Abstract
Background
The aim of this study was to explore effects of microRNA-200c regulating TGF-β/Smad3 pathway by targeting Zeb1 on the occurrence and development of hypospadias and to evaluate the relationship between microRNA-200c and occurrence of hypospadias.
Material/Methods
Pregnant rats with a gestational age of 12 days were allocated into 2 groups; one received gavage of DEHP-contained soybean oil (1 ml/day, 8 days; Group A) and the other had gavage of normal soybean oil (1 ml/day, 8 days; Group B). Baby rats with hypospadias from Group A were assigned to the model group (n=20) and healthy baby rats from Group B were assigned to the control group (n=20). Real-time quantitative polymerase chain reaction (qRT-PCR), immunohistochemistry and Western blot analysis were performed to detect microRNA-200c, Zeb1, TGF-β, and Smad3 mRNA and protein expressions in the model group (n=20) and the control group (n=20). The relationship between microRNA-200c and Zeb1 was detected using a dual-luciferase reporter gene experiment. After the in vitro intervention experiment in fetal rat penises, Western blot was used to detect the expression of Zeb1, TGF-β, and Smad3.
Results
In the model group, microRNA-200c was expressed at a low level, and microRNA-200c expression in control group was 2.1 times higher than in the model group (P<0.05). When compared with the control group, mRNA expressions, protein expressions, and positive rates of Zeb1, TGF-β, and Smad3 were higher in the model group (all P<0.01). Luciferase gene report determined that Zeb1 is a target gene of microRNA-200c. The in vitro intervention experiment in fetal rat penises found that a high concentration of microRNA-200c inhibited hypospadias occurrence by suppressing the expression of Zeb1, TGF-β, and Smad3.
Conclusions
MicroRNA-200c was expressed in hypospadias penis tissues at low levels and was negatively correlated with Zeb1 expression. MicroRNA-200c up-regulated Zeb1 expression to regulate the TGF-β/Smad3 pathway, which led to the occurrence of hypospadias.
MeSH Keywords: Afferent Pathways, Hypospadias, MicroRNAs
Background
Hypospadias, defined as a malformation of the penis due to incomplete development of the ventral part of the penis [1], is one of most common congenital anomalies, affecting between 1 in 200 to 1 in 300 live male births [2]. A study has suggested a progressive increase in incidence of hypospadias in recent years and found an association between living in urban areas and hypospadias [3]. According to severity, hypospadias is classified into 3 degrees: the urethra opens on the anterior portion of the penis; openings on the midshaft of the penis; and posterior penile, penoscrotal, scrotal and perineal openings [4]. With the further study of hypospadias, we found the etiology is directly related to the following factors: genetic factors; epidermal growth factor (EGF); and environmental factors, including extra estrogen and progestin [5,6]. With the increasing use of molecular biology, several molecular methods have been used in the treatment of hypospadias, such as abnormal genetic transcription by gene site-directed mutation [7] and regulation of the expression and function of related target protein by microRNA overexpression [8], which indicate a new direction for hypospadias treatment.
MicroRNA (miRNA) is a small (21–25 nucleotides long) non-coding, single-stranded RNA derived from a 70-nucleotide-long precursor that controls gene expression by targeting mRNA transcripts, leading to its translational repression or degradation [9]. Several studies have demonstrated the important role of miRNA in cancers such as colorectal cancer, breast cancer, and gastric cancer [10,11]. Several studies have reported that miRNA-200 (miR-200c), a member of the miRNA-200 family, plays a critical role in the metastatic behavior of colorectal cancer; increasing expression of miR-200c results in the negative regulation of Zeb1 gene, which in turn regulates E-cadherin and vimentin expression to trigger epithelial-to-mesenchymal transition (EMT) [12,13]. In 2008, Korpal found that miR-200c can resist and reverse TGF-β1-induced EMT [14]. Another study confirmed that TGF-β1 has a vital role in the occurrence and development of urethral scar fibrosis [15]. Previously, Smad3 was found to be a downstream signal transduction factor of TGF-β1 [16]. These studies suggest that miRNA-200c plays a critical role in the pathogenesis of hypospadias. Therefore, the aim of this study was to evaluate the expression of miR-200c in hypospadias and normal penis tissues to determine the target gene of miR-200c and its related signal pathway in the occurrence of hypospadias.
Material and Methods
Ethics statement
The study design was reviewed and approved by the Ethics Committee of the First People’s Hospital of Yulin. This study was conducted in accordance with International Animal Convention.
Study subjects
The healthy and qualified rats used this study were provided by the Experimental Animal Centre of the First People’s Hospital of Yulin and the rats were housed as female-male pairs. The day on which sperm was detected on a vaginal smear was designated as gestation day (GD) 0. A total of 40 health rats on GD 12 were randomly selected and divided into 2 groups: the Di (2-ethylhexyl) phthalate (DEHP) experimental group (group A, n=20) and the soybean oil control group (group B, n=20). DEHP (500 mg/kg) dissolved in 1 ml of soybean oil was intragastrically administered to pregnant rats at 8:00 AM from GD 12 to GD 19, while the control group was only given an equivalent volume of soybean oil [17]. On postnatal day (PND) 1, the male newborn rats were counted and weighted, and their anogenital distances (AGD) were measured under a dissecting microscope. On PND 70, male offspring were intraperitoneally anesthetized with 10% chloral hydrate. After anesthesia, the length, curvature, and urethral opening position of each rat’s penis were examined to identify the presence of hypospadias.
A total of 20 male rats with hypospadias was assigned to the model group and 20 normal male rats were assigned to the control group. All rats were aged PND 70. After processing by liquid nitrogen, penis tissues were kept at −80°C. Half of the tissue specimen of each rat was used to detect microRNA-200c, and the other half underwent immunohistochemistry detection after 4% polyoxymethylene fixation and paraffin embedding.
Detecting the mRNA expressions of microRNA-200c, Zeb1, TGF-β, and Smad3 by quantitative real-time polymerase chain reaction (qRT-PCR)
A total of 50 mg frozen tissues were obtained to extract the total RNA using the miRNeasy Mini Kit (Qiagen, Valencia, CA). After 5 μl of the RNA sample was diluted 20 times with RNAase-free ultrapure water, the quality and purity of RNA were measured with optical density (OD) of 260 nm and 280 nm. OD260/OD280 between 1.7 and 2.1, which indicates higher purity of RNA, indicated qualification for inclusion in the following experiment. Briefly, total RNA was reverse-transcribed into cDNA templates using PCR amplification, and primers were synthesized by Invitrogen (Guangzhou) Trading Co. Ltd. The qRT-PCR was performed using 7500HT fluorogenic quantitative PCR (ABI Co.) at conditions of: pre-denaturation at 95°C for 10 min, 40 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 20 s, and extension at 72°C for 34 s). All primers are shown in Table 1. U6 snRNA was considered as an internal reference for miR-200c and GAPDH as internal reference for Zeb1, TGF-β, and Smad3. 2−ΔΔCt showed the ratio of genetic expressions in the experimental group and control group, and the formula was as follows: ΔΔCT=ΔCtexperimental group – ΔCtcontrol group; ΔCt=Ct miRNA – Ct U6. Here, Ct is the number of amplification cycles when the real-time fluorescence intensity of the reaction reaches the set threshold and the amplification increases as logarithmic phase.
Table 1.
Relative primer sequences.
| Gene | Forward primers: 5′-3′ | Reverse primers: 5′-3′ |
|---|---|---|
| mir-200c | ACACTCCAGCTGGGTAATACTGCC | TGGTGTCGTGCAGTCG |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
| Zeb1 | GGCACAACCAAAATCATCATCAA | GCAGTCCTCACACAAAAGGCAAG |
| TGF-β | TACCAACTACTGCTTCAGCTCCAC | GTTGTGTTGGTTGTAGAGGGCA |
| Smad3 | GAGAAACCAGTGACCACCAGATG | TAGGAGATGGAGCACCAGAAGG |
| GAPDH | GGAAATCCCATCACCATCT | GGACTCCACGACGTACTCA |
Detecting the expression of Zeb1, TGF-β, and Smad3 by immunohistochemistry
Two-step method was performed to detect the expressions of Zeb1, TGF-β, and Smad3 by PV-9000. After de-paraffinization in graded ethyl alcohol to distilled water, serial sections (4 μm) of each paraffin-embedded tissue chip were retrieved with antigens using a microwave, cooled naturally at room temperature, and washed by distilled water twice and phosphate-buffered (PBS) twice. After being treated with 3% hydrogen peroxide to block endogenous peroxidase, sections were incubated with primary antibodies (monoclonal rabbit anti-rat ZEB1, 1:100; rabbit anti-rat TGF-β, 1:100; rabbit anti-rat Smad 3, 1:100; Beijing Bioss Co.) overnight at 4°C, followed by 20-min incubation with polymerase adjuvant at room temperature, and 30-min incubation with horseradish peroxidase-labelled secondary antibodies (goat anti-rabbit ZEB1, 1:500; goat anti-rabbit TGF-β, 1:500; goat anti-rabbit Smad 3, 1:500; Beijing Bioss Co.) at room temperature. Finally, the sections were visualized by addition of a buffered diaminobenzidine (DAB) and then counterstained with hematoxylin and sealed. Primary antibody was replaced by PBS as negative control, and normal penis tissues were used as positive control. Zeb1 protein was mainly expressed in the cell nucleus as diffuse yellow granules, while TGF-β and Smad3 were expressed in the cell nucleus of penis matrix and vascular endothelial cells of subcutaneous tissue. The percentage of positive cells was assigned to one of the following categories: 1=1~10%; 2=11~25%; 3=26~50%; and 4=51~75%; 5=76%100%. The relative intensity of positive signals was classified into 3 grades with 1~3 scores: 1 (weakly stained, stronger than background, negative); 2 (moderately stained, light yellow, weakly positive); 3 (intensely stained, yellow, positive). These 2 scores were added and divided by 2 to produce the final score for each section (negative=1~2.5; positive=3~4) [18].
Detecting the expression of Zeb1, TGF-β, and Smad3 by western blot
After cells were washed 3 times in pre-chilled PBS, cultured cells were lysed in lysis buffer in a culture bottle (100 μl/50 ml) and placed on ice for 30 min, followed by a 10-min centrifugation at 12 000 rpm at 4°C. Then, supernatant was absorbed and placed in a 0.5-ml centrifuge tube separately and saved at −20°C. PBS was used to dilute 2 μg/μl BSA protein standards into 20 μg/μl, 15 μg/μl, 10 μg/μl, 5 μg/μl, 2.5 μg/μl, and 0 μg/μl. Based on the number of samples, we calculated the amount of BCA detection reagent, and blended solution A and solution B (BCA kit, Pierce Co.) at a 50:1 ratio.
A total of 2 μl protein was diluted with 18 μl of double-distilled water and each sample was divided and placed into 2 wells. After 200 ul of detection liquid were added to 96-well plates, diluted protein standards and detecting samples (10 ul/well) were added in detection wells with light and even shaking, followed by 30-min incubation at 37°C and then cooling to room temperature. The absorption value of samples was obtained using a full-automatic microplate reader (Beijing Nuoya Instrument Co.) at 490 nm, and the protein concentration of samples was calculated with a standard curve. All samples were saved at −70°C.
In a cold chamber at 4°C, electrophoresis began at 60 V and then at 120 V for 1–2 h as soon as protein went into separation gel. The wet-transfer method was used to transfer protein onto PVDF membrane for 2 h in the 4°C cold chamber. After PVDF membrane was removed, samples were sealed with 5% nonfat dried milk Tris-buffered saline tween (TBST) for 1–2 h at room temperature. Then, the samples were incubated with primary antibodies for Zeb1, TGF-β, and Smad3 overnight at 4°C, and washed with TBST (10 min ×3). The cells were then incubated with secondary antibodies at room temperature for 1 h and washed 3 times in TBST (10 min ×3). After the samples were exposed and fixed to X-ray film, chemiluminescent assay was performed to detect protein bands, followed by data analysis.
Cell culture
Penis tissues were rinsed with normal saline, placed in an Erlenmeyer flask (0.25% trypsin and 0.02% EDTA solution), sealed, and kept at 4°C for 18 h. The skin graft was transferred to a petri dish, following by separating and discarding the epidermis. The dermis graft was rinsed with DMEM, cut into pieces, and digested with collagenase. After 1-h incubation at 37°C and repeated shaking for cell scattering, the digestive solution was filtered through a 100-mesh screen to collect cell suspensions, following by centrifugation. The supernatant was then discarded, and the cell cluster was made into a cell suspension using DMEM medium, following by counting and adjusting the cell density. The cells were cultured in DMEM supplemented with 10% fetal bovine serum in 5% CO2 and 95% humidity at 37°C. The culture cells grew as a monolayer and were subjected to routine cell passage.
Luciferase reporter gene experiment
We designed respectively mutant sequence and wild sequence, that Zeb1 has a deletion in 3′UTR for miR-200c binding, to insert reporter plasmids. In hypospadias penile tissue cells, miR-200c mimics were co-transfected with wild (Wt-miR-200c/Zeb1) and mutant (Mut-miR-200c/Zeb1) recombinant plasmids. DNA was extracted with TIANamp Genomic DNA Kit (TIANGEN) according to the manufacture’s instruction. With constructing luciferase reporter vector, luciferase activity of samples was measured using Dual-Luciferase Reporter Assay System (E1910) (Promega Co.). After 48-hour transfection, the former medium was removed, and the cells were washed with PBS twice and lysed in 100 μl passive lysis buffer (PLB) with 15-min shaking at room temperature so as to collect cell lysis buffer.
The program was set as 2 s pre-reading, 10 s reading and 100 μl sample size (LARII and Stop & Glo® Reagent). After the pre-made LARII, Stop & Glo ® Reagent and luminescent tube or plate with cell lysis buffer (20 μl per sample) was added in bioluminescence detector, program was conducted till the end of fluorescence reading and data were recorded.
In-vitro intervene experiment of fetal rat penis
The pregnant rats were killed by cervical dislocation at GD 12.5. The fetal rats were removed under sterile condition, washed with sterile PBS, and the abdominal cavity was dissected to identify male fetuses by finding testes. Cross-cutting along the penis root, the full penis was located 0.4 μm above the microporous membrane as ventral side downward, followed by adding 1.5 ml BjGb medium in each well until the penis was in a gas-liquid interface. The experiments were classified as the experimental group (200 μM mono-2-ethylhexyl phthalate (MEHP), in vivo metabolite of DEHP, supplemented with a high dose of miR-200c); the control group (200 μM MEHP, in vivo metabolite of DEHP); and the blank group (no treatment). Each group was incubated with 5% CO2 and saturation humidity at 37°C and the medium was replaced every 24 h. After 72-h incubation, the expression of Zeb1, TGF-β, and Smad3 mRNAs and proteins were measured.
Statistical analysis
All data were analyzed using SPSS 21.0 software (SPSS Inc, Chicago, IL, USA) and are expressed as mean ± standard deviation (SD). Measurement data were analyzed by t test, the chi-square test was performed to analyze enumeration data, and correlation analysis was analyzed using Pearson correlation method. P<0.05 was considered statistically significant.
Results
Modeling results of hypospadias in rats
In the DEHP experiment group, 20 pregnant rats had 127 baby rats. Of these 127, 67 were male newborns, of which 26 had hypospadias (an occurrence rate of 20.5%). In the soybean oil control group, 20 pregnant rats had 103 baby rats, of which 50 newborns were males with no hypospadias. There were no significant differences in the number of newborns or their sex ratios (both P>0.05). When compared with the soybean oil control group, the occurrence rate of hypospadias in the DEHP experiment group was significantly higher (P<0.05), indicating the success in modeling (Table 2). For normal male rats, the urethra opens at the top of the penis with complete foreskin, and micturition is linear when stimulated. Male rats with hypospadias have an abnormal opening at the junction between the penis and the perinea or ventral side, with ventral foreskin division.
Table 2.
Comparison between experiment group (DEHP-induced hypospadias) and control group.
| Group | Pregnant rats (n) | Newborns (n) | Male newborns (n) | Occurrence of hypospadias (n) | AGD (cm) | Occurrence rate of hypospadias (%) |
|---|---|---|---|---|---|---|
| Experiment group | 20 | 127 | 67 | 26 | 0.181* | 20.5 |
| Control group | 20 | 103 | 50 | 0 | 0.208 | 0 |
AGD – anogential distance;
compare with control group, P<0.001
The expression of microRNA-200c
When compared with the control group, qRT-PCR detection results showed that the expression of microRNA-200c in penis tissues was lower in the model group. The expression quantity of microRNA-200c was 2.1 times higher than in the model group (t=2.09, P<0.01) (Figure 1).
Figure 1.
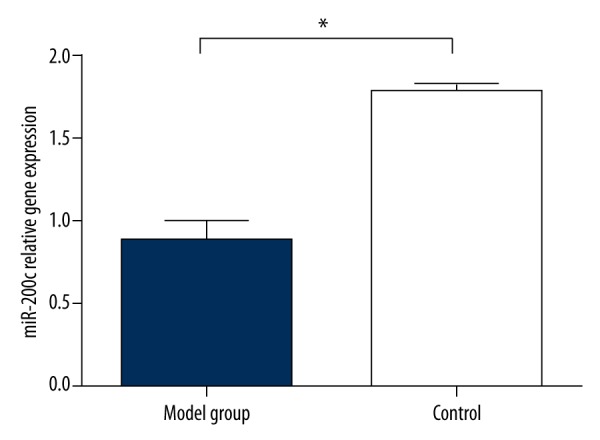
Differences in the expression level of microRNA-200c in penis tissues between model group and control group, * compared with control group, P<0.01.
The mRNA expression levels of Zeb1, TGF-β and Smad3
qPT-PCR was performed to detect the expression of Zeb1 mRNA, TGF-β, and Smad3. The results (Figure 2) showed that expression level of Zeb1 mRNA in penis tissues in the model group was 1.3±0.12, which was significantly higher than the 0.67±0.06 in the control group (t=2.04, P<0.01). In the model group, the expression level of TGF-β mRNA in penis tissues was 1.45±0.13, which was significantly higher than the 1.08±0.10 in the control group (t=2.03, P<0.01). When compared with the control group (0.78±0.07), the expression level of Smad3 mRNA in penis tissues was much higher (1.25±0.08) (t=2.03, P<0.01).
Figure 2.
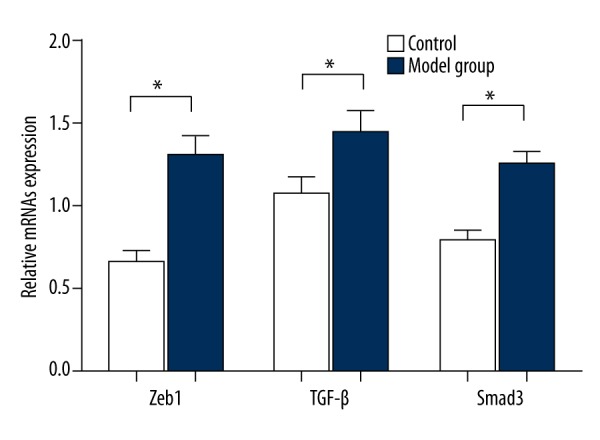
Comparisons of the expression levels of Zeb1, TGF-β, and Smad3 mRNAs in penis tissues between control group and model group, * compared with control group, P<0.01.
The protein expression levels of Zeb1, TGF-β, and Smad3
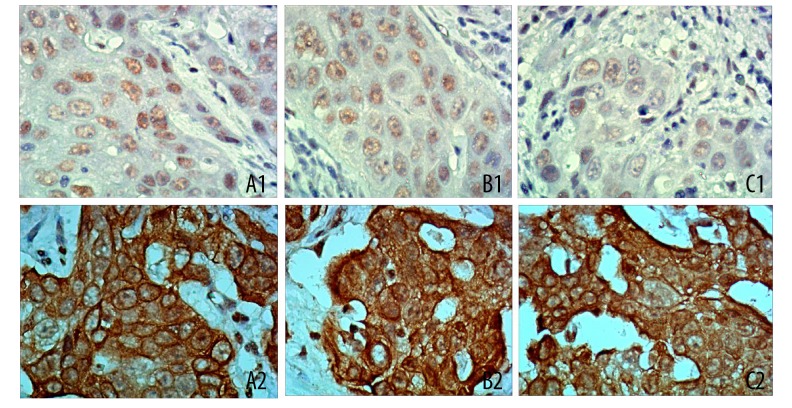
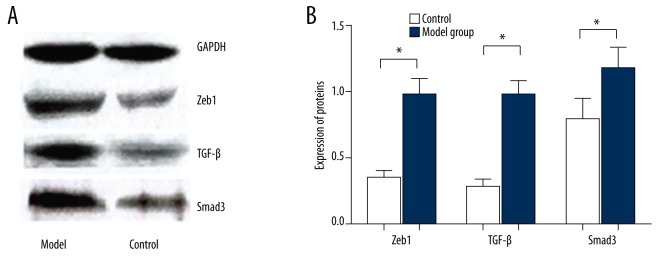
Immunohistochemical results demonstrated that Zeb1 protein was mainly positively expressed in cell nucleus as diffuse yellow granules, while TGF-β and Smad3 proteins were positively expressed in the cell nuclei of penis matrix and vascular endothelial cells of subcutaneous tissues (Figure 3). As shown in Table 3, the positive expression rate of Zeb1, TGF-β, and Smad3 in penis tissues in the model group was significantly higher than in the control group. Moreover, Western blot results (Figure 4) indicated that the expression of Zeb1 in penis tissues in the model group was 0.97±1.23, which was significantly higher than the 0.35±0.05 in the control group (P<0.01). When compared with the control group (0.29±0.04), the expression of TGF-β in penis tissues in the model groups was much higher (0.98±0.10) (P<0.01). In the model group, the expression of Smad3 in penis tissues was 1.15±0.16, which was significantly higher than the 0.78±0.16 found in the control group (P<0.01).
Figure 3.
Immunohistochemistry results on the expressions of Zeb1, TGF-β, and Smad3 (200×). (A1) weakly positive expression of Zeb1 in normal penis tissues; (A2) strongly positive expression of Zeb1 in penis tissues with hypospadias; (B1) weakly positive expression of TGF-β in normal penis tissues; (B2) strongly positive expression of TGF-β in penis tissues with hypospadias; (C1) weakly positive expression of Smad3 in normal penis tissues; (C2) strongly positive expression of Smad3 in penis tissues with hypospadias.
Table 3.
The expression status of Zeb1, TGF-β and Smad3 proteins.
| N | Zeb1 | TGF-β | Smad3 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Negative | Positive | Positive rate (%) | Negative | Positive | Positive rate (%) | Negative | Positive | Positive rate (%) | ||
| Model group | 20 | 3 | 17 | 85 | 1 | 19 | 95 | 2 | 18 | 90 |
| Control group | 20 | 18 | 2 | 10 | 18 | 2 | 10 | 17 | 3 | 15 |
| χ2 | 22.56 | 28.97 | 22.56 | |||||||
| P | P<0.001 | P<0.001 | P<0.001 | |||||||
Figure 4.
(A, B) The Western blot results on expression levels of Zeb1, TGF-β, and Smad3 proteins in model group and control group, * compared with control group, P<0.01.
Correlation analysis of microRNA-200c with Zeb1, TGF-β, and Smad3
As shown in Table 4, Pearson correlation analysis results demonstrated that the miRNA-200c expression level and the Zeb1 expression level were negatively correlation (r=−0.991, P<0.01), but the Zeb1 expression level was positively correlated with the expression levels of TGF-β and Smad3 (r=0.999, P<0.001; r=0.995, P<0.01).
Table 4.
Correlation of the microRNA-200c expression with the expression of Zeb1, TGF-β and Smad3 in model group.
| Indicator | microRNA-200c | Zeb 1 | TGF-β | Smad3 |
|---|---|---|---|---|
| microRNA-200c | 1.000 | −0.991* | −0.990* | −0.985* |
| Zeb 1 | −0.991* | 1.000 | 0.999* | 0.995* |
| TGF-β | −0.990* | 0.999* | 1.000 | 0.994* |
| Smad3 | −0.985* | 0.995* | 0.994* | 1.000 |
P<0.01, significant correlation between two indicators.
Verifying the target gene of miR-200c
The online prediction software Target Scan was used to determine the target site for Zeb1 to bind miR-200c and the sequences in 3′-UTR where Zeb1 mRNA binds miR-200c (Figure 5A). In order to prove miR-200c was targeting Zeb1, we designed a mutant sequence and a wild sequence that is a 3′ UTR deletion in Zeb1 for miR-200 binding, to insert reporter plasmid. In hypospadias penile tissue cells, miR-200c mimics were co-transfected with wild (Wt-miR-200c/Zeb1) and mutant (Mut-miR-200c/Zeb1) recombinant plasmids. The luciferase activity detection results showed that miR-200c mimics had no significant effects on the luciferase activity intensity in the mutant (Mut-miR-200c/Zeb1) reporter plasmid group; however, in the wild (Wt-miR-200c/Zeb1) reporter plasmid group, the luciferase activity intensity was decreased (P<0.05) (Figure 5B).
Figure 5.
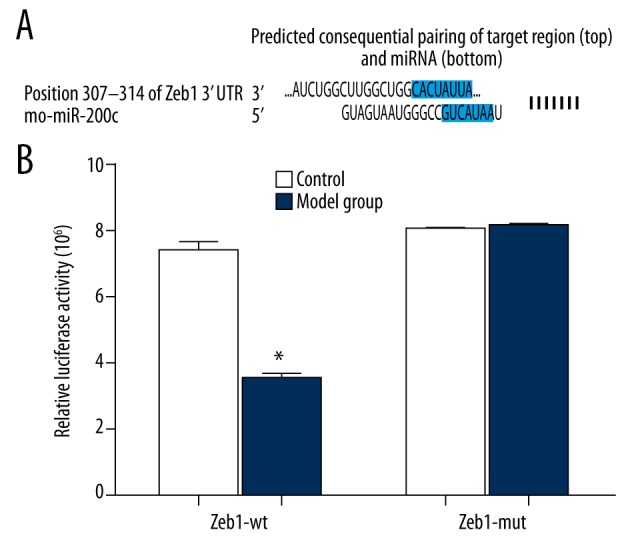
The Target Scan software identified that miR-200c directly targets Zeb1. (A) Gene comparison of miR-200c and Zeb1 3′UTR; (B) Dual-luciferase reporter gene activity assay. * P<0.01 in comparison with the control group.
In vitro intervention results of fetal rats
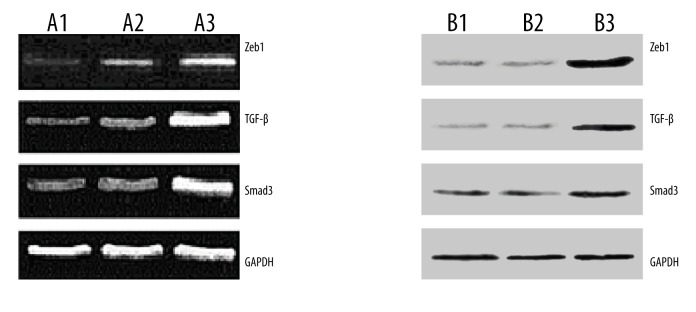
After 72-h in vitro incubation of penis tissues, expressions of Zeb1, TGF-β, and Smad3 mRNAs were quantified (Figure 6A), showing that the largest expression quantity was in the control group, but the expression quantity in the experiment group was close to that in the blank group. Western blot analysis was performed to detect the expression of Zeb1, TGF-β, and Smad3 proteins. Taking GAPDH as an internal reference, Zeb1, TGF-β, and Smad3 expressions were highly expressed in the control group and the expression results in the experiment group were similar to that in the blank group (Figure 6B). The results indicated that the high dose of miRNA-200c inhibited the expressions of Zeb 1, TGF-β, and Smad3 in penis tissues and reduced the occurrence and severity of hypospadias. The results also identified that Zeb1, TGF-β, and Smad3 were highly expressed in hypospadias penis tissues.
Figure 6.
In vitro intervention results in fetal rats. (A1) expression of Zeb1, TGF-β, and Smad3 mRNAs in the blank group; (A2) expression of Zeb1, TGF-β, and Smad3 mRNAs in the experimental group, which was similar to the blank group; (A3) high expression of Zeb1, TGF-β, and Smad3 mRNAs in the control group (GAPDH as internal reference). (B1) the expression of Zeb1, TGF-β, and Smad3 proteins in penis tissues in blank group; (B2) the expression of Zeb1, TGF-β, and Smad3 proteins in penis tissues in the experimental group, similar to the blank group; (B3) the high expression of Zeb1, TGF-β, and Smad3 proteins in penis tissues in the control group.
Discussion
There is currently no clear understanding of the complex etiology of hypospadias. By intragastrically administering pregnant rats with DEHP to produce baby rats with hypospadias, we found microRNA-200c was under-expressed in hypospadias tissues, and more importantly, the expression of Zeb1 was negatively correlated with miR-200c. As a member of the miRNA-200 family, related research involving miR-200c has concentrated on its functions in tumors. Recently, studies reported that miRNA-200 families were up-regulated in different tumor cells and able to inhibit tumor cell invasion and migration [10,19,20]. One study showed that microRNA-200c was negatively correlated with Zeb1 protein expression in pancreatic cancer [21]. In colorectal cancer, miR-200c inhibits Zeb1 protein expression [12]. Moreover, miR-200c inhibits Zeb1 protein expression on cell membranes so as to promote cancer cell invasion [22]. Furthermore, it was found that miR-200c can resist and reverse TGF-β by suppressing Zeb1 and Zeb2 activities [14]. Together, these data suggest that miR-200c is negatively correlated with Zeb1 protein expression in various diseases. This further confirms that reduction of miR-200c and increase of Zeb1 protein expression in hypospadias promotes the occurrence and development of hypospadias.
By detecting the expression of Zeb1, TGF-β, and Smad3 at the protein level, we found the expression level of Zeb1, TGF-β, and Smad3 in the model group was significantly higher than that in the control group. In vitro experiment results showed that high levels of miR-200c can reduce the expression of Zeb1, TGF-β, and Smad3 so as to inhibit the occurrence of hypospadias. A recent study reported that miR-200c inhibited activity of Smad pathway and secretion of TGF-β in keloid fibroblasts [23]. Another study reported that the development of fetal rat genitalia used urethral epithelium as a molecular signal center, which was regulated by TGF-β1 [7]. As the potent chemokine of fibroblasts, TGF-β1 can inhibit proliferations of most cells and induce epithelial cell apoptosis through autocrine and paracrine functioning in inflammatory and repair cells directly or indirectly, individually or synergistically, synchronously or asynchronously [24]. Moreover, a study found that phosphorylation of Smad3 necessarily involves TGF-β1 because only phosphorylated Smad3 can enter into the nucleus from the cytoplasm and then regulate the expression of the specific downstream gene, which triggers a series of biological events, including promoting fibroblast collagen synthesis, reducing collagen degradation, and adhering cells to extracellular matrix [25]. These findings suggest that TGF-β and Smad3 promote the occurrence of hypospadias, leading to the inhibition of growth of urethral epithelial cells and apoptosis, and that inhibitory expression of TGF-β and Smad3 play a vital role in healing hypospadias.
However, the present study has its own limitations. Besides the small number of experiment subjects, this study did not completely explain the relationship between DEPH and miRNA-200c in pregnant rats. Furthermore, it is still unknown whether high doses of DEPH cause the low expression of miRNA-200c, leading to hypospadias. Finally, this study did not explore the relationship of different DEHP doses with degree of hypospadias and miRNA-200c expression quantity.
Conclusions
This study indicates that miR-200c is expressed at low levels in hypospadias penis tissues and is negatively correlated with Zeb1 protein expression. Furthermore, miRNA-200c might up-regulate Zeb1 gene and protein expression to up-regulate TGF-β/Smad3 pathway level, suggesting that miRNA-200c is correlated with hypospadias occurrence and degree.
Footnotes
Competing interests
None.
Source of support: Departmental sources
References
- 1.Kalfa N, Philibert P, Werner R, et al. Minor hypospadias: the “tip of the iceberg” of the partial androgen insensitivity syndrome. PLoS One. 2013;8:e61824. doi: 10.1371/journal.pone.0061824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winston JJ, Meyer RE, Emch ME. Geographic analysis of individual and environmental risk factors for hypospadias births. Birth Defects Res A Clin Mol Teratol. 2014;100:887–94. doi: 10.1002/bdra.23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, Mao M, Dai L, et al. Time trends and geographic variations in the prevalence of hypospadias in China. Birth Defects Res A Clin Mol Teratol. 2012;94:36–41. doi: 10.1002/bdra.22854. [DOI] [PubMed] [Google Scholar]
- 4.Akin Y, Ercan O, Telatar B, et al. Hypospadias in Istanbul: incidence and risk factors. Pediatr Int. 2011;53:754–60. doi: 10.1111/j.1442-200X.2011.03340.x. [DOI] [PubMed] [Google Scholar]
- 5.Kalfa N, Sultan C, Baskin LS. Hypospadias: etiology and current research. Urol Clin North Am. 2010;37:159–66. doi: 10.1016/j.ucl.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Kalfa N, Philibert P, Baskin LS, Sultan C. Hypospadias: interactions between environment and genetics. Mol Cell Endocrinol. 2011;335:89–95. doi: 10.1016/j.mce.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Li J, Willingham E, Baskin LS. Gene expression profiles in mouse urethral development. BJU Int. 2006;98:880–85. doi: 10.1111/j.1464-410X.2006.06435.x. [DOI] [PubMed] [Google Scholar]
- 8.Tang F. Small RNAs in mammalian germline: Tiny for immortal. Differentiation. 2010;79:141–46. doi: 10.1016/j.diff.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Rios JM. [The big world of the small RNAs]. Rev Latinoam Microbiol. 2006;48:73–78. [in Spanish] [PubMed] [Google Scholar]
- 10.Zaravinos A. The regulatory role of microRNAs in EMT and cancer. J Oncol. 2015;2015:865816. doi: 10.1155/2015/865816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang L, Guo F, Huo B, et al. Expression and clinical significance of the microRNA-200 family in gastric cancer. Oncol Lett. 2015;9:2317–24. doi: 10.3892/ol.2015.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hur K, Toiyama Y, Takahashi M, et al. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2013;62:1315–26. doi: 10.1136/gutjnl-2011-301846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mongroo PS, Rustgi AK. The role of the miR-200 family in epithelial-mesenchymal transition. Cancer Biol Ther. 2010;10:219–22. doi: 10.4161/cbt.10.6312548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li C, Xu YM, Li HB. Preliminary experimental study of urethral reconstruction with tissue engineering and RNA interference techniques. Asian J Androl. 2013;15:430–33. doi: 10.1038/aja.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ungefroren H, Groth S, Sebens S, et al. Differential roles of Smad2 and Smad3 in the regulation of TGF-beta1-mediated growth inhibition and cell migration in pancreatic ductal adenocarcinoma cells: Control by Rac1. Mol Cancer. 2011;10:67. doi: 10.1186/1476-4598-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, He DW, Zhang DY, et al. Di(2-ethylhexyl) phthalate (DEHP) increases transforming growth factor-beta1 expression in fetal mouse genital tubercles. J Toxicol Environ Health A. 2008;71:1289–94. doi: 10.1080/15287390802114915. [DOI] [PubMed] [Google Scholar]
- 18.Konno R, Yamakawa H, Utsunomiya H, et al. Expression of survivin and Bcl-2 in the normal human endometrium. Mol Hum Reprod. 2000;6:529–34. doi: 10.1093/molehr/6.6.529. [DOI] [PubMed] [Google Scholar]
- 19.Elson-Schwab I, Lorentzen A, Marshall CJ. MicroRNA-200 family members differentially regulate morphological plasticity and mode of melanoma cell invasion. PLoS One. 2010;5(10) doi: 10.1371/journal.pone.0013176. pii: e13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhlmann S, Zhang JD, Schwager A, et al. miR-200bc/429 cluster targets PLCgamma1 and differentially regulates proliferation and EGF-driven invasion than miR-200a/141 in breast cancer. Oncogene. 2010;29:4297–306. doi: 10.1038/onc.2010.201. [DOI] [PubMed] [Google Scholar]
- 21.Ma C, Ding YC, Yu W, et al. microRNA-200c overexpression plays an inhibitory role in human pancreatic cancer stem cells by regulating epithelial-mesenchymal transition. Minerva Med. 2015;106:193–202. [PubMed] [Google Scholar]
- 22.Burk U, Schubert J, Wellner U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–89. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee JW, Zoumalan RA, Valenzuela CD, et al. Regulators and mediators of radiation-induced fibrosis: Gene expression profiles and a rationale for Smad3 inhibition. Otolaryngol Head Neck Surg. 2010;143:525–30. doi: 10.1016/j.otohns.2010.06.912. [DOI] [PubMed] [Google Scholar]
- 24.Gordon KJ, Blobe GC. Role of transforming growth factor-beta superfamily signaling pathways in human disease. Biochim Biophys Acta. 2008;1782:197–228. doi: 10.1016/j.bbadis.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 25.Jagadeesan J, Bayat A. Transforming growth factor beta (TGFbeta) and keloid disease. Int J Surg. 2007;5:278–85. doi: 10.1016/j.ijsu.2006.04.007. [DOI] [PubMed] [Google Scholar]