVVPIYN | 1.45 | Low or undetectable | Non |
| 242 | 1 | MDLSIVVPI | 0.88 | Low or undetectable | Antigen |
The peptides from H. influenzae type b, S. pneumoniae, and N. meningitidis serogroup A and W that passed proteasomal cleavage prediction, TAP binding efficiency, and antigenicity prediction were used for the prediction of HLA class I binding using IEDB resource. They are as follows: DLALLLLGL and MQYGDKTTF from H. influenzae type b; LDTRVKRPK and MKEQNTLEI from S. pneumoniae; ECTEGEPDY, GECTEGEPD, NGECTEGEP, and QDMLIPINF from N. meningitidis serogroup A; and SIVVPIYNV, DLSIVVPIY, and MDLSIVVPI from N. meningitidis serogroup W (Table 3).
Table 3.
Candidates of T-cell epitope for MHC class I.
| EPITOPE INTERACTION WITH HLA CLASS I (IC50) |
|---|
| Haemophilus influenzae type b |
| DLALLLLGL HLA-B*15:02(41.04), HLA-C*12:03(196.43), HLA-C*14:02(280.94), HLA-C*03:03(329.31), HLA-A*02:01(492.16) |
| MQYGDKTTF HLA-C*03:03(13.23), HLA-B*15:01(23.07), HLA-C*12:03(54.60), HLA-A*32:01(59.83), HLA-A*02:06(142.74), HLA-C*14:02(165.05), HLA-B*48:01(198.88), HLA-B*35:01(239.65), HLA-B*15:02(260.75), HLA-B*18:01(409.30), HLA-B*39:01(438.93), HLA-B*53:01(442.47) |
| Streptococcus pneumoniae |
| LDTRVKRPK HLA-C*03:03(19.84), HLA-C*12:03(49.91), HLA-C*14:02(109.05) |
| MKEQNTLEI HLA-C*12:03(29.19), HLA-C*05:01(221.14), HLA-C*14:02(243.01), HLA-C*03:03(294.18), HLA-B*15:02(387.46) |
| Neisseria meningitidis serogroup A |
| ECTEGEPDY HLA-C*12:03(34.29), HLA-C*03:03(35.78), HLA-C*14:02(148.46), HLA-C*07:02(275.53), HLA-B*15:02(279.40), HLA-B*35:01(296.40) |
| GECTEGEPD HLA-C*12:03(34.06), HLA-C*03:03(53.90) |
| NGECTEGEP HLA-C*12:03(26.38), HLA-C*03:03(90.70), HLA-C*14:02(90.91), HLA-C*07:02(191.94), HLA-C*05:01(280.98) QDMLIPINF HLA-C*12:03(54.85), HLA-C*14:02(61.18), HLA-B*15:02(309.19), HLA-C*03:03(336.98) |
| Neisseria meningitidis serogroup W |
| SIVVPIYNV HLA-C*12:03(15.46), HLA-A*68:02(66.46), HLA-C*15:02(141.72), HLA-A*02:06(172.01), HLA-C*03:03(264.61), HLA-A*02:01(264.92) |
| DLSIVVPIY HLA-C*12:03(48.00), HLA-C*07:02(196.87), HLA-C*03:03(197.97), HLA-C*14:02(205.88), HLA-B*15:02(298.01), HLA-A*30:02(319.49), HLA-B*18:01(431.56), HLA-A*29:02(433.27), HLA-C*05:01(434.18) |
| MDLSIVVPI HLA-C*03:03(17.40), HLA-C*12:03(48.00), HLA-A*32:01(61.26), HLA-A*32:01(90.56), HLA-C*14:02(95.20), HLA-B*53:01(387.15), HLA-B*40:02(389.82), HLA-A*68:02(406.06). |
The analysis in stabilized matrix method-based IEDB MHC I prediction tool retrieved several possible MHC I alleles that could interact well with the epitopes from four bacteria (Table 3). Each predicted MHC I allele was given an IC50 value that indicates affinity between epitope and MHC I molecule. A lower IC50 value indicates higher affinity toward MHC molecules. A peptide showing an IC50 value lower than 50, 500, and 5000 nM is associated with high affinity, intermediate affinity, and low affinity toward MHC class I molecule, respectively. Moreover, a peptide is categorized as a binder if it has IC50 lower than 500 nM and is categorized as a nonbinder if IC50 is equal to or more than 500 nM.37 In this study, the peptides were selected if they possess antigenicity, IC50 value lower than 500 nM, and more than 5 bonds with HLA class I.16 Our study identified four antigenic peptides (MQYGDKTTF, MKEQNTLEI, ECTEGEPDY, and DLSIVVPIY) as candidates of T-cell epitope for MHC class I that could interact with HLA-B*15:02, HLA-C*03:03, and HLA-C*14:02 (Table 3). Of these three types mentioned, HLA-C*03:03 has the best interaction with the selected epitope candidates. Therefore, HLA-C*03:03 structure was chosen as a model for molecular docking study of HLA peptides.
Identification of MHC class II epitope
MHC class II molecules present antigenic peptides to stimulate cellular and humoral immunity through the actions of helper T-lymphocytes. Identification of MHC class II-restricted epitope is very important in designing epitope-based vaccine.
There are four T-cell epitopes, namely, YPMAMM-WRNASNRAI, TLQMTLLGIVPNLNK, ETSLHHIP-GISNYFI, and SLLYILEKNAEMEFD, generated for MHC class II from IEDB prediction. Each of these epitopes similarly binds to three types of HLA class II, HLA-DRB1*01:01, HLA-DRB1*04:04, and HLA-DRB1*11:01. However, HLA-DRB1*11:01 has the strongest affinity with the predicted epitopes. Therefore, this type of HLA will be used as a model in molecular docking simulation to predict the strength of association between HLA and epitopes (Table 4).
Table 4.
Candidates of T-cell epitope for MHC class II.
| EPITOPE PREDICTION | INTERACTION WITH HLA CLASS II (IC50) | ANTIGENICITY |
|---|---|---|
| Haemophilus influenzae type b | ||
| YGDKTTFKQSLAIQG | HLA-DRB1*09:01(55.00), HLA-DRB1*01:01(60.00), HLA-DRB5*01:01(69.00), HLA-DRB1*07:01 (102.00), HLA-DRB1*04:04(147.00), HLA-DRB1*11:01(163.00), HLA-DRB1*04:05(178.00) | Antigen |
| GDKTTFKQSLAIQGR | HLA-DRB1*09:01(55.00), HLA-DRB1*01:01(55.00), HLA-DRB5*01:01(65.00), HLA-DRB1*07:01(102.00), HLA-DRB1*04:04(147.00), HLA-DRB1*11:01(158.00), HLA-DRB1*04:05(176.00) | Antigen |
| GLVMVKNFSKGVEPQ | HLA-DRB1*13:02(35.00), HLA-DRB5*01:01(86.00), HLA-DRB1*01:01(143.00), HLA-DRB1*15:01(262.00), HLA-DRB1*07:01(266.00), HLA-DRB1*11:01(305.00), HLA-DRB1*09:01(470.00) | Non |
| YPMAMMWRNASNRAI | HLA-DRB1*01:01(83.00), HLA-DRB1*11:01(116.00), HLA-DRB1*07:01(117.00), HLA-DRB1*13:02(127.00), HLA-DRB5*01:01(235.00), HLA-DRB1*04:01(317.00), HLA-DRB1*04:04(349.00), HLA-DRB1*09:01(373.00) | Antigen |
| MAMMWRNASNRAIGS | HLA-DRB1*01:01(87.00), HLA-DRB1*07:01(124.00), HLA-DRB1*13:02(129.00), HLA-DRB1*11:01(188.00), HLA-DRB5*01:01(239.00), HLA-DRB1*04:01(320.00), HLA-DRB1*09:01(392.00) | Antigen |
| PMAMMWRNASNRAIG | HLA-DRB1*01:01(88.00), HLA-DRB1*07:01(124.00), HLA-DRB1*13:02(130.00), HLA-DRB1*11:01(189.00), HLA-DRB5*01:01(244.00), HLA-DRB1*04:01(326.00), HLA-DRB1*09:01(395.00), HLA-DRB1*04:04(473.00) | Antigen |
| AMMWRNASNRAIGSI | HLA-DRB1*01:01(88.00), HLA-DRB1*07:01(124.00), HLA-DRB1*13:02(130.00), HLA-DRB1*11:01(189.00), HLA-DRB5*01:01(244.00), HLA-DRB1*04:01(326.00), HLA-DRB1*09:01(395.00) | Non |
| MMWRNASNRAIGSIS | HLA-DRB1*01:01(106.00), HLA-DRB1*07:01(119.00), HLA-DRB1*13:02(156.00), HLA-DRB1*11:01(218.00), HLA-DRB5*01:01(308.00), HLA-DRB1*04:01(360.00), HLA-DRB1*09:01(402.00) | Non |
| Streptococcus pneumoniae | ||
| ISITRVSDVTTLEEA | HLA-DRB1*07:01(243.00), HLA-DRB1*01:01(302.00), HLA-DRB1*08:02(403.00), HLA-DRB1*04:04(457.00), HLA-DRB1*04:01(499.00) | Non |
| DTLQMTLLGIVPNLN | HLA-DRB1*04:04(10.00), HLA-DRB1*01:01(60.00), HLA-DRB1*04:05(79.00), HLA-DRB1*04:01(182.00), HLA-DRB1*07:01(269.00), HLA-DRB1*15:01(290.00), HLA-DRB4*01:01(333.00), HLA-DRB5*01:01(364.00) | Antigen |
| TLQMTLLGIVPNLNK | HLA-DRB1*04:04(10.00), HLA-DRB1*01:01(59.00), HLA-DRB1*04:05(78.00), HLA-DRB1*04:01(163.00), HLA-DRB5*01:01(163.00), HLA-DRB1*15:01(252.00), HLA-DRB4*01:01(342.00), HLA-DRB1*08:02(378.00), HLA-DRB1*07:01(465.00), HLA-DRB1*11:01(473.00) | Antigen |
| Neisseria meningitidis serogroup A | ||
| HIHKTNISKAQSNIS | HLA-DRB1*01:01(171.00), HLA-DRB1*07:01(200.00), HLA-DRB1*04:01(312.00), HLA-DRB4*01:01(481.00), HLA-DRB1*13:02(492.00) | Antigen |
| ETSLHHIPGISNYFI | HLA-DRB1*01:01(98.00), HLA-DRB1*04:04(158.00), HLA-DRB1*15:01(205.00), HLA-DRB1*04:05(258.00), HLA-DRB1*11:01(306.00), HLA-DRB1*07:01(308.00), HLA-DRB1*08:02(498.00) | Antigen |
| IETSLHHIPGISNYF | HLA-DRB1*01:01(134.00), HLA-DRB1*04:04(242.00), HLA-DRB1*04:05(277.00), HLA-DRB1*11:01(319.00), HLA-DRB1*07:01(491.00) | Non |
| NKFRSLDDIAVTGYL | HLA-DRB1*01:01(29.00), HLA-DRB1*04:01(201.00), HLA-DRB1*09:01(219.00), HLA-DRB1*04:05(325.00), HLA-DRB1*04:04 (440.00) | Antigen |
| LHNKFRSLDDIAVTG | HLA-DRB1*01:01(30.00), HLA-DRB1*04:01(203.00), HLA-DRB1*09:01(230.00), HLA-DRB1*04:05(303.00), HLA-DRB1*04:04(438.00) | Antigen |
| FFNFEYIVKKLNNQN | HLA-DRB1*11:01(18.00), HLA-DRB5*01:01(68.00), HLA-DRB1*04:04(207.00), HLA-DRB1*04:05(230.00), HLA-DRB1*01:01(323.00), HLA-DRB1*08:02(500.00) | Antigen |
| YKPDFNSDATSTSRF | HLA-DRB1*04:01(19.00), HLA-DRB1*04:05(218.00), HLA-DRB1*01:01(236.00), HLA-DRB3*01:01(390.00), HLA-DRB1*07:01(429.00), HLA-DRB1*04:04(499.00) | Antigen |
| KPDFNSDATSTSRFL | HLA-DRB1*04:01(19.00), HLA-DRB1*04:05(206.00), HLA-DRB1*01:01(207.00), HLA-DRB1*07:01(295.00), HLA-DRB3*01:01(461.00) | Antigen |
| EGEPDYLNGARNANT | HLA-DRB1*01:01(100.00), HLA-DRB1*04:04(124.00), HLA-DRB1*04:01(256.00), HLA-DRB1*04:05(355.00) | Antigen |
| MFILNNRKWRKLKRD | HLA-DRB1*11:01(81.00), HLA-DRB1*03:01(142.00), HLA-DRB5*01:01(206.00), HLA-DRB1*13:02(373.00). | Non |
| Neisseria meningitidis serogroup A | ||
| ARNTGIKNSNGKYIV | HLA-DRB1*13:02(11.00), HLA-DRB1*07:01(99.00), HLA-DRB1*09:01(257.00), HLA-DRB5*01:01(283.00), HLA-DRB1*01:01(474.00) | Antigen |
| RNTGIKNSNGKYIVF | HLA-DRB1*13:02(11.00), HLA-DRB1*07:01(95.00), HLA- DRB1*09:01(248.00), HLA-DRB5*01:01(278.00), HLA-DRB1*01:01(483.00) | Antigen |
| EARNTGIKNSNGKYI | HLA-DRB1*13:02(12.00), HLA-DRB1*07:01(105.00), HLA-DRB1*09:01(266.00), HLA-DRB5*01:01(295.00) | Antigen |
| SLLYILEKNAEMEFD | HLA-DRB1*01:01(64.00), HLA-DRB1*04:01(123.00), HLA-DRB5*01:01(133.00), HLA-DRB1*04:04(161.00), HLA-DRB1*11:01(264.00), HLA-DRB1*04:05 (310.00) | Antigen |
| LLYILEKNAEMEFDR | HLA-DRB1*01:01(70.00), HLA-DRB1*04:01(128.00), HLA-DRB5*01:01(136.00), HLA-DRB1*04:04(172.00), HLA-DRB1*04:05(404.00) | Non |
| KSLLYILEKNAEMEF | HLA-DRB1*01:01(53.00), HLA-DRB1*04:01(118.00), HLA-DRB5*01:01(130.00), HLA-DRB1*04:04(147.00), HLA-DRB1*11:01(220.00), HLA-DRB1*04:05(237.00), HLA-DRB1*15:01(293.00), HLA-DRB1*12:01(392.00) | Non |
Coverage population prediction of class I and class II epitopes
An epitope will evoke a response only in individuals that express an MHC molecule capable of binding that particular epitope. However, human MHC (HLA) alleles are highly polymorphic and different types of HLA are expressed differently in the different population. Therefore, ensuring broad population coverage by selecting epitopes with different HLA binding specificities is an important consideration in designing epitope-based vaccine.31 An epitope is said to show good coverage if its value is approaching 100% or close to 100%.16 In this study, the average population coverage is 80%. Maximum coverage (94%) was found in the population of East Asia and North America, followed by Europe (93%). On the opposite side, the lowest coverage was showed in the population of Central America. While for Indonesian population, the selected epitopes showed more than half population coverage (Table 5).
Table 5.
Prediction of population coverage.
| POPULATION | COVERAGE (%) |
|---|---|
| East Asia | 94 |
| Northeast Asia | 84 |
| South Asia | 85 |
| Southeast Asia | 83 |
| Southwest Asia | 67 |
| Europe | 93 |
| East Africa | 78 |
| West Africa | 83 |
| Indonesia | 65 |
| Central Africa | 78 |
| North Africa | 84 |
| South Africa | 63 |
| West Indies | 81 |
| North America | 94 |
| Central America | 53 |
| South America | 81 |
| Oceania | 75 |
| Average | 80 |
Molecular docking
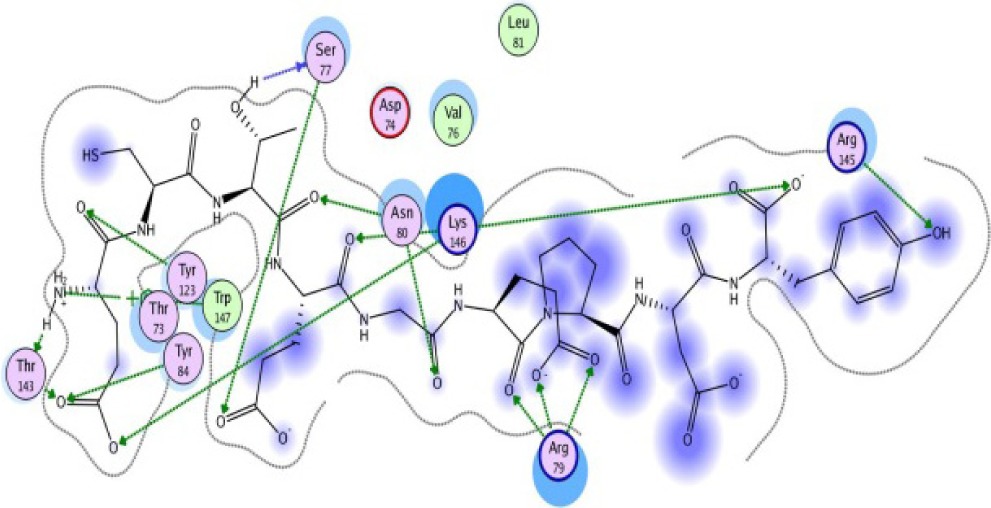
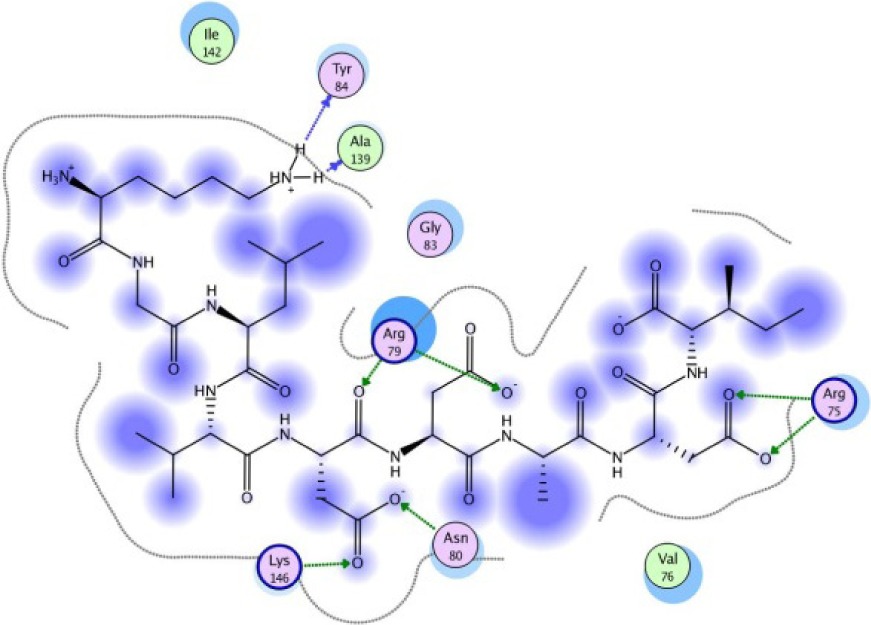
A binding interaction between epitopes and HLA alleles was assessed using MOE. The 3D structures of epitopes were predicted using PEP-FOLD26 and prepared using MOE, which includes wash, partial charge, and energy minimization. MHC I-restricted epitope and HLA C*03:03 formed a stable HLA–peptide complex with lower ΔGbinding than standards (KGLVDDADI), as presented in Table 6. The more negative ΔGbinding value, the stronger the interaction between the epitope and HLA. Apart from the ΔGbinding value, the interaction between epitope and HLA C*03:03 can also be studied by analyzing the hydrogen bond between them. Table 6 showed that eight hydrogen bonds were present in MQYGDKTTF–HLA class I, DLSIVVPIY–HLA class I, and standard (KGLVDDADI)–HLA class I complexes, which involved six amino acid residues, while MKEQNTLEI–HLA and ECTEGEPDY–HLA complexes formed 13 and 14 hydrogen bonds, respectively. Despite the same number of hydrogen bonds in two epitope–HLA class I complexes and standard–HLA class I complex, the ΔGbinding value for each of the complexes is different. This is due to the inequality of hydrogen bonds, which depends on the atom distances and angles. Moreover, the estimation of ΔGbinding also takes into account the contribution from other noncovalent interactions such as electrostatic solvation, hydrophobic interaction, rotational entropy, and translational entropy.38,39
Table 6.
Molecular docking simulation of MHC class I-restricted epitope with HLA-C*03:03.
| FREE ENERGY AND 3D EPITOPE | EPITOPE INTERACTION WITH HLA C*03:03 |
|---|---|
| MQYGDKTTF −14.43 kcal/mol 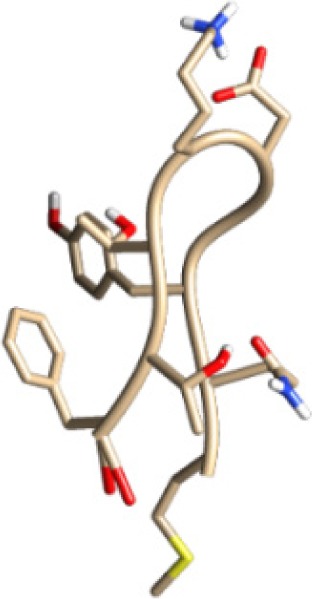
|
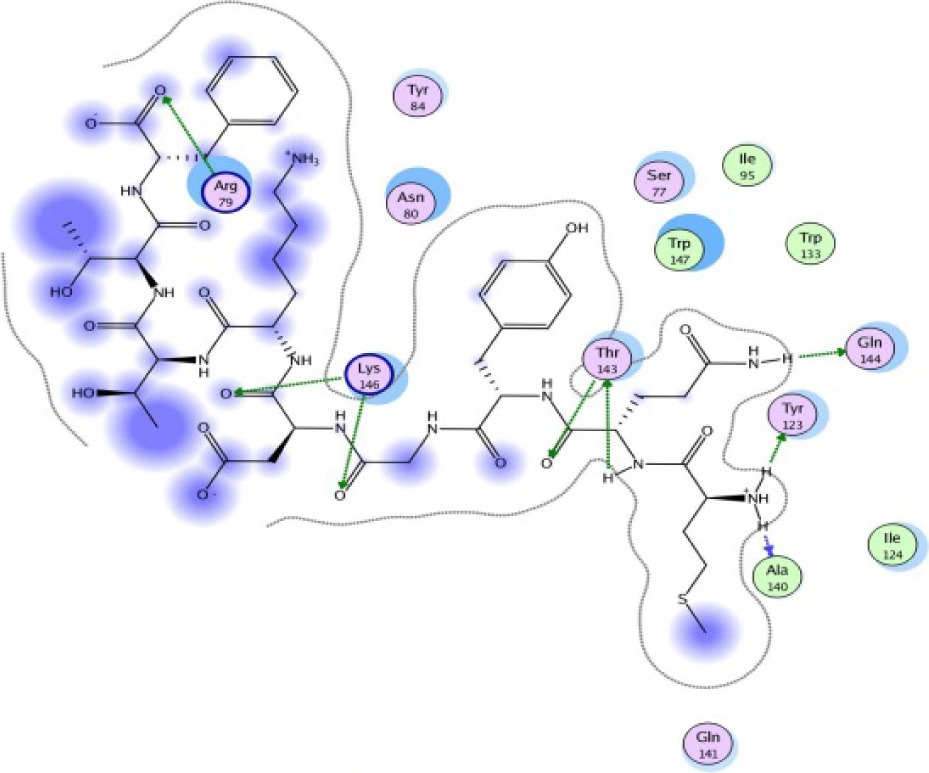
|
| MKEQNTLEI −17.46 kcal/mol 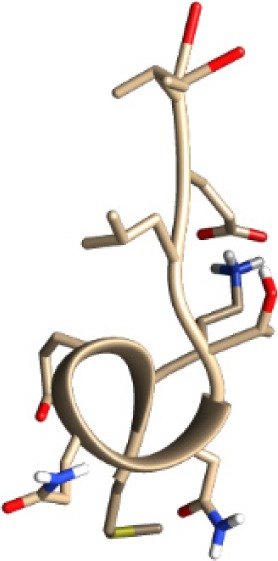
|
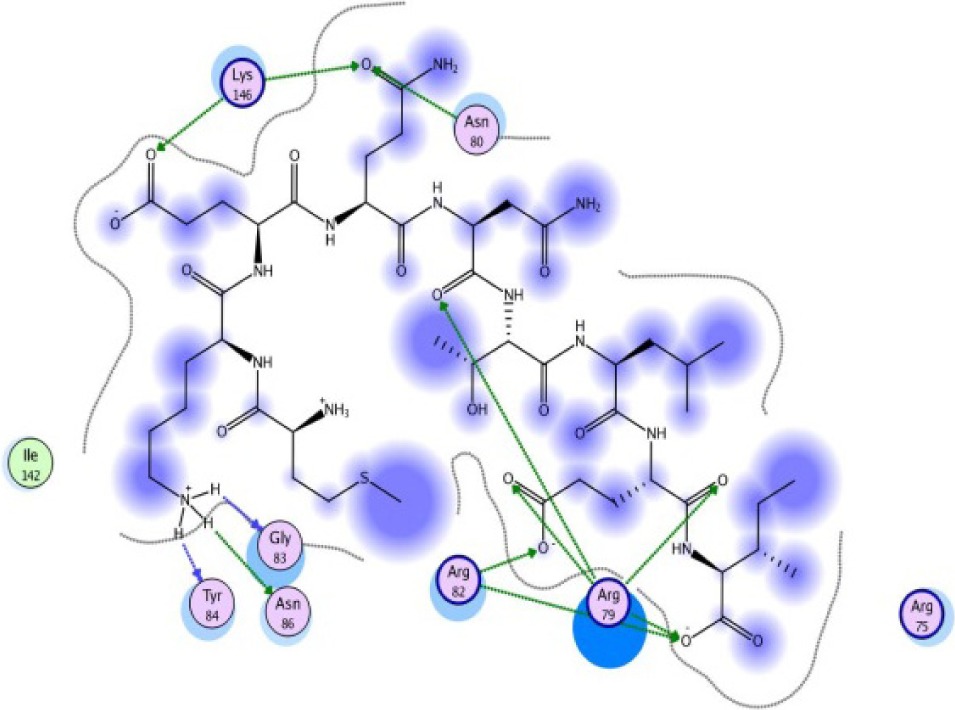
|
| ECTEGEPDY −16.28 kcal/mol 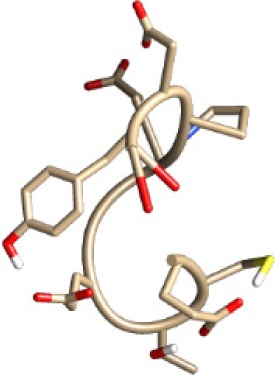
|
|
| DLSIVVPIY −16.88 kcal/mol 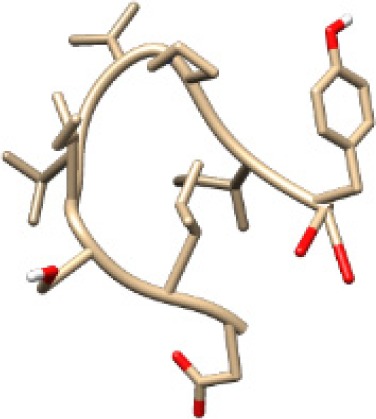
|

|
| KGLVDDADI −13.14 kcal/mol 
|
|
Notes:
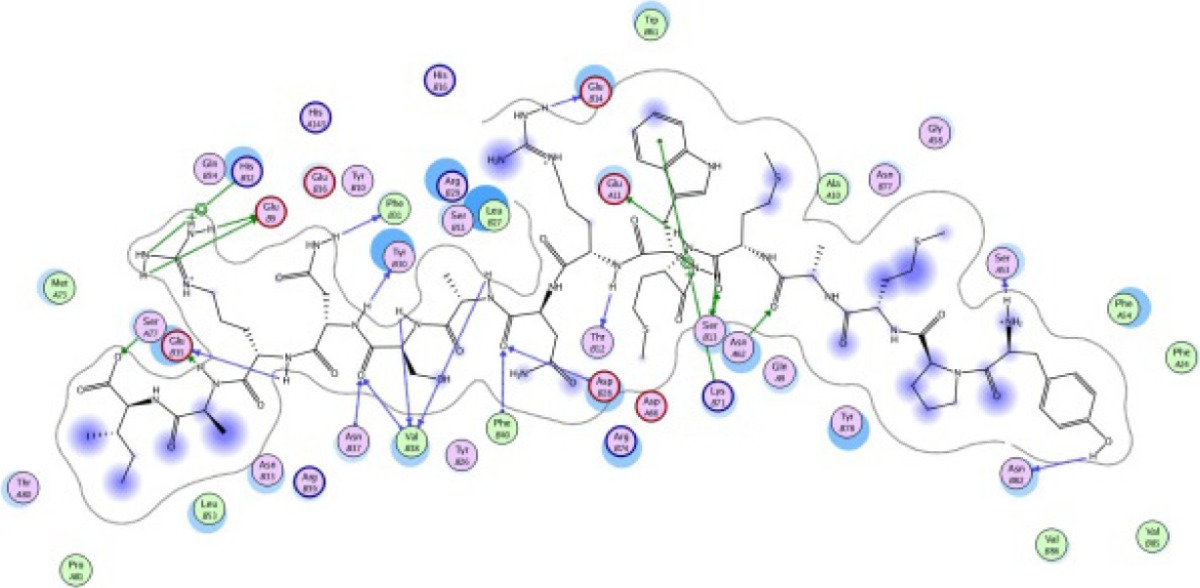
The same result was observed for the molecular docking simulation between MHC class II-restricted epitope and HLA-DRB1*11:01 (Table 7). The complex between selected epitopes and HLA class II showed more negative free energy of binding than the standard–HLA class II complex. The ETSLHHIPGISNYFI–HLA-DRB1*11:01 complex has the lowest ΔGbinding value of all complexes (−60.16 kcal/mol). The most favored binding orientation between each epitope and HLA class II molecule is displayed in Table 7.
Table 7.
Molecular docking simulation of MHC class II-restricted epitope with HLA-DRB1*11:01.
| FREE ENERGY AND 3D EPITOPE | EPITOPE INTERACTION WITH HLA DRB1*11:01 |
|---|---|
| YPMAMMWRNASNRAI −30.54 kcal/mol 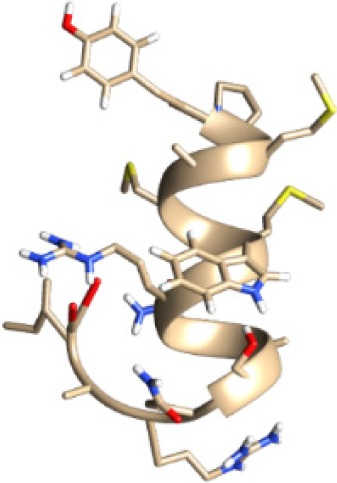
|
|
| TLQMTLLGIVPNLNK −25.61 kcal/mol 
|
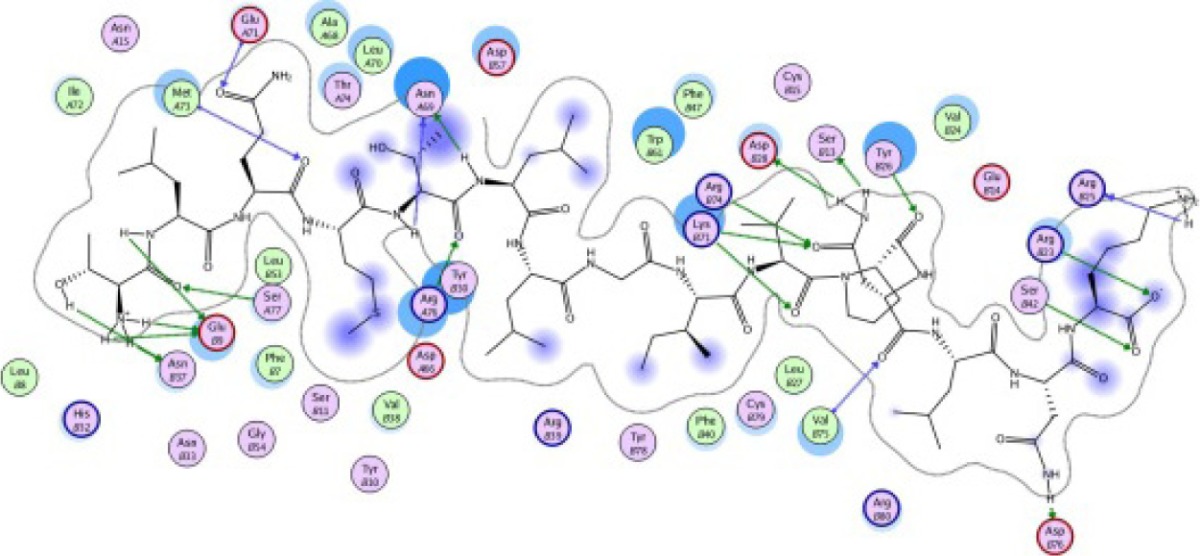
|
| ETSLHHIPGISNYFI −60.16 kcal/mol 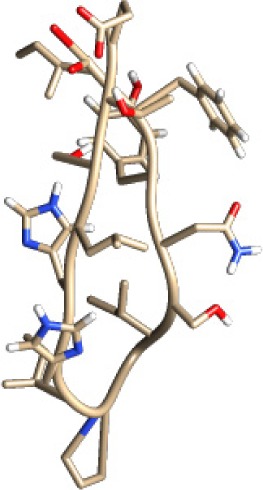
|
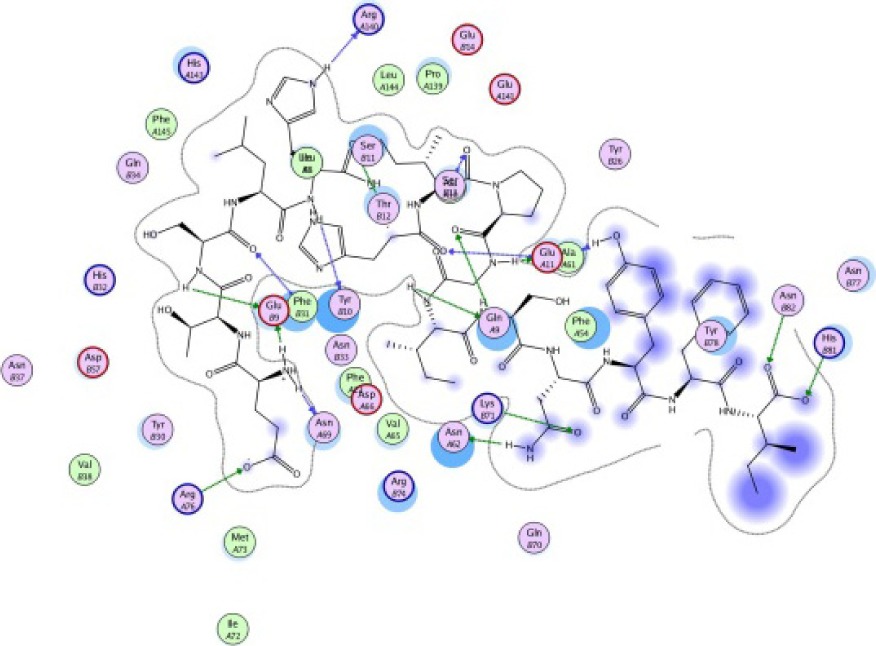
|
| SLLYILEKNAEMEFD −25.55 kcal/mol 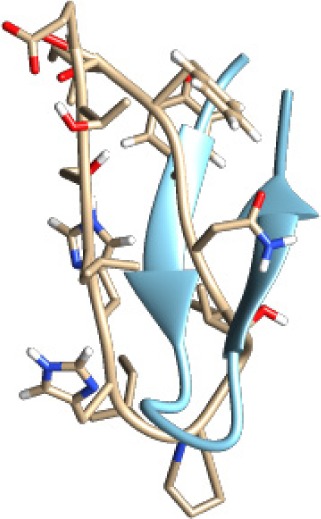
|
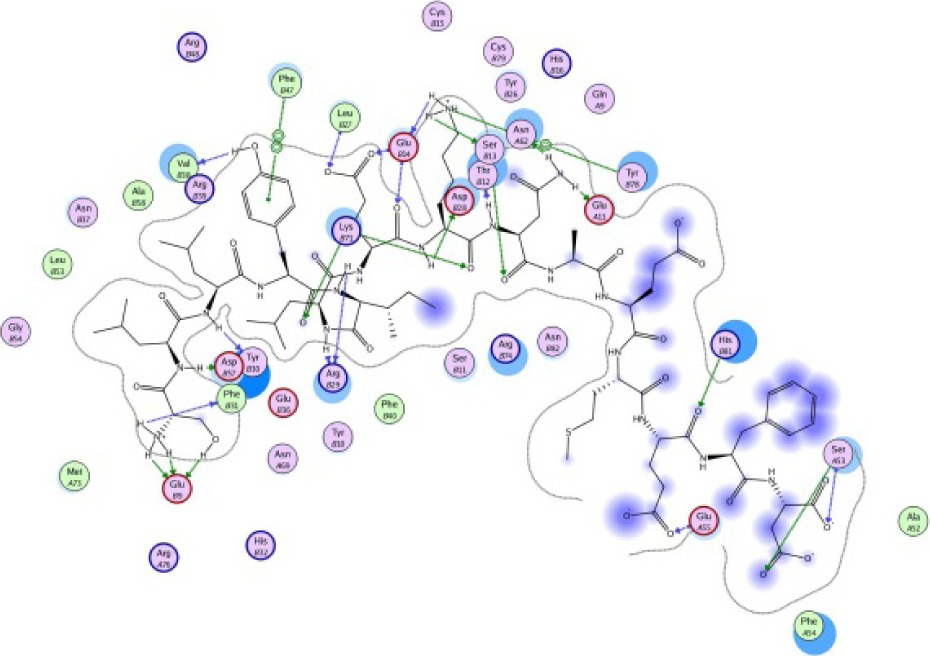
|
| FMILPIFNV −24.76 kcal/mol 
|
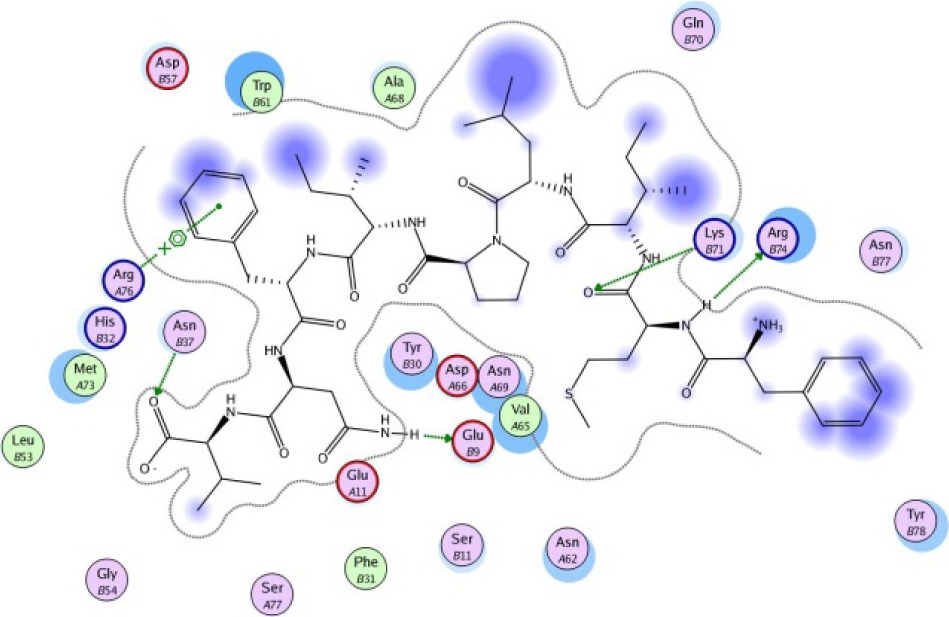
|
Notes:

Discussion
The main objective of epitope prediction is to design a molecule that can replace an antigen in the process of eliciting a relevant immune response. Designed molecules are favorable to use in vaccine production because they are cost-effective and noninfectious in contrast to whole pathogen organisms, which might possess risks to researchers or experimental subjects (animals and humans). This study incorporates immunoinformatics approach to reducing time- and cost-consuming hit and trial sets of wet laboratory experiments. This approach is used for the prediction of antigenic determinants in the capsular protein sequence of H. influenzae type b, S. pneumoniae, and N. meningitidis serogroup A and W.
According to the prediction result of IEDB and molecular docking study, the peptides that passed several criteria of probable epitope such as possessing antigenicity, binder attribute, and good affinity with HLA molecules are MQYGDKTTF, MKEQNTLEI, ECTEGEPDY, and DLSIVVPIY for MHC class I and YPMAMMWRNASNRAI, TLQMTLLGIVPNLNK, ETSLHHIPGISNYFI, and SLLYILEKNAEMEFD for MHC class II. These peptides also passed proteasomal cleavage and TAP binding efficiency prediction, which are of main concerns in designing good epitopes for vaccine candidates. Molecular docking study has been widely used in computer-aided drug design. However, it is now applied to investigate the epitope candidates that could bind MHC class I and class II molecules. Computational immunology is now considered to contribute to vaccine design in the way computational chemistry contributes to drug design.35 The algorithms for epitopes identification served by IEDB and molecular docking study have increased the overall efficiency in epitope discovery for vaccine research.
These epitopes also showed good population coverage (80% in average) and reached above average values in East Asia, North America, and Europe population. The high value of population coverage is needed to minimize the complexity of putting different epitopes in the development of vaccine.31 The predicted epitopes can be synthesized for further in vitro and in vivo assays.
Conclusion
We have predicted numerous antigenic peptides from the capsular protein sequence of H. influenzae type b, S. pneumoniae, and N. meningitidis serogroup A and W, which would be beneficial for effective vaccine development against meningococcal diseases. Results indicated that MQYGDKTTF, MKEQNTLEI, ECTEGEPDY, and DLSIVVPIY are potential vaccine candidates that have considerable binding with MHC class I alleles, while YPMAMMWRNASNRAI, TLQMTLLGIVPNLNK, ETSLHHIPGISNYFI, and SLLYILEKNAEMEFD are the potential candidates for MHC class II-restricted T-cell epitopes. These epitopes also had low energy minimization values that favored the stability of the epitope–MHC allele complex. However, experiments using model animals should be performed to verify their suitability to be included in a vaccine formulation against meningococcal diseases.
Acknowledgments
The authors thank Directorate of Research and Community Engagement, University of Indonesia, for providing facilities for this research.
Abbreviations
- MHC
major histocompatibility complex
- HLA
human leukocyte antigen
- DRB
antigen D-related beta chain
- RCSB
Research Collaboratory for Structural Bioinformatics
Footnotes
ACADEMIC EDITOR: Laurence Fitzhenry, Deputy Editor in Chief
PEER REVIEW: Three peer reviewers contributed to the peer review report. Reviewers’ reports totaled 420 words, excluding any confidential comments to the academic editor.
FUNDING: The authors thank DRPM Universitas Indonesia, PITTA 2016 for funding this research. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert single-blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Supervised this research: USFT and HZ. Worked on the technical details: AM. Gave technical assistance to the whole process: AAP. All the authors are responsible for writing this manuscript.
REFERENCES
- 1.Morley SL, Pollard AJ. Vaccine prevention of meningococcal disease, coming soon? Vaccine. 2002;20:666–687. doi: 10.1016/s0264-410x(01)00410-8. [DOI] [PubMed] [Google Scholar]
- 2.Bekairy AM, Harbi S, Alkatheri AM, Dekhail S, Swaidan L, Khalidi N. Bacterial meningitis: an update review. Afr J Pharm Pharmacol. 2014;8(18):469–478. [Google Scholar]
- 3.Lingappa JR, Al-rabeah AM, Hajjeh R, et al. Serogroup W-135 meningococcal disease during the Hajj, 2000. Emerg Infect Dis. 2003;9(6):665–671. doi: 10.3201/eid0906.020565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quagliarello VJ, Scheld WM. Treatment of bacterial meningitis. N Engl J Med. 1997;336(10):708–716. doi: 10.1056/NEJM199703063361007. [DOI] [PubMed] [Google Scholar]
- 5.Cooper B, Detora L. Menveo®: a novel quadrivalent conjugate vaccine against. Expert Rev Vaccines. 2011;10(1):21–33. doi: 10.1586/erv.10.147. [DOI] [PubMed] [Google Scholar]
- 6.Paulo S, Infectious P, Division D, Hospital SL, Paulo S, Vaccines N. Epidemiology and prevention of meningococcal disease: a critical appraisal of vaccine policies. Expert Rev Vaccines. 2012;10(12):1717–1730. doi: 10.1586/erv.11.159. [DOI] [PubMed] [Google Scholar]
- 7.Nolan T, Lambert S, Roberton D, et al. A novel combined Haemophilus influenzae type b-Neisseria meningitidis serogroups C and Y-tetanus-toxoid conjugate vaccine is immunogenic and induces immune memory when co-administered with DTPa-HBV-IPV and conjugate pneumococcal vaccines in infants. Vaccine. 2007;25(51):8487–8499. doi: 10.1016/j.vaccine.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 8.Tan LK, Carlone GM, Borrow R. Advances in the development of vaccines against Neisseria meningitidis. N Engl J Med. 2010;362(16):1511–1520. doi: 10.1056/NEJMra0906357. [DOI] [PubMed] [Google Scholar]
- 9.Pizza M, Scarlato V, Masignani V, et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287(5459):1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 10.Panatto D, Amicizia D, Lai PL, Cristina ML, Domnich A, Gasparini R. New versus old meningococcal group B vaccines: how the new ones may benefit infants & toddlers. Indian J Med Res. 2013;138(6):835–846. [PMC free article] [PubMed] [Google Scholar]
- 11.Lazer D, Kennedy R, King G, Vespignani A. The parable of Google flu: traps in big data analysis. Science. 2014;343(6167):1203–1205. doi: 10.1126/science.1248506. [DOI] [PubMed] [Google Scholar]
- 12.Oyarzun P, Kobe B. Recombinant and epitope-based vaccines on the road to the market and implications for vaccine design and production. Hum Vaccin Immunother. 2015;12(3):763–767. doi: 10.1080/21645515.2015.1094595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munikumar M, Priyadarshini IV, Pradhan D, Umamaheswari A, Vengamma B. Computational approaches to identify common subunit vaccine. Interdiscip Sci. 2013;5(2):155–164. doi: 10.1007/s12539-013-0161-1. [DOI] [PubMed] [Google Scholar]
- 14.Shah K, Chaubey P, Mishra N. Bioinformatics approach for screening and modeling of putative T cell epitopes from Por B protein of Neisseria meningitides as vaccine constructs. Indian J Biotechnol. 2010;9(October):351–359. [Google Scholar]
- 15.Tambunan U, Sipahutar F, Parikesit A, Kerami D. Vaccine design for H5N1 based on B- and T-cell epitope predictions. Bioinform Biol Insights. 2016;2016(10):27. doi: 10.4137/BBI.S38378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakib MS, Islam MR, Hasan AK, Nabi AH. Prediction of epitope-based peptides for the utility of vaccine development from fusion and glycoprotein of nipah virus using in silico approach. Adv Bioinforma. 2014;2014:402492. doi: 10.1155/2014/402492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar A, Jain A, Verma SK. Screening and structure-based modeling of T-cell epitopes of marburg virus NP, GP and VP40: an immunoinformatic approach for designing peptide-based vaccine. Trends Bioinforma. 2013;6(1):10–16. [Google Scholar]
- 18.Ingale AG. Epitopes identification for vaccine design and structural aspects of dengue virus 3 envelope protein. Biochem Physiol Open Access. 2014;3(2):1–7. [Google Scholar]
- 19.Jain A, Tripathi P, Shrotriya A, Chaudhary R, Singh A. In silico analysis and modeling of putative T cell epitopes for vaccine design of Toscana virus. 3 Biotech. 2015;5(4):497–503. doi: 10.1007/s13205-014-0247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Liu H, Yang J, Chou K. Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids. 2007;33(3):423–428. doi: 10.1007/s00726-006-0485-9. [DOI] [PubMed] [Google Scholar]
- 21.Doytchinova IA, Flower DR. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nussbaum AK, Kuttler C, Hadeler KP, Rammensee HG, Schild H. PAProC: a prediction algorithm for proteasomal cleavages available on the WWW. Immunogenetics. 2001;53(2):87–94. doi: 10.1007/s002510100300. [DOI] [PubMed] [Google Scholar]
- 23.Bhasin M, Lata S, Raghava GPS. TAPPred prediction of TAP-binding peptides in antigens. Methods Mol Biol. 2007;409:381–386. doi: 10.1007/978-1-60327-118-9_28. [DOI] [PubMed] [Google Scholar]
- 24.Bhasin M, Raghava GPS. Analysis and prediction of affinity of TAP binding peptides using cascade SVM. Protein Sci. 2004;13(3):596–607. doi: 10.1110/ps.03373104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vita R, Overton JA, Greenbaum JA, et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43(Database issue):D405–D412. doi: 10.1093/nar/gku938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thévenet P, Shen Y, Maupetit J, Guyon F, Derreumaux P, Tufféry P. PEP-FOLD: an updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 2012;40(Web Server issue):W288–W293. doi: 10.1093/nar/gks419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molecular Operating Environment (MOE), 2009.10. Chemical Computing Group Inc; 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7: 2016. [Google Scholar]
- 28.Pettersen EF, Goddard TD, Huang CC, et al. UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 29.Buus S, Brunak S, Kes C, Saxova P. Predicting proteasomal cleavage sites: a comparison of available methods. Int Immunol. 2003;15(7):781–787. doi: 10.1093/intimm/dxg084. [DOI] [PubMed] [Google Scholar]
- 30.Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bui HH, Sidney J, Dinh K, Southwood S, Newman MJ, Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics. 2006;7:153. doi: 10.1186/1471-2105-7-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tambunan USF, Zahroh H, Utomo BB, Parikesit AA. Screening of commercial cyclic peptide as inhibitor NS5 methyltransferase of dengue virus through molecular docking and molecular dynamics simulation. Bioinformation. 2014;10(1):23–27. doi: 10.6026/97320630010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tambunan USF, Zahroh H, Parikesit AA, Idrus S, Kerami D. Screening analogs of β-OG pocket binder as fusion inhibitor of dengue virus 2. Drug Target Insights. 2015;9:33–49. doi: 10.4137/DTI.S31566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ranjbar MM, Ghorban K, Alavian SM, et al. GB virus C/hepatitis G virus envelope glycoprotein E2: computational molecular features and immunoinformatics study. Hepat Mon. 2013;13(12):e15342. doi: 10.5812/hepatmon.15342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patronov A, Doytchinova I. T-cell epitope vaccine design by immunoinformatics. Open Biol. 2013;3(1):120139. doi: 10.1098/rsob.120139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang GL, Petrovsky N, Kwoh CK, August JT, Brusic V. PRED(TAP): a system for prediction of peptide binding to the human transporter associated with antigen processing. Immunome Res. 2006;2(1):3. doi: 10.1186/1745-7580-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh R, Singh S, Sharma PK, et al. Helper T cell epitope-mapping reveals MHC-peptide binding affinities that correlate with T helper cell responses to pneumococcal surface protein A. PLoS One. 2010;5(2):e9432. doi: 10.1371/journal.pone.0009432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kitchen D, Decornez H, Furr J, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov. 2004;3(11):935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 39.Corbeil CR, Williams CI, Labute P. Variability in docking success rates due to dataset preparation. J Comput Aided Mol Des. 2012;26(6):775–786. doi: 10.1007/s10822-012-9570-1. [DOI] [PMC free article] [PubMed] [Google Scholar]


